Abstract
Polymicrobial biofilms play important roles in oral and systemic infections. The oral plaque bacterium Streptococcus gordonii is known to attach to the hyphal cell wall of the fungus Candida albicans to form corn-cob like structures in biofilms. However, the role of C. albicans in formation of polymicrobial biofilms is not completely understood. The objective of this study was to determine the role of C. albicans transcription factors in regulation of polymicrobial biofilms and antibiotic tolerance of S. gordonii. The proteins secreted by C. albicans and S. gordonii in mixed planktonic cultures were determined using mass spectrometry. Antibiotic tolerance of S. gordonii to ampicillin and erythromycin was determined in mixed cultures and mixed biofilms with C. albicans. Additionally, biofilm formation of S. gordonii with C. albicans knock-out mutants of 45 transcription factors that affect cell wall integrity, filamentous growth and biofilm formation was determined. Furthermore, these mutants were also screened for antibiotic tolerance in mixed biofilms with S. gordonii. Analysis of secreted proteomes resulted in the identification of proteins being secreted exclusively in mixed cultures. Antibiotic testing showed that S. gordonii had significantly increased survival in mixed planktonic cultures with antibiotics as compared to single cultures. C. albicans mutants of transcription factors Sfl2, Brg1, Leu3, Cas5, Cta4, Tec1, Tup1, Rim101 and Efg1 were significantly affected in mixed biofilm formation. Also mixed biofilms of S. gordonii with mutants of C. albicans transcription factors, Tec1 and Sfl2, had significantly reduced antibiotic tolerance as compared to control cultures. Our data indicates that C. albicans may have an important role in mixed biofilm formation as well as antibiotic tolerance of S. gordonii in polymicrobial biofilms. C. albicans may play a facilitating role than being just an innocent bystander in oral biofilms and infections.
Keywords: Interkingdom interactions, Polymicrobial biofilms, Phenotypic switch, Streptococcus gordonii, Ampicillin resistance, Candida albicans, Polymicrobial biofilms, Antibiotic tolerance
Introduction
Candida albicans can be found as a commensal in the majority of healthy humans in the oral, vaginal, and gastrointestinal mucosae (Kim & Sudbery, 2011). However, C. albicans is also the most common fungal pathogen in humans. It turns opportunistic and causes candidiasis in immunocompromised patients, denture wearers and the elderly (Muzyka & Epifanio, 2013). It is found in over 90% of human fungal infections and is a major etiologic factor in hospital-acquired infections (Azie et al., 2012). With an increasing number of individuals immunocompromised from organ transplantation, chemotherapy, HIV infection, and medical device implantation, infections caused by C. albicans are becoming an increasing cause of concern. Candidemia may occur if the fungus is able to disseminate into the bloodstream causing symptoms similar to bacterial septicemia with extremely high mortality rates (Pfaller & Diekema, 2007).
Fungal infections occurring in the mouth, such as oral candidiasis, can become a nidus for disseminated infection (Odds, 1987). C. albicans is unique in that it is pleiomorphic and is able to grow in multiple morphological forms. It can exist as yeast (unicellular), hyphae, pseudohyphae and chlamydiospores. The transition from yeast to hyphae is stimulated by multiple factors, including the presence of serum, neutral pH, high CO2, N-acetylglucosamine, 37 °C, and when within biofilms (Kim & Sudbery, 2011). The ability of C. albicans to phenotypically switch from yeast to hyphal form is associated with virulence, even though both forms are found in infections (Calderone & Fonzi, 2001). Candida infections involve formation of a biofilm, which is a structured microbial community adherent to a surface and embedded within a matrix of extracellular polymeric substance. Biofilms in the oral cavity are found on the surface of teeth and dentures. Typically, the dental plaque biofilm is polymicrobial in nature and has Candida albicans in close association with several bacteria including Streptococci, which are the pioneer bacteria for dental plaque (Jenkinson, Lala & Shepherd, 1990; Ranjan & Dongari-Bagtzoglou, 2018; Xu, Jenkinson & Dongari-Bagtzoglou, 2014).
Streptococcal species most frequently found in the oral cavity, with the exception of Streptococcus mutans, the etiologic agent of dental caries, are generally considered beneficial and non-pathogenic (Xu, Jenkinson & Dongari-Bagtzoglou, 2014; Kuramitsu et al., 2007). These commensal bacteria can colonize multiple sites in the oral cavity, including keratinized and non-keratinized oral mucosa, the tongue, periodontal pockets, and teeth. A subgroup of Streptococcus sp., named the mitis group, includes: Streptococcus gordonii, Streptococcus oralis, Streptococcus mitis, Streptococcus parasanguinis, and Streptococcus sanguinis. The mitis group comprises over 60% of the identifiable oral microbiota (Syed & Loesche, 1978) and dominates the composition of the buccal mucosa in healthy individuals (Diaz et al., 2012). The mitis group is a research focus because of their ability to initiate polymicrobial biofilms in the oral cavity (Whitmore & Lamont, 2011).
C. albicans and S. gordonii are known to colonize the same habitats within the oral cavity. Streptococcus sp. has been verified to form corn-cob like structures around C. albicans in dental plaque (Zijnge et al., 2010). Their interactions may therefore be important to commensal colonization in health as well as in pathogenicity. There are several examples of interkingdom interactions between these two microorganisms. C. albicans secretes a quorum-sensing molecule, farnesol, that represses hyphae formation at high concentrations (Hornby et al., 2001). However, when C. albicans and S. gordonii are cultured together, hyphae formation is induced overriding the effect of farnesol (Bamford et al., 2009). Also S. gordonii, as with other Streptococci, produces lactic acid that may serve as an energy source for C. albicans, suggesting another synergistic interkingdom interaction (Jenkinson, Lala & Shepherd, 1990). Additionally, S. gordonii was found to have a higher affinity for C. albicans compared to other mitis bacteria (Holmes, Gopal & Jenkinson, 1995). Bacterial cell wall polypeptides SspA and SspB were identified as adhesion molecules in this interaction (Holmes, McNAB & Jenkinson, 1996). C. albicans hyphal wall proteins Als3 and Hwp1 have been identified as receptors for these bacterial polypeptides (Nobbs, Vickerman & Jenkinson, 2010). Attachment between these organisms may enable a variety of cell–cell interactions to occur.
Past studies have shown that C. albicans and S. gordonii grown together in mixed biofilms have a greater biomass than when cultured in a monospecies biofilm (Xu, Jenkinson & Dongari-Bagtzoglou, 2014; Bamford et al., 2009; Dutton et al., 2014). It has been shown that their interactions induce changes in gene expression that lead to differences in growth, virulence, and drug susceptibility (Dutton et al., 2015). Additionally, a more recent study has shown that polymicrobial biofilms of C. albicans and S. gordonii have increased resistance to antibiotics (Montelongo-Jauregui et al., 2016). In this study, our primary objective was to understand the functions of C. albicans in polymicrobial biofilm formation with S. gordonii and antibiotic resistance. In order to better understand the proteomic interactions between these microorganisms we determined secreted proteins unique to planktonic co-cultures of C. albicans and S. gordonii. We further aimed to demonstrate differences in biofilm formation and anitbiotic tolerance in single species versus mixed biofilms. Additionally, we performed screening of several mutants of C. albicans transcription factors that affect cell wall integrity to study regulation of polymicrobial biofilm formation and antibiotic tolerance.
Materials and Methods
Strains and culturing conditions
C. albicans wild type strain SC5314 and transcription factor knock-out mutant strains (Homann plates) were obtained from the fungal genetics stock center (FGSC). The genetic background and creation of the transcription factor knock-out mutants has been described previously (Homann et al., 2009). The C. albicans wild type strain SC5314 and C. albicans transcription factor mutant strains were cultured from frozen stock in liquid Yeast Nitrogen Base (YNB) medium (Amresco , OH) with 2% Glucose (Amresco, OH) as a carbon source supplemented with a complete amino acid supplement mixture (CSM) (MP Biomedicals, OH) and grown overnight at 30 °C with shaking at 225 rpm. The Streptococcus gordonii wild type strain Challis CH1 was cultured from frozen stock in liquid Tryptic Soy Broth plus 0.3% Yeast Extract (TSBY) medium (Difco, BD Diagnostics, MD) and grown statically overnight at 37 °C in a candle jar. Solid media was prepared by adding 1.5% agar (Alfa Aesar, MA). Both C. albicans and S. gordonii were also cultured from frozen stock in a mixture of 50% YNB and 50% TSBY in their respective growth conditions where indicated. C. albicans cell concentration was monitored with a hemocytometer and S. gordonii concentration was monitored by measuring the optical density at 600nm (OD600).
Effect of pH on C. albicans Growth
An overnight culture of wild type C. albicans in YNB was inoculated from frozen stock. The overnight was used to inoculate 20 ml of five different mixtures of YNB and TSBY: 100% YNB, 75% YNB/25% TSBY, 50% YNB/50% TSBY, 25% YNB/75% TSBY, and 100% TSBY. The pH of each of these mixtures was measured prior to inoculation. The cultures were grown at 30 °C with shaking at 225 rpm. Cell concentrations were taken at 0, 2, 4, and 6 h. A fresh overnight culture of C. albicans was used to inoculate five, 20 ml volumes of YNB that had each been adjusted to the pH of one of the YNB/TSBY mixtures used previously. Cultures were grown at 30 °C with shaking and cell concentrations were taken at 0, 2, 4, and 6 h. These cultures served as controls to make sure that alterations in the growth of C. albicans were due to the change in media rather than just a change in pH.
Secreted proteome analysis of co-cultures of C. albicans and S. gordonii
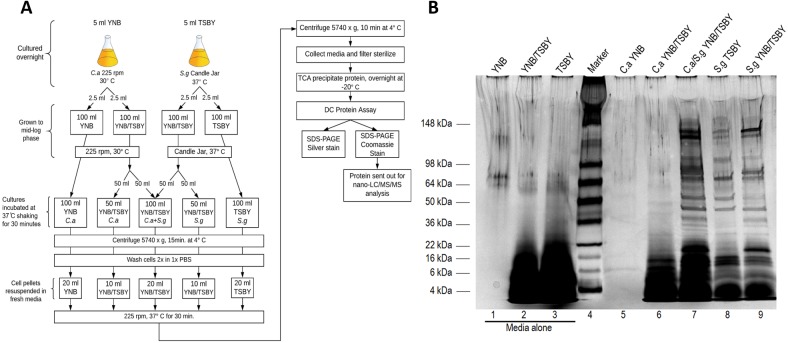
Portions (2.5 ml) of a wild type C. albicans overnight culture grown in YNB at 30 °C with shaking were used to inoculate 100 ml of YNB and 100 ml of YNB/TSBY (50/50). Portions (2.5 ml) of a S. gordonii overnight culture grown in TSBY statically at 37 °C in a candle jar were used to inoculate 100 ml of TSBY and 100 ml of YNB/TSBY. The two C. albicans cultures were grown at 30 °C with shaking and the two S. gordonii cultures were grown statically at 37 °C in a candle jar until they reached mid-log phase (approximately 5 × 107 cells/ml and OD600 = 0.6 respectively) for a total of four different cultures. Upon reaching desired cell concentration, 50 ml of the C. albicans culture grown in YNB/TSBY (50/50) was combined with 50 ml of the S. gordonii culture that was grown in the same type of media for a total volume of 100 ml in a new container to make a fifth culture group. The cells from all five groups were harvested by centrifugation (5,470×g, 15 min). The cell pellets were washed twice with cold 1× PBS. The cell pellets from the cultures with a 100 ml volume prior to centrifugation (C. albicans in YNB, S. gordonii in TSBY, and the YNB/TSBY combined co-culture) were re-suspended a fresh 20ml volume of their respective original medias. The cell pellets from the cultures with a 50 ml volume prior to centrifugation (the remainders of the single cultures of C. albicans and S. gordonii grown in YNB/TSBY left over from the creation of the co-culture group) were re-suspended in a fresh 10 ml volume of YNB/TSBY. All five cultures were then allowed to grow at 37 °C with shaking for 30 min. After the incubation period, the cells were removed from the media by centrifugation (5,470× g, 10 min.) followed by filtration using 0.2 µm syringe filters (Corning, NY). The proteins in the media were collected for analysis by TCA precipitation using a final concentration of 12.5% TCA (Amresco, OH) and 50% acetone (Fisher Scientific, ON). Proteins were allowed to precipitate for 24 h at −20 °C. In addition to the spent culture media samples, TCA precipitations of each fresh media (YNB, TSBY, or YNB/TSBY) were included as controls to see what protein was coming from the media alone. The precipitated proteins were collected by centrifugation (10,510×g, 10 min.) and washed twice with cold 100% acetone. All samples were re-suspended in a small volume of 1× PBS after allowing the acetone to evaporate completely in a flow hood and subjected to a DC Protein Assay (Bio-Rad, CA) to determine protein concentration. The protein samples were then subjected to SDS-PAGE. 9 µg of protein of each sample were loaded onto a 4 to 15% Mini-Protean TGX gel (Bio-Rad, CA) with the exceptions of the YNB media only control and the C. albicans sample that was grown in YNB. For these samples, as much as possible was loaded since there is no protein coming from the YNB itself and C. albicans secretes very little protein when grown in YNB. The proteins were visualized using a silver stain kit (Bio-Rad, CA). An additional gel was run for the preparation of samples for mass spectrometric (MS) analysis. 9 µg of each sample were loaded onto a SDS-PAGE gel and subjected to a brief electrophoresis step of 5 min so that the dye front only traveled a short distance into the gel. The gel was then stained with Coomassie Brilliant Blue (Amresco, OH). The protein bands from the co-culture and the single cultures of C. albicans and S. gordonii (all grown in YNB/TSBY) were cut out of the gel after de-staining and sent for nano-LC/MS/MS analysis at the Fred Hutchinson Cancer Research Center (FHCRC), Seattle, WA. Samples from two repeats of the experiment were sent for MS analysis. Please see Fig. 1A for a flow chart of methods.
Figure 1. C. albicans and S. gordonii secreted proteomes in planktonic cultures.
(A) Schematic of culture conditions and proteomic analysis of secreted proteins. (B) Silver-stained 4–15% gradient SDS-PAGE gel of TCA precipitated secreted protein from single and co-cultures of 30 minutes only. C. albicans was grown in yeast nitrogen base (YNB) and a mixture of yeast nitrogen base and tryptic soy broth with yeast extract (YNB/TSBY) media. C. albicans and S. gordonii co-culture was grown in YNB/TSBY. S. gordonii was grown in TSBY and YNB/TSBY media. Lanes 1, 2, and 3 contain TCA precipitated proteins from media alone. Proteins 22 kDa and smaller are likely from the TSBY due to the undefined nature of the media. There is no protein from the YNB alone (lane 1) as it is a completely synthetic media with no added protein. Image taken with Gel Doc XR+ and viewed with Image Lab Software (Bio-Rad, USA).
Assessment of ampicillin tolerance in planktonic cultures
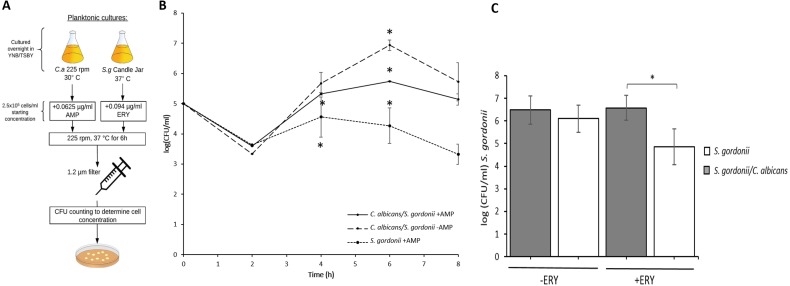
Overnight cultures of wild type C. albicans and S. gordonii were inoculated from frozen stocks in YNB/TSBY (50/50) and grown in their respective preferred growth conditions overnight. The concentrations of the overnights were determined and then used to inoculate three different groups, all in a 10 ml total volume of YNB/TSBY (50/50), at a concentration of 1 × 105 cells/ml. The test group was a co-culture of C. albicans and S. gordonii supplemented with ampicillin (G-Biosciences, MO) at a concentration of 0.125 µg/ml. The positive control group was a single culture of S. gordonii supplemented with ampicillin. The negative control group was a co-culture of C. albicans and S. gordonii with no antibiotic added. The ampicillin concentration used was based on a MIC value range found in literature (Etienne, Gruer & Fleurette, 1984; Nemoto et al., 2013). All cultures were grown at 37 °C with shaking. Samples were taken at 2, 4, 6, and 8 h and filtered through a 1.2 µm syringe filter (Whatman, GE Healthcare Life Sciences, IL) to remove C. albicans. The filtrate was diluted and plated on TSBY agar plates in triplicate. Plates were incubated overnight at 37 °C in a candle jar. Colony forming units (CFU) were counted the next day to determine CFU/ml. Raw data was statistically analyzed utilizing ANOVA performed using the default stats package in R Core Team v3.1.2 with results adjusted post-hoc with Tukey’s test. Please see Fig. 2A for a flow chart of methods.
Figure 2. C. albicans provides S. gordonii antibiotic tolerance in planktonic co-cultures.
(A) Schematic of culture conditions and antibiotic treatment. (B) CFUs (Mean+/-SD) of S. gordonii in mono and dual cultures with C. albicans, in the presence or absence of ampicillin. At 4 h, a significant difference was found between the positive control (S. gordonii. + ampicillin) and negative control (C. albicans. + S. gordonii, no ampicillin) and between the test group (C. albicans + S. gordonii. + ampicillin) and positive control group. No significant differences between the groups were found at 8 h. Statistical significance was observed between all groups at 6 h ( p < 0.05) using ANOVA with results adjusted post-hoc using Tukey’s Test. (C) CFUs (Mean ±SD) of S. gordonii in mono and dual cultures with C. albicans, with and without erythromycin at 6 h time point. There was no significant difference between S. gordonii survival with and without erythromycin when C. albicans was present. However, significantly increased killing was observed when S. gordonii was present alone. Three replicates were used for each experimental group and a minimum of two repeats were performed for each experiment. Statistical analysis was done using Student’s t-Test (p < 0.05).
Assessment of erythromycin tolerance in planktonic cultures
Overnight cultures of wild type C. albicans and S. gordonii were inoculated from frozen stocks in YNB/TSBY and grown in their respective preferred growth conditions overnight. The concentrations of the overnights were determined and used to inoculate six different cultures in a total volume of 10 ml YNB/TSBY. Four cultures served as controls. The first two cultures were grown without antibiotic and include a single culture of S. gordonii and a co-culture of C. albicans and S. gordonii. The second pair of cultures, single culture of S. gordonii and a co-culture, were supplemented with ampicillin (0.0625 µg/ml). The last pair of cultures, single culture of S. gordonii and a co-culture, were supplemented with erythromycin (0.094µg/ml) (Bancescu et al., 2004). All six cultures were grown at 37 °C with shaking. Samples were taken at 2, 4, 6, and 8 h and filtered through a 1.2 µm syringe filter (Whatman, GE Healthcare Life Sciences, IL) to remove C. albicans. The filtrate was diluted and plated on TSBY agar plates in triplicate. Plates were incubated overnight at 37 °C in a candle jar. Colony forming units (CFU) were counted the next day to determine CFU/ml. Raw data was statistically analyzed using Student’s t-test. Please see Fig. 2A for a flow chart of methods.
Analysis of dual species biofilm formation of C. albicans with S. gordonii
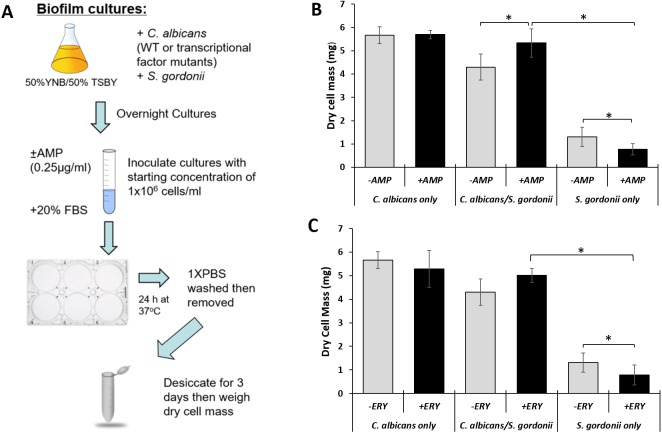
Analysis of dual species biofilms was done as described previously (Mancuso et al., 2018). Briefly, overnight cultures of wild type C. albicans and S. gordonii strains were inoculated from frozen stocks in YNB/TSBY and grown in their respective growth conditions overnight. The concentrations of the overnights were determined and used to inoculate several different cultures with the starting concentration of 1 × 106 cells/ml for each organism in a 6 ml total volume of YNB/TSBY supplemented with 20% FBS. The first two cultures were grown without antibiotic and include a single culture of S. gordonii and a co-culture of C. albicans and S. gordonii. The second pair of cultures, single culture of S. gordonii and a co-culture, were supplemented with ampicillin (0.25 µg/ml). The last pair of cultures, single culture of S. gordonii and a co-culture, were supplemented with erythromycin (0.376 µg/ml). Each culture was transferred to uncoated 6-well polystyrene culture plates (2 ml/well) and incubated statically for 24 h at 37 °C. After incubation, the media was carefully removed and the biofilms were washed once with 1x PBS. The biofilms were removed with additional 1x PBS and transferred to pre-weighed microfuge tubes. The samples were centrifuged to pellet the cells and remove most of the liquid to facilitate drying. The sample tubes were opened and placed in a desiccator jar with anhydrous calcium chloride used as the desiccant. Dry cell mass was determined after 3 days using an analytical weighing scale. Raw data was statistically analyzed using Student’s t-test. Please see Fig. 3A for a flow chart of methods.
Figure 3. C. albicans provides S. gordonii antibiotic tolerance in biofilm co-cultures.
(A) Schematic of biofilm culture conditions, antibiotic treatment and measurement of biofilm weight following dessication. (B) Dry weight measurement (Mean ± SD) of S. gordonii and C. albicans biofilms with and without ampicillin at 24 h time point. There was no significant difference between dry weights of biofilms with and without ampicillin when C. albicans was present. However, significantly reduced biofilm dry weight was observed for single species S. gordonii biofilms with ampicillin. Statistical analysis was done by Student’s t-Test (p < 0.05). (C) Dry weight measurement (Mean ± SD) of S. gordonii and C. albicans biofilms with and without erythromycin at 24 h time point. There was no significant difference between dry weights of biofilms with and without erythromycin when C. albicans was present. However, significantly reduced biofilm dry weight was observed for single species S. gordonii biofilms with erythromycin. Three replicates were used for each experimental group and a minimum of two repeats were performed for each experiment. Statistical analysis was done using Student’s t-Test (p < 0.05).
Dual species biofilm formation and ampicillin tolerance
Biofilms were prepared as described previously (Mancuso et al., 2018). Overnight cultures of wild type and transcription factor mutant C. albicans strains and the wild type Challis CH1 S. gordonii strain were inoculated from frozen stocks in YNB/TSBY and grown in their respective growth conditions overnight. The concentrations of the overnight cultures were determined and then used to inoculate several different cultures with the starting concentration of 1 × 106 cells/ml for each organism in a total volume of 6 ml of YNB/TSBY supplemented with 20% Fetal Bovine Serum (FBS) (Seradigm, VWR, GA). There were three groups and two cultures in each group. One culture for each group was grown with ampicillin (0.25 µg/ml) and the other was grown without ampicillin. The first group was a co-culture of wild type C. albicans and S. gordonii, the second was a co-culture of one of the 45 C. albicans transcriptional regulator mutants from the Homann knockout set and S. gordonii, and the third was a single culture of S. gordonii. Each culture was transferred to uncoated 6-well polystyrene culture plates (2 ml/well) and incubated statically for 24 h at 37 °C. After incubation, the media was carefully removed and the biofilms were washed once with 1 ×PBS. The biofilms were removed with additional 1x PBS and transferred to pre-weighed microfuge tubes. The samples were centrifuged to pellet the cells and remove most of the liquid to facilitate drying. The sample tubes were opened and placed in a desiccator jar with anhydrous calcium chloride used as the desiccant. Dry cell mass was determined after 3 days. Please see Fig. 3A for a flow chart of methods.
Scanning electron microscopy (SEM) of biofilms
Biofilms were prepared as described previously (Mancuso et al., 2018). Briefly, overnight cultures of the wild type C. albicans and S. gordonii strains were inoculated from frozen stocks in YNB/TSBY (50/50) and grown in their respective growth conditions overnight. The concentrations of the overnight cultures were determined and then used to inoculate several different cultures with the starting concentration of 1 × 106 cells/ml for each organism in a total volume of 4 ml of YNB/TSBY (50/50) supplemented with 20% FBS. Two, dual-species biofilm cultures with both organisms were set up. Mono-species biofilm cultures for S. gordonii and C. albicans, were also set up. 2 ml/well of each culture was transferred to uncoated 6-well polystyrene culture plates (Falcon, Corning, NY) containing FBS-coated glass microscope slide (Globe Scientific, NJ) pieces for the biofilms to grow on for use in SEM imaging. The culture plates were incubated statically for 24 h at 37 °C. After incubation, the media was carefully removed from the biofilm cultures. The biofilms were fixed with cold 2.5% glutaraldehyde (Electron Microscopy Sciences, PA) in 1×PBS at 4 °C for 20 min. 1×PBS was added to wash the biofilms and left on for 10 min. The biofilms were dehydrated with a series of ethanol washes lasting 5 min each including 30, 50, 70, and 90% ethanol v/v in water with two final washes of 100% ethanol. Biofilms were not allowed to dry out until the final drying step that included incubating in the chemical drying agent, hexamethyldisilazane (HMDS) (Acros Organics, Fisher Scientific, ON), for 5 min and then removal. The biofilms were then allowed to air dry completely. The biofilm samples were coated with evaporated carbon at high vacuum (Denton 502 Evaporator). SEM images were acquired with a Hitachi SU70 FESEM at 2.0 KeV using the lower detector and a 45° tilt.
Results
C. albicans and S. gordonii secreted proteomes in planktonic co-cultures
In order to culture C. albicans and S. gordonii in mixed planktonic and biofilm cultures, the independent growth of C. albicans in several combinations of yeast nitrogen base (YNB) and tryptic soy broth yeast extract (TSBY) was tested. A culture medium with YNB/TSBY (50/50) was found to be the most conducive for growth of C. albicans with optimal cell concentrations (Fig. S1). Additionally, the growth of C. albicans under various pH conditions was examined and pH 7 was found to be optimal (Fig. S2). S. gordonii is normally grown under static conditions. However, the co-cultures were to be cultured with shaking for C. albicans. Hence, we tested whether there was any difference in the cultures for S. gordonii in YNB/TSBY (50/50) for static and shaking conditions and found no significant differences (Fig. S3). For proteomic analysis of secreted proteins co-cultures of C. albicans and S. gordonii were prepared as depicted in the schematic in Fig. 1A. The isolated secreted proteins from the illustrates the On the silver-stained SDS-PAGE gel (Fig. 1B), the lanes for the medium alone show that proteins with a molecular weight of 22 kDa and smaller are most likely proteins from the TSBY medium. No proteins from the YNB medium formed obvious identifiable bands as it is synthetic media. S. gordonii, regardless of which medium it grew in, exhibited more secreted protein compared to C. albicans. Some proteins, like the 50-kDa protein for S. gordonii, were present in higher quantity in the 100% TSBY medium versus the YNB/TSBY (50/50) medium. There appeared to be more proteins released in co-culture (lane 7) and this is simply due to cumulative secretion of proteins by both microorganisms. However, it is unclear which proteins belong to which organism on the gel. The majority of these proteins may possibly belong to S. gordonii since it produced more proteins in YNB/TSBY (50/50) medium than C. albicans did in the same medium. All lanes were equally loaded with the exception of YNB alone since it is a synthetic media (lane 1) and the C. albicans single culture grown in YNB since it secretes very little protein when grown in this media (lane 5).
The first nano-LC/MS/MS analysis identified 85 proteins in the C. albicans single culture (all samples were obtained YNB/TSBY (50/50)). A total of 237 proteins were identified in the S. gordonii single culture. A total of 348 proteins were identified in the co-culture, with 80 of those belonging to C. albicans and 268 belonging to S. gordonii. The second MS analysis identified 208 proteins in the C. albicans single culture. A total of 468 proteins were identified in the S. gordonii single culture. A total of 539 proteins were identified in the co-culture, with 137 of those belonging to C. albicans and 402 belonging to S. gordonii. The MS analyses used three genomic databases to rule out protein contamination from other sources (such as the media) and to verify that the proteins did belong to either C. albicans or S. gordonii. The results from the first and second MS analyses were combined. Proteins present in both the first and second analyses for C. albicans in single culture were identified, as they were for S. gordonii in single culture. Proteins present in both the first and second analyses for C. albicans and S. gordonii in co-culture were also identified. Thirteen proteins from C. albicans were identified in the single culture. One protein was identified for S. gordonii in single culture. In co-culture, four proteins belonged to C. albicans and one protein belonged to S. gordonii, for a total of 5 proteins identified in co-culture (Table 1).
Table 1. Secreted proteome of C. albicans and S. gordonii interactions in planktonic cultures.
Proteomic analysis was performed by nano-LC/MS/MS analysis. Proteins identified in two separate analyses have been listed in the table.
| Candida albicans | Streptococcus gordonii | |
|---|---|---|
| Single culture only proteins | Translation initiation factor | Conserved hypothetical protein TIGR00096 |
| Cytosolic ribosomal protein L12 | ||
| Elongation factor 2 | ||
| Elongation factor 3 | ||
| Adenosine kinase | ||
| Hypothetical protein CaO19.10988 | ||
| Likely cytosolic ribosomal protein L3 | ||
| Galactose/glucose transporter | ||
| Hypothetical protein CaO19.6673 | ||
| Sphingolipid long chain base-responsive protein LSP1 | ||
| Ribosomal protein 10 | ||
| Co-culture only proteins | Hypothetical protein 76573724 | Ethanol-active dehydrogenase/acetaldehyde-active reductase |
| Hypothetical protein CAWG_05665 | ||
| SNF-2 family ATP dependent chromatin remodeling factor snf21 | ||
| Conserved hypothetical protein 238879991 |
Antibiotic tolerance in planktonic co-cultures and dual species biofilms
In order to assess the role of C. albicans in resistance to ampicillin in planktonic co-cultures with S. gordonii, they were cultured in YNB/TSBY (50/50) with ampicillin (0.25 µg/ml) and CFU/ml were analyzed every 2 h for 8 h using a protocol described in Fig. 2A. As seen in Fig. 2B, S. gordonii growth for all groups deceased for all groups at 2 h, then increased at 4 and 6 h before finally decreasing at 8 h. At 4 h, a significant difference was found between the test (C. albicans + S. gordonii + ampicillin) and negative control (C. albicans +S. gordonii - ampicillin) groups. At 6 h, all groups were significantly different, and at 8 h no significant differences were found between any of the groups. To test whether tolerance of co-cultures extends to other antibacterial agents, the cultures were incubated with erythromycin for 6 h and S. gordonii CFUs were analyzed. The data show that in co-cultures S. gordonii CFUs were significantly higher indicating protection from erythromycin (Fig. 2C). This finding suggests that C. albicans has a protective effect on S. gordonii when it is grown in the presence of antibacterial agents. This tolerance to antibiotics was also observed for dual species biofilms of C. albicans and S. gordonii (Fig. 3). For the dual species biofilms with antibiotics, biofilm formation was more than the no antibiotic group (Fig. 3B & Fig. 3C). In fact, for the ampicillin group there was a significantly increased biofilm formation for dual species. There is a possibility that this increase in biofilm formation by weight in the antibiotic group could be because of an increase in proliferation of C. albicans in the biofilm while S. gordonii struggles to grow under antibiotic stress. This may be further supported by the finding that mono-species biofilm formation is at similar levels to the antibiotic treated dual species biofilms. These data indicate that dual species biofilms may be more tolerant to ampicillin and erythromycin.
Binding and coaggregation of C. albicans and S. gordonii
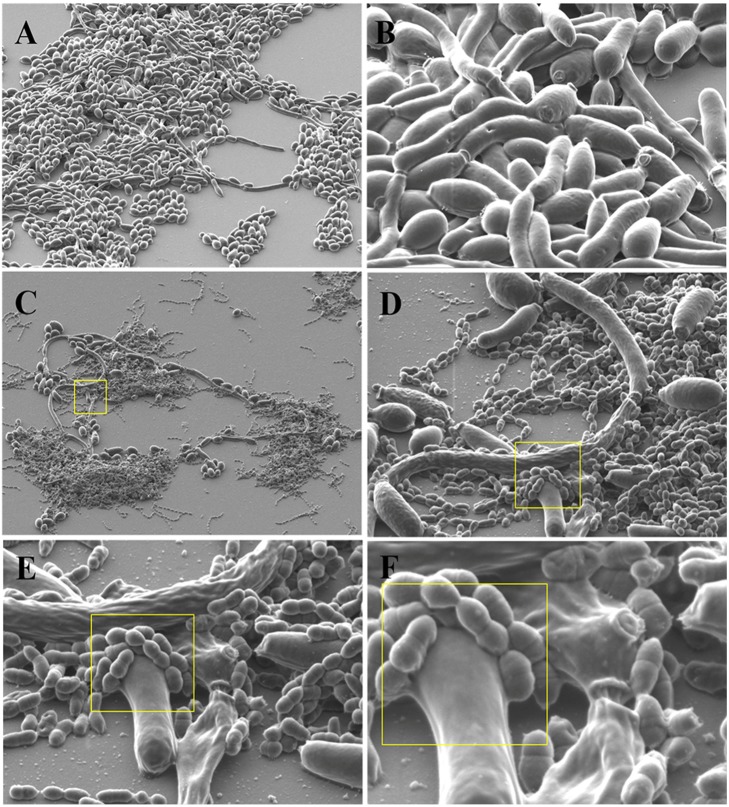
SEM analysis of early biofilms cultured in YNB/TSBY (50/50) with 10% fetal bovine serum (FBS) revealed coaggregation of C. albicans and S. gordonii (Fig. 4). S. gordonii cells were found to be growing comfortably on C. albicans yeast and hyphae and appeared to be firmly attached to the candida cell wall (Figs. 4C–4D).
Figure 4. Binding and coaggregation of S. gordonii and C. albicans in early biofilms.
Scanning Electron Microscopy (SEM)images were acquired with a Hitachi SU70 FESEM at 2.0 KeV using the lower detector and a 45° tilt. (A) C. albicans biofilm at 1,000× magnification. (B) C. albicans biofilm at 5,000× magnification. (C) C. albicans and S. gordonii dual species biofilm at 1,000× magnification. (D) C. albicans and S. gordonii dual species biofilm at 5,000× magnification. (E) C. albicans and S. gordonii dual species biofilm at 10,000× magnification. (F) C. albicans and S. gordonii dual species biofilm at 20,000× magnification.
C. albicans cell wall integrity transcription factors affect dual species biofilm formation with S. gordonii as well as antibacterial tolerance
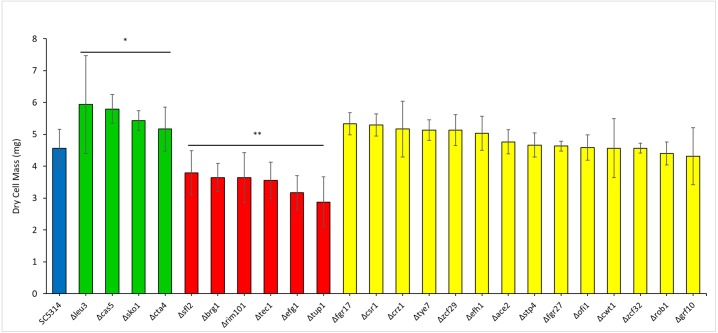
In order to assess polymicrobial biofilm formation C. albicans cell wall integrity transcription factor mutants (Fig. 5) were cultured as dual species biofilms with S. gordonii. The dry weight of the resulting biofilms was compared with those formed by WT C. albicans with S. gordonii. Interestingly, mutants of transcription factors leu3, cas5, cta4 and sko1 formed significantly heavier biofilms while mutants of sfl2, brg1, tec1, tup1, efg1 and rim101 formed significantly lighter biofilms (Fig. 5). Additionally, dual species biofilms of S, gordonii and sfl2, tec1 and efg1 transcription factor mutants of C. albicans also displayed reduced tolerance to antibiotics.
Figure 5. Transcriptional regulators affect the ability of C. albicans to form polymicrobial biofilms with S. gordonii.
Dry weights were measured for biofilms of C. albicans transcription factor Knock-out mutants with S. gordonii and compared with biofilms of WT C. albicans and S. gordonii. Statistical analysis was done using Student’s t-Test (p < 0.05).
Discussion
Biofilms are composed of microbial organisms embedded in extracellular matrix. In a C. albicans biofilm it has been documented that 55% of the dry weight of the extracellular matrix is composed of proteins (Zarnowski, Sanchez & Andes, 2016). It has also been found that there is a striking resemblance in glycoproteins that are part of the extracellular matrix of the biofilm as well as supernatants of planktonic cultures (Thomas, Bachmann & Lopez-Ribot, 2006). Thus it is very likely that glycoproteins secreted during planktonic cultures will end up in the extracellular matrix during growth as biofilms (Pierce et al., 2017). Moreover, in polymicrobial biofilms, proteins secreted by microorganisms may play important roles in interactions. Hence our proteomic analysis of secreted proteins in dual planktonic cultures of C. albicans and S. gordonii is justified and may represent the proteins being secreted in their biofilms as well. For determining optimal culture conditions for dual cultures, we tested the growth of C. albicans in mixed media at various concentrations and found that TSBY/YNB (50/50) was the most optimal for growth (Fig. S1). The pH of our various media mixtures ranged from 5.4 to 7.02, acidic to neutral, however, this did not seem to significantly impact the growth of C. albicans (Fig. S2). Since we planned to study mixed cultures of C. albicans with S. gordonii, which prefers TSBY medium, we chose TSBY/YNB (50/50) medium without pH adjustment as our co-culture media. S. gordonii is usually cultured in static conditions in the laboratory. However, the dual planktonic cultures were to be under shaking conditions for optimal growth of C. albicans. Hence, we compared growth of S. gordonii in TSBY/YNB (50/50) medium at static and shaking conditions and found no significant differences (Fig. S3). Although S. gordonii appeared to produce more protein bands in both single and co-cultures compared to C. albicans, when samples were analyzed using nano-LC/MS/MS, more differentially secreted protein bands were identified for C. albicans (Table 1). However, C. albicans may have produced more protein bands if the co-cultures had been allowed to incubate for more than the allotted 30 min, which only minimally offers eukaryotes time to translate protein. The proteomic analysis data presented in this manuscript is purely qualitative and the protein bands may not represent individual proteins but may also be processed proteins and thus future quantitative Mass Spectrometry analysis will be needed to ascertain the quantity and identity of those protein bands. Furthermore, limitations of proteomic analysis in this study include the limited number of repetitions (n = 2), and the variable environments used for culture conditions. Many of the proteins that were differentially secreted appeared to be non-classical, i.e., without a signal peptide. This has been observed in past studies of proteomic analysis as well (Chaffin et al., 1998; Maddi, Bowman & Free, 2009; Nombela, Gil & Chaffin, 2006). One of these differentially secreted proteins non-classical protein, SNF2-family ATP dependent chromatin remodeling factor Snf21, is essential for cell viability and is needed for mitotic progression (Yamada et al., 2008). There is evidence that this complex regulates the expression of a gene that promotes adhesion for cell-to-cell contact, biofilm formation and the dimorphic switch (Barrales et al., 2012). Since this protein appears in co-culture, it would be interesting to investigate if this protein was present because genes related to adhesion with S. gordonii were being upregulated. Three different hypothetical proteins without identified functions unique to C. albicans were also found in the co-culture only. These proteins of unknown function will be a source of interest for future studies as they may have possible implications in increased biofilm formation and virulence for either, or both, C. albicans and S. gordonii. There are multiple proteins identified in the single culture only for C. albicans. These include translation initiation factor, likely cytosolic ribosomal protein L12, elongation factor 2 (EF-2) and 3 (EF-3), likely cytosolic ribosomal protein L3, and ribosomal protein 10. EF-3 is unique to all fungi (Ross-Smith et al., 1995). EF-2 and EF-3 are both necessary for protein synthesis. EF-2 is encoded by one single gene (Mendoza et al., 1999), therefore, the presence of both EF-2 and EF-3 requires the activation of more than one gene. The genes that encode these proteins are on different chromosomes; EF-3 is encoded by a gene on chromosome 5 (Myers, Fonzi & Sypherd, 1992). Examples of proteins that may have been located in the cell wall and been either released from the cell wall or secreted include galactose/glucose transporter and sphingolipid long chain base-responsive protein LSP1. Adenosine kinase was identified, which is a cellular protein integral to multiple cellular functions. Additional unknown proteins that were identified that may be a source of interest for future studies include four hypothetical proteins. Only one protein was identified in co-culture that was unique to S. gordonii: ethanol-active dehydrogenase/acetaldehyde-active reductase. The increase in this protein may indicate an increase in cellular glycolysis. Only one protein, a conserved hypothetical protein, was identified in single culture that was unique to S. gordonii. This protein of unknown function may be a source of interest for future studies.
Fungi and bacteria in biofilms show increased resistance to antibiotics, including ampicillin (Donlan & Costerton, 2002). Past studies show that C. albicans can enhance antibacterial resistance/tolerance of gram positive bacteria like Staphylococcus aureus (Scheres & Krom, 2016). In that study, it was shown that attachment of S. aureus to C. albicans hyphae resulted in decreased susceptibility of the bacteria to antibiotic treatment. Furthermore, it was found that co-inoculation of S. aureus with C. albicans resulted in a more widespread infection as compared to either microorganism alone (Scheres & Krom, 2016). Similarly, another study of biofilms of S. gordonii with C. albicans found that there was enhanced resistance to antibiotics for dual species biofilms as opposed to individual biofilms (Montelongo-Jauregui et al., 2016). Based on this information, we expected that the interaction between planktonic C. albicans and S. gordonii in planktonic cultures could result in antibiotic tolerance to ampicillin and erythromycin which were routinely used for treating oral infections (Etienne, Gruer & Fleurette, 1984; Nemoto et al., 2013). The MIC concentration for ampicillin for Streptococci has been well established (Nemoto et al., 2013). In this reference the 0.25 microgram/ml has been determined as a concentration of antibiotic susceptibility for Streptococci (Nemoto et al., 2013). Thus we have utilized the above concentration for ampicillin. For Erythromycin, a range of 0.16–4 microgram/mL has been provided as MIC as described previously (Bancescu et al., 2004). We initially did some quick screening experiments to determine the Mic of 0.375 microgram/mL as ideal for our experiments.
S. gordonii killing was more prominent when incubated with ampicillin, however protection from ampicillin was observed by significantly increased survival of S. gordonii when C. albicans was present in the culture (Fig. 2B). This protection from ampicillin was observed at the 4, 6 and 8 h time points but it was statistically significant at 6 h. Overall, our data suggest that C. albicans provides a survival advantage to S. gordonii in the presence of ampicillin. Furthermore, this antibiotic protection in plaktonic cultures was also observed with other antibiotics such as erythromycin (Fig. 2C). We further show that dual species biofilms of C. albicans and S. gordonii are more resistant to ampicillin and erythromycin (Fig. 3). This is consistent and confirmatory with other studies (Montelongo-Jauregui et al., 2016).
Coaggregation between C. albicans and S. gordonii has been documented utilizing confocal microscopy and electron microscopy as well as light microscopy (Zijnge et al., 2010). As such coaggregation typically occurs with S. gordonii cells bound to hyphae of C. albicans in a biofilm environment (Zijnge et al., 2010). Our SEM experiments of early biofilms, using our unique culture media and conditions, confirm the binding of S. gordonii to C. albicans hyphae as well as yeast form cells (Fig. 4). In our analysis, the bacterial cells appear to be bound firmly, as if fused to the cell wall of C. albicans (Fig. 4). The induction of hyphae has been thought to increase the surface area and protein adhesins available for increased S. gordonii to C. albicans cell-to-cell binding (Bamford et al., 2015). This binding can play a role in biofilm formation and virulence (Bamford et al., 2009). Based on this information we hypothesized that C. albicans cell wall integrity had a role in the dual species biofilm formation with S. gordonii. To test this we screened knock-out mutants of 45 C. albicans transcription factors (Table 2), that regulate key functions like cell to cell adherence, hyphae formation, biofilm formation and cell membrane maintenance, for dual species biofilm formation with S. gordonii. Among the 45 transcription factors, we found that knock-outs of four factors - Leu3, Cta4, Cas5 and Sko1 had increased biofilm formation as compared to WT dual species biofilms, indicating that these factors negatively regulated dual species biofilms (Fig. 5). On the other hand, we found that knock-outs of six factors –Sfl2, Brg1, Tec1, Tup1, Efg1 and Rim101 had reduced dual species biofilm formation as compared to WT dual species biofilms, indicating that these factors positively regulated dual species biofilms (Fig. 5). Some of these transcription factor mutants are inherently affected in biofilm formation. Hence, we also measured biofilm formation of these mutant strains in mono-species biofilms (Fig. S4). Cas5 and Cta4 mutants had significantly increased while Sfl2, Tec1 and Rim101 had significantly decreased mono-species biofilm formation indicating that even the dual species biofilm formation could be affected by their inherent abnormalities in biofilm formation (Fig. S4). However, the remaining transcription factor mutants were unaffected in dual species biofilm formation, by their inherent abnormalities in mono-species biofilm formation (Fig. S4).
Table 2. List of C. albicans transcription factor knock-out mutants that were used in this study.
The mutant strains were sourced from the Hommann plates obtained from Fungal Genetics Stock Center (FGSC).
| Transcription Factors | Functions | |
|---|---|---|
| SFL2, GRF10, BRG1, OFI1, ZCF29, EFH1, BAS1, ZCF3, CSR1, ASH1, FGR17, TUP1, RCA1 | Involved in regulation of filamentous growth | |
| LEU3, CAS5, ZCF31, TRY4, ACE2, MRR2, TRY6, UGA33, SUC1, ZCF39, CZF1, BCR1 | Required for yeast cell adherence | |
| FGR27 | Involved in yeast cell adherence and filamentous growth | |
| TYE7, ZCF32, UGA32, CUP9, CTA4, ZAC7, OPI1, INO4, ROB1, GZF3, TEC1, SSN6, RIM101, CPH2, EFG1 | Required for biofilm formation, involved in regulating hyphal growth | |
| CWT1, SKO1 | Involved in cell wall architecture | |
| STP4 | Induced in core caspofungin response, colony morphology-related gene regulation by SSN6 | |
| CRZ1 | Role in maintenance of membrane integrity |
Negative regulators of dual species biofilm formation
Leu3 is a Zn(II)2Cys6 transcription factor which functions in branched chain aminoacid synthesis. Leu3 is induced by Mnl1 under weak acid stress conditions (Ramsdale et al., 2008). A Leu3 homozygous null mutant is viable (Finkel et al., 2012). Leu3 is required for positive regulation of cell adhesion in a monospecies biofilm. Additionally Leu3 is also required for cell adhesion to a silicone substrate (Finkel et al., 2012). Cta4 is a Zn(II)2Cys6 transcription factor which is induced under weak acid stress conditions (Bamford et al., 2015). It is repressed upon adherence to polystyrene (Finkel et al., 2012). Cta4 virulence is reduced in a mouse model (Chiranand et al., 2007). Cas5 is a zinc finger transcription factor that plays a critical role in cell wall integrity response. It is required for the induction of caspofungin responsive genes in C. albicans including ECM33 and CRH11 (Chiranand et al., 2007). Cas5 is also required for virulence in mouse and drosophila infection models (Chamilos et al., 2009). Sko1 is a bZip transcription factor that is involved in cell wall damage response (Rauceo et al., 2008). It is known to repress yeast to hyphal transformation and regulate oxidative stress response in C. albicans (Alonso-Monge et al., 2010). Regulation of oxidative and osmotic stress response by Sko1 occurs in a Hog1 dependent manner (Enjalbert et al., 2006).
Positive regulators of dual species biofilm formation
Sfl2 is a transcription factor involved in regulation of morphogenesis. Null mutant of Sfl2 has decreased hyphal growth and is required for infection in a reconstituted epithelial infection model (Spiering et al., 2009). Additionally, Sfl2 regulates transcriptional response dependent on sensing carbon dioxide levels (Tao et al., 2017). Brg1 is a transcription factor that plays a role in transcriptional response of hypha specific genes. Hence the null mutant has a decreased hyphal formation and virulence in mice (Cleary et al., 2012). Additionally, Brg1 null mutants have decreased biofilm formation in spider medium (Lin et al., 2013). The reduced biofilms of Brg1 mutants is observed for both conventional and pheromone-induced biofilms indicating its important role in cell cohesion (Lin et al., 2013). Tec1 is a TEA/ATTS transcription factor that is predominantly expressed in the hyphae. Tec1 deletion leads to reduced hypha formation in vitro, but normal hypha formation in vivo (Schweizer et al., 2000). Tec1 has been shown to regulate Bcr1 which is required for biofilm formation. Bcr1 induces several cell surface proteins and adhesins that are expressed during hyphal development (Nobile & Mitchell, 2005). Tup1 is a transcriptional repressor of filamentous growth. Tup1 null mutant grows only as filaments under various conditions. Tup1 null mutants produce 17-fold increased farnesol in biofilms (Nett Jeniel et al., 2009). Tup1 represses hypha specific genes and cell wall adhesins and is regulated by Tor1 (Bastidas, Heitman & Cardenas, 2009). Efg1 is a basic-helix-loop-helix (bHLH) transcription factor. It is a major transcriptional regulator that is involved in hyphal morphogenesis (Stoldt, 1997). Efg1 has a central role as a downstream component of protein kinase A (PKA) in the regulation of yeast to hypha transition (Bockmühl & Ernst, 2001). Rim101 is a zinc finger transcription factor that is involved in regulation of yeast to hypha transition in response to pH changes (Davis et al., 2000). Specifically, Rim101 induces alkaline responsive genes while repressing acid responsive genes (Baek, Martin & Davis, 2006). Additionally, null mutants of Rim101 are defective in virulence in a mouse model of disseminated candidiasis (Davis et al., 2000).
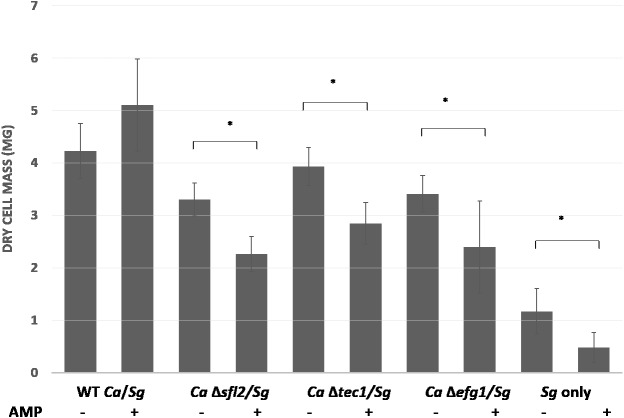
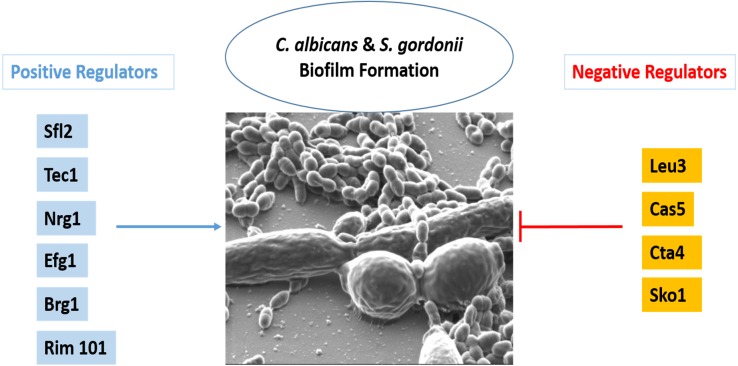
The C. albicans transcription factors which may negatively regulate dual species biofilm formation with S. gordonii are mainly involved in candida cell–cell adhesion, adhesion to a surface or cell wall integrity. It is reasonable that repression of candida cell to cell adhesion may be needed in order to promote cell adhesion with S. gordonii for a mixed biofilm formation. Thus the functions of transcription factors especially Leu3 and Cta4 might be important for adherence of C. albicans with S. gordonii. Cell wall integrity or cell wall damage response usually occurs in response to stress and is mediated by the Cek1 pathway in C. albicans (Román et al., 2015). Cell wall integrity is very important for cell survival and hence several transcription factors regulate this key function. Thus it is possible that although Cas5 and Sko1 play a role in cell wall integrity and damage response they might not be required for the biofilm formation with S. gordonii due to compensatory effect by other transcription factors. On the other hand, the transcription factors Sfl2, Brg1, Tec1, Tup1, Efg1 and Rim101 are mainly required for yeast to hyphal morphogenesis and thus also play critical roles in candida biofilm formation. A reduced biofilm formation of mutants of these transcription factors with S. gordonii indicates that hypha formation by C. albicans is critical for robust biofilm formation. Furthermore, we also screened the knock-out mutants of all 45 transcription factors in order to identify putative regulators of ampicillin resistance in dual species biofilms with S. gordonii. We observed that knock-out mutants of Sfl2, Efg1 and Tec1 demonstrated significantly decreased biofilm formation with S. gordonii in the presence of ampicillin as compared to control biofilms indicating a reduced antibiotic resistance (Fig. 6). This data indicates a role for these transcription factors in antibiotic resistance of dual species biofilms (Fig. 7).
Figure 6. C. albicans transcriptional regulators Sfl2, Tec1 and Efg1 affect ampicillin resistance in polymicrobial biofilms with S. gordonii.
Dry weights were measured for biofilms of C. albicans transcription factor Knock-out mutants with S. gordonii and compared with biofilms of WT C. albicans and S. gordonii in the presence or absence of ampicillin. Statistical analysis was done using Student’s t-Test (p < 0.05).
Figure 7. Transcriptional regulation of C. albicans and S. gordonii biofilms.
Please see Table 2 for functions related to the transcription factors in this figure.
Conclusions
Differential protein secretion occurs in dual species planktonic cultures of C. albicans and S. gordonii indicating a possible way of interaction between the two microorganisms. Additionally, antibiotic resistance to ampicillin and erythromycin was observed in both planktonic and biofilm cultures of C. albicans and S. gordonii. Binding between these oral microorganisms in early biofilms was confirmed using SEM analysis. Additionally, C. albicans transcription factors play a key role in dual species biofilm formation and ampicillin resistance of S. gordonii. This study can serve as a template for other studies investigating the relationship of C. albicans with bacteria in polymicrobial biofilms.
Supplemental Information
Biofilms were cultured for 24h in YNB with serum as described in methods. Biofilms were dried and weighed. WT (SC5314) strain was used as control.
Acknowledgments
We acknowledge David Beery for his work on the statistical analysis for the ampicillin resistance experiment. We acknowledge Dr. Peter Bush for assistance in scanning electron microscopy. We would like to thank Dr. Amarpreet Sabharwal for assisting in preparation of the manuscript for submission. We acknowledge Jason Chwirut for preparation of figures for the manuscript.
Funding Statement
This work was supported by State University of New York Start up funding. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
Abhiram Maddi is an Academic Editor for PeerJ.
Author Contributions
Jennifer Chinnici conceived and designed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, approved the final draft.
Lisa Yerke conceived and designed the experiments, contributed reagents/materials/analysis tools, prepared figures and/or tables, approved the final draft.
Charlene Tsou conceived and designed the experiments, contributed reagents/materials/analysis tools, authored or reviewed drafts of the paper, approved the final draft.
Sujay Busarajan conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, approved the final draft.
Ryan Mancuso conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, authored or reviewed drafts of the paper, approved the final draft.
Nishanth D. Sadhak conceived and designed the experiments, performed the experiments, analyzed the data, prepared figures and/or tables, approved the final draft.
Jaewon Kim conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, prepared figures and/or tables, authored or reviewed drafts of the paper, approved the final draft.
Abhiram Maddi conceived and designed the experiments, authored or reviewed drafts of the paper, approved the final draft, submitting the manuscript.
Data Availability
The following information was supplied regarding data availability:
Maddi, Abhiram (2019): Raw Data for PeerJ manuscript 38361. figshare. Dataset. https://doi.org/10.6084/m9.figshare.9862856.v1.
References
- Alonso-Monge et al. (2010).Alonso-Monge R, Román E, Arana DM, Prieto D, Urrialde V, Nombela C, Pla J. The Sko1 protein represses the yeast-to-hypha transition and regulates the oxidative stress response in Candida albicans. Fungal Genetics and Biology. 2010;47(7):587–601. doi: 10.1016/j.fgb.2010.03.009. [DOI] [PubMed] [Google Scholar]
- Azie et al. (2012).Azie N, Neofytos D, Pfaller M, Meier-Kriesche HU, Quan SP, Horn D. The PATH (Prospective Antifungal Therapy) Alliance® registry and invasive fungal infections: update 2012. Diagnostic Microbiology and Infectious Disease. 2012;73(4):293–300. doi: 10.1016/j.diagmicrobio.2012.06.012. [DOI] [PubMed] [Google Scholar]
- Baek, Martin & Davis (2006).Baek Y-U, Martin SJ, Davis DA. Evidence for novel pH-dependent regulation of Candida albicans Rim101, a direct transcriptional repressor of the cell wall β-Glycosidase Phr2. Eukaryotic Cell. 2006;5(9):1550–1559. doi: 10.1128/ec.00088-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford et al. (2009).Bamford CV, d’Mello A, Nobbs AH, Dutton LC, Vickerman MM, Jenkinson HF. Streptococcus gordonii modulates Candida albicans biofilm formation through intergeneric communication. Infection and Immunity. 2009;77(9):3696–3704. doi: 10.1128/iai.00438-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford et al. (2015).Bamford CV, Nobbs AH, Barbour ME, Lamont RJ, Jenkinson HF. Functional regions of Candida albicans hyphal cell wall protein Als3 that determine interaction with the oral bacterium Streptococcus gordonii. Microbiology. 2015;161(1):18–29. doi: 10.1099/mic.0.083378-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancescu et al. (2004).Bancescu G, Dumitriu S, Bancescu A, Defta C, Pana M, Ionescu D, Alecu S, Zamfirescu M. Susceptibility testing of Streptococcus mitis group isolates. The Indian Journal of Medical Research. 2004;119:257–261. [PubMed] [Google Scholar]
- Barrales et al. (2012).Barrales RR, Korber P, Jimenez J, Ibeas JI. Chromatin modulation at the FLO11 promoter of Saccharomyces cerevisiae by HDAC and Swi/Snf complexes. Genetics. 2012;191(3):791–803. doi: 10.1534/genetics.112.140301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastidas, Heitman & Cardenas (2009).Bastidas RJ, Heitman J, Cardenas ME. The protein kinase Tor1 regulates adhesin gene expression in Candida albicans. PLOS Pathogens. 2009;5(2):e1000294. doi: 10.1371/journal.ppat.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockmühl & Ernst (2001).Bockmühl DP, Ernst JF. A potential phosphorylation site for an A-type kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans. Genetics. 2001;157(4):1523–1530. doi: 10.1093/genetics/157.4.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone & Fonzi (2001).Calderone RA, Fonzi WA. Virulence factors of Candida albicans. Trends in Microbiology. 2001;9(7):327–335. doi: 10.1016/s0966-842x(01)02094-7. [DOI] [PubMed] [Google Scholar]
- Chaffin et al. (1998).Chaffin WL, López-Ribot JL, Casanova M, Gozalbo D, Martínez JP. Cell wall and secreted proteins of Candida albicans: identification, function, and expression. Microbiology and Molecular Biology Reviews. 1998;62(1):130–180. doi: 10.1128/mmbr.62.1.130-180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamilos et al. (2009).Chamilos G, Nobile Clarissa J, Bruno Vincent M, Lewis Russell E, Mitchell Aaron P, Kontoyiannis Dimitrios P. Candida albicans Cas5, a regulator of cell wall integrity, is required for virulence in murine and toll mutant fly models. The Journal of Infectious Diseases. 2009;200(1):152–157. doi: 10.1086/599363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiranand et al. (2007).Chiranand W, McLeod I, Zhou H, Lynn JJ, Vega LA, Myers H, Yates 3rd JR, Lorenz MC, Gustin MC. CTA4 Transcription factor mediates induction of nitrosative stress response in Candida albicans. Eukaryotic Cell. 2007;7(2):268–278. doi: 10.1128/ec.00240-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary et al. (2012).Cleary IA, Lazzell AL, Monteagudo C, Thomas DP, Saville SP. BRG1 and NRG1 form a novel feedback circuit regulating Candida albicans hypha formation and virulence. Molecular Microbiology. 2012;85(3):557–573. doi: 10.1111/j.1365-2958.2012.08127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis et al. (2000).Davis D, Edwards JE, Mitchell AP, Ibrahim AS. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infection and Immunity. 2000;68(10):5953–5959. doi: 10.1128/iai.68.10.5953-5959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz et al. (2012).Diaz PI, Dupuy AK, Abusleme L, Reese B, Obergfell C, Choquette L, Dongari-Bagtzoglou A, Peterson DE, Terzi E, Strausbaugh LD. Using high throughput sequencing to explore the biodiversity in oral bacterial communities. Molecular Oral Microbiology. 2012;27(3):182–201. doi: 10.1111/j.2041-1014.2012.00642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlan & Costerton (2002).Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clinical Microbiology Reviews. 2002;15(2):167–193. doi: 10.1128/cmr.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton et al. (2014).Dutton LC, Nobbs AH, Jepson K, Jepson MA, Vickerman MM, Aqeel Alawfi S, Munro CA, Lamont RJ, Jenkinson HF. O-Mannosylation in Candida albicans enables development of interkingdom biofilm communities. mBio. 2014;5(2):e00911–e009114. doi: 10.1128/mbio.00911-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton et al. (2015).Dutton LC, Paszkiewicz KH, Silverman RJ, Splatt PR, Shaw S, Nobbs AH, Lamont RJ, Jenkinson HF, Ramsdale M. Transcriptional landscape of trans-kingdom communication between Candida albicans and Streptococcus gordonii. Molecular Oral Microbiology. 2015;31(2):136–161. doi: 10.1111/omi.12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enjalbert et al. (2006).Enjalbert B, Smith DA, Cornell MJ, Alam I, Nicholls S, Brown AJ, Quinn J. Role of the Hog1 stress-activated protein kinase in the global transcriptional response to stress in the fungal pathogen Candida albicans. Molecular Biology of the Cell. 2006;17(2):1018–1032. doi: 10.1091/mbc.e05-06-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne, Gruer & Fleurette (1984).Etienne J, Gruer LD, Fleurette J. Antibiotic susceptibility of streptococcal strains associated with infective endocarditis. European Heart Journal. 1984;5(Suppl C):33–37. doi: 10.1093/eurheartj/5.suppl_c.33. [DOI] [PubMed] [Google Scholar]
- Finkel et al. (2012).Finkel JS, Xu W, Huang D, Hill EM, Desai JV, Woolford CA, Nett JE, Taff H, Norice CT, Andes DR, Lanni F, Mitchell AP. Portrait of Candida albicans adherence regulators. PLOS Pathogens. 2012;8(2):e1002525. doi: 10.1371/journal.ppat.1002525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, Gopal & Jenkinson (1995).Holmes AR, Gopal PK, Jenkinson HF. Adherence of Candida albicans to a cell surface polysaccharide receptor on Streptococcus gordonii. Infection and Immunity. 1995;63(5):1827–1834. doi: 10.1128/iai.63.5.1827-1834.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, McNAB & Jenkinson (1996).Holmes AR, McNAB R, Jenkinson HF. Candida albicans binding to the oral bacterium Streptococcus gordonii involves multiple adhesin-receptor interactions. Infection and Immunity. 1996;64(11):4680–4685. doi: 10.1128/iai.64.11.4680-4685.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann et al. (2009).Homann OR, Dea J, Noble SM, Johnson AD. A phenotypic profile of the Candida albicans regulatory network. PLOS Genetics. 2009;5(12):e1000783. doi: 10.1371/journal.pgen.1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornby et al. (2001).Hornby JM, Jensen EC, Lisec AD, Tasto JJ, Jahnke B, Shoemaker R, Dussault P, Nickerson KW. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Applied and Environmental Microbiology. 2001;67(7):2982–2992. doi: 10.1128/aem.67.7.2982-2992.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson, Lala & Shepherd (1990).Jenkinson H, Lala H, Shepherd M. Coaggregation of Streptococcus sanguis and other streptococci with Candida albicans. Infection and Immunity. 1990;58(5):1429–1436. doi: 10.1128/iai.58.5.1429-1436.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim & Sudbery (2011).Kim J, Sudbery P. Candida albicans, a major human fungal pathogen. The Journal of Microbiology. 2011;49(2):171–177. doi: 10.1007/s12275-011-1064-7. [DOI] [PubMed] [Google Scholar]
- Kuramitsu et al. (2007).Kuramitsu HK, He X, Lux R, Anderson MH, Shi W. Interspecies interactions within oral microbial communities. Microbiology and Molecular Biology Reviews. 2007;71(4):653–670. doi: 10.1128/mmbr.00024-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin et al. (2013).Lin CH, Kabrawala S, Fox EP, Nobile CJ, Johnson AD, Bennett RJ. Genetic control of conventional and pheromone-stimulated biofilm formation in Candida albicans. PLOS Pathogens. 2013;9(4):e1003305. doi: 10.1371/journal.ppat.1003305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddi, Bowman & Free (2009).Maddi A, Bowman SM, Free SJ. Trifluoromethanesulfonic acid–based proteomic analysis of cell wall and secreted proteins of the ascomycetous fungi Neurospora crassa and Candida albicans. Fungal Genetics and Biology. 2009;46(10):768–781. doi: 10.1016/j.fgb.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso et al. (2018).Mancuso R, Chinnici, Tsou C, Busarajan S, Maddi A. Candida albicans cell wall glycosidases Dfg5 and Dcw1 are required for biofilm formation and Hog-1 signaling. PeerJ Preprints. 2018:e26526v1. doi: 10.7717/peerj.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza et al. (1999).Mendoza A, Serramıá Ma J, Capa L, Garcıá-Bustos JF. Translation elongation factor 2 is encoded by a single essential gene in Candida albicans. Gene. 1999;229(1–2):183–191. doi: 10.1016/s0378-1119(99)00024-4. [DOI] [PubMed] [Google Scholar]
- Montelongo-Jauregui et al. (2016).Montelongo-Jauregui D, Srinivasan A, Ramasubramanian AK, Lopez-Ribot JL. An in vitro model for oral mixed biofilms of Candida albicans and Streptococcus gordonii in synthetic saliva. Frontiers in Microbiology. 2016;7:686. doi: 10.3389/fmicb.2016.00686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzyka & Epifanio (2013).Muzyka BC, Epifanio RN. Update on oral fungal infections. Dental Clinics. 2013;57(4):561–581. doi: 10.1016/j.cden.2013.07.002. [DOI] [PubMed] [Google Scholar]
- Myers, Fonzi & Sypherd (1992).Myers KK, Fonzi WA, Sypherd PS. Isolation and sequence analysis of the gene for translation elongation factor 3 from Candida albicans. Nucleic Acids Research. 1992;20(7):1705–1710. doi: 10.1093/nar/20.7.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto et al. (2013).Nemoto H, Nomura R, Ooshima T, Nakano K. Distribution of amoxicillin-resistant oral streptococci in dental plaque specimens obtained from Japanese children and adolescents at risk for infective endocarditis. Journal of Cardiology. 2013;62(5):296–300. doi: 10.1016/j.jjcc.2013.05.008. [DOI] [PubMed] [Google Scholar]
- Nett Jeniel et al. (2009).Nett Jeniel E, Lepak Alexander J, Marchillo K, Andes David R. Time course global gene expression analysis of an in vivo candida biofilm. The Journal of Infectious Diseases. 2009;200(2):307–313. doi: 10.1086/599838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs, Vickerman & Jenkinson (2010).Nobbs AH, Vickerman MM, Jenkinson HF. Heterologous expression of Candida albicans cell wall-associated adhesins in Saccharomyces cerevisiae reveals differential specificities in adherence and biofilm formation and in binding oral Streptococcus gordonii. Eukaryotic Cell. 2010;9(10):1622–1634. doi: 10.1128/ec.00103-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile & Mitchell (2005).Nobile CJ, Mitchell AP. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Current Biology. 2005;15(12):1150–1155. doi: 10.1016/j.cub.2005.05.047. [DOI] [PubMed] [Google Scholar]
- Nombela, Gil & Chaffin (2006).Nombela C, Gil C, Chaffin WL. Non-conventional protein secretionin yeast. Trends in Microbiology. 2006;14(1):15–21. doi: 10.1016/j.tim.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Odds (1987).Odds FC. Candida infections: an overview. CRC Critical Reviews in Microbiology. 1987;15(1):1–5. doi: 10.3109/10408418709104444. [DOI] [PubMed] [Google Scholar]
- Pfaller & Diekema (2007).Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clinical Microbiology Reviews. 2007;20(1):133–163. doi: 10.1128/cmr.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce et al. (2017).Pierce CG, Vila T, Romo JA, Montelongo-Jauregui D, Wall G, Ramasubramanian A, Lopez-Ribot JL. The Candida albicans biofilm matrix: composition, structure and function. Journal of Fungi. 2017;3(1):14. doi: 10.3390/jof3010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsdale et al. (2008).Ramsdale M, Selway L, Stead D, Walker J, Yin Z, Nicholls SM, Crowe J, Sheils EM, Brown AJ. MNL1 regulates weak acid–induced stress responses of the fungal pathogen Candida albicans. Molecular Biology of the Cell. 2008;19(10):4393–4403. doi: 10.1091/mbc.e07-09-0946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan & Dongari-Bagtzoglou (2018).Ranjan A, Dongari-Bagtzoglou A. Tipping the balance: C. albicans adaptation in polymicrobial environments. Journal of Fungi. 2018;4(3):112. doi: 10.3390/jof4030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauceo et al. (2008).Rauceo JM, Blankenship JR, Fanning S, Hamaker JJ, Deneault JS, Smith FJ, Nantel A, Mitchell AP. Regulation of the Candida albicans cell wall damage response by transcription factor Sko1 and PAS kinase Psk1. Molecular Biology of the Cell. 2008;19(7):2741–2751. doi: 10.1091/mbc.e08-02-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Román et al. (2015).Román E, Alonso-Monge R, Miranda A, Pla J. The Mkk2 MAPKK regulates cell wall biogenesis in cooperation with the Cek1-Pathway in Candida albicans. PLOS ONE. 2015;10(7):e0133476. doi: 10.1371/journal.pone.0133476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Smith et al. (1995).Ross-Smith N, Tan P, Belfield G, Tuite MF. Translational elongation factor 3 (Ef-3): a study of its structural and functional divergence in fungi. Biochemical Society Transactions. 1995;23(1):132. doi: 10.1042/bst023132s. [DOI] [PubMed] [Google Scholar]
- Scheres & Krom (2016).Scheres N, Krom BP. Methods in molecular biology. Springer; New York: 2016. Staphylococcus–Candida interaction models: antibiotic resistance testing and host interactions; pp. 153–161. [DOI] [PubMed] [Google Scholar]
- Schweizer et al. (2000).Schweizer A, Rupp S, Taylor BN, Röllinghoff M, Schröppel K. The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Molecular Microbiology. 2000;38(3):435–445. doi: 10.1046/j.1365-2958.2000.02132.x. [DOI] [PubMed] [Google Scholar]
- Spiering et al. (2009).Spiering MJ, Moran GP, Chauvel M, Maccallum DM, Higgins J, Hokamp K, Yeomans T, d’Enfert C, Coleman DC, Sullivan DJ. Comparative transcript profiling of Candida albicans and Candida dubliniensis Identifies SFL2, a C. albicans gene required for virulence in a reconstituted epithelial infection model. Eukaryotic Cell. 2009;9(2):251–265. doi: 10.1128/ec.00291-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoldt (1997).Stoldt VR. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. The EMBO Journal. 1997;16(8):1982–1991. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed & Loesche (1978).Syed SA, Loesche WJ. Bacteriology of human experimental gingivitis: effect of plaque age. Infection and Immunity. 1978;21(3):821–829. doi: 10.1128/iai.21.3.821-829.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao et al. (2017).Tao L, Zhang Y, Fan S, Nobile CJ, Guan G, Huang G. Integration of the tricarboxylic acid (TCA) cycle with cAMP signaling and Sfl2 pathways in the regulation of CO2 sensing and hyphal development in Candida albicans. PLOS Genetics. 2017;13(8):e1006949. doi: 10.1371/journal.pgen.1006949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, Bachmann & Lopez-Ribot (2006).Thomas DP, Bachmann SP, Lopez-Ribot JL. Proteomics for the analysis of the Candida albicans biofilm lifestyle. Proteomics. 2006;6(21):5795–5804. doi: 10.1002/pmic.200600332. [DOI] [PubMed] [Google Scholar]
- Whitmore & Lamont (2011).Whitmore SE, Lamont RJ. The pathogenic persona of community-associated oral streptococci. Molecular Microbiology. 2011;81(2):305–314. doi: 10.1111/j.1365-2958.2011.07707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Jenkinson & Dongari-Bagtzoglou (2014).Xu H, Jenkinson HF, Dongari-Bagtzoglou A. Innocent until proven guilty: mechanisms and roles of Streptococcus-Candida interactions in oral health and disease. Molecular Oral Microbiology. 2014;29(3):99–116. doi: 10.1111/omi.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada et al. (2008).Yamada K, Hirota K, Mizuno K-I, Shibata T, Ohta K. Essential roles of Snf21, a Swi2/Snf2 family chromatin remodeler, in fission yeast mitosis. Genes & Genetic Systems. 2008;83(5):361–372. doi: 10.1266/ggs.83.361. [DOI] [PubMed] [Google Scholar]
- Zarnowski, Sanchez & Andes (2016).Zarnowski R, Sanchez H, Andes DR. Large-scale production and isolation of Candida biofilm extracellular matrix. Nature Protocols. 2016;11(12):2320–2327. doi: 10.1038/nprot.2016.132. [DOI] [PubMed] [Google Scholar]
- Zijnge et al. (2010).Zijnge V, Van Leeuwen MBM, Degener JE, Abbas F, Thurnheer T, Gmür R, Harmsen HJ. Oral biofilm architecture on natural teeth. PLOS ONE. 2010;5(2):e9321. doi: 10.1371/journal.pone.0009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Biofilms were cultured for 24h in YNB with serum as described in methods. Biofilms were dried and weighed. WT (SC5314) strain was used as control.
Data Availability Statement
The following information was supplied regarding data availability:
Maddi, Abhiram (2019): Raw Data for PeerJ manuscript 38361. figshare. Dataset. https://doi.org/10.6084/m9.figshare.9862856.v1.