Abstract
Synthetic gene drives may soon be used to suppress or eliminate populations of disease vectors, pathogens, invasive species, and agricultural pests. Recent proposals have focused on using Z-linked gene drives to control species with ZW sex determination, which include Lepidopteran pests, parasitic trematodes, and cane toads. These proposals include Z-linked ‘W-shredders’, which would suppress populations by cleaving the W chromosome and causing females to produce only sons, as well as Z-linked female-sterilizing gene drives. Here, I use eco-evolutionary simulations to evaluate the potential of some proposed Z-linked gene drives, and to produce recommendations regarding their design and use. The simulations show that W-shredders are likely to be highly effective at eradicating populations provided that resistance to W-shredding cannot evolve. However, W-shredder alleles can invade populations from very low frequencies, making it difficult to eliminate specific populations while leaving nearby populations untouched; this issue may restrict their possible uses.
Keywords: sex chromosomes, gene drives, population control, schistosomiasis, selfish genes
1. Introduction
Developments in genetic engineering will soon make it feasible to alter or eliminate populations of disease vectors, pathogens, agricultural pests, and invasive species using ‘gene drives’ [1–6]. Gene drives cause particular alleles (usually transgenes) to propagate through populations via a range of mechanisms, including gene conversion, poison-antidote systems, segregation distortion, and genetic incompatibility [7–9]. For example, CRISPR-Cas9 gene editing can be used to create a transgene that will be transmitted to almost 100% of the offspring of heterozygous individuals instead of the usual 50%; this type of gene drive functions by inducing a double-stranded DNA break at the homologous wild-type locus, which is then repaired using the transgene as a template. Gene drives are often categorized into two types: replacement drives, which aim to spread a human-beneficial allele throughout a population (e.g. a mosquito allele that interferes with the transmission of malaria [1,10]), and suppression drives, which reduce the size of a population (potentially to extinction). Suppression drives typically work by using non-Mendelian inheritance to spread alleles that cause lethality or sterility [2,5,11], or skew the offspring sex ratio—typically towards males [12–16].
Recent theoretical papers have investigated the feasibility, efficacy, and potential negative consequences of various types of gene drives. For example, Noble et al. [6] showed that the basic version of a CRISPR-Cas9 gene drive might be highly invasive and could rapidly spread to fixation across whole species, which is often an undesirable outcome. Conversely, other models have concluded that gene drives are likely to fail if populations can evolve resistance to their effects [17,18]. The issue of resistance is compounded because the standard implementation of a CRISPR-Cas9 gene drive (but perhaps not updated versions; [4,5,18,19]) tends to create its own resistance alleles, e.g. when the double-stranded break induced by Cas9 is repaired using an alternative DNA repair pathway (non-homologous end joining; NHEJ) instead of homology-directed repair [1–3,18,20]. Given the potential safety, ethical, and sociopolitical concerns surrounding gene drives, some models have focused on gene drives that would go extinct after a time [15,21,22], would stay confined to particular populations [11,22], and/or could be reversed once they have spread [23].
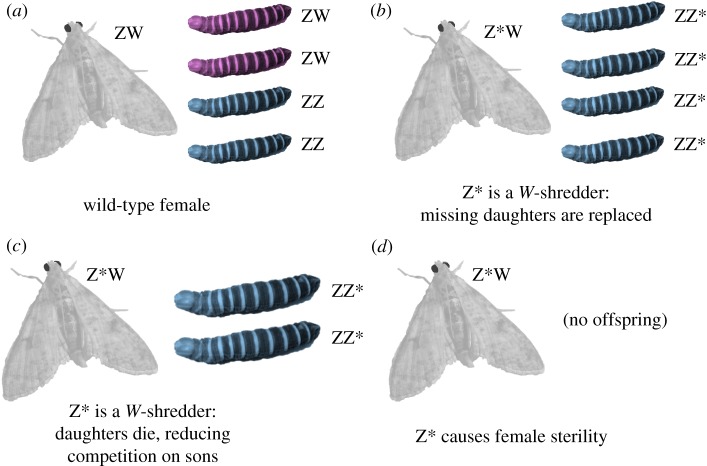
This paper focuses on the evolutionary dynamics of Z-linked suppression gene drives. The simulation is inspired by proposals for various types of Z-linked gene drives by Kevin Esvelt and colleagues, as well as ongoing efforts to develop these Z drives (see www.sculptingevolution.org; at the time of writing, these ideas have not been published elsewhere). Various Z-linked suppression drives proposed by Esvelt and colleagues are shown schematically in figure 1. Depending on its design, mode of action, and the biology of the target species, Z chromosomes carrying the drive allele (denoted Z*) might enjoy a transmission advantage in Z*W females (figure 1b, and perhaps also 1c), and optionally also in Z*Z males. Esvelt et al. focus on using Z drives to control the Schistosoma trematodes responsible for schistosomiasis, though Z drives could theoretically be used to control any organism with female-heterogametic sex determination, such as Lepidopteran agricultural pests, invasive populations of cane toads Bufo marinus [24], or even invasive birds.
Figure 1.
Some hypothetical Z-linked suppression drives considered in this study. Panel (a) illustrates normal inheritance of sex chromosomes in a wild-type ZW female (assumed to be mated to a wild-type ZZ male; not shown): the offspring sex ratio is even. (b) The female carries a W-shredder allele (Z*) that kills gametes or offspring early enough that missing daughters are replaced with more Z*-bearing sons. (c) The lost daughters are not replaced, though their absence increases the survival probability of the sons somewhat (shown by their larger size), causing super-Mendelian inheritance of the Z* allele. Lastly, panel (d) shows a Z-linked female-sterilizing allele (e.g. an allele that cleaves the W chromosome or a female-essential gene in somatic cells); since it is strongly disadvantageous in females, such an allele would go extinct unless it benefits from gene drive in heterozygous males. (Online version in colour.)
A Z-linked gene drive could suppress populations by biasing gametogenesis in females, for example, by inducing double-stranded DNA breaks in the W chromosome in order to inactivate it; such a gene drive would be a ‘W-shredder’, analogous to the X- and Y-shredders under development for XY species [12,13,15,16,25,26]. Females carrying the gene drive would thus produce relatively few viable W-bearing eggs, and therefore produce mainly drive-carrying sons. Esvelt et al. point out that the evolutionary dynamics of the drive will depend on the fitness of drive carriers relative to wild types, the timing of W-shredding (e.g. in germ cells, ova, or zygotes), and the ecology of the target species. For example, some W-shredder designs might allow drive females to produce roughly the same number of (mostly male) offspring as a wild-type female provided that the W chromosome is destroyed early enough in oogenesis/development that the lost daughters can be replaced by sons (figure 1b). Alternatively, drive-carrying females might produce half the number of offspring (or fewer), e.g. if the drive works by destroying all ova or offspring that carry a W chromosome, and this loss is not compensated by reduced competition on the surviving offspring. Esvelt et al. also proposed that one could suppress populations using a Z-linked locus that caused sterility or lethality in females, either by shredding the W in somatic tissues, or by spreading some other allele that harms females but not males. If this female-harming allele were capable of gene drive in males, it could perhaps reach high enough frequencies to suppress the population. The W-shredder could be designed to also cause gene drive in males. Male gene drive could be accomplished using ‘standard’ CRISPR-Cas9 gene conversion, whereby the driving Z allele would convert the wild-type locus using homing endonuclease activity followed by homology-directed repair, causing heterozygous males to produce mostly drive-carrying sperm. Esvelt et al. note that male gene drive might not be necessary, since a Z-linked locus that prevents transmission of the W may already enjoy a transmission advantage (figure 1b,c).
Here, I present an evolutionary simulation that can accommodate all of these proposed Z-linked drives. I aimed to test which properties of the gene drive and the ecology of the target species are critical to determining the likelihood and speed of extinction. For example, the gene drive will presumably spread faster if it can bias transmission in both sexes, but perhaps a simpler female-only drive would be adequate. Also, since the population will become more male-biased as the gene drive invades, eco–evo feedback might affect the outcome of the drive release in non-intuitive ways. For example, alleles that prevent W-shredding might have an especially large fitness advantage relative to resistance alleles against ‘standard’ CRISPR drives [17,18], since the former restore a normal sex ratio as well as defusing the harmful effect of the driver on individual fitness. Moreover, the change in sex ratio could affect the ecology and evolution of the population, particularly if males and females contribute differentially to density-dependent population growth [27,28], or have different dispersal rates [29]. The model incorporates the possibility that Z-linked resistant-to-drive alleles are sometimes created by NHEJ in heterozygote males, to test whether resistance is just as problematic as for autosomal drives [1–3,18,20].
2. Methods
A full description of the simulation is provided as electronic supplementary material. In brief, I simulate a finite population of dioecious diploids with ZW sex determination that inhabits k discrete habitat patches arranged linearly in a ring. The aim of the simulation is to identify properties of the gene drive and the ecology of the target species that influence the outcome of a single release of nrelease homozygote males carrying a Z-linked gene drive allele, Z*. The drive allele causes biased inheritance and/or reduced fecundity in females, and optionally also causes biased inheritance in heterozygous males (e.g. via gene conversion). Each generation proceeds in discrete steps: birth, dispersal between patches, breeding within patches, and death of the parental generation. The equilibrium population size was roughly 10 000 in all simulations upon release of the gene drive, and the main outcomes of interest are the likelihood and speed of extinction. The simulation was written in R v. 3.4.0 and run on the Spartan cluster at the University of Melbourne; table 1 lists the simulation parameters that were manipulated to study their effects.
Table 1.
List of variables, and their corresponding parameter(s) in the model, which were varied in order to study their effects on extinction.
| variable | parameter(s) |
|---|---|
| strength of gene drive in females (e.g. W-shredding) | pshred |
| strength of gene drive in males (e.g. gene conversion) | pconv |
| cost of gene drive allele to female fecundity | cf |
| cost of gene drive allele to male mating success | cm |
| frequency of W-linked resistance mutations | μW |
| frequency of Z-linked resistance mutations and NHEJ | μZ and pnhej |
| frequency of autosomal resistance alleles | μA and μB |
| patchiness of the population | k |
| dispersal rate of males and females | xm and xf |
| global versus local density-dependence of female fecundity | ψ |
| contribution of males relative to females in density-dependence | δ |
| number of gene drive carrier males released | nrelease |
| release strategy: all in one patch, or global | — |
| fecundity of females at low population densities | r |
| shape of density dependence | α |
Each male carries two diploid autosomal loci (termed A/a and B/b) and one diploid Z-linked locus, while females carry both autosomal loci, a single allele at the Z-linked locus, plus a W chromosome. There are three possible Z-linked alleles: the drive allele (Z*), a wild-type allele (Z+) that is vulnerable to gene drive in Z*Z+ males, and a resistant allele (Zr) that is immune to gene drive in Z*Zr males. Similarly, there are two types of W chromosome: a wild-type W that is vulnerable to shredding by the Z* allele (W+), and an immune variant (Wr). The autosomal alleles A and B are dominant ‘trans-acting’ resistance alleles that confer immunity to W-shredding and gene conversion, respectively. The Z* allele imposes a cost cf on the fecundity of female carriers, and a cost cm on the mating success of male carriers. The resistance alleles Wr, Zr, A, and B are assumed to be cost-free. Setting cf = 1 allows simulation of a female-sterilizing Z-linked drive (figure 1d).
Females carrying Z* (and no A or Wr alleles) produce Z-bearing gametes and W-bearing gametes, and thus produce >50% sons when pshred > 0. Secondly, Z*Z+ males produce gametes carrying the Z* allele, gametes carrying the Z+ allele, and gametes carrying the Zr allele. Thus, gene conversion occurs in males if pconv > 0, meaning that the Z* allele is over-represented in the gametes of these three male genotypes. The parameter pnhej represents the creation of resistance alleles via NHEJ.
Female fecundity depends on the local and/or global density and fitness of other females, and the density of males, via functions involving five parameters that control the carrying capacity (K), maximum possible fecundity (r), the shape of the density-dependence function (α), the relative effect of males and females on density (δ), and the scale of density-dependence (local or global; ψ). These parameters allow the model to capture a range of possible life histories suitable for different organisms that could be suppressed with W-shredders. For example, maximum fecundity r is lower for birds than for cane toads, and δ might be lower in Lepidoptera than trematodes, since male trematodes consume space/resources inside the host (and males are much larger than females), while male Lepidoptera do not consume one of the main limited resources that females need (i.e. host plants on which to lay their eggs). Female and male offspring disperse to other patches with probabilities xf and xm, respectively, allowing for variable and sex-specific gene flow between patches. Dispersal was either local or global (i.e. to a neighbouring patch or a random patch).
3. Results
(a). Three illustrative simulation runs
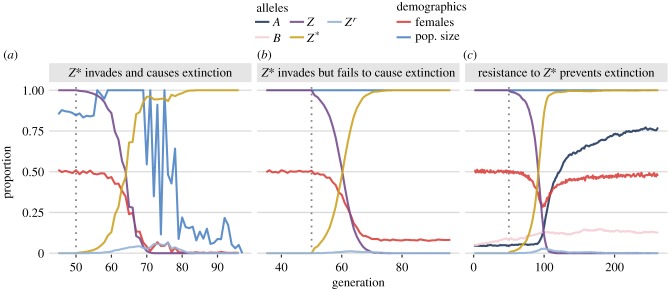
Figure 2 shows three contrasting simulation runs. In figure 2a, the release of Z*Z* males at generation 50 caused the Z* allele to invade and fix, causing rapid extinction due to a lack of females. The simulation run in figure 2a assumed that Z* causes near-complete W-shredding, Z* benefits from gene drive in males, that Z* has moderate fitness costs to carriers, and there is no resistance to W-shredding (electronic supplementary material, table S1).
Figure 2.
Three illustrative runs of the simulation, showing evolution in response to the release of males carrying a W-shredder at generation 50 (dotted line). (a) The driving Z* allele fixed very quickly, causing population extinction through a shortage of females. (b) The Z* allele fixed but did not cause extinction, because carrier females continued to produce some daughters due to incomplete W-shredding. (c) The Z* allele invaded, which selected for the W-shredding resistance allele A, causing Z* to go extinct by removing its transmission advantage. The population size N is shown as a fraction of its maximum value of 10 000. Electronic supplementary material, table S1 lists the parameter spaces used for these three runs. (Online version in colour.)
In figure 2b, Z* went to fixation but failed to cause extinction, due to incomplete W-shredding by the Z* allele. In this run, pshred = 0.84 (electronic supplementary material, table S1), meaning that 8% of the offspring of Z*W+ mothers are female. The invasion of Z* altered the population sex ratio, but did not cause extinction (or even a decline in population size).
Lastly, figure 2c shows a case where the invasion of Z* was reversed by the evolution of autosomal resistance alleles. Following the release of the Z* allele, the initially rare autosomal resistance allele A (which prevents W-shredding) rapidly increased in frequency, disarming the effects of Z* in females and returning the population-level sex ratio to 50 : 50, even though the Z* allele reached fixation. Incidentally, the resistant allele A was favoured over a because the male-biased population sex ratio created by Z* favours the production of daughters, and AA and Aa females produce more daughters than aa females in populations where Z* is present.
(b). Effects of each parameter on the evolution of a W-shredder
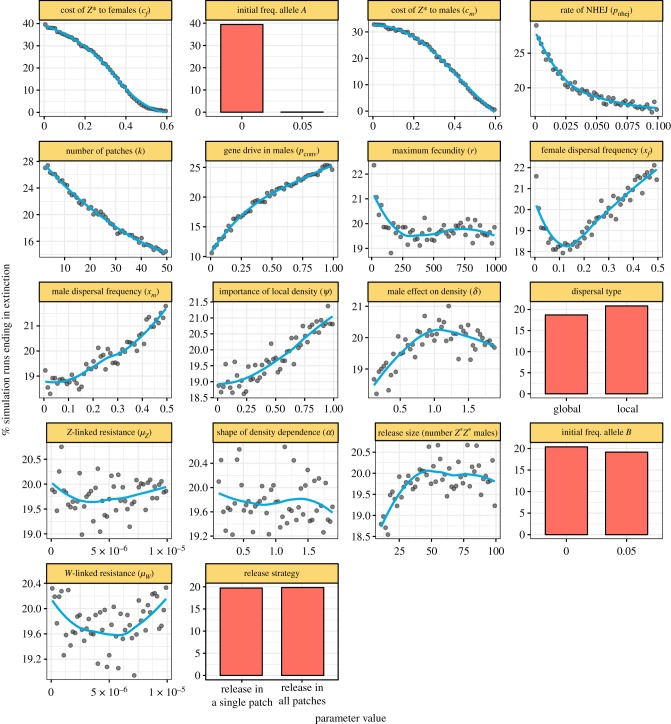
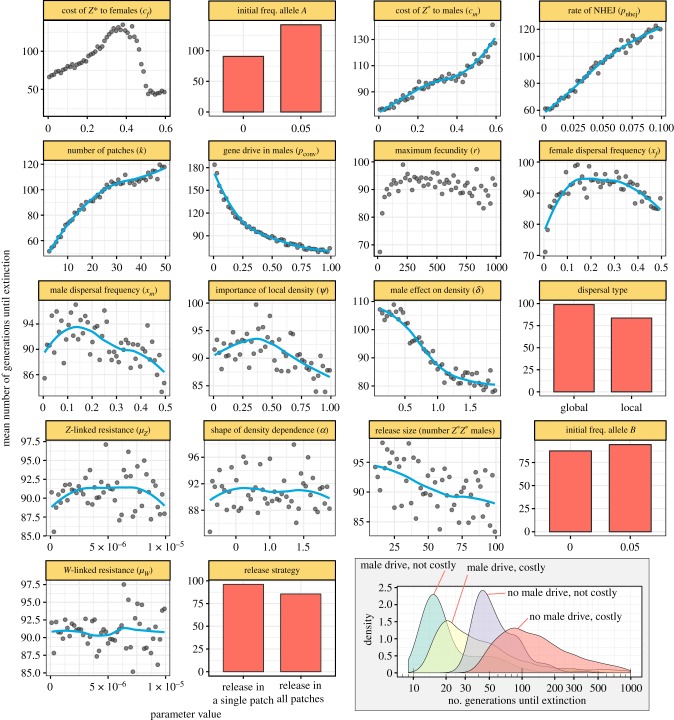
Figure 3 shows the effects of each parameter for simulations of a Z-linked W-shredder allele that also potentially causes gene drive in Z*Z males. Figure 3 assumes complete W-shredding (pshred = 1), because initial trials revealed that extinction almost never occurred if pshred < 0.95 (electronic supplementary material, figure S1). This result confirms that a W-shredder needs to effectively prevent female carriers from producing daughters in order to reliably cause extinction. Otherwise, all the parameters were allowed to vary within pre-specified limits using Latin hypercube sampling. Thus, the relationships in figure 3 show the expected effect of the focal parameter on extinction probability, while the other parameters vary independently within the ranges shown on the x-axis of figure 3. Figure 4 is similar to figure 3, except that it plots the average time until extinction (for the subset of runs in which extinction actually occurred).
Figure 3.
The plot shows the effect of each model parameter on the percentage of simulation runs in which the population went extinct, for simulations of a Z-linked W-shredder that completely prevents the production of daughters by females (pshred = 1; see also electronic supplementary material, figure S1). The plot was generated by sampling evenly from the complete parameter space 721 587 times using Latin hypercube sampling, such that each plot shows the marginal effect of one parameter while the other parameters vary independently across their possible ranges (shown by the relevant x-axis scale). Continuous parameters were grouped into 51 bins (each containing approx. 14 400 runs, 2% of the total), and the lines were fitted using LOESS (locally estimated scatterplot smoothing). Note that the y-axis scales to the range of the data, and the panels are ordered by this range (showing the approximate relative importance of each parameter). (Online version in colour.)
Figure 4.
The main plot shows the average number of generations until extinction, among the subset of runs in which extinction occurred. The density plot in the lower right shows the distribution of the response variable across all the parameter spaces in which the Z* allele was costly (defined as cf > 0.05 and/or cm > 0.05) or not costly (cf < 0.05 and cm < 0.05), and in which the Z* allele benefitted from strong gene drive in males (pconv > 0.95) or not (pconv < 0.95). Other details are the same as for figure 3. (Online version in colour.)
(i). Gene drive parameters
Extinction probability declined sharply as the costs of the gene drive to female fecundity (cf) and male mating success (cm) increased, suggesting that one should work to minimize such costs when developing a W-shredder. Consistent with this conclusion, time until extinction increased monotonically with the cost of male mating success cm, but interestingly, the relationship between cf and the speed of extinction was hump-shaped. W-shredders with no costs in females caused extinction quickly due to their more rapid spread, while W-shredders that carried a sufficiently high cost to females actually resulted in faster extinction (though note that extinction rarely occurred when cf was high), since female fecundity is critical to population growth. On balance, the model suggests that W-shredders should be designed to have minimal impact of the fitness of carriers, though reassuringly (from a design standpoint), extinction still occurred frequently even if carriers were 10–20% less fit than wild types.
Another very important parameter was the presence/absence of autosomal resistance alleles that prevent W-shredding from occurring: extinction almost never happened when such alleles were present. Perhaps surprisingly, when I allowed wild-type W+ chromosomes to mutate to a resistant type, ensuring the population almost always contained a small number of resistant Wr chromosomes, the probability of extinction was essentially unaffected. The difference in outcome relative to the A allele may have to do with the reduced population size of the W chromosome, which is limited to females, whose population size drops very rapidly once the Z* allele invades (figure 2).
Extinction was substantially more likely when the W-shredder was also capable of gene drive in Z*Z males. Although male gene drive was non-essential for the W-shredder to cause extinction, efficient male-acting drive greatly reduced the number of generations required for extinction to occur (see inset of figure 4). Similarly, resistance to the male-acting component of the gene drive reduced the extinction probability (and increased the time until extinction), especially when resistance alleles were created by NHEJ rather than arising randomly by mutation (presumably because the latter causes the resistance alleles to appear in the same patches as the drive allele, where they immediately enjoy a selective advantage).
The number of Z*Z* males released into the population had little effect on extinction probability: releasing 20 individuals was almost as likely to cause extinction as releasing 100. This result reflects the classic population genetics result that the fixation probability of a strongly selected allele (such as a gene drive) does not depend on its frequency (only on selection and population structure; reviewed in [30]). Additionally, releasing the drive-carrying males throughout the meta-population was equally likely to cause extinction as releasing them all in one patch. As well as suggesting that multi-site release programmes might not be strictly necessary, this result implies that Z-linked gene drives are very likely to spread between populations connected by gene flow, as previously found for autosomal drives [6].
(ii). Ecological drive parameters
The most important ecological parameter was the number of patches into which the population was divided. Spatially structured populations were harder to drive extinct, presumably because the population can persist in habitat patches where the drive allele is absent, and because migrants can recolonize patches where local extinction has occurred. Since patches containing the gene drive will produce fewer migrants (due to their imbalanced sex ratio), migrants that recolonize empty patches are more likely to carry wild-type or drive-resistant alleles. The time until extinction increased with the patchiness of the population, with diminishing returns.
Interestingly, the migration rate had a sex-specific relationship with extinction probability. Extinction became increasingly likely as the male migration rate (xm) increased, since the drive allele is primarily carried by males due to its effect on the sex ratio. Conversely, the effect of the female migration rate (xf) was U-shaped. With very high female migration rates, population structure is much reduced, ensuring that the drive allele quickly reaches any patch that lacks it. With low female migration rates, the gene drive causes local extinctions faster than patches can be recolonized by dispersal. Intermediate rates of female dispersal allow for the creation of long-lasting refugia from the drive allele, and also allow for some recolonization, resulting in a reduced probability of extinction.
Unsurprisingly, extinction was more likely when the maximum possible fecundity of each female (r) was lower. However, the decline was not linear, and the results suggest that W-shredders are capable of eliminating even those species in which a handful of surviving females can rapidly repopulate. The results also suggest that extinction was slightly more probable when female fecundity was determined primarily by local rather than global density (ψ). This is because local density can remain high (and thus, per-female fecundity can remain low) even in meta-populations that are declining due to the spread of the Z* allele in some of their sub-populations.
The parameter δ, which represents sex differences in ecological niche use and behaviour, had a hump-shaped relationship with extinction probability. Low δ indicates that female fecundity is only weakly affected by the density of males, and this scenario was associated with lower extinction probabilities. This is because the gene drive creates male-biased populations, boosting female fecundity for any given population size, and helping to stave off extinction. Conversely when δ is high, male density has a stronger negative effect on female fecundity, which slightly reduced the probability of extinction (perhaps because patches containing many drive alleles are male-biased, and thus send out comparatively few drive-carrying migrants when δ is high). Extinction was most likely for intermediate values of δ. By contrast, the time until extinction declined monotonically with δ, since high values of δ mean that male-biased populations produce even fewer offspring.
Finally, the parameter α, which controls the shape of the relationship between female fecundity and population density, had a weak negative effect on the likelihood or speed of extinction. Setting α < 1 means that female fecundity declines at a decelerating rate as population density increases, such that per-female fecundity only approaches its maximum possible value once the population is heavily depleted, making extinction more likely.
(c). Well-designed W-shredders cause rapid extinction
The inset of figure 4 shows the frequency distribution for time until extinction. With a well-designed W-shredder and favourable ecological conditions, the Z* allele caused extinction in as few as nine generations. When considering only those simulations in which pshred = 1 (i.e. drive females produce no daughters) and the fitness of drive carriers was within 5% of the wild type, extinction occurred within 20 generations for the majority of the parameter space shown in figures 3 and 4, illustrating that a well-designed W-shredder is likely to succeed in controlling species with very different ecologies.
(d). Female-sterilizing Z-linked gene drive
For runs in which the Z* allele caused females to become sterile rather than biasing the offspring sex ratio (i.e. cf = 1), extinction never occurred (in 1 559 817 simulation runs). This was true even when all other parameters besides cf took values that make extinction more likely, e.g. when the Z* allele was transmitted to 100% of the offspring of male heterozygotes, and when Z* did not harm male fitness. The reason for this result is that the fitness gains from gene drive in males were not compensated by a complete loss of fitness in females, at least for the ecological conditions considered here. I hypothesize that a female-sterilizing Z* allele might be capable of causing extinction if Z* males were continuously released into the population, but this possibility is outside the scope of the model implemented here.
4. Discussion
The model shows that W-shredders are, in principle, very effective at eliminating populations, particularly if they fully prevent females from producing daughters (pshred ≈ 1) and if resistance to W-shredding cannot evolve. The results have implications for the design of Z-linked W-shredders and female-sterilizing suppression drives, and help to identify ecological parameters that are (and are not) important to the outcome of a gene drive release.
A major design consideration is whether to engineer W-shredders that are also capable of gene drive in males, e.g. by including guide RNAs that target the Z as well as the W chromosome, to allow gene conversion in male heterozygotes. Foregoing male drive could simplify the design of W-shredders since they would only need to target the W chromosome (and not also the Z), particularly because male-acting gene conversion drives seem more challenging to develop than female-acting ones in some taxa (due to sex differences in DNA repair; [31]). In the model, W-shredders very often caused extinction even without male gene drive (i.e. when pconv ≈ 0), provided that individuals carrying the W-shredder had comparable fitness to wild types, and that carrier females produce very few daughters (as in figure 1b). Conversely, if W-shredder females had low fecundity (around half that of a wild type, or below; figure 1c) or produced some daughters, male gene drive tended to be essential for extinction to occur, or at least for extinction to occur rapidly enough to be useful. Although male gene drive was not always needed for extinction to occur, it did substantially reduce the number of generations until extinction. Therefore, I conclude that it would almost certainly be worth the effort to incorporate a male-acting component when developing a W-shredder, especially in species with longer generation times.
Another aim when designing W-shredders should be to ensure that female carriers produce as few daughters as possible (ideally none; represented in the model as pshred = 1), while producing a large number of drive-carrying sons (ideally as many as the total offspring produced by non-carriers; cf = 0). This implies that one should aim to design a construct that cleaves the W chromosome early in gametogenesis or offspring development, to increase the chance that the number of surviving progeny produced by each female is unaffected. Expression of Cas9 and cleavage of the W should also be restricted to the tissues where it is needed (e.g. the germline), to minimize fitness losses due to the loss of the W in somatic cells, or detrimental effects of Cas9. For some species, this may be as simple as placing the W-shredder under the control of a promoter such as nanos [32,33], assuming that females are able to replace lost W-bearing oocytes before they are provisioned with limiting resources. Even if the lost daughters are not replaced with additional sons, the Z* allele might still exhibit super-Mendelian inheritance across generations, because Z*-bearing males will experience reduced competition due to the deaths of their sisters (somewhat like the selfish genetic element Medea; [34]). In Lepidoptera, juvenile density is often strongly negatively correlated with survival, and there are various maternally transmitted endosymbionts that drive through populations by killing males to lessen competition on their infected sisters (e.g. [35,36]); these observations suggest that W-shredder alleles might drive through Lepidopteran populations even if Z*W females produced half as many viable eggs, though male gene drive would certainly help the invasion. The model also highlighted that strong fitness costs to males (cm) hinder the spread of the drive allele, though it seems unlikely that a W-shredder allele would have major costs in males.
The W-shredding mechanism should also be designed in a way that makes it difficult for W-linked or trans-acting resistance to shredding to evolve. One way to do this would be to use a single-guide RNA that targets W-specific sequences that are present in multiple copies, or to use multiple guide RNAs that target multiple W-linked sequences [32,37]; the former is similar to the X-shredder developed for Anopheles mosquitos, which cleaves repetitive, X-linked ribosomal genes [13]. By designing gene drives with multiple targets, the reference sequence must evolve multiple changes to acquire resistance to cleavage, which is relatively unlikely. To ensure that the targets of cleavage do not become resistant as a result of indels induced by NHEJ, one can ensure that the guide RNA’s target lies within an essential gene where an indel would be selectively disadvantageous, preventing resistant alleles from accumulating in the population [32,37]. This may not be necessary if the W-shredder targets many W-linked loci, but it is an important design consideration for the male gene drive component of the W-shredder, because the evolution of Z-linked resistance completely nullified the usefulness of male gene drive in the simulation (echoing [18]). Recent work demonstrated the feasibility of arrays containing many guide RNAs separated by spacers [38], suggesting it may soon be easier to create gene drives with multiple guide RNAs. A final way of developing resistance is to ‘disarm’ the endonuclease responsible for W-shredding and gene conversion, for example, by producing a protein that inhibits Cas9, or through evolution of gene regulatory processes that cause the Cas9 to no longer be expressed at the right time. There appears to be little data on whether this type of resistance is possible or likely, though models such as the present one indicate that they could certainly impede population suppression efforts.
The model also indicated that extinction does not require the release of large numbers of individuals: releasing just 20 Z* males was often enough to eliminate a spatially structured metapopulation of 10 000 individuals in a few generations. On the one hand, this is advantageous because W-shredders would be cheap and easy to deploy once they are developed, and they are likely to extirpate whole metapopulations even if gene flow between sites is quite weak. However, such high invasiveness is not always desirable, because it makes the gene drive very difficult to restrict to a particular area. This could limit the usefulness of W-shredders to control species like Lepidoptera and birds, where one may wish to eradicate only invasive or agriculturally damaging populations, while leaving other populations untouched. Modifications to gene drive design—such as the self-limiting ‘daisy drive’ system—might someday address this important concern [21,22].
The model found several ecological variables that affect the likelihood of extinction, but reassuringly the effects on extinction probability were somewhat minor, and I found no ecological parameter values that made extinction impossible. Populations in which females can produce many offspring (high r) were harder to drive extinct, though the effect of r reached an asymptote around r = 250, such that even extremely fecund organisms could be controlled using W-shredders: this is because fecundity does not matter if the population is all-male. Patchy populations were harder to drive extinct, though this could probably be mitigated by releasing gene drive individuals in larger numbers, over a greater range, over multiple releases. Dispersal was a double-edged sword: weak gene flow meant that the gene drive spread more slowly through the metapopulation, while high dispersal rates allow for re-colonization of patches in which the gene drive had caused extinction. Demographic parameters such as the scale of competition (ψ) and the shape of density-dependence (α) mattered little. Perhaps surprisingly, the relative importance of male density to population growth (δ) had only a weak effect: extinction was a little more likely when the presence of males suppresses female fecundity just as much (or more so) as the presence of other females. However, extinction still occurred when female fecundity depended only weakly on male density (again, probably because female fecundity becomes unimportant as females begin to disappear from the population).
Finally, I note that W-shredders might in general be easier to develop than X-shredders. Efforts to develop an X-shredder in Anopheles mosquitos were initially hindered, because the I-PpoI protein used to cleave the X was paternally transmitted to the embryo inside sperm, causing all of the drive male’s offspring to die (not just the daughters) due to cleavage of the maternally inherited X in the male’s sons. Although this technical issue was later addressed [13], such intergenerational effects would not hinder a W-shredder since the W chromosome is unique to females (provided that the W-shredding protein was not expressed in males, or at least was absent from their sperm). Additionally, W-shredders might sometimes be easier to develop than gene drives that work by deleting genes that are essential to female (but not male) fitness (e.g. [15]). This is because one could design a prototype W-shredder based only on sequence data from the sex chromosomes, while identifying genes with female-specific fitness effects requires more detailed data (e.g. expression profiling or knockout studies) that are unavailable for most taxa.
Supplementary Material
Acknowledgements
I thank the organizers (Anna Lindholm and Tom Price), funding bodies (European Society for Evolutionary Biology; Swiss National Science Foundation), and attendees of the 2018 ESEB Progress Meetings in Evolutionary Biology, which provided the impetus for this paper. I also thank Kevin Esvelt and colleagues for describing their ongoing research on a personal webpage; their ideas were instrumental to this paper.
Data accessibility
A website presenting all R scripts used to the run the simulation and analyse the data can be found at https://lukeholman.github.io/W_shredder/.
Competing interests
I declare I have no conflict of interests.
Funding
This project was stimulated by an ESEB Progress Meetings in Evolutionary Biology meeting, funded by grants from ESEB (European Society for Evolutionary Biology) and from the Swiss National Science Foundation.
References
- 1.Gantz VM, Jasinskiene N, Tatarenkova O, Fazekas A, Macias VM, Bier E, James AA. 2015. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl Acad. Sci. USA 112, E6736–E6743. ( 10.1073/pnas.1521077112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hammond A. et al. 2016. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechnol. 34, 78–83. ( 10.1038/nbt.3439) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Z, Pan Q, Gendron P, Zhu W, Guo F, Cen S, Wainberg MA, Liang C. 2016. CRISPR/Cas9-derived mutations both inhibit HIV-1 replication and accelerate viral escape. Cell Rep. 15, 481–489. ( 10.1016/j.celrep.2016.03.042) [DOI] [PubMed] [Google Scholar]
- 4.Prowse TA, Cassey P, Ross JV, Pfitzner C, Wittmann TA, Thomas P. 2017. Dodging silver bullets: good CRISPR gene-drive design is critical for eradicating exotic vertebrates. Proc. R. Soc. B 284, 20170799 ( 10.1098/rspb.2017.0799) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kyrou K, Hammond AM, Galizi R, Kranjc N, Burt A, Beaghton AK, Nolan T, Crisanti A. 2018. A CRISPR–Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat. Biotechnol. 36, 1062–1066. ( 10.1038/nbt.4245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noble C, Adlam B, Church GM, Esvelt KM, Nowak MA. 2018. Current CRISPR gene drive systems are likely to be highly invasive in wild populations. eLife 7, e33423 ( 10.7554/eLife.33423) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindholm AK. et al. 2016. The ecology and evolutionary dynamics of meiotic drive. Trends Ecol. Evol. 31, 315–326. ( 10.1016/j.tree.2016.02.001) [DOI] [PubMed] [Google Scholar]
- 8.Champer J, Buchman A, Akbari OS. 2016. Cheating evolution: engineering gene drives to manipulate the fate of wild populations. Nat. Rev. Genet. 17, 146–159. ( 10.1038/nrg.2015.34) [DOI] [PubMed] [Google Scholar]
- 9.Oberhofer G, Ivy T, Hay BA. 2019. Cleave and Rescue, a novel selfish genetic element and general strategy for gene drive. Proc. Natl Acad. Sci. USA 116, 6250–6259. ( 10.1073/pnas.1816928116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marshall JM, Akbari OS. 2015. Gene drive strategies for population replacement. In Genetic control of malaria and dengue (ed. Adelman ZN.), pp. 169–200. London, UK: Academic Press. [Google Scholar]
- 11.Maselko M, Heinsch SC, Das S, Smanski MJ. 2018. Genetic incompatibility combined with female-lethality is effective and robust in simulations of Aedes aegypti population control. bioRxiv, p. 316406 ( 10.1101/316406) [DOI] [Google Scholar]
- 12.Windbichler N, Papathanos PA, Crisanti A. 2008. Targeting the X chromosome during spermatogenesis induces Y chromosome transmission ratio distortion and early dominant embryo lethality in Anopheles gambiae. PLoS Genet. 4, e1000291 ( 10.1371/journal.pgen.1000291) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galizi R, Doyle LA, Menichelli M, Bernardini F, Deredec A, Burt A, Stoddard BL, Windbichler N, Crisanti A. 2014. A synthetic sex ratio distortion system for the control of the human malaria mosquito. Nat. Commun. 5, 3977 ( 10.1038/ncomms4977) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beaghton A, Beaghton PJ, Burt A. 2017. Vector control with driving Y chromosomes: modelling the evolution of resistance. Malar. J. 16, 286 ( 10.1186/s12936-017-1932-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burt A, Deredec A. 2018. Self-limiting population genetic control with sex-linked genome editors. Proc. R. Soc. B 285, 20180776 ( 10.1098/rspb.2018.0776) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papathanos PA, Windbichler N. 2018. Redkmer: an assembly-free pipeline for the identification of abundant and specific X-chromosome target sequences for X-shredding by CRISPR endonucleases. CRISPR J. 1, 88–98. ( 10.1089/crispr.2017.0012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drury DW, Dapper AL, Siniard DJ, Zentner GE, Wade MJ. 2017. CRISPR/Cas9 gene drives in genetically variable and nonrandomly mating wild populations. Sci. Adv. 3, e1601910 ( 10.1126/sciadv.1601910) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Unckless RL, Clark AG, Messer PW. 2017. Evolution of resistance against CRISPR/Cas9 gene drive. Genetics 205, 827–841. ( 10.1534/genetics.116.197285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esvelt KM, Smidler AL, Catteruccia F, Church GM. 2014. Emerging technology: concerning RNA-guided gene drives for the alteration of wild populations. Elife 3, e03401 ( 10.7554/eLife.03401) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gantz VM, Bier E. 2015. The mutagenic chain reaction: a method for converting heterozygous to homozygous mutations. Science 348, 442–444. ( 10.1126/science.aaa5945) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Min J, Noble C, Najjar D, Esvelt KM. 2017. Daisyfield gene drive systems harness repeated genomic elements as a generational clock to limit spread. BioRxiv, p. 104877 ( 10.1101/104877) [DOI] [Google Scholar]
- 22.Noble C. et al. 2019. Daisy-chain gene drives for the alteration of local populations. Proc. Natl Acad. Sci. USA 116, 8275–8282. ( 10.1073/pnas.1716358116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vella MR, Gunning CE, Lloyd AL, Gould F. 2017. Evaluating strategies for reversing CRISPR-Cas9 gene drives. Sci. Rep. 7, 11038 ( 10.1038/s41598-017-10633-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abramyan J, Ezaz T, Graves JAM, Koopman P. 2009. Z and W sex chromosomes in the cane toad (Bufo marinus). Chromosome Res. 17, 1015–1024. ( 10.1007/s10577-009-9095-1) [DOI] [PubMed] [Google Scholar]
- 25.North A, Burt A, Godfray HCJ. 2013. Modelling the spatial spread of a homing endonuclease gene in a mosquito population. J. Appl. Ecol. 50, 1216–1225. ( 10.1111/1365-2664.12133) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prowse TA, Adikusuma F, Cassey P, Thomas P, Ross JV. 2019. A Y-chromosome shredding gene drive for controlling pest vertebrate populations. eLife 8, e41873 ( 10.7554/eLife.41873) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rankin DJ, Kokko H. 2007. Do males matter? The role of males in population dynamics. Oikos 116, 335–348. ( 10.1111/oik.2007.116.issue-2) [DOI] [Google Scholar]
- 28.Li XY, Kokko H. 2019. Intersexual resource competition and the evolution of sex-biased dispersal. Front. Ecol. Evol. 7, 111 ( 10.3389/fevo.2019.00111) [DOI] [Google Scholar]
- 29.Li XY, Kokko H. 2019. Sex-biased dispersal: a review of the theory. Biol. Rev. 94, 721–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patwa Z, Wahl LM. 2008. The fixation probability of beneficial mutations. J. R. Soc. Interface 5, 1279–1289. ( 10.1098/rsif.2008.0248) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grunwald HA, Gantz VM, Poplawski G, Xu XRS, Bier E, Cooper KL. 2019. Super-Mendelian inheritance mediated by CRISPR–Cas9 in the female mouse germline. Nature 566, 105–109. ( 10.1038/s41586-019-0875-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Champer J, Liu J, Oh SY, Reeves R, Luthra A, Oakes N, Clark AG, Messer PW. 2018. Reducing resistance allele formation in CRISPR gene drive. Proc. Natl Acad. Sci. USA 115, 5522–5527. ( 10.1073/pnas.1720354115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z, Niu B, Ji D, Li M, Li K, James AA, Tan A, Huang Y. 2018. Silkworm genetic sexing through W chromosome-linked, targeted gene integration. Proc. Natl Acad. Sci. USA 115, 8752–8756. ( 10.1073/pnas.1810945115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hay BA, Chen CH, Ward CM, Huang H, Su JT, Guo M. 2010. Engineering the genomes of wild insect populations: challenges, and opportunities provided by synthetic Medea selfish genetic elements. J. Insect. Physiol. 56, 1402–1413. ( 10.1016/j.jinsphys.2010.05.022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiggins F, Hurst G, Jiggins C, VD Schulenburg J, Majerus M. 2000. The butterfly Danaus chrysippus is infected by a male-killing Spiroplasma bacterium. Parasitology 120, 439–446. ( 10.1017/S0031182099005867) [DOI] [PubMed] [Google Scholar]
- 36.Jiggins FM. 2003. Male-killing Wolbachia and mitochondrial DNA: selective sweeps, hybrid introgression and parasite population dynamics. Genetics 164, 5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oberhofer G, Ivy T, Hay BA. 2018. Behavior of homing endonuclease gene drives targeting genes required for viability or female fertility with multiplexed guide RNAs. Proc. Natl Acad. Sci. USA 115, E9343–E9352. ( 10.1073/pnas.1805278115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurata M. et al. 2018. Highly multiplexed genome engineering using CRISPR/Cas9 gRNA arrays. PLoS ONE 13, e0198714 ( 10.1371/journal.pone.0198714) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
A website presenting all R scripts used to the run the simulation and analyse the data can be found at https://lukeholman.github.io/W_shredder/.