Abstract
To create artificial cartilage in vitro, mimicking the function of native extracellular matrix (ECM) and morphological cartilage‐like shape is essential. The interplay of cell patterning and matrix concentration has high impact on the phenotype and viability of the printed cells. To advance the capabilities of cartilage bioprinting, we investigated different ECMs to create an in vitro chondrocyte niche. Therefore, we used methacrylated gelatin (GelMA) and methacrylated hyaluronic acid (HAMA) in a stereolithographic bioprinting approach. Both materials have been shown to support cartilage ECM formation and recovery of chondrocyte phenotype. We used these materials as bioinks to create cartilage models with varying chondrocyte densities. The models maintained shape, viability, and homogenous cell distribution over 14 days in culture. Chondrogenic differentiation was demonstrated by cartilage‐typical proteoglycan and type II collagen deposition and gene expression (COL2A1, ACAN) after 14 days of culture. The differentiation pattern was influenced by cell density. A high cell density print (25 × 106 cells/mL) led to enhanced cartilage‐typical zonal segmentation compared to cultures with lower cell density (5 × 106 cells/mL). Compared to HAMA, GelMA resulted in a higher expression of COL1A1, typical for a more premature chondrocyte phenotype. Both bioinks are feasible for printing in vitro cartilage with varying differentiation patterns and ECM organization depending on starting cell density and chosen bioink. The presented technique could find application in the creation of cartilage models and in the treatment of articular cartilage defects using autologous material and adjusting the bioprinted constructs size and shape to the patient. © 2019 The Authors. Journal of Biomedical Materials Research Part B: Applied Biomaterials published by Wiley Periodicals, Inc. J Biomed Mater Res Part B: Appl Biomater 107B:2649–2657, 2019.
Keywords: bioprinting, stereolithography, photopatterning, articular cartilage, tissue engineering, biomaterial, gelatin, hyaluronic acid
INTRODUCTION
Three‐dimensional (3D) biological environments are needed to model complex cell–cell and cell–matrix interactions, the foundation for basic biological life. Tissue engineering focuses on the modeling and recreation of these biological niches. With the adaption of new technologies, more and more detailed in vitro cultures are created, surpassing simple 2D cell cultures through microphysiological cultivation in sophisticated organ‐on‐a‐chip co‐cultures.1 Solid freeform manufacturing (SFM) is making a big contribution to this motion.2 Technologies, which have their origin in rapid prototyping, are applied and adapted to biotechnological workflows. The idea is to utilize these technologies to create detailed models of human tissue or organs.3 Especially 3D printing is fueling this evolution. 3D printing with biological material, coined bioprinting, allows the deposition of biological materials such as cells, biopolymers, and chemokines in three‐dimensional space. Different technologies from SFM are applied to bioprinting—inkjet printing, extrusion, laser‐induced forward transfer (LIFT), or magnetic levitation are used to produce 3D objects.4, 5 Although bioprinting is coming of age, a complete organ is not printable yet, but small functional units are possible.6 Due to the modularity of the human body, these bricks can resemble the function of a complete organ. Thus, small 3D organ models can be used to study cell–cell or cell–matrix interactions, organ neogenesis, transplantational studies, or drug development. It is important to create these small bricks as precise and detailed as possible to imitate the in vivo situation. Subsequently in vivo situations might be modeled in vitro.
With the presented bottom–up stereolithographic bioprinting technology, precise printing of multiple materials within one print run is possible. Thus, different layers and gradients inside a single object can be created. In addition, different cell types can be deposited which allows the printing of complex objects. Furthermore, multiple objects can be bioprinted simultaneously in the projection area without time delay.7
In this work, we investigated the close interplay between cell patterning, matrix choice, and initial cell density for cartilage bioprinting. Cartilage is the ideal model to test this technology due to its biological properties. It has a rather simple composition, but the chondrocytes require a sophisticated extracellular matrix (ECM).8, 9 To create well‐defined 3D in vitro cartilage of different compositions, we used stereolithography, as this technology offers a mechanically gentle printing procedure, by incorporating the cells in the bioink by light exposure.10, 11, 12, 13 Depending on the used bioprinting technique, parameters such as cell pattern, matrix stiffness, shape, porosity, and so on, have to be adapted to the desired model structure.13 A substantial contribution to the creation of the in vitro cell niche as organ model is made by the used bioinks.14
In this study, we used gelatin and hyaluronic acid as base matrices for the bioink development. Both materials have been shown to support cartilage ECM formation and maintenance of chondrocyte phenotype.15 By modifying the materials with acrylic groups, highly structured, cell‐laden hydrogels can be formed by stereolithographic bioprinting. Hyaluronic acid was found to improve cell viability in bioprinted constructs.16 Methacrylated gelatin was found to enhance chondrogenesis.17
Aside from technical parameters for stereolithographic bioprinting to create an in vitro cartilage, we investigated the survival of chondrocytes and the development of cartilage‐like ECM. We varied matrices and densities of cells to demonstrate technical possibilities and the potential to influence biological properties of tissue that is essentially emulating human biology. For analysis of biocompatibility, we made use of porcine chondrocytes that have been shown to model substantial aspects of cartilage physiology and pathology such as osteoarthritis in vitro.18
METHODS
Chondrocyte isolation and propagation
Articular cartilage slices were harvested from the medial and lateral femoral condyle of domestic pigs (n = 3, 6–12 months old). Chondrocytes were isolated according to a protocol previously published.19 Cartilage slices were incubated for 19 h in stirred flasks containing RPMI 1640 medium (Merck, Darmstadt, Germany) supplemented with 10% fetal bovine serum (FBS, v/v), 100 U/mL penicillin and 100 μg/mL streptomycin, 333.3 U/mL collagenase II (all Merck), 1 U/mL collagenase P (Roche Diagnostics, Mannheim, Germany), and 33.3 U/mL hyaluronidase (Sigma‐Aldrich, Steinheim, Germany). After incubation, cell suspensions were strained through a nylon mesh with 100 μm pore diameter (Becton Dickinson, Heidelberg, Germany), washed in Hanks solution (Merck), resuspended in propagation medium (RPMI 1640, 10% FBS, penicillin/streptomycin as above), and seeded at 104 cells/cm2 in standard cell culture flasks (passage 0). Medium was changed completely 3 times a week. Once the cells reached confluence, they were detached with trypsin (Merck) and seeded as above.
Chondrocytes of passage 2 from three different individuals were equally pooled and used for construction of 3D cultures. Constructs were maintained for 14 days serum‐free in DMEM high glucose (Merck) supplemented with ITS+1, 0.1 μM dexamethasone, 1 mM sodium pyruvate, 0.17 mM l‐ascorbate‐2‐phosphate; 0.35 mM l‐prolin (all Sigma‐Aldrich), 100 U/mL penicillin, 100 μg/mL streptomycin, and 10 ng/mL transforming growth factor beta 3 (TGF‐β3, Peprotech, Rocky Hill, NJ). Medium (1 mL per construct) was changed completely three times a week.
Bioink preparation
Bioinks based on gelatin and hyaluronic acid were synthesized as previously described resulting in methacrylated gelatin (GelMA) and methacrylated hyaluronic acid (HAMA).20, 21 In short, a 10 wt % gelatin (porcine skin Type B, ~300 bloom, Sigma) or 2 wt % hyaluronic acid (Alfa Aesar, ~120 kDa) were dissolved in PBS at 50°C and 5°C, respectively.22 Twenty‐fold excess methacrylic anhydride (Sigma) was added and reaction continued for 3 h (24 h for hyaluronic acid). After reaction, the product was dialyzed against distilled water. Products were freeze dried and lyophilized for precise bioink preparation. The degree of methacrylation was found at 60% (GelMA) and 14% (HAMA) via NMR‐spectroscopy. The photoinitiator lithium phenyl‐2,4,6‐trimethylbenzoylphosphinate was used at 0.1 wt % in all bioinks.23 For the bioprinting process, cells were mixed with bioink solutions containing the photoinitiator to form a bioink cell suspension ready for photopolymerization. Thereby, the final polymer concentration is diluted. GelMA was printed at 5 wt % and HAMA was printed at 1 wt %. Both bioinks were used with two different cell densities for comparison. 5 × 106 cells/mL (0.4 × 106 cells/construct) and 25 × 106 cells/mL (2 × 106 cells/construct) were defined as low and high cell density, respectively.
Microfabrication of cartilage equivalents via bioprinting
Microfabrication of tissue equivalents was performed by a DLP‐based bioprinting process as visualized in Figure 1. Prior to the bioprinting, a 3D model of the tissue construct was created using computer‐aided design (CAD) software (Rhinoceros 5, McNeel Europe) and processed by the bioprinter for fabrication. The bioink cell suspension was prepared by mixing cells with GelMA or HAMA [Figure 2(A)]. Tissue constructs were fabricated layer by layer in a stereolithographic process as illustrated in Figure 2. During the printing process, each layer of the tissue construct was photopolymerized directly onto the print‐head holding a carrier membrane by blue light illumination (385–405 nm) for 30 s each. The carrier membrane is used to easily remove the printed constructs from the printer for subsequent cultivation of the tissue models. Each model consists of three layers (~300 μm per layer) resulting in a model height of ~1 mm and a model diameter of 8 mm. After printing, the constructs are detached from the bioprinter and placed in a 24‐well plate filled with cell culture medium for cultivation. Tissue constructs were incubated at 37°C at 5% CO2 over 14 days cultivation time. Two time points, 1 day after printing and 14 days after printing, were chosen for analysis in triplicates unless stated otherwise. Figure 2E shows the photograph of the printed cartilage model.
Figure 1.
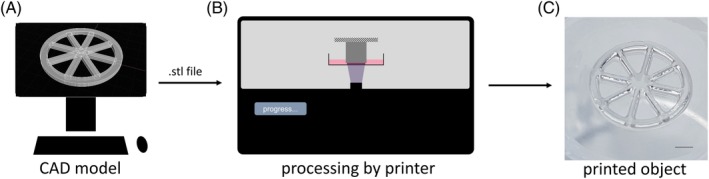
Schematic: from CAD model to printed object. A: A 3D model is designed with a CAD software and exported as stereolithography (STL) file. B: The file is fed to the bioprinter and interpreted. Layers of defined thickness (here ~300 μm) are created from the 3D file and processed for the DLP projection. C: The object is printed layer‐by‐layer ready for cultivation afterward, scale bar is 2 mm.
Figure 2.

Schematic of bioprinting process. A: Below the print‐head, a bioink reservoir filled with the bioink cell suspension is located. The suspension is prepared in advance. B: A digital light processing (DLP) unit projects each layer of the 3D model through the bottom of the bioink reservoir onto the print‐head. C: With each layer, the print‐head adjusts its position to the transparent projection screen to a defined height. D: After the last layer is printed, the print‐head drives to its initial position (E). The printed tissue can be detached from the printer and it is ready for cultivation. A photograph illustrates the finished print. Scale bar is 1 mm.
Histological and immunohistochemical analysis
To document ECM formation, histological and immunohistochemical stainings were performed on 8 μm cryosections obtained from bioink‐cell constructs embedded in optimal cutting temperature (OCT) compound (Sakura Finetek, Alphen aan den Rijn, Netherlands). Cartilage‐typical sulfated glycosaminoglycans (GAG) were stained with 0.7% Safranin O in 66% ethanolic solution, and cell nuclei were counterstained with 0.2% Fast Green FCF (Sigma‐Aldrich) in 0.3% acetic acid. Additionally, cartilage‐specific type II collagen was detected immunohistochemically with polyclonal rabbit anti‐porcine type II collagen antibodies (Acris Antibodies, Herford, Germany). Rabbit IgG (DAKO, Hamburg, Germany) served as control. EnVision++ horseradish peroxidase (HRP) rabbit Kit (DAKO) was used for antibody detection, and nuclei were counterstained with hematoxylin (DAKO). Stainings were photodocumented using a light microscope (Axio 10, Zeiss, Jena, Germany) equipped with the ProgRes speed XT core 5 camera and ProgRes CapturePro 2.10 software (both Jenoptik, Jena, Germany).
Collagen type II‐stained sections were used to determine the cell number inside the construct. Nuclei of five representative areas (each 0.25 mm2) were manually counted (Supporting Information, File 1). Cell number per construct was calculated according to the method published by Aherne considering section thickness and diameter of cells.24
Test of viability
Whole constructs were examined for viability involving propidium iodide/fluorescein diacetate staining (PI/FDA; Sigma‐Aldrich) after 14 days of maintenance. Washing steps were done using PBS (Merck). The staining was performed first in FDA solution (3 μg/mL; 15 min, 37°C) and subsequently in PI staining solution (100 μg/mL; 2 min; RT). For microscopy, an Olympus CKX41 combined with a reflected fluorescence microscopy system was used (Olympus, Hamburg, Germany; camera and software as above).
RNA preparation and real‐time rtPCR analysis
For total RNA from construct and monolayer cultures, one construct or an equivalent of detached and pooled cells (from three different individuals) were snap‐frozen and stored at −80°C. Frozen samples were transferred to TriReagent (Sigma‐Aldrich) and mechanically homogenized. Subsequently, 1‐bromo‐3‐chloro‐propane (Sigma‐Aldrich) was admixed followed by centrifugation for 45 min at 13,000g. The aqueous phase was collected and nucleic acids were precipitated by the addition of an equal volume of ice‐cold isopropanol. After 30 min of incubation, precipitated nucleic acids were collected and resolved in RNA isolation buffer (RLT, Qiagen, Hilden, Germany). Further purification was performed according to a protocol for animal tissues of the RNeasy Mini Kit (Qiagen).
For real‐time rtPCR analysis, RNA was reverse transcribed (rt) using the iScript cDNA synthesis kit (Bio‐Rad, München, Germany). Real‐time rtPCR was performed in technical triplicates in 96‐well plates (Becton Dickinson) on a iCycler (Bio‐Rad) using expression assays for TaqMan probes and primer sets (order no. in parentheses): collagen type II alpha 1 (COL2A1, Ss03373344_g1), collagen type I alpha 1 (COL1A1, Ss003373341_g1), aggrecan (ACAN, SS03373387_S1). To normalize the samples, the expression of GAPDH (glyceraldehyde‐3‐phosphate dehydrogenase, SS03375435_u1) was used. Marker gene expression is given as a percentage related to GAPDH expression applying the efficiency corrected Δ‐Ct method.25
Statistical analysis
The significance level was determined with the independent two‐sample t‐test statistics of the Excel 2007 software package (Microsoft). Normality distribution was checked applying the Anderson−Darling test,26 and equal variance of compared sample groups was tested applying the f test27 In all groups, signals were normally distributed. If the equal variance test was not passed, Welch's t test was applied.28
RESULTS
Maintenance of construct specifications in cell culture
Bioprinted constructs were cultivated in chondrogenic medium for 14 days. The construct's spherical shape and its adhesion to the carrier membrane were maintained over the whole culture time in cell‐free gelatin and HA as well in low, high cell density constructs [Figure 3(A–F)]. The diameter of cell‐free constructs was found constant with 8 mm in GelMA and 7.9 mm in HAMA‐based construct after 14 days of culture. Cell‐laden cultures tended to shrink by 12% in average in diameter after culture (Supporting Information, File 2). Histological stainings demonstrated that cells were homogenously distributed in all types of constructs (Figure 3). In accordance with intended cell density, the number of cells in low cell density constructs [Figure 3(H,K)] was apparently lower compared to the high cell density constructs [Figure 3(I,L)]. Determination of cell number on tissue sections demonstrated that low density constructs contained 0.39–0.44 × 106 cells and high density constructs contained 2.1–2.2 × 106 cells confirming visual observations (Supporting Information, File 1).
Figure 3.
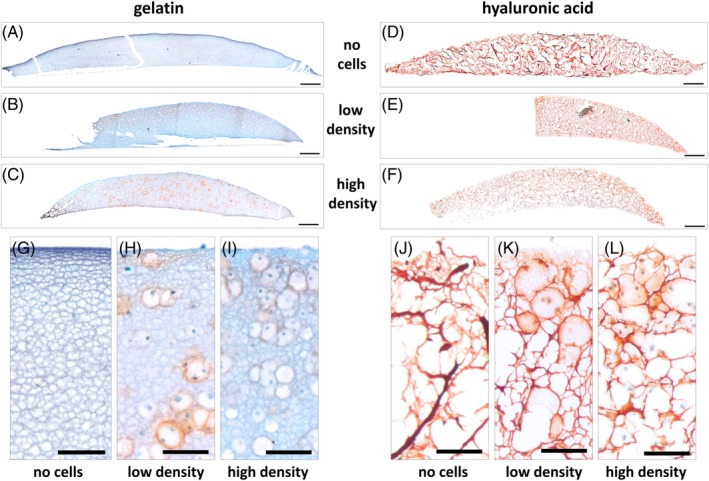
Detection of proteoglycans in constructs after 14 days of culture. Safranin O stain of tissue section documented the presence of proteoglycans in red. Nuclei and other ECM/bioink appear green to blue by Fast Green counterstain. Porcine chondrocytes were embedded in (A,B,C,G,H,I) gelatin and (D,E,F,J,K,L) hyaluronic acid bioinks with low (B,E,H,K) and high (C,F,I,L) cell density using the Cellbricks Bioprinting technique. (A,D,G,J) Cell‐free constructs cultured for 14 days. (A–F) Scale bar is 500 μm; (G–L) Scale bar is 100 μm.
Enabling viability of cells
Live/dead staining of whole constructs demonstrated that the vast majority of cells remained vital in all types of constructs after 14 days of culture [Figure 4(A–D)]. Only a small proportion of cell (approximately <5%) was found dead at this late stage of cell culture.
Figure 4.

Vitality of cells in constructs after 14 days of culture. Fluorescent microscopy images of PI/FDA stained constructs showing living cells in green and dead cells in red: (A,C) cells in bioprinted gelatin; (B,D) cells in bioprinted hyaluronic acid; (A,B) low density = 5 mio cells/mL; (C,D) high density = 25 mio cells/mL. Scale bar = 250 μm.
Detection of a cartilage‐like proteoglycans
In gelatin‐based constructs, the deposition of sulfated proteoglycans was indicated by positive (red) Safranin O staining. Whereas in low‐density cultures, formed proteoglycans appeared more evenly distributed [Figure 3(B)], in high cell density cultures, this ECM was found more pronounced at the (outer) surface, facing the medium, compared to (inner) areas close to the carrier membrane [Figure 3(C)]. The ECM is predominantly deposited closely adjacent to cells visible as a red ring around the cell nucleus in histological stainings [Figure 3(B,C)].
Assessment of proteoglycan formation in HAMA‐based constructs by Safranin O staining was only possible to a limited extent, as HA itself was intensively stained [Figure 3(D)]. In accordance with gelatin, surface areas of high cell density constructs appeared to contain more ECM compared to inner areas [Figure 3(F)]. Likewise, ring‐like structures around cell nuclei displayed a more intensive staining [Figure 3(K,L)] than cell‐free areas [Figure 3(J)]. Alcian blue stainings of proteoglycans confirmed the abovementioned observations (Supporting Information, File 3).
Formation of cartilage‐typical collagen type II
In high cell density constructs made of both GelMA and HAMA, the formation of cartilage‐specific collagen type II was detected after 14 days of culture [Figure 5(C,F)]. ECM‐forming chondrocytes were predominantly found at the surface area, but also in deeper zones closer to the carrier membrane [Figure 5(I,L)].
Figure 5.

Detection of cartilage‐specific collage type II in constructs after 14 days of culture. Collagen type II immunostaining stain of tissue section documented the presence of proteoglycans in red. Nuclei and other ECM/bioink appear blue to purple by hematoxylin counterstain. Porcine chondrocytes were embedded in (A,B,C,G,H,I) gelatin and (D,E,F,J,K,L) hyaluronic acid bioinks with low (B,E,H,K) and high (C,F,I,L) cell density using the Cellbricks bioprinting technique. (A,D,G,J) Cell‐free constructs cultured for 14 days. (A–F) Scale bar = 500 μm; (G–L) Scale bar = 100 μm.
In low cell density constructs, collagen type II was not detected in HAMA‐based constructs [Figure 5(K)], but succeeded in GelMA in only a few cells close to the surface [Figure 5(H)].
Induction of a chondrocyte‐like gene expression pattern
The gene expression patterns of chondrocytes embedded in gelatin and HA constructs at low and high cell density (day 14) were compared with the patterns of monolayer chondrocytes (Figure 6). The induction of collagen type IIα1 (COL2A1) and aggrecan (ACAN) expression was remarkable higher in high cell density than in low cell density in both gelatin and HA constructs (p < 0.01). In contrast to histological findings, in low cell density constructs the gene expression of COL2A1 and ACAN was higher in HA than in gelatin constructs, but only significant for ACAN. Distinct differences between gelatin and HA were also observed for collagen type Iα1 (COL1A1) expression. In both low (p < 0.05) and high cell density (p < 0.001), the expression of this dedifferentiation gene was found higher in gelatin compared to HA.
Figure 6.

Quantitative gene expression analysis in constructs after 14 days of culture. Relative expression of cartilage‐specific collagen type II (COL2A1) and aggrecan (ACAN) as well as dedifferentiation/bone‐related collagen type I (COL1A1) was analyzed in constructs of gelatin (Gel) and hyaluronic acid (HA) with cell densities of 5 × 106 cells/mL (low) and 25 × 106 cells/mL (high) compared to the expression of monolayer (ML) chondrocytes. Glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) was used as reference gene. Bars represent SD; *p < 0.05, **p < 0.01, ***p < 0.001, above column: low versus high.
DISCUSSION
This study focused on the feasibility to create and maintain the biological function of an articular cartilage tissue model for in vitro research purposes produced by stereolithographic bioprinting technology. The creation and reproduction of the model with gelatin‐ and HA‐based bioinks was successful. Thereby, gelatin was used at 5 wt % and HA at 1 wt % as these concentrations have shown to be well manageable in our daily stereolithographic bioprinting routine. The complexity of HA solution preparation, for example at varying temperatures, different molecule sizes or polymer concentrations, probably influences the effects of the hydrogel on the cells. Therefore, we only followed methods published by Smeds et al. for HAMA synthesis from HA solution.22 We demonstrated the stability in shape and dimension for both bioinks over 14 days under cell culture conditions. It was possible to encapsulate chondrocytes with different cell densities that were maintained over culture time in low and high cell number constructs, respectively. Intended cell density was reached with minor deviations between 3% and 10%, and construct's size (diameter) was found deviated <1%. High viability and the ability to print with varying cell number have been demonstrated shortly after printing as well as after 14 days of maintenance in vitro. Cell‐laden constructs tended to slightly shrink during culture, which has been shown by other authors as a result of tissue remodeling through matrix–cell interaction29 or matrix degradation and synthesis.30 Gelatin and HA are prone to such cell‐driven modifications through their natural origin, being advantageous for cell differentiation processes such as chondrocyte redifferentiation observed in this study.
In this study, we printed one cartilage model per printing procedure to test feasibility and biological relevance. In contrast to extrusion‐based bioprinting, where each layer needs to be drawn by the x–y matrix, the digital light processing (DLP) projection‐based printing technology allows to print multiple objects simultaneously without time delay.7 Ideally, simultaneous production of many constructs at a time can be adapted in future applications. This results in high reproducibility and comparability between replicates.
The culture in GelMA and HAMA hydrogels facilitated the redifferentiation of monolayer‐expanded chondrocytes demonstrated by cartilage‐typical proteoglycan and cartilage‐specific type II collagen deposition as well as cartilage marker gene expression (COL2A1, ACAN) after 14 days of culture. Two weeks is a time that most 3D in vitro models require to develop an acceptable redifferentiated phenotype (if dedifferentiated by monolayer expansion) and to create sufficient ECM for in vitro testing.19, 31, 32 On histological level, for both materials, a different pattern of differentiation depending on the cell density was observed. Whereas low cell density (5 × 106 cells/mL) constructs displayed a more homogenous distribution of proteoglycans and collagen type II, constructs with high cell density (25 × 106 cells/mL) displayed the formation of zones with an higher extent of proteoglycan distribution in layers closer to the culture medium. As oxygen limitation normally induces enhanced proteoglycan synthesis in chondrocytes,33 the lower proteoglycan content in deeper zones is more likely associated with lower nutrients availability such as glucose.34 So, influencing cell density is an expedient feature of the applied bioprinting technique, as a sophisticated modeling of native cartilage structures requires the generation of zones of different ECM components and cell numbers.35 For example, native articular cartilage is characterized by zonal segmentation, ranging from a collagen‐rich layer with higher cell density in the superficial zone and a zone rich of proteoglycans with fewer cells in deeper layers close to the subchondral bone36 constituting the unique biomechanical properties such as high resilience to shear force at the cartilage surface and to compression in the deep zone.37 In high‐density and conventional matrix‐assisted cell culture models, the initial homogenous distribution is normally altered by cells. This act of self‐organization is time‐consuming,38 so that the maintenance of a steady state and, thus reproducibility, is only possible within restrictive specifications.
On gene expression level differences in quality of differentiation between GelMA‐ and HAMA‐based constructs were detected. Whereas the expression of COL1A1 in HAMA‐based constructs was on a level similar to monolayer‐expanded chondrocytes, the expression was remarkably higher in GelMA‐based constructs. High expressions of COL1A1 are typical for in vitro culture using TGF‐beta as chondroinductor.39 Its induction is closely associated with dedifferentiating chondrocytes and loss of chondrocyte phenotype.40 Therefore, the GelMA component seems to promote a more premature chondrocyte phenotype compared to HAMA. Most studies on bioprinted cartilage use extrusion‐based technologies. In contrast to many studies summarized by Wu et al.,41 we achieved bioprinting with cell densities exceeding reported ranges of 2 × 107 cells/mL.42 Furthermore, the presented stereolithographic approach facilitates the production of constructs that are highly customizable (Supporting Information, File 4).
Comparing different bioprinting technologies, stereolithography showed promising results concerning feasibility and scalability for future applications. Extrusion‐based bioprinting technologies are easily capable of creating constructs with dimensions in the range of centimeters. Laser‐based printing technologies such as LIFT result in very high precision. Both technologies are more time consuming the larger, the printed objects are or the more objects are to be printed. The presented stereolithographic bioprinting technology combines high resolution with the ability to print multiple objects at a time.
For applications that require a high initial mechanical stability of the constructs, the materials used in its present configuration (GelMA, HAMA) are not recommended. Parameters such as degree of methacrylation, concentration of gelatin and HA, and UV light exposure were adjusted to allow flexible spatial definition of the construct and survival of the cells.
In many use cases, high initial load bearing capabilities are not necessarily required. For example, established clinical applications in cartilage repair make use of chondrocytes, which are encapsulated in fibrin within a polyglycol acid carrier fleece43 or are applied as spheroids,44 both posing only minimal initial mechanical stability. The load‐bearing features of the transplant develop in vivo promoted by environmental factors such as mechanical force, oxygen tension, and synovial fluid constituents.45 Mimicking those conditions during in vitro culture supports the generation of a mechanically robust cartilage models and is also mandatory for constructs composed of biodegradable materials to maintain stability, as the chondrocyte actively drive matrix turnover and tissue remodeling.46, 47
Most studies on bioprinted cartilage use extrusion‐based technologies, only one stereolithographic techniques is available so far.41 Zhu et al. have reported the successful fabrication of in vitro cartilage based on mesenchymal progenitor cells, which have been differentiated toward a chondrocytic phenotype with TGF‐beta in GelMA.48 In this study, we have demonstrated for the first time the utilization of methacrylated hyaluronic acid in a stereolithographic bioprinting approach to generate a viable cartilage‐like tissue in vitro. Furthermore, we were able to encapsulate cells in two different materials (GelMA, HAMA) using the same bioprinter setting opening the perspective for combination (mixtures) and zonal stratification (layering) of bioinks to generate more sophisticated cartilage constructs.
CONCLUSION
Using GelMA and HAMA in a stereolithographic bioprinting approach, we were able to create in vitro cartilage models with different cell concentrations, which allowed recovery of chondrocyte differentiation status over the course of 14 days. Based on these results, future experiments focus on the combination of bioinks to amplify the power of both materials, as cartilage shows a three‐dimensional zonal structure with different matrix composition and rigidity. Therefore, multi material bioprinting seems to be an ideal technology to create an in vitro model with blended bioinks. The bioprinted articular cartilage constructs could extend a previously published bone marrow model49 to a complete tissue engineered in vitro femoral head describing joint diseases like osteoarthritis. Furthermore, bioprinting technology becomes crucial in enhancing tissue models mimicking human in vivo organ interaction. Such models find increasing application in a number of sophisticated micro physiological systems used to solve the drug‐testing dilemma.50, 51 Despite the usage as an in vitro organ model, bioprinted cartilage based on gelatin and hyaluronic acid could potentially find clinical application in repairing cartilage defects using patient‐specific cells incorporated in the printed constructs. Size and shape of the printed hydrogel could be adjusted according to the defects dimensions.
Supporting information
Supporting Information File 1 Determination of construct cell number. Collagen type II immunostainings that have been counterstained with hematoxylin were used for the verification of cell number inside the construct after 14 days of cell culture. Gelatin‐ (A,B) and HA‐based (C,D) with low (A,C) and high cell density (B,D) were analyzed. Cell nuclei of five representative areas (each 0.5 mm × 0.5) were manually counted (E, area 1–5). The average was used to calculate the cell number according to a method published by Aherne in 1967 (PMID 5582822). The cell number per area volume (N A) was estimated from the mean count (n), area (A = 0.25 mm2), section thickness (t = 8 μm), and the estimated diameter of the cell nuclei (d = 6 μm) with N A = n/(A × (d + t)). Neglecting the minimal volume loss of the construct observed during culture (V K = 50 μL), the cell number per construct was calculated through N K = V K/V A × N A with V A = A × (d + t). Bar = 500 μm.
Supporting Information Figure 2 Measurements of diameter. Intact (A–C) or symmetrically interpolated (D, if not complete) sections were used to determine the diameter of the constructs (E). Sections were chosen that showed highest diameter. Measuring the height was not possible as it could not be assured that cutting plane was exactly at right angle to the construct's plane.
Supporting Information Figure 3 Detection of proteoglycans in constructs after 14 days of culture. Alcian blue stain of tissue section documented the presence of proteoglycans in blue. Nuclei and other ECM/bioink appear pink to red by Nuclear Fast Red counterstain. Porcine chondrocytes were embedded in (A, B, E, F) gelatin and (C, D, G, H) hyaluronic acid bioinks with low (A, C, E, G) and high (B, D, F, H) cell density using the Cellbricks Bioprinting technique. (A–D) Bar = 500 μm; (E–H) Bar = 100 μm. Alcian blue staining method: Detection of proteoglycan by Alcian blue was performed on 8 μm cryosections as described in the main text with 1% Alcian blue 8GX (Sigma) in 3% acetic acid (pH 2.5). Cell nuclei were counterstained with Nuclear Fast Red aluminum sulphate solution (ready to use, Roth).
Supporting Information Figure 4 Bioprinted constructs. These are examples of bioprinted cartilage models. (A) Bioprinted form of meniscal structure, (B) except of meniscal structure. (C, D) A defined defect was printed within the meniscal topography, here marked by yellow arrows. All scale bars represent 1 cm.
ACKNOWLEDGMENTS
JPK, SH, and ME are employees of TransTissue Technologies GmbH (TTT). TTT develops products and cartilage treatment strategies in the field of regenerative medicine. TL and AT are employees of Cellbricks GmbH engineering human tissue models via bioprinting technology. TD, MS, and RL declare no conflict of interest. The authors are very grateful to Samuel Vetterlein for his excellent technical assistance.
Lam T, Dehne T, Krüger JP, Hondke S, Endres M, Thomas A, Lauster R, Sittinger M, Kloke L. 2019. Photopolymerizable gelatin and hyaluronic acid for stereolithographic 3D bioprinting of tissue‐engineered cartilage. J Biomed Mater Res Part B. 2019:107B:2649–2657.
REFERENCES
- 1. Maschmeyer I, Lorenz AK, Schimek K, Hasenberg T, Ramme AP, Hübner J, Lindner M, Drewell C, Bauer S, Thomas A, Sambo NS, Sonntag F, Lauster R, Marx U. A four‐organ‐chip for interconnected long‐term co‐culture of human intestine, liver, skin and kidney equivalents. Lab Chip 2015;15(12):2688–2699. [DOI] [PubMed] [Google Scholar]
- 2. Mandrycky C, Wang Z, Kim K, Kim DH. 3D bioprinting for engineering complex tissues. Biotechnol Adv 2016;34(4):422–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bajaj P. 3D biofabrication strategies for tissue engineering and regenerative medicine. Annu Rev Biomed Eng 2014;16:247–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kang T‐Y, Hong JM, Jung JW, Kang H‐W, Cho D‐W. Construction of large‐volume tissue mimics with 3D functional vascular networks. PLoS One 2016;11(5):e0156529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ozbolat IT, Hospodiuk M. Current advances and future perspectives in extrusion‐based bioprinting. Biomaterials 2016;76:321–343. [DOI] [PubMed] [Google Scholar]
- 6. Billiet T, Vandenhaute M, Schelfhout J, Van Vlierberghe S, Dubruel P. A review of trends and limitations in hydrogel‐rapid prototyping for tissue engineering. Biomaterials 2012;33(26):6020–6041. [DOI] [PubMed] [Google Scholar]
- 7. Grix T, Ruppelt A, Thomas A, Amler A‐K, Noichl B, Lauster R, Kloke L. Bioprinting perfusion‐enabled liver equivalents for advanced organ‐on‐a‐chip applications. Genes (Basel) 2018;9(4):176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bryant SJ, Durand KL, Anseth KS. Manipulations in hydrogel chemistry control photoencapsulated chondrocyte behavior and their extracellular matrix production. J Biomed Mater Res A 2003;67(4):1430–1436. [DOI] [PubMed] [Google Scholar]
- 9. Bajaj P, Chan V, Jeong JH, Zorlutuna P, Kong H, Bashir R. 3‐D biofabrication using stereolithography for biology and medicine. Conf Proc IEEE Eng Med Biol Soc 2012;2012:6805–6808. [DOI] [PubMed] [Google Scholar]
- 10. Melchels FPW, Feijen J, Grijpma DW. A review on stereolithography and its applications in biomedical engineering. Biomaterials 2010;31(24):6121–6130. [DOI] [PubMed] [Google Scholar]
- 11. Skoog S a, Goering PL, Narayan RJ. Stereolithography in tissue engineering. J Mater Sci Mater Med 2014;25(3):845–856. [DOI] [PubMed] [Google Scholar]
- 12. Han L‐H, Mapili G, Chen S, Roy K. Projection microfabrication of three‐dimensional scaffolds for tissue engineering . J Manuf Sci Eng 2008;130(2):021005. [Google Scholar]
- 13. Chan V, Zorlutuna P, Jeong JH, Kong H, Bashir R. Three‐dimensional photopatterning of hydrogels using stereolithography for long‐term cell encapsulation. Lab Chip 2010;10(16):2062–2070. [DOI] [PubMed] [Google Scholar]
- 14. Klotz BJ, Gawlitta D, Rosenberg AJWP, Malda J, Melchels FPW. Gelatin‐methacryloyl hydrogels: Towards biofabrication‐based tissue repair. Trends Biotechnol 2016;34(5):394–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Levett P a, Melchels FPW, Schrobback K, Hutmacher DW, Malda J, Klein TJ. Chondrocyte redifferentiation and construct mechanical property development in single‐component photocrosslinkable hydrogels. J Biomed Mater Res A 2014;102(8):2544–2553. [DOI] [PubMed] [Google Scholar]
- 16. Souness A, Zamboni F, Walker GM, Collins MN. Influence of scaffold design on 3D printed cell constructs. J Biomed Mater Res B Appl Biomater 2018;106(2):533–545. [DOI] [PubMed] [Google Scholar]
- 17. Murphy C, Kolan K, Li W, Semon J, Day D, Leu M. 3D bioprinting of stem cells and polymer/bioactive glass composite scaffolds for bone tissue engineering. Int J Biporinting 2017;3:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schlichting N, Dehne T, Mans K, Endres M, Stuhlmüller B, Sittinger M, Kaps C, Ringe J. Suitability of porcine chondrocyte micromass culture to model osteoarthritis in vitro. Mol Pharm 2014;11(7):2092–2105. [DOI] [PubMed] [Google Scholar]
- 19. Lübke C, Ringe J, Krenn V, Fernahl G, Pelz S, Kreusch‐Brinker R, Sittinger M, Paulitschke M. Growth characterization of neo porcine cartilage pellets and their use in an interactive culture model. Osteoarthr Cartil 2005;13(6):478–487. [DOI] [PubMed] [Google Scholar]
- 20. Van Den Bulcke a I, Bogdanov B, De Rooze N, Schacht EH, Cornelissen M, Berghmans H. Structural and rheological properties of methacrylamide modified gelatin hydrogels. Biomacromolecules 2000;1(1):31–38. [DOI] [PubMed] [Google Scholar]
- 21. SMEDS K, PFISTER‐SERRES A, HATCHELL D, GRINSTAFF M. Synthesis of a novel polysaccharide hydrogel. J Macromol Sci A Pure Appl Chem 1999;36(7&8):981–989. [Google Scholar]
- 22. Smeds K a, Grinstaff MW. Photocrosslinkable polysaccharides for in situ hydrogel formation. J Biomed Mater Res 2001;54(1):115–121. [DOI] [PubMed] [Google Scholar]
- 23. Fairbanks BD, Schwartz MP, Bowman CN, Anseth KS. Photoinitiated polymerization of PEG‐diacrylate with lithium phenyl‐2,4,6‐trimethylbenzoylphosphinate: Polymerization rate and cytocompatibility. Biomaterials 2009;30(35):6702–6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aherne W. Methods of counting discrete tissue components in microscopical sections. J R Microsc Soc 1967;87(3):493–508. [DOI] [PubMed] [Google Scholar]
- 25. Pfaffl MW. A new mathematical model for relative quantification in real‐time RT‐PCR. Nucleic Acids Res 2001;29(9):e45–e445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Anderson TW, Darling DA. Asymptotic theory of certain "goodness of fit" criteria based on stochastic processes. Ann Math Stat Jun. 1952;23(2):193–212. [Google Scholar]
- 27. Box GEP. Non‐normality and tests on variances. Biometrika Dec. 1953;40(3/4):318–335. [Google Scholar]
- 28. Welch BL. The generalization of student's ’ problem when several different population variances are involved. Biometrika 1947;34(1):28–35. [DOI] [PubMed] [Google Scholar]
- 29. Poveda‐Reyes S, Moulisova V, Sanmartín‐Masiá E, Quintanilla‐Sierra L, Salmerón‐Sánchez M, Ferrer GG. Gelatin—hyaluronic acid hydrogels with tuned stiffness to counterbalance cellular forces and promote cell differentiation. Macromol Biosci 2016;16:1311–1324. [DOI] [PubMed] [Google Scholar]
- 30. Lu P, Takai K, Weaver VM, Werb Z. Extracellular matrix degradation and remodeling in development and disease. Cold Spring Harb Perspect Biol 2011;3(12):1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Endres M, Neumann K, Schröder SEA, Vetterlein S, Morawietz L, Ringe J, Sittinger M, Kaps C. Human polymer‐based cartilage grafts for the regeneration of articular cartilage defects. Tissue Cell Oct. 2007;39(5):293–301. [DOI] [PubMed] [Google Scholar]
- 32. Dehne T, Karlsson C, Ringe J, Sittinger M, Lindahl A. Chondrogenic differentiation potential of osteoarthritic chondrocytes and their possible use in matrix‐associated autologous chondrocyte transplantation. Arthritis Res Ther 2009;11(5):R133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schrobback K, Klein TJ, Crawford R, Upton Z, Malda J, Leavesley DI. Effects of oxygen and culture system on in vitro propagation and redifferentiation of osteoarthritic human articular chondrocytes. Cell Tissue Res 2012;347(3):649–663. [DOI] [PubMed] [Google Scholar]
- 34. Heywood HK, Bader DL, Lee D a. Glucose concentration and medium volume influence cell viability and glycosaminoglycan synthesis in chondrocyte‐seeded alginate constructs. Tissue Eng 2006;12(12):3487–3496. [DOI] [PubMed] [Google Scholar]
- 35. Stockwell R. The cell density of human articular and costal cartilage. J Anat 1967;101(Pt 4):753–763. [PMC free article] [PubMed] [Google Scholar]
- 36. Poole a R. What type of cartilage repair are we attempting to attain? J Bone Joint Surg Am 2003;85–A(Suppl):40–44. [DOI] [PubMed] [Google Scholar]
- 37. Fox AJS, Wanivenhaus F, Rodeo SA. The basic science of the patella: Structure, composition, and function. J Knee Surg 2012;25(2):127–141. [DOI] [PubMed] [Google Scholar]
- 38. Dawson E, Mapili G, Erickson K, Taqvi S, Roy K. Biomaterials for stem cell differentiation. Adv Drug Deliv Rev 2008;60(2):215–228. [DOI] [PubMed] [Google Scholar]
- 39. Baugé C, Cauvard O, Leclercq S, Galéra P, Boumédiene K. Modulation of transforming growth factor beta signalling pathway genes by transforming growth factor beta in human osteoarthritic chondrocytes: Involvement of Sp1 in both early and late response cells to transforming growth factor beta. Arthritis Res Ther 2011;13(1):R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Benya PD, Shaffer JD. Dedifferentiated chondrocytes reexpress the differentiated collagen phenotype when cultured in agarose gels. Cell 1982;30(1):215–224. [DOI] [PubMed] [Google Scholar]
- 41. Wu Y, Kennedy P, Bonazza N, Yu Y, Dhawan A, Ozbolat I. Three‐dimensional bioprinting of articular cartilage: A systematic review. Cartilage 2018. 10.1177/1947603518809410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ren X, Wang F, Chen C, Gong X, Yin L, Yang L. Engineering zonal cartilage through bioprinting collagen type II hydrogel constructs with biomimetic chondrocyte density gradient. BMC Musculoskelet Disord 2016;17(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Erggelet C, Kreuz PC, Mrosek EH, Schagemann JC, Lahm A, Ducommun PP, Ossendorf C. Autologous chondrocyte implantation versus ACI using 3D‐bioresorbable graft for the treatment of large full‐thickness cartilage lesions of the knee. Arch Orthop Trauma Surg 2010;130(8):957–964. [DOI] [PubMed] [Google Scholar]
- 44. Becher C, Laute V, Fickert S, Zinser W, Niemeyer P, John T, Diehl P, Kolombe T, Siebold R, Fay J. Safety of three different product doses in autologous chondrocyte implantation: Results of a prospective, randomised, controlled trial. J Orthop Surg Res 2017;12(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gaut C, Sugaya K. Critical review on the physical and mechanical factors involved in tissue engineering of cartilage. Regen Med 2015;10(5):665–679. [DOI] [PubMed] [Google Scholar]
- 46. Roberts S, Hollander AP, Caterson B, Menage J, Richardson JB. Matrix turnover in human cartilage repair tissue in autologous chondrocyte implantation. Arthritis Rheum Nov. 2001;44(11):2586–2598. [DOI] [PubMed] [Google Scholar]
- 47. Lammi MJ, Piltti J, Prittinen J, Qu C. Challenges in fabrication of tissue‐engineered cartilage with correct cellular colonization and extracellular matrix assembly. Int J Mol Sci Sep. 2018;19:2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhu W, Cui H, Boualam B, Masood F, Flynn E, Rao RD, Zhang Z‐Y, Zhang LG. 3D bioprinting mesenchymal stem cell‐laden construct with core–shell nanospheres for cartilage tissue engineering. Nanotechnology May 2018;29(18):185101. [DOI] [PubMed] [Google Scholar]
- 49. Sieber S, Wirth L, Cavak N, Koenigsmark M, Marx U, Lauster R, Rosowski M. Bone marrow‐on‐a‐chip: Long‐term culture of human hematopoietic stem cells in a 3D microfluidic environment. J Tissue Eng Regen Med 2017;12:479–489. [DOI] [PubMed] [Google Scholar]
- 50. Dehne E‐M, Hasenberg T, Marx U. The ascendance of microphysiological systems to solve the drug testing dilemma. Futur Sci OA 2017;3:FSO185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Marx U, Andersson TB, Bahinski A, Beilmann M, Cassee FR, Cirit M, Daneshian M, Frey O, Gaertner C, Giese C, Griffith L, Hartung T, Heringa MB, Hoeng J, De Jong WH, Kuehnl J, Leist M, Luch A, Maschmeyer I, Roth A. Biology‐inspired microphysiological system approaches to solve the prediction dilemma of substance testing. ALTEX 2016;33:1–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information File 1 Determination of construct cell number. Collagen type II immunostainings that have been counterstained with hematoxylin were used for the verification of cell number inside the construct after 14 days of cell culture. Gelatin‐ (A,B) and HA‐based (C,D) with low (A,C) and high cell density (B,D) were analyzed. Cell nuclei of five representative areas (each 0.5 mm × 0.5) were manually counted (E, area 1–5). The average was used to calculate the cell number according to a method published by Aherne in 1967 (PMID 5582822). The cell number per area volume (N A) was estimated from the mean count (n), area (A = 0.25 mm2), section thickness (t = 8 μm), and the estimated diameter of the cell nuclei (d = 6 μm) with N A = n/(A × (d + t)). Neglecting the minimal volume loss of the construct observed during culture (V K = 50 μL), the cell number per construct was calculated through N K = V K/V A × N A with V A = A × (d + t). Bar = 500 μm.
Supporting Information Figure 2 Measurements of diameter. Intact (A–C) or symmetrically interpolated (D, if not complete) sections were used to determine the diameter of the constructs (E). Sections were chosen that showed highest diameter. Measuring the height was not possible as it could not be assured that cutting plane was exactly at right angle to the construct's plane.
Supporting Information Figure 3 Detection of proteoglycans in constructs after 14 days of culture. Alcian blue stain of tissue section documented the presence of proteoglycans in blue. Nuclei and other ECM/bioink appear pink to red by Nuclear Fast Red counterstain. Porcine chondrocytes were embedded in (A, B, E, F) gelatin and (C, D, G, H) hyaluronic acid bioinks with low (A, C, E, G) and high (B, D, F, H) cell density using the Cellbricks Bioprinting technique. (A–D) Bar = 500 μm; (E–H) Bar = 100 μm. Alcian blue staining method: Detection of proteoglycan by Alcian blue was performed on 8 μm cryosections as described in the main text with 1% Alcian blue 8GX (Sigma) in 3% acetic acid (pH 2.5). Cell nuclei were counterstained with Nuclear Fast Red aluminum sulphate solution (ready to use, Roth).
Supporting Information Figure 4 Bioprinted constructs. These are examples of bioprinted cartilage models. (A) Bioprinted form of meniscal structure, (B) except of meniscal structure. (C, D) A defined defect was printed within the meniscal topography, here marked by yellow arrows. All scale bars represent 1 cm.


