Abstract
Variation across dog breeds presents a unique opportunity to investigate the evolution and biological basis of complex behavioural traits. We integrated behavioural data from more than 14 000 dogs from 101 breeds with breed-averaged genotypic data (n = 5697 dogs) from over 100 000 loci in the dog genome. We found high levels of among-breed heritability for 14 behavioural traits (the proportion of trait variance attributable to genetic similarity among breeds). We next identified 131 single nucleotide polymorphisms associated with breed differences in behaviour, which were found in genes that are highly expressed in the brain and enriched for neurobiological functions and developmental processes, suggesting that they may be functionally associated with behavioural differences. Our results shed light on the heritability and genetic architecture of complex behavioural traits and identify dogs as a powerful model in which to address these questions.
Keywords: dog, breed, behaviour, evolution, heritability, genetics
1. Introduction
Genetic and phenotypic variation across dog breeds provides a unique opportunity for investigating questions about the evolution and biological basis of complex traits. For example, studies of breed differences have led to major advances in our understanding of the genetics of diseases, including cancer, metabolic disorders and blindness [1], as well as the genetic underpinnings of morphological traits, such as body mass, coat type and coloration [2]. Despite rapid progress in these areas, we still know little about the biological bases of breed differences in behaviour (reviewed in [3]). For instance, the extent to which phenotypic similarities are predicted by genetic relatedness among breeds remains unknown. Furthermore, even less is known about the genetic architecture of these behavioural traits. Are they largely polygenic—as is the case for most complex traits—or instead predominantly influenced by a handful of loci [2,4]?
Dogs provide a powerful model to address questions about behavioural evolution, due to their simplified genetic architecture resulting from population bottlenecks during domestication and strong diversifying selection during subsequent breed evolution [5–7]. The majority of variance among modern breeds has probably resulted from the repeated crossing of novel phenotypes, which—originating from a limited pool of genetic variation—has nonetheless given rise to extraordinary phenotypic diversity. In addition to these practical advantages, dogs exhibit complex cognitive and behavioural phenotypes, some of which exhibit striking parallels to traits in humans [8–13]. For example, common genetic mechanisms contribute to individual differences in social behaviour in dogs and humans, with relevance to understanding behavioural syndromes such the hypersociability found in Williams syndrome [14]. However, research to date has been conducted with small sample sizes from a restricted number of breeds, limiting our ability to make broader inferences about the evolution and biological basis of behavioural diversity across breeds [15,16].
In the current study, we combined a behavioural dataset of more than 14 000 dogs from 101 breeds (electronic supplementary material, table S1) with breed-typical genotypic data from over 100 000 loci across the dog genome. Using these data, we quantified the extent to which breed differences in behaviour are attributable to genetic similarity among breeds, and identified genomic regions associated with breed differences in behaviour.
First, we hypothesized that if diversifying selection in dogs has led to genetically based breed differences in behaviour, then phenotypic similarity among breeds should be attributable to genetic similarity among breeds. In quantitative genetics, this proportion of phenotypic variance attributable to additive genetic factors is typically quantified as (narrow-sense) heritability (h2). Heritability estimates are population-specific, and can vary across time, environments and between populations, due to population differences in both genetic and environmental variance [17]. When considering variance across breeds, we can make use of the heritability concept while acknowledging some potentially important differences from studies of heritability in randomly interbreeding populations.
Although dogs have been bred by humans for millennia, the formalization of modern breeds, as defined by closed genetic pools, occurred only 200–300 years ago [18]. Consequently, most modern breeds are characterized by limited genetic diversity, and members of the same breed can be reliably assigned to a single breed-specific clade [18–20]. From an evolutionary perspective, variance across breeds reflects diversifying selection, which has occurred rapidly through selective breeding by humans. Thus, we hypothesized that due to relatively high levels of among-breed (relative to within-breed) variance, heritability estimates across breeds would far exceed those obtained within breeds. To disambiguate within- from across-breed measures of heritability (acknowledging fundamental differences in the reference populations for these studies), we use the term ‘among-breed heritability’.
Second, if breed differences in behaviour are highly heritable, we expected that models predicting breed-average behavioural scores as a function of breed-average allele frequency would identify loci contributing to phenotypic variance. We hypothesized that if the genes implicated in these analyses contribute to variance in behaviour across breeds, that they should be highly expressed in the brain, and play functional roles in nervous system processes with relevance to behaviour.
2. Methods
(a). Data sources
(i). Genetic data
We used genetic data from two large-scale genotyping analyses: Hayward et al. [4] and Parker et al. [19]. Both genetic datasets were generated using the Illumina Canine HD SNP chip (Illumina, San Diego, CA) which includes a set of 172 000 SNPs from across the dog genome [21]. Using these data, we estimated genetic relationships among breeds using identity-by-state (IBS) matrices generated in PLINK v.1.9, using the default settings (plink—file datafile—cluster—matrix [22]. From these matrices (one for each genetic dataset), we then generated breed-average IBS matrices by calculating the mean IBS within breeds and between pairs of breeds. The off-diagonal elements in these matrices represent the proportion of single nucleotide polymorphisms (SNPs) that are (on average) identical between each pairwise combination of breeds. The Hayward et al. dataset included 12 143 additional custom markers, and in total included 160 727 SNPs with a minor allele frequency (MAF) greater than 0.01 [4]. The Parker et al. dataset included 150 131 SNPs with a MAF > 0.01 and was obtained directly from the authors. The dataset from Parker et al. [19] contains individuals sampled in Hayward et al. [4], which were removed from the Parker et al. data in order to obtain independent genetic datasets for analysis. The resulting datasets included a mutually exclusive set of 4342 dogs from Hayward et al. [4] and 1355 dogs from Parker et al. [19]. Median inter-SNP distances were 5829 bases for the Parker et al. data and 6606 bases for the Hayward et al. data. For heritability analyses, breed-level IBS matrices were multiplied by an individual-level incidence matrix in order to generate an individual-level IBS matrix. This approach allowed us to incorporate data at the individual level within a mixed-model framework (electronic supplementary material). To avoid the assumption that members of the same breed were clonal, pairwise within-breed IBS values were set to the average IBS value between members of that breed.
(ii). Behavioural data
Behavioural data were obtained from the Canine Behavioral Assessment and Research Questionnaire (C-BARQ) Database at the University of Pennsylvania (https://vetapps.vet.upenn.edu/cbarq/). The C-BARQ is a widely used behavioural assessment tool for dogs with established reliability characteristics and demonstrated construct validity [23–26]. It comprises 14 factors or subscales extracted by factor analysis from responses to 78 behavioural items that are scored either in terms of severity or frequency (table 1). All C-BARQ items are presented as five-point ordinal rating scales with a score of ‘0’ corresponding to ‘absent/never’ and a score of ‘4’ corresponding to ‘severe/always’. C-BARQ factor scores are calculated by averaging all the item scores within each factor [23]. The current sample was derived from 29 656 C-BARQ entries for pet dogs collected from dog owners between 2005 and 2016. These data were filtered to include only pure-bred dogs (owner report), and further restricted to breeds represented in the genetic data sources described above (n = 14 020; electronic supplementary material, table S1). To ensure representative samples, we included only breeds with at least 25 observations per breed for analysis. Our final behavioural dataset linked to the Parker et al. [19] genetic data included 12 806 dogs from 86 breeds, and our final behavioural dataset linked to the Hayward et al. [4] genetic data included 13 907 dogs from 98 breeds.
Table 1.
Description of C-BARQ factors.
| C-BARQ factor (type) | definition (number of questionnaire items) |
|---|---|
| trainability (frequency) | willingness to attend to the owner, obey simple commands, fetch objects, respond positively to correction, and ignore distracting stimuli (8) |
| stranger aggression (severity) | threatening or aggressive responses to strangers approaching or invading the dog's or owner's personal space, territory or home range (10) |
| owner aggression (severity) | threatening or aggressive responses to the owner or other household members when challenged, handled, stared at, stepped over, or approached while in possession of food or objects (8) |
| dog aggression (severity) | threatening or aggressive responses when approached directly by unfamiliar dogs (4) |
| dog rivalry (severity) | aggressive or threatening responses to other familiar dogs in the same household (4) |
| dog fear (severity) | fearful or wary responses when approached directly by unfamiliar dogs (4) |
| stranger fear (severity) | fearful or wary responses when approached directly by unfamiliar people (4) |
| non-social fear (severity) | fearful or wary responses to sudden or loud noises, traffic, and unfamiliar objects and situations (6) |
| touch sensitivity (severity) | fearful or wary responses to potentially painful or uncomfortable procedures, including bathing, grooming, nail-clipping, and veterinary examinations (4) |
| separation problems (frequency) | vocalizing and/or destructive behaviour when separated from the owner, including autonomic signs of anxiety—restlessness, loss of appetite, trembling and excessive salivation (8) |
| excitability (frequency) | reaction to potentially exciting or arousing events, such as going for walks or car trips, doorbells, arrival of visitors or the owner arriving home; difficulty settling down after such events (6) |
| attachment/attention-seeking (frequency) | maintains close proximity to the owner or other members of the household, solicits affection or attention, becomes agitated when the owner gives attention to third parties (6) |
| chasing (severity + frequency) | pursues cats, birds and/or other small animals, given the opportunity (4) |
| energy (frequency) | level of energetic, boisterous and/or playful behaviour (2) |
(iii). Heritability analyses
Heritability was estimated using efficient mixed model association (EMMA) [27,28]. For comparison to a more conventional approach, we conducted supplementary analyses implementing the animal model [29] with Markov chain Monte Carlo (MCMC) linear mixed models [30]. Although both approaches are mixed models for estimation of the phenotypic variance attributable to (additive) genetic and environmental effects, they differ in the computations through which variance components are estimated. Specifically, EMMA employs restricted maximum likelihood (REML) to efficiently estimate variance components whereas variance components are numerically optimized in the MCMC framework. EMMA models were fitted using the NAM R package [31], and MCMC models were fitted using the MCMCglmm R package [30]. The equations for these mixed models, and additional details, are included in the electronic supplementary material.
To estimate heritability for each behavioural trait, we employed a resampling procedure to reduce computational demands and assess a distribution of results across random subsamples across breeds. The resampling procedure was performed as follows: (i) randomly sample 25 individuals from each breed without replacement; (ii) estimate heritability in this subsample. The mean value across resampling was used as the final heritability estimate. For MCMC modelling, each model used a 1000 iteration burn-in, followed by a 10-iteration thinning interval across 9000 subsequent iterations. For each MCMC model, we retained the mean heritability value from the posterior distribution. Results from EMMA and MCMC heritability models were highly similar (electronic supplementary material). To assess statistical significance of heritability estimates, we performed the resampling procedure but randomly permuted the trait values across breeds at each iteration.
To test whether these behaviours arose from selection, rather than genetic drift due to the likely genetic bottleneck early in breeding, we conducted Qst − Fst analysis [32,33]. We approximated Qst using Pst across all 14 behaviours (after controlling for breed-average body weight) using the R package Pstat, and calculated confidence intervals using 1000 bootstrap iterations. We calculated genome-wide Fst using all SNPs in the Parker et al. data, under the reasonable assumption that majority of the sites would be neutrally evolving.
(iv). Genome-wide association study
We assessed associations between SNPs and the behavioural traits using EMMA [28]. Specifically, we modelled breed-average behavioural scores as a function of breed-average allele frequency and a polynomial term for the log of breed-average body weight. EMMA models were fitted using the EMMREML R package [34]. We included a second-order polynomial term for log body weight because preliminary analyses revealed nonlinear associations between breed-average weight and behaviour that were best captured using a polynomial term. We only included SNPs with a median MAF across breeds of at least 0.05 (Hayward et al.: 127 970 SNPs; Parker et al. 110 096 SNPs). Genome-wide association study (GWAS) analyses were conducted separately with each genetic dataset, and the resulting p-values were combined across datasets using meta-analysis (Fisher's method), first at the level of the SNP (109 780 overlapping SNPs), and then at the level of the gene. Combined p-values were Bonferroni corrected for identification of genes associated with behavioural traits, and false discovery rate (FDR) corrected for the enrichment analyses described below [35]. Our primary analyses were restricted to SNPs located in genes, but we report supplemental analyses that associated SNPs with the nearest gene within 20 kb (electronic supplementary material). To derive gene-level p-values from the GWAS, we combined p-values from multiple SNPs associated with the same gene using meta-analysis (Fisher's method).
(v). Enrichment analyses
To infer the biological relevance of genes implicated in the GWAS, we conducted enrichment analyses using gene ontology (GO), and tests of tissue-specific gene expression. GO enrichment analyses test whether a set of genes identified in an analysis are disproportionately related to specific functions (grouped within three broad domains involving cellular components, molecular functions, or biological processes). Because we were interested in processes related to behaviour, we conducted the GO analysis using the biological process domain. GO analyses were implemented using a Fisher exact test and the ‘weight01’ algorithm in the topGO R package [36,37], with ENSEMBL gene identifiers mapped to GO terms using the biomaRt R package [38,39]. Genes were included as significant if the FDR corrected p-value for the gene was less than or equal to 0.05. Our primary GO analyses were restricted to gene-level p-values derived from meta-analysis of SNPs in the gene, but additional analyses using SNPs within 20Kb of the nearest gene are provided in the electronic supplementary material.
Tissue enrichment analyses investigated whether the genes identified through meta-analysis disproportionately included genes with tissue-specific expression (e.g. brain, heart, liver, etc.). These analyses were conducted using the TissueEnrich R package [40]. Data on tissue-specific gene expression in dogs were compiled from a study of gene expression in 10 tissues from a sample of four dogs [41]. Microarray expression data were averaged across dogs (two beagles and two mixed-breed dogs), and across probes in cases where genes were mapped to more than one probe. Tissue-specific genes were defined using the algorithm from Uhlén et al. [42], with a gene classified as ‘tissue-specific’ if it was 3× more highly expressed in a particular tissue compared to all other tissues. Because dogs and humans share similar tissue enrichment patterns [41,43], we also conducted enrichment analyses using human gene expression data from a larger sample of individuals and tissues [42].
3. Results
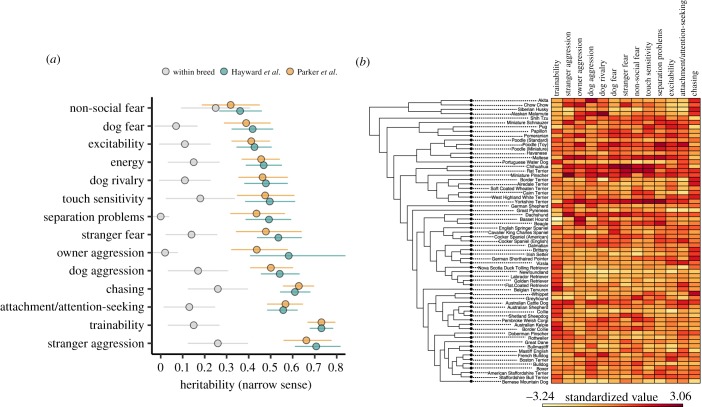
We found that a large proportion of behavioural variance across breeds (among-breed heritability) is attributable to genetic factors (figure 1a). The mean among-breed heritability was 0.51 ± 0.12 (s.d.) across all 14 traits (range: h2 = 0.27–0.77), and significantly higher than the null expectation in all cases (permutation tests, p < 0.001). These estimates are also significantly higher than those in previous studies assessing heritability of the same traits in large within-breed samples (mean difference = 0.37, 95% CI = 0.44–0.30; t13 = −12.25, p < 0.001; [44], but see [45]). Incorporating among-breed variance thus yields h2 estimates that are on average, five times higher (range = 1.3–25.5 times higher) than traditional within-breed estimates, which could be due to limited genetic and phenotypic variation within breeds. Although heritability estimates at these different scales stem from fundamentally different underlying populations (randomly interbreeding versus highly structured populations), the statistical component of estimating variance ratios is similar in both cases, suggesting that a larger fraction of phenotypic variance across breeds is explained by genetic factors compared with typical within-breed studies.
Figure 1.
Within- and among-breed heritability estimates, and breed-level behavioural data. (a) Heritability (h2) estimates (proportion of variance attributable to genetic factors) for 14 behavioural traits. Genotypic variation accounts for five times more variance in analyses across versus within breeds (within-breed estimates compiled from Ilska et al. [44]). Points for Hayward et al. [4] and Parker et al. [19] reflect the results of analyses with independent genetic datasets. Error bars reflect the 95% confidence intervals. (b) Heatmap of breed-average behavioural scores plotted alongside a cladogram of breed relatedness from Parker et al. [19]. (Online version in colour).
Interestingly, the traits with the highest among-breed heritability were trainability (h2 = 0.73), stranger-directed aggression (h2 = 0.68), chasing (h2 = 0.62) and attachment and attention-seeking (h2 = 0.56), which is consistent with the hypothesis that these behaviours have been important targets of selection during the formation of modern breeds [3]. Indeed, the overall patterns of breed differences align closely with genetic clades corresponding to functional breed groups (figure 1b). Qst − Fst analysis revealed that all 14 behavioural traits were under strong positive selection, and could not be explained by genetic drift alone (electronic supplementary material, figure S1).
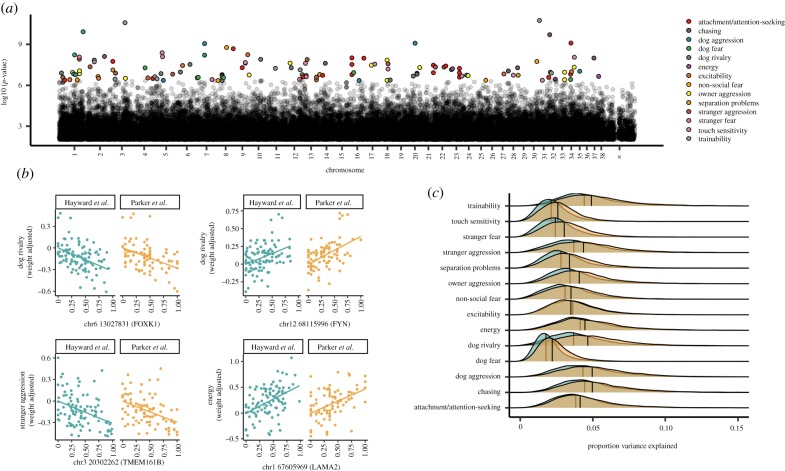
To identify specific loci associated with breed differences in behaviour, we conducted a GWAS across breeds. Following the approach used previously to identify genomic regions associated with morphological [2] and athletic variation between breeds [46], we modelled breed-average behavioural scores as a function of breed-average allele frequencies, controlling for relatedness among breeds using EMMA [28]. To confirm that our approach was sensitive to known genetic associations with morphological traits, we first conducted a GWAS for body mass using owner-reported body weights from the same dataset used for our behavioural analysis. These analyses identified SNPs in IGF1 and SMAD2 as the top autosomal hits associated with body mass, replicating the results of previous studies [2,47,48]. The top SNPs associated with body mass were located in the same region of IGF1 implicated in previous studies, and explained approximately 50% of variation in body mass across breeds, consistent with earlier reports [2,47]. For our analysis of behavioural traits, we also implemented a control for possible inflation of p-values due to cryptic population stratification by correcting p-values using the approach described by Amin et al. [49]. Each behavioural trait was modelled twice: once with each of the independent genetic datasets. The effects of the same single-nucleotide polymorphisms (SNPs) on each behaviour were strongly correlated across the two datasets (median r = 0.77, range: r = 0.68–0.82; electronic supplementary material, figure S2), and the strength of this association increased to a Pearson correlation of 0.93 when we included SNPs significantly associated with a trait at a nominal p ≤ 0.01 (electronic supplementary material, figure S3). This suggested that our results were not subject to variation in the representation of breeds or genotyping platforms of a specific study. We, therefore, used Fisher's combined probability test to combine the p-values for each shared SNP across the two datasets.
Overall, we identified 131 unique SNPs that were significantly associated with at least one of the 14 behavioural traits following Bonferroni correction (figure 2; electronic supplementary material, figure S4). Forty per cent of these SNPs (n = 52) were located within a gene—none of which encoded for changes in the amino acid sequence of the protein (see electronic supplementary material, for analyses using other distance thresholds for mapping SNPs to genes). On average, the top SNP explained 15% of variance in the behavioural trait (range: PVE = 0.06–0.25; figure 2; electronic supplementary material, table S2). Thus, while we identify multiple variants with moderately large effects, the variance explained by individual SNPs is far less than that explained by additive variation across the genome (heritability) suggesting that as in humans, behavioural traits in dogs are highly polygenic. However, the variance explained by the top SNPs in our analysis across breeds was, on average, more than five times higher than that from within-breed association studies using the C-BARQ [44]. Importantly, estimates of variation explained by SNPs with the lowest significance values may be subject to inflation (known as the ‘winner's curse’) [50]. However, given that we expect this to be true for both within- and across-breed studies, our finding that a relatively larger proportion of variance is attributable to single SNP differences across breeds is unlikely to be driven by inflation of effect sizes only in our study.
Figure 2.
Genetic associations with breed differences in behaviour. (a) Manhattan plot showing SNPs associated with behavioural traits (colour coded) after Bonferroni correction. Individual Manhattan plots for each trait are shown in electronic supplementary material, figure S4. (b) Behavioural trait values, corrected for body weight, as a function of allele frequency. Plots show data for 4 SNPs in genes with sequence or expression differences between foxes artificially bred for tameness or aggression. (c) Distributions of the proportion variance explained (PVE) by SNPs associated with behavioural traits at p ≤ 0.05, after correction for the FDR. Turquoise points and distributions: Hayward et al. [4] genetic data; yellow points and distributions: Parker et al. [19] genetic data. (Online version in colour).
To examine if these variants may be linked to behaviour-relevant genes, we further derived gene-level associations using a meta-analytic approach (electronic supplementary material). Many of the gene-level associations with dog behavioural traits (electronic supplementary material, table S3) include (i) candidate domestication genes, (ii) genes mapped to phenotypes implicated in domestication, (iii) genes implicated in behavioural differences between foxes bred for tameness or aggression and (iv) genes that underwent positive selection in both human evolution and dog domestication (electronic supplementary material, table S4). For example, breed differences in stranger-directed aggression were associated with SNPs in GRM8, a gene that has undergone positive selection in both humans and dogs [51]. This gene encodes a glutamate receptor, one of the major excitatory neurotransmitters in the central nervous system. Importantly, a reduction in stranger-directed aggression is thought to be the unifying factor in (early) dog domestication and human self-domestication [52], making GRM8 an interesting candidate gene for these processes. Similarly, PDE7B, which is differentially expressed in the brains of tame and aggressive foxes [53], has been identified as a target of selection during domestication, and is highly expressed in the brain [54], where it functions in dopaminergic pathways [55]. In our analyses, SNPs in this gene were associated with breed differences in aggression, which is consistent with data from experimentally bred foxes, as well as hypotheses that selection against aggression was the primary evolutionary pressure during initial domestication events [56–58].
The gene–trait associations identified in our study also align closely with similar associations in human populations (electronic supplementary material, table S5). For example, breed differences in aggression are associated with multiple genes that have been linked to aggressive behaviour in humans. Molecular associations with breed differences in energy (frequency of energetic, boisterous and playful behaviour) include genes previously linked to resting heart rate, daytime rest, and sleep duration in humans. Lastly, breed differences in fear were associated with genes linked to temperament and startle response in humans, and several of the genes implicated in breed differences in trainability have been previously associated with intelligence and information processing speed in humans.
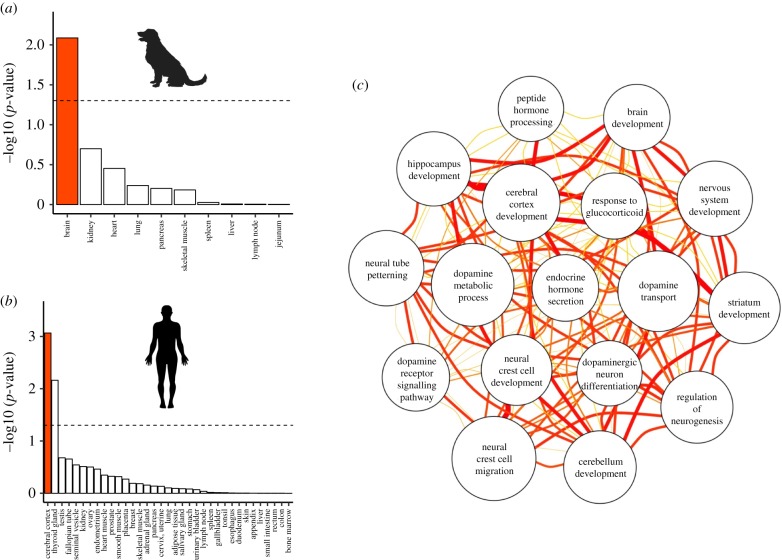
If the variants in genes identified in our analyses make major contributions to behaviour and cognition we expected that the associated genes should be (i) involved in biological processes related to nervous system development and function, and (ii) primarily expressed in the brain. Indeed, we found that behaviour-associated genes (as identified through meta-analysis) were enriched for numerous nervous system processes (figure 3; electronic supplementary material, table S6). These processes include neurogenesis, neuron migration and differentiation, axon and dendrite development, and regulation of neurotransmitter transport and release. We also identified GO terms related to key developmental processes implicated in domestication [60,61], including neural tube development, neural crest cell differentiation and migration, and the development and organization of brain structures including the hippocampus, cerebral cortex and striatum (electronic supplementary material, table S6). On a trait-specific level, we identified several interesting links between dog behaviour and putative biological mechanisms implicated in the regulation of these behaviours. For example, dopaminergic pathways play important roles in mammalian social attachment [62,63], and GO terms relating to dopamine transport and metabolism were significantly enriched in genes associated with breed differences in attachment and attention-seeking. Gene-level associations and enriched GO terms from analyses mapping SNPs to the nearest gene within 20 Kb are shown in electronic supplementary material, tables S7 and S8.
Figure 3.
Genes containing behaviour-associated SNPs are highly expressed in the brain and associated with gene ontology terms relating to brain development and function. (a) Enrichment tests using dog gene expression to identify tissue-specific genes. (b) Enrichment tests using human gene expression to identify tissue-specific genes. Bars reflect the −log10 (p-value) from a hypergeometric test for tissue-specific gene enrichment. The dashed line indicates −log10 (p = 0.05) and the results for brain tissue are highlighted in red. (c) Network plot for a subset of gene ontology terms relating to brain development and function that were associated with breed differences in behaviour. Edge colours and line widths reflect Resnik's similarity scores between GO terms, as implemented in NaviGO [59]. Wider and redder lines reflect greater similarity between nodes. Node sizes are inversely proportional to p-values from enrichment tests. (Online version in colour).
To examine if the genes identified through GWAS are predominantly expressed in the brain, we analysed tissue-specific enrichment for the set of genes with SNPs that were significantly associated with any of the 14 behavioural traits. We used two datasets of tissue-specific expression: (i) gene expression across 10 tissues from a sample of four dogs [41], and (ii) human gene expression from larger samples of individuals across a greater range of tissues (n = 35) [42]. In both datasets, we found that the genes containing SNPs associated with dog behaviour are significantly more likely to be expressed in the brain (figure 3; hypergeometric test: dog tissue: p < 0.001, human tissue: p < 0.001; see electronic supplementary material, and figure S5 for similar results including SNPs nearby, but not necessarily in, genes). Together, this suggests that the SNPs identified in our GWAS may affect behavioural processes by altering expression in genes that are highly expressed in the brain.
4. Discussion
The vast phenotypic diversity and simplified genetic architecture of dog breeds has led to major advances in our understanding of complex traits relevant to morphology and disease. Our findings suggest that dog breeds also provide a powerful and highly tractable model for questions about the evolution and genetic basis of behavioural traits. Breed differences in behaviour covary strongly with relatedness between breeds, and for several traits, genotype accounts for more than 50% of behavioural variation across breeds—up to 25× higher than heritability estimates from genetic studies within breeds. Although among-breed analyses draw on genotypic and phenotypic variation in a fundamentally different population than heritability studies with randomly interbreeding populations, both analyses address the proportion of phenotypic variance within a study population that is attributable to genetic factors. While the h2 estimates from our study are high relative to within-breed studies, they are perhaps unsurprising given intense diversifying selection among breeds. Specifically, if different breeds or breed groups have been selected for particular behavioural traits—or if behavioural traits are genetically correlated with other traits under selection—then we would expect that a large fraction of among-breed phenotypic variance should be attributable to genetic factors.
Given sufficient among-breed heritability, we expected that using a genome-wide approach it should be possible to identify specific loci associated with breed differences in behaviour. Our GWAS revealed that SNPs associated with breed differences in behaviour tend to fall in genes that are disproportionately expressed in the brain and involved in pathways related to the development and expression of behaviour and cognition. Therefore, although we cannot infer causal effects of specific SNPs, the genes we identified have potential to influence behavioural processes through altered expression in the brain, or contributions to other biological processes involving the nervous system. In addition, the variants associated with breed differences in behaviour are found in genes with sequence or brain-expression differences in foxes artificially bred for tameness or aggression, and several of the genotype–phenotype associations we found align closely with similar associations in human populations. Collectively, these findings suggest that these genes may play important roles in modulating behaviour across species (e.g. [64]).
Because we aimed to integrate behavioural data from large samples across more than a hundred breeds, the genotypic and phenotypic data in our study were not collected from the same subjects, but rather aggregated across independent datasets. Although this approach does not provide resolution at the level of the individual, both the genetic and behavioural datasets were collected from large representative samples, and our findings were robust across resampling and independent genetic datasets. This approach has also been implemented successfully in other trait-mapping studies in dogs, and our GWAS for body mass with the current dataset replicated well-established findings regarding loci linked to variation in size across breeds, as well as the proportion of variance attributable to these variants. Thus, while future work incorporating genotypic and phenotypic data from the same subjects will be important for finer-resolution trait-mapping, our findings are robust at the breed level.
Lastly, more than most model organisms, dogs also exhibit a suite of cognitive and behavioural traits that make them a powerful model for many aspects of human social behaviour and cognition [11,65]. These similarities are hypothesized to result from convergent evolution, due to similar selective pressures in human evolution and dog domestication [8,66]. Many of the gene–trait associations in the current study are consistent with similar findings from human populations (electronic supplementary material, table S5), suggesting the possibility of common mechanisms contributing to behavioural variation in both dogs and humans. Thus, the combination of phenotypic diversity across breeds, the expression of complex traits shared with humans and the comparative simplicity of trait-mapping in this species make dogs an invaluable organism for questions about the genetic bases of complex behaviours.
Supplementary Material
Supplementary Material
Acknowledgements
We thank X. Zhou and E. Karlsson for helpful comments regarding analysis and A. Boyko and H. Parker for discussion of data from their laboratories. We thank Michael Morrissey and one anonymous reviewer for helpful comments on an earlier draft of this manuscript.
Data accessibility
Genetic data used in these analyses are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.266k4 [67] and GEO accession nos. GSE90441, GSE83160, GSE70454 and GSE96736. C-BARQ data are available from serpell@vet.upenn.edu.
Authors' contributions
E.L.M., N.S.-M. and J.A.S. designed the studies. E.L.M. and N.S.-M. analysed the data. E.L.M., N.S.-M., B.M.v. and J.A.S. wrote the paper.
Competing interests
The authors declare no competing interests.
Funding
N.S.-M. was supported by grant no. R00AG051764 from the National Institute of Aging.
References
- 1.Sutter NB, Ostrander EA. 2004. Dog star rising: the canine genetic system. Nat. Rev. Genet. 5, 900–910. ( 10.1038/nrg1492) [DOI] [PubMed] [Google Scholar]
- 2.Boyko AR, et al. 2010. A simple genetic architecture underlies morphological variation in dogs. PLoS Biol. 8, e1000451 ( 10.1371/journal.pbio.1000451) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serpell JA, Duffy DL. 2014. Dog breeds and their behavior. In Domestic dog cognition and behavior (ed. A Horowitz), pp. 31–57. Berlin, Germany: Springer. [Google Scholar]
- 4.Hayward JJ, et al. 2016. Complex disease and phenotype mapping in the domestic dog. Nat. Commun. 7, 10460 ( 10.1038/ncomms10460) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ostrander EA, Kruglyak L. 2000. Unleashing the canine genome. Genome Res. 10, 1271–1274. ( 10.1101/gr.155900) [DOI] [PubMed] [Google Scholar]
- 6.Lindblad-Toh K, et al. 2005. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438, 803 ( 10.1038/nature04338) [DOI] [PubMed] [Google Scholar]
- 7.Wayne RK, Ostrander EA. 1999. Origin, genetic diversity, and genome structure of the domestic dog. Bioessays 21, 247–257. () [DOI] [PubMed] [Google Scholar]
- 8.MacLean EL, Herrmann E, Suchindran S, Hare B. 2017. Individual differences in cooperative communicative skills are more similar between dogs and humans than chimpanzees. Anim. Behav. 126, 41–51. ( 10.1016/j.anbehav.2017.01.005) [DOI] [Google Scholar]
- 9.MacLean EL. 2016. Unraveling the evolution of uniquely human cognition. Proc. Natl Acad. Sci. USA 113, 6348–6354. ( 10.1073/pnas.1521270113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miklósi Á. 2014. Dog behaviour, evolution, and cognition. Oxford, UK: Oxford University Press. [Google Scholar]
- 11.Hare B, Woods V. 2013. The genius of dogs: how dogs are smarter than you think. New York, NY: Dutton Adult. [Google Scholar]
- 12.Nagasawa M, Mitsui S, En S, Ohtani N, Ohta M, Sakuma Y, Onaka T, Mogi K, Kikusui T. 2015. Oxytocin-gaze positive loop and the coevolution of human–dog bonds. Science 348, 333–336. ( 10.1126/science.1261022) [DOI] [PubMed] [Google Scholar]
- 13.Kaminski J, Marshall-Pescini S. 2014. The social dog: behavior and cognition. San Diego, CA: Elsevier. [Google Scholar]
- 14.vonHoldt BM, et al. 2017. Structural variants in genes associated with human Williams-Beuren syndrome underlie stereotypical hypersociability in domestic dogs. Sci. Adv. 3, e1700398 ( 10.1126/sciadv.1700398) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zapata I, Serpell JA, Alvarez CE. 2016. Genetic mapping of canine fear and aggression. BMC Genomics 17, 572 ( 10.1186/s12864-016-2936-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Rooy D, Arnott ER, Early JB, McGreevy P, Wade CM.. 2014. Holding back the genes: limitations of research into canine behavioural genetics. Canine Genet. Epidemiol. 1, 1 ( 10.1186/2052-6687-1-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Visscher PM, Hill WG, Wray NR. 2008. Heritability in the genomics era—concepts and misconceptions. Nat. Rev. Genet. 9, 255 ( 10.1038/nrg2322) [DOI] [PubMed] [Google Scholar]
- 18.Parker HG, et al. 2004. Genetic structure of the purebred domestic dog. Science 304, 1160–1164. ( 10.1126/science.1097406) [DOI] [PubMed] [Google Scholar]
- 19.Parker HG, Dreger DL, Rimbault M, Davis BW, Mullen AB, Carpintero-Ramirez G, Ostrander EA. 2017. Genomic analyses reveal the influence of geographic origin, migration, and hybridization on modern dog breed development. Cell Rep. 19, 697–708. ( 10.1016/j.celrep.2017.03.079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.vonHoldt BM, et al. 2010. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature 464, 898 ( 10.1038/nature08837) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lequarré A-S, et al. 2011. LUPA: a European initiative taking advantage of the canine genome architecture for unravelling complex disorders in both human and dogs. Vet. J. 189, 155–159. ( 10.1016/j.tvjl.2011.06.013) [DOI] [PubMed] [Google Scholar]
- 22.Purcell S, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genetics 81, 559–575. ( 10.1086/519795) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu Y, Serpell JA. 2003. Development and validation of a questionnaire for measuring behavior and temperament traits in pet dogs. J. Am. Vet. Med. Assoc. 223, 1293–1300. ( 10.2460/javma.2003.223.1293) [DOI] [PubMed] [Google Scholar]
- 24.van den Berg SM, Heuven H, van den Berg L, Duffy DL, Serpell JA. 2010. Evaluation of the C-BARQ as a measure of stranger-directed aggression in three common dog breeds. Appl. Anim. Behav. Sci. 124, 136–141. ( 10.1016/j.applanim.2010.02.005) [DOI] [Google Scholar]
- 25.Duffy DL, Serpell JA. 2012. Predictive validity of a method for evaluating temperament in young guide and service dogs. Appl. Anim. Behav. Sci. 138, 99–109. ( 10.1016/j.applanim.2012.02.011) [DOI] [Google Scholar]
- 26.Jakuba T, et al. 2013. Differences in evaluation of a dog's temperament by individual members of the same household. Soc. Anim. 21, 582–589. ( 10.1163/15685306-12341314) [DOI] [Google Scholar]
- 27.Zhou X, Stephens M. 2012. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 44, 821–824. ( 10.1038/ng.2310) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E. 2008. Efficient control of population structure in model organism association mapping. Genetics 178, 1709–1723. ( 10.1534/genetics.107.080101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson AJ, et al. 2010. An ecologist's guide to the animal model. J. Anim. Ecol. 79, 13–26. ( 10.1111/j.1365-2656.2009.01639.x) [DOI] [PubMed] [Google Scholar]
- 30.Hadfield JD. 2010. MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J. Stat. Softw. 33, 1–22. ( 10.18637/jss.v033.i02)20808728 [DOI] [Google Scholar]
- 31.Xavier A, Xu S, Muir WM, Rainey KM. 2015. NAM: association studies in multiple populations. Bioinformatics 31, 3862–3864. ( 10.1093/bioinformatics/btv448) [DOI] [PubMed] [Google Scholar]
- 32.Leinonen T, McCairns RS, O'hara RB, Merilä J. 2013. Q ST–F ST comparisons: evolutionary and ecological insights from genomic heterogeneity. Nat. Rev. Genet. 14, 179 ( 10.1038/nrg3395) [DOI] [PubMed] [Google Scholar]
- 33.Brommer J. 2011. Whither PST? The approximation of QST by PST in evolutionary and conservation biology. J. Evol. Biol. 24, 1160–1168. ( 10.1111/j.1420-9101.2011.02268.x) [DOI] [PubMed] [Google Scholar]
- 34.Akdemir D, Godfrey OU. 2015. EMMREML: Fitting mixed models with known covariance structures. R package version 3.
- 35.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300. ( 10.1111/j.2517-6161.1995.tb02031.x) [DOI] [Google Scholar]
- 36.Alexa A, Rahnenfuhrer J. 2010. topGO: enrichment analysis for gene ontology. R package version 2.
- 37.Alexa A, Rahnenführer J.. 2009. Gene set enrichment analysis with topGO. See http://bioconductor.uib.no/2.7/bioc/vignettes/topGO/inst/doc/topGO.pdf.
- 38.Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, Huber W. 2005. BioMart and bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440. ( 10.1093/bioinformatics/bti525) [DOI] [PubMed] [Google Scholar]
- 39.Durinck S, Spellman PT, Birney E, Huber W. 2009. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 4, 1184 ( 10.1038/nprot.2009.97) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jain A, Tuteja G. 2018. TissueEnrich: A tool to calculate tissue-specific gene enrichment.
- 41.Briggs J, Paoloni M, Chen Q-R, Wen X, Khan J, Khanna C. 2011. A compendium of canine normal tissue gene expression. PLoS ONE 6, e17107 ( 10.1371/journal.pone.0017107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uhlén M, et al. 2015. Tissue-based map of the human proteome. Science 347, 1260419 ( 10.1126/science.1260419) [DOI] [PubMed] [Google Scholar]
- 43.Li Y, Vonholdt BM, Reynolds A, Boyko AR, Wayne RK, Wu D-D, Zhang Y-P. 2013. Artificial selection on brain-expressed genes during the domestication of dog. Mol. Biol. Evol. 30, 1867–1876. ( 10.1093/molbev/mst088) [DOI] [PubMed] [Google Scholar]
- 44.Ilska J, Haskell MJ, Blott SC, Sánchez-Molano E, Polgar Z, Lofgren SE, Clements DN, Wiener P. 2017. Genetic characterization of dog personality traits. Genetics 206, 1101–1111. ( 10.1534/genetics.116.192674) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liinamo A-E, van den Berg L, Leegwater PA, Schilder MB, van Arendonk JA, van Oost BA. 2007. Genetic variation in aggression-related traits in golden retriever dogs. Appl. Anim. Behav. Sci. 104, 95–106. ( 10.1016/j.applanim.2006.04.025) [DOI] [Google Scholar]
- 46.Kim J, Williams FJ, Dreger DL, Plassais J, Davis BW, Parker HG, Ostrander EA. 2018. Genetic selection of athletic success in sport-hunting dogs. Proc. Natl Acad. Sci. USA 115, E7212–E7E21. ( 10.1073/pnas.1800455115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sutter NB, et al. 2007. A single IGF1 allele is a major determinant of small size in dogs. Science 316, 112–115. ( 10.1126/science.1137045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones P, Chase K, Martin A, Davern P, Ostrander EA, Lark KG. 2008. Single-nucleotide-polymorphism-based association mapping of dog stereotypes. Genetics 179, 1033–1044. ( 10.1534/genetics.108.087866) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amin N, Van Duijn CM, Aulchenko YS.. 2007. A genomic background based method for association analysis in related individuals. PLoS ONE 2, e1274 ( 10.1371/journal.pone.0001274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wray NR, Yang J, Hayes BJ, Price AL, Goddard ME, Visscher PM. 2013. Pitfalls of predicting complex traits from SNPs. Nat. Rev. Genet. 14, 507 ( 10.1038/nrg3457) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang G-D, et al. 2013. The genomics of selection in dogs and the parallel evolution between dogs and humans. Nat. Commun. 4, 1860 ( 10.1038/ncomms2814) [DOI] [PubMed] [Google Scholar]
- 52.Hare B. 2017. Survival of the friendliest: homo sapiens evolved via selection for prosociality. Annu. Rev. Psychol. 68, 155–186. ( 10.1146/annurev-psych-010416-044201) [DOI] [PubMed] [Google Scholar]
- 53.Hekman JP, et al. 2018. Anterior pituitary transcriptome suggests differences in ACTH release in tame and aggressive foxes. G3: Genes, Genomes, Genetics. 8, 859–873. ( 10.1534/g3.117.300508) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Freedman AH, et al. 2016. Demographically-based evaluation of genomic regions under selection in domestic dogs. PLoS Genet. 12, e1005851 ( 10.1371/journal.pgen.1005851) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.De Gortari P, Mengod G. 2010. Dopamine D1, D2 and mu-opioid receptors are co-expressed with adenylyl cyclase 5 and phosphodiesterase 7B mRNAs in striatal rat cells. Brain Res. 1310, 37–45. ( 10.1016/j.brainres.2009.11.009) [DOI] [PubMed] [Google Scholar]
- 56.Trut LN. 1999. Early canid domestication: the farm-fox experiment: foxes bred for tamability in a 40-year experiment exhibit remarkable transformations that suggest an interplay between behavioral genetics and development. Am. Sci. 87, 160–169. ( 10.1511/1999.2.160) [DOI] [Google Scholar]
- 57.Belyaev DK. 1979. Destabilizing selection as a factor in domestication. J. Hered. 70, 301–308. ( 10.1093/oxfordjournals.jhered.a109263) [DOI] [PubMed] [Google Scholar]
- 58.Hare B, Tomasello M. 2005. Human-like social skills in dogs? Trends Cogn. Sci. 9, 439–444. ( 10.1016/j.tics.2005.07.003) [DOI] [PubMed] [Google Scholar]
- 59.Wei Q, Khan IK, Ding Z, Yerneni S, Kihara D. 2017. NaviGO: interactive tool for visualization and functional similarity and coherence analysis with gene ontology. BMC Bioinf. 18, 177 ( 10.1186/s12859-017-1600-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilkins AS, Wrangham RW, Fitch WT. 2014. The ‘domestication syndrome’ in mammals: a unified explanation based on neural crest cell behavior and genetics. Genetics 197, 795–808. ( 10.1534/genetics.114.165423) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pendleton AL, Shen F, Taravella AM, Emery S, Veeramah KR, Boyko AR, Kidd JM. 2018. Comparison of village dog and wolf genomes highlights the pivotal role of the neural crest in dog domestication. bioRxiv. 118794 ( 10.1101/118794) [DOI] [PMC free article] [PubMed]
- 62.Insel TR. 1997. A neurobiological basis of social attachment. Am. J. Psychiatry. 154, 726 ( 10.1176/ajp.154.6.726) [DOI] [PubMed] [Google Scholar]
- 63.Carter CS. 1998. Neuroendocrine perspectives on social attachment and love. Psychoneuroendocrinology 23, 779–818. ( 10.1016/S0306-4530(98)00055-9) [DOI] [PubMed] [Google Scholar]
- 64.Noh HJ, et al. 2017. Integrating evolutionary and regulatory information with a multispecies approach implicates genes and pathways in obsessive-compulsive disorder. Nat. Commun. 8, 774 ( 10.1038/s41467-017-00831-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miklósi A, Topál J, Csányi V. 2007. Big thoughts in small brains? Dogs as a model for understanding human social cognition. Neuroreport 18, 467–471. ( 10.1097/WNR.0b013e3280287aae) [DOI] [PubMed] [Google Scholar]
- 66.MacLean EL, Hare B. 2015. Dogs hijack the human bonding pathway. Science 348, 280–281. ( 10.1126/science.aab1200) [DOI] [PubMed] [Google Scholar]
- 67.Hayward Jessica J. et al 2016. Data from: Complex disease and phenotype mapping in the domestic dog Dryad Digital Repository. ( 10.5061/dryad.266k4) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Hayward Jessica J. et al 2016. Data from: Complex disease and phenotype mapping in the domestic dog Dryad Digital Repository. ( 10.5061/dryad.266k4) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Genetic data used in these analyses are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.266k4 [67] and GEO accession nos. GSE90441, GSE83160, GSE70454 and GSE96736. C-BARQ data are available from serpell@vet.upenn.edu.