Summary
Chronic lymphocytic leukaemia (CLL) is the most common leukaemia in Western countries but very rare in Asia. Peripheral blood or bone marrow mononuclear cells obtained at initial diagnosis from 194 patients with CLL were analysed to determine the ethnic difference in genetic abnormalities. Mutated IGHV was detected in 71·2% of Taiwanese CLL and IGHV3‐23 was the most frequently used gene. Stereotyped BCR was present in 18·3% with subset 8 being the most frequent. All cases with subset 8 belonged to IGHV 4‐39 and were exclusively associated with un‐mutated IGHV and poor outcome. Mutation frequencies of SF3B1 (9·7%), NOTCH1 (8·6%), BIRC3 (1·1%), ATM (16·9%) or TP53 (8·1%), and frequencies of cytogenetic abnormalities including trisomy 12 (18·6%), del(17p) (10·4%), del(13q) (43·7%) and IGH translocation (10·1%) were comparable to those reported from Western countries, except del(11q) (6·9%) which was lower in our patients. Patients with un‐mutated IGHV, subset 8, disrupted TP53, trisomy 12, and SF3B1 mutations had a worse outcome compared to patients without these mutations. In conclusion, IGHV3‐23 usage, stereotyped subset 8 and lower frequency of del(11q) show an ethnicity‐dependent association in Taiwanese CLL patients.
Keywords: chronic lymphocytic leukaemia, IGHV, ATM, TP53, BCR stereotype
Chronic lymphocytic leukaemia (CLL) is a disease characterized by the proliferation and accumulation of morphologically mature lymphocytes expressing CD5, CD20 and CD23, together with low expression of surface IgM (Rozman & Montserrat, 1995). CLL is the most common leukaemia in Western countries (Rozman & Montserrat, 1995; Morton et al, 2006) but its incidence is very low in Asia (Weiss, 1979). In CLL, mutational status of the immunoglobulin heavy‐chain variable (IGHV) gene, which is widely accepted as one of the most reliable predictors of clinical outcome, can be divided into two subgroups with prognostic relevance: mutated IGHV associated with indolent clinical course and un‐mutated IGHV, with a progressive disease course even in the patients with early stage disease (Damle et al, 1999; Hamblin et al, 1999; Oscier et al, 2002).
The reported frequencies of mutated IGHV ranged from 42·4% to 64·2% in the West (Duke et al, 2003; Tobin et al, 2004; Agathangelidis et al, 2012) compared to 60~78·8% in Oriental patients with CLL (Nakahashi et al, 2009; Xia et al, 2015; Marinelli et al, 2016). In addition to IGHV mutational status, IGHV repertoire analysis showed that IGHV families or IGHV gene usage had geographically biased predispositions. In the West, the most predominant IGHV family was IGHV3 followed by IGHV1 (Hamblin et al, 1999; Duke et al, 2003; Tobin et al, 2004). In contrast, Asian cohorts were dominated by IGHV3 followed by IGHV4 (Chen et al, 2008; Hojjat‐Farsangi et al, 2009; Nakahashi et al, 2009; Marinelli et al, 2016) except in one recent small series study from Taiwan, which described a very low frequency of IGHV4 (Wu et al, 2017).
Another important characteristic of the IGHV repertoire is the expression of stereotyped B‐cell receptors (BCRs). Homologous stereotyped Complementarity‐Determining region 3 (CDR3) within BCRs were identified in 20~30% of CLL cases, which has been suggested to be involved in the pathogenesis of CLL and could be subdivided into 19 major subsets and other minor subsets (Stamatopoulos et al, 2007; Agathangelidis et al, 2012; Rani et al, 2016). Notably, the frequency of the usage of BCR subsets showed great variation among different geographic areas (Marinelli et al, 2016).
In addition to IGHV mutational status, chromosomal aberrations and gene mutations are other important prognostic markers of CLL. Patients with 17p deletion [del(17p)] had inferior survival whereas those with del(13q) or trisomy 12 had better outcomes, and patients carrying del(11q) had an intermediate survival (Dohner et al, 2000). CLL patients with TP53, NOTCH1, SF3B1, ATM or BIRC3 mutations had poor prognosis (Foa et al, 2013; Puiggros et al, 2014). A substantial difference in the frequencies of gene mutations was observed between Caucasian and Asian patients (Xia et al, 2015). In the present study, we sought to investigate the untreated diagnostic samples of a larger cohort of CLL patients in Taiwan for the frequencies of IGHV mutational status, IGHV usage, BCR stereotype, chromosomal aberrations and gene mutations, as well as correlate the genetic abnormalities with their outcomes.
Materials and methods
Patients and samples
Between July 1991 and December 2017, 194 patients with CLL were consecutively diagnosed and were regularly followed‐up at a single tertiary referral centre in Taiwan. Diagnosis of CLL was based on the International Workshop of CLL‐National Cancer Institute (IWCLL‐NCI) criteria (Hallek et al, 2008). All patients met the criteria of ≧ 5 × 109/l monoclonal B cells in the peripheral blood with expression of CD5, CD20 and CD23 by flow cytometry. Peripheral blood or bone marrow mononuclear cells (MNC) were obtained from the diagnostic sample of each patient by Ficoll‐Hypaque density gradient centrifugation and freshly frozen at −80°C until testing. The study was approved by the Institutional Review Board of Chang Gung Memorial Hospital (105‐1282C).
gDNA extraction, cDNA synthesis, PCR amplification, and sequencing
gDNA and total RNA were extracted from peripheral blood or bone marrow MNC and complementary DNA (cDNA) was synthesized. To determine IGHV usage, clonal IGH rearrangements were amplified from cDNA using two primer pairs: (i) seven leader primers and an IgM/IgG primer (Fais et al, 1998); (ii) seven leader primers and a JH primer (McCarthy et al, 2003). In three cases with no cDNA as template, targets were amplified from gDNA. PCR products were purified and sequenced in both directions, and then aligned to the closest matched germline gene by using the IMGT/V‐QUEST analysis software (IMGT; http://www.imgt.org/, Montpellier, France). Sequences with a germline identity of 98% or higher were considered un‐mutated IGHV, and those with less than 98% identity as mutated IGHV (Hamblin et al, 1999). The IGHV CDR3 of each sequence was also analysed by IMGT analysis software. For the clustering analysis, sequences were applied to ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) and then successive filtering was carried out on the basis of previously proposed criteria (Darzentas et al, 2010). All IGHV sequences were also evaluated using the online tool ARResT/AssignSubsets (http://tools.bat.infspire.org/arrest/assignsubsets) (Bystry et al, 2015). Novel clusters identified in this study but not defined (Agathangelidis et al, 2012) were assigned a number preceded by the word ‘Cluster’.
Fluorescence in situ hybridisation in CLL
Locus‐specific probes including TP53 (17p13)/MPO (17q22) (Kreatech, Amsterdam, Netherlands) for del(17p), D13S319/13q34 (Vysis, Des Plaines, IL, USA) for del(13q), ATM (11q22)/GLI1 (12q13) (Kreatech) for del(11q)/trisomy 12 and IGH breakapart (Vysis) for IGH translocation were used.
Analysis of ZAP70 expression by flow cytometry
Fluorescently labelled antibodies to CD5‐peridinin chlorophyll protein (PerCP)‐cyanine (Cy) 5·5 and CD19‐allophycocyanin (APC) were obtained from Becton Dickinson (San Jose, CA, USA). ZAP70‐Alexa Fluor 488 and Mouse IgG1 Alexa Fluor 488 antibodies were purchased from Caltag (Buckingham, UK). Frozen or fresh MNC were stained by adding CD5‐PerCP‐Cy5·5 and CD19‐APC, permeabilised with 8E reagent, which was kindly provided by Prof. Dario Campana at National Singapore University, and followed by ZAP70‐Alexa Fluor 488 staining. Cells were then analysed with a BD FACS Aria III flow cytometer and FACS Diva Software (Becton Dickinson, San Jose, CA, USA). The percentage of B cells positive for ZAP70 was determined by gating the CD19/CD5 population. The threshold was set at 20%, as described previously (Crespo et al, 2003; Richardson et al, 2006).
Mutational analysis using next generation sequencing
Ion AmpliSeq primer pools for TP53 (exons 2–11), NOTCH1 (exons 28–34), SF3B1 (exons 11–16), BIRC3 (exons 3–9) and ATM (whole coding exons) were used to amplify the targets. Library was constructed by using Ion AmpliSeq™ Library kit (Life Technologies, Carlsbad, CA, USA) and sequenced on the Ion Torrent PGM (Life Technologies) machine. Mutations were then analysed with the Variant Caller software offered by the Torrent Server. Sanger sequencing or pyroseqencing was used to validate the mutations.
Statistical analysis
Patients were followed until initiation of CLL‐specific treatment or death or end of follow‐up, defined as treatment‐free survival (TFS), and until death or end of follow‐up, defined as overall survival (OS). All statistical analyses were carried out using the Sigmaplot statistical package (Systat Software Inc., San Jose, CA, USA). Categorical variables were compared using Fisher exact test. Multivariate analysis was done by Cox proportional hazard regression. Survival curves were constructed by Kaplan–Meier estimate and differences were evaluated by log rank test. Two‐tailed P values less than 0·05 were considered as statistically significant.
Results
IGHV usage
Three of 194 patients with no reliable or reproducible clonal IGHV were excluded from IGHV analysis. Based on a cut‐off of 2% deviation from the germline sequence, 136 out of 191 patients (71·2%) had IGHV mutated gene sequences, and the remaining 55 had sequences (28·8%) that belonged to the un‐mutated subgroup. The most frequently expressed IGHV family was IGHV3 (n = 102, 53·4%), followed by IGHV4 (n = 49, 25·7%), IGHV1 (n = 26, 13·6%), IGHV2 (n = 7, 3·7%), IGHV5 (n = 4, 2·1%), IGHV6 (n = 2, 1·0%) and IGHV7 (n = 1, 0·5%) (Fig 1A). For mutational status of IGHV region among the most common IGHV families, the vast majority of IGHV1 expressing‐cases showed more un‐mutated (16/26; 61·5%) compared to IGHV3 (20/102; 19·6%, P < 0·0001) or IGHV4 families (14/49; 28·6%, P < 0·0071) (Fig 1A). The IGHV mutational status in the most frequent IGHV usage subtypes is shown in Fig 1B. IGHV3‐23 (n = 20, 10·5%) was the most used, followed by IGHV3‐7 (n = 18, 9·4%), IGHV3‐74 (n = 14, 7·3%), IGHV4‐59 (n = 13, 6·8%), IGHV3‐30 (n = 12, 6·3%), IGHV4‐34 (n = 11, 5·8%), IGHV4‐39 (n = 11, 5·8%) and IGHV1‐69 (n = 11, 5·8%). IGHV3‐23 (19·6%), IGHV3‐7 (17·6%) and IGHV3‐74 (13·7%) constituted 51·0% of all IGHV3 cases, which were highly associated with mutated IGHV status (95·0%, 94·4% and 78·6%). IGHV4‐59 (26·5%), IGHV4‐34 (22·4%) and IGHV4‐39 (22·4%) constituted 71·4% of all IGHV4‐expressing cases, in which mutated cases were 76·9%, 90·9%, and 27·3%, respectively. In the IGHV1 family, IGHV1‐69 (42·3%) was the most frequent IGHV1‐expressing subtype with a mutated frequency of 9·1%.
Figure 1.
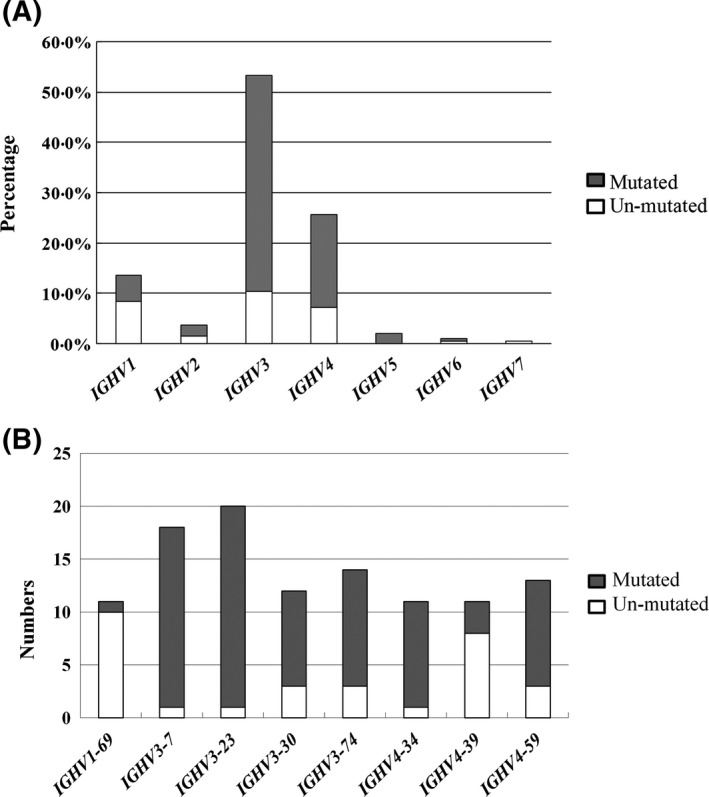
(A) Distribution of IGHV usage in 191 chronic lymphocytic leukaemia patients; (B) Number of mutated IGHV in the most frequent subtypes.
Stereotyped BCR
Thirty‐five of 191 patients (18·3%) had homologous IGHV CDR3 (stereotyped BCR), 17 of them (8·9%) were assigned to the seven defined major subsets, i.e. subset 8 (n = 8), subset 1 (n = 3), subset 2 (n = 2), and one each for subset 5, subset 12, subset 14 and subset 77. Thirteen of 17 cases could be assigned to a previously identified subset using ARResT/AssignSubsets tool (Bystry et al, 2015). Four sequences belonged to three minor subsets, including subsets 7 (n = 2), 13 (n = 1) and 98 (n = 1), and the remaining seven clusters, which could not be allocated to the known subsets, were numbered as Clusters 1–7 with two each for the seven clusters.
Table 1 shows the subsets or clusters of the 35 patients carrying stereotyped BCR, their IGHV subtypes and mutational status, CDR3 length, and amino acid sequence. Un‐mutated IGHV gene showed strong association with stereotyped BCR compared with non‐stereotyped BCR group (21 of 35 vs. 34 of 156; P < 0·0001). Clusters 1, 2, 3, 6 and subset 2 were composed mainly of mutated IGHV sequences whereas subsets 1, 7 and 8, and Clusters 5 and 7 were composed of un‐mutated sequences. For IGHV CDR3 length, the median amino acid (aa) length of the 191 CLL patients was 14 aa (range: 6–37). Un‐mutated IGHV‐CLL patients had a significantly longer IGHV CDR3 (median aa: 19, range: 9–37) than mutated IGHV‐CLL patients (median aa: 13, range: 6–27) (P < 0·0001).
Table 1.
IGHV CDR3 cluster distribution in 35 CLL patients with stereotyped BCR.
| Subset or cluster | Case no. | IGHV gene | Mutational status | CDR3 length | Amino acid sequence |
|---|---|---|---|---|---|
| Subset | |||||
| 1 | 53 | IGHV7‐4 | UM | 13 | AREQWLVLPYFDY |
| 1 | 95 | IGHV1‐3 | UM | 13 | AREQWLVRVYFDY |
| 1 | 211 | IGHV1‐8 | UM | 13 | ARVQWLVLDYFDY |
| 2 | 131 | IGHV3‐30 | M | 9 | ARDSYGMDV |
| 2 | 202 | IGHV3‐21 | M | 9 | ASDRNGMDV |
| 5 | 312 | IGHV1‐69 | UM | 21 | ARVKARGVITSLYYYYYYMDV |
| 8 | 23 | IGHV4‐39 | UM | 19 | ARRDGYSSSWYQRENWFDP |
| 8 | 79 | IGHV4‐39 | UM | 19 | ARRVGYSSSWYSHDNWFDP |
| 8 | 93 | IGHV4‐39 | UM | 19 | ARTAGYSSSWYSSYNWFDP |
| 8 | 130 | IGHV4‐39 | UM | 19 | ARRVGYSSSWYSTHNWFDP |
| 8 | 151 | IGHV4‐39 | UM | 19 | ARLVGYSSSWYGPYNWFDP |
| 8 | 230 | IGHV4‐39 | UM | 19 | ARGLGYSSSWYGVYNWFDP |
| 8 | 241 | IGHV4‐39 | UM | 18 | ASLNGYSSSWHSNNWFDP |
| 8 | 249 | IGHV4‐39 | UM | 18 | AKASGYSSSWYGSNWFDP |
| 12 | 214 | IGHV1‐46 | UM | 19 | ARDSYYYDSSGYYSGFFDY |
| 14 | 336 | IGHV4‐34 | M | 10 | ARGGLRRADP |
| 77 | 326 | IGHV4‐34 | M | 14 | ARGADTTGWNAFDY |
| 7 | 178 | IGHV4‐59 | UM | 24 | ARSWARDYDFWSGHRPAYYYYMDV |
| 7 | 343 | IGHV4‐59 | UM | 23 | ARATTYYDFWSGYSPYYYYYMDV |
| 13 | 288 | IGHV4‐59 | M | 18 | ARDYYCSGGTCFDWFSDL |
| 98 | 59 | IGHV3‐30 | UM | 25 | ATSVPTYYDFWSGLGDYYYYYGMDV |
| Cluster | |||||
| 1 | 275 | IGHV3‐30 | M | 12 | ANSADYGDRFDY |
| 1 | 284 | IGHV3‐74 | M | 12 | ASAGDYGDYADY |
| 2 | 159 | IGHV3‐7 | M | 11 | ARDQHRQAYNY |
| 2 | 182 | IGHV3‐7 | M | 11 | ARDQHRQAYNY |
| 3 | 238 | IGHV5‐10 | M | 16 | ARQRYYFGSGSSPMDV |
| 3 | 308 | IGHV5‐51 | M | 16 | ARQRYNFGSLLSQVDF |
| 4 | 36 | IGHV3‐7 | M | 11 | AKDGTKYSFDY |
| 4 | 180 | IGHV3‐43 | UM | 12 | AKDGSSGYLVDY |
| 5 | 133 | IGHV3‐74 | UM | 20 | ARDSGGYSYGIYYYYYGMDV |
| 5 | 298 | IGHV3‐30 | UM | 22 | ARDSTYYYDSSGYYYYYYGMDV |
| 6 | 165 | IGHV3‐74 | M | 11 | AGGEGGQCLDS |
| 6 | 306 | IGHV3‐74 | M | 11 | ARDEGGQCLDY |
| 7 | 104 | IGHV2‐5 | UM | 18 | AHSPAETLIAAPVGYFDY |
| 7 | 164 | IGHV2‐5 | UM | 18 | AHSPAETLIAAPVGYFDY |
The sequences in bold were identified by ARResT/AssignSubsets (Bystry et al, 2015).
BCR, B‐cell receptor; CDR3, complementarity‐determining region 3; CLL, chronic lymphocytic leukaemia; M, mutated; UM, un‐mutated.
Cytogenetic and genetic lesions
Cytogenetic abnormalities were detected in 80 of 183 (43·7%) for del(13q), 35 of 188 (18·6%) for trisomy 12, 19 of 183 (10·4%) for del(17p), 13 of 188 (6·9%) for del(11q) and 18 of 179 (10·1%) for IGH translocation. The mutational status of TP53, NOTCH1 and SF3B1 was analysed in 186 patients with a frequency of 8·1%, 8·6% and 9·7%, respectively; 31 of 183 patients (16·9%) had ATM mutations, and 2 of 183 patients (1·1%) had BIRC3 mutations. The results of gene mutations in CLL patients are summarized in Fig 2. Of the 183 patients with mutational status available for all 5 genes, 12 patients had 2 concurrent mutations: 5 co‐existing SF3B1and ATM mutations; 2 each co‐existing SF3B1 and NOTCH1, and ATM and TP53 mutations; one each co‐existing NOTCH1 and ATM, NOTCH1 and BIRC3, and SF3B1 and TP53 mutations. NOTCH1 and TP53 mutations were mutually exclusive. The remaining patients had only one mutation. Correlations of gene mutations with cytogenetic abnormalities demonstrated that TP53 mutations were closely associated with del(17p) compared with no del(17p) (42·1% vs. 4·4%; P < 0·0001), and ATM mutations correlated with del(11q) compared with no del(11q) (53·8% vs. 14·4%; P = 0·0019).
Figure 2.
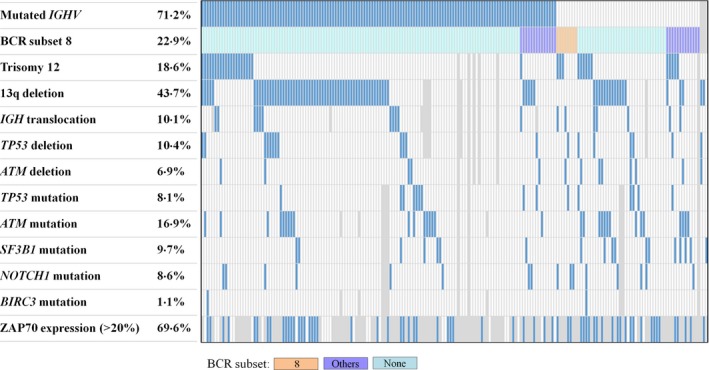
Correlation among the genetic lesions in 194 patients with chronic lymphocytic leukaemia. Areas not examined are indicated in grey. BCR, B‐cell receptor.
Among the gene mutations analysed, SF3B1 mutations were significantly associated with un‐mutated IGHV (10 of 52 vs. 7 of 131; P = 0·0083) and absent for stereotyped BCR subset 8. There was no correlation between mutated genes and ZAP70 > 20% but a higher frequency of un‐mutated IGHV in patients with ZAP70 > 20% (P = 0·0064) was observed: 25 (39·7%) of the 63 patients with ZAP70 > 20% had un‐mutated IGHV compared with 3 (10·7%) of the 28 patients with ZAP70 ≦ 20% had un‐mutated IGHV. Although no correlation of SF3B1, NOTCH1 or TP53 mutations with stereotyped BCR subsets was observed, we found TP53‐mutated cases had a higher frequency of IGHV1‐69 usage compared with TP53‐unmutated cases (3 of 15 vs. 8 of 168; P = 0·0495).
Prognostic relevance of cytogenetic/genetic abnormalities
With a median follow‐up of 49·6 months, the median TFS and OS was 24·4 months [95% confidence interval (CI): 13·6–35·2] and 110·0 months (95% CI: 82·3–136·7), respectively. The impact of cytogenetic abnormalities on outcomes was analysed. Patients with cytogenetic lesions had a comparable TFS among all groups (P = 0·215) or between any two different groups except that patients with trisomy 12 had a shorter TFS than those with del(13q) (median, 19·4 months vs. 30·0 months, P = 0·028). In contrast, there was a significant difference in OS among different cytogenetic groups (P = 0·001). The median OS of patients with del(13q), del(11q), trisomy 12, del(17p) and normal karyotypes was 154·6 months, 88·5 months, 80·5 months, 44·3 months and 95·8 months, respectively. Patients with trisomy 12 had an inferior OS compared to those with del(13q) (P = 0·0001), or those with normal karyotypes (P = 0·037).
Prognostic impact of the genetic abnormalities in CLL is shown in Table 2. By univariate analysis, TFS was significantly shorter in patients with un‐mutated IGHV (median, 7·7 months vs. 36·2 months; P = 0·001), subset 8 (median, 5·1 months vs. 28·0 months; P = 0·001), trisomy12 (median, 19·4 months vs. 27·7 months; P = 0·044), TP53 disruption (TP53 mutations and/or 17p deletions) (median, 10·6 months vs. 30·0 months, P = 0·038), SF3B1 mutations (median, 1·7 months vs. 32·7 months, P = 0·001), and ZAP70 > 20% (median, 13·1 months vs. 84·5 months, P = 0·009). OS was significantly worse in patients with un‐mutated IGHV (median, 63·0 months vs. 144·2 months; P < 0·0001), subset 8 (median, 41·1 months vs. 114·4 months; P = 0·001), trisomy 12 (median, 80·5 months vs. 153 months; P = 0·001), negative for del(13q) (median, 88·1 months vs. 154·6 months, P = 0·016), TP53 disruption (median, 44·3 months vs. 123·0 months, P = 0·003) and SF3B1 mutations (median, 58·0 months vs. 126·3 months, P = 0·0001). In multivariate analysis (Table 3), the independent predicators for inferior TFS included SF3B1 mutations [Hazard ratio (HR) = 2·942, 95% CI: 1·217–7·114; P = 0·017], trisomy12 (HR = 1·997, 95% CI: 1·009–3·951; P = 0·047), and a borderline level for TP53 disruption (HR = 1·876, 95% CI: 0·902–3·904; P = 0·092) and ZAP70 > 20% (HR = 2·070, 95% CI: 0·984–4·356; P = 0·055). Independent predicators for inferior OS included un‐mutated IGHV (mutated IGHV: HR = 0·487, 95% CI: 0·262–0·907; P = 0·023), trisomy 12 (HR = 2·301, 95% CI: 1·150–3·951; P = 0·007), TP53 disruption (HR = 3·667, 95% CI: 2·025–6·639; P < 0·0001), SF3B1 mutations (HR = 2·786, 95% CI: 1·239–6·267; P = 0·013) and at a borderline significant level for BCR subset 8 (HR = 2·307, 95% CI: 0·896–5·938; P = 0·083) (Table 3).
Table 2.
Risk factors that influence the outcome of 194 Taiwanese CLL patients.
| Feature | Treatment‐free survival (months) | Overall survival (months) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | P | Positive | Negative | P | |||||
| N | Median (95% CI) | N | Median (95% CI) | N | Median (95% CI) | N | Median (95% CI) | |||
| Mutated IGHV | 129 | 36·2 (21·0–51·4) | 51 | 7·7 (0·8–14·6) | 0·001 | 136 | 144·2 (112·6–175·8) | 55 | 63·0 (44·4–81·6) | <0·0001 |
| BCR subset 8 | 7 | 5·1 (0·0–15·6) | 176 | 28·0 (15·9–40·1) | 0·001 | 8 | 41·1 (22·4–59·8) | 186 | 114·4 (84·9–143·7) | 0·001 |
| Trisomy 12 | 32 | 19·4 (5·6–33·2) | 145 | 27·7 (12·7–42·7) | 0·044 | 35 | 80·5 (35·4–125·6) | 153 | 144·2 (86·4–202·0) | 0·001 |
| 13q deletion | 75 | 30·0 (14·5–45·5) | 98 | 14·3 (7·2–21·4) | 0·193 | 80 | 154·6 (120·8–188·4) | 103 | 88·1 (57·4–118·8) | 0·016 |
| IGH translocation | 15 | 22·8 (0·0–58·1) | 155 | 33·7 (14·1–33·3) | 0·795 | 18 | 175·3 | 161 | 110·0 (69·6–150·4) | 0·430 |
| TP53 disruption | 26 | 10·6 (3·0–18·2) | 142 | 30·0 (14·3–45·7) | 0·038 | 26 | 44·3 (8·8–79·8) | 152 | 123·0 (76·8–169·2) | 0·003 |
| ATM disruption | 45 | 16·7 (5·8–27·6) | 134 | 28·6 (10·0–46·7) | 0·254 | 47 | 95·8 (57·8–133·8) | 143 | 114·4 (81·5–147·3) | 0·961 |
| SF3B1 mutation | 17 | 1·7 (0·0–6·8) | 158 | 32·7 (18·8–46·6) | 0·001 | 18 | 58·0 (32·4–83·6) | 168 | 126·3 (80·6–172·0) | 0·0001 |
| NOTCH1 mutation | 14 | 13·6 (11·2–16·0) | 161 | 30·0 (15·5–44·5) | 0·092 | 16 | 57·7 (37·7–77·7) | 170 | 110 (80·1–139·9) | 0·340 |
| BIRC3 mutation | 2 | 0·3 | 170 | 27·7 (14·4–41·0) | 0·144 | 2 | 4·3 | 181 | 110·0 (82.‐137·8) | 0·070 |
| ZAP70 expression (>20%) | 62 | 13·1 (6·7–19·5) | 28 | 84·5 | 0·009 | 64 | 96·8 (45·2–148·4) | 28 | 173·6 | 0·141 |
BCR, B‐cell receptor; CI, confidence interval; CLL, chronic lymphocytic leukaemia; N; number of cases.
Table 3.
Survival by multivariate analysis of risk factors in Taiwanese CLL patients.
| Feature | Treatment‐free survival | Overall survival | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate | Multivariate | Univariate | Multivariate | |||||||||
| HR | 95% CI | P value | HR | 95% CI | P value | HR | 95% CI | P value | HR | 95% CI | P value | |
| Mutated IGHV | 0·517 | 0·347–0·769 | 0·001 | 0·870 | 0·433–1·747 | 0·695 | 0·386 | 0·242–0·617 | <0·0001 | 0·487 | 0·262–0·907 | 0·023 |
| BCR subset 8 | 3·386 | 1·556–7·367 | 0·002 | 1·444 | 0·394–5·300 | 0·579 | 3·643 | 1·645–8·065 | 0·001 | 2·307 | 0·896–5·938 | 0·083 |
| Trisomy 12 | 1·613 | 1·005–2·587 | 0·048 | 1·997 | 1·009–3·951 | 0·047 | 2·339 | 1·370–3·992 | 0·002 | 2·301 | 1·150–3·951 | 0·007 |
| 13q deletion | 0·776 | 0·528–1·140 | 0·196 | 0·561 | 0·348–0·904 | 0·018 | 0·801 | 0·469–1·369 | 0·417 | |||
| TP53 disruption | 1·689 | 1·021–2·794 | 0·041 | 1·876 | 0·902–3·904 | 0·092 | 2·313 | 1·320–4·054 | 0·003 | 3·667 | 2·025–6·639 | <0·0001 |
| SF3B1 mutation | 2·715 | 1·504–4·904 | 0·001 | 2·942 | 1·217–7·114 | 0·017 | 3·148 | 1·695–5·846 | 0·0003 | 2·786 | 1·239–6·267 | 0·013 |
| ZAP70 expression (>20) | 2·421 | 1·219–4·809 | 0·012 | 2·070 | 0·984–4·356 | 0·055 | 2·185 | 0·752–6·353 | 0·151 | |||
BCR, B‐cell receptor; CI, confidence interval; CLL, chronic lymphocytic leukaemia; HR, hazard ratio.
We divided the patients into four genetic groups based on TP53 disruption and IGHV mutational status. Patients with wild‐type TP53 and mutated IGHV had a longer TFS compared to those with TP53 disruption and un‐mutated IGHV, those with wild‐type TP53 and un‐mutated IGHV, and those with TP53 disruption and mutated IGHV (P = 0·001) (Fig 3A). As shown in Fig 3B, patients with wild‐type TP53 and mutated IGHV had a superior OS compared with those with wild‐type TP53 and un‐mutated IGHV (P < 0·0001). OS of patients with TP53 disruption and mutated IGHV was also significantly different from patients with TP53 disruption and un‐mutated IGHV (P = 0·040). A significant difference was also observed between mutated IGHV patients with and without TP53 disruption (P = 0·002), and between un‐mutated IGHV patients with and without TP53 disruption (P = 0·005). Patients with wild‐type TP53 and mutated IGHV had the longest OS among the four groups (P < 0·0001).
Figure 3.
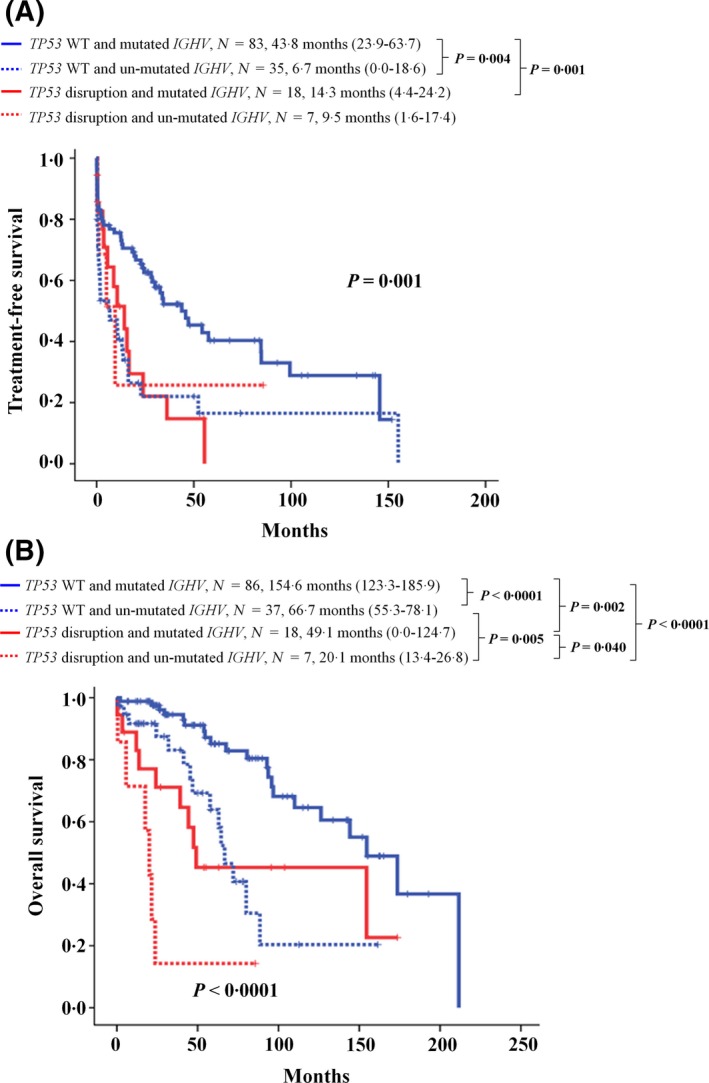
Kaplan–Meier estimates of (A) treatment‐free survival and (B) overall survival according to TP53 disruption and IGHV mutational status in Taiwanese chronic lymphocytic leukaemia patients.
Discussion
A frequency of 71·2% mutated IGHV genes in our Taiwanese patients with CLL was comparable with a Japanese study (78·8%) (Nakahashi et al, 2009) and a study from Tianjin, China (75·3%) (Marinelli et al, 2016). As shown in Table SI, the frequency of IGHV hypermutation was higher in Asian countries. The distribution of IGHV family usage in our cohort followed an order of IGHV3>IGHV4>IGHV1, which was similar to that observed in other Asian cohorts (Nakahashi et al, 2009; Marinelli et al, 2016) but different from IGHV3>IGHV1>IGHV4 hierarchy in the West (Agathangelidis et al, 2012; Marinelli et al, 2016). Our findings confirmed that IGHV gene repertoire in CLL is geographically heterogeneous. The present study showed that the frequency of mutated IGHV was higher in IGHV3 and IGHV4 families than that of un‐mutated IGHV whereas IGHV1 family carried predominantly un‐mutated IGHV, which was in line with the studies of Caucasians (Duke et al, 2003; Karan‐Djurasevic et al, 2012; Rani et al, 2016). Notably, IGHV3‐23 was most frequently expressed in the present study and another Taiwanese study (Wu et al, 2017), which were different from all other Asian countries in which IGHV4‐34 was the most frequently expressed gene in Japan and China (Nakahashi et al, 2009; Marinelli et al, 2016). On the contrary, the most frequently used gene in the Western studies was IGHV1‐69 (Agathangelidis et al, 2012; Marinelli et al, 2016) (Table SI).
Based on the stringent criteria for the analysis of stereotype in IGHV CDR3 amino acid sequences (Darzentas et al, 2010), the expression frequency of stereotyped BCR was lower than those observed in Western studies (Stamatopoulos et al, 2007; Agathangelidis et al, 2012; Marinelli et al, 2016) (18·3% vs. 25–30%). The length of IGHV CDR3 in the mutated CLL patients was significantly smaller than that in the un‐mutated CLL cases in the present study, as well as in other Asian or Western countries (Bianchi et al, 2010; Agathangelidis et al, 2012; Marinelli et al, 2016; Rani et al, 2016), suggesting that mutated IGHV and smaller CDR3 length might work together to affect the antibody‐binding pocket for antigen. According to the studies listed in Table SI, there was a reverse correlation of the frequency of patients with stereotyped BCR and mutated IGHV. The higher frequency of mutated IGHV might be attributed to the lower frequency of stereotyped BCR in our Taiwanese cohort.
We also observed that the frequency of major subsets of stereotyped BCR was lower in Taiwan and China compared with that in the West. Among the reported 19 major subsets, subset 8 was the most frequent subset in our patients, which was the least frequent subset in the Western cohort (Agathangelidis et al, 2012; Marinelli et al, 2016). These also supported an ethnicity‐dependent association of IGHV usage. In addition, previous studies showed that stereotyped BCRs were of prognostic relevance: subset 1 or 2 was associated with poor outcome in the Western studies (Maura et al, 2011; Strefford et al, 2013; Baliakas et al, 2014; Del Giudice et al, 2014), whereas patients with subset 4 might not require treatment (Rani et al, 2016). Both the present study and that reported by Marinelli et al. (2016) observed that patients with subset 8 had an unfavourable survival.
The cytogenetic and mutational analyses were performed at initial diagnosis (untreated samples) in all our patients. The frequencies of cytogenetic abnormalities in the present study were in line with those reported from European descendants with CLL at initial diagnosis, except for del(11q). The incidence of del(11q) (6·9%) was slightly lower in our series than that in Western studies (10–25%) (Dohner et al, 2000; Amare et al, 2013). A lower frequency of del(11q) was also observed in a Chinese study (9·5%) (Xu et al, 2008), a Korean study (12·5%) (Yoon et al, 2014) and another Taiwanese study (11%) (Wu et al, 2017). These data suggested that del(11q) was relatively lower in Asia compared with that in the West.
The mutation rate of TP53 (8·1%) in the present study was comparable to 8% of 166 diagnostic CLL samples while the mutation frequency was 15% in 307 CLL patients of all stages in a report from China (Xia et al, 2015) and Western studies (Takahashi et al, 2018; Leeksma et al, 2019). The frequency of 8·1% for TP53 mutations was lower than 20·5% of a small Taiwanese cohort (Wu et al, 2017). It is plausible that different times of sample collection from untreated patients was one of the major reasons to explain these discrepancies. The relationship among molecular alterations in our study was mostly consistent with the reported data, including that NOTCH1 mutations occurred exclusively without TP53 mutations (Rossi et al, 2012) and un‐mutated IGHV was frequently associated with SF3B1 mutations (Xia et al, 2015). However, more IGHV‐mutated patients in our cohort had ZAP70 expression compared with that of reported data (Wiestner et al, 2003). The higher frequency of more than 70% of IGHV hypermutation in our CLL patients might partly explain the lower correlation between un‐mutated IGHV and ZAP70 expression in our cohort. In addition, we also failed to find associations of un‐mutated IGHV with TP53 or NOTCH1 mutations (Xia et al, 2015) and the correlation of stereotyped BCR subsets with any gene mutations (Sutton et al, 2016). Notably, the present result showed that TP53 mutations were especially common in cases using IGHV1‐69, suggesting that IGHV usage rather than subsets of stereotyped BCR correlated with gene mutations.
Our patients were consecutively diagnosed and were regularly followed‐up in a single institution, a tertiary referral centre; however, in our health care system, every patient could visit the tertiary centre directly without following the referral procedure. As the prevalence of CLL is very low and no standard protocol or guideline was available in the earlier years, it was possible that physicians might give oral alkylating agent (chlorambucil) to asymptomatic patients, making the TFS shorter. We believe that OS, rather than the TFS, is more representative of outcome in our CLL cohort. The survival of our patients was inferior to that reported by previous studies from Western countries (Hamblin et al, 1999; Dohner et al, 2000), to which we have compared our series with regard to the distribution of clinical stages; no differences were observed. The worse survival and earlier progression of Asian CLL patients were also previously reported from South Asia (India, Pakistan or Bangladesh), China and Taiwan (Gunawardana et al, 2008; Wu et al, 2013; Marinelli et al, 2016), suggesting that the inferior outcome of the present series compared with that of Western patients might be also attributed to ethnic differences. The underlying mechanisms require further investigation. In agreement with the previous studies, our results showed that patients with del(13q) had the most favourable OS and those with del(17p) had the most unfavourable OS (Dohner et al, 2000; Rossi et al, 2013; Fischer et al, 2016). However, the results of the impact of del(11q) or trisomy 12 on outcome were conflicting among different series (Rossi et al, 2013; Hernandez et al, 2015; Fischer et al, 2016; Gonzalez‐Gascon et al, 2016). Our results showed that there was no effect on TFS in patients with trisomy 12 or del(11q) but a shorter OS was found in patients with trisomy 12 compared with that of del(11q) patients, and a shorter OS compared with patients without four types of cytogenetic lesions. Three (8·6%) of the 35 patients with trisomy 12 carried a poor risk factor of BCR subset 8 compared with 5 (3·3%) of the 150 patients without trisomy 12 carrying BCR subset 8, which might partly explain one of the reasons for the poor outcome of patients with trisomy 12. However, it will be necessary to confirm this association by enrolling more cases with BCR subset 8. In addition, we showed that patients without TP53 disruption and un‐mutated IGHV had the longest TFS and OS compared with patients with other groups of different combination of the two unfavourable factors of outcome, which was also reported in other studies (Xia et al, 2015; Fischer et al, 2016).
To summarise, in a relatively large Taiwanese cohort of CLL, we showed a high frequency of mutated IGHV (71·2%), low frequency of stereotyped BCR (18·3%), the most frequent usage of IGHV3‐23 gene, lower frequency of del(11q) (6·9%) and the most frequent stereotyped BCR subset 8. Our results showed that IGHV features and occurrence of del(11q) are ethnicity‐dependent associated. In addition, un‐mutated IGHV, TP53 disruption, trisomy 12, and SF3B1 mutations were independent predictors for inferior OS.
Authorship contributions
LY designed and supervised the study; MC, H, PN, JH, YM, MC, TC, CY, and LY provided patients’ samples and their clinical data; LY and YJ developed the methodology, LY and YJ analysed and interpreted the data, YJ and LY wrote the manuscript.
Disclosure of conflict of interest
The authors have no conflict interest.
Supporting information
Table SI. Comparative analysis of IGHV features of Taiwanese CLL patients with other studies.
Acknowledgements
This work was supported by Chang Gung Memorial Hospital (CMRPG3E0271, CMRPG3E0272, and CMRPG3E0273). We thank Mr. Tung‐Huei Lin for statistical analysis and Ms. Ting‐Yu Huang for secretarial assistance.
References
- Agathangelidis, A. , Darzentas, N. , Hadzidimitriou, A. , Brochet, X. , Murray, F. , Yan, X.J. , Davis, Z. , van Gastel‐Mol, E.J. , Tresoldi, C. , Chu, C.C. , Cahill, N. , Giudicelli, V. , Tichy, B. , Pedersen, L.B. , Foroni, L. , Bonello, L. , Janus, A. , Smedby, K. , Anagnostopoulos, A. , Merle‐Beral, H. , Laoutaris, N. , Juliusson, G. , di Celle, P.F. , Pospisilova, S. , Jurlander, J. , Geisler, C. , Tsaftaris, A. , Lefranc, M.P. , Langerak, A.W. , Oscier, D.G. , Chiorazzi, N. , Belessi, C. , Davi, F. , Rosenquist, R. , Ghia, P. & Stamatopoulos, K. (2012) Stereotyped B‐cell receptors in one‐third of chronic lymphocytic leukemia: a molecular classification with implications for targeted therapies. Blood, 119, 4467–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amare, P.S. , Gadage, V. , Jain, H. , Nikalje, S. , Manju, S. , Mittal, N. , Gujral, S. & Nair, R. (2013) Clinico‐pathological impact of cytogenetic subgroups in B‐cell chronic lymphocytic leukemia: experience from India. Indian Journal of Cancer, 50, 261–267. [DOI] [PubMed] [Google Scholar]
- Baliakas, P. , Hadzidimitriou, A. , Sutton, L.A. , Minga, E. , Agathangelidis, A. , Nichelatti, M. , Tsanousa, A. , Scarfò, L. , Davis, Z. , Yan, X.J. , Shanafelt, T. , Plevova, K. , Sandberg, Y. , Vojdeman, F.J. , Boudjoghra, M. , Tzenou, T. , Chatzouli, M. , Chu, C.C. , Veronese, S. , Gardiner, A. , Mansouri, L. , Smedby, K.E. , Pedersen, L.B. , van Lom, K. , Giudicelli, V. , Francova, H.S. , Nguyen‐Khac, F. , Panagiotidis, P. , Juliusson, G. , Angelis, L. , Anagnostopoulos, A. , Lefranc, M.P. , Facco, M. , Trentin, L. , Catherwood, M. , Montillo, M. , Geisler, C.H. , Langerak, A.W. , Pospisilova, S. , Chiorazzi, N. , Oscier, D. , Jelinek, D.F. , Darzentas, N. , Belessi, C. , Davi, F. , Rosenquist, R. , Ghia, P. & Stamatopoulos, K. (2014) Clinical effect of stereotyped B‐cell receptor immunoglobulins in chronic lymphocytic leukaemia: a retrospective multicentre study. The Lancet Haematology, 1, e74–e84. [DOI] [PubMed] [Google Scholar]
- Bianchi, S. , Moreno, P. , Landoni, A.I. , Naya, H. , Oppezzo, P. , Dighiero, G. , Gabus, R. & Pritsch, O. (2010) Immunoglobulin heavy chain V‐D‐J gene rearrangement and mutational status in Uruguayan patients with chronic lymphocytic leukemia. Leukaemia & Lymphoma, 51, 2070–2078. [DOI] [PubMed] [Google Scholar]
- Bystry, V. , Agathangelidis, A. , Bikos, V. , Sutton, L.A. , Baliakas, P. , Hadzidimitriou, A. , Stamatopoulos, K. & Darzentas, N. (2015) ARResT/AssignSubsets: a novel application for robust subclassification of chronic lymphocytic leukemia based on B cell receptor IG stereotypy. Bioinformatics, 31, 3844–3846. [DOI] [PubMed] [Google Scholar]
- Chen, L. , Zhang, Y. , Zheng, W. , Wu, Y. , Qiao, C. , Fan, L. , Xu, W. & Li, J. (2008) Distinctive IgVH gene segments usage and mutation status in Chinese patients with chronic lymphocytic leukemia. Leukemia Research, 32, 1491–1498. [DOI] [PubMed] [Google Scholar]
- Crespo, M. , Bosch, F. , Villamor, N. , Bellosillo, B. , Colomer, D. , Rozman, M. , Marce, S. , Lopez‐Guillermo, A. , Campo, E. & Montserrat, E. (2003) ZAP‐70 expression as a surrogate for immunoglobulin‐variable‐region mutations in chronic lymphocytic leukemia. New England Journal of Medicine, 348, 1764–1775. [DOI] [PubMed] [Google Scholar]
- Damle, R.N. , Wasil, T. , Fais, F. , Ghiotto, F. , Valetto, A. , Allen, S.L. , Buchbinder, A. , Budman, D. , Dittmar, K. , Kolitz, J. , Lichtman, S.M. , Schulman, P. , Vinciguerra, V.P. , Rai, K.R. , Ferrarini, M. & Chiorazzi, N. (1999) Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood, 94, 1840–1847. [PubMed] [Google Scholar]
- Darzentas, N. , Hadzidimitriou, A. , Murray, F. , Hatzi, K. , Josefsson, P. , Laoutaris, N. , Moreno, C. , Anagnostopoulos, A. , Jurlander, J. , Tsaftaris, A. , Chiorazzi, N. , Belessi, C. , Ghia, P. , Rosenquist, R. , Davi, F. & Stamatopoulos, K. (2010) A different ontogenesis for chronic lymphocytic leukemia cases carrying stereotyped antigen receptors: molecular and computational evidence. Leukemia, 24, 125–132. [DOI] [PubMed] [Google Scholar]
- Dohner, H. , Stilgenbauer, S. , Benner, A. , Leupolt, E. , Krober, A. , Bullinger, L. , Dohner, K. , Bentz, M. & Lichter, P. (2000) Genomic aberrations and survival in chronic lymphocytic leukemia. New England Journal of Medicine, 343, 1910–1916. [DOI] [PubMed] [Google Scholar]
- Duke, V.M. , Gandini, D. , Sherrington, P.D. , Lin, K. , Heelan, B. , Amlot, P. , Mehta, A.B. , Hoffbrand, A.V. & Foroni, L. (2003) V(H) gene usage differs in germline and mutated B‐cell chronic lymphocytic leukemia. Haematologica, 88, 1259–1271. [PubMed] [Google Scholar]
- Fais, F. , Ghiotto, F. , Hashimoto, S. , Sellars, B. , Valetto, A. , Allen, S. , Schulman, P. , Vinciguerra, V.P. , Rai, K. , Rassenti, L.Z. , Kipps, T.J. , Dighiero, G. , Schroeder, H.W.J. , Ferrarini, M. & Chiorazzi, N. (1998) Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. Journal of Clinical Investigation, 102, 1515–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, K. , Bahlo, J. , Fink, A.M. , Goede, V. , Herling, C.D. , Cramer, P. , Langerbeins, P. , von Tresckow, J. , Engelke, A. , Maurer, C. , Kovacs, G. , Herling, M. , Tausch, E. , Kreuzer, K.A. , Eichhorst, B. , Bottcher, S. , Seymour, J.F. , Ghia, P. , Marlton, P. , Kneba, M. , Wendtner, C.M. , Dohner, H. , Stilgenbauer, S. & Hallek, M. (2016) Long‐term remissions after FCR chemoimmunotherapy in previously untreated patients with CLL: updated results of the CLL8 trial. Blood, 127, 208–215. [DOI] [PubMed] [Google Scholar]
- Foa, R. , Del Giudice, I. , Guarini, A. , Rossi, D. & Gaidano, G. (2013) Clinical implications of the molecular genetics of chronic lymphocytic leukemia. Haematologica, 98, 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Giudice, I. , Chiaretti, S. , Santangelo, S. , Tavolaro, S. , Peragine, N. , Marinelli, M. , Ilari, C. , Raponi, S. , Messina, M. , Nanni, M. , Mauro, F.R. , Piciocchi, A. , Bontempi, K. , Rossi, D. , Gaidano, G. , Guarini, A. & Foà, R. (2014) Stereotyped subset #1 chronic lymphocytic leukemia: a direct link between B‐cell receptor structure, function, and patients’ prognosis. American Journal of Hematology, 89, 74–82. [DOI] [PubMed] [Google Scholar]
- Gonzalez‐Gascon, Y.M.I. , Hernandez‐Sanchez, M. , Rodriguez‐Vicente, A.E. , Sanzo, C. , Aventin, A. , Puiggros, A. , Collado, R. , Heras, C. , Munoz, C. , Delgado, J. , Ortega, M. , Gonzalez, M.T. , Marugan, I. , de la Fuente, I. , Recio, I. , Bosch, F. , Espinet, B. , Gonzalez, M. , Hernandez‐Rivas, J.M. & Hernandez, J.A. (2016) A high proportion of cells carrying trisomy 12 is associated with a worse outcome in patients with chronic lymphocytic leukemia. Hematological Oncology, 34, 84–92. [DOI] [PubMed] [Google Scholar]
- Gunawardana, C. , Austen, B. , Powell, J.E. , Fegan, C. , Wandroo, F. , Jacobs, A. , Pratt, G. & Moss, P. (2008) South Asian chronic lymphocytic patinets have more rapid disease progression in comparison to White patients. British Journal of Haematology, 142, 606–609. [DOI] [PubMed] [Google Scholar]
- Hallek, M. , Cheson, B.D. , Catovsky, D. , Caligaris‐Cappio, F. , Dighiero, G. , Döhner, H. , Hillmen, P. , Keating, M.J. , Montserrat, E. , Rai, K.R. & Kipps, T.J. (2008) Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute‐Working Group 1996 guidelines. Blood, 111, 5446–5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblin, T.J. , Davis, Z. , Gardiner, A. , Oscier, D.G. & Stevenson, F.K. (1999) Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood, 94, 1848–1854. [PubMed] [Google Scholar]
- Hernandez, J.A. , Hernandez‐Sanchez, M. , Rodriguez‐Vicente, A.E. , Grossmann, V. , Collado, R. , Heras, C. , Puiggros, A. , Martin, A.A. , Puig, N. , Benito, R. , Robledo, C. , Delgado, J. , Gonzalez, T. , Queizan, J.A. , Galende, J. , de la Fuente, I. , Martin‐Nunez, G. , Alonso, J.M. , Abrisqueta, P. , Luno, E. , Marugan, I. , Gonzalez‐Gascon, I. , Bosch, F. , Kohlmann, A. , Gonzalez, M. , Espinet, B. & Hernandez‐Rivas, J.M. (2015) A low frequency of losses in 11q chromosome is associated with better outcome and lower rate of genomic mutations in patients with chronic lymphocytic leukemia. PLoS ONE, 10, e0143073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojjat‐Farsangi, M. , Jeddi‐Tehrani, M. , Razavi, S.M. , Sharifian, R.A. , Mellstedt, H. , Shokri, F. & Rabbani, H. (2009) Immunoglobulin heavy chain variable region gene usage and mutational status of the leukemic B cells in Iranian patients with chronic lymphocytic leukemia. Cancer Science, 100, 2346–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karan‐Djurasevic, T. , Palibrk, V. , Kostic, T. , Spasovski, V. , Nikcevic, G. , Srzentic, S. , Colovic, M. , Colovic, N. , Vidovic, A. , Antic, D. , Mihaljevic, B. , Pavlovic, S. & Tosic, N. (2012) Mutational status and gene repertoire of IGHV‐IGHD‐IGHJ rearrangements in Serbian patients with chronic lymphocytic leukemia. Clinical Lymphoma Myeloma and Leukemia, 12, 252–260. [DOI] [PubMed] [Google Scholar]
- Leeksma, A.C. , Taylor, J. , Wu, B. , Gardner, J.R. , He, J. , Nahas, M. , Gonen, M. , Alemayehu, W.G. , Te Raa, D. , Walther, T. , Hullein, J. , Dietrich, S. , Claus, R. , de Boer, F. , de Heer, K. , Dubois, J. , Dampmann, M. , Durig, J. , van Oers, M.H.J. , Geisler, C.H. , Eldering, E. , Levine, R.L. , Miller, V. , Mughal, T. , Lamanna, N. , Frattini, M.G. , Heaney, M.L. , Zelenetz, A. , Zenz, T. , Abdel‐Wahab, O. & Kater, A.P. (2019) Clonal diversity predicts adverse outcome in chronic lymphocytic leukemia. Leukemia, 33, 390–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinelli, M. , Ilari, C. , Xia, Y. , Del Giudice, I. , Cafforio, L. , Della Starza, L. , Raponi, S. , Mariqlia, P. , Bonina, S. , Yu, Z. , Yang, W. , Qiu, L. , Chan, T. , Piciocchi, A. , Kwong, Y.L. , Tse, E. , Li, J. , Guarini, A. , Xu, W. & Foà, R. (2016) Immunoglobulin gene rearrangements in Chinese and Italian patients with chronic lymphocytic leukemia. Oncotarget, 7, 20520–20531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maura, F. , Cutrona, G. , Fabris, S. , Colombo, M. , Tuana, G. , Agnelli, L. , Matis, S. , Lionetti, M. , Gentile, M. , Recchia, A.G. , Di Raimondo, F. , Musolino, C. , Ilariucci, F. , Di Renzo, N. , Pesce, E. , Molica, S. , Federico, M. , Cortelezzi, A. , Morabito, F. , Ferrarini, M. & Neri, A. (2011) Relevance of stereotyped B‐cell receptors in the context of the molecular, cytogenetic and clinical features of chronic lymphocytic leukemia. PLoS ONE, 6, e24313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, H. , Wierda, W.G. , Barron, L.L. , Cromwell, C.C. , Wang, J. , Coombes, K.R. , Rangel, R. , Elenitoba‐Johnson, K.S. , Keating, M.J. & Abruzzo, L.V. (2003) High expression of activation‐induced cytidine deaminase (AID) and splice variants is a distinctive feature of poor‐prognosis chronic lymphocytic leukemia. Blood, 101, 4903–4908. [DOI] [PubMed] [Google Scholar]
- Morton, L.M. , Wang, S.S. , Devesa, S.S. , Hartge, P. , Weisenburger, D.D. & Linet, M.S. (2006) Lymphoma incidence patterns by WHO subtype in the United States, 1992–2001. Blood, 107, 265–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahashi, H. , Tsukamoto, N. , Hashimoto, Y. , Koiso, H. , Yokohama, A. , Saitoh, T. , Uchiumi, H. , Handa, H. , Murakami, H. , Nojima, Y. & Karasawa, M. (2009) Characterization of immunoglobulin heavy and light chain gene expression in chronic lymphocytic leukemia and related disorders. Cancer Science, 100, 671–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oscier, D.G. , Gardiner, A.C. , Mould, S.J. , Glide, S. , Davis, Z.A. , Ibbotson, R.E. , Corcoran, M.M. , Chapman, R.M. , Thomas, P.W. , Copplestone, J.A. , Orchard, J.A. & Hamblin, T.J. (2002) Multivariate analysis of prognostic factors in CLL: clinical stage, IGVH gene mutational status, and loss or mutation of the p53 gene are independent prognostic factors. Blood, 100, 1177–1184. [PubMed] [Google Scholar]
- Puiggros, A. , Blanco, G. & Espinet, B. (2014) Genetic abnormalities in chronic lymphocytic leukemia: where we are and where we go. BioMed Research International, 2014, 435983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani, L. , Mathur, N. , Gogia, A. , Vishnubhatla, S. , Kumar, L. , Sharma, A. , Dube, D. , Kaur, P. & Gupta, R. (2016) Immunoglobulin heavy chain variable region gene repertoire and B‐cell receptor stereotypes in Indian patients with chronic lymphocytic leukemia. Leukaemia & Lymphoma, 57, 2389–2400. [DOI] [PubMed] [Google Scholar]
- Richardson, S.J. , Matthews, C. , Catherwood, M.A. , Alexander, H.D. , Carey, B.S. , Farrugia, J. , Gardiner, A. , Mould, S. , Oscier, D. , Copplestone, J.A. & Prentice, A.G. (2006) ZAP‐70 expression is associated with enhanced ability to respond to migratory and survival signals in B‐cell chronic lymphocytic leukemia (B‐ALL). Blood, 107, 3584–3592. [DOI] [PubMed] [Google Scholar]
- Rossi, D. , Rasi, S. , Fabbri, G. , Spina, V. , Fangazio, M. , Forconi, F. , Marasca, R. , Laurenti, L. , Bruscaggin, A. , Cerri, M. , Monti, S. , Cresta, S. , Famà, R. , De Paoli, L. , Bulian, P. , Gattei, V. , Guarini, A. , Deaglio, S. , Capello, D. , Rabadan, R. , Pasqualucci, L. , Dalla‐Favera, R. , Foà, R. & Gaidano, G. (2012) Mutations of NOTCH1 are an independent predictor of survival in chronic lymphocytic leukemia. Blood, 119, 521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi, D. , Rasi, S. , Spina, V. , Bruscaggin, A. , Monti, S. , Ciardullo, C. , Deambrogi, C. , Khiabanian, H. , Serra, R. , Bertoni, F. , Forconi, F. , Laurenti, L. , Marasca, R. , Dal‐Bo, M. , Rossi, F.M. , Bulian, P. , Nomdedeu, J. , Del Poeta, G. , Gattei, V. , Pasqualucci, L. , Rabadan, R. , Foa, R. , Dalla‐Favera, R. & Gaidano, G. (2013) Integrated mutational and cytogenetic analysis identifies new prognostic subgroups in chronic lymphocytic leukemia. Blood, 121, 1403–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozman, C. & Montserrat, E. (1995) Chronic lymphocytic leukemia. New England Journal of Medicine, 333, 1052–1057. [DOI] [PubMed] [Google Scholar]
- Stamatopoulos, K. , Belessi, C. , Moreno, C. , Boudjograh, M. , Guida, G. , Smilevska, T. , Belhoul, L. , Stella, S. , Stavroyianni, N. , Crespo, M. , Hadzidimitriou, A. , Sutton, L. , Bosch, F. , Laoutaris, N. , Anagnostopoulos, A. , Montserrat, E. , Fassas, A. , Dighiero, G. , Caligaris‐Cappio, F. , Merle‐Beral, H. , Ghia, P. & Davi, F. (2007) Over 20% of patients with chronic lymphocytic leukemia carry stereotyped receptors: Pathogenetic implications and clinical correlations. Blood, 109, 259–270. [DOI] [PubMed] [Google Scholar]
- Strefford, J.C. , Sutton, L.A. , Baliakas, P. , Agathangelidis, A. , Malčíková, J. , Plevova, K. , Scarfó, L. , Davis, Z. , Stalika, E. , Cortese, D. , Cahill, N. , Pedersen, L.B. , di Celle, P.F. , Tzenou, T. , Geisler, C. , Panagiotidis, P. , Langerak, A.W. , Chiorazzi, N. , Pospisilova, S. , Oscier, D. , Davi, F. , Belessi, C. , Mansouri, L. , Ghia, P. , Stamatopoulos, K. & Rosenquist, R. (2013) Distinct patterns of novel gene mutations in poor‐prognostic stereotyped subsets of chronic lymphocytic leukemia: the case of SF3B1 and subset #2. Leukemia, 27, 2196–2199. [DOI] [PubMed] [Google Scholar]
- Sutton, L.A. , Young, E. , Baliakas, P. , Hadzidimitriou, A. , Moysiadis, T. , Plevova, K. , Rossi, D. , Kminkova, J. , Stalika, E. , Pedersen, L.B. , Malcikova, J. , Agathangelidis, A. , Davis, Z. , Mansouri, L. , Scarfò, L. , Boudjoghra, M. , Navarro, A. , Muggen, A.F. , Yan, X.J. , Nguyen‐Khac, F. , Larrayoz, M. , Panagiotidis, P. , Chiorazzi, N. , Niemann, C.U. , Belessi, C. , Campo, E. , Strefford, J.C. , Langerak, A.W. , Oscier, D. , Gaidano, G. , Pospisilova, S. , Davi, F. , Ghia, P. , Stamatopoulos, K. & Rosenquist, R. (2016) Different spectra of recurrent gene mutations in subsets of chronic lymphocytic leukemia harboring stereotyped B‐cell receptors. Haematologica, 101, 959–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, K. , Hu, B. , Wang, F. , Yan, Y. , Kim, E. , Vitale, C. , Patel, K.P. , Strati, P. , Gumbs, C. , Little, L. , Tippen, S. , Song, X. , Zhang, J. , Jain, N. , Thompson, P. , Garcia‐Manero, G. , Kantarjian, H. , Estrov, Z. , Do, K.A. , Keating, M. , Burger, J.A. , Wierda, W.G. , Futreal, P.A. & Ferrajoli, A. (2018) Clinical implications of cancer gene mutations in patients with chronic lymphocytic leukemia treated with lenalidomide. Blood, 131, 1820–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin, G. , Thunberg, U. , Karlsson, K. , Murray, F. , Laurell, A. , Willander, K. , Enblad, G. , Merup, M. , Vilpo, J. , Juliusson, G. , Sundstrom, C. , Soderberg, O. , Roos, G. & Rosenquist, R. (2004) Subsets with restricted immunoglobulin gene rearrangement features indicate a role for antigen selection in the development of chronic lymphocytic leukemia. Blood, 104, 2879–2885. [DOI] [PubMed] [Google Scholar]
- Weiss, N.S. (1979) Geographical variation in the incidence of the leukemias and lymphomas. National Cancer Institute Monograph, 1979, 139–142. [PubMed] [Google Scholar]
- Wiestner, A. , Rosenwald, A. , Barry, T.S. , Wright, G. , Davis, R.E. , Henrickson, S.E. , Zhao, H. , Ibbotson, R.E. , Orchard, J.A. , Davis, Z. , Sterler‐Stevenson, M. , Raffeld, M. , Arthur, D.C. , Marti, G.E. , Wilson, W.H. , Hamblin, T.J. , Oscier, D.G. & Staudt, L.M. (2003) ZAP‐70 expression identfies a chronic lymphocytic leukemia subtype with unmutated immunoglobulin genes, inferior clinical outcome, and distinct gene expression profile. Blood, 101, 4944–4951. [DOI] [PubMed] [Google Scholar]
- Wu, S.J. , Chiang, C.J. , Lin, C.T. , Tien, H.F. & Lai, M.S. (2013) Improving but inferior suvival in patinets with chronic lymphocutic leukemia in Taiwan: a population‐based study, 1990–2003. PLoS ONE, 8, e62930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S.J. , Lin, C.T. , Agathangelidis, A. , Lin, L.I. , Kuo, Y.Y. , Tien, H.F. & Ghia, P. (2017) Distinct molecular genetics of chronic lymphocytic leukemia in Taiwan: clinical and pathogenetic implications. Haematologica, 102, 1085–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, Y. , Fan, L. , Wang, L. , Gale, R.P. , Wang, M. , Tian, T. , Wu, W. , Yu, L. , Chen, Y.Y. , Xu, W. & Li, J.Y. (2015) Frequencies of SF3B1, NOTCH1, MYD88, BIRC3 and IGHV mutations and TP53 disruptions in Chinese with chronic lymphocytic leukemia: disparities with Europeans. Oncotarget, 6, 5426–5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, W. , Li, J.Y. , Wu, Y.J. , Yu, H. , Shen, Q.D. , Li, L. , Fan, L. & Qiu, H.X. (2008) Prognostic significance of ATM and TP53 deletions in Chinese patients with chronic lymphocytic leukemia. Leukemia Research, 32, 1071–1077. [DOI] [PubMed] [Google Scholar]
- Yoon, J.H. , Kim, Y. , Yahng, S.A. , Shin, S.H. , Lee, S.E. , Cho, B.S. , Eom, K.S. , Kim, Y.J. , Lee, S. , Kim, H.J. , Min, C.K. , Kim, D.W. , Lee, J.W. , Min, W.S. , Park, C.W. , Lim, J. , Kim, Y. , Han, K. , Kim, M. & Cho, S.G. (2014) Validation of Western common recurrent chromosomal aberrations in Korean chronic lymphocytic leukaemia patients with very low incidence. Hematological Oncology, 32, 169–177. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table SI. Comparative analysis of IGHV features of Taiwanese CLL patients with other studies.


