Abstract
A common genetic variant in the gene encoding the protein tyrosine phosphatase nonreceptor type 22 (PTPN22 C1858T) has been linked to a wide range of autoimmune disorders. Although a B cell-intrinsic role in promoting disease has been reported, the mechanism(s) through which this variant functions to alter the pre-immune B cell repertoire remains unknown. Using a series of polyclonal and transgenic self-reactive models harboring the analogous mutation in murine Ptpn22, we show evidence for enhanced BCR, BAFFR and CD40 co-receptor programs, leading to broadly enhanced positive selection of B cells at two discrete checkpoints in the bone marrow and spleen. We further identified a bias for selection of B cells into the follicular (FM) vs. marginal zone (MZ) B cell compartment. Using a biomarker to track a self-reactive heavy chain in peripheral blood, we found evidence of similarly enhanced positive selection in human carriers of the PTPN22 C1858T variant. Our combined data supports a model whereby the risk variant augments the BCR and co-receptor programs throughout B cell development promoting enrichment of self-reactive specificities into the FM compartment, thereby likely increasing the risk for seeding of autoimmune B cell responses.
INTRODUCTION
The protein tyrosine phosphatase nonreceptor type 22 gene (PTPN22) encodes for the phosphatase, LYP, (or PEP in mice) which functions as a negative regulator of antigen receptor (AgR) signaling through its direct modulation of Src-family kinases (1). A genetic variant in PTPN22 (C1858T; encoding LYP-R620W) is a major risk factor for a number of autoimmune disorders including type 1 diabetes (T1D), systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), Graves’ disease, and others (2-6). To model this variant in vivo, we previously generated knock-in mice with the analogous risk allele on a mixed 129/Sv and C57BL/6J background. Expression of the Ptpn22 variant significantly altered lymphocyte function and led to the development of systemic autoimmunity (7).
Although the PTPN22 risk variant promotes disease via its impact on multiple cell lineages, B cells appear to be particularly important for this process (7,8). Notably, the disorders associated with PTPN22 risk variant are characterized by high titers of disease-specific pathogenic autoantibodies (9). While autoantibodies may result from B and/or T cell-driven processes, our group found that B cell-intrinsic Ptpn22 variant expression was sufficient to promote autoimmunity (7). The conclusion that altered B cell tolerance may potentiate similar risks in human subjects arose from the observation that transitional B cells were increased in both human and murine carriers of the risk variant (7,10). Lending further support to this idea, increased proportions of self-reactive B cells were identified at two checkpoints during human B cell development based on analyses of cells isolated from the peripheral blood of healthy subjects with the risk allele (11).
Taken together, these data suggest that the PTPN22 variant plays an important role in shaping the pre-immune B cell repertoire in at-risk individuals and in murine models; however, several key questions remain that warrant further study. First, one major unresolved issue is whether the variant confers a gain- vs. loss-, or alternatively an altered-, functional activity. Indeed, a range of contradictory findings with respect to the impact of the variant on AgR signals have been observed in human and murine studies (reviewed in (12)). The studies to date have relied upon in vitro stimulated cells, thus direct ex vivo analysis of AgR signaling is needed. Secondly, other than the BCR signaling pathway, it is unclear whether additional networks are impacted by PTPN22 variant. Of particular relevance are the BAFFR and CD40 co-receptor pathways, given their importance in regulating B cell tolerance and known crosstalk with the BCR signaling program (13-16). Lastly, a more complete understanding of how the PTPN22 variant shapes the specificities selected into the mature, naïve B cell compartments might help to predict the risk for subsequent aberrant activation of such cells in autoimmune individuals.
In the current study, we use a series of murine models, in association with a rigorous assessment of the naïve repertoire, to track the development and selection of B cells expressing the Ptpn22 risk variant. Murine studies included mice homozygous for the non-risk allele (Ptpn22CC) and heterozygous (Ptpn22CT) or homozygous (Ptpn22TT) risk allele animals intercrossed with various selection models. To reduce potential impacts from additional genetic modifiers, we used Ptpn22 variant and controls backcrossed onto the non-autoimmune C57BL/6J background. In parallel, a flow-based assay tracking a self-reactive heavy chain (HC) was used to monitor peripheral B cell selection in human carriers with the variant. Our combined results suggest the Ptpn22 variant augments the coordinate BCR, BAFFR, and CD40 programs throughout B cell development, leading to altered tolerance at discrete checkpoints in the bone marrow and periphery. These events promoted enhanced positive selection of transitional B cells, with an unexpected bias for self-reactive specificities into the FM compartment. Healthy human subjects expressing the risk variant exhibited a reduced proportion of transitional B cells utilizing a specific, self-reactive heavy-chain family, findings most consistent with broadly enhanced positive selection for developing B cells with a range of self-reactive specificities. Our collective data add to the understanding of B cell-mediated autoimmunity, suggesting that allelic variants that enhance the BCR and/or key co-receptor pathways preferentially skew self-reactive B cells into the follicular B cell compartment, thereby increasing the probability of subsequent events that trigger autoimmune germinal center responses.
MATERIALS AND METHODS
Mice
Ptpn22CC (Ly 5.1 and Ly 5.2 lines), Ptpn22TT, Nur77-GFP Tg, BAFFR−/−, CD40−/−, CD40L−/−, MD4 Tg, mHEL, sHEL, μMT, and 125 Tg (VH125 and VK125 lines) mice were maintained in the specific pathogen-free animal facility of Seattle Children’s Research Institute and handled according to Institutional Animal Care and Use Committee-approved protocols. Ptpn22TT knock in mice were generated as previously described (7) and backcrossed to C57BL/6J for 10 generations before crossing to Nur77-GFP Tg, MD4, 125 Tg (VH125) or 125 Tg (VK125). The experimental mice contained 1 copy of Nur77-GFP, 1 copy of MD4, or 1 copy each of VH125 and VK125 transgenes.
High-throughput BCR sequencing
Murine FM and MZ B cell populations were bulk sorted and genomic DNA was extracted for survey-depth sequencing of the IgH locus (Adaptive Biotechnologies, Seattle WA). Adaptive Biotechnologies’ ImmunoSEQ Illumina-based sequencing platform was used to identify productive templates for assignment of IgH V and J genes and to determine CDR3 boundaries (defined as including the first base of the codon for the conserved cysteine in the V gene through the last base of the codon for the conserved residue in the J gene). Average hydrophobicity scores (GRAVY) were calculated using http://www.gravy-calculator.de/. Diversity scores (reciprocal Simpson Index) were calculated as described (17). The total number of productive templates generated can be found in Supplemental Table I. Data are representative of two independent experiments.
Human subjects and sample preparation
Frozen PBMCs from age and sex-matched healthy subjects screened for PTPN22 1858 were obtained from the Benaroya Research Institute Immune Mediated Disease Registry. Subjects included PTPN22 C/C (n=35), C/T (n=35) and T/T (n=3). For flow cytometry, single-cell PBMC suspensions were incubated with fluorescently labeled Abs (see below) for 20 min at 4°C in staining buffer, and data collected on an LSRII (BD) and analyzed using FlowJo software (Tree Star). Subject information can be found in Supplemental Table II. Data are representative of three independent experiments.
Reagents and Abs
Anti-murine Abs used in these studies include: B220 (RA3-6B2), CD24 (M1/69), CD21 (7E9), CD23 (B3B4), CD4 (RM4-4) and CD44 (IM7) from Biolegend; IgMa (DS-1) and CD40 (3/23) from BD; Ly 5.1 (A20), Ly 5.2 (104), Gr1 (RB6-8C5), CD11b (M1/70), CD3 (17A2), BAFFR (eBio7H22-E16), CD93 (AA4.1), CD62L (MEL-14) and CD40L (MR1) from eBiosciences; IgM (1B4B1) and IgD (11-26) from Southern Biotech; recombinant human insulin conjugated to biotin from Fitzgerald Industries; Streptavadin (S-868) from Life Technologies. Anti-human Abs used include: CD19 (HIB19) and IgM (MHM-88) from Biolegend; CD27 (O323) from eBiosciences; CD10 (HI10a), CD24 (ML5), CD38 (HIT2), IgD (IA6-2), and BAFFR (IIC1) from BD; anti–human FITC-conjugated 9G4 antibody (18).
Flow cytometry and cell sorting
Murine single-cell BM and splenocyte suspensions were incubated with fluorescently labeled Abs for 30 min at 4°C in staining buffer, and data collected on an LSRII (BD) and analyzed using FlowJo software (Tree Star). Cell sorting was performed using an Aria II (BD); sort purities were >90% in all experiments.
Single-cell BCR cloning
Murine single FM and MZ cells were FACS sorted into 96-well plates. BCRs were cloned from the cDNA of single cells and used to generate mAbs using methods previously described (16,19). Data are representative of at least two independent experiments.
ELISA tests
All monoclonal antibody ELISAs were performed with each mAb first normalized to a standard dilution ranging from 10ng/ml to 10ug/ml. Individual FM and MZ mAbs were tested for reactivity with insulin (Fitzgerald Industries International), MDA-LDL (20P-MD L-105; Academy Bio-Medical), dsDNA (Sigma-Aldrich), phosphorylcholine (PC)-12 (Sigma-Aldrich) and smRNP (ATR01-10; Arotech Diagnostics Limited) as previously described (18). Abs were considered reactive if the observed OD at the highest mAb concentration (10ug/ml) was greater than a threshold value set at 0.5 OD. Anti-HEL Abs in MD4 chimeras were measured by incubating serum on HEL pre-coated plates, followed by detection with anti-IgMa conjugated to biotin (DS-1) and streptavidin-HRP. Serum BAFF levels were measured using BAFF/BlyS Quantikine ELISA kit (R&D Systems). Data are representative of two independent experiments.
Competitive Chimera BM transplantations
BM was harvested from donor Ptpn22CC (Ly 5.1), Ptpn22TT (Ly 5.2) and μMT (Ly 5.1/Ly 5.2) mice and single-cell suspensions were mixed at a 10:10:80 ratio for retro-orbital injection of 5 × 106 cells into lethally irradiated (900 cGy) Ptpn22CC (Ly 5.1/Ly 5.2) recipients. Resulting BM chimeras were sacrificed 12-14 wks post-transplantation. Data are representative of five independent experiments.
HEL BM transplantations
BM was harvested from donor Ptpn22CC MD4 or Ptpn22TT MD4 mice and made into single-cell suspensions, where 5 × 106 cells were retro-orbitally injected into lethally irradiated (900 cGy) mHEL or sHEL recipients. Resulting BM chimeras were sacrificed 8-12 wks post-transplantation. Data are representative of two independent experiments.
Quantitative PCR
RNA was isolated from sorted cells (purity >90% for all samples) using the AllPrep DNA/RNA Micro Kit (QIAGEN) and converted into cDNA by reverse transcription (Maxima Reverse Transcriptase; Thermo Scientific). Real-time PCR was performed using a CFX96 real-time PCR detection system (Bio-Rad) with iTaq universal SYBR Green supermix (Bio-Rad). Ratios were calculated using the comparative CT method with β2-microglobulin as an endogenous control. Primers used were as follows: β2M FP, 5’-CTTCAGTCGTCAGCATGGCTCG-3’; β2M RP, 5’-GCAGTTCAGTATGTTCGG CTTCCC-3’; BAFFR FP, 5’-CT GAGGCTGCAGAGCTGTC-3’; BAFFR RP, 5’-GGTGAGAAACTGCGTGTCCT-3’; CD40 FP, 5’-CTGCATGGTGTCTTTGCCT-3’; CD40 RP, 5’-GCCATCGTGGAGGTACTGTT-3’; PIM2 FP, 5’-CTTTCGAGGCC GATAACCGA-3’; PIM2 RP, 5’-GATGGCCACCTGACGTCTAT-3’; A1 FP, 5’-CCTGGCTGAGCACTACCTTCA-3’; A1 RP, 5’-CTGCATGCTTGGCTTGGA-3’; Notch2 FP, 5’-TTCGTGTCCCCCAGGCACCC-3’; Notch2 RP, 5’-AATCCGGT CCACGCACTGGC-3’; Deltex1 FP, 5’-CGGACATTTGAGACCCACTT-3’; Deltex1 RP, 5’-CCACTTTCAAGGAGGGAGAA-3’; Hes1 FP, 5’-GAGAAGAGGCGAA GGGCAAGAAT-3’; Hes1 RP, 5’-GAGGTGACTTCACAGTCA-3’.
In vitro stimulations
To allow for receptor internalization, total splenocytes were cultured in RPMI media alone or stimulated with 0.1μg/ml recombinant murine BAFF (R&D) or 0.1μg/ml mouse sCD40L (Southern-Biotech) at 37°C for 5-60min in pre-warmed media. To prevent receptor internalization and thus assess the potential impact of ligand-mediated competition with BAFFR or CD40 antibody staining, similar stimulations were performed on ice for a similar timecourse in pre-chilled media in the presence of sodium azide (0.2%). All cells were immediately washed post-stimulation with ice-cold PBS in a pre-chilled centrifuge and incubated with fluorescently labeled B220, BAFFR, and CD40 Abs for 30 min on ice in staining buffer. After washing, cells were immediately fixed in 2% PFA with data collected on an LSRII (BD) and analyzed using FlowJo software (Tree Star). Data are representative of two independent experiments.
Statistical analysis
The p-values were calculated using the two-tailed Student t test or Paired t test where appropriate (GraphPad software). Differences were considered significant when p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), and p < 0.0001 (****).
RESULTS
Ptpn22 variant increases BCR and co-receptor signaling programs in developing B cells
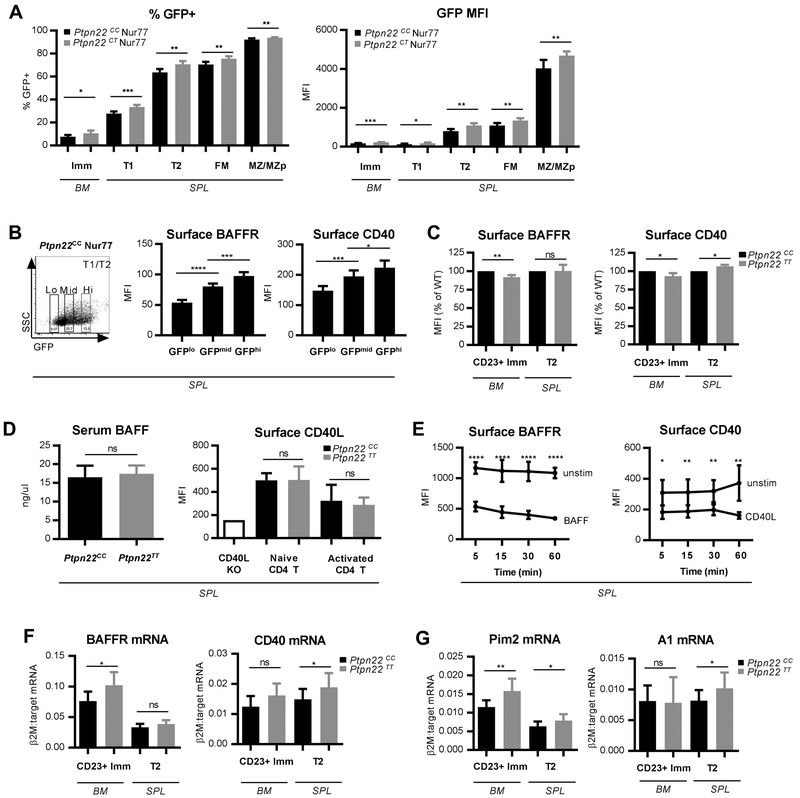
Previous in vitro stimulation studies revealed evidence for subtly enhanced BCR signaling in bulk splenic B cells isolated from Ptpn22 variant mice (7). This change in signaling, however, was only evident in cells pre-stimulated with a TLR ligand and correlated with an upregulation in PEP expression. In order to assess the potential impact of BCR signaling within unmanipulated B cells in vivo, we utilized the Nur77-GFP Tg reporter strain. In this model, endogenous BCR signals activate a wide spectrum of GFP expression under control of the Nur77 regulatory region, consistent with self-antigen-mediated in vivo BCR signaling (20). For these studies, we assessed Ptpn22 variant mice with a single copy of the variant crossed to the Nur77-GFP Tg model (Ptpn22CT Nur77 Tg), as we were unable to generate pups with two transgene copies (likely due to co-inheritance with the randomly inserted Nur77 transgene). Compared to control animals, B cells from Ptpn22CT Nur77 Tg mice exhibited a higher frequency of GFP+ cells as well as a higher GFP MFI beginning at the immature stage in the bone marrow and continuing in all transitional and naïve B cell subsets in the periphery (Fig. 1A and Supplemental Fig. 1A). These findings suggest that the Ptpn22 variant promotes a greater proportion of self-reactive B cells to survive tolerance mechanisms and enter the periphery, consistent with an enhanced BCR signaling program mediating these events.
FIGURE 1. Ptpn22 variant B cells exhibit enhanced antigen-mediated BCR, BAFFR and CD40 programs.
(A) The Nur77 GFP Tg model was used to assess in vivo antigen-mediated BCR signaling. Frequency of GFP+ cells (left) and mean fluorescent intensity (MFI) of GFP (right) in specific B cell subsets in 12wk Ptpn22CC Nur77 (n=6) and Ptpn22CT Nur77 (n=6) mice. See Supplemental Fig. 1A for representative GFP histograms and Supplemental Fig. 2A-B for details of B cell subset gating. (B) Representative gating of GFPlo, GFPmid and GFPhi (left panel); and MFI of BAFFR (middle panel) and CD40 (right panel) on T1/T2 cells (gated B220+ CD24hi CD21lo-mid) across varying GFP intensities in 15wk Ptpn22CC Nur77 (n=6) mice. (C) MFI of surface BAFFR (left panel) and CD40 (right panel) in bone marrow CD23+ immature and splenic T2 B cells in 10-12wk Ptpn22CC (n=9) and Ptpn22TT (n=9) mice. See Supplemental Fig. 2B-C for gating. (D) Serum BAFF levels (left panel) in 8-12wk Ptpn22CC (n=10) and Ptpn22TT (n=11) mice; surface CD40L MFI (right panel) of CD4+ naïve (CD44− CD62L+) and activated (CD44+ CD62L−) T cell subsets in 8-11wk Ptpn22CC (n=12) and Ptpn22TT (n=12) mice. CD40L knockout mouse used as a negative staining control. (E) Splenocytes from 10wk Ptpn22CC (n=6) mice were stimulated with either soluble BAFF (0.1 ug/ml) or CD40L (0.1 ug/ml) for the indicated times at 37 degrees and MFI of surface BAFFR (left panel) and CD40 (right panel) of B220+ cells were analyzed by FACs. Similar stimulations were performed at 4 degrees in the presence of sodium azide (0.2%) to prevent receptor internalization and test for possible competition between ligand and staining antibodies (Supplemental Fig. 1C). (F and G) Quantitative PCR of CD23+ immature and T2 cells sorted from 8-10wk Ptpn22CC (n=8 samples for BM, 11 samples for SPL) and Ptpn22TT (n=6 samples for BM, 11 samples for SPL) mice; mRNA levels of BAFFR (F, left panel), CD40 (F, right panel), Pim2 (G, left panel), and A1 (G, right panel) relative to β2-microglobulin. See Supplemental Fig. 2B-C for gating. All data are representative of at least two independent experiments. Error bars show SD. Statistical analysis was performed using Student’s t test: * p<0.05; ** p<0.01; *** p<0.001; **** p<0.0001.
Recent studies suggest that BCR signals coordinate with both BAFF family receptors and CD40 to shape the mature, naive B cell repertoire (21). BCR signaling promotes the BAFFR program through modulation of receptor transcript levels (22), as well as providing p100 substrate for induction of the alternative NFκB pathway (23). We therefore determined whether BCR signaling in vivo correlated with surface expression levels of BAFFR and CD40. Using the Nur77-GFP Tg model, we found that higher GFP expression in transitional cells indeed correlates with higher expression of both BAFFR and CD40 (Fig. 1B), suggesting a direct and/or indirect role for antigen-mediated BCR signaling in regulating sensitivity to these co-receptor signals.
We also compared BAFFR and CD40 surface levels across bone marrow and splenic subsets in control Ptpn22CC mice in order to determine the developmental stages most likely to be impacted by co-receptor signals. Prior reports have identified increased levels of BAFFR on the CD23+ subset of immature BM B cells, a population comprising ~20% of all immature IgM+ cells (gated as B220+ IgM+ AA4.1+ CD23+) (21,23-26). Therefore, we adopted a similar gating strategy in the BM. As shown in Supplemental Fig. 1B, CD23+ immature BM and splenic late transitional (T2) B cells (gated as B220+ CD24hi CD21mid) expressed higher levels of BAFFR and CD40 relative to their earlier developmental counterparts. Thus, we focused our subsequent analysis of co-receptor studies on these specific subsets in control and risk variant mice.
Based on the increased BCR signal observed in Ptpn22CT Nur77 Tg mice (Fig. 1A) and correlation of BCR signal strength with co-receptor expression (Fig. 1B), we hypothesized that Ptpn22TT animals may exhibit enhanced BAFFR and/or CD40 signaling in CD23+ immature and T2 cells. Consistent with this prediction, T2 cells had greater surface CD40 levels in Ptpn22TT mice (Fig. 1C). In contrast, surface levels of both BAFFR and CD40 were reduced in Ptpn22TT immature CD23+ B cells compared to controls (Fig. 1C). Because serum BAFF and surface CD40L levels on T cell subsets did not differ between control and Ptpn22TT animals (Fig. 1D), reduced receptor levels could not be explained by reduced ligand availability. Instead, a potential explanation for the paradoxical reduction in co-receptor surface levels on Ptpn22TT CD23+ immature B cells was enhanced receptor internalization. Consistent with this possibility, we first demonstrated that stimulation of control splenic B cells with either BAFF or soluble CD40L is sufficient to reduce surface expression levels of BAFFR or CD40, respectively (Fig. 1E; Supplemental Fig. 1C). Similar to our findings, BAFFR levels were reported to decline following ex vivo BAFF stimulation of human B cells (27). Therefore, to determine whether reduced levels of these co-receptors on risk variant BM CD23+ B cells reflected increased signaling, we assessed transcript levels using quantitative PCR in sort-purified, CD23+ and T2 B cells isolated from 8-10wk control and Ptpn22TT mice. Consistent with an enhanced co-receptor signaling program, Ptpn22TT immature and transitional cells exhibited greater levels of BAFFR and CD40 transcripts, respectively (Fig. 1F).
To more directly parse out the interconnected BCR and co-receptor signaling programs, we measured transcript levels of the alternative NFκB-pathway target, Pim2 - responsive to co-receptor signals, and the alternative and classical NFκB-pathway target, A1 – responsive to the BCR and co-receptor signals. (26,28-30). Pim2 transcripts were substantially increased at both stages of development in Ptpn22TT mice (Fig. 1G), while A1 was significantly increased only in the periphery. Collectively, these results demonstrate that the Ptpn22 variant coordinately increases both antigen-mediated, BCR signaling and the BAFFR and CD40 co-receptor programs. While enhanced BCR signals mediated by the risk variant likely impacts both co-receptor programs (directly and/or indirectly) during this developmental window, our findings suggest that the variant may exert a greater role on BAFFR in the BM and on CD40 in the periphery.
Ptpn22 risk variant expression alters B cell tolerance checkpoints at two discrete stages
The hen egg lysozyme (HEL) and anti-HEL (MD4) BCR transgenic mouse models have been used extensively to study negative selection of self-reactive B cells (31,32). In these studies, MD4 B cells express high-affinity BCRs specific to the neo-self antigen, HEL. Upon development in membrane-bound HEL (mHEL) expressing mice, MD4 B cells receive a strong BCR signal and are deleted or undergo receptor editing before entry into the periphery (33,34). In contrast, exposure to soluble HEL (sHEL) elicits a weaker BCR signal in MD4 B cells that allows them to enter the periphery, where they exhibit features of functional anergy, including down-regulation of surface IgM and an inability to secrete anti-HEL antibodies (35).
To evaluate whether the Ptpn22 variant modulates either deletion or anergy of self-reactive B cells, we created mixed BM chimeras using Ptpn22CC MD4 Tg or Ptpn22TT MD4 Tg mice as donors for transplantation into lethally irradiated mHEL (Fig. 2A) and sHEL recipient mice (Fig. 2B). We found that Ptpn22TT MD4 Tg B cells developing in either mHEL or sHEL recipients exhibit comparable deletion (Fig. 2C-E, gating as in Supplemental Fig. 2D) and anergy (Fig. 2F-H) as their Ptpn22CC MD4 Tg counterparts.
FIGURE 2. Intact negative selection of self-reactive Ptpn22 variant B cells in a high-affinity BCR transgenic model.
Mixed BM chimeras utilizing the donor MD4 (anti-HEL Ig transgenic) and either (A) membrane-bound (m) or (B) soluble (s) HEL recipients were used to assess deletion and anergy, respectively. (C-E) mHEL recipients reconstituted with either Ptpn22CC MD4 (n=6) or Ptpn22TT MD4 (n=6) BM were analyzed 2-3 months after transplant. Reconstitution of donor MD4 B cell subsets in the (C) BM and (D) SPL of mHEL recipients were analyzed by FACS. (E) HEL-specific serum antibody levels from mHEL recipients. (F-H) sHEL recipients reconstituted with either Ptpn22CC MD4 (n= 12) or Ptpn22TT MD4 (n= 12) BM were analyzed 2-3 months after transplant. (F) Reconstitution of donor MD4 B cells in the SPL of sHEL recipients. (G) MFI of MD4 anti-HEL transgene (IgMa) in sHEL recipients, gated on B220+ splenic B cells. (H) HEL-specific serum antibody levels from sHEL recipients. See Supplemental Fig. 2D for gating. All data represents at least two independent experiments. Error bars show SD. Statistical analysis was performed using Student’s t test.
While negative selection mechanisms thus appeared intact in Ptpn22 variant animals, it remained possible that the subtly enhanced BCR signal in Ptpn22TT mice was masked by the high-affinity signal utilized in the HEL models (which express anti-HEL BCRs originally generated from post-germinal center cells). We therefore generated mixed BM chimeras to assess the development of Ptpn22CC versus Ptpn22TT variant B cells within a competitive, polyclonal setting. Congenically marked Ptpn22CC (Ly5.1), Ptpn22TT (Ly5.2), and B-cell deficient Ptpn22CC (μMT; Ly5.1/5.2) donors were mixed at a 10:10:80 ratio for transplantation into lethally irradiated Ptpn22CC (Ly5.1/5.2) recipients. The addition of μMT BM allowed us to assess B-intrinsic effects of Ptpn22 variant expression on B cell selection, since the majority (~90%) of all non-B cells would express wildtype Ptpn22CC, whereas B cells were mixed evenly between those expressing Ptpn22CC and Ptpn22TT (Fig. 3A).
FIGURE 3. Competitive advantage of murine Ptpn22 variant B cells in mixed BM chimeras.
(A) B cell-intrinsic Ptpn22CC and Ptpn22TT competitive BM chimeras created by transfer of congenically marked 80% μMT (Ly 5.1/5.2) + 10% Ptpn22CC (Ly5.1) + 10% Ptpn22TT (Ly5.2) BM into lethally irradiated Ptpn22CC mice (Ly5.1/5.2). Chimeras sacrificed three months after transplant (n=28). (B) Representative gating of B cell and splenic monocyte (B220− CD3− Gr1− CD11b+) subsets by FACS. (C) Frequency of Ptpn22CC vs. Ptpn22TT vs. host + μMT cells in each subset. Data represents five independent experiments. See Supplemental Fig. 2A-B for gating. Error bars show SD. Statistical analysis was performed using Student’s t test: *** p<0.001; **** p<0.0001.
Chimeras were sacrificed at 3 months post-transplant to evaluate the frequencies of Ptpn22CC (Ly5.1) and Ptpn22TT (Ly5.2) B cells at each stage of development. Equivalent levels of donor cell engraftment were observed in splenic monocytes based upon congenic marker expression (Fig. 3B-C). The Pre+Pro B cell fraction also exhibited equal contributions from each genotype, suggesting that variant expression does not alter early B cell development (Fig. 3B-C). In contrast, Ptpn22TT B cells exhibited a competitive advantage at the BM immature and peripheral early transitional (T1) and late transitional (T2) stages (Fig. 3B-C). Notably, while this competitive advantage was maintained in mature FM B cells, it was lost in MZ B cells (Fig. 3B-C). The enhanced competition at the immature/transitional stage, followed by discrepant competition into FM vs. MZ B cell compartments, is consistent with a model whereby the Ptpn22 risk variant alters B cell tolerance at two discrete checkpoints in the bone marrow and periphery.
Ptpn22 variant selectively promotes B cells into the FM compartment
To better understand the differential impact of Ptpn22 risk variant expression on mature B cell subsets, we decided to test whether FM versus MZ cell fate was altered. Given the role that BCR signal strength has been proposed to play in determining cell fate (36), we predicted that the increased BCR signal in Ptpn22TT mice might promote a preferential bias into the FM compartment. Previous studies in Ptpn22 TT mice at >12 weeks of age (in a mixed 129/Sv and C57BL/6J background) revealed normal proportions of mature subsets (7), a finding reproduced in the current study in the C57BL/6J background (data not shown). Because the murine MZ B cell compartment is established during the initial ~12 weeks of life, we determined whether earlier assessment of splenic subsets in Ptpn22TT animals might reveal subtle differences in FM vs. MZ fate. Indeed, comparing the proportions of splenic peripheral subsets during an 8.5-11 week interval of age, we observed a significant increase in FM and concomitant decrease in the proportion of marginal zone precursor (MZp) and MZ B cells in Ptpn22TT mice compared to controls (Fig. 4A). Consistent with an alteration in Notch2 dependent signals required for MZ B cell development (37,38), analysis of MZ precursor cells sorted from Ptpn22CC vs. Ptpn22TTmice at 8.5 weeks of age revealed reduced Notch2 and Notch target gene transcript levels. In addition, a reduction in Hes1 transcripts was also observed in sorted T2 cells (Fig. 4B).
FIGURE 4. Preferential selection of murine Ptpn22 variant B cells into FM compartment.
(A) 8.5-11wk Ptpn22CC (n=9) and Ptpn22TT (n=10) mice were analyzed for splenic B cell subsets by FACS. (B) Quantitative PCR of B cell subsets sorted from 8.5wk Ptpn22CC (n=6) and Ptpn22TT (n=6) mice; mRNA levels of Notch2 (left panel), Deltex1 (center panel) and Hes1 (right panel) relative to β2-microglobulin. (C) The 125 Tg (anti-insulin Ig transgenic) model was used to track insulin-specific B cells in the periphery. 9-16wk Ptpn22CC 125 Tg (n= 11) and Ptpn22TT 125 Tg (n=11) mice were analyzed for total splenic B cell subsets by FACS. All data represents at least two independent experiments. See Supplemental Fig. 2B for gating. Error bars show SD. Statistical analysis was performed using Student’s t test: * p<0.05; ** p<0.01; *** p<0.001; **** p<0.0001.
While these polyclonal studies suggested widespread selection bias, these data did not assess whether self-reactive B cells in particular were affected. To track the selection of B cells with self-reactivity relevant to an autoimmune disorder associated with the PTPN22 risk variant, we crossed Ptpn22TTmice to the 125 Tg model. In this model, ~95% of peripheral B cells express a fixed H and L chain (identified as IgMa+ by FACS) and are insulin-reactive (39). Using FACs analysis of splenic B cell subsets, we found that Ptpn22TT 125 Tg mice similarly exhibited a marked reduction in the proportion of insulin-reactive MZ B cells, and a subtle (though not statistically significant) increase in FM B cells compared to controls (Fig. 4C). Taken together with data demonstrating skewed mature compartments in a polyclonal setting, these findings support the idea that variant protein expression promotes antigen-mediated selection of self-reactive peripheral B cells, biasing their entry into the FM over MZ compartment.
FM and MZ naïve repertoires are differentially skewed in Ptpn22 variant mice
Given the enhanced positive selection and differential selection of transitional cells into the FM and MZ compartments in Ptpn22TT mice, we hypothesized that these findings might correlate with skewing of the naïve repertoires. We therefore performed a detailed assessment of the naïve repertoire of Ptpn22TT mice using combined approaches: high throughput sequencing (HTS) of the BCR H chain, and single cell BCR cloning and assessment of BCR self-reactivity.
We first sought to validate IgH HTS as an appropriate platform for reading out distinct CDR3 profiles between FM and MZ subsets. While useful in many respects, prior sequencing studies have been limited by their restriction to a single VH family, gating strategies that included transitional cells, and/or analysis restricted to the BALB/c background (40-42). To expand on these findings, we bulk sorted FM and MZ B cells from wildtype Ptpn22CC mice on the C57BL/6 background for high-throughput sequencing of the IgH locus (obtaining a total of 120,805 – 142,910 productive sequences for each sample; Supplemental Table I). Our studies revealed broadly altered differences in CDR3 length and composition between mature, naïve FM and MZ subsets (Fig. 5). Consistent with published data, we found that MZ B cells had increased usage for JH2 family (Fig. 5A), shorter average CDR3s (Fig. 5B), a greater proportion of CDR3s lacking N nucleotides (Fig. 5C), and slightly reduced hydrophobicity (Fig. 5D) compared to FMs. This high-throughput approach further revealed novel differences, including broadly altered VH family (Fig. 5E) and amino acid usage (Fig. 5F) between subsets, decreased N2 insertions (Fig. 5G), and reduced diversity (Fig. 5H) in MZ cells. Notably, we found no difference in charged amino acid usage between subsets (data not shown), in contrast to prior studies (40,42).
FIGURE 5. High-throughput sequencing reveals broadly altered CDR3 characteristics between murine FM and MZ subsets.
Follicular (FM) and marginal zone (MZ) cells were sorted from 11-14wk Ptpn22CC mice (n=4 per subset) for high throughput BCR IgH chain sequencing. Nonproductive templates were excluded from analysis. (A) JH family usage. (B) CDR3 length. (C) Templates lacking N insertions (between both V-D and D-J junctions). (D) Hydrophobic amino acids (includes: F, I, W, L, V, M, Y, C, A). (E) VH family usage. (F) Amino acid usage. (G) N2 insertions (between D-J junction). (H) Diversity index. See Supplemental Fig. 2B for gating. See Supplemental Table I for numbers of sequences analyzed. Data represents two independent experiments. Error bars show SD. Statistical analysis was performed using Student’s t test: * p<0.05; ** p<0.01; *** p<0.001; **** p<0.0001.
Armed with an ability to read out distinct CDR3 profiles between mature, naïve subsets using IgH HTS, we next expanded our sequencing studies to include FM and MZ subsets sorted from Ptpn22TT mice (obtaining a total of 119,459 – 142,910 productive sequences for each sample; Supplemental Table I). Surprisingly, we found that the CDR3 profiles in naïve subsets derived from Ptpn22 variant mice did not differ markedly from controls. We identified only minor differences in VH usage and N1 insertions in Ptpn22CC vs. Ptpn22TT MZ B cells (Fig. 6). Thus, while an HTS approach was suitable for identifying unique CDR3 characteristics between FM and MZ B cells, it was for the most part unable to detect any impact of the Ptpn22 variant on the BCR repertoire.
FIGURE 6. Control and murine Ptpn22 variant FM and MZ B cells exhibit indistinguishable IgH CDR3 profiles.
Follicular (FM) and marginal zone (MZ) subsets were FACS sorted from 11-14wk Ptpn22CC (n=4) and Ptpn22TT (n=4) mice for high throughput BCR IgH chain sequencing. Nonproductive templates were excluded from analysis. (A) FM VH and (B) FM JH family usage. (C) MZ VH and (D) MZ JH family usage. (E) CDR3 length. (F) N1 insertions (between V-D junction). See Supplemental Fig. 2B for gating. See Supplemental Table I for numbers of sequences analyzed. Data represents two independent experiments. Error bars show SD. Statistical analysis was performed using Student’s t test: * p<0.05.
One possible explanation, supported by our polyclonal studies (Fig. 3-4), is that the Ptpn22 variant impacts a broad range of BCR specificities, leading to a setting where individual CDR3 features might appear largely unchanged. Differences that may exist in a subpopulation, such as in self-reactive cells, could be lost in a high-throughput approach. Therefore, we next turned to single cell BCR cloning for a more targeted assessment of the naïve repertoire. Using established methods (16,19), BCRs were cloned from single FM and MZ cells sorted from Ptpn22CC and Ptpn22TT mice, producing a total of 273 recombinant mAbs. We assessed antigen specificity using ELISAs for a range of self-antigens, including insulin, malondialdehyde (MDA)-LDL, dsDNA, phosphorylcholine (PC-12) and smRNP. Strikingly, for each self-antigen tested, the proportion of Ptpn22TT mAbs considered reactive (reaching an OD threshold greater than 0.5) was approximately doubled in FM cells. In striking contrast, we observed the opposite impact in MZ B cells with the proportion of Ptpn22TT mAbs considered reactive reduced nearly in half compared to control mAbs (Fig. 7; ELISA curves in Supplemental Fig. 1D-E). The proportions of polyreactive clones exhibited similar differential skewing, with higher and lower proportions found in the FM and MZ compartments, respectively, of variant mice (Fig. 7). As predicted by our tolerance studies, we conclude that Ptpn22 variant mice exhibit a subset-specific skewing of the naïve repertoire, with a preferential bias for self-reactive BCRs within the FM over MZ compartment.
FIGURE 7. Ptpn22 variant mice exhibit an increased proportion of self-reactive FM B cells but fewer self-reactive MZ B cells.
Follicular (FM) and marginal zone (MZ) cells were FACS sorted from 8-12wk Ptpn22CC (n=5) and Ptpn22TT (n=5) mice for single cell BCR cloning of mAbs (n=70 Ptpn22CC FM, n=71 Ptpn22TT FM, n=67 Ptpn22CC MZ, n=65 Ptpn22TT MZ mAbs). Proportions of (A) FM and (B) MZ mAbs reactive to self-antigens (insulin, MDA-LDL, dsDNA, phosphorylcholine [PC-12], smRNP) or polyreactive (reactive to three or more of above self-antigens) by ELISA assay. See Supplemental Fig. 2B for gating and Supplemental Fig. 1D-E for ELISA curves. Data represents two independent experiments.
Carriers of the PTPN22 variant exhibit evidence for broadly enhanced positive selection
The increased self-reactivity of the FM compartment in Ptpn22TT mice closely mirrors the increase in self-reactivity observed in circulating naïve B cells of healthy human subjects with the PTPN22 risk variant (11). In an effort to better understand the signaling events that drive the selection of autoreactive B cells in PTPN22 variant carriers, we turned to a flow based assay based on tracking of a self-reactive heavy chain (HC). We chose the VH4-34 family as a candidate for study due to its well-documented polyreactivity towards a range of self-antigens including B cells, RBCs, and dsDNA (43-45), as well as its relatively high frequency within the transitional and naïve mature B cell compartments in healthy subjects. In addition, previous work from our laboratory and others have documented enrichment of VH4-34 family expressing transitional and mature B cells in autoimmune settings, thought to be driven through modulations in dual BCR and TLR signals (18,46). Based on these previous studies, we anticipated that healthy PTPN22 risk variant carriers might exhibit an increased proportion of VH4-34+ B cells (as detected by the anti-idiotypic mAb, 9G4), as would be consistent with a naïve repertoire skewed towards autoreactivity prior to disease development. Surprisingly, in contrast to this prediction, we found that healthy PTPN22 risk subjects had fewer 9G4+ B cells across all peripheral blood compartments compared to healthy non-risk controls, including most notably within the transitional subset (Fig. 8A-B; cells gated as in Supplemental Fig. 2E).
FIGURE 8. Healthy subjects with the PTPN22 risk variant exhibit broadly enhanced positive selection.
(A-B) PBMCs from age and sex-matched healthy subjects screened for PTPN22 1858 C/C (n=34), C/T (n=34), and T/T (n=3) genotypes were analyzed by FACS for VH4-34 family (9G4+) B cells. (A) Representative histograms quantifying 9G4+ B cells. (B) Percentage of 9G4+ cells across peripheral B cell subsets. (C-D) PBMCs from age and sex-matched healthy subjects screened for PTPN22 1858 C/C (n=18) and C/T (n=18) genotypes were analyzed by FACS for surface BAFFR MFI of total B cells (C) and 9G4− and 9G4+ gated B cells (D). See Supplemental Fig. 2E for gating. See Supplemental Table II for subject information. All data represents at least two independent experiments. Error bars show SD. Statistical analysis was performed using Student’s t test (B) or paired t test (C): * p<0.05; ** p<0.01.
One possible explanation for the reduced frequency of 9G4+ B cells in PTPN22 risk variant subjects, supported by our murine studies, is that globally enhanced positive selection across multiple specificities might reduce the relative proportion of BCRs utilizing this specific HC family. In partial support of this idea, CD40 levels and signaling activity were previously reported to be increased in PTPN22 risk variant subjects (11). To test whether BAFFR levels were similarly increased, and might reflect events similar to our observations in the murine model, we next compared surface BAFFR levels in PTPN22 non-risk vs. risk subjects. Strikingly, the MFI of BAFFR expression was increased in all peripheral B cell subsets in risk subjects (Fig. 8C). The elevated BAFFR levels in risk subjects were unlikely to reflect differences in available cytokine, as we have previously reported that non-risk and risk subjects exhibit similar serum levels of BAFF (10) However, it remained possible that the reduced frequency of self-reactive 9G4+ B cells in risk subjects might reflect an impaired ability compete for available BAFF. To test for this possibility, we compared surface BAFFR levels among 9G4+ (representing predominantly self-reactive BCRs) or 9G4− peripheral B cell subsets (representing a more heterogeneous population) in both non-risk and risk subjects. Strikingly, relative to non-risk subjects, PTPN22CT subjects exhibited a global increase (or trend for increase) in BAFFR levels across all populations, including 9G4− and 9G4+ cells within the transitional, naive and IgM memory B cell compartments (Fig. 8D). Overall, these data imply that in risk subjects, BCR and/or other coordinate signals promote increased BAFFR expression, thereby permitting such cells to compete more effectively for BAFF family survival and differentiation signals.
DISCUSSION
Our findings in the Ptpn22TT murine models indicate that while negative selection mechanisms are intact, enhanced BCR and/or co-receptor signaling programs promote increased positive selection of transitional B cells (including cells with a range of self-reactive specificities) into the follicular mature B cell compartment. Moreover, the counter selection of VH4-34+ expressing cells in human carriers, coupled with global increases in BAFFR and CD40 co-receptor expression described here and in a previous study (11), respectively, support a similar role for the risk variant in human B cell selection. Our combined datasets imply that the risk variant facilitates a subtle yet widespread increase in positive selection signaling programs throughout BM and transitional B cell development. These combined events ultimately allows for a broader range of autoreactive B cells to compete for survival into the mature, naïve B cell compartment. While additional work is required to definitively assess the specific biochemical impact(s) of PTPN22 C1858T variant on BCR and/or co-receptor signaling pathways in developing B cells, our findings strongly suggest that similar mechanisms alter central and peripheral B cell tolerance in both murine and human immune development.
As described earlier, multiple studies in murine models and human subjects support the idea that PTPN22 variant alters B cell tolerance mechanisms (7,10,11). However, it remained unknown whether this defect was B cell-intrinsic. To eliminate the potential impact of disrupted T cell tolerance and homeostasis present in Ptpn22 variant mice and human subjects (7,12,47), our BM chimera studies restricted variant expression predominantly to B cells. Our finding that Ptpn22TT expressing B cells exhibit a competitive advantage at key developmental checkpoints in the bone marrow and periphery highlight the importance of B cell-intrinsic signaling pathways in regulating these events. Consistent with this, we found an identical competitive advantage for variant expressing B cells in competitive chimeras depleted of CD4 T cells (data not shown).
To better address the key question regarding how the Ptpn22 risk variant alters BCR signaling in murine B cells, we utilized the newly described Nur77-GFP model (20). This model has the advantage of permitting assessment of BCR signaling directly ex vivo at discrete stages of B cell development. Our results indicate that risk variant expression promotes a greater proportion of B cells to survive tolerance mechanisms throughout development, mediated in part through stronger BCR signals. Taken as a whole, these data provide compelling evidence that the Ptpn22 variant modulates key survival programs dependent upon BCR signals in vivo, and that engagement with self-antigen in the periphery can promote these events.
Although BCR signaling serves as a master regulator of B cell tolerance, its synergy with key co-receptor pathways, predominantly BAFFR and CD40, ultimately determines the developmental fate of a given B cell (21,48). Each of these co-receptor pathways, in turn, play an important role in promoting positive selection in the periphery (13-16). In addition, a growing body of work, including our own, suggests they serve a similar function in the bone marrow (16,23,25,49). Our finding that Ptpn22TT mice exhibit slightly augmented BCR and co-receptor signals is consistent with subtle signaling changes whereby the variant promotes greater positive selection throughout immature and naïve B cell development.
While the complexity of receptor crosstalk (21) prevented us from fully dissecting the relative contributions of the BCR versus co-receptor signals in mediating selection, our studies demonstrate how subtle fluctuations in these signaling networks can influence the mature repertoire, and may do so in distinct ways. For instance, enhanced positive selection observed in Ptpn22 risk variant mice differs from other autoimmune settings. Unlike models in which a transgenic self-reactive BCR (50) or a specific, self-reactive BCR family (18) facilitates positive selection at the T2 stage via clonal expansion, the Ptpn22 risk variant instead impacts a broad range of BCR specificities during BM and splenic transitional B cell development, allowing multiple self-reactive B cells to compete for BAFF and CD40L signals. Thus, while highly specific VH family skewing is evident in both mice and humans with defects in the Wiskott-Aldrich syndrome (WAS) gene – an autoimmune setting of augmented BCR and TLR signals – (18), we did not observe major differences in either IgH CDR3 profiles or VH family use in Ptpn22TT mature B cells. We further propose that the broad impact of the Ptpn22 variant limited the relative expansion of individual (including self-reactive) VH families. Although the implication of these subtle distinctions in dual BCR/TLR vs. BCR/BAFFR/CD40-mediated positive selection in distinct models remains to be seen (including implications likely beyond autoimmunity), they nevertheless illustrate the usefulness of our integrated approach in assessing the naïve repertoire.
This integrated approach also helped us identify an unexpected bias for selection into the FM compartment of Ptpn22TT mice, including of several self-reactive specificities. Several lines of evidence led to this idea: First, a larger proportion of FM relative to MZ cells was observed in both polyclonal and insulin-specific Ptpn22TT murine models, and marginal zone precursors exhibited a significant decrease in Notch2 expression and in Notch target gene transcripts. Secondly, and consistent with a preferential bias for selection into the FM compartment, our single cell BCR cloning studies revealed an increased proportion of self-reactive and polyreactive BCRs within the FM compartment. In parallel, we observed the opposite finding in the MZ with decreases in both self-reactive and polyreactive specificities in individual MZ B cells. These observations support the conclusion that the Ptpn22 risk variant skews the naïve FM B cell compartment towards self-reactivity, while revealing an additional novel role for restricting MZ development and/or fate. While the mechanistic basis for these surprising observations with respect to MZ B cell development remain unclear, these findings align with recent studies in which BCR and Notch2 signaling exhibit crosstalk that critically regulates MZ lineage commitment (51).
Notably, while we were unable to directly study human splenic B cell subsets, we made progress in translating our murine findings to human subjects. Our observations provide an alternative interpretation of the prevailing model for how the PTPN22 variant impacts human B cell development. As noted above, previous human studies suggest that PTPN22 C1858T carriers exhibit an attenuated BCR signal, promoting relaxed negative selection and subsequent enrichment of self-reactive B cells within the naïve compartment (10,11,52,53). Interestingly, these previous studies have proposed that hypo-responsive BCR signals in risk subjects leads to both increased numbers of self-reactive, new-emigrant (transitional) B cells (10,53), and a seemingly paradoxical aberrant activation of these cells, as demonstrated by elevated BCR target genes involved in cell activation, proliferation, and survival (11). Most notably, previous work described higher levels of both CD40 transcripts and expression and increased CD40 signaling in risk variant new emigrant B cells (11). Thus, while the interpretation differs from our conclusions, these earlier findings indicate that healthy subjects with the risk allele exhibit an increase in the CD40 co-receptor program in transitional B cells. Consistent with these observations, our murine studies revealed intact negative selection and evidence for augmented BCR and/or BAFFR and CD40 co-receptor signals in Ptpn22 variant mice, leading to broadly enhanced positive selection and preferential skewing for several self-reactive specificities into the FM compartment. Thus, while the end result appears the same, explanations for how a greater proportion of self-reactive B cells enter the naïve repertoire differ with respect to previous human and our current murine studies.
As a means to begin to test whether our findings of broadly enhanced positive selection in murine models also applied to human carriers, we utilized a flow-based assay to track the selection of a polyreactive VH family in peripheral blood (VH4-34; identified as 9G4+) (46). Our observation that healthy PTPN22 variant subjects had fewer 9G4+ B cells, yet exhibited increased BAFFR levels in all peripheral B subsets, including both 9G4+ and 9G4− cell populations, is most consistent with a model whereby positive selection is globally enhanced, thereby reducing the relative contribution of this single family VH family. Finally, an additional (but not mutually exclusive) interpretation for the reduced proportion of 9G4+ B cells in PTPN22 carriers, is that this may reflect impaired MZ fate/development – an untested yet intriguing possibility given our murine findings of impaired Notch2 signaling, as well as the presumed enrichment of 9G4+ B cells into an MZ-like B cell subset within human peripheral blood (18,46).
In summary, our collective murine and human data provides an alternative model for how the PTPN22 C1858T variant promotes self-reactivity into the naïve B cell repertoire, and consequently, is likely to increase the probability of triggering autoimmune B cell responses in at-risk individuals. Our studies highlight the importance of synergistic BCR and co-receptor signaling pathways in regulating these events, and in doing so, identifies novel pathways for future study.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank J. Thomas for providing 125 Tg mice; A. Weiss for Nur77-GFP Tg mice; D. Hamm (Adaptive Biotechnologies) for expert assistance with sequencing analysis; S. Khim and K. Sommer for animal and technical assistance. In addition, we thank the Benaroya Research Institute Clinical Core and subjects that participated in the BRI immune-mediated disease biorepository.
This work was supported by NHLBI, NIDDK and NIAID of the National Institutes of Health under award numbers: R01HL075453 (DJR), R01A1084457 (DJR), R01A1071163 (DJR), DP3DK097672 (DJR and JHB), DP3DK111802 (DJR), and DP3DK097672-01S1 (GM). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Additional support provided by the Benaroya Family Gift Fund (DJR) and by an HHMI/NIH Molecular Medicine Training Grant (GM). The authors have no conflicting financial interests.
Abbreviations used in this article:
- FM
follicular mature
- MZ
marginal zone
- AgR
antigen receptor
- BAFFR
B-cell activating factor receptor
- CD40
cluster of differentiation 40
- HEL
hen egg lysozyme
- IgH
BCR heavy chain
- CDR3
complementarity determining region 3
- BM
bone marrow
- MDA-LDL
malondialdehyde low-density lipoprotein
- PC
phosphorylcholine
- smRNP
Smith and nuclear ribonucleoprotein
- β2M
beta-2-microglobulin
- MFI
mean fluorescent intensity
- T2
late transitional
- BAFF
B-cell activating factor
- MZp
marginal zone precursor
- HTS
high-throughput sequencing
REFERENCES
- 1.Rhee I, and Veillette A. 2012. Protein tyrosine phosphatases in lymphocyte activation and autoimmunity. Nat. Rev. Immunol 13: 439–447. [DOI] [PubMed] [Google Scholar]
- 2.Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, MacMurray J, Meloni GF, Lucarelli P, Pellecchia M, Eisenbarth GS, Comings D, and Mustelin T. 2004. A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet. 36: 337–338. [DOI] [PubMed] [Google Scholar]
- 3.Kyogoku C, Ortmann WA, Lee A, Selby S, Carlton VEH, Chang M, Ramos P, Baechler EC, Batliwalla FM, Novitzke J, Williams AH, Gillett C, Rodine P, Graham RR. Ardlie KG, Gaffney PM, Moser KL, Petri M, Begovich AB, Gregersen PK, and Behrens TW. 2004. Genetic Association of the R620W Polymorphism of Protein Tyrosine Phosphatase PTPN22 with Human SLE. Am J Hum Gen. 75: 504–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Begovich AB, Carlton VEH, Honigberg LA, Schrodi SJ, Chokkalingam AP, Alexander HC, Ardlie KG, Huang Q, Smith AM, Spoerke JM, Conn MT, Chang M, Chang SP, Saiki RK, Catanese JJ, Leong DU, Garcia VE, McAllister LB, Jeffery DA, Lee AT, Batliwalla F, Remmers E, Criswell LA, Seldin MF, Kastner DL, Amos CI, Sninsky JJ, and Gregersen PK. 2004. A Missense Single-Nucleotide Polymorphism in a Gene Encoding a Protein Tyrosine Phosphatase (PTPN22) Is Associated with Rheumatoid Arthritis. Am J Hum Gen. 75: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Velaga MR, Wilson V, Jennings CE, Owen CJ, Herington S, Donaldson PT, Ball SG, James RA, Quinton Q, Perros P, and Pearce SHS. 2004. The Codon 620 Tryptophan Allele of the Lymphoid Tyrosine Phosphatase (LYP) Gene Is a Major Determinant of Graves’ Disease. J. Clin Endocrinol Metab 89: 5862–5865. [DOI] [PubMed] [Google Scholar]
- 6.Hermiston ML, Zikherman J, and Zhu JW. 2009. CD45, CD148, and Lyp/Pep: critical phosphatases regulating Src family kinase signaling networks in immune cells. Immunol Rev. 228: 288–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai X, James RG, Habib T, Singh S, Jackson S, Khim S, Moon RT, Liggitt D, Wolf-Yadlin A, Buckner JH, and Rawlings DJ. 2013. A disease-associated PTPN22 variant promotes systemic autoimmunity in murine models. J Clin Invest. 123: 2024–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Shaked I, Stanford SM, Zhou W, Curtsinger JM, Mikulski S, Shaheen ZR, Cheng G, Sawatzke K, Campbell AM, Auger JL, Bilgic H, Shoyama FM, Schmeling DO, Balfour HH, Hasegawa K, Chan AC, Corbett JA, Binstadt BA, Mescher MF, Ley K, Bottini N, and Peterson EJ. 2013. The Autoimmunity-Associated Gene PTPN22 Potentiates Toll-like Receptor-Driven, Type 1 Interferon-Dependent Immunity. Immunity. 39: 111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng J, Ibrahim S, Petersen F, and Yu X. 2012. Meta-analysis reveals an association of PTPN22 C1858T with autoimmune diseases, which depends on the localization of the affected tissue. Genes Immun. 13: 641–652. [DOI] [PubMed] [Google Scholar]
- 10.Habib T, Funk A, Rieck M, Brahmandam A, Dai X, Panigrahi AK, Prak ETL, Meyer-Bahlburg A, Sanda S, Greenbaum C, Rawlings DJ, and Buckner JH. 2011. Altered B Cell Homeostasis Is Associated with Type I Diabetes and Carriers of the PTPN22 Allelic Variant. J. Immunol 188: 487–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menard L, Saadoun D, Isnardi I, Ng Y-S, Meyers G, Massad C, Price C, Abraham C, Motaghedi R, Buckner JH, Gregersen PK, and Meffre E. 2011. The PTPN22 allele encoding an R620W variant interferes with the removal of developing autoreactive B cells in humans. J. Clin. Invest 121: 3635–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rawlings DJ, Dai X, and Buckner JH. 2015. The Role of PTPN22 Risk Variant in the Development of Autoimmunity: Finding Common Ground between Mouse and Human. J. Immunol 194: 2977–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mackay F, and Schneider P. 2009. Cracking the BAFF code. Nat. Rev. Immunol 9: 491–502. [DOI] [PubMed] [Google Scholar]
- 14.Sater RA, Sandel PC, and Monroe JG. 1998. B cell receptor-induced apoptosis in primary transitional murine B cells: signaling requirements and modulation by T cell help. Int Immunol. 10: 1673–1682. [DOI] [PubMed] [Google Scholar]
- 15.Khan WN, Nilsson A, Mizoguchi E, Castigli E, Forsell J, Bhan AK, Geha R, Sideras P, and Alt FW. 1997. Impaired B cell maturation in mice lacking Bruton’s tyrosine kinase (Btk) and CD40. Int Immunol. 9: 395–405. [DOI] [PubMed] [Google Scholar]
- 16.Schwartz MA, Kolhatkar NS, Thouvenel C, Khim S, and Rawlings DJ. 2014. CD4+ T cells and CD40 participate in selection and homeostasis of peripheral B cells. J. Immunol 193: 3492–3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruggiero E, Nicolay JP, Fronza R, Arens A, Paruzynski A, Nowrouzi A, Urenden G, Lulay C, Schneider S, Goerdt S, Glimm H, Krammer PH, Schmidt M, and von Kalle C. 2015. High-resolution analysis of the human T-cell receptor repertoire. Nat Commun. 6: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolhatkar NS, Brahmandam A, Thouvenel CD, Becker-Herman S, Jacobs HM, Schwartz MA, Allenspach EJ, Khim S, Panigrahi AK, Prak ETL, Torgerson TR, Sanz I, and Rawlings DJ. 2015. Altered BCR and TLR signals promote enhanced positive selection of autoreactive transitional B cells in Wiskott-Aldrich syndrome. J. Exp. Med 212: 1663–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tiller T, Busse CE, and Wardemann H. 2009. Cloning and expression of murine Ig genes from single B cells. J. Immunol. Methods 350: 183–193. [DOI] [PubMed] [Google Scholar]
- 20.Zikherman J, Parameswaran R, and Weiss A. 2013. Endogenous antigen tunes the responsiveness of naive B cells but not T cells. Nature 489: 160–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rawlings DJ, Metzler G, Wray-Dutra M, Jackson SW. 2017. Altered B cell signalling in autoimmunity. Nat. Rev. Immunol Advance online publication April 10, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith SH, and Cancro MP. 2003. Cutting Edge: B Cell Receptor Signals Regulate BLyS Receptor Levels in Mature B Cells and Their Immediate Progenitors. J. Immunol 170: 5820–5823. [DOI] [PubMed] [Google Scholar]
- 23.Stadanlick JE, Kaile M, Karnell FG, Scholz JL, Miller JP, Quinn III WJ, Brezski RJ, Treml LS, Jordan KA, Monroe JG, Sen R, and Cancro MP. 2008. Tonic B cell antigen receptor signals supply an NF-κB substrate for prosurvival BLyS signaling. Nat Immunol. 9:1379–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindsley RC, Thomas M, Srivastava B, Allman D. 2007. Generation of peripheral B cells occurs via two spatially and temporally distinct pathways. Blood. Blood 10:2521–2528. [DOI] [PubMed] [Google Scholar]
- 25.Rowland SL, Leahy KF, Halverson R, Torres RM, Pelanda R. 2010. BAFF Receptor Signaling Aids the Differentiation of Immature B Cells into Transitional B Cells following Tonic BCR Signaling. J. Immunol 185:4570–4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu BL, Harless SM, Lindsley RC, Hilbert DM, Cancro MP. 2002. Cutting Edge: BLyS Enables Survival of Transitional and Mature B Cells Through Distinct Mediators. J. Immunol 168: 5993–5993. [DOI] [PubMed] [Google Scholar]
- 27.Barbosa RR, Silva SL, Silva SP, Melo AC, Pereira-Santos MC, Barata JT, Hammarstrom , Cascalho M, Sousa AE. 2014. Reduced BAFF-R and Increased TACI Expression in Common Variable Immunodeficiency. J Clin Immunol 34: 573–583. [DOI] [PubMed] [Google Scholar]
- 28.Woodland RT, Fox CJ, Schmidt MR, Hammerman PS, Opferman JT, Korsmeyer SJ, Hilbert DM, and Thompson CB. 2008. Multiple signaling pathways promote B lymphocyte stimulator dependent B-cell growth and survival. Blood 111: 750–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hatada EN, Do RKG, Orlofsky A, Liou HC, Prystowsky M, MacLennan ICM, Caamano J, and Chen-Kiang S. 2003. NF- B1 p50 Is Required for BLyS Attenuation of Apoptosis but Dispensable for Processing of NF- B2 p100 to p52 in Quiescent Mature B Cells. J. Immunol 171: 761–768. [DOI] [PubMed] [Google Scholar]
- 30.Craxton A, Chuang PI, Shu G, Harlan JM, and Clark EA. 2000. The CD40-Inducible Bcl-2 Family Member A1 Protects B Cells from Antigen Receptor-Mediated Apoptosis. Cell Immunol. 200: 56–62. [DOI] [PubMed] [Google Scholar]
- 31.Shlomchik MJ 2008. Sites and Stages of Autoreactive B Cell Activation and Regulation. Immunity 28: 18–28. [DOI] [PubMed] [Google Scholar]
- 32.Cambier JC, Gauld SB, Merrell KT, and Vilen BJ. 2007. B-cell anergy: from transgenic models to naturally occurring anergic B cells? Nat. Rev. Immunol 7: 633–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hartley SB, Crosbie J, Brink R, Kantor AB, Basten A, Goodnow CC. 1991. Elimination from peripheral lymphoid tissues of self-reactive B lymphocytes recognizing membrane-bound antigens. Nature 353: 765–769. [DOI] [PubMed] [Google Scholar]
- 34.Pelanda R, Torres RM. 2012. Central B-Cell Tolerance: Where Selection Begins. Cold Spring Harb Perspect Biol 4: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodnow CC, Crosbie J, Adelstein S, Lavoie TB, Smith-Gill SJ, Brink RA, Pritchard-Brisco H, Wotherspoon JS, Loblay RH, Raphael K, Trent RJ, and Basten A. 1988. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature 334: 676–682. [DOI] [PubMed] [Google Scholar]
- 36.Pillai S, and Cariappa A. 2009. The follicular versus marginal zone B lymphocyte cell fate decision. Nat. Rev. Immunol. 9: 767–777. [DOI] [PubMed] [Google Scholar]
- 37.Saito T, Chiba S, Ichikawa M, Kunisato A, Asai T, Shimizu K, Yamaguchi T, Yamamoto G, Seo S, Kumano K, Nakagami-Yamaguchi E, Hamada Y, Aizawa S, and Hirai H. 2003. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity 18: 675–685. [DOI] [PubMed] [Google Scholar]
- 38.Witt CM, Won WJ, Hurez V, Klug CA. 2003. Notch2 Haploinsufficiency Results in Diminished B1 B Cells and a Severe Reduction in Marginal Zone B Cells. J. Immunol 171: 2783–2788. [DOI] [PubMed] [Google Scholar]
- 39.R Rojas M, Hulbert C, Thomas JW. 2001. Anergy and not clonal ignorance determines the fate of B cells that recognize a physiological autoantigen. J. Immunol 166: 3194–3200. [DOI] [PubMed] [Google Scholar]
- 40.Schelonka RL, Tanner J, Zhuang Y, Gartland GL, Zemlin M, Schroeder HW. 2007. Categorical selection of the antibody repertoire in splenic B cells. Eur J Immunol. 37: 1010–1021. [DOI] [PubMed] [Google Scholar]
- 41.Carey JB, Moffatt-Blue CS, Watson LC, Gavin AL, Feeney AJ. 2008. Repertoire-based selection into the marginal zone compartment during B cell development. J. Exp. Med 205:2043–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ippolito GC, Schelonka RL, Zemlin M, Ivanov II, Kobayashi R, Zemlin C, Gartland GL, Nitschke L, Pelkonen J, Fujihashi K, Rajewsky K, and Schroeder HW Jr. 2006. Forced usage of positively charged amino acids in immunoglobulin CDR-H3 impairs B cell development and antibody production. J. Exp. Med 203: 1567–1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cappione AJ, Pugh-Bernard AE, Anolik JH, Sanz I. 2004. Lupus IgG VH4.34 Antibodies Bind to a 220-kDa Glycoform of CD45/B220 on the Surface of Human B Lymphocytes. J. Immunol 172: 4298–4307. [DOI] [PubMed] [Google Scholar]
- 44.Jenks SA, Palmer EM, Marin EY, Hartson L, Chida AS, Richardson C, and Sanz I. 2013. 9G4+ Autoantibodies Are an Important Source of Apoptotic Cell Reactivity Associated With High Levels of Disease Activity in Systemic Lupus Erythematosus. Arthritis Rheum. 65: 3165–3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richardson C, Chida AS, Adlowitz D, Silver L, Fox E, Jenks SA, Palmer E, Wang Y, Heimburg-Molinaro J, Li Q-Z, Mohan C, Cummings R, Tipton C, and Sanz I. 2013. Molecular Basis of 9G4 B Cell Autoreactivity in Human Systemic Lupus Erythematosus. J. Immunol 191: 4926–4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pugh-Bernard AE, Silverman GJ, Cappione AJ, Villano ME, Ryan DH, Insel RA, and Sanz I. 2001. Regulation of inherently autoreactive VH4-34 B cells in the maintenance of human B cell tolerance. J. Clin. Invest 108: 1061–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang J, Zahir N, Jiang Q, Miliotis H, Heyraud S, Meng X, Dong B, Xie G, Qiu F, Hao Z, McCulloch CA, Keystone EC, Peterson AC, and Siminovitch KA. 2011. The autoimmune disease–associated PTPN22 variant promotes calpain-mediated Lyp/Pep degradation associated with lymphocyte and dendritic cell hyperresponsiveness. Nat. Genet 43: 902–907. [DOI] [PubMed] [Google Scholar]
- 48.Cancro MP 2009. Signalling crosstalk in B cells: managing worth and need. Nat. Rev. Immunol 9: 657–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Castigli E, Young F, Carossino AM, Alt FW, and Geha RS. 1996. CD40 expression and function in murine B cell ontogeny. Int. Immunol 8 :405–411. [DOI] [PubMed] [Google Scholar]
- 50.Meyer-Bahlburg A, Andrews SF, Yu KOA, Porcelli SA, and Rawlings DJ. 2008. Characterization of a late transitional B cell population highly sensitive to BAFF-mediated homeostatic proliferation. J. Exp. Med 205: 155–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hammad H, Vanderkerken M, Pouliot P, Deswarte K, Toussaint W, Vergote K, Vandersarren L, Janssens S, Ramou I, Savvides SN, Haigh JJ, Hendriks R, Kopf M, Craessaerts K, de Strooper B, Kearney JF, Conrad DH, and Lambrecht BN. 2017. Transitional B cells commit to marginal zone B cell fate by Taok3-mediated surface expression of ADAM10. Nat. Immunol 9: 1–10. [DOI] [PubMed] [Google Scholar]
- 52.Rieck M, Arechiga A, Onengut-Gumuscu S, Greenbaum C, Concannon P, Buckner JH. 2007. Genetic Variation in PTPN22 Corresponds to Altered Function of T and B Lymphocytes. J. Immunol 179: 4704–4710. [DOI] [PubMed] [Google Scholar]
- 53.Schickel J-N, Kuhny M, Baldo A, Bannock JM, Massad C, Wang H, Katz N, Oe T, Menard L, Soulas-Sprauel P, Strowig T, Flavell R, and Meffre E. 2016. PTPN22 inhibition resets defective human central B cell tolerance. Sci Immunol 1: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.