Colour is a naturally important characteristic in plant biology. It is also an important trait for fruits and vegetables. Three major groups of pigments—betalains, flavonoids and carotenoids—are responsible for pigmentation in plants. Carotenoids, the most widely distributed pigment, display diverse colours, ranging from yellow and orange to deep red. The first step of carotenogenesis is the dimerization of two molecules of geranylgeranyl pyrophosphate (GGPP) to phytoene by the enzyme phytoene synthase (PSY). This is the rate‐limiting step in the carotenoid biosynthesis route and has long been considered a ‘bottleneck’ in the pathway. Previous plant biotechnology studies have enhanced the carotenoid levels to increase the nutritional value of crops and fruits by conventional breeding and genetic engineering. In the past two decades, on the purpose to alleviate the public health problem of vitamin A deficiency (VAD) in large number of countries, the Crt B (for phytoene synthase), Crt I (for phytoene desaturase) and Crt Y genes (for lycopene ß‐cyclase) of bacteria were singly or in combination overexpressed in various crops to generate provitamin A‐rich foods. For example, the Golden rice (Ye et al., 2000), Brassica golden seed (Shewmaker et al., 1999), Golden potato tuber (Diretto et al., 2010), maize of high carotenoid endosperm (Aluru et al., 2008) and tomato (Fraser et al., 2002) were successfully enhanced with the predominant accumulation of ß‐carotene. In addition, the RNAi‐mediated technology to down‐regulate DET 1 (De‐etiolated 1 gene, a negative regulator of light signal transduction in tomato) expression and to obtain the increasing carotenoid level in tomato has also been carried out (Davuluri et al., 2005). With regard to the flower colour alteration using genetic engineering, considerable progress has been made in the past 20 years. However, those works were concentrated on flavonoid modification, such as blue rose, carnation and petunia (Tanaka et al., 2010). Thus far, the carotenoid modification to change flower colour was only reported on transgenic chrysanthemum, by knockdown of CCD4 to generate vivid yellow flower from white flower (Ohmiya, 2009). Next to the success of transgenic chrysanthemum, silencing of phytoene synthase by RNAi to reduce carotenoid content in Oncidium floral tissues is performed and achieved in this paper.
Oncidium orchids are native to Middle and South America. Oncidium hybrids are the second most popular varieties of orchids in the orchid industry, next to the Phalaenopsis spp. The past breeding efforts have produced many commercial cultivars such as Oncidium Sharry Baby, Sweet Sugar, Gower Ramsey and Honey Angel, which are widely available in the current global market. Among these cultivars, Onc. Gower Ramsey and Onc. Honey Angel have been the most popular cultivars in the commercial cut‐flower market in Asian countries. The current Onc. Gower Ramsey and Honey Angel have the drawbacks of reproductive sterility, which makes conventional breeding difficult to generate new variety. Although several somatic mutants thus far have been occurred in tissue‐culture propagation process (Chiou et al., 2010), their floral characters were difficult to meet customer favourites, for example, the two somaclonal mutant varieties—White Jade and Sunkist. The former was caused by the natural mutation of methylation effect on OgCCD1 (Carotenoid Cleavage Dioxygenase 1, catabolizing carotenoid metabolites) promoter. The elevated expression level of OgCCD1 resulted in white floral colour, but with negative side effect of tiny, sparse florets and short vast life; the latter cause by the down‐regulation of OgHYB and OgZEP in carotenoid biosynthetic pathway exhibited the un‐delightful dark brownish florets (β‐carotene accumulation). Efforts were taken to overexpress OgCCD1 gene to obtain white‐colour blooms with good phenotype characters and the results were negative (data not shown). Therefore, an attempt to obtain a white‐flowered Oncidium variety was designed by blocking the carotenoid biosynthesis pathway in the yellow‐coloured flowers of an Oncidium hybrid, such as Honey Angel. Herewith, we constructed a chimeric hairpin‐RNA (hpRNA) transfer‐DNA (T‐DNA) vector targeting PSY mRNA (Figure 1a). After blast analysis on the five PSY nucleotide sequences from other four monocot plant species (rice, wheat, maize, Phalaenopsis) in NCBI data bank, the 150‐bp cDNA region (+51~ +200 bp in ORF) of the OgPSY gene (accession number: FJ859989; refer to Chiou et al., 2010), containing a 22‐bp conserved sequence of 100% homology across corresponding complementary DNA (cDNA) sequences, was chosen for RNAi targeting. Pchrc, a previously identified promoter region (1.5 kb) of the CHRC gene with specific expression in floral tissues (Chiou et al., 2008), was employed to drive the DNA segment of chimeric hairpin‐RNA. The hpRNA T‐DNA vector was delivered into protocorm‐like body (PLB) of WT (Onc. Honey Angel) by Agrobacterium‐mediated transformation. The transformed PLB cultures, exhibiting an approximately 20% transformation efficiency, were screened under hygromycin (30 ppm), regenerated to form seedlings and then planted in pots for two years. After flowering, 50 transgenic orchids harbouring the PSY‐RNAi construct were obtained. These transgenic orchids—approximately 60% of which showed the RNAi effect—displayed whitish to white florets. Subsequently, two independent lines were selected for further study. One displayed a yellowish‐white bloom designated as ‘MF‐1’, and the other showed white blooms were designated as ‘MF‐5’ (Figure 1b). Unlike the previous work employing CaMV 35S promoter, it caused the transgenic Oncidium orchids growth‐arrested and non‐flowering (Liu et al., 2014). The current two transgenic lines were very similar to WT orchids in terms of growth rate and phenotypic traits, such as floral size, floral stem, floret number and leaf morphology. Comparing to the former somaclonal mutant variety, White Jade, which displayed tiny, few florets in floral stem and early wilting (Chiou et al., 2010), it suggests that RNAi technology is an effective and under‐controllable genetic manipulation. Upon analysis of carotenoid levels in floral tissues, MF‐1 and MF‐5 were found to contain 40% of 9‐cis‐violaxanthin levels relative to WT orchid. Further, while MF‐1 contained 30% of neoxanthin, violaxanthin and β‐carotene levels relative to the recipient, MF‐5 contained only 1–3% of these compounds (Figure 1c).
Figure 1.
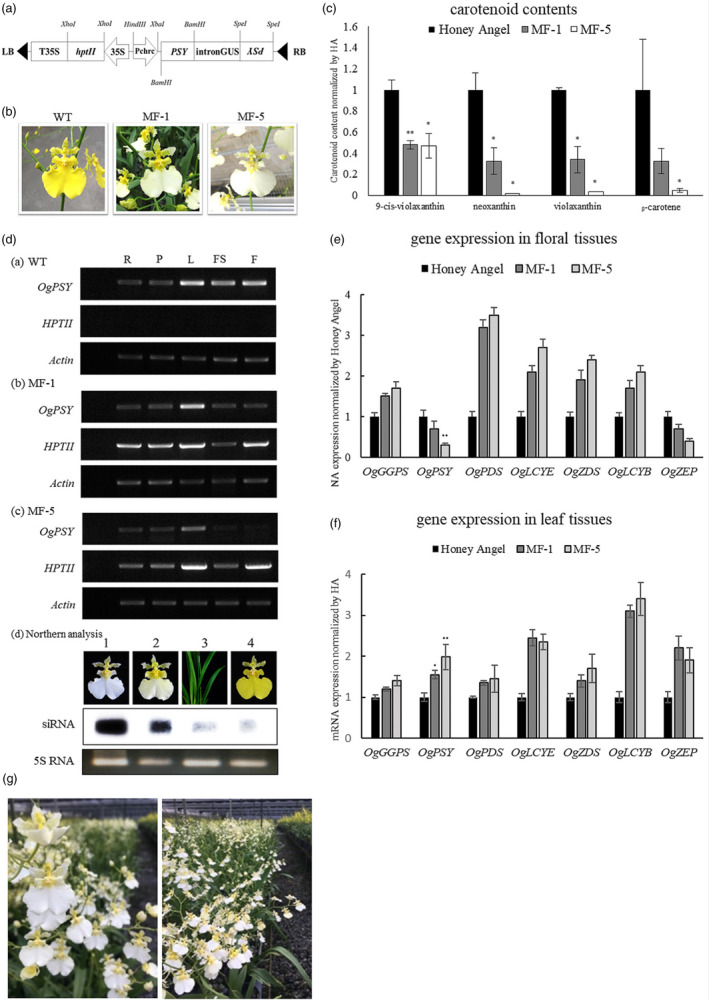
RNAi construct transformation, generation of white transgenic Oncidium varieties, and characterization of carotenoid compounds and relevant gene expression. (a) Schematic representation of hpRNA silencing vectors. Two 150‐bp PSY ORF fragments were inversely fused to both ends of the spacer DNA fragment, GUS intron (750 bp), and driven by Pchrc promoter. (b) Photographs of the transgenic Oncidium orchids harbouring PSY‐RNAi constructs. From left to right: WT orchid (yellow), transgenic orchid MF‐1 (yellowish white); and transgenic orchid MF‐5 (white). (c) Carotenoid levels in the floral tissues of the three Oncidium orchids. (*significantly different compared to control at P < 0.05, **significantly different compared to control at P < 0.01, by t tests, n = 2) (d) Analysis of OgPSY RNA expression in transgenic orchids. Semi‐quantitative real‐time PCR analysis of OgPSY expression in WT orchid; transgenic orchid MF‐1; and transgenic orchid MF‐5. R, root; P, pseudobulb; L, leaf; FS, flower stem; F, floral tissues; HPTII, hygromycin phosphotransferase II gene. Northern analysis of siRNA for the floral tissues of MF‐5 (lane 1), MF‐1 (lane 2), leaves of MF‐5 (lane 3) and the WT orchid (lane 4). The α‐32P‐labelled 18‐bp RNA probe, 5′‐GUUAGAUACACACCACGU‐3′, was used to hybridize. (e) and (f) Real‐time quantitative PCR analysis of the expression patterns of genes involved in carotenoid biosynthesis in floral tissues and in leaf tissues. (*significantly different compared to control at P < 0.05, **significantly different compared to control at P < 0.01, by t tests, n = 3) (g) The white transgenic orchid plants (MF‐5) grown in a greenhouse. They displayed the stable traits.
As shown in Figure 1d, WT orchid showed high OgPSY (designation given to the PSY gene of Oncidium orchids) expression levels in floral tissues, leaves and flower stems; in comparison, the OgPSY expression levels in MF‐1 were lower, and those in MF‐5 were the least among them (Figure 1d). Meanwhile, HPTII was constitutively expressed in MF‐1 and MF‐5, while no expression was observed in WT orchid (Figure 1d). Moreover, northern blot analysis, performed to estimate the extent of OgPSY silencing in the floral tissues of both transgenic orchid lines, demonstrated the presence of OgPSY‐derived small interfering RNAs (siRNAs) in the floral tissues of MF‐1 and MF‐5 (Figure 1d—northern analysis). It showed more extensive disruption of OgPSY expression in MF‐5 than in MF‐1. Upon comparing the OgPSY‐derived siRNA levels in the floral and leaf tissues of MF‐5 (Figure 1d—northern analysis, lane 1& 3), although there seemed little siRNA levels existing in the leaves and WT, it could demonstrate that the Pchrc promoter was activated specifically in petal tissues. The data clearly indicated the successful silencing of PSY expression specifically in the floral tissues of the white orchid varieties. We also analysed the mRNA levels of the carotenoid biosynthesis pathway genes in floral tissues by Q‐PCR. The data demonstrated that the carotenoid pathway genes exhibited enhanced expression level, except for OgPSY, which was down‐regulated to 70% in MF‐1 and 20% in MF‐5 (Figure 1e). This enhanced expression can be explained as a compensation effect for the flux in the entire carotenoid pathway. Upon analysing the expression patterns in leaf tissues, it was interesting to note that all carotenoid pathway genes in the transgenic orchids exhibited higher expression levels than in the WT (Figure 1f). Although the gene silencing was not targeted to the leaf, the side effects were present.
The phenotypic stability of a product is a priority for commercialization purposes. Because the Oncidium hybrid has lost pollen fertility for sexual reproduction, asexual multiplication by tissue culture is the sole way for propagating its seedlings. In this study, we multiplied the MF‐1 and MF‐5 plants from the P0 stage by culturing tiny meristematic bud tissues on half‐strength Murashige & Skoog (MS) medium containing 30 ppm hygromycin. Through PLB and shooting steps, we were able to produce two thousand P1 seedlings of each individual line after 2 years. Seedlings were grown at a temperature of 30°C/25°C (day/night), and the light intensity was controlled at the PPFD (photosynthetic photon flux density) of approximately 300 μmol/m2/s. Bloom colour was used as the main parameter for assessing phenotypic stability. Throughout the 2‐year cultivation period, the P1 plants displayed stable white pigments in their flowers, approximately 85% and 88% in MF‐1 and MF‐5, respectively. Further, we selected the best one from the P1 population and propagated them by tissue culture again. We then evaluated the phenotypic traits of the P2 population over another 2‐year period. As shown in Figure 1g, the P2 plants of both lines produced approximately 100% whitish/white florets, suggesting that the RNAi‐mediated suppression of OgPSY expression was stably inherited in this population through consecutive selection. The results of analysis of OgPSY expression revealed that the OgPSY mRNA levels in the P2 plants were as low as those in the P0 transgenic orchids and lower than those in WT (data not shown). These results strongly confirmed the genetic stability of the RNAi‐mediated transformation. In conclusion, our work demonstrates that down‐regulation of PSY expression by RNAi technology under the regulation of a tissue‐specific promoter, Pchrc, is effective to modify the flower colour specifically in floral tissues.
Acknowledgements
This work was financially supported by the Ministry of Science and Technology, Taiwan, under Project No.105C1402 to Dr. Kai‐Wun Yeh.
References
- Aluru, M. , Xu, Y. , Guo, R. , Wang, Z.G. , Li, S.S. , White, W. , Wang, K. et al. (2008) Generation of transgenic maize with enhanced provitamin A content. J. Exp. Bot. 59, 3551–3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou, C.Y. , Wu, K. and Yeh, K.W. (2008) Characterization and promoter activity of chromoplast specific carotenoid associated gene (CHRC) from Oncidium Gower Ramsey. Biotechnol. Lett. 30, 1861–1866. [DOI] [PubMed] [Google Scholar]
- Chiou, C.Y. , Pan, H.A. , Chuang, Y.N. and Yeh, K.W. (2010) Differential expression of carotenoid‐related genes determines diversified carotenoid coloration in floral tissues of Oncidium cultivars. Planta 232, 937–948. [DOI] [PubMed] [Google Scholar]
- Davuluri, G.R. , Tuinen, A.V. , Fraser, P.D. , Manfredonia, A. , Newman, R. , Burgess, D. and Brummell, D.A. (2005) Fruit‐specific RNAi‐mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nat. Biotechnol. 23, 890–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diretto, G. , Al‐Babili, S. , Tavazza, R. , Scossa, F. , Papacchioli, V. , Migliore, M. , Beyer, P. et al. (2010) Transcriptional‐metabolic networks in beta‐carotene‐enriched potato tubers: the long and winding road to the golden phenotype. Plant Physiol. 154, 899–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser, P.D. , Romer, S. , Shipton, C.A. , Mills, P.B. , Kiano, J.W. , Misawa., N. , Drake, R.G. et al. (2002) Evaluation of transgenic tomato plants expressing an additional phytoene synthase in a fruit‐specific manner. Proc. Natl Acad. Sci. USA, 99, 1092–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J.X. , Chiou, C.Y. , Shen, C.H. , Chen, P.J. , Liu, Y.C. , Jian, C.D. and Yeh, K.W. (2014) RNA interference‐based gene silencing of phytoene synthase impairs growth, carotenoids, and plastid phenotype in Oncidium hybrid orchid. SpringerPlus 3, 478–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmiya, A. .(2009) Carotenoid cleavage dioxygenases and their apocarotenoid products in plants. Plant Biotechnol. 26, 351–358. [Google Scholar]
- Shewmaker, C.K. , Sheehy, J.A. , Daley, M. , Colburn, S. and Ke, D.Y. (1999) Seed‐specific overexpression of phytoene synthase: increase in carotenoids and other metabolic effects. Plant J. 20, 401–412. [DOI] [PubMed] [Google Scholar]
- Tanaka, Y. , Brugliera, F. , Kalc, G. , Senior, M. , Dyson, B. , Nakamura, N. , katsumoto, Y. et al. (2010) Flower color modification by engineering of the flavonoid biosynthetic pathway: practical perspectives. Biosci. Biotechnol. Biochem. 74, 1760–1769. [DOI] [PubMed] [Google Scholar]
- Ye, X.D. , Babilli, S.A. , Kloti, A. , Zhang, J. , Lucca, P. , Beyer, P. and Potrykus, I. (2000) Engineering the provitamin A (β‐carotene) biosynthetic pathway into (carotenoid‐free) rice endosperm. Science 287, 303–305. [DOI] [PubMed] [Google Scholar]


