SUMMARY
In collective cell migration, directional protrusions orient cells in response to external cues, which requires coordinated polarity among the migrating cohort. However, the molecular mechanism has not been well defined. Drosophila border cells (BCs) migrate collectively and invade via the confined space between nurse cells, offering an in vivo model to examine how group polarity is organized. Here, we show that the front/back polarity of BCs requires Rap1, hyperactivation of which disrupts cluster polarity and induces misoriented protrusions and loss of asymmetry in the actin network. Conversely, hypoactive Rap1 causes fewer protrusions and cluster spinning during migration. A forward genetic screen revealed that downregulation of the Hippo (Hpo) pathway core components hpo or mats enhances the Rap1V12-induced migration defect and misdirected protrusions. Mechanistically, association of Rap1V12 with the kinase domain of Hpo suppresses its activity, which releases Hpo signaling-mediated suppression of F-actin elongation, promoting cellular protrusions in collective cell migration.
Graphical Abstract
In Brief
In collective cell movement, coordinated polarity among the migrating cohort is required for directional protrusions in response to external cues. Chang et al. show that such group polarity requires Rap1 activity, which promotes cellular protrusions and gives cells an advantage in competing to lead the cluster.
INTRODUCTION
Cellular polarization is important in a variety of biological events, including cell division, cell fate determination, vesicle trafficking, and migration. Migrating cells extend directional protrusions, driven by actin polymerization, to form lamellipodia, filopodia, or a combination of both in response to external chemical or mechanical cues (Krause and Gautreau, 2014; Rørth, 2011). Over recent decades, studies on single cells such as Dictyostelium and mammalian neutrophils have elucidated how chemical signaling, Rho small GTPase, and the cytoskeleton network polarize cells during chemotaxis (Artemenko et al., 2014; Kamp et al., 2016). When a chemoattractant binds to its receptor, differential recruitment of downstream effectors to the front or back of cells establishes polarity, which orchestrates the forward protrusions and rear retraction that result in directional movement. Suppression of actin polymerization renders cells spherical even in the presence of external cues, although asymmetrically localized signaling components are still engaged at the front of the cell (Janetopoulos et al., 2005; Wang et al., 2014). Thus, molecules that connect membrane receptors to the cytoskeleton network are crucial for transducing external stimuli and for integrating signals to create biased protrusions. However, how adhering cells acquire group polarity to migrate collectively remains less clear, despite a great number of structural components having been identified (Haeger et al., 2015; Mayor and Etienne-Manneville, 2016; Nelson, 2016).
To address this topic, we took advantage of a group of migratory cells called border cells (BCs), which are derived from the follicular epithelium and migrate collectively within the egg chamber during Drosophila oogenesis (Prasad et al., 2015). Four to six motile cells are recruited by a pair of polar cells to form a cluster. and these extend protrusions that guide the cluster through the confined space between nurse cells to reach the oocyte (Figure 1A; Cai et al., 2014; Duchek et al., 2001; McDonald et al., 2006). Blockage of guidance cues by expressing dominant-negative forms of receptor tyrosine kinase, PVR (plaetelet-derived growth factor [PDGF]/vascular endothelial growth factor [VEGF] receptor) and EGFR (epidermal growth factor receptor), results in numerous protrusions toward all directions (Prasad and Montell, 2007). The receptors for these guidance cues stimulate the Rac-dependent F-actin assembly that promotes protrusions (Duchek et al., 2001). Here we report another player involved in the polarity system, a small GTPase, Rap1, which promotes directional protrusions in BCs. Expression of the constitutively active form of Rap1 (Rap1V12) in BCs disrupted the front/back polarity of entire clusters, causing misoriented protrusions and evenly distributed phalloidin staining. In contrast, expression of dominant-negative Rap1 (Rap1N17) decreased the protrusion number, causing clusters to spin during migration. Random induction of Rap1V12 in one or two BCs caused them to preferentially assume leading positions in migrating clusters, whereas cells expressing Rap1N17 were more frequently found at the rear. To uncover genes involved in Rap1-dependent polarization, we conducted a forward genetic screen and identified two Hpo pathway members, hpo and mats (mob as tumor suppressor). Hypoactivity of these genes not only enhanced Rap1V12-induced migration defects but also increased protrusion number in both the forward and backward directions. Hpo encodes a Ser/Thr kinase and possesses a linker domain conjoining the N-terminal kinase and C-terminal SARAH (Sav/Rassf/Hpo) domains. Activation of Hpo can be initiated by autophosphorylation via homodimerization of the SARAH domain, which subsequently activates other core components to transduce the kinase cascade (Creasy et al., 1996; Jin et al., 2012; Meng et al., 2016; Ni et al., 2015). We provide biochemical evidence to show that Rap1V12 binds to the Hpo kinase domain to hinder its activation. Moreover, Rap1V12 increases the expression of Enabled (Ena, Drosophila homolog of vasodilator-stimulated phosphoroprotein [VASP]), an actin filament elongation factor that promotes BC protrusion but is inactivated by Hpo signaling. Taken together, our results suggest that, at the leading edge of migrating BCs, activated Rap1 binds to Hpo, disinhibiting Ena and promoting protrusions.
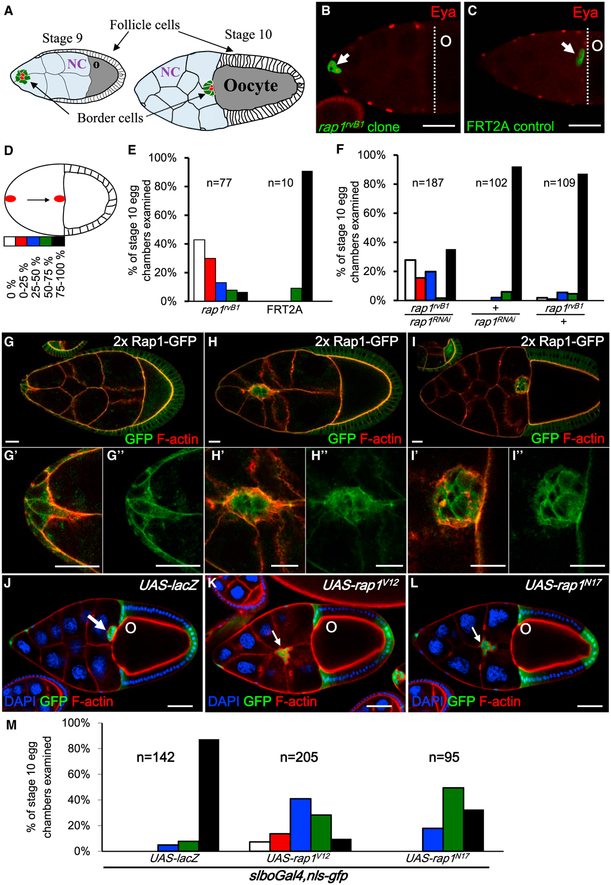
Figure 1. Rap1 Is Required for BC Migration.
(A) Schematic drawings of stage 9 and 10 egg chambers (posterior to the right). BCs (green), surrounding the polar cell pair (red), move through the nurse cells (NCs, light blue) posteriorly and reach the oocyte at stage 10 (gray).
(B and C) Stage 10 egg chambers are stained with anti-Eyes absent (Eya) (red) to mark follicle cells, including BCs (white arrows). The GFP-labeled BC cluster with the rap1rvB1 mutation (green) remains attached at the anterior end (B), but the GFP-labeled FRT2A control clone (green) reaches the oocyte border (dotted line) (C).
(D) Schematic showing the migration path divided into five sessions to quantify migration delay.
(E and F) Quantification of the migration defect caused by rap1rvB1 mutation (E) or by rap1 RNAi knockdown (F).
(G–I) Rap1-GFP (green) shows endogenous expression domains of Rap1 in egg chambers through stages 9 to 10.
(J–L) GFP labels the slboGal4 domains, including BCs (white arrows). Nuclei of follicle cells and nurse cells were visualized by DAPI. Phalloidin staining (red) shows F-actin in egg chambers expressing UAST-lacZ, UAST-rap1V12, and UAST-rap1N17.
(M) Quantification of migration defects for the indicated genotypes. n represents the numbers of stage 10 egg chambers examined.
Scale bars, 50 μm
RESULTS
Rap1 Is Required for BC Migration
Rap1 belongs to the small GTPase family, members of which are active when bound to guanosine triphosphate (GTP) and inactive when bound to guanosine diphosphate (GDP). They are regulated by guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) (Cherfils and Zeghouf, 2013). In Drosophila, Rap1, its downstream effector, and several GEFs have been linked to adhesion complex formation, integrin signaling, hemocyte migration, and tissue morphogenesis (Boettner et al., 2003; Huelsmann et al., 2006; Knox and Brown, 2002; Siekhaus et al., 2010; Wang et al., 2013). To investigate Rap1 function in BCs, GFP-labeled rap1rvB1 homozygous mutant cells were generated in egg chambers and showed impaired motility (Figure 1B). We assessed the extent of the migration defect by quantifying where BCs stopped along the migration path, which we divided into five sections, as shown in Figure 1D. Of the rap1rvB1 mutant BC clusters, 43% (n = 77) stalled at the anterior tip of the egg chamber (Figure 1E), whereas none of the wild-type control cells did so (Figures 1C and 1E). Knockdown of rap1 by UAS-rap1RNAi expression in BCs barely affected motility but, in combination with one copy of mutant rap1rvB1dramatically delayed migration by 65% (n = 187) (Figure 1F). The expression pattern of rap1-rap1::gfp under endogenous rap1 promoter control (Knox and Brown, 2002) revealed that Rap1-GFP was mainly localized at the follicle cell membrane but co-localized with F-actin at the leading edge during BC migration (Figures 1G–1I). We further drove expression of UAS-rap1V12 (Boettner et al., 2003) in BCs to study Rap1 function and found that 90% of BCs failed to reach the oocyte border (Figures 1K and 1M), but only about 10% of control cells exhibited migration delay (Figures 1J and 1M). Attenuation of Rap1 activity by expressing UAS-rap1N17 also blocked cell migration, with only 33% of clusters reaching their destination (Figures 1L and 1M). Despite research attention on Rap1 focusing on single-cell migration in Drosophila macrophages, Dictyostelium, or epithelial invagination during embryonic development (Boettner et al., 2003; Choi et al., 2013; Huelsmann et al., 2006; Lee and Jeon, 2012; O’Keefe et al., 2012; Schurmans et al., 2015; Siekhaus et al., 2010; Wang et al., 2014), our observations demonstrate that collective cell movement requires tightly regulated Rap1 activity.
Polarized Actomyosin Is Regulated by Rap1
To investigate how Rap1 affects cell motility, we examined rap1 mutant cells by phalloidin staining of F-actin. F-actin was enriched at the periphery of wild-type BCs and accumulated at the leading edge of the clusters during migration (Figures S1A–1F). However, in GFP-labeled rap1rvB1 mutant clones, F-actin was reduced and lacked front end accumulation, unlike neighboring wild-type cells (Figures S1G–S1G”). Evenly distributed F-actin staining was also observed under expression of Rap1N17 (Figures S1H and H’). Conversely, Rap1V12-expressing BCs presented high levels of uniformly distributed phalloidin staining throughout the entire cluster (Figures S1I and S1I’) rather than leading edge enrichment as in the controls (Figures S1B and S1E). To further test the requirement for Rap1 activity to induce F-actin-mediated protrusions, Drosophila S2 cells were transfected with gfp (Figure S2A), rap1 (Figure S2B), rap1V12 (Figure S2C), or rap1N17 (Figure S2D) and stained with phalloidin to analyze their morphologies. S2 cells are derived from a macrophage lineage and are spherical under standard culture conditions. In Rap1V12-transfected cells, 71% formed filopodium-like protrusions, but only about one-third of wild-type and Rap1 or Rap1N17 overexpression cells exhibited such structures (Figure S2E). Time-lapse movies further demonstrate the vigorous dynamics of Rap1V12-induced long and multiple protrusions (Movie S1). Our results suggest that Rap1 regulates F-actin expression and the dynamics of actin-rich protrusions in cells.
Several lines of evidence show that guidance signals trigger protrusion by activating Rac-dependent actin polymerization (Duchek et al., 2001; McDonald et al., 2006; Wang et al., 2010), but little is known about how front/back cluster polarity is established to maintain collective migration. Therefore, we employed UAS-gfp::act (Verkhusha et al., 1999) to monitor actin dynamics by fixed-sample analysis or time-lapse imaging to further unravel the effect of Rap1 on actin-dependent protrusions. In wild-type BCs, GFP::Act was enriched at the leading edge, where membrane protrusions extended toward the oocyte (Figures 2A, 2B, 2D and 2E; Figure 3A; Movie S2; Kim et al., 2011; Poukkula et al., 2014). However, rap1V12 overexpression disrupted front/back polarity, resulting in protrusions projecting in all directions (Figures 2J and 2K and 3B; Movie S3), occasionally causing the cluster to disperse (Figures 2M–2O). In contrast, in Rap1N17-expressing BCs, fewer protrusions were observed (Figures 2G–2I), and the location of GFP::Actin enrichment continuously changed during migration (Figure 3C; Movie S4). Our time-lapse movies further support the idea that Rap1 is required for directional polarization of F-actin-driven protrusions.
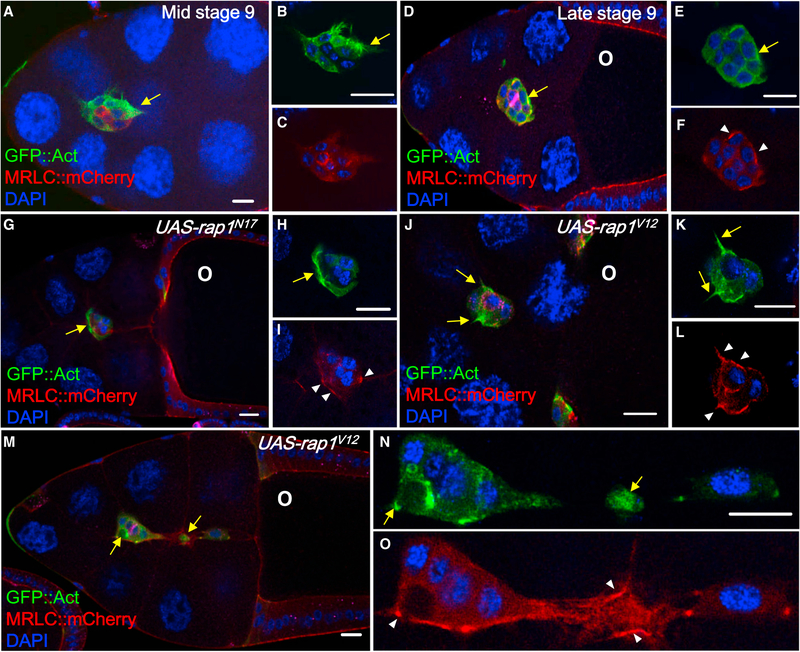
Figure 2. Rap1 Regulates the Distribution of Actomyosin in BCs.
(A–F) DAPI (blue) is used to label nuclei. MRLC::mCherry (red) and slboGal4 > UAS-gfp::act (green) are applied to analyze actin dynamics and myosin contraction, respectively, in mid-stage 9 (A–C) and late stage 9 BCs that have completed 75%–100% of the migration path (D–F). Wild-type BCs display one or two protrusions projecting toward the oocyte (O) (A, B, D, and E) as well as a few speckles of MRLC::mCherry enriched in the center of migrating clusters (A and C) or at the cell margin membrane (D and F).
(G–O) Expression of UAS-rap1N17 (G–I) or UAS-rapV12 (J–O) disrupts the polarized patterns of both reporters.
Anti-FasIII (magenta) staining serves as a marker of polar cells (D). O indicates the oocyte. The scale bars in (A), (G) and (M) indicate 20 μm, and the others indicate 10 μm.
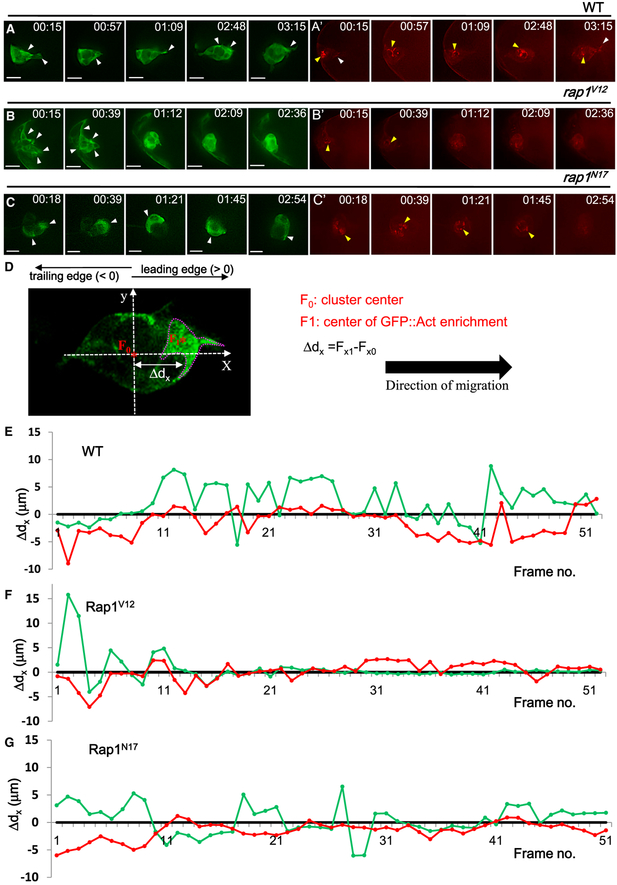
Figure 3. Live Imaging Analysis of Rap1-Regulated Actomyosin Distribution.
(A–C) Time-lapse imaging is used to monitor the dynamic behavior of BCs expressing UAS-gfp::act (green) and MRLC::mCherry (red) under wild-type (A), UAS-rap1V12 (B), and UAS-rap1N17 expression (C).
(D) A live imaging frame taken from a BC cluster demonstrates our definition for the centers of the migrating cluster (F0) and the fluorescence-enriched region (F1; circled with a dotted line, center marked by an asterisk), which are tracked frame by frame using the NIS-Elements AR program (Nikon). Ddx is defined as the distance between the x coordinates of these two points (Fx0 and Fx1), the values of which were plotted against sequential GFP::Act and MRLC-mCherry signals (E–G). In wild-type BCs, actin and myosin distributions are polarized (E). However, BCs expressing UAS-rap1V12 (F) or UAS-rap1N17 (G) lack these polarized patterns.
Scale bars, 10 μm.
In addition to actin filament assembly, periodic myosin contraction is also indispensable for driving protrusions in cycles of extension and withdrawal. Although BCs are known to move collectively, how contractile activity is organized in clusters remains unclear. We used MRLC-mCherry (myosin II regulatory light chain, named spaghetti squash [sqh] in Drosophila) (Munjal and Lecuit, 2014; Rauzi et al., 2010), which accumulates at sites of active contraction, to investigate the distribution of myosin activity during BC movement. In migrating clusters, MRLC accumulated at the interface of border/polar cells (Figures 2A and 2C). Time-lapse video further revealed periodic accumulation and dispersal of MRLC-mCherry in the centers of clusters (Figure 3A’, yellow arrowheads; Movie S2), suggesting that the contractile force was pulled inward collectively but restricted at the junctions between border/polar cells. In addition, MRLC-mCherry was also found to localize on the membrane margins of BCs during migration (Figures 2D and 2F, white arrowheads). However, aggregated signals were decreased in Rap1V12expressing BCs (Figures 2J and 2L, white arrowheads; Figure 3B’; Movie S3) or Rap1N17-expressing BCs (Figures 2G and 2I, white arrowheads; Figure 3C’; Movie S4), but the membrane distribution of MRLC-mCherry was retained and co-localized with GFP::Act (Figures 2I, 2L, and 2O, white arrowheads). Based on these analyses, we propose that polarized actomyosin, which is regulated by Rap1 activity, is essential for BC migration.
Tracking and Comparison of Actomyosin Polarity
To analyze the dynamic pattern of the actomyosin signal in BCs, we used an auto-tracking program to examine its localization and fluorescence intensity frame-by-frame in time-lapse videos. For each frame, the x coordinates of the cluster center (Fx0) and the center of enriched fluorescent signal (Fx1) were recorded, and the distance between these two coordinates (Ddx; Figure 3D)was used to plot a sequential graph of GFP::Act and MRLCmCherry (Figures 3E–3G). When the enriched signal resided at the leading edge, Δdx was defined as positive; on the contrary ΔdX was treated as negative. According to these criteria, we observed wild-type BCs maintaining distinct polarity of actin/myosin during migration. Before detachment, the center of the GFP::Act-enriched domain was 2 μm to 6 μm away from the cluster center, and the MRLC-mCherry signal was predominantly localized at the back of BCs (Figure 3E, frames 1–11). Gradually, the aggregated spots of MRLC-mCherry moved toward the center of migrating clusters (Figure 3E, frames 11–21). After detachment, BCs exhibited a polarized signal pattern, with GFP::Act at the front and MRLC-mCherry close to the center or at the rear of the clusters, until they approached the oocyte (Figure 3E, frames 21–51). Rap1V12expressing cells lacked this polarized pattern (Figure 3F), and the signal intensity was increased over the entire cluster so that the cluster center and that of the GFP-enriched area were barely distinguishable. A similar non-polarized MRLC-mCherry signal was obtained for Rap1N17 expression (Figure 3G), but the GFP::Act signal fluctuated considerably (Figure 3G), reflecting how frequently the GFP::Act-enriched region switches positions during migration (Movie S4). Periodic protrusion and retraction, generated by actin polymerization and myosin contraction, is apparently essential to drive the cluster forward. Because disruption of Rap1 activity not only interrupts this rhythm but also impedes cell mobility, we conclude that Rap1 is responsible for the maintenance of actomyosin polarity in BC migration.
Rap1 Genetically Interacts with Hpo Signaling in BC Migration
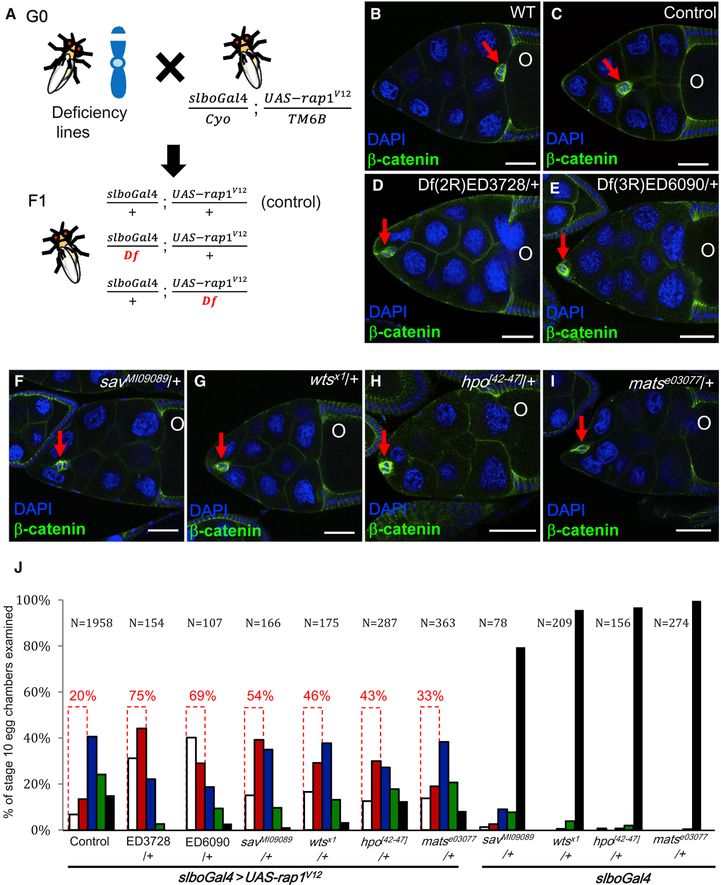
To shed light on the molecular mechanism by which Rap1 regulates actomyosin polarity, we performed a forward genetic screen for genes involved in the Rap1V12-induced migration defect. We crossed 158 Drosophila deficiency lines with slboGal4 > UAS-rap1V12 to look for enhancers or suppressors of the migration defect (Figure 4A). To determine the penetrance of the Rap1V12-induced migration defect, we examined 1,958 stage 10 egg chambers and found that 20% of their clusters failed to pass one-quarter of the migration path, and only about 15% completed three-quarters of it. Thus, we chose BCs stalled in the first quarter of the migration path as the criterion for conducting the modifier screen (Figure 4J, dotted boxes). Because the SD for this group of BCs was 8%, a frequency of more than 28% of BCs stalling at this stage of migration was defined as enhancement. Based on this criterion, 38 deficiency lines displayed modified phenotypes, and the top 14 candidates enhanced the migration defect ~1.8- to 3-fold compared with Rap1V12 expression alone. Two of the 14 lines, Df(2R)ED3728 and Df(3R)ED6090, drew our attention because they both represent a core component of the Hpo (Hpo) pathway, specifically hpo and mats (Figures 4D and 4E). Because recent findings indicate that Hpo signaling is central to transduction of mechanical and cytoskeletal cues in collective cell movement (Gaspar and Tapon, 2014; Lucas et al., 2013; Sansores-Garcia et al., 2011), we further investigated the relationship between Rap1 and Hpo in BCs. Although 20% of clusters failed to pass one-quarter of the migration path in Rap1V12-expressing flies, loss of one allele of hpo or mats escalated the migration defect to 43% and 33% of clusters, respectively (Figures 4H–4J). This outcome is consistent with the results of our screen, wherein a 75% and 69% migration delay was evident for the Df(2R)ED3728 and Df(3R)ED6090 lines, respectively (Figures 4D and 4E). Furthermore, lack of one copy of other key members of the Hpo pathway, warts (wts) or salvador (sav), also greatly intensified the Rap1V12-induced defective phenotype to 46% and 54% of clusters, respectively (Figures 4F, and 4G, and 4J). However, in no case did the hemizygote alone affect migration (Figure 4J). Upon activation of Hpo signaling, the downstream effector Yorkie (yki) is phosphorylated by Wts and retained in the cytoplasm. Conversely, unphosphorylated Yki is translocated into the nucleus to activate downstream genes (Chen et al., 2015; Huang et al., 2005; Ren et al., 2010). Therefore, we expressed constitutively active Yki (phosphorylation sitemutated forms) to mimic the hypoactivity of Hpo signaling (Oh and Irvine, 2008), but, consistent with a previous report (Lin et al., 2014), neither the wild-type nor the Yki constitutively active forms delayed migration (Figure S3E). More importantly, loss of yki does not impair BC motility, indicating that endogenous yki is not required for BC migration (Lin et al., 2014), even though overexpression of hyperactive Yki exacerbated the Rap1V12-induced migration defect (Figures S3A–S3E). Hence, we conclude that Rap1 interacts with non-canonical Hpo signaling to regulate BC migration. Next we examined whether loss of the Hpo upstream components expanded (ex), kibra (Kib), and merlin (mer) affected the Rap1V12-dependent migration defect. However, none of these upstream components had a significant effect (Figures S3F–S3J). These results suggest that the core kinase components of the Hpo pathway interact with Rap1 to control BC migration.
Figure 4. Downregulation of Hpo Signaling Enhances the Rap1V12-Induced Migration Defect.
(A) Schematic overview of the genetic screen for mutant loci that affect Rap1V12-induced migration defects.
(B–I) Confocal micrographs of stage 10 egg chambers stained with anti-b-catenin (green) and DAPI (blue) to label follicle cells with the indicated genotypes. Red arrows pinpoint BC clusters. Two deficiency lines, Df(2R)ED3728 and Df(3R)ED6090, each of which uncovers one component of the Hpo pathway, enhance the UAS-rap1V12-induced migration defect (D and E). BCs expressing rap1V12 stall at the anterior part of egg chambers upon loss of one allele of indicated Hpo pathway members (F–I). O indicates oocytes.
(J) Quantification of the degree of BC migration in the indicated genotypes. Dotted boxes indicate the data used for the screen. Scale bars, 50 μm.
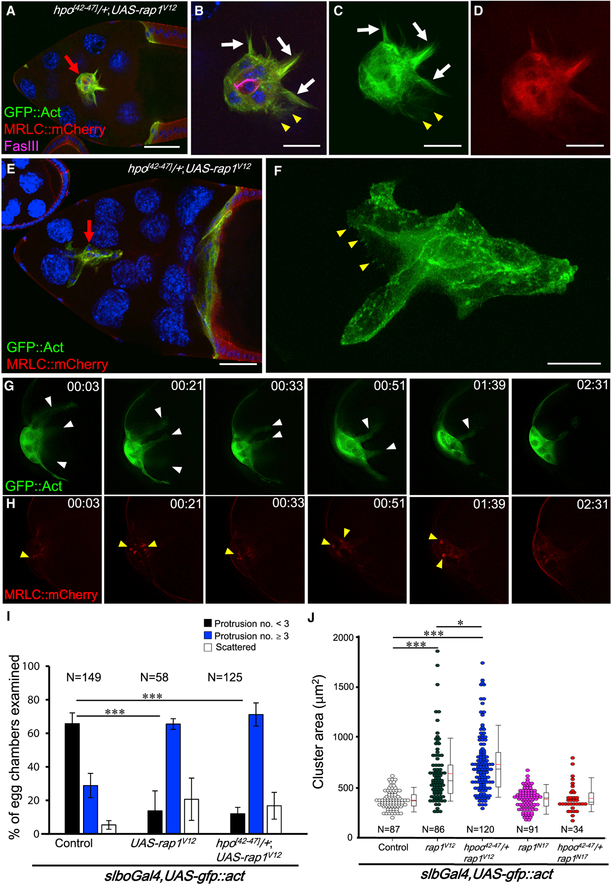
Hpo signaling has been shown to polarize actin polymerization in BCs (Lucas et al., 2013), so we tested whether downregulation of hpo affects actin-mediated protrusion or exacerbates the disruption of actomyosin polarity arising from Rap1V12 overexpression. In wild-type egg chambers, 62% of BC clusters presented one or two long protrusions, and only 33% hosted three or more during migration (Figures 2A and 2B, 3A, and 5I). A combination of one allele of hpo[42–47] with Rap1V12 overexpression gave rise to multiple and spiny protrusions in 70% of BC clusters (Figures 5A–5D and 5G), which substantially intensified the phenotype resulting from Rap1V12 alone (66%) (Figure 5I). Consistently, we observed long persistent protrusions extending in all directions under live imaging (Figure 5G, white arrowheads; Movie S5). Moreover, upon simultaneous overexpression of Rap1V12 and downregulation of Hpo, BCs gradually dissociated and became scattered because of misoriented protrusions pulling the cluster apart (Figures 5E and 5F), which, consequently, may have resulted in loss of the inner contractile force, as manifested by aggregation of MRLC::mCherry (Figures 5D and 5H; Movie S5). Further analysis revealed that the average area of wild-type BC clusters was 377 μm2; i.e., much smaller than that of rap1V12-expressing cells (639 μm2). Downregulation of hpo further enlarged the cluster area to 730 μm2 (Figure 5J). Accordingly, our results suggest that Hpo functions as a negative regulator of Rap1-mediated actin polymerization in BCs.
Figure 5. Increased Protrusion Number and Cluster Size Caused by rap1V12 Overexpression under hpo Hemizygosity.
(A–F) 3D confocal micrographs of BCs. The UAS-gfp::act (green) transgene is driven by slboGal4, and MRLC::mCherry (red) serves as readout for contractile force exertion during migration. Nuclei are labeled by DAPI (blue), and anti-FasIII (magenta) is used to label polar cells (A and B). Rap1V12 overexpression gives rise to brush-like protrusions (yellow arrowheads) and multiple long protrusions (white arrows) in the hpo[42–47]/+ background (A–D). Lamellipodium-like membrane extension is also observed in scattered BCs (E and F).
(G and H) Time-lapse micrographs show actin protrusions (green, G) and MRLC-mCherry accumulation (red, H) in hpo[42–47]-hemizygous BCs expressing UAS-rap1V12.
(I) Quantification of protrusion number and cluster scattering in BCs with specific genotypes.
(J) Quantification of spreading areas of BC clusters.
*p < 0.05, ***p < 0.001. n is the total number of egg chambers examined. Scale bars, 50 μm in (A) and (E) and 20 μm in (B)–(D) and (F). The error bars represent SD.
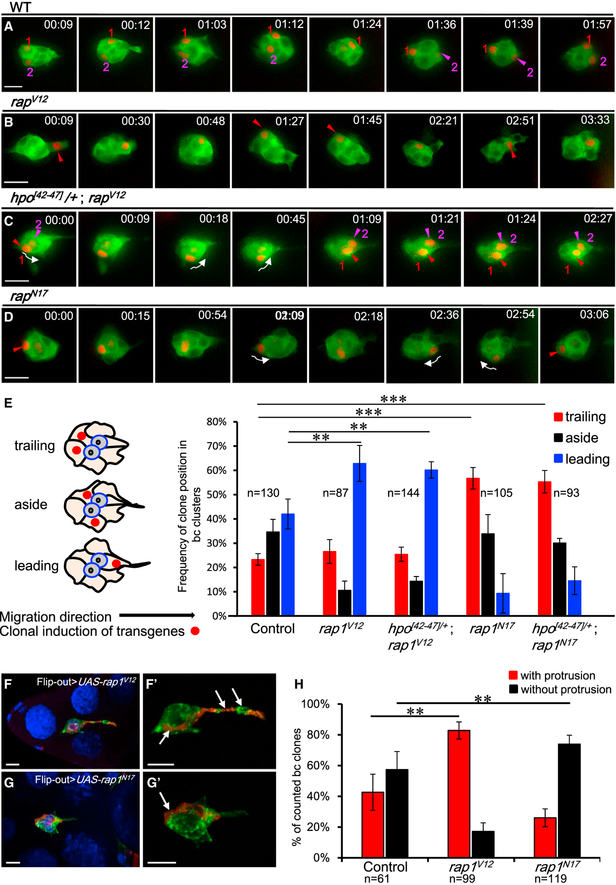
Forward extension is thought to be indispensable for collective movement of BCs, so we were greatly interested in examining whether the Rap1 activity that regulates protrusion assembly also promotes directed migration. To do so, we exploited live imaging technology to analyze the migration behavior of individual BCs whose Rap1 activity was genetically modified with the indicated transgenes via a clonal induction system (Ito et al., 1997). To exclude the effect of delamination impairment caused by dysregulated Rap1 activity, only BCs completely detached from the anterior epithelium were selected for the following experiments. In the time-lapse video (Figure 6A; Movie S6), two randomly selected and RFP (red fluorescence protein)-labeled wild-type BCs continuously alternated their positions during migration, seemingly in competition to reach the oocyte first. Such dynamic competition is consistent with a previous report (Prasad and Montell, 2007) and can be easily appreciated from our plot of the migratory track (Figure S4B). The cell with high Rap1 activity tended to take the lead position, with the exception of a period (frames 20–49) when the migratory group underwent tumbling (Figures 6B and S4B; Movie S7). In an interesting case, one pair of BCs moved in parallel from the back to the front of the migrating cluster under Rap1 hyperactivity in combination with hpo hemizygosity (Figures 6C and S4B; Movie S8). Conversely, blocking Rap1 activity using Rap1N17 impaired forward migration, making a single cell mainly stay at the back of the cluster (Figures 6D and S4B; Movie S9). A 192-min movie reveals that only 10 frames spanning 27 min show occupancy of the cell in the front of the cluster (Movie S9, 02:27 to 03:04) without forward protrusion (Figure 6D, 2:36). These data indicate that the directional mobility of individual BCs greatly depends on Rap1 activity. To further assess our live imaging observations, we conducted a detailed quantification analysis on fixed samples in which the positions of clonally marked BCs were counted as leading, trailing, or aside (Figure 6E). Consistently, 63% of scored Rap1V12 cells resided in the leading edge of the cluster (i.e., higher than that of control cells [42%]), and the trailing edge frequency greatly increased from 23% in control cells to 57% in Rap1N17 cells (Figure 6E). Note that neither of these phenotypes was affected by loss of one hpo allele. We further tested whether the positional advantage was associated with cellular extension by simultaneously labeling actin protrusions and induced clones with RFP::Act. Our clonal analysis revealed that 83% of Rap1V12-expressing clones extended protrusions (Figures 6F and 6H) and that 74% of Rap1N17 clones did not exhibit protrusions (Figure 6G and 6H). Consequently, distribution biases and protrusion preferences demonstrate that greater Rap1 activity rendered BCs better at competing for a frontal position in the cluster and extending protrusions at the leading edge. Together, our results suggest an instructive role for Rap1 in promoting forward movement of individual cells, which may contribute to the persistence and directionality of the migrating cohort.
Figure 6. Rap1 Activity Promotes Directional Migration in Individual BCs.
(A–D) Fluorescence micrographs are exported from time-lapse videos of BC clusters (posterior to the right). Live imaging is applied to trace the migratory behavior of BCs marked by GFP::Act, and RFP is used to label cells randomly expressing the indicated transgenes. See Movies S6, S7, S8, and S9. Wild-type cells continuously alter their positions during migration (A). In rap1V12 overexpression (B), cells more frequently remain at the front of the cluster, whereas rap1N17-expressing cells spend more time at the back (D). In hpo hemizygous egg chambers, two rap1V12-expressing BCs migrate from the back to the font in parallel (C). Arrows indicate the direction of cell movement.
(E) Quantification of the frequency of clone distribution in BC clusters. The left schematic illustrates the definition for BC positions within the cluster.
(F–H) Rap1 activity promotes cell protrusion. 3D confocal micrographs of BCs expressing rap1v12 (F) or rap1N17 (G) (marked by arrows and RFP::Act [red]). GFP::Actin was used to mark BCs and to quantify the frequency of protrusions extending from Flip-out clones (H).
***p < 0.001, **p < 0.01. n means the number of egg chambers examined. Scale bars, 10 μm. The error bars represent SD.
To decipher the molecular mechanism of how Rap1 facilitates cell migration, we tested whether dysregulation of Rap1 overrides the Rac activity that controls protrusion-dependent movement in BCs (Wang et al., 2010) using Rac fluorescence resonance energy transfer (RacFRET). There was a stronger RacFRET signal at the front of migrating clusters in wild-type cells, but this asymmetry was disrupted upon overexpression of Rap1V12 or Rap1N17 (causing increased or decreased Rac activity, respectively) (Figure S5), indicating that Rap1 can act through Rac to activate actin polymerization. In addition, we scrutinized the expression of Ena, which promotes actin elongation but is negatively regulated by Hpo signaling (Lafuente et al., 2004; Lucas et al., 2013). Under Rap1V12 overexpression, clusters presented a 25% increase in Ena immunostaining intensity (Figures S5C–S5E and S5G), which was further enhanced to 75% in clusters lacking one hpo allele (Figures S5F and S5G). Epistasis analysis further demonstrated that a reduction of Ena expression by incorporating one null allele of ena (ena210) greatly suppressed the Rap1V12-induced migration defect in comparison with Rap1V12 alone (Figure S5H). Hence, we conclude that Rap1 acts like a positive regulator of Ena and that the Hpo pathway exerts the opposite effect.
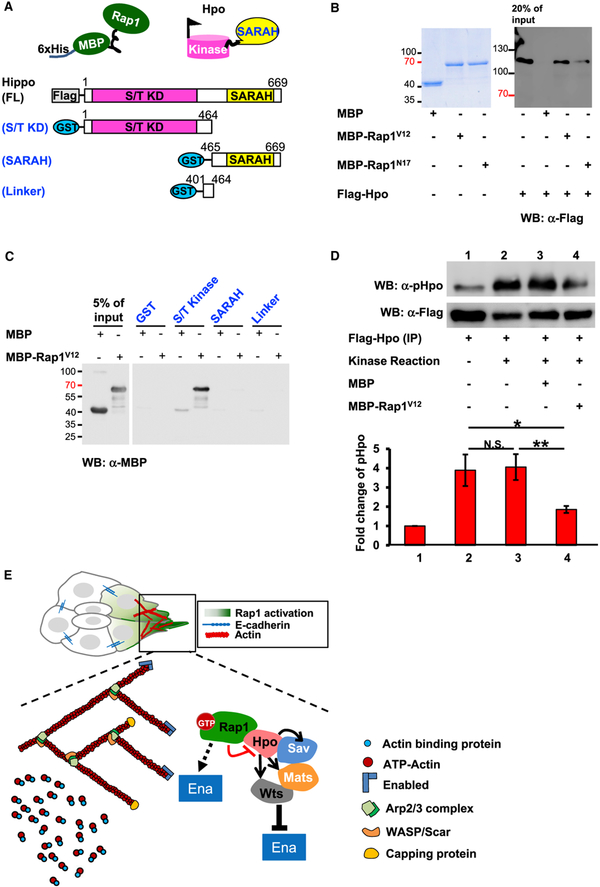
To further unveil how Rap1 and Hpo work together during BC migration, we performed a pull-down assay to test for possible interactions between these two proteins. We found that FLAGHpo bound to bacterially expressed Rap1 proteins fused with 6xHis-MBP (maltose binding protein) (Figures 7A and 7B). This association was stronger for Rap1V12 than Rap1N17 (Figure 7B). Hpo and its homologs, the mammalian Ste20-like kinases 1/2 (MST1/2), are highly conserved and share the same genomic structure, featuring an N-terminal kinase domain, a linker domain, and a C-terminal SARAH domain (Figure 7A; Meng et al., 2016). To determine which region is responsible for the Rap1-Hpo interaction, we generated hpo deletion constructs for a glutathione S-transferase (GST) pull-down assay (Figure 7A). Domain analysis indicates a direct association of Rap1V12 with the N-terminal kinase domain of Hpo (Figure 7C). Hpo/MST1/2 operates at the center of the Hpo pathway, integrating and transducing upstream stimuli with downstream effectors in a process that requires kinase activation via autophosphorylation of the phosphorylation loop at Thr195 (Hpo), Thr183 (MST1), and Thr180 (MST2) (Glantschnig et al., 2002; Jin et al., 2012; Meng et al., 2016). Because Hpo genetically and physically interacts with Rap1, we next tested whether Rap1V12 affects Hpo activity (as measured by autophosphorylation levels) and performed an in vitro kinase assay with purified recombinant proteins as substrates. Quantitative immunoblotting with an antibody against Hpo-pThr195 revealed an ~50% reduction in Hpo autophosphorylation in the presence of Rap1V12 (Figure 7D), suggesting that Rap1V12 antagonizes Hpo kinase activity. Based on our genetic and biochemical data, we propose that activation of Rap1 brings about a stronger interaction between it and Hpo, which impedes Hpo activation, and this, in turn, relieves the suppression of Ena by Hpo signaling, promoting actin polymerization (Figure 7E).
Figure 7. Rap1 Interacts with Hpo to Suppress Its Kinase Activity.
(A) A schematic diagram of full-length Hpo and its deletion constructs used in this study.
(B) SDS-PAGE gel stained with Coomassie blue showing the bacterially expressed MBP fusion proteins. Rap1 recombinant proteins are incubated in cell lysate containing FLAG-Hpo, and the precipitated complexes were subjected to western blotting (WB) with anti-FLAG antibody.
(C) WB with anti-MBP antibody revealing which of the Hpo domains pulls down MBP-Rap1V12.
(D) Hpo autophosphorylation is analyzed by immunoblot with anti-Hpo-pThr195 antibody, and the optical density ratio is normalized with FLAG-Hpo protein (α-FLAG). *p < 0.5, **p < 0.01. N.S. means not significant.
(E) Model of the Rap1-Hpo pathway in BC polarization. At the front of migratory clusters, GTP-bound Rap1 binds to Hpo to prevent its activation, relieving Ena/VASP suppression and promoting actin polymerization to advance BC migration. The error bars represent SD.
DISCUSSION
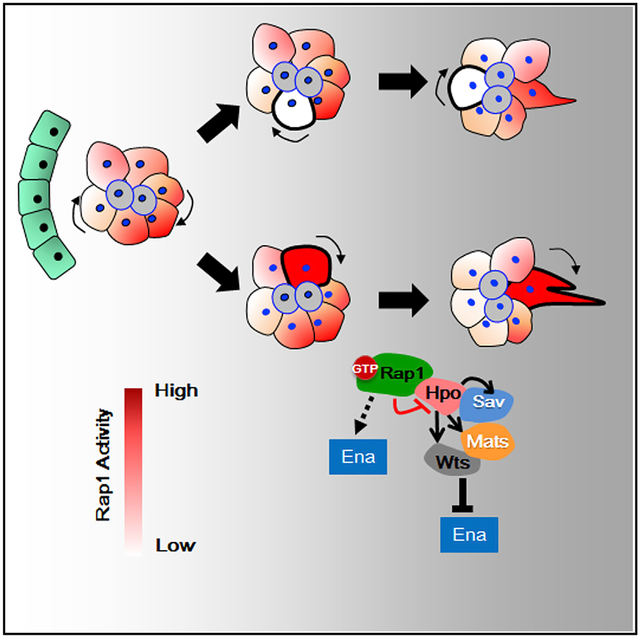
How cells induce group polarity to form directional protrusions is a long-standing and intensively studied question in collective cell migration. In particular, which molecular mechanism induces individual cells to compete for the leading position or to communicate with other members of the migratory cluster has yet to be established (Cai et al., 2014; Ellison et al., 2016; Ramel et al., 2013; Rappel, 2016; Xiang et al., 2016). Our finding that a Rap1 and Hpo complex polarizes actin protrusion provides a mechanism underlying group polarity in collective cell migration. First, the polarity of front end-enriched actin protrusions and inward contraction directed by non-muscle myosin II are continuously retained during migration. Rap1 hyperactivity disrupts the front/rear polarity of BCs, resulting in excessive misoriented protrusions. Furthermore, individual cells expressing Rap1V12 gain an advantage in protrusion extension and take the lead in the migrating cluster, whereas Rap1N17-expressing cells behave in the opposite way. As a consequence, BCs compete to move forward, making the cluster migrate in that direction. Thus, upon higher Rap1 activity, the “winner” cell extends protrusions and moves to the front, whereas the others, having lower activity, are outcompeted and lag behind. Second, our forward genetic screen reveals that hypoactivity of Hpo signaling enhances the Rap1V12-induced migration defect and protrusion numbers in either the backward or forward direction. Most intriguingly, our pull-down assays illustrate a strong physical interaction between Rap1V12 and Hpo protein, which significantly reduces Hpo activity. Suppression of Hpo signaling has long been demonstrated to cause abnormal F-actin polymerization, which, in turn, has been linked to enormous organ size in vertebrates and invertebrates (Fernández et al., 2011; Gaspar and Tapon, 2014; Sansores-Garcia et al., 2011; Yu et al., 2015). Manipulation of the actin cytoskeleton or stiffness of the extracellular matrix also affects nuclear localization of YAP (Yes-associated protein; i.e., the mammalian homolog of Yki; Aragona et al., 2013), which supports the role of Hpo signaling in sensing the actin architecture or local environment. Therefore, our results link Rap1 to the Hpo pathway and unfold a mechanism through which group polarity and protrusions are organized by attenuating Hpo signaling activity at the leading edge of a migrating cluster (Figure 7E).
In general, actin-based extension occurs at the front of motile cells and, subsequently, is followed by myosin II-mediated contraction, resulting in rhythmic cycles during locomotion (Lecuit et al., 2011). In external cue-directed cell movement, chemotaxis molecules bind to membrane receptors to activate Rho GTPase and its downstream effectors (Wiskott-Aldrich syndrome protein [WASP] family proteins), which interact with the Arp2/3 complex to initiate actin polymerization (Arthur et al., 2004; Kamp et al., 2016; Krause and Gautreau, 2014). Previous studies in the social ameba Dictyostelium discoideum uncovered that, in response to chemoattractant stimulation, spatially activated Rap1 binds to RacGEF1 at the front of cells to activate Rac1-dependent actin polymerization (Arthur et al., 2004; Liu et al., 2016; Mun and Jeon, 2012). Accumulated evidence suggests that, during single cell migration, Rap1 is a vital part of the polarity system that accounts for the asymmetric actin cytoskeleton. However, in terms of group migration, whether Rap1 plays the same role had not been investigated until our study, in which we demonstrate the conserved function of Rap1 in polarizing individual cells to move during collective migration.
Hpo signaling was first identified as being responsible for cell growth in Drosophila, but it has since been shown to be conserved in humans and to be pivotal in various fundamental aspects of biology, including embryonic development, tissue homeostasis, and disease (Meng et al., 2016; Pan, 2010; Wu et al., 2003). Intensive genetic screens and biochemical studies have explored the complexity of the Hpo network, but the mechanism of Hpo regulation remains less understood. The Hpo pathway can be initiated by phosphorylation of the activation loop of Hpo/MST, which is accomplished by transphosphorylation by Tao-1/TAO kinase or autophosphorylation via dimerization of the SARAH domains (Boggiano et al., 2011; Glantschnig et al., 2002; Jin et al., 2012; Poon et al., 2011). Active Hpo/MST2 subsequently autophosphorylates its linker domain to create docking sites for Mats/Mob1, whose interaction with Hpo/MST2 enables it to relay the kinase cascade to Warts/large tumor suppressor kinases 1/2 (LATS1/2) (Meng et al., 2016; Ni et al., 2015). Our results suggest a role for Rap1 as a Hpo signaling suppressor, impairing Hpo activation by binding to the Hpo kinase domain. It would be of great interest to investigate whether a similar mechanism operates in other systems, such as cancer metastasis, and which signals/stimuli activate Rap1 in such contexts.
In Drosophila, Hpo is activated by multiple upstream scaffold proteins such as Kib, Mer, and Ex, but our screen did not reveal any of them to have genetic interaction with Rap1 in BCs. We also examined whether Rap1V12 affects Yki transcriptional activity using a well-characterized reporter (ex-lacZ; Hamaratoglu et al., 2006) but observed no alteration in BCs even in combination with one hpo mutant allele (data not shown). In fact, neither wild-type nor constitutively active Yki alone delayed migration (Figure S3E). In light of our work and other independent research showing that depletion of yki does not affect BC migration (Lin et al., 2014), we conclude that Rap1 functions in the Hpo pathway independent of yki to induce biased protrusions and facilitate BC migration. Interestingly, although yki has no role in normal BC migration, we observed a strong genetic interaction between Rap1V12 and wild-type or hyperactive yki, which only arose in double gain-of-function scenarios (Figures S3B–S3E). This finding may represent a potential mechanism by which Yki/YAP participates in Rap1-mediated migration in an oncogenic context.
Over the past decade, several upstream regulators of Hpo signaling have been identified, including adhesion junction proteins, actin-binding proteins, molecules determining apical/basal/planar cell polarity, and proteins involved in cell matrix attachment. Most of these regulators are not required to regulate the core kinase activity of Hpo through Kib, Mer, or Ex. For example, loss of capping protein (which restricts actin polymerization) leads to a reduction of Hpo signaling activity and tissue outgrowth (Fernández et al., 2011; Sansores-Garcia et al., 2011). F-actin-destabilizing treatment in NIH 3T3 cells enhances Mst1 activity, which is sufficient to stabilize p21, a key cell cycle regulator (Densham et al., 2009). This evidence relating to actin cytoskeleton integrity and cell morphology has led to Hpo signaling being proposed as a mechanosensor for monitoring the local environment. However, it remains unclear whether Hpo core kinase components are directly regulated by actin architecture or whether additional adaptor proteins, such as actinbinding proteins or membrane-associated proteins, are required to transduce mechanical cues. Rap1 has been implicated in E-cadherin-dependent morphogenesis and integrin-mediated cell attachment in several tissues. Our results might imply that the stress generated from BC invasion through nurse cells can be transduced to interact with Rap1 through the membrane-tethering cadherin complex, the integrin/focal adhesion complex, or even a membrane-associated protein, suppressing Hpo activity and promoting protrusion.
Rap1 has been reported to interact with the mammalian Hpo, Mst1, through RapL; i.e., the downstream effector of Rap1 that regulates T lymphocyte polarization and migration (Ebisuno et al., 2010; Katagiri et al., 2006). Both these latter processes require the kinase activity of Mst1, activation of which relies on recruitment of this protein to the leading edge by Rap1 and RapL (Katagiri et al., 2006). Combined with our own evidence presented here, the association of Rap1 with Hpo appears to be an evolutionarily conserved mechanism for advancing cellular motion, but the effect of Rap1 on Hpo/MST1 varies in different contexts. Here we reveal a role for Rap1; rather than acting as a positive regulator of Hpo, it inhibits Hpo suppression of the actin polymerization involved in cell cluster migration.
In summary, our study provides cellular and molecular insights into how Rap1 regulates directional protrusions to achieve collective cell movement. In BCs, the intermolecular autophosphorylation essential for Hpo activation is hampered by its association with activated Rap1, which suppresses Hpo signaling activity and, thereby, promotes cellular protrusions.
MATERIALS AND METHODS
Drosophila Strains and Genetics
The following Drosophila strains were used in this study. slboGal4 was used to express transgenes, and UAS-lacZ served as a control in gain-of-function analysis. UAS-rap1V12 and UAS-rap1N17 were gifts from Dr. Ulrike Gaul. rap1rvB1 mutant stock was obtained from Dr. Nick Brown. Fly stock P[hsp70-flp],UAS-mCD8gfp;tubGal80,FRT2A and c306Gal4;rap1rvB1 were used to generate mutant clones to observe homozygous mutant phenotypes of rap1rvB1 in BCs. The following fly lines were obtained from the Bloomington Drosophila Stock Center: UAS-rfp::act, UAS-gfp::act, DrosDel kits of chromosome II, wtsx1, mats03077, savMI09089, exe1, merlin4, kibraEY03746, UAS-yki, UAS-ykiS168A and UAS-ykiS111A,S168A,S250A, and pUAS-dMST.FLAG/TM2 (dMST encodes Drosophila Hpo). UAS-rap1RNAiv20761 was obtained from Vienna Drosophila Resource Center (VDRC). Mutant hpo[42–47] was provided by Dr. Madhuri Kango-Singh. For visualizing myosin II contractile activity, Dr. Thomas Lecuit kindly provided sqh-MRLC::mCherry transgenic flies that expressed MRLC fused with mCherry under endogenous promoter control. slbo-gfp::act was provided by Dr. Mohit Prasad. rap1-rap1::gfp was obtained from Dr. Nick Brown. The P[hsp70-flp];AyGal4,UAS-rfp/TM6B flies used in a clonal induction technique (Flip-out) were provided by Dr. Henry Y. Sun.
Immunohistochemistry
Female ovarioles were dissected in S2 cell medium and then fixed on ice by standard protocol. For BC protrusion analysis, dissection was completed within 10 min, followed by room temperature fixation for 18 min. The following primary antibodies from the Developmental Studies Hybridoma Bank (DSHB) were used for immunostaining: mouse anti-Enabled, mouse anti-Armadillo, mouse anti-Eyes Absent, and mouse anti-Fasciclin. Alexa 488, Alexa 568, or Alexa 647 (Molecular Probes) secondary antibody was used at a dilution of 1:400. DAPI and phalloidin were used to detect nuclear DNA and F-actin. Images were obtained under Zeiss LSM 780-Meta confocal microscopy and Zeiss Observer D1. ZEN 2010 software and Axio Vision SE64 Rel. 4.8.2 software were used for image analyses.
Egg Chamber Culture for Time-Lapse Imaging Analysis
Our time-lapse imaging conditions are a modification of previously published culture conditions (Prasad et al., 2007). After dissection, egg chambers were cultured in a glass-bottomed dish (ibidi). Images were taken every 3 min by Zeiss Observer D1 microscopy and then processed using Axio Vision SE64Rel.4.8.2 software. All time-lapse movies were processed with the deconvolution algorithm of the AutoQuant X3 software. To track the dynamic behavior of GFP::Actin and MRLC-mCherry, NIS-Elements AR (Nikon) was employed to detect and record target signals frame by frame.
Clonal Induction (Flip-Out)
Female flies were subjected to transient heat shock (37°C) for 3 min. For live imaging, heat-shocked flies were kept at 18°C for 1 day and then switched to 25°C overnight before dissection. For fixed sample analysis, heat-shocked flies were kept at 18°C for 1 day and then switched to 29°C overnight before dissection.
Plasmid DNA Construction
For biochemical analysis, rap1, rap1V12, and rap1N17 DNA was amplified by PCR and cloned into the pAc5.1/V5-His A vector (Invitrogen) for expression in S2 cells. To generate MBP fusion proteins, rap1 genes were cloned into the pDB-6xHis-MBP vector. Different domain deletion constructs of Hpo were generated to express GST fusion proteins. The following constructs were amplified by PCR and cloned into the pGEX-5X-3 vector for GST fusion protein expression: Hpo (S/T kinase) (1,392 bp), Hpo SARAH (618 bp), and Hpo linker (192 bp).
S2 Cell Culture and Protrusion Dynamics Analysis
Drosophila S2 cells and S2-Mt-GFP-Act5C cells that express GFP::Act fusion protein were cultured in S2 cell medium at 25°C. Plasmid DNA, pAc5.1-rap1, pAc5.1-rap1V12, or pAc5.1-rap1N17 was transfected into cells to analyze protrusion dynamics. Each movie lasts 15 min, and the time interval is 30 s per frame.
Pull-Down Assays for Rap1 and Hpo
DNA constructs pDB-6xHis-MBP-Rap1, pDB-6xHis-MBP-Rap1V12, and pDB-6xHis-MBP-Rap1N17 were applied to express fusion proteins. Bacterial fusion proteins were purified by Ni2+ affinity chromatography. The expression of UAS-dMST-flag (UAS-flag-hpo) in larvae was induced by a Flip-out system. Fifty pairs of eye imaginal discs and fat bodies expressing UAS-dMST-flag were lysed with 50 mL lysis buffer. The cell lysate was further diluted with 550 mL immunoprecipitation (IP) buffer for the MBP pull-down assay. Protein extracts were incubated with bacterially expressed Rap1 proteins at 4°C for 3 hr and then washed with buffer before SDS-PAGE analysis. Mouse anti-FLAG antibody was used for western blot detection of FLAG-Hpo. GST-Hpo fusion proteins were prepared by a standard protocol. MBP-Rap1 proteins and GST-Hpo fusion proteins were incubated in binding buffer and then washed five times with 13 PBS buffer before SDS-PAGE analysis. Antibody against MBP was applied to detect MBP-Rap1 by western blotting.
In Vitro Kinase Assay
FLAG-Hpo proteins expressed by 75 pairs of eye imaginal discs and fat bodies were prepared and then immunoprecipitated by anti-FLAG M2 magnetic beads. FLAG-Hpo-bound beads were washed and then incubated with 1 μg MBP or MBP-Rap1V12 in 500 μL kinase buffer at 30°C for 4.5 hr. Immunoblotting was performed according to a standard protocol with mouse anti-phospho-MST1 (Thr183)/MST2 (Thr180).
Supplementary Material
Highlights.
Polarized actomyosin protrusions in border cells are regulated by Rap1
Genetic screen reveals that Rap1 interacts with Hpo pathway in collective cell migration
Activated Rap1 binds to Hpo to suppress its autophosphorylation
Rap1 activation relieves suppression of Ena by the Hpo pathway to promote actin assembly
ACKNOWLEDGMENTS
We thank Madhuri Kango-Singh, Thomas Lecuit, Ulrike Gaul, Nick Brown, the Fly Core in Taiwan, and the Bloomington Stock Center for providing fly stocks. We are grateful to Jocelyn A. McDonald, Y. Henry Sun, Chang-Shi Chen, and Shian-Jang Yan for their valuable suggestions. We appreciate the Instrument Development Center of NCKU for access to and technical support for a Carl Zeiss LSM 780 laser-scanning microscope. This work was supported by grants to A.C.-C.J. by the Ministry of Science and Technology, Taiwan, ROC (MOST 106–2311-B-006–002, MOST 105–2311-B-006–003, and MOST104–2311-B-006–002) and the Headquarters of University Advancement at the National Cheng Kung University granted by the Ministry of Education, Taiwan, ROC.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, five figures, and nine movies and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.01.080.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, and Piccolo S (2013). A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 154, 1047–1059. [DOI] [PubMed] [Google Scholar]
- Artemenko Y, Lampert TJ, and Devreotes PN (2014). Moving towards a paradigm: common mechanisms of chemotactic signaling in Dictyostelium and mammalian leukocytes. Cell. Mol. Life Sci 71, 3711–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur WT, Quilliam LA, and Cooper JA (2004). Rap1 promotes cell spreading by localizing Rac guanine nucleotide exchange factors. J. Cell Biol 167, 111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner B, Harjes P, Ishimaru S, Heke M, Fan HQ, Qin Y, Van Aelst L, and Gaul U (2003). The AF-6 homolog canoe acts as a Rap1 effector during dorsal closure of the Drosophila embryo. Genetics 165, 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boggiano JC, Vanderzalm PJ, and Fehon RG (2011). Tao-1 phosphorylates Hippo/MST kinases to regulate the Hippo-Salvador-Warts tumor suppressor pathway. Dev. Cell 21, 888–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D, Chen SC, Prasad M, He L, Wang X, Choesmel-Cadamuro V, Sawyer JK, Danuser G, and Montell DJ (2014). Mechanical feedback through E-cadherin promotes direction sensing during collective cell migration. Cell 157, 1146–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Zhang N, Xie R, Wang W, Cai J, Choi KS, David KK, Huang B, Yabuta N, Nojima H, et al. (2015). Homeostatic control of Hippo signaling activity revealed by an endogenous activating mutation in YAP. Genes Dev. 29, 1285–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherfils J, and Zeghouf M (2013). Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev 93, 269–309. [DOI] [PubMed] [Google Scholar]
- Choi W, Harris NJ, Sumigray KD, and Peifer M (2013). Rap1 and Canoe/afadin are essential for establishment of apical-basal polarity in the Drosophila embryo. Mol. Biol. Cell 24, 945–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creasy CL, Ambrose DM, and Chernoff J (1996). The Ste20-like protein kinase, Mst1, dimerizes and contains an inhibitory domain. J. Biol. Chem 271, 21049–21053. [DOI] [PubMed] [Google Scholar]
- Densham RM, O’Neill E, Munro J, König I, Anderson K, Kolch W, and Olson MF (2009). MST kinases monitor actin cytoskeletal integrity and signal via c-Jun N-terminal kinase stress-activated kinase to regulate p21Waf1/Cip1 stability. Mol. Cell. Biol 29, 6380–6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchek P, Somogyi K, Jékely G, Beccari S, and Rørth P (2001). Guidance of cell migration by the Drosophila PDGF/VEGF receptor. Cell 107, 17–26. [DOI] [PubMed] [Google Scholar]
- Ebisuno Y, Katagiri K, Katakai T, Ueda Y, Nemoto T, Inada H, Nabekura J, Okada T, Kannagi R, Tanaka T, et al. (2010). Rap1 controls lymphocyte adhesion cascade and interstitial migration within lymph nodes in RAPL-dependent and -independent manners. Blood 115, 804–814. [DOI] [PubMed] [Google Scholar]
- Ellison D, Mugler A, Brennan MD, Lee SH, Huebner RJ, Shamir ER, Woo LA, Kim J, Amar P, Nemenman I, et al. (2016). Cell-cell communication enhances the capacity of cell ensembles to sense shallow gradients during morphogenesis. Proc. Natl. Acad. Sci. USA 113, E679–E688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández BG, Gaspar P, Brás-Pereira C, Jezowska B, Rebelo SR, and Janody F (2011). Actin-Capping Protein and the Hippo pathway regulate F-actin and tissue growth in Drosophila. Development 138, 2337–2346. [DOI] [PubMed] [Google Scholar]
- Gaspar P, and Tapon N (2014). Sensing the local environment: actin architecture and Hippo signalling. Curr. Opin. Cell Biol 31, 74–83. [DOI] [PubMed] [Google Scholar]
- Glantschnig H, Rodan GA, and Reszka AA (2002). Mapping of MST1 kinase sites of phosphorylation. Activation and autophosphorylation. J. Biol. Chem 277, 42987–42996. [DOI] [PubMed] [Google Scholar]
- Haeger A, Wolf K, Zegers MM, and Friedl P (2015). Collective cell migration: guidance principles and hierarchies. Trends Cell Biol. 25, 556–566. [DOI] [PubMed] [Google Scholar]
- Hamaratoglu F, Willecke M, Kango-Singh M, Nolo R, Hyun E, Tao C, Jafar-Nejad H, and Halder G (2006). The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol 8, 27–36. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, and Pan D (2005). The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122, 421–434. [DOI] [PubMed] [Google Scholar]
- Huelsmann S, Hepper C, Marchese D, Knöll C, and Reuter R (2006). The PDZ-GEF dizzy regulates cell shape of migrating macrophages via Rap1 and integrins in the Drosophila embryo. Development 133, 2915–2924. [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, and Yamamoto D (1997). The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development 124, 761–771. [DOI] [PubMed] [Google Scholar]
- Janetopoulos C, Borleis J, Vazquez F, Iijima M, and Devreotes P (2005). Temporal and spatial regulation of phosphoinositide signaling mediates cytokinesis. Dev. Cell 8, 467–477. [DOI] [PubMed] [Google Scholar]
- Jin Y, Dong L, Lu Y, Wu W, Hao Q, Zhou Z, Jiang J, Zhao Y, and Zhang L (2012). Dimerization and cytoplasmic localization regulate Hippo kinase signaling activity in organ size control. J. Biol. Chem 287, 5784–5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp ME, Liu Y, and Kortholt A (2016). Function and Regulation of Heterotrimeric G Proteins during Chemotaxis. Int. J. Mol. Sci 17, E90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri K, Imamura M, and Kinashi T (2006). Spatiotemporal regulation of the kinase Mst1 by binding protein RAPL is critical for lymphocyte polarity and adhesion. Nat. Immunol 7, 919–928. [DOI] [PubMed] [Google Scholar]
- Kim JH, Cho A, Yin H, Schafer DA, Mouneimne G, Simpson KJ, Nguyen KV, Brugge JS, and Montell DJ (2011). Psidin, a conserved protein that regulates protrusion dynamics and cell migration. Genes Dev. 25, 730–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox AL, and Brown NH (2002). Rap1 GTPase regulation of adherens junction positioning and cell adhesion. Science 295, 1285–1288. [DOI] [PubMed] [Google Scholar]
- Krause M, and Gautreau A (2014). Steering cell migration: lamellipodium dynamics and the regulation of directional persistence. Nat. Rev. Mol. Cell Biol 15, 577–590. [DOI] [PubMed] [Google Scholar]
- Lafuente EM, van Puijenbroek AA, Krause M, Carman CV, Freeman GJ, Berezovskaya A, Constantine E, Springer TA, Gertler FB, and Boussiotis VA (2004). RIAM, an Ena/VASP and Profilin ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion. Dev. Cell 7, 585–595. [DOI] [PubMed] [Google Scholar]
- Lecuit T, Lenne PF, and Munro E (2011). Force generation, transmission, and integration during cell and tissue morphogenesis. Annu. Rev. Cell Dev. Biol 27, 157–184. [DOI] [PubMed] [Google Scholar]
- Lee MR, and Jeon TJ (2012). Cell migration: regulation of cytoskeleton by Rap1 in Dictyostelium discoideum. J. Microbiol 50, 555–561. [DOI] [PubMed] [Google Scholar]
- Lin TH, Yeh TH, Wang TW, and Yu JY (2014). The Hippo pathway controls border cell migration through distinct mechanisms in outer border cells and polar cells of the Drosophila ovary. Genetics 198, 1087–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Lacal J, Veltman DM, Fusetti F, van Haastert PJ, Firtel RA, and Kortholt A (2016). A Ga-Stimulated RapGEF Is a Receptor-Proximal Regulator of Dictyostelium Chemotaxis. Dev. Cell 37, 458–472. [DOI] [PubMed] [Google Scholar]
- Lucas EP, Khanal I, Gaspar P, Fletcher GC, Polesello C, Tapon N, and Thompson BJ (2013). The Hippo pathway polarizes the actin cytoskeleton during collective migration of Drosophila border cells. J. Cell Biol 201, 875–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor R, and Etienne-Manneville S (2016). The front and rear of collective cell migration. Nat. Rev. Mol. Cell Biol 17, 97–109. [DOI] [PubMed] [Google Scholar]
- McDonald JA, Pinheiro EM, Kadlec L, Schupbach T, and Montell DJ (2006). Multiple EGFR ligands participate in guiding migrating border cells. Dev. Biol 296, 94–103. [DOI] [PubMed] [Google Scholar]
- Meng Z, Moroishi T, and Guan KL (2016). Mechanisms of Hippo pathway regulation. Genes Dev. 30, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun H, and Jeon TJ (2012). Regulation of actin cytoskeleton by Rap1 binding to RacGEF1. Mol. Cells 34, 71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munjal A, and Lecuit T (2014). Actomyosin networks and tissue morphogenesis. Development 141, 1789–1793. [DOI] [PubMed] [Google Scholar]
- Nelson CM (2016). Collective migration in tissues. Mol. Biol. Cell 27, 877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L, Zheng Y, Hara M, Pan D, and Luo X (2015). Structural basis for Mob1-dependent activation of the core Mst-Lats kinase cascade in Hippo signaling. Genes Dev. 29, 1416–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Keefe DD, Gonzalez-Niño E, Edgar BA, and Curtiss J (2012). Discontinuities in Rap1 activity determine epithelial cell morphology within the developing wing of Drosophila. Dev. Biol 369, 223–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, and Irvine KD (2008). In vivo regulation of Yorkie phosphorylation and localization. Development 135, 1081–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D (2010). The hippo signaling pathway in development and cancer. Dev. Cell 19, 491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon CL, Lin JI, Zhang X, and Harvey KF (2011). The sterile 20-like kinase Tao-1 controls tissue growth by regulating the Salvador-Warts-Hippo pathway. Dev. Cell 21, 896–906. [DOI] [PubMed] [Google Scholar]
- Poukkula M, Hakala M, Pentinmikko N, Sweeney MO, Jansen S, Mattila J, Hietakangas V, Goode BL, and Lappalainen P (2014). GMF promotes leading-edge dynamics and collective cell migration in vivo. Curr. Biol 24, 2533–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad M, and Montell DJ (2007). Cellular and molecular mechanisms of border cell migration analyzed using time-lapse live-cell imaging. Dev. Cell 12, 997–1005. [DOI] [PubMed] [Google Scholar]
- Prasad M, Jang AC, Starz-Gaiano M, Melani M, and Montell DJ (2007). A protocol for culturing Drosophila melanogaster stage 9 egg chambers for live imaging. Nat. Protoc 2, 2467–2473. [DOI] [PubMed] [Google Scholar]
- Prasad M, Wang X, He L, Cai D, and Montell DJ (2015). Border Cell Migration: A Model System for Live Imaging and Genetic Analysis of Collective Cell Movement. Methods Mol. Biol 1328, 89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel D, Wang X, Laflamme C, Montell DJ, and Emery G (2013). Rab11 regulates cell-cell communication during collective cell movements. Nat. Cell Biol 15, 317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappel WJ (2016). Cell-cell communication during collective migration. Proc. Natl. Acad. Sci. USA 113, 1471–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauzi M, Lenne PF, and Lecuit T (2010). Planar polarized actomyosin contractile flows control epithelial junction remodelling. Nature 468, 1110–1114. [DOI] [PubMed] [Google Scholar]
- Ren F, Zhang L, and Jiang J (2010). Hippo signaling regulates Yorkie nuclear localization and activity through 14-3-3 dependent and independent mechanisms. Dev. Biol 337, 303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rørth P (2011). Whence directionality: guidance mechanisms in solitary and collective cell migration. Dev. Cell 20, 9–18. [DOI] [PubMed] [Google Scholar]
- Sansores-Garcia L, Bossuyt W, Wada K, Yonemura S, Tao C, Sasaki H, and Halder G (2011). Modulating F-actin organization induces organ growth by affecting the Hippo pathway. EMBO J. 30, 2325–2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurmans S, Polizzi S, Scoumanne A, Sayyed S, and Molina-Ortiz P (2015). The Ras/Rap GTPase activating protein RASA3: from gene structure to in vivo functions. Adv. Biol. Regul 57, 153–161. [DOI] [PubMed] [Google Scholar]
- Siekhaus D, Haesemeyer M, Moffitt O, and Lehmann R (2010). RhoL controls invasion and Rap1 localization during immune cell transmigration in Drosophila. Nat. Cell Biol 12, 605–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhusha VV, Tsukita S, and Oda H (1999). Actin dynamics in lamellipodia of migrating border cells in the Drosophila ovary revealed by a GFP-actin fusion protein. FEBS Lett. 445, 395–401. [DOI] [PubMed] [Google Scholar]
- Wang X, He L, Wu YI, Hahn KM, and Montell DJ (2010). Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat. Cell Biol 12, 591–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YC, Khan Z, and Wieschaus EF (2013). Distinct Rap1 activity states control the extent of epithelial invagination via a-catenin. Dev. Cell 25, 299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MJ, Artemenko Y, Cai WJ, Iglesias PA, and Devreotes PN (2014). The directional response of chemotactic cells depends on a balance between cytoskeletal architecture and the external gradient. Cell Rep. 9, 1110–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Huang J, Dong J, and Pan D (2003). hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell 114, 445–456. [DOI] [PubMed] [Google Scholar]
- Xiang W, Zhang D, and Montell DJ (2016). Tousled-like kinase regulates cytokine-mediated communication between cooperating cell types during collective border cell migration. Mol. Biol. Cell 27, 12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhao B, and Guan KL (2015). Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell 163, 811–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.