ABSTRACT
Background
A high-quality reference genome is an essential tool for applied and basic research on arthropods. Long-read sequencing technologies may be used to generate more complete and contiguous genome assemblies than alternate technologies; however, long-read methods have historically had greater input DNA requirements and higher costs than next-generation sequencing, which are barriers to their use on many samples. Here, we present a 2.3 Gb de novo genome assembly of a field-collected adult female spotted lanternfly (Lycorma delicatula) using a single Pacific Biosciences SMRT Cell. The spotted lanternfly is an invasive species recently discovered in the northeastern United States that threatens to damage economically important crop plants in the region.
Results
The DNA from 1 individual was used to make 1 standard, size-selected library with an average DNA fragment size of ∼20 kb. The library was run on 1 Sequel II SMRT Cell 8M, generating a total of 132 Gb of long-read sequences, of which 82 Gb were from unique library molecules, representing ∼36× coverage of the genome. The assembly had high contiguity (contig N50 length = 1.5 Mb), completeness, and sequence level accuracy as estimated by conserved gene set analysis (96.8% of conserved genes both complete and without frame shift errors). Furthermore, it was possible to segregate more than half of the diploid genome into the 2 separate haplotypes. The assembly also recovered 2 microbial symbiont genomes known to be associated with L. delicatula, each microbial genome being assembled into a single contig.
Conclusions
We demonstrate that field-collected arthropods can be used for the rapid generation of high-quality genome assemblies, an attractive approach for projects on emerging invasive species, disease vectors, or conservation efforts of endangered species.
Background
In September 2014, Lycorma delicatula (Hemiptera: Fulgoridae), commonly referred to as the spotted lanternfly, was first detected in the United States in Berks County, Pennsylvania. L. delicatula is a highly polyphagus phloem-feeding insect native to Asia that is documented to feed upon >65 plant species [1,2]. Because this insect was an invasive that damaged grapevines and tree fruit in South Korea in the mid-2000s [3, 4], its potential to cause economic damage was known. Shortly after it was detected in the USA, the Pennsylvania Department of Agriculture established a quarantine zone surrounding the site of first detection. The invasion likely began with a shipment of stone that harbored egg masses, as L. delicatula lays inconspicuous egg masses seemingly indiscriminately on a wide variety of surfaces (e.g., tree bark, automobiles, railcars, shipping pallets), contributing to the potential for abrupt and distant spread. Since that time, the L. delicatula quarantine zone has expanded from an area of 50 mi2 to >9,400 mi2, and as of the time of publication, has spread throughout southeastern Pennsylvania, encompassing 13 counties, with detections in surrounding states. Entering the fall, during the period of mass adult flights, the spread of this insect will likely increase further. While this pest has huge potential for spread and increased impact, essentially nothing is known at the genomic level about this species or any Fulgorid species, and there is a need to develop resources rapidly for this pest to support development of management and control practices.
A high-quality genome as a foundation to understand arthropod biology can be a powerful tool to combat invasions and disease-carrying vectors, aid in conservation, and many other fields (e.g., see [5–8]). To this end, large-scale initiatives are underway to comprehensively catalog the genomes of many arthropod species, including the i5K initiative aiming to sequence and analyze the genomes of 5,000 arthropod species [9–11] associated with the Darwin Tree of Life Project [12] and the Earth BioGenome Project [13]. Within the context of the Earth BioGenome Project, the United States Department of Agriculture Agricultural Research Service (USDA-ARS) Ag100Pest initiative is focused on rapidly deciphering the genomes of 100 insect species destructive to crops and livestock, projected to have profound bioeconomic impacts to agriculture and livestock industries, as well as habitat and species conservation. Despite many hemipterans being both direct pests as well as vectors of plant diseases, overall, genomic resources are lacking in this order relative to other insect groups, with the exception of the Aphidoidea [14].
Arthropod genome assembly projects face unique challenges stemming from their small body size and high heterozygosity. Owing to the limited quantities of genomic DNA that can be extracted from a small-bodied animal, researchers may pool multiple individuals, such as by generating next-generation sequencing libraries of different insert sizes, each from a different individual [10,14], or by pooling multiple individuals for a single long-read sequencing library from an iso-female laboratory strain [15–19] or laboratory colony [5,20]. Pooling introduces multiple haplotypes into the sample and complicates the assembly and curation process [20], and while this issue may be ameliorated by inbreeding, it is not always an option for organisms that cannot be cultured in the laboratory. Moreover, genomic regions with high heterozygosity tend to be assembled into more fragmented contigs [21], so computational methods specifically developed for heterozygous samples are needed [22–24]. Recently, high-quality long-read assemblies have been published for a single diploid mosquito (Anopheles coluzzii) [25] and a single haploid honeybee (Apis mellifera) [7]. Despite both species having relatively small genomes (<300 Mb), multiple Pacific Biosciences (PacBio) SMRT Cells were needed for sufficient sequencing coverage (N = 3 for mosquito, N = 29 for bee).
Here, we demonstrate the sequencing and high-quality de novo assembly of a 2.25 Gb genome from a single, field-collected spotted lanternfly (Lycorma delicatula, NCBI:txid130591) insect, requiring only 1 sequencing library and 1 SMRT Cell sequencing run on the Sequel II System. The genome assembly is highly contiguous, complete, and accurate, and it resolves the maternal and paternal haplotypes over 60% of the genome. In addition to the lanternfly genome, the assembly immediately provided complete genomes from 2 of the organism's bacterial endosymbionts. The approach outlined here can be applied to field-collected arthropods or other taxa for which the rapid generation of high-quality contig-level genome assemblies is critical, such as for invasive species or for conservation efforts of endangered species.
Results
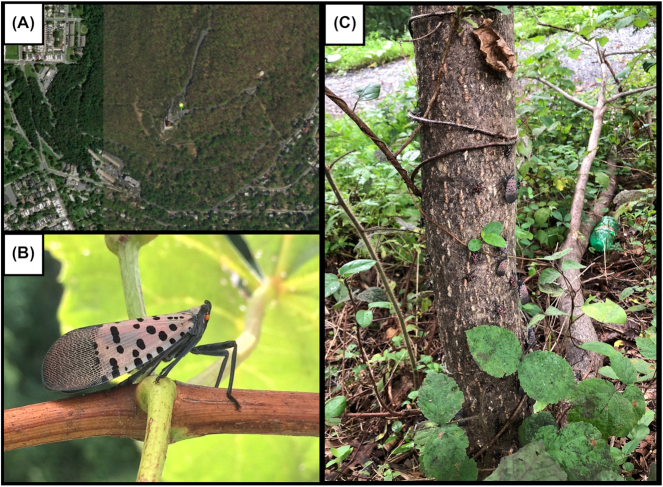
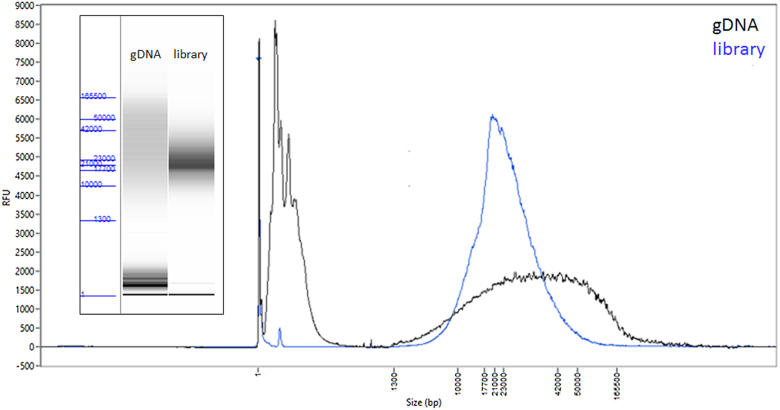
We extracted DNA from a single female L. delicatula collected from the main trunk of Ailanthus altissima (tree of heaven) in Reading, Berks County, Pennsylvania, USA (N 40 20.189, W 75 54.283) on 26 August 2018 (Fig. 1). L. delicatula is known to harbor several endosymbionts in specialized bacteriocytes, predominantly in the distal end of the insect abdomen; to avoid a high proportion of these symbionts in the sequencing, DNA was extracted from the head and thorax regions of the insect only (see Materials and Methods for details). While more recently developed single arthropod assemblies have significantly lowered DNA input requirements [25], here the amount of extracted genomic DNA was more plentiful because of the relatively larger size, allowing for sufficient DNA for a standard library preparation with size selection, resulting in a ∼20 kb average insert size sequencing library (Fig. 2). The library was sequenced on the Sequel II System with 1 SMRT Cell 8M, yielding 131.6 Gb of total sequence contained in 5,639,857 reads, with a polymerase read length N50 of 41.7 kb and insert (subread) length N50 of 22.3 kb (Fig. S1).
Figure 1.
w. (A) Location of specimen collection (green marker), near the Reading Pagoda on Mt. Penn (Reading, Berks County, Pennsylvania, USA [40.33648 N, 75.90471 W]); (B) adult female Lycorma delicatula; (C) the host Ailanthus altissima tree (tree of heaven) from which the female adult sample was collected on 26 August 2018. Late nymph stage and adults can be seen covering the trunk of this host tree.
Figure 2.
Lycorma delicatula input DNA and resulting library. FEMTO Pulse traces and “gel” images (inset) of the genomic DNA (gDNA) input (black) and the final library (blue) before sequencing.
The genome was assembled with FALCON-Unzip, a diploid assembler that captures haplotype variation in the sample [22]. A single subread per zero-mode waveguide (ZMW) was used in assembly for a total of 82.4 Gb of sequence (36-fold coverage for a 2.3 Gb genome). Reads longer than 8 kb were selected as “seed reads” for pre-assembly, a process of error correction using alignment and consensus calling with the PacBio data. Pre-assembled reads totaled 55.5 Gb of sequence (24-fold) with mean (N50) read length of 10.8 kb (15.2 kb) (Fig. S1). The draft FALCON assembly consisted of 5,158 contigs with N50 length of 1.38 Mb and total assembly size of 2.43 Gb. We screened this draft assembly for bacterial symbiont or contaminant DNA (see Methods) and identified 2 contigs originating from microbial symbionts, Sulcia muelleri and Vidania fulgoroideae, respectively, 2 known bacterial symbionts of planthoppers [26]. These contigs were removed from the final curated assembly and analyzed separately (see below).
The FALCON-Unzip module was applied to phase and haplotype-resolve the assembly. The unzipped assembly was then polished twice to increase base-level accuracy of the contigs. The first polishing round used phased reads that were assigned to haplotypes during FALCON-Unzip. The second round of polishing with Arrow used all subreads mapped to the concatenated primary contigs plus haplotigs. For both polishing rounds, all subreads were used, including multiple passes from a single library molecule. The resulting assembly consisted of 4,209 primary contigs comprising 2.40 Gb with contig N50 of 1.42 Mb. A total of 1.25 Gb of the assembly “unzipped” into 10,103 haplotigs of mean (N50) length 76.9 kb (152 kb) (Table 1).
Table 1.
Spotted lanternfly de novo genome assembly stats for the FALCON-Unzip and curated assemblies
| Assembly version | FALCON-Unzip | Curated assembly |
|---|---|---|
| Primary assembly size | 2.395 Gb | 2.252 Gb |
| Number of primary contigs | 4,209 | 2,927 |
| Contig length N50 | 1.423 Mb | 1.520 Mb |
| Haplotig assembly size (proportion of primary length) | 1.249 Gb (52%) | 1.349 Gb (60%) |
| Number of haplotigs | 10,103 | 10,652 |
| Haplotig N50 | 185.5 kb | 178.1 kb |
| BUSCO complete | 96.8% | 96.8% |
| BUSCO duplicate | 3.3% | 2.4% |
Assembly contiguity and BUSCO completeness stats are shown after FALCON-Unzip, and after curation to recategorize duplicated haplotypes in the primary contigs and removal of repetitive and redundant haplotigs and bacterial contigs. For complete BUSCO stats see Table S1.
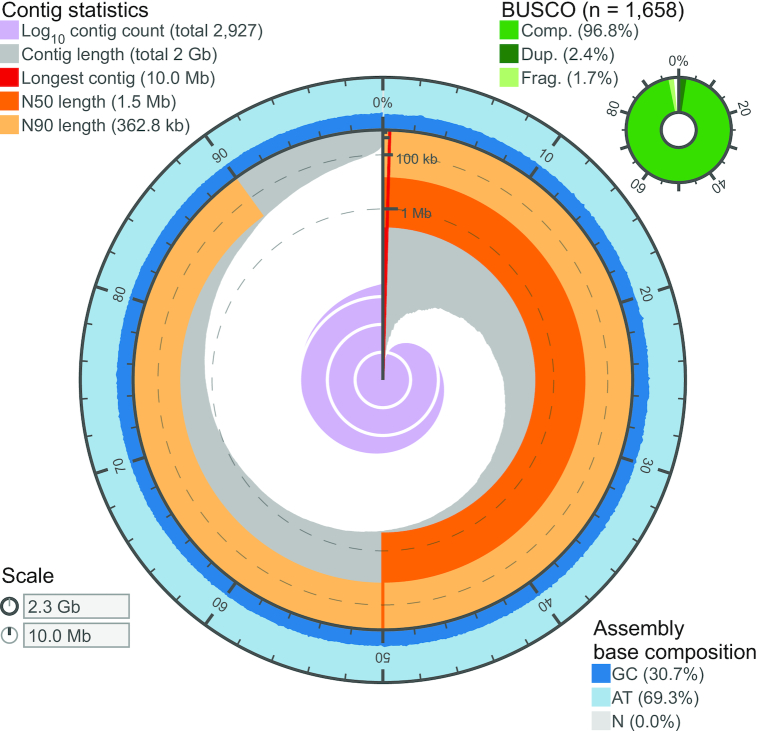
While FALCON-Unzip is designed to resolve haplotypes in non-inbred organisms, some homologous regions of the genome with high heterozygosity may be assembled on separate primary contigs. Our goal was to generate a haploid reference sequence, so we performed additional curation to both recategorize duplicated haplotypes from the primary set as haplotigs and remove repetitive, artifactual, and redundant haplotigs (see Methods). The final curated assembly consisted of 2,927 primary contigs of total length 2.252 Gb with contig N50 1.520 Mb. The alternate haplotypes spanned 60% of the primary contig length: 10,652 haplotigs comprised a total of 1.349 Gb with an N50 length of 178.1 kb (Fig. S1). A visualization of the assembly contiguity and completeness was generated using assembly-stats [27] and is presented in Fig. 3 and Table 1.
Figure 3.
Assembly visualization. The contiguity and completeness of the L. delicatula genome assembly is visualized as a circle, with the full circle representing the full assembly length of ∼2.3 Gb. The longest contig was 10.0 Mb, and the assembly has uniform GC content throughout, with very few contigs <50 kb in length.
Despite attempting to avoid bacteriocyte-associated internal symbionts by excluding the abdomen during DNA extraction, 2 contigs that were identified as circular and of microbial origin were present in the assembly. Contig 001940F is a complete representation of the Candidatus S. muelleri obligate symbiont, 212,195 bp in length with a guanine-cytosine (GC) content of 23.8% and sequenced at ∼46.6× coverage of subreads. A second contig 5193, designated to be circular in the FALCON assembly, was identified as a complete representation of the Candidatus V. fulgoroideae obligate symbiont genome. This genome was 126,523 bp in length with a GC content of 19.15%. Contig names are relative to the FALCON assembly prior to running Unzip, which is available in the supporting dataset on the Ag Data Commons (see Availability of Supporting Data and Materials). More details on these symbionts will be provided in an future publication.
We assessed additional aspects of genome assembly completeness and sequence accuracy with analysis of conserved genes, and an orthogonal method to estimate the genome size using read coverage depth. First, using the “insecta_odb9” BUSCO gene set collection [28], we observed that >96% of the 1,658 genes were complete and >96% occurred as single copies (Tables 1 and S1). Concordantly with the recategorization of initial primary contigs into haplotigs by Purge Haplotigs, the percentage of duplicated genes decreased from 3.3% to 2.4%. As an additional evaluation, we aligned to the primary assembly the core Drosophila melanogaster CEGMA gene set, resulting in 416 alignments (91%) and a mean alignment length of 86%, and with >96.6% of alignments showing no frame shift–inducing indels. Using read coverage (see Fig. S2 and Materials and Methods), we see a single unimodal coverage peak in the primary haploid assembly and also generated a genome size estimate of ∼2.75 Gb, which is slightly larger than our curated primary assembly, consistent with telomeric, centromeric, and ribosomal DNA satellite regions being refractory to genome assembly [29–31].
Discussion and Conclusions
We sequenced and assembled a high-quality reference genome for a single wild-caught spotted lanternfly (Lycorma delicatula), a Fulgorid planthopper species invasive in the northeastern USA. Previous planthopper genome projects required 100–5,000 inbred individuals and ≥16 different sequencing libraries [32–34] (Table 2). We generated long-read sequence data sufficient for de novo assembly from a single sequencing library, run on 1 PacBio SMRT Cell. Despite the fact that the genome of our planthopper species is 2–4 times larger compared with the 3 previous described planthopper genomes, it is 13–63 times more contiguous. The new workflow presented here improves on many aspects of previous approaches for generating arthropod genome assemblies, and the genomes of their endosymbionts. These include sample (i) collection strategies, (ii) library preparation efforts and sequencing time, (iii) assembly considerations, and (iv) endosymbiont genome capture and are discussed in detail below.
Table 2.
Comparison of the spotted lanternfly genome assembly with previously described planthopper species assemblies, highlighting the improvements with regard to the required number of insect individuals, sequencing libraries, assembly sizes, and contiguity qualities
| Species | Nilaparvata lugens (2014)a | Sogatella furcifera (2017)b | Laodelphax striatellus (2017)c | Lycorma delicatula (this work) |
|---|---|---|---|---|
| Number of individuals (source) | ∼5,000 (F13 from inbred line) | ∼120 (F6 from inbred line) | ∼100 (F22 from inbred line) | 1 (field-collected) |
| Number of sequencing libraries | 16 (+fosmid libraries) | 17 | 47 | 1 |
| Assembly size | 1.14 Gb | 0.72 Gb | 0.54 Gb | 2.25 Gb |
| Contig N50 | 24 kb | 71 kb | 118 kb | 1,520 kb |
Collection strategies
The strategy of performing single-insect genome assemblies has several advantages. First, it dispenses with the requirement of inbred laboratory colonies, which may take months or even years to establish, can be expensive to maintain, and are impractical or impossible for many species. Second, by sampling field-collected animals, genetic variation can be more accurately characterized for local populations, without the risk of adaptation to laboratory culture [35] or loss of heterozygosity [36]. For invasive pests, methods for artificial rearing often do not exist and there is a desire to rapidly generate foundational data on these pests, so direct sequencing of wild specimens is advantageous. The ability to generate genomes de novo from field-collected arthropods makes high-quality genomes accessible for many more species. This approach also enables comprehensive comparisons of genetic diversity within and between populations without the bias from previous single reference-based studies [16] and allows generation of a diploid genome assembly that more closely captures the organism's biology [23].
Library preparation and sequencing
The methods described here for DNA extraction, library preparation, and sequencing are straightforward and rapid, using established kits and leveraging the higher throughput of the Sequel II System to generate sufficient sequencing coverage with just 1 SMRT Cell and 30 hours of sequencing run time. The need for multiple libraries from several individuals or pool fractions, or for covering different insert size ranges is eliminated. These improvements potentially allow a genome project, with infrastructure optimization, to be completed in <1 week (estimating 1 day each for DNA extraction, library preparation, sequencing, and data analysis) and can be carried out by individual laboratories rather than requiring large consortia that were typical of previous genome assembly efforts. All steps in the workflow are amenable to automation to accommodate larger sample numbers in a high-throughput manner. The rapid nature of the workflow will allow not only for the generation of a single reference-grade genomic resource but for the comprehensive genomic monitoring of species before or throughout a field season, and for rapid testing of intervention strategies.
Assembly
An additional advantage to single-insect assemblies is that genome assembly for a diploid sample is algorithmically simpler than for a sample of many pooled individuals, each of which may contribute up to 2 unique haplotypes. Several de novo assembly methods are available for diploid samples [22,23,37] and have been broadly applied taxonomically [5,38]. Recent work indicates that assembly of high-heterozygosity samples is more accurate than for inbred samples when parental data can be used to partition long-read sequence data by haplotype, an approach called trio-binning [23,39]. When trio samples are not available, long-range contact data may be leveraged in combination with long-read assemblies to enhance haplotype phasing [40]. This represents a reversal in the paradigm for high-quality references in insect genomics [23], where one now should target outcrossed or highly heterozygous (wild) individuals, rather than inbreeding to reduce polymorphism and avoid complications caused by heterozygosity that may arise using previous assembly methods. While, in the past, PacBio-only assemblies may have contained a significant number of insertion/deletion (indel) errors detected in gene models [41], advances in sequencing chemistry, read lengths, and improved polishing methods (such as use of all subreads from each sequencing ZMW) translate to similar consensus accuracy compared to the highest quality published human genome generated from single-molecule data [39,41,42]. We anticipate further improvements of the genome assembly upon collection and integration of RNA-sequencing data, and the application of scaffolding methods.
Endosymbionts and metagenomic approaches for symbiosis
Although our method of DNA extraction was intended to avoid structures in the lanternfly that house bacterial symbionts, our results included the complete genomes of 2 known planthopper endosymbionts, S. muelleri and V. fulgoroideae. Early work by Müller revealed that the cells (bacteriocytes) housing endosymbionts in planthoppers are organized into organs, or bacteriomes, and that these structures often display complex morphologies and occupy a variety of positions within an insect's abdomen [43]. Dissections of L. delicatula reveal the presence of complex, string-like bacteriome structures positioned around the alimentary canal that are large enough to be visible to the naked eye. As such, it is not surprising that some bacteriome tissue was included with the thorax as it was separated from the abdomen for extraction. Despite the attempt to avoid these symbionts, their complete genomes were recovered at sufficient coverage to be assembled into single contigs from a host-targeted DNA extraction. This approach allows for high-quality assemblies in a metagenomic context, with the long reads and robust assembly strategy allowing for clear discrimination of the microbial symbionts. This dramatically simplifies strategies for symbiont sequencing: rather than dissection and pooling of bacteriocytes from the host, a shotgun metagenomics strategy can be used to not only recover the symbiont genome but also a draft reference of the host, at a similar cost to targeted methods. Additional follow-up shotgun approaches could yield discovery of novel or unexpected microbes associated with the host.
Genomic applications for control
High-quality reference genomes for L. delicatula and its associated endosymbionts represent invaluable resources for this dangerous invasive, about which little is known of its basic biology. Because obligate symbionts in phloem-feeding insects typically provide nutritional benefit to their hosts [44,45], the symbiont genomes offer insight into nutritional requirements and basic metabolic functioning of L. delicatula. They also offer additional potential opportunities for control. For example, obligate endosymbionts are typically vertically transferred from female to offspring transovarially. In L. delicatula, development of the female reproductive system appears to require substantial time and resources. Females typically eclose as adults in late July and feed voraciously over several months and accumulate abdominal mass, before mass flights and egg laying in October–November. During this time, bacterial symbionts must proliferate and get transferred to developing ovarioles. This may present a time window for potential disruption of symbiont transmission, which would represent a control strategy that is highly specific to L. delicatula. Alternatively, RNA inhibition (RNAi) strategies used for control often target highly conserved genes in the insect's genome that perform vital cellular functions. Inhibition of one of these core gene functions is lethal to the insect. Targeting such highly conserved genes, however, reduces the species specificity of this approach. Obligate bacterial endosymbionts, however, only occur within the host insects with whom they have coevolved over tens to hundreds of millions of years and, as such, provide highly species-specific genomic targets for control with RNAi [46]. Additionally, the assembly presented here is being used immediately as a foundation for population genetic studies to track the movement and potentially source new detections of this invader as its range expands in years to come.
Conclusions
The genome assembly presented here can be used as a foundation for further assembly and curation efforts with long-range scaffolding technologies such as Bionano Genomics [47,48] and/or Hi-C [20, 49–51] to generate a reference-quality, chromosome-scale genome scaffold representation. Similarly, full-length RNA-sequencing (Iso-Seq) [52,53] or other RNA-sequencing data types can be applied, with the assembly serving as a mapping reference, for gene and other functional element annotation. While these follow-up efforts are currently underway in our laboratories, we wanted to make this initial, high-quality draft genome assembly available to provide immediate resources to the scientific community working to battle this invasive pest and restrict its expansion and impact on the breadth of tree fruit, nut, and horticultural crops that are at risk. Furthermore, this approach, of 1 insect, 1 library, and 1 PacBio Sequel II SMRT Cell, can hopefully be used as a model for pursuing high-quality, single-individual diploid assemblies across organisms that were previously unobtainable using other approaches.
Materials and Methods
Sample collection and DNA isolation
A cohort of L. delicatula females were collected off the trunk of their preferred host A. altissima (tree of heaven) in Reading, Berks County, Pennsylvania, USA (40.33648 N, 75.90471 W) on 26 August 2018. Individuals were snap-frozen in liquid nitrogen in the field and stored at −80°C until processing. L. delicatula were extracted individually, by first cutting off the abdomen, and grinding the head and thorax in liquid nitrogen to a powder. High molecular weight DNA was extracted using a modification of a “salt-out” protocol [54]. Briefly, the ground material was resuspended in 1.8 mL of lysis buffer (10 mM Tris-HCl, 400 mM NaCl, and 100 mM EDTA, pH 8.0) and 120 µL of 10% sodium dodecyl sulfate (SDS) and 300 uL of Proteinase K solution (1 mg/mL Proteinase K, 1% SDS, and 4 mM EDTA, pH 8.0) was added. The sample was incubated overnight at 37°C. To remove RNA, 40 µL of 20 mg/mL RNAse A was added and the solution was incubated at room temperature for 15 minutes. A total of 720 µL of 5 M NaCl was added and mixed gently through inversion. The sample was centrifuged at 4°C at 1500g for 20 minutes. A wide-bore pipette tip was then used to transfer the supernatant, avoiding any precipitated protein material, to a new tube and DNA was precipitated through addition of 3.6 mL of 100% ethanol. The DNA was pelleted at 4°C at 6,250g for 15 minutes, and all ethanol was decanted from the tube. The DNA pellet was allowed to dry and then was resuspended in 150 µL of TE. Initial quality and quantity of DNA was determined using a Qubit fluorometer and evaluating DNA on a 1% agarose genome on a Pippin Pulse using a 14-hour 5–80 kb separation protocol. DNA was sent to Pacific Biosciences (Menlo Park, California) for library preparation and sequencing.
Library preparation and sequencing
Genomic DNA quality was evaluated using the FEMTO Pulse automated pulsed-field capillary electrophoresis instrument (Agilent Technologies, Wilmington, DE) and showed a DNA smear, with majority >20 kb (Fig. 2), appropriate for SMRTbell library construction without shearing.
One SMRTbell library was constructed using the SMRTbell Express Template Prep kit 2.0 (Pacific Biosciences). Briefly, 5 µg of the genomic DNA was carried into the first enzymatic reaction to remove single-stranded overhangs followed by treatment with repair enzymes to repair any damage that may be present on the DNA backbone. After DNA damage repair, ends of the double-stranded fragments were polished and subsequently tailed with an A-overhang. Ligation with T-overhang SMRTbell adapters was performed at 20°C for 60 minutes. Following ligation, the SMRTbell library was purified with 1X AMPure PB beads. The size distribution and concentration of the library were assessed using the FEMTO Pulse and dsDNA BR reagents Assay kit (Thermo Fisher Scientific, Waltham, MA). Following library characterization, 3 µg was subjected to a size selection step using the BluePippin system (Sage Science, Beverly, MA) to remove SMRTbells ≤15 kb. After size selection, the library was purified with 1X AMPure PB beads. Library size and quantity were assessed using the FEMTO Pulse (Fig. 2), and the Qubit Fluorometer and Qubit dsDNA HS reagents Assay kit.
Sequencing primer v2 and Sequel II DNA Polymerase were annealed and bound, respectively, to the final SMRTbell library. The library was loaded at an on-plate concentration of 30 pM using diffusion loading. SMRT sequencing was performed using a single 8M SMRT Cell on the Sequel II System with Sequel II Sequencing Kit, 1800-minute movies, and Software v6.1.
Assembly
Data were assembled with FALCON-Unzip [22] using pb-falcon version 0.2.6 from the bioconda pb-assembly metapackage version 0.0.4 with the following configuration:
genome_size = 2 500 000 000; seed_coverage = 30; length_cutoff = -1; length_cutoff_pr = 10 000; pa_daligner_option = -e0.8 -l1000 -k18 -h70 -w8 -s100; ovlp_daligner_option = -k24 -h1024 -e.92 -l1000 -s100; pa_HPCdaligner_option = -v -B128 -M24; ovlp_HPCdaligner_option = -v -B128 -M24; pa_HPCTANmask_option = -k18 -h480 -w8 -e.8 -s100; pa_HPCREPmask_option = -k18 -h480 -w8 -e.8 -s100; pa_DBsplit_option = -x500 -s400; ovlp_DBsplit_option = -s400; falcon_sense_option = –output-multi –min-idt 0.70 –min-cov 3 –max-n-read 100 –n-core 4; overlap_filtering_setting = –max-diff 100 –max-cov 200 –min-cov 3 –n-core 24; polish_include_zmw_all_subreads = true
The assembly was polished once as part of the FALCON-Unzip workflow and a second time by mapping all subreads to the concatenated reference with pbmm2 v1.1.0 (“pbmm2 align $REF $BAM $MOVIE.aln.bam –sort -j 48 -J 48”) and consensus calling with Arrow with gcpp v 0.0.1-e2ea76a (“gcpp -j 4 -r $REF -o $OUT.$CONTIG.fasta $BAM -w “$W””). Both tools are available through bioconda [55]. We screened the primary assembly for duplicate haplotypes using Purge Haplotigs (bioconda v1.0.4) [56]. Purge Haplotigs identifies candidate haplotigs in the primary contigs using PacBio read coverage depth and contig alignments. To determine the coverage thresholds, we mapped only the unique subreads to the primary contigs rather than all subreads. This resulted in more distinct modes in the coverage histogram (data not shown). A fasta file of unique subreads was generated with the command “python -m falcon_kit.mains.fasta_filter median movie.subreads.fasta > movie.median.fasta”, which is available in the pb-assembly software. We used coverage thresholds of 5, 25, and 10 and default parameters except “-s 90” (diploid coverage maximum for auto-assignment of contigs as suspect haplotigs). We recategorized 1,269 primary contigs as haplotigs (total length 141.8 Mb) and discarded 12 as artifactual (total length 869 kb) and 201 as repeats (total length 19.1 Mb). A perl script [57] was used to rename the haplotigs using the FALCON-Unzip nomenclature so that each haplotig can be easily associated with a primary contig. Following renaming, we aligned each haplotig to its associate primary contig, chained sub-alignments in 1 dimension, and removed redundant haplotigs whose alignment to the primary was completely contained within another haplotig [40]. This process removed 518 haplotigs totaling 22.6 Mb.
Contaminant and symbiont screening
All primary contigs from the draft FALCON assembly were searched using DIAMOND BLASTx against the NCBI nr database (downloaded 8 April 2019) [58], and the subsequent hits were used to assign taxonomic origin of each contig using a least common ancestor assignment for each contig utilizing MEGAN 6.15.2 Community Edition with the longReads LCA Algorithm and readCount assignment mode [59]. Any contigs that were identified as microbial were flagged and removed from the final assembly. To avoid assignment of contigs as microbial when a microbial gene may have horizontally transferred to the insect, any potentially microbial contigs were screened for presence of BUSCO insect genes and retained if a BUSCO was present on the contig.
Genome assembly evaluation
To assess the completeness of the curated assembly, we searched for conserved, single copy genes using BUSCO (Benchmarking Universal Single-Copy Orthologs, BUSCO, RRID:SCR_015008) v3.0.2 [28] with the “insecta_odb9” database. In addition, we evaluated assembly completeness and accuracy against the Drosophila melanogaster CEGMA gene set [60], using a previously described script [61]. A visualization of the assembly contiguity and completeness was generated using assembly-stats [] and are presented in Fig. 3 and Table 1.
We also applied an orthogonal method to estimate the genome size by dividing the total base pairs of unique subreads (82.4 Gb) by the modal read coverage (30-fold, Fig. S2) of the PacBio data. This calculation is possible because PacBio data has minimal sequencing bias across DNA content and sequence complexity [62, 63]. Unique subreads were mapped to the curated primary assembly (“minimap2 -ax map-pb $REF $QRY –secondary = no” [64], read depth was estimated with “bedtools genomecov” [65], and a histogram was visualized in R [66].
Availability of Supporting Data and Materials
Raw data and final assembly for this project were submitted to NCBI under BioProject PRJNA540533; sample is described in BioSample SAMN11546444; and the SRA accession for raw PacBio subreads (fastq formatted) is SRR9005207. Supporting data for this article have been submitted to the AgDataCommons, including polished FALCON assembly, polished FALCON-Unzip assembly, final curated assembly and placement file, microbial symbiont assemblies, and associated metadata [67]. Additional supporting data and materials are also available in the GigaScience GigaDB database [68].
Additional Files
Figure S1: Cumulative distribution of subread lengths for Sequel II 8M SMRT Cell of 15-kb size-selected library. Data were bioinformatically filtered prior to assembly to remove reads shorter than 500 bp and retain 1 subread per library molecule (see Materials and Methods).
Figure S2: Coverage depth histogram. PacBio reads mapped to curated primary contigs shows unimodal coverage with peak centered at 30-fold.
Table S1: Full summary from BUSCO analysis of primary contigs, using the “insecta_odb9” gene set (total = 1,658), after different stages of assembly and curation.
Chris Wheat, PhD -- 6/27/2019 Reviewed
Shanlin Liu -- 7/9/2019 Reviewed
Shanlin Liu -- 8/28/2019 Reviewed
Abbreviations
BLAST: Basic Local Alignment Search Tool; BUSCO: Benchmarking Universal Single-Copy Orthologs; CEGMA: Core Eukaryote Gene Mapping Approach; EDTA: ethylenediaminetetraacetic acid; Gb: gigabase pairs; GC: guanine-cytosine; gDNA: genomic DNA; kb: kilobase pairs; Mb: megabase pairs; MEGAN: MEtaGenome ANalyzer; NCBI: National Center for Biotechnology Information; PacBio: Pacific Biosciences; RNAi: RNA inhibition; SDS: sodium dodecyl sulfate; SMRT: single-molecule real-time; SRA: Sequence Read Archive; USDA-ARS: United States Department of Agriculture Agricultural Research Service; ZMW: zero-mode waveguide.
Competing interests
S.B.K., C.C.L., P.B., and J.K. are full-time employees at Pacific Biosciences, a company developing single-molecule sequencing technologies.
Funding
Funding for A.K.C., B.C., B.S., K.H., and S.M.G. provided by USDA-ARS. Funding to J.U. from USDA APHIS-PPQ Cooperative Agreement #AP18PPQS&T00C221, USDA NIFA Hatch Funding #1004464, and College of Agriculture, Penn State University. Computational analyses were performed on the USDA-ARS Moana HPC (Hilo, Hawaii) and the USDA-ARS CERES HPC (Ames, Iowa) supported by USDA-ARS as well as other HPC systems. This project is a component of the Ag100Pest Genomics Initiative at USDA-ARS. Map image was created using ArcGIS® software by Esri with imagery in the public domain (USDA FSA). USDA is an equal opportunity employer. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA.
Authors' contributions
S.B.K. performed assembly and curation. S.M.G. performed genomic extraction and assembly curation. J.U. performed sample collection. P.B. and C.C.L. performed library preparation and sequencing. J.K., S.B.K., S.M.G., and J.U. wrote the manuscript. S.B.K., P.B., A.K.C., B.C., B.S., K.H., J.K., and S.M.G. conceived of and designed the project.
ACKNOWLEDGEMENTS
We thank Q. Wu (University of Science and Technology of China) and F. Cui (Institute of Zoology, Chinese Academy of Sciences) for sharing technical details about their previous genome assembly studies. We thank Angela Kauwe for assistance in the wetlab at USDA-ARS Hilo and Erica Smyers for providing the photograph for Fig. 1B.
References
- 1. Dara SK, Barringer L, Arthurs SP. Lycorma delicatula (Hemiptera: Fulgoridae): A new invasive pest in the United States. J Integr Pest Manag. 2015;6(1):20. [Google Scholar]
- 2. Parra G, Moylett H, Bulluck R. Technical working group summary report: Spotted lanternfly, Lycorma delicatula (White, 1845). 2018. http://agriculture.pa.gov/Plants_Land_Water/PlantIndustry/Entomology/spotted_lanternfly/research/Documents/SLF%20TWG%20Report%20020718%20final.pdf. Accessed 27 Apr 2019. [Google Scholar]
- 3. Han JM, Kim H, Lim EJ, et al.. Lycorma delicatula (Hemiptera: Auchenorrhyncha: Fulgoridae: Aphaeninae) finally, but suddenly arrived in Korea. Entomol Res. 2008;38:281–6. [Google Scholar]
- 4. Kim JG, Lee E-H, Seo Y-M, et al.. Cyclic behavior of Lycorma delicatula (Insecta: Hemiptera: Fulgoridae) on host plants. J Insect Behav. 2011;24:423–35. [Google Scholar]
- 5. Matthews BJ, Dudchenko O, Kingan SB, et al.. Improved reference genome of Aedes aegypti informs arbovirus vector control. Nature. 2018;563:501–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McKenna DD, Scully ED, Pauchet Y, et al.. Genome of the Asian longhorned beetle (Anoplophora glabripennis), a globally significant invasive species, reveals key functional and evolutionary innovations at the beetle–plant interface. Genome Biol. 2016;17:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wallberg A, Bunikis I, Pettersson OV, et al.. A hybrid de novo genome assembly of the honeybee, Apis mellifera, with chromosome-length scaffolds. BMC Genomics. 2019,20:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang K, Li P, Gao Y, et al.. De novo genome assembly of the white-spotted flower chafer (Protaetia brevitarsis). Gigascience. 2019;8(4), doi: 10.1093/gigascience/giz019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. i5K Consortium. The i5K Initiative: Advancing arthropod genomics for knowledge, human health, agriculture, and the environment. J Hered. 2013;104:595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thomas GWC, Dohmen E, Hughes DST, et al.. The genomic basis of arthropod diversity. bioRxiv. 2018;382945. [Google Scholar]
- 11. Poelchau M, Childers C, Moore G, et al.. The i5k Workspace@NAL—enabling genomic data access, visualization and curation of arthropod genomes. Nucleic Acids Res. 2015;43:D714–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Darwin Tree of Life Project. https://www.sanger.ac.uk/news/view/genetic-code-66000-uk-species-be-sequenced. Accessed 16 Apr 2019. [Google Scholar]
- 13. Lewin HA, Robinson GE, Kress WJ, et al.. Earth BioGenome Project: Sequencing life for the future of life. Proc Natl Acad Sci U S A. 2018;115:4325–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Panfilio KA, Vargas Jentzsch IM, Benoit JB, et al.. Molecular evolutionary trends and feeding ecology diversification in the Hemiptera, anchored by the milkweed bug genome. Genome Biol. 2019;20:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Berlin K, Koren S, Chin C-S, et al.. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat Biotechnol. 2015;33:623. [DOI] [PubMed] [Google Scholar]
- 16. Chakraborty M, Emerson JJ, Macdonald SJ, et al.. Structural variants exhibit allelic heterogeneity and shape variation in complex traits. bioRxiv. 2018:419275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chakraborty M, VanKuren NW, Zhao R, et al.. Hidden genetic variation shapes the structure of functional elements in Drosophila. Nat Genet. 2018;50:20–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim KE, Peluso P, Babayan P, et al.. Long-read, whole-genome shotgun sequence data for five model organisms. Sci Data. 2014;1:140045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miller DE, Staber C, Zeitlinger J, et al.. Highly contiguous genome assemblies of 15 Drosophila species generated using nanopore sequencing. G3 (Bethesda). 2018;8:3131–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ghurye J, Koren S, Small ST, et al.. A chromosome-scale assembly of the major African malaria vector Anopheles funestus. Gigascience. 2019;8(6), doi: 10.1093/gigascience/giz063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Holt RA, Subramanian GM, Halpern A, et al.. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–49. [DOI] [PubMed] [Google Scholar]
- 22. Chin CS, Peluso P, Sedlazeck FJ, et al.. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods. 2016;13:1050–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Koren S, Rhie A, Walenz BP, et al.. De novo assembly of haplotype-resolved genomes with trio binning. Nat Biotechnol. 2018;36:1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vinson JP, Jaffe DB, O'Neill K, et al.. Assembly of polymorphic genomes: Algorithms and application to Ciona savignyi. Genome Res. 2005;15:1127–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kingan SB, Heaton H, Cudini J, et al.. A high-quality de novo genome assembly from a single mosquito using PacBio sequencing. Genes. 2019;10:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Urban JM, Cryan JR. Two ancient bacterial endosymbionts have coevolved with the planthoppers (Insecta: Hemiptera: Fulgoroidea). BMC Evol Biol. 2012;12:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Challis R. rjchallis/assembly-stats 17.02. Zenodo 2014, doi: 10.5281/zenodo.322347. [DOI] [Google Scholar]
- 28. Waterhouse RM, Seppey M, Simao FA, et al.. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol. 2018;35(3): 543–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chang C-H, Larracuente AM. Heterochromatin-enriched assemblies reveal the sequence and organization of the Drosophila melanogaster Y chromosome. Genetics. 2019;211:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Doležel J, Čížková J, Šimková H, et al.. One major challenge of sequencing large plant genomes is to know how big they really are. Int J Mol Sci. 2018;19:3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wright FA, Lemon WJ, Zhao WD, et al.. A draft annotation and overview of the human genome. Genome Biol. 2001;2:research0025.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang L, Tang N, Gao X, et al.. Genome sequence of a rice pest, the white-backed planthopper (Sogatella furcifera). Gigascience. 2017;6(1), doi: 10.1093/gigascience/giw004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xue J, Zhou X, Zhang C-X, et al.. Genomes of the rice pest brown planthopper and its endosymbionts reveal complex complementary contributions for host adaptation. Genome Biol. 2014;15:521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhu J, Jiang F, Wang X, et al.. Genome sequence of the small brown planthopper, Laodelphax striatellus. GigaScience. 2017;6(12), doi: 10.1093/gigascience/gix109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hoffmann AA, Ross PA. Rates and patterns of laboratory adaptation in (mostly) insects. J Econ Entomol. 2018;111:501–9. [DOI] [PubMed] [Google Scholar]
- 36. Nowak C, Vogt C, Diogo JB, et al.. Genetic impoverishment in laboratory cultures of the test organism Chironomus riparius. Environ Toxicol Chem. 2007;26:1018–22. [DOI] [PubMed] [Google Scholar]
- 37. Weisenfeld NI, Kumar V, Shah P, et al.. Direct determination of diploid genome sequences. Genome Res. 2017;27:757–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Low WY, Tearle R, Bickhart DM, et al.. Chromosome-level assembly of the water buffalo genome surpasses human and goat genomes in sequence contiguity. Nat Commun. 2019;10:260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wenger AM, Peluso P, Rowell WJ, et al.. Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat Biotechnol. 2019, doi: 10.1038/s41587-019-0217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kronenberg ZN, Rhie A, Koren S, et al.. Extended haplotype phasing of de novo genome assemblies with FALCON-Phase. bioRxiv. 2019:327064, doi: 10.1101/327064. [DOI] [Google Scholar]
- 41. Watson M, Warr A. Errors in long-read assemblies can critically affect protein prediction. Nat Biotechnol. 2019;37:124–6. [DOI] [PubMed] [Google Scholar]
- 42. Koren S, Walenz BP, Berlin K, et al.. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27:722–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Müller HJ. Die Symbiose der Fulgoroiden (Homoptera Cicadina). Zoologica 1940, 98(1):1-110. [Google Scholar]
- 44. Bennett GM, Moran NA. Small, smaller, smallest: The origins and evolution of ancient dual symbioses in a phloem-feeding insect. Genome Biol Evol. 2013;5:1675–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Douglas AE. Phloem-sap feeding by animals: Problems and solutions. J Exp Bot. 2006;57:747–54. [DOI] [PubMed] [Google Scholar]
- 46. Chung SH, Jing X, Luo Y, et al.. Targeting symbiosis-related insect genes by RNAi in the pea aphid-Buchnera symbiosis. Insect Biochem Mol Biol. 2018;95:55–63. [DOI] [PubMed] [Google Scholar]
- 47. Jiao Y, Peluso P, Shi J, et al.. Improved maize reference genome with single-molecule technologies. Nature. 2017;546:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kronenberg ZN, Fiddes IT, Gordon D, et al.. High-resolution comparative analysis of great ape genomes. Science. 2018;360:eaar6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bickhart DM, Rosen BD, Koren S, et al.. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat Genet. 2017;49:643–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. VanBuren R, Wai CM, Pardo J, et al.. Exceptional subgenome stability and functional divergence in allotetraploid teff, the primary cereal crop in Ethiopia. bioRxiv. 2019:580720, doi: 10.1101/580720. [DOI] [Google Scholar]
- 51. McKernan K, Helbert Y, Kane LT et al.. Cryptocurrencies and zero mode wave guides: An unclouded path to a more contiguous Cannabis sativa L. genome assembly. OSF Preprints. 2018, doi: 10.17605/OSF.IO/N98GP. [DOI] [Google Scholar]
- 52. Workman RE, Myrka AM, Wong GW, et al.. Single-molecule, full-length transcript sequencing provides insight into the extreme metabolism of the ruby-throated hummingbird Archilochus colubris. Gigascience. 2018;7(3), 10.1093/gigascience/giy009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhou Y, Zhao Z, Zhang Z, et al.. Isoform sequencing provides insight into natural genetic diversity in maize. Plant Biotechnol J. 2019;17:1473–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. DNA Extraction from Single Insects. https://support.10xgenomics.com/de-novo-assembly/sample-prep/doc/demonstrated-protocol-dna-extraction-from-single-insects. Accessed 9 Mar 2019. [Google Scholar]
- 55. Bioconda. https://github.com/PacificBiosciences/pbbioconda. Accessed 9 Mar 2019. [Google Scholar]
- 56. Roach MJ, Schmidt SA, Borneman AR. Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinformatics. 2018;19:460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. adapt_PurgeHaplotigs_for_FALCONPhase. https://github.com/skingan/adapt_PurgeHaplotigs_for_FALCONPhase. Accessed 9 Mar 2019. [Google Scholar]
- 58. Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59. [DOI] [PubMed] [Google Scholar]
- 59. Huson DH, Beier S, Flade I, et al.. MEGAN community Edition - Interactive Exploration and analysis of large-scale microbiome sequencing data. PLoS Comput Biol. 2016;12:e1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Korf Lab. http://korflab.ucdavis.edu/datasets/cegma/core_genome/D.melanogaster.aa. Accessed 9 Mar 2019. [Google Scholar]
- 61. Korlach J, Gedman G, Kingan SB, et al.. De novo PacBio long-read and phased avian genome assemblies correct and add to reference genes generated with intermediate and short reads. GigaScience. 2017;6(10), doi: 10.1093/gigascience/gix085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Chaisson MJP, Wilson RK, Eichler EE. Genetic variation and the de novo assembly of human genomes. Nat Rev Genet. 2015;16:627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ross MG, Russ C, Costello M, et al.. Characterizing and measuring bias in sequence data. Genome Biol. 2013;14:R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Quinlan AR. BEDTools: The Swiss-Army tool for genome feature analysis. Curr Protoc Bioinformatics. 2014;47:11.12.11–11.12.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. https://www.r-project.org/. [Google Scholar]
- 67. Kingan S, Urban J, Lambert C. Data from: A high-quality genome assembly from a single, field-collected spotted lanternfly (Lycorma delicatula) using the PacBio Sequel II System. Ag Data Commons. 2019. 10.15482/USDA.ADC/1503745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kingan SB, Urban J, Lambert CC, et al.. Supporting data for “A high-quality genome assembly from a single, field-collected spotted lanternfly (Lycorma delicatula) using the PacBio Sequel II System.”. GigaScience Database. 2019. 10.5524/100650. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Chris Wheat, PhD -- 6/27/2019 Reviewed
Shanlin Liu -- 7/9/2019 Reviewed
Shanlin Liu -- 8/28/2019 Reviewed