Summary
Apple exhibits S‐RNase‐mediated self‐incompatibility. Although the cytotoxic effect of S‐RNase inside the self‐pollen tube has been studied extensively, the underlying defence mechanism in pollen tube in Rosaceae remains unclear. On exposure to stylar S‐RNase, plant defence responses are activated in the pollen tube; however, how these are regulated is currently poorly understood. Here, we show that entry of both self and non‐self S‐RNase into pollen tubes of apple (Malus domestica) stimulates jasmonic acid (JA) production, in turn inducing the accumulation of MdMYC2 transcripts, a transcription factor in the JA signalling pathway widely considered to be involved in plant defence processes. MdMYC2 acts as a positive regulator in the pollen tube activating expression of MdD1, a gene encoding a defence protein. Importantly, MdD1 was shown to bind to the RNase activity sites of S‐RNase leading to inhibition of enzymatic activity. This work provides intriguing insights into an ancient defence mechanism present in apple pollen tubes where MdD1 likely acts as a primary line of defence to inhibit S‐RNase cytotoxicity prior to self/non‐self recognition.
Keywords: Malus domestica, MdD1, pollen tube growth, plant defence, S‐RNase
Introduction
Plant reproduction requires reciprocal recognition and communication events between the pollen and the stigma/style cells (Boavida et al., 2005; Higashiyama, 2010; Suzuki, 2009; Thomas and Noni, 2013). While many flowering plants exhibit intraspecific reproductive barrier that is widely found to prevent self‐fertilization in order to promote cross‐breeding, this process is called self‐incompatibility (SI) (Chen et al., 2018; Franklin‐Tong, 2008; Li et al., 2018; Nettancourt, 2001; Takayama and Isogai, 2005). To date, a number of genes involved in SI system between pollen and pistil have been identified, among these many plant species from the Rosaceae, Plantaginaceae and Solanaceae family exhibit S‐RNase‐based gametophytic self‐incompatibility (GSI), which is the most commonly occurring and has been centrally studied (Anderson et al., 1989; Broothaerts et al., 2004; De et al., 2012; Roalson and Mccubbin, 2003; Sassa et al., 1992, 1994). Apple (Malus x domestica), which belongs to S‐RNase‐based GSI in Rosaceae, is one of the most important horticultural crops in the world. In the GSI system, S‐RNase is female determinant. During fertilization, S‐RNase is transported into pollen tubes by ABCF protein and recognized by S‐locus F‐box protein (SLF/SFBB), which is the male determinant (Anderson et al., 1989; Hua et al., 2008; Mcclure, 2004; Meng et al., 2014; Qiao et al., 2004; Sijacic et al., 2004; Wei et al., 2014).
S‐RNases are style‐specific secreted ribonuclease glycoproteins, which act both as recognition proteins and as highly specific cytotoxins. The critical role of S‐RNase is the rejection of self‐pollen. In GSI response, both self and non‐self pollen tube could grow into pistil. In compatible pollen tubes, S‐RNase subsequently degrades via the ubiquitin‐26S‐proteasome pathway, while in incompatible pollen tubes, S‐RNase exhibits cytotoxicity characteristic and has been attributed to its degradation of pollen rRNA, resulting in the arrest of protein synthesis (Gray et al., 1991).
Recent studies in Rosaceae mainly focus on the downstream signalling cascade likely occurs in SI in order to terminate incompatible pollen tubes growth, such as changes of calcium concentration, actin cytoskeleton depolymerization, burst out of reactive oxygen and tRNA aminoacylation in pollen tubes (Di et al., 2010; Li et al., 2018; Wang and Zhang, 2011; Wang et al., 2009). Indeed, in general, when an extracellular protein enters a cell, a series of processes will be triggered, including defence response and signal transduction (Kwak et al., 2014; Sawamoto et al., 1996; Wan et al., 2009). In response to undesirable invading substances, plants could activate a wide range of defences which can reduce potential cellular damage (Erb et al., 2012; Schuman and Baldwin, 2015; Wu et al., 2010). Taken together, as S‐RNase possesses cytotoxicity, it seems that S‐RNase may trigger signalling pathways or defence responses following its entry into the pollen tube. Furthermore, SI and the plant innate immunity systems are considered to have common pathways, and Nasrallah (2005) proposed that SI originates from an ancient plant defence system so that there are many common pathways between SI and plant defence; for instance, programmed cell death is caused by RNase‐based cytotoxicity (Dangl and Jones, 2001; Geitmann et al., 2004; Jones and Dangl, 2006; Jordan et al., 2000; Nasrallah, 2005; Penn and Potts, 1999; Schuman and Baldwin, 2015).
Previous reports have demonstrated that cysteine‐rich peptides (CRPs), comprising various subgroups of defence molecules, regulate a wide range of reproductive processes and defence in plants, including pollen tube growth and guidance, gamete activation and pathogen responses (Bircheneder and Dresselhaus, 2016; Qu et al., 2015; Satohiro et al., 2009; Takeuchi and Higashiyama, 2016; Tavormina et al., 2015). Moreover, in GSI system of poppy (Papaver rhoeas), stigma S‐determinant (PrsS) encodes a secreted CRP, is a stigma determinant in poppies and could activate downstream signalling events when it interacted with incompatible pollen (Foote et al., 1994; Wheeler et al., 2010). In addition, Brassica families display another SI system, SCR, a S‐locus CRP cysteine‐rich peptide, that is a pollen determinant in Brassica and structurally similar to defensin (Kachroo and Nasrallah, 2002; Schopfer et al., 1999; Suzuki et al., 1999; Takayama et al., 2001). These findings propose that CRPs act as signalling substance triggering varies responses in SI process. Furthermore, previous studies have shown that defence genes, such as defensins or γ‐thionins, could be regulated by jasmonic acid (JA), which is known to be important for plant defence and fertility (Ahmad et al., 2016; Davis et al., 1969; Devoto and Turner, 2003; Farmer and Ryan, 1990; Ishiguro et al., 2001; Melo et al., 2002; MuradoğLu et al., 2010; Nakata and Ohme‐Takagi, 2013; Song et al., 2005; Thomma et al., 1998; Wasternack and Hause, 2013; Wasternack et al., 2013; Yildiz and Yilmaz, 2002; Yuan and Zhang, 2015). Thus, the SI reaction is more likely to trigger the defence response in pollen tube in Rosaceae species. To date, it has not been reported that plant hormones or CRPs participate in pollen tube growth during the SI response in apple.
In GSI system in the Rosaceae, both self and non‐self S‐RNase could be transported into pollen tubes, and then, recognition occurs in the tube (Meng et al., 2014). However, there has been no evidence of the molecular events that occur between entry of S‐RNase into pollen tubes and its recognition. A key question is whether non‐self S‐RNase also affects pollen tube growth in a negative way before its targeted degradation? Are there endogenous response mechanisms which combat the effects of S‐RNase during the early stages of pollen tube growth in the style? Thus, we propose the feasibility that a plant defence response may be triggered by self/non‐self S‐RNase in the pollen tube. Here, we focus on the S‐RNase‐induced reaction in pollen tube before self/non‐self discrimination. We report that apple S‐RNase triggers the accumulation of JA in both self and non‐self pollen tubes, resulting in the deployment of defence protein, MdD1. We also provide evidence that the MdD1 protein inhibits S‐RNase activity by interacting with its active site, ensuring normal pollen tube growth prior to self/non‐self recognition via the SI mechanism. Based on these results, we propose that MdD1 acts as a primary defence protein in apple pollen tubes facilitating compatible pollen tube growth in the presence of S‐RNase.
Results
S‐RNase increases the concentration of JA in both compatible and incompatible apple pollen tubes
As the SI reaction primarily occurs in the pollen tube, we established an in vitro pollen tube culture assay to study SI in apple. For self‐pollination‐induced (SPI) treatment, equal amounts of recombinant S‐RNase proteins (S 1 ‐RNase and S 2 ‐RNase) matching the ‘Ralls Janet’ pollen S haplotype (S 1 S 2 ) were added to the pollen germination media at a range of concentrations. For cross‐pollination‐induced (CPI) treatment, equal amounts of recombinant S‐RNase proteins (S 3 ‐RNase and S 9 ‐RNase) were added to the pollen germination media at a range of concentrations. The results showed that pollen tube lengths were found to be significantly reduced when the total concentration of self S‐RNase (S 1 ‐RNase and S 2 ‐RNase) was 30 μg/mL. In contrast, pollen tubes were not significantly suppressed when the concentration of non‐self S‐RNase (combination of S 3 ‐RNase and S 9 ‐RNase) was 30 μg/mL. Furthermore, pollen tube growth was inhibited by both self and non‐self S‐RNases when these proteins were added at a concentration of 45 μg/mL, indicating that high protein concentration of S‐RNase could not distinguish self and non‐self S‐RNase treatment, while inactivated S‐RNase, which was denatured by boiling, failed to induce significant inhibition of pollen tube growth (Figure S1). This result clearly demonstrates that SI was induced by adding recombinant S 1 +S 2 ‐RNase proteins to a final total concentration of 30 μg/mL in the pollen culture medium and that self and non‐self reactions can be distinguished in vitro.
To investigate whether self or non‐self S‐RNase influences JA content changes in pollen tubes, we used high‐performance liquid chromatography (HPLC) and observed a significant increase of JA levels in apple pollen tubes under the treatments of S 1 +S 2 ‐RNase (self) and S 3 +S 9 ‐RNase (non‐self) (Figure 1a). These results suggested that the JA content could be induced by both self and non‐self S‐RNase. And there was no significant difference in JA level between the self and non‐self S‐RNase treatments. Given that MYC2 has previously been identified as a key TF involved in the JA signal transduction pathway (Kazan and John, 2013), in order to further investigate the regulation mechanisms of JA response to S‐RNase in apple pollen tubes, we identified the MdMYC2 cDNA sequence from RNA extracted from ‘Ralls Janet’ pollen tubes. MdMYC2 is predicted to encode a protein with bHLH‐MYC TF N‐terminal structural domain (Figure S2). Moreover, based on protein sequence alignment and crystal structure superimposition analysis, MdMYC2 polypeptide is composed of helix–loop–helix domains, as is characteristic of the specific DNA‐binding proteins (Figure 1b, Figure S2 and Table S1). Combining the result of subcellular localization, MdMYC2‐GFP fluorescence indicated that MdMYC2 is localized in the nucleus (Figure S3b), indicating that MdMYC2 act as a TF in apple pollen tube. The expression of MdMYC2 was evaluated in different organs of apple, the results showed that MdMYC2 transcript abundance ubiquitously expressed in all organs (Figure S3a). In addition, qRT‐PCR revealed that expression of MdMYC2 in cultured ‘Ralls Janet’ pollen tubes increases significantly after both self and non‐self S‐RNase treatments (Figure 1c). Furthermore, the expression of MdMYC2 was also induced by MeJA treatment in pollen tube (Figure 1c), indicating that it participates in JA pathway. No difference was observed in the expression of MdMYC2 between samples of pollen tubes that had been treated with the JA biosynthesis inhibitor, DIECA (sodium diethyldithiocarbamate), and samples of pollen tubes that had been treated with S 1 +S 2 ‐RNase/S 3 +S 9 ‐RNase after DIECA treatment (Figure 1c). These results suggested that the expression of MdMYC2 may be induced in pollen tubes by both self and non‐self S‐RNase via the JA signalling pathway.
Figure 1.
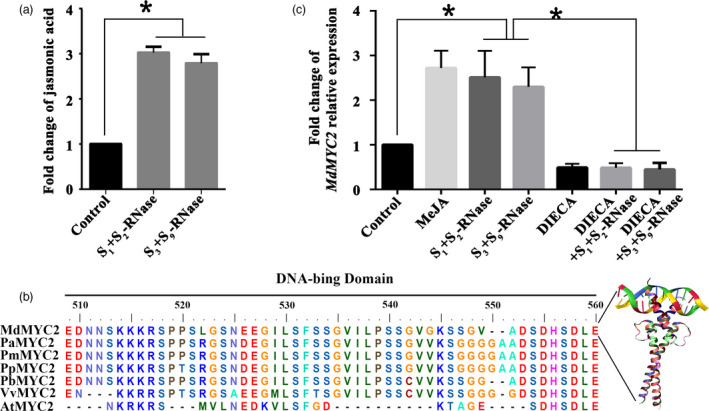
S‐RNase induces the JA‐MdMYC2 signalling pathway in pollen tubes. (a) Pollen tubes from ‘Ralls Janet’ were treated with 30 μg/mL of S 1 +S 2 ‐RNase and S 3 +S 9 ‐RNase, respectively, and then collected the pollen tubes. Fold change of jasmonic acid content in ‘Ralls Janet’ pollen tubes treated with 30 μg/mL of S 1 +S 2 ‐RNase and S 3 +S 9 ‐RNase. Control, untreated pollen tubes. (b) Comparison of the deduced amino acid sequence of MdMYC2 with MYC2 protein sequences from other plant species. The DNA‐binding domain is shown as a black bar above the alignment. AtMYC2 is from Arabidopsis thaliana, PaMYC2 is from Prunus avium, PmMYC2 is from Prunus mume, PpMYC2 is from Prunus persica, PbMYC2 is from Pyrus bretschneideri, and VvMYC2 is from Vitis vinfera. (c) Fold change of MdMYC2 relative expression. qRT‐PCR analysis of MdMYC2 expression in pollen tubes following various treatments. Pollen tubes were treated with 30 μg/mL of S 1 +S 2 ‐RNase and S 3 +S 9 ‐RNase, 30 μm methyl jasmonate 30 μm (MeJA), sodium diethyldithiocarbamate (DIECA) and 30 μg/mL of S 1 +S 2 ‐RNase and S 3 +S 9 ‐RNase after DIECA treatment. Control, untreated pollen tubes. The final data were normalized to the expression in the untreated pollen tubes (control). Values are means + SD of three biological replicates. Asterisks indicate significantly different values (*P < 0.05).
MdD1 is a downstream factor in the MdMYC2 response to S‐RNase in apple pollen tubes
In order to identify the downstream gene targets of MdMYC2, we performed a chromatin immunoprecipitation sequencing (ChIP‐seq) assay using anti‐MdMYC2 antibody to pull down the genomic fragments bound by MdMYC2 in vivo in apple (Figure S4). Based on the binding pattern analysis, we found that G‐box is a MdMYC2 binding motif (Figure 2a). Using a distance to transcription start site between −1000 and +100 bp, a total of 114 candidate genes were identified, 30 of which were found to be expressed in pollen by qRT‐PCR (Figure 2b, Table S3). Using quantitative PCR, we identified three genes from the 30 pollen‐expressed candidate genes that responded to self or non‐self S‐RNase in a MdMYC2‐dependent manner (Figure 2b). Of these, a gene, in which plaza number is MD00G040950, exhibits strongly up‐regulated expression in pollen tubes in response to both self and non‐self S‐RNases. Furthermore, we silenced MdMYC2 in pollen tube to analysis whether MD00G040950 was regulated by MdMYC2. The result was shown that the expression was significantly down‐regulated when MdMYC2 was silenced by antisense oligonucleotide (as‐ODN) (Figure 2c). Therefore, we chose MD00G040950 for further research. Structure analysis showed that MD00G040950 is predicted to encode a protein that has sequence homology to γ‐thionins, and to possess a signal peptide (Figure 2d and Table S1). Phylogenetic analysis of homologs from other species revealed the closest relationship between MD00G040950 and pear (Pyrus) defence proteins (Figure S5b), so we renamed the corresponding gene, MdD1. qRT‐PCR analysis showed that MdD1 was ubiquitously expressed in most organs of apple, with the highest expression in pollen (Figure S5a). Particle bombardment of growing apple pollen tubes was performed with a construct containing the MdD1 fused to green fluorescent protein (GFP), and heterologously expressed MdD1‐GFP in maize protoplasts to investigate the subcellular localization of MdD1. In both cases, MdD1 was observed to localize mainly in the cytoplasm (Figure S5c, d).
Figure 2.
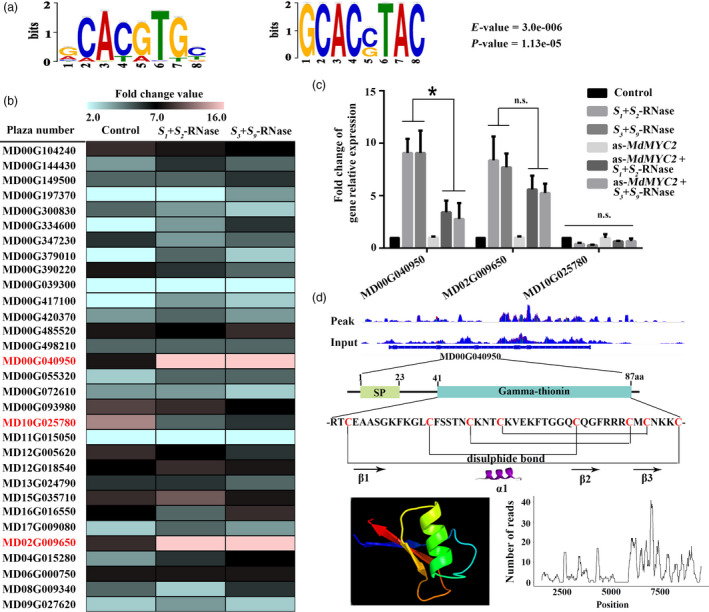
MdD1 is a downstream factor in the MdMYC2 response to S‐RNase in apple pollen tubes. (a) MdMYC2 binding elements. Potential MdMYC2 binding motifs as determined by MEME analysis. (b) The candidate gene expression levels were investigated by qRT‐PCR. Expression of candidate genes which expressed in pollen tube under S 1 +S 2 ‐RNase and S 3 +S 9 ‐RNase‐treated pollen tubes. Screening target genes, which strongly up‐regulated expression in pollen tubes, in response to both S 1 +S 2 ‐RNase and S 3 +S 9 ‐RNase. (c) Expression of MdMYC2 target gene levels was investigated by qRT‐PCR in S 1 +S 2 ‐RNase and S 3 +S 9 ‐RNase treated with or without MdMYC2 silencing using antisense oligonucleotide MdMYC2 (as‐MdMYC2). (d) Structure analysis of MdD1.
To confirm the regulation of the MdD1 promoter by MdMYC2, MdMYC2 binding to the MdD1 promoter was examined by yeast one‐hybrid (Y1H) analysis. Various fragments of the promoter were tested, and this revealed that MdMYC2 bound to a fragment containing the G‐box motif and a MeJA responsiveness motif (Figure 3a). To confirm that MdMYC2 bound to the promoter of MdD1 in vivo, a chromatin immunoprecipitation (ChIP)‐PCR assay was performed. The results showed that MdMYC2 significantly enhanced the PCR‐based detection of the MdD1 promoter, indicating that MdMYC2 binds to the MdD1 promoter in vivo (Figure 3b). In addition, using a GUS (β‐glucuronidase) transactivation assay in tobacco leaves involving co‐transformation with the 35S:MdMYC2 and pMdMdD1:GUS constructs, we saw that the activity of the MdD1 promoter was enhanced by MdMYC2 and further enhanced by jasmonate (MeJA) treatment (Figure 3c). It was also observed that MdD1 expression was up‐regulated by both self and non‐self S‐RNase treatments. The expression of MdD1 was also induced by MeJA treatment in pollen tube (Figure 3d), indicating that it participates in JA pathway. No difference was observed in the expression of MdD1 between samples of pollen tubes treated with DIECA and samples of pollen tubes treated with S 1 +S 2 ‐RNase/S 3 +S 9 ‐RNase after DIECA treatment, or between samples of pollen tubes that were MdMYC2 silenced by as‐ODN and S 1 +S 2 ‐RNase/S 3 +S 9 ‐RNase treated after silencing MdMYC2 (Figure 3d). Additionally, a sense oligonucleotide (s‐ODN) assay and S 1 +S 2 ‐RNase/S 3 +S 9 ‐RNase‐treated tubes after s‐ODN‐MdMYC2 were run as controls (Figure S6). These results suggest that the expression of MdD1 in apple pollen tubes is regulated by both self and non‐self S‐RNase through JA‐MdMYC2 signalling.
Figure 3.
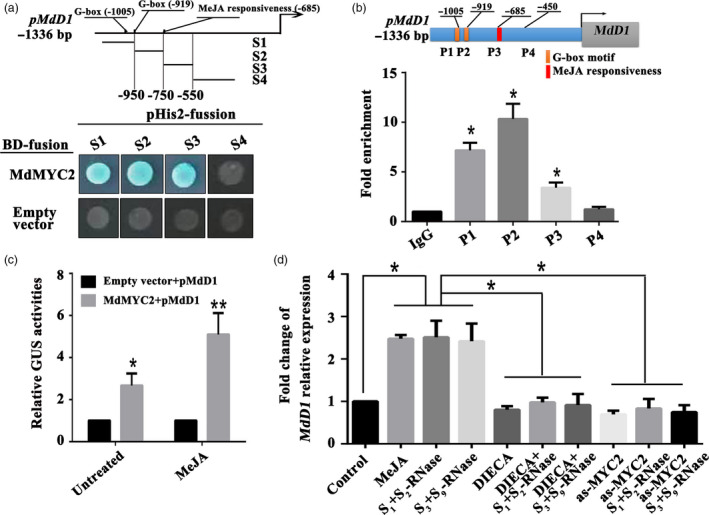
S‐RNase induces MdD1 expression via the JA‐MdMYC2 signalling pathway in pollen tubes. (a) Yeast one‐hybrid (Y1H) analysis showing that MdMYC2 binds to the MdD1 promoter fragment containing the G‐box motif and MeJA responsiveness motif. The promoter of MdD1 was divided into four fragments (S1‐S4). S1 and S2 fragment contain G‐box motif, respectively; S3 fragment contains MeJA responsiveness motif; S4 fragment without the binding motif as a negative control. X‐α‐gal was used as a screening marker. The empty vector and the MdD1 promoter were used as negative controls. These experiments were repeated three times. (b) Chromatin immunoprecipitation (ChIP)‐PCR showing the in vivo binding of MdMYC2 to the MdD1 promoter. Chromatin samples were extracted from pollen tubes and precipitated with an anti‐MdMYC2 antibody. Eluted DNA was used to amplify the sequences neighbouring the G‐box by qPCR. Four regions (P1–P4) were examined. The ChIP assay was repeated three times, and the enriched DNA fragments in each ChIP were used as one biological replicate for qPCR. Data are the mean ± SEM. *P < 0.05. (c) β‐glucuronidase (GUS) reporter activity analysis, showing that MdMYC2 activates the MdD1 promoter and transcriptional activity was significantly enhanced under MeJA treatment. The MdMYC2 effector vector, together with the reporter vector containing the MdD1 promoter, was co‐injected into tobacco leaves to analyse the GUS activity. The empty vector as control, together with the reporter vector containing the MdD1 promoter, was co‐injected into tobacco. Three independent transfection experiments were performed. Data are the mean values ± SEM. *P < 0.05, **P < 0.01. (d) Fold change of MdD1 relative expression by qRT‐PCR. The following pollen tube treatments were used: methyl jasmonate (MeJA) and the inhibitor (DIECA for MeJA); a treatment with the above inhibitors followed by S‐RNase treatment; a treatment with S‐RNase with or without MdMYC2 silencing. Pollen tubes without any treatment were used as controls. Data from three biological replicates were combined using a linear mixed‐effects model. The final data were normalized to the expression in the untreated pollen tubes (control). Data are the mean ± SEM. *P < 0.05.
MdD1 inhibits S‐RNase activity
As MdD1 could be triggered by both self and non‐self S‐RNase, to investigate the function of MdD1 in pollen tubes, its expression was silenced in ‘Ralls Janet’ pollen tubes by using an antisense oligonucleotide assay. A significant reduction in MdD1 transcript and corresponding protein levels were confirmed by qRT‐PCR and immunoblot analysis, respectively (Figure 4a). The growth of MdD1‐silenced pollen tubes was significantly inhibited by a treatment with 15 μg/mL S 1 +S 2 ‐RNase, while growth was not significantly inhibited in the untransformed pollen tubes at this concentration (Figure 4b). Furthermore, we also observed that MdD1‐silenced pollen tube was significantly inhibited by 30 μg/mL non‐self S‐RNase (S 3 +S 9 ‐RNase), while growth was not inhibited by the pollen tubes without silencing MdD1 in pollen tubes at this concentration. Thus, down‐regulation of MdD1 in pollen tubes appears to enhance the effectiveness of S‐RNase inhibition of pollen tube growth, suggesting that MdD1 may function as an inhibitor of RNase enzymatic activity. To test this hypothesis, purified recombinant His‐S‐RNase and GST‐MdD1 protein were combined in an S‐RNase activity assay using yeast RNA as a substrate and S‐RNase activity was found to decrease with increasing MdD1 concentration (Figure 4c). This trend was also seen when different recombinants S 3 +S 9 ‐RNase, S 2 +S 3 ‐RNase or S 1 +S 9 ‐RNase were incubated with MdD1 (Figure S7). We then treated pollen tube cultures with S 1 +S 2 ‐RNase and different concentrations of the MdD1 protein and found that the S‐RNase‐mediated inhibition of pollen tube growth was significantly reduced in the presence of MdD1 (Figure 4d). As non‐self S‐RNase could inhibit the MdD1‐silenced pollen tubes, we speculate that the MdD1 response to S‐RNase might occur before non‐self S‐RNase ubiquitination and subsequent proteasomal degradation in pollen tubes. In order to verify this hypothesis, pollen tubes were treated with the 26S proteasome inhibitor MG132, to block the 26S proteasome‐mediated ubiquitin pathway, and then added non‐self S‐RNase (S 3 +S 9 ‐RNase). The result showed that MdD1 expression remained unchanged regardless of degradation of non‐self S‐RNase. This indicates that the MdD1 response to both self and non‐self S‐RNase in pollen tubes occurs before non‐self S‐RNase recognition (Figure S8).
Figure 4.
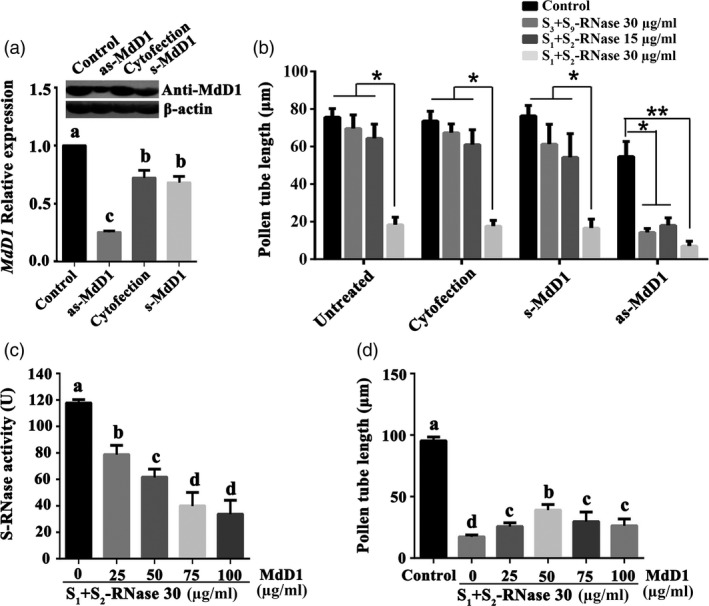
MdD1 inhibits S‐RNase activity. (a) MdD1 expression level was investigated by qRT‐PCR. The pollen tubes were treated with as‐MdD1, s‐MdD1 and cytofection, respectively. as‐MdD1 is phosphorothioated antisense oligodeoxynucleotide of MdD1, s‐MdD1 is phosphorothioated sense oligodeoxynucleotide of MdD1, cytofection is transfection agent buffer, and control is the untreated pollen tubes. Data from three biological replicates were combined using a linear mixed‐effects model. The final data were normalized to the expression levels in untreated pollen tubes (control). (b) Pollen tube length in differently treated samples. (c) Inhibition of S‐RNase activity by various amounts of MdD1. Recombinant GST‐tagged MdD1 protein and His‐tagged S‐RNase protein were expressed in E. coli. S‐RNase (30 μg/mL) protein was incubated with different concentration MdD1 protein, respectively. S‐RNase activity was measured using torula yeast RNA as the substrate. Data are the mean ± SEM. *P < 0.05. (d) Pollen tube length in differently treated samples. Data are the mean ± SEM. At least 60 pollen tubes were measured.
MdD1 interacts with S‐RNase
According to the phenotype of pollen tube growth which has been shown that MdD1 could inhibit S‐RNase activity in pollen tube, in order to investigate the mechanism of MdD1 inhibiting S‐RNase activity, we first investigate whether MdD1 has protease activity that could degrade S‐RNase directly? We purified recombinant GST‐MdD1 and His‐S 1 ‐RNase, respectively. The MdD1 and S 1 ‐RNase were incubated together with or without ATP, and Western blot analysis was performed to evaluate the change of S 2 ‐RNase concentration. No difference was observed in the concentration between ATP‐treated and nontreated control group (Figure S9), indicating that MdD1 has no protease activity to degrade S‐RNase directly.
As MdD1 could not function as protease, we speculated that MdD1 act as a protease inhibitor that could interact with S‐RNase directly. To test this hypothesis, we verified the interaction between MdD1 and S‐RNase. A yeast two‐hybrid (Y2H) assay was employed to determine whether an interaction occurred between MdD1 and different S‐RNase (S 1 ‐, S 2 ‐, S 3 ‐ and S 9 ‐RNase) haplotypes. MdD1 was found to directly interact in a non‐haplotype‐specific manner with S‐RNase (Figure 5a, Figure S10). A pull‐down assay using purified recombinant polyhistidine‐tagged S‐RNase (His‐S‐RNase) and glutathione S‐transferase‐tagged MdD1 (GST‐MdD1) confirmed this interaction (Figure S11). Additional evidence of interaction between MdD1 and S‐RNase in both pollen tube and apple leaves was obtained using bimolecular fluorescence complementation (BiFC). As shown in Figure 5c and d, fluorescence was clearly observed for the S‐RNase‐YFPn and MdD1‐YFPc combination. Meanwhile, as shown in Figure 5e, we performed Western bolt to test the expression of S‐RNase and MdD1 in pollen tube and apple leaves during the BiFC assay. Furthermore, when a competitor (S‐RNase) was present YFP fluorescence was weakened (Figure S12). Lastly, we performed a semi‐co‐immunoprecipitation (Co‐IP) using GST‐MdD1 purified from E. coli and incubated it with a total protein extract from the style of ‘Ralls Janet’, using an anti‐S‐RNase antibody to detect S‐RNase binding to MdD1. The result showed that MdD1 interacted with S‐RNase in the crude protein extract (Figure 5b).
Figure 5.
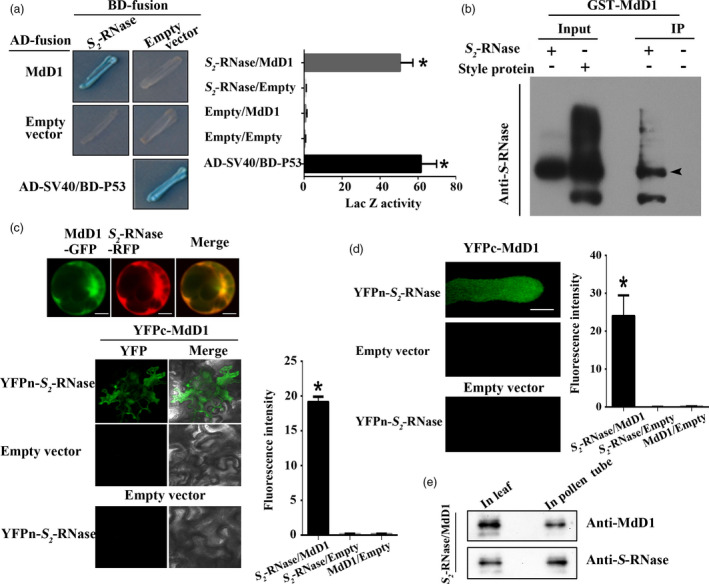
MdD1 interacts with S‐RNase.(a) The interactions between MdD1 and S‐RNase were analysed using a yeast two‐hybrid (Y2H) assay. The coding sequence of MdD1 was ligated into the pGADT7 vector (AD, activation domain), and the coding sequence of S 2 ‐RNase was ligated into the pGBKT7 vector (BD, binding domain). X‐α‐gal was used as a screening marker. The SV40 and P53 genes were used as the positive control, and the empty AD and BD vectors as the negative control. Blue plaques indicate the interaction between two proteins. (b) A pull‐down analysis of the interaction between MdD1 and S 2 ‐RNase. Purified His‐S 2 ‐RNase was used as bait against purified GST‐MdD1. Bound proteins were examined using an anti‐GST antibody. GST was used as a negative control. (b) In vitro immunoprecipitation (IP) assay for binding between S 2 ‐RNase from apple styles and the GST‐MdD1 fusion protein. The band detected by the S‐RNase antibody in the precipitated protein sample indicates the interaction between MdD1 and S 2 ‐RNase. (c) Fusions of MdD1‐GFP (green fluorescent protein) and S 2 ‐RNase‐RFP (red fluorescent protein) were co‐transformed into maize protoplasts. Bimolecular fluorescence complementation (BiFC) assays showed the interaction of MdD1 and S 2 ‐RNase. MdD1‐YFPc and S 2 ‐RNase‐YFPn were co‐injected into apple leaves. (d) BiFC assays showed the interaction of MdD1 and S 2 ‐RNase in pollen tube. Scale bars = 10 μm. (e) Western blot was performed to analysis the expression of MdD1 and S‐RNase in pollen tube and apple leaves. These experiments were repeated three times.
MdD1 inhibits the activity of S‐RNase by binding to the RNase activity sites of S‐RNase
As MdD1 inhibits S‐RNase activity and also directly interacts with S‐RNase, we investigated whether MdD1 inhibits S‐RNase activity via interaction with the active sites of S‐RNase. The active sites of S‐RNase have been identified as two histidine residues, which are located at amino acids 60 and 116 in segments C2 and C3 of the protein (Matsuura et al., 2001). To investigate the mechanism by which MdD1 inhibits S‐RNase activity, a Y2H assay was used to test the binding of each of four fragments (C1C2Hv, HvC3C4C5, C2HvC3 and C4C5) of the S‐RNase to MdD1 (Figure 6a). We found that MdD1 interacted with the fragments containing the active sites of S‐RNase (Figure 6b and Figure S13), and this was confirmed using a BiFC assay in pollen tube and apple leaves. As shown in Figure 6c and d, fluorescence was clearly observed for combinations that included MdD1‐YFPc together with S‐RNase‐YFPn fragments that containing the active sites. By contrast, no fluorescence was observed for the other combinations tested suggesting that MdD1 likely interacts with the active sites of S‐RNase in vivo (Figure 6c and d).
Figure 6.
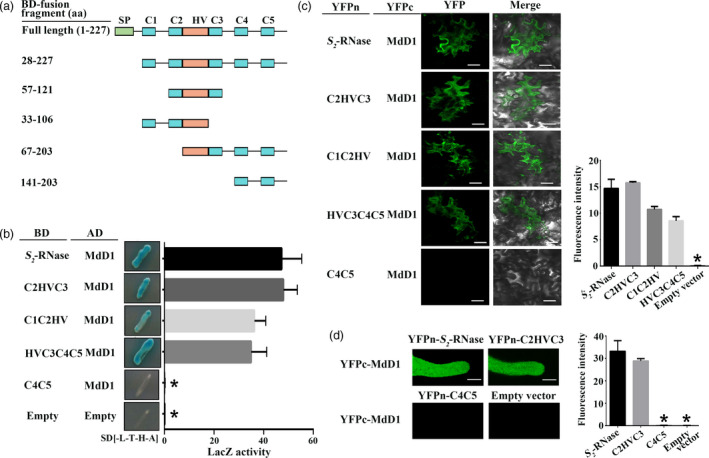
MdD1 interacts with the RNase activity site of S‐RNase. (a) A segment model of S‐RNase. The sequence of S‐RNase was divided into four fragments according to the RNase activity site of S‐RNase. (b) Yeast two‐hybrid (Y2H) assay showing the interactions between MdD1 and different fragments of S 2 ‐RNase as described in (a). The active sites of S 2 ‐RNase are localized in the C2 and C3 domains, and S 2 ‐RNase was divided into four fragments. (c) Bimolecular fluorescence complementation (BiFC) assay showing the interactions between MdD1 and different fragments of the S 2 ‐RNase. Scale bars = 10 μm. (d) BiFC assay showing the interactions between MdD1 and different fragments of the S 2 ‐RNase in pollen tube. Scale bars = 10 μm.
To test whether MdD1 does indeed interact with the active sites of S‐RNase, one, or both, of the active site histidine residues was mutated to aspartic acid (D) generating lines H60D and H116D, or H60D/H116D, respectively (Figure 7a). Ribonuclease activity assays confirmed that the mutated S‐RNase had lost its ability to degrade RNA (Figure 7b). Furthermore, the ability of the mutated S‐RNase to inhibit pollen tube growth was abolished (Figure 7c). Meanwhile, we confirmed that the mutated S‐RNase could not trigger the up‐regulation of MdD1 in pollen tube (Figure 7d). A Y2H assay clearly demonstrated that MdD1 was not able to bind S‐RNase when the active sites were mutated (Figure 7e). Furthermore, a BiFC assay carried out in pollen tube and apple leaf cells also confirmed that MdD1 was unable to interact with S‐RNase when both active site residues were mutated, although a limited interaction was detected when only histidine 60 or histidine 116 was individually mutated (Figure 7f and g). Using a Co‐IP assay, we also observed that the MdD1 protein was immuno‐precipitated by an anti‐MdD1 antibody from extracts from the S‐RNase‐cultured pollen tubes, but not from extracts from the mutated S‐RNase‐cultured pollen tubes (Figure 7h). Taken together, these results suggest that MdD1 directly interacts with the active sites of S‐RNase and inhibits its activity following S‐RNase entry into the pollen tube.
Figure 7.
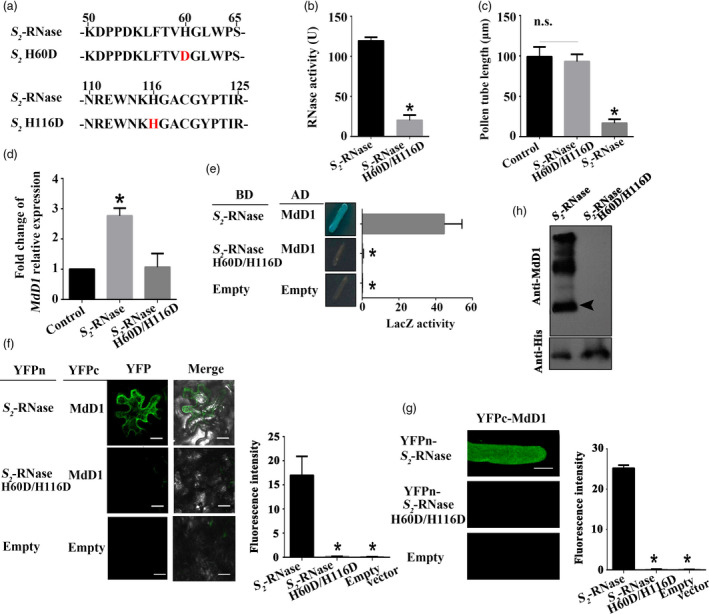
MdD1 inhibits the activity of S‐RNase by binding to its active sites. (a) A segment model of mutant S‐RNase. The active sites of S 2 ‐RNase are two histidine (H) residues located at amino acid 60 and 116. (b) Measurement of the ribonuclease activity of S 2 ‐RNase or mutated S 2 ‐RNase indicated that the mutated S 2 ‐RNase lost most of their ability to degrade RNA. Data are the mean ± SE. n = 4 for biological replicates. *, P < 0.05. t‐test. (c) Measurement of ribonuclease activity of original or mutated S‐RNase indicated that the mutated S‐RNase lost most of the ability to degrade RNA. Data are the mean values ± SEM. *P < 0.05. (d) Measurement of pollen tube length (mutated S‐RNase could not inhibit pollen tube growth). Control, untreated pollen tubes; NS, no significant difference. Data are the mean ± SEM. At least 60 pollen tubes from three pistils were measured. (e) Yeast two‐hybrid (Y2H) assay showing that MdD1 interacts with the active sites of S 2 ‐RNase. The active sites of S 2 ‐RNase are two histidine (H) residues located at amino acid 60 and 116. Each of the histidine was mutated to an aspartic acid (D) residue, and the interactions between these S 2 ‐RNases and MdD1 were investigated. The sequences corresponding to the mature peptides of MdD1 were ligated into the pGADT7 vector (AD, activation domain), and differing mutant fragments of S 2 ‐RNase were ligated into the pGBKT7 vector (BD, binding domain). X‐α‐gal was used as a screening marker. The SV40 and P53 genes were used as the positive control, and AD and BD vectors as the negative control. Blue plaques indicate the interaction between two proteins. (f) Bimolecular fluorescence complementation (BiFC) assay showing that MdD1 was not able to interact with mutated S 2 ‐RNase. MdD1‐YFPc and the various mutant fragments of S 2 ‐RNase‐YFPn were co‐injected into apple leaves. Scale bars = 10 μm. (g) BiFC assay showing that MdD1 was not able to interact with mutated S 2 ‐RNase in pollen tube. Scale bars = 10 μm. (h) In vivo immunoprecipitation (IP) assay showing that MdD1 was not able to interact with mutated S 2 ‐RNase in growing pollen tubes. The band detected by the MdD1 antibody in the precipitated protein sample indicates the interaction between MdD1 and S 2 ‐RNase.
Discussion
Previous studies have demonstrated that SI and immunity share the same pathway (Kao and Tsukamoto, 2004; Mcclure and Franklin‐Tong, 2006, Thomas and Franklin‐Tong, 2004). SI has a lot in similarity with plant immunity, including defence against invaders. Furthermore, both SI and immunity could reject the unwanted substance or cells (Sanabria et al., 2008). Consistent with this notion, we found that both self and non‐self S‐RNase promoted the accumulation of JA in apple pollen tubes, which in turn stimulated the expression of MdMYC2 and its target, a defensin gene, MdD1 (Figure 3). These results imply that entry of S‐RNase into pollen tubes may induce a defence response pathway.
A study by Lush and Clarke showed that Nicotiana alata pollen tubes challenged with self S‐RNases are capable of recovering from inhibition if the top of the incompatible pistil is grafted onto a compatible pistil, indicating that pollen rejection may occur through a triggered mechanism, rather than a directly executed process (Lush and Clarke, 1997). Indeed, in general, when an extracellular protein enters a cell, more complex processes are initiated for it to exert its function with signal transduction or defence reactions often being involved (Sawamoto et al., 1996; Wang et al., 2009). During pollination, style‐specific determinants may trigger signalling pathways or defence responses in the pollen, suggesting similarities between defence signalling and SI (Baxter et al., 2014; Thomas and Franklin‐Tong, 2004; Wang et al., 2010; Wilkins et al., 2011). Several studies have shown that ROS can induce the accumulation of plant hormones (Baxter et al., 2014; Xu et al., 2015), and JA plays important roles in regulating plant defence, such as responding to ROS signalling and in regulating various developmental processes, including fertility and reproduction (Rao et al., 2000; Wasternack, 2007; Wasternack et al., 2013; Yuan and Zhang, 2015). In this study, we found that S‐RNase stimulated an increase in JA content (Figure 1a), suggesting that JA signalling plays a role in the pollen tube response to both self and non‐self S‐RNase.
MYC2 is one of the most important transcription factors in JA signalling (Kazan and John, 2013), and several studies have demonstrated that MYC family genes participate in pollen development (Figueroa and Browse, 2012; Kazan and John, 2013); nevertheless, their specific role remains unclear. Here, we found that S‐RNase‐induced MdMYC2 expression is dependent on the JA signalling pathway in apple pollen tubes (Figure 1c), suggesting that a similar molecular mechanism exists in both plant defence and S‐RNase‐based SI in apple. Consistent with these results, a study by Shi et al. (2017) on S‐RNase‐based SI in pear found that expression of genes involved in the ‘plant hormone signal transduction pathway’ and ‘plant–pathogen interaction pathway’ was significantly enriched in self‐ and cross‐pollinations (Shi et al., 2017). Furthermore, previous reports have indicated that MYC2 can bind G‐box‐related motifs (Gangappa and Chattopadhyay, 2013; Kazan and John, 2013), and in this study, we identified a new signalling component, MdD1, using a ChIP‐seq approach (Figure 2). The G‐box promoter motif of MdD1 was shown to bind MdMYC2 in both Y1H and ChIP‐PCR assays (Figure 3 a,b). Further, a GUS activity analysis revealed that MdMYC2 promotes the activity of the MdD1 promoter in response to MeJA (Figure 3c).
MdD1 is a defensin gene belonging to the γ‐thionin subfamily (Pelegrini and Franco, 2005). It is predicted to encode a secreted cysteine‐rich peptide (CRP), many of which have been reported to play roles in pollination and defence against pathogen infection (Bircheneder and Dresselhaus, 2016; Marshall et al., 2011). For example, in the Brassicaceae (Cruciferae), the pollen coat‐derived ligand S‐locus cysteine‐rich/S‐locus protein 11 (SCR/SP11) is a member of the defensin‐like subfamily of CRPs (Takayama et al., 2001). During SI, SCR/SP11, which acts as the male determinant, binds a membrane‐spanning serine/threonine receptor kinase to activate a signalling cascade, leading to the arrest of pollen tube growth (Kachroo and Nasrallah, 2002). Furthermore, AtPCP‐Bs, also small secreted CRPs, are key regulators of the hydration ‘checkpoint’ in establishment of pollen–stigma compatibility in Arabidopsis thaliana (Wang et al., 2016). In poppy (Papaver rhoeas), the S‐determinant PrsS, a CRP that is secreted by the stigma, interacts with a pollen tube surface receptor S‐determinant, PrpS, inducing increased levels of Ca2+ and ROS (Franklin‐Tong et al., 2002; Wheeler et al., 2010). Additionally, a number of CRPs have been identified as signalling molecules, including pollen‐derived LAT52, a member of the Kunitz trypsin inhibitor‐like subfamily of CRPs that plays a role in tomato (Solanum lycopersicum) pollen tube germination (Tang et al., 2002). Another type of secreted CRP that directly binds to the tip of the growing pollen tube is the short‐range pollen tube‐attracting LURE peptides (Okuda and Higashiyama, 2010; Okuda et al., 2009; Takeuchi and Higashiyama, 2016). Taken together, these studies indicate that CRPs play important roles throughout the entire process of pollen tube development and growth.
Although defensins are a major class of the CRP family, there is no evidence to date that they participate in defence‐related processes in pollen tubes. MdD1 is a defensin gene belonging to the γ‐thionin subfamily. Thionins are an important component of the plant nonspecific defence system, and they can inhibit the growth of many pathogenic microorganisms (Pelegrini and Franco, 2005). At present, most thionins isolated from plants have broad‐spectrum antifungal or bacterial characteristics (Loeza‐Ángeles et al., 2008). In this study, we found that MdD1 could be induced by both self and non‐self S‐RNase suggesting this is a defensive response mounted in both compatible and incompatible tubes (Figure 3d). This adds weight to the hypothesis that MdD1 is a component of an ancient defence system acting as a first line of defence in pollen tubes following perception of RNase. Interestingly, the silencing of MdD1 in pollen tubes strengthened the inhibition of both self/non‐self S‐RNase‐treated pollen tubes (Figure 4), our data also proved that MdD1 inhibited S‐RNase activity by interacting with its active site (Figure 6). These results suggest that MdD1 may play a protective role during the early stage after entry of S‐RNase by reducing the RNase activity of S‐RNase in pollen tube. Furthermore, in this work, we found that there was no change of MdD1 expression between MG132‐treated and nontreated pollen tubes, indicating that MdD1 functions prior to self/non‐self recognition. Importantly, both self and non‐self S‐RNase has the potential to exhibit cytotoxicity via RNA degradation in pollen tubes early in the pollen–pistil interaction, and both of them are considered as a kind of invasion of exogenous substances in pollen tubes. It has been reported that the self‐recognition in SI is familiar to immunologists and the plant innate immune response is operated by a two‐phase pathogen recognition mechanism (Jones and Dangl, 2006). Additionally, both recognition and defence responses are early reaction in plant–pathogen interactions. In lichen symbionts, the recognition‐related genes were triggered only after physical contact; in the meanwhile, the defence‐related genes showed significant changes in early stage (Athukorala and Piercey‐Normore, 2015). In this study, we found that MdD1 was triggered by both self and non‐self S‐RNase before self/non‐self recognition. While this may be desirable in a self‐pollination, cross‐pollen tube growth would also be inhibited with the potential for activation of further downstream processes leading to death of the pollen tube via PCD. Thus, MdD1 likely acts as primary defence factor during the SI process preventing cellular damage from S‐RNase, which in turn would guarantee normal pollen tube growth before self/non‐self recognition. In conclusion, our study has revealed for the first time that a plant hormone‐mediated defence mechanism induced by S‐RNase in pollen tubes interfaces with gametophytic SI in the Rosaceae. MdD1, identified in this study as a key component of this system, acts to inhibit both self and non‐self S‐RNase activity during the initial phases of pollen tube growth in the style. This work adds a new dimension to studies on SI and makes some important links between SI and plant defence that previously have not been explored.
Experimental procedures
Plant materials
Pollen, styles, leaves, sepals, filaments and petals were collected from Malus domestica cv. ‘Ralls Janet’ (S 1 S 2 ) and stored at −80 °C until further use.
Pollen tube growth and in vitro bioassay for apple SI
‘Ralls Janet’ pollen (S 1 S 2 ) was cultured in liquid germination medium (10% (w/v) sucrose, 0.01% (w/v) H3BO3 and 0.015% (w/v) CaCl2 (pH 5.8)) at 23 °C for 40 min in darkness. For the SI treatment in vitro, the nucleotide sequences encoding mature S 1 ‐ or S 2 ‐RNase peptides were amplified and ligated into the pEASY‐E1 vector (Transgene, Beijing). The transformation of the resulting plasmids into Escherichia coli BL21 and the recombinant His‐S‐RNase proteins were purified as previously described (Meng et al., 2014). Inactivation of purified S‐RNase was achieved through treatment of the sample in a boiling water bath. Primer sequences are shown in Supplemental Table S2.
Measurement of JA in pollen tubes
‘Ralls Janet’ pollen was cultured in liquid germination medium for 40 min in darkness at 23 °C, the recombinant S‐RNase proteins were added into the medium for 20 min, and then, the pollen tubes were collected. As a control, pollen tubes were grown in germination media without recombinant S‐RNase proteins being present. JA levels were measured by the Zoonbio Biotechnology Company in Nan Jing using high‐performance liquid chromatography (HPLC). After S‐RNase treatment, the pollen tubes were ground into powder with liquid nitrogen. 1 g of pollen tube powder was collected, and 10 mL of isopropanol/hydrochloric acid was added to the powder, which was shaken at 4 °C for 30 min. After that, 20 mL of dichloromethane was added and then shaken at 4 °C for 30 min. Samples were centrifuged for 5 min at 14,000 rpm and take out the lower organic phase, while the organic phase was dried with nitrogen and dissolved with 150 μL of methanol (0.1% formic acid) in dark. After filtered with membrane of 0.22 μm in diameter, the obtained sample was detected by HPLC‐MS/MS. The standard solutions of JA with gradient of 0.1, 0.5, 1, 5, 20, 50 and 200 ng/mL were prepared with methanol (0.1% formic acid) as solvent. Chromatographic column was Agilent ZORBAX SB‐C18 reversed‐phase column (2.1 × 150, 3.5 μm). Column temperature was 30 °C. Mobile phase: A:B = (methanol/0.1% formic acid): (water/0.1% formic acid). The sample volume used for testing is 2 μL. The mass spectrometry conditions were as follows: air curtain gas was 15 psi, spray voltage was 4500 V, atomization pressure was 65 psi, auxiliary pressure was 70 psi, and atomization temperature was 400 °C.
RNA isolation, RT‐PCR and qRT‐PCR analysis
Total RNA was extracted from pollen tubes and other tissues using the SV Total RNA Isolation System (Promega), according to the manufacturer's instructions. To eliminate genomic DNA contamination, the RNA was treated with DNase I (Takara Bio) for 20 min. First‐strand cDNA was synthesized from total RNA using an RNA PCR Kit (Takara Bio). qRT‐PCRs (20 μL volume containing 1.5 μL cDNA as the template) were performed using an ABI 7500 Real‐Time PCR System (Applied Biosystems), following the manufacturer's instructions and using SYBR Premix Ex Taq (Perfect Real Time; Takara Bio). The qRT‐PCR was conducted with three biological replicates, and each sample was analysed at least in triplicate and normalized using MdACTIN (forward primer 5′‐GTCAGTACCGTGGGAGGGTA‐3′ and reverse primer 5′‐ ACCTCTCGCATGCTAAGC‐3′) as an internal control. Transcription levels were assessed using the 2–▵▵CT method (Schmittgen and Livak, 2008). The primer sequences used for qRT‐PCR analyses are listed in Additional file 13: Table S2.
Bioinformatics analysis
Phylogenetic analyses were performed using MEGA version 5 and the neighbour‐joining method with 1000 bootstrap replicates. The transcript factor binding motif analysis was performed using MEME (http://meme-suite.org/meme_4.11.1/).
ChIP‐seq and ChIP‐PCR assays
The CDS region of MdMYC2 was cloned from cDNA of pollen tubes of ‘Ralls Janet’. The PCR product was ligated into the pEASY‐E1 vector (Transgene, Beijing). The transformation of the resulting plasmids into Escherichia coli BL21 (DE3) and the recombinant His‐MdMYC2 proteins were purified as previously described (Meng et al., 2014). Primer sequences are shown in Supplemental Table S2. The MdMYC2 antibody was prepared by the ABclone Company in Wu Han. Both ChIP‐seq and ChIP‐PCR assays were completed by the ABclone Company in Wu Han. Total DNA was extracted from apple pollen tubes. The ChIP‐seq assay was performed by Illumina HiSeq 2500 sequencing platform.
Y1H assay
The coding sequence of MdMYC2 (2097 bp) was ligated into the pGADT7 vector. A MdD1 promoter fragment (30 bp) was ligated into the pHIS2 vector (Clontech). All constructs were transformed into yeast strain AHY187 by the lithium acetate method. The Y1H assay was conducted as described according to the manufacturer's instructions. All primers used are listed in Additional file 13: Table S2.
GUS analysis
The MdD1 promoter sequence (1336 bp) was ligated into the pCambia1305 vector to generate the GUS reporter construct. The coding sequence of MdMYC2 was ligated into the pBI121 vector to generate the effector construct. Co‐transfection of the reporter and effector constructs into tobacco leaves was performed as described by Li (Li et al., 2016). The expression level of GUS was quantified using qRT‐PCR, and this value was used to represent GUS activity levels. The qRT‐PCR assay was described above. The PCR primers used are listed in Additional file 13: Table S2.
Y2H assays
The Matchmaker GAL4 Two‐hybrid System (Clontech) was used for the Y2H assays. The coding sequence of MdD1 was cloned into the pGADT7 (Clontech) vector, while the coding sequences of S 1 ‐, S 2 ‐, S 3 ‐ and S 9 ‐RNase were cloned into the pGBKT7 vector. All primers used are listed in Additional file 13: Table S2. The GAL4 Y2H assay was performed according to the manufacturer's instructions. At least three independent experiments were performed.
S‐RNase was divided into four fragments according to its active sites. All primers used are listed in Supplemental Table S2. According to the active sites of S‐RNase, we then mutated one, or both, of the histidine residues of the predicted S‐RNase active sites to aspartic acid (D) to generate the lines H60D and H116D, or H60D/H116D, respectively. All primers used are listed in Additional file 13: Table S2.
Pull‐down assays
The coding sequence of MdD1 was cloned into the expression vector, pGEX4T‐1. The GST‐MdD1 fusion construct was transformed into Escherichia coli BL21 (DE3), and the recombinant protein, GST‐MdD1, was expressed and purified as previously described (Meng et al., 2014). The sequences corresponding to the mature peptides of S 1‐, S 2‐, S 3‐ and S 9‐RNase peptides were cloned into the pEXSY‐E1 vector (Transgene, Beijing). The PCR primers used are listed in Additional file 13: Table S2. The recombinant His‐S1‐RNase, His‐S2‐RNase, His‐S3‐RNase and His‐S9‐RNase proteins were expressed and purified as previously described (Meng et al., 2014).
For the pull‐down assay, the purified fusion protein, GST‐MdD1, was incubated with each of the four His‐S‐RNase recombinant proteins and applied to amylase resin, as previously described by Yuan (2014). Proteins were detected by Western blot with anti‐GST and anti‐His as previously described (Meng et al., 2014).
BiFC assays
The full‐length MdD1 and S 1‐, S 2‐, S 3‐ and S 9‐RNase cDNA sequences were cloned into vectors harbouring a yellow fluorescent protein (YFP) coding sequence to generate either N‐terminal or C‐terminal fusions (Meng et al., 2014; a). The PCR primers used are listed in Supplemental Table S2. The resulting constructs were transiently expressed in apple leaves as previously described (Meng et al., 2014). The YFP fluorescence was imaged 5 days after transformation using an Olympus BX61 confocal laser scanning microscope. The excitation wavelength for YFP fluorescence was 488 nm, and emission fluorescence was detected at 500–542 nm.
Transient expression of MdD1 in apple pollen and maize protoplasts
The full‐length cDNA of MdD1 was cloned in‐frame into the pEZS‐NL vector. The PCR primers used are listed in Supplemental Table S1. MdD1 was transiently expressed in ‘Ralls Janet’ pollen as a green fluorescent protein (GFP)‐fusion driven by the LAT52 pollen‐specific promoter. The MdD1‐GFP construct was transformed into pollen by particle bombardment, and GFP expression was visualized 2 h after transformation using fluorescence microscopy (Olympus BX61). To determine the subcellular localization of MdD1 and S‐RNases in maize protoplasts, the full‐length cDNAs of MdD1 and S 2 ‐RNase were cloned into the pEZS‐NL vector driven by the cauliflower mosaic virus promoter (35S). The constructs 35S‐MdD1‐GFP and 35S‐S2‐RNase‐RFP (red fluorescent protein) were transiently expressed in maize protoplasts. Maize protoplast preparation and transformation were performed as previously described (Han et al., 2015). The transformed protoplasts were imaged for GFP/RFP fluorescence using an Olympus BX61 confocal laser scanning microscope. For excitation of the fluorescent proteins, an argon ion laser and the following wavelengths were used: 488 nm for GFP and 543 nm for RFP. Fluorescence was detected at 493–542 nm for GFP and 578–625 nm for RFP.
Protein extraction from pollen tubes and immunoblotting
Pollen tubes were collected from liquid medium by centrifugation at 3500 g and washed three times with cold PBS buffer (0.1 m Na2HPO4 and 0.1 m NaH2PO4, pH 7.0). Pollen proteins were extracted as previously described (Rudd et al., 1996). Antibodies against the MdD1 recombinant proteins (anti‐MdD1) were raised in rabbits (CW Biotech). The immunoblot analyses were performed as described by Yuan (Meng et al., 2014).
Immunoprecipitation assay
For the in vitro immunoprecipitation of MdD1 and S‐RNase, the recombinant protein, GST‐MdD1, was expressed and purified as previously described (Meng et al., 2014). GST‐MdD1 was incubated with Glutathione Sepharose 4B (GE Healthcare), but do not wash from the resin. Pistil proteins were extracted as described (Wang et al., 2006). The pistil crude proteins were incubated with GST‐MdD1 protein which bond to Glutathione Sepharose 4B for 2 h at 4 °C. We then washed with cold PBS buffer (0.1 mol/L Na2HPO4 and 0.1 mol/L NaH2PO4, pH 7.0) for five times in order to take out the proteins which do not bind with GST‐MdD1 protein. For immunoblot analyses, the GST‐MdD1 protein bound to the beads were resuspended in PBS buffer and separated by 12% SDS‐PAGE gels, and then transferred to nitrocellulose membranes (Bio‐Rad) using Trans‐Blot Turbo (Bio‐Rad). Proteins were detected by Western blot with anti‐S‐RNase antibody. The SDS‐PAGE and Western blot assay were described by Yuan (Meng et al., 2014).
Antisense oligo experiments
An antisense oligonucleotide experiment was performed essentially as described by Moutinho et al. (2001). A phosphorothioated antisense oligodeoxynucleotide (as‐ODN) and its sense control (s‐ODN) were synthesized (Taihe Biotechnology Co., Ltd.) to correspond to bp 115–178 of the MdD1 open reading frame and to bp 719‐738 of the MdMYC2 open reading frame, in order to down‐regulate expression of the genes. The antisense and sense sequences are listed in Additional file 12: Table S1. Hydrated pollen tubes were transfected as described above, and pollen tube lengths were measured 40 min after adding S‐RNase into the culture medium. For transfection, 200 μL (10 pmol) of oligonucleotides (as‐ODN or s‐ODN), 280 μL cytofectin buffer and 40 μL cytofectin were pre‐mixed and added immediately to 2 mL germination medium. As a control, cytofectin alone was used to treat the pollen.
S‐RNase activity analysis
S‐RNase activity was measured using torula yeast RNA as the substrate. Recombinant GST‐tagged MdD1 protein and His‐tagged S‐RNase protein were expressed in E. coli. S‐RNase protein (30 μg/mL) was incubated with different concentration of MdD1 protein, respectively. The reaction buffer (490 mL), containing 0.1 m imidazole hydrochloric acid (HCl) (pH 7.0), 0.1 m KCl and 2 mg RNA, was incubated at 37 °C for 10 min before the addition of the enzyme solution (10 mL). After a further incubation at 37°C for 20 min, 100 mL of stop solution (25% perchloric acid, 0.75% (w/v) lanthanum acetate) was added. Following a 30‐min incubation on ice, the solution was centrifuged at 14 000 g for 5 min. The absorbance of the supernatant at 260 nm was measured with a Hitachi U‐2000 spectrophotometer. Data shown are the means ± SEM of three independent biological replicates.
S‐RNase degradation analysis
Recombinant GST‐tagged MdD1 protein and His‐tagged S‐RNase protein were expressed in E. coli. S‐RNase protein (30 μg/mL) was incubated with equal amount of MdD1 protein in the condition with or without ATP. After 60 min of incubation, MdD1 antibody and S‐RNase antibody were used to detect changes in protein content.
Accession numbers
Sequence data from this article can be found in the GenBank/NCBI data libraries under the following accession numbers: NP_001315873.1 (MdMYC2); D50837 (S 1 ‐RNase); U12199 (S 2 ‐RNase); U12200 (S 3 ‐RNase); and U19793 (S 9 ‐RNase).
Conflict of interest
The authors declare no conflict of interest.
Author contributions
T.L., Z.G. and D.M. designed the research; J.D. gave critical assessment of the manuscript; Z.G., Q.Y., W.L., H.Y., J.Y. and C.L. performed the experiments; Z.G., Q.Y. and Q.C. analysed the data; Z.G., J.D., D.M., T.L. and W.L. wrote the paper with input from all the other authors.
Supporting information
Figure S1 Pollen tube growth in control pollen, or those treated with inactive S‐RNase, or different concentrations of S 1 +S 2 ‐RNase and S 3 +S 9 ‐RNase.
Figure S2 Sequence alignments of the deduced amino acid sequences of MdMYC2.
Figure S3 The expression analysis of MdMYC2.
Figure S4 Genome‐wide mapping of MdMYC2 performed by ChIP‐Seq.
Figure S5 The expression analysis of MdD1.
Figure S6 MdD1 relative expression in pollen tube.
Figure S7 MdD1 causes a decrease in S‐RNase activity in different S‐haplotype combinations.
Figure S8 MdD1 relative expression.
Figure S9 S‐RNase could not be degraded by MdD1.
Figure S10 Yeast two‐hybrid analysis of the physical interaction between the mature peptide of MdD1 and the mature peptides of S 1‐, S 3 ‐, and S 9 ‐RNase.
Figure S11 A pull‐down analysis of the interaction between MdD1 and S 1 ‐, S 3 ‐, S 9 ‐RNase.
Figure S12 Bimolecular fluorescence complementation (BiFC) assay showing the interactions between MdD1 and S 1 ‐, S 3 ‐, and S 9 ‐RNase.
Figure S13 Yeast two‐hybrid (Y2H) assay showing the interactions between MdD1 and different S 1 ‐, S 3 ‐, S 9 ‐RNase fragments.
Table S1 Sequences of proteins listed in Table S1.
Table S2 Primers and oligonucleotides used in this study.
Table S3 The candidate genes found by ChIP‐seq.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (Grant Number 2018YFD1000107) and the National Natural Science Foundation of China (31630066, 31572096 and 31372035).
References
- Ahmad, P. , Rasool, S. , Gul, A. , Sheikh, S.A. , Akram, N.A. , Ashraf, M. , Kazi, A.M. et al. (2016) Jasmonates: multifunctional roles in stress tolerance. Front Plant Sci. 7, 813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, M.A. , Mcfadden, G.I. , Bernatzky, R. , Atkinson, A. , Orpin, T. , Dedman, H. , Tregear, G. et al. (1989) Sequence variability of three alleles of the self‐incompatibility gene of Nicotiana alata . Plant Cell, 1, 483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athukorala, S.N.P. and Piercey‐Normore, M.D. (2015) Recognition‐ and defense‐related gene expression at 3 resynthesis stages in lichen symbionts. Can. J. Microbiol. 61, 1–12. [DOI] [PubMed] [Google Scholar]
- Baxter, A. , Mittler, R. and Suzuki, N. (2014) ROS as key players in plant stress signalling. J. Exp. Bot. 65, 1229–1240. [DOI] [PubMed] [Google Scholar]
- Bircheneder, S. and Dresselhaus, T. (2016) Why cellular communication during plant reproduction is particularly mediated by CRP signalling. J. Exp. Bot. 67, 4849–4861. [DOI] [PubMed] [Google Scholar]
- Boavida, L.C. , Vieira, A.M. , Becker, J.D. and Feijó, J.A. (2005) Gametophyte interaction and sexual reproduction: how plants make a zygote. Int. J. Dev. Biol. 49, 615–632. [DOI] [PubMed] [Google Scholar]
- Broothaerts, W. , Keulemans, J. and Nerum, I.V. (2004) Self‐fertile apple resulting from S‐RNase gene silencing. Plant Cell Rep. 22, 497–501. [DOI] [PubMed] [Google Scholar]
- Chen, J. , Wang, P. , Graaf, B.H.J.D. , Zhang, H. and Wu, J. (2018) Phosphatidic acid counteracts S‐RNase signaling in pollen by stabilizing the actin cytoskeleton. Plant Cell, 30, 1023–1039. tpc.00021.02018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl, J.L. and Jones, J.D. (2001) Plant pathogens and integrated defence responses to infection. Nature, 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Davis, B.D. , Dulbecco, R. , Eisen, H.N. , Ginsberg, H.S. and Wood, W.B. Jr . (1969) Microbiology and Immunology Principles of Microbiology and Immunology. BioScience 19, 82–82. [Google Scholar]
- De, F.P. , Dondini, L. and Sanzol, J. (2012) Molecular bases and evolutionary dynamics of self‐incompatibility in the Pyrinae (Rosaceae). J. Exp. Bot. 63, 4015–4032. [DOI] [PubMed] [Google Scholar]
- Devoto, A. and Turner, J.G. (2003) Regulation of Jasmonate‐mediated Plant Responses in Arabidopsis. Ann. Bot. 92, 329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di, S.A. , Del, D.S. , Verderio, E. , Hargreaves, A.J. , Scarpellini, A. , Cai, G. , Cresti, M. et al. (2010) An extracellular transglutaminase is required for apple pollen tube growth. Biochem. J. 429, 261–271. [DOI] [PubMed] [Google Scholar]
- Erb, M. , Meldau, S. and Howe, G.A. (2012) Role of phytohormones in insect‐specific plant reactions. Trends Plant Sci. 17, 250–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer, E.E. and Ryan, C.A. (1990) Interplant communication: airborne methyl jasmonate induces synthesis of proteinase inhibitors in plant leaves. Proc. Natl Acad. Sci. USA 87, 7713–7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa, P. and Browse, J. (2012) The Arabidopsis JAZ2 promoter contains a G‐Box and thymidine‐rich module that are necessary and sufficient for jasmonate‐dependent activation by MYC transcription factors and repression by JAZ proteins. Plant Cell Physiol. 53, 330–343. [DOI] [PubMed] [Google Scholar]
- Foote, H.C. , Ride, J.P. , Franklin‐Tong, V.E. , Walker, E.A. , Lawrence, M.J. and Franklin, F.C. (1994) Cloning and expression of a distinctive class of self‐incompatibility (S) gene from Papaver rhoeas L. Proc. Natl Acad. Sci. USA, 91, 2265–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin‐Tong, V.E. , (ed). (2008) Self‐incompatibility in papaver rhoeas: progress in understanding mechanisms involved in regulating self‐incompatibility in papaver. In Self‐Incompatibility in Flowering Plants: Evolution, Diversity, and Mechanisms, pp. 37‐258. Berlin, Heidelberg: Springer. [Google Scholar]
- Franklin‐Tong, V.E. , Holdaway‐Clarke, T.L. , Straatman, K.R. , Kunkel, J.G. and Hepler, P.K. (2002) Involvement of extracellular calcium influx in the self‐incompatibility response of Papaver rhoeas . Plant J. 29, 333–345. [DOI] [PubMed] [Google Scholar]
- Gangappa, S.N. and Chattopadhyay, S. (2013) MYC2 differentially regulates GATA‐box containing promoters during seedling development in Arabidopsis. Plant Signal. Behav. 8, e25679. 10.4161/psb.25679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geitmann, A. , Franklintong, V.E. and Emons, A.C. (2004) The self‐incompatibility response in Papaver rhoeas pollen causes early and striking alterations to organelles. Cell Death Differ. 11, 812–822. [DOI] [PubMed] [Google Scholar]
- Gray, J.E. , Mcclure, B.A. , Bönig, I. , Anderson, M.A. and Clarke, A.E. (1991) Action of the style product of the self‐incompatibility gene of nicotiana alata (S‐RNase) on in Vitro‐grown pollen tubes. Plant Cell, 3, 271–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, Y. , Dang, R. , Li, J. , Jiang, J. , Zhang, N. , Jia, M. , Wei, L. et al. (2015) Sucrose nonfermenting1‐related protein KINASE2.6, an ortholog of open Stomata1, is a negative regulator of strawberry fruit development and ripening. Plant Physiol. 167, 915–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiyama, T. (2010) Peptide signaling in pollen‐pistil interactions. Plant Cell Physiol. 51, 177–189. [DOI] [PubMed] [Google Scholar]
- Hua, Z.H. , Fields, A. and Kao, T. (2008) Biochemical models for S‐RNase‐based self‐incompatibility. Mol. Plant, 1, 575–585. [DOI] [PubMed] [Google Scholar]
- Ishiguro, S. , Kawaioda, A. , Ueda, J. , Nishida, I. and Okada, K. (2001) The defective in anther Dehiscence1 Gene encodes a novel Phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in arabidopsis. Plant Cell, 13, 2191–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.D. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Jordan, N.D. , Franklin, F.C. and Franklin‐Tong, V.E. (2000) Evidence for DNA fragmentation triggered in the self‐incompatibility response in pollen of Papaver rhoeas. Plant J. 23, 471–479. [DOI] [PubMed] [Google Scholar]
- Kachroo, A. and Nasrallah, J.B. (2002) Self‐incompatibility in the Brassicaceae: receptor‐ligand signaling and cell‐to‐cell communication. Plant Cell, 14(Suppl), S227–S238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao, T.H. and Tsukamoto, T. (2004) The molecular and genetic bases of S‐RNase‐based self‐incompatibility. Plant Cell, 16, S72–S83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan, K. and John, M. (2013) MYC2: the master in action. Mol. Plant 6, 686–703. [DOI] [PubMed] [Google Scholar]
- Kwak, Y.D. , Hendrix, B.J. and Sugaya, K. (2014) Secreted type of amyloid precursor protein induces glial differentiation by stimulating the BMP/Smad signaling pathway. Biochem. Biophys. Res. Comm. 447, 394–399. [DOI] [PubMed] [Google Scholar]
- Li, T. , Jiang, Z. , Zhang, L. , Tan, D. , Wei, Y. , Yuan, H. , Li, T. et al. (2016) Apple (Malus domestica) MdERF2 negatively affects ethylene biosynthesis during fruit ripening by suppressing MdACS1 transcription. Plant J. 88, 735–748. [DOI] [PubMed] [Google Scholar]
- Li, W. , Meng, D. , Gu, Z. , Yang, Q. , Yuan, H. , Li, Y. , Chen, Q. et al. (2018) Apple S‐RNase triggers inhibition of tRNA aminoacylation by interacting with a soluble inorganic pyrophosphatase in growing self'ollen tubes in vitro. New Phytol. 218, 579–593. [DOI] [PubMed] [Google Scholar]
- Loeza‐Ángeles, H. , Sagrero‐Cisneros, E. , Lara‐Zárate, L. , Villagómez‐Gómez, E. , López‐Meza, J.E. and Ochoa‐Zarzosa, A. (2008) Thionin Thi2.1 from Arabidopsis thaliana expressed in endothelial cells shows antibacterial, antifungal and cytotoxic activity. Biotech. Lett., 30, 1713. [DOI] [PubMed] [Google Scholar]
- Lush, W.M. and Clarke, A.E. (1997) Observations of pollen tube growth in Nicotiana alata and their implications for the mechanism of self‐incompatibility. Sex. Plant Reprod. 10, 27–35. [Google Scholar]
- Marshall, E. , Costa, L.M. and Gutierrezmarcos, J. (2011) Cysteine‐rich peptides (CRPs) mediate diverse aspects of cell‐cell communication in plant reproduction and development. J. Exp. Bot. 62, 1677–1686. [DOI] [PubMed] [Google Scholar]
- Matsuura, T. , Sakai, H. , Unno, M. , Ida, K. , Sato, M. , Sakiyama, F. and Norioka, S. (2001) Crystal structure at 1.5a resolution of pyrus pyrifolia pistil ribonuclease responsible for gametophytic self‐incompatibility. J. Biol. Chem. 48, 45261‐45269. [DOI] [PubMed] [Google Scholar]
- Mcclure, B. (2004) S‐RNase and SLF determine S‐haplotype‐specific pollen recognition and rejection. Plant Cell, 16, 2840–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcclure, B.A. and Franklin‐Tong, V. (2006) Gametophytic self‐incompatibility: understanding the cellular mechanisms involved in “self” pollen tube inhibition. Planta, 224, 233–245. [DOI] [PubMed] [Google Scholar]
- Melo, F.R. , Rigden, D.J. , Franco, O.L. , Mello, L.V. , Ary, M.B. , Mf, G.D.S. and Jr, B.C. (2002) Inhibition of trypsin by cowpea thionin: characterization, molecular modeling, and docking. Proteins‐Struct. Funct. Bioinformat. 48, 311–319. [DOI] [PubMed] [Google Scholar]
- Meng, D. , Gu, Z. , Li, W. , Wang, A. , Yuan, H. , Yang, Q. and Li, T. (2014) Apple MdABCF assists in the transportation of S‐RNase into pollen tubes. Plant J. for Cell Mol. Biol. 78, 990–1002. [DOI] [PubMed] [Google Scholar]
- Moutinho, A. , Camacho, L. , Haley, A. , Pais, M. , Trewavas, A. and Malho, R. (2001) Antisense perturbation of protein function in living pollen tubes. Sex. Plant Reprod. 14, 101‐104. [Google Scholar]
- MuradoğLu, F. , Yıldız, K. and Balta, F. (2010) Methyl jasmonate influences of pollen germination and pollen tube growth of apricot (Prunus armeniaca L.). Yüzüncü Yıl University J. Agric. Sci. 20, 183–188. [Google Scholar]
- Nakata, M. and Ohme‐Takagi, M. (2013) Two bHLH‐type transcription factors, JA‐ASSOCIATED MYC2‐LIKE2 and JAM3, are transcriptional repressors and affect male fertility. Plant Signal. Behav. 8, e26473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah, J.B. (2005) Recognition and rejection of self in plant self‐incompatibility: comparisons to animal histocompatibility. Trends Immunol. 26, 412–418. [DOI] [PubMed] [Google Scholar]
- Nettancourt, D.D. (2001) The Genetics of Self‐Incompatibility.
- Okuda, S. and Higashiyama, T. (2010) Pollen tube guidance by attractant molecules: LUREs. Cell Struct. Funct. 35, 45–52. [DOI] [PubMed] [Google Scholar]
- Okuda, S. , Tsutsui, H. , Shiina, K. , Sprunck, S. , Takeuchi, H. , Yui, R. , Kasahara, R.D. et al. (2009) Defensin‐like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature, 458, 357–361. [DOI] [PubMed] [Google Scholar]
- Pelegrini, P.B. and Franco, O.L. (2005) Plant gamma‐thionins: novel insights on the mechanism of action of a multi‐functional class of defense proteins. Int. J. Biochem. Cell Biol. 37, 2239–2253. [DOI] [PubMed] [Google Scholar]
- Penn, D.J. and Potts, W.K. (1999) The evolution of mating preferences and major histocompatibility complex genes. Am. Nat. 153, 145–164. [DOI] [PubMed] [Google Scholar]
- Qiao, H. , Wang, H. , Zhao, L. , Zhou, J. , Huang, J. , Zhang, Y. and Xue, Y. (2004) The F‐box protein AhSLF‐S2 physically interacts with S‐RNases that may be inhibited by the ubiquitin/26S proteasome pathway of protein degradation during compatible pollination in Antirrhinum. Plant Cell, 16, 582–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, L.J. , Li, L. , Lan, Z. and Dresselhaus, T. (2015) Peptide signalling during the pollen tube journey and double fertilization. J. Exp. Bot. 66, 5139–5150. [DOI] [PubMed] [Google Scholar]
- Rao, M.V. , Lee, H. , Creelman, R.A. , Mullet, J.E. and Davis, K.R. (2000) Jasmonic acid signaling modulates ozone‐induced hypersensitive cell death. Plant Cell, 12, 1633–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roalson, E.H. and Mccubbin, A.G. (2003) S‐RNases and sexual incompatibility: structure, functions, and evolutionary perspectives. Mol. Phylogenet. Evol. 29, 490–506. [DOI] [PubMed] [Google Scholar]
- Rudd, J.J. , Franklin, F. , Lord, J.M. and Franklintong, V.E. (1996) Increased Phosphorylation of a 26‐kD Pollen Protein Is Induced by the Self‐Incompatibility Response in Papaver rhoeas. Plant Cell, 8, 713–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanabria, N. , Goring, D. , Nürnberger, T. and Dubery, I. (2008) Self/nonself perception and recognition mechanisms in plants: a comparison of self‐incompatibility and innate immunity. New Phytol. 178, 503–514. [DOI] [PubMed] [Google Scholar]
- Sassa, H. , Hirano, H. and Ikehashi, H. (1992) Self‐Incompatibility‐Related RNases in Styles of Japanese Pear (Pyrus serotina Rehd.). Plant Cell Physiol. 33, 811–814. [Google Scholar]
- Sassa, H. , Mase, N. , Hirano, H. and Ikehashi, H. (1994) Identification of self‐incompatibility‐related glycoproteins in styles of apple (Malus×domestica). Tag.theoretical & Applied Genetics.theoretische Und Angewandte Genetik, 89, 201–205. [DOI] [PubMed] [Google Scholar]
- Satohiro, O. , Hiroki, T. , Keiko, S. , Stefanie, S. , Hidenori, T. , Ryoko, Y. , Kasahara, R.D. et al. (2009) Defensin‐like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature, 458, 357–361. [DOI] [PubMed] [Google Scholar]
- Sawamoto, K. , Okabe, M. , Tanimura, T. , Mikoshiba, K. , Nishida, Y. and Okano, H. (1996) The Drosophila secreted protein Argos regulates signal transduction in the Ras/MAPK pathway. Dev. Biol. 178, 13–22. [DOI] [PubMed] [Google Scholar]
- Schmittgen, T.D. and Livak, K.J. (2008) Analyzing real‐time PCR data by the comparative CT method. Nat. Protoc. 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Schopfer, C.R. , Nasrallah, M.E. and Nasrallah, J.B. (1999) The male determinant of self‐incompatibility in Brassica. Science 286, 1697–1700. [DOI] [PubMed] [Google Scholar]
- Schuman, M.C. and Baldwin, I.T. (2015) The layers of plant responses to insect herbivores. Annu. Rev. Entomol. 61, 373–394. [DOI] [PubMed] [Google Scholar]
- Shi, D. , Chao, T. , Wang, R. , Chao, G. , Xiao, W. , Shi, H. , Jin, J. et al. (2017) Transcriptome and phytohormone analysis reveals a comprehensive phytohormone and pathogen defence response in pear self‐/cross‐pollination. Plant Cell Rep. 36, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sijacic, P. , Wang, X. , Skirpan, A.L. , Wang, Y. , Dowd, P.E. , Mccubbin, A.G. , Huang, S. et al. (2004) Identification of the pollen determinant of S‐RNase‐mediated self‐incompatibility. Nature, 429, 302–305. [DOI] [PubMed] [Google Scholar]
- Song, X. , Wang, J. , Wu, F. , Li, X. , Teng, M. and Gong, W. (2005) cDNA cloning, functional expression and antifungal activities of a dimeric plant defensin SPE10 from Pachyrrhizus erosus seeds. Plant Mol. Biol. 57, 13–20. [DOI] [PubMed] [Google Scholar]
- Suzuki, G. (2009) Recent progress in plant reproduction research: the story of the male gametophyte through to successful fertilization. Plant Cell Physiol. 50, 1857–1864. [DOI] [PubMed] [Google Scholar]
- Suzuki, G. , Kai, N. , Hirose, T. , Fukui, K. , Nishio, T. , Takayama, S. , Isogai, A. et al. (1999) Genomic organization of the s locus: identification and characterization of genes in SLG/SRK region of S9 haplotype of Brassica campestris (syn. rapa). Genetics, 153, 391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama, S. and Isogai, A. (2005) Self‐incompatibility in plants. Annu. Rev. Plant Biol. 56, 467–489. [DOI] [PubMed] [Google Scholar]
- Takayama, S. , Shimosato, H. , Shiba, H. , Funato, M. , Che, F.S. , Watanabe, M. , Iwano, M. et al. (2001) Direct ligand‐receptor complex interaction controls Brassica self‐incompatibility. Nature, 413, 534–538. [DOI] [PubMed] [Google Scholar]
- Takeuchi, H. and Higashiyama, T. (2016) Tip‐localized receptors control pollen tube growth and LURE sensing in Arabidopsis. Nature, 531, 245–248. [DOI] [PubMed] [Google Scholar]
- Tang, W. , Ezcurra, I. , Muschietti, J. and Mccormick, S. (2002) A cysteine‐rich extracellular protein, LAT52, interacts with the extracellular domain of the pollen receptor kinase LePRK2. Plant Cell, 14, 2277–2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavormina, P. , De, C.B. , Nikonorova, N. , De, S.I. and Cammue, B.P. (2015) The plant peptidome: an expanding repertoire of structural features and biological functions. Plant Cell, 27, 2095–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, S.G. and Franklin‐Tong, V.E. (2004) Self‐incompatibility triggers programmed cell death in Papaver pollen. Nature, 429, 305–309. [DOI] [PubMed] [Google Scholar]
- Thomas, D. and Noni, F.T. (2013) Male‐female crosstalk during pollen germination, tube growth and guidance, and double fertilization. Mol. Plant 6, 1018–1036. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P. , Eggermont, K. , Penninckx, I.A. , Mauch‐Mani, B. , Vogelsang, R. , Cammue, B.P. and Broekaert, W.F. (1998) Separate jasmonate‐dependent and salicylate‐dependent defense‐response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl Acad. Sci. USA, 95, 15107–15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, B. , Wang, X. , Zhou, Y. , Zhang, X. , Huo, K. and Han, Z. (2009) C120RF39, a novel secreted protein with a typical amidation processing signal. Biosci. Rep. 30, 1–10. [DOI] [PubMed] [Google Scholar]
- Wang, C. and Zhang, S. (2011) A cascade signal pathway occurs in self‐incompatibility of Pyrus pyrifolia . Plant Signal. Behav. 6, 420–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W. , Vignani, R. , Scali, M. and Cresti, M. (2006) A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomic analysis. Electrophoresis, 27, 2782–2786. [DOI] [PubMed] [Google Scholar]
- Wang, C.L. , Xu, G.H. , Jiang, X.T. , Chen, G. , Wu, J. , Wu, H.Q. and Zhang, S.L. (2009) S‐RNase triggers mitochondrial alteration and DNA degradation in the incompatible pollen tube of Pyrus pyrifolia in vitro . Plant J. 57, 220–229. [DOI] [PubMed] [Google Scholar]
- Wang, C.L. , Wu, J. , Xu, G.H. , Gao, Y.B. , Chen, G. , Wu, J.Y. , Wu, H.Q. et al. (2010) S‐RNase disrupts tip‐localized reactive oxygen species and induces nuclear DNA degradation in incompatible pollen tubes of Pyrus pyrifolia . J. Cell Sci. 123, 4301–4309. [DOI] [PubMed] [Google Scholar]
- Wang, L. , Clarke, L.A. , Eason, R.J. , Parker, C.C. , Qi, B. , Scott, R.J. and Doughty, J. (2016) PCP‐B class pollen coat proteins are key regulators of the hydration checkpoint in Arabidopsis thaliana pollen–stigma interactions. New Phytol. 213, 764–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C. (2007) Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, Growth and Development. Ann. Bot. 100, 681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C. and Hause, B. (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111, 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack, C. , Forner, S. , Strnad, M. and Hause, B. (2013) Jasmonates in flower and seed development. Biochimie, 95, 79–85. [DOI] [PubMed] [Google Scholar]
- Wei, L. , Fan, J. , Li, J. , Song, Y. , Li, Q. , Zhang, Y.E. and Xue, Y. (2014) SCFSLF‐mediated cytosolic degradation of S‐RNase is required for cross‐pollen compatibility in S‐RNase‐based self‐incompatibility in Petunia hybrida . Front. Genet. 5, 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, M.J. , Vatovec, S. and Franklintong, V.E. (2010) The pollen S‐determinant in Papaver: comparisons with known plant receptors and protein ligand partners. J. Exp. Bot. 61, 2015–2025. [DOI] [PubMed] [Google Scholar]
- Wilkins, K.A. , Bancroft, J. , Bosch, M. , Ings, J. , Smirnoff, N. and Franklintong, V.E. (2011) ROS and NO mediate actin reorganization and programmed cell death in the Self‐Incompatibility response of Papaver. Plant Physiol. 156, 404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J. , Shang, Z. , Wu, J. , Jiang, X. , Moschou, P.N. , Sun, W. , Roubelakis‐Angelakis, K.A. et al. (2010) Spermidine oxidase‐derived H2O2 regulates pollen plasma membrane hyperpolarization‐activated Ca2+‐permeable channels and pollen tube growth. Plant J. 63, 1042–1053. [DOI] [PubMed] [Google Scholar]
- Xu, E.J. , Vaahtera, L. and Brosche, M. (2015) Roles of defense hormones in the regulation of ozone‐induced changes in gene expression and cell death. Mol. Plant 8, 1776–1794. [DOI] [PubMed] [Google Scholar]
- Yildiz, K. and Yilmaz, H. (2002) Effect of jasmonic acid, ACC and ethephon on pollen germination in strawberry. Plant Growth Regul. 38, 145–148. [Google Scholar]
- Yuan, Z. and Zhang, D. (2015) Roles of jasmonate signalling in plant inflorescence and flower development. Curr. Opin. Plant Biol. 27, 44–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Pollen tube growth in control pollen, or those treated with inactive S‐RNase, or different concentrations of S 1 +S 2 ‐RNase and S 3 +S 9 ‐RNase.
Figure S2 Sequence alignments of the deduced amino acid sequences of MdMYC2.
Figure S3 The expression analysis of MdMYC2.
Figure S4 Genome‐wide mapping of MdMYC2 performed by ChIP‐Seq.
Figure S5 The expression analysis of MdD1.
Figure S6 MdD1 relative expression in pollen tube.
Figure S7 MdD1 causes a decrease in S‐RNase activity in different S‐haplotype combinations.
Figure S8 MdD1 relative expression.
Figure S9 S‐RNase could not be degraded by MdD1.
Figure S10 Yeast two‐hybrid analysis of the physical interaction between the mature peptide of MdD1 and the mature peptides of S 1‐, S 3 ‐, and S 9 ‐RNase.
Figure S11 A pull‐down analysis of the interaction between MdD1 and S 1 ‐, S 3 ‐, S 9 ‐RNase.
Figure S12 Bimolecular fluorescence complementation (BiFC) assay showing the interactions between MdD1 and S 1 ‐, S 3 ‐, and S 9 ‐RNase.
Figure S13 Yeast two‐hybrid (Y2H) assay showing the interactions between MdD1 and different S 1 ‐, S 3 ‐, S 9 ‐RNase fragments.
Table S1 Sequences of proteins listed in Table S1.
Table S2 Primers and oligonucleotides used in this study.
Table S3 The candidate genes found by ChIP‐seq.


