Summary
Drought is an abiotic stress that affects plant growth, and lipids are the main economic factor in the agricultural production of oil crops. However, the molecular mechanisms of drought response function in lipid metabolism remain little known. In this study, overexpression (OE) of different copies of the drought response genes LEA3 and VOC enhanced both drought tolerance and oil content in Brassica napus and Arabidopsis. Meanwhile, seed size, membrane stability and seed weight were also improved in OE lines. In contrast, oil content and drought tolerance were decreased in the AtLEA3 mutant (atlea3) and AtVOC‐RNAi of Arabidopsis and in both BnLEA‐RNAi and BnVOC‐RNAi B. napus RNAi lines. Hybrids between two lines with increased or reduced expression (LEA3‐OE with VOC‐OE, atlea3 with AtVOC‐RNAi) showed corresponding stronger trends in drought tolerance and lipid metabolism. Comparative transcriptomic analysis revealed the mechanisms of drought response gene function in lipid accumulation and drought tolerance. Gene networks involved in fatty acid (FA) synthesis and FA degradation were up‐ and down‐regulated in OE lines, respectively. Key genes in the photosynthetic system and reactive oxygen species (ROS) metabolism were up‐regulated in OE lines and down‐regulated in atlea3 and AtVOC‐RNAi lines, including LACS9, LIPASE1, PSAN , LOX2 and SOD1. Further analysis of photosynthetic and ROS enzymatic activities confirmed that the drought response genes LEA3 and VOC altered lipid accumulation mainly via enhancing photosynthetic efficiency and reducing ROS. The present study provides a novel way to improve lipid accumulation in plants, especially in oil production crops.
Keywords: LEA3, VOC , Brassica napus, drought tolerance, lipid accumulation, photosynthetic efficiency, reactive oxygen species
Introduction
Drought is an important abiotic stress (Zhu, 2002) that can affect the morphological, physiological and biochemical characteristics of plants (Suzuki et al., 2014). With the loss of cellular homeostasis under drought, an accumulation of toxic metabolites injures plant cells (Miller et al., 2010; Xiong and Zhu, 2002). In addition, increased production of reactive oxygen species (ROS) could cause further damage to plant tissue (Miller et al., 2010).
Some progress has been made in understanding how plants respond to drought stress, and some drought‐responsive genes have been identified in plants (Zhang, 2015; Zhao et al., 2016). Significant drought stress can induce transcription of dehydration‐responsive element‐binding proteins (DREBs) in plants and then activate genes that are involved in detoxification, water and ion movement and chaperone functions, including LEA (Shinozaki and Yamaguchi‐Shinozaki, 2007; Wang et al., 2003; Xiong and Zhu, 2002; Yoshida et al., 2014). In this model, LEA genes are part of a category that could function in the protection of membranes and proteins. Previous studies have suggested that LEA‐type proteins could act as water‐binding molecules, playing important roles in macromolecule and membrane stabilization and in ion sequestration (Chakrabortee et al., 2012; Wang et al., 2003). Most LEA proteins are hydrophilic and are not thought to have a stable secondary structure (Bremer et al., 2017; Hincha and Thalhammer, 2012; Tunnacliffe et al., 2010). These structural characteristics are related to the prediction of their function in response to desiccation stress (Candat et al., 2014; Chakrabortee et al., 2007). VOC proteins are members of an enzyme superfamily with a common mechanistic attribute enabled by conserved active site residues. Glyoxalase I (GLYI) is a major member of the VOC family. It is a metalloenzyme that participates in the glyoxalase system, which has been reported to be a major pathway for the detoxification of methylglyoxal (MG) in living organisms. GLYI can use one molecule of glutathione (GSH) to convert MG to S‐D‐lactoylglutathione and functions in abiotic stress response (Mustafiz et al., 2014; Singla‐Pareek et al., 2003).
Lipids are essential for plants and humans; they provide not only the energy for metabolic processes but also a structural basis for cell membranes (Okazaki and Saito, 2014). In Arabidopsis thaliana and Brassica napus, oil is mainly stored in seeds as triacylglycerols (TAGs), together with starch and storage proteins (Graham, 2008). TAG biosynthesis is achieved through the coordinated action of multiple pathways in various subcellular compartments. The synthetic process could be briefly classified into different phases in plant cells (Chapman and Ohlrogge, 2012). Some genes in these TAG biosynthetic pathways were identified. Diacylglycerol acyltransferases (DGATs) are well known in TAG biosynthesis in plants. DGAT1 is required for TAG accumulation in Arabidopsis; disruption of the DGAT1 gene could result in a 20%–40% reduction in seed oil in Arabidopsis, while overexpression of DGAT1 in vegetative tissues or seeds could lead to TAG accumulation (Yen et al., 2008; Zhang et al., 2009).
In plants, lipids also participate in signalling pathways that respond to environmental cues (Okazaki and Saito, 2014). In recent years, it was revealed that some enzymes involved in lipid metabolism could play a part in abiotic stress tolerance. For example, freezing tolerance was decreased with disruption of AtDGAT1 (Arisz et al., 2018). A lipase, designated as HEAT INDUCIBLE LIPASE1 (HIL1), induces the catabolism of monogalactosyldiacylglycerol (MGDG) under heat stress in Arabidopsis leaves (Higashi et al., 2018). Whether abiotic stress response genes can also function in lipid metabolism remains unclear. In our previous study, it was revealed that the LEA and VOC proteins were highly expressed in B. napus plants that had high oil content (Gan et al., 2013). We also found that most of the BnLEA and BnVOC genes had higher expression levels under drought stress in B. napus with high oil content (Liang et al., 2016, 2017). These results suggested that drought‐stress‐responsive genes might have a relationship with lipid metabolism.
Brassica napus (AACC, 2n = 38) is an allopolyploid species with a triplicated genome structure and many duplicated genes (Chalhoub et al., 2014). Rapeseed is the third largest oilseed crop in the world, and many areas where it is planted are affected by drought. Thus, it is important to further study the exact roles of genes involved in oil accumulation and response to drought stress conditions. In the present study, oil content and drought resistance were analysed in LEA3 and VOC transgenic and mutant plants in Arabidopsis and rapeseed. RNA‐seq analysis revealed that FA synthesis was up‐regulated and FA degradation was down‐regulated in OE lines. In addition, in the photosynthetic system, chlorophyll synthesis was up‐regulated in OE lines and down‐regulated in atlea3 and AtVOC‐RNAi. Hybrids between either OE lines or RNAi lines had corresponding effects on drought tolerance and oil content in B. napus. Finally, we confirmed that LEA3 and VOC play an important role in lipid accumulation, mainly by enhancing photosynthetic efficiency and reducing ROS. The present research reveals that stress response genes could not only function in abiotic stress but also enhance oil accumulation in oil crops.
Results
LEA3 and VOC Arabidopsis transgenic lines could enhance drought tolerance and lipid accumulation under drought treatment
To investigate the role of LEA3s and VOCs in oil metabolism, different copies of BnLEA3 and BnVOC and the AtLEA3 and AtVOC genes were cloned into plant expression vectors, and two kinds of vectors were constructed. One kind of vector contained the 35S CaMV promoter, and the other contained the glycinin promoter. These two vector types could be used to confer overexpression in whole plants and in only the seeds of transgenic lines, respectively (Figures S1, S2). These twenty OE vectors were all transformed into Arabidopsis. The red fluorescent protein (DsRed) gene was used to mark the transgenic seeds (Figure S1B).
As the LEA and VOC genes were both associated with drought stress, the drought tolerance of the LEA3‐OE and VOC‐OE transgenic lines was first validated under drought treatment (Figure S1C). The T‐DNA insertion SALK Arabidopsis mutant (atlea3) (Figures 1a, S2) of the AtLEA3 gene and the RNA interference line of AtVOC (AtVOC‐RNAi) (Figures 2a, S2) were used to further investigate the role of AtLEA3 and AtVOC in drought tolerance and lipid accumulation. After being confirmed as homozygous by PCR and RT‐PCR, the atlea3 materials were planted under normal conditions and subjected to drought treatment after flowering. The drought tolerances of the atlea3 and AtVOC‐RNAi plants were analysed. The atlea3 plants showed much weaker growth under drought treatment; they had lower relative water contents (RWCs) (Figure 1n), chlorophyll contents (Figure 1l), and leaf temperatures (Figure 1k) and higher anthocyanin contents (Figure 1o) and H2O2 accumulations (Figure 1c). The growth of the AtVOC‐RNAi plants was also affected by drought stress (Figure 2b–g). The AtVOC‐RNAi plants exhibited decreased drought tolerance with changes in the relevant physiological indexes compared with the wild‐type (WT) and empty vector controls (Figure 2k, l, n, o). A structural analysis of the seeds and leaves supported the conclusion of weaker drought tolerance (Figure 2e–g). The oil content of mature seeds in the mutant lines (24.5%) was 14.9% lower than that of WT (28.8%) under drought conditions (Figure 1i). Under normal conditions, the oil content of mature seeds in the mutant lines (27.5%) was 10.7% lower than that of WT (30.8%). C18:2 was also significantly affected by drought treatment in atlea3 (Figure 1j). However, the oil content of the AtVOC‐RNAi lines was only slightly decreased in normal conditions and reduced by 5.3%–13.9% in the drought treatment (Figure 2h); a decrease in C18:0 was also observed (Figure 2i). Meanwhile, the thousand seed weights (TSWs) and seed sizes of atlea3 and AtVOC‐RNAi were reduced by 10.6% (19.2 mg) and 8.7% (19.7 mg) compared with those of the control under drought conditions, respectively (Figures 1m, 2m). The expression of genes involved in lipid accumulation was down‐regulated in atlea3 and AtVOC‐RNAi (Figures 1h, 2j). These results suggest that knockout of AtLEA3 and knockdown of AtVOC could affect gene expression related to lipid accumulation.
Figure 1.

Drought tolerance and oil content of the atlea3 mutant and complementation of the full‐length AtLEA3 gene into atlea3. (a): Schematic diagram of the T‐DNA insertion mutant used in this study (atlea3). (b): Phenotype of the atlea3 mutant, HB (complementary vector transformed into atlea3) and WT under normal, drought and rewatering conditions. (c): DAB staining of WT (i), atlea3 (ii) and HB (iii). The number below indicates the quantitative data of H2O2 content, and unit is nmol/g FW. (d and e): Seed and leaf sections of WT, atlea3 and HB under drought stress. Bar = 500 μm. (f): SEM observation of leaves of WT, atlea3 and HB under drought conditions. Bar = 100 μm. (g): Images of mutant, HB and WT plants in a thermal imaging system under drought conditions. (h): Relative expression levels of genes in the TAG synthetic pathway in HB and atlea3 under drought conditions. LPAT: lysophosphatidic acid acyltransferase, LPP: lipid phosphate phosphatase, DGAT: diacylglycerol acyltransferase, PDAT: phospholipid:diacylglycerol acyl transferase, CCT: choline‐phosphate:CTP cytidylyltransferase, AAPT: aminoalcohol‐phosphotransferase. (i): Oil contents of the HB lines and mutants under drought conditions. (j): FA compositions of the HB lines and mutants under drought conditions. (k–o): Leaf temperatures, chlorophyll a + b contents, RWCs, TSWs and anthocyanin contents of the HB lines and mutants under drought conditions. All the results are represented as the mean ± standard deviation (STD; n = 3). Statistically significant differences were determined using a two‐tailed paired Student's t‐test compared with WT plants under similar conditions, and the results are indicated by **P < 0.01 and *P < 0.05.
Figure 2.

AtVOC‐RNAi lines have lower oil production and lower drought tolerance. (a): Schematic diagram of the RNA interference vector built in this study. (b): Phenotype of AtVOC‐RNAi and the empty vector and WT controls. (c): Thermal images of AtVOC‐RNAi and WT under drought conditions. (d): DAB staining of WT (i) and AtVOC‐RNAi (ii). Bar = 1 cm. The number below indicates the quantitative data of H2O2 content, and unit is nmol/g FW (e and f): Seed and leaf sections of WT and AtVOC‐RNAi under drought stress. Bar = 500 μm. (g): SEM observation of leaves of WT and AtVOC‐RNAi under drought conditions. Bar = 100 μm. (h): Oil content changes in the AtVOC‐RNAi lines. (i): FA composition of the AtVOC‐RNAi lines. (j): Relative expression levels of genes in the TAG synthetic pathway in AtVOC‐RNAi under drought stress. LPAT, lysophosphatidic acid acyltransferase; LPP, lipid phosphate phosphatase; DGAT, diacylglycerol acyltransferase; PDAT, phospholipid:diacylglycerol acyl transferase; CCT, choline‐phosphate; CTP, cytidylyltransferase; AAPT, aminoalcohol‐phosphotransferase. (k–o): Physiological indexes and TSWs of RNAi and WT under drought conditions. All the results are represented as the mean ± standard deviation (STD; n = 3). Statistically significant differences were determined using a two‐tailed paired Student's t‐test compared with WT plants under similar conditions, and the results are indicated by **P < 0.01 and *P < 0.05.
The full‐length AtLEA3 gene was cloned, and a complementation vector was constructed and then transformed into the lea3 mutant (Figure 1). Compared with that of the mutant, the oil contents of the homozygous transgenic materials (AtLEA‐HBs) were rescued, with increases of 11.1%–25.3% and 12.9%–19.3% under drought and normal growth conditions, respectively (Figures 1b, S3), while the C18:1 contents of seeds in the AtLEA‐HBs were lower than those of atlea3 (Figure 1j). In addition, the growth of the AtLEA3‐HBs under drought stress was recovered (Figure 1). These results suggested that loss or repression of these drought response genes could affect drought tolerance, lead to decreased oil content and reduce oil production.
Overall, these results indicated that overexpression of LEA3 and VOC could enhance plant drought tolerance and maintain normal oil accumulation under drought conditions.
Overexpression of the BnLEA3, BnVOC, AtLEA3 and AtVOC genes could increase oil content and seed size in transgenic Arabidopsis under normal conditions
The oil content of transgenic lines in generation T3 was analysed, and the average oil contents of seeds in the BnLEA3.1‐Gly‐OE, BnLEA3.2‐Gly‐OE, BnLEA3.3‐Gly‐OE, BnLEA3.4‐Gly‐OE and AtLEA‐Gly‐OE transgenic lines were 34.2%–36.1%, 9.6%–15.7% higher than those of the control (CK, oil content: 31.2%; Figure 3). The maximum oil content in some transgenic lines was increased by 25% (Table S1). The seed oil contents of BnLEA3‐35S‐OE and AtLEA3‐35S‐OE were similar to those of LEA3‐Gly‐OE (Figure 3d), and the oil contents of BnLEA3.2‐35S‐OE and BnLEA3.3‐35S‐OE were as high as 34%–36.4%, an average increase of 18.3% (Table S1). The average oil contents of transgenic seeds from the BnVOC1‐Gly‐OE, BnVOC2‐Gly‐OE, BnVOC3‐Gly‐OE, BnVOC4‐Gly‐OE and AtVOC‐Gly‐OE lines increased from 10.8% to 23.9% (Figure 3d). The BnVOC‐35S‐OE and AtVOC‐35S‐OE lines also showed increased oil contents from 14% to 22.6% (Figure 3d). These results suggested that the LEA3 and VOC genes take part in oil accumulation and help to increase seed oil content.
Figure 3.
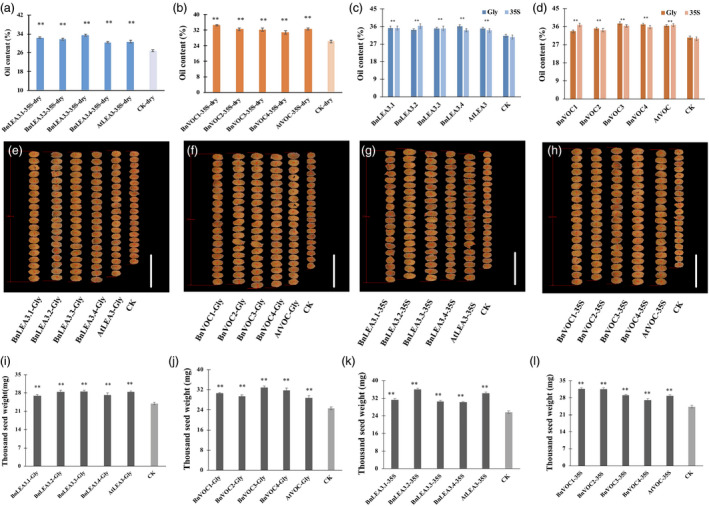
Overexpression of the BnLEA3, BnVOC , AtLEA3 and AtVOC genes resulted in higher oil content, seed size and seed weight. (a and b): Oil contents of transgenic Arabidopsis seeds under drought conditions. (c and d): Oil contents of transgenic Arabidopsis seeds under normal conditions. (e–h): Seed sizes of LEA‐ and VOC‐OE lines (Gly and 35S) and CK. Bar = 1 mm. (i–l): TSWs of LEA‐ and VOC‐OE lines (Gly and 35S) and CK. The data represent the mean and standard deviation (STD) of three biological replicates (n = 3). Statistically significant differences were determined using a two‐tailed paired Student's t‐test compared with WT plants under similar conditions, and the results are indicated by **P < 0.01.
The sizes of the transgenic OE seeds were also measured. The seeds in the transgenic lines were larger than those of CK without morphological malformation (Figure 3e–h). Further analysis revealed that the TSW of LEA‐OE seeds was inconsistently increased by 4–9 mg. The LEA‐Gly‐OE lines showed a greater improvement in TSW than the LEA‐35S‐OE lines (Figure 3i, k). Compared with CK, the average TSWs of the BnLEA3‐Gly‐OE and AtLEA3‐Gly‐OE lines increased by 11.1%–19.1%, reaching 27–28.6 mg (Figure 3i). The TSWs of the BnVOC‐Gly‐OE lines ranged from 30.6 mg to 32.95 mg, with an average of 32 mg, which was higher than that of the BnVOC‐35S‐OE lines (TSWs: 27–31 mg; Figure 3j, l). These results indicated that overexpression of the LEA3 and VOC genes could also improve seed size.
Overexpression and RNA interference of the BnLEA3 and BnVOC genes affect oil content, seed size and drought tolerance in B. napus
To investigate the performance of the BnLEA3 and BnVOC genes in B. napus, overexpression vectors containing four BnLEA3s (BnLEA‐OE) and four BnVOCs (BnVOC‐OE) and the corresponding RNA interference (BnLEA‐RNAi and BnVOC‐RNAi) vectors were constructed (Figures 4a, 2a) and then introduced into B. napus. The transgenic plants were confirmed by qPCR (Figure S4), Southern blotting and Western blotting (Figure 4b and c). The relative expression levels of the LEA and VOC genes in the OE and RNAi lines were 8‐ to 21‐fold higher and lower, respectively (Figure S4).
Figure 4.

Transgenic B. napus showed improved drought tolerance. (a): Schematic diagram of the OE vector transformed into B. napus in this study. (b): Southern blotting was used to confirm the stable integration of the transgene in B. napus. (c): Western blotting was performed to check the expression of protein in WT (CK) and transgenic plants. (d): Phenotypes of transgenic plants and WT plants in the drought treatment. Bar = 5 cm. (e): Thermal images of transgenic and WT B. napus under drought conditions. (f): DAB staining of transgenic and WT B. napus under drought conditions. The number below indicates the quantitative data of H2O2 content, and unit is nmol/g FW. All the results are represented as the mean ± standard deviation (STD; n = 3).
Under drought stress, the BnLEA‐OE and BnVOC‐OE lines showed higher drought tolerance than the WT, BnLEA‐RNAi and BnVOC‐RNAi lines (Figure 4d). In addition, the RWCs and leaf temperatures greatly varied between the OE and RNAi lines and WT. The RWCs of BnLEA‐OE, BnVOC‐OE, BnLEA‐RNAi and BnVOC‐RNAi were 75.28%, 68.27%, 40.28% and 44.14%, respectively. The leaf temperatures of BnLEA‐OE and BnVOC‐OE were 20.5 and 19.9 °C, which were higher than those of WT (17.9 °C), BnLEA‐RNAi (16.8 °C) and BnVOC‐RNAi (16.1 °C) (Figure 4e). Further analysis showed that H2O2 accumulated more in the RNAi lines (Figure 5f). These results suggested that the BnLEA and BnVOC genes could also increase drought tolerance in B. napus.
Figure 5.

Transgenic B. napus showed changes in oil content and seed size. (a and b): Oil contents of transgenic B. napus lines. (c): TEM observation of transgenic and WT seeds. Bar = 10 μm. (d): Seed sizes of transgenic plants, hybrids (H‐35S: OE hybrids; H‐RNAi: RNAi hybrids) and WT. (e): Average oil contents of transgenic and hybrid plants under drought conditions. (f): TSWs of transgenic and hybrid plants. All the results are represented as the mean ± standard deviation (STD; n = 3). Statistically significant differences were determined using a two‐tailed paired Student's t‐test compared with WT plants under similar conditions, and the results are indicated by **P < 0.01.
Under normal condition, the oil content of WT was 37.6%, while the oil contents of the BnVOC‐OE and BnLEA‐OE lines were 43%–49.8% and 42%–48%, increases of 14.3%–32.9% and 11.7%–27.6%, respectively (Figure 5a and b). In contrast, the BnLEA‐RNAi and BnVOC‐RNAi lines had an average of 13.2%–15.9% decrease in oil content. In drought conditions, the oil contents of all lines were decreased, but those of the OE lines (above 30%) were higher than those of WT (27%). The oil content of BnLEA‐RNAi and BnVOC‐RNAi decreased more obviously, with an oil content of only 22%–23% (Figures 5e, S5). The changes in FA composition were mainly an increase in C18:1 and a decrease in C18:2 (Figure S6).
Further observation showed that the BnVOC‐OE and BnLEA‐OE lines had relatively large seed sizes with higher average TSWs (6.74 and 6.31 g, respectively) than WT. The seeds of the RNAi lines were much smaller than those of WT, with reduced average TSWs (only 1.9 and 1.78 g, respectively; Figure 5c and d). TEM observation showed that the seeds of both OE lines contained more oil bodies and less protein. In addition, the seeds of the RNAi lines showed fewer oil bodies, which were loosely arranged in the cells (Figure 5c). These results indicated that BnLEAs and BnVOCs play an important role in lipid accumulation and promote oil production.
Transcriptome analyses of BnLEA‐OE, AtLEA‐OE and atlea3 seeds reveal distinct patterns of gene activity and regulatory networks affecting oil metabolism
RNA‐seq analysis was performed to explore the regulatory mechanisms active in seeds under drought stress. To investigate the function of the LEA and VOC genes in drought response and drought tolerance, two types of drought treatment were performed: short‐term drought treatment to observe the response when plants initially encountered drought and long‐term drought treatment to observe tolerance when plants were under prolonged drought stress (Table 1). In total, 8 groups of lines were subjected to transcriptome analysis, including BnLEASL, AtLEASL, mLEASL, BnLEASS, AtLEASS and mLEASS. WTSL and WTSS were used as controls (the first letter ‘S’ indicates a seed sample, and for the second letter, ‘L’ and ‘S’ represent the long‐term and short‐term drought treatments, respectively; the initial ‘m’ indicates mutant). The gene sets that were activated in these genotypes in response to drought were identified (Figure 6b). In the short‐term drought treatment, compared with WTSS, 1676, 2707 and 695 differentially expressed genes (DEGs) were detected in AtLEASS, BnLEASS and mLEASS (Figure 6c and d). Further analysis revealed that 163 DEGs were the same in all these lines. In the long‐term drought treatment, only 80, 638 and 7 DEGs were found in AtLEASL, BnLEASL and mLEASL, respectively. These results suggested that there were large, coordinated shifts in gene expression at the early stage of drought stress in the OE lines and mutants.
Table 1.
RNA‐seq analysis codes for the drought treatments combining the genotype and sample tissues
| Code | Genotype | Drought treatment | Sample tissue |
|---|---|---|---|
| AtLEALL | Over expression of AtLEA | Long‐term | Leaves |
| BnLEALL | Over expression of BnLEA | Long‐term | Leaves |
| AtVOCLL | Over expression of AtVOC | Long‐term | Leaves |
| BnVOCLL | Over expression of BnVOC | Long‐term | Leaves |
| mLEALL | Mutant of AtLEA | Long‐term | Leaves |
| AtVOCRNAiLL | RNAi of AtVOC | Long‐term | Leaves |
| WTLL | Wild type | Long‐term | Leaves |
| AtLEALS | Over expression of AtLEA | Short‐term | Leaves |
| BnLEALS | Over expression of BnLEA | Short‐term | Leaves |
| AtVOCLS | Over expression of AtVOC | Short‐term | Leaves |
| BnVOCLS | Over expression of BnVOC | Short‐term | Leaves |
| mLEALS | Mutant of AtLEA | Short‐term | leaves |
| AtVOCRNAiLS | RNAi of AtVOC | Short‐term | Leaves |
| WTLS | Wild type | Short‐term | Leaves |
| AtLEASL | Over expression of AtLEA | Long‐term | Silique |
| BnLEASL | Over expression of BnLEA | Long‐term | Silique |
| AtVOCSL | Over expression of AtVOC | Long‐term | Silique |
| BnVOCSL | Over expression of BnVOC | Long‐term | Silique |
| mLEASL | Mutant of AtLEA | Long‐term | Silique |
| AtVOCRNAiSL | RNAi of AtVOC | Long‐term | Silique |
| WTSL | Wild type | Long‐term | Silique |
| AtLEASS | Over expression of AtLEA | Short‐term | Silique |
| BnLEASS | Over expression of BnLEA | Short‐term | Silique |
| AtVOCSS | Over expression of AtVOC | Short‐term | Silique |
| BnVOCSS | Over expression of BnVOC | Short‐term | Silique |
| mLEASS | Mutant of AtLEA | Short‐term | Silique |
| AtVOCRNAiSS | RNAi of AtVOC | Short‐term | Silique |
| WTSS | Wild type | Short‐term | Silique |
Figure 6.

RNA‐seq analysis of BnLEA‐OE, AtLEA‐OE, atlea3 and WT under short‐ and long‐term drought stress. (a and b): Cluster heatmaps of DEGs in the leaves (a) and seeds (b) of plants differentially expressing LEA under drought conditions. (c–f): Venn diagrams showing the numbers of DEGs in different genotypes of Arabidopsis (P ≦ 0.05, false discovery rate < 0.05). (g–j): Significantly enriched GO terms (coloured in red and yellow) of the DEGs in the seeds of differential LEA expression lines in the long‐term (g) and short‐term (i) drought treatments and the corresponding leaves in the long‐term (j) and short‐term (h) drought treatments. (k and l): Major enriched KEGG pathways and the expression of the related genes in the leaves (k) and seeds (l) of plants differentially expressing LEA.
To examine the biological processes encoded within differentially expressed gene sets, the Gene Ontology (GO) enrichment function of SeqEnrich was used (Figure 6g, i). DNA replication and cell proliferation were significantly up‐regulated in both OE lines under short‐term drought; many genes showed increased expression, and cell growth processes were the most strongly up‐regulated. In atlea3, genes related to response to oxygen‐containing compound and abiotic stimuli were up‐regulated in the short‐term drought treatment (Figure 6g, i). These results suggested that other abiotic stress response pathways were still able to respond to the threat of drought without the AtLEA3 gene. In the long‐term drought treatment, genes related to extracellular region, regulation of endosperm development and lipid transport were mainly up‐regulated in the OE lines, and response to oxygen‐containing compound and stimulus genes were down‐regulated in the mutant lines. These results suggest that, because overexpression of LEA3 genes can increase the expression of genes involved in lipid‐related processes, a loss of LEA3 might lead to desiccation tolerance in long‐term drought. To further identify potential regulatory networks, Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was performed (Figure 6l). In the LEA‐OE lines, FA biosynthesis was enriched and FA degradation was depleted. This result indicated that LEA overexpression could contribute to oil accumulation.
Transcriptome changes in LEA‐OE and atlea3 identified that photosynthetic carbon fixation confers drought tolerance in leaves
To study the gene expression changes of leaves under drought conditions, the leaves of BnLEA‐OE, AtLEA‐OE, atlea3 and WT under drought treatment were subjected to RNA‐seq analysis (BnLEALL, AtLEALL mLEALL, WTLL, BnLEALS, AtLEALS mLEALS, WTLS: the first letter ‘L’ indicates a leaf sample, and for the second letter, ‘L’ and ‘S’ indicate the long‐term and short‐term drought treatments, respectively; Figure 6a). Compared to WT, 98 DEGs were commonly found in BnLEALL, AtLEALL and mLEALL (Figure 6f), but only 6 DEGs were common among these three genotypes in the short‐term drought treatment (AUXIN ASSOCIATED GENE 2, leucine‐rich repeat family protein, transmembrane amino acid transporter family protein, transposable_element_gene, AT5G38005 and RIBOSOMAL RNA26S; Figure 6e). These results demonstrated that more genes were differentially expressed in leaves in the long‐term drought treatment than in the short‐term drought treatment. The GO analysis showed that genes associated with response to oxygen‐containing compound, stress and stimulus were affected in OE lines in the early stage. Chloroplast part and plastid part were enriched in OE lines in the long‐term drought treatment. However, cutin biosynthetic process and structural constituent of the cell wall were reduced in the mutant in the short‐term drought treatment. Response to oxygen‐containing compound was significantly enriched in the mutant in the long‐term drought treatment (Figure 6h, j). These results indicated that the LEA‐OE lines could quickly enrich biological processes involved in stress response in the early stages of drought and keep chloroplasts and other parts of the cell stable under long‐term drought. In atlea3, this genetic response to stress was delayed, and the plants lacked the ability to stabilize their organelles. KEGG analyses showed that carbon fixation in photosynthetic organisms and chlorophyll metabolism were up‐regulated in the OE lines (Figure 6k), which indicated that LEA could confer greater photosynthetic capacity under drought stress.
Transcriptome analyses of BnVOC‐OE, AtVOC‐OE and AtVOC‐RNAi lines reveal VOC function in FA degradation in seeds
To identify the potential mechanism of VOC action in drought treatment, RNA‐seq of seeds in the OE and RNAi transgenic lines was conducted in two drought treatments (BnVOCSL, AtVOCSL, AtVOCRNAiSL, BnVOCSS, AtVOCSS, AtVOCRNAiSS: the first letter ‘S’ indicates a seed sample; for the second letter, ‘L’ and ‘S’ represent the long‐term and short‐term drought treatments, respectively; Figure 7b). In the short‐term drought treatment, 41 and 786 common DEGs in these three genotypes in the early stage and late stage were identified compared with WT, respectively (Figure 7c and d). These results suggest that VOC genes tend to affect late seed development under drought conditions. The GO analysis of BnVOCSS showed enrichment in response to stress and oxygen‐containing compound. In AtVOCSS, auxin metabolic process was enriched. In AtVOCRNAiSS, DNA replication was significantly enriched. In the long‐term drought treatment, genes involved in seed maturation, response to stimulus, lipid localization, seed oil body biogenesis and lipid storage were active in the BnVOC‐OE and AtVOC‐OE lines. However, response to abiotic stimulus, oxygen‐containing compound and water were down‐regulated in AtVOCRNAiSL (Figure 7g–h).
Figure 7.

Transcriptome analysis of plants differentially expressing VOC and WT plants under short‐ and long‐term drought stress. (a and b): Cluster heatmaps of DEGs in the leaves (a) and seeds (b) of plants differentially expressing VOC under drought conditions. (c–f): Venn diagrams showing the numbers of DEGs in different genotypes of Arabidopsis (P ≦ 0.05, false discovery rate < 0.05). (g–j): Significantly enriched GO terms (coloured in red and yellow) of the DEGs in the seeds of differential VOC expression lines in the long‐term (g) and short‐term (h) drought treatments and the corresponding leaves in the long‐term (j) and short‐term (i) drought treatments. (k and l): Major enriched KEGG pathways and the expression of the related genes in the seeds (k) and leaves (l) of plants differentially expressing VOC.
Kyoto Encyclopedia of Genes and Genomes analysis revealed that the VOC genes take an active part in FA degradation and FA elongation and that many genes involved in FA degradation were down‐regulated in the VOC‐OE lines. The expression levels of lipid metabolism genes in the AtVOC‐RNAi lines showed opposite trends to those in the AtVOC‐OE lines. These results demonstrated that VOC could function in lipid metabolism by reducing FA degradation (Figure 7k).
Glyoxylate metabolism is significantly affected in the leaves of VOC transgenic plants
To further investigate the role of the VOC gene in response to drought, RNA‐seq analysis was performed with the leaves of VOC transgenic plants in short‐ and long‐term drought treatment (BnVOCLL, AtVOCLL, AtVOCRNAiLL, BnVOCLS, AtVOCLS, AtVOCRNAiLS: the first letter ‘L’ after the gene/construct name indicates a leaf sample, and for the second letter, ‘L’ and ‘S’ indicate the long‐term and short‐term drought treatments, respectively; Figure 7a). Compared with WTLL, 44 common DEGs were found in BnVOCLL, AtVOCLL and AtVOCRNAiLL, but only 2 common DEGs were found in BnVOCLS, AtVOCLS and AtVOCRNAiLS (AT1G15380: GLYI4; AT3G59930: a defensin‐like (DEFL) family protein; Figure 7e and f). These results indicated that genes affected by VOC were mainly expressed in the long‐term drought treatment. The GO categories enriched in BnVOCLS and AtVOCLS were mainly ribonucleotide biosynthetic process and response to ROS. In AtVOCRNAiLS, response to hormone was enriched. In the long‐term drought treatment, the BnVOCLL lines showed enrichment of response to oxygen‐containing compound and chloroplast part. The GO categories enriched in AtVOCLL and AtVOCRNAiLL were mainly photosynthesis and light reaction (Figure 7i and j). These results suggested that BnVOC and AtVOC differed under both short‐ and long‐term drought conditions but consistently affected light reactions and hormones. KEGG pathway analysis showed that glyoxylate and dicarboxylate metabolism and plant hormone signal transduction were enriched in OE transgenic leaves (Figure 7l). The BnVOC‐OE and AtVOC‐OE lines contained more up‐regulated genes in the glyoxylate pathway than the AtVOC‐RNAi lines. These results indicated that glyoxylate metabolism and oxygen‐related metabolic process are affected by VOC genes.
Hybrids of LEA and VOC transgenic lines had altered oil content, seed size and drought tolerance in B. napus and Arabidopsis
As mentioned above, both LEA and VOC could improve oil content and stress tolerance under drought conditions, but these genes had different regulatory mechanisms. To further investigate whether they could work together to strengthen drought tolerance or oil production under drought conditions, we hybridized BnLEA‐OE with BnVOC‐OE (H‐35S) and BnLEA‐RNAi with BnVOC‐RNAi (H‐RNAi), and also cross BnLEA‐OE with BnVOC‐RNAi in B. napus. We also crossed the transgenic Arabidopsis BnLEA‐35S line with BnVOC‐35S (H‐Bn‐OE), AtLEA‐35S with AtVOC‐35S (H‐At‐OE) and atlea3 with AtVOC‐RNAi (H‐mi) in Arabidopsis.
In B. napus, the hybrids of OE and RNAi showed nearly 10% changes in oil content (highest in H‐35S: 51.15%; lowest in H‐RNAi: 27.23%) under normal conditions (Figure 5). After drought treatment, the growth of the hybrids was affected, and H‐35S showed better drought tolerance than the H‐RNAi lines (Figure 8a, d, e). Under drought conditions, the oil content of H‐RNAi was only 18.26%; in contrast, H‐35S had an oil content of 44.07%. The TSW of H‐35S could be improved to 10.21 g. The average TSW of the H‐RNAi lines was greatly diminished to only 1.16 g under drought conditions (Figure 5f). Scanning electron microscope (SEM) observation found that the oil bodies of the hybrids showed corresponding variations: the number of oil bodies in H‐35S was increased, and the oil bodies were closely arranged. In contrast, the protein bodies were fewer than in WT. In contrast, the seeds of the H‐RNAi lines contained fewer oil bodies and more protein bodies than those of WT (Figure 8g).
Figure 8.

Hybrids of LEA and VOC in B. napus and Arabidopsis. (a): Performance of Arabidopsis hybrids in the drought treatment. (b): Thermal images of Arabidopsis hybrids in the drought treatment. (c and d): DAB staining of Arabidopsis (c) and B. napus (d) hybrids in the drought treatment. The number below indicates the quantitative data of H2O2 content, and unit is nmol/g FW. (E): Performance of B. napus hybrids in the drought treatment. Bar = 5 cm. (f): Thermal images of B. napus hybrids in the drought treatment. (g): SEM observations of B. napus hybrids in the drought treatment. Bar = 5 μm. (h): Seed sizes of Arabidopsis hybrids. (i): Oil contents of Arabidopsis hybrids. (j): TSWs of Arabidopsis hybrids. (k and l): Relative expression levels of key genes in transgenic and hybrid B. napus under normal conditions (k) and drought conditions (l). All the results are represented as the mean ± standard deviation (STD; n = 3). Statistically significant differences were determined using a two‐tailed paired Student's t‐test compared with WT plants under similar conditions, and the results are indicated by **P < 0.01.
In Arabidopsis, the H‐Bn‐OE lines had a higher oil content than H‐At‐OE and H‐mi under both normal and drought conditions. The oil content of H‐At‐OE was 31.8%, while the oil content of H‐mi was only 21.9% under drought conditions (Figure 8i). The FA composition was also altered under drought conditions (Figure S7). The TSW of H‐Bn‐OE was increased by 28.8%, and the H‐mi was decreased by 27.52% under drought conditions (Figure 8j). Further analysis showed that the drought tolerance of H‐Bn‐OE and H‐At‐OE was much better than that of H‐mi (Figure 8a–c). The seeds of the OE hybrids and RNAi/mutant hybrids had larger and smaller seed sizes, respectively, in both B. napus and Arabidopsis than those of CK (Figure 8h). The expression of key genes in the regulatory pathways of FA metabolism, oxidation and photosynthesis in the hybrids and transgenic lines showed that the hybrids had higher expression levels of positive‐effect genes and lower expression levels of negative‐effect genes in drought response and lipid accumulation (Figure 8k and l). The hybrid combination of LEA‐OE and VOC‐RNAi shows the similar level with WT. These results demonstrated that LEA and VOC have different mechanisms in response to drought, and their effects could be reinforced or weakened in hybrids.
Transgenic and hybrid OE and RNAi B. napus lines maintain altered photosynthetic machinery under drought stress
The transcriptome results showed that the LEA and VOC genes could affect photosynthesis and FA metabolism in the transgenic lines and hybrids of LEA‐OE and VOC‐OE B. napus lines. In addition, the BnLEA‐OE and BnVOC‐OE lines and the OE hybrids had better drought tolerance and higher oil contents than WT. Various determinants of photosynthesis were measured in the drought treatment (Figure 9). The BnLEA‐OE and BnVOC‐OE transgenic lines and the 35S hybrid lines all showed higher chlorophyll content, higher photosynthetic rate and higher RWC than WT under drought conditions. Other parameters in these transgenic and hybrid plants further supported their ability to maintain better photosynthetic capacity under drought stress. On the other hand, the BnLEA‐RNAi, BnVOC‐RNAi, and RNAi hybrids all had lower chlorophyll contents and photosynthetic rates. These results indicated that the LEA and VOC genes could help to improve photosynthetic efficiency under drought conditions.
Figure 9.

Photosynthetic efficiency of transgenic B. napus under drought stress. (a–f): Comparison of various photosynthetic and physiological parameters of WT and transgenic oilseed rape under drought conditions. (a) Transpiration rate, (b) stomatal conductance, (c) chlorophyll content, (d) Fv/Fm, (e) photosynthetic rate and (f) RWC were measured. All the results are represented as the mean ± standard deviation (STD; n = 3). Statistically significant differences were determined using a two‐tailed paired Student's t‐test compared with WT plants under similar conditions, and the results are indicated by **P < 0.01 and *P < 0.05.
Combining the regulatory pathway information gathered from the many transcriptome results and experiments shown above, we concluded that the LEA genes could enhance oil accumulation mainly via improved photosynthetic efficiency. As shown in Figure 11, we can deduce that the LEA‐OE lines contained more chlorophyll and had a higher photosynthetic rate, ensuring sucrose synthesis under drought stress, which provided energy and an upstream substrate for FA metabolism. Second, LEA affected many relative proteins during FA metabolism in the plastid and endoplasmic reticulum, such as LACS9, acyl carrier proteins to enhance FA biosynthesis, and LOX and LIPASE to decrease lipid oxidation and degradation. Third, the LEA‐OE lines had altered levels of heavy metal‐associated proteins, SnRK and plant lipid transfer proteins (LTP), which could increase plant cell integrity by reducing metal toxicity, reducing stomatal aperture and enhancing membrane stability.
Reduced ROS level, oxidation system activity and enhanced MG tolerance conferred drought tolerance on BnVOC transgenic and hybrid B. napus
The identification of enriched redox‐related GO terms and DEGs prompted us to study additional differences in the redox system between the VOC lines and WT under drought stress. VOC genes play an important role in MG detoxification reactions, and MG tolerance was confirmed by experiments. The results of prokaryotic expression showed that VOC proteins could confer MG tolerance in E. coli (Figure S8). Under drought stress, the contents of H2O2 in the B. napus BnVOC‐OE and BnVOC‐RNAi lines were reduced and increased, respectively, compared with those of WT. MG, a cytotoxic by‐product of the glycolytic pathway, also had a lower content in the BnVOC‐OE lines.
In the glyoxalase system, GLY I and glyoxalase II (GLY II) catalyse the MG detoxification reactions. The main subfamily of VOCs is GLY I. It was found that the activity of GLY I significantly increased, and the activity of GLY II also improved in the BnVOC‐OE lines (Figure 10). In addition, the BnVOC‐OE lines had a lower content of malondialdehyde (MDA, a product of lipid peroxidation). GSH and oxidized glutathione (GSSG) contents were also changed under drought conditions. Furthermore, lipoxygenase (LOX) and superoxide dismutase (SOD) activities were increased in the BnVOC‐RNAi lines and decreased in the BnVOC‐OE lines. Catalase (CAT) activity was inversely correlated with those of SOD and LOX. The results for the H‐35S and H‐RNAi hybrids were consistent with those for the OE and RNAi lines (Figure 11). Based on the results above and the transcriptome results, the molecular pathway of VOC in drought conditions was deduced (Figure 11). First, VOC proteins could definitely reduce MG toxicity through glyoxylate metabolism with GSH. VOC proteins also contributed to lower levels of ROS synthesis (H2O2). Finally, lipid peroxidation and membrane damage were reduced.
Figure 10.

Activity of the oxidation system and glyoxalase system of BnVOC transgenic B. napus under drought stress. (a–k): GLY I activity (a), GLY II activity (b), MG content (c), H2O2 content (d), MDA content (e), GSH content (f), GSSG content (g), GSH/GSSG ratio (h), LOX activity (i), CAT activity (j) and SOD activity (k) in transgenic and hybrid B. napus under drought stress. All the results are represented as the mean ± standard deviation (STD; n = 3). Statistically significant differences were determined using a two‐tailed paired Student's t‐test compared with WT plants under similar conditions, and the results are indicated by **P < 0.01 and *P < 0.05.
Figure 11.

Model of LEA and VOC functional regulation in plants. (a): Schematic representation of the influence of LEA genes on FA metabolic and catabolic biosynthetic pathways under drought stress. Photosynthetic efficiency, membrane stability and lipid synthesis and transportation are improved under drought conditions, resulting in higher seed oil contents in B. napus and Arabidopsis. The plant hormone signalling pathways lead to stomatal closure, improved membrane stability and reduced metal ion toxicity, conferring drought tolerance in OE leaves. HMA, heavy metal‐associated domain protein; G6P, glucose‐6‐phosphate; ACBP, Acyl‐CoA binding proteins; ER, endoplasmic reticulum; ACP, acyl carrier protein; LACS9, long‐chain acyl‐CoA synthetase 9; HSP, heat shock factor protein. (b): The effect of VOC genes on the ROS system reduces H2O2 and MG contents, which improves oil content and drought tolerance by reducing lipid peroxidation and membrane damage. ABF, ABRE binding factors; ABA, abscisic acid; PP2C, protein phosphatase 2C; SNRK2, SNF‐related kinase 2; CRK, cysteine‐rich receptor‐like kinase; hv, irradiation with light; PS I, photosystem I; GPOX, glutathione peroxidase; GR, glutathione reductase.
Discussion
Drought tolerance was the first obvious change in the LEA and VOC differential expression lines. Stomata regulate the flow of gases and transpiration by limiting water loss in different environments (Hetherington and Woodward, 2003). In this study, the stomata in the LEA and VOC‐OE lines tended to be closed under drought conditions, as observed in other plants (He et al., 2018; Prasch et al., 2015; Ullah et al., 2017). To study the lipid changes associated with drought stress, the oil content of transgenic plants under drought conditions was also analysed. The oil content of WT was only 25.3%–26.6% (Figure 3a and b), a reduction of 14.1%–18.3% compared with the control, under drought stress. However, the oil contents of the LEA3‐OE and VOC‐OE lines were 30.9%–34.6% (Figure 3a) and 30.7%–33.5% (Figure 3b), respectively. These results suggested that LEA3 and VOC could stabilize oil accumulation under drought stress. The morphology of the OE and WT plants under drought stress was observed by paraffin sectioning and SEM analysis (Figure S9). The OE seeds developed normally, while the WT seeds became wizened under drought stress. Meanwhile, the leaves of WT were more wrinkled than the OE leaves, and stomatal closure was more pronounced in the OE leaves. These results indicated that LEA3 and VOC can maintain normal seed and leaf development under drought stress. The enhanced drought tolerance of the LEA3‐35S‐OE and VOC‐35S‐OE lines was also reflected by physiological index analysis. The chlorophyll contents and RWCs of the LEA3‐35S‐OE and VOC‐35S‐OE lines were significantly higher than those of WT after drought treatment, while their anthocyanin contents and H2O2 accumulations were lower than those of WT (Figure S1D‐I). The temperature of the leaf surface is a phenotype that cannot be observed by the naked eye, but it could be investigated with infrared thermal imaging technology. This experiment revealed that LEA3‐35S‐OE and VOC‐35S‐OE plants achieved a stable state enabling their leaves to remain at a suitable temperature under drought stress (Figure S1D, G). To explore whether the differential sensitivities of the LEA3‐35S‐OE and VOC‐35S‐OE lines are associated with the expression levels of drought‐responsive marker genes, the relative expression levels of RD29A, SnRK2.6 and RbohD were further determined by qRT‐PCR in WT and transgenic plants. The results showed that the expression levels of these genes in the OE lines were higher than those in WT (Figure S1F). RD29A, RbohD and SnRK2.6 are drought responsive and have positive functions in drought tolerance (Ding et al., 2015). RD29A is a drought‐inducible gene with a promoter that contains two major cis‐acting elements, which are involved in stress‐inducible gene expression (Shinozaki and Yamaguchi‐Shinozaki, 2007). RbohD is involved in the stress response and ROS metabolic process, and it is required for hydrogen sulphide‐induced stomatal closure (Ding et al., 2015; He et al., 2018; Scuffi et al., 2018). SnRK2.6 is a protein kinase that functions in the gene regulatory pathway of ABA signal transduction and can be involved in plant drought tolerance through mediating phosphorylation (Ding et al., 2015; Kobayashi et al., 2005). Stomatal closure is a defensive action that is induced by ABA and accompanied by the production of H2O2 (He et al., 2018; Li et al., 2017; Zhang et al., 2001). The gene expression changes in the ABA pathway in the present study were consistent with this (Figures 6, 7, 8). OsbZIP23 was reported as a central regulator of ABA signalling and drought resistance in rice (Zong et al., 2016). Under drought stress, ZIP family gene expression and reduced H2O2 levels were identified in the BnLEA‐OE and VOC‐OE lines. These results support the hypothesis that ABA signal transduction could confer drought tolerance.
LEA genes are activated by DREBs (Battaglia et al., 2008; Lee et al., 2012; Morran et al., 2011). The expression of DREB in the differential LEA expression lines, as revealed by RNA‐seq analysis, was consistent with the known functions of LEA in Arabidopsis, wheat and barley (Morran et al., 2011; Sakuma et al., 2006). The products of LEA genes are often quite hydrophobic and are involved in the direct protection of the cell from stress by increasing membrane stability and processing proteins (Battaglia et al., 2008; Chakrabortee et al., 2007; Goyal et al., 2005; Tolleter et al., 2007). In transgenic B. napus, the membrane of BnLEA‐35S was more stable under drought conditions. In the differential LEA expression lines, AT2G39030 (N‐ACETYLTRANSFERASE ACTIVITY 1, NATA1) and AT5G24780 (VEGETATIVE STORAGE PROTEIN 1, ATVSP1) were the common DEGs in long‐term drought treatment (Figures 6d, S6). NATA1 acts as an ornithine N‐delta‐acetyltransferase, leading to the formation of N‐delta‐acetylornithine (Lou et al., 2016). ATVSP1 acts as an acid phosphatase similar to soya bean vegetative storage proteins, and it can be induced by wounding and jasmonic acid (Courteaux et al., 2013). These results indicate a gene expression trend similar to that of WTSL, but some key genes were still regulated in the late stage of drought stress in seeds. Many lipases were down‐regulated in the LEA‐OE lines. In addition, lipoxygenase genes were down‐regulated; for instance, AT3G45140 (LOX2) and AT1G72520 (LOX4) were down‐regulated in BnLEASS and AtLEASS, respectively. These results suggested that lipid degradation was down‐regulated in the seeds of the LEA‐OE lines. To further investigate transcript coexpression in the drought treatment, a K‐means clustering algorithm and network analysis were used to identify expressed transcripts with patterns of gene activity and to group genes with similar expression profiles (Figures S10, S11). These findings demonstrate that LEA can be active and play an important role in drought stress.
Methylglyoxal is a highly cytotoxic and mutagenic compound known to arrest growth (Singla‐Pareek et al., 2003). In plants, MG has been found to accumulate to higher levels under abiotic stress (Kaur et al., 2014). The glyoxalase pathway has been shown to be associated with abiotic stress adaptation, and GLY I and GLY II can function in salinity adaptation (Ghosh et al., 2014; Mustafiz et al., 2014; Singla‐Pareek et al., 2003). In B. napus, MG was significantly reduced by BnVOC in the drought treatment because of its enhancement of GLY I and GLY II activities (Figure 11). These results suggest that the glyoxalase system can function in drought stress and that increased expression of VOC can augment drought tolerance in B. napus.
In this study, the oil contents of the LEA‐OE and VOC‐OE lines were also increased. Microscopic observation of the seeds indicated that the LEA and VOC genes might have a function in enabling relatively normal seed development under drought stress. LEA proteins could bind to the cell membrane and maintain its integrity (Goyal et al., 2005). The results of microscopic observation in the present study show that LEA‐OE seeds had stable and thick membranes, which can maintain normal lipid synthesis in plant cells, especially under drought stress conditions. The complement of unsaturated FAs was found to be changed in the BnLEA‐OE and BnVOC‐OE lines compared with WT (Figure S12). The FA composition of transgenic plants subjected to drought was also changed (Figure S12), which indicated that overexpression of BnLEA and BnVOC could lead to changes in FA composition. LTPs are defined as proteins that facilitate/accelerate lipid exchange between membranes in vitro (D'Angelo et al., 2008). Our results showed that LTPs were down‐ and up‐regulated in the AtVOC‐RNAi and OE lines, respectively. This phenomenon could lead to more stable membrane structures under drought conditions and more efficient lipid fluxes in the OE lines. In the FA biosynthetic pathway, chloroplastic acetylcoenzyme A carboxylase 1 (CAC1), CAC2, transferase, 3‐ketoacyl‐acyl carrier protein synthase 1 (KAS1), an NAD(P)‐binding protein, hydroxyacyl‐[acyl carrier protein] dehydratase, MOD1, LACS9, and LACS1 were all up‐regulated in the seeds of the LEA‐OE lines (Figure 8). Notably, each step of the main FA biosynthetic pathway (CAC → AT2G30200 → KAS1 → AT1G24360 → AT5G10160 → MOD1 → LACS) had at least one up‐regulated gene in these OE lines (Figure S13). LACS can esterify free FAs to acyl‐CoAs, which is a key activation step that is necessary for the utilization of FAs by most lipid metabolic enzymes (Browse, 2002). The transcriptome and qPCR analyses of the hybrids showed that the expression of LACS was affected, suggesting that the final step of FA biosynthesis could be more efficient in the OE lines. Many LEA proteins localized to the cytosol and plastids, with most being able to diffuse into the nucleus. LEA are thus expected to establish interactions with various cellular membranes under stress conditions (Candat et al., 2014). The FAs were mainly synthesized in plastids, and membranes were important for cell stability in stress. On the other hand, events downstream of FA metabolism, especially FA degradation and lipid peroxidation, were down‐regulated. This reduced expression of genes related to FA degradation and lipid peroxidation also contributed to FA accumulation.
Reactive oxygen species usually act as major signalling molecules in plant hormone response pathways (Bright et al., 2006; Li et al., 2017). Among ROS, H2O2 is a versatile molecule that participates in the response to abiotic stress (Quan et al., 2008; Reczek and Chandel, 2015). High concentrations of H2O2 lead to cell injury (Dietz et al., 2016; Huang et al., 2016). In the present study, H2O2 contents were decreased in the OE lines and OE hybrids. Drought‐responsive genes contributed to these lower concentrations of H2O2, which could reduce cell injury in plants under drought stress. The content of MDA usually represents the level of lipid peroxidation. VOC genes might mitigate lipid peroxidation, which could also affect lipid production. Low concentrations of H2O2 and MDA were also found in other abiotic treatments in B. napus (Farooq et al., 2016; Nouairi et al., 2009). The activities of enzymes related to ROS metabolism (CAT, SOD) showed corresponding changes under drought stress (Figure 11). This result suggested that VOC could affect genes encoding the enzymes involved in ROS metabolism. MG and GSH are also involved in ROS metabolism, and the relative contents of these compounds were altered in the differential VOC expression lines of B. napus (Figure 10). The subcellular location of VOC was the nucleus, cell membrane, and cytosol (Figure S14), which indicate that they can function in MG and ROS metabolism. GLY I and GLY II can catalyse the MG detoxification with GSH (Singla‐Pareek et al., 2003). In the present study, the activities of the GLY I and GLY II enzymes and the contents of GSH in the differential VOC expression lines of transgenic B. napus indicated that VOC could function in MG metabolism. In addition, GLY I plays a key role in glyoxylate metabolism. The glyoxylate cycle could influence the final lipid accumulation in the plant glyoxysome via peroxidation. Under other abiotic stresses, glyoxylate metabolism in the seeds of different B. napus lines was also found to change (Zhou et al., 2018). The results of this study suggest that VOC plays a role in reducing ROS, which can lead to enhanced drought tolerance and lipid accumulation in B. napus.
Photosynthesis provides sucrose, which is the upstream substrate for FA biosynthesis (Durrett et al., 2008). Photosynthetic efficiency and photosynthesis‐related pathways were found to be up‐regulated in the OE transgenic lines and 35S hybrids of B. napus under drought stress. Previous studies have revealed that photosynthesis plays an important role in responses to abiotic stress (Garg et al., 2002; Ghosh et al., 2014; He et al., 2018; Munns and Tester, 2008). Under drought conditions, plants need to maintain their internal water balance and enable photosynthesis. In the present study, the transgenic B. napus lines maintained a balance between photosynthetic capability and osmotic loss by fine‐tuning stomatal closure, transpiration rate, chlorophyll content, RWC and photosynthetic rate (Figures 1, 8, S1, S7). The relative expression of photosystem genes was found to correspond to the expression level of the genes of interest. In addition, the chlorophyll contents of the OE lines were higher than those of the RNAi lines and WT. The changes in chlorophyll contents were consistent with the transcriptome analysis, in which the genes that were involved in chlorophyll synthesis were up‐regulated and down‐regulated in the OE and RNAi lines, respectively. For example, the important genes CHLM, GUN5, CRD1 and CHLG were all up‐regulated in the LEA‐OE and VOC‐OE lines (Chen et al., 2016, 5; Lin et al., 2014; Schlicke et al., 2014; Van Wilder et al., 2009). This maintenance of chlorophyll synthesis in the leaves of the OE lines could contribute to the enhanced photosynthesis in the drought treatment. The higher carbon fixation levels and elevated capacity for photosynthesis also contribute to FA accumulation. Previously, an integrated omics analysis of B. napus and Glycine max also showed that some genes involved in photosynthesis were related to lipid metabolism (Zhang et al., 2018). Thus, the enhanced photosynthetic efficiency in the LEA‐OE and VOC‐OE lines could confer drought tolerance and contribute to lipid accumulation.
In summary, we functionally identified different copies of drought response genes from B. napus and Arabidopsis. The LEA and VOC genes could contribute to drought tolerance and oil content in both B. napus and Arabidopsis. Hybridization of lines overexpressing these two genes showed reinforced performance in drought tolerance and oil production. The mechanism by which LEA and VOC improve oil accumulation is mainly by enhancing photosynthetic efficiency and reducing ROS under drought stress. Therefore, these drought response genes could not only function in drought stress but also improve oil accumulation. The present results provide novel insights into the relationship between oil accumulation and drought resistance and provide a new way to improve oil production in the breeding of oil crops.
Experimental procedures
Plant materials and growth conditions
The WT Arabidopsis used was Columbia ecotype, and B. napus cv. Westar was used as WT B. napus. The T‐DNA insertion mutant of the atlea3 gene was obtained from the Arabidopsis Biological Resource Center and then identified by PCR. Arabidopsis was planted, and the growth conditions were established according to a previous study (Zhang et al., 2013). The B. napus seeds were similarly sterilized and grown under the same conditions. After growing for 4 weeks, the B. napus plants were transplanted to fields until harvest.
Vector constructs and plant transformation
Four gene copies of BnLEA3 and BnVOC from B. napus were acquired and named BnLEA3.1, BnLEA3.2, BnLEA3.3, BnLEA3.4, BnVOC1, BnVOC2, BnVOC3, and BnVOC4 (GenBank Nos. KP315905, KP315906, KP315907, KP315908, KP315909, KP315910, KP315911, KP315912), respectively. These copies were selected because their expression levels were relatively stable and higher in seeds and leaves during drought, based on our previous studies (Liang et al., 2016, 2017). In addition, they were located in four different A or C subgenomes of B. napus. The homologous AtLEA3 and AtVOC genes (AT1G02820, AT1G07645) in Arabidopsis were cloned. The pGEX and pCAMBIA1305 vectors were used for prokaryotic expression and for functional complementation in the atlea3 mutant, respectively. These plasmids were transformed into Arabidopsis and B. napus using Agrobacterium GV3101 with the floral dip method and the B. napus transformation method (Cardoza and Stewart, 2003). The DsRed marker was used for the selection of transformant seeds, which were also PCR confirmed (Zhang et al., 2013).
RNA isolation and RT‐qPCR analysis
Gene‐specific primers were designed using Primer 5.0 (Table S2). A housekeeping gene (ACTIN) that is constitutively expressed in B. napus and Arabidopsis was used as a reference for normalization and analysed using three biological replicates for the qPCR analysis. RNA isolation and RT‐qPCR analysis were performed according to a previous study (Liang et al., 2016). Twenty DEGs that were observed in FA biosynthesis, photosynthesis, ROS metabolism and other random genes were selected for RT‐qPCR. As shown in Figure S11, the RT‐qPCR results generally matched the RNA‐seq results.
Generation of transgenic plants and confirmation through southern hybridization and Western blotting
The presence of the transgene and its expression were first confirmed by RT‐qPCR and then by Southern blotting and Western blotting. For Southern blotting, genomic DNA that tested positive by PCR was digested by the HindIII enzyme, blotted and probed by using the hygromycin gene following the standard protocol (Mustafiz et al., 2014). For Western blotting, FLAG was used as the tag, and ACTIN was used as the housekeeping protein. The next steps were performed according to the protocol (Lagarde et al., 1996).
Drought treatment of the experimental materials
In Arabidopsis, 15% PEG for 9–10 h and withholding water for 10–15 days (RWC of soil was <40%) were used for the short‐term and long‐term drought treatments, respectively. The leaf samples were collected from plants that were 4 weeks (short‐term) and 5 weeks (long‐term) after germination. The silique samples were collected from plants 6 weeks (short‐term) and 7 weeks (long‐term) after flowering. The drought response marker genes (SnRK2.6, RbohD and RbohF) started to be up‐regulated in short‐term drought and reached peak levels in long‐term drought (Tran et al., 2004; Zheng et al., 2016). In B. napus, water was withheld from the plants 10 days after flowering for 15 days.
3,3′‐Diaminobenzidine (DAB) staining and anthocyanin measurements
DAB staining was performed as described previously (He et al., 2018), and the anthocyanin measurements were conducted according to a previous report (Gou et al., 2011).
Morphological observation and analysis
For SEM, TEM and light observation, the samples were fixed and prepared according to the previously published methods (Dolan et al., 1993; Gan et al., 2013; Girard et al., 2017) and observed using a scanning electron microscope (Hitachi S‐4700, Japan), a transmission electron microscope (Hitachi t7700, Japan), and a light microscope (Olympus BX53, Japan), respectively. The leaf surface temperature images were taken using a Thermal Imager (Fluke Ti400) and analysed using Fluke SmartView Infrared Analysis Software. The seed sizes were scanned and measured by a crop scanning test system (Wanshen SC‐G, China).
Oil content and FA analysis of experimental materials
The lipids of the seeds were extracted by the protocol described in a previous study, and the oil contents were measured as the proportion of total oil content to seed dry weight. The change percentages were measured as change percentage = (oil content of sample‐oil content of CK)/oil content of CK *100 (Maisonneuve et al., 2010; Zhang et al., 2013). The extracted FAs were analysed by GC/MS using an Agilent 6850 gas chromatograph with frame ionization detection (FID) on a polar column (Agilent, DB‐23, 30 m by 0.25 mm i.d., 0.25 μm film).
Library generation for high‐throughput sequencing and bioinformatics analyses
Method S1 shows details of the RNA sequencing and bioinformatics analyses (Figures [Link], [Link], [Link], [Link], [Link], [Link], Tables S3 and S4).
Measurement of MG, H2O2, MDA, GSSG and GSH concentrations
The contents of MG, GSH, GSH, H2O2 and MDA were measured using a plant MG ELISA kit (Nanjing Jin Yibai, JEB~14422, China), a Reduced Glutathione (GSH) Assay Kit (Solarbio BC1170, China), an Oxidized Glutathione (GSSG) Assay Kit (Solarbio BC1180, China) or according to Shi's and Basu's studies (Basu et al., 2001; Shi et al., 2014;).
Enzyme assays
For the enzyme assays, the plant samples were prepared and quantified using previously published protocols (Doderer et al., 1992; Shi et al., 2014; Singla‐Pareek et al., 2003).
Measurement of photosynthetic parameters
An infrared gas analyser (Li‐COR 6400, Agilent, Lincoln, Nebraska, USA) was used to detect photosynthetic parameters.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
ML designed the research and wrote the article. YL and LG performed the vector constructions, transformation, GCMS, and data analysis and wrote the article. KK, YL and JG conducted bioinformatics analyses and discussed the results. SN, JX, SS, LX and SL performed the crossing, fieldwork and oil extractions. LG and YY conducted the drought treatment. JX, SL and BW performed the enzyme assays and MDA, MG, H2O2, GSSG and GSH and measurements.
Supporting information
Figure S1 Enhanced drought tolerance in LEA3‐OE and VOC‐OE lines of Arabidopsis.
Figure S2 Identification of mutant atlea3 and gene expression of transgenic Arabidopsis.
Figure S3 Oil contents of the atlea3 mutant and HB lines.
Figure S4 qPCR of LEA and VOC genes in B. napus.
Figure S5 Oil reduction comparison.
Figure S6 FA compositions of OE, RNAi and hybrid seeds in B. napus.
Figure S7 FA compositions of hybrid Arabidopsis.
Figure S8 Prokaryotic expression of VOC genes in E. coli.
Figure S9 Microscopic observations of OE and WT seeds and leaves in Arabidopsis under drought conditions.
Figure S10 K‐means clusters and RT‐qPCR confirmation of DEGs.
Figure S11 Network of DEGs in LEA and VOC transgenic seeds under drought conditions.
Figure S12 FA compositions of OE and WT seeds in Arabidopsis.
Figure S13 Change folds of gene in the FA synthesis in LEA‐relative lines.
Figure S14 Subcellular localization of VOC.
Table S1 Detailed oil and FA contents of transgenic Arabidopsis overexpressing LEA and VOC.
Table S2 Primers used in RNA‐seq confirmation
Table S3 Detailed RNA‐seq information.
Table S4 Abbreviations used in this study.
Method S1 RNA‐seq analysis methods.
Acknowledgements
This work was supported by the National Key Research and Development Program (2016YFD0100200, 2016YFD0101300), National Natural Science Foundation of China (31871656) and the National Program on Key Basic Research Project (2015CB150205). We appreciate Weimeng Sha, Efan Wang, Chenfei Shang, Hai Chen for their help in preparing plant materials. We also acknowledge the support of the Research Core Facilities for Life Science in HUST.
References
- Arisz, S.A. , Heo, J.‐Y. , Koevoets, I.T. , Zhao, T. , van Egmond, P. , Meyer, J. , Zeng, W. , et al. (2018) Diacylglycerol acyltransferase 1 contributes to freezing tolerance. Plant Physiol. 177, 1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu, U. , Good, A.G. and Taylor, G.J. (2001) Transgenic Brassica napus plants overexpressing aluminium‐induced mitochondrial manganese superoxide dismutase cDNA are resistant to aluminium. Plant, Cell Environ. 24, 1269–1278. [Google Scholar]
- Battaglia, M. , Olvera‐Carrillo, Y. , Garciarrubio, A. , Campos, F. and Covarrubias, A.A. (2008) The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 148, 6–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer, A. , Wolff, M. , Thalhammer, A. and Hincha, D.K. (2017) Folding of intrinsically disordered plant LEA proteins is driven by glycerol‐induced crowding and the presence of membranes. FEBS J. 284, 3765. [DOI] [PubMed] [Google Scholar]
- Bright, J. , Desikan, R. , Hancock, J. , Weir, I. and Neill, S. (2006) ABA‐induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J. 45, 113–122. [DOI] [PubMed] [Google Scholar]
- Browse, J.A. (2002) Arabidopsis contains nine long‐chain acyl‐coenzyme a synthetase genes that participate in fatty acid and glycerolipid metabolism. Plant Physiol. 129, 1710–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candat, A. , Paszkiewicz, G. , Neveu, M. , Gautier, R. , Logan, D.C. , Avelange‐Macherel, M.‐H. and Macherel, D. (2014) The ubiquitous distribution of late embryogenesis abundant proteins across cell compartments in Arabidopsis offers tailored protection against abiotic stress. Plant Cell, 26, 3148–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoza, V. and Stewart, C. (2003) Increased Agrobacterium‐mediated transformation and rooting efficiencies in canola (Brassica napus L.) from hypocotyl segment explants. Plant Cell Rep. 21, 599–604. [DOI] [PubMed] [Google Scholar]
- Chakrabortee, S. , Boschetti, C. , Walton, L.J. , Sarkar, S. , Rubinsztein, D.C. and Tunnacliffe, A. (2007) Hydrophilic protein associated with desiccation tolerance exhibits broad protein stabilization function. Proc. Natl Acad. Sci. 104, 18073–18078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabortee, S. , Tripathi, R. , Watson, M. , Schierle, G.S.K. , Kurniawan, D.P. , Kaminski, C.F. and Tunnacliffe, A. (2012) Intrinsically disordered proteins as molecular shields. Mol. BioSyst. 8, 210–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub, B. , Denoeud, F. , Liu, S. , Parkin, I.A. , Tang, H. , Wang, X. , Chiquet, J. et al. (2014) Plant genetics. Early allopolyploid evolution in the post‐Neolithic Brassica napus oilseed genome. Science, 345, 950–953. [DOI] [PubMed] [Google Scholar]
- Chapman, K.D. and Ohlrogge, J.B. (2012) Compartmentation of triacylglycerol accumulation in plants. J. Biol. Chem. 287, 2288–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S.‐T. , He, N.‐Y. , Chen, J.‐H. and Guo, F.‐Q. (2016) Identification of core subunits of photosystem II as action sites of HSP21, which is activated by the GUN5‐mediated retrograde pathway in Arabidopsis. Plant J. 89, 1106–1118. [DOI] [PubMed] [Google Scholar]
- Courteaux, B. , Profizi, C. , Clément, C. , Gérard, C. , Baillieul, F. , Rabenoelina, F. , Le Hénanff, G. et al. (2013) Grapevine NAC1 transcription factor as a convergent node in developmental processes, abiotic stresses, and necrotrophic/biotrophic pathogen tolerance. J. Exp. Bot. 64, 4877–4893. [DOI] [PubMed] [Google Scholar]
- D'Angelo, G. , Vicinanza, M. and De Matteis, M.A. (2008) Lipid‐transfer proteins in biosynthetic pathways. Curr. Opin. Cell Biol. 20, 360–370. [DOI] [PubMed] [Google Scholar]
- Dietz, K.‐J. , Mittler, R. and Noctor, G. (2016) Recent progress in understanding the role of reactive oxygen species in plant cell signaling. Plant Physiol. 171, 1535–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, S. , Zhang, B. and Qin, F. (2015) Arabidopsis RZFP34/CHYR1, a ubiquitin E3 ligase, regulates stomatal movement and drought tolerance via SnRK2.6‐mediated phosphorylation. Plant Cell, 27, 3228–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doderer, A. , Kokkelink, I. , Veen, S. , van der Valk, B.E. , Schram, A. and Douma, A.C. (1992) Purification and characterization of two lipoxygenase isoenzymes from germinating barley. Biochim. Biophys. Acta, 1120, 97–104. [DOI] [PubMed] [Google Scholar]
- Dolan, L. , Janmaat, K. , Willemsen, V. , Linstead, P. , Poethig, S. , Roberts, K. and Scheres, B. (1993) Cellular organisation of the Arabidopsis thaliana root. Development, 119, 71. [DOI] [PubMed] [Google Scholar]
- Durrett, T.P. , Benning, C. and Ohlrogge, J. (2008) Plant triacylglycerols as feedstocks for the production of biofuels. Plant J. 54, 593–607. [DOI] [PubMed] [Google Scholar]
- Farooq, M.A. , Gill, R.A. , Islam, F. , Ali, B. , Liu, H. , Xu, J. , He, S. , et al. (2016) Methyl jasmonate regulates antioxidant defense and suppresses arsenic uptake in Brassica napus L. Front. Plant Sci. 7, 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan, L. , Zhang, C.Y. , Wang, X.D. , Wang, H. , Long, Y. , Yin, Y.T. , Li, D.R. et al. (2013) Proteomic and comparative genomic analysis of two Brassica napus lines differing in oil content. J. Proteome Res. 12, 4965–4978. [DOI] [PubMed] [Google Scholar]
- Garg, A. , Kim, J. , Owens, T. , Ranwala, A. , Do Choi, Y. , Kochian, L. and Wu, R. (2002) Trehalose accumulation in rice plants confers high tolerance levels to different abiotic stresses. Proc. Natl Acad. Sci. 99, 15898–15903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh, A. , Pareek, A. , Sopory, S.K. and Singla‐Pareek, S.L. (2014) A glutathione responsive rice glyoxalase II, OsGLYII‐2, functions in salinity adaptation by maintaining better photosynthesis efficiency and anti‐oxidant pool. Plant J. 80, 93–105. [DOI] [PubMed] [Google Scholar]
- Girard, I.J. , Tong, C. , Becker, M.G. , Mao, X. , Huang, J. , de Kievit, T. , Fernando, W.G.D. et al. (2017) RNA sequencing of Brassica napus reveals cellular redox control of Sclerotinia infection. J. Exp. Bot. 68, 5079–5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou, J. , Felippes, F.F. , Liu, C. , Weigel, D. and Wang, J. (2011) Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156‐Targeted SPL transcription factor. Plant Cell, 23, 1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal, K. , Walton, L. and Tunnacliffe, A. (2005) LEA proteins prevent protein aggregation due to water stress. Biochem. J. 388, 151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, I.A. (2008) Seed storage oil mobilization. Annu. Rev. Plant Biol. 59, 115–142. [DOI] [PubMed] [Google Scholar]
- He, F. , Wang, H.L. , Li, H.G. , Su, Y. , Li, S. , Yang, Y. , Feng, C.H. , et al. (2018) PeCHYR1, a ubiquitin E3 ligase from Populus euphratica, enhances drought tolerance via ABA‐induced stomatal closure by ROS production in Populus. Plant Biotechnol. J. 16, 1514‐1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetherington, A.M. and Woodward, F.I. (2003) The role of stomata in sensing and driving environmental change. Nature, 424, 901–908. [DOI] [PubMed] [Google Scholar]
- Higashi, Y. , Okazaki, Y. , Takano, K. , Myouga, F. , Shinozaki, K. , Knoch, E. , Fukushima, A. et al. (2018) HEAT INDUCIBLE LIPASE1 remodels chloroplastic monogalactosyldiacylglycerol by liberating α‐linolenic acid in Arabidopsis leaves under heat stress. Plant Cell, 30, 1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hincha, D.K. and Thalhammer, A. (2012) LEA proteins: IDPs with versatile functions in cellular dehydration tolerance. Biochem. Soc. Trans. 40, 1000–1003. [DOI] [PubMed] [Google Scholar]
- Huang, S. , Van Aken, O. , Schwarzlaender, M. , Belt, K. and Millar, A.H. (2016) The roles of mitochondrial reactive oxygen species in cellular signaling and stress response in plants. Plant Physiol. 171, 1551–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur, C. , Ghosh, A. , Pareek, A. , Sopory, S.K. and Singla‐Pareek, S.L. (2014) Glyoxalases and stress tolerance in plants. Biochem. Soc. Trans. 42, 485–499. [DOI] [PubMed] [Google Scholar]
- Kobayashi, Y. , Murata, M. , Minami, H. , Yamamoto, S. , Kagaya, Y. , Hobo, T. , Yamamoto, A. et al. (2005) Abscisic acid‐activated SNRK2 protein kinases function in the gene‐regulation pathway of ABA signal transduction by phosphorylating ABA response element‐binding factors. Plant J. 44, 939–949. [DOI] [PubMed] [Google Scholar]
- Lagarde, D. , Basset, M. , Lepetit, M. , Conejero, G. , Gaymard, F. , Astruc, S. and Grignon, C. (1996) Tissue‐specific expression of Arabidopsis AKT1 gene is consistent with a role in K+ nutrition. Plant J. 9, 195–203. [DOI] [PubMed] [Google Scholar]
- Lee, S.‐C. , Lim, M.‐H. , Yu, J.‐G. , Park, B.‐S. and Yang, T.‐J. (2012) Genome‐wide characterization of the CBF/DREB1 gene family in Brassica rapa. Plant Physiol. Biochem. 61, 142–152. [DOI] [PubMed] [Google Scholar]
- Li, J. , Li, Y. , Yin, Z. , Jiang, J. , Zhang, M. , Guo, X. , Ye, Z. et al. (2017) OsASR5 enhances drought tolerance through a stomatal closure pathway associated with ABA and H2O2 signalling in rice. Plant Biotechnol. J. 15, 183–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Y. , Xiong, Z. , Zheng, J. , Xu, D. , Zhu, Z. , Xiang, J. , Gan, J. et al. (2016) Genome‐wide identification, structural analysis and new insights into late embryogenesis abundant (LEA) gene family formation pattern in Brassica napus . Sci. Rep. 6, 24265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Y. , Wan, N. , Cheng, Z. , Mo, Y. , Liu, B. , Liu, H. , Raboanatahiry, N. et al. (2017) Whole‐genome identification and expression pattern of the vicinal oxygen chelate family in Rapeseed (Brassica napus L.). Front. Plant Sci. 8, 745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y. , Lee, T. , Tanaka, A. and Charng, Y. (2014) Analysis of an Arabidopsis heat‐sensitive mutant reveals that chlorophyll synthase is involved in reutilization of chlorophyllide during chlorophyll turnover. Plant J. 80, 14–26. [DOI] [PubMed] [Google Scholar]
- Lou, Y.‐R. , Bor, M. , Yan, J. , Preuss, A.S. and Jander, G. (2016) Arabidopsis NATA1 acetylates putrescine and decreases defense‐related hydrogen peroxide accumulation. Plant Physiol. 171, 1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve, S. , Bessoule, J.‐J. , Lessire, R. , Delseny, M. and Roscoe, T.J. (2010) Expression of rapeseed microsomal lysophosphatidic acid acyltransferase isozymes enhances seed oil content in Arabidopsis. Plant Physiol. 152, 670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, G. , Suzuki, N. , Ciftci‐Yilmaz, S. and Mittler, R. (2010) Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant, Cell Environ. 33, 453–467. [DOI] [PubMed] [Google Scholar]
- Morran, S. , Eini, O. , Pyvovarenko, T. , Parent, B. , Singh, R. , Ismagul, A. , Eliby, S. et al. (2011) Improvement of stress tolerance of wheat and barley by modulation of expression of DREB/CBF factors. Plant Biotechnol. J. 9, 230–249. [DOI] [PubMed] [Google Scholar]
- Munns, R. and Tester, M. (2008) Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. [DOI] [PubMed] [Google Scholar]
- Mustafiz, A. , Ghosh, A. , Tripathi, A.K. , Kaur, C. , Ganguly, A.K. , Bhavesh, N.S. , Tripathi, J.K. et al. (2014) A unique Ni2+‐dependent and methylglyoxal‐inducible rice glyoxalase I possesses a single active site and functions in abiotic stress response. Plant J. 78, 951–963. [DOI] [PubMed] [Google Scholar]
- Nouairi, I. , Ben Ammar, W. , Ben Youssef, N. , Ben Miled, D.D. , Ghorbal, M.H. and Zarrouk, M. (2009) Antioxidant defense system in leaves of Indian mustard (Brassica juncea) and rape (Brassica napus) under cadmium stress. Acta Physiol. Plant. 31, 237–247. [Google Scholar]
- Okazaki, Y. and Saito, K. (2014) Roles of lipids as signaling molecules and mitigators during stress response in plants. Plant J. 79, 584–596. [DOI] [PubMed] [Google Scholar]
- Prasch, C.M. , Ott, K.V. , Bauer, H. , Ache, P. , Hedrich, R. and Sonnewald, U. (2015) Beta‐amylase1 mutant Arabidopsis plants show improved drought tolerance due to reduced starch breakdown in guard cells. J. Exp. Bot. 66, 6059–6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan, L. , Zhang, B. , Shi, W. and Li, H. (2008) Hydrogen peroxide in plants: a versatile molecule of the reactive oxygen species network. J. Integr. Plant Biol. 50, 2–18. [DOI] [PubMed] [Google Scholar]
- Reczek, C.R. and Chandel, N.S. (2015) ROS‐dependent signal transduction. Curr. Opin. Cell Biol. 33, 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma, Y. , Maruyama, K. , Osakabe, Y. , Qin, F. , Seki, M. , Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2006) Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought‐responsive gene expression. Plant Cell, 18, 1292–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlicke, H. , Hartwig, A.S. , Firtzlaff, V. , Richter, A.S. , Glässer, C. , Maier, K. , Finkemeier, I. et al. (2014) Induced deactivation of genes encoding chlorophyll biosynthesis enzymes disentangles tetrapyrrole‐mediated retrograde signaling. Mol. Plant, 7, 1211–1227. [DOI] [PubMed] [Google Scholar]
- Scuffi, D. , Nietzel, T. , Di Fino, L.M. , Meyer, A.J. , Lamattina, L. , Schwarzländer, M. , Laxalt, A.M. et al. (2018) Hydrogen sulfide increases production of NADPH oxidase‐dependent hydrogen peroxide and phospholipase D‐derived phosphatidic acid in guard cell signaling. Plant Physiol. 176, 2532–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, H. , Ye, T. , Zhu, J.‐K. and Chan, Z. (2014) Constitutive production of nitric oxide leads to enhanced drought stress resistance and extensive transcriptional reprogramming in Arabidopsis. J. Exp. Bot. 65, 4119–4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2007) Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58, 221–227. [DOI] [PubMed] [Google Scholar]
- Singla‐Pareek, S.L. , Reddy, M.K. and Sopory, S.K. (2003) Genetic engineering of the glyoxalase pathway in tobacco leads to enhanced salinity tolerance. Proc. Natl Acad. Sci. 100, 14672–14677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, N. , Rivero, R.M. , Shulaev, V. , Blumwald, E. and Mittler, R. (2014) Abiotic and biotic stress combinations. New Phytol. 203, 32–43. [DOI] [PubMed] [Google Scholar]
- Tolleter, D. , Jaquinod, M. , Mangavel, C. , Passirani, C. , Saulnier, P. , Manon, S. , Teyssier, E. et al. (2007) Structure and function of a mitochondrial late embryogenesis abundant protein are revealed by desiccation. Plant Cell, 19, 1580–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, L.‐S.P. , Nakashima, K. , Sakuma, Y. , Simpson, S.D. , Fujita, Y. , Maruyama, K. , Fujita, M. et al. (2004) Isolation and functional analysis of arabidopsis stress‐inducible NAC transcription factors that bind to a drought‐responsive cis‐element in the early responsive to dehydration stress 1 Promoter. Plant Cell, 16, 2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunnacliffe, A. , Hincha, D.K. , Leprince, O. and Macherel, D. (2010) LEA proteins: versatility of form and function. In Dormancy and Resistance in Harsh Environments ( Esther, L. , Joan, C. , Melody, C. ), pp. 91–108. Berlin: Springer. [Google Scholar]
- Ullah, A. , Sun, H. , Yang, X. and Zhang, X. (2017) Drought coping strategies in cotton: increased crop per drop. Plant Biotechnol. J. 15, 271–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wilder, V. , De Brouwer, V. , Loizeau, K. , Gambonnet, B. , Albrieux, C. , Van Der Straeten, D. , Lambert, W.E. et al. (2009) C1 metabolism and chlorophyll synthesis: the Mg‐protoporphyrin IX methyltransferase activity is dependent on the folate status. New Phytol. 182, 137–145. [DOI] [PubMed] [Google Scholar]
- Wang, W. , Vinocur, B. and Altman, A. (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta, 218, 1–14. [DOI] [PubMed] [Google Scholar]
- Xiong, L. and Zhu, J.K. (2002) Molecular and genetic aspects of plant responses to osmotic stress. Plant, Cell Environ. 25, 131–139. [DOI] [PubMed] [Google Scholar]
- Yen, C.L.E. , Stone, S.J. , Koliwad, S. , Harris, C. and Farese, R.V. (2008) DGAT enzymes and triacylglycerol biosynthesis. J. Lipid Res. 49, 2283–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, T. , Mogami, J. and Yamaguchi‐Shinozaki, K. (2014) ABA‐dependent and ABA‐independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 21, 133–139. [DOI] [PubMed] [Google Scholar]
- Zhang, B.H. (2015) MicroRNA: a new target for improving plant tolerance to abiotic stress. J. Exp. Bot. 66, 1749–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Miao, Y. , An, G. , Zhou, Y. , Shangguan, Z. , Gao, J. and Song, C. (2001) K+ channels inhibited by hydrogen peroxide mediate abscisic acid signaling in Vicia guard cells. Cell Res. 11, 195–202. [DOI] [PubMed] [Google Scholar]
- Zhang, M. , Fan, J.L. , Taylor, D.C. and Ohlrogge, J.B. (2009) DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell, 21, 3885–3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C. , Iskandarov, U. , Klotz, E.T. , Stevens, R.L. , Cahoon, R.E. , Nazarenus, T.J. , Pereira, S.L. et al. (2013) A thraustochytrid diacylglycerol acyltransferase 2 with broad substrate specificity strongly increases oleic acid content in engineered Arabidopsis thaliana seeds. J. Exp. Bot. 64, 3189–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. , Dunwell, J.M. and Zhang, Y.‐M. (2018) An integrated omics analysis reveals molecular mechanisms that are associated with differences in seed oil content between Glycine max and Brassica napus. BMC Plant Biol. 18, 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Chan, Z.L. , Gao, J.H. , Xing, L. , Cao, M.J. , Yu, C.M. , Hu, Y.L. et al. (2016) ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl Acad. Sci. 113, 1949–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Y. , Ding, Y. , Sun, X. , Xie, S. , Wang, D. , Liu, X. , Su, L. et al. (2016) Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in Arabidopsis. J. Exp. Bot. 67, 1703–1713. [DOI] [PubMed] [Google Scholar]
- Zhou, L. , Yan, T. , Chen, X. , Li, Z. , Wu, D. , Hua, S. and Jiang, L. (2018) Effect of high night temperature on storage lipids and transcriptome changes in developing seeds of oilseed rape. J. Exp. Bot. 69, 1721–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J.K. (2002) Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong, W. , Tang, N. , Yang, J. , Peng, L. , Ma, S. , Xu, Y. , li, G. et al. (2016) Feedback regulation of ABA signaling and biosynthesis by a bZIP transcription factor targets drought‐resistance‐related genes. Plant Physiol. 171, 2810–2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Enhanced drought tolerance in LEA3‐OE and VOC‐OE lines of Arabidopsis.
Figure S2 Identification of mutant atlea3 and gene expression of transgenic Arabidopsis.
Figure S3 Oil contents of the atlea3 mutant and HB lines.
Figure S4 qPCR of LEA and VOC genes in B. napus.
Figure S5 Oil reduction comparison.
Figure S6 FA compositions of OE, RNAi and hybrid seeds in B. napus.
Figure S7 FA compositions of hybrid Arabidopsis.
Figure S8 Prokaryotic expression of VOC genes in E. coli.
Figure S9 Microscopic observations of OE and WT seeds and leaves in Arabidopsis under drought conditions.
Figure S10 K‐means clusters and RT‐qPCR confirmation of DEGs.
Figure S11 Network of DEGs in LEA and VOC transgenic seeds under drought conditions.
Figure S12 FA compositions of OE and WT seeds in Arabidopsis.
Figure S13 Change folds of gene in the FA synthesis in LEA‐relative lines.
Figure S14 Subcellular localization of VOC.
Table S1 Detailed oil and FA contents of transgenic Arabidopsis overexpressing LEA and VOC.
Table S2 Primers used in RNA‐seq confirmation
Table S3 Detailed RNA‐seq information.
Table S4 Abbreviations used in this study.
Method S1 RNA‐seq analysis methods.


