Abstract
Single cell molecular profiling technologies are gaining rapid traction, but the manual process by which resulting cell types are typically annotated is labor-intensive and rate-limiting. We describe Garnett, an algorithm and accompanying software for rapidly annotating cell types in scRNA-seq and scATAC-seq datasets, based on an interpretable, hierarchical markup language of cell type-specific genes. Garnett successfully classifies cell types in tissue and whole organism datasets, as well as across species.
Single-cell transcriptional profiling (scRNA-seq) is a powerful means of cataloging the myriad cell types present in multicellular organisms1. The computational steps of constructing a cell atlas typically include unsupervised clustering of cells based on their gene expression profiles, followed by annotation of known cell types amongst the resulting clusters2,3. For the latter task, there are at least four key challenges. First, cell type annotation is labor intensive, requiring extensive literature review of cluster-specific genes4. Second, any revision to the analysis (e.g. additional data, parameter adjustment) necessitates manual reevaluation of all previous annotations. Third, annotations are not easily transferred between datasets generated by independent groups on related tissues, resulting in wasteful repetition of effort. Finally, annotation labels are typically ad hoc; although ontologies of cell types exist5,6, we lack tools for systematically applying these ontologies to scRNA-seq data. Collectively, these challenges hinder progress towards a consensus framework for cell types and the features that define them.
Towards addressing these challenges, we devised Garnett (Figure 1A). Garnett consists of four components. First, Garnett defines a markup language for specifying cell types using the genes that they specifically express. The markup language is hierarchical in that a cell type can have subtypes (e.g. CD4+ and CD8+ are subsets of T cells). Second, Garnett includes a parser that processes the markup file together with a single-cell dataset, identifying representative cells bearing markers that unambiguously identify them as one of the cell types defined in the file. Third, Garnett trains a classifier that recognizes additional cells as belonging to each cell type based on their similarity to representative cells, similar to an approach that our groups recently developed for annotating a single cell mouse atlas of chromatin accessibility7. Importantly, Garnett does not require that cells be organized into clusters, but it can optionally extend classifications to additional cells using either its own internal clustering routines or those of other tools. Finally, Garnett provides a method for applying a markup file together with a classifier trained on one dataset to rapidly annotate additional datasets.
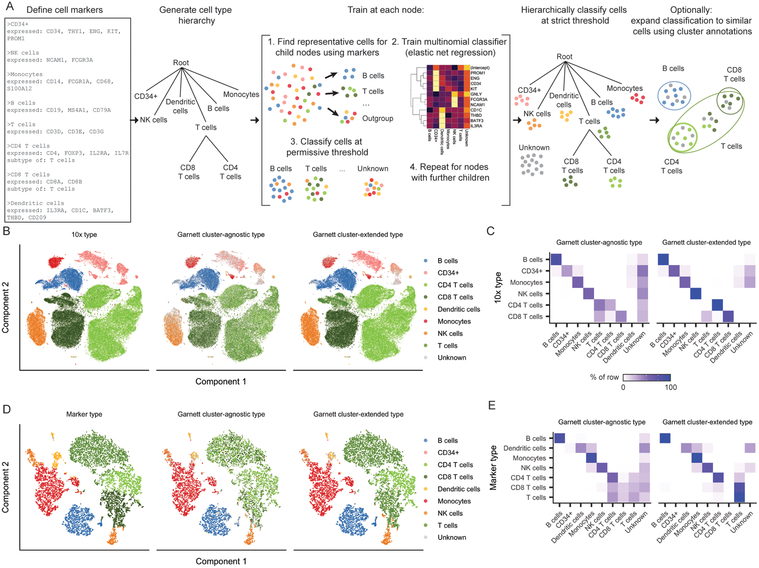
Figure 1. Garnett accurately classifies peripheral blood mononuclear cells.
A) Overview of Garnett algorithm. See Methods for algorithmic details. Briefly, Garnett takes as input a marker file that defines cell types using marker genes, and builds a cell type hierarchy that can include cell subtypes. Next, Garnett trains a classifier using elastic net multinomial regression17 at each node beginning at the root of the tree by comparing cell type representative cells. Lastly, Garnett hierarchically classifies all cells and optionally provides a second cluster-extended classification. B) t-SNE plots of 10x Genomics’ 100,000 cell PBMC dataset (n = 94,571 cells). The first panel is colored by cell type based on FACS sorting, the second panel is colored by cluster-agnostic cell type according to Garnett classification, and the third panel is colored by the Garnett cluster-extended type, which labels cells based on the composition of their cluster or community. C) A heatmap of data in (B) comparing the labels based on FACS (rows) with the cluster-agnostic (left) and cluster-extended (right) cell type assignments by Garnett (columns). Color represents the percent of cells of a certain FACS type labeled each type by Garnett. D) t-SNE plots of 10x Genomics V2 chemistry applied to PBMCs from a healthy donor (n = 8,381 cells). The first panel is colored by type determined manually using known gene markers. The second and third panels are colored by Garnett cluster-agnostic and cluster-extended cell type assignments by a classifier trained on the data shown in panels (B) and (C). E) Similar to panel (C), a heatmap of data in (D).
We tested Garnett on a benchmark scRNA-seq dataset comprising 94,571 immunophenotyped peripheral blood mononuclear cells (PBMCs), generated with the 10X Chromium platform8. Garnett requires ≥1 marker gene for each cell type. To classify the PBMCs, we populated a marker file including each of the expected cell types using literature-based markers. As a supervised method, Garnett’s accuracy will be dependent on these markers, so we devised a measure of each marker’s usefulness for the purposes of Garnett classification (see Methods). We used this quality metric to exclude poorly scoring markers (ambiguity > 0.5) before proceeding with classification (Supplementary Figure 1A).
Garnett assigned 71% (3% incorrect, 26% unclassified) of cells to the correct type (“cluster-agnostic type”), with 34% of T cells also receiving a correct subtype classification (41% not subclassified, 23% unclassified, 2% incorrect) (Figure 1B–C). Cells remaining unlabeled were comparably distributed amongst immunophenotypes. Moreover, by expanding cell type assignments to nearby cells using Louvain clustering9 (“cluster-extended type”), correct assignments increased to 94% (2% incorrect, 4% unclassified), with 91% of T cells also receiving a correct subtype classification (8% not subclassified, <1% unclassified, <1% incorrect).
We next evaluated Garnett’s ability to classify data not seen during training by analyzing PBMCs that were profiled to a higher molecular depth with a different library preparation method and a different Chromium system (“V2”) (Supplementary Figure 1B). Because these cells were unsorted, we manually assigned cell types to clusters based on classic markers (Supplementary Figure 1C–D). Although trained on sparser molecular data from a different method and instrument, classification accuracy remained high, with 80% (3% incorrect, 17% unclassified) of cells correctly labeled with cluster-agnostic type and 95% (3% incorrect, 2% unclassified) with cluster-extended type (Figure 1D–E). Of note, when trained on these more deeply profiled V2 cells, Garnett also accurately classified the more sparsely profiled V1 cells (83% correct with cluster-agnostic type and 95% correct with cluster-extended) (Supplementary Figure 1E–G). We furthermore used the PBMC datasets to explore the limits of Garnett and found that the algorithm was robust to rare and missing cell types and low data quality (Supplementary Figures 2 and 3, Supplementary Note).
To assess Garnett’s ability to catalog cell types in complex solid tissues, we analyzed lung tissue data from two recently reported cell atlases, the Mouse Cell Atlas (MCA)3 and Tabula Muris (TM)2. We defined a single hierarchy of expected lung cell types based on those studies and compiled marker genes from literature to recognize them in each dataset (marker files are Supplementary Files, consensus cell type names in Supplementary Table 1). Overall, Garnett’s classifications agreed with both the MCA (58% correct, 29% unclassified with cluster-agnostic type; 65% correct, 23% unclassified with cluster-extended type; Supplementary Figure 4A–B) and TM (71% correct, 22% unclassified with cluster-agnostic type; 87% correct, 8% unclassified with cluster-extended type; Supplementary Figure 4C–D) annotations, which were derived by manual inspection of genes enriched in each cluster. Moreover, a Garnett model trained on the MCA accurately classified the TM cells and vice versa (trained on MCA: 82% correct, 5% unclassified with cluster-agnostic type; 86% correct, 2% unclassified with cluster-extended type; trained on TM: 46% correct, 30% unclassified with cluster-agnostic type; 56% correct, 21% unclassified with cluster-extended type; Supplementary Figures 4E–H).
We next sought to evaluate whether Garnett was similarly useful for annotating single-cell chromatin accessibility (scATAC-seq) datasets, which we have generally found to be more challenging to manually annotate than scRNA-seq datasets. We and colleagues recently used regularized, multinomial regression to classify clusters of cells based on chromatin accessibility7. We adapted Garnett to classify cells based on scATAC-seq-derived “gene activity scores”, a measure of open chromatin around each gene10. Applying it to our recent scATAC-seq atlas of the mouse7, Garnett labeled 39% of cells concordantly with our previous assignments (cluster-extended; 22% incorrect; 39% unclassified) (Supplementary Figure 5). A caveat is that the marker file was informed by our previous literature-based annotation of the dataset by a related method, but these analyses nonetheless illustrate the potential of Garnett to enable the rapid annotation of not only scRNA-seq but also scATAC-seq datasets.
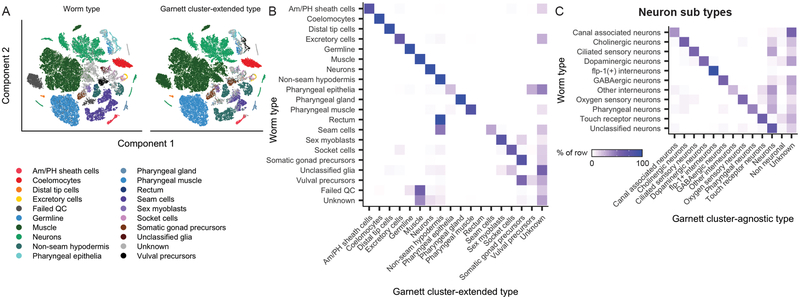
We next applied Garnett to the task of discriminating all cell types of a whole animal, L2 stage C. elegans11. We defined a cell hierarchy that discriminated 29 major cell types, as well as subtypes of neurons, using the marker genes from the original study. Of cells previously assigned, Garnett labeled 87% of cells concordantly for major cell type (cluster-extended; 8% incorrect; 5% unclassified), with rectum cells being frequently mislabeled as non-seam hypodermis (Figure 2A–B, Supplementary Figures 6–7). Of 4,186 neurons originally assigned subtypes, 53% were subtyped correctly, and a further 18% were labeled as neurons of unknown subtype (cluster-agnostic; 8% incorrect) (Figure 2C). These analyses demonstrate Garnett can scale to classifying the cell types found in a whole animal.
Figure 2. Garnett can discriminate among cell types across a whole animal, across species and between normal and pathological tissue.
Garnett classification results for sci-RNA-seq data from whole C. elegans, published in ref11. A) t-SNE plots of the whole worm dataset (n = 42,035 cells). First panel is colored by published type from ref11, second panel colored by the major (top level) Garnett cluster-extended classification. Garnett cluster-agnostic type is available in Supplementary Figure 7. B) Heatmap comparing the reported cell types versus the Garnett cluster-extended cell types. Color represents the percent of cells of a certain reported type labelled as each type by Garnett. C) Heatmap comparing the reported neuron subtypes versus the Garnett cluster-agnostic neuron subtypes.
To evaluate how Garnett would perform on a complex system with a deep hierarchy, we generated a 4-level classifier with 144 cell definitions for mouse nervous system based on the data and taxonomy presented in ref12 (Supplementary Figure 8). We found Garnett performed very well at the higher levels, but often under-classified cells at the lower, more specific levels (e.g., classifying a cerebellum neuron as a neuron) (Supplementary Figure 9A–E; Supplementary Figure 10). The size and complexity of this hierarchy facilitated exploration of the properties of markers chosen by the elastic-net regression to discriminate amongst cell types. Garnett tended to select genes that were more highly expressed and more specific than other genes (Supplementary Figure 11).
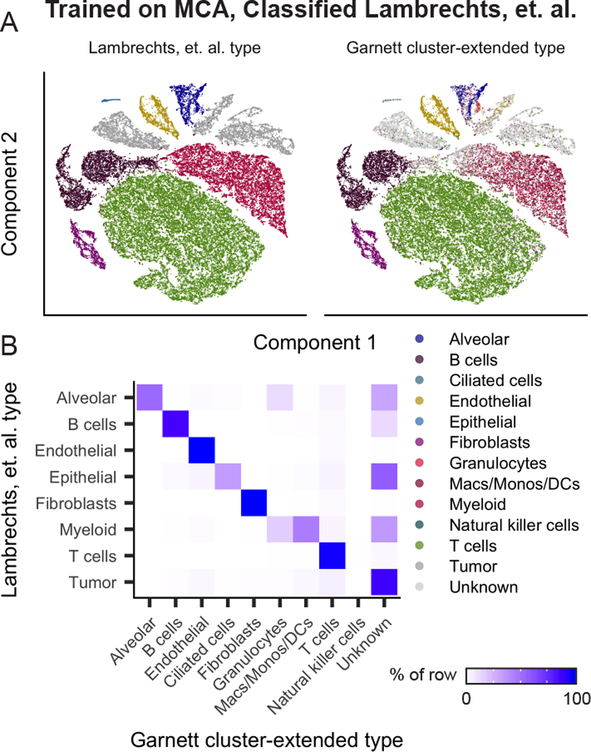
Finally, as tissue-specific gene expression patterns are largely conserved across vertebrates13, we wondered whether Garnett models trained on mouse data could be used to classify human cell types. We applied the Garnett model trained on the MCA lung dataset to scRNA-seq data from human lung tumors14 (Figure 3A–B, Supplementary Figure 12, Supplementary Table 1). Over 92% of alveolar, B cells, T cells, epithelial (ciliated) cells, endothelial cells, and fibroblasts were accurately assigned by the Garnett MCA model. Of the 9,756 cells annotated as myeloid14, Garnett labeled 44% as monocyte/macrophage/dendritic cell and a further 16% granulocytes, leaving 34% unclassified. 22% of the dataset was labeled “unknown”, of which 55% were identified as tumor cells in the original study. As expected, given that they are not represented in the original marker file nor in the MCA lung dataset, 88% of all cells annotated as tumor cells in the original study were labeled as “unknown” by Garnett. These analyses demonstrate that Garnett has the potential to operate across related species, and is not necessarily confounded by the presence of pathological cell states when trained on normal healthy tissue.
Figure 3. Garnett accurately classifies across species and distinguishes normal and pathological tissue.
A) Garnett cluster-extended results for human lung tumors from ref14 classified based on a Garnett classifier trained on lung cells from the Mouse Cell Atlas. t-SNE plots of the human lung tumor dataset (n = 52,698 cells). First panel is colored by published type from ref14, second panel colored by the Garnett cluster-extended classification. B) Heatmap comparing the reported cell types versus the Garnett cluster-extended cell types from panel D. Color represents the percent of cells of a certain reported type labelled as each type by Garnett.
Cell type annotation is a critical and rate-limiting step in cell type atlas construction, as illustrated by recent studies that resorted to labor-intensive, ad hoc literature review to achieve this end2,3,7,11,12,15 Garnett is an algorithm and accompanying software that automates and standardizes the process of classifying cells based on marker genes. While other algorithms for automated cell type assignment have been published3,16 we believe that Garnett’s ease-of-use and lack of requirement of pre-classified training datasets will make it an asset for future cell type annotation. One existing method, scMCA, trained a model using Mouse Cell Atlas data that can be applied to newly sequenced mouse tissues. scMCA reported slightly higher accuracy than Garnett3, likely owing to a training procedure that relies on manual annotation of cell clusters. . But a key distinction is that the hierarchical marker files on which Garnett is based are interpretable to biologists and explicitly relatable to the existing literature. Furthermore, together with these markup files, Garnett classifiers trained on one dataset are easily shared and applied to new datasets, and are robust to differences in depth, methods, and species.
We anticipate the potential for an “ecosystem” of Garnett marker files and pre-trained classifiers that: 1) enable the rapid, automated, reproducible annotation of cell types in any newly generated dataset. 2) minimize redundancy of effort, by allowing for marker gene hierarchies to be easily described, compared, and evaluated. 3) facilitate a systematic framework and shared language for specifying, organizing, and reaching consensus on a catalog of molecularly defined cell types. To these ends, in addition to releasing the Garnett software, we have made the marker files and pre-trained classifiers described in this manuscript available at a wiki-like website that facilitates further community contributions, together with a web-based interface for applying Garnett to user datasets (https://cole-trapnell-lab.github.io/garnett).
Online Methods
Garnett
Garnett is designed to simplify, standardize, and automate the classification of cells by type and subtype. To train a new model with Garnett, the user must specify a cell hierarchy of cell types and subtypes, which may be organized into a tree of arbitrary depth; there is no limit to the number of cell types allowed in the hierarchy. For each cell type and subtype, the user must specify at least one marker gene that is taken as positive evidence that the cell is of that type. Garnett includes a simple language for specifying these marker genes, in order to make the software more accessible to users unfamiliar with statistical regression. Negative marker genes, i.e. taken as evidence against a cell being of a given type, can also be specified. In addition, Garnett includes tools for selecting and checking the quality of markers. Garnett uses the marker information provided to select cells that are then used to train a regression-based classifier, similar to the approach taken in ref7. After a classifier is trained, it can be applied to other single cell datasets run on the same or different platforms. Algorithmic details are provided below.
Constructing marker files
Garnett uses a marker file to allow users to specify cell type definitions. These definitions are then used to choose representative cells from each cell type to use when training the classifier. Full details describing the syntax of the marker file are provided as part of the software package. Briefly, the marker file consists of a series of cell type entries, beginning with a cell type name, followed by lists of expressed markers and metadata. In addition, cell types can be specified to be a subtype of another defined type, i.e. hierarchical definitions. Marker files also have the capability to hold literature references for the chosen marker genes that are then included as metadata in the classifier.
Because only markers that are expressed specifically in a given cell type are useful for Garnett classification’ we also provide functions for assessing the value of each of the provided marker genes. These functions estimate the number of cells that a given marker nominates for their cell type, the number of cells that become “ambiguously” nominated to multiple cell types in a given level of the hierarchy when the marker is included, and an overall marker score G, defined as:
where a is the fraction of cells of the cells nominated by the given marker that are made ambiguous by that marker, p is a small pseudocount, b is the number of cells nominated by the marker, and n is the total number of cells nominated for that cell type. In addition to estimating these values, Garnett will plot a diagnostic chart to aid the user in choosing markers (e.g. Supplementary Figure 1A).
Training the classifier
Garnett’s first step in training a cell type classifier is to choose representative cells to train on. Let M be an m by n matrix of input gene expression data. First, Mi,j is normalized by size factor (the geometric mean of the total UMIs expressed for each cell j) to adjust for read depth, resulting in a normalized m by n matrix N. In addition, the gene IDs of the expression data are converted to Ensembl IDs using correspondence tables from a Bioconductor AnnotationDbi-class18 package. Next, the input marker file is parsed and the gene IDs are also converted to Ensembl IDs as above. Finally, a tree representation of the marker file is constructed, with any designated subtypes placed as children of the parent cell type in the tree. In addition to the tree, a dataset-wide size factor is generated and saved to the tree to allow normalization to new datasets for later classification (see classifying cells section below).
For each parent node in the tree, the following steps are taken: First, cells are scored as “expressed” or “not expressed” for each of the provided markers and an aggregate marker score is derived for each cell type for each cell (details on scoring below). Next, any metadata or hard expression cutoffs are applied to exclude a subset of cells from consideration. Lastly, outgroup samples are chosen (see below). After choosing the training sample, the classifier is trained (see below), and a preliminary classification is made in order to further train downstream nodes.
Aggregated marker scores
We devised an aggregated marker scoring system to address two challenges of single-cell RNA-seq data for the purposes of identifying representative cell types based on markers. The first challenge when choosing cells is that of differing levels of expression of different markers. If a lowly expressed but specific marker is found in a cell profile, this is better evidence of cell type than a highly expressed and less specific marker. To address this, we use the term frequency-inverse document frequency19 (TF-IDF) transformation when generating aggregate marker scores. The TF-IDF transformed matrix is defined by,
where Ni,j is the m by n normalized gene expression matrix defined above.
The second challenge we addressed in our aggregate marker score calculation was that highly expressed genes have been known to leak into the transcriptional profiles of other cells. For example, in samples including hepatocytes, albumin transcripts are often found in low copy numbers in non-hepatocyte profiles. To address this, we assign a cutoff above which a gene is considered expressed in that cell. To determine this cutoff we use a heuristic measure defined as
where ci is the gene cutoff for gene i and qi is the 95th percentile of T for gene i. Any gene i in cell j with a value Ti,j below ci is set to 0 for the purposes of generating aggregated marker scores.
After these transformations, the aggregated marker score is defined by a simple sum of the genes defined as markers in the cell marker file,
where Sc,j is the aggregated score for cell type c and cell j, and Gc is the list of marker genes for cell type c. Cells in the 75th percentile and above for aggregated marker score S in only 1 cell type are chosen as good representatives. Any metadata specifications (e.g. the requirement that a cell type have come from a particular tissue), provided in the marker file are then used to exclude cells and generate a final training dataset.
Choosing outgroup cells
When choosing outgroup samples for training, we wanted to make sure that the outgroup set is not dominated by the most abundant cell type. So instead, we cluster a random subset of potential outgroup cells and choose equal numbers of random cells from each cluster to make up the outgroup. Specifically, we first calculate the first 50 principal components using principal components analysis (PCA) as implemented by the irlba20 R package. Next, we calculate jaccard coefficients on a k-nearest-neighbors (kNN) graph generated using k = 20. Lastly, we generate clusters using Louvain community detection on the resulting cell-cell map of jaccard coefficients. A random set of cells from each resulting community is then combined to create the outgroup.
Training with GLMnet
The classifier is trained on the normalized expression matrix N for cells chosen as representatives and for all genes expressed in greater than 5% of cells in at least one training set and not expressed in the 90th percentile of TF-IDF transformed expression in all cell types. This last filter prevents ubiquitously expressed genes from being chosen as features. The classifier is trained using genes as features and cells as observations with a grouped multinomial elastic net regularized (alpha = 0.3) generalized linear model using the package GLMnet21 in R. Observations are weighted by the geometric mean of the counts in each of the training groups. The GLMnet regularization parameter λ is chosen using 3-fold cross validation. Genes provided in the marker files are required to be included in the model not regularized.
Classifying cells
Because we wished to be able to use pre-trained classifiers to classify cells across datasets and platforms, we include a dataset size factor D for the training data with the classifier object. D is the geometric mean of the total read counts per cell divided by the median number of genes expressed above zero per cell. Formally, D is defined by
where g is the number of genes expressed above zero per cell. When applying an existing classifier to a new dataset, we can then transform the new expression data, an m′ by n′ matrix M′, to the scale of the training data using D,
where g′ is the number of genes expressed above zero per cell in the new data.
After normalization, gene IDs for the new dataset are also converted to Ensembl IDs. At each internal node in the classifier, the multinomial model for that node is applied to the data, the output probabilities of each class are normalized by dividing by the maximum probability for each cell, and the ratio of the top scoring cell type to the second best scoring cell type is calculated. If this odds ratio is greater than the user-specified rank probability ratio (in this paper and by default, we use 1.5), the top type is assigned, otherwise the cell type is set to “Unknown”. Optionally, Garnett will add a second set of classifications which classify an entire cluster of cells if: greater than 90% of assigned cells within a cluster are the same type and greater than 5% of all cells in the cluster are classified (not “Unknown”), and greater than 5 cells in the cluster are classified. Cluster labels can be provided by the user or generated by Garnett using Louvain community detection in the top 50 principal components of the expression matrix.
10x Genomics Peripheral Blood Mononuclear Cells (PBMCs)
10x PBMC datasets from both version 1 (V1) and version 2 (V2) chemistry were downloaded from the 10x Genomics website. The V1 cells are a combination of each of the pure cell type populations isolated by 10x Genomics using FACS sorting (CD14+ Monocytes, CD19+ B cells, CD34+ cells, CD4+ Helper T cells, CD4+/CD25+ Regulatory T cells, CD4+/CD45RA+/CD25- Naive T cells, CD4+/CD45RO+ Memory T cells, CD56+ Natural killer cells, CD8+ Cytotoxic T cells and CD8+/CD45RA+ Naive cytotoxic T cells) preprocessed using CellRanger 1.1.0 and published in Zheng et. al. The V2 cells are the V2 chemistry distributed demonstration dataset labelled “8k PBMCs from a healthy donor”, preprocessed using CellRanger 2.1.0. Markers for PBMCs were those often cited in the literature. Using Garnett’s marker scoring system, we excluded the markers with high ambiguity (>0.5). The final PBMC marker file used is available as Supplementary Data File 1. Garnett classification for V1 and V2 was run using default parameter values defined in the preceding sections. For testing the limits of Garnett (Supplementary Figure 2), we used a 1-level marker file with no T cell subtypes defined. To downsample the reads in training and classification datasets (Supplementary Figures 2D–E), we used the downsampleMatrix function from the DropletUtils22,23 R package which uses sampling without replacement per cell so that the total reads in that cell is reduced by the specified proportion. For the T cell ablation experiment (Supplementary Figure 2G–H) we removed the T cells from the matrix and also set all values of the T cell marker genes to zero.
Tabula muris and mouse cell atlas MCA lung analysis
The Tabula muris FACS dataset from ref2 was downloaded from their figshare website. The MCA dataset from ref3 was downloaded from their figshare website. For the purposes of this analysis, only data derived from lung tissue from both datasets were used. To facilitate comparisons between each of the lung datasets used, a set of consensus cell type names were used as described in Table 1. The marker file used is available as Supplementary Data File 2. Garnett classification was run using default parameter values for both datasets.
sci-ATAC-seq analysis
The sci-ATAC-seq data was downloaded from the website associated with ref7. The input to Garnett was the previously calculated Cicero gene activity scores presented in the original publication. The final marker file used is available as Supplementary Data File 3. Garnett classification was run using default parameter values.
Worm analysis
The worm data was downloaded from the website associated with ref11. Markers were those used by the original publication to identify cell types. Using Garnett’s marker scoring system, we excluded the markers with high ambiguity (Supplementary Figure 6). The final marker file used is available as Supplementary Data File 4. Garnett classification was run using default parameter values.
Human lung tumor analysis
The human lung tumor data was downloaded from the ArrayExpress database entry associated with ref14. Because expression data were log-transformed, we first exponentiated the expression data before classification. To allow for cross-species classification, we first converted the human expression data to mouse gene labels by creating a correspondence table using the biomaRt hsapiens_gene_ensembl and mmusculus_gene_ensembl databases. Only unique rows (one-to-one correspondences) were used. Ultimately 15,336 of the original 22,180 human genes could be converted to mouse labels including 89 percent of the genes in the MCA classifier with non-zero coefficients. The final marker file used is available as Supplementary Data File 5. Garnett classification was run using default parameter values.
Mouse nervous system analysis
The mouse nervous system data and potential marker genes were downloaded from the website associated with ref12. The final marker file used is available as Supplementary Data File 6.
Data Availability
No new data was generated for this study. All data used in this study is publicly available.
Software Availability
Garnett is an R package available through github.
Supplementary Material
Acknowledgements
We gratefully acknowledge S. Tapscott, W. Noble, and D. Witten as well as members of the Shendure and Trapnell labs, particularly A. Hill, for their advice. Z. Pliner named the software. This work was supported by the following funding: NIH grant U54DK107979 to JS and CT; NIH grant DP2HD088158, RC2DK114777 and R01HL118342 to CT; NIH grants DP1HG007811 and R01HG006283 to JS; and the Paul G. Allen Frontiers Group to JS and CT. JS is an Investigator of the Howard Hughes Medical Institute. CT is partly supported by an Alfred P. Sloan Foundation Research Fellowship. HAP was supported by the National Science Foundation Graduate Research Fellowship under Grant No. (DGE-1256082).
Footnotes
Competing Financial Interests Statement
The authors declare no competing interests.
REFERENCES
- 1.Svensson V, Vento-Tormo R & Teichmann SA Exponential scaling of single-cell RNA-seq in the past decade. Nat. Protoc 13, 599–604 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Tabula Muris Consortium et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han X et al. Mapping the Mouse Cell Atlas by Microwell-Seq. Cell 173, 1307(2018). [DOI] [PubMed] [Google Scholar]
- 4.Zhang X et al. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 47, D721–D728 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diehl AD et al. The Cell Ontology 2016: enhanced content, modularization, and ontology interoperability. J. Biomed. Semantics 7, 44(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bard J, Rhee SY & Ashburner M An ontology for cell types. Genome Biol. 6, R21(2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cusanovich DA et al. A Single-Cell Atlas of In Vivo Mammalian Chromatin Accessibility. Cell 174, 1309–1324.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng GXY et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8, 14049(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levine JH et al. Data-Driven Phenotypic Dissection of AML Reveals Progenitor-like Cells that Correlate with Prognosis. Cell 162, 184–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pliner HA et al. Cicero Predicts cis-Regulatory DNA Interactions from Single-Cell Chromatin Accessibility Data. Mol. Cell 71, 858–871.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao J et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeisel A et al. Molecular Architecture of the Mouse Nervous System. Cell 174, 999–1014.e22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Merkin J, Russell C, Chen P & Burge CB Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science 338, 1593–1599 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lambrechts D et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat. Med 24, 1277–1289 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Rosenberg AB et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 360, 176–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alavi A, Ruffalo M, Parvangada A, Huang Z & Bar-Joseph Z A web server for comparative analysis of single-cell RNA-seq data. Nat. Commun 9, 4768(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou H & Hastie T Regularization and variable selection via the elastic net. J. R. Stat. Soc. Series B Stat. Methodol 67, 301–320 (2005). [Google Scholar]
- 18.Carlson M, Falcon S, Pages H & Li N AnnotationDbi: Annotation Database Interface. R package version 1, [Google Scholar]
- 19.Jones KS A STATISTICAL INTERPRETATION OF TERM SPECIFICITY AND ITS APPLICATION IN RETRIEVAL. Journal of Documentation 28, 11–21 (1972). [Google Scholar]
- 20.Baglama J, Reichel L & Lewis BW irlba: Fast truncated singular value decomposition and principal components analysis for large dense and sparse matrices. R package version 2, (2017). [Google Scholar]
- 21.Friedman J, Hastie T & Tibshirani R glmnet: Lasso and elastic-net regularized generalized linear models. R package version 1, (2009). [Google Scholar]
- 22.Lun A et al. Distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. doi: 10.1101/234872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Griffiths JA, Richard AC, Bach K, Lun ATL & Marioni JC Detection and removal of barcode swapping in single-cell RNA-seq data. Nat. Commun 9, 2667(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No new data was generated for this study. All data used in this study is publicly available.