Abstract
Type III polyketide synthases (PKSs) produce an incredibly diverse group of plant specialized metabolites with medical importance despite their structural simplicity compared with the modular type I and II PKS systems. The type III PKSs use homodimeric proteins to construct the molecular scaffolds of plant polyketides by iterative condensations of starter and extender CoA thioesters. Ever since the structure of chalcone synthase (CHS) was disclosed in 1999, crystallographic and mutational studies of the type III PKSs have explored the intimate structural features of these enzyme reactions, revealing that seemingly minor alterations in the active site can drastically change the catalytic functions and product profiles. New structures described in this review further build on this knowledge, elucidating the detailed catalytic mechanism of enzymes that make curcuminoids, use extender substrates without the canonical CoA activator, and use noncanonical starter substrates, among others. These insights have been critical in identifying structural features that can serve as a platform for enzyme engineering via structure-guided and precursor-directed engineered biosynthesis of plant polyketides. In addition, we describe the unique properties of the recently discovered “second-generation” type III PKSs that catalyzes the one-pot formation of complex molecular scaffolds from three distinct CoA thioesters or from “CoA-free” substrates, which are also providing exciting new opportunities for synthetic biology approaches. Finally, we consider post-type III PKS tailoring enzymes, which can also serve as useful tools for combinatorial biosynthesis of further unnatural novel molecules. Recent progress in the field has led to an exciting time of understanding and manipulating these fascinating enzymes.
Keywords: enzyme, enzyme mechanism, enzyme structure, biosynthesis, natural product biosynthesis
Introduction
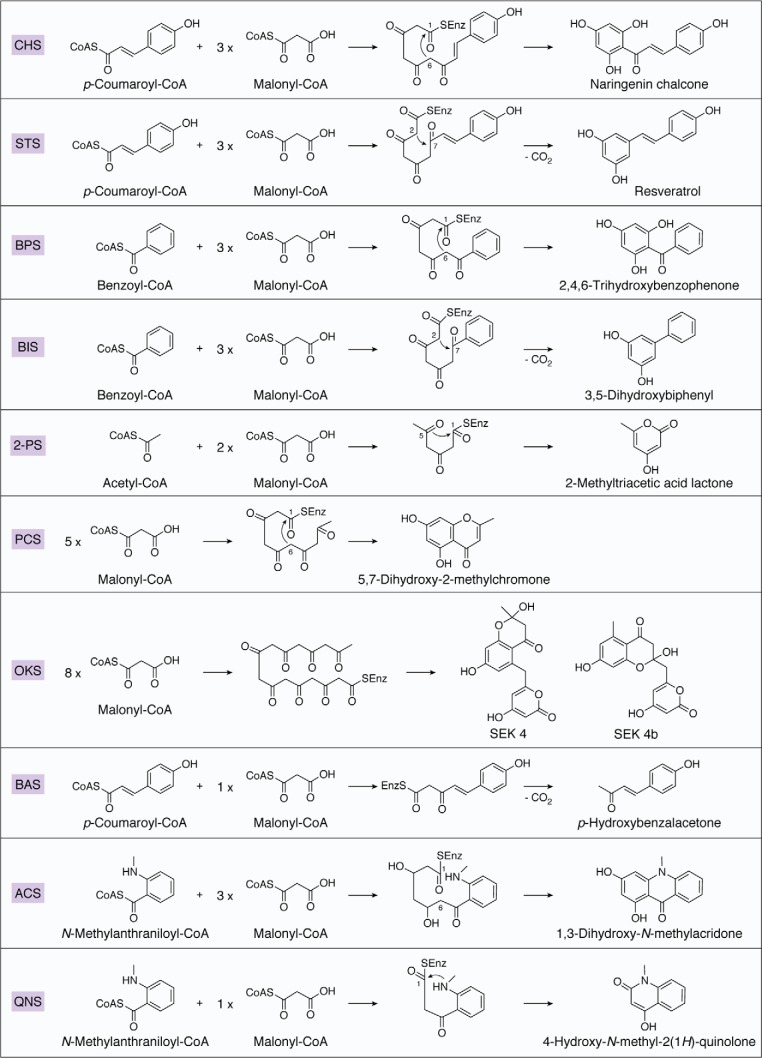
Plant polyketides are medicinally important plant specialized metabolites that have been used as medicines, foods, pigments, and so on. Very interestingly, the core structures of these structurally and chemically diverse plant polyketides (flavonoids, chromones, stilbenes, phloroglucinols, xanthones, acridones, quinolones, diarylheptanoids, etc.) are produced by type III PKS3-superfamily enzymes by iterative condensations of starter and extender acyl-CoA thioesters. Thus, in contrast to bacterial modular type I and dissociated type II PKS megaenzyme systems with multiple catalytic domains, such as ketoacyl synthase, acyltransferase, and acyl carrier protein (1, 2), the structurally simple type III PKSs are homodimeric proteins of ∼40-kDa subunits, which play a pivotal role in the remarkable structural diversification of polyketide backbone assemblies (3–12). Each independent ketoacyl synthase domain in the type III PKS commonly maintains a “Cys-His-Asn” catalytic triad to catalyze the tethering of a CoA-linked starter substrate to the active-site Cys, which is followed by carbon–carbon (C–C) bond formation via decarboxylative Claisen-type condensation with an extender substrate (normally malonyl-CoA). The resulting poly-β-ketide intermediate usually undergoes further linear carbon chain extensions through iterative thioester bond cleavage and new C–C bond formation. Most of the enzyme-tethered linear polyketide intermediates are then cyclized by intramolecular Claisen and/or aldol condensation(s) and/or lactonization to yield various molecular scaffolds within the single active site of the enzyme.
The primary sequences and substrate and product specificities of the selected type III PKSs are summarized in Figs. S1 and 1. Since the discovery in 1983 of the chalcone synthase (chs) gene from parsley (Petroselinum hortense) that encodes the enzyme responsible for the biosynthesis of flavonoids (13), more than 20 functionally different plant type III PKSs, which share 30–95% amino acid sequence identity with each other, have been cloned and characterized (3–12, 14–17). This includes stilbene synthase (STS) (18–21), biphenyl synthase (BIS) (22, 23), benzophenone synthase (BPS) (23–27), 2-pyrone synthase (28, 29), pentaketide synthase (30), octaketide synthase (31–33), and benzalacetone synthase (BAS) (34, 35). The diverse functions of the plant type III PKSs result from the differences in (i) the selection of the substrates, (ii) the number of condensations, and (iii) the cyclization reactions.
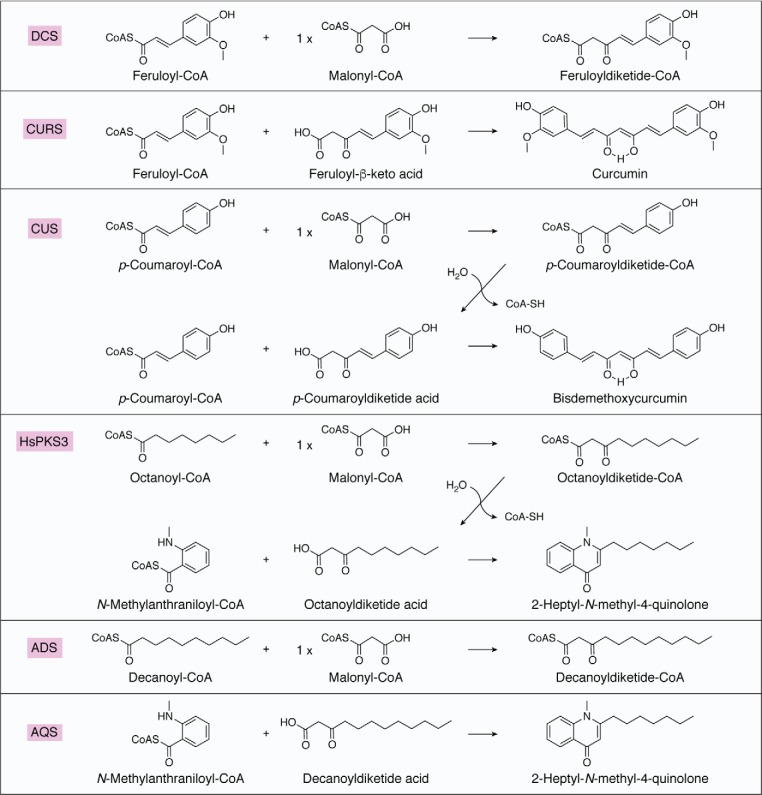
In certain cases, multiple functionally distinct type III PKS enzymes are involved in the biosynthesis, as exemplified by the collaboration between diketide-CoA synthase (DCS) and curcumin synthase (CURS) for the formation of curcumin in Curcuma longa (36, 37). In other cases, a single enzyme catalyzes the one-pot synthesis of curcumin and 2-alkylquinolone, as exemplified by curcuminoid synthase (CUS) from Oryza sativa (38) and PKS3 from Huperzia serrata (HsPKS3) (16), respectively (Fig. 2).
Figure 2.
Formations of curcumin by DCS and CURS, bisdemethoxycurcumin by CUS, 2-heptyl-N-methyl-4-quinolone by HsPKS3, and 2-heptyl-N-methyl-4-quinolone by ADS and AQS.
The type III PKSs share a common αβαβα structural fold, and each monomer contains a functionally independent active site in the cleft between the lower and upper domains (Fig. 3) (10, 17, 23, 39–57). The Cys-His-Asn catalytic triad sits between the active-site cavity and the “CoA-binding tunnel” that is connected to the protein surface. Structural comparisons of the active site have revealed the various features that contribute to the diverse catalytic functions of the type III PKS enzymes. Specifically, the dissimilarities in the volumes and orientations of residues 197, 256, 338, 215, and 265 (numbering according to Medicago sativa CHS2 (39), with the first solved crystal structure among these enzymes) (Fig. S1) are thought to control the polyketide chain length, substrate selectivity, and ring construction reactions. The “aldol-switch” hydrogen bond network of Ser338–H2O–Thr132–Glu192, observed in the crystal structures of the STSs from Pinus sylvestris and Arachis hypogaea, guides the course of the decarboxylative aldol-type cyclization of the enzyme-bound tetraketide intermediate to generate the stilbene scaffold instead of the chalcone-forming Claisen-type cyclization in the CHSs (42, 43).
Figure 3.

Crystal structure of M. sativa CHS. A, overall structure. Each monomer is colored white or blue. The catalytic Cys164 is represented by a black sphere. The Met137 residues, which form a partial wall of the active-site cavity in the opposing monomer, are represented by cyan stick models. Black arrows denote the entrance of the CoA-binding tunnel of each monomer. B, close-up view of the catalytic triad. Cys164, His303, and Asn338 are represented by black sticks. The CoA-SH molecules are depicted by green stick models.
Because plant type III PKS-superfamily enzymes share such high sequence identity, it is extremely difficult to predict their catalytic functions from the amino acid sequences. Indeed, we have often seen that only a small modification of the active site, even single amino acid substitutions, drastically changes the catalytic functions of the enzymes and their product profiles. Elucidations of the structural bases for the functionally distinct plant type III PKSs are thus prerequisite for understanding the structure–function relationships of the enzymes and for further manipulation of their catalytic machineries (5, 8, 58).
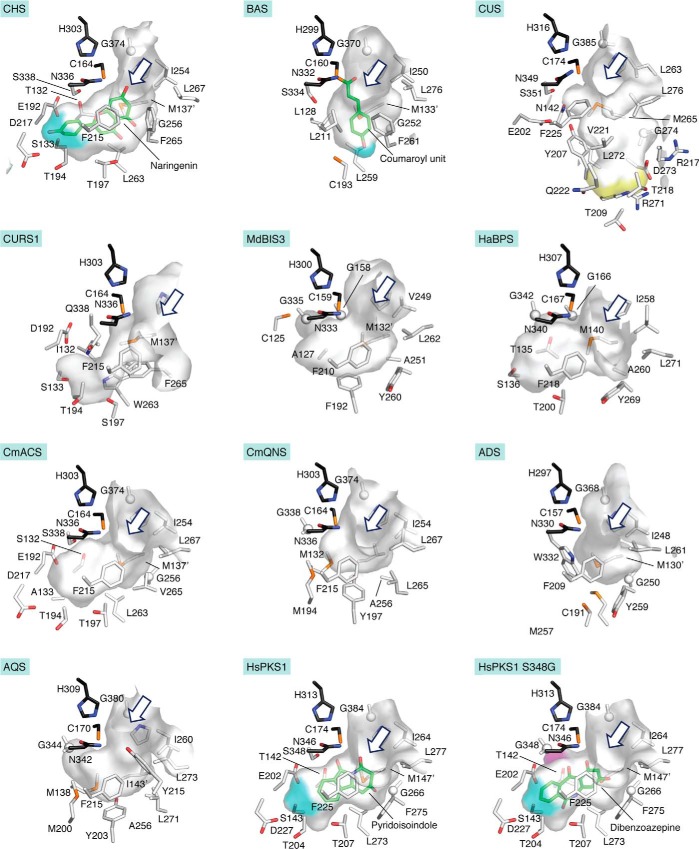
At present, the crystal structures of 16 functionally distinct plant type III PKSs have been experimentally defined. This short article is an update for the previous review in 2010 (5) and highlights the structural characteristics of CUS (51), CURS1 (53), BIS3 from Malus domestica (23), BPS from Hypericum androsaemum (23), acridone synthase (ACS) and quinolone synthase (QNS) from Citrus microcarpa (56), alkyldiketide-CoA synthase (ADS) and alkylquinolone synthase (AQS) from Evodia rutaecarpa (10), HsPKS1 (55), and CHSL1 from Marchantia paleacea (MpCHSL1) (57). The active-site architectures of M. sativa CHS, BAS, CUS, CURS1, CmACS, CmQNS, ADS, AQS, M. domestica BIS3 (MdBIS3), H. androsaemum BPS (HaBPS), HsPKS1, and CHSL1 are summarized in Fig. 4. These type III PKS structures provided important information on how these enzymes regulate their substrates specificities. In particular, the structures of CUS provided insight into the mechanism of the unique one-pot formation of curcuminoids (51), whereas those of CURS1 and AQS revealed their unusual preference for “CoA-free” diketide acids as extender substrates (10, 53). Moreover, this review also introduces recently reported “second-generation” type III PKSs, such as HsPKS3 and Atropa belladonna pyrrolidine ketide synthase (AbPYKS), which catalyze a one-pot formation of alkylquinolone scaffolds from three distinct CoA thioesters (16) and construction of tropane alkaloid scaffolds from CoA-free substrates (12), respectively. These enzymes not only significantly expanded the catalytic repertoire of the type III PKS enzymes but also provided further opportunities for the engineered biosynthesis to create new molecules that are structurally different from those of the canonical type III PKS products.
Figure 4.
Active-site cavities observed in the crystal structures of M. sativa CHS (PDB entry 1BI5), Rheum palmatum BAS (PDB entry 3A5Q), O. sativa CUS (PDB entry 3OIT), C. longa CURS1 (PDB entry 3OV2), MdBIS3 (PDB entry 5W8Q), HaBPS (PDB entry 5UCO), CmACS (PDB entry 3WD7), CmQNS (PDB entry 3WD8), E. rutaecarpa ADS (PDB entry 5WX3), E. rutaecarpa AQS (PDB entry 5WX4), and HsPKS1 (PDB entry 3AWK) and in the model structure of the HsPKS1 S348G mutant enzyme. The sizes and shapes of the active-site cavities are represented by surface models. The catalytic triads are represented by black stick models. The bottoms of the coumaroyl-binding pocket in M. sativa CHS, the corresponding pockets in HsPKS1 and its mutant, and the alternative coumaroyl-binding pocket in R. palmatum BAS are highlighted by cyan surfaces. The bottom of the downward-expanding pocket in CUS is indicated by a yellow surface. The expanded wall of the HsPKS1 S348G mutant is highlighted by a pink surface. The entrances to the active-site cavities are shown by blue arrows. In the M. sativa CHS and R. palmatum BAS crystal structures, the complexed naringenin and coumaroyl unit, respectively, are shown with green stick models. Three-dimensional structures of pyridoisoindole (2-hydroxybenzo[f]pyrido[2,1-a]isoindole-4,6-dione) and dibenzoazepine (1,3-dihydroxy-5H-dibenzo[b,e]azepine-6,11-dione), shown as green stick models, are docked into the active-site cavities of the WT and S348G mutant HsPKS1 enzymes, respectively. All active-site cavities in this figures were rendered using PyMOL.
Finally, this review also covers the structural features of post-type III PKS tailoring enzymes, such as a novel plant polyketide cyclase from Cannabis sativa, olivetolic acid cyclase (OAC). This is so far the only known plant polyketide cyclase that directly accepts a linear poly-β-ketide intermediate, which is expected to be a useful tool for combinatorial biosynthesis for further production of unnatural novel molecules for drug discovery.
Curcuminoid synthase
CUS was discovered in the O. sativa genome as an unusual type III PKS that catalyzes the one-pot formation of the C6-C7-C6 diarylheptanoid scaffold of bisdemethoxycurcumin. CUS first condenses one molecule of p-coumaroyl-CoA with one malonyl-CoA to produce the C6-C5 p-coumaroyldiketide-CoA intermediate, which is followed by hydrolysis to yield the corresponding β-keto acid. Finally, condensation of the β-keto acid with the second p-coumaroyl-CoA affords the C6-C7-C6 diarylheptanoid (38) (Fig. 2). In CUS, the active-site residues Thr132, Thr197, Gly256, and Phe265 in CHS are characteristically replaced with Asn142, Tyr207, Met265, and Gly274, respectively (Fig. S1). As reported previously, the first three residues differ in a number of functionally divergent type III PKSs and are thought to control the substrate and product specificities of the enzyme reactions (4, 5, 41). The gatekeeper Phe265, positioned at the junction between the active-site cavity and the CoA-binding tunnel, guides the substrate loaded within the active-site cavity (39).
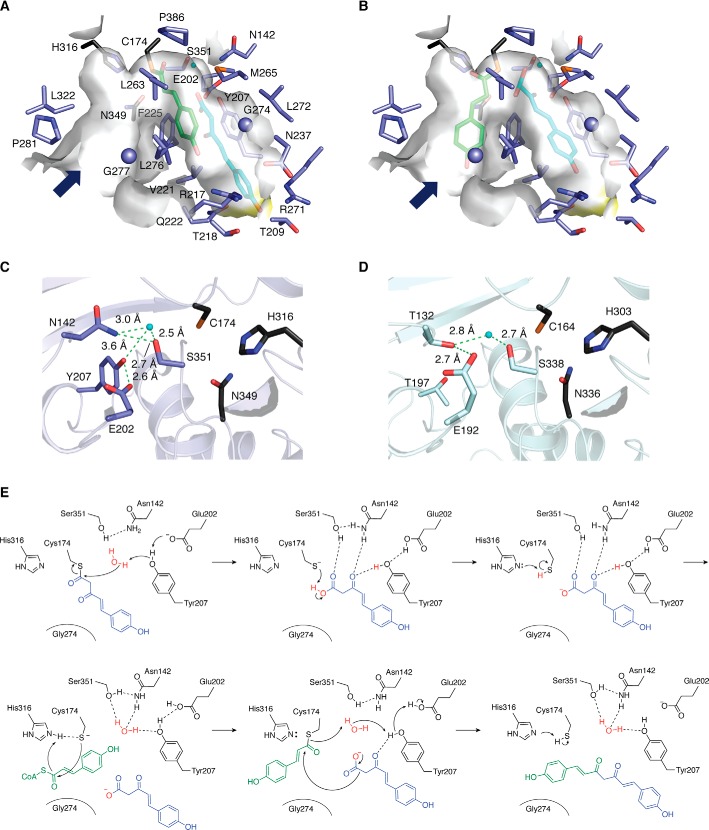
The crystal structure of CUS revealed that the overall structure is analogous to those of other type III PKSs, whereas the aforementioned residues in CUS form a large and unique downward expansion of the active site (Fig. 4) (51). Thus, in the CUS active site, the Asn142 and Tyr207 substitutions cause the loss of the “coumaroyl-binding pocket” of CHS, whereas the large-to-small replacement of Phe265 in CHS with Gly274 in CUS not only enlarges the active-site entrance but also forms the unusual downward expanding pocket. The Met265 side chain protrudes toward the catalytic center and partially alters the active-site architecture. Docking simulations predicted that the active site of CUS is sufficient to accommodate both the p-coumaroyldiketide acid and the second coumaroyl unit (Fig. 5A). The wide active-site entrance of CUS may facilitate the reorientation of the coumaroyl unit tethered at the catalytic Cys174 from the active-site cavity to the CoA-binding tunnel (Fig. 5B). Another noticeable structural feature is the presence of the unique Ser351–Asn142–H2O–Tyr207–Glu202 hydrogen bond network adjacent to the catalytic Cys174, which is similar to the aldol switch observed in the active sites of STSs (Fig. 5, C and D) (42, 43).
Figure 5.
Close-up views of the active-site cavity and the CoA-binding tunnel of O. sativa CUS. A and B, three-dimensional models of the coumaroyl monoketide covalently bound to the catalytic Cys174 (green stick model) and the p-coumaroyldiketide acid (cyan stick model) within the active-site cavity (A) and those within the CoA-binding tunnel and the active-site cavity (B), respectively. C and D, the hydrogen bond networks of O. sativa CUS (C) and P. sylvestris STS (D). The catalytic triads are represented by black stick models. The water molecules and hydrogen bonds are represented by cyan spheres and green dotted lines, respectively. E, proposed catalytic mechanism for the formation of bisdemethoxycurcumin by CUS.
These structural data suggested that CUS utilizes unique catalytic machinery for the one-pot formation of the C6-C7-C6 diarylheptanoid scaffold of bisdemethoxycurcumin. Thus, the synthesis likely involves (i) thioester bond cleavage of the enzyme-bound diketide via nucleophilic attack of a water molecule activated by the hydrogen bond network, (ii) the conversion of the diketide acid into an anion by proton abstraction by Cys174, (iii) the subsequent loading of the second p-coumaroyl-CoA to the catalytic Cys followed by reorientation into the CoA-binding tunnel, and (iv) a final decarboxylative condensation of the diketide acid kept in the active-site cavity with the second coumaroyl unit (Fig. 5E). Interestingly, the CUS M265L and G274F mutants accepted 4-hydroxyphenylpropionyl-CoA as a starter substrate to produce an unnatural novel product, tetrahydrobisdemethoxycurcumin (51). Homology models predicted that the M265L and G274F substitutions narrow the active-site entrance, thereby altering the substrate preference to accept the more flexible 4-hydroxyphenylpropionyl-CoA over p-coumaroyl-CoA to yield the new product. These findings suggested not only the importance of the downward expanding active-site architecture for the unique one-pot enzyme reaction but also a strategy to expand the substrate scope for further generation of unnatural novel molecules.
Diketide synthase and curcumin synthase
In some cases, the collaboration of two functionally distinct type III PKSs generates one natural product scaffold. DCS and CURS from C. longa are the first reported type III PKSs that collaborate to produce curcumin (36, 37) (Fig. 2). Thus, DCS first condenses feruloyl-CoA with malonyl-CoA to yield feruloyldiketide-CoA. After hydrolysis, CURS1 catalyzes the coupling of the C6-C5 feruloyldiketide acid with another feruloyl-CoA to construct the C6-C7-C6 diarylheptanoid scaffold. Interestingly, the C. longa plant also expresses CURS2 and CURS3, which accept both feruloyl-CoA and p-coumaroyl-CoA as substrates.
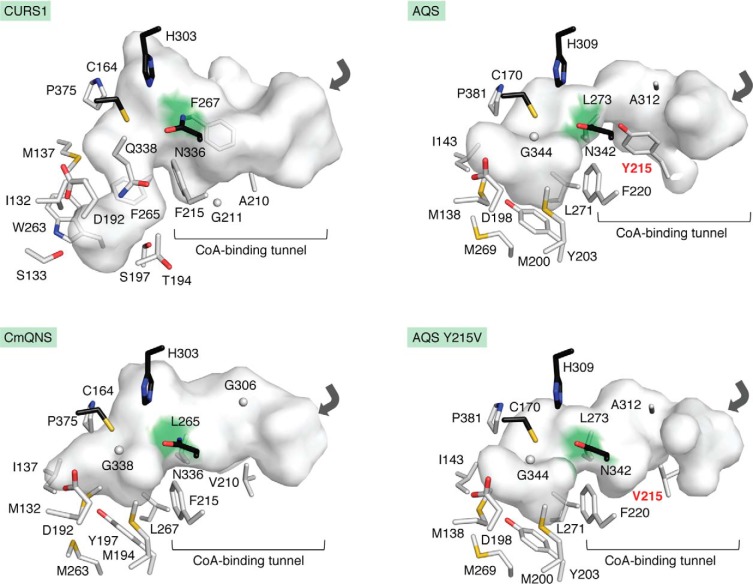
The crystal structure of CURS1 revealed that the active-site cavity is much narrower than that of M. sativa CHS because of the characteristic Gln338 and the reorientation of the gatekeeper Phe265 (Fig. 6). As a result, CURS1 creates a unique hydrophobic pocket with Gly211, Phe215, Phe265, and Phe267 in the CoA-binding tunnel. Furthermore, analyses of the G211F and G211W mutants, which drastically decreased the curcuminoid-forming activity in vitro, and docking studies supported the proposal that this pocket allows the bulky hydrophobic feruloyldiketide acid to be loaded into the catalytic center, thereby forming the C6-C7-C6 curcuminoid scaffold by the decarboxylative condensation with feruloyl-CoA. In contrast, the three-dimensional structure of DCS has yet to be solved. Notably, in DCS, the conserved Ser338 next to the catalytic Cys164 is replaced with the bulky and negatively charged Glu337, which may explain the substrate and product specificities of DCS.
Figure 6.
Comparison of the active sites of C. longa CURS1, E. rutaecarpa AQS, E. rutaecarpa QNS, and the AQS Y215V mutant. The catalytic triads are represented by black stick models. The junctions between the CoA-binding tunnel and the active-site cavity are highlighted with green surfaces. The entrances to the CoA-binding tunnels are shown with gray arrows. Tyr215 and the mutated Val215 are highlighted with red designations.
Biphenyl synthase and benzophenone synthase
BIS and BPS are type III PKSs that select CoA thioesters of benzoate as the starter substrate to catalyze three condensations with malonyl-CoA (Fig. 1). As a result of the decarboxylative C2–C7 aldol-type cyclization, BIS produces 3,5-dihydroxybiphenyl, which is the core scaffold of the biphenyl and dibenzofuran phytoalexins (22–23) (Fig. 1). BIS has been characterized in Sorbus aucuparia and M. domestica (22) and exploited for the precursor-directed biosynthesis of 4-hydroxycoumarin, which is the immediate precursor of the synthetic anticoagulants (59, 60). In contrast, BPS catalyzes C1–C6 Claisen-type cyclization to yield 2,4,6-trihydroxybenzophenone, which becomes the backbone of the xanthones, guttiferones, and sampsoniones that are predominantly found in the closely related Hypericaceae and Clusiaceae families (Fig. 1) (23–27). BPS has been identified in H. androsaemum, H. perforatum, and Garcinia mangostana (26, 27). In BPS, Thr135 in the active-site cavity is thought to control the Claisen-type cyclization, as supported by the dominant production of phenylpyrone by the T135L mutant of H. androsaemum BPS (61). The triple L263M/F265Y/S338V mutant of H. androsaemum CHS, which shares 60% amino acid sequence identity with BPS, preferentially accepted benzoyl-CoA over p-coumaroyl-CoA as the starter substrate (25). Notably, in all known benzoate-specific type III PKSs, the CHS active-site residues Phe265 and Ser338 are uniquely substituted with Tyr and Gly, respectively.
Figure 1.
Typical reactions catalyzed by plant type III PKSs. 2-PS, 2-pyrone synthase; PCS, pentaketide synthase; OKS, octaketide synthase.
The crystal structures of MdBIS3 and HaBPS have been solved by Noel and co-workers (23) along with that of M. domestica CHS2. MdBIS3 and HaBPS utilize an alternate, novel active-site pocket behind the catalytic Cys, instead of the coumaroyl-binding pocket in CHS, which facilitates the selective binding of the smaller benzoate starter (Fig. 4). The benzoate-binding pocket in MdBIS3 is formed by Cys125, Ala127, Ala157, Gly158, Glu187, Gly335, and Ala336. Correspondingly, Thr135, Gly166, Glu195, Gly342, and Ser343 form a similar pocket in HaBPS. These substantial changes are mainly caused by the single substitution of Thr197 in M. domestica CHS with Phe192 in MdBIS3 and by the replacement of Ser338 of M. domestica CHS2 with Gly342 in HaBPS. As a result, BIS and BPS create a hydrophobic surface for the interaction with the small hydrophobic CoA thioesters of benzoates. Interestingly, the size of the active-site cavity of MdBIS3 is dramatically reduced by the replacement of Thr197 in CHS with Phe192, as compared with that of HaBPS (Fig. 4). Thus, these structural features were suggested to be the decisive factors for the substrate and product specificities of the BIS and BPS enzyme reactions. Notably, MdBIS3 lacks the aldol-switch hydrogen bond network underlying the aldol-type cyclization reaction. Although water molecules that may function as the nucleophile were observed in the MdBIS3 active site, further detailed analyses of the enzyme reaction are required.
Acridone synthase
Type III PKSs that catalyze the C–N bond-forming reaction have also been isolated from Rutaceae plants, which are rich producers of acridone and quinolone alkaloids. The ACSs from C. microcarpa (56) and Ruta graveolens (62, 63) catalyze the iterative condensations of three malonyl-CoAs with N-methylanthraniloyl-CoA followed by Claisen-type and C–N bond-forming cyclization reactions to generate 1,3-dihydroxy-N-methylacridone, which is the core skeleton of the acridone alkaloids (Fig. 1). Interestingly, ACSs can also accept p-coumaroyl-CoA as the starter substrate to produce naringenin chalcone in vitro (56, 63–67).
In the ACSs, the active-site residues in CHS, Thr132, Ser133, and Phe265, are substituted with Ser, Ala, and Val, respectively (Fig. S1). These residues determine the selection of N-methylanthraniloyl-CoA as the starter substrate, which was confirmed by the full conversion of the R. graveolens ACS S132T/A133S/V265F mutant activity to that of CHS (64). Furthermore, our structural studies of C. microcarpa ACS (CmACS) revealed that the F265V substitution, together with the conformational changes of Phe215 and Leu267, forms a wider active-site entrance, which accounts for the bulky starter substrate preference of ACS (Fig. 4) (56). Interestingly, the crystal structures of BAS (49) and the M. sativa CHS F215S mutant (65), which both also accept the bulky N-methylanthraniloyl-CoA and anthraniloyl-CoA as starters, revealed similar expanded active-site entrances. The large-to-small T132S/S133A substitutions in CmACS also alter the shape and size of the active site. The structural studies thus suggested that the ACSs utilize a catalytic mechanism similar to those of the CHSs, but due to the wider active-site entrance, the ACSs prefer the N-methylanthraniloyl-CoA starter to generate the acridone scaffold.
Quinolone synthase
The QNSs from Aegle marmelos (AmQNS) and C. microcarpa (CmQNS) generate the core skeletons of the quinolone alkaloids (56, 68). Both enzymes catalyze the decarboxylative condensation of malonyl-CoA with N-methylanthraniloyl-CoA and the C–N bond-forming cyclization to yield 4-hydroxy-2(1H)-quinolone (Fig. 2). AmQNS also produces acridone and benzalacetone from N-methylanthraniloyl-CoA and p-coumaroyl-CoA, respectively (Fig. 2). The characteristic Ser132/Ala133/Val265 replacements in ACS are also maintained in AmQNS (Fig. S1). In contrast, CmQNS specifically produces 4-hydroxy-N-methylquinolone from N-methylanthraniloyl-CoA starter and malonyl-CoA extender. Furthermore, CmQNS does not accept p-coumaroyl-CoA as a substrate but accepts benzoyl-CoA and hexanoyl-CoA starters to produce the corresponding triketide lactones (56). Interestingly, half of the conserved active-site residues, such as Thr132, Met137, Ser133, Thr194, Thr197, Gly256, Phe265, and Ser338 in M. sativa CHS, are uniquely altered with Met, Ile, Ala, Met, Tyr, Ala, Leu, and Gly in CmQNS, respectively (Fig. S1).
The crystal structure of CmQNS revealed that the F265L replacement and the conformational changes of Phe215 and Leu267 result in the formation of an unusually wide active-site entrance that favors the N-methylanthraniloyl-CoA substrate (Fig. 4) (56). Furthermore, these characteristic residues make the active-site cavity of CmQNS significantly smaller than that of ACS, thus preventing the binding of a long and large substrate, such as p-coumaroyl-CoA (Fig. 4) (49). The unique active-site architecture of CmQNS is mostly attributed to Met132, Met194, and Tyr197. Indeed, the CmQNS Y197A mutant was able to accept the p-coumaroyl-CoA starter to produce a triketide lactone, along with benzalacetone (Fig. 2) (56). Interestingly, the CmACS S132M, T194M, and T197Y mutants lost the acridone-forming activity and afforded 4-hydroxy-N-methyl-2(1H)-quinolones as the single product from N-methylanthraniloyl-CoA (56). Moreover, these mutants no longer accepted p-coumaroyl-CoA to produce chalcone, tetraketide, and triketide lactones.
Alkyldiketide-CoA synthase and alkylquinolone synthase
In contrast to QNS, ADS and AQS are functionally distinct type III PKSs that collaboratively generate the core skeleton of 2-alkylquinolone alkaloids in the Rutaceae medicinal plant E. rutaecarpa (10), in a manner similar to those described for DCS and CURS in curcumin biosynthesis. Steady-state kinetics analyses suggested that ADS could efficiently catalyze the condensation of malonyl-CoA with decanoyl-CoA to yield decanoylalkyldiketide-CoA, whereas AQS preferentially catalyzes the coupling of a decanoyldiketide acid with N-methylanthraniloyl-CoA to produce 2-nonylquinolone via C–N bond formation (Fig. 2). In vitro analyses revealed that ADS also accepts octanoyl-CoA and lauroyl-CoA as starter substrates, whereas AQS accepts alkyldiketide acids with alkyl lengths ranging from hexanoyl to hexadecanoyl as the extender substrates.
In E. rutaecarpa ADS, the CHS active-site residues, Thr132, Thr194, Thr197, Leu263, Phe265, and Ser338, are altered to Ser, Met, Cys, Met, Tyr, and Trp, respectively (Fig. S1). Furthermore, the crystal structure of ADS revealed that the bulky side chains of Trp332 and Met257 form the narrow and shallow active-site cavity, as compared with that of the functionally closest BAS (Fig. 4) (10). In addition, Trp332 dramatically narrows the active-site entrance. Mutation experiments suggested that Trp332 and Cys191 are important for the preference of the fatty acyl-CoA starter substrate. The active-site cavity volume allows the accommodation of up to a 10-carbon-length enzyme-bound monoketide intermediate.
In contrast, the active site of E. rutaecarpa AQS is quite similar to that of the quinolone-forming CmQNS (10), although AQS has a much narrower CoA-binding tunnel as compared with that of CmQNS (Fig. 6). This unique architecture is caused by the substitution of Val in CmQNS with Tyr215 in AQS. As a result, AQS selects an alkyldiketide acid as the preferred extender substrate, in sharp contrast to the functionally closest CURS1 (53), which has a characteristic hydrophobic pocket formed by Gly211, Phe215, Phe265, and Phe267 in the CoA-binding tunnel for the accommodation of the aromatic diketide extender (Fig. 6). Indeed, unlike the WT AQS, the Y215V mutant efficiently accepts benzoyl-CoA and p-coumaroyl-CoA as starter substrates (8). The crystal structure confirmed the expanded CoA-binding tunnel of the AQS Y215V mutant (Fig. 6). Hence, the highly homologous active-site architecture to that of CmQNS and the presence of the narrow CoA-binding tunnel control the substrate and product specificities of the AQS enzyme reaction, in a manner similar to that of CURS1. These findings have clarified the structural basis for the versatile catalytic repertoires of the type III PKSs in the biogenesis of various plant metabolites.
Notably, in the Gram-negative bacterium Pseudomonas aeruginosa, three β-ketoacyl carrier protein synthase III (FabH)-like enzymes, PqsB, -C, and -D, which share 19% amino acid sequence similarities with ADS and AQS, and an additional thioesterase, PqsE, are involved in the 2-alkylquinolone biosynthesis. This is different from the synthesis in plants and proceeds in the following order: (i) the condensation of anthraniloyl-CoA and malonyl-CoA to produce 2-aminobenzoyldiketide-CoA, (ii) the hydrolysis of 2-aminobenzoyldiketide-CoA to yield 2-aminobenzoyldiketide acid, and (iii) finally the tail-to-tail condensation of 2-aminobenzoyldiketide acid with a fatty acyl-CoA to generate 2-heptyl-4-quinolone (69–71). Thus, the comparative studies also provide new insight into the functional and evolutionary implications of the biosynthesis of 2-alkylquinolones in plants and bacteria.
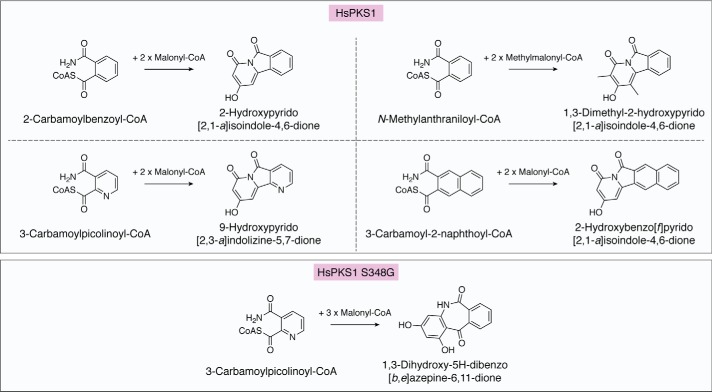
PKS1 from H. serrata
HsPKS1, from the medicinal moss H. serrata, exhibits extraordinarily promiscuous substrate specificity and catalytic versatility to produce numerous aromatic polyketides in vitro (55, 67). For example, HsPKS1 efficiently catalyzes the formation of naringenin chalcone and 1,3-dihydroxy-N-methylacridone from p-coumaroyl-CoA and N-methylanthraniloyl-CoA, respectively. A precursor-directed biosynthesis using N-containing nonphysiological synthetic substrates successfully generated unnatural novel alkaloids, such as 2-hydroxypyrido[2,1-a]isoindole-4,6-dione, 1,3-dimethyl-2-hydroxypyrido[2,1-a]isoindole-4,6-dione, and 9-hydroxypyrido[2,3-a]indolizine-5,7- dione, with the 6.5.6-fused ring system (Fig. 7). HsPKS1 also accepted the bulky naphthalene-containing 3-carbamoyl-2-naphthoyl-CoA as a starter substrate with good efficiency to yield 2-hydroxybenzo[f]pyrido[2,1-a]isoindole-4,6-dione, with the 6.6.5.6-fused tetracyclic ring system. Notably, this product has a structure similar to that of camptothecin, a well-known DNA topoisomerase I inhibitor from Camptotheca acuminata and Nothapodytes foetida (72).
Figure 7.
Enzymatic formation of pyridoisoindole and benzoazepine alkaloids by WT HsPKS1 and its S348G mutant.
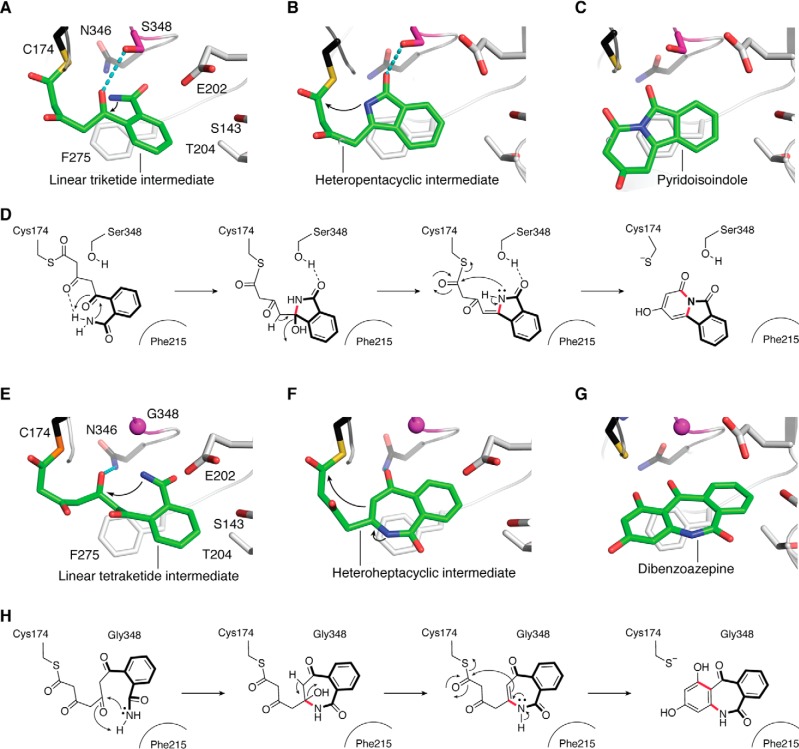
The crystal structure of HsPKS1 complexed with CoA-SH revealed that the cavity volume of the active site is comparable with that of M. sativa CHS, but the active-site entrance is wider due to the conformational change of the gatekeeper Phe275 (Fig. 4) (55). The promiscuous substrate specificity and catalytic versatility of HsPKS1 are thus principally attributed to this widening of the active-site entrance. Docking studies with linear and partially cyclized triketide intermediates tethered to the catalytic Cys174 suggested that the hydroxyl group of Ser348, next to the catalytic Cys, first forms a hydrogen bond with the C5 carbonyl oxygen of the linear intermediate and then bonds with the amide oxygen of the partially cyclized intermediate (Fig. 8, A and B). These findings suggested that Ser348 could be the crucial residue governing the mechanisms of the polyketide chain elongation and both N–C5 and N–C1 cyclizations to generate the pyridoisoindole scaffolds (Fig. 8, C and D).
Figure 8.
Proposed mechanism for the formation of 2-hydroxybenzo[f]pyrido[2,1-a]isoindole-4,6-dione by WT HsPKS1 and 1,3-dihydroxy-5H-dibenzo[b,e]azepine-6,11-dione by the HsPKS1 S348G mutant. A–C, model structures of HsPKS1 complexed with the linear triketide intermediate (A), the heteropentacyclic intermediate (B), and 2-hydroxybenzo[f]pyrido[2,1-a]isoindole-4,6-dione (pyridoisoindole) (C). The catalytic Cys174 and Asn346 are depicted by black stick models. Hydrogen bonds between Ser348 (purple stick model) and the intermediate are represented by cyan dotted lines. D, schematic representation of the proposed mechanism for the formation of pyridoisoindole. E–G, model structures of HsPKS1 S348G complexed with the linear tetraketide intermediate (E), the heteroheptacyclic intermediate (F), and 1,3-dihydroxy-5H-dibenzo[b,e]azepine-6,11-dione (dibenzoazepine) (G). H, schematic representation of the proposed mechanism for the formation of dibenzoazepine. The catalytic Cys174 and Asn346 are depicted by black stick models. Ser348 and Gly348 are highlighted by purple stick models. Hydrogen bonds between Ser348 and the intermediate are represented by cyan dotted lines.
The structure-guided HsPKS1 S348G mutant further produced the B-ring–expanded, unnatural 6.7.6-fused tricyclic novel dibenzoazepine alkaloid 1,3-dihydroxy-5H-dibenzo[b,e]azepine-6,11-dione from 2-carbamoylbenzoyl-CoA by three condensations with malonyl-CoA (Fig. 7) (55). The S348G substitution is thought to increase the space adjacent to the catalytic Cys174 for the polyketide chain elongation, leading to N–C5 and C6–C1 tandem cyclizations of the linear intermediate to afford the bulkier tetracyclic product with the 6.7.6-fused tricyclic ring system (Fig. 8, E–H). The incorporation of the basic nitrogen atom and the structure-based mutagenesis thus dramatically expanded the catalytic repertoire of the type III PKS enzyme reactions.
Chalcone synthase L1 from M. paleacea
Four type III PKSs, MpCHSL1, MpCHSL2, MpCHSL3, and MpCHS, were identified from the liverwort M. paleacea, which is a rich source of phenolic compounds, including bibenzyls and flavonoids (57). An in vitro analysis utilizing p-coumaroyl-CoA and dihydro-p-coumaroyl-CoA as starter substrates revealed that MpCHS efficiently catalyzes the formation of naringenin chalcone and phloretin. In contrast, the MpCHSLs preferentially produced lactonized α-pyrones, coumaroyl triacetic acid lactone (CTAL), and dihydro-CTAL. These assays also indicated that the MpCHSLs have the same catalytic functions. The crystal structure of MpCHSL1 confirmed the high homology to other CHS enzymes. MpCHSL1 shares not only the regular catalytic triad but also the other residues lining the catalytic cavity, Thr132, Ser133, Met137, Thr194, Gly216, Ile254, Gly256, Ser338, and Pro375, in MsCHS (Fig. S1) (57). In MpCHSL1, the nonsequence-conserved α-helix (residues 278–295) leads to the formation of a larger active site that affords more space for the cyclization reaction. The lack of the aldol switch in MpCHSL1 further suggested that it is catalytically closer to CHS than STS.
PKS3 from H. serrata
On a different note, the HsPKS3 from H. serrata is another unique type III PKS that can generate a series of unnatural 2-alkylquinolones and 1,3-diketones, including curcumin, via the formation of a diketide acid from acyl-CoA and malonyl-CoA followed by the tail-to-tail coupling of a second acyl-CoA and the diketide acid (Fig. 2). For example, the one-pot formation of 2-heptyl-N-methyl-4-quinolone was achieved by the condensation of octanoyl-CoA, malonyl-CoA, and N-methylanthraniloyl-CoA (16). The conspicuous substrate promiscuity of HsPKS3 also facilitated the synthesis of a more diverse group of 2-substituted quinolones and 1,3-diketones.
Considering the product profile of the enzyme reactions, HsPKS3 is likely to employ a catalytic mechanism similar to that of CUS. A primary sequence comparison of HsPKS3 with those of the other type III PKSs indicates that most of the active-site residues of CHS are well-conserved in this enzyme, except for only three residues, Thr132, Ser133, and Leu263, in CHS, which are characteristically substituted with Ser, Asn, and Met in HsPKS3. To fully understand the catalytic mechanism of HsPKS3, further crystallographic studies are required.
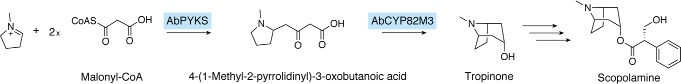
Pyrrolidine ketide synthase
Tropane alkaloids, such as scopolamine, are pharmaceutically important natural products. The biosynthesis of tropane alkaloids has not been entirely elucidated and thus has remained enigmatic for a long time. However, in 2018, it was demonstrated that a novel type III PKS, AbPYKS, is involved in the biosynthesis of tropinone, which is the core skeleton of the tropane alkaloids (12). Remarkably, AbPYKS is able to utilize a CoA-free substrate, N-methyl-Δ1-pyrrolinium cation, as the starter substrate with two malonyl-CoAs to form 4-(1-methyl-2-pyrrolidinyl)-3-oxobutanoic acid, which is then cyclized by the cytochrome P450 monooxygenase AbCYP82M3 to generate the tropinone scaffold (Fig. 9). A sequence analysis indicated that the catalytic triad common to all type III PKSs is also conserved, as Cys166, His305, and Asn338. Thus, it is likely that AbPYKS forms 4-(1-methyl-2-pyrrolidinyl)-3-oxobutanoic acid in a manner similar to those of the other type III PKSs.
Figure 9.
Proposed biosynthetic pathway for the formation of tropinone and scopolamine, catalyzed by AbPYKS.
However, many questions still remain to be answered. Is the N-methyl-Δ1-pyrrolinium cation also tethered to the catalytic Cys? When and how is the pyrrolinium cation loaded onto the catalytic center? Does the loading of the pyrrolinium cation affect the decarboxylative condensation with malonyl-CoA to form the proposed 2-(1-methylpyrrolidinyl)-acetyl-CoA? Is AbPYKS able to control the absolute configuration of the product, and if so, what is the mechanism? To fully understand the noncanonical catalytic mechanism of AbPYKS, further functional and structural analyses are required. The discovery of AbPYKS greatly expanded the catalytic repertoire of the type III PKS enzymes and provided further opportunities for the engineered biosynthesis of plant polyketides.
Triketide synthase and olivetolic acid cyclase
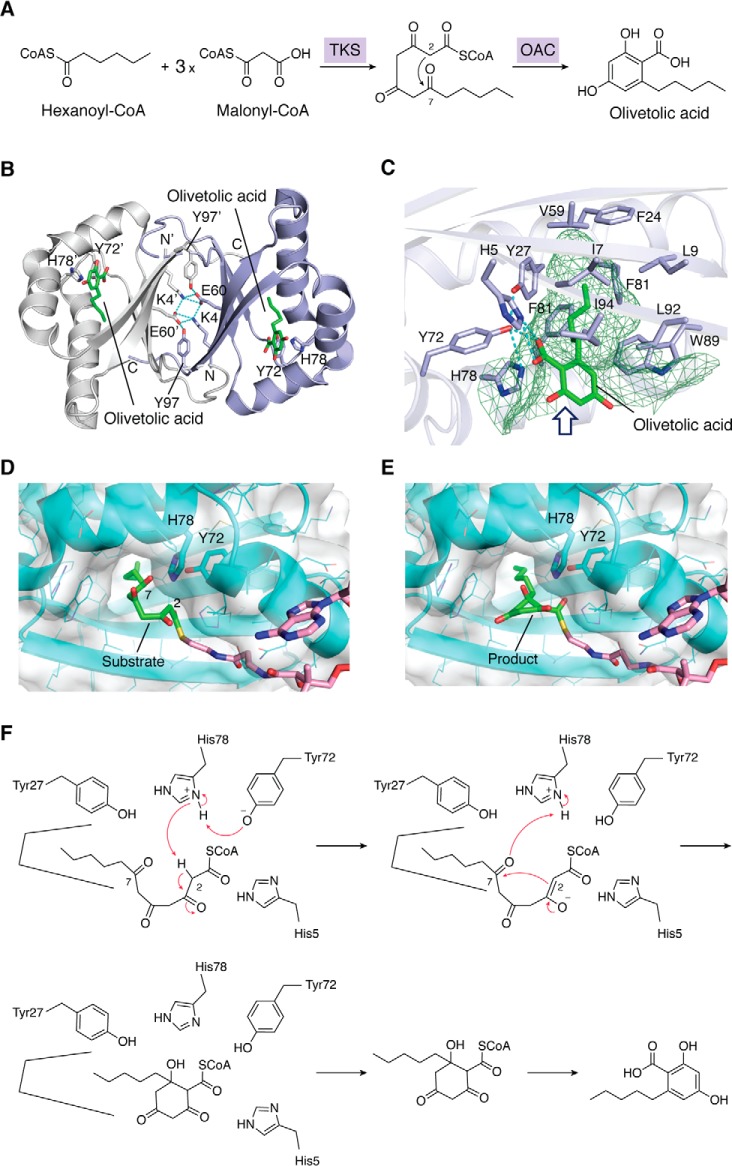
The ring construction is one of the crucial diversification steps in plant polyketide biosynthesis. Although polyketide chain elongation and cyclization generally proceed within a single active site of a type III PKS enzyme, the formation of olivetolic acid, the biosynthetic precursor of cannabinoids in C. sativa, requires OAC for the cyclization of the linear poly-β-keto intermediate provided by a type III PKS (73–75). Thus, tetraketide synthase first produces the linear pentyltetra-β-ketide-CoA from the hexanoyl-CoA starter, and then OAC catalyzes the C2–C7 aldol cyclization of this linear tetraketide to form the aromatic ring of olivetolic acid. This is similar to the chalcone reductase enzyme reaction, and the OAC does not require any cofactors (Fig. 10A).
Figure 10.
Proposed biosynthetic pathway for the formation of olivetolic acid and structure of OAC. A, the formation of olivetolic acid by tetraketide synthase (TKS) and OAC. B, overall binary structure of OAC complexed with olivetolic acid. Each monomer is colored white or purple. Olivetolic acid molecules are shown as green stick models. Hydrogen bonds forming the dimer interface between two monomers are represented with cyan dotted lines. C, close-up view of the active-site cavity. The size and shape of the active-site cavity are represented with green mesh. D and E, docking model of pentyltetra-β-ketide-CoA (substrate) (D) and product (E) in OAC. F, the proposed mechanism catalyzed by OAC to generate olivetolic acid.
Structurally, OAC is a dimeric protein composed of 101-amino-acid-residue monomers, with the dimeric α + β barrel (DABB) fold (Fig. 10B) (73). OAC shares 17% identity with tetracenomycin F2 cyclase (TcmI), a bacterial polyketide cyclase from Streptomyces glaucescens (76). The OAC crystal structures revealed a unique active-site architecture with a 9-Å-long pentyl-binding pocket, which is a narrow hydrophobic pocket in the interior of each monomer (Fig. 10, B and C) that binds the pentyl moiety of the olivetolic acid (77, 78). Furthermore, His78, which forms a hydrogen network with Tyr72, protrudes toward the C1 and C6 positions of the olivetolic acid molecule, thus facilitating the C2–C7 aldol-type cyclization of the substrate. Structure-based site-directed mutagenesis studies confirmed the crucial roles of the pentyl-binding pocket, for locking the pentyl moiety of the substrate, and the Tyr72–His78 catalytic dyad, located near the active-site entrance, which functions as an acid/base catalyst. Indeed, docking studies suggested that His78 is located in the proximity of the C2 carbon and the C7 carbonyl oxygen for the aldol-type cyclization, whereas Tyr72 recognizes the side chain of His78 and the thioester carbonyl oxygen of the substrate (Fig. 10, D and E).
However, the OAC structure lacks the pockets or tunnels that are usually associated with the binding of the CoA moiety of the substrate. This suggested that the CoA moiety is exposed to the solvent or bound on the protein surface. Furthermore, the absence of water molecules, candidate residues, and metal ions suggested that OAC lacks thioesterase and aromatase activities. Nonetheless, these analyses confirmed that OAC employs a unique catalytic mechanism that utilizes acid/base chemistry for the biogenesis of olivetolic acid (Fig. 10F).
Perspective
Due to the unusually versatile catalytic potential to accept a wide range of substrates to generate incredibly structurally diverse and chemically different molecular scaffolds, the type III PKS-superfamily enzymes have served as an excellent platform for synthetic biology. Recent progress on the structural studies of functionally diverse type III PKSs has further opened the door toward understanding their detailed catalytic mechanisms and engineering to modify their functions. As exemplified by the synthesis of the 6.7.6-fused tricyclic dibenzoazepine alkaloid (55), combination of the precursor-directed biosynthesis with rationally designed synthetic substrate analogues, including CoA-free substrates such as N-methyl-Δ1-pyrrolinium cation, and the structure-guided reconstruction of the active-site architecture will further significantly expand the catalytic repertoire of the type III PKS enzymes.
One of the future directions is extender unit engineering. Although most of the structure-guided engineering of the type III PKSs has been achieved by focusing on size and shape of the active-site central cavity, the fact that the extender substrate specificity is controlled by the CoA-binding tunnel architecture in AQS and CURS provides another strategy for further development of novel biocatalysts. In addition, CUS and HsPKS3, which catalyze the noncanonical one-pot formation of curcumin and 2-alkylquinolone scaffold, respectively, are also attractive targets. In particular, it is remarkable that the latter enzyme couples two different acyl units with one malonate-derived carbon unit in between, thus coupling three distinct acyl-CoA thioesters.
Significant progress has been made during the past 10 years. With the growing number of the three-dimensional structures of the type III PKSs, it is now possible to manipulate the starter substrate specificity and the number of the malonyl-CoA condensations. The next question is how to control the cyclization reactions of the enzyme-tethered poly-β-keto intermediates to generate the desired molecular scaffold. The interactions with modification enzymes, such as the novel plant polyketide cyclase OAC, will also present further opportunities for engineering. However, there are many questions that still remain unanswered. What is the timing of the thioester bond cleavage during cyclization reactions of the poly-β-ketide intermediate? How does the active-site chaperone the highly reactive poly-β-keto intermediates and guide the course of the enzyme reaction to afford specific products? In some cases, cyclization might even occur spontaneously without elaborate enzymic assistance. Detailed structural analyses of these enzymes will certainly answer these questions and provide strategies for the further manipulation of the type III PKS enzyme reactions for future engineered biosynthesis to create novel unnatural molecules for drug discovery.
Supplementary Material
This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (Japan Society for the Promotion of Science (JSPS) KAKENHI Grants JP16H06443, JP16H06442, and JP19H04649) and Japan Science and Technology Agency Strategic International Collaborative Research Program (JST SICORP) Grant JPMJSC1701. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Fig. S1.
- PKS
- polyketide synthase
- Ab
- A. belladonna
- ADS
- alkyldiketide-CoA synthase
- ACS
- acridone synthase
- Am
- A. marmelos
- AQS
- alkylquinolone synthase
- BAS
- benzalacetone synthase
- BIS
- biphenyl synthase
- BPS
- benzophenone synthase
- CHS
- chalcone synthase
- CHSL
- chalcone synthase–like
- Cm
- C. microcarpa
- CTAL
- coumaroyl triacetic acid lactone
- CURS
- curcumin synthase
- CUS
- curcuminoid synthase
- DCS
- diketide-CoA synthase
- HsPKS
- H. serrata type III polyketide synthase
- Ha
- H. androsaemum
- Md
- M. domestica
- Mp
- M. paleacea
- OAC
- olivetolic acid cyclase
- PYKS
- pyrrolidine ketide synthase
- QNS
- quinolone synthase
- STS
- stilbene synthase.
References
- 1. Smith S., and Tsai S. C. (2007) The type I fatty acid and polyketide synthases: a tale of two megasynthases. Nat. Prod. Rep. 24, 1041–1072 10.1039/b603600g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hertweck C., Luzhetskyy A., Rebets Y., and Bechthold A. (2007) Type II polyketide synthases: gaining a deeper insight into enzymatic teamwork. Nat. Prod. Rep. 24, 162–190 10.1039/B507395M [DOI] [PubMed] [Google Scholar]
- 3. Schröder J. (1999) The chalcone-stilbene synthase-type family of condensing enzymes, in Comprehensive Natural Products Chemistry (Sankawa U., ed) pp. 749–771, Elsevier, Oxford, UK [Google Scholar]
- 4. Austin M. B., and Noel J. P. (2003) The chalcone synthase superfamily of type III polyketide synthases. Nat. Prod. Rep. 20, 79–110 10.1039/b100917f [DOI] [PubMed] [Google Scholar]
- 5. Abe I., and Morita H. (2010) Structure and function of the chalcone synthase superfamily of plant type III polyketide syntheses. Nat. Prod. Rep. 27, 809–838 10.1039/b909988n [DOI] [PubMed] [Google Scholar]
- 6. Morita H., Abe I., and Noguchi H. (2010) Plant type III polyketide synthases, in Comprehensive Natural Products II: Chemistry and Biology (Liu H.-W., and Mander L., eds) pp. 171–225, Elsevier, Oxford, UK [Google Scholar]
- 7. Abdel-Rahman I. A., Beuerle T., Ernst L., Abdel-Baky A. M., Desoky E. el-D., Ahmed A. S., and Beerhues L. (2013) In vitro formation of the anthranoid scaffold by cell-free extracts from yeast-extract-treated Cassia bicapsularis cell cultures. Phytochemistry 88, 15–24 10.1016/j.phytochem.2013.01.001 [DOI] [PubMed] [Google Scholar]
- 8. Lim Y. P., Go M. K., and Yew W. S. (2016) Exploiting the biosynthetic potential of type III polyketide synthases. Molecules 21, E806 10.3390/molecules21060806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pietiäinen M., Kontturi J., Paasela T., Deng X., Ainasoja M., Nyberg P., Hotti H., and Teeri T. H. (2016) Two polyketide synthases are necessary for 4-hydroxy-5-methylcoumarin biosynthesis in Gerbera hybrida. Plant J. 87, 548–558 10.1111/tpj.13216 [DOI] [PubMed] [Google Scholar]
- 10. Matsui T., Kodama T., Mori T., Tadakoshi T., Noguchi H., Abe I., and Morita H. (2017) 2-Alkylquinolone alkaloid biosynthesis in the medicinal plant Evodia rutaecarpa involves collaboration of two novel type III polyketide synthases. J. Biol. Chem. 292, 9117–9135 10.1074/jbc.M117.778977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Taura F., Iijima M., Yamanaka E., Takahashi H., Kenmoku H., Saeki H., Morimoto S., Asakawa Y., Kurosaki F., and Morita H. (2016) A novel class of plant type III polyketide synthase involved in orsellinic acid biosynthesis from Rhododendron dauricum. Front. Plant Sci. 7, 1452 10.3389/fpls.2016.01452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bedewitz M. A., Jones A. D., D'Auria J. C., and Barry C. S. (2018) Tropinone synthesis via an atypical polyketide synthase and P450-mediated cyclization. Nat. Commun. 9, 5281 10.1038/s41467-018-07671-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reimold U., Kröger M., Kreuzaler F., and Hahlbrock K. (1983) Coding and 3′ non-coding nucleotide sequence of chalcone synthase mRNA and assignment of amino acid sequence of the enzyme. EMBO J. 2, 1801–1805 10.1002/j.1460-2075.1983.tb01661.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matsuzawa M., Katsuyama Y., Funa N., and Horinouchi S. (2010) Alkylresorcylic acid synthesis by type III polyketide synthases from rice Oryza sativa. Phytochemistry 71, 1059–1067 10.1016/j.phytochem.2010.02.012 [DOI] [PubMed] [Google Scholar]
- 15. Kim S. Y., Colpitts C. C., Wiedemann G., Jepson C., Rahimi M., Rothwell J. R., McInnes A. D., Hasebe M., Reski R., Sterenberg B. T., and Suh D. (2013) Physcomitrella PpORS, basal to plant type III polyketide synthases in phylogenetic trees, is a very long chain 2-oxoalkylresorcinol synthase. J. Biol. Chem. 288, 2767–2777 10.1074/jbc.M112.430686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang J., Wang X. H., Liu X., Li J., Shi X. P., Song Y. L., Zeng K. W., Zhang L., Tu P. F., and Shi S. P. (2016) Synthesis of unnatural 2-substituted quinolones and 1,3-diketones by a member of type III polyketide synthases from Huperzia serrata. Org. Lett. 18, 3550–3553 10.1021/acs.orglett.6b01501 [DOI] [PubMed] [Google Scholar]
- 17. Wang X., Zhang Z., Dong X., Feng Y., Liu X., Gao B., Wang J., Zhang L., Wang J., Shi S., and Tu P. (2017) Identification and functional characterization of three type III polyketide synthases from Aquilaria sinensis calli. Biochem. Biophys. Res. Commun. 486, 1040–1047 10.1016/j.bbrc.2017.03.159 [DOI] [PubMed] [Google Scholar]
- 18. Schröder G., Brown J. W., and Schröder J. (1988) Molecular analysis of resveratrol synthase. cDNA, genomic clones and relationship with chalcone synthase. Eur. J. Biochem. 172, 161–169 10.1111/j.1432-1033.1988.tb13868.x [DOI] [PubMed] [Google Scholar]
- 19. Melchior F., and Kindl H. (1990) Grapevine stilbene synthase cDNA only slightly differing from chalcone synthase cDNA is expressed in Escherichia coli into a catalytically active enzyme. FEBS Lett. 268, 17–20 10.1016/0014-5793(90)80961-H [DOI] [PubMed] [Google Scholar]
- 20. Fliegmann J., Schröder G., Schanz S., Britsch L., and Schröder J. (1992) Molecular analysis of chalcone and dihydropinosylvin synthase from Scots pine (Pinus sylvestris), and differential regulation of these and related enzyme activities in stressed plants. Plant Mol. Biol. 18, 489–503 10.1007/BF00040665 [DOI] [PubMed] [Google Scholar]
- 21. Wang W., Wan S. B., Zhang P., Wang H. L., Zhan J. C., and Huang W. D. (2008) Prokaryotic expression, polyclonal antibody preparation of the stilbene synthase gene from grape berry and its different expression in fruit development and under heat acclimation. Plant Physiol. Biochem. 46, 1085–1092 10.1016/j.plaphy.2008.07.005 [DOI] [PubMed] [Google Scholar]
- 22. Liu B., Raeth T., Beuerle T., and Beerhues L. (2007) Biphenyl synthase, a novel type III polyketide synthase. Planta 225, 1495–1503 10.1007/s00425-006-0435-5 [DOI] [PubMed] [Google Scholar]
- 23. Stewart C. Jr, Woods K., Macias G., Allan A. C., Hellensc R. P., and Noel J. P. (2017) Molecular architectures of benzoic acid-specific type III polyketide synthases. Acta Crystallogr. D Struct. Biol. 73, 1007–1019 10.1107/S2059798317016618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Beerhues L. (1996) Benzophenone synthase from cultured cells of Centaurium erythraea. FEBS Lett. 383, 264–266 10.1016/0014-5793(96)00265-7 [DOI] [PubMed] [Google Scholar]
- 25. Liu B., Falkenstein-Paul H., Schmidt W., and Beerhues L. (2003) Benzophenone synthase and chalcone synthase from Hypericum androsaemum cell cultures: cDNA cloning, functional expression, and site-directed mutagenesis of two polyketide synthases. Plant J. 34, 847–855 10.1046/j.1365-313X.2003.01771.x [DOI] [PubMed] [Google Scholar]
- 26. Schmidt W., and Beerhues L. (1997) Alternative pathways of xanthone biosynthesis in cell cultures of Hypericum androsaemum L. FEBS Lett. 420, 143–146 10.1016/S0014-5793(97)01507-X [DOI] [PubMed] [Google Scholar]
- 27. Nualkaew N., Morita H., Shimokawa Y., Kinjo K., Kushiro T., De-Eknamkul W., Ebizuka Y., and Abe I. (2012) Benzophenone synthase from Garcinia mangostana L. Pericarp. Phytochemistry 77, 60–69 10.1016/j.phytochem.2012.02.002 [DOI] [PubMed] [Google Scholar]
- 28. Helariutta Y., Elomaa P., Kotilainen M., Griesbach R. J., Schröder J., and Teeri T. H. (1995) Chalcone synthase-like genes active during corolla development are differentially expressed and encode enzymes with different catalytic properties in Gerbera hybrida (Asteraceae). Plant Mol. Biol. 28, 47–60 10.1007/BF00042037 [DOI] [PubMed] [Google Scholar]
- 29. Eckerman S., Schröder G., Schmidt J., Strack D., Edrada R. A., Helariutta Y., Elomaa P., Kotilainen M., Kilpeläinen I., Proksch P., Teeri T. H., and Schröder J. (1998) New pathway to polyketides in plants. Nature 396, 387–390 10.1038/24652 [DOI] [Google Scholar]
- 30. Abe I., Utsumi Y., Oguro S., Morita H., Sano Y., and Noguchi H. (2005) A plant type III polyketide synthase that produces pentaketide chromone. J. Am. Chem. Soc. 127, 1362–1363 10.1021/ja0431206 [DOI] [PubMed] [Google Scholar]
- 31. Abe I., Oguro S., Utsumi Y., Sano Y., and Noguchi H. (2005) Engineered biosynthesis of plant polyketides: chain length control in an octaketide-producing plant type III polyketide synthase. J. Am. Chem. Soc. 127, 12709–12716 10.1021/ja053945v [DOI] [PubMed] [Google Scholar]
- 32. Karppinen K., Hokkanen J., Mattila S., Neubauer P., and Hohtola A. (2008) Octaketide-producing type III polyketide synthase from Hypericum perforatum is expressed in dark glands accumulating hypericins. FEBS J. 275, 4329–4342 10.1111/j.1742-4658.2008.06576.x [DOI] [PubMed] [Google Scholar]
- 33. Mizuuchi Y., Shi S. P., Wanibuchi K., Kojima A., Morita H., Noguchi H., and Abe I. (2009) Novel type III polyketide synthases from Aloe arborescens. FEBS J. 276, 2391–2401 10.1111/j.1742-4658.2009.06971.x [DOI] [PubMed] [Google Scholar]
- 34. Abe I., Takahashi Y., Morita H., and Noguchi H. (2001) Benzalacetone synthase: a novel polyketide synthase that plays a crucial role in the biosynthesis of phenylbutanones in Rheum palmatum. Eur. J. Biochem. 268, 3354–3359 10.1046/j.1432-1327.2001.02255.x [DOI] [PubMed] [Google Scholar]
- 35. Zheng D., and Hrazdina G. (2008) Molecular and biochemical characterization of benzalacetone synthase and chalcone synthase genes and their proteins from raspberry (Rubus idaeus L.). Arch. Biochem. Biophys. 470, 139–145 10.1016/j.abb.2007.11.013 [DOI] [PubMed] [Google Scholar]
- 36. Katsuyama Y., Kita T., Funa N., and Horinouchi S. (2009) Curcumonoid biosynthesis by two type III polyketide synthases in the herb Curcuma longa. J. Biol. Chem. 284, 11160–11170 10.1074/jbc.M900070200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Katsuyama Y., Kita T., and Horinouchi S. (2009) Identification and characterization of multiple curcumin synthases from the herb Curcuma longa. FEBS Lett. 583, 2799–2803 10.1016/j.febslet.2009.07.029 [DOI] [PubMed] [Google Scholar]
- 38. Katsuyama Y., Matsuzawa M., Funa N., and Horinouchi S. (2007) In vitro synthesis of curcuminoids by type III polyketide synthase from Oryza sativa. J. Biol. Chem. 282, 37702–37709 10.1074/jbc.M707569200 [DOI] [PubMed] [Google Scholar]
- 39. Ferrer J.-L., Jez J. M., Bowman M. E., Dixon R. A., and Noel J. P. (1999) Structure of chalcone synthase and the molecular basis of plant type III polyketide biosynthesis. Nat. Struct. Biol. 6, 775–784 10.1038/11553 [DOI] [PubMed] [Google Scholar]
- 40. Go M. K., Wongsantichon J., Cheung V. W. N., Chow J. Y., Robinson R. C., and Yew W. S. (2015) Synthetic polyketide enzymology: platform for biosynthesis of antimicrobial polyketides. ACS Catal. 5, 4033–4042 10.1021/acscatal.5b00477 [DOI] [Google Scholar]
- 41. Jez J. M., Austin M. B., Ferrer J., Bowman M. E., Schröder J., and Noel J. P. (2000) Structural control of polyketide formation in plant-specific polyketide synthases. Chem. Biol. 7, 919–930 10.1016/S1074-5521(00)00041-7 [DOI] [PubMed] [Google Scholar]
- 42. Austin M. B., Bowman M. E., Ferrer J.-L., Schröder J., and Noel J. P. (2004) An aldol-switch discovered in stilbene synthase mediates cyclization specificity of type III polyketide synthase. Chem. Biol. 11, 1179–1194 10.1016/j.chembiol.2004.05.024 [DOI] [PubMed] [Google Scholar]
- 43. Shomura Y., Torayama I., Suh D. Y., Xiang T., Kita A., Sankawa U., and Miki K. (2005) Crystal structure of stilbene synthase from Arachis hypogaea. Proteins 60, 803–806 10.1002/prot.20584 [DOI] [PubMed] [Google Scholar]
- 44. Morita H., Kondo S., Abe T., Noguchi H., Sugio S., Abe I., and Kohno T. (2006) Crystallization and preliminary crystallographic analysis of a novel plant type III polyketide synthase that produces pentaketide chromone. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62, 899–901 10.1107/S174430910602968X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Morita H., Kondo S., Oguro S., Noguchi H., Sugio S., Abe I., and Kohno T. (2007) Structural insight into chain length control and product specificity of pentaketide chromone synthase from Aloe arborescens. Chem. Biol. 14, 359–369 10.1016/j.chembiol.2007.02.003 [DOI] [PubMed] [Google Scholar]
- 46. Morita H., Kondo S., Kato R., Wanibuchi K., Noguchi H., Sugio S., Abe I., and Kohno T. (2007) Crystallization and preliminary crystallographic analysis of an octaketide-producing plant type III polyketide synthase. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63, 947–949 10.1107/S1744309107047069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shi S. P., Wanibuchi K., Morita H., Endo K., Noguchi H., and Abe I. (2009) Enzymatic formation of unnatural novel chalcone, stilbene, and benzophenone scaffolds by plant type III polyketide synthase. Org. Lett. 11, 551–554 10.1021/ol802606w [DOI] [PubMed] [Google Scholar]
- 48. Morita H., Tanio M., Kondo S., Kato R., Wanibuchi K., Noguchi H., Sugio S., Abe I., and Kohno T. (2008) Crystallization and preliminary crystallographic analysis of a plant type III polyketide synthase that produces benzalacetone. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64, 304–306 10.1107/S1744309108006775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Morita H., Shimokawa Y., Tanio M., Kato R., Noguchi H., Sugio S., Kohno T., and Abe I. (2010) A structure-based mechanism for benzalacetone synthase from Rheum palmatum. Proc. Natl. Acad. Sci. U.S.A. 107, 669–673 10.1073/pnas.0909982107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Morita H., Wanibuchi K., Kato R., Sugio S., and Abe I. (2010) Expression, purification and crystallization of a plant type III polyketide synthase that produces curcuminoid. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 948–950 10.1107/S1744309110023572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Morita H., Wanibuchi K., Nii H., Kato R., Sugio S., and Abe I. (2010) Structural basis for the one-pot formation of the diarylheptanoid scaffold by curcuminoid synthase from Oryza sativa. Proc. Natl. Acad. Sci. U.S.A. 107, 19778–19783 10.1073/pnas.1011499107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Miyazono K., Um J., Imai F. L., Katsuyama Y., Ohnishi Y., Horinouchi S., and Tanokura M. (2011) Crystal structure of curcuminoid synthase CUS from Oryza sativa. Proteins 79, 669–673 10.1002/prot.22888 [DOI] [PubMed] [Google Scholar]
- 53. Katsuyama Y., Miyazono K., Tanokura M., Ohnishi Y., and Horinouchi S. (2011) Structural and biochemical elucidation of mechanism for decarboxylative condensation of β-keto acid by curcumin synthase. J. Biol. Chem. 286, 6659–6668 10.1074/jbc.M110.196279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Morita H., Kondo S., Kato R., Wanibuchi K., Noguchi H., Sugio S., Abe I., and Kohno T. (2007) Crystallization and preliminary crystallographic analysis of an acridone-producing novel multifunctional type III polyketide synthase from Huperzia serrata. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63, 576–578 10.1107/S1744309107026164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Morita H., Yamashita M., Shi S. P., Wakimoto T., Kondo S., Kato R., Sugio S., Kohno T., and Abe I. (2011) Synthesis of unnatural alkaloid scaffolds by exploiting plant polyketide synthase. Proc. Natl. Acad. Sci. U.S.A. 108, 13504–13509 10.1073/pnas.1107782108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mori T., Shimokawa Y., Matsui T., Kinjo K., Kato R., Noguchi H., Sugio S., Morita H., and Abe I. (2013) Cloning and structure-function analyses of quinolone- and acridone-producing novel type III polyketide synthases from Citrus microcarpa. J. Biol. Chem. 288, 28845–28858 10.1074/jbc.M113.493155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yu H. N., Liu X. Y., Gao S., Sun B., Zheng H. B., Ji M., Cheng A. X., and Lou H. X. (2018) Structural and biochemical characterization of the plant type III polyketide synthases of the liverwort Marchantia paleacea. Plant Physiol. Biochem. 125, 95–105 10.1016/j.plaphy.2018.01.030 [DOI] [PubMed] [Google Scholar]
- 58. Wanibuchi K., Morita H., Noguchi H., and Abe I. (2011) Enzymatic formation of an aromatic dodecaketide by engineered plant polyketide synthase. Bioorg. Med. Chem. Lett. 21, 2083–2086 10.1016/j.bmcl.2011.01.135 [DOI] [PubMed] [Google Scholar]
- 59. Liu B., Raeth T., Beuerle T., and Beerhues L. (2010) A novel 4-hydroxycoumarin biosynthetic pathway. Plant Mol. Biol. 72, 17–25 10.1007/s11103-009-9548-0 [DOI] [PubMed] [Google Scholar]
- 60. Lin Y., Shen X., Yuan Q., and Yan Y. (2013) Microbial biosynthesis of the anticoagulant precursor 4-hydroxycoumarin. Nat. Commun. 4, 2603 10.1038/ncomms3603 [DOI] [PubMed] [Google Scholar]
- 61. Klundt T., Bocola M., Lütge M., Beuerle T., Liu B., and Beerhues L. (2009) A single amino acid substitution converts benzophenone synthase into phenylpyrone synthase. J. Biol. Chem. 284, 30957–30964 10.1074/jbc.M109.038927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lukacin R., Springob K., Urbanke C., Ernwein C., Schröder G., Schröder J., and Matern U. (1999) Native acridone synthases I and II from Ruta graveolens L. form homodimers. FEBS Lett. 448, 135–140 10.1016/S0014-5793(99)00355-5 [DOI] [PubMed] [Google Scholar]
- 63. Springob K., Lukacin R., Ernwein C., Gröning I., and Matern U. (2000) Specificities of functionally expressed chalcone and acridone synthases from Ruta graveolens. Eur. J. Biochem. 267, 6552–6559 10.1046/j.1432-1327.2000.01746.x [DOI] [PubMed] [Google Scholar]
- 64. Lukacin R., Schreiner S., and Matern U. (2001) Transformation of acridone synthase to chalcone synthase. FEBS Lett. 508, 413–417 10.1016/S0014-5793(01)03061-7 [DOI] [PubMed] [Google Scholar]
- 65. Jez J. M., Bowman M. E., and Noel J. P. (2002) Expanding the biosynthetic repertoire of plant type III polyketide synthases by altering starter molecule specificity. Proc. Natl. Acad. Sci. U.S.A. 99, 5319–5324 10.1073/pnas.082590499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lukacin R., Schreiner S., Silber K., and Matern U. (2005) Starter substrate specificities of wild-type and mutant polyketide synthases from Rutaceae. Phytochemistry 66, 277–284 10.1016/j.phytochem.2004.11.023 [DOI] [PubMed] [Google Scholar]
- 67. Wanibuchi K., Zhang P., Abe T., Morita H., Kohno T., Chen G., Noguchi H., and Abe I. (2007) An acridone-producing novel multifunctional type III polyketide synthase from Huperzia serrata. FEBS J. 274, 1073–1082 10.1111/j.1742-4658.2007.05656.x [DOI] [PubMed] [Google Scholar]
- 68. Resmi M. S., Verma P., Gokhale R. S., and Soniya E. V. (2013) Identification and characterization of a type III polyketide synthase involved in quinolone alkaloid biosynthesis from Aegle marmelos correa. J. Biol. Chem. 288, 7271–7281 10.1074/jbc.M112.429886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dulcey C. E., Dekimpe V., Fauvelle D. A., Milot S., Groleau M. C., Doucet N., Rahme L. G., Lépine F., and Déziel E. (2013) The end of an old hypothesis: the Pseudomonas signaling molecules 4-hydroxy-2-alkylquinolines derive from fatty acids, not 3-ketofatty acids. Chem. Biol. 20, 1481–1491 10.1016/j.chembiol.2013.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Drees S. L., Li C., Prasetya F., Saleem M., Dreveny I., Williams P., Hennecke U., Emsley J., and Fetzner S. (2016) PqsBC, a condensing enzyme in the biosynthesis of the Pseudomonas aeruginosa quinolone signal: crystal structure, inhibition, and reaction mechanism. J. Biol. Chem. 291, 6610–6624 10.1074/jbc.M115.708453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Drees S. L., and Fetzner S. (2015) PqsE of Pseudomonas aeruginosa acts as pathway-specific thioesterase in the biosynthesis of alkylquinolone signaling molecules. Chem. Biol. 22, 611–618 10.1016/j.chembiol.2015.04.012 [DOI] [PubMed] [Google Scholar]
- 72. Lorence A., and Nessler C. L. (2004) Camptothecin, over four decades of surprising findings. Phytochemistry 65, 2735–2749 10.1016/j.phytochem.2004.09.001 [DOI] [PubMed] [Google Scholar]
- 73. Gagne S. J., Stout J. M., Liu E., Boubakir Z., Clark S. M., and Page J. E. (2012) Identification of olivetolic acid cyclase from Cannabis sativa reveals a unique catalytic route to plant polyketides. Proc. Natl. Acad. Sci. U.S.A. 109, 12811–12816 10.1073/pnas.1200330109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Taura F., Tanaka S., Taguchi C., Fukamizu T., Tanaka H., Shoyama Y., and Morimoto S. (2009) Characterization of olivetol synthase, a polyketide synthase putatively involved in cannabinoid biosynthetic pathway. FEBS Lett. 583, 2061–2066 10.1016/j.febslet.2009.05.024 [DOI] [PubMed] [Google Scholar]
- 75. Taguchi C., Taura F., Tamada T., Shoyama Y., Shoyama Y., Tanaka H., Kuroki R., and Morimoto S. (2008) Crystallization and preliminary X-ray diffraction studies of polyketide synthase-1 (PKS-1) from Cannabis sativa. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64, 217–220 10.1107/S1744309108003795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Thompson T. B., Katayama K., Watanabe K., Hutchinson C. R., and Rayment I. (2004) Structural and functional analysis of tetracenomycin F2 cyclase from Streptomyces glaucescens: a type II polyketide cyclase. J. Biol. Chem. 279, 37956–37963 10.1074/jbc.M406144200 [DOI] [PubMed] [Google Scholar]
- 77. Yang X., Matsui T., Mori T., Taura F., Noguchi H., Abe I., and Morita H. (2015) Expression, purification, and crystallization of a plant polyketide cyclase from Cannabis sativa. Acta Crystallogr. F Struct. Biol. Commun. 71, 1470–1474 10.1107/S2053230X15020385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yang X., Matsui T., Kodama T., Mori T., Zhou X., Taura F., Noguchi H., Abe I., and Morita H. (2016) Structural basis for olivetolic acid formation by a polyketide cyclase from Cannabis sativa. FEBS J. 283, 1088–1106 10.1111/febs.13654 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.