ABSTRACT
CKLF-like MARVEL transmembrane domain containing 6 (CMTM6) plays a crucial role in the stability of the programmed death-ligand 1 (PD-L1). However, there has been no previous study of CMTM6 in non-small cell lung cancer (NSCLC) and its association with PD-L1 has not been confirmed. The aim of this study was to investigate the expression of CMTM6 and PD-L1 and to confirm their predictive roles for anti-PD-1 therapy in non-small cell lung cancer. CMTM6 and PD-L1 immunohistochemical expressions were evaluated in 35 advanced, treatment-refractory NSCLC patients who received PD-1 inhibitor therapy. The correlation between CMTM6 and PD-L1 expression was also determined based on immunohistochemistry and RNA-sequencing data obtained from The Cancer Genome Atlas (TCGA) database. CMTM6 expression was positively correlated with PD-L1 expression in immunohistochemical data (Pearson’s r = 0.342 and p = .044). A positive correlation was also identified in the mRNA expression data. Using receiver operating characteristic curves, the levels of CMTM6 and PD-L1 expression which provided the best distinguishing point between responder versus non-responder to PD-1 inhibitors were 70 and 75 H-scores, respectively. The patients in the PD-1 inhibitor responder group had higher CMTM6 expressions in univariate logistic regression analysis (odds ratio (OR) = 5.333, p = .037). However, PD-L1 expression was not associated with response to PD-1 inhibitor (p = .288). In multivariate analysis, CMTM6 was also found to be an independent predictor of the response to PD-1 inhibitors (OR = 6.226, p = .032). CMTM6 expression can be a promising predictor useful for therapeutic decision-making regarding PD-1 inhibitors.
KEYWORDS: Non-small cell lung cancer, CMTM6, PD-L1, biomarker
Introduction
In the field of lung cancer, monoclonal antibodies targeting the programmed death 1 (PD-1) receptor and its ligand (PD-L1) have demonstrated clinical responses and survival improvement and the PD-1 inhibitors, nivolumab and pembrolizumab, have been approved by the FDA for the treatment of advanced non-small cell lung cancer (NSCLC).1–3 Despite the impressive treatment outcomes for PD-1/PD-L1 inhibitors in NSCLC, it is noteworthy that only 15–20% of patients respond to the therapy. Previous results have highlighted the need for better predictive biomarkers.4
Although high PD-L1 expression has been associated with higher objective response rates to PD-1/PD-L1 inhibitors,1,5 conflicting results have also been reported. Durable responses to PD-1/PD-L1 inhibitors have been reported in NSCLC patients with low or no PD-L1 expression.6,7 Various predictors, including tumor-infiltrating lymphocytes,8 tumor mutational burden,9 and immune-related gene signatures,10 have been reported to supplement the predictive role of PD-L1, however, the predictors have not been validated yet.
More recently, two studies showed that CKLF-like MARVEL transmembrane domain containing 6 (CMTM6) was identified as an important regulator of the PD-L1 protein.11,12 Inhibition of CMTM6 expression resulted in impaired PD-L1 protein expression in all human tumor cell types, including lung cancer.12 The depletion of CMTM6 greatly reduced the inhibition of tumor-specific T cell activity in vitro and in vivo.11 PD-L1 relies on the CMTM6 to efficiently inhibit T cells and these researches on CMTM6 provide a new way of blocking the PD-L1 pathway. However, there has been no previous study of CMTM6 in lung cancer and its association with PD-L1 has not been confirmed.
The present work was conducted to determine whether CMTM6 affected the response to anti-PD-1 therapy in NSCLC patients. In addition, correlation analyses of CMTM6 and PD-L1 were performed on immunohistochemistry and web-based mRNA expression data.
Results
Patient demographics
The demographic data of patients included in this study are reported in Table 1. Although most samples were pulmonary specimens, three samples were obtained from metastatic lesions (two from soft tissue and one from a lymph node). Nineteen patients (54.3%) received pembrolizumab and 16 patients (45.7%) received nivolumab. In our study, all patients were treated with anti-PD-1 as a second-or-higher line of treatment because they were refractory to conventional chemotherapy, radiation therapy, and targeted therapy. Nineteen patients (54.3%) were sorted into the responder group, while 16 patients (45.7%) were classified into the non-responder group. Four EGFR-mutated patients were treated with tyrosine kinase inhibitor therapy before PD-1 blockade therapy. There was no ALK-rearranged patient.
Table 1.
Demographic and clinical characteristics of patients.
| Variable | Number (%) |
|---|---|
| Age, median (range) (years) | 67 (40–85) |
| Male sex | 28 (80.0%) |
| Smoking history | 20 (69.0%) |
| Histologic subtype | |
| Adenocarcinoma | 15 (42.9%) |
| Squamous cell carcinoma | 12 (34.3%) |
| Pleomorphic carcinoma | 4 (11.4%) |
| NSCLC, NOS | 4 (11.4%) |
| Clinical stage at diagnosis | |
| III | 9 (25.7%) |
| IV | 26 (74.3%) |
| Genetic alteration status | |
| EGFR-mutated | 4 (11.4%) |
| ALK-rearranged | 0 (0%) |
| Wild type | 31 (88.6%) |
| Type of PD-1 blockade | |
| Nivolumab | 16 (45.7%) |
| Pembrolizumab | 19 (54.3%) |
| PD-L1 expression | |
| Low (< H-score 75) | 10 (28.6%) |
| High (≥ H-score 75) | 25 (71.4%) |
| Response to PD-1 blockade | |
| Responder | 19 (54.3%) |
| Non-responder | 16 (45.7%) |
Smoking history was collected for 29 patients.
EGFR test was performed in 28 patients.
ALK test was performed in 31 patients.
Abbreviations: epidermal growth factor receptor; EGFR, non-small-cell lung cancer-not otherwise specified; NSCLC, NOS, programmed cell death protein 1, PD-1; programmed death-ligand 1, PD-L1.
Correlation between CMTM6 and PD-L1 in immunohistochemistry and mrna expression
We performed correlation analyses between CMTM6 and PD-L1 expression in the immunohistochemical data. CMTM6 and PD-L1 immunohistochemical data showed normal distributions (Kolmogorov-Smirnov test p = .117 and p = .642, respectively). Therefore, we used Pearson’s correlation coefficient for the analysis. CMTM6 immunohistochemical expression was positively correlated with PD-L1 immunohistochemical expression (Pearson’s r = 0.342 and p = .044, Figure 1A). Next, we performed correlation analyses between CMTM6 and PD-L1 expression in the mRNA data. CMTM6 and PD-L1 mRNA expression were not normally distributed in adenocarcinoma and squamous cell carcinomas (Kolmogorov-Smirnov test p < .001 for CMTM6 and PD-L1 mRNA expression in adenocarcinoma and squamous cell carcinoma). Therefore, we used Spearman’s correlation coefficient for the analysis. CMTM6 mRNA expression was positively correlated with PD-L1 mRNA expression in adenocarcinoma and squamous cell carcinoma (Spearman’s rho = 0.21 and p < .001, Figure 1B; and Spearman’s rho = 0.13 and p = .005, Figure 1C).
Figure 1.
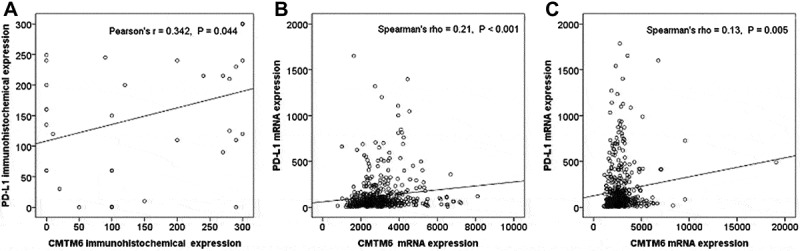
Correlation analyses between the CMTM6 and PD-L1 expression. (A) Correlation between CMTM6 and PD-L1 immunohistochemical expression. (B) Correlation between CMTM6 and PD-L1 mRNA expression in lung adenocarcinoma. (C) Correlation between CMTM6 and PD-L1 mRNA expression in lung squamous cell carcinoma.
We performed correlation analyses between CMTM6 and PD-L1 mRNA expression in other type carcinomas using TCGA, PanCancer Atlas data. In 28 solid tumors, 18 solid tumors showed a significant positive correlation between CMTM6 and PD-L1 mRNA expression (Supplementary Table 1).
Associations between CMTM6, PD-L1, clinicopathologic parameters, and response to PD-1 inhibitors
ROC curves for CMTM6 or PD-L1 according to PD-1 inhibitor responses were generated to determine the appropriate cutoff values. The area under the curve was 0.715 for CMTM6 at a CMTM6 value of 70, corresponding to the maximum joint sensitivity and specificity on the ROC curve (84% sensitivity and 50% specificity, Figure 2A). The area under the curve was 0.618 for PD-L1 at a PD-L1 value of 75, corresponding to the maximum joint sensitivity and specificity on the ROC curve (79% sensitivity and 38% specificity, Figure 2B).
Figure 2.
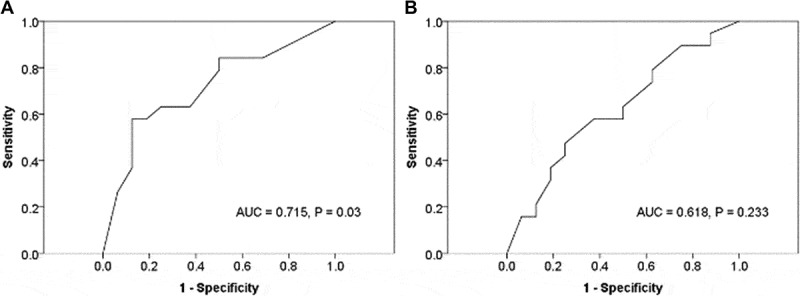
Receiver operating characteristic (ROC) and area under the curve (AUC) for CMTM6 (A) and PD-L1 (B).
We analyzed the effects of CMTM6, PD-L1, and clinicopathologic factors on the response to PD-1 inhibitors. In univariate analysis, CMTM6 was found to be a predictor of the response to PD-1 inhibitors. (p = .037, odds ratio (OR) = 5.333, Table 2). However, PD-L1 and clinicopathologic factors were not predictors of the response to PD-1 inhibitors. Age and sex were clinically important variables, therefore, they were included in the final multivariate analysis. In multivariate analysis, CMTM6 was also found to be an independent predictor of the response to PD-1 inhibitors (p = .032, OR = 6.226, Table 2). CMTM6 as continuous value was also significant predictor of clinical response to PD-1 blockade in univariate analysis (p = .02, OR = 1.007). In multivariate analysis, CMTM6 as continuous value was also a significant independent predictor (p = .027, OR = 1.007).
Table 2.
Univariate and multivariate logistic regression analysis for predicting clinical response to PD-1 blockade.
| Univariate analysis |
Multivariate analysis |
|||||
|---|---|---|---|---|---|---|
| covariate | OR | 95%CI | P-value† | OR | 95%CI | P-value† |
| Age (≥65 years vs.<65 years) | 2.204 | 0.567–8.571 | 0.254 | 2.654 | 0.585–12.04 | 0.206 |
| Sex (male vs. female) | 1.788 | 0.106–2.999 | 0.499 | 0.443 | 0.069–2.854 | 0.392 |
| Smoking history (+ vs. –) | 1.875 | 0.382–9.197 | 0.438 | |||
| Presence of EGFR mutation (+ vs. -) | 0.200 | 0.018–2.225 | 0.190 | |||
| Type of PD-1 blockade (Nivolumab vs. Pembrolizumab) |
1.157 | 0.304–4.404 | 0.831 | |||
| PD-L1 (≥75 vs.<75) | 2.250 | 0.504–10.05 | 0.288 | |||
| CMTM6 (≥70 vs.<70) | 5.333 | 1.104–25.76 | 0.037 | 6.226 | 1.175–32.99 | 0.032 |
Abbreviations: confidence interval; CI, epidermal growth factor receptor; EGFR, odd ratio; OR, programmed cell death protein 1, PD-1; programmed death-ligand 1, PD-L1
Prognostic significance of CMTM6, PD-L1, and clinicopathologic parameters
Responders to PD-1 inhibitors had a better overall survival rate than non-responders to PD-1 inhibitors (p = .015) (Figure 3A). Patients with high CMTM6 protein expression had better overall survival than patients with low CMTM6 protein expression but were not statistically significant (p = .349) (Figure 3B). High PD-L1 protein expression was also correlated with better overall survival with borderline statistical significance (p = .059) (Figure 3C).
Figure 3.
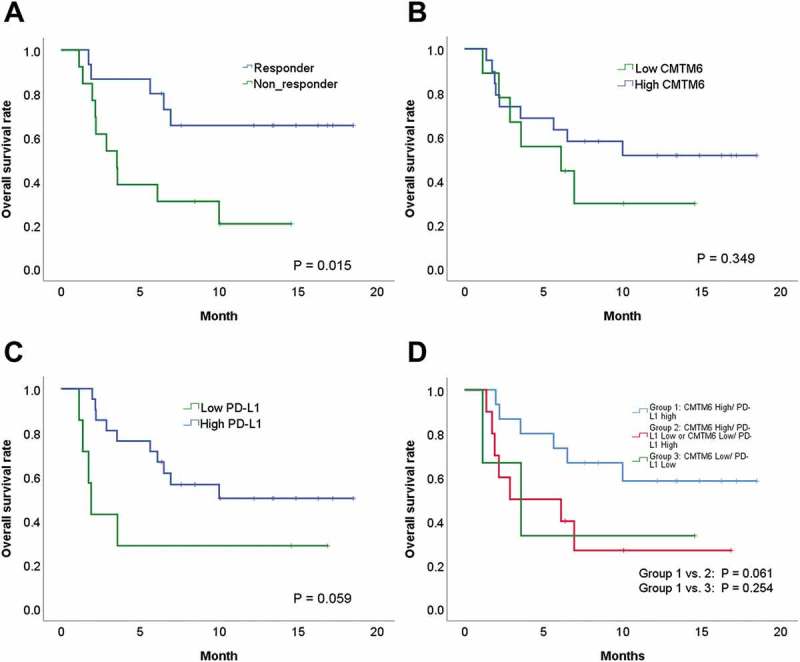
Comparison of survival rates according to CMTM6 and PD-L1 expression. (A) Overall survival (OS) and the clinical response to PD-1 inhibitors. (B) OS and CMTM6. (C) OS and PD-L1. (D) OS, CMTM6 and PD-L1.
Next, we combined CMTM6 and PD-L1 expression for survival analysis. Patients with high expression of both CMTM6 and PD-L1 expression had longer overall survival than patients with high CMTM6 and low PD-L1 and patients with low CMTM6 and high PD-L1 (p = .061) (Figure 3D). Patients with high expression of both CMTM6 and PD-L1 expression also had longer overall survival than patients with low CMTM6 and low PD-L1 (p = .254) (Figure 3D). However, statistical significance was not reached in the above analyzes.
Discussion
Our study made several novel findings. First, we found that CMTM6 expression was an independent predictive factor of PD-1 inhibitor response and reported the cutoff value for CMTM6 expression to distinguish the responder and non-responder groups. Second, CMTM6 expression was positively correlated with PD-L1 expression in immunohistochemical and mRNA expression data.
Intratumoral PD-L1 expression is regulated by multiple mechanisms, including the phosphoinositide 3-kinase (PI3K) and mitogen-activated protein kinase (MAPK) or Akt pathways, transcriptional factors signal transducer and activator of transcription 3 (STAT3), hypoxia-inducible factor-1 (HIF1), and nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB).13 PD-L1 expression on tumor cells is transient and expression disappears quickly. Tumor-associated macrophages (TAM) or IFN-gamma help maintain PD-L1 expression.14 Intratumoral heterogeneity of PD-L1 expression has been frequently observed in NSCLC.15 For these reasons, tumors not expressing detectable levels of PD-L1 can also respond to PD-1 inhibitors. Therefore, another predictor to supplement PD-L1 is needed.
The novel chemokine-like factor (CKLF)-like Marvel Transmembrane Domain-containing gene family (CMTM) consists of 8 members (CMTM1-8).16 CMTM3 and CMTM5 are associated with tumorigenesis.17,18 CMTM6 is involved in the DNA methylation of hepatocellular carcinoma19 and CMTM6 overexpression is associated with prognosis in glioma20 and hepatocellular carcinoma.21 Recently, two studies reported that CMTM6 regulated antitumor immunity by maintaining the expression of PD-L1.11,12 Mezzadra et al. reported that CMTM6 was associated with PD-L1 protein. It reduced PD-L1 ubiquitination and increased its protein half-life in a melanoma cell line.12 Burr et al. also reported that CMTM6 coexisted with PD-L1 in the plasma membrane and recycling endosomes, where it prevented PD-L1 lysosomal degradation in a breast cancer cell line.11 CMTM6 depletion, via the reduction of PD-L1, significantly promoted tumor-specific T cell activity in vitro and in vivo.11 Furthermore, inhibition of CMTM6 provided a survival benefit in a mouse model of cancer.11 Since PD-L1 is a transient protein, CMTM6 plays a critical role in the survival of PD-L1 because it increases the stability of PD-L1.
Mamessier et al. reported that the group with CMTM6 high and PD-L1 high was associated with shorter overall survival in mRNA expression data of pancreatic adenocarcinomas.22 However, the group with CMTM6 high and PD-L1 high showed better metastasis-free survival in mRNA expression data of triple-negative primary breast cancers.22 Our results also showed that the group with CMTM6 high and PD-L1 high showed better overall survival. Responders to PD-1 inhibitors were more likely to survive in our results. Therefore, interaction between CMTM6 and PD-L1 increases response to PD-1 inhibitors. Increased response to PD-L1 inhibitors may contribute to improved survival.
In present study, 54.3% of the patients were in the responder group. Our study’s response rate is somewhat higher than other studies. Garon et al. reported that the objective response rate was 19.4% in all PD-L1-positive NSCLC treated with pembrolizumab and the response rate was 45.2% in groups with ≥ 50% PD-L1 expression.1 Gettinger et al. revealed that the objective response rate was 23% in all advanced NSCLC treated with Nivolumab.2 In study of Gettinger et al, the objective response rate was 50% in tumors with ≥ 50% PD-L1 expression. In our study, 71.4% of patients showed high expression of PD-L1 (≥ H-score 75). High PD-L1 expression rate may have affected the response rate.
This study had several limitations. First, our study was a retrospective design and had a relatively small sample size. Second, in logistic regression analyses, overfitting a model is an important problem. To avoid overfitting our model, we should perform validation tests with independent data. However, our sample number was not enough to perform the validation test. Another large scale validation test should be performed to confirm our results. Third, three samples were obtained from metastatic lesions. In these cases, CMTM6 expression in primary tumors was not accurately reflected. Fourth, we used CMTM6 immunohistochemical scoring to determine whether CMTM6 affected the response to anti-PD-1 therapy in patients with NSCLC. However, immunohistochemical methods have weaknesses in standardization and reproducibility.23 We used a fallopian tube as a positive control and an automated immunohistochemistry platform to improve reproducibility. We used H-scores for immunohistochemical scoring. The H-score method has been used as a standard for immunohistochemical scoring and has been shown to be highly reproducible among pathologists.24,25 Fifth, we did not include other potentially important predictors of response to PD-1 inhibitors including tumor mutational burden,9 tumor-infiltrating lymphocytes8 and the predictive expression signatures10 in multivariate logistic analysis.
In conclusion, we examined CMTM6 and PD-L1 expression in 35 pretreatment NSCLC samples from PD-1 inhibitor-treated patients. High CMTM6 expression was identified as an independent factor predicting the response to PD-1 inhibitors. In addition, CMTM6 expression was positively correlated with PD-L1 in immunohistochemical and mRNA expression data. Based on our findings, CMTM6 expression might become a promising predictor for therapeutic decision-making.
Materials and methods
Patients
We enrolled advanced non-small cell lung cancer patients treated with PD-1 blockade therapy at our institution who had pretreatment tumor tissue available for study and evaluated their drug response from 2016 to 2018. A total of 35 patients with NSCLC were consecutively collected. This retrospective study was approved by the Institutional Review Board of Ajou University School of Medicine. Informed consent was waived due to the retrospective nature of the study. This study’s involvement with human subjects complies with the Declaration of Helsinki.
Age, sex, smoking history, histologic subtype, clinical stage at diagnosis, and genetic alteration status were obtained from the medical records. Patients treated with PD-1 blockade therapy were classified into the responder group (complete response, partial response, or stable disease) or the non-responder group (progression disease) according to RECIST criteria version 1.1.26
Immunohistochemical staining and scoring of CMTM6
All H&E slides were carefully reviewed by one pathologist (YWK) to determine tumor subtype according to the 2015 World Health Organization Classification of Lung Tumors.27 Clinical TNM staging was recorded according to the eighth edition of the TNM classification.
Immunohistochemical staining of CMTM6 was carried out with a Benchmark XT automatic IHC staining device (Ventana Medical Systems, Tucson, AZ, USA). The samples were incubated with antibody against CMTM6 (dilution 1:200, polyclonal, GeneTex, Irvine, CA, USA). We used fallopian tissue as a positive control for CMTM6.28 CMTM6 expression was identified in the membranous or cytoplasmic areas of the fallopian tube and CMTM6 expression was not identified in normal lung tissue (Supplementary Figure 1A and B).
The intensity of the CMTM6 was also evaluated on a four-point intensity scale: 0 (no staining), 1 (faint staining = light yellow), 2 (moderate staining = yellow-brown), and 3 (strong staining = brown) (Figure 4A and 4B). Percentages (0–100%) of cytoplasmic or membranous expression of CMTM6 were also evaluated. We used the H-scores to interpret the CMTM6 stains.29 H-score = [1 × (% cells 1+) + 2 × (% cells 2+) + 3 × (% cells 3+)]. H-scores (0–300) were obtained by multiplying the intensity by the percentage of positive cells.
Figure 4.
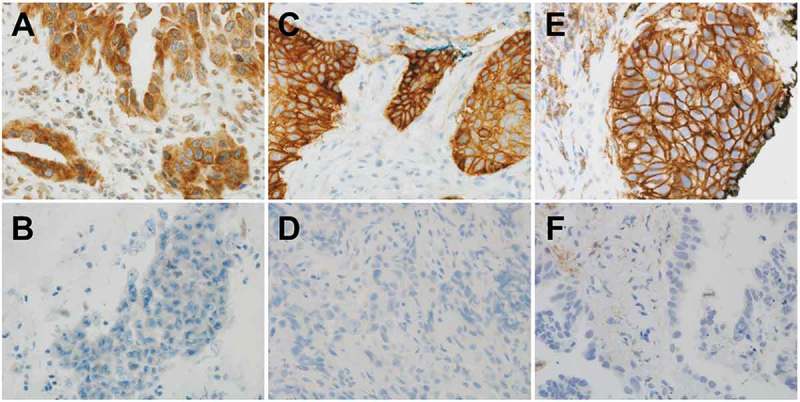
CMTM6 or PD-L1 expression in non-small cell carcinoma. (A) Positive cytoplasmic or membranous CMTM6 expression in tumor cells. (B) Negative CMTM6 expression in tumor cells. (C) Positive membranous SP263 PD-L1 expression in tumor cells. (D) Negative SP263 PD-L1 expression in tumor cells. (E) Positive membranous 22C3 PD-L1 expression in tumor cells. (F) Negative 22C3 PD-L1 expression in tumor cells.
Immunohistochemical staining and scoring of PD-L1
We performed 22C3 and/or SP263 assays before PD-1 inhibitor treatment for all patients. Five of the 35 (14.3%) specimens were tested for both SP263 and 22C3, 16 (45.7%) for only SP263, and 14 (40%) for only 22C3. The SP263 assay was performed with the OptiView DAB Immunohistochemical Detection Kit on a VENTANA BenchMark ULTRA instrument and the 22C3 assay was performed on the Dako Link-48 platform as recommended by the manufacturer30 (Figure 4C-F). We used H-scores to interpret the PD-L1 stains.29 When both the 22C3 and SP263 tests were performed, the mean value of the 22C3 and SP263 tests was used as the H-Score.
Web-based mrna profiling
The mRNA expression data of 510 lung adenocarcinoma patients and 484 lung squamous cell carcinoma patients were downloaded from The Cancer Genome Atlas (TCGA) cBioportal (http://cbioportal.org).31
Statistical analyses
Categorical variables were compared using chi-squared tests. We used Pearson’s correlation analysis to describe the correlation between quantitative variables with a normal distribution. We used Spearman’s correlation analysis to describe the correlation between quantitative variables without a normal distribution. The probability of clinical benefit from a PD-1 inhibitor based on clinicopathologic variables was examined by univariate and multivariate logistic regression analyses. The cutoff values for CMTM6 and PD-L1 were determined by receiver operating curve (ROC) analysis. We defined overall survival as the interval between the date that therapy started and the date of death from any cause or the date of last follow-up. The overall survival difference between the cohorts was assessed using the log-rank test. IBM SPSS Statistics for Windows (Version 25.0. Armonk, NY) was used for all analyses and a p-value of less than 0.05 was considered statistically significant.
Funding Statement
No funding
Abbreviations
CKLF-like MARVEL transmembrane domain containing 6, CMTM6; hypoxia-inducible factor-1, HIF1; mitogen-activated protein kinase, MAPK; non-small cell lung cancer, NSCLC; nuclear factor kappa-light-chain-enhancer of activated B cells, NF-κB; phosphoinositide 3-kinase, PI3K; programmed death 1, PD-1; programmed death-ligand 1, PD-L1; receiver operating characteristic curve, ROC; signal transducer and activator of transcription 3, STAT3; The Cancer Genome Atlas, TCGA; tumor-associated macrophages, TAM
Ethics approval and consent to participate
The human cancer tissues used in this study were approved by the Institutional Review Board of Ajou University School of Medicine. This study’s involvement with human subjects complies with the Declaration of Helsinki.
Consent for publication
This retrospective study was approved by the Institutional Review Board of Ajou University School of Medicine. Informed consent was waived due to the retrospective nature of the study.
Competing interests
The authors declare that they have no conflict of interest
Supplementary Material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L, Carcereny E., et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med. 2015;372:28–2018. doi: 10.1056/NEJMoa1501824 [DOI] [PubMed] [Google Scholar]
- 2.Gettinger S, Rizvi NA, Chow LQ, Borghaei H, Brahmer J, Ready N, Gerber DE, Shepherd FA, Antonia S, Goldman JW, Juergens R A., et al. Nivolumab monotherapy for first-line treatment of advanced non-small-cell lung cancer. J Clin Oncol. 2016;34:7–2980. doi: 10.1200/jco.2016.66.9929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herbst RS, Baas P, Kim DW, Felip E, Perez-Gracia JL, Han JY, Molina J, Kim JH, Arvis CD, Ahn MJ, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet. 2016;387:50–1540. doi: 10.1016/s0140-6736(15)01281-7 [DOI] [PubMed] [Google Scholar]
- 4.Shukuya T, Carbone DP.. Predictive markers for the efficacy of anti-PD-1/PD-L1 antibodies in lung cancer. J Thorac Oncol. 2016;11:88–976. doi: 10.1016/j.jtho.2016.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brahmer J, Reckamp KL, Baas P, Crino L, Eberhardt WE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE, Holgado E, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373:35–123. doi: 10.1056/NEJMoa1504627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mino-Kenudson M. Programmed cell death ligand-1 (PD-L1) expression by immunohistochemistry: could it be predictive and/or prognostic in non-small cell lung cancer? Cancer Biol Med. 2016;13:70–157. doi: 10.20892/j.issn.2095-3941.2016.0009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Km Kerr, Ms Tsao, Ag Nicholson, Yatabe Y, Wistuba II, Fr Hirsch. Programmed death-ligand 1 immunohistochemistry in lung cancer: in what state is this art? J Thorac Oncol. 2015;10:9–985. doi: 10.1097/jto.0000000000000526 [DOI] [PubMed] [Google Scholar]
- 8.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, West A N., et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:71–568. doi: 10.1038/nature13954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, Stephens PJ, Daniels GA, Kurzrock R.. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16:608–2598. doi: 10.1158/1535-7163.mct-17-0386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prat A, Navarro A, Pare L, Reguart N, Galvan P, Pascual T, Martinez A, Nuciforo P, Comerma L, Alos L, Pardo N. et al Immune-related gene expression profiling after PD-1 blockade in non-small cell lung carcinoma, head and neck squamous cell carcinoma, and melanoma. Cancer Res. 2017;77:50–3540. doi: 10.1158/0008-5472.can-16-3556 [DOI] [PubMed] [Google Scholar]
- 11.Burr ML, Sparbier CE, Chan YC, Williamson JC, Woods K, Beavis PA, Lam EYN, Henderson MA, Bell CC, Stolzenburg S, Gilan O. et al CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature. 2017;549:5–101. doi: 10.1038/nature23643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mezzadra R, Sun C, Jae LT, Gomez-Eerland R, de Vries E, Wu W, Logtenberg MEW, Slagter M, Rozeman EA, Hofland I, Broeks A. et al Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature. 2017;549:10–106. doi: 10.1038/nature23669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Jiang CC, Jin L, Zhang XD.. Regulation of PD-L1: a novel role of pro-survival signalling in cancer. Ann Oncol. 2016;27:16–409. doi: 10.1093/annonc/mdv615 [DOI] [PubMed] [Google Scholar]
- 14.Noguchi T, Ward JP, Gubin MM, Arthur CD, Lee SH, Hundal J, Selby MJ, Graziano RF, Mardis ER, Korman AJ, Schreiber R D.. Temporally distinct PD-L1 expression by tumor and host cells contributes to immune escape. Cancer Immunol Res. 2017;5:17–106. doi: 10.1158/2326-6066.cir-16-0391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura S, Hayashi K, Imaoka Y, Kitamura Y, Akazawa Y, Tabata K, Groen R, Tsuchiya T, Yamasaki N, Nagayasu T, Fukuoka J., Deb S.. Intratumoral heterogeneity of programmed cell death ligand-1 expression is common in lung cancer. PLoS One. 2017;12:e0186192. doi: 10.1371/journal.pone.0186192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu J, Wu QQ, Zhou YB, Zhang KH, Pang BX, Li L, Sun N, Wang HS, Zhang S, Li WJ, Zheng W., et al. Cancer research advance in CKLF-like MARVEL transmembrane domain containing member family (review). Asian Pac J Cancer Prev. 2016;17:4–2741 [PubMed] [Google Scholar]
- 17.Yuan W, Liu B, Wang X, Li T, Xue H, Mo X, Yang S, Ding S, Han W.. CMTM3 decreases EGFR expression and EGF-mediated tumorigenicity by promoting Rab5 activity in gastric cancer. Cancer Lett. 2017;386:77–86. doi: 10.1016/j.canlet.2016.11.015 [DOI] [PubMed] [Google Scholar]
- 18.Xu G, Dang C.. CMTM5 is downregulated and suppresses tumour growth in hepatocellular carcinoma through regulating PI3K-AKT signalling. Cancer Cell Int. 2017;17:113. doi: 10.1186/s12935-017-0485-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yafune A, Kawai M, Itahashi M, Kimura M, Nakane F, Mitsumori K, Shibutani M.. Global DNA methylation screening of liver in piperonyl butoxide-treated mice in a two-stage hepatocarcinogenesis model. Toxicol Lett. 2013;222:295–302. doi: 10.1016/j.toxlet.2013.08.006 [DOI] [PubMed] [Google Scholar]
- 20.Guan X, Zhang C, Zhao J, Sun G, Song Q, Jia W.. CMTM6 overexpression is associated with molecular and clinical characteristics of malignancy and predicts poor prognosis in gliomas. EBioMedicine. 2018;35:43–233. doi: 10.1016/j.ebiom.2018.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu X, Qi G, Li C, Bei C, Tan C, Zhang Y, Shi W, Zeng W, Kong J, Fu Y, Tan S.. Expression and Clinical Significance of CMTM6 in Hepatocellular Carcinoma. DNA Cell Biol. 2019;38:7–193. doi: 10.1089/dna.2018.4513 [DOI] [PubMed] [Google Scholar]
- 22.Mamessier E, Birnbaum DJ, Finetti P, Birnbaum D, Bertucci F.. CMTM6 stabilizes PD-L1 expression and refines its prognostic value in tumors. Ann Transl Med. 2018;6:54. doi: 10.21037/atm.2017.11.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Hurley G, Sjostedt E, Rahman A, Li B, Kampf C, Ponten F, Gallagher WM, Lindskog C.. Garbage in, garbage out: a critical evaluation of strategies used for validation of immunohistochemical biomarkers. Mol Oncol. 2014;8:98–783. doi: 10.1016/j.molonc.2014.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aviles-Salas A, Muniz-Hernandez S, Maldonado-Martinez HA, Chanona-Vilchis JG, Ramirez-Tirado LA, HernaNdez-Pedro N, Dorantes-Heredia R, Rui ZMJM, Motola-Kuba D, Arrieta O.. Reproducibility of the EGFR immunohistochemistry scores for tumor samples from patients with advanced non-small cell lung cancer. Oncol Lett. 2017;13:20–912. doi: 10.3892/ol.2016.5512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Detre S, Saclani Jotti G, Dowsett M.. A “quickscore” method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J Clin Pathol. 1995;48:8–876 10.1136/jcp.48.9.876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:47–228. doi: 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 27.Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JH, Beasley MB, Chirieac LR, Dacic S, E Duhig, Flieder DB, et al. The 2015 world health organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol. 2015;10:60–1243. doi: 10.1097/jto.0000000000000630 [DOI] [PubMed] [Google Scholar]
- 28.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I. et al Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- 29.McCarty KS Jr., Szabo E, Flowers JL, Cox EB, Leight GS, Miller L, Konrath J, Soper JT, Budwit DA, Creasman WT, et al. Use of a monoclonal anti-estrogen receptor antibody in the immunohistochemical evaluation of human tumors. Cancer Res. 1986;46:4244s–8s [PubMed] [Google Scholar]
- 30.Munari E, Rossi G, Zamboni G, Lunardi G, Marconi M, Sommaggio M, Netto GJ, Hoque MO, Brunelli M, Martignoni G, Haffner M C. et al PD-L1 assays 22C3 and SP263 are not interchangeable in non-small cell lung cancer when considering clinically relevant cutoffs: an interclone evaluation by differently trained pathologists. Am J Surg Pathol. 2018;42:9–1384. doi: 10.1097/pas.0000000000001105 [DOI] [PubMed] [Google Scholar]
- 31.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, Omberg L. et al An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173:400–16.e11. doi: 10.1016/j.cell.2018.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


