Abstract
Escherichia coli serotype O9a provides a model for export of lipopolysaccharide (LPS) O-antigen polysaccharide (O-PS) via ABC transporters. In O9a biosynthesis, a chain-terminator enzyme, WbdD, caps the nonreducing end of the glycan with a methylphosphate moiety and thereby establishes chain-length distribution. A carbohydrate-binding module (CBM) in the ABC transporter recognizes terminated glycans, ensuring that only mature O-PS is exported and incorporated into LPS. Here, we addressed two questions arising from this model. Are both residues in the binary terminator necessary for termination and export? And is a terminal methylphosphate moiety sufficient for export of heterologous glycans? To answer the first question, we uncoupled WbdD kinase and methyltransferase activities. WbdD mutants revealed that although the kinase activity is solely responsible for chain-length regulation, both activities are essential for CBM recognition and export. Consistent with this observation, a saturation transfer difference NMR experiment revealed a direct interaction between the CBM and the terminal methyl group. To determine whether methylphosphate is the sole determinant of substrate recognition by the CBM, we exploited Klebsiella pneumoniae O7, whose O-PS repeat-unit structure differs from O9a, but, as shown here, offers the second confirmed example of a terminal methylphosphate serving in substrate recognition. In vitro and in vivo experiments indicated that each CBM can bind the O-PS only with the native repeat unit, revealing that methylphosphate is essential but not sufficient for substrate recognition and export. Our findings provide important new insight into the structural determinants in a prototypical quality control system for glycan assembly and export.
Keywords: lipopolysaccharide (LPS), glycolipid, ABC transporter, membrane transport, glycobiology, bacterial outer membrane, carbohydrate-binding module (CBM), glycan export, O-antigen polysaccharide
Introduction
Lipopolysaccharide (LPS)5 is the major glycolipid of the Gram-negative bacterial outer membrane (1). It is almost always essential for cellular viability and is important for pathogenicity. The tripartite structure of LPS consists of the outer membrane–embedded component, lipid A (also known as endotoxin), a core oligosaccharide, and a structurally variable and immunogenic sugar polymer termed O-antigen (O-PS) (2). In human pathogens, O-PS is one of the first molecules encountered by the host, and it provides protection to the bacterium from immune processes such as the complement system (3). For resistance to serum killing, the length of the O-PS is a critical factor in promoting a successful defense, with longer O-PS providing a more effective barrier (4, 5). Loss of O-PS chain-length regulation has resulted in loss of serum resistance and susceptibility to leukocytes in some Gram-negative bacteria (5). There are many different O-PS structures with the diversity driven by selective environmental factors, such as host immune response and bacteriophages that exploit O-PSs as receptors (6).
Many O-PSs are assembled by a mechanism that requires an ATP-binding cassette (ABC) transporter to export lipid-linked O-PS intermediates from the site of synthesis in the cytosol to the periplasm (7, 8), where the LPS molecule is completed before being translocated to the cell surface by the Lpt (LPS transport) machinery (9, 10). The ABC transporter provides an important role in determining the chain-length distribution of the O-PS products, with two main mechanisms being described. In the simpler process exhibited by Klebsiella pneumoniae serotype O2a (11), the O-PS is polymerized to completion within the cytosol by a biosynthetic enzyme complex. The chain-length distribution is controlled by the relative activities of a complex of glycosyltransferase (GT) enzymes and the ABC transporter. The O2a ABC transporter does not possess strict O-PS specificity, and it can export polymers with diverse repeat-unit structures, but polymerization and export are obligatorily coupled (11, 12). The more intricate Escherichia coli serotype O9a O-PS assembly system incorporates an additional mechanism that imposes a stricter level of control over chain length. This requires the installation of a chain-terminating residue that creates an export signal recognized by a carbohydrate-binding module (CBM) attached to the ABC transporter. In this system, polymerization and export can be temporally uncoupled in vitro (11). The structural requirements for glycan recognition by the CBM provide the focus of this study.
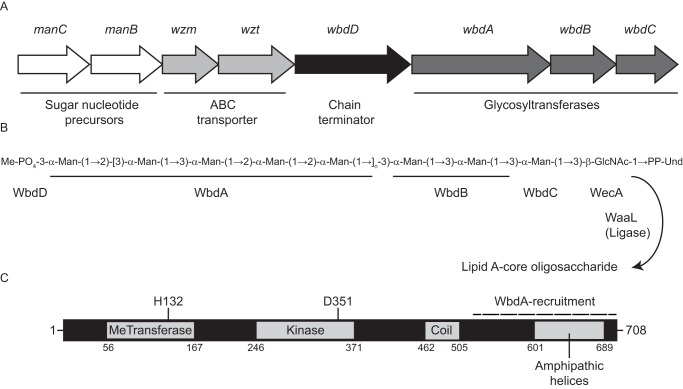
During O-PS biosynthesis in both systems, the 55-carbon polyisoprene carrier lipid, undecaprenol phosphate (Und-P) serves as the acceptor for the shared initiating phosphoglycosyltransferase enzyme, named WecA (2). In serotype O9a, the resulting Und-PP-GlcNAc product is then extended by serotype-specific mannosyltransferases to create the O9a O-PS backbone (Fig. 1). Central to polymerization is WbdA, a dual-domain GT polymerase that catalyzes formation of alternating pairs of α1–2- and α1–3-linkages (13, 14). WbdA operates in a distributive (rather that processive) manner, where the growing chain is released from the enzyme at each step (14, 15). The length of the O9a O-PS is dictated by WbdD. The C terminus of WbdD contains a membrane-anchoring helix and a region that recruits WbdA into an active membrane-bound heterocomplex (Fig. 1C) (14, 16). The N terminus of WbdD possesses kinase and methyltransferase catalytic sites that add a phosphomethyl moiety to the nonreducing terminus of the O-PS, blocking further extension by WbdA (17–19). The physical separation of the chain-terminating domain from the membrane by an extended coiled-coil ensures that only O-PS of a minimum length can be capped (20). In addition to operating as a molecular ruler, the coiled-coil structure facilitates multimerization of functional WbdD trimers. The relative abundance of WbdA and WbdD also affects the distribution of O-PS lengths, and this is explained by the variable geometry model that describes the outcome of changes in the stoichiometries of the WbdA-WbdD heterocomplexes (21).
Figure 1.
Structure and biosynthesis of the E. coli O9a O-PS. A, organization of the O-PS synthesis (wb*) operon. B, structure of the Und-PP–linked O9a biosynthetic intermediate. The square brackets delineate the tetrasaccharide O-PS repeat unit. Man, d-mannose; GlcNAc, d-GlcNAc. C, domain organization of the dual kinase methyltransferase WbdD. The coiled-coil domain acts as a molecular ruler separating WbdA, which is recruited to its interaction site at the C terminus of WbdD (indicated), from the chain-terminating catalytic sites of WbdD.
Essential to this chain-length regulation strategy is a quality control mechanism on the ABC transporter protein complex, which ensures that only terminated (and chain-regulated) O-PS is exported for assembly on the cell surface. O-PS ABC transporters are composed of homodimers of the transmembrane domain protein (Wzm) and the nucleotide-binding domain protein (Wzt). In E. coli O9a and related assembly systems, Wzt possesses a C-terminal CBM, which is specific for its cognate O-PS (12, 22, 23). Binding of O-PS by the CBM is a prerequisite for transport, and removal or mutation of the CBM abrogates export (23). The Aquifex aeolicus Wzm-Wzt ABC transporter is currently the only solved structure for this class of exporter, and it shares similarity with E. coli O9a Wzm-Wzt (24). Although the A. aeolicus structure was solved without the cognate CBM, it has allowed the development of mechanistic models for O-PS transport. Key mechanistic predictions were tested by examining the phenotypes of the corresponding mutations in E. coli O9a Wzm-Wzt. In vitro data shows that the CBM stimulates ATP turnover by Wzm-Wzt in proteoliposomes (24), suggesting a more complex role that is not confined to substrate recognition. Notably, constructs where the O9a CBM and nucleotide-binding domain are artificially separated retain export function in vivo (23).
Similar chain termination/export strategies exist in other species and/or serotypes. For example, O-PS chain regulation is achieved by the addition of a methyl group in E. coli O8 or a sugar (3-deoxy-d-manno-oct-2-ulosonic acid; Kdo) in K. pneumoniae O12 (25). The involvement of both phosphate and methyl residues in chain-length regulation in E. coli O9a appears unnecessarily complicated; in principle, a single terminating residue should suffice. Given the pivotal role the E. coli O9a O-PS system plays as an influential prototype for bacterial glycan export, it is essential to understand the details and limitations of substrate recognition and export. This provides the goal of the current study.
Results
WbdD kinase activity is solely responsible for arresting O9a O-PS polymerization
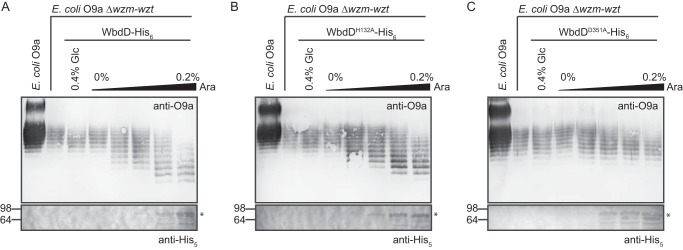
The modal distribution of O9a-PS chain lengths is established by the coiled-coil domain, which physically separates the WbdD termination enzymes from membrane-anchored WbdA (20). Previous work established that WbdD halts WbdA-mediated O-PS extension by installation of a phosphomethyl moiety to the nonreducing terminus, where new backbone residues are added (17, 20, 21). Logically, the addition of a single terminating residue to the site of O-PS extension should be sufficient for regulation of chain-length distribution. To confirm this hypothesis and rule out any unanticipated requirements for a functional methyltransferase, we exploited the observation that overexpression of a functional WbdD terminator decreases the average chain length of O-PS (21). Mathematical modeling supports the hypothesis that O-PS chain length responds to altered stoichiometry within the WbdA:WbdD heterocomplex, with the resulting changes in geometry of the complex altering access of the glycan to termination catalytic sites (21). The O-PS phenotype is readily assessed by SDS-PAGE profiles of Und-PP–linked O9a intermediates in whole-cell lysates. These intermediates are not visible in silver-stained SDS-PAGE but are readily visualized in immunoblots using O-PS–specific antibodies (11, 12, 26).
These experiments were performed in a manA Δwzm-wzt::aphA-3 background, where O-PS production is conditionally dependent on the addition of d-mannose to the growth medium (17). This is necessary to avoid any unexpected effects on viability of the transformants, known to occur when O9a export is eliminated (22). After SDS-PAGE of whole-cell lysates, O9a intermediates were detected using O9a-specific antibodies (Fig. 2). Prior in vitro experiments verified that WbdDH132A and WbdDD351A lack methyltransferase and kinase activities, respectively (19). Each variant was expressed from a pBAD promoter by titrating the l-arabinose inducer, and the expected increase in protein production was confirmed by exploiting the His6 tag for Western immunoblotting. As shown previously (21), increasing levels of WT WbdD resulted in an inducer-dependent reduction in the average O-PS intermediate chain-length (Fig. 2A). The profiles of O9a intermediates in cells overexpressing the methyltransferase mutant (WbdDH132A) were qualitatively similar to those with WT WbdD (Fig. 2B). In contrast, the kinase mutant (WbdDD351A) had no effect on the chain-length distribution of O-PS intermediates (Fig. 2C). These results show that the addition of the activity of the kinase is sufficient to determine O-PS chain length in vivo and that the kinase and methyltransferase activities can be uncoupled.
Figure 2.
The kinase domain of WbdD is sufficient for chain termination in vivo. Levels of WbdD-His6 (expressed from pWQ470) (A) and its mutants with inactivated methyltransferase (His132 → Ala; pWQ829) (B) or kinase (Asp351 → Ala; pWQ830) (C) catalytic sites were titrated in E. coli CWG638 manA Δwzm-wzt::aphA-3. The addition of 0.4% d-glucose was used to repress the pBAD promoter, whereas varying concentrations of l-arabinose induced expression of the WbdD variants. To overcome second site mutations occurring during prolonged growth in the absence of functional O-PS export (17), a manA mutant was used, making O9a O-PS biosynthesis conditional on the addition of mannose to the growth medium at the beginning of the experiment. Und-PP–linked O-PS intermediates (accumulating in the absence of transport) were detected in proteinase K–digested whole-cell lysates by SDS-PAGE and immunoblotting with O9a-specific antibodies (top panels). The control lane is authentic O9a LPS from the manA strain (CWG634). The bottom panels show the detection of WbdD-His6 variants by immunoblotting with anti-His5 antibodies.
The terminating methyl group is essential for export by Wzm-Wzt
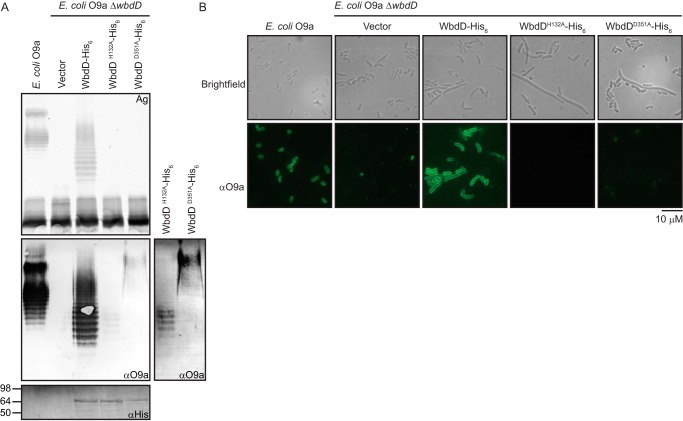
To evaluate whether O9a O-PS lacking normal nonreducing terminal modification could be transported across the inner membrane, an E. coli O9a manA ΔwbdD mutant was transformed with plasmids expressing the WbdD mutant variants. Export activity was monitored by silver-stained LPS profiles in whole-cell lysates separated by SDS-PAGE (Fig. 3A). Whereas plasmids expressing WT WbdD restored the production of O-PS–containing LPS visible in silver-stained gels, only O-PS–deficient LPS was evident in cells expressing either WbdDH132A or WbdDD351A. However, as expected, small amounts of Und-PP–linked O-PS were still detected by Western immunoblotting in cells expressing WbdDH132A or WbdDD351A. Whereas the WbdDH132A variant produced a banding pattern resembling the O-PS chain-length profile of the WT LPS, the WbdDD351A O-PS was of a higher molecular weight, as anticipated from the lack of chain-length regulation described above.
Figure 3.
The complete O-antigen–terminating moiety is essential for export of O9a O-PS and cell-surface exposure. E. coli O9a manA ΔwbdD (CWG900) cells were transformed with plasmids expressing WbdD-His6 (pWQ470) and its variants, WbdDH132A-His6 (pWQ829) or WbdDD351A-His6 (pWQ830), and examined by SDS-PAGE and immunoblotting (A) or immunofluorescence microscopy (B). Protein expression was induced with 0.02% l-arabinose. E. coli O9a manA (CWG634) provided the positive control. A, the top panel displays the LPS profile of proteinase K–treated whole-cell lysates resolved by SDS-PAGE and stained with silver. The middle panel is an immunoblot using polyclonal primary antibodies directed against O9a (note that immunoblotting detects both Und-PP-linked and lipid A-linked O-PS). Due to the high signal from complete LPS compared with Und-PP–linked intermediates in equivalent amounts of cells, a longer exposure of the lanes containing the Und-PP–linked intermediates is shown on the right. The bottom panel is a Western immunoblot using antibodies directed against the C-terminal His6 tags on WbdD to confirm protein expression. B, immunofluorescence microscopy of the corresponding cells, using O9a-specific antibodies to detect surface-exposed O9a O-PS. Brightfield and immunofluorescence microscopy experiments were performed on formalin-fixed whole cells, probed with an anti-O9a rabbit antiserum and an FITC-conjugated secondary antibody.
To confirm export status, immunofluorescence microscopy was performed to determine whether O-PS was present on the cell surface of the transformed mutants. O-PS was probed with a primary antiserum directed against the O9a antigen and a secondary antibody conjugated with FITC (Fig. 3B). Images of the parent O9a strain and the manA ΔwbdD mutant transformed with the WT wbdD gene displayed robust signal from surface-exposed O9a O-PS. In contrast, manA ΔwbdD cells transformed with vector or either mutant wbdD gene generated no immunofluorescence, consistent with the data in Fig. 3A. Brightfield images show that cells accumulating intracellular O-PS intermediates possess morphological defects (typically elongated cells), as reported previously (22). Taken together, these results indicated that the ABC transporter could not export either the nonmethylated O-PS or the nonterminated (nonphosphorylated and nonmethylated) O-PS.
The carbohydrate-binding module on Wzt is unable to recognize nonmethylated O9a O-PS
Previous work demonstrated that the C-terminal domain of Wzt functions as a CBM (23). In that study, an in vitro pulldown approach revealed the CBM binds its cognate O-PS, but not O-PS lacking the terminal methylphosphate or O-PS from E. coli O8 possessing a polymannose structure (with a repeat-unit structure different from that of O9a) capped with a terminal methyl group. To determine whether the requirement for O-PS methylation for export is dictated by the CBM-binding step, purified LPSs lacking either terminal phosphomethyl or methyl modifications were tested using the same pulldown approach. LPS molecules with chemically distinct nonreducing termini were generated using a strategy exploited previously to generate nonterminated K. pneumoniae O12 LPS (12). This method takes advantage of the ability of the K. pneumoniae O2a ABC transporter to export heterologous O-PS intermediates. This transporter lacks a CBM and has no specificity for either the repeat-unit structure or terminal modifications of O-PS substrates (11, 27). It can therefore export LPS molecules with varying terminal modifications. LPS species were purified from E. coli O9a ΔwbdD transformed with plasmids to express the K. pneumoniae O2a transporter and appropriate WbdD variants.
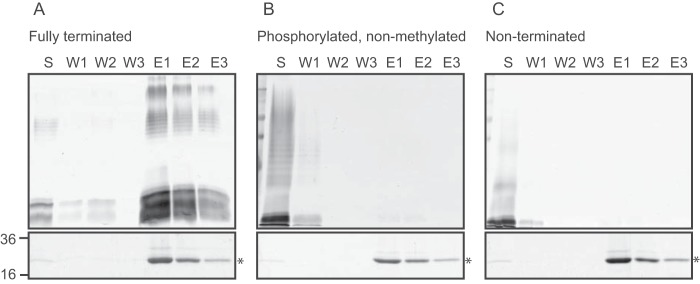
The pulldown experiments follow retention (or not) of LPS by His6-tagged CBM using nickel magnetic beads (Fig. 4). In the control experiments with WT O9a LPS, a considerable amount of LPS eluted in fractions containing CBM as reported previously (23). However, when CBM was incubated with either nonmethylated or nonmethylated/nonphosphorylated LPS, no binding was evident, indicating that the methyl group is essential for recognition by the CBM, and the phosphate group alone is insufficient. The inability of the CBM to bind improperly terminated O-PS is entirely consistent with the transport defects described above. These data support the notion that the impaired transport reflects altered substrate recognition but cannot exclude additional defects in later transport stages resulting from a terminally exposed phosphate.
Figure 4.
The Wzt CBM does not recognize nonmethylated LPS. Pulldown experiments were performed using purified E. coli O9a CBM (Wzt-C-His6) and one of three LPS species: WT O9a (A), nonmethylated O9a (B), or nonterminated O9a (C). Reaction mixtures consisting of 200 μg of CBM and 200 μg of LPS were mixed with nickel magnetic beads and collected using a magnet. The samples are unbound material in the supernatant (S) and material released by sequential washes of the beads in the absence (W1–W3) or presence of imidazole (E1–E3). Aliquots of the collected samples were subjected to SDS-PAGE. LPS was detected by silver staining (top panels) and protein was detected by simply blue staining (bottom panels).
Direct interaction between the nonreducing terminal methylphosphate and the CBM
During the characterization of the O9a CBM, residues in a putative binding pocket were identified that abrogated O-PS binding in vitro or had adverse effects on export in vivo (23). A saturation transfer difference (STD) NMR experiment (28) was used to provide more insight into the interaction and to formally verify the direct role of the methyl group in WztO9a-C:O9a binding. Similar experiments have been performed previously to precisely define the interaction sites of antibodies with glycans by “epitope mapping” (e.g. see Refs. 29–31). The method relies on the application of precise radiofrequency pulses to selectively saturate the NMR signals of the protein; this saturation is then transferred to ligands that transiently bind the protein, attenuating the NMR signals of ligand protons at and near the binding site. To better observe small (>5%) attenuations, STD spectra are collected as the difference of an “on-resonance” spectrum (i.e. saturation pulses applied on the protein signals) and an “off-resonance” reference spectrum (i.e. saturation pulses applied far away from any NMR signals). STD spectra therefore display only signals corresponding to ligand protons that are at and near the protein-binding site. To address complications arising from saturation in the presence of larger ligands (30), a saturation transfer double difference spectrum (STDD) (32) is obtained by subtracting an STD spectrum obtained from a sample without protein from the protein-ligand STD spectra.
Serotype O9a O-PS was purified, and its structure was confirmed by comparison against the reported NMR data (13, 33) (data not shown). For STD NMR experiments, two samples were prepared. The first sample contained an approximately 100-fold excess of O9a polysaccharide mixed with WztO9a-C, whereas the second sample contained O9a polysaccharide without protein. Saturation times were tested at 1, 2, 3, and 6 s (Fig. 5A). It was determined that 3 s was sufficient for magnetization transfer, and data were collected. The most intense signal from the obtained STDD spectrum (aside from a differencing artifact arising from the Tris buffer) was a doublet at δH 3.63 ppm with coupling constant J = 11 Hz (Fig. 5B), which precisely corresponds to the chemical shift of the methyl protons on the nonreducing terminal methylphosphate group from O9a O-PS (33). These findings offer clear and direct data for the interaction of the nonreducing terminal methyl group with WztO9a-C and are entirely consistent with its requirement for WztO9a-C-binding in LPS pulldown experiments as well as its requirement for display of the O9a polysaccharide on the cell surface.
Figure 5.
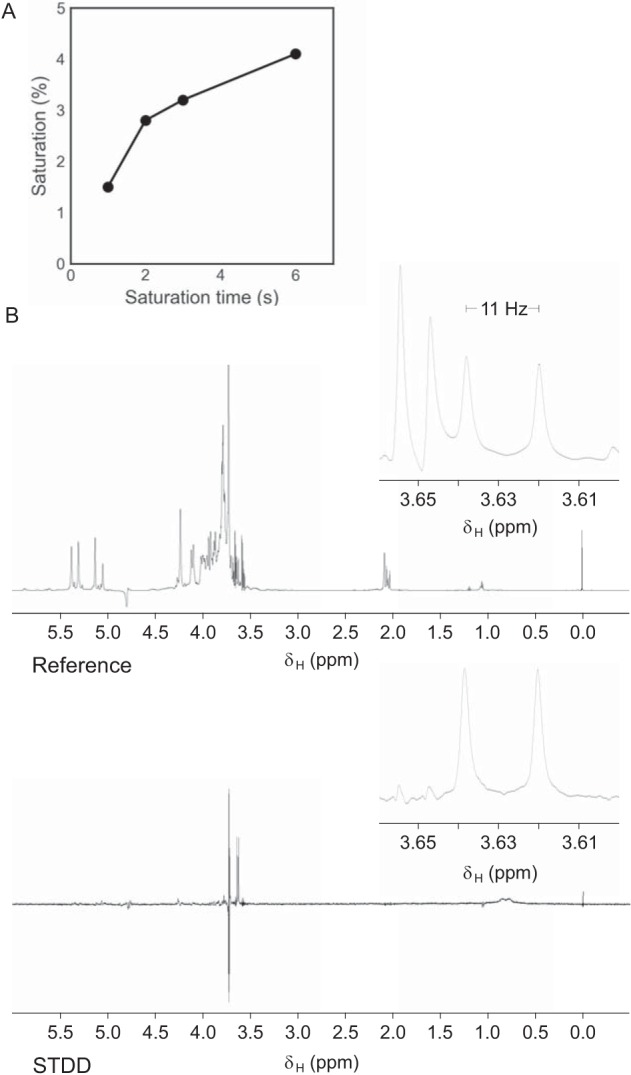
WztO9a-C binds to the terminal methylphosphate group of the O9a polysaccharide. A, time dependence of magnetization transfer (STDD%) of the terminal methylphosphate methyl protons for four saturation time points. B, 1H NMR spectra of off-resonance (top) and STDD (bottom) experiments after 3-s saturation. Panels to the right display the region between 3.60 and 3.66 ppm.
Identification of a terminal methylphosphate moiety in the K. pneumoniae O7 O-PS
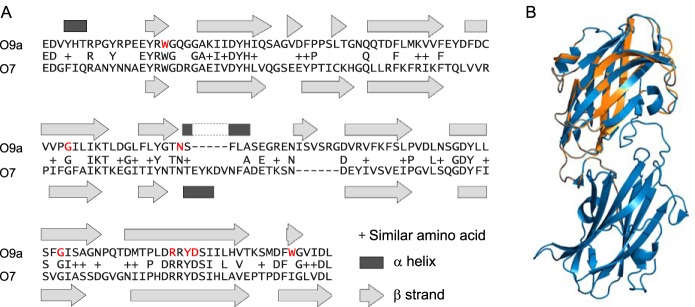
The data above reveal the importance of the methylphosphate group for substrate recognition and export but do not reveal whether the terminal modification is sufficient by itself for these processes. To address this question, we examined the ability of the O9a CBM to bind an O-PS possessing a terminal methylphosphate group but a different repeat-unit structure. This was provided by K. pneumoniae O7, whose detailed structure has not been reported previously. Examination of the K. pneumoniae O7 wb* locus (Fig. S1) revealed features consistent with chain-terminating ABC transporter-dependent O-PS biosynthesis. WztO7 possesses a proposed CBM domain sharing 34%/50% identity/similarity (E = 7e−26) with the CBM of WztO9a (Fig. 6A). A tertiary structure prediction of WztO7-C using Phyre2 (34) indicates structural homology between the C-terminal domains (Fig. 6B). Furthermore, residues equivalent to those essential for WztO9a-C O-PS binding, Trp-286, Gly-333, Asn-348, Gly-387, Arg-402, Tyr-404, and Asp-405 (23), are found in WztO7-C. These similarities between the CBMs suggested that they recognize similar ligand structures, and therefore the O7 O-PS likely possesses a methylphosphate nonreducing terminal modification.
Figure 6.
Alignment of WztO9a-C (residues 270–423) to WztO7-C (residues 303–455). A, pairwise sequence alignment was performed by Blast (A). The indicated secondary structure elements are from the WztO9a-C crystal structure (Protein Data Bank entry 2R5O) and a Phyre2 model of WztO7-C. The residues coloured red are important for O-PS binding by WztO9a-C (23). B, cartoon representation overlay of WztO7-C model (orange) with WztO9a-C crystal structure (blue). The structural overlay was generated using PyMOL (Schrödinger, LLC).
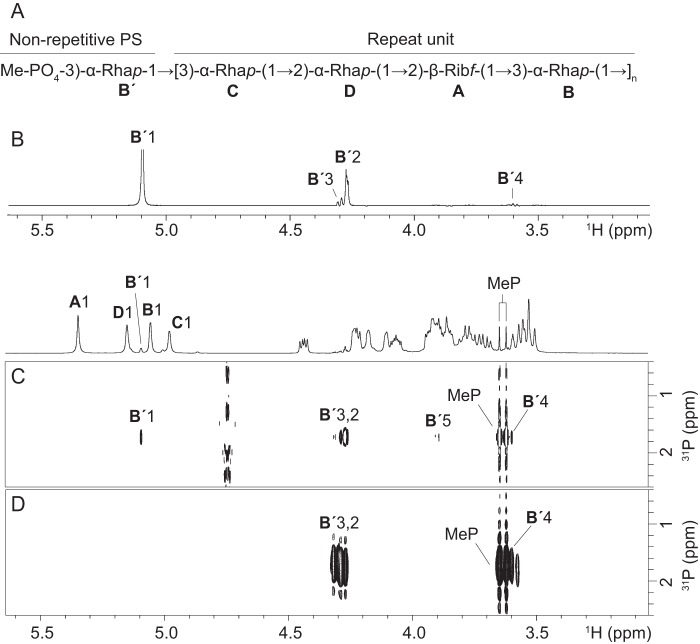
The O7 O-PS has a tetrasaccharide repeat unit [→2-α-l-Rhap-(1→2)-β-d-Ribf-(1→3)-α-l-Rhap-(1→3)-α-l-Rhap-(1→] (35). The previously reported structural study of O7 O-PS (32) was performed without the use of NMR spectroscopy and did not exclude the possibility that the anomeric configuration of one of the sugar residues could be reversed. Additionally, the possibility of terminal modification of O-PS chain was not investigated. We used 1H, 13C, and 31P NMR spectroscopy, including two-dimensional 1H,1H correlation spectroscopy (COSY), total correlation spectroscopy (TOCSY), NOE spectroscopy (NOESY), heteronuclear single-quantum coherence (HSQC), heteronuclear multiple bond correlation (HMBC), and HMQC-TOCSY experiments to unequivocally determine the structure of the repeat unit and the nonreducing terminus of the O-PS. NMR spectra of O-PS contained the signals for four major spin systems, which were assigned to a Ribf residue (A) and three Rhap residues (B, C, and D) (Table 1, Fig. 7A, and Figs. S2–S4). The β-anomeric configuration of Rib was confirmed by the position of the Rib C-1 signal at δ 108.2 (compare published data (36) of δ 103.1 and 108.0 for methyl glycosides of α- and β-Ribf, respectively). The α-configuration of all three Rha residues were inferred from the H-5 and C-5 chemical shifts at δH 3.76–3.90 and δC 70.4–70.7 (compare published data (37) of δH 3.86, δC 69.1 for α-Rhap, δH 3.39, δC 72.8 for β-Rhap). These data, together with the sequence of the sugar residues determined by HMBC and NOESY (Fig. S2) experiments, confirmed the structure of O7 O-PS reported earlier (Fig. 7A).
Table 1.
1H and 13C NMR chemical shifts of K. pneumoniae 264(1) serotype O7 O-PS
| Sugar residue | H-1 C-1 | H-2 C-2 | H-3 C-3 | H-4 C-4 | H-5 (5a,b) C-5 | H-6 C-6 |
|---|---|---|---|---|---|---|
| Repeat unit | ||||||
| →2)-β-d-Ribf-(1→ | ||||||
| A | 5.35 | 4.22 | 4.44 | 4.07 | 3.87, 3.71 | |
| 108.2 | 81.9 | 71.0 | 83.8 | 63.0 | ||
| →3)-α-d-Rhap-(1→ | ||||||
| B | 5.06 | 4.24 | 3.91 | 3.53 | 3.90 | 1.31 |
| 103.3 | 71.1 | 79.9 | 72.3 | 70.4 | 18.0 | |
| →3)-α-d-Rhap-(1→ | ||||||
| C | 4.98 | 4.18 | 3.85 | 3.57 | 3.80 | 1.29 |
| 103.2 | 71.1 | 79.4 | 72.5 | 70.7 | 17.9 | |
| →2)-α-d-Rhap-(1→ | ||||||
| D | 5.15 | 4.10 | 3.93 | 3.53 | 3.76 | 1.32 |
| 100.9 | 79.3 | 71.0 | 73.4 | 70.7 | 18.0 | |
| Non-reducing enda | ||||||
| →3)-α-d-Rhap-(1→ | ||||||
| B′ | 5.09 | 4.27 | 4.29 | 3.60 | 3.90 | 1.31 |
| 103.2 | 70.7 | 76.7 | 72.2 | 70.4 | 18.0 |
a The signals for the methylphosphate group are at δH 3.63, δC 54.3, and δP 1.75.
Figure 7.
K. pneumoniae O7 O-PS is terminated with a methylphosphate group on C3 of the nonreducing terminal rhamnose residue. A, the determined repeat unit and nonreducing terminal structure of the O7 O-PS. Rha, l-rhamnose; Rib, d-ribose. B, part of the one-dimensional selective TOCSY spectrum obtained after irradiation of B′ H-1 at δ 5.09. 1H,31P HMQC-TOCSY (C) and 1H,31P HMBC (D) spectra demonstrate correlations between phosphate and methyl protons as well as protons within the B′ spin system. MeP, methylphosphate. The corresponding part of the 1H NMR spectrum is shown along the horizontal axis.
In addition to the signals for sugar residues, the NMR spectra contained a characteristic sharp signal at δH/δC 3.63/54.3 with JH,P = 11 Hz, which was previously assigned to a methylphosphate group at the nonreducing terminus of E. coli O9a O-PS (33). The presence of a methylphosphate group was confirmed by correlation between methyl protons and phosphate at δH/δP 3.64/1.75 observed in the 1H,31P HMBC spectrum (Fig. 7D). Furthermore, a 1H,31P HMQC-TOCSY experiment showed correlation between phosphate and H-1 to H-5 of a minor α-Rhap spin system denoted as B′. The selective one-dimensional TOCSY (Fig. 7B) and NOESY experiments (not shown) confirmed the assignment of B′ H-1 to H-6 signals. A relatively low-field position of the signal for B′ C-3, together with strong HMBC correlation between phosphate and B′ H-3, demonstrate that the methylphosphate group is attached at position O3 of B′. The linkage between B′ and the next sugar residue could not be determined unambiguously, although the HMBC correlation at δH/δC 5.09/79.3 indicates that B′ is linked to another Rha residue. Thus, the NMR spectroscopic analysis confirmed the structure of the O7 repeat unit reported earlier (35) and established the second known example of a methylphosphate O-PS terminal modification.
The methylphosphate group is necessary but not sufficient for CBM binding and transport
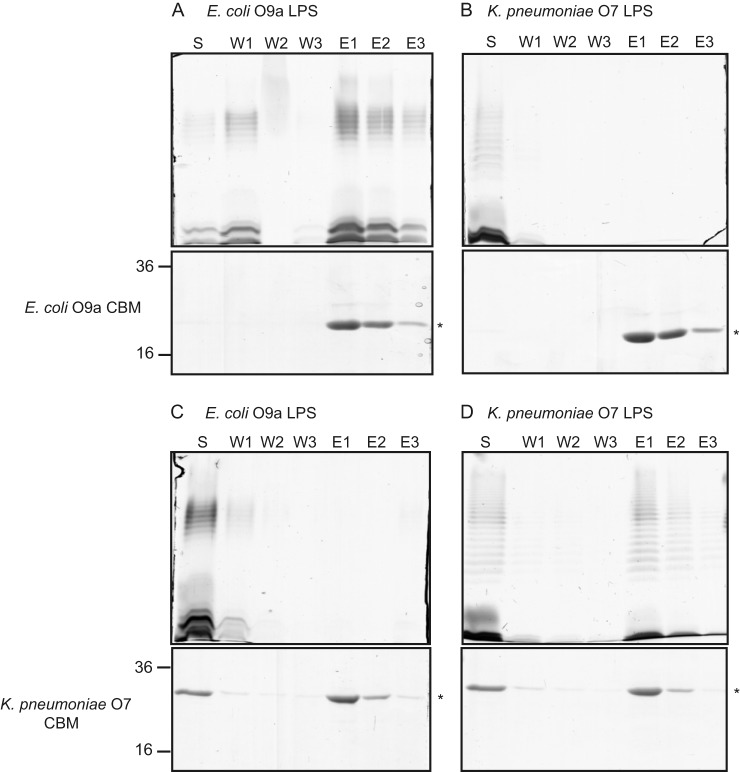
To examine whether the nonreducing terminal modification is the sole contributor to WztO9a-C O-PS recognition, we performed a pulldown experiment using the E. coli O9a CBM with K. pneumoniae O7 LPS (Fig. 8, A and B). No O7 LPS was detected in elution fractions, indicating that the CBM did not bind O7 LPS. To validate these results, the reciprocal experiments were done using purified WztO7-C. Evident from the silver stain, the O7 CBM was only able to bind its cognate LPS (Fig. 8, C and D). Finally, in vivo mutant complementation experiments were performed in E. coli O9a mutants lacking a functional O-PS ABC transporter (manA Δwzm-wzt::aphA-3) (Fig. 9). Expression of the K. pneumoniae O7 ABC transporter in this mutant could not restore translocation of the O-PS across the inner membrane, as evidenced by the absence of banding on silver-stained SDS-polyacrylamide gels, consistent with results from in vitro pulldown experiments. Because the O7 and O9a LPS structures contain the same nonreducing terminal modification, the lack of O-PS binding and export must be due to differences in the main-chain polymer.
Figure 8.
WztO9a-C and WztO7-C can only bind LPS from the native serotype. Purified WztO9a-C (O9a CBM; A and B) and WztO7-C (O7 CBM; C and D) were incubated with the O9a and O7 LPSs. Reaction mixtures consisting of 200 μg of CBM and 200 μg of LPS were mixed with nickel magnetic beads and collected using a magnet. The samples are unbound material in the supernatant (S) and material released by sequential washes of the beads in the absence (W1–W3) or presence of imidazole (E1–E3). Aliquots of the collected samples were subjected to SDS-PAGE. LPS was detected by silver staining (top panels), and protein was detected by simply blue staining (bottom panels).
Figure 9.
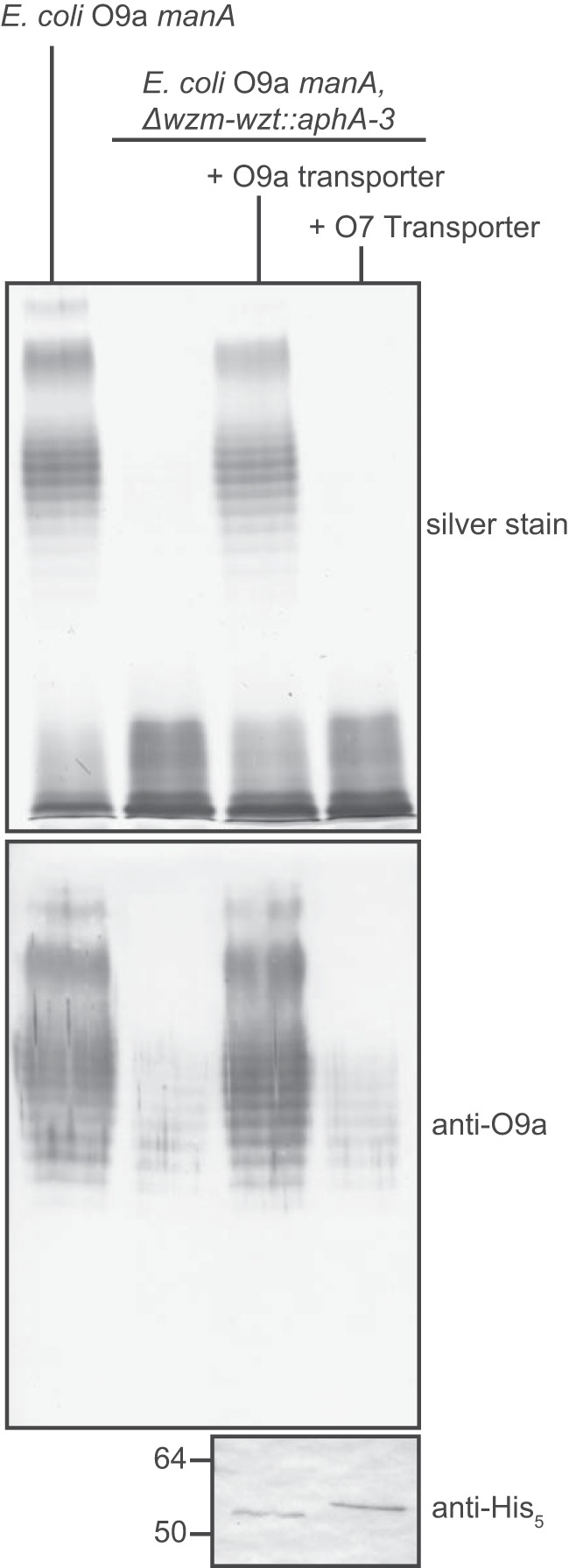
The K. pneumoniae O7 ABC transporter is unable to export the E. coli O9a O-PS. Overnight cultures of E. coli manA (CWG634; O9a O-PS control) and E. coli manA Δwzm-wzt::aph3-A (CWG638) transformed with plasmids encoding either the O9a (pWQ332) or O7 (pWQ1014) transporter (Wzm-Wzt-His6) were subcultured for 3 h in medium supplemented with 0.1% d-mannose and 0.1% l-arabinose to induce O-PS biosynthesis and transporter expression, respectively. LPS profiles were examined in proteinase K–treated whole-cell lysates using silver-stained SDS-polyacrylamide gels (top). α-His5 Western blots of whole cell lysates were used to confirm transporter expression (bottom). O-PS present in lanes 2 and 4 of the Western blot is a result of nonexported O-PS, evident from its lack of silver staining.
Discussion
The E. coli O9a O-PS biosynthesis and export system represents an elegant model for regulating polymer chain length distribution. By combining a molecular ruler with polymerization-termination and exporter-recognition mechanisms, the cell ensures an appropriate O-PS length distribution to optimize survival within the host. This system has become an important and influential prototype for similar strategies involved in the assembly of a variety of O-PS and other cell-surface glycans from Gram-negative and Gram-positive bacteria. The experimental platform for investigating substrate engagement by the E. coli O9a transporter therefore has broad relevance in microbial glycobiology.
Comparable systems have been identified in E. coli O8 and K. pneumoniae O12 and in S-layer glycoprotein assembly in Geobacillus stearothermophilus NRS2004/3a, Geobacillus tepidamans GS5-97T, and Aneurinibacillus thermoaerophilus L420-91T (27). In most examples, a single terminal residue (and presumed termination-export signal) is used. Examples include an O-methyl group on E. coli O8 O-PS and on the S-layer glycans from G. stearothermophilus NRS2004/3a (38) and A. thermophilus L420-91T (39), as well as a Kdo on K. pneumoniae O12 (25). An exception is the addition of GlcNAc and N-acetylmuramic acid residues onto separate positions of the final rhamnose in G. tepidimans GS5-97T (40), but only one of these residues is positioned to directly impede chain extension. The overall strategy for the biosynthesis of the terminally methylated d-rhamnose polymer in G. stearothermophilus NRS2004/3a is remarkably similar to that used for the methylated d-mannose E. coli O8 antigen (41). In contrast, E. coli O9a utilizes a binary terminator, consisting of a phosphate and an O-methyl group (18, 25). An identical structure terminates the related mannans produced by E. coli O9, K. pneumoniae O3a/O3b/O3c (42), and Hafnia alvei PCM1223 (33). This reflects lateral gene transfer of the locus, followed by diversification of the repeat-unit structure by mutations in wbdA. K. pneumoniae O7 uses the same methylphosphate terminating moiety to cap a different repeat-unit structure.
Despite the similar termination chemistry, the K. pneumoniae O7-biosynthesis gene cluster does not encode a homolog of the E. coli O9a WbdD protein. Instead, a candidate methyltransferase domain is part of a large multidomain predicted protein (Orf6) that also contains two candidate glycosyltransferases (belonging to family GT2) and a coiled-coil region (Fig. S1). This multidomain format is reminiscent of the K. pneumoniae O12 WbbB protein, where the terminating CMP-Kdo–dependent glycosyltransferase is separated by a coiled-coil domain from the glycosyltransferase modules that polymerize the repeat-unit glycan (43, 44). The kinase involved in the O7 termination reaction has not been identified, but, by analogy to WbdD, we anticipate this domain being located in proximity to the methyltransferase. Orf6 does possess an extensive region (47 kDa) downstream of the methyltransferase that could not be assigned by a homology search, and this may provide kinase activity. Because the identity of the kinase is not central to this study and is not required for data interpretation, it was not pursued here.
We demonstrated that the WbdD kinase domain was sufficient for chain length determination. However, the quality control mechanism conferred by the CBM shows an additional dependence on methyltransferase activity. STDD NMR experiments demonstrated that WztO9a-C interacts directly with the methyl substituent at the nonreducing terminus, and this likely provides the most prominent hydrophobic interaction involved in O9a polysaccharide recognition by Wzt. The O9a CBM crystal structure has been solved, and the binding pocket was characterized using site-directed mutagenesis in conjugation with transport and CBM binding experiments (23). Several residues were necessary for substrate binding. Aromatic residues (Trp-286 and Tyr-404) presumably contribute via π-stacking with the monosaccharide rings, whereas polar residues (Asn-348, Arg-402, and Asp-405) may interact with sugar hydroxyl moieties or the terminal unit. Because these are conserved between WztO9a and WztO7, additional side chains distinct to each homologue must provide the noncovalent interactions critical for serotype specificity. However, these are not sufficient for robust productive binding in the absence of any terminal modification, as seen here and in previous experiments using in vitro–synthesized O9a glycan with or without complete terminal modification (23). STDD NMR experiments implicated the terminal methyl group as an interaction group with the CBM. Unfortunately, these experiments could not identify additional hydrophobic interactions arising from the main polymer, due to inefficient magnetization transfer. Furthermore, STD approaches are not suitable for identification of electrostatic or polar interactions. Interactions contributed by the main-chain polymer may be predominantly polar, arising from hydroxyl groups, and these are not detected because of the fast exchange of hydroxyl protons with solvent. It is likely that the phosphate moiety on the O9a glycan provides contact points within the binding pocket (e.g. through electrostatic interaction with the critical Arg-402 residue) (23) and therefore still contributes to recognition of the fully modified glycan. The extensive binding pocket likely accommodates main-chain sugars that contribute to the binding interface.
The O9a and O7 O-PS structures contain identical termination groups on the C-3 position of the nonreducing terminal sugar of their given polysaccharides. Because WztO9a-C did not interact with O7 LPS during pulldown experiments, we conclude that one or more monosaccharide units provide functional groups that contribute to direct interactions with the CBM. Cocrystal structures with terminated O-PS analogues are essential to conclusively verify structural features related to O-PS specificity, but, to date, we have been unable to obtain structures of CBM:O-PS complexes for a detailed view of the interactions.
Here, we focus solely on the role of the terminal modification in the substrate recognition process. How this interfaces with substrate engagement by the transporter proper remains to be resolved. The ultimate goal of this and related studies is to recapitulate the export process with purified proteins and substrates in proteoliposomes, and, in this context, resolution of the chemical determinants for export is a critical step toward this objective. However, why E. coli O9a has evolved and maintained a more complex strategy, compared with the simple methylation process in the related E. coli O8, remains unanswered.
Experimental procedures
Strains, plasmids, and growth conditions
The bacterial strains and plasmids used in this study are described in Table 2. Cultures were grown in lysogeny broth (LB) at 37 °C with aeration and were supplemented with 100 μg ml−1 of ampicillin, d-glucose (0.4% w/v), d-mannose (0.4% w/v), or l-arabinose (0.0002–0.2% (w/v)) where appropriate, unless otherwise specified.
Table 2.
List of strains and plasmids
| Strain/plasmid name | Description | Source |
|---|---|---|
| Strain | ||
| E. coli E69 | Serotype O9a:K30 | F. Orskov |
| K. pneumoniae 264(1) | Serotype O7:K67 | F. Orskov |
| CWG28 | E. coli E69 derivative; trp his lac rpsL cpsK30; Smr | Ref. 52 |
| CWG634 | CWG28 derivative; manA; Smr, Tcr | Ref. 17 |
| CWG635 | CWG634 derivative; wbdD::aacC1; Smr, Tcr, Gmr | Ref. 17 |
| CWG638 | CWG634 derivative; Δwzm-wzt::aphA-3; Smr Tcr Kmr | Ref. 17 |
| CWG900 | CWG634 derivative; ΔwbdD; Smr, Tcr | Ref. 16 |
| CWG1412 | CWG635 derivative; Δwzm-wzt::aphA-3; Smr Tcr Kmr, Gmr | This study |
| Plasmid | ||
| pBAD24 | Plasmid vector with l-arabinose-inducible promoter; Apr | Ref. 53 |
| pKD3 | Source of chloramphenicol resistance cassette; Cmr | Ref. 54 |
| pWQ470 | pBAD24 derivative containing His6-wbdD | Ref. 16 |
| pWQ829 | pWQ470 derivative containing His6-wbdDH132A | Ref. 19 |
| pWQ830 | pWQ470 derivative containing His6-wbdDD351A | Ref. 19 |
| pWQ831 | pWQ470 derivative containing His6-wbdDY16FA | Ref. 19 |
| pWQ391 | pBAD24 derivative containing a fragment including wzm-wzt from K. pneumoniae O2a and the Cmr cassette from pKD3 cloned into the ScaI site; Cmr | Ref. 11 |
| pWQ1013 | pBAD24 derivative containing the open-reading frame encoding His6-Wzt-C (amino acids 268–464) from K. pneumoniae O7 cloned between the NheI and NcoI sites | This study |
| pWQ1014 | pBAD24 derivative containing the open reading frame encoding Wzm-Wzt-His6 tag from K. pneumoniae O7 cloned between the NheI and NcoI sites | This study |
| pWQ332 | pBAD24 derivative containing E. coli O9a Wzm-Wzt-His6 | Ref. 22 |
| pWQ424 | pBAD24 derivative containing E. coli O9a Wzt-C-His10 | Ref. 23 |
DNA methods
KOD Hot Start DNA polymerase (Novagen) was used for cloning DNA fragments. Oligonucleotide primers were obtained from Sigma and are listed in Table S1. Amplified products were purified using a GeneJET purification kit (Thermo Fisher Scientific), and restriction endonuclease digests were performed using enzymes from New England Biolabs. Restriction sites used are indicated in Table 1. Plasmids were confirmed by restriction digests and by DNA sequencing performed by the Advanced Analysis Centre, University of Guelph.
Construction of E. coli CWG1412
E. coli CWG1412 (manA wbdD::aacC1 Δwzm-wzt::aphA-3) was constructed using the same primers and protocol as described previously for construction of CWG638 (17). However, CWG635 was used as the parent strain instead of CWG634.
Genomic DNA sequencing
Whole-genome shotgun sequence data for K. pneumoniae 264(1) was generated using the Solexa paired-end sequencing platform (Illumina) and genomic DNA prepared using the Illumina sample preparation kit. De novo assembly of sequencing reads (∼100× coverage) was performed with Velvet (45). The sequence of the gene cluster for O7 O-PS biosynthesis has been deposited in NCBI (GenBankTM accession no. MN173773).
SDS-PAGE and immunoblotting
To prepare whole-cell lysates for SDS-PAGE and Western immunoblotting, bacterial cells (1 A600 nm unit equivalent) were collected by centrifugation, solubilized in SDS-PAGE loading buffer, and heated to 100 °C for 10 min prior to electrophoresis. For LPS analysis, the whole-cell lysates were treated with proteinase K (Invitrogen) for 1 h at 55 °C (46). SDS-PAGE was performed using 12% acrylamide resolving gels in Tris-glycine buffer (47). LPS was visualized by staining with a silver nitrate solution (48), and protein was detected using SimplyBlue SafeStain (Invitrogen).
For immunoblot analyses, material resolved by SDS-PAGE was transferred to nitrocellulose membranes (Protran; PerkinElmer Life Sciences). O9a antigen was detected using polyclonal antibodies raised in rabbits (17). Alkaline phosphatase–conjugated goat anti-rabbit was used as a secondary antibody (Cedar Lane Laboratories). Hexahistidine-tagged proteins were detected using mouse-anti-His5 mAb (Qiagen) and alkaline phosphatase-conjugated goat anti-mouse secondary antibody (CedarLane Laboratories). Immunoblots were developed with nitro blue tetrazolium and 5-bromo-4-chloro-3-indoyl phosphate (Roche Applied Science).
Immunofluorescence microscopy
Overnight cultures grown in LB, 0.4% d-glucose were subcultured 1:100 into fresh LB, glucose. After 4 h, cells were resuspended in fresh medium containing 0.4% d-mannose and 0.02% l-arabinose to facilitate WbdD synthesis, and growth was continued for an additional 1 h before cells were harvested. Microscopy was performed on formaldehyde-fixed whole cells as described previously (17), using rabbit-anti-O9a primary antibody and FITC-conjugated goat anti-rabbit secondary antibody (Thermo Fisher Scientific).
Wzt-C purification
WztO9a-C was purified as described previously (23). Briefly, E. coli Top10 harboring pWQ424 was grown at 37 °C with aeration to mid-exponential growth phase and induced with 0.3% l-arabinose for an additional 2 h. Cells were collected by centrifugation, resuspended in buffer A (25 mm Tris, pH 7.5, containing 100 mm NaCl), and lysed by sonication. Cleared lysate was applied to 1-ml columns of nickel affinity resin. Resin was washed with buffer A containing 20 mm imidazole, and protein was eluted using buffer A containing 200 mm imidazole. WztO7-C was purified from E. coli Top10 harboring pWQ1013, using the same growth and induction conditions. WztO7-C was also purified by nickel-affinity chromatography, but buffer B (25 mm Bis-Tris, pH 7.0, 250 mm NaCl) was used.
LPS purification
WT E. coli O9a LPS was purified from overnight cultures of E. coli CWG28 and E69 for pulldown experiments and STDD NMR experiments, respectively. WT K. pneumoniae O7 LPS was purified from K. pneumoniae 264(1). LPS containing nonterminated O-PS was prepared from E. coli CWG1412 (manA wbdD::aacC1 Δwzm-wzt::aphA-3), which lacks innate termination and transport activities, transformed with pWQ391, which provides the ABC transporter from K. pneumoniae O2a. This transporter operates independently of O-PS repeat-unit structure and does not require a terminating moiety (11, 12). LPS lacking a nonreducing terminal O-PS modification was purified from E. coli CWG1412 transformed with pWQ391 and either pWQ830 (WbdDD351A) or pWQ831 (wbdDY16F). The WbdDY16F variant lacks methyltransferase activity (19). l-Arabinose (0.1%) was added to induce the recombinant K. pneumoniae O2a ABC transporter. After 30 min of growth, d-mannose (0.4%) was added to initiate O-PS expression, and the cultures were grown for an additional 2 h. LPS was purified using the hot water phenol method (49). After dialysis of the extraction mixture against water to remove the phenol, proteins and nucleic acids were precipitated using cold, aqueous TCA and dialyzed until neutral. To separate O7 LPS from capsule, LPS was sedimented by ultracentrifugation for 18 h at 100,000 × g in 20 mm NaOAc, pH 7.0.
O-PS for NMR analysis was isolated by acid hydrolysis of LPS at 100 °C in 2% (v/v) acetic acid, followed by centrifugation at 13,000 × g to remove lipid precipitate. The carbohydrate-containing supernatant was fractionated on a Sephadex G-50 superfine column (2.5 × 75 cm) in 50 mm pyridinium acetate buffer (pH 4.5) at a flow rate of 0.6 ml min−1. Elution was monitored with a Smartline 2300 refractive index detector (Knauer). The O9a O-PS–containing fraction was acidic (potentially due to contaminating capsule; data not shown), so the polysaccharide mixture was applied to a DEAE Fast Flow (GE Healthcare) column and eluted with a 5–500 mm sodium phosphate gradient at pH 6.3 monitored using absorbance spectroscopy at a wavelength of 208 nm (50). The O9a polysaccharide-containing fractions were pooled and buffer-exchanged by chromatography on a Sephadex G-50 superfine column equilibrated with water.
Binding assays
In vitro binding assays were performed as described previously (12, 23). 1-ml reaction volumes of buffer B (25 mm Bis-Tris, pH 7.0, 250 mm NaCl) containing 200 μg of LPS and 200 μg of Wzt-C were incubated on a rotary shaker for 30 min at room temperature. This mixture was added to 50 μl of PureProteome nickel magnetic beads (Millipore), which were pre-equilibrated with buffer B, and incubated for 30 min at room temperature on a rotary shaker. The beads were collected with a magnet and washed three times with 500 μl of buffer B. Protein was eluted stepwise from the beads using three washes with 100 μl of buffer B containing 500 mm imidazole.
STDD NMR
NMR studies were performed at the University of Guelph Advanced Analysis Centre. The chemical shifts are referenced to 3-trimethylsilylpropanoate-2,2,3,3-d4 (δH 0, δC −1.6) added as an internal standard. The Bruker TopSpin 3.2 program was used to acquire and process the NMR data.
STD NMR samples contained 22.2 mg/ml (estimated 2.2 mm) O9a polysaccharide and 0.022 mm WztO9a-C (omitted for the “double difference” sample) in 25 mm Tris, pH 7.4, 100 mm NaCl. Deuterium oxide was added to 10% (v/v). NMR spectra were collected on a Bruker AVANCE III 600-MHz spectrometer equipped with a 5-mm TCI cryoprobe. The sample temperature was regulated at 298.2 ± 1 K. STD spectra (28) were acquired using the Bruker pulse sequence stddiffesgp, which suppresses the water resonance via an excitation sculpting scheme (51). WztO9a-C was saturated with a train of Gaussian-shaped pulses of 25 ms each (calibrated to the correct power level for a 90° Gaussian pulse), applied at 0.67 ppm for the “on-resonance” spectrum and at −11.66 ppm for the “off-resonance” spectrum. Four different irradiation times of 1, 2, 3, and 6 s were employed, and the total prescan delay (i.e. including the irradiation time) was kept constant at 7 s. The acquisition time was 1.7 s. STD data were obtained by subtracting the on-resonance FID from the off-resonance FID using the stdsplit macro provided by the Bruker software program TopSpin.
The frequency-selective pulse employed in the excitation sculpting scheme is not infinitely narrow and can cause undesired attenuation of nearby ligand resonances (e.g. anomeric protons). In the default implementation, the frequency-selective pulse duration is 2 ms, which resulted in the undesired suppression of the anomeric proton resonance at 5.04 ppm to 6% of its nominal intensity. For the experiments presented in this paper, the frequency-selective pulse duration was increased to 5 ms; the reduction in the corresponding pulse bandwidth restored the intensity of the 5.04-ppm anomeric proton to 80% of the nominal value.
When performed on larger ligands, the STD experiment can result in partial saturation of the ligand. To correct for this, a set of “ligand-only” STD experiments were performed on a second sample not containing WztO9a-C (but otherwise identical to the sample described above), and the resulting “ligand-only” STD data were subtracted from the STD data of O9a in the presence of WztO9a-C to produce the STDD spectra reported (32).
Elucidation of the K. pneumoniae O7 O-PS structure
O-PS samples were lyophilized twice in 99.9% D2O for deuterium exchange and then analyzed in 99.9% D2O at 30 °C. Data were collected using a Bruker Avance III 600-MHz spectrometer equipped with a 5-mm TCI cryoprobe, with the exception of 31P-based NMR experiments, performed on a Bruker 400-MHz Avance III spectrometer equipped with a 5-mm broadband Prodigy cryoprobe. Mixing times of 100 and 200 ms were used in TOCSY and NOESY experiments, respectively. The HMBC experiment was optimized for the JH,C coupling constant of 8 Hz and a JH,P coupling constant of 11 Hz. Mixing times of 100 and 80 ms were used for selective TOCSY and 1H,31P HMQC-TOCSY, respectively.
Author contributions
E. M., S. D. K., and C. W. conceived the study. E. M., S. D. K., and S. A. performed the experiments. B. L. provided genome sequence data. E. M., S. D. K., S. A., B. R. C., O. G. O., and C. W. analyzed and interpreted the data. E. M., S. D. K., S. A., O. G. O., and C. W. prepared the initial manuscript draft, and all authors provided input for the final version.
Supplementary Material
Acknowledgments
We are grateful to Dr. Vladimir Ladizhansky for helpful discussions concerning NMR experiments, Gregory Whitfield for purifying WztO9a-C and LPS, and Lu Wang and Xi Gou for genomic sequence data.
This research was supported by funding from the Natural Sciences and Engineering Research Council of Canada (to C. W.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Table S1 and Figs. S1–S4.
- LPS
- lipopolysaccharide
- O-PS
- O-antigeni polysaccharide
- ABC
- ATP-binding cassette
- Lpt
- LPS transport
- GT
- glycosyltransferase
- CBM
- carbohydrate-binding module
- Und-P
- undecaprenol phosphate
- Kdo
- 3-deoxy-d-manno-oct-2-ulosonic acid
- STD
- saturation transfer difference
- STDD
- saturation transfer double difference
- COSY
- correlation spectroscopy
- TOCSY
- total correlation spectroscopy
- NOESY
- nuclear Overhauser effect spectroscopy
- HSQC
- heteronuclear single-quantum coherence
- HMBC
- heteronuclear multiple-bond correlation
- LB
- lysogeny broth
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
References
- 1. Whitfield C., and Trent M. S. (2014) Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 83, 99–128 10.1146/annurev-biochem-060713-035600 [DOI] [PubMed] [Google Scholar]
- 2. Raetz C. R. H., and Whitfield C. (2002) Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700 10.1146/annurev.biochem.71.110601.135414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lerouge I., and Vanderleyden J. (2002) O-antigen structural variation: mechanisms and possible roles in animal/plant-microbe interactions. FEMS Microbiol. Rev. 26, 17–47 10.1111/j.1574-6976.2002.tb00597.x [DOI] [PubMed] [Google Scholar]
- 4. Grossman N., Schmetz M. A., Foulds J., Klima E. N., Jimenez-Lucho V. E., Leive L. L., Joiner K. A., and Jiminez V. (1987) Lipopolysaccharide size and distribution determine serum resistance in Salmonella montevideo. J. Bacteriol. 169, 856–863 10.1128/jb.169.2.856-863.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burns S. M., and Hull S. I. (1998) Comparison of loss of serum resistance by defined lipopolysaccharide mutants and an acapsular mutant of uropathogenic Escherichia coli O75:K5. Infect. Immun. 66, 4244–4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mostowy R. J., and Holt K. E. (2018) Diversity-generating machines: genetics of bacterial sugar-coating. Trends Microbiol. 26, 1008–1021 10.1016/j.tim.2018.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Greenfield L. K., and Whitfield C. (2012) Synthesis of lipopolysaccharide O-antigens by ABC transporter-dependent pathways. Carbohydr. Res. 356, 12–24 10.1016/j.carres.2012.02.027 [DOI] [PubMed] [Google Scholar]
- 8. Liston S. D., Mann E., and Whitfield C. (2017) Glycolipid substrates for ABC transporters required for the assembly of bacterial cell-envelope and cell-surface glycoconjugates. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 1862, 1394–1403 10.1016/j.bbalip.2016.10.008 [DOI] [PubMed] [Google Scholar]
- 9. Okuda S., Sherman D. J., Silhavy T. J., Ruiz N., and Kahne D. (2016) Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat. Rev. Microbiol. 14, 337–345 10.1038/nrmicro.2016.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sperandeo P., Martorana A. M., and Polissi A. (2017) The lipopolysaccharide transport (Lpt) machinery: A nonconventional transporter for lipopolysaccharide assembly at the outer membrane of Gram-negative bacteria. J. Biol. Chem. 292, 17981–17990 10.1074/jbc.R117.802512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kos V., Cuthbertson L., and Whitfield C. (2009) The Klebsiella pneumoniae O2a antigen defines a second mechanism for O antigen ATP-binding cassette transporters. J. Biol. Chem. 284, 2947–2956 10.1074/jbc.M807213200 [DOI] [PubMed] [Google Scholar]
- 12. Mann E., Mallette E., Clarke B. R., Kimber M. S., and Whitfield C. (2016) The Klebsiella pneumoniae O12 ATP-binding Cassette (ABC) transporter recognizes the terminal residue of its O-antigen polysaccharide substrate. J. Biol. Chem. 291, 9748–9761 10.1074/jbc.M116.719344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Greenfield L. K., Richards M. R., Li J., Wakarchuk W. W., Lowary T. L., and Whitfield C. (2012) Biosynthesis of the polymannose lipopolysaccharide O-antigens from Escherichia coli serotypes O8 and O9a requires a unique combination of single- and multiple-active site mannosyltransferases. J. Biol. Chem. 287, 35078–35091 10.1074/jbc.M112.401000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liston S. D., Clarke B. R., Greenfield L. K., Richards M. R., Lowary T. L., and Whitfield C. (2015) Domain interactions control complex formation and polymerase specificity in the biosynthesis of the Escherichia coli O9a antigen. J. Biol. Chem. 290, 1075–1085 10.1074/jbc.M114.622480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Greenfield L. K., Richards M. R., Vinogradov E., Wakarchuk W. W., Lowary T. L., and Whitfield C. (2012) Domain organization of the polymerizing mannosyltransferases involved in synthesis of the Escherichia coli O8 and O9a lipopolysaccharide O-antigens. J. Biol. Chem. 287, 38135–38149 10.1074/jbc.M112.412577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Clarke B. R., Greenfield L. K., Bouwman C., and Whitfield C. (2009) Coordination of polymerization, chain termination, and export in assembly of the Escherichia coli lipopolysaccharide O9a antigen in an ATP-binding cassette transporter-dependent pathway. J. Biol. Chem. 284, 30662–30672 10.1074/jbc.M109.052878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Clarke B. R., Cuthbertson L., and Whitfield C. (2004) Nonreducing terminal modifications determine the chain length of polymannose O antigens of Escherichia coli and couple chain termination to polymer export via an ATP-binding cassette transporter. J. Biol. Chem. 279, 35709–35718 10.1074/jbc.M404738200 [DOI] [PubMed] [Google Scholar]
- 18. Clarke B. R., Richards M. R., Greenfield L. K., Hou D., Lowary T. L., and Whitfield C. (2011) In vitro reconstruction of the chain termination reaction in biosynthesis of the Escherichia coli O9a O-polysaccharide: the chain-length regulator, WbdD, catalyzes the addition of methyl phosphate to the non-reducing terminus of the growing glycan. J. Biol. Chem. 286, 41391–41401 10.1074/jbc.M111.295857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hagelueken G., Huang H., Clarke B. R., Lebl T., Whitfield C., and Naismith J. H. (2012) Structure of WbdD: a bifunctional kinase and methyltransferase that regulates the chain length of the O antigen in Escherichia coli O9a. Mol. Microbiol. 86, 730–742 10.1111/mmi.12014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hagelueken G., Clarke B. R., Huang H., Tuukkanen A., Danciu I., Svergun D. I., Hussain R., Liu H., Whitfield C., and Naismith J. H. (2015) A coiled-coil domain acts as a molecular ruler to regulate O-antigen chain length in lipopolysaccharide. Nat. Struct. Mol. Biol. 22, 50–56 10.1038/nsmb.2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. King J. D., Berry S., Clarke B. R., Morris R. J., and Whitfield C. (2014) Lipopolysaccharide O antigen size distribution is determined by a chain extension complex of variable stoichiometry in Escherichia coli O9a. Proc. Natl. Acad. Sci. U.S.A. 111, 6407–6412 10.1073/pnas.1400814111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cuthbertson L., Powers J., and Whitfield C. (2005) The C-terminal domain of the nucleotide-binding domain protein Wzt determines substrate specificity in the ATP-binding cassette transporter for the lipopolysaccharide O-antigens in Escherichia coli serotypes O8 and O9a. J. Biol. Chem. 280, 30310–30319 10.1074/jbc.M504371200 [DOI] [PubMed] [Google Scholar]
- 23. Cuthbertson L., Kimber M. S., and Whitfield C. (2007) Substrate binding by a bacterial ABC transporter involved in polysaccharide export. Proc. Natl. Acad. Sci. U.S.A. 104, 19529–19534 10.1073/pnas.0705709104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bi Y., Mann E., Whitfield C., and Zimmer J. (2018) Architecture of a channel-forming O-antigen polysaccharide ABC transporter. Nature 553, 361–365 10.1038/nature25190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vinogradov E., Frirdich E., MacLean L. L., Perry M. B., Petersen B. O., Duus JØ., Whitfield C. (2002) Structures of lipopolysaccharides from Klebsiella pneumoniae: eluicidation of the structure of the linkage region between core and polysaccharide O chain and identification of the residues at the non-reducing termini of the O chains. J. Biol. Chem. 277, 25070–25081 10.1074/jbc.M202683200 [DOI] [PubMed] [Google Scholar]
- 26. Mann E., Ovchinnikova O. G., King J. D., and Whitfield C. (2015) Bacteriophage-mediated glucosylation can modify lipopolysaccharide O-antigens synthesized by an ATP-binding cassette (ABC) transporter-dependent assembly mechanism. J. Biol. Chem. 290, 25561–25570 10.1074/jbc.M115.660803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cuthbertson L., Kos V., and Whitfield C. (2010) ABC transporters involved in export of cell surface glycoconjugates. Microbiol. Mol. Biol. Rev. 74, 341–362 10.1128/MMBR.00009-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mayer M., and Meyer B. (1999) Characterization of ligand binding by saturation transfer difference NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 38, 1784–1788 [DOI] [PubMed] [Google Scholar]
- 29. van de Weerd R., Berbís M. A., Sparrius M., Maaskant J. J., Boot M., Paauw N. J., de Vries N., Boon L., Baba O., Cañada F. J., Geurtsen J., Jiménez-Barbero J., and Appelmelk B. J. (2015) A murine monoclonal antibody to glycogen: characterization of epitope-fine specificity by saturation transfer difference (STD) NMR spectroscopy and its use in mycobacterial capsular α-glucan research. Chembiochem 16, 977–989 10.1002/cbic.201402713 [DOI] [PubMed] [Google Scholar]
- 30. Kooistra O., Herfurth L., Lüneberg E., Frosch M., Peters T., and Zähringer U. (2002) Epitope mapping of the O-chain polysaccharide of Legionella pneumophila serogroup 1 lipopolysaccharide by saturation-transfer-difference NMR spectroscopy. Eur. J. Biochem. 269, 573–582 10.1046/j.0014-2956.2001.02684.x [DOI] [PubMed] [Google Scholar]
- 31. Mayer M., and Meyer B. (2001) Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J. Am. Chem. Soc. 123, 6108–6117 10.1021/ja0100120 [DOI] [PubMed] [Google Scholar]
- 32. Claasen B., Axmann M., Meinecke R., and Meyer B. (2005) Direct observation of ligand binding to membrane proteins in living cells by a saturation transfer double difference (STDD) NMR spectroscopy method shows a significantly higher affinity of integrin αIIbβ3 in native platelets than in liposomes. J. Am. Chem. Soc. 127, 916–919 10.1021/ja044434w [DOI] [PubMed] [Google Scholar]
- 33. Kubler-Kielb J., Whitfield C., Katzenellenbogen E., and Vinogradov E. (2012) Identification of the methyl phosphate substituent at the non-reducing terminal mannose residue of the O-specific polysaccharides of Klebsiella pneumoniae O3, Hafnia alvei PCM 1223 and Escherichia coli O9/O9a LPS. Carbohydr. Res. 347, 186–188 10.1016/j.carres.2011.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kelley L. A., Mezulis S., Yates C. M., Wass M. N., and Sternberg M. J. E. (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lindberg B., Lönngren J., Nimmich W., and Rudén U. (1973) Structural studies on the Klebsiella O group 7 lipopolysaccharide. Acta Chem. Scand. 27, 3787–3790 10.3891/acta.chem.scand.27-3787 [DOI] [PubMed] [Google Scholar]
- 36. Bock K., and Pedersen C. (1983) Carbon-13 nuclear magnetic resonance spectroscopy of monosaccharides. Adv. Carbohydr. Chem. Biochem. 41, 27–66 10.1016/S0065-2318(08)60055-4 [DOI] [Google Scholar]
- 37. Jansson P. E., Kenne L., and Widmalm G. (1989) Computer-assisted structural analysis of polysaccharides with an extended version of casper using 1H- and 13C-N.M.R. data. Carbohydr. Res. 188, 169–191 10.1016/0008-6215(89)84069-8 [DOI] [PubMed] [Google Scholar]
- 38. Schäffer C., Wugeditsch T., Kählig H., Scheberl A., Zayni S., and Messner P. (2002) The surface layer (S-layer) glycoprotein of Geobacillus stearothermophilus NRS 2004/3a: analysis of its glycosylation. J. Biol. Chem. 277, 6230–6239 10.1074/jbc.M108873200 [DOI] [PubMed] [Google Scholar]
- 39. Schäffer C., Müller N., Christian R., Graninger M., Wugeditsch T., Scheberl A., and Messner P. (1999) Complete glycan structure of the S-layer glycoprotein of Aneurinibacillus thermoaerophilus GS4–GS97. Glycobiology 9, 407–414 10.1093/glycob/9.4.407 [DOI] [PubMed] [Google Scholar]
- 40. Kählig H., Kolarich D., Zayni S., Scheberl A., Kosma P., Schäffer C., and Messner P. (2005) N-acetylmuramic acid as capping element of α-d-fucose-containing S-layer glycoprotein glycans from Geobacillus tepidamans GS5–97T. J. Biol. Chem. 280, 20292–20299 10.1074/jbc.M501724200 [DOI] [PubMed] [Google Scholar]
- 41. Steiner K., Novotny R., Werz D. B., Zarschler K., Seeberger P. H., Hofinger A., Kosma P., Schäffer C., and Messner P. (2008) Molecular basis of S-layer glycoprotein glycan biosynthesis in Geobacillus stearothermophilus. J. Biol. Chem. 283, 21120–21133 10.1074/jbc.M801833200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guachalla L. M., Stojkovic K., Hartl K., Kaszowska M., Kumar Y., Wahl B., Paprotka T., Nagy E., Lukasiewicz J., Nagy G., and Szijártó V. (2017) Discovery of monoclonal antibodies cross-reactive to novel subserotypes of K. pneumoniae O3. Sci. Rep. 7, 6635 10.1038/s41598-017-06682-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Williams D. M., Ovchinnikova O. G., Koizumi A., Mainprize I. L., Kimber M. S., Lowary T. L., and Whitfield C. (2017) Single polysaccharide assembly protein that integrates polymerization, termination, and chain-length quality control. Proc. Natl. Acad. Sci. U.S.A. 114, E1215–E1223 10.1073/pnas.1613609114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ovchinnikova O. G., Mallette E., Koizumi A., Lowary T. L., Kimber M. S., and Whitfield C. (2016) Bacterial β-Kdo glycosyltransferases represent a new glycosyltransferase family (GT99). Proc. Natl. Acad. Sci. U.S.A. 113, E3120–E3129 10.1073/pnas.1603146113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zerbino D. R., and Birney E. (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hitchcock P. J., and Brown T. M. (1983) Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154, 269–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- 48. Tsai C. M., and Frasch C. E. (1982) A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119, 115–119 10.1016/0003-2697(82)90673-X [DOI] [PubMed] [Google Scholar]
- 49. Westphal O. (1965) Bacterial lipopolysaccharide-extraction with phenol water and further application of procedure. Methods Carbohydr. Chem. 1, 83–91 [Google Scholar]
- 50. Fedonenko Y. P., Zatonsky G. V., Konnova S. A., Zdorovenko E. L., and Ignatov V. V. (2002) Structure of the O-specific polysaccharide of the lipopolysaccharide of Azospirillum brasilense Sp245. Carbohydr. Res. 337, 869–872 10.1016/S0008-6215(02)00061-7 [DOI] [PubMed] [Google Scholar]
- 51. Hwang T. L., and Shaka A. J. (1995) Water suppression that works: excitation sculpting using arbitrary waveforms and pulsed field gradients. J. Magn. Reson. Ser. A 112, 275–279 10.1006/jmra.1995.1047 [DOI] [Google Scholar]
- 52. Whitfield C., Schoenhals G., and Graham L. (1989) Mutants of Escherichia coli O9:K30 with altered synthesis and expression of the capsular K30 antigen. J Gen Microbiol. 135, 2589–2599 [DOI] [PubMed] [Google Scholar]
- 53. Guzman L. M., Belin D., Carson M. J., and Beckwith J. (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130 10.1128/jb.177.14.4121-4130.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Datsenko K. A., and Wanner B. L. (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 97, 6640–6645 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.