Abstract
Protein ubiquitylation is one of the most prevalent post-translational modifications (PTM) within cells. Ubiquitin modification of target lysine residues typically marks substrates for proteasome-dependent degradation. However, ubiquitylation can also alter protein function through modulation of protein complexes, localization or activity, without impacting protein turnover. Taken together, ubiquitylation imparts critical regulatory control over nearly every cellular, physiological, and pathophysiological process. Affinity purification techniques coupled with quantitative mass spectrometry have been robust tools to identify PTMs on endogenous proteins. A peptide antibody-based affinity approach has been successfully utilized to enrich for and identify endogenously ubiquitylated proteins. These antibodies recognize the Lys-ϵ-Gly-Gly (diGLY) remnant that is generated following trypsin digestion of ubiquitylated proteins, and these peptides can then be identified by standard mass spectrometry approaches. This technique has led to the identification of >50,000 ubiquitylation sites in human cells and quantitative information about how many of these sites are altered upon exposure to diverse proteotoxic stressors. In addition, the diGLY proteomics approach has led to the identification of specific ubiquitin ligase targets. Here we provide a detailed method to interrogate the ubiquitin-modified proteome from any eukaryotic organism or tissue.
Keywords: Ubiquitin, Proteomics, diGLY, Affinity Purification, Mass Spectrometry
1. Introduction
Protein post-translational modifications (PTMs) impart critical regulatory control on nearly every cellular process. PTMs diversify the proteome to such a degree that we will likely never realize the full extent of proteome complexity. Common post-translational modifications include phosphorylation [1–6], glycosylation [7–11], ubiquitylation [12–20], nitrosylation [21], methylation [22], acetylation [23], and lipidation [24], all of which impact normal cell biology as well as pathogenesis. Therefore, identifying and understanding the function of individual PTMs is critical to understand cellular homeostasis.
PTMs modify a subpopulation of proteins and only a small portion of the total proteome, therefore identification relies on the development of efficient and robust enrichment approaches. A number of different strategies to enrich for ubiquitylated proteins have been developed, including utilizing overexpression of epitope-tagged versions of ubiquitin followed by affinity-based enrichment of ubiquitylated proteins [12,25]. Another strategy uses purified ubiquitin binding domains from select proteins to enrich for endogenously ubiquitylated proteins [26]. Each of these techniques has several advantages and drawbacks which have been reviewed previously [27–29]. While each approach can identify putative ubiquitylated proteins, they are less effective in identifying the precise sites of ubiquitylation on those proteins.
Since the initial peptide sequencing of ubiquitin and the demonstration that ubiquitin-modifies lysine residues on substrates, it was appreciated that trypsin-digestion of a ubiquitylated protein would generate peptides containing a characteristic diglycine(diGLY)-modified lysine residue that could be used to identify ubiquitylation sites. In fact, the existence of the remnant ubiquitin diGLY residue was first reported on histone H2A by A24 (later renamed ubiquitin) by Goldknopf and Busch in 1977 [30]. These diGLY-modified peptides were then easily distinguished by “bottom-up” mass spectrometry approaches, identifying ubiquitin-modified proteins along with the exact site of modification. However, the low abundance of ubiquitylated peptides compared to linear non-modified peptides in a whole proteome digest, make identification of diGLY-modified peptides difficult without enrichment steps. In 2003, Peng, J. et.al first attempted to study the ubiquitin-modified proteome based on the existence of the known ubiquitin remnant diGLY motif on ubiquitylated substrate peptides upon trypsinolysis [12]. This report pressed the need for development of new tools to capture these modifications.
Global analysis of the ubiquitin-modified proteome using mass spectrometry gained momentum when initial studies reported the development of antibody-based enrichment strategies that utilized antibodies capable of binding diGLY-modified peptides [18]. Thereafter, the generation of more robust ubiquitin remnant diGLY motif-specific antibodies allowed for the identification of more than 10,000 ubiquitylated (diGLY) peptides [19] [20]. It is critical to note that the C-terminal protein sequences of the ubiquitin-like proteins, Nedd8 and ISG15, are like ubiquitin and will leave identical diGLY-modified peptides upon trypsinolysis of NEDDylated or ISGylated proteins. As such, identification of a diGLY-modified peptide does not, on its own, unequivocally identify a protein as being ubiquitylated. However, studies have shown that ~95% of all diGLY-peptides identified using the diGLY-antibody enrichment approach arise from ubiquitylation versus neddylation or ISGylation [19].
The diGLY proteomics approach now has become an indispensable tool to systematically interrogate protein ubiquitylation with site-level resolution [31–33]. The use of quantitative mass spectrometry approaches coupled with diGLY-antibody affinity enrichment steps have led to a more complete understanding of both the breadth of ubiquitylation, and global alterations in protein ubiquitylation in response to an increasing variety of cell stimuli and stressors [34–43]. This approach has also proven useful to identify substrates for specific ubiquitin ligases [19,38,41,43–49]. Another advantage of the diGLY-antibody affinity approach is that it can be applied toward the identification of ubiquitylated proteins from any human or murine primary tissue or with any eukaryote [37,40,50–54].
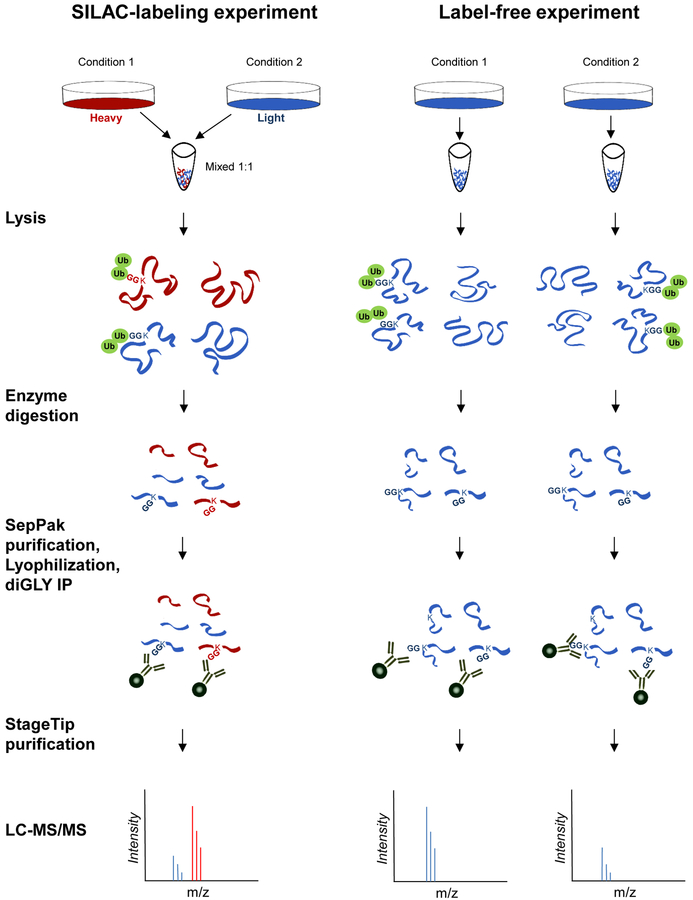
Here, we describe a SILAC-based quantitative proteomic method to identify differential ubiquitylation between two samples (Figure 1). SILAC-based approaches are not required, and label-free as well as chemical isobaric-labeling approaches can also be used (see Note 2). Additional enrichment and/or fractionation steps can be used to improve the depth of the enrichment of diGLY-modified peptides depending on the exact experimental question [18,20,35,39,41,42,48,51,53–58]. Here, we limit our method description to a simple one-pot affinity strategy without extensive post-lysis or post-digestion fractionation steps.
Figure 1 – Workflow Schematic for diGLY Proteomics.
2. Materials
2.1. Cell Culture Media
The media may vary according to the cells used for the experiment. Here, we use standard DMEM which can be used with a variety of human cell types commonly grown in culture.
DMEM lacking lysine and arginine: Thermo Fisher – catalog #88364 (see Note 1).
Heavy lysine (K8): Cambridge Isotope laboratories - L-LYSINE:2HCL (13C6, 99%; 15N2, 99%) – catalog #CNLM-291-H-PK
Heavy arginine (R10): Cambridge Isotope laboratories - L-ARGININE: HCL (13C6, 99%; 15N4, 99%)- catalog #CNLM-539-H-PK
Dialyzed Fetal Bovine Serum: Thermo Fisher – catalog # 26400044
L-Lysine Hydrochloride (for control “light” media)
L-Arginine Hydrochloride (for control “light” media)
Penicillin-Streptomycin
Recipe for Stable Isotope Labelling:
Light Media: Remove 50mL of DMEM media from the 500mL bottle and add 50mL of thawed dialyzed Fetal Bovine Serum (FBS). Dissolve 150mg of LLysine-2HCl (light) and 85mg of L-Arginine-HCl (light) using 1mL of DMEM media. Mix thoroughly and add it to the Light media. Add Pen/Strep to avoid contamination during the cell culture.
Heavy Media: Remove 50mL of DMEM media from the 500mL bottle and add 50mL of thawed dialyzed Fetal Bovine Serum (FBS). Dissolve 150mg of 13C6-15N2 L-Lysine-2HCl (Heavy) and 85mg of 13C6-15N4 L L-Arginine-HCl (Heavy) using 1mL of DMEM media. Mix thoroughly and add it to the Light media. Add Pen/Strep to avoid contamination during the cell culture.
2.2. Cell/tissue Lysis Buffer
Lysis Buffer composition:
8M Urea
150mM NaCl
50mM Tris-HCl, pH 8
Complete Protease Inhibitor- Roche - catalog #5056489001
1mM Sodium Fluoride (NaF)
1mM β-glycerophosphate (β-Gly)
1mM Sodium Orthovanadate (NaV)
5mM N-Ethylmaleimide (NEM): dissolve in ethanol and prepare fresh before addition to the lysis buffer (see Notes 3-5).
2.3. Reagents for Protein Digestion
LysC protease enzyme (Wako, catalog #125–02543, 2AU): For 2AU vial of LysC, resuspend in 400μL of HPLC grade water for a final concentration of 0.005AU/μL, approximately 2mg/mL. Make 20μL aliquots in 0.5mL tubes and store at −80°C.
Trypsin protease enzyme (Sigma, catalog #T1426 TPCK treated): Prepare the stock concentration of 0.1mg/mL in a 50mM Ammonium bicarbonate buffer and store at −80°C (see Note 7).
1M CaCl2 stock solution
2.4. Reagents for peptide desalting
SepPak™ tC18 reverse phase column, (Waters - catalog #WAT036815): For 30mg of a protein digest, a 500mg SepPak™ tC18 column is recommended [59]. This protocol uses 3cc volume capacity cartridges (see Note 8). If using the other cartridge sizes, the volume of solutions should be adapted accordingly (i.e., for 6cc, tC18 cartridge, use 6mL of the volume of the solution during each wash step)
100% Acetonitrile (ACN): Use HPLC-grade Acetonitrile for preparing the solutions.
50% Acetonitrile (ACN), 0.5% Acetic acid (HAcO)
0.1% Trifluoroacetic acid (TFA)
0.4% Trifluoroacetic acid (TFA)
0.5% Acetic Acid (HAcO)
2.5. Reagents for diGLY IP
Ubiquitin Remnant Motif (K-Ɛ-GG) Antibody or PTMScan® Ubiquitin Remnant Motif (K-Ɛ-GG) Kit: (Cell Signaling Technologies, catalog #5562). (see Note 9)
Protein-A Plus Ultralink™ Resin (Thermo Fisher, Pierce, catalog # 53142).
Phosphate Buffered Saline.
HPLC-grade water.
Immunoaffinity Purification (IAP) Buffer: The composition of 10X IAP buffer is 500mM MOPS-NaOH, pH 7.5, 100mM Na2HPO4, 500mM NaCl. IAP buffer can be stored at 4°C for up to 1 month. We typically make the 10X IAP fresh each time.
2.6. Reagents for antibody cross-linking (optional)
Cross-linking buffer (0.2M Triethanolamine, pH 8.2 (F.W.149.19, d=1.124g/mL)
Cross-linking agent (Dimethyl pimelidate dihydrochloride - DMP): Prepare fresh
Blocking buffer (0.1M Ethanolamine pH 8.2 (M.W.61.08g/mol)
Antibody cross-linked beads storage buffer (0.1% Tween-20, 0.02% Sodium azide in PBS)
2.7. Reagents for Stage-tip peptide purification
Stage Tips, Empore™ C18-embeded membrane (Sigma-Aldrich - catalog #2215-C18): Punch a small hole through the four layers of Empore C18 material discs and mount it at the narrow end of an unautoclaved 200μL pipet tip.
Methanol, HPLC grade.
80% Acetonitrile (ACN), 5% Formic acid (FA).
5% Formic acid.
5% Acetonitrile (ACN), 5% Formic acid (FA).
2.8. Equipment
Sonicator.
37°C incubator.
Lyophilizer.
Speed Vacuum Centrifuge
nano-HPLC.
Mass Spectrometer.
3. Methods
3.1. Cell Culture
We typically use eight 15cm dishes of HCT116 cells per experiment (4 plates for heavy-labeled cells, 4 plates for light-labeled cells). This corresponds to ~2×108 cells and ~30mg of total protein upon lysis (see Note 10). The heavy cell population is typically treated with a cell stressor or is altered in other ways (e.g. knockout of a E3 ligase gene).
3.2. Cell/Tissue Harvest and Lysis
Once the cells are ready to harvest, remove the cells from 15cm dishes with 4mL of 0.25% trypsin EDTA (cell culture grade) per dish.
Add 30mL of cold SILAC media (add appropriate heavy or light media depending upon the treatment condition) to a single dish.
Resuspend the cells and move to the next dish to collect all the heavy or light cell populations. Alternatively, the cells can be scraped and collected in 5mL cold PBS for each dish.
Collect the separated heavy and light cells by centrifugation (300 RCF, 4°C, 5 min). Decant and discard the supernatant.
Resuspend the cell pellets in 20mL of cold PBS. Make sure the cell pellet is well resuspended to get the accurate cell count.
Count the cells using hemocytometer or by using an automated cell counter. Mix equal amounts of heavy and light cells (by cell number) into a single 50mL tube. Save approximately 100uL or more of the unmixed heavy and light cell populations to perform Western blot analysis if necessary (see Note 11).
Collect the combined heavy and light cells by centrifugation (300 RCF, 4°C, 5 min). Decant and discard the supernatant.
Cell pellets can be stored at −80°C at this stage.
Add 1.5 to 2mL of freshly prepared urea lysis buffer (depending on the pellet size to be mixed) to the frozen cell pellets. As the cell pellet thaws, resuspend the pellet completely (see Notes 4,6).
Sonicate with a 30% intensity (8mV) setting using a microtip-fitted sonicator. Sonicate three times for 10 seconds each with 30 second rest on ice between the cycles.
Centrifuge the cell lysate at 20,000 RCF for 15 min at 4°C to pellet the insoluble material.
Collect and transfer the supernatant to a new 15mL conical tube.
3.3. Protein Digestion
Determine the protein concentration of the cell lysates using an appropriate total protein detection assay method (i.e., BCA or Bradford assay).
Dilute the lysate with equal volume of lysis buffer without urea or protease inhibitors to bring the final urea concentration to 4M.
Add LysC to the lysates at the final concentration of 10ng/μL.
Incubate at 37°C for 1–2h with end-over-end rotation.
Dilute the LysC digested samples to 1M urea final concentration using 50mM Tris-HCl, pH 8
Add CaCl2 to final concentration of 1mM
Add Sigma trypsin (T1426 TPCK treated) at the ratio of 1:100; enzyme: substrate.
Incubate overnight at 37°C.
Stop the digestion by adding TFA to a final concentration of 0.4%. Verify the pH is less than 2.0 using pH determination strips. Add enough TFA to bring down the pH if in case it is above 2.
Centrifuge at 300 RCF for 15 min at room temperature. Collect the supernatant.
3.4. Peptide desalting
The peptide desalting steps are done essentially as described previously [59]. Wash and condition the C18 SepPak™ cartridge 3 times using 3mL of ACN for 3cc capacity cartridges (use vacuum) (see Note 8).
Wash with 3mL of 50% ACN and 0.5% HAcO to clear any unwanted material bound to the cartridges (use vacuum).
Equilibrate 3 times with 3mL of 0.1% TFA (use vacuum).
Load the digested cell lysates in 0.4% TFA (gravity flow).
Wash and desalt 4 times with 3mL of 0.1% TFA (use vacuum).
Wash (to remove TFA) with 1mL of 0.5% HAcO (gravity flow).
Elute the peptides bound to the C18 cartridges 2 times with 3mL of 50% ACN, 0.5% HAcO (gravity flow). Use a new 15cm conical tube to collect the peptide eluate. To increase the throughput, the C18 cartridge can be placed in a 15mL conical tube and spun at 200 RCF to increase the flow rate (see Note 8).
At this point, collect ~250μg of the digested and desalted peptides in a separate tube to be used for analysis of the total proteome by mass spectrometry.
Freeze the eluate with liquid N2 or store at −80°C for 2–4h and lyophilize for a minimum of 2 days. The lyophilized peptides appear as white (sometimes yellowish) fluffy powder. The lyophilization step should remove all the residual acid from the peptide sample and the peptides should be completely dry prior to diGLY IP.
3.5. Affinity purification with diGLY antibody
The subsequent steps of the diGLY IP should be done on ice or in the cold room.
To prepare the antibody/protein A resin, pre-equilibrate the protein-A beads by washing 5 times in 1mL of cold PBS followed by 2 washes in 2X IAP buffer. One IP reaction uses 20μL of dry resin (40 μL of the 1:1 slurry is needed) (see Note 9).
After the final wash, resuspend the resin as a 1:1 slurry in 2X IAP buffer.
Add 60μg of the diGLY antibody per 20μL of prepared protein-A beads and couple overnight (end-over-end rotation at 4°C). Alternatively, the antibody can be cross-linked to the resin (e.g. using Pierce crosslink IP Kit #26147) (see Note 12).
Resuspend the lyophilized peptides in 1.3mL of 2X IAP buffer. Shaking and sonication can help dissolve peptides. Check pH of solution (see Note 13). The pH should be around 7–7.5 (use the pH paper and compare peptide solution to 2X IAP buffer alone).
Clear solution by centrifugation, 30 min at 20,000 RCF at 4°C (A sizable pellet may be present but most of the peptides will be in solution).
Transfer the peptide solution to the tube with the antibody-coupled beads and incubate for 1–2h rotating at 4°C.
Centrifuge at 300 RCF, 4oC, and collect the supernatant. Save the supernatant for additional IP reactions or as a backup if IP fails (see Note 14).
Wash the beads 4 times with 1mL of 2X IAP buffer. Rotate at 4°C for 10 min between each wash.
Wash with 1mL cold HPLC-grade water. Rotate at 4°C for 10 min.
Elution 1: Add 55μL of 5% formic acid, mix (tap bottom of tube lightly) and let it stand at room temperature for 10 min. Collect eluate in a new 1.5mL Eppendorf tube.
Elution 2: Add 45μL of 5% formic acid, mix, incubate 10 min at room temperature and combine it with the first elution.
3.6. Antibody cross-linking (optional)
The cross-linking of the ubiquitin remnant Lys-ϵ-Gly-Gly motif specific antibody has been reported to increase the number of detectable diGLY sites due to the reduced deleterious effects that antibody-derived contaminants have on the enrichment [55,60]. The cross-linking of antibody is an optional step to save the antibody reagents for subsequent immune-enrichments. The shelf life of the cross-linked antibody is not well-studied (see Note 12). The cross-linking procedure is adapted and modified from a protocol available from New England BioLabs.
After diGLY antibody coupling to the Protein-A beads (as per diGLY IP protocol procedure), wash 2 times with PBS. Resuspend resin as a 1:1 slurry with PBS.
Add 1mL of cross-linking buffer to the Protein-A immobilized antibody and resuspend thoroughly. Mix with end-over-end rotation at room temperature for 10 min.
Centrifuge at 300 RCF and remove the supernatant.
Repeat steps 2–3 one more time.
Resuspend in 1mL cross linking buffer containing 25mM DMP. Mix thoroughly and incubate at room temperature for 45 min with agitation.
Centrifuge at 300 RCF and remove the supernatant.
Add 1mL blocking buffer and resuspend completely.
Centrifuge at 300 RCF and remove the supernatant.
Add 1mL blocking buffer, resuspend and incubate for 1h at room temperature with agitation.
Centrifuge at 300 RCF and remove the supernatant.
Wash the beads with 1mL PBS.
Repeat PBS wash 2 more times.
After the final PBS wash, proceed with the diGLY protocol, add the reconstituted peptides in 1.3mL 2X IAP buffer, check the pH (it should be around 7).
After the final elution (as per diGLY protocol) of the peptides in 5% formic acid, wash the beads with 1mL, 5% formic acid 3 times, and then with 1mL PBS 5 times.
Resuspend the beads in 100μL PBS, 0.1% Tween-20, 0.02% sodium azide for long-term storage at 4°C.
3.7. Stage-tip purification of peptides
Samples are cleaned up for mass spectrometry analysis using in-house prepared C18 Stage Tips, as described previously [61].
Puncture a hole in the cap of a 1.5 mL microcentrifuge tube (unautoclaved) to allow for placement of a 200μ micropipette. Condition the stage tip containing 4 layers of Empore C18-embeded membrane with 40μL of methanol. Centrifuge at 550 RCF for 1 min (see Note 15).
Wash stage tip with 40μL of 80% acetonitrile, 5% formic acid. Centrifuge at 550 RCF for 1 min.
Wash three times with 40μL of 5% Formic acid. Centrifuge at 550 RCF for 1 min.
Load the diGLY IP eluate on the stage tip. Centrifuge at 550 RCF for 1 min. Pass the IP eluate through the tip twice.
Wash three times with 40μL of 5% Formic acid. Centrifuge at 550 RCF for 1 min.
Transfer the stage tip to the new punctured 1.5 mL microcentrifuge tube.
Elute the peptides with 40μL of 80% Acetonitrile, 5% Formic acid. Repeat two times.
Evaporate the solvent using a speed vacuum centrifuge.
Resuspend the dried peptides in 14μL of 5% Formic acid, 5% Acetonitrile. Centrifuge the tubes. Transfer the peptides to an autosampler vial or other mass spec compatible vial. Centrifuge the sample again. (sample can be stored in 4°C for short-term or at −20°C for long-term storage before using it for MS analysis).
3.8. Mass-spectrometry analysis
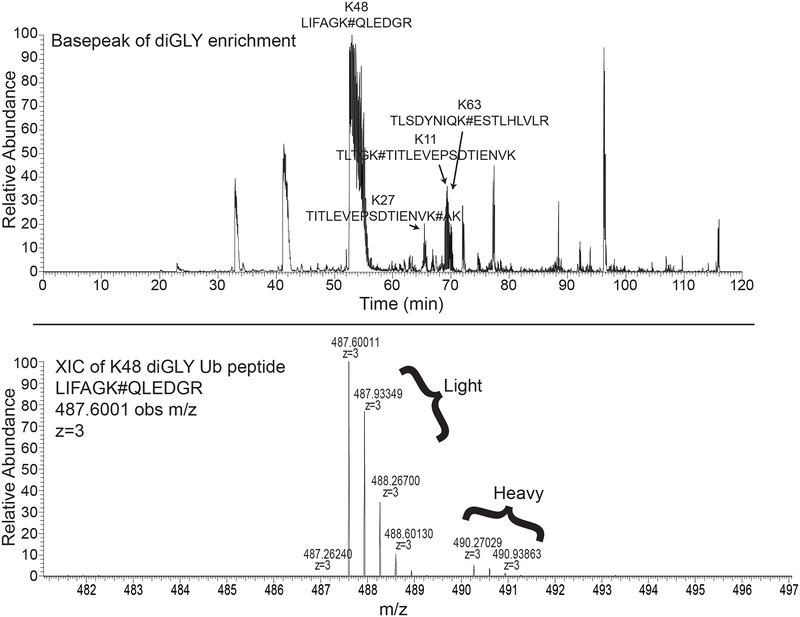
The choice of the nano-HPLC and the mass spectrometer depends on the availability of the instrument for the user. High resolution, high speed instruments are ideal for acquiring data of high quality and quantity. Here, the information is provided for the EASY-nLC1000 and the Q-Exactive Mass Spectrometer instrument (Thermo Fisher) (see Note 16). Solvent A contains 0.1% Formic acid in water and solvent B contains 0.1% Formic acid in Acetonitrile. We typically inject 3μL of the sample per run, and run each sample in triplicate (Figure 2).
Figure 2 – Example of typical base peak ion chromatogram for a diGLY-antibody enriched sample.
Top) Heavy (K8-only) labeled cells were treated with a ubiquitin E1 enzyme inhibitor and the proteasome inhibitor MG132. Heavy cells were mixed with light cells treated with MG132 only prior to cell lysis and subsequent sample processing for diGLY-based proteomic analysis. Shown is the base peak ion chromatogram for the diGLY-antibody enriched sample. Arrows signify prominent ion peaks that correspond to a subset of peptides from abundant ubiquitin-ubiquitin linkages.
Bottom) Extracted ion chromatogram for the K48 linked ub peptide (z+3). The sequence of the peptide is shown with # representing a diGLY-modified lysine residue. Note that the ion intensity of the heavy ion pair is significantly lower than the corresponding light pair due to E1 enzyme inhibition.
LC-MS-MS Parameters:
Pack a fused silica column (15cm, 100μm ID) with C18 material- Dr. Maisch GmbH, part #r119.aq.001 (porous spherical silica: 1.9μM; pore diameter to surface area: 120A0 /300m2/g; %carbon: 15%) for the in-line reverse phase chromatography.
We run a 2h gradient and instrument method for diGLY-enriched samples.
Peptides are eluted from the C18 column with the following gradient. 100 min, 2–30% ACN gradient; 5 min, 30–60% ACN gradient; 5 min, 60–95% ACN gradient; 5 min 95–0% gradient; with a final 5 min, isocratic step in solvent A. Total run time of 120 min at a flow rate of 250 nL/min.
Collect the MS/MS data in a data dependent fashion using a top 10 method with a full MS mass range from 300–1750 m/z, 70,000 resolution, and an AGC target of 3e6.
Set the MS2 scans to trigger on when an ion intensity threshold of 1e5 reaches with a maximum injection time of 250ms.
Set the normalized collision energy setting to 22 for peptides fragmentation.
A dynamic exclusion time of 40 seconds is used and the peptide match setting is disabled.
Singly charged ions, charge states above 8, and unassigned charge states are excluded.
Peptide and Protein Identification and Quantification:
Subsequent instrument files containing the raw MS/MS data can be processed with a variety of MS data processing pipelines including the Trans-Proteomic pipeline, MaxQuant, or vendor-specific software packages.
Use the suitable search algorithm such as SEQUEST, OMSSA or MASCOT to search MS/MS spectra against a concatenated target-decoy database comprised of forward and reversed sequences from the appropriate sequence database.
Typical search parameters used are as follows: 10 ppm precursor ion tolerance and 0.01 Da fragment ion tolerance; Trypsin (1 1 KR P) is set as the enzyme; allow up to three missed cleavages; dynamic modifications of 15.99491 Da on methionine (oxidation), and 114.04293 Da on lysine (diGLY), and 42.010564 Da on peptide N-term (acetylation), and static modification of 125.047679 Da on cysteines (NEM alkylation).
Filter the peptide matches to a peptide false discovery rate of 1% (see Notes 17,18).
Perform appropriate quantitative measurement (MS1 precursor area or max signal comparison, peptide counting, or MS2-based quantification). Exclude all peptides with a C-terminal diGLY-modified lysine residue.
4. Notes
SILAC media using only heavy lysine can be used as well. All peptides of interest (i.e., ubiquitylated peptides) will have a lysine residue which will carry the heavy label. Thus, usage of heavy arginine is not required. However, we routinely use input peptides to the diGLY-IP to measure changes in total protein abundance. For this, labeling with both lysine and arginine can be helpful although not essential. If lysine-only labeling is used, we discard all non-lysine containing peptides during quantitative analysis of both the ubiquitin-modified and total proteome.
A SILAC-based approach can be used for experiments in Saccharomyces cerevisiae as well [36,37,48,56]. Label-free quantitative approaches have also been used to examine the ubiquitin-modified proteome with good success. Further, tandem-mass-tagged approaches that label peptides after diGLY-antibody-based enrichment has been used to quantify changes to the ubiquitin-modified proteome [53]. However, special care must be taken with both label-free and post-IP labeling approaches to ensure consistent lysis, sample digestion, and IP conditions across both control and experimental samples. Small deviations in sample handling between experiments can result in quantitative differences between peptide amounts resulting in sample handling rather than for biological reasons.
All reagents used should be of highest mass spectrometry grade quality.
Urea lysis buffer should be prepared fresh before every experiment.
N-Ethylmaleimide (NEM) is a cysteine alkylating agent that will inhibit cellular thiol-dependent deubiquitylating enzymes upon lysis. Addition of fresh NEM (or other cysteine alkylating agents) is critical as omission can lead to loss of ubiquitylated peptides upon lysis (even in 8M urea). NEM is readily soluble in ethanol and it should be prepared fresh before addition to the lysis buffer.
Try to use the minimum possible volume of lysis buffer for cell lysis so when the samples are diluted to decrease the urea concentration, prior to lysC and trypsin digestion, the protein concentration remains high enough for efficient digestion.
We typically do not use the highest quality mass spectrometry grade trypsin during sample preparation for subsequent diGLY enrichment. The recommended starting protein amount is more than 20mg and use of mass spectrometry grade trypsin to facilitate proper digestion of this amount of protein would add additional reagent costs to the experiment.
The SepPak™ cartridges are available with varying volume holding capacities. The 3cc tC18 cartridge allows for convenient nesting on 15mL conical tubes for centrifugation in the last step of the peptide desalting procedure to extract maximum volume from the cartridge.
Lucerna also sells a diGLY-antibody (catalog # 30–1000) that has been successfully used in many studies [20,34,51,62]. However, the clear majority of published studies have used the Cell Signaling antibody, and we have used it exclusively [19,35–44,46–58,60,63–65]. As such, the subsequent protocol is based on our experience using the diGLY antibody available from Cell-Signaling Technologies. The Ubiquitin Remnant Motif (K-Ɛ-GG) Antibody, when purchased as part of the PTM-scan kit, typically arrives already conjugated to the resin. We prefer to use the unconjugated antibody without the kit. As such, this protocol includes the procedure for antibody coupling to the beads (both with and without crosslinking). If you are using the antibody pre-coupled to the resin as part of the PTM-scan kit, then skip to step 4 in the affinity purification section.
We recommend using ~30mg of starting material (tissue lysate, worm extract, etc.). Other studies report using less starting material with good outcomes; we have obtained good data from as little as 10mg. However, 30mg of starting material is recommended to obtain the depth of coverage of ubiquitin-modified peptides usually desired.
Heavy and light-labeled samples can also be mixed 1:1 according to total protein after lysis.
Previous studies have reported greater numbers of ubiquitin-modified peptides identified after crosslinking the antibody to the resin [55] (Table 1). We have not observed this result and have obtained similar results from un-crosslinked and crosslinked resins. We have successfully reused crosslinked resin after acid elution as long as the resin is reused within 2–3 weeks.
After the lyophilized peptides are resuspended in the 2X IAP buffer, it is very important to check the pH of the dissolved peptides. If the pH is below 7, it indicates that trace amounts of TFA and acetic acid used in the desalting steps are still present in the sample and the lyophilization was not efficient. To avoid such a case, it is recommended to lyophilize the sample for more than 2 days.
To identify and quantify larger numbers of diGLY-modified peptides, we have previously done sequential IPs from a single sample (i.e., the flow through of the first diGLY IP is applied to a second round of IP using a new aliquot of antibody-coupled resin). While this can achieve a greater number of identified peptides, it comes at the cost of using more diGLY antibody per experiment.
During the stage tip purification of the peptides, make sure that the C18 material does not dry down during all the centrifugation steps. Adjust the centrifugation speed if required.
Individual LC and mass spectrometer settings will need to be adjusted based on the instrumentation available. The settings listed here are meant to serve as an example and are not to be considered the only suitable instrument method for data collection (Figure 2).
We typically observe that between 30–80% of all identified peptides contain a diGLY-modified lysine residue. Enrichment is typically higher when using samples treated with a proteasome inhibitor.
We typically identify ~10,000 total diGLY peptides (4000 unique peptides) in a single sample shot in triplicate. Treatment of cells with proteasome inhibitor will result in a greater number of identified peptides (25,000 total diGLY-modified peptides, 10,000 unique peptides) (Figure 2, Table 1).
Table 1 –
Table describing results of key previous published diGLY studies.
| Published study PMID (ref #) | 12872131 (12) |
20639865 (18) |
21139048 | 21987572 (45) |
21906983 (19) |
21890473 (20) |
21963094 (44) |
| Authors | Peng, J. et.al. | Xu, G. et.al. | Danielsen, J.M. et.al. | Lee, KA., et.al. | Kim, W. et.al. | Wagner, S.A. et.al. | Emanuele, M. J., et.al. |
| Year | 2003 | 2010 | 2011 | 2011 | 2011 | 2011 | 2011 |
| Tissues/Cells | S. cerevisiae | HEK293T cells | U2OS, HEK293T cells | HeLa TREx cells | HCT116, HEK293T | HEK293T | HeLa cells |
| Labeling | SILAC | SILAC | SILAC | SILAC | SILAC | SILAC | |
| Ub-sites Identified | 110 | 374 | 753 | 1,800 | 19,000 | 11,054 | 9,957 |
| Ub-Proteins Identified | 72 | 236 | 471 | 900 | 5,000 | 4,273 | 2,814 |
| IP/Pre-IP Enrichment | Ni-NTA affinity chromatography of His-tagged Ub | 1) Ni-NTA affinity chromatography of His-tagged Ub and 2) diGLY IP | Streptavidin affinity chromatography of Strep-HA-tagged Ub | diGLY-antibody IP (antibody 3925), Cell Signaling Technology | diGLY-antibody IP Cell Signaling Technology | diGLY-antibody IP (Lucerna) | PTMScan Ubiquitin Remnant Antibody Beads (Cell Signaling Technology) diGLY-antibody IP |
| PostEnrichment fractionation | SCX | IEF | |||||
| Remarks | First report of Ub-modified peptide identification using the signature diGLY remnant for identification by MS | Reported the use of diGLY affinity purification strategy for immuno-enrichment of the diGLY modified peptides | Study reports the promiscuity of Lysine ubiquitylation at the site level | Study used peptide and protein level approaches to enrich for ubiquitinated proteins in the presence and absence of the HRD1 ubiquitin ligase | Reported the identification and quantification of a large number of diGLY-modified peptides and their response to proteasome inhibition and translation inhibition. Also identified substrates for CRL ligases. | Reported the identification and quantification of a large number of diGLY-modified peptides and their response to proteasome inhibition. Characterized possible crosstalk with acetylation | Identified proteins regulated by CRL ligases |
| Published study PMID (ref #) | 22871113 (50) |
23000965 (34) |
22505724 (35) |
22790023 (51) |
22817900 (63) |
23266961 (55) |
23749301 (56) |
23716602 (36) |
| Authors | Na, C. H., et.al. | Povlsen, L. K., et.al. | Udeshi, N. D., et.al. | Wagner, S. A., et.al. | Beltrao, P., et.al. | Udeshi, N. D., et.al. | Swaney, D. L., et.al. | Ng, A. H., et.al. |
| Year | 2012 | 2012 | 2012 | 2012 | 2012 | 2013 | 2013 | 2013 |
| Tissues/Cells | Rat brain tissue | Human U2OS cells | Jurkat E6–1 cells | Tissues from C57BL/6 mice | S. cerevisiae | Jurkat E6–1 cells | S. cerevisiae | S. cerevisiae |
| Labeling | SILAC | SILAC | SILAC | SILAC | SILAC | |||
| Ub-sites Identified | 1,786 | 6,700 | 3,300 peptides | 20,085 | 2,500 | 20,004 sites, 3,143 peptides | 5,465 | 1,321 |
| Ub-Proteins Identified | 921 | 1,307 | 240 | |||||
| IP/Pre-IP Enrichment | 1) SCX, 2) PTMScan Ubiquitin Remnant Antibody Beads (Cell Signaling Technology) |
diGLY IP (Lucerna, clone GX41) | 1) SCX, 2) PTMScan Ubiquitin Remnant Antibody Beads (Cell Signaling Technology) |
diGLY-antibody IP (clone GX41) and PTMScan Ubiquitin Remnant Antibody Beads (Cell Signaling Technology), diGLY-antibody IP | 1) HisTag purification of His-tagged Ub and 2) PTMScan Ubiquitin Remnant Antibody Beads (Cell Signaling Technology) | 1) Basic pH reversed-phase separation pre-IP, 2) PTMScan Ubiquitin Remnant Antibody Beads-cross linked with DMP (Cell Signaling Technology) diGLY-antibody IP |
A) i) cobalt-NTA affinity chromatography for His-tag purification, ii) PTMScan Ubiquitin Remnant Antibody Beads (Cell Signaling Technology) B) i) SCX, ii) PTMScan Ubiquitin Remnant Antibody Beads (Cell Signaling Technology) | PTMScan Ubiquitin Remnant Antibody Beads (Cell Signaling Technology) |
| PostEnrichment fractionation | SCX | |||||||
| Remarks | Study utilized the antibody enrichment strategy for analyzing the ubiquitome from rat brain | Study demonstrated the widespread ultraviolet-regulated ubiquitylation of known DDR components and many proteins not previously implicated in this response | Examined the effects of proteasome inhibition and deubiquitinase inhibition by PR-619 on ubiquitination sites | Study demonstrated the enrichment of endogenous ubiquitylated peptides from murine tissue lysates and their subsequent identification by mass spectrometry |
This study identified 2,500 ubiquitylation sites for S. cerevisiae | Study described a number of enhancements to the diGLY-based enrichment method | Study developed two methods to identify protein isoforms that are both phosphorylated and ubiquitylated in the yeast Saccharomyces cerevisiae, identifying 466 proteins with 2,100 phosphorylation sites co-occurring with 2,189 ubiquitylation sites | Study shows that upon heat shock the distinct populations of structured and intrinsically disordered proteins are prone to ubiquitylation |
| Published study PMID (ref #) | 23503661 (46) |
23749302 (57) |
25093938 | 25078903 (48) |
24961812 (48) |
25147182 (64) |
25278611 (58) |
| Authors | Sarraf, S. A., et.al. | Mertins, P., et.al. | Akimov, V. et.al. | Tong, Z., et.al. | Iesmantavicius, V., et.al. | Thompson, J. W., et.al. | Theurillat, J. P., et.al. |
| Year | 2013 | 2013 | 2014 | 2014 | 2014 | 2014 | 2014 |
|
Tissues/ Cells |
HCT116PARKIN,HeLaPARKIN, HCT116, SH-SY5Y | HeLa S3 cells | StUbEx-stable expressing HeLa cells | S. cerevisiae | S. cerevisiae | BT-549 cells stably transduced with an inducible HUWE1 shRNA | LHMAR cells |
| Labeling | SILAC | SILAC | SILAC | SILAC | SILAC | SILAC | SILAC |
| Ub-sites Identified | 6,934 sites in HCT116PARKIN; 3,286 sites in HeLaPARKIN; 6,149 sites in HCT116; 3,450 sites in SH-SY5Y cells | 15,408 | 3,116 | 2,299 | A) 2,735 peptides in condition 1 and 2,806 peptides in condition 2; B) 6,614 peptides in MG132 treated cells and 4,157 peptides in DMSO treated cells | 7,181 Ub peptides | |
| Ub-Proteins Identified | 1,993 proteins in HCT116PARKIN; 1,220 proteins in HeLaPARKIN; 1,927 proteins in HCT116; 1,329 proteins in SH-SY5Y cells | 3,400 | 1,111 | ||||
| IP/Pre-IP Enrichment | diGLY antibody IP (Cell Signaling Technology) | 1) Basic pH reversed-phase separation pre-IP 2) PTMScan Ubiquitin Remnant Antibody Beads (Cell Signaling Technology) |
Ni-Sepharose beads | 1) Ni-NTA affinity chromatography of His-tagged Ub 2) PTMScan Ubiquitin Remnant Antibody Beads (Cell Signaling Technology) | PTMScan Ubiquitin Remnant Antibody Beads (Cell Signaling Technology) | A) PTMScan Ubiquitin Remnant Antibody Beads (Cell Signaling Technology) B) i) bRPLC, ii) PTMScan Ubiquitin Remnant Antibody Beads | 1) Basic pH reversed-phase (bRP) chromatography, 2) diGLY antibody IP (Cell Signaling Technology) crosslinked using DMP |
| PostEnrichment fractionation | pH gradient fractionation | SCX | SCX | ||||
| Remarks | Study elucidated the ubiquitylation site specificity and topology of PARKIN-dependent target modification in response to mitochondrial depolarization. | Reported the integrated analysis of protein expression, phosphorylation, ubiquitylation and acetylation by serial enrichment | Used the StUbEx strategy to identify Ub-modified peptides | diGLY proteomics approach identified 3,116 ubiquitylation sites, including 10 sites in Tul1 candidate substrates. | Study reported the parallel quantification of ubiquitylation, phosphorylation, and proteome changes in rapamycin-treated yeast cells. | Study implemented an inducible loss of function approach in combination with quantitative diGly proteomics to find novel Huwe1 substrates | Study analyzed changes in the ubiquitin landscape induced by prostate cancer-associated mutations of SPOP, an E3 ubiquitin ligase substrate-binding protein. |
| Published study PMID (ref #) | 26038114 (62) |
26051181 (39) |
26051182 (40) |
26131937 (41) |
27211868 (65) |
27185884 (42) |
27667366 (53) |
28665616 (54) |
| Authors | Satpathy, S., et.al. | Elia, A. E., et.al. | Higgins, R., et.al. | Kronke, J., et.al. | Fiskin, E., et.al. | Gendron, J. M., et.al. | Rose, C. M., et.al. | Sap, K. A., et.al. |
| Year | 2015 | 2015 | 2015 | 2015 | 2016 | 2016 | 2016 | 2017 |
|
Tissues/ Cells |
A20 cells | HeLa cells | HCT116 cells Drosophila S2 cells S. cerevisiae | KG-1 cells | human HCT116 and HeLa cells infected with Salmonella or left untreated | HCT116 cells | HTC116 cells, mice tissues | Drosophila melanogaster Schneider’s line 2 cells |
| Labeling | SILAC | SILAC | SILAC | SILAC | SILAC | SILAC | Ten-plex tandem mass tags (TMTs) | SILAC |
| Ub-sites Identified | Quantified 6,059 | 33,500 | 5,893 | 13,061 | 11,606 | 9,000 | 16,842 in HCT116 cells, 16,925 in mice tissues | 14,018 |
| Ub-Proteins Identified | 3,824 | 3,000 | ||||||
| IP/Pre-IP Enrichment | diGLY-antibody IP (Lucerna, clone GX41) | 1) SCX (for nuclear extracts) 2) diGLY-antibody IP (Cell Signaling Technology) | diGLY antibody IP (Cell Signaling Technologies) | 1) Basic pH reversed-phase separation pre-IP 2) PTMScan Ubiquitin Remnant Antibody Beads (Cell Signaling Technology) |
diGLY antibody IP (Cell Signaling Technology) | 1) Cell fractionation using differential centrifugation, 2) diGLY antibody IP (Cell Signaling Technology) | 1) diGLY antibody IP (Cell Signaling Technology) 2) Basic pH reversed-phase separation | 1) Fractionation by HILIC, 2) PTMScan Ubiquitin Remnant Antibody Beads (Cell Signaling Technology) |
| PostEnrichment fractionation | SCX | |||||||
| Remarks | Study illustrated the power of multilayered proteomic analyses for discovering novel BCR signaling. | Systematically examined alterations to the ub-modified proteome upon induction of DNA damage responses. | Study identified the evolutionarily conserved, site-specific regulatory ubiquitylation of 40S ribosomal proteins upon UPR activation and translation inhibition. | Study demonstrate that lenalidomide induces the ubiquitination of casein kinase 1A1 (CK1a) by the E3 ubiquitin ligase CUL4-RBX1-DDB1-CRBN | Study observed ~5%−10% of all quantified diGLY sites being over 2-fold regulated, corresponding to Salmonella invasion-induced changes in multiple cellular processes. | Study demonstrated that combining ub-proteomics with subcellular fractionation can effectively separate degradative and regulatory ubiquitylation events on distinct protein populations | Study reports TMT-multiplexing strategy to quantify ubiquitin-modified peptides and reveals PINK1- and PARKIN- dependent ubiquitylation events during early and late mitophagy | Study reports ub- modified proteome changes upon proteasome inactivation both by chemical inhibitors and by dsRNA-mediated knockdown of specific subunits in Drosophila S2 cells |
Acknowledgements
We thank Marilyn Leonard and Danielle Garshott for providing a critical reading of this manuscript and Ruoyu (Lulu) Li for assistance in making figure 1. This work was supported the NIH (DP2-GM119132, PGM085764) (E.J.B).
5. References
- 1.Mann M, Ong S-E, Grønborg M, Steen H, Jensen ON, Pandey A (2002) Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends in biotechnology 20 (6):261–268 [DOI] [PubMed] [Google Scholar]
- 2.Steen H, Küster B, Fernandez M, Pandey A, Mann M (2001) Detection of tyrosine phosphorylated peptides by precursor ion scanning quadrupole TOF mass spectrometry in positive ion mode. Analytical chemistry 73 (7):1440–1448 [DOI] [PubMed] [Google Scholar]
- 3.Thalassinos K, Grabenauer M, Slade SE, Hilton GR, Bowers MT, Scrivens JH (2008) Characterization of phosphorylated peptides using traveling wave-based and drift cell ion mobility mass spectrometry. Analytical chemistry 81 (1):248–254 [DOI] [PubMed] [Google Scholar]
- 4.Carr SA, Huddleston MJ, Annan RS (1996) Selective detection and sequencing of phosphopeptides at the femtomole level by mass spectrometry. Analytical biochemistry 239 (2):180–192 [DOI] [PubMed] [Google Scholar]
- 5.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM (2002) Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nature biotechnology 20 (3):301–305 [DOI] [PubMed] [Google Scholar]
- 6.Ptacek J, Devgan G, Michaud G, Zhu H (2005) Global analysis of protein phosphorylation in yeast. Nature 438 (7068):679. [DOI] [PubMed] [Google Scholar]
- 7.Zhang H, Xiao-jun L, Martin DB, Aebersold R (2003) Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nature biotechnology 21 (6):660. [DOI] [PubMed] [Google Scholar]
- 8.Dell A, Morris HR (2001) Glycoprotein structure determination by mass spectrometry. Science 291 (5512):2351–2356 [DOI] [PubMed] [Google Scholar]
- 9.Apweiler R, Hermjakob H, Sharon N (1999) On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochimica et Biophysica Acta (BBA)-General Subjects 1473 (1):4–8 [DOI] [PubMed] [Google Scholar]
- 10.Brownlee M, Michael (1995) Advanced protein glycosylation in diabetes and aging. Annual review of medicine 46 (1):223–234 [DOI] [PubMed] [Google Scholar]
- 11.Spiro RG (2002) Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12 (4):43R–56R [DOI] [PubMed] [Google Scholar]
- 12.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP (2003) A proteomics approach to understanding protein ubiquitination. Nat Biotechnol 21 (8):921–926. doi: 10.1038/nbt849 [DOI] [PubMed] [Google Scholar]
- 13.Scheffner M, Nuber U, Huibregtse JM (1995) Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature 373 (6509):81. [DOI] [PubMed] [Google Scholar]
- 14.Kirkpatrick DS, Denison C, Gygi SP (2005) Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics. Nat Cell Biol 7 (8):750–757. doi: 10.1038/ncb0805-750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu P, Peng J (2006) Dissecting the ubiquitin pathway by mass spectrometry. Biochim Biophys Acta 1764 (12):1940–1947. doi: 10.1016/j.bbapap.2006.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J (2009) Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137 (1):133–145. doi: 10.1016/j.cell.2009.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bennett EJ, Rush J, Gygi SP, Harper JW (2010) Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell 143 (6):951–965. doi: 10.1016/j.cell.2010.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu G, Paige JS, Jaffrey SR (2010) Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat Biotechnol 28 (8):868–873. doi: 10.1038/nbt.1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW, Gygi SP (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell 44 (2):325–340. doi: 10.1016/j.molcel.2011.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, Choudhary C (2011) A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics 10 (10):M111. 10.1074/mcp.M111.013284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH (2001) Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nature cell biology 3 (2):193. [DOI] [PubMed] [Google Scholar]
- 22.Clarke S (1993) Protein methylation. Current opinion in cell biology 5 (6):977–983 [DOI] [PubMed] [Google Scholar]
- 23.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325 (5942):834–840 [DOI] [PubMed] [Google Scholar]
- 24.Ichimura Y, Kirisako T, Takao T, Satomi Y (2000) A ubiquitin-like system mediates protein lipidation. Nature 408 (6811):488. [DOI] [PubMed] [Google Scholar]
- 25.Mayor T, Deshaies RJ (2005) Two-step affinity purification of multiubiquitylated proteins from Saccharomyces cerevisiae. Methods Enzymol 399:385–392. doi: 10.1016/S0076-6879(05)99026-5 [DOI] [PubMed] [Google Scholar]
- 26.Aillet F, Lopitz-Otsoa F, Hjerpe R, Torres-Ramos M, Lang V, Rodríguez MS (2012) Isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. Ubiquitin Family Modifiers and the Proteasome: Reviews and Protocols:173–183 [DOI] [PubMed] [Google Scholar]
- 27.Iconomou M, Saunders DN (2016) Systematic approaches to identify E3 ligase substrates. Biochem J 473 (22):4083–4101. doi: 10.1042/BCJ20160719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beaudette P, Popp O, Dittmar G (2016) Proteomic techniques to probe the ubiquitin landscape. Proteomics 16 (2):273–287. doi: 10.1002/pmic.201500290 [DOI] [PubMed] [Google Scholar]
- 29.Ordureau A, Munch C, Harper JW (2015) Quantifying ubiquitin signaling. Mol Cell 58 (4):660–676. doi: 10.1016/j.molcel.2015.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldknopf IL, Busch H (1977) Isopeptide linkage between nonhistone and histone 2A polypeptides of chromosomal conjugate-protein A24. Proceedings of the National Academy of Sciences 74 (3):864–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bustos D, Bakalarski CE, Yang Y, Peng J, Kirkpatrick DS (2012) Characterizing ubiquitination sites by peptide-based immunoaffinity enrichment. Mol Cell Proteomics 11 (12):1529–1540. doi: 10.1074/mcp.R112.019117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sylvestersen KB, Young C, Nielsen ML (2013) Advances in characterizing ubiquitylation sites by mass spectrometry. Curr Opin Chem Biol 17 (1):49–58. doi: 10.1016/j.cbpa.2012.12.009 [DOI] [PubMed] [Google Scholar]
- 33.Carrano AC, Bennett EJ (2013) Using the ubiquitin-modified proteome to monitor protein homeostasis function. Molecular & Cellular Proteomics 12 (12):3521–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Povlsen LK, Beli P, Wagner SA, Poulsen SL, Sylvestersen KB, Poulsen JW, Nielsen ML, Bekker-Jensen S, Mailand N, Choudhary C (2012) Systems-wide analysis of ubiquitylation dynamics reveals a key role for PAF15 ubiquitylation in DNA-damage bypass. Nat Cell Biol 14 (10):1089–1098. doi: 10.1038/ncb2579 [DOI] [PubMed] [Google Scholar]
- 35.Udeshi ND, Mani D, Eisenhaure T, Mertins P, Jaffe JD, Clauser KR, Hacohen N, Carr SA (2012) Methods for quantification of in vivo changes in protein ubiquitination following proteasome and deubiquitinase inhibition. Molecular & Cellular Proteomics 11 (5):148–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ng AH, Fang NN, Comyn SA, Gsponer J, Mayor T (2013) System-wide analysis reveals intrinsically disordered proteins are prone to ubiquitylation after misfolding stress. Mol Cell Proteomics 12 (9):2456–2467. doi: 10.1074/mcp.M112.023416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iesmantavicius V, Weinert BT, Choudhary C (2014) Convergence of ubiquitylation and phosphorylation signaling in rapamycin-treated yeast cells. Molecular & Cellular Proteomics 13 (8):1979–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kronke J, Udeshi ND, Narla A, Grauman P, Hurst SN, McConkey M, Svinkina T, Heckl D, Comer E, Li X, Ciarlo C, Hartman E, Munshi N, Schenone M, Schreiber SL, Carr SA, Ebert BL (2014) Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 343 (6168):301–305. doi: 10.1126/science.1244851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elia AE, Boardman AP, Wang DC, Huttlin EL, Everley RA, Dephoure N, Zhou C, Koren I, Gygi SP, Elledge SJ (2015) Quantitative Proteomic Atlas of Ubiquitination and Acetylation in the DNA Damage Response. Mol Cell 59 (5):867–881. doi: 10.1016/j.molcel.2015.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Higgins R, Gendron JM, Rising L, Mak R, Webb K, Kaiser SE, Zuzow N, Riviere P, Yang B, Fenech E, Tang X, Lindsay SA, Christianson JC, Hampton RY, Wasserman SA, Bennett EJ (2015) The Unfolded Protein Response Triggers Site-Specific Regulatory Ubiquitylation of 40 S Ribosomal Proteins. Mol Cell 59 (1):35–49. doi: 10.1016/j.molcel.2015.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kronke J, Fink EC, Hollenbach PW, MacBeth KJ, Hurst SN, Udeshi ND, Chamberlain PP, Mani DR, Man HW, Gandhi AK, Svinkina T, Schneider RK, McConkey M, Jaras M, Griffiths E, Wetzler M, Bullinger L, Cathers BE, Carr SA, Chopra R, Ebert BL (2015) Lenalidomide induces ubiquitination and degradation of CK1alpha in del(5q) MDS. Nature 523 (7559):183–188. doi: 10.1038/nature14610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gendron JM, Webb K, Yang B, Rising L, Zuzow N, Bennett EJ (2016) Using the Ubiquitin-modified Proteome to Monitor Distinct and Spatially Restricted Protein Homeostasis Dysfunction. Mol Cell Proteomics 15 (8):2576–2593. doi: 10.1074/mcp.M116.058420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sundaramoorthy E, Leonard M, Mak R, Liao J, Fulzele A, Bennett EJ (2017) ZNF598 and RACK1 Regulate Mammalian Ribosome-Associated Quality Control Function by Mediating Regulatory 40 S Ribosomal Ubiquitylation. Mol Cell 65 (4):751–760 10.1016/j.molcel.2016.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emanuele MJ, Elia AE, Xu Q, Thoma CR, Izhar L, Leng Y, Guo A, Chen YN, Rush J, Hsu PW, Yen HC, Elledge SJ (2011) Global identification of modular cullin-RING ligase substrates. Cell 147 (2):459–474. doi: 10.1016/j.cell.2011.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee KA, Hammerle LP, Andrews PS, Stokes MP, Mustelin T, Silva JC, Black RA, Doedens JR (2011) Ubiquitin ligase substrate identification through quantitative proteomics at both the protein and peptide levels. J Biol Chem 286 (48):41530–41538. doi: 10.1074/jbc.M111.248856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarraf SA, Raman M, Guarani-Pereira V, Sowa ME, Huttlin EL, Gygi SP, Harper JW (2013) Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 496 (7445):372–376. doi: 10.1038/nature12043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fang NN, Chan GT, Zhu M, Comyn SA, Persaud A, Deshaies RJ, Rotin D, Gsponer J, Mayor T (2014) Rsp5/Nedd4 is the major ubiquitin ligase that targets cytosolic misfolded proteins upon heat-stress. Nature cell biology 16 (12):1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tong Z, Kim M-S, Pandey A, Espenshade PJ (2014) Identification of candidate substrates for the Golgi Tul1 E3 ligase using quantitative diGly proteomics in yeast. Molecular & Cellular Proteomics 13 (11):2871–2882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garzia A, Jafarnejad SM, Meyer C, Chapat C, Gogakos T, Morozov P, Amiri M, Shapiro M, Molina H, Tuschl T (2017) The E3 ubiquitin ligase and RNA-binding protein ZNF598 orchestrates ribosome quality control of premature polyadenylated mRNAs. Nature Communications 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Na CH, Jones DR, Yang Y, Wang X, Xu Y, Peng J (2012) Synaptic protein ubiquitination in rat brain revealed by antibody-based ubiquitome analysis. J Proteome Res 11 (9):4722–4732. doi: 10.1021/pr300536k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagner SA, Beli P, Weinert BT, Scholz C, Kelstrup CD, Young C, Nielsen ML, Olsen JV, Brakebusch C, Choudhary C (2012) Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Mol Cell Proteomics 11 (12):1578–1585. doi: 10.1074/mcp.M112.017905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buckley SM, Aranda-Orgilles B, Strikoudis A, Apostolou E, Loizou E, Moran-Crusio K, Farnsworth CL, Koller AA, Dasgupta R, Silva JC, Stadtfeld M, Hochedlinger K, Chen EI, Aifantis I (2012) Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell 11 (6):783–798. doi: 10.1016/j.stem.2012.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rose CM, Isasa M, Ordureau A, Prado MA, Beausoleil SA, Jedrychowski MP, Finley DJ, Harper JW, Gygi SP (2016) Highly Multiplexed Quantitative Mass Spectrometry Analysis of Ubiquitylomes. Cell Syst 3 (4):395–403 10.1016/j.cels.2016.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sap KA, Bezstarosti K, Dekkers DHW, Voets O, Demmers JAA (2017) Quantitative Proteomics Reveals Extensive Changes in the Ubiquitinome after Perturbation of the Proteasome by Targeted dsRNA-Mediated Subunit Knockdown in Drosophila. J Proteome Res 16 (8):2848–2862. doi: 10.1021/acs.jproteome.7b00156 [DOI] [PubMed] [Google Scholar]
- 55.Udeshi ND, Svinkina T, Mertins P, Kuhn E, Mani D, Qiao JW, Carr SA (2013) Refined preparation and use of anti-diglycine remnant (K-ε-GG) antibody enables routine quantification of 10,000 s of ubiquitination sites in single proteomics experiments. Molecular &Cellular Proteomics 12 (3):825–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swaney DL, Beltrao P, Starita L, Guo A, Rush J, Fields S, Krogan NJ, Villén J (2013) Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nature methods 10 (7):676–682 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mertins P, Qiao JW, Patel J, Udeshi ND, Clauser KR, Mani DR, Burgess MW, Gillette MA, Jaffe JD, Carr SA (2013) Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat Methods 10 (7):634–637. doi: 10.1038/nmeth.2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Theurillat JP, Udeshi ND, Errington WJ, Svinkina T, Baca SC, Pop M, Wild PJ, Blattner M, Groner AC, Rubin MA, Moch H, Prive GG, Carr SA, Garraway LA (2014) Prostate cancer. Ubiquitylome analysis identifies dysregulation of effector substrates in SPOP-mutant prostate cancer. Science 346 (6205):85–89. doi: 10.1126/science.1250255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Villen J, Gygi SP (2008) The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc 3 (10):1630–1638. doi: 10.1038/nprot.2008.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Udeshi ND, Mertins P, Svinkina T, Carr SA (2013) Large-scale identification of ubiquitination sites by mass spectrometry. Nat Protoc 8 (10):1950–1960. doi: 10.1038/nprot.2013.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rappsilber J, Mann M, Ishihama Y (2007) Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nature protocols 2 (8):1896. [DOI] [PubMed] [Google Scholar]
- 62.Satpathy S, Wagner SA, Beli P, Gupta R, Kristiansen TA, Malinova D, Francavilla C, Tolar P, Bishop GA, Hostager BS, Choudhary C (2015) Systems-wide analysis of BCR signalosomes and downstream phosphorylation and ubiquitylation. Mol Syst Biol 11 (6):810. doi: 10.15252/msb.20145880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beltrao P, Albanese V, Kenner LR, Swaney DL, Burlingame A, Villen J, Lim WA, Fraser JS, Frydman J, Krogan NJ (2012) Systematic functional prioritization of protein posttranslational modifications. Cell 150 (2):413–425. doi: 10.1016/j.cell.2012.05.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thompson JW, Nagel J, Hoving S, Gerrits B, Bauer A, Thomas JR, Kirschner MW, Schirle M, Luchansky SJ (2014) Quantitative Lys--Gly-Gly (diGly) proteomics coupled with inducible RNAi reveals ubiquitin-mediated proteolysis of DNA damage-inducible transcript 4 (DDIT4) by the E3 ligase HUWE1. J Biol Chem 289 (42):28942–28955. doi: 10.1074/jbc.M114.573352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fiskin E, Bionda T, Dikic I, Behrends C (2016) Global Analysis of Host and Bacterial Ubiquitinome in Response to Salmonella Typhimurium Infection. Mol Cell 62 (6):967–981. doi: 10.1016/j.molcel.2016.04.015 [DOI] [PubMed] [Google Scholar]