Abstract
Introduction
Type 1 diabetes (T1D) is the second most common autoimmune disease among children. There is evidence suggesting that dysbiosis of some gut colonizing bacteria are associated with the pathogenesis of T1D. However, these studies are still controversial and a systematic review was conducted to evaluate the association between gut microbiota and T1D.
Methods
A systematic search was carried out in Medline (Via Pubmed) and Embase from January 2000 to January 2019 for all original cross-sectional, cohort, case–control or nested case–control studies investigating the association between gut microbiota and T1D.
Results
Of 568 articles identified, 26 studies met the inclusion criteria. The total population study of these articles consists of 2600 children (under 18 years old) and 189 adults. Among the included studies, 24 articles confirmed the association between gut microbiota dysbiosis and T1D. The most common bacterial alterations in T1D patients included Bacteroides spp., Streptococcus spp., Clostridium spp., Bifidobacterium spp., Prevotella spp., Staphylococcus spp., Blautia spp., Faecalibacterium spp., Roseburia spp., and Lactobacillus spp.
Conclusion
Our study showed a significant association between alterations in intestinal microbial composition and T1D; however, in some articles, it is not clear which one happens first. Investigation of altered gut microbiota can help in the early detection of T1D before seropositivity. Targeted microbiome modulation can be a novel potential therapeutic strategy.
Keywords: Type 1 diabetes, Microbiota, Dysbiosis
Introduction
Type 1 diabetes (T1D) is the second most common autoimmune disease among children. It is accompanied by many complications and has life-long morbidity [1]. The incidence of T1D is increasing universally and accounts for 5–10% of all diabetic morbidity [2]. T1D is a chronic autoimmune inflammatory process that affects insulin-producing beta cells of the pancreas, results in less insulin production [3]. Destruction of 90% of beta cells is a critical point that clinical manifestations emerge [4]. Because of the early onset of disease and chronicity, T1D is of great importance. Previous animal and human studies have shown the role of genetic factors like human leukocyte antigen (HLA) DQ and DRB in the pathogenesis of disease but recent studies propose the significant role of environmental factors such as gut colonizing bacteria [5]. Gut microbiota has an important role in the regulation of metabolism, systemic and local immunity [6]. From birth to age 3, gut microbiota undergoes a lot of changes and the microbiota composition of a 3-year-old child is similar to that of an adult [7]. The most important factors affecting gut microbiota include the type of delivery [8], breastfeeding [9] or bottle feeding, maternal microbiota composition, mother’s diet during pregnancy and the western diet [5, 10–13], contact with peers, environment, and use of antibiotics [14–17]. Gut dysbiosis, an imbalance of the microbial communities, can be associated with metabolic disorders, obesity, insulin resistance, Type 2 diabetes (T2D), inflammatory bowel disease, celiac disease and immunity dysfunction [18–20]. Lately, there is evidence suggesting the correlation between dysbiosis and pathogenesis of T1D [21]. However, these studies are still controversial and need further investigation; Thus, we carried out a systematic review about the association between gut microbiota and T1D according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses statement [22].
Materials and methods
Search strategy
A systematic search was carried out in Medline (Via Pubmed) and Embase from January 2000 to January 2019. Medical Subject Headings (MeSH) were “gastrointestinal microbes”, “dysbiosis”, “gut microbiota”, “gut bacteria”, “gut microbes” combined with “type 1 diabetes mellitus”. Lists of references of selected articles and relevant review articles were hand-searched to identify further studies. Only studies written in English were selected.
Study selection
Two reviewers independently performed the review of titles and abstracts and chose those fitting selection criteria for full-text evaluation. Discrepancies were discussed with a third reviewer. All original cross-sectional, cohort, case–control or nested case–control studies investigating the association between gut microbiota and T1D patients were considered. The following articles were excluded: animal studies, case reports, reviews, and editorials.
Data extraction
The following variables were extracted: first author; year of publication; study duration, type of study, country/ies where the study was conducted; the number of cases with T1D; age; gender; microbiota analysis technique; modifications of intestinal microbiota and modifications of biochemical and immunological factors. Data were independently collected by two authors.
Results
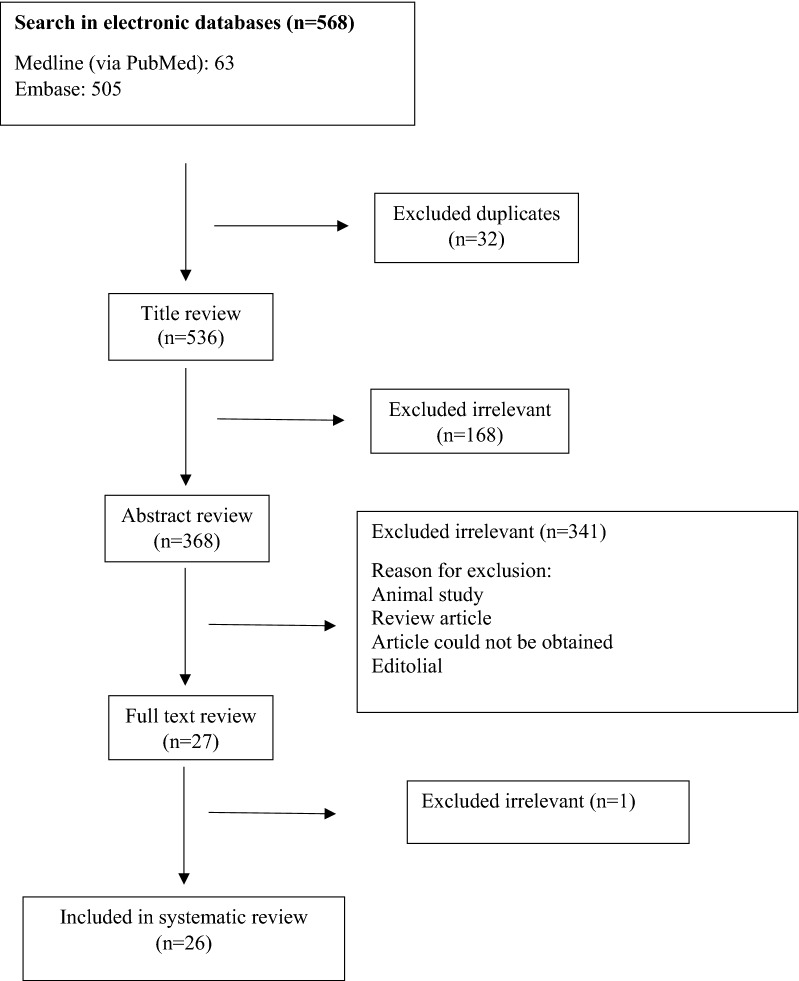
The selection process of articles is shown in Fig. 1. Twenty-six articles were included and classified into 16 case–control studies [18, 21, 23–36], 6 cohort studies [37–42], 2 cross-sectional studies [5, 43] and 2 nested case–control studies [44, 45]. Four of these studies were conducted in the USA, three in Italy, three in Finland, two in China, two in Spain and others in Netherland, Germany, Turkey, UK, Portugal, Poland, Russia, Mexico, Brazil, Australia, Czech Federation, and France. The population of these articles consists of 2600 children (under 18 years old) and 189 adults. The most applied techniques for detection and assessment of gut microbiota in stool samples where PCR, real-time quantitative PCR, 16s rRNA sequencing, microarray analysis, proteomics and quantitative cultures of stool samples (Table 1).
Fig. 1.
Flow chart of study selection for inclusion in the systematic review
Table 1.
Characteristics of included studies
| Authors | Year | Country | Type of study | Study population (control and case) | Age (mean) | Microbiota analysis technique |
|---|---|---|---|---|---|---|
| Rozanova et al. [41] | 2002 | Russia | Cohort | 38 T1D | 3 years | Not mentioned |
| Brown et al. [23] | 2011 | Finland | Case–control | 8 T1D | Children | DNA sequencing |
| Giongo et al. [31] | 2011 | USA | Case–control | Control: 4, case: 4 | 5 months | 16s rRNA sequencing |
| Murri et al. [26] | 2013 | Spain | Case–control | Control: 16, case: 16 | 7 years | Real time quantitative PCR |
| Richardson et al. [37] | 2014 | Finland | Cohort | Control: 47, case: 29 | 2 years | 16s rRNA sequencing |
| de Goffau et al. [24] | 2014 | Netherlands | Case–control | Control: 27, case: 28 | 3 years | Microarray analysis |
| Endesfelder et al. [38] | 2014 | Germany | Cohort | Control: 22, case: 22 | 19.5 months | 16s rRNA sequencing |
| Mejia et al. [30] | 2014 | Mexico | Case–control | Control: 8, case: 21 | 12.5 years | 16s rRNA pyrosequencing |
| Soyucen et al. [28] | 2014 | Turkey | Case–control | Control: 35, case: 35 | 10 years | Quantitative cultures on selective and non-selective media |
| Kostic et al. [39] | 2015 | Finland | Cohort | Control: 22, case: 11b | Infants | 16s rRNA sequencing |
| Alkanani et al. [5] | 2015 | USA | Cross-sectional | Control: 23, casea: 88 | 11 years | 16s rRNA sequencing |
| Cui et al. [18] | 2016 | China | Case–control | Control: 15, case: 15 | 11 years | 16s rRNA sequencing |
| Maffeis et al. [25] | 2016 | Italy | Case–control | Control: 10, case: 10 | 11 years | Semi-quantitative PCR |
| Stewart et al. [29] | 2017 | UK | Case–control | Control: 10, case: 10 | 27 years | 16s rRNA sequencing |
| Pinto et al. [40] | 2017 | Portugal | Cohort | Control: 3, case: 3 | 9 years | Real time quantitative PCR |
| Pellegrini et al. [27] | 2017 | Italy | Case–control | Control: 35c, case: 19 | 36 years | Real time quantitative PCR |
| Traversi et al. [21] | 2017 | Italy | Case–control | Control: 13, case: 13 | 8 years | Real time quantitative PCR |
| Gao et al. [42] | 2018 | France | Cohort | 33 genetically predisposed to T1D | 1.5 years | 16s rRNA sequencing |
| Vatanen et al. [44] | 2018 | USA | Nested case–control | Control: 415, case: 368d | 3 month | 16s rRNA sequencing |
| Stewart et al. [45] | 2018 | USA | Nested case–control | 903 children | 24.5 month | 16s rRNA sequencing |
| Huang et al. [32] | 2018 | China | Case–control | Control: 10, case: 12 | 23.5 | 16s rRNA sequencing |
| Gavin et al. [43] | 2018 | Australia | Cross-sectional | Control: 22, case: 69e | 10.8 | Proteomics and 16S rRNA sequencing |
| Leiva-Gea et al. [33] | 2018 | Spain | Case–control | Control: 13, case: 15 T1D | 12.6 | 16S rRNA pyrosequencing |
| Higuchi et al. [34] | 2018 | Brazil | Case–control | Control: 28, case: 20 | 23.1 | 16s rRNA sequencing |
| Salamon et al. [35] | 2018 | Poland | Case–control | Control: 23, case: 22 | 42.5 | 16s rRNA sequencing |
| Cinek et al. [36] | 2018 | Czech federation | Case–control | Control: 103, case: 73f | 11.8 | 16s rRNA sequencing |
a35 new-onset patients; 21 seropositive; 32 seronegative FDRs (first degree relatives)
bSeroconverters
c16 healthy control; 19 gut inflammatory disease as the second control
d267 seroconverters and 101 diagnosed with T1D
e23 recent onset type 1 diabetes; 17 islet autoantibody–positive subjects; 29 low-risk autoantibody-negative subjects
fAzerbaijan: 19, Jordan: 20, Nigeria: 14, Sudan: 20
Gut microbiota and type 1 diabetes
Twenty-four out of twenty-six articles confirmed the association of T1D and gut microbiota dysbiosis. In one study alterations could not be attributable to T1D [23] and one of the articles is only a preliminary study and doesn’t have any obvious conclusion yet [21] (Table 2). The most common bacterial alterations in T1D patients group versus healthy individuals included Bacteroides spp., Streptococcus spp., Clostridium spp., Bifidobacterium spp., Prevotella spp., Staphylococcus spp., Blautia spp., Faecalibacterium spp., Roseburia spp., and Lactobacillus spp. Details indicating the altered bacteria are shown in Table 3.
Table 2.
Association of gut microbiota and type 1 diabetes
| Authors | Year | Country | Type of study | Association between diabetes and microbiome |
|---|---|---|---|---|
| Rozanova et al. [41] | 2002 | Russia | Cohort | Yes |
| Brown et al. [23] | 2011 | Finland | Case–control | May |
| Giongo et al. [31] | 2011 | USA | Case–control | Yes |
| Murri et al. [26] | 2013 | Spain | Case–control | Yes |
| Richardson et al. [37] | 2014 | Finland | Cohort | Yes |
| de Goffau et al. [24] | 2014 | Netherlands | Case–control | Yes |
| Endesfelder et al. [38] | 2014 | Germany | Cohort | Yes |
| Mejia et al. [30] | 2014 | Mexico | Case–control | Yes |
| Soyucen et al. [28] | 2014 | Turkey | Case–control | Yes |
| Kostic et al. [39] | 2015 | Finland | Cohort | Yes |
| Alkanani et al. [5] | 2015 | USA | Cross-sectional | Yes |
| Cui et al. [18] | 2016 | China | Case–control | Yes |
| Maffeis et al. [25] | 2016 | Italy | Case–control | Yes |
| Stewart et al. [29] | 2017 | UK | Case–control | Yes |
| Pinto et al. [40] | 2017 | Portugal | Cohort | Yes |
| Pellegrini et al. [27] | 2017 | Italy | Case–control | Yes |
| Traversi et al. [21] | 2017 | Italy | Case–control | Not mentioned |
| Gao et al. [42] | 2018 | France | Cohort | Yes |
| Vatanen et al. [44] | 2018 | USA | Nested case–control | Yes |
| Stewart et al. [45] | 2018 | USA | Nested case–control | Yes |
| Huang et al. [32] | 2018 | China | Case–control | Yes |
| Gavin et al. [43] | 2018 | Australia | Cross-sectional | Yes |
| Leiva-Gea et al. [33] | 2018 | Spain | Case–control | Yes |
| Higuchi et al. [34] | 2018 | Brazil | Case–control | Yes |
| Salamon et al. [35] | 2018 | Poland | Case–control | Yes |
| Cinek et al. [36] | 2018 | Czech federation | Case–control | Yes |
Table 3.
Intestinal microbiota modifications
| Authors | Intestinal microbiota modifications |
|---|---|
| Rozanova et al. [41] | |
| Increased | Lactose-negative Enterobacteriaceae, Klebsiella spp., Enterococcus spp., Candida spp., Clostridium spp., Staphylococcus epidermidis |
| Decreased | Bifidobacterium spp., Lactobacillus spp., Escherichia coli |
| Brown et al. [23] | |
| Increased | Bacteroides spp., Veillonella spp., Alistipes spp. |
| Decreased | Prevotella spp., Akkermansia spp. |
| Giongo et al. [31] | |
| Increased | Bacteroidetes |
| Decreased | Firmicutes |
| Murri et al. [26] | |
| Increased | Clostridium spp., Bacteroides spp. and Veillonella spp. |
| Decreased | Lactobacillus spp., Bifidobacterium spp., Blautia coccoides, Eubacterium rectale, Prevotella spp., lactic acid-producing bacteria, butyrate-producing bacteria and mucin-degrading bacteria |
| Richardson et al. [37] | |
| Increased | Bacteroides dorei, Bacteroides vulgatus |
| Decreased | – |
| de Goffau et al. [24]a | |
| Increased | Streptococcus mitis, Bacteroidetes |
| Decreased | – |
| de Goffau et al. [24]b | |
| Increased | Non butyrate producing species of Clostridium cluster 14a, Clostridium stercorarium |
| Decreased | – |
| Endesfelder et al. [38]c | |
| Increased | Enterococcus spp., Sarcina spp., Prevotella spp., Corynebacterium spp. |
| Decreased | – |
| Endesfelder et al. [38]d | |
| Increased | Barnesiella spp., Candidatus Nardonella |
| Decreased | Staphylococcus spp., Nocardioides spp. |
| Mejia et al. [30] | |
| Increased | Bacteroides spp. |
| Decreased | Prevotella spp. |
| Soyucen et al. [28] | |
| Increased | Enterobacteriaceae, Candida albicans |
| Decreased | Bifidobacterium spp. |
| Kostic et al. [39] | |
| Increased | – |
| Decreased | Coprococcus eutactus, Dialister invisus |
| Alkanani et al. [5] | |
| Increased | Lactobacillus spp., Staphylococcus spp. |
| Decreased | Prevotellaceae |
| Cui et al. [18] | |
| Increased | Blautia spp., Haemophilus spp., Lachnospira spp., Intestinimonas spp., Dialister spp., Micrococcales spp. |
| Decreased | Pasteurella spp., Caulobacterales spp. |
| Maffeis et al. [25] | |
| Increased | Dialister invisus, Globicatella sanguinis, Bifidobacterium longum |
| Decreased | – |
| Stewart et al. [29] | |
| Increased | Actinomyces spp. |
| Decreased | – |
| Pinto et al. [40] | |
| Increased | Eubacterium rectale, Faecalibacterium prausnitzzi, Bacteroides dorei, Bacteroides uniformis |
| Decreased | Collinsella aerofaciens, Coprococcus Comes, Clostridium spp., Bifidobacterium adolescentis, Bifidobacterium longum infantis, Ruminococcus spp., Collinsella spp. |
| Pellegrini et al. [27] | |
| Increased | Firmicutes |
| Decreased | Clostridium spp., Bacteroidetes, Proteobacteria |
| Traversi et al. [21] | |
| Increased | Bacteroides clarus, Alistipes obesi, Bifidobacterium longus, Methanobrevibacter Smithii |
| Decreased | Bacteroides coprophilus, Bacteroides dorei, Fusicatenibacter saccharivorans, Bacteroides vulgatus, Bacteroides oleiciplenus, Firmicutes |
| Gao et al. [42] | This study emphasizes on interactions between gut microbiota rather than quantitative changes |
| Vatanen et al. [44] | |
| Increased | Bifidobacterium pseudocatenulatum, Roseburia hominis, Alistipes shahii |
| Decreased | Streptococcus thermophilus, Lactococcus lactis |
| Stewart et al. [45] | |
| Increased | – |
| Decreased | Ruminococcus spp., Lactococcus spp., Streptococcus spp., Akkermansia spp. |
| Huang et al. [32] | |
| Increased | Bacteroidetes/Firmicutes ratio, Porphyromonadaceae |
| Decreased | Ruminococcus spp., Veillonella spp., Phascolarctobacterium spp., Fusobacterium spp., Paenibacillaceae |
| Gavin et al. [43] | |
| Increased | Bacteroides spp., Prevotella spp. |
| Decreased | Alistipes spp., Ruminococcus spp., Barnesiella spp., Clostridium spp., Dorea spp., Faecalibacterium Prausnitzii |
| Leiva-Gea et al. [33] | |
| Increased | Bacteroides spp., Rikenellaceae, Ruminococcus spp., Veillonella spp., Enterobacteriaceae, Blautia spp., Streptococcus spp., Prevotellaceae, Sutterella spp. |
| Decreased | Bifidobacterium spp., Roseburia spp., Faecalibacterium spp., Lachnospira spp., Anaerostipes spp., Actinobacteria, Proteobacteria, Firmicutes |
| Higuchi et al. [34] | |
| Increased | Bacteroides spp., Alistipes spp., Prevotella spp. |
| Decreased | – |
| Salamon et al. [35] | |
| Increased | Akkermansia spp., Ruminococcus spp., Bacteroides spp., Blautia spp. |
| Decreased | Lachnospira spp., Faecalibacterium spp., Bifidobacterium spp., Coprococcus spp., Collinsella spp., Dorea spp. |
| Cinek et al. [36] | |
| Increased | Escherichia coli |
| Decreased | Eubacterium spp., Roseburia spp., Haemophilus spp., Clostridium clusters IV and XIVa |
aIn cases with < 2.9 years
bIn cases with > 2.9 years
cEigenvector centrality (EC) at age 0.5 years
dEC at age 2 years
The relationship between intestinal microbiota and HbA1C, inflammatory mediators and serum zonulin level
Some articles reported evidence of an association between HbA1C level and bacterial groups such as Blautia spp. count and Firmicutes:Bacteroidetes ratio (F:B ratio) [18, 33]. Murri et al. [26] in 2013 by designing a case–control study noticed the HbA1C level affected Clostridium spp. positively and F:B ratio negatively in both uni and multivariant statistical analysis. Univariate statistical analysis also showed that Bifidobacterium spp. and Lactobacillus spp. may affect HbA1C levels [26]. On the contrary, Alkanani et al. [5] reported that bacterial alterations in the case group were not associated with the HbA1C level.
There is an elevated level of TNF-α expression in lamina propria of intestinal biopsy in T1D patients in comparison with healthy individuals [27]. Higuchi et al. [34] reported a negative correlation between TNF plasma level and Proteobacteria and Clostridiaceae abundance. In another study, increment of Bacteroides spp. and decrement of Roseburia spp. was correlated with TNF-α level [33].
Interleukin-6 has an important correlation with Ruminococcaceae abundance as reported by the Higuchi et al. study [34]. Increase of Bacteroides spp. and decrease of Roseburia spp. abundance is correlated with serum IL-6 level [33].
According to Leiva-Gea et al. [33] an increase in Bacteroides spp. and Veillonella spp. and decrease in Bifidobacterium spp., Roseburia spp. and Faecalibacterium spp. was associated with serum IL-1β; in addition, an increase of Streptococcus spp. and decrease of Bifidobacterium spp. was reported related to serum IL-10 and IL-13 levels.
Serum zonulin level has a significant role in the pathogenesis of T1D. Leiva-Gea et al. [33] showed that an increase in Bacteroides spp. and Veillonella spp. and decrease in Faecalibacterium spp. and Roseburia spp. was correlated with an increased serum zonulin level.
Discussion
We reviewed 26 articles, twenty-four of them approved a straight correlation between microbiota and diabetes; however, most of them didn’t clarify if microbiota induces T1D or T1D changes gut microbiome. The articles were screened according to the type of gut microbiota and correlation with T1D as explained below: one article mentioned that microbiome alteration occurs after diabetes [26], two articles studied microbiota as a therapeutic agent on T1D [35, 41], seven articles just showed the differences in gut microbiota of healthy and diabetic people and didn’t discuss the type of relation [21, 24, 29, 32, 34, 36, 40], finally fourteen articles suggested the exact mechanism that leads to autoimmunity by the change in gut microbiome [5, 18, 25, 27, 28, 31, 33, 37, 39, 42–46] (one article was just in the abstract form and we couldn’t read the details [30]).
Different mechanisms have been suggested about the role of gut microbiota in the pathogenesis of T1D. These mechanisms are mainly derived from the 14 articles mentioned above. In more details, the following points can be noticed:
In patients with T1D, some bacteria increase mucin degradation, results in reduced integrity and increased permeability of intestinal mucosa that leads to bacterial penetration [47]. The penetration of bacteria into intestinal mucosa leads to stimulation of the immune system and production of antibodies against them [47]. Cross-reaction of these antibodies and surface antigens of pancreatic beta cells, as well as T cell cross-reactivity results in the destruction of beta cells and formation of T1D [47].
Butyrate is one of the most important byproducts of microbiota metabolisms and plays an important role in colonic T-reg induction, down-regulation of pro-inflammatory macrophages and integrity enhancement of gut barriers through increasing mucin production [48, 49].
Zonulin is a protein that can be assumed as an important indicator of mucosal integrity and gut permeability [33]. This protein modulates intercellular junctions and macromolecular passage through them [33]. Some bacterial groups can alter mucosal integrity by affecting zonulin; increase in Bacteroides spp. and Veillonella spp. or decrease in Faecalibacterium spp. and Roseburia spp. correlates with increased serum zonulin levels in T1D patients [33]. However, according to Leiva-Gea et al. [33] the impaired gut permeability in T1D patients can be more attributed to the binding of Veillonella to colonic crypt cells rather than change in zonulin levels. Lactate produced by Veillonella is pushed to the luminal surface and weakens tight junctions [33].
Gut microbiota ingests and ferment fibers and produce short-chain fatty acids (SCFA) [50–52]. SCFAs enter the blood circulation and modulate T-reg differentiation; thus autoimmunity is prevented [5, 53–55].
With keeping these mechanisms in mind, now we are going to discuss the known attributable mechanism of some highlighted bacteria in more details:
Bacteroidetes, Firmicutes, Proteobacteria, and Actinobacteria were of great importance in our reviewed articles.
Genera Bacteroides and Prevotella are two important subgroups in Phylum Bacteroides that were increased in most of the T1D patient’s samples and can affect gut microbial composition by several mechanisms. Succinate and acetate are the main byproducts of anaerobic metabolism in this phylum that compromise epithelial tight junctions, decrease gut mucosal integrity, block T-reg differentiation and activates inflammatory pathways [25, 32, 33].
These bacteria also produce Glutamic acid decarboxylase (GAD) which can stimulate GAD autoimmunity by molecular mimicry [24, 32].
Phylum Actinobacteria including genus Bifidobacterium is butyrate-producing taxa that have anti-inflammatory effects and augments gut barrier by cytokine modulation [33]. This bacteria also induces T-reg development that results in immune response suppression by regulation of IL-10 production [18].
The third important phylum, Firmicutes, consists of eight notable subgroups: Veillonella, Roseburia, Ruminococcus, Lactobacillus, Blautia, Streptococcus, Faecalibacterium, and Staphylococcus.
Association of T1D and Veillonella is controversy. Kostic et al. [39] reported a decrease in Veillonella in T1D patients and proposed the following mechanism: reduced level of lithocholic acid results in stimulation of gut inflammation by an increased level of reactive oxygen species, reactive nitrogen species and nuclear factor-kB (NF-kB) activity in epithelial cells. Sphingomyelin increment also inhibits NK-T cell function that prevents inflammation [39].
Ruminococcaceae are butyrate-producing taxa that were reported declined in some studies and increased in others. The mechanism of reduced Ruminococcaceae in T1D patients is the same as Veillonella reduction mechanism [33, 39].
Faecalibacterium and Roseburia have anti-inflammatory effects, their presence may augment gut barrier function by modulating cytokine production and butyrate synthesis [24, 33, 36]. These genera have decreased in almost all patient samples.
Genus Blautia is also butyrate-producing taxa that have declined in most of the reviewed articles and plays an important role in blood glucose regulation, lipid metabolism and regulation of T-cell differentiation [18, 33]. There is also evidence of its increment in literature.
Genus Lactobacillus eliminates peroxidase radicals by superoxide dismutase and peroxidase enzymes thus provide a suitable condition for Bifidobacterium reproduction [28]. Lactobacillus down-modulate inflammation and previous studies have demonstrated that dendritic cells co-cultured with species of lactobacilli induce polarization of T-reg cells [5, 56, 57].
Staphylococcaceae may stimulate the growth of Bifidobacterium, Clostridium and Bacteroides which results in augmentation of neonate’s gut maturation [5, 58]. Streptococcaceae produce GAD so they have the same effects as Bacteroides [24, 59].
Limitations
Limitations of our study that complicates interpretation of results can be listed as below: various geographical areas studied have an effect on diet of patients and controls; diversity of microbiota analysis techniques; colonizing microbiome and genetic susceptibility to T1D, some studies considered HLA as a genetic predisposing factor in the selection process of case and control individuals whereas others have ignored this point; different study design (e.g. some studies noticed seropositive group and seronegative FDRs in addition to T1D patients and healthy individuals while others just compared T1D patients with healthy individuals) [5, 25, 39, 44, 45]; various statistical analysis methods and different levels of p-value significance were reported in reviewed articles, however, in this study we used only statistical significant findings from the articles included.
Suggestions
According to our study, we suggest new therapeutic and diagnostic strategies that need further clinical trials for assessing their effectiveness:
Use of prebiotics, probiotics and fecal microbial transplantation to modulate gut microbiome; e.g. a probiotic mixture of certain bacteria (mentioned in “Results”) can reduce HbA1c level so it can be considered as a complementary strategy for T1D management.
In order to detect early evidence of dysbiosis and prevention of T1D progression, serial stool exams in genetically susceptible children can be done by using a specific kit that semi-quantitatively compares microbiota composition of healthy control and suspected individual. In designing the kit, Firmicutes: Bacteroidetes ratio should be considered because it has been reported decreased in all the reviewed studies.
Conclusions
Our study showed a significant association between alterations in intestinal microbial composition and T1D; however, in some articles, it is not clear which one happens first. Investigation of altered gut microbiota can help in the early detection of T1D before seropositivity against classical autoantigens. Targeted microbiome modulation can be a novel potential therapeutic strategy.
Acknowledgements
This study is related to the project No. 1397/68977 From Student Research Committee, Shahid Beheshti University of Medical Sciences, Tehran, Iran. We also appreciate “Student Research Committee” and “Research and Technology Chancellor” in Shahid Beheshti University of Medical Sciences for their financial support of this study.
Authors’ contributions
All authors had made considerable contributions to the present study. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
All data were included.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Parnian Jamshidi, Saba Hasanzadeh and Azin Tahvildari equally first authors
Contributor Information
Yeganeh Farsi, Email: yeganehfarsi@sbmu.ac.ir.
Mahta Arbabi, Email: mahtaarbabi@sbmu.ac.ir.
Mohammad Javad Nasiri, Email: mj.nasiri@hotmail.com.
References
- 1.Mejia-Leon ME, Barca AM. Diet, microbiota and immune system in type 1 diabetes development and evolution. Nutrients. 2015;7(11):9171–9184. doi: 10.3390/nu7115461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pettitt DJ, Talton J, Dabelea D, Divers J, Imperatore G, Lawrence JM, et al. Prevalence of diabetes in US youth in 2009: the SEARCH for diabetes in youth study. Diabetes Care. 2014;37(2):402–408. doi: 10.2337/dc13-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Notkins AL, Lernmark Å. Autoimmune type 1 diabetes: resolved and unresolved issues. J Clin Invest. 2001;108(9):1247–1252. doi: 10.1172/JCI14257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Islam ST, Srinivasan S, Craig ME. Environmental determinants of type 1 diabetes: a role for overweight and insulin resistance. J Paediatr Child Health. 2014;50(11):874–879. doi: 10.1111/jpc.12616. [DOI] [PubMed] [Google Scholar]
- 5.Alkanani AK, Hara N, Gottlieb PA, Ir D, Robertson CE, Wagner BD, et al. Alterations in intestinal microbiota correlate with susceptibility to type 1 diabetes. Diabetes. 2015;64(10):3510–3520. doi: 10.2337/db14-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121–141. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5(7):e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, Chen YY, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137(5):1716–1724. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turnbaugh PJ, Backhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3(4):213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 13.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1(6):6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blaser MJ, Falkow S. What are the consequences of the disappearing human microbiota? Nat Rev Microbiol. 2009;7(12):887–894. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, et al. The long-term stability of the human gut microbiota. Science. 2013;341(6141):1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knip M, Siljander H. The role of the intestinal microbiota in type 1 diabetes mellitus. Nat Rev Endocrinol. 2016;12(3):154–167. doi: 10.1038/nrendo.2015.218. [DOI] [PubMed] [Google Scholar]
- 17.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158(4):705–721. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi C-J, Zhang Q, Yu M, Xu J-P, Zheng J, Wang T, et al. Imbalance of fecal microbiota at newly diagnosed type 1 diabetes in chinese children. Chin Med J. 2016;129(11):1298–1304. doi: 10.4103/0366-6999.182841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valdes AM, Walter J, Segal E, Spector TD. Role of the gut microbiota in nutrition and health. BMJ. 2018;361:k2179. doi: 10.1136/bmj.k2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parekh PJ, Balart LA, Johnson DA. The influence of the gut microbiome on obesity, metabolic syndrome and gastrointestinal disease. Clin Transl Gastroenterol. 2015;6(6):e91. doi: 10.1038/ctg.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Traversi D, Rabbone I, Ignaccolo MG, Carletto G, Racca I, Vallini C, et al. Gut microbiota diversity and T1DM onset: preliminary data of a case-control study. Hum Microbiome J. 2017;5:11–13. doi: 10.1016/j.humic.2017.11.002. [DOI] [Google Scholar]
- 22.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151(4):264–269. doi: 10.7326/0003-4819-151-4-200908180-00135. [DOI] [PubMed] [Google Scholar]
- 23.Brown CT, Davis-Richardson AG, Giongo A, Gano KA, Crabb DB, Mukherjee N, et al. Gut microbiome metagenomics analysis suggests a functional model for the development of autoimmunity for type 1 diabetes. PLoS ONE. 2011;6(10):e25792. doi: 10.1371/journal.pone.0025792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Goffau MC, Fuentes S, van den Bogert B, Honkanen H, de Vos WM, Welling GW, et al. Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia. 2014;57(8):1569–1577. doi: 10.1007/s00125-014-3274-0. [DOI] [PubMed] [Google Scholar]
- 25.Maffeis C, Martina A, Corradi M, Quarella S, Nori N, Torriani S, et al. Association between intestinal permeability and faecal microbiota composition in Italian children with beta cell autoimmunity at risk for type 1 diabetes. Diabetes Metab Res Rev. 2016;32(7):700–709. doi: 10.1002/dmrr.2790. [DOI] [PubMed] [Google Scholar]
- 26.Murri M, Leiva I, Gomez-Zumaquero JM, Tinahones FJ, Cardona F, Soriguer F, et al. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med. 2013;11(1):1–12. doi: 10.1186/1741-7015-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pellegrini S, Sordi V, Bolla AM, Saita D, Ferrarese R, Canducci F, et al. Duodenal mucosa of patients with type 1 diabetes shows distinctive inflammatory profile and microbiota. J Clin Endocrinol Metab. 2017;102(5):1468–1477. doi: 10.1210/jc.2016-3222. [DOI] [PubMed] [Google Scholar]
- 28.Soyucen E, Gulcan A, Aktuglu-Zeybek AC, Onal H, Kiykim E, Aydin A. Differences in the gut microbiota of healthy children and those with type 1 diabetes. Pediatr Int. 2014;56(3):336–343. doi: 10.1111/ped.12243. [DOI] [PubMed] [Google Scholar]
- 29.Stewart CJ, Nelson A, Campbell MD, Walker M, Stevenson EJ, Shaw JA, et al. Gut microbiota of type 1 diabetes patients with good glycaemic control and high physical fitness is similar to people without diabetes: an observational study. Diabet Med. 2017;34(1):127–134. doi: 10.1111/dme.13140. [DOI] [PubMed] [Google Scholar]
- 30.Mejia-Leon M, Petrosino J, Ajami N, Domínguez-Bello M, Calderon de la Barca A. HLA DQ/DR prevalence and microbiota disturbance in northwestern Mexican children with type 1 diabetes (1118.3) FASEB J. 2014;28(1_supplement):1118.3. [Google Scholar]
- 31.Giongo A, Gano KA, Crabb DB, Mukherjee N, Novelo LL, Casella G, et al. Toward defining the autoimmune microbiome for type 1 diabetes. ISME J. 2011;5(1):82–91. doi: 10.1038/ismej.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Y, Li SC, Hu J, Ruan HB, Guo HM, Zhang HH, et al. Gut microbiota profiling in Han Chinese with type 1 diabetes. Diabetes Res Clin Pract. 2018;141:256–263. doi: 10.1016/j.diabres.2018.04.032. [DOI] [PubMed] [Google Scholar]
- 33.Leiva-Gea I, Sanchez-Alcoholado L, Martin-Tejedor B, Castellano-Castillo D, Moreno-Indias I, Urda-Cardona A, et al. Gut microbiota differs in composition and functionality between children with type 1 diabetes and MODY2 and healthy control subjects: a case–control study. Diabetes Care. 2018;41(11):2385–2395. doi: 10.2337/dc18-0253. [DOI] [PubMed] [Google Scholar]
- 34.Higuchi BS, Rodrigues N, Gonzaga MI, Paiolo JCC, Stefanutto N, Omori WP, et al. Intestinal dysbiosis in autoimmune diabetes is correlated with poor glycemic control and increased interleukin-6: a pilot study. Front Immunol. 2018;9:1689. doi: 10.3389/fimmu.2018.01689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salamon D, Sroka-Oleksiak A, Kapusta P, Szopa M, Mrozinska S, Ludwig-Slomczynska AH, et al. Characteristics of gut microbiota in adult patients with type 1 and type 2 diabetes based on nextgeneration sequencing of the 16S rRNA gene fragment. Pol Arch Intern Med. 2018;128(6):336–343. doi: 10.20452/pamw.4246. [DOI] [PubMed] [Google Scholar]
- 36.Cinek O, Kramna L, Mazankova K, Odeh R, Alassaf A, Ibekwe MU, et al. The bacteriome at the onset of type 1 diabetes: a study from four geographically distant African and Asian countries. Diabetes Res Clin Pract. 2018;144:51–62. doi: 10.1016/j.diabres.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 37.Davis-Richardson AG, Ardissone AN, Dias R, Simell V, Leonard MT, Kemppainen KM, et al. Bacteroides dorei dominates gut microbiome prior to autoimmunity in Finnish children at high risk for type 1 diabetes. Front Microbiol. 2014;5:678. doi: 10.3389/fmicb.2014.00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Endesfelder D, Engel M, Zu Castell W. Gut immunity and type 1 diabetes: a mélange of microbes, diet, and host interactions? Curr Diabetes Rep. 2016;16(7):60. doi: 10.1007/s11892-016-0753-3. [DOI] [PubMed] [Google Scholar]
- 39.Kostic AD, Gevers D, Siljander H, Vatanen T, Hyotylainen T, Hamalainen AM, et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe. 2015;17(2):260–273. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinto E, Anselmo M, Calha M, Bottrill A, Duarte I, Andrew PW, et al. The intestinal proteome of diabetic and control children is enriched with different microbial and host proteins. Microbiology. 2017;163(2):161–174. doi: 10.1099/mic.0.000412. [DOI] [PubMed] [Google Scholar]
- 41.Rozanova GN, Voevodin DA, Stenina MA, Kushnareva MV. Pathogenetic role of dysbacteriosis in the development of complications of type 1 diabetes mellitus in children. Bull Exp Biol Med. 2002;133(2):164–166. doi: 10.1023/A:1015503006854. [DOI] [PubMed] [Google Scholar]
- 42.Gao X, Huynh B-T, Guillemot D, Glaser P, Opatowski L. Inference of significant microbial interactions from longitudinal metagenomics data. Front Microbiol. 2018;9:2319. doi: 10.3389/fmicb.2018.02319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gavin PG, Mullaney JA, Loo D, Cao KL, Gottlieb PA, Hill MM, et al. Intestinal metaproteomics reveals host-microbiota interactions in subjects at risk for type 1 diabetes. Diabetes Care. 2018;41(10):2178–2186. doi: 10.2337/dc18-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vatanen T, Franzosa EA, Schwager R, Tripathi S, Arthur TD, Vehik K, et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature. 2018;562(7728):589–594. doi: 10.1038/s41586-018-0620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stewart CJ, Ajami NJ, O’Brien JL, Hutchinson DS, Smith DP, Wong MC, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018;562(7728):583–588. doi: 10.1038/s41586-018-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Endesfelder D, Zu Castell W, Ardissone A, Davis-Richardson AG, Achenbach P, Hagen M, et al. Compromised gut microbiota networks in children with anti-islet cell autoimmunity. Diabetes. 2014;63(6):2006–2014. doi: 10.2337/db13-1676. [DOI] [PubMed] [Google Scholar]
- 47.Cole DK, Bulek AM, Dolton G, Schauenberg AJ, Szomolay B, Rittase W, et al. Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity. J Clin Invest. 2016;126(6):2191–2204. doi: 10.1172/JCI85679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27(2):104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 49.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504(7480):446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 50.Brestoff JR, Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14(7):676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40(3):235–243. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 52.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous clostridium species. Science. 2011;331(6015):337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Izcue A, Coombes JL, Powrie F. Regulatory lymphocytes and intestinal inflammation. Annu Rev Immunol. 2009;27:313–338. doi: 10.1146/annurev.immunol.021908.132657. [DOI] [PubMed] [Google Scholar]
- 55.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341(6145):569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bron PA, van Baarlen P, Kleerebezem M. Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nat Rev Microbiol. 2011;10:66. doi: 10.1038/nrmicro2690. [DOI] [PubMed] [Google Scholar]
- 57.Smits HH, Engering A, van der Kleij D, de Jong EC, Schipper K, van Capel TM, et al. Selective probiotic bacteria induce IL-10-producing regulatory T cells in vitro by modulating dendritic cell function through dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin. J Allergy Clin Immunol. 2005;115(6):1260–1267. doi: 10.1016/j.jaci.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 58.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118(2):511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 59.Garcia E, Lopez R. Streptococcus pneumoniae type 3 encodes a protein highly similar to the human glutamate decarboxylase (GAD65) FEMS Microbiol Lett. 1995;133(1–2):113–118. doi: 10.1016/0378-1097(95)00346-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data were included.