Abstract
Endoplasmic reticulum glycoprotein folding quality control (ERQC) and ER-associated degradation (ERAD) preside over cellular glycoprotein secretion and maintain steady glycoproteostasis. When cells turn malignant, cancer cell plasticity is affected and supported either by point mutations, preferential isoform selection, altered expression levels, or shifts to conformational equilibria of a secreted glycoprotein. Such changes are crucial in mediating altered extracellular signalling, metabolic behavior, and adhesion properties of cancer cells. It is therefore conceivable that interference with ERQC and/or ERAD can be used to selectively damage cancers. Indeed, inhibitors of the late stages of ERAD are already in the clinic against cancers such as multiple myeloma. Here, we review recent advances in our understanding of the complex relationship between glycoproteostasis and cancer biology and discuss the potential of ERQC and ERAD modulators for the selective targeting of cancer cell plasticity.
1. Introduction
Plasticity, an intrinsic characteristic of healthy cells in biological contexts as varied as embryonal development [1], tissue development and repair [2], adaptation to injury [3], and wound healing [4], is also central to cancer initiation, progression, and metastasis. The proteins establishing and maintaining cancer plasticity are good anticancer drug targets in the fight against cancer initiation, progression, and therapy resistance itself [5]. Plasticity of cancer cells relies heavily on glycoproteins that traverse the secretory pathway, such as cell surface receptors and signalling molecules released in the extracellular medium [6, 7]. These secreted glycoproteins respond to and steer changes in the surroundings of a cancer cell, and contribute to tumour immunity [8], tumour growth and cancer cell division, adhesion and metastasis.
The reliance of cancer cells on secreted glycoproteins begs the question as to whether the endoplasmic reticulum glycoprotein folding quality control (ERQC) and/or endoplasmic reticulum associated degradation (ERAD) systems (together with the parallel misfolding-associated protein secretion system, MAPS [9]) could constitute potential anti-cancer targets. It is conceivable that ERQC/ERAD would make attractive targets for the treatment of cell malignancies [10], in that the fitness of the cancer cells, particularly those bearing a high secretory burden such as multiple myeloma cells [11], is critically dependent on functional integrity of the endoplasmic reticulum (ER), which in turn relies on ERQC/ERAD as ER stress-attenuating mechanisms.
The therapeutic value of pharmacological chaperones (small molecules specifically stabilising a misfolded glycoprotein as it traverses the ER) is already well established in a number of congenital glycoprotein misfolding endocrine and metabolic disorders [12], further supporting the idea that therapeutic modulation of ER glycoprotein folding and degradation systems could also be successfully applied to cancer treatment, at least in cases where ERQC-assisted glycoprotein folding and ERAD play a major role.
Importantly, while pharmacological chaperones are designed to bind individual misfolded glycoproteins, any drug targeting a specific ERQC/ERAD component would affect folding of all glycoproteins that are dependent on it for their folding/degradation. Given the unique and central role of ERQC/ERAD in the fate of hundreds of secreted glycoproteins, and remembering that plasticity of different cancers depends on different subsets of secreted glycoproteins, ERQC/ERAD modulating drugs may have the potential to represent broad-spectrum anti-cancer agents.
Of course, like any strategy aimed at inhibition/modulation of basic cell housekeeping machineries, molecules developed to interfere with ERQC/ERAD have the potential to be toxic to healthy cells as well as cancerous ones. In addition, ERQC/ERAD inhibition could lead to increased levels of prematurely secreted misfolded glycoproteins (a scenario akin to the opening of an “ER Pandora's box”).
In this review article, we explore the evidence suggesting that the ability of cancer cells to create and spread tumours around the body, to resist current therapies, and to recur post-treatment, hinges vitally on ERQC/ERAD. We review our current understanding of how ERQC/ERAD preserve ER glycoproteostasis and discuss how we may harness the molecular detail so far established on these systems in order to develop new broad-spectrum anti-cancer therapeutics.
2. Materials and Methods
2.1. Homology Modelling
The HHPred server [13] was used to align the protein sequences with the ones of orthologues of known structure and create homology models with MODELLER [14]. The transmembrane helix of MmMOGS (mouse GCS1, UniProt Q80UM7, MOGS_MOUSE, residues 42–62) was homology modelled on PDB entry 1HH0, residues A20-A40. The C-terminal part of human calnexin (UniProt P27824, CALX_HUMAN, residues 461–484) was homology modelled using PDB ID 6A69, residues B223–B246. The C-terminal part of human Sep15 (UniProt O60613, SEP15_HUMAN, residues 46–134) was homology modelled using PDB ID 2A4H, residues A11-A99. The human ER UDPase (UniProt O75356, ENTP5_HUMAN, residues 22–404) was homology modelled using PDB ID 5U7W, residues A4–A412. The human UDP-Glc transporter (putatively identified as UniProt P78383, S35B1_HUMAN, residues 9–321; although a recently published paper reports ATP/ADP antiporter activity [15]) was homology modelled on the basis of PDB ID 5OGE, residues C16–C333. The ER lumenal domain of human EDEM1 (UniProt Q92611, EDEM1_HUMAN, residues 126–587) was homology modelled on the basis of PDB ID 1X9D A84-A535; its N-terminal transmembrane part, residues 1–34, was homology modelled on the basis of PDB ID 5MRW, residues E57–E90. The HRD1/HRD3 complex was modelled by docking the crystal structures (PDB IDs 5V6P and 5V7V) in the cryo-EM map for the complex (Electron Microscopy Data Bank ID EMD-8638 [16]), using Chimera [17]. All protein structure figures were made with PyMol [18].
2.2. ERQC/ERAD and Cancer
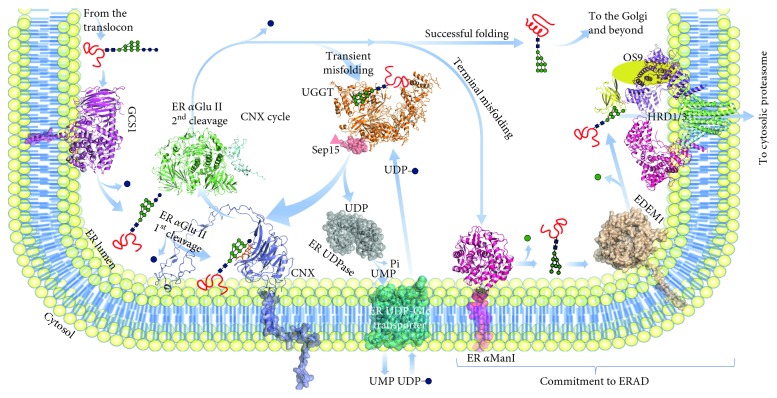
Glycoproteins traversing the secretory pathway of eukaryotic cells reach their cellular or extracellular destinations after folding in the ER [19]. To deal with the constant challenge of protein misfolding in the ER, eukaryotic cells have evolved the ERQC system, centred around the calnexin cycle [20]. Collectively, ERQC components (left-hand side of Figure 1) identify, retain in the ER, and aid folding of misfolded glycoproteins on the way down the secretory pathway. ERQC surveys glycoprotein folding, prevents premature glycoprotein secretion, and is integrated with the adaptive stress response [10]. ERQC proteins either reside in the ER lumen or are inserted in/associated with the ER membrane. A second ER-resident machinery called endoplasmic reticulum-associated degradation (ERAD, right-hand side of Figure 1) comprises proteins that commit terminally misfolded glycoproteins to demannosylation, retrotranslocation to the cytoplasm, and ubiquitination, ultimately targeting them to cytoplasmic proteasomes. Both ERQC and ERAD support cells in their effort to fine tune the rate of glycoprotein synthesis and entry into the ER to match the ER folding capacity (glycoprotein homeostasis or glycoproteostasis) [21].
Figure 1.
Structural view of ERQC/ERAD. The ER trajectory of the folding stages of a glycoprotein, bound to a glycan of variable composition, is followed through its interactions with ERQC/ERAD components, either ER-lumenal or ER-membrane associated. Proteins whose structures have been described in the literature for at least one eukaryotic species are in cartoon representation, while homology models of proteins of unknown structure are in semitransparent surface representation. Proteins for which no orthologue of known structure exists are represented as arbitrary 2D shapes. The red squiggly symbol represents the polypeptidic part of a glycoprotein on its way to folding (either incompletely folded, transiently misfolded, or terminally misfolded), while the red helix and two-stranded-β-sheet symbol represents a glycoprotein that has successfully attained its native fold. The symbols of the monosaccharides in the glycan are from [151]: blue squares, N-acetyl glucosamine (GlcNAc); green circles, mannose (Man); blue circles, glucose (Glc). The symbolic N-linked glycan on the glycoprotein is not to scale with the proteins, and the proteins themselves are not represented to exact relative scale to one another. The nascent glycoprotein in the upper-left corner has just been synthesised into the ER by a translocon-associated ribosome. After translation/translocation, the polypeptide has been glycosylated by oligosaccharyl-transferase (OST, not shown) and carries a GlcNAc2Man9Glc3 glycan. The nascent glycoprotein is first deglucosylated by GCS1 (purple, mouse GCS1, PDB ID 5MHF, on the left), which leaves a GlcNAc2Man9Glc2 glycan; a further deglucosylation step by ER αGluII (mouse GCS2, green and cyan, PDB ID 5F0E) produces a GlcNAc2Man9Glc1 glycan, through which the glycoprotein can associate with the lectin domain of either calnexin (CNX, indigo, dog calnexin, PDB ID 1JHN) or calreticulin (not shown) and the associated foldases/chaperones (not shown). A second cleavage by ER αGluII leaves a GlcNAc2Man9 glycan on the glycoprotein, which can bind no longer to the ER lectins and if folded is free to progress to the Golgi and beyond (upper right corner). If the glycoprotein fold is not native yet, the misfolded glycoprotein is recognised and reglucosylated by UGGT (orange, CtUGGT, PDB ID 5NV4), using UDP-Glc as a source of glucose. The resulting GlcNAc2Man9Glc1 glycoprotein can reassociate with the CNX/CRT to profit from further attempts at chaperone/foldase-assisted folding. The UGGT partner protein Sep15 is in salmon pink (triangle: Nterm domain; surface: Cterm domain). The UDP molecule produced by UGGT-mediated reglucosylation is hydrolysed by a ER UDPase (grey) to inorganic phosphate (Pi) and UMP—the latter being antiported back to the cytoplasm by an ER-membrane-resident ER UDP-Glc transporter (cyan), in exchange for a molecule of UDP-Glc. Terminally misfolded glycoproteins are first demannosylated by ER αManI (magenta, PDB ID 1X9D) and then by EDEM1 (wheat), to yield a GlcNAc2Man5–7 glycan. The mannose-6-phosphate receptor homology (MRH) domain of OS-9 (yellow) binds the demannosylated glycan and recruits the terminally misfolded glycoprotein to the ERAD retrotranslocon assembled around the HRD1/HRD3 ubiquitin ligase (magenta, violet, green and cyan, yeast structures, PDB IDs 5V6P and 5V7V, on the right). A cytoplasmic proteasome then degrades the terminally misfolded ubiquitinated glycoprotein (not shown).
Malignant cells are deprived of nutrients and their protein synthesis is dysregulated, so that they are especially prone to ER stress. The latter results from protein misfolding within the ER, and it has profound effects on cancer cells' proliferation and survival [22]. It is therefore not surprising that ERQC and ERAD play a key role in cancer biology. Yet, the complexity of ER glycoproteostasis, coupled with the galaxy of cancer cell phenotypes, makes it nontrivial to predict if the activity of a specific ERQC/ERAD component helps or hinders establishment and progression of a specific type of cancer. Indeed, ER quality control and degradation systems have been suggested to represent a double-edged sword that may aid progression as well as prevention of cancer cell growth in a context-dependent manner [23].
Table 1 lists a number of ERQC/ERAD components and association of their expression levels with cancer patient survival in the Human Protein Atlas (HPA) [24, 25], as evidenced by Kaplan–Meier survival plots [26] derived from cancer tissue images. Quite a few of these ERQC/ERAD components have been identified as unfavourable prognostic markers in cancer studies. We also list the frequency of somatic mutations detected in the same genes, as reported by the Catalogue of Somatic Mutations in Cancer (COSMIC), the world's largest source of manually curated somatic mutation information relating to human cancers [27]. Other useful resources are the database of therapeutic vulnerability of cancer [28], lists of oncogenes [29], and the tumour suppressor gene TUSON ranking [30] (https://bioinfo.uth.edu/TSGene/ [40, 41]) in the cancer cell metabolism genes database [31], but in the interest of simplicity, we did not compile values from these online sources in Table 1.
Table 1.
ERQC/ERAD components and association of their expression levels with cancer patient survival in the Human Protein Atlas (HPA) [24, 25].
| Protein/gene and UniProt entry/name | Prognosis upon overexpression (Human Protein Atlas, p < 0.001) | Frequency of somatic mutations in cancer (COSMIC) |
|---|---|---|
| GCS1/MOGS, Q13724/MOGS_HUMAN | Unfavourable prognosis in renal, liver, and colorectal cancers | 191/47211 (0.4%) |
| ER αGlu II α subunit/GANAB, Q14697/GANAB_HUMAN | Unfavourable prognosis in liver and urothelial cancers | 254/47211 (0.5%) |
| ER αGlu II β subunit/PRKCSH, P14314/GLU2B_HUMAN | Unfavourable prognosis in renal cancer | 191/47211 (0.4%) |
| UGGT1/UGGT1, Q9NYU2/UGGG1_HUMAN | Unfavourable prognosis in renal cancer | 333/47297 (0.7%) |
| UGGT2/UGGT2, Q9NYU1/UGGG2_HUMAN | Unfavourable prognosis in lung and liver cancers | 406/47212 (0.8%) |
| Sep15/Sep15, O60613/SEP15_HUMAN | Unfavourable prognosis in liver, head, and neck cancers but favourable prognosis in colorectal cancer | 17/47187 (0.04%)) |
| Calnexin/CANX, P27824/CALX_HUMAN | Favourable prognosis in colorectal cancer but unfavourable in thyroid cancer | 151/47211 (0.3%) |
| Calreticulin/CALR, P27797/CALR_HUMAN | Favourable prognosis in ovarian cancer but unfavourable in renal cancer | 4344/81169 (5.3%) |
| ER UDPase, O75356, ENTP5_HUMAN | Favourable prognosis in renal cancer | 110/47209 (0.2%) |
| ER αMan I, Q9UKM7, MA1B1_HUMAN | Unfavourable prognosis in liver cancer | 178/47354 (0.4%) |
| EDEM1, Q92611, EDEM1_HUMAN | N/A | 141/47255 (0.3%) |
| EDEM2, Q9BV94 EDEM2_HUMAN | Unfavourable prognosis in renal cancer | 162/35626 (0.4%) |
| EDEM3, Q9BZQ6 EDEM3_HUMAN | Unfavourable prognosis in renal cancer | 231/35629 (0.6%) |
| ERDJ5, Q8IXB1, DJC10_HUMAN | Favourable prognosis in endometrial cancer but unfavourable in renal and thyroid cancers | 205/47347 (0.4%) |
| HRD1, Q86TM6, SYVN1_HUMAN | Favourable prognosis in head and neck cancer | 143/47298 (0.3%) |
| OS-9/, Q13438/OS9_HUMAN | N/A | 188/47797 (0.4%) |
The HPA correlation (p < 0.001) between high levels of expression of the protein with the survival rates of cancer patients is reported, together with the frequency of somatic mutations detected in the same genes, as per the Catalogue of Somatic Mutations in Cancer (COSMIC) [27]. For comparison, the tumour suppressors TP53 and CDKN2A have mutation frequencies of 25% (40416/160297) and 6% (6067/100370), respectively.
In the following paragraphs, we briefly review some of the published evidence of direct cancer association for a selected subset of ERQC/ERAD components, before examining the second-order involvement of ERQC/ERAD with cancer, through their regulation of folding and degradation of specific cancer-associated secreted glycoproteins.
2.3. ERQC and Cancer
ER α-glucosidase I (GCS1, in purple on the left-hand side in Figure 1) directly interacts with subunits of the ER-membrane-associated oligosaccharyl transferase (OST) [30, 31], in agreement with what was observed for the yeast orthologues [32, 33]. GCS1 acts as the porter at the ERQC one-way entrance door, removing the outer glucose (Glc) residue from the Glc3Man9GlcNAc2 N-linked glycan transferred by OST to a nascent glycoprotein. With this cleavage, ER Glu I generates diglucosylated glycoproteins, i.e., glycoproteins carrying Glc2Man9GlcNAc2 N-linked glycans. This kind of glycan in turn is necessary for the first interaction with the second major ERQC player, ER αGlu II: without the ER αGlu I-mediated Glc cleavage, glycoproteins cannot interact with ER αGlu II nor enter ERQC [32, 33]. A direct role for diglucosylated glycans in ERQC has also been hypothesised in conjunction with malectin, the ER lectin that binds them specifically [34]. Genetic defects in MOGS, the gene encoding GCS1, cause rare congenital disorder of glycosylation type IIb (CDG-IIb) and confer decreased susceptibility to infections due to viruses whose life cycle depends on the host cell's calnexin cycle [35]. The Human Protein Atlas (HPA) [24, 25] reports unfavourable prognoses in human renal, liver, and colorectal cancers overexpressing the MOGS gene (see Table 1).
ER α-glucosidase II (ER αGlu II, in green and cyan in Figure 1) acts as an usher, mediating both entry and exit of a glycoprotein into the cycle [36]. Entry into ERQC is conditional on ER αGlu II-mediated removal of the terminal Glc from the Glc2Man9GlcNAc2 glycan, enabling recruitment of the resulting monoglucosylated glycoprotein to the ER lectins calnexin and calreticulin, and the associated oxido-reductases, isomerases, and foldases. The same ER αGlu II eventually removes the remaining Glc from the Glc1Man9GlcNAc2 glycan, preventing further association with the ER lectins, thus freeing a glycoprotein from the refolding end of ERQC [37]. The noncatalytic ER αGlu II β subunit likely mediates association with the client glycoprotein glycan via its C-terminal mannose 6-phosphate receptor homology (MRH) domain, and it contains the ER-retrieval motif localising ER αGlu II to the ER [38]. Overexpression of ER αGlu II β subunit (ER αGlu II β) in different tumour tissues has been reported [40, 41]. More recently, it has been suggested that activation of ERQC through ER αGlu II can help tumour cells to escape from autophagy and apoptosis [42]. A study of molecular chaperones regulating the invasion phenotype of head and neck cancer (HNC) established that loss of the tumour suppression function of the ER αGlu II α subunit contributed to aggressive cancers [39].
Calnexin (CNX, ER-membrane inserted, in violet in Figure 1) and calreticulin (CRT, ER lumenal and soluble) are ERQC lectins with a specificity for mono-glucosylated glycans (Glc1Man9GlcNAc2). They recruit monoglucosylated glycoproteins to oxido-reductases, isomerases, and foldases, effectively constituting the refolding end of the calnexin cycle. In one lung cancer study, low levels of CNX contributed to poor prognosis: in a cell-culture model, targeted depletion of calnexin reduced cancer proliferation, invasion, and migration [44]. CNX expression positively correlates with metastasis of breast cancer to the brain [45]. CNX was also significantly upregulated in oral squamous cell carcinoma, and its levels correlated with poor prognosis in patients affected by this tumour [46].
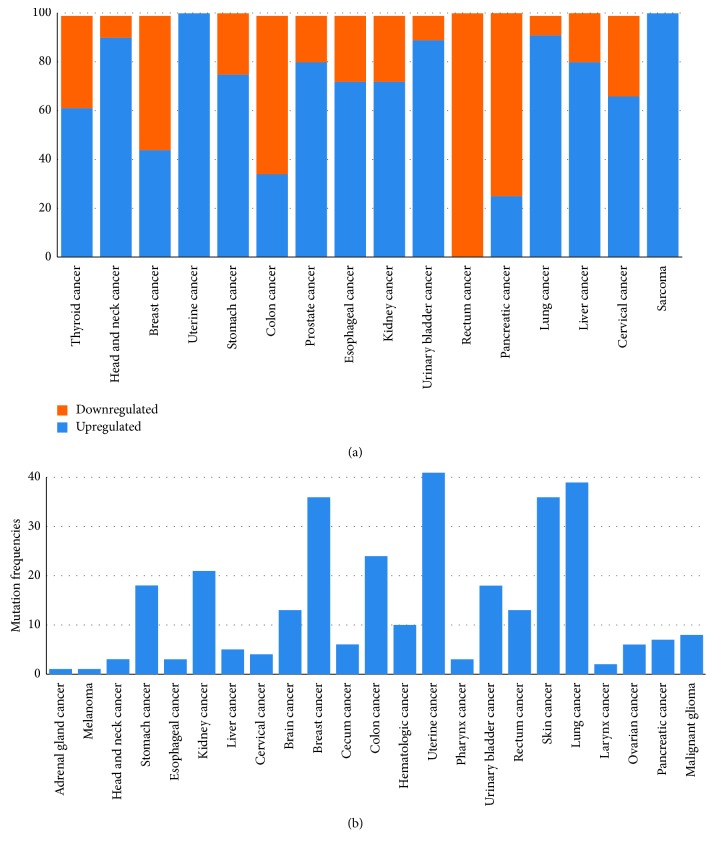
UGGT (UDP-glucose glycoprotein glucosyltransferase) is the ERQC checkpoint, detecting misfolded glycoproteins and reglucosylating them in order to enable further rounds of association with CNX/CRT, beyond the initial one(s) afforded by the OST-transferred N-glycan(s) after the initial ER αGlu II cleavages [40]. In higher vertebrates, there are two UGGT isoforms, UGGT1 and UGGT2. Although UGGT2 was initially reported not to reglucosylate UGGT1 misfolded glycoprotein clients [41], this isoform is also competent in reglucosylating synthetic glycoproteins carrying high-mannose glycans [42, 43], suggesting that UGGT1 and UGGT2 evolved to act on different subsets of glycoprotein clients. The mechanism by which UGGT recognises and selectively reglucosylates misfolded glycoproteins remains unclear. The observation that UGGT bears demannosylated glycans that are the hallmark of ERAD [44, 45] is compatible with the hypothesis that UGGT may recognise misfolded glycoproteins via an intrinsically misfolded domain (“it takes one to know one”), as observed for the mouse ERAD mannosidase [46]. Despite the centrality of UGGT to eukaryotic glycoprotein secretion, only a few bona fide UGGT glycoprotein clients are known [47–52], and the full lists of clients of the two isoforms (“UGGT-omes”) remain to be compiled. The Human Protein Atlas (HPA) [24, 25] reports unfavourable prognoses in renal cancers and lung and liver cancers overexpressing UGGT1 or UGGT2, respectively. A majority of cancers are reported to overexpress the UGGT1 gene (see Figure 2(a)), and a few cancer types report a significant rate of mutations in the same gene (see Figure 2(b))—although without functional data, it is difficult to assess if they are likely to impair or enhance protein function. No studies have directly tested the role of UGGT in cancer plasticity.
Figure 2.
(a) Percentage of upregulation and downregulation of the UGGT1 gene in a number of cancers from RNA-seq experiments on bulk cancer tissues. (b) Mutation frequencies in various cancer types in the same gene (from the Q9NYU2 entries in BioMuta and BioXpress at the OncoMX server [152]).
Sep15 (aka Selenoprotein F, Selenof) is a 15 kDa protein which in humans (but not in fruit fly, mosquito, zebrafish or rat) contains a selenocysteine residue [53]. Selenium has been implicated in cancer prevention [54], but the mechanism and possible involvement of selenoproteins in this process are not well understood. Based on the fact that abnormal glycoprotein folding and secretion were observed in conjunction with Sep15 deficiency, it has been proposed that it may have an important role in the ER maturation of N-glycosylated proteins [55], in particular M-immunoglobulins [56]. Sep15 mitigates oxidative stress and apoptosis [57]. Its C-terminal domain (residues 46–134) folds as a thioredoxin-like domain [58]; the N-terminal domain (residues 1–45), whose fold is not easily predictable from sequence, likely mediates Sep15 nanomolar association with UGGT1 [59]. Indeed, Sep15 enhances UGGT1-mediated reglucosylation of IL-8 and crambin containing mispaired disulphides [42, 43], suggesting that the Sep15 redox potential may have evolved to selectively reduce/isomerise disulphides in nonnative over native environments. A number of studies point to a role of Sep15 in cancer aetiology. The Sep15 coding gene is located in a highly mutated region of chromosome 1, and several mutations and deletions of Sep15 coding gene are involved in cancer progression and tumorigenesis [53]. The expression levels of Sep15 were investigated in various cancer models: downregulation of the protein was found in hepatocarcinomas and colorectal, gastric, and prostate cancers [53, 54, 60, 61]. On the other hand, decreased expression of Sep15 reduces proliferation and growth of liver and colon cancer cell lines, pointing to a role of Sep15 in tumour progression [60, 62–64]. Single-nucleotide polymorphisms in the Sep15 gene have been studied in conjunction with differential levels of selenocysteine insertion [65] and susceptibility to lung and breast cancer [66–68], highlighting the need for a stratified medicine approach in the development of Sep15 modulators as anticancer therapeutics.
Supply of UDP-glucose to the ER is thought to be mediated by an ER-transmembrane UDP-Glc/UMP antiporter (in cyan in Figure 1), in analogy with other sugar nucleotides synthesised in the cytoplasm and transported to the ER or to the Golgi by specific antiporters. Sugar nucleotide/nucleotide monophosphate antiporters (or nucleotide sugar transporters, NST for short) are a subclass of the solute carrier transporter family of molecules that have been proposed as potential targets for digestive system neoplasms [69]. Until recently, and on the basis of sequence homology to known NSTs [70], the putative gene encoding the human UDP-Glc/UMP antiporter was the solute carrier family 35 member B1 aka SLC35B1 or UGTrel1 (UniProt P78383, S35B1_HUMAN). Intriguingly, deletion of the ER-localised members of the NST family in Schizosaccharomyces pombe produces phenotypes similar to the deletion of the UGGT gene, but even when combined with disruption of all known NST genes whose products have an unknown location, loss of genes encoding known ER NSTs did not obliterate UDP-Glc ER entrance [71]. Last year, a study characterised SLC35B1 as an ATP/ADP antiporter [15]. These observations combined now support the hypothesis that UDP-Glc entrance into the yeast ER may not follow the classical NST antiport mechanism.
Whichever the source of ER UDP-glucose, once UGGT has transferred a Glc molecule from UDP-Glc to a misfolded glycoprotein glycan, a molecule of UDP is produced, which would inhibit UGGT [72]. As is the case for other nucleoside diphosphates produced by sugar transferases [73], an ER-specific UDPase (NTPD5, UniProt O75356, ENTP5_HUMAN, in grey in Figure 1) hydrolyses the ER UDP pool to UMP [72, 74]. NTPD5 may mediate some of the cancer-related phenotypes associated with AKT1 activation: NTPD5 is upregulated in cell lines and primary human tumor samples with active AKT and, together with cytidine monophosphate kinase-1 and adenylate kinase-1, is part of an ATP hydrolysis cycle that converts ATP to AMP, resulting in the cancer-associated compensatory increase in aerobic glycolysis known as the Warburg effect [75]. Many studies have correlated dysregulation of the expression of the ER UDPase with a range of cancers, explaining why the enzyme has been proposed as a potential target for anticancer therapy [76–80].
2.4. ERAD in Cancer
Just as the N-linked glycan is used by ERQC to add/remove the glucose whose presence/absence marks a misfolded glycoprotein for ER retention/progression to the Golgi, ERAD mannosidases remove mannose residues from the N-linked glycan, flagging a terminally misfolded glycoprotein for degradation [81]. In particular, trimming of N-glycans by ERAD mannosidases generates Man6GlcNAc2 and Man5GlcNAc2 (M6 and M5) glycans, with three main consequences [82]: (i) removal of the outer Man residues on branch A precludes reentry of the glycoprotein molecule in the calnexin cycle; (ii) the trimmed M5-6 structures bind to the lectins OS-9 and XTP-3B [83], targeting the glycoprotein to retrotranslocation by the SEL1L/HRD1 ERAD dislocon complex; and (iii) the trimmed species are selected against ER-to-Golgi transport [84]. Unlike ERQC, where the glucose residue can be put back on the N-linked glycan by UGGT and the cycle glucose-on/glucose-off repeated, no ERAD mannosyl-transferase is known, so after the first steps of ERAD-mediated demannosylation, a glycoprotein is irretrievably dispatched to degradation [85].
Correct identification of misfolded secretory glycoproteins and their degradation by ERAD are crucial for cellular health and survival. ERAD processing is not stochastic: ERAD glycan trimming is selectively accelerated on misfolded glycoprotein [82]. Without functional ERAD, misfolded glycoproteins accumulate, the ER is stressed, and the unfolded protein response (UPR) ensues. While the early UPR response tries to increase the production of molecular chaperones involved in protein folding, prolonged stress activates UPR arms steering the cell towards apoptosis.
High growth rate, impaired ATP generation, hypoxia, hypoglycemia, and specific mutations perturb cancer cells' ER homeostasis [86, 87] and may also induce UPR [88]. This in turn can lead to cell death. ERAD unwittingly (but effectively) helps cancer cells by conferring them tolerance to glycoproteotoxic stress. Indeed, survival under chronic ER stress is a feature of aggressive cancers [89], and tumour cells attempt survival by hijacking ERAD [90]. For these reasons, terminal ERAD components inhibitors have been proposed as targets to specifically impair the survival of cancer cells [22, 91]. Blocking ERAD can also trigger cellular apoptosis [92].
The ERAD components acting early in the pathway are the endoplasmic reticulum degradation-enhancing mannosidases (EDEM), committing misfolded glycoproteins to degradation. To date, no EDEM-specific inhibitors are known, and the effects of EDEM inhibition/deletion on cancer cells have not been investigated, although the generic α-mannosidase inhibitor kifunensine [93] increased adhesion of breast cancer cells to endothelial cells [94] and 1-deoxymannojirimycin (another broad-spectrum mannosidase inhibitor) induced cellular ER stress in a human hepatocarcinoma cell line [95].
ER mannosyl-oligosaccharide 1,2-α-mannosidase (ER αMan I, UniProt Q9UKM7, MA1B1_HUMAN, in pink in Figure 1) is an 80 kDa enzyme with a short cytoplasmic tail, a single transmembrane α helix localising it to the ER membrane, and an ER lumenal mannosidase domain, initially believed to selectively remove only the middle arm terminal α(1,2)-linked D-mannose residue from the oligomannose Man9GlcNAc2 N-linked glycan [96], for which it has an affinity of 0.4 mM [97]. More recent in vitro and in cellula data highlight that ER αMan I can in fact remove all four α(1,2)-linked D-mannose residues from the glycan, although it does have a preference for the one on arm B [82, 98–100]. A crystal structure of human ER αMan I in complex with a glycan has revealed the structural basis for its substrate recognition and catalysis [101]. A conserved motif within the 3′UTR of ER αMan I is a target of miR-125b, a microRNA frequently downregulated in numerous types of cancers, including hepatocellular carcinoma (HCC), with the expression of ERManI significantly elevated in HCC, as measured by immunohistochemistry in a liver disease spectrum tissue microarray [102].
ER ManIA aka mannosyl-oligosaccharide 1,2-alpha-mannosidase IA (ER ManIA, UniProt P33908, MA1A1_HUMAN)— originally annotated as resident in the Golgi—has been shown to colocalise with ER αMan I in quality control vesicles (QCVs) and is also implicated in targeting to ERAD [103]. In cancer, the enzyme levels showed impact on degradation of cell-surface glycoprotein involved in cell-cell adhesion and metastasis: reduced ER ManIA expression or mannosidase inhibition lead to a significantly increased adhesion of breast cancer cells to endothelial cells [94]. Conversely, ER ManIA was downregulated in metastatic hepatocellular cancer (HCC) cell lines and orthotopic xenograft tumours, in comparison with nonmetastatic HCC controls [104].
ER degradation enhancing mannosidases (EDEMs) target misfolded glycoproteins for degradation [105] by cleaving α(1,2) mannoses from the glycan and exposing Man α(1,6) bonded residues [106]. There are two degradation-enhancing α(1,2) mannosidases (MNS4 and MNS5) in Arabidopsis thaliana [107] and three EDEMs (EDEM1, 2 and 3) in mammals. Human EDEM1 (UniProt Q92611, EDEM1_HUMAN, in wheat brown in Figure 1) is a 74 kDa enzyme inserted in the ER membrane via an N-terminal trans-membrane helix. EDEM3 (UniProt Q9BZQ6, EDEM3_HUMAN) is also ER-localised, because it carries an ER retrieval sequence at its C-terminus. EDEM2 (UniProt Q9BV94, EDEM2_HUMAN) lacks both an ER retrieval sequence and a transmembrane region [81] so its ER localisation is less certain [108]. EDEM1 overexpression can trigger ERAD in absence of ER αMan I [109]. Unlike ER αMan I, which is active even on isolated glycans, EDEM1 is more active on misfolded human glycoprotein substrates [109–112], similarly to what was observed in yeast [113, 114]. A mouse EDEM1 N-terminal region predicted to be intrinsically disordered accelerates ERAD of tyrosinase misfolded mutants [46], suggesting that misfold can be used to recognise misfold (again, as may be the case for UGGT, one hypothesis is that “it takes one to know one”). In agreement with this model is the observation that EDEM1 may be itself subjected to ERAD [115]. The Human Protein Atlas (HPA) [24, 25] reports unfavourable prognoses in renal cancers overexpressing EDEM2 and EDEM3. A somatic variant of EDEM1 (N198I), which loses one of its five N-linked glycans, was found to confer a selective advantage to hepatocellular carcinoma cells [116].
In mouse, EDEM1 has been found in association with the ERDJ5 protein disulfide isomerase (PDI) [117, 118], and the same interaction was observed between human EDEM1 and the human ERAD PDIs ERDJ5 [119] and TXNDC11 [112]. To date, no structure of a EDEM : PDI complex exists. It is likely that ERAD PDIs thioredoxin-like (TRXL) domains confer them the ability to process misfolded glycoproteins in the presence of nonnative disulfide bridges: if this is the case, ERAD PDI-mediated reduction of nonphysiological disulfide bridges may help the retrograde transport of misfolded substrates through the retrotranslocation channel [117]. High levels of ERDJ5 and TXNDC11 are unfavourable prognostic markers in renal and thyroid cancers and glioma, respectively [24, 25]. Knockdown of ERDJ5 by RNA interference in neuroectodermal tumour cells increases the apoptotic response to fenretinide [120]. These data make the case for selective ERDJ5 and/or TXNDC11 modulators as novel chemotherapeutic targets. On the other hand, high levels of TXNDC11 or ERDJ5 were a favourable prognostic marker in endometrial cancer [24, 25], and overexpression of ERDJ5 sensitizes neuroblastoma cells to ER stress-induced apoptosis [121], so it is clear that inhibiting this PDI may not work against some cancers. Similar results were observed for TXNDC11, whose elevated levels of expression correlated with suppression of tumour-promoting genes [122].
Once demannosylated by EDEMs, misfolded glycoproteins in the ER lumen and membrane are recruited by the osteosarcoma 9 (OS-9) and XTP3B ERAD lectins [123] which direct them to the ER membrane-bound complexes assembled around E3 ubiquitin ligases [124–126]. Both OS-9 and XTP3B are localised in the ER lumen [123]. OS-9 and XTP3B specifically recognise Man α(1,6)-Man α(1,6)-Man residues on the processed C-arm of the N-linked glycan [127]. XTP3B also inhibits the degradation of nonglycosylated proteins [83]. Yet again, different studies report opposite roles of the ERAD lectins in different cancers. For example, OS-9 is highly upregulated in osteosarcoma [128] and XTP3B was found to be critical for metastatic properties of human lung cancer cell lines [129], while a long noncoding RNA suppresses pancreatic ductal adenocarcinoma (PDAC) cell invasion by increasing both mRNA and protein levels for OS-9 [130].
The ERAD E3 ubiquitin-protein ligases accept ubiquitin specifically from an ER-associated E2 ligase and transfer it to glycoprotein substrates that need degradation [131]. After ubiquitination, the p97 (aka VCP) ATPase helps feeding substrates to a cytosolic proteasome [132]. Most of these ERAD ubiquitin-protein ligases are poorly characterised, and only few targets for each of them, for example, for the ERAD E3 enzymes HRD1 and MARCH6 [125, 133], have been identified. HRD1 protects cells from ER stress-induced apoptosis [134], and its upregulation promotes cell migration and invasion in colon cancer [135]. Another ERAD E3 enzyme, AMFR (aka gp78), mediates tumour invasion and metastasis functioning as a receptor for the GPI/autocrine motility factor [136]. Modulation of components of HRD1 partners [137] has been proposed as a novel point of intervention for cancer therapies, although there is published evidence that HRD1 suppresses the growth and metastasis of breast cancer cells [138] and the decrease of HRD1 expression contributed to tamoxifen resistance in breast cancer [139] (the latter by promoting the degradation of S100A8, a divalent metal ion binding protein involved in the chemistry-drug resistance in many tumours).
2.5. Glycoproteins and Antiancer Strategies Focussing on ERQC/ERAD Modulation
Cancer cells survive by adjusting to ER stress, and a number of studies in the literature have pointed out that components of the ERQC/ERAD machineries may constitute anticancer therapeutic targets [120, 121]. The centrality of ERQC/ERAD to glycoproteostasis would potentially endow such compounds with broad-spectrum activity, but depending on their glycoprotein secretory burden, different cancers will vary in their sensitivity to strategies that interfere with ER stress and glycoprotein folding and degradation [10]. Importantly, as is the case for any drug that interferes with basic cellular pathways, ERQC/ERAD modulators are potentially toxic to healthy cells as well. For these reasons, the most promising use for ERQC/ERAD modulators will likely be in combination with existing chemotherapeutics. For example, inhibition of homeostatic ER stress responses enhances apoptosis induced by oxidative stress-inducing drugs acting through the ER stress pathway [120].
Any attempt to develop ERQC/ERAD modulators as anticancer therapeutics would want to aim at ER stress-mediated selective killing of malignant cells without imposing significant damage to surrounding healthy cells. To be selective in aid of anticancer therapy, any ERQC/ERAD inhibitor of this kind needs to exploit different folding requirements of specific glycoproteins in cancerous vs. healthy cells. Amongst the many glycoprotein-dependent strategies used specifically by cancer cells are the expression of tumour-specific glycoprotein isoforms (with patterns of alternative splicing of mRNAs differing between tumour and normal tissues from which they are derived [140]); tumour-specific glycoprotein conformations [141]; upregulation of membrane-embedded drug transporters mediating chemotherapic multidrug resistance [142]; and expression of surface adhesion glycoproteins involved in tissue penetration and/or metastasis in leukemic cells [143] and solid malignancies [144]. Cancer cells also rely extensively on receptor tyrosine kinases (RTK): these glycoproteins are important in squamous cell carcinomas, breast/pancreas/prostate adenocarcinomas, and malignant gliomas. Indeed, nanomolar concentration of tunicamycin, a well-known inhibitor of N-glycosylation, reduces protein levels of at least four RTKs involved in tumour cell proliferation and survival [27–29].
Glycoproteins are also central to cancer immunotherapy [145, 146]: therapeutic anticancer antibodies, their cell-surface receptors, most of their epitopes [145], and complement components [147] are all glycoproteins. Many glycoproteins also underpin cancers' lack of response to immunotherapy response [12]. Drugs altering glycoprotein secretion/degradation will alter a patient's glycosecretome, including the surface antigens targeted by immunotherapy monoclonal antibodies, Fcγ receptors (FcγRs), and components of the complement system. Indeed, recent evidence has implicated polymorphisms of FcγR in the efficacy of monoclonal antibody- (mAb-) mediated therapy [148]. As the molecular basis for the opposite effects between inhibitory vs. activating FcγR resides in different intracellular phosphotyrosyl-based motifs [149], the folding/degradation requirements of different FcγRs may differ. Unfortunately, we have only partially uncovered the roles played by ERQC/ERAD during anticancer mAb therapy and, in particular, the folding and stability of cancer-specific surface glycoprotein epitopes, FcγRs and complement components [8]: the hypothesis that drugs that selectively impair glycoprotein folding and degradation may aid cancer immunotherapy remains to be tested.
3. Conclusions
A large number of published studies have highlighted the dependency of a number of cancers on specific ERQC/ERAD components, but the lack of specific inhibitors of the components in both pathways has hampered proper characterisation of the roles played by ERQC/ERAD in cancer biology. Even if such specific inhibitors were available, in order to make a convincing case for ERQC/ERAD as valid anticancer targets, several aspects of ERQC/ERAD biology in healthy and cancer cells need to be better elucidated.
For example, only a few bona fide glycoprotein clients of ERQC/ERAD are known [150] and none of the glycoproteins with proven roles in cancer biology have been tested for their dependency on ERQC/ERAD. As the checkpoint enzymes of both machineries are likely to be critical ones, useful first pieces of knowledge towards gauging the potential of ERQC/ERAD as anticancer targets would be the lists of substrates of UGGTs and EDEMs (which collectively we call “UGGT-omes”/“EDEM-omes”), in healthy cells and in their corresponding cancer counterparts.
Other important open questions involve the degrees of redundancy and interplay between ERAD and ERQC checkpoints (again, UGGTs and EDEMs) in deciding the fate of a specific misfolded glycoprotein. Whether there is a general mechanism by which the dilemma ER retention vs. secretion is solved or whether different individual glycoproteins are taken care by the ERQC and/or the ERAD branch to different extents during their lifetime in the ER still remains to be elucidated. The extents to which specific cancers tip the EQRC/ERAD balance for glycoproteins that are crucial to their survival will be of course one of the next big questions to answer, ultimately helping in each case to make choices between ERQC vs. ERAD modulation for the most effective anticancer prescription of this kind.
Last but not least, when it comes to toxicity, although evidence of ER-retention and/or ER associated degradation exists for a few cancer-associated glycoproteins, we do not know which EQRC/ERAD clients would risk premature and unwanted secretion in healthy cells (a scenario we dubbed the “ER Pandora's box”) upon administration of an ERQC/ERAD modulator. Thus, the relative toxicity of such drugs to healthy vs. cancer cells is difficult to predict. Targeting ERQC/ERAD may well prove a broad-spectrum spanner in the plasticity works of cancer cells, but—as it often happens with cancer biology—winning this battle will require a better understanding of the roles that these machineries play in cells at various stages of the cell cycle (in healthy cells as well as in cancer tissues). Only then may ERQC/ERAD inhibitors reach the clinic, adding to the expanding arsenal of anticancer therapeutics.
Acknowledgments
P. R. is the recipient of a LISCB Wellcome Trust ISSF award (grant reference 204801/Z/16/Z). G. T. is funded by a Wellcome Trust Seed Award in Science to P. R. (grant reference 214090/Z/18/Z). A. L. is the recipient of an Italian Government Ph.D. Studentship. Helpful discussions with Armando Parodi, Cecilia D'Alessio, Nicole Zitzmann, Dominic S. Alonzi, John L. Kiappes, Alessandro T. Caputo, Juan I. Blanco Capurro, Carlos Modenutti, Marcelo Martí, Ariel Orellana, Gerardo Lederkremer, Simon Spiro, and Johan C. Hill on the topic are gratefully acknowledged.
Disclosure
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Conflicts of Interest
The authors declare no conflicts of interest.
Authors' Contributions
Gábor Tax and Andrea Lia contributed equally to this work.
References
- 1.Ourednick J. Stem Cell Biology: Development and Plasticity. Vol. 1049. Ames, Iowa, USA: New York Academy of Sciences; 2005. [PubMed] [Google Scholar]
- 2.Long M. W. Stem cell plasticity: molding the future of tissue development and repair. Blood Cells, Molecules and Diseases. 2001;27(3):586–589. doi: 10.1006/bcmd.2001.0432. [DOI] [PubMed] [Google Scholar]
- 3.Harrison O. J., Linehan J. L., Shih H.-Y., et al. Commensal-specific T cell plasticity promotes rapid tissue adaptation to injury. Science. 2019;363(6422) doi: 10.1126/science.aat6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dekoninck S., Blanpain C. Stem cell dynamics, migration and plasticity during wound healing. Nature Cell Biology. 2019;21(1):18–24. doi: 10.1038/s41556-018-0237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta P. B., Pastushenko I., Skibinski A., Blanpain C., Kuperwasser C. Phenotypic plasticity: driver of cancer initiation, progression, and therapy resistance. Cell Stem Cell. 2019;24(1):65–78. doi: 10.1016/j.stem.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanahan D., Weinberg R. A. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 7.Hanahan D., Weinberg R. A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Eggleton P., Michalak M., Bremer E. Editorial: endoplasmic reticulum and its role in tumor immunity. Frontiers in Oncology. 2015;5(5):p. 252. doi: 10.3389/fonc.2015.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Volkmar N., Fenech E., Christianson J. C. New MAPS for misfolded proteins. Nature Cell Biology. 2016;18(7):724–726. doi: 10.1038/ncb3381. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y., Ye Y. Proteostasis regulation at the endoplasmic reticulum: a new perturbation site for targeted cancer therapy. Cell Research. 2011;21(6):867–883. doi: 10.1038/cr.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar S. K., Rajkumar V., Kyle R. A., et al. Multiple myeloma. Nature Reviews Disease Primers. 2017;3(1):p. 17046. doi: 10.1038/nrdp.2017.47. [DOI] [PubMed] [Google Scholar]
- 12.Simoes-Correia J., Figueiredo J., Oliveira C., et al. Endoplasmic reticulum quality control: a new mechanism of E-cadherin regulation and its implication in cancer. Human Molecular Genetics. 2008;17(22):3566–3576. doi: 10.1093/hmg/ddn249. [DOI] [PubMed] [Google Scholar]
- 13.Zimmermann L., Stephens A., Nam S.-Z., et al. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. Journal of Molecular Biology. 2018;430(15):2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 14.Sali A., Potterton L., Yuan F., van Vlijmen H., Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23(3):318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- 15.Klein M.-C., Zimmermann K., Schorr S., et al. AXER is an ATP/ADP exchanger in the membrane of the endoplasmic reticulum. Nat Commun. 2018;9(1):p. 3489. doi: 10.1038/s41467-018-06003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schoebel S., Mi W., Stein A., et al. Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3. Nature. 2017;548(7667):352–355. doi: 10.1038/nature23314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pettersen E. F., Goddard T. D., Huang C. C., et al. UCSF Chimera?A visualization system for exploratory research and analysis. Journal of Computational Chemistry. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 18.DeLano W. L. The case for open-source software in drug discovery. Drug Discovery Today. 2005;10(3):213–217. doi: 10.1016/s1359-6446(04)03363-x. [DOI] [PubMed] [Google Scholar]
- 19.Zappa F., Failli M., De Matteis M. A. The Golgi complex in disease and therapy. Current Opinion in Cell Biology. 2018;50:102–116. doi: 10.1016/j.ceb.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 20.Lamriben L., Graham J. B., Adams B. M., Hebert D. N. N-Glycan-based ER molecular chaperone and protein quality control system: the calnexin binding cycle. Traffic. 2016;17(4):308–326. doi: 10.1111/tra.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hebert D. N., Lamriben L., Powers E. T., Kelly J. W. The intrinsic and extrinsic effects of N-linked glycans on glycoproteostasis. Nature Chemical Biology. 2014;10(11):902–910. doi: 10.1038/nchembio.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clarke H. J., Chambers J. E., Liniker E., Marciniak S. J. Endoplasmic reticulum stress in malignancy. Cancer Cell. 2014;25(5):563–573. doi: 10.1016/j.ccr.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 23.Kim H., Bhattacharya A., Qi L. Endoplasmic reticulum quality control in cancer: friend or foe. Seminars in Cancer Biology. 2015;33:25–33. doi: 10.1016/j.semcancer.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uhlen M., Fagerberg L., Hallstrom B. M., et al. Tissue-based map of the human proteome. Science. 2015;347(6220):p. 1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 25.Bairi El K., Amrani M., Afqir S. The Human Cancer Pathology Atlas: an open-access source for more than 900,000 Kaplan-Meier plots and 5 million cancer tissue images. Biomed Pharmacother. 2017;96:1438–1439. doi: 10.1016/j.biopha.2017.10.063. [DOI] [PubMed] [Google Scholar]
- 26.Sedgwick P. How to read a Kaplan-Meier survival plot. BMJ. 2014;349(4):p. g5608. doi: 10.1136/bmj.g5608. [DOI] [PubMed] [Google Scholar]
- 27.Forbes S. A., Beare D., Boutselakis H., et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Research. 2017;45(D1):D777–D783. doi: 10.1093/nar/gkw1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aksoy B. A., Demir E., Babur Ö., et al. Prediction of individualized therapeutic vulnerabilities in cancer from genomic profiles. Bioinformatics. 2014;30(14):2051–2059. doi: 10.1093/bioinformatics/btu164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Higgins M. E., Claremont M., Major J. E., Sander C., Lash A. E. CancerGenes: a gene selection resource for cancer genome projects. Nucleic Acids Research. 2007;35:D721–D726. doi: 10.1093/nar/gkl811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davoli T., Xu A. W., Mengwasser K. E., et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell. 2013;155(4):948–962. doi: 10.1016/j.cell.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim P., Cheng F., Zhao J., Zhao Z. ccmGDB: a database for cancer cell metabolism genes. Nucleic Acids Research. 2016;44(D1):D959–D968. doi: 10.1093/nar/gkv1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Praeter C. M., Gerwig G. J., Bause E., et al. A novel disorder caused by defective biosynthesis of N-linked oligosaccharides due to glucosidase I deficiency. The American Journal of Human Genetics. 2000;66(6):1744–1756. doi: 10.1086/302948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gallo G. L., Valko A., Aramburu S. I., et al. Abrogation of glucosidase I-mediated glycoprotein deglucosylation results in a sick phenotype in fission yeasts: model for the human MOGS-CDG disorder. Journal of Biological Chemistry. 2018 doi: 10.1074/jbc.RA118.004844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tannous A., Pisoni G. B., Hebert D. N., Molinari M. N-linked sugar-regulated protein folding and quality control in the ER. Seminars in Cell and Developmental Biology. Elsevier Ltd. 2014:1–11. doi: 10.1016/j.semcdb.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sadat M. A., Moir S., Chun T.-W., et al. Glycosylation, hypogammaglobulinemia, and resistance to viral infections. New England Journal of Medicine. 2014;370(17):1615–1625. doi: 10.1056/nejmoa1302846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hammond C., Braakman I., Helenius A. Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proceedings of the National Academy of Sciences. 1994;91(3):913–917. doi: 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caputo A. T., Alonzi D. S., Kiappes J. L., et al. Advances in Experimental Medicine and Biology. 8. Vol. 1062. Singapore: Springer; 2018. Structural insights into the broad-spectrum antiviral target endoplasmic reticulum alpha-glucosidase II; pp. 265–276. [DOI] [PubMed] [Google Scholar]
- 38.Caputo A. T., Alonzi D. S., Marti L., et al. Structures of mammalian ER α-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals. Proceedings of the National Academy of Sciences. 2016;113(32):E4630–E4638. doi: 10.1073/pnas.1604463113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chiu C.-C., Lin C.-Y., Lee L.-Y., et al. Molecular chaperones as a common set of proteins that regulate the invasion phenotype of head and neck cancer. Clinical Cancer Research. 2011;17(14):4629–4641. doi: 10.1158/1078-0432.ccr-10-2107. [DOI] [PubMed] [Google Scholar]
- 40.Parodi A. J., Caramelo J. J., D’Alessio C. Handbook of Glycosyltransferases and Related Genes. Tokyo, Japan: Springer; 2014. UDP-glucose: glycoprotein glucosyltransferase 1,2 (UGGT1,2) pp. 15–30. [Google Scholar]
- 41.Arnold S. M., Kaufman R. J. The noncatalytic portion of human UDP-glucose:glycoprotein glucosyltransferase I confers UDP-glucose binding and transferase function to the catalytic domain. Journal of Biological Chemistry. 2003;278(44):43320–43328. doi: 10.1074/jbc.m305800200. [DOI] [PubMed] [Google Scholar]
- 42.Takeda Y., Seko A., Hachisu M., et al. Both isoforms of human UDP-glucose:glycoprotein glucosyltransferase are enzymatically active. Glycobiology. 2014;24(4):344–350. doi: 10.1093/glycob/cwt163. [DOI] [PubMed] [Google Scholar]
- 43.Ito Y., Takeda Y., Seko A., Izumi M., Kajihara Y. Functional analysis of endoplasmic reticulum glucosyltransferase (UGGT): synthetic chemistry’s initiative in glycobiology. Seminars in Cell & Developmental Biology. 2015;41:90–98. doi: 10.1016/j.semcdb.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 44.Daikoku S., Seko A., Ito Y., Kanie O. Glycan structure and site of glycosylation in the ER-resident glycoprotein, uridine 5′-diphosphate-glucose: glycoprotein glucosyltransferases 1 from rat, porcine, bovine, and human. Biochemical and Biophysical Research Communications. 2014;451(3):356–360. doi: 10.1016/j.bbrc.2014.07.095. [DOI] [PubMed] [Google Scholar]
- 45.Daikoku S., Seko A., Son S.-H., Suzuki K., Ito Y., Kanie O. The relationship between glycan structures and expression levels of an endoplasmic reticulum-resident glycoprotein, UDP-glucose: glycoprotein glucosyltransferase 1. Biochemical and Biophysical Research Communications. 2015;462(1):58–63. doi: 10.1016/j.bbrc.2015.04.105. [DOI] [PubMed] [Google Scholar]
- 46.Marin M. B., Ghenea S., Spiridon L. N., Chiritoiu G. N., Petrescu A.-J., Petrescu S.-M. Tyrosinase degradation is prevented when EDEM1 lacks the intrinsically disordered region. PLoS One. 2012;7(8) doi: 10.1371/journal.pone.0042998.e42998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Molinari M., Galli C., Vanoni O., Arnold S. M., Kaufman R. J. Persistent glycoprotein misfolding activates the glucosidase II/UGT1-driven calnexin cycle to delay aggregation and loss of folding competence. Molecular Cell. 2005;20(4):503–512. doi: 10.1016/j.molcel.2005.09.027. [DOI] [PubMed] [Google Scholar]
- 48.Pankow S., Bamberger C., Calzolari D., et al. ∆F508 CFTR interactome remodelling promotes rescue of cystic fibrosis. Nature. 2015;528(7583):510–516. doi: 10.1038/nature15729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Labriola C., Cazzulo J. J., Parodi A. J. Trypanosoma cruzi calreticulin is a lectin that binds monoglucosylated oligosaccharides but not protein moieties of glycoproteins. Molecular Biology of the Cell. 1999;10(5):1381–1394. doi: 10.1091/mbc.10.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin H., Yan Z., Nam K. H., Li J. Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Molecular Cell. 2007;26(6):821–830. doi: 10.1016/j.molcel.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li J., Zhao-Hui C., Batoux M., et al. Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proceedings of the National Academy of Sciences. 2009;106(37):15973–15978. doi: 10.1073/pnas.0905532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pearse B. R., Tamura T., Sunryd J. C., Grabowski G. A., Kaufman R. J., Hebert D. N. The role of UDP-Glc:glycoprotein glucosyltransferase 1 in the maturation of an obligate substrate prosaposin. The Journal of Cell Biology. 2010;189(5):829–841. doi: 10.1083/jcb.200912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumaraswamy E., Malykh A., Korotkov K. V., et al. Structure-expression relationships of the 15-kDa selenoprotein gene. Journal of Biological Chemistry. 2000;275(45):35540–35547. doi: 10.1074/jbc.m004014200. [DOI] [PubMed] [Google Scholar]
- 54.Apostolou S., Klein J. O., Mitsuuchi Y., et al. Growth inhibition and induction of apoptosis in mesothelioma cells by selenium and dependence on selenoprotein SEP15 genotype. Oncogene. 2004;23(29):5032–5040. doi: 10.1038/sj.onc.1207683. [DOI] [PubMed] [Google Scholar]
- 55.Ren B., Liu M., Ni J., Tian J. Role of selenoprotein F in protein folding and secretion: potential involvement in human disease. Nutrients. 2018;10(11):p. 1619. doi: 10.3390/nu10111619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yim S. H., Everley R. A., Schildberg F. A., et al. Role of Selenof as a gatekeeper of secreted disulfide-rich glycoproteins. Cell Reports. 2018;23(5):1387–1398. doi: 10.1016/j.celrep.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Labunskyy V. M., Yoo M.-H., Hatfield D. L., Gladyshev V. N. Sep15, a thioredoxin-like selenoprotein, is involved in the unfolded protein response and differentially regulated by adaptive and acute ER stresses. Biochemistry. 2009;48(35):8458–8465. doi: 10.1021/bi900717p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ferguson A. D., Labunskyy V. M., Fomenko D. E., et al. NMR structures of the selenoproteins Sep15 and SelM reveal redox activity of a new thioredoxin-like family. Journal of Biological Chemistry. 2006;281(6):3536–3543. doi: 10.1074/jbc.m511386200. [DOI] [PubMed] [Google Scholar]
- 59.Zhu J., Wang G., Li C., et al. Maize Sep15-like functions in endoplasmic reticulum and reactive oxygen species homeostasis to promote salt and osmotic stress resistance. Plant Cell Environ. 2018;96(11):p. 1106. doi: 10.1111/pce.13507. [DOI] [PubMed] [Google Scholar]
- 60.Irons R., Tsuji P. A., Carlson B. A., et al. Deficiency in the 15-kDa selenoprotein inhibits tumorigenicity and metastasis of colon cancer cells. Cancer Prevention Research. 2010;3(5):630–639. doi: 10.1158/1940-6207.capr-10-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lan X., Xing J., Gao H., et al. Decreased expression of selenoproteins as a poor prognosticator of gastric cancer in humans. Biological Trace Element Research. 2017;178(1):22–28. doi: 10.1007/s12011-016-0908-8. [DOI] [PubMed] [Google Scholar]
- 62.Short S. P., Williams C. S. Selenoproteins in tumorigenesis and cancer progression. Advances in Cancer Research. 2017;136:49–83. doi: 10.1016/bs.acr.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsuji P. A., Naranjo-Suarez S., Carlson B. A., Tobe R., Yoo M.-H., Davis C. D. Deficiency in the 15 kDa selenoprotein inhibits human colon cancer cell growth. Nutrients. 2011;3(9):805–817. doi: 10.3390/nu3090805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bang J., Huh J. H., Na J.-W., et al. Cell proliferation and motility are inhibited by G1 phase Arrest in 15-kDa selenoprotein-deficient chang liver cells. Molecules and Cells. 2015;38(5):457–465. doi: 10.14348/molcells.2015.0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu Y. J., Korotkov K. V., Mehta R., et al. Distribution and functional consequences of nucleotide polymorphisms in the 3′-untranslated region of the human Sep15 gene. Cancer Research. 2001;61(5):2307–2310. [PubMed] [Google Scholar]
- 66.Jablonska E., Gromadzinska J., Sobala W., Reszka E., Wasowicz W. Lung cancer risk associated with selenium status is modified in smoking individuals by Sep15 polymorphism. European Journal of Nutrition. 2008;47(1):47–54. doi: 10.1007/s00394-008-0696-9. [DOI] [PubMed] [Google Scholar]
- 67.Mohammaddoust S., Salehi Z., Saeidi Saedi H. SEPP1 and SEP15 gene polymorphisms and susceptibility to breast cancer. British Journal of Biomedical Science. 2018;75(1):36–39. doi: 10.1080/09674845.2017.1333241. [DOI] [PubMed] [Google Scholar]
- 68.Watrowski R., Castillo-Tong D. C., Fabjani G., Schuster E., Fischer M., Zeillinger R. The 811 C/T polymorphism in the 3′ untranslated region of the selenoprotein 15-kDa (Sep15) gene and breast cancer in Caucasian women. Tumor Biology. 2016;37(1):1009–1015. doi: 10.1007/s13277-015-3847-7. [DOI] [PubMed] [Google Scholar]
- 69.Xie J., Zhu X. Y., Liu L. M., Meng Z. Q. Solute carrier transporters: potential targets for digestive system neoplasms. Cancer Management and Research. 2018;10:153–166. doi: 10.2147/cmar.s152951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Orellana A., Moraga C., Araya M., Moreno A. Overview of nucleotide sugar transporter gene family functions across multiple species. Journal of Molecular Biology. 2016;428(16):3150–3165. doi: 10.1016/j.jmb.2016.05.021. [DOI] [PubMed] [Google Scholar]
- 71.Bredeston L. M., Marino-Buslje C., Mattera V. S., Buzzi L. I., Parodi A. J., D’Alessio C. The conundrum of UDP-Glc entrance into the yeast ER lumen. Glycobiology. 2017;27(1):64–79. doi: 10.1093/glycob/cww092. [DOI] [PubMed] [Google Scholar]
- 72.Trombetta E. S., Helenius A. The EMBO Journal. 12. Vol. 18. EMBO Press; 1999. Glycoprotein reglucosylation and nucleotide sugar utilization in the secretory pathway: identification of a nucleoside diphosphatase in the endoplasmic reticulum; pp. 3282–3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hadley B., Maggioni A., Ashikov A., Day C. J., Haselhorst T., Tiralongo J. Structure and function of nucleotide sugar transporters: current progress. Computational and Structural Biotechnology Journal. 2014;10(16):23–32. doi: 10.1016/j.csbj.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mulero J. J., Yeung G., Nelken S. T., Ford J. E. CD39-L4 is a secreted human apyrase, specific for the hydrolysis of nucleoside diphosphates. Journal of Biological Chemistry. 1999;274(29):20064–20067. doi: 10.1074/jbc.274.29.20064. [DOI] [PubMed] [Google Scholar]
- 75.Fang M., Shen Z., Huang S., et al. The ER UDPase ENTPD5 promotes protein N-glycosylation, the Warburg effect, and proliferation in the PTEN pathway. Cell. 2010;143(5):711–724. doi: 10.1016/j.cell.2010.10.010. [DOI] [PubMed] [Google Scholar]
- 76.Páez J. G., Recio J. A., Rouzaut A., Notario V. Identity between the PCPH proto-oncogene and the CD39L4 (ENTPD5) ectonucleoside triphosphate diphosphohydrolase gene. International Journal of Oncology. 2001;19(6):1249–1254. doi: 10.3892/ijo.19.6.1249. [DOI] [PubMed] [Google Scholar]
- 77.Recio J. A., Páez J. G., Sanders S., Kawakami T., Notario V. Partial depletion of intracellular ATP mediates the stress-survival function of the PCPH oncoprotein. Cancer Research. 2002;62(9):2690–2694. [PubMed] [Google Scholar]
- 78.Tirado O. M., Mateo-Lozano S., Sanders S., Dettin L. E., Notario V. The PCPH oncoprotein antagonizes the proapoptotic role of the mammalian target of rapamycin in the response of normal fibroblasts to ionizing radiation. Cancer Research. 2003;63(19):6290–6298. [PubMed] [Google Scholar]
- 79.Villar J., Arenas M. I., MacCarthy C. M., Blánquez M. J., Tirado O. M., Notario V. PCPH/ENTPD5 expression enhances the invasiveness of human prostate cancer cells by a protein kinase cδ-dependent mechanism. Cancer Research. 2007;67(22):10859–10868. doi: 10.1158/0008-5472.can-07-2041. [DOI] [PubMed] [Google Scholar]
- 80.Villar J., Quadri H. S., Song I., Tomita Y., Tirado O. M., Notario V. PCPH/ENTPD5 expression confers to prostate cancer cells resistance against cisplatin-induced apoptosis through protein kinase C -mediated bcl-2 stabilization. Cancer Research. 2009;69(1):102–110. doi: 10.1158/0008-5472.can-08-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hebert D. N., Molinari M. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiological Reviews. 2007;87(4):1377–1408. doi: 10.1152/physrev.00050.2006. [DOI] [PubMed] [Google Scholar]
- 82.Shenkman M., Lederkremer G. Z. Compartmentalization and selective tagging for disposal of misfolded glycoproteins. Trends in Biochemical Sciences. 2019 doi: 10.1016/j.tibs.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 83.van der Goot A. T., Pearce M. M. P., Leto D. E., Shaler T. A., Kopito R. R. Redundant and antagonistic roles of XTP3B and OS9 in decoding glycan and non-glycan degrons in ER-associated degradation. Molecular Cell. 2018;70(3):516–530. doi: 10.1016/j.molcel.2018.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kamiya Y., Kamiya D., Yamamoto K., Nyfeler B., Hauri H.-P., Kato K. Molecular basis of sugar recognition by the human L-type lectins ERGIC-53, VIPL, and VIP36. Journal of Biological Chemistry. 2008;283(4):1857–1861. doi: 10.1074/jbc.m709384200. [DOI] [PubMed] [Google Scholar]
- 85.Parodi A. J. Role of N-oligosaccharide endoplasmic reticulum processing reactions in glycoprotein folding and degradation. Biochemical Journal. 2000;348(1):1–13. doi: 10.1042/bj3480001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shuda M., Kondoh N., Imazeki N., et al. Activation of the ATF6, XBP1 and grp78 genes in human hepatocellular carcinoma: a possible involvement of the ER stress pathway in hepatocarcinogenesis. Journal of Hepatology. 2003;38(5):605–614. doi: 10.1016/s0168-8278(03)00029-1. [DOI] [PubMed] [Google Scholar]
- 87.Yoo Y. S., Han H. G., Jeon Y. J. Unfolded protein response of the endoplasmic reticulum in tumor progression and immunogenicity. Oxidative Medicine and Cellular Longevity. 2017;2017:p. 2969271. doi: 10.1155/2017/2969271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tsai Y. C., Weissman A. M. The unfolded protein response, degradation from the endoplasmic reticulum, and cancer. Genes and Cancer. 2010;1(7):764–778. doi: 10.1177/1947601910383011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chen X., Iliopoulos D., Zhang Q., et al. XBP1 promotes triple-negative breast cancer by controlling the HIF1α pathway. Nature. 2014;508(7494):103–107. doi: 10.1038/nature13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Croft A., Tay K. H., Boyd S. C., et al. Oncogenic activation of MEK/ERK primes melanoma cells for adaptation to endoplasmic reticulum stress. Journal of Investigative Dermatology. 2014;134(2):488–497. doi: 10.1038/jid.2013.325. [DOI] [PubMed] [Google Scholar]
- 91.Lee A.-H., Iwakoshi N. N., Anderson K. C., Glimcher L. H. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proceedings of the National Academy of Sciences. 2003;100(17):9946–9951. doi: 10.1073/pnas.1334037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bagheri-Yarmand R., Sinha K. M., Li L., et al. Combinations of tyrosine kinase inhibitor and ERAD inhibitor promote oxidative stress-induced apoptosis through ATF4 and KLF9 in medullary thyroid cancer. Molecular Cancer Research. 2019;17(3):751–760. doi: 10.1158/1541-7786.mcr-18-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Elbein A. D., Tropea J. E., Mitchell M., Kaushal G. P. Kifunensine, a potent inhibitor of the glycoprotein processing mannosidase I. Journal of Biological Chemistry. 1990;265(26):15599–15605. [PubMed] [Google Scholar]
- 94.Legler K., Rosprim R., Karius T., et al. Reduced mannosidase MAN1A1 expression leads to aberrant N-glycosylation and impaired survival in breast cancer. British Journal of Cancer. 2018;118(6):847–856. doi: 10.1038/bjc.2017.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lu Y., Xu Y.-Y., Fan K.-Y., Shen Z.-H. 1-Deoxymannojirimycin, the α1,2-mannosidase inhibitor, induced cellular endoplasmic reticulum stress in human hepatocarcinoma cell 7721. Biochemical and Biophysical Research Communications. 2006;344(1):221–225. doi: 10.1016/j.bbrc.2006.03.111. [DOI] [PubMed] [Google Scholar]
- 96.Gonzalez D. S., Karaveg K., Vandersall-Nairn A. S., Lal A., Moremen K. W. Identification, expression, and characterization of a cDNA encoding human endoplasmic reticulum mannosidase I, the enzyme that catalyzes the first mannose trimming step in mammalian Asn-linked oligosaccharide biosynthesis. Journal of Biological Chemistry. 1999;274(30):21375–21386. doi: 10.1074/jbc.274.30.21375. [DOI] [PubMed] [Google Scholar]
- 97.Tremblay L. O., Herscovics A. Cloning and expression of a specific human 1,2-mannosidase that trims Man9GlcNAc2 to Man8GlcNAc2 isomer B during N-glycan biosynthesis. Glycobiology. 1999;9(10):1073–1078. doi: 10.1093/glycob/9.10.1073. [DOI] [PubMed] [Google Scholar]
- 98.Romero P. A., Vallée F., Howell P. L., Herscovics A. Mutation of arg 273 to leu alters the specificity of the yeast N -glycan processing class I α1,2-mannosidase. Journal of Biological Chemistry. 2000;275(15):11071–11074. doi: 10.1074/jbc.275.15.11071. [DOI] [PubMed] [Google Scholar]
- 99.Vallée F., Karaveg K., Herscovics A., Moremen K. W., Howell P. L. Structural basis for catalysis and inhibition ofn-glycan processing class I α1,2-mannosidases. Journal of Biological Chemistry. 2000;275(52):41287–41298. doi: 10.1074/jbc.m006927200. [DOI] [PubMed] [Google Scholar]
- 100.Vallée F., Lipari F., Yip P., Sleno B., Herscovics A., Howell P. L. Crystal structure of a class I α1,2-mannosidase involved in N-glycan processing and endoplasmic reticulum quality control. The EMBO Journal. 2000;19(4):581–588. doi: 10.1093/emboj/19.4.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xiang Y., Karaveg K., Moremen K. W. Substrate recognition and catalysis by GH47 α-mannosidases involved in Asn-linked glycan maturation in the mammalian secretory pathway. Proceedings of the National Academy of Sciences. 2016;113(49):E7890–E7899. doi: 10.1073/pnas.1611213113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pan S., Cheng X., Chen H., et al. ERManI is a target of miR-125b and promotes transformation phenotypes in hepatocellular carcinoma (HCC) PLoS One. 2013;8(8) doi: 10.1371/journal.pone.0072829.e72829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ogen-Shtern N., Avezov E., Shenkman M., Benyair R., Lederkremer G. Z. Mannosidase IA is in quality control vesicles and participates in glycoprotein targeting to ERAD. Journal of Molecular Biology. 2016;428(16):3194–3205. doi: 10.1016/j.jmb.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 104.Liu T., Zhang S., Chen J., et al. Guan X.-Y., editor. The transcriptional profiling of glycogenes associated with hepatocellular carcinoma metastasis. PLoS One. 2014;9(9):p. e107941. doi: 10.1371/journal.pone.0107941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hosokawa N., Wada I., Hasegawa K., et al. A novel ER -mannosidase-like protein accelerates ER-associated degradation. EMBO Reports. 2001;2(5):415–422. doi: 10.1093/embo-reports/kve084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ermonval M., Kitzmuller C., Mir A. M., Cacan R., Ivessa N. E. N-glycan structure of a short-lived variant of ribophorin I expressed in the MadIA214 glycosylation-defective cell line reveals the role of a mannosidase that is not ER mannosidase I in the process of glycoprotein degradation. Glycobiology. 2001;11(7):565–576. doi: 10.1093/glycob/11.7.565. [DOI] [PubMed] [Google Scholar]
- 107.Hüttner S., Veit C., Vavra U., et al. Arabidopsis class I α-mannosidases MNS4 and MNS5 are involved in endoplasmic reticulum-associated degradation of misfolded glycoproteins. The Plant Cell. 2014;26(4):1712–1728. doi: 10.1105/tpc.114.123216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mast S. W., Diekman K., Karaveg K., Davis A., Sifers R. N., Moremen K. W. Human EDEM2, a novel homolog of family 47 glycosidases, is involved in ER-associated degradation of glycoproteins. Glycobiology. 2005 Apr;15(4):421–436. doi: 10.1093/glycob/cwi014. [DOI] [PubMed] [Google Scholar]
- 109.Ron E., Shenkman M., Groisman B., Izenshtein Y., Leitman J., Lederkremer G. Z. Bypass of glycan-dependent glycoprotein delivery to ERAD by up-regulated EDEM1. Molecular Biology of the Cell. 2011;22(21):3945–3954. doi: 10.1091/mbc.e10-12-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Molinari M., Calanca V., Galli C., Lucca P., Paganetti P. Role of EDEM in the release of misfolded glycoproteins from the calnexin cycle. Science. 2003;299(5611):1397–1400. doi: 10.1126/science.1079474. [DOI] [PubMed] [Google Scholar]
- 111.Sokołowska I., Piłka E. S., Sandvig K., Węgrzyn G., Słomińska-Wojewódzka M. Hydrophobicity of protein determinants influences the recognition of substrates by EDEM1 and EDEM2 in human cells. BMC Molecular and Cell Biology. 2015;16(1):p. 1. doi: 10.1186/s12860-015-0047-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shenkman M., Ron E., Yehuda R., Benyair R., Khalaila I., Lederkremer G. Z. Mannosidase activity of EDEM1 and EDEM2 depends on an unfolded state of their glycoprotein substrates. Communications Biology. 2018;1(1):p. 172. doi: 10.1038/s42003-018-0174-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gauss R., Kanehara K., Carvalho P., Ng D. T. W., Aebi M. A complex of Pdi1p and the mannosidase Htm1p initiates clearance of unfolded glycoproteins from the endoplasmic reticulum. Molecular Cell. 2011;42(6):782–793. doi: 10.1016/j.molcel.2011.04.027. [DOI] [PubMed] [Google Scholar]
- 114.Liu Y.-C., Fujimori D. G., Weissman J. S. Htm1p-Pdi1p is a folding-sensitive mannosidase that marks N-glycoproteins for ER-associated protein degradation. Proceedings of the National Academy of Sciences. 2016;113(28):E4015–E4024. doi: 10.1073/pnas.1608795113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sun S., Shi G., Han X., et al. Sel1L is indispensable for mammalian endoplasmic reticulum-associated degradation, endoplasmic reticulum homeostasis, and survival. Proceedings of the National Academy of Sciences. 2014;111(5):E582–E591. doi: 10.1073/pnas.1318114111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Papaioannou A., Higa A., Jégou G., et al. Alterations of EDEM 1 functions enhance ATF 6 pro‐survival signaling. The FEBS Journal. 2018;285(22):4146–4164. doi: 10.1111/febs.14669. [DOI] [PubMed] [Google Scholar]
- 117.Ushioda R., Hoseki J., Araki K., Jansen G., Thomas D. Y., Nagata K. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321(5888):569–572. doi: 10.1126/science.1159293. [DOI] [PubMed] [Google Scholar]
- 118.Määttänen P., Gehring K., Bergeron J. J. M., Thomas D. Y. Protein quality control in the ER: the recognition of misfolded proteins. Seminars in Cell and Developmental Biology. 2010;21(5):500–511. doi: 10.1016/j.semcdb.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 119.Lamriben L., Oster M. E., Tamura T., et al. EDEM1’s mannosidase-like domain binds ERAD client proteins in a redox-sensitive manner and possesses catalytic activity. Journal of Biological Chemistry. 2018;293(36):13932–13945. doi: 10.1074/jbc.ra118.004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Corazzari M., Lovat P. E., Armstrong J. L., et al. Targeting homeostatic mechanisms of endoplasmic reticulum stress to increase susceptibility of cancer cells to fenretinide-induced apoptosis: the role of stress proteins ERdj5 and ERp57. British Journal of Cancer. 2007;96(7):1062–1071. doi: 10.1038/sj.bjc.6603672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Thomas C. G., Spyrou G. ERdj5 sensitizes neuroblastoma cells to endoplasmic reticulum stress-induced apoptosis. Journal of Biological Chemistry. 2009;284(10):6282–6290. doi: 10.1074/jbc.m806189200. [DOI] [PubMed] [Google Scholar]
- 122.Cho S. Y., Kim S., Son M.-J., et al. Clinical significance of the thioredoxin system and thioredoxin-domain-containing protein family in hepatocellular carcinoma. Digestive Diseases and Sciences. 2019;64(1):123–136. doi: 10.1007/s10620-018-5307-x. [DOI] [PubMed] [Google Scholar]
- 123.Christianson J. C., Shaler T. A., Tyler R. E., Kopito R. R. OS-9 and GRP94 deliver mutant α1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nature Cell Biology. 2008;10(3):272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kikkert M., Doolman R., Dai M., et al. Human HRD1 is an E3 ubiquitin ligase involved in degradation of proteins from the endoplasmic reticulum. Journal of Biological Chemistry. 2004;279(5):3525–3534. doi: 10.1074/jbc.m307453200. [DOI] [PubMed] [Google Scholar]
- 125.Zattas D., Berk J. M., Kreft S. G., Hochstrasser M. A conserved C-terminal element in the yeast Doa10 and human MARCH6 ubiquitin ligases required for selective substrate degradation. Journal of Biological Chemistry. 2016;291(23):12105–12118. doi: 10.1074/jbc.m116.726877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Carvalho P., Goder V., Rapoport T. A. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 2006;126(2):361–373. doi: 10.1016/j.cell.2006.05.043. [DOI] [PubMed] [Google Scholar]
- 127.Satoh T., Chen Y., Hu D., Hanashima S., Yamamoto K., Yamaguchi Y. Structural basis for oligosaccharide recognition of misfolded glycoproteins by OS-9 in ER-associated degradation. Molecular Cell. 2010;40(6):905–916. doi: 10.1016/j.molcel.2010.11.017. [DOI] [PubMed] [Google Scholar]
- 128.Kimura Y., Nakazawa M., Yamada M. Cloning and characterization of three isoforms of OS-9 cDNA and expression of the OS-9 gene in various human tumor cell lines. Journal of Biochemistry. 1998;123(5):876–882. doi: 10.1093/oxfordjournals.jbchem.a022019. [DOI] [PubMed] [Google Scholar]
- 129.Yanagisawa K., Konishi H., Arima C., et al. Novel metastasis-related gene CIM functions in the regulation of multiple cellular stress-response pathways. Cancer Research. 2010;70(23):9949–9958. doi: 10.1158/0008-5472.can-10-1055. [DOI] [PubMed] [Google Scholar]
- 130.Sun Y.-W., Chen Y.-F., Li J., et al. A novel long non-coding RNA ENST00000480739 suppresses tumour cell invasion by regulating OS-9 and HIF-1α in pancreatic ductal adenocarcinoma. British Journal of Cancer. 2014;111(11):2131–2141. doi: 10.1038/bjc.2014.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mehnert M., Sommer T., Jarosch E. ERAD ubiquitin ligases. Bioessays. 2010;32(10):905–913. doi: 10.1002/bies.201000046. [DOI] [PubMed] [Google Scholar]
- 132.Christianson J. C., Ye Y. Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nature Structural and Molecular Biology. 2014;21(4):325–335. doi: 10.1038/nsmb.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ye Y., Baek S.-H., Ye Y., Zhang T. Proteomic characterization of endogenous substrates of mammalian ubiquitin ligase Hrd1. Cell and Bioscience. 2018;8:p. 46. doi: 10.1186/s13578-018-0245-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kaneko M., Ishiguro M., Niinuma Y., Uesugi M., Nomura Y. Human HRD1 protects against ER stress-induced apoptosis through ER-associated degradation. FEBS Letters. 2002;532(1-2):147–152. doi: 10.1016/s0014-5793(02)03660-8. [DOI] [PubMed] [Google Scholar]
- 135.Tan X., He X., Fan Z. Upregulation of HRD1 promotes cell migration and invasion in colon cancer. Molecular and Cellular Biochemistry. 2018;16:p. 359. doi: 10.1007/s11010-018-3447-0. [DOI] [PubMed] [Google Scholar]
- 136.Fang S., Ferrone M., Yang C., Jensen J. P., Tiwari S., Weissman A. M. The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proceedings of the National Academy of Sciences. 2001;98(25):14422–14427. doi: 10.1073/pnas.251401598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Schulz J., Avci D., Queisser M. A., et al. Conserved cytoplasmic domains promote Hrd1 ubiquitin ligase complex formation for ER-associated degradation (ERAD) Journal of Cell Science. 2017;130(19):3322–3335. doi: 10.1242/jcs.206847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Xu Y.-M., Wang H.-J., Chen F., et al. HRD1 suppresses the growth and metastasis of breast cancer cells by promoting IGF-1R degradation. Oncotarget. 2015;6(40):42854–42867. doi: 10.18632/oncotarget.5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang Y., Guo A., Liang X., et al. HRD1 sensitizes breast cancer cells to Tamoxifen by promoting S100A8 degradation. Oncotarget. 2017;8(14):23564–23574. doi: 10.18632/oncotarget.15797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Slansky J. E., Spellman P. T. Alternative splicing in tumors—A path to immunogenicity? New England Journal of Medicine. 2019;380(9):877–880. doi: 10.1056/nejmcibr1814237. [DOI] [PubMed] [Google Scholar]
- 141.Garrett T. P. J., Burgess A. W., Gan H. K., et al. Antibodies specifically targeting a locally misfolded region of tumor associated EGFR. Proceedings of the National Academy of Sciences. 2009;106(13):5082–5087. doi: 10.1073/pnas.0811559106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kumar A., Jaitak V. Natural products as multidrug resistance modulators in cancer. European Journal of Medicinal Chemistry. 2019;176:268–291. doi: 10.1016/j.ejmech.2019.05.027. [DOI] [PubMed] [Google Scholar]
- 143.Santamaria S., Delgado M., Kremer L., Garcia-Sanz J. A. Will a mab-based immunotherapy directed against cancer stem cells be feasible? Frontiers in Immunology. 2017;8:p. 1509. doi: 10.3389/fimmu.2017.01509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Menke-van der Houven van Oordt C. W., Gomez-Roca C., van Herpen C., et al. First-in-human phase I clinical trial of RG7356, an anti-CD44 humanized antibody, in patients with advanced, CD44-expressing solid tumors. Oncotarget. 2016;7(48):80046–80058. doi: 10.18632/oncotarget.11098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Scott A. M., Wolchok J. D., Old L. J. Antibody therapy of cancer. Nature Reviews Cancer. 2012;12(4):278–287. doi: 10.1038/nrc3236. [DOI] [PubMed] [Google Scholar]
- 146.Dekker N. R. P., Stigt J. A., Visser O., Netters F. J. S., Koornstra R. H. T., de Groot J. W. B. Immunotherapy for cancer: from medical breakthrough to application in daily practice. Nederlands Tijdschrift voor Geneeskunde. 2019;163 [PubMed] [Google Scholar]
- 147.Bordron A., Bagacean C., Tempescul A., et al. Complement system: a neglected pathway in immunotherapy. Clinical Reviews in Allergy & Immunology. 2019;8(1):p. 569. doi: 10.1007/s12016-019-08741-0. [DOI] [PubMed] [Google Scholar]
- 148.Kaifu T., Nakamura A. Polymorphisms of immunoglobulin receptors and the effects on clinical outcome in cancer immunotherapy and other immune diseases: a general review. International Immunology. 2017;29(7):319–325. doi: 10.1093/intimm/dxx041. [DOI] [PubMed] [Google Scholar]
- 149.Sibéril S., Dutertre C.-A., Fridman W.-H., Teillaud J.-L. FcγR: the key to optimize therapeutic antibodies? Critical Reviews in Oncology/Hematology. 2007;62(1):26–33. doi: 10.1016/j.critrevonc.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 150.Roversi P., Marti L., Caputo A. T., et al. Interdomain conformational flexibility underpins the activity of UGGT, the eukaryotic glycoprotein secretion checkpoint. Proceedings of the National Academy of Sciences. 2017;114(32):8544–8549. doi: 10.1073/pnas.1703682114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.McNicholas S., Agirre J. Glycoblocks: a schematic three-dimensional representation for glycans and their interactions. Acta Crystallographica Section D Structural Biology. 2017;73(2):187–194. doi: 10.1107/s2059798316013553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Dingerdissen H. M., Torcivia-Rodriguez J., Hu Y., Chang T.-C., Mazumder R., Kahsay R. BioMuta and BioXpress: mutation and expression knowledgebases for cancer biomarker discovery. Nucleic Acids Research. 2018;46(D1):D1128–D1136. doi: 10.1093/nar/gkx907. [DOI] [PMC free article] [PubMed] [Google Scholar]