Short abstract
Background
Duchenne muscular dystrophy is a muscle wasting disease caused by dystrophin gene mutations resulting in dysfunctional dystrophin protein. Autophagy, a proteolytic process, is impaired in dystrophic skeletal muscle though little is known about the effect of dystrophin deficiency on autophagy in cardiac muscle. We hypothesized that with disease progression autophagy would become increasingly dysfunctional based upon indirect autophagic markers.
Methods
Markers of autophagy were measured by western blot in 7-week-old and 17-month-old control (C57) and dystrophic (mdx) hearts.
Results
Counter to our hypothesis, markers of autophagy were similar between groups. Given these surprising results, two independent experiments were conducted using 14-month-old mdx mice or 10-month-old mdx/Utrn± mice, a more severe model of Duchenne muscular dystrophy. Data from these animals suggest increased autophagosome degradation.
Conclusion
Together these data suggest that autophagy is not impaired in the dystrophic myocardium as it is in dystrophic skeletal muscle and that disease progression and related injury is independent of autophagic dysfunction.
Keywords: mdx, utrophin, heart, Duchenne muscular dystrophy
Introduction
Duchenne muscular dystrophy (DMD) is a muscle wasting disease caused by the absence of functional dystrophin protein. Dystrophin anchors the dystrophin-associated glycoprotein complex (DGC), which connects the extracellular matrix to the actin cytoskeleton. This sarcolemmal architecture is necessary for the DGC to function as a conduit of force transduction, to provide stability to the sarcolemma, and participate in intracellular signaling.1 In addition to sarcolemmal instability,2 dystrophin deficiency results in secondary cellular dysfunctions3–9 and ultimately leads to loss of motor function, respiratory failure, and cardiac failure.
Autophagy functions as a homeostatic mechanism, in which lysosomes degrade autophagosomal cargo.10,11 Upon stimulation of autophagy, nucleation of a phagophore begins and matures to become an autophagosome, which packages tagged proteins, organelles, and other cellular components. Autophagosomes then fuse with lysosomes creating an autophagolysosome, in which the autophagosomal cargo is degraded. We recently demonstrated that in dystrophic skeletal muscle degradation of autophagosomes is impaired due, at least in part, to lysosomal insufficiency.12 In other investigations, rapamycin was used to inhibit mTOR in the mdx model, which increased autophagic flux and partially corrected disease-related losses in skeletal and cardiac muscle function.13,14 That stimulation of autophagy in the dystrophic myocardium preserved function suggests that autophagy is impaired by dystrophin deficiency. Other models of cardiac injury have shown that dysfunctional autophagy contributes to disease-related injury,15–17 although in limb-girdle muscular dystrophy dysfunctional autophagy was not apparent.18 Importantly, the extent to which autophagy changes with progressive cardiac injury in DMD is unknown. Consequently, the purpose of this investigation was to determine the extent to which degradation of autophagosomes is altered by disease progression in dystrophic cardiac muscle. We hypothesized that degradation of autophagosomes would become increasingly dysfunctional with disease progression.
Methods
Animal treatment
All procedures were approved by the Institutional Animal Care and Use Committees at Iowa State University or Auburn University. In Experiment 1, 7 wk and 17 mo C57 (7 wk n = 5; 17 mo n = 3) and mdx (7 wk n = 8; 17 mo n = 8) male mice were sedated with tribromoethanol (0.15 ml/g) and sacrificed by cervical dislocation. In an independent experiment (Experiment 2), 14 mo C57 and mdx mice (n = 7 per group) were sedated with isoflurane and sacrificed by cardiac excision. Before eight weeks of age, cardiac function has been found to be normal in mdx mice but progresses to cardiomyopathy by 10 months of age19; therefore, 7 wk old mice were used to model early disease progression and 14–17 mo mice were used to model late disease progression. In Experiment 3, 10 mo male C57 and dystrophin deficient mice heterozygous for a utrophin mutation (mdx/Utrn±) (n = 8 per group) were treated as in Experiment 2. As these mice also suffer from decreased utrophin abundance, they have a more severe pathology compared to age-matched mdx mice. Excised hearts from all mice were frozen and stored at −80°C for subsequent analyses. Previously published data from this animal cohort include a detailed histopathological examination of hearts for the C57, mdx, and mdx/Utrn± mice.20,21 Data regarding skeletal muscle from these mice have also been previously published,22,23 including the effect of disease progression on markers of autophagy.12
Biochemistry
Protein from hearts was isolated and subjected to western blotting as previously described.12,20,23 Briefly, cardiac muscle was powdered using a dry ice-chilled metal mortar and pestle, protein extracted and concentration measured, and loaded onto a 4–20% gradient gel (Lonza). After separation and transfer, membranes were stained with Ponceau stain to verify equal loading, blocked in 5% milk made in Tris buffered saline with 0.2% Tween20 (TBST) and incubated in primary antibody overnight at 4°C. Membranes were washed and secondary antibodies were applied for 1 h at room temperature. Dilutions of primary and secondary antibodies were used as follows from Cell Signaling Technology Inc. (Danvers, MA, USA), unless otherwise noted: AMP-activated protein kinase (AMPK) (product no. 5832, Primary (P) 1:1000 5% milk in TBST, Secondary (S) 1:2000 2.5% milk in TBST), phosphorylated (p) AMPK (T172) (product no. 2535, P 1:1000 in TBST, S 1:2000 5% milk in TBST), Unc-51 Like Autophagy Activating Kinase 1 (ULK) (product no. 8054, P 1:500 in TBST, S 1:500 in TBST), p-ULK1 (S555) (product no. 5869, P 1:500 1% milk in TBST, S 1:500 in TBST), phosphoinositide 3-kinases (PI3K) Class III (product no. 3358, P 1:1000 5% milk in TBST, S 1:1000 5% milk in TBST), beclin 1 (product no. 3495, P 1:750 5% milk in TBST, S 1:1000 in TBST), p-beclin 1 (S93) (product no. 14717, P 1:500 1% milk in TBST, S 1:500 in TBST), autophagy-related protein (ATG) 5 (product no. 9980, P 1:1000 5% milk in TBST, S 1:2000 5% milk in TBST), ATG5/12 (product no. 4180, P 1:1000 1% milk in TBST, S 1:2000 5% milk in TBST), ATG7 (product no. 2631, P 1:1000 in TBST, S1:2000 in TBST), light chain 3 (LC3) (product no. 12741, P 1:500 5% milk in TBST, S 1:2000 5% milk in TBST), sequestosome 1 (SQSTM1, p62) (Abcam, Cambridge, UK) (product no. EPR4844, P 1:500 5% milk in TBST, S 1:1000 5% milk in TBST). Species specific secondary antibodies were obtained from Cell Signaling Technology Inc. (product no. 7074S, Danvers, MA, USA). Clarity™ (BioRad, Hercules, CA, USA) enhanced chemiluminescence was applied to each membrane for approximately 7 min, and then films were developed in a dark room to visualize protein bands. Protein bands were quantified using Carestream densitometry software (Carestream Health, Inc., New Haven, CT).
qPCR
Quantitative PCR was used to evaluate Spstm1 (p62) transcript abundance. RNA was extracted from snap frozen, powdered tissue using TriZol (15596018, ThermoScientific) and RNeasy purification kit (Qiagen), then cDNA was reverse transcribed using QuantiTect Reverse Transcriptase Kit (205310, Qiagen). Manufacturers’ protocols were followed except for use of random hexamers (51-0118-01, IDT Premade Primers) instead of the RT primer mix provided (Qiagen). Sqsmt1 (p62) was evaluated (Forward – 5′tgtggtgggaactcgctataa-3′, Reverse – 5′-cagcggctatgagagaagctat-3′) and normalized to 18s (Forward – 5′-ctctagataacctcgggccg-3′, Reverse – 5′-gtcgggagtgggtaatttgc-3′).
Statistics
In Experiment 1, a two-way analysis of variance was used to compare differences between 7 wk and 17 mo C57 and mdx mice groups with age and disease status as main effects followed by a Newman–Keul post hoc test when indicated. In Experiments 2 and 3, a Student’s t-test was used to compare differences between groups. Significance was established at P < 0.05, a priori.
Results
Experiment 1: 7 wk and 17 mo C57 and mdx mice
In skeletal muscle from these mice we previously reported impaired degradation of autophagosomes due, at least in part, to a lysosomal insufficiency.12 To assess the effects of disease severity on markers of autophagy in dystrophic hearts, markers of activation, autophagosome formation, and autophagosome degradation were measured. Relative protein abundance and phosphorylation of upstream autophagy activator AMPK at threonine 172 (p-AMPK (T172)) was similar between groups as was the ratio of phosphorylated to total AMPK protein (Figure 1). ULK1 protein abundance was decreased as a function of dystrophin deficiency compared to healthy hearts (Figure 2). Phosphorylation of ULK1 at serine 555 (p-ULK1 (S555)) was similar between groups as was p-ULK1 (S555)/ULK1 ratio. After activation by ULK1, beclin 1 binds with PI3K class III to initiate phagophore formation.24 Relative protein abundance of PI3K class III, beclin 1, and p-beclin 1 (S93)/beclin 1 ratio were similar between groups, though relative abundance of p-beclin 1 increased as a function of age.
Figure 1.
Activation of autophagy. AMPK is similar between C57 and mdx mice at both 7 wk and 17 mo of age. Significance was established at p < 0.05. White bars – C57 7 wk (n = 4–5), gray bars – mdx 7 wk (n = 7–8), white bars with black hatching – C57 17 mo (n = 3), black bars – mdx 17 mo (n = 8).
Figure 2.
Initiation of autophagy. Markers of autophagic initiation were similar between all groups. Significance was established at p < 0.05. White bars – C57 7 wk (n = 4–5), gray bars – mdx 7 wk (n = 7–8), white bars with black hatching – C57 17 mo (n = 3), black bars – mdx 17 mo (n = 8). # indicates significant main effect of age, † indicates significant main effect of disease.
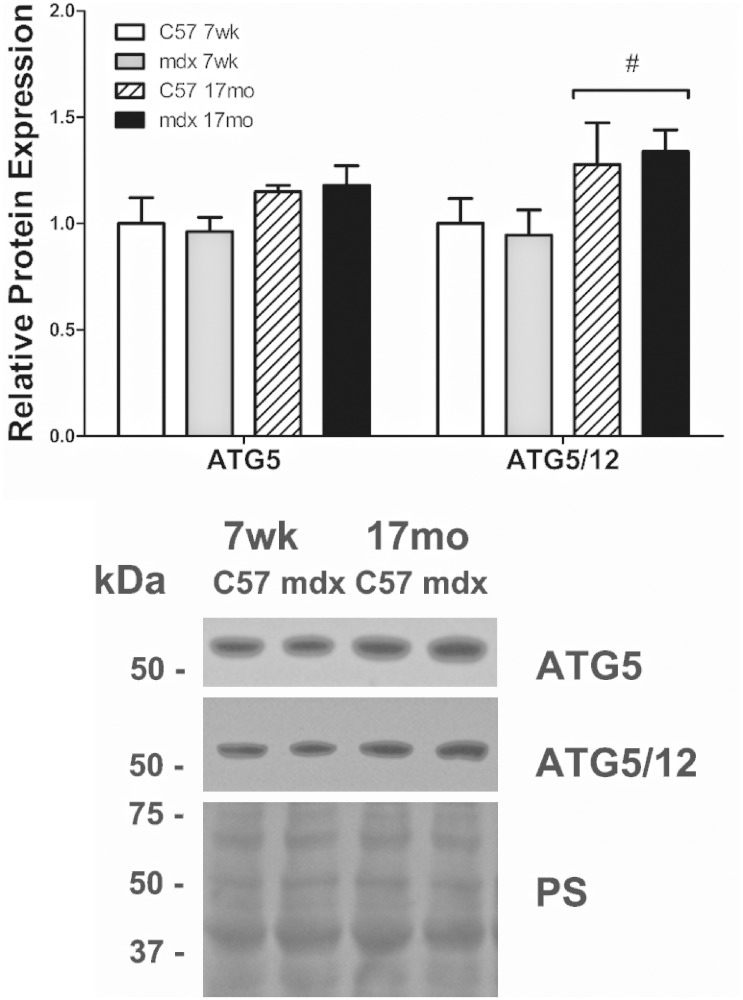
The maturation of a phagophore to an autophagosome is regulated by ATG proteins. Specifically, ATG5 and 12 form a complex that ultimately binds with ATG16L. ATG5 protein abundance was similar between groups and abundance of the ATG5/12 complex increased as a function of age but was independent of disease status (Figure 3). The lipidation of LC3I to LC3II suggests movement toward the formation of mature autophagosomes, thus the ratio of LC3II/I was used as a marker for autophagic maturation. LC3I, LC3II, and LC3II/I were similar between healthy and diseased groups, though LC3II protein abundance and LC3II/I ratio increased as a function of age (Figure 4). Lastly, protein abundance of p62, an inverse correlate of autophagic degradation,25 was similar between groups. Further, p62 transcript expression was measured and was also similar between groups.
Figure 3.
Autophagosome formation. ATG proteins involved in autophagosome formation were similar between all groups, suggesting no alteration in autophagosome formation in dystrophic cardiac muscle regardless of age. Significance was established at p < 0.05. White bars – C57 7 wk (n = 4–5), gray bars – mdx 7 wk (n = 7–8), white bars with black hatching – C57 17 mo (n = 3), black bars – mdx 17 mo (n = 8). # indicates significant main effect of age.
Figure 4.
Autophagic degradation. (a,b) Markers of autophagosome degradation were similar between mdx and age-matched controls. (c) Transcript abundance of Sqstm1, which encodes p62, was similar between all groups. Significance was established at p < 0.05. White bars – C57 7 wk (n = 4–5), gray bars – mdx 7 wk (n = 7–8), white bars with black hatching – C57 17 mo (n = 3), black bars – mdx 17 mo (n = 8). # indicates significant main effect of age.
Experiment 2: 14 mo C57 and mdx mice
We previously reported increased histological damage and declining cardiac function in dystrophic hearts20 as well as substantial impairments to limb and diaphragmatic function in these animals.22,23 Biochemical findings from 14 mo hearts largely recapitulated findings from Experiment 1. Protein markers of autophagosome maturation, ATG5 and 7, were similar between groups. LC3I was also similar between groups; however, LC3II, a marker of autophagosome abundance, was decreased by 56% in mdx mice compared to C57 raising the possibility of increased degradation of autophagosomes (Table 1). p62 protein abundance was similar between groups.
Table 1.
Autophagy in 14 mo C57 and mdx mice.
| C57 | mdx | p-Value | |
|---|---|---|---|
| beclin 1 | 1.00 ± 0.04 | 0.95 ± 0.11 | 0.71 |
| ATG5 | 1.00 ± 0.03 | 1.00 ± 0.07 | 0.98 |
| ATG7 | 1.00 ± 0.03 | 1.02 ± 0.07 | 0.81 |
| LC3I | 1.00 ± 0.04 | 0.97 ± 0.13 | 0.83 |
| LC3II | 1.00 ± 0.20 | 0.44 ± 0.08a | 0.02 |
| LC3II/I | 1.00 ± 0.22 | 0.59 ± 0.20 | 0.19 |
| p62 | 1.00 ± 0.02 | 0.99 ± 0.03 | 0.76 |
Note: Markers of autophagy were largely similar except LC3II, a marker of autophagosome maturation, which was significantly decreased. Mean ± SEM.
aSignificantly different from C57 controls (p < 0.05). C57 14 mo (n = 7) and mdx 14 mo (n = 7).
Experiment 3: 10 mo C57 and mdx/Utrn± mice
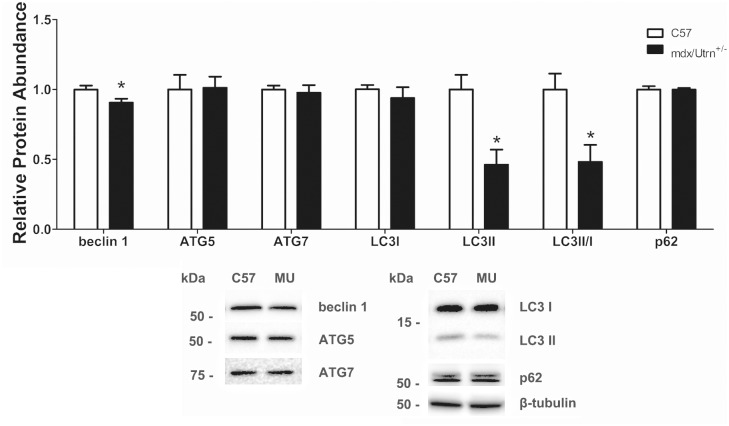
A common criticism of the mdx model is its mild phenotype. While our investigation deliberately used mice early in disease progression (7 wk) and with evident cardiomyopathy (14 and 17 mo), we also assessed autophagy in a more severe disease model that is dystrophin deficient and heterozygous for a utrophin mutation (mdx/Utrn±). We previously found that hearts from 10-month-old mdx/Utrn± mice are severely damaged and demonstrate cardiac function decline compared to age-matched C57 mice.21 Notably, beclin 1 protein abundance was decreased by 10% in mdx/Utrn± compared to C57 mice but ATG5 and ATG7 were similar between groups (Figure 5). Relative protein abundance of LC3I was similar between groups but LC3II and LC3II/I were significantly reduced by 50% suggestive of increased degradation of autophagosomes. Lastly, p62 protein abundance was similar between groups.
Figure 5.
Autophagy in 10 mo C57 and mdx/Utrn± mice. Markers of autophagosome maturation (LC3II and LC3II/I) were significantly decreased while p62, a marker of autophagosome degradation, was similar. Significance was established at p < 0.05. White bars – C57 10 mo (n = 8) and black bars – mdx/Utrn± 10 mo (n = 8). * indicates significantly different from C57 controls.
Discussion
DMD is caused by the absence of dystrophin protein and collapse of the DGC. Dystrophic muscle cells are susceptible to contraction-induced injury resulting in secondary effects such as sarcolemma injury and cell degeneration.26 The application of advanced respiratory support therapies now prolongs life in those with DMD to the extent that cardiomyopathy is an emergent threat facing these patients. Recent evidence indicates that several cardioprotective drugs27 and nutraceuticals.20,21,28 attenuate the severity of cardiomyopathy. Recently, autophagy has been identified as a target for pharmacological manipulation as autophagy is blunted in dystrophic skeletal muscle serving to limit removal of protein aggregates and damaged organelles.12,14 Pleiotropic therapeutics, such as mTOR inhibitors, which may, among other effects, activate autophagy, have successfully decreased disease severity in dystrophic skeletal and cardiac muscle13,14; however, the role of autophagy in progressive cardiomyopathy is unknown. We hypothesized that with advancing cardiac injury autophagy would exhibit markers of progressive dysfunction.
On the whole, markers of autophagy were similar between healthy and diseased hearts at 7 wk and 17 mo of age, suggesting that autophagy is independent of disease-related injury to the myocardium. As these results were quite unexpected and there were numerical changes in several key variables, particularly with advanced disease, we carried out two additional confirmatory experiments. In the first confirmatory experiment using 14 mo mice, LC3II protein abundance decreased and p62 was similar in diseased hearts compared to healthy, raising the possibility that degradation of autophagosomes is enhanced in dystrophic cardiac muscle as autophagosomes are being degraded rapidly resulting in decreased LC3II. Further supporting this notion, LC3II abundance was decreased without alteration in p62 in 10 mo mdx/Utrn± mice compared to healthy. Given findings from Experiments 2 and 3, data from Experiment 1, in which LC3II and LC3II/I ratio were numerically decreased without a change in p62, could be supportive of increased degradation of autophagosomes, particularly in aged mice. Collectively these findings are interesting and raise novel insights into the nuanced role of autophagy and disease progression in dystrophic hearts.
Based on our preliminary interpretation of these findings, the reader should be aware that conclusions are tempered to match study design limitations. Specifically, the current approach relied upon archived samples, and as such, measures of autophagic flux were not immediately feasible. An alternative interpretation of these findings does not exclude the possibility that autophagy is impaired in dystrophic hearts. If autophagy were impaired, a reduction in LC3II could reflect decreased autophagosome formation and reduced efficiency of autophagosome degradation as suggested by similar p62. However, published findings likely do not support the notion of decreased autophagic flux in that a well-designed study focused on dystrophic hearts clearly delineated that LC3II increased following inhibition of autophagic degradation with chloroquine, indicative of increased flux.29 In combination with outcomes in the current investigation, these findings suggest an increased flux accompanies disease progression. Indeed, careful scrutiny of work by Kang et al. indicates that autophagy increased following isolation of cardiomyocytes, which the authors attributed to cellular stress caused specifically by isolation.29 An important extension of this point is that despite increased autophagy as a result of dystrophinopathy, cells were able to further increase autophagy suggesting it could be responsive to therapeutics intended to stimulate autophagy. In support, stimulation of autophagy via inhibition of mTOR was cardioprotective in 18 mo mdx mice.13
Our first experiment also provides a limited opportunity to consider the effect of aging on markers of autophagy in healthy hearts. Here, we found that p-beclin and the ATG12/5 complex were increased as a function of age suggesting increased activation of autophagy. We also discovered increased LC3II and LC3II/I without a corresponding change in p62. Accumulation of LC3II can be due to either an increase in formation of autophagosomes, or a failure to degrade autophagosomes, or a combination of the two. As LC3II increased without a corresponding increase in p62 it is suggestive of increased degradation of autophagosomes in hearts from 17 mo mice compared to 7 wk old mice. Such findings are consistent with aging studies using pathway biomarkers of mice of similar age30; however, in older mice autophagic dysfunction is apparent.31–34
In total, and counter to our hypothesis, indices of autophagic dysregulation were not evident in dystrophic hearts examined in these experiments. We interpret this finding to be particularly important in that the emergent understanding of autophagy in various cardiomyopathies, including dystrophic heart failure, is more nuanced and contextual than initially thought.16,17 Indeed, findings from the current investigation include hearts from several mouse cohorts that exhibited significant evidence of histochemical and physiological pathology.20,21 As such, these initial experiments to probe autophagy in dystrophic hearts suggest that despite disease progression, markers of autophagic activation and degradation appear to be independent of disease-related cardiomyopathy in the dystrophic myocardium. Moreover, our collective evidence is interpreted within the current literature,29 which suggests autophagy is enhanced in dystrophin-deficient hearts. Given the pathologic intracellular environment of a dystrophin-deficient fiber and that (nonspecific) autophagy activators provided therapeutic benefits,13 cellular activators of autophagy may represent druggable targets leading to cardioprotection.
Declaration of conflicting interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical approval
All procedures were approved by the Institutional Animal Care and Use Committees at Iowa State University or Auburn University.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported in part by Duchenne Alliance in grants to JTS and JCQ. HRS was supported in part by Parent Project Muscular Dystrophy (Grant 01297), Ryan’s Quest, and Michael’s Cause. MBH was supported in part by NIH 5P20GM113125-03.
Guarantor
JTS
Contributorship
All authors were involved in study design, data analysis, and data interpretation. All authors revised the manuscript and approved the final manuscript.
References
- 1.Allen DG, Whitehead NP, Froehner SC. Absence of dystrophin disrupts skeletal muscle signaling: roles of Ca2+, reactive oxygen species, and nitric oxide in the development of muscular dystrophy. Physiol Rev 2016; 96: 253–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petrof BJ, Shrager JB, Stedman HH, et al. Dystrophin protects the sarcolemma from stresses developed during muscle contraction. Proc Natl Acad Sci U S A 1993; 90: 3710–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alderton JM, Steinhardt RA. Calcium influx through calcium leak channels is responsible for the elevated levels of calcium-dependent proteolysis in dystrophic myotubes. J Biol Chem 2000; 275: 9452–9460. [DOI] [PubMed] [Google Scholar]
- 4.Godin R, Daussin F, Matecki S, et al. Peroxisome proliferator-activated receptor γ coactivator 1-α gene transfer restores mitochondrial biomass and improves mitochondrial calcium handling in post-necrotic mdx mouse skeletal muscle. J Physiol 2012; 590: 5487–5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morris CA, Selsby JT, Morris LD, et al. Bowman–Birk inhibitor attenuates dystrophic pathology in mdx mice. J Appl Physiol 2010; 109: 1492–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Selsby JT. Increased catalase expression improves muscle function in mdx mice. Exp Physiol 2011; 96: 194–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moulin M, Ferreiro A. Muscle redox disturbances and oxidative stress as pathomechanisms and therapeutic targets in early-onset myopathies. Semin Cell Dev Biol 2017; 64: 213–223. [DOI] [PubMed] [Google Scholar]
- 8.Williams IA, Allen DG. Intracellular calcium handling in ventricular myocytes from mdx mice. Am J Physiol Heart Circ Physiol 2007; 292: H846–H855. [DOI] [PubMed] [Google Scholar]
- 9.Kaspar RW, Allen HD, Montanaro F. Current understanding and management of dilated cardiomyopathy in Duchenne and Becker muscular dystrophy. J Am Acad Nurse Pract 2009; 21: 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sandri M, Coletto L, Grumati P, et al. Misregulation of autophagy and protein degradation systems in myopathies and muscular dystrophies. J Cell Sci 2013; 126: 5325–5333. [DOI] [PubMed] [Google Scholar]
- 11.Zhang J. Teaching the basics of autophagy and mitophagy to redox biologists – mechanisms and experimental approaches. Redox Biol 2015; 4: 242–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spaulding HR, Kelly EM, Quindry JC, et al. Autophagic dysfunction and autophagosome escape in the mdx mus musculus model of Duchenne muscular dystrophy. Acta Physiol (Oxf) 2018; 222. [DOI] [PubMed]
- 13.Bibee KP, Cheng YJ, Ching JK, et al. Rapamycin nanoparticles target defective autophagy in muscular dystrophy to enhance both strength and cardiac function. FASEB J 2014; 28: 2047–2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pal R, Palmieri M, Loehr JA, et al. Src-dependent impairment of autophagy by oxidative stress in a mouse model of Duchenne muscular dystrophy. Nat Commun 2014; 5: 4425. DOI: 10.1038/ncomms5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi S, Liang Q. Autophagy and mitophagy in diabetic cardiomyopathy. Biochim Biophys Acta 2015; 1852: 252–261. [DOI] [PubMed] [Google Scholar]
- 16.Yan Y, Finkel T. Autophagy as a regulator of cardiovascular redox homeostasis. Free Radic Biol Med 2017; 109: 108–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottlieb RA, Mentzer RM. Autophagy: an affair of the heart. Heart Fail Rev 2013; 18: 575–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraysse B, Nagi SM, Boher B, et al. Ca2+ overload and mitochondrial permeability transition pore activation in living delta-sarcoglycan-deficient cardiomyocytes. Am J Physiol Cell Physiol 2010; 299: C706–C713. [DOI] [PubMed] [Google Scholar]
- 19.Quinlan JG, Hahn HS, Wong BL, et al. Evolution of the mdx mouse cardiomyopathy: physiological and morphological findings. Neuromuscul Disord 2004; 14: 491–496. [DOI] [PubMed] [Google Scholar]
- 20.Ballmann C, Denney T, Beyers RJ, et al. Long term dietary quercetin enrichment as a cardioprotective countermeasure in mdx mice. Exp Physiol 2017; 102: 635–649. [DOI] [PubMed] [Google Scholar]
- 21.Ballmann C, Denney TS, Beyers RJ, et al. Lifelong quercetin enrichment and cardioprotection in Mdx/Utrn± mice. Am J Physiol Heart Circ Physiol 2017; 312: H128–H140. [DOI] [PubMed] [Google Scholar]
- 22.Selsby JT, Ballmann CG, Spaulding HR, et al. Oral quercetin administration transiently protects respiratory function in dystrophin-deficient mice. J Physiol 2016; 594: 6037–6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spaulding HR, Ballmann CG, Quindry JC, et al. Long-term quercetin dietary enrichment partially protects dystrophic skeletal muscle. PloS One 2016; 11: e0168293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nazarko VY, Zhong Q. ULK1 targets Beclin-1 in autophagy. Nat Cell Biol 2013; 15: 727–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bjorkoy G, Lamark T, Brech A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 2005; 171: 603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fayssoil A, Abasse S, Silverston K. Cardiac involvement classification and therapeutic management in patients with Duchenne muscular dystrophy. J Neuromuscul Dis 2017; 4: 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rafael-Fortney JA, Chimanji NS, Schill KE, et al. Early treatment with lisinopril and spironolactone preserves cardiac and skeletal muscle in Duchenne muscular dystrophy mice. Circulation 2011; 124: 582–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ballmann C, Hollinger K, Selsby JT, et al. Histological and biochemical outcomes of cardiac pathology in mdx mice with dietary quercetin enrichment. Exp Physiol 2015; 100: 12–22. [DOI] [PubMed] [Google Scholar]
- 29.Kang C, Badr MA, Kyrychenko V, et al. Deficit in PINK1/PARKIN-mediated mitochondrial autophagy at late stages of dystrophic cardiomyopathy. Cardiovasc Res 2018; 114: 90–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boyle AJ, Shih H, Hwang J, et al. Cardiomyopathy of aging in the mammalian heart is characterized by myocardial hypertrophy, fibrosis and a predisposition towards cardiomyocyte apoptosis and autophagy. Exp Gerontol 2011; 46: 549–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoshino A, Mita Y, Okawa Y, et al. Cytosolic p53 inhibits Parkin-mediated mitophagy and promotes mitochondrial dysfunction in the mouse heart. Nat Commun 2013; 4: 2308. [DOI] [PubMed] [Google Scholar]
- 32.Ren J, Yang L, Zhu L, et al. Akt2 ablation prolongs life span and improves myocardial contractile function with adaptive cardiac remodeling: role of Sirt1-mediated autophagy regulation. Aging Cell 2017; 16: 976–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hua Y, Zhang Y, Ceylan-Isik AF, et al. Chronic Akt activation accentuates aging-induced cardiac hypertrophy and myocardial contractile dysfunction: role of autophagy. Basic Res Cardiol 2011; 106: 1173–1191. [DOI] [PubMed] [Google Scholar]
- 34.Taneike M, Yamaguchi O, Nakai A, et al. Inhibition of autophagy in the heart induces age-related cardiomyopathy. Autophagy 2010; 6: 600–606. [DOI] [PubMed] [Google Scholar]