Abstract
The mitogen-activated protein kinase (MAPK) MoMkk1 governs the cell-wall integrity (CWI) pathway in rice blast fungus Magnaporthe oryzae. To understand the underlying mechanism, we have identified MoSsb1 as one of the MoMkk1-interacting proteins. MoSsb1 is a stress-seventy subfamily B (Ssb) protein homolog, sharing high amino acid sequence homology with the 70-kDa heat shock proteins (Hsp70s). Hsp70 are a family of conserved and ubiquitously expressed chaperones that regulate protein biogenesis by promoting protein folding, preventing protein aggregation, and controlling protein degradation. We found that MoSsb1 regulates the synthesis of nascent polypeptide chains and this regulation is achieved by being in complex with other members of Hsp70s MoSsz1 and 40-kDa Hsp40 MoZuo1. MoSsb1 is important for the growth, conidiation, and full virulence of the blast fungus and this role is also shared by MoSsz1 and MoZuo1. Importantly, MoSsb1, MoSsz1, and MoZuo1 are all involved in the regulation of the CWI MAPK pathway by modulating MoMkk1 biosynthesis. Our studies reveal novel insights into how MoSsb1, MoSsz1, and MoZuo1 affect CWI signaling that is involved in regulating growth, differentiation, and virulence of M. oryzae and highlight the conserved functional mechanisms of heat-shock proteins in pathogenic fungi.
The 70-kilodalton heat shock proteins (Hsp70s or DnaK) are a family of conserved ubiquitously expressed heat-shock proteins that play various important functions in both prokaryotic and eukaryotic cells (Li and Srivastava 2004). They function as molecular chaperones to promote protein folding, prevent protein aggregation, and regulate protein degradation by reversibly binding and releasing short hydrophobic stretches of unfolding polypeptide substrates (Benjamin and McMillan 1998; Borges and Ramos 2005; Mashaghi et al. 2016; Mayer and Bukau 2005; Nelson et al. 1992; Preissler and Deuerling 2012; Walter and Buchner 2002). The budding yeast Saccharomyces cerevisiae has 14 Hsp70s, of which nine exist in the cytosolic form, three are localized to the endoplasmic reticulum (ER), and two are found in mitochondria (Boorstein et al. 1994). In M. oryzae, seven Hsp70s have been identified by sequence alignments, namely MoSsa1 (MGG_06958), MoSsb1 (MGG_11513), MoSse1 (MGG_06065), MoSsz1 (MGG_02842), MoKar2 (MGG_02503), MoLhs1 (MGG_06648), and MoSsc1 (MGG_04191).
Stress-seventy subfamily B (Ssb) proteins are the members of cytosolic Hsp70s identified only in fungi (Nelson et al. 1992). Their structure is typical of Hsp70s, containing an N-terminal ATPase domain connected via a linker to the C-terminal substrate-binding domain. Ssb proteins directly interact with translating ribosomes and form cross-link products with nascent polypeptides to assist newly made polypeptides exiting the ribosomal tunnel. The loss of Ssb causes misfolding of newly synthesized proteins and the accumulation of protein aggregates (Hanebuth et al. 2016; Nelson et al. 1992; Peisker et al. 2010; Raue et al. 2007). Hsp70s together with the corresponding 40-kDa Hsp40s also regulate polypeptide substrate modifications. The C-terminal peptide-binding domain of Hsp70s is responsible for the dynamic interaction with unfolding substrate polypeptides (Finka et al. 2015; Lopez-Buesa et al. 1998). The substrate binding and release are modulated by the activity of its N-terminal ATPase domain, which is stimulated by the corresponding J-domain of Hsp40s (Cyr et al. 1994; Qiu et al. 2006). In S. cerevisiae, the Hsp40 zuotin (Zuo1) is a partner of Ssb proteins (Yan et al. 1998) involved in stimulating Ssb ATPase activity when in complex with an atypical Hsp70, Ssz1 (Hundley et al. 2002; Walsh et al. 2004). Ssz1 and Zuo1 form a stable ribosome-associated complex to recruit cytosolic Hsp70s to nascent polypeptides (Gautschi et al. 2001; Hanebuth et al. 2016; Huang et al. 2005; Rakwalska and Rospert 2004; Zhang et al. 2014). Together, Ssb, Ssz1, and Zuo1 function as a triad to facilitate de novo protein folding (Gautschi et al. 2002; Koplin et al. 2010; Zhang et al. 2017). In human, Hsp70L1 (analogous to Ssz1) is identified as the partner for human MPP11 (analogous to Zuo1), and they also function as ribosome-associated complex to function together (Hundley et al. 2005; Otto et al. 2005).
Mitogen-activated protein kinases (MAPK) are a family of protein kinases that regulate proliferation, gene expression, differentiation, mitosis, cell survival, and apoptosis in a wide variety of organisms (Avruch 2007; Manning et al. 2002). In M. oryzae, the cell-wall integrity (CWI) MAPK cascade plays important roles in vegetative growth, conidiation, the response to extracellular stresses, and virulence. In response to internal cues and environmental challenges, MoMck1 transmits a signal to the MAPK kinase MoMkk1, which activates the MAPK MoMps1 via protein phosphorylation. Consequently, MoMps1 phosphorylates transcription factors, such as MoSwi6, to regulate the nuclear expression of genes involved in cell-wall biosynthesis and cell-cycle progression (Jeon et al. 2008; Xu et al. 1998; Yin et al. 2016).
We have previously found that a conserved component of the MAPK cascade, MoMkk1, is important not only for the maintenance of CWI but also for aerial hyphal growth, conidiation, and virulence of M. oryzae (Yin et al. 2016). To further explore the functional mechanism of MoMkk1, we searched for proteins that interact with MoMkk1 and identified MoSsb1 as a Hsp70 homolog, using coimmunoprecipitation (co-IP). As the relationships and functions of Ssb proteins and their cochaperones were clearly illustrated in S. cerevisiae but not as thoroughly in M. oryzae or other pathogenic fungi, we sought to establish or confirm such functional links. We have provided evidence that MoSsb1 in complex with MoSsz1 and MoZuo1 contributes to the growth, conidiation, and pathogenicity of the blast fungus, likely by modulating MoMkk1 biosynthesis to impact CWI signaling.
RESULTS
Identification of MoSsb1 as a MoMkk1-interacting protein.
As we have previously demonstrated, MoMkk1 is an integral component of the CWI MAPK cascade that is critical for aerial hyphal growth, conidiation, and pathogenicity of M. oryzae (Yin et al. 2016). To determine the mechanism by which MoMkk1 regulates these biological processes, we performed a protein co-IP assay to identify proteins that interact with MoMkk1. The MoMKK1-S construct was generated by linking the S-tag sequence to the C terminal of the MoMKK1 with its native promoter and transforming it into the wild-type Guy11 strain. The presence of a 56-kDa band indicative of the fusion protein in the recipient strain was demonstrated by Western blot analysis. We used anti-S beads to enrich the MoMkk1-S-interacting proteins prior to the analysis by mass spectrometry. A series of proteins that potentially interact with MoMkk1 and function in the CWI MAPK pathway were identified (Supplementary Table S1). Among proteins with a high peptide-occurring frequency, we identified MoSsb1 (MGG_11513), containing 614 amino acids and sharing 70% amino acid sequence identities with yeast Ssb1 and Ssb2 proteins, as an interacting protein of MoMkk1. Further phylogenetic analysis revealed that MoSsb1 is also highly homologous to other Ssb proteins from various organisms (Supplementary Fig. S1).
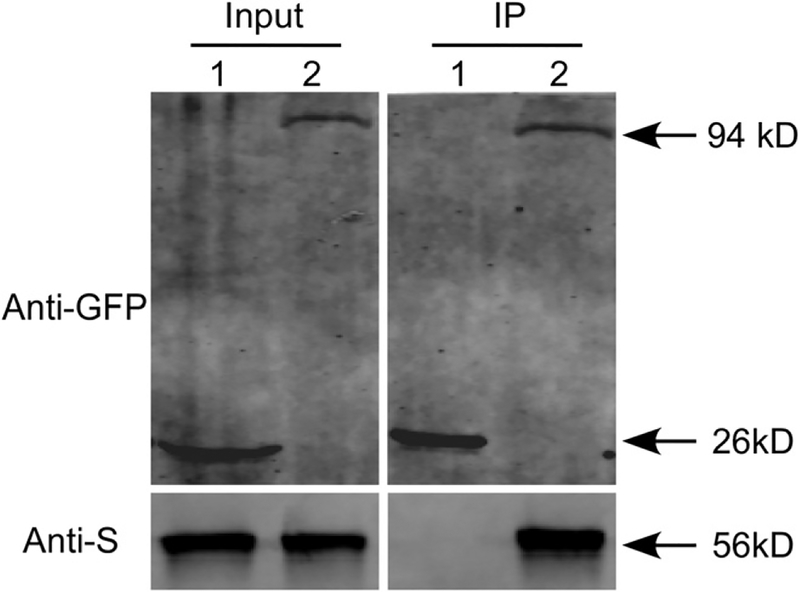
To verify the interaction between MoSsb1 and MoMkk1, the co-IP, yeast two-hybrid, and bimolecular fluorescence complementation (BiFC) assays were all performed. The MoSsb1-MoMkk1 interaction could be established by the co-IP assay (Fig. 1) but not by yeast two-hybrid and BiFC (Supplementary Fig. S2).
Fig. 1.
Coimmunoprecipitation assay between MoSsb1 and MoMkk1. The following strains were grown in liquid complete medium for 48 h. 1 = Guy11 containing green fluorescent protein (GFP) and MoMkk1-S; 2 = Guy11 containing MoSsb1-GFP and MoMkk1-S. Total proteins extracted from the strains were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblots were incubated with anti-GFP and anti-S antibodies as indicated. Samples were pulled down using anti-GFP beads and were then detected with the anti-S antibody.
MoSsb1 is required for vegetative growth, conidiation, and virulence.
To characterize the function of MoSsb1, two ΔMossb1 mutants were identified and confirmed (Supplementary Fig. S3A and B). In addition, the MoSSB1-GFP construct with the native promoter of MoSSB1 was introduced into the ΔMossb1 mutant to obtain two complemented ΔMossb1/MoSSB1 strains. Since both the two mutant and the two complemented strains exhibited similar phenotypes, we selected one each for further analysis. On complete medium (CM), minimal medium (MM), oatmeal agar medium (OM), and straw decoction and corn (SDC) agar medium, colonies of the ΔMossb1 mutant were significantly smaller with very compact mycelial mass (Table 1; Fig. 2A). Considering MoSsb1 is a HSP, we also tested the growth of the ΔMossb1 mutant at 20, 25, 28, and 30°C. Because of severe growth reduction, the test could not establish a role of MoSsb1 in heat-shock responses (Supplementary Fig. S4).
Table 1.
Comparison of mycological characteristics among Guy11, mutant strains, and complemented strains
| Growth (cm)a | Conidiation (× 104/cm2)b | Abnormal conidia rate (%) | Germination rate (%)c | Appressorium formation (%)d | ||||
|---|---|---|---|---|---|---|---|---|
| Strain | CM | MM | OM | SDC | ||||
| Guy11 | 5.33 ± 0.1a | 4.07 ± 0.1a | 4.73 ± 0.1a | 4.43 ± 0.1a | 21.2 ± 1.6a | 2.7 ± 1.2a | 95.3 ± 2.0a | 93.0 ± 2.0a |
| ΔMossb1 | 1.63 ± 0.1b | 1.43 ± 0.1b | 1.47 ± 0.1b | 1.53 ± 0.1b | 11.5 ± 1.1b | 79.3 ± 4.9b | 93.0 ± 1.2a | 88.7 ± 1.2a |
| ΔMossb1/MoSSB1V443F | 1.63 ± 0.2b | N/A | N/A | 1.57 ± 0.1b | N/A | N/A | N/A | N/A |
| ΔMossb1/MoSSB1 | 5.23 ± 0.1a | 4.00 ± 0.0a | 4.70 ± 0.0a | 4.30 ± 0.0a | 19.2 ± 0.3a | 2.6 ± 1.3a | 96.6 ± 2.1a | 94.2 ± 2.4a |
| ΔMossz1 | 1.43 ± 0.1b | 1.40 ± 0.0b | 1.23 ± 0.1b | 1.57 ± 0.1b | 8.1 ± 1.8b | 85.3 ± 0.5b | 82.3 ± 1.5a | 89.3 ± 1.5a |
| ΔMossz1/MoSSZ1 | 5.30 ± 0.1a | 4.03 ± 0.1a | 4.63 ± 0.1a | 4.37 ± 0.1a | 21.1 ± 1.3a | 2.9 ± 2.1a | 93.6 ± 1.5a | 92.8 ± 1.5a |
| ΔMozuo1 | 1.10 ± 0.0b | 1.23 ± 0.1b | 0.67 ± 0.1b | 0.80 ± 0.0b | 3.4 ± 0.3b | 82.7 ± 3.7b | 89.3 ± 3.5a | 89.3 ± 3.5a |
| ΔMozuo1/MoZUO1 | 5.23 ± 0.1a | 4.03 ± 0.1a | 4.63 ± 0.1a | 4.30 ± 0.1a | 21.0 ± 0.9a | 2.7 ± 1.5a | 94.3 ± 3.1a | 93.6 ± 1.2a |
Colony diameter was measured after 7 days of growth at 28°C. CM = complete medium, MM = minimal medium, OM = oatmeal agar medium, and SDC = straw decoction and corn.
Quantification of the conidial production of the indicated strains from SDC cultures.
Percentage of conidial germination on artificial surface at 24 h postinoculation (hpi).
Percentage of appressorium formation on artificial surface at 24 hpi. ± standard deviation was calculated from three repeated experiments and asterisks indicate statistically significant differences (Duncan’s new multiple range test, P < 0.01). N/A = not assayed.
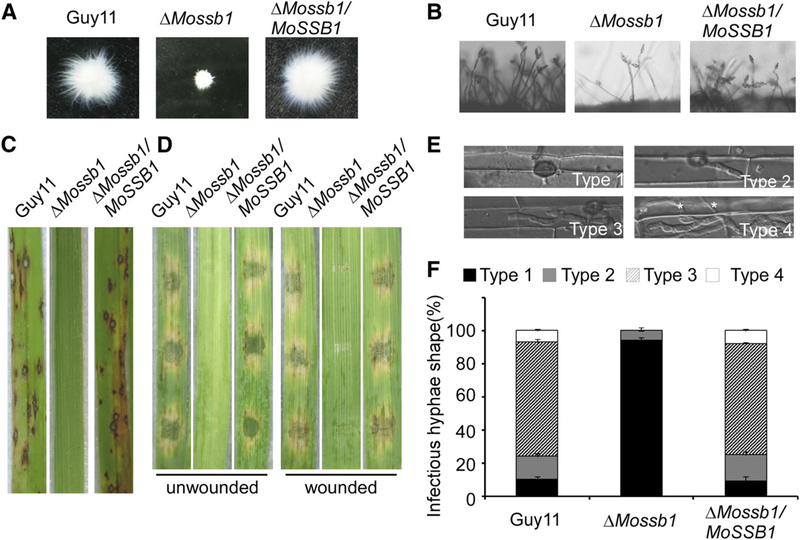
Fig. 2.
MoSsb1 is important for vegetative growth, asexual development and pathogenesis in Magnaporthe oryzae. A, The wild-type strain Guy 11, ΔMossb1 mutant, and the complemented strain were inoculated in liquid complete medium for 48 h at 28°C and were then photographed. B, Conidia formation was observed under a light microscope after induction of conidiation onto coverslips. C, Rice (Oryza sativa cv. CO39) seedlings were inoculated with conidial suspension (5 × 104 spores/ml). Typical leaves were photographed at 7 days postinoculation (dpi). D, Detached unwounded and wounded barley (Hordeum vulgare cv. Four-arris) leaves were drop-inoculated with conidial suspension and were photographed at 5 dpi. E and F, Detailed observation and statistical analysis of invasive growth in rice sheath cells at 36 hpi. Appressorium penetration sites (n = 100) were checked and the invasive hyphae (IH) were rated from type 1 to 4. Asterisks indicate IH extended to surrounding cells. All the experiments were performed three times with similar results.
Conidia are important in the spread of rice blast. Following 10 days of growth on SDC medium, conidiation was reduced in the ΔMossb1 mutant, as compared with controls (Table 1; Fig. 2B). Nearly 80% of the conidia produced by the ΔMossb1 mutant were abnormal when compared with the <5% produced by wild type and complemented strains (Table 1; Supplementary Fig. S5). In addition, when the ΔMossb1 mutant was crossed with the tester strain TH3 (MAT1–1), no perithecia were observed after 3 weeks (Supplementary Fig. S6), suggesting that MoSsb1 is indispensable for sexual reproduction.
To examine if MoSsb1 is required for pathogenicity, we inoculated conidial suspensions of the wild type, ΔMossb1 mutant, and complemented strains on CO-39, a susceptible rice cultivar. The ΔMossb1 mutant produced few lesions, whereas the control strains produced many typical lesions at 7 days postinoculation (dpi) (Fig. 2C). Similar results were obtained when conidial suspensions were drop-inoculated on detached unwounded and wounded leaves of the barley cultivar Fourarris at 5 dpi (Fig. 2D). To explain the reduced pathogenicity, the ΔMossb1 mutant was observed for germination and appressorium formation. The ΔMossb1 mutant displayed delayed germination and appressorium formation initially, but there were no significant differences at 24 h (Table 1; Supplementary Fig. S7). We then examined the penetration and invasive hyphal growth in rice sheath cells and found that about 80% of the invasive hyphae by the wild type and complemented strains were types 3 and 4, at 36 h postinoculation (hpi). By contrast, more than 95% of the appressorium did not form invasive hyphae in the ΔMossb1 mutant and tested as type 1 (Fig. 2E and F). These results all indicated that MoSsb1 plays a crucial role in the vegetative growth, conidiation, and virulence of M. oryzae.
MoSsb1, MoSsz1, and MoZuo1 function as a complex.
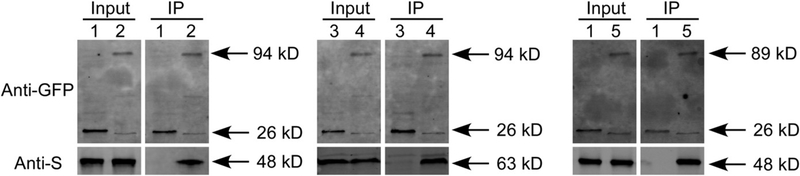
In S. cerevisiae, the Ssb proteins function only in the presence of the Hsp40 Zuo1, which functions by forming a stable heterodimer with Ssz1, an atypical Hsp70 (Boorstein et al. 1994; Gautschi et al. 2001). To examine whether MoSsb1 functions through the similar mechanism, gene loci MGG_02842 and MGG_10565, encoding Ssz1 and Zuo1 homologs, were identified and named MoSsz1 and MoZuo1, respectively. We tested the interactions among these three proteins using co-IP, yeast two-hybrid, and BiFC assays. Again, the co-IP assay but not the yeast two-hybrid and BiFC assays showed that MoSsb1, MoSsz1, and MoZuo1 each interacted with the other proteins (Fig. 3, Supplementary Fig. S8). This indicated that MoSsb1, MoSsz1, and MoZuo1 could form a complex and function in a fashion similar to that of yeast proteins.
Fig. 3.
Coimmunoprecipitation assays among MoSsb1, MoSsz1, and MoZuo1. The following strains were grown in liquid complete medium for 48 h. 1 = Guy11 containing green fluorescent protein (GFP) and MoZuo1-S; 2 = Guy11 containing MoSsb1-GFP and MoZuo1-S; 3 = Guy11 containing GFP and MoSsz1-S; 4 = Guy11 containing MoSsb1-GFP and MoSsz1-S; 5 = Guy11 containing MoSsz1-GFP and MoZuo1-S. Total proteins extracted from the strains were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblots were incubated with anti-GFP and anti-S antibodies as indicated. Samples were pulled down using anti-GFP beads and were then detected with anti-S antibodies.
MoSsz1 and MoZuo1 are important for vegetative growth, conidiation, and virulence.
To test the functions of MoSsz1 and MoZuo1, ΔMossz1 and ΔMozuo1 mutants were identified and confirmed. Again, the MoSSZ1-GFP and MoZUO1-GFP constructs with native promoters were introduced into, ΔMossz1 and ΔMozuo1 mutants, and we obtained complemented strains, respectively. The ΔMossz1 and ΔMozuo1 mutants produced significantly reduced colonies on all four media (Table 1) and mycelia masses were also very compact (Fig. 4A). An experiment to test the response to improper temperatures, including 20, 25, 28, and 30°C, was also carried out, and the results were also inconclusive in supporting that MoSsz1 and MoZuo1 are involved in heat shock response.
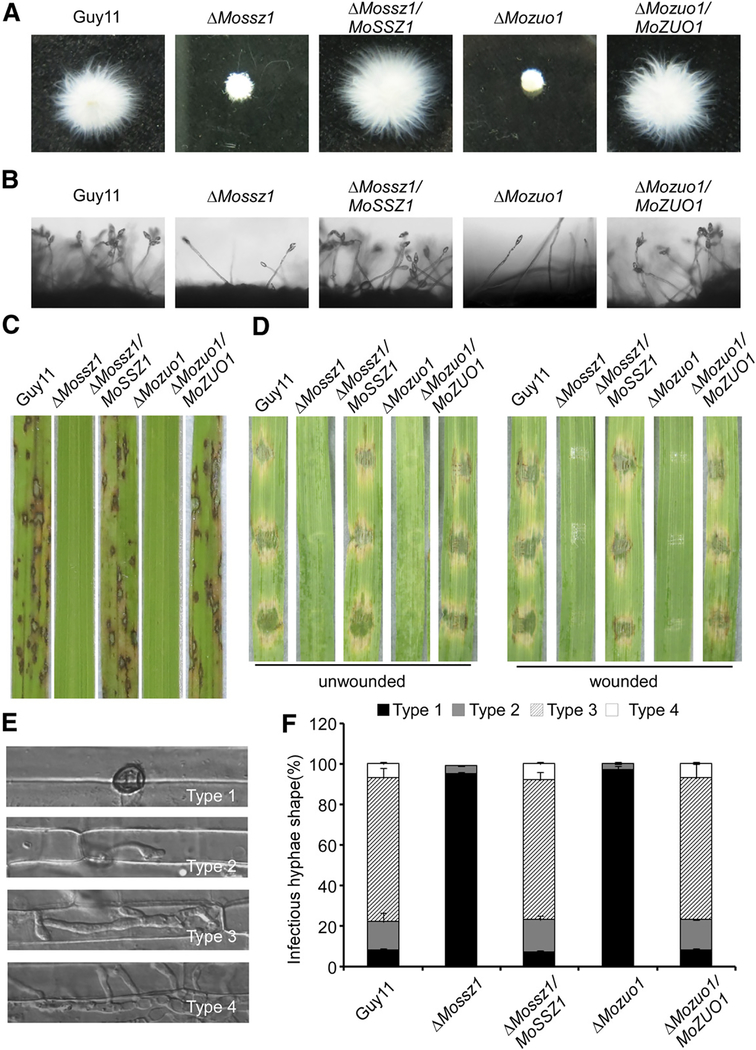
Fig. 4.
MoSsz1 and MoZuo1 are also important for vegetative growth, asexual development, and pathogenesis in Magnaporthe oryzae. A, Guy 11, ΔMossz1, ΔMossz1/MoSSZ1, ΔMozuo1, and ΔMozuo1/MoZUO1 were inoculated in liquid complete medium for 48 h at 28°C and were then photographed. B, Conidia formation was observed under a light microscope. C, Rice (Oryza sativa cv. CO39) seedlings were inoculated with conidial suspension (5 × 104 spores/ml). Typical leaves were photographed at 7 days postinoculation (dpi). D, Detached unwounded and wounded barley (Hordeum vulgare cv. Four-arris) leaves were drop-inoculated with conidial suspension and were photographed at 5 dpi. E and F, Detailed observation and statistical analysis of invasive growth in rice sheath cells at 36 h postinoculation. Appressorium penetration sites (n = 100) were checked and the invasive hyphae (IH) were rated from type 1 to 4. Asterisks indicate IH extended to surrounding cells. All the experiments were performed three times with similar results.
Conidia counts and microscopic observations revealed that the ΔMossz1 and ΔMozuo1 mutants had defects in conidia formation (Table 1; Fig. 4B). The percentage of abnormal conidia exceeded 80% in the ΔMossz1 and ΔMozuo1 mutants, compared with <5% in the wild type and complemented strains (Table 1). In addition, the ΔMossz1 and ΔMozuo1 strains were also defective in mating with no perithecia found after 3 weeks when crossed with the tester strain TH3, similar to the ΔMossz1 strains.
The pathogenicity assay showed that the ΔMossz1 and ΔMozuo1 strains failed to generate any lesions on rice leaves, detached unwounded, and wounded barley leaves (Fig. 4C and D). Germination and appressorium formation were also delayed in the ΔMozuo1 and ΔMozuo1 mutants compared with the wild type and complemented strains (Table 1). To elaborate these observations, we examined the penetration and invasive hyphal growth in rice sheath cells. For the ΔMossz1 and ΔMozuo1 mutants, more than 95% of the appressorium did not achieve invasive hyphal growth and rested at type 1 at 36 hpi, in contrast to 80% and types 3 and 4 invasive hyphal found in the wild-type and complemented strains (Fig. 4E and F). These results indicated that both MoSsz1 and MoZuo1 play important roles in the vegetative growth, conidiation, and virulence of M. oryzae. These characters are very similar to those of ΔMossb1, consistent with the hypothesis that MoSsb1, MoSsz1, and MoZuo1 form a complex to mediate the same cellular processes.
MoSsb1, MoSsz1, and MoZuo1 are critical for CWI.
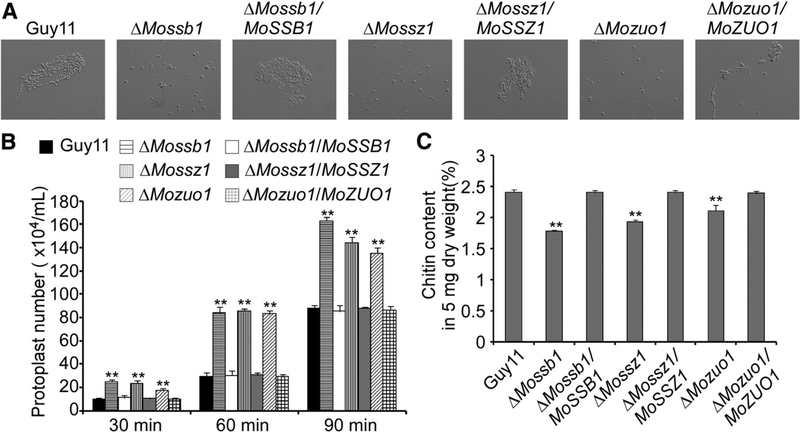
To examine the roles of MoSsb1, MoSsz1, and MoZuo1 in cell-wall integrity, we performed a protoplast release assay and a chitin content test. The protoplast release assay involves counting of the protoplasts following mycelium treatment with cell wall–degrading enzymes. Protoplast release was faster by the three mutants compared with the wild-type and corresponding complemented strains (Fig. 5A and B). As chitin is an important cell-wall component (Roncero 2002), we examined whether these three proteins are involved in regulating chitin biosynthesis. We measured the chitin content using the fluorometric Morgan-Elson method and found that the chitin contents of ΔMossb1, ΔMossz1, and ΔMozuo1 were significantly reduced (Fig. 5C). These two findings indicated the presence of a compromised cell-wall structure in these mutants and that MoSsb1, MoSsz1, and MoZuo1 are important for CWI signaling of M. oryzae.
Fig. 5.
MoSsb1, MoSsz1, and MoZuo1 are required for cell-wall integrity. A, Light microscopic examination of protoplast release after treatment with cell wall–degrading enzymes for 1 h at 30°C. B, Data statistics of protoplast release assay. Asterisks indicate significant differences at P < 0.01. The experiment was performed three times with similar results. C, N-acetylglucosamine (GlcNAc) determination by the fluorimetric Morgan-Elson method showed significantly decreased chitin content in the three mutants. Asterisks indicate significant differences at P < 0.01, according to Duncan’s range test.
MoSsb1, MoSsz1, and MoZuo1 are involved in regulating MoMps1 phosphorylation.
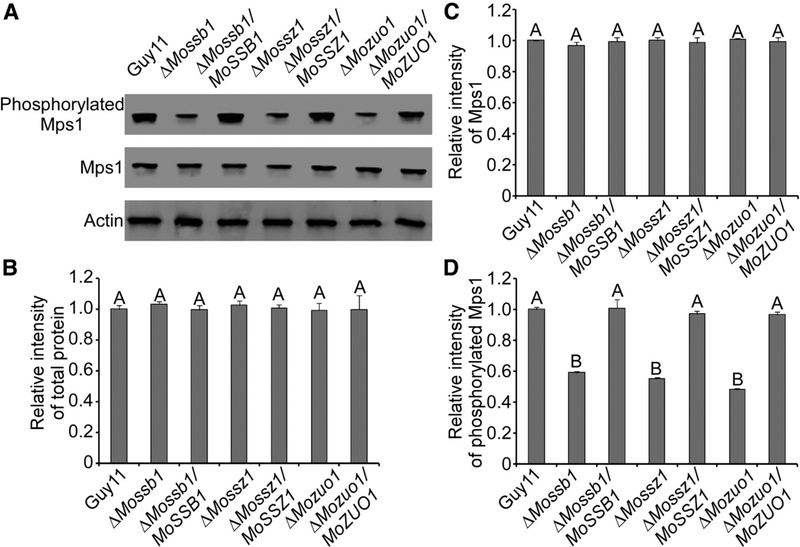
As MoMkk1 is an important component of the MAPK cascade, the interaction between MoSsb1 and MoMkk1 suggests that MoSsb1 or the MoSsb1-MoSsz1-MoZuo1 complex may play a function in MAPK signaling. To test this hypothesis, we examined if MoSsb1, MoSsz1, and MoZuo1 are related to MoMps1 phosphorylation, using antibodies specific to MoMps1 and phosphorylated MoMps1. We found that phosphorylation of MoMps1 was significantly reduced in the ΔMossb1, ΔMossz1, and ΔMozuo1 mutants when compared with the wild type and corresponding complemented strains (Fig. 6). This finding may provide an explanation, at least partially, for the reduced CWI, appressorium penetration, and invasive growth exhibited by these three mutants.
Fig. 6.
MoSsb1, MoSsz1, and MoZuo1 are involved in the regulation of MoMps1 phosphorylation. A, Immunoblots were performed with total proteins from hyphae of Guy11, ΔMossb1, ΔMossz1, ΔMozuo1, and respective complemented strains. The signal corresponding to phosphorylated Mps1 was detected by binding of the antiphospho-p44/42 antibody, with the Mpk1 and actin antibodies used as controls. MoMps1 phosphorylations in the three mutants were significantly reduced. B to D, The Western blotting bands detected with antiphosphorylated-Mps1 were quantified with the ODYSSEY infrared imaging system (application software version 2.1). The intensity of the phosphorylated Mps1 band for each strain is relative to that of total protein and Mps1. Bars denote standard errors from three independent experiments. Values on the bars followed by the same letter were not significantly different at P = 0.05.
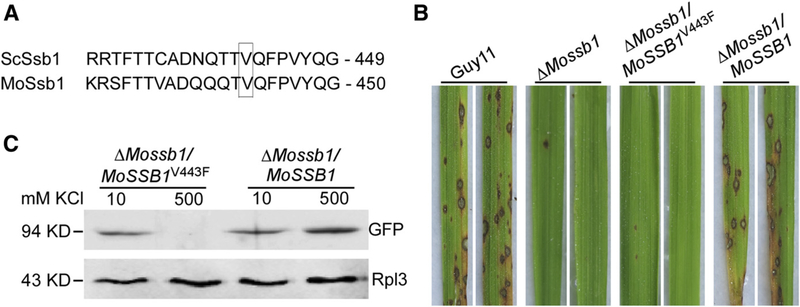
MoSsb1V443 is important for pathogenicity and the interaction with nascent polypeptides.
Ssb proteins are thought to directly bind to nascent polypeptides to affect their modifications during protein synthesis (Peisker et al. 2010). During the initial stage when nascent polypeptides combine with the ribosome, the interaction between Ssb proteins and the nascent polypeptides can be tested by determining the quantity of Ssb proteins on the ribosome (Pfund et al. 2001). Based on findings by Pfund and colleagues (2001), MoSsb1 valine-443 is likely a key site for ribosome binding (Fig. 7A). To test this, we introduced the MoSSB1V443F-GFP construct into the ΔMossb1 mutant and obtained the ΔMossb1/MoSSB1V443F strain. In pathogenicity and growth tests, no lesions were found on rice leaves infected by ΔMossb1/MoSSB1V443F, whereas many lesions were found on leaves infected with the wild type and complemented strains (Fig. 7B). The growth of the ΔMossb1/MoSSB1V443F was also significantly reduced compared with controls (Table 1). These results were all similar to those of the ΔMossb1 mutant.
Fig. 7.
MoSsb1 valine-443 is important for the pathogenicity and its interaction with the nascent polypeptides. A, Comparison of Ssb1 sequences in Magnaporthe oryzae (accession number XP_003718330.1) with those in Saccharomyces cerevisiae (accession number NP_010052.1) and identification of valine-443 of MoSsb1 as a key site for binding to nascent polypeptides. B, Conidia suspensions (5 × 104 spores/ml) of the Guy11, ΔMossb1 mutant, point mutation strain ΔMossb1/MoSSB1V443F, and complemented strain ΔMossb1/MoSSB1 were sprayed on rice seedlings, respectively. Typical leaves were photographed at 7 days postinoculation. These experiments were performed three times with similar results. C, Extracts were run through sucrose cushions and ribosome pellets were resuspended in buffer containing the indicated amount of KCl. Pellets were incubated for 25 min on ice and were then run through sucrose cushions of the indicated concentration of KCl. Then, pellets were resuspended, were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and were immunoblotted with antibodies against green fluorescent protein and the ribosomal protein L3 (Rpl3).
To further test the interaction between MoSsb1 and the nascent polypeptides, we extracted ribosome proteins from the ΔMossb1/MoSSB1V443F and complemented ΔMossb1/MoSSB1 strains. Western blotting analysis showed that the amount of MoSsb1V443F precipitating together with the ribosome in 500 mM KCl was significantly lower when compared with to that precipitating in 10 mM KCl, whereas the two protein samples of MoSsb1V443F were approximately equal (Fig. 7C). This indicated that the interaction between the point mutation MoSsb1 and nascent polypeptides is salt-sensitive and that valine-443 of MoSsb1 is important for this interaction.
MoSsb1, MoSsz1, and MoZuo1 are required to maintain the amount of MoMkk1 protein.
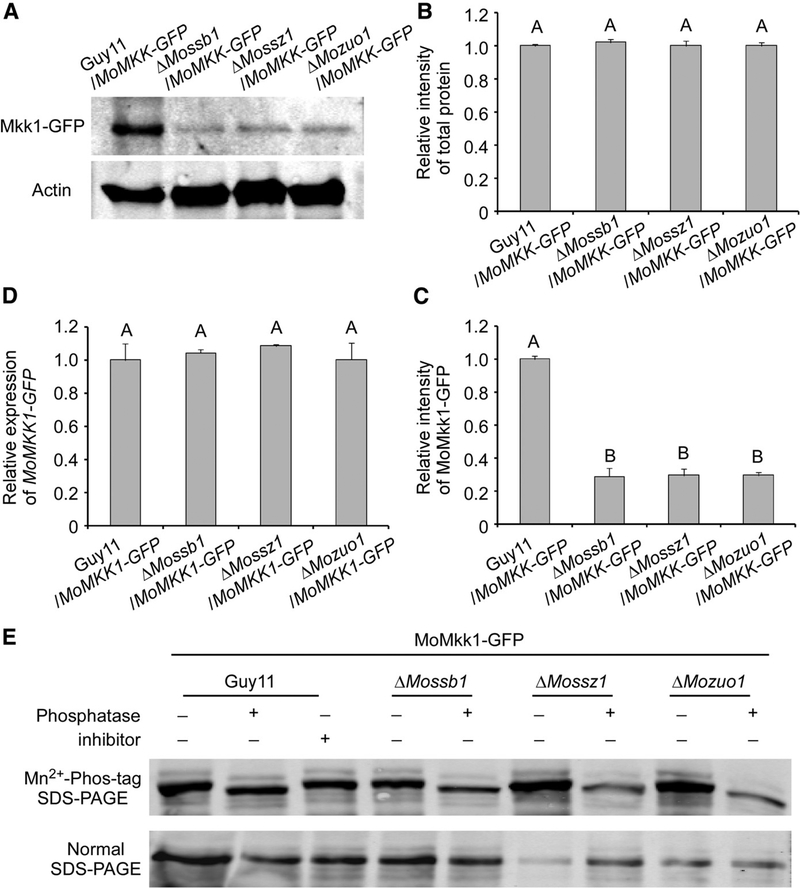
We found that MoSsb1, MoSsz1, and MoZuo1 did not influence the amount of the MoMps1 protein but, rather, its phosphorylation levels (Fig. 6). Given the functions of MoMkk1 in transferring the signaling cue to MoMps1 and of the MoSsb1-MoSsz1-MoZuo1 complex in folding and maintaining nascent polypeptides, we speculated that the MoSsb1-MoSsz1-MoZuo1 complex was involved in the synthesis of MoMkk1. Therefore, we determined the amount of the MoMkk1 protein in the ΔMossb1, ΔMossz1, and ΔMozuo1 mutants. We first transformed the MoMKK1-GFP construct into three mutant strains as well as the wild-type strain. Western blot analysis of total protein extracts revealed that the amount of MoMkk1-GFP was significantly lower in the three mutants compared with the wild-type strain (Fig. 8A to C). To rule out the possibility that the MoSsb1-MoSsz1-MoZuo1 complex controls the level of MoMKK1-GFP transcription, we used quantitative reverse transcription-polymerase chain reaction (qRT-PCR), which ruled out this possibility (Fig. 8D). We also analyzed MoMkk1-GFP from the wild-type strain, ΔMossb1, ΔMossz1, and ΔMozuo1 mutants in gels containing Phos-tag, which retards the mobility of phosphoproteins. We found that the wild-type strain and three mutants had similar mobility, implying that the MoSsb1-MoSsz1-MoZuo1 complex does not regulate MoMkk1 via protein phosphorylation when assisting the synthesis and maintenance of MoMkk1 (Fig. 8E). These results indicated that MoMkk1 is a potential client of the MoSsb1-MoSsz1-MoZuo1 complex.
Fig. 8.
MoSsb1, MoSsz1, and MoZuo1 are required for the maintenance of protein amounts of MoMkk1. A, Guy11, ΔMossb1, ΔMossz1, and ΔMozuo1 strains containing MoMkk1-GFP (green fluorescent protein) were grown in liquid complete medium for 48 h. Total proteins extracted from the strains were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblots were incubated with anti-GFP. Protein amounts of MoMkk1-GFP in DMossb1, ΔMossz1, and ΔMozuo1 were significantly reduced. B and C, The Western blotting bands detected with anti-GFP were quantified with the ODYSSEY infrared imaging system (application software version 2.1). The intensity of the MoMkk1-GFP band for each strain is relative to that of total protein. Bars denote standard errors from three independent experiments. Values on the bars followed by the same letter were not significantly different at P = 0.05. D, Transcription analysis of MoMKK1-GFP in Guy11 and three mutants containing MoMkk1-GFP using quantitative reverse transcription-polymerase chain reaction. E, MoMkk1-GFP from the wild-type strain, ΔMossb1, ΔMossz1, and ΔMozuo1 mutants cell extracts treated with alkaline phosphatase or phosphatase inhibitors were subjected to Phos-tag SDS-PAGE and normal SDS-PAGE followed by immunoblotting with anti-GFP.
DISCUSSION
Hsp are cytoplasmic and organellar proteins playing important biological functions. Several Hsps located in the ER have been identified in filamentous fungi. In Ustilago maydis, Dnj1 was characterized as part of the conserved cellular response to ER stress important in pathogenicity (Lo Presti et al. 2016). In M. oryzae, the proper processing of secreted proteins, including effectors, by chaperone MoLhs1 in the ER is required for successful colonization and for determining host-pathogen compatibility via gene-for-gene interactions (Yi et al. 2009). Other than those two findings, fungal Hsp in other organelles or the cytosol remain poorly understood. Here, we identified three cytosolic Hsps, including a typical Hsp70 (MoSsb1), an atypical Hsp70 (MoSsz1), and a Hsp40 (MoZuo1) from M. oryzae and showed that all three proteins play key roles in the regulation of pathogenicity due, in part, to their roles in attenuating CWI signaling. We also identified the putative mechanism by which they act, via their influence on the CWI MAPK pathway, which is essential for appressorium formation and colonization in M. oryzae.
After nascent polypeptides emerge from the ribosome, growing chains assume an aggregation-prone and partially folded state and require ribosome-associated chaperones to maintain translational fidelity. In yeast, Ssb1, Ssz1, and Zuo1 form a complex and function as a triad to facilitate de novo protein folding (Hartl and Hayer-Hartl 2009). In M. oryzae, a similar mechanism also exists for MoSsb1, MoSsz1, and MoZuo1, based on that ΔMossb1, ΔMossz1, and ΔMozuo1 mutants exhibit similar growth and development phenotypes, their interactions with each other, and the functional dissection between them and MoMkk1-mediated CWI signaling.
Hsp70s and Hsp40s function together to activate the modification of unfolding polypeptides (Bukau and Horwich 1998; Kelley 1998; Qiu et al. 2006). Ssb1-Zuo1 is one such Hsp70-Hsp40 pair in which the conserved J domain of Zuo1 stimulates the N-terminal ATPase domain of Ssb1 and assists Ssb1 to bind with nascent polypeptides (Yan et al. 1998). We speculate that MoSsb1-MoZuo1 functions in the same way in M. oryzae. However, we could only establish this interaction through co-IP but not yeast two-hybrid and BiFC. This may suggest that additional conditions or components may be required for the interaction. In the interaction between Zuo1 and Ssz1, the stable ribosome-associated complex promotes the substrate binding of Ssb1. However, unlike typical Hsp70s represented by Ssa1 and Ssb1, Ssz1 has not been demonstrated to be capable of directly binding to unfolding polypeptides (Gautschi et al. 2001; Huang et al. 2005). We thought that the MoSsz1-MoZuo1 interaction model could be similar to MoSsb1-MoZuo1, but the mechanism may be different, as an uncharacterized domain or other factors may account for the interaction. In addition, we hypothesized that interactions of MoSsb1-MoZuo1 and MoSsz1-MoZuo1 may be transient and they occur only in the presence of suitable substrates in vivo. Previous studies showed that MoDnm1 did not couple directly with MoFis1 and that MoMdv1 functions as an adaptor linking MoDnm1 with MoFis1 in M. oryzae (Zhong et al. 2016). Given the interaction between MoSsb1 and MoSsz1 based on the co-IP assay, we speculated that MoZuo1 could function as an adaptor protein linking MoSsb1 to MoSsz1.
As demonstrated in our studies, the MoSsb1-MoSsz1-MoZuo1 complex regulates the phosphorylation of MoMps1, but it does not affect the amount of MoMps1 protein. In the MoMck1-MoMkk1-MoMps1 pathway, the changes in MoMkk1 influence the phosphorylation of MoMps1, and the activation of MoMkk1 can rescue Momps1 deletional defects in M. oryzae (Yin et al. 2016). In view of these findings, we hypothesized that the reduced amount of MoMkk1 protein leads to a decrease in signaling from MoMkk1 to MoMps1, and the defects of MoMkk1 in the phosphorylation of MoMps1 lead to a decrease in the levels of MoMps1 phosphorylation. In additional experiments, we found that the amount of MoMkk1 protein was significantly reduced in the ΔMossb1, ΔMossz1, and ΔMozuo1 mutants. Considering the interaction between MoMkk1 and MoSsb1 and the function of the MoSsb1-MoSsz1-MoZuo1 complex in binding with nascent polypeptides to mediate the synthesis of nascent proteins, we are confident that the reduced amount of MoMkk1 protein accounts for the decrease in phosphorylated MoMps1 that results in a defect in CWI signaling. As we could establish the interaction between MoSsb1 and MoMkk1 by co-IP but not by yeast two-hybrid and BiFC, we also speculate that MoMkk1 interacts with MoSsb1 transiently and conditionally in vivo. MoMkk1 interacting with MoSsb1 may be very brief or MoSsb1 functions as chaperone only in the presence of MoSsz1 and MoZuo1. MoMkk1 could also be a substrate of MoSsb1. When the nascent peptides of MoMkk1 appear in the exit of a ribosomal tunnel, MoSsb1 could bind with them and pull them out of the ribosome for further synthesis and maturity.
Serving as classical chaperones, Ssb proteins and their cochaperones play key roles not only in de novo protein folding but also in the transport of proteins to appropriate cellular locations. Recent work showed that the translocation of MoYvh1, a protein phosphatase, into the nucleus following oxidative stress is dependent on MoSsb1 and MoSsz1 and this translocation promotes infection (Liu et al. 2018). Moreover, in addition to being a classical chaperone, Ssb proteins are multifunctional proteins involved in the regulation of cellular signaling processes. In yeast, Ssb proteins also bind Akt/PKB serine/threonine protein kinase Sch9 posttranslationally and are required for the TORC1-dependent phosphorylation of Sch9 at T737 to control ribosome biogenesis (Mudholkar et al. 2017). In M. oryzae, it appears that one of the functions by the MoSsb1-MoSsz1-MoZuo1 complex is to impact CWI signaling by reducing the protein amount of MoMkk1 rather than the phosphorylation levels of MoMkk1 protein. Such a role for Ssb proteins remains to be explored in M. oryzae.
In summary, we revealed that MoSsb1 together with MoSsz1 and MoZuo1 regulates the vegetative growth, conidiation, mating, and pathogenicity of the rice blast fungus. MoSsb1, MoSsz1, and MoZuo1 form a complex that binds to likely unfolding MoMkk1 polypeptides and affects its biosynthesis, thereby participating in CWI signaling and exerting their regulatory functions. Our studies reveal the conserved functional mechanisms of Hsps in fungi and provide new insights into how MoSsb1, MoSsz1, and MoZuo1 affect CWI signaling involved in growth, differentiation, and virulence of M. oryzae.
MATERIALS AND METHODS
Strains and culture conditions.
The M. oryzae Guy11 strain was used as wild type for transformation in this study. All strains were maintained on CM agar plates at 28°C (Talbot et al. 1993). Mycelia were harvested from liquid CM and were used for DNA and RNA extractions. For vegetative growth, small agar blocks were cut from the edge of 7-day-old cultures and were placed onto fresh media (CM, MM, OM, and SDC) followed by incubation in the dark at 28°C. For conidiation, strain blocks were maintained on SDC (100 g of straw, 40 g of corn powder, 15 g of agar in 1 liter of distilled water) agar media (Zhang et al. 2011b) at 28°C for 7 days in the dark, followed by constant illumination with mycelia removed for 3 days.
Targeted gene deletion and complementation.
The gene-deletion mutants were generated using the standard one-step gene replacement strategy. First, two fragments with 1.0 kb of sequences flanking the targeted gene were amplified with primer pairs by PCR. The resulting PCR products of MoSSB1, MoSSZ1, and MoZUO1 were digested with restriction endonucleases and were ligated with the hygromycin resistance cassette (HPH) released from pCX62. Finally, the recombinant inserts were sequenced. The 3.4-kb fragments, which include the flanking sequences and the HPH cassette, were amplified and were transformed into Guy11 protoplasts. Putative mutants were first screened by PCR and later confirmed by Southern blot analysis. The complement fragments, which contain the entire gene-coding region and its native promoter region, was amplified by PCR with primers and inserted into pYF11 (bleomycin resistance) to complement the mutant strains.
Protoplast release assay and measurement of the chitin content.
Protoplasts were prepared and transformed as described (Sweigard et al. 1992). The number of the released protoplasts from the strains were counted and were analyzed statistically after incubation for 30, 60, and 90 min. The protoplast release of the indicated strains was observed under a light microscope and was photographed after 60 min.
Chitin (N-acetylglucosamine [GlcNAc]) content was analyzed as described previously (Ram and Klis 2006; Song et al. 2010). First, mycelial samples were freeze-dried and, then, 5 mg of dried mycelia were resuspended in 1 ml of 6% KOH and were heated at 80°C for 90 min. Samples were centrifuged (16,000 × g, 10 min) and pellets were washed with phosphate buffered saline in three cycles of centrifugation and suspension (16,000 × g, 10 min) before final suspension in 0.5 ml of McIlvaine’s buffer (pH = 6). An aliquot of 100 ml (13 units) of Streptomyces plicatus chitinase (Sigma, St. Louis) was added and was incubated for 16 h at 37°C with gentle mixing; 100-ml samples were then combined with 100 ml of 0.27 M sodium borate (pH = 9), were heated for 10 min at 100°C with final addition of 1 ml of freshly diluted (1: 10) Ehrlich’s reagent (10 g of p-dimethylaminobenzaldehyde in 1.25 ml of HCl and 8.75 ml of glacial acetic acid). After incubation at 37°C for 20 min, 1 ml of the sample was transferred to a 2.5 ml plastic cuvette (Greiner, Frickenhausen, Germany) and absorbance at 585 nm was recorded. Standard curves were prepared with GlcNAc (Sigma). The experiment was repeated three times. The primers used in this study are listed in Supplementary Table S2.
qRT-PCR analysis.
For qRT-PCR, total RNA was reverse transcribed into first-strand cDNA using the oligo (dT) primer and M-MLV reverse transcription (Invitrogen, Carlsbad, CA, U.S.A.). qRT-PCR assays were performed following previously established procedures (Guo et al. 2010). qRT-PCR was performed with three independent pools of tissues in three sets of experimental replicates.
co-IP.
To confirm the interactions of MoSsb1, MoSsz1, and MoZuo1 in vivo, the three pairs of constructs were introduced by cotransformation into protoplasts of the wild-type strain as follows: i) Guy11 containing gtrrm fluorescent protein (GFP) and MoZuo1-S; ii) Guy11 containing MoSsb1-GFP and MoZuo1-S; iii) Guy11 containing GFP and MoSsz1-S; iv) Guy11 containing MoSsb1-GFP and MoSsz1-S; v) Guy11 containing MoSsz1-GFP and MoZuo1-S. Strains i and iii are negative controls. Total proteins were isolated from transformants expressing both fusion proteins and were incubated with anti-GFP affinity beads. Proteins bound to resins were eluted after a series of washing steps, according to manufacture instructions. Western blots of total proteins and elution from resins were detected with anti-GFP and anti-S antibodies, using the ECL Supersignal system (Pierce, Rockford, IL, U.S.A.). The same methods were applied to confirm the interaction between MoSsb1 and MoMkk1.
Extraction of ribosomal proteins.
Ribosomal proteins were extracted from mycelia as previously described by Lo et al. (2009). Briefly, fungal strains were cultured on solid CM medium, first, for 7 days at 28°C, and an approximately 1 × 1 mm squares of agar containing the culture were inoculated in liquid CM and were grown for another 2 days. Mycelia were filtered through Miracloth, were blotted dry, and were ground into powder in liquid nitrogen with a mortar and a pestle. A total of 5 g of mycelium was mixed with 15 ml of first extraction buffer I (0.1 M natrium aceticum, 10 mM Tris-HCl, 10 mM MgCl2 with 0.07% β-mercaptoethanol) and were incubated at 4°C for 2 h. Samples were centrifuged at 5,000 × g for 30 min at 4°C, repeated once before discarding the pellets. The upper phase was centrifuged at 96,000 × g at 4°C for 2 h, and the pellets were dissolved in a second extraction buffer (20 mM Tris, pH = 7.5, 6 mM MgCl2, 10% glycerol, 0.1% NP-40, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 μM leupeptin, and 1 μM pepstatin A). Protein extracts (2.5 ml) were overlaid on 7.5 ml of 1 M sucrose in 20 mM Tris, 8 mM MgCl2, and 100 mM KCl in 10-ml ultracentrifuge tubes (Beckman Coulter, Brea, CA, U.S.A.). Samples were centrifuged again at 96,000 × g at 4°C for 2 h. Finally, the pellets were dissolved in dissolution buffer (5 M urea, 2 M thiourea, 2% CHAPS, 2% SB3–10, 40 mM Tris, 5 mM Mercaptoethanol).
Western blot analysis of protein phosphorylation.
The mutants and wild-type Guy11 strains were cultured in liquid CM for 2 days and, then, the mycelia of each strain were harvested. Total proteins were isolated from vegetative hyphae as described by Bruno et al. (2004). The intensity of the signal corresponding to phosphorylated Mps1 was detected by binding of an antiphospho-p44/42 MAPK antibody (Cell Signaling Technology, Boston), with the Mpk1 antibody (N-terminal anti-Mpk1) from Santa Cruz Biotechnology (Santa Cruz, CA, U.S.A.) used as a control. Each experiment was repeated three times.
Phos-tag analysis.
The MoMKK1-GFP fusion constructs were transferred into their respective wild-type strain, ΔMossb1, ΔMossz1, and ΔMozuo1 mutants. The positive transformants were cultured in liquid CM for 48 h. For protein isolation, about 150 to 200 mg of mycelia were ground into powder in liquid nitrogen and were resuspended in 1 ml of extraction buffer (10 mM Tris-HCl [pH = 7.5], 150 mM NaCl, 0.5 mM EDTA, 0.5% NP-40) to which 1 mM PMSF, 10 μl of protease inhibitor cocktail (Sigma), and 10 μl of phosphatase inhibitor cocktail 3 (Sigma) had to be freshly added. For the preparation of the phosphatase-treated cell lysates, the phosphatase inhibitor cocktail was omitted for 2.5 U per milliliter of alkaline phosphatase (final concentration [P6774; Sigma]) and the sample was incubated for 1 h with the addition of 1 mM MgCl2 (37°C). Then, the samples were resolved on 8% sodium dodecyl sulfate-polyacrylamide gels prepared with 50 μM acrylamide-pendant Phos-tag ligand and 100 μM MnCl2 according to the instructions provided by the Phos-tag Consortium. Gels were electrophoresed at 80 V/gel for 3 to 6 h. Prior to transfer, gels were first equilibrated two times in transfer buffer containing 5 mM EDTA for 20 min and, then, in transfer buffer without EDTA for 10 min. Protein transfer from the Mn2+-Phos-tag acrylamide gel to the polyvinylidene diflouride membrane was performed overnight at 80 V at 4°C, and then, the membrane was analyzed by Western blotting (using anti-GFP antibodies).
Virulence assays.
Virulence assays were performed as described (Zhang et al. 2011a). Conidia were harvested from 10-day-old SDC agar cultures, were filtered through one-layer Miracloth, and were resuspended by 0.2% (wt/vol) gelatin solution to a concentration of 5 × 104 spores/ml. For detached barley assay, leaves from 7-day-old barley (H. vulgare cv. Four-arris) seedlings were drop-inoculated with three droplets (20 μl) of conidial suspension. Photographs were taken 5 days after incubation, alternating light and dark at 28°C. For the rice seedling spraying assay, two-week-old seedlings of rice (O. sativa cv. CO-39) were sprayed with a 5-ml conidial suspension of each treatment. Inoculated plants were kept in a growth chamber at 28°C and in the dark for the first 24 h, followed by a 12-h light and 12-h dark cycle. Lesion formation was checked and photographed 7 days after inoculation.
For microscopic observation of penetration and invasive hyphae expansion, a conidial suspension (1 × 105 spores/ml) was inoculated in the inner leaf sheath. After incubation for 30 or 48 h at 28°C, the inner leaf sheath cuticle cells were observed under Zeiss Axio Observer A1 inverted microscope.
Supplementary Material
Funding:
This research was supported by National Natural Science Foundation of China (grant number 31470248 to X. Zheng), the Fundamental Research Funds for the Central Universities number KYTZ201604 and KYT201805, and Innovation Team Program for Jiangsu Universities (2017). The Wang lab research was supported by the United States National Institutes of Health grant AI121451.
Footnotes
The e-Xtra logo stands for “electronic extra” and indicates that eight supplementary figures and two supplementary tables are published online.
LITERATURE CITED
- Avruch J 2007. MAP kinase pathways: The first twenty years. Biochim. Biophys. Acta 1773:1150–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin IJ, and McMillan DR 1998. Stress (heat shock) proteins: Molecular chaperones in cardiovascular biology and disease. Circ. Res 83:117–132. [DOI] [PubMed] [Google Scholar]
- Boorstein WR, Ziegelhoffer T, and Craig EA 1994. Molecular evolution of the HSP70 multigene family. J. Mol. Evol 38:1–17. [DOI] [PubMed] [Google Scholar]
- Borges JC, and Ramos CH 2005. Protein folding assisted by chaperones. Protein Pept. Lett 12:257–261. [DOI] [PubMed] [Google Scholar]
- Bruno KS, Tenjo F, Li L, Hamer JE, and Xu JR 2004. Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea. Eukaryot. Cell 3:1525–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B, and Horwich AL 1998. The Hsp70 and Hsp60 chaperone machines. Cell 92:351–366. [DOI] [PubMed] [Google Scholar]
- Cyr DM, Langer T, and Douglas MG 1994. DnaJ-like proteins: Molecular chaperones and specific regulators of Hsp70. Trends Biochem. Sci 19:176–181. [DOI] [PubMed] [Google Scholar]
- Finka A, Sharma SK, and Goloubinoff P 2015. Multi-layered molecular mechanisms of polypeptide holding, unfolding and disaggregation by HSP70/HSP110 chaperones. Front. Mol. Biosci 2:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautschi M, Lilie H, Fünfschilling U, Mun A, Ross S, Lithgow T, Rücknagel P, and Rospert S 2001. RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and zuotin. Proc. Natl. Acad. Sci. U.S.A 98:3762–3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautschi M, Mun A, Ross S, and Rospert S 2002. A functional chaperone triad on the yeast ribosome. Proc. Natl. Acad. Sci. U.S.A 99: 4209–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Guo W, Chen Y, Dong S, Zhang X, Zhang H, Song W, Wang W, Wang Q, Lv R, Zhang Z, Wang Y, and Zheng X 2010. The basic leucine zipper transcription factor Moatf1 mediates oxidative stress responses and is necessary for full virulence of the rice blast fungus Magnaporthe oryzae. Mol. Plant-Microbe Interact 23:1053–1068. [DOI] [PubMed] [Google Scholar]
- Hanebuth MA, Kityk R, Fries SJ, Jain A, Kriel A, Albanese V, Frickey T, Peter C, Mayer MP, Frydman J, and Deuerling E 2016. Multivalent contacts of the Hsp70 Ssb contribute to its architecture on ribosomes and nascent chain interaction. Nat. Commun 7:13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, and Hayer-Hartl M 2009. Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol 16:574–581. [DOI] [PubMed] [Google Scholar]
- Huang P, Gautschi M, Walter W, Rospert S, and Craig EA 2005. The Hsp70 Ssz1 modulates the function of the ribosome-associated J-protein Zuo1. Nat. Struct. Mol. Biol 12:497–504. [DOI] [PubMed] [Google Scholar]
- Hundley H, Eisenman H, Walter W, Evans T, Hotokezaka Y, Wiedmann M, and Craig E 2002. The in vivo function of the ribosome-associated Hsp70, Ssz1, does not require its putative peptide-binding domain. Proc. Natl. Acad. Sci. U.S.A 99:4203–4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundley HA, Walter W, Bairstow S, and Craig EA 2005. Human Mpp11 J protein: Ribosome-tethered molecular chaperones are ubiquitous. Science 308:1032–1034. [DOI] [PubMed] [Google Scholar]
- Jeon J, Goh J, Yoo S, Chi MH, Choi J, Rho HS, Park J, Han SS, Kim BR, Park SY, Kim S, and Lee YH 2008. A putative MAP kinase kinase kinase, MCK1, is required for cell wall integrity and pathogenicity of the rice blast fungus, Magnaporthe oryzae. Mol. Plant-Microbe Interact 21:525–534. [DOI] [PubMed] [Google Scholar]
- Kelley WL 1998. The J-domain family and the recruitment of chaperone power. Trends Biochem. Sci 23:222–227. [DOI] [PubMed] [Google Scholar]
- Koplin A, Preissler S, Ilina Y, Koch M, Scior A, Erhardt M, and Deuerling E 2010. A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J. Cell Biol 189:57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, and Srivastava P 2004. Heat-shock proteins. Curr. Protoc. Immunol 1 (Appendix):1T. [DOI] [PubMed] [Google Scholar]
- Liu X, Yang J, Qian B, Cai Y, Zou X, Zhang H, Zheng X, Wang P, and Zhang Z 2018. MoYvh1 subverts rice defense through functions of ribosomal protein MoMrt4 in Magnaporthe oryzae. PLoS Pathog 14: e1007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo KY, Li Z, Wang F, Marcotte EM, and Johnson AW 2009. Ribosome stalk assembly requires the dual-specificity phosphatase Yvh1 for the exchange of Mrt4 with P0. J. Cell Biol 186:849–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Presti L, López Díaz C, Turrà D, Di Pietro A, Hampel M, Heimel K, and Kahmann R 2016. A conserved co-chaperone is required for virulence in fungal plant pathogens. New Phytol 209:1135–1148. [DOI] [PubMed] [Google Scholar]
- Lopez-Buesa P, Pfund C, and Craig EA 1998. The biochemical properties of the ATPase activity of a 70-kDa heat shock protein (Hsp70) are governed by the C-terminal domains. Proc. Natl. Acad. Sci. U.S.A 95:15253–15258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning G, Whyte DB, Martinez R, Hunter T, and Sudarsanam S 2002. The protein kinase complement of the human genome. Science 298:1912–1934. [DOI] [PubMed] [Google Scholar]
- Mashaghi A, Bezrukavnikov S, Minde DP, Wentink AS, Kityk R, Zachmann-Brand B, Mayer MP, Kramer G, Bukau B, and Tans SJ 2016. Alternative modes of client binding enable functional plasticity of Hsp70. Nature 539:448–451. [DOI] [PubMed] [Google Scholar]
- Mayer MP, and Bukau B 2005. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell. Mol. Life Sci 62:670–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudholkar K, Fitzke E, Prinz C, Mayer MP, and Rospert S 2017. The Hsp70 homolog Ssb affects ribosome biogenesis via the TORC1-Sch9 signaling pathway. Nat. Commun 8:937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, and Craig EA 1992. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell 71:97–105. [DOI] [PubMed] [Google Scholar]
- Otto H, Conz C, Maier P, Wölfle T, Suzuki CK, Jenö P, Rücknagel P, Stahl J, and Rospert S 2005. The chaperones MPP11 and Hsp70L1 form the mammalian ribosome-associated complex. Proc. Natl. Acad. Sci. U.S.A 102:10064–10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisker K, Chiabudini M, and Rospert S 2010. The ribosome-bound Hsp70 homolog Ssb of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1803:662–672. [DOI] [PubMed] [Google Scholar]
- Pfund C, Huang P, Lopez-Hoyo N, and Craig EA 2001. Divergent functional properties of the ribosome-associated molecular chaperone Ssb compared with other Hsp70s. Mol. Biol. Cell 12:3773–3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissler S, and Deuerling E 2012. Ribosome-associated chaperones as key players in proteostasis. Trends Biochem. Sci 37:274–283. [DOI] [PubMed] [Google Scholar]
- Qiu XB, Shao YM, Miao S, and Wang L 2006. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci 63:2560–2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakwalska M, and Rospert S 2004. The ribosome-bound chaperones RAC and Ssb1/2p are required for accurate translation in Saccharomyces cerevisiae. Mol. Cell. Biol 24:9186–9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram AF, and Klis FM 2006. Identification of fungal cell wall mutants using susceptibility assays based on Calcofluor white and Congo red. Nat. Protoc 1:2253–2256. [DOI] [PubMed] [Google Scholar]
- Raue U, Oellerer S, and Rospert S 2007. Association of protein biogenesis factors at the yeast ribosomal tunnel exit is affected by the translational status and nascent polypeptide sequence. J. Biol. Chem 282:7809–7816. [DOI] [PubMed] [Google Scholar]
- Roncero C 2002. The genetic complexity of chitin synthesis in fungi. Curr. Genet 41:367–378. [DOI] [PubMed] [Google Scholar]
- Song W, Dou X, Qi Z, Wang Q, Zhang X, Zhang H, Guo M, Dong S, Zhang Z, Wang P, and Zheng X 2010. R-SNARE homolog MoSec22 is required for conidiogenesis, cell wall integrity, and pathogenesis of Magnaporthe oryzae. PLoS One 5:e13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigard JA, Chumley FG, and Valent B 1992. Disruption of a Magnaporthe grisea cutinase gene. Mol. Gen. Genet 232:183–190. [PubMed] [Google Scholar]
- Talbot NJ, Ebbole DJ, and Hamer JE 1993. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell 5:1575–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh P, Bursać D, Law YC, Cyr D, and Lithgow T 2004. The J-protein family: Modulating protein assembly, disassembly and trans-location. EMBO Rep 5:567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter S, and Buchner J 2002. Molecular chaperones—Cellular machines for protein folding. Angew. Chem. Int. Ed. Engl 41:1098–1113. [DOI] [PubMed] [Google Scholar]
- Xu JR, Staiger CJ, and Hamer JE 1998. Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc. Natl. Acad. Sci. U.S.A 95:12713–12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Schilke B, Pfund C, Walter W, Kim S, and Craig EA 1998. Zuotin, a ribosome-associated DnaJ molecular chaperone. EMBO J 17: 4809–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M, Chi MH, Khang CH, Park SY, Kang S, Valent B, and Lee YH 2009. The ER chaperone LHS1 is involved in asexual development and rice infection by the blast fungus Magnaporthe oryzae. Plant Cell 21:681–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Tang W, Wang J, Liu X, Yang L, Gao C, Zhang J, Zhang H, Zheng X, Wang P, and Zhang Z 2016. Phosphodiesterase MoPdeH targets MoMck1 of the conserved mitogen-activated protein (MAP) kinase signalling pathway to regulate cell wall integrity in rice blast fungus Magnaporthe oryzae. Mol. Plant Pathol 17:654–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liu K, Zhang X, Tang W, Wang J, Guo M, Zhao Q, Zheng X, Wang P, and Zhang Z 2011b. Two phosphodiesterase genes, PDEL and PDEH, regulate development and pathogenicity by modulating intracellular cyclic AMP levels in Magnaporthe oryzae. PLoS One 6:e17241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Xue C, Kong L, Li G, and Xu JR 2011a. A Pmk1-interacting gene is involved in appressorium differentiation and plant infection in Magnaporthe oryzae. Eukaryot. Cell 10:1062–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ma C, Yuan Y, Zhu J, Li N, Chen C, Wu S, Yu L, Lei J, and Gao N 2014. Structural basis for interaction of a cotranslational chaperone with the eukaryotic ribosome. Nat. Struct. Mol. Biol 21: 1042–1046. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Sinning I, and Rospert S 2017. Two chaperones locked in an embrace: Structure and function of the ribosome-associated complex RAC. Nat. Struct. Mol. Biol 24:611–619. [DOI] [PubMed] [Google Scholar]
- Zhong K, Li X, Le X, Kong X, Zhang H, Zheng X, Wang P, and Zhang Z 2016. MoDnm1 dynamin mediating peroxisomal and mitochondrial fission in complex with MoFis1 and MoMdv1 is important for development of functional appressorium in Magnaporthe oryzae. PLoS Pathog 12:e1005823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.