Abstract
Background:
Arsenic exposure affects people worldwide, including in Bangladesh. Arsenic exposure increases the risk of cancer and other chronic diseases, and one potential mechanism of arsenic toxicity is epigenetic dysregulation.
Objective:
We assessed associations between arsenic exposure and genome-wide DNA methylation measured at baseline among 396 Bangladeshi adults participating in the Health Effects of Arsenic Longitudinal Study (HEALS) who were exposed by drinking naturally contaminated well water.
Methods:
Methylation in whole blood DNA was measured at using the Illumina InfiniumMethylationEPIC (EPIC) array. To assess associations between arsenic exposure and CpG methylation, we used linear regression models adjusted for covariates and surrogate variables (SVs) (capturing unknown technical and biologic factors). We attempted replication and conducted a meta-analysis using an independent dataset of from 400 Bangladeshi individuals with arsenical skin lesions.
Results:
We identified 34 CpGs associated with creatinine-adjusted urinary arsenic []. Sixteen of these CpGs annotated to the array, and 10 associations were replicated (). The top two CpGs annotated upstream of the ABR gene (cg01912040, cg10003262). All urinary arsenic–associated CpGs were also associated with arsenic concentration measured in drinking water (). Meta-analysis ( samples) identified 221 urinary arsenic–associated CpGs (). The arsenic-associated CpGs from the meta-analysis were enriched in non-CpG islands and shores () and depleted in promoter regions (). Among the arsenic-associated CpGs (), we observed significant enrichment of genes annotating to the reactive oxygen species pathway, inflammatory response, and tumor necrosis factor () signaling via nuclear factor kappa-B () hallmarks ().
Conclusions:
The novel and replicable associations between arsenic exposure and DNA methylation at specific CpGs observed in this work suggest that epigenetic alterations should be further investigated as potential mediators in arsenic toxicity and as biomarkers of exposure and effect in exposed populations. https://doi.org/10.1289/EHP3849
Introduction
Arsenic exposure from naturally contaminated drinking water is a global public health concern that affects people worldwide (Naujokas et al. 2013), and in Bangladesh (Flanagan et al. 2012). Prior to remediation efforts, of the drinking water wells in Bangladesh contained water with inorganic arsenic concentrations above , the World Health Organization (WHO) recommended maximum level of exposure (Ahsan et al. 2000; Anawar et al. 2002; Van Geen et al. 2002). Inorganic arsenic is classified as a group I carcinogen by the International Agency for Research on Cancer, and long-term exposure is associated with risk for nonmelanoma skin (Karagas et al. 2015), lung (Lamm et al. 2015), and bladder cancer (Gamboa-Loira et al. 2017; Lamm et al. 2015) as well as arsenical skin lesions (a hallmark of chronic arsenic exposure). Chronic environmental arsenic exposure also increases the risk of other chronic diseases, including cardiovascular diseases (Moon et al. 2017; Navas-Acien et al. 2005) and respiratory disease (Sanchez et al. 2018) and overall mortality among Bangladeshi adults (Argos et al. 2010).
Arsenic is not considered to be directly genotoxic, and potential mechanisms of arsenic toxicity include induction of oxidative stress and inflammation, inhibition of DNA repair, and epigenetic dysregulation, including alteration of DNA methylation. DNA methylation is characterized by the addition of a methyl group to a cytosine nucleotide, frequently located within CpG sites in the genome. DNA methylation (and the underlying chromatin state it represents) provides an additional level of transcriptional regulation and has an important role in maintaining genomic stability. Exposure to environmental toxicants, like arsenic, can induce changes in both global and gene-specific DNA methylation (Cortessis et al. 2012; Martin and Fry 2018). Some potential mechanisms by which arsenic could disrupt DNA methylation include interaction with methylation or chromatin maintenance machinery, depletion of cofactors involved in DNA methylation synthesis, interaction with transcription factor binding sites (TFBS), and alteration of the inflammatory and oxidative environment of the cell (Bailey and Fry 2014; Ren et al. 2011). Arsenic-induced alterations of DNA methylation may be an important component in the mechanism of arsenic toxicity and carcinogenesis.
While several studies have examined prenatal arsenic exposure and genome-wide DNA methylation (Cardenas et al. 2015a, 2015b; Green et al. 2016; Kile et al. 2014; Koestler et al. 2013; Rojas et al. 2015), only four studies have examined arsenic exposure and genome-wide DNA methylation in adults (Ameer et al. 2017; Argos et al. 2015; Liu et al. 2014; Seow et al. 2014). Argos et al. identified four urinary arsenic–associated CpGs and three blood arsenic–associated CpGs (Bonferroni threshold of ) in Bangladeshi adults (), with a wide range of environmental arsenic exposure and diagnosed with skin lesions (Argos et al. 2015). Liu et al. identified 22 differentially methylated CpGs () in adults () from the United States with low arsenic exposure compared with adults in Bangladesh (Liu et al. 2014). Ameer et al. identified six urinary arsenic–associated CpGs [] among Andean women from Argentina () with a wide range of environmental exposure, similar to Bangladesh (Ameer et al. 2017). Seow et al. conducted a pilot study comparing genome-wide DNA methylation between 10 arsenical skin lesions cases and 10 lesion-free controls from Bangladesh, but no significant associations with methylation were observed (Seow et al. 2014). These prior studies differed with respect to study population, sample size, exposure assessment and level, disease status, and statistical approaches, likely contributing to the variability in CpGs identified and patterns of genome-wide methylation observed (Argos 2015). Prior studies used the Illumina array, while the newer Illumina InfiniumMethylationEPIC (EPIC) array measures methylation at approximately 850,000 CpGs, including of the CpGs on the array as well as substantial coverage of CpGs in enhancer regions and CpG shores (Pidsley et al. 2016). By including these additional sites, the EPIC array enables evaluation of methylation within distal regulatory regions of the genome that may be more susceptible to environmental exposures, like arsenic, since methylation at enhancers is more variable and dynamic and not as well preserved (Jones 2012).
In this study, we assess the association between exposure to arsenic measured in urine (prior to arsenic mitigation efforts) and genome-wide DNA methylation assessed using the EPIC array in 396 adults from the Health Effects of Arsenic Longitudinal Study (HEALS). HEALS is a population-based cohort of arsenic-exposed Bangladeshi adults established to assess the health effects associated with consumption of arsenic-contaminated water (Ahsan et al. 2006a). We validate arsenic-associated CpGs by examining their association with arsenic concentration in drinking water and replicate observed associations in an independent cohort of 400 Bangladeshi adults. We then conduct a meta-analysis of both cohorts to identify additional putative arsenic-associated CpGs.
Methods
Study Population
HEALS was initiated to prospectively investigate the health outcomes associated with chronic arsenic exposure through consumption of groundwater in a sample of Bangladeshi adults with homogenous ethnic and sociocultural characteristics () and little to no genetic admixture in Araihazar, Bangladesh (Pierce et al. 2012). This study has been described previously (Ahsan et al. 2006a). HEALS participants were recruited between October 2000 and May 2002. Participants were sampled from married couples between ages 18–75 y who resided in study area for at least 5 y. Trained study physicians (blinded to arsenic exposure) conducted in-person interviews, clinical evaluations, and skin lesion assessment, and collected urine and blood samples using structured protocols (Ahsan et al. 2006b). Participants in this epigenome-wide association study (EWAS) () were randomly selected from the random subcohort recently included in a genome-wide association study (Pierce et al. 2012).
Data from the Bangladesh Vitamin E and Selenium Trial (BEST) were used to replicate associations observed in HEALS. BEST is a factorial randomized chemoprevention trial evaluating the effects of vitamin E and selenium dietary supplemental and skin cancer risk among individuals with arsenical skin lesions from rural central Bangladesh (Argos et al. 2013). BEST participants were recruited between August 2006 and August 2009 and were between 25–65 y old, resided in the study area, and had arsenical skin lesions. Arsenic exposure was assessed in urine, and whole blood was collected at baseline for all BEST participants. Genome-wide methylation was previously measured on the array for 400 BEST participants, and the results were reported by Argos et al. (2015). The study protocols were approved by the Institutional Review Boards of the University of Chicago, Columbia University, and the Bangladesh Medical Research Council. Informed consent was obtained from all participants.
Exposure Assessment
Arsenic was measured in both urine and water for each HEALS participant. At baseline, each participant identified the primary well used as their main source of drinking water. Spot urine samples were obtained from each participant at baseline. Arsenic in urine and water was measured using graphite furnace atomic absorption spectrometry (AAnalyst 600 spectrometer; Perkin Elmer) in a single laboratory (Trace Metal Core Laboratory at Columbia University), and the limit of detection for this method was and for urine and water, respectively (Nixon et al. 1991). For arsenic in well water, any samples that were below the limit of detection were reanalyzed using inductively coupled plasma mass spectrometry with a detection limit of (Cheng et al. 2004). Urine creatinine was measured by a colorimetric Sigma Diagnostics Kit (Sigma). Total urinary arsenic was divided by urine creatinine to obtain creatinine-adjusted urine arsenic ( creatinine).
DNA Methylation
DNA was extracted from clotted blood using FlexiGene DNA Kit (Qiagen). We then bisulfite converted of DNA using EZ-96 DNA Methylation™ Kit (Zymo Research). All samples were then prepared and analyzed in accordance with the manufacturer guidelines and protocol for the Infinium MethylationEPIC array (Illumina). The EPIC array measured methylation at 866,895 CpGs. We removed CpGs with a detection in one or more samples () or missing methylation in of samples (). Cross-reactive CpGs were removed (), as well as CpGs annotating to a single-nucleotide polymorphism (SNP) or within a single base pair extension () (Pidsley et al. 2016). We removed CpGs annotating to the X and Y chromosomes to avoid potential gender bias in methylation patterns as well as non-CpG and SNP probes on the array (). The total CpGs included in the analysis were 771,192.
One sample from HEALS was removed due to mismatched sex. After imputing beta values for missing CpG methylation using k-nearest neighbors method () (Troyanskaya et al. 2001), the dataset was normalized using the beta mixture quantile (BMIQ) method. BMIQ is an assumption-free approach to adjust for type I/II probe bias (Teschendorff et al. 2013). We applied the ComBat function in R to adjust for batch effect due to plate (Johnson et al. 2007; Leek et al. 2012). This batch effect was removed after reviewing the principal components analysis of the batch-adjusted data. All processing of DNA methylation array data and analyses were conducted in R 3.4.1.
Association Analyses
All statistical analyses were performed using logit transformed beta values and M-values {log2[beta/(1 − beta)]}, unless noted. The range of beta values is 0 (0% methylated) to 1 (100% methylated), while M-values range from to . We selected covariates a priori and adjusted for age (continuous variable), sex, smoking status (categorized as never, former, and current smoker based on self-report), and body mass index (BMI) category [categorized as normal (reference), underweight, overweight, obese, and unknown]. We urinary and water arsenic to reduce the influence of outlying values; arsenic exposure was also modeled as an untransformed continuous and an ordinal variable across integer-coded quartiles [first (), second (), third (), fourth ()]. We applied surrogate variable analysis (SVA) to adjust for unknown biologic and technical effects (Leek and Storey 2007; Leek et al. 2012). SVA has been demonstrated to perform stably (McGregor et al. 2016) and optimally identify informative CpGs in genome-wide methylation studies (Kaushal et al. 2017), and it was chosen over a cell type–adjusted approach (Houseman et al. 2012) as our primary analysis approach since it produced a lower genomic inflation factor. We estimated surrogate variables (SVs) using a permutation-based approach (Buja and Eyuboglu 1992). For each CpG, we analyzed the association with arsenic using linear regression adjusting for SVs and covariates in limma in R (Ritchie et al. 2015) and computed the genomic inflation factor () for EWAS (Aulchenko et al. 2007; van Iterson et al. 2017). A Bonferroni threshold was determined at , our threshold for epigenome-wide testing. The FDR p-values were computed using the Benjamini-Hochberg method (Hochberg and Benjamini 1990). -tests were applied to compare the distribution of arsenic-associated CpGs to the remaining CpGs across genomic features. UCSC genome (http://genome.ucsc.edu/), FANTOM5 (Andersson et al. 2014), and ENCODE (Thurman et al. 2012) annotations were provided by Illumina and used to annotate and assign genomic features to CpGs based on human reference genome GRCh37. If CpGs were assigned TSS200 or TSS1500 (at least 200 and 1500 base pairs upstream of transcriptional start site), we annotated these CpGs as being located in a promoter region. DNase I hypersensitive sites (DHS) and TFBS regions were indicated by evidence from ENCODE (Thurman et al. 2012) while enhancer regions were indicated by supporting evidence from FANTOM5 (Andersson et al. 2014). In a sensitivity analysis, we also analyzed our data using a reference-based approach, estimating six cell type proportions (monocytes, B-cells, granulocytes, CD8T, CD4T, and natural killer cells) from a whole blood reference panel (Houseman et al. 2012). For the reference-based approach, we performed linear regression adjusting for estimated cell type proportions and covariates using limma.
Differential Methylation Regional Analysis
We examined associations between urinary arsenic in HEALS and differentially methylated regions (DMRs) using DMRcate (Peters et al. 2015). DMRcate is agnostic to the direction of the association for each CpG and all annotations except those related to the spatial location (i.e., chromosome number and map position). DMRs are then defined by agglomerating CpG locations with an adjusted p-value below a selected threshold (based on FDR) that are at most nucleotides from each other. Each DMR is assigned a minimum FDR (minFDR) that is from the lowest adjusted p-value among the CpG locations contained within it. We searched for DMRs using a bandwidth of 1,000 nucleotides () and scaling factor of 2 for the bandwidth () (recommended parameters for and EPIC arrays). We restricted to DMRs that contained at least two CpGs, and applied a stringent cutoff threshold of . The genomic region for the top three DMRs are visualized using coMET (Martin et al. 2015).
Gene Set Enrichment Analyses
To evaluate the annotation of urinary arsenic–associated CpGs to pathways and biologic processes from the discovery analysis and the meta-analysis, we conducted gene set enrichment analyses (GSEA) using the gometh function in missMethyl (Phipson et al. 2016). Gometh accounts for the potential bias in GSEA due to number of CpGs per gene by computing prior probabilities (Geeleher et al. 2013) and evaluates enrichment using a hypergeometric test. We tested enrichment among arsenic-associated CpGs among the top 500 arsenic-associated CpGs and arsenic-associated CpGs below FDR of 0.05 within gene sets from the KEGG pathways (https://www.genome.jp/kegg/) ( pathways) and hallmark gene set collection ( sets), concise sets curated from multiple founder gene sets and gene expression datasets (Liberzon et al. 2015). Our justification for examining enrichment among the top 500 arsenic-associated CpGs was that it was difficult to examine genomic enrichments among a limited number of arsenic-associated CpGs at an , and arsenic likely has numerous biologic effects on the epigenome that might not reach epigenome-wide significance. We extracted results with since our analysis was exploratory and underpowered to detect enrichment at more stringent significance thresholds.
Meta-Analysis
We conducted a meta-analysis of genome-wide methylation and urinary arsenic using data from HEALS () and the BEST cohort (). Using the same procedure applied to the HEALS EPIC array data, we preprocessed, BMIQ normalized, estimated SVs, and analyzed the BEST array data. Among the overlapping CpGs in HEALS and BEST (), the association estimate and p-value for each CpG were obtained from BEST and HEALS and meta-analyzed using the sample size based approach in METAL software (Willer et al. 2010).
Gene Expression
Gene expression data were only available for the BEST participants included in this study and have been previously described (Argos et al. 2015; Gao et al. 2015). Briefly, gene expression was measured using the Illumina HT-12 v4 BeadChip according to manufacturer’s protocol. The chip includes 37,231 probes and covers 31,335 genes. The gene expression values were quantile normalized and then . COMBAT was used to adjust for batch effect (Johnson et al. 2007). We extracted gene expression probes corresponding to genes that annotated to our arsenic-associated CpGs identified in the meta-analysis for 371 BEST participants with expression data. To assess the associations between gene expression and methylation, we ran Pearson correlations and linear models adjusted for age and sex, and extracted the direction and p-value for each association.
Local Methylation Quantitative Trait Loci (cis-mQTL) Analyses of Arsenic-Associated CpGs
HEALS participants were genotyped on the Illumina HumanCytoSNP-12 v2.1 array with 299,140 markers. We used MaCH software (Li et al. 2010) to conduct genotype imputation using 1,000 genomes reference haplotypes (version 5; 1000G Phase 3). We examined cis associations between SNPs and CpGs within a 1-megabase window ( upstream and downstream of the CpG) using genotype dosages and matrixeQTL software (Shabalin 2012). The model was adjusted for age, sex, and first four methylation principal components (PCs). We summarized the total SNPs tested for each CpG and number that reached FDR of 0.01, and reported the genomic information for the lead SNP (smallest p-value) and distance from CpG.
Replication Analysis for Meta-Analysis Arsenic-Associated CpGs among Andean Women
We attempted replication of the urinary arsenic–associated CpGs identified the meta-analysis () using data from an independent population of Andean women from Argentina that have been previously published (Ameer et al. 2017). Methylation was measured on the array for 93 women. For each CpG, the association between methylation (M-values) and urinary arsenic was examined and adjusted for age, coca usage (yes/no), and estimated fractions of granulocyte and natural killer cells. We extracted the direction of association and p-value for 217 of our 221 CpGs with available data from the Ameer et al. (2017) analysis.
Results
Our HEALS subcohort used for the discovery EWAS () was randomly drawn from previously genotyped participants in HEALS (also randomly selected). The median age of our subcohort was 36.5 y [interquartile range (IQR): 30.0, 45.0 y] (Table 1), and there were more women (58%) than men (42%). Current smokers comprised 34% of the HEALS subcohort. At baseline, 76% of our participants consumed water from arsenic-contaminated hand-pumped tube wells with estimated arsenic concentrations above the WHO guideline of , and 50% of our participants consumed water with concentrations above the national Bangladesh standard of . The median urinary arsenic was creatinine (IQR: 113.5, ).
Table 1.
Subcohort descriptive statistics [median (25th, 75th percentiles) or n (%)] for HEALS (discovery) and BEST (validation).
| HEALS cohort () | HEALS subcohort (discovery; ) | BEST subcohort (validation; ) | |
|---|---|---|---|
| Age (years) | 36.0 (29.0,45.0) | 36.5 (30.0, 45.0) | 44.0 (35.0, 50.3) |
| Urinary arsenic () | 199.0 (106.0, 352.0) | 201.5 (113.5, 350.0) | 137.3 (76.2, 394.4) |
| Water arsenic () | 60.0 (12.0,147.0) | 50.5 (11.0, 124.8) | Not measured |
| Sex | |||
| Male | 4,855 (43.3%) | 167 (42.2%) | 212 (53.0%) |
| Female | 6,369 (56.7%) | 229 (57.8%) | 188 (47.0%) |
| Smoking | |||
| Never | 7,204 (64.2%) | 240 (60.6%) | 251 (62.8%) |
| Former | 743 (6.6%) | 21 (5.3%) | 40 (10.0%) |
| Current | 3,271 (29.1%) | 135 (34.1%) | 109 (27.3%) |
| Unknown | 6 (0.1%) | 0 (0.0%) | 0 (0.0%) |
| BMI () | |||
| Normal (18.5–22.9) | 5,067 (45.1%) | 175 (44.2%) | 176 (44.0%) |
| Underweight () | 4,421 (39.4%) | 154 (38.9%) | 150 (37.5%) |
| Overweight (23.0–29.9) | 1,575 (14.0%) | 61 (15.4%) | 73 (18.3%) |
| Obese () | 80 (0.7%) | 5 (1.3%) | 1 (0.3%) |
| Unknown | 81 (0.7%) | 1 (0.3%) | 0 (0.0%) |
Note: BEST, Bangladesh Vitamin E and Selenium Trial; BMI, body mass index; HEALS, Health Effects of Arsenic Longitudinal Study.
Urinary arsenic–associated DNA Methylation in HEALS
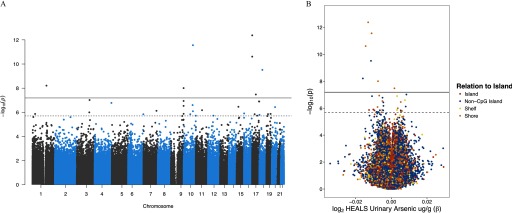
We conducted an EWAS of urinary arsenic (creatinine adjusted) using data on 771,192 CpGs measured on the EPIC array among 396 HEALS participants (Figure 1; Figure S1; Excel Table S1). After adjustment for covariates and SVs (), 34 CpGs were associated with urinary arsenic at an FDR of 0.05 () (Table 2; Figure S2). From the analysis of beta values (range 0 to 1), the association estimates across these 34 CpGs ranged from 0.016 decrease to a 0.008 increase in methylation per doubling of urinary arsenic. Decreased methylation was associated with increased arsenic exposure for 23 of the 34 associated CpGs (). Arsenic-associated CpGs () were predominantly located in CpG shores () and non-CpG islands () (Figure 1B). Seven CpGs that passed Bonferroni-threshold () were located upstream of the ABR (ABR activator of RhoGEF and GTPase) gene on chromosome 17 (cg01912040, cg10003262), in the SEMA4G (semaphorin 4G) gene body on chromosome 10 (cg05962511), in the MAPRE2 (microtubule associated protein RP/EB family member 2) gene body (cg17420142), in the GBAP1 (glucosylceramidase beta pseudogene 1) gene body (cg06466147), in the NSMF gene body (cg09082427), and in the NBR1 (NBR1 autophagy cargo receptor) 5′ untranslated region (UTR) (cg04193083).
Figure 1.
Genome-wide associations between urinary arsenic concentration and CpG site–specific methylation in the Health Effects of Arsenic Longitudinal Study (HEALS). Using the EPIC () array, the association between urinary arsenic (creatinine adjusted) and methylation was evaluated at 771,192 CpG sites across 396 individuals from HEALS. (A) Manhattan plot of the chromosomal location and p-value for each CpG–arsenic association. (B) Volcano plot presenting association estimate and p-value for each CpG–arsenic association. Colors correspond to CpG relationship to island. In (A) and (B), solid and dashed lines designate the Bonferroni threshold () and false discovery rate (FDR) 0.05 threshold (), respectively.
Table 2.
CpGs associated with urinary arsenic () discovered in HEALS (EPIC array).
| HEALS (discovery, EPIC) | BESTd (validation 450 K) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Chr | Position | CpG Location | Nearest gene | Feature | Distance (basepairs) | Mean (SD)a | b | c | Mean (SD)a | b | ||
| cg01912040 | 17 | 1106553 | Shore | ABR | Upstream | 15,937 | 0.73 (0.05) | 0.72 (0.06) | |||||
| cg05962511 | 10 | 102730022 | Shore | SEMA4G | Body | 732 | 0.35 (0.05) | 0.34 (0.05) | |||||
| cg10003262 | 17 | 1106589 | Shore | ABR | Upstream | 15,973 | 0.34 (0.06) | 0.26 (0.05) | |||||
| cg17420142 | 18 | 32702783 | Non-CpG island | MAPRE2 | Body | 81,169 | 0.59 (0.05) | — | — | — | |||
| cg06466147 | 1 | 155188982 | Non-CpG island | GBAP1 | Body | 19,459 | 0.62 (0.07) | — | — | — | |||
| cg09082427 | 9 | 140349184 | Shore | NSMF | Body | 4,602 | 0.75 (0.05) | — | — | — | |||
| cg04193083 | 17 | 41323562 | Shore | NBR1 | 5'UTR | 316 | 0.04 (0.02) | 0.002 | 0.03 (0.01) | 0.000 | |||
| cg19534475 | 3 | 141632139 | Non-CpG island | ATP1B3 | Body | 36,669 | 0.47 (0.04) | 0.008 | — | — | — | ||
| cg12608784 | 9 | 140349197 | Shore | NSMF | Body | 4,589 | 0.69 (0.05) | — | — | — | |||
| cg14891900 | 17 | 76341204 | Shelf | SOCS3 | Downstream | 14,954 | 0.91 (0.01) | 0.003 | 0.93 (0.01) | ||||
| cg11308227 | 17 | 79202435 | Non-CpG island | ENTHD2 | 3'UTR | 10,456 | 0.54 (0.06) | — | — | — | |||
| cg13832772 | 4 | 186283800 | Non-CpG island | SNX25 | Body | 41,913 | 0.87 (0.03) | 0.94 (0.02) | 0.000 | ||||
| cg15108641 | 10 | 99263320 | Shelf | UBTD1 | Body | 4,552 | 0.35 (0.06) | 0.008 | — | — | — | ||
| cg09658504 | 9 | 140349188 | Shore | NSMF | Body | 4,598 | 0.47 (0.04) | — | — | — | |||
| cg05438461 | 15 | 40401720 | Shore | BMF | Promoter | 632 | 0.20 (0.03) | 0.005 | — | — | — | ||
| cg10663081 | 20 | 36837543 | Non-CpG island | KIAA1755 | Downstream | 51,631 | 0.47 (0.03) | — | — | — | |||
| cg08759026 | 11 | 69061454 | Non-CpG island | MYEOV | Promoter | 159 | 0.24 (0.04) | 0.27 (0.04) | |||||
| cg11644394 | 7 | 144148615 | Non-CpG island | TPK1 | Downstream | 419 | 0.62 (0.06) | 0.008 | — | — | — | ||
| cg05428706 | 10 | 102730130 | Shore | SEMA4G | Body | 840 | 0.79 (0.04) | 0.76 (0.04) | |||||
| cg12865207 | 3 | 138669373 | Shore | FOXL2NB | Body | 3,297 | 0.15 (0.03) | 0.004 | — | — | — | ||
| cg09183146 | 16 | 1429863 | Island | UNKL | Promoter | 34,842 | 0.26 (0.05) | 0.24 (0.05) | |||||
| cg04622454 | 9 | 140349128 | Shore | NELF | Body | 4,658 | 0.63 (0.04) | 0.57 (0.04) | |||||
| cg06381803 | 19 | 46119475 | Island | EML2 | Body | 29,300 | 0.41 (0.07) | 0.40 (0.07) | |||||
| cg02772605 | 1 | 28912323 | Shelf | SNHG12 | Upstream | 3957 | 0.88 (0.02) | 0.003 | 0.89 (0.02) | 0.000 | |||
| cg10283165 | 19 | 17375666 | Non-CpG island | USHBP1 | Promoter | 61 | 0.39 (0.05) | — | — | — | |||
| cg04891961 | 17 | 27939900 | Island | ANKRD13B | Body | 19,373 | 0.49 (0.07) | 0.47 (0.08) | 0.002 | ||||
| cg12746706 | 6 | 169276508 | Non-CpG island | SMOC2 | Upstream | 207,834 | 0.86 (0.03) | 0.94 (0.01) | |||||
| cg02330195 | 10 | 73342047 | Non-CpG island | CDH23 | Body | 142,463 | 0.67 (0.04) | 0.005 | — | — | — | ||
| cg08077890 | 18 | 157838 | Shore | USP14 | Promoter | 645 | 0.77 (0.04) | 0.006 | — | — | — | ||
| cg20433952 | 17 | 55607898 | Non-CpG island | MSI2 | Body | 273,524 | 0.51 (0.04) | 0.52 (0.04) | |||||
| cg10185759 | 12 | 60366859 | Non-CpG island | SLC16A7 | Downstream | 283,741 | 0.46 (0.04) | — | — | — | |||
| cg05646745 | 10 | 135172466 | Shore | FUOM | Promoter | 937 | 0.73 (0.04) | 0.005 | — | — | — | ||
| cg22345623 | 9 | 125050297 | Non-CpG island | MRRF | Body | 17,154 | 0.78 (0.05) | — | — | — | |||
| cg13480898 | 19 | 10195914 | Shore | C19orf66 | Promoter | 892 | 0.73 (0.05) | 0.76 (0.04) | |||||
Note: Results from the discovery analysis are shown for CpGs associated with urinary arsenic (creatinine adjusted) below false discovery rate (FDR) of 0.05 from models adjusted for age, sex, BMI category, smoking status, and 27 SVs. The p-values are obtained from the analyses using methylation expressed as M-values as the outcome variable, and reported association estimates () are from analysis with methylation expressed as beta values as the outcome variable (for easier interpretability). Gene assignments are from UCSC gene annotation provided by Illumina. The replication results from BEST are also presented for the CpGs measured on both the EPIC and arrays. Extended results for arsenic-associated CpGs with are presented in Excel Table S1. —, no data; BEST, Bangladesh Vitamin E and Selenium Trial; BMI, body mass index; Chr, chromosome; HEALS, Health Effects of Arsenic Longitudinal Study; SD, standard deviation; SV, surrogate variable.
Mean and SD of beta values at each CpG (range: 0 to 1) in HEALS and BEST.
and are association estimate and p-value from covariate and SV-adjusted analysis model.
is p-value from the analysis model of urinary arsenic and adjusted for covariates and estimated cell type proportions.
Results from the replication analysis in BEST are shown for urinary arsenic–CpG associations from models adjusted for age, sex, BMI category, smoking status, and 24 SVs.
The results from the urinary arsenic analysis were compared with results from analyses where urinary arsenic was modeled as an untransformed continuous or an ordinal quartile (integer-coded) variable. Across arsenic exposure models, the genomic inflation was similar, and all untransformed continuous and ordinal quartile arsenic-associated CpGs () overlapped with arsenic-associated CpGs (; ) (Table S1). Among the 34 arsenic-associated CpGs identified (), 31 were significantly associated with urinary arsenic () prior to adjustment for covariates and SVs. Of the 34 arsenic-associated CpGs identified from the SV model, 68% () and 97% () were also associated with urinary arsenic at FDR of 0.05 and , respectively, in the cell type–adjusted model. When we examined the associations between urinary arsenic and estimated cell type proportions, urinary arsenic was associated with decreased CD4T percentage [ (0.2); ] and increased granulocyte percentage [ (0.4); ] after adjustment for age, sex, smoking status, and BMI (Table S2), suggesting that urinary arsenic was modestly associated with variation in estimated cell composition.
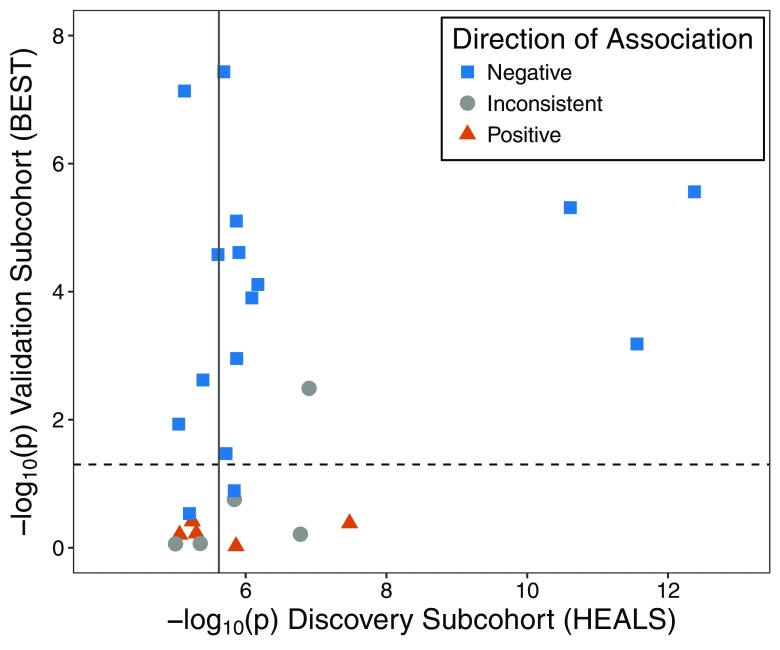
Among the 34 urinary arsenic–associated CpGs () discovered in HEALS, we attempted replication for the 16 CpGs present on the array using previously measured methylation data from 400 BEST participants. Ten CpGs replicated with consistent direction of association in BEST () (Table 2). Among the urinary arsenic–associated CpGs discovered in HEALS at the threshold (), 26 CpGs were measured in BEST (Figure 2; Excel Table S1). Among these 26 CpGs, 15 (58%) were associated with urinary arsenic (), and 14 (93%) were directionally consistent in BEST.
Figure 2.
Associations between urinary arsenic and CpG methylation discovered in the Health Effects of Arsenic Longitudinal Study (HEALS) and tested for replication in the Bangladesh Vitamin E and Selenium Trial (BEST). Using results from BEST, we attempted to validate 26 arsenic-associated CpGs that annotated to array among the 67 CpGs identified using the EPIC array in HEALS (). The BEST results consisted of associations between urinary arsenic and methylation evaluated for 390,810 CpGs using data on 400 BEST participants with existing array data. Horizontal dashed line represents , and solid vertical line corresponds to . Direction of association summarizes whether results from the cohorts are both positive (triangle), both negative (square), or inconsistent (directions of association differ) (circle). The corresponding results data are presented in Table 2 and Excel Table S1.
Water Arsenic–Associated DNA Methylation in HEALS
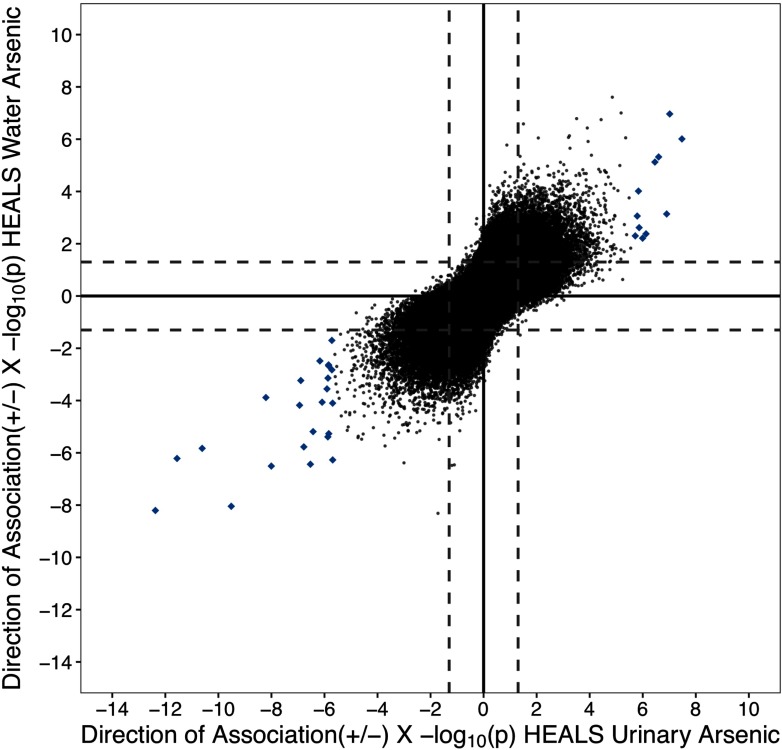
To examine whether urinary arsenic–associated CpGs were consistent across different exposure assessments of arsenic, we examined arsenic in drinking water and its association with genome-wide methylation. Both water and urinary arsenic were measured in HEALS, and the correlation between the two measures was 0.61 () (Figure S3). Four CpGs passed a Bonferroni threshold () when we assessed associations with water arsenic (Figures S4; Figure S5; Excel Table S2). At an FDR of 0.05, 24 water arsenic–associated CpGs were identified. Among the urinary arsenic–associated CpGs (, ), all 34 CpGs were associated with water arsenic (i.e., and consistent direction), and eight were associated at a FDR of 0.05. When we examined all CpG methylation and arsenic associations across the epigenome for both water and urine, the correlation between the and direction of association was 0.67 (), mirroring the correlation we observed between the two exposures (Figure 3).
Figure 3.
Comparison between the associations of urinary arsenic and water arsenic with genome-wide methylation in the Health Effects of Arsenic Longitudinal Study (HEALS). The direction of association and for each CpG–arsenic association are plotted for urinary (x-axis) and water (y-axis) (both ). Solid lines and dashed lines designate and , respectively. CpGs highlighted in blue diamonds denote urinary arsenic–associated sites []. Pearson’s correlation between all urinary and water arsenic CpG associations was 0.67 ().
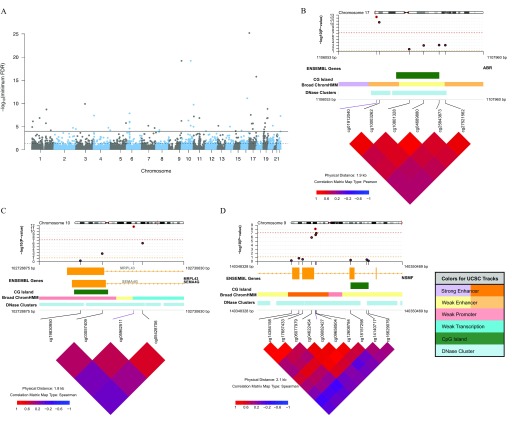
Arsenic-Associated Regions in HEALS
To identify regions of DNA methylation associated with urinary arsenic, we searched for DMRs using the DMRcate method. We identified 45 DMRs with (Figure 4A; Table S3). Eight CpGs individually associated with urinary arsenic () annotated to six of these DMRs based on region start and end locations (see CpG location in Table 2). The average methylation decreased with increased urinary arsenic in 64.4% () of these DMRs. Most DMRs (82.2%; ) contained CpGs annotated to a gene; however, we also identified eight intergenic DMRs. Among the DMRs that annotated to a gene, the DMRs most frequently spanned promoters (), gene bodies (), and 5′UTRs (). The top three DMRs were a) upstream of ABR on chromosome 17 (), overlapping an enhancer region (Figure 4B); b) in the body of SEMA4G on chromosome 10 (), overlapping a weak promoter (Figure 4C); and c) a region within the NSMF gene on chromosome 9 (), overlapping an enhancer region (Figure 4D). All three regions spanned a DNase cluster region. The mean effect size related to the arsenic-associated change in DNA methylation (beta values) across the DMR was for ABR, for SEMA4G, and for NSMF.
Figure 4.
Differentially methylated regions (DMRs) associated with urinary arsenic in the Health Effects of Arsenic Longitudinal Study (HEALS). Using 771,192 CpGs from the EPIC array, DMRs were identified using DMRcate ( base pairs and ). (A) Manhattan plot of the minimum false discovery rate (minFDR) for the CpG within all regions identified across EPIC array (no significance threshold set) ( regions). Solid and dashed lines designate minFDR of and minFDR of 0.05, respectively. (B–D) coMET plots of top three DMRs associated with urinary arsenic. Genomic annotations to gene, CpG islands, chromatin regulation, and DNase clusters are shown below each plot and are from UCSC CpG Island, UCSC DNase Cluster, and the Broad UCSC ChromatinHMM tracks (http://genome.ucsc.edu/, see coMET software documentation for further information and color coding) (Martin et al. 2015).
Enrichment of Arsenic-Associated CpGs within Genomic Features and Gene Sets
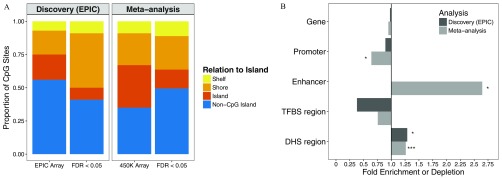
We examined the enrichment of arsenic-associated CpGs within promoters, enhancers, TFBS, and DHS regions. Arsenic-associated CpGs () were enriched in shores and depleted in islands compared with CpGs not associated with arsenic () [see “Discovery (EPIC)” in Figure 5A]. We observed no enrichment/depletion of arsenic-associated CpGs in gene regions, promoters, TFBS, and enhancers. Arsenic-associated CpGs were 29% more likely to be annotated to DHS regions () [see “Discovery (EPIC)” in Figure 5B].
Figure 5.
Enrichment of arsenic-associated CpGs among genomic features at . (A) Locational (with relation to island) distribution of arsenic-associated CpGs () from the Health Effects of Arsenic Longitudinal Study (HEALS) discovery analysis on EPIC array ( CpGs) and meta-analysis of HEALS and the Bangladesh Vitamin E and Selenium Trial (BEST) ( CpGs) compared with distribution of CpGs on entire EPIC () and () array, respectively. Colors correspond to CpG relationship to island. (B) Fold enrichment (FE) is plotted for each genomic feature comparing urinary arsenic–associated CpGs below FDR of 0.05 to the remaining CpGs. CpGs annotating to a promoter region were defined as those annotating to TSS200 or TSS1500. Abbreviations correspond to transcription factor binding site (TFBS) and DNase I hypersensitive site (DHS). P-values were obtained from comparing distribution of CpGs above and below FDR of 0.05 for each genomic feature [ (*), (**), and (***)].
GSEA was applied to the urinary arsenic–associated CpGs discovered in HEALS. No gene sets were enriched among the genes annotated to the arsenic-associated CpGs at FDR of 0.05 (21 unique genes). We assessed enrichment of hallmark gene sets among the genes that annotated to our top 500 arsenic-associated CpGs (348 unique genes) (see “HEALS” in Table 3) and observed enrichment for genes annotated to the tumor necrosis factor () signaling via hallmark (seven genes; ), cholesterol homeostasis (four genes; ), and angiogenesis (three genes; ). Among KEGG pathways, the hematopoietic cell lineage pathway was significantly enriched (four genes; ) (Table S4).
Table 3.
Results from gene set enrichment analysis of hallmark gene set collection ( sets) among the urinary arsenic–associated CpGs identified in the HEALS discovery analysis () and meta-analysis of HEALS and BEST ().
| Top 500 arsenic-associated CpGsa | Arsenic-associated CpGs below b | ||||
|---|---|---|---|---|---|
| n | Genes | p | Genes | p | |
| HEALS | |||||
| Angiogenesis | 36 | 3 | — | — | |
| Cholesterol homeostasis | 74 | 4 | — | — | |
| signaling via | 200 | 7 | — | — | |
| META-ANALYSIS | |||||
| Allograft rejection | 200 | 9 | 4 | ||
| 105 | 6 | 3 | |||
| Reactive oxygen species pathway | 49 | 3 | 2 | ||
| Inflammatory response | 200 | 6 | 3 | ||
| signaling via | 200 | 7 | 4 | ||
| Estrogen response early | 200 | 10 | — | — | |
| UV response up-regulated | 158 | 7 | — | — | |
| MTORC1 signaling | 200 | — | — | 4 | |
| Oxidative phosphorylation | 200 | — | — | 3 | |
| MYC targets V1 | 200 | — | — | 3 | |
| Apical junction | 200 | — | — | 4 | |
Note: Gene annotations for arsenic-associated CpGs are provided as gene set input for the gometh function. Results are presented for hallmark gene sets ( sets) with at least two genes from arsenic-associated CpG gene set and enrichment (). Background gene set for is 19,246 genes, and EPIC is 23,234 genes. The table reports total number of genes in set (n), genes identified in pathway from CpG gene set, and p-value from CpG bias-corrected hypergeometric test. —, no data; BEST, Bangladesh Vitamin E and Selenium Trial; FDR, false discovery rate; HEALS, Health Effects of Arsenic Longitudinal Study; , tumor necrosis factor .
Top 500 arsenic-associated CpGs, annotated to 348 unique genes in the HEALS discovery analysis (EPIC array), 338 unique genes in the meta-analysis ( array).
Arsenic-associated CpGs (), annotated to 21 unique genes in the discovery analysis, 153 unique genes in the meta-analysis.
Meta-Analysis of Urinary Arsenic and Genome-Wide DNA Methylation in HEALS and BEST
We conducted a meta-analysis of 390,810 CpGs measured in both HEALS and BEST (see Figure S6 and Figure S7 for full results) to more robustly identify urinary arsenic–associated CpGs. This meta-analysis identified 41 CpGs passing the Bonferroni threshold () (Figure 6; Table 4). At an FDR of 0.05, 221 urinary arsenic–associated CpGs were identified, and all had consistent direction of association in each study () (Excel Table S3). Among the 41 arsenic-associated CpGs (), 34 (82.9%) were negatively associated with increased exposure, and this pattern of hypomethylation with increased arsenic persisted among associated CpGs with FDR of 0.05 (, 76.9%) (Table 4). Our top two arsenic-associated CpGs were located upstream of ABR (cg01912040, cg10003262). The remaining CpGs of the top 10 CpGs annotated to the upstream region of SEMA4G (cg05962511), C19orf66 promoter (cg13480898), EFNA1 (ephrin A1) gene body (cg07207669), SQSTM1 (sequestosome 1) 5’UTR (cg01225779), EML2 (echinoderm 486 microtubule associated protein like 2) gene body (cg06381803), UNKL (unk like zinc finger) TSS200 (cg09183146), MYEOV (myeloma overexpressed) promoter (cg08759026), and SPSB1 (splA/ryanodine receptor domain and SOCS box containing 1) 5’UTR (cg17489312).
Figure 6.
Epigenome-wide meta-analysis of associations between urinary arsenic and DNA methylation in Bangladeshi adults using data from the Health Effects of Arsenic Longitudinal Study (HEALS) and the Bangladesh Vitamin E and Selenium Trial (BEST). Using data from EPIC (HEALS; ) and (BEST; ) arrays, 390,810 CpG sites were meta-analyzed using METAL [summary statistics from covariate- and surrogate variable (SV)–adjusted model were provided as input]. (A) Q-Q plot of the observed distribution of meta-analysis p-values. The red line represents expected distribution of p-values under the null, and corresponds to the genomic inflation factor (see “Methods” section). (B) Manhattan plot of the location of each CpG and p-value for each CpG–arsenic association. Solid and dashed lines designate the Bonferroni threshold () and false discovery rate (FDR) 0.05 threshold (), respectively.
Table 4.
CpGs Associated with urinary arsenic () from meta-analysis of HEALS () and BEST ().
| Name | Chr | Position | CpG location | Nearest gene | Feature | Distance (basepairs) | Direction of association | p |
|---|---|---|---|---|---|---|---|---|
| cg01912040 | 17 | 1106553 | Shore | ABR | Upstream | 15,937 | ||
| cg10003262 | 17 | 1106589 | Shore | ABR | Upstream | 15,973 | ||
| cg05962511 | 10 | 102730022 | Shore | SEMA4G | Body | 732 | ||
| cg13480898 | 19 | 10195914 | Shore | C19orf66 | Promoter | 892 | ||
| cg07207669 | 1 | 155102388 | Shore | EFNA1 | Body | 2,039 | ||
| cg01225779 | 5 | 179238472 | Shelf | SQSTM1 | 5'UTR | 4,148 | ||
| cg06381803 | 19 | 46119475 | Island | EML2 | Body | 29,300 | ||
| cg09183146 | 16 | 1429863 | Island | UNKL | Promoter | 34,842 | ||
| cg08759026 | 11 | 69061454 | Non-CpG island | MYEOV | Promoter | 159 | ||
| cg17489312 | 1 | 9376039 | Non-CpG island2 | SPSB1 | 5'UTR | 23,098 | ||
| cg00472758 | 16 | 2552820 | Shelf | TBC1D24 | 3'UTR | 4,581 | ||
| cg05428706 | 10 | 102730130 | Shore | SEMA4G | Inside | 840 | ||
| cg05425326 | 16 | 58439361 | Non-CpG island | GINS3 | 3'UTR | 13,063 | ||
| cg26435149 | 3 | 55605611 | Non-CpG island | ERC2 | 3'UTR | 896,780 | ||
| cg17393635 | 19 | 49843565 | Island | CD37 | Body | 4,888 | ||
| cg13223043 | 1 | 26492308 | Shore | FAM110D | Upstream | 3,189 | ||
| cg03871754 | 17 | 79320652 | Island | TMEM105 | Upstream | 16,178 | ||
| cg19240637 | 2 | 7172297 | Island | RNF144A | Body | 114,774 | ||
| cg07782285 | 19 | 13085442 | Non-CpG island | DAND5 | 3'UTR | 5,118 | ||
| cg26390598 | 21 | 41032396 | Non-CpG island | B3GALT5 | 5'UTR | 3,142 | ||
| cg04622454 | 9 | 140349128 | Shore | NELF | Body | 4,658 | ||
| cg22959742 | 10 | 13913931 | Non-CpG island | FRMD4A | Body | 136,229 | ||
| cg00281776 | 2 | 209224225 | Shore | PTH2R | Promoter | 344 | ||
| cg12261095 | 19 | 7764345 | Non-CpG island | FCER2 | Body | 2,687 | ||
| cg14718533 | 10 | 33355576 | Non-CpG island | ITGB1 | Upstream | 108,283 | ||
| cg04459545 | 19 | 17375685 | Non-CpG island | USHBP1 | Promoter | 61 | ||
| cg14145338 | 9 | 139649039 | Non-CpG island | LCN8 | Body | 3,692 | ||
| cg04920032 | 12 | 50262986 | Non-CpG island | FAIM2 | 3'UTR | 34,774 | ||
| cg18413900 | 12 | 58160989 | Shore | CYP27B1 | Promoter | 13 | ||
| cg01757312 | 13 | 112720565 | Island | SOX1 | Promoter | 1,348 | ||
| cg05816193 | 6 | 26018127 | Shelf | HIST1H1A | Promoter | 87 | ||
| cg07367302 | 1 | 19967428 | Shelf | MINOS-NBL1 | Body | 43,957 | ||
| cg02306995 | 3 | 122635049 | Shelf | SEMA5B | Body | 111,610 | ||
| cg24318728 | 17 | 39649283 | Non-CpG island | KRT36 | Downstream | 484 | ||
| cg04875062 | 1 | 17305562 | Shore | MFAP2 | 5'UTR | 2,519 | ||
| cg13764516 | 9 | 139648911 | Non-CpG island | LCN8 | 3'UTR | 3,820 | ||
| cg23050300 | 1 | 3281321 | Non-CpG island | PRDM16 | Body | 295,579 | ||
| cg18050715 | 13 | 97996992 | Shore | MBNL2 | Body | 69,106 | ||
| cg04826368 | 6 | 27130208 | Non-CpG island | HIST1H2AH | Upstream | 14,867 | ||
| cg08596618 | 1 | 24275885 | Non-CpG island | SRSF10 | Downstream | 31,068 | ||
| cg27092191 | 16 | 31884699 | Shore | ZNF267 | Promoter | 380 |
Note: Results from meta-analysis are shown CpGs associated with urinary arsenic (creatinine adjusted) (). Meta-analysis was conducted using the METAL software, and summary statistics from the covariate- and surrogate variable (SV)–adjusted models for HEALS and BEST were provided as input. Direction of association indicated by arrows for HEALS and BEST, and downward and upward arrows correspond to inverse and positive association with increasing arsenic exposure. UCSC gene annotation provided by Illumina. BEST, Bangladesh Vitamin E and Selenium Trial; Chr, chromosome; HEALS, Health Effects of Arsenic Longitudinal Study.
Among the BEST participants, gene expression data was available for 26 of the 28 genes that annotated to our urinary arsenic–associated CpGs () (Table S5). At a threshold of , methylation was positively correlated with gene expression for four genes: RNF144A (ring finger protein 144A) (cg19240637; ), C19orf66 (cg13480898; ), SEMA5B (semaphorin 5B) (cg02306995; ), and NELF (negative elongation factor) (cg04622454; ). Inverse correlations between methylation and expression were observed for five CpGs in four genes: EML2 (cg06381803; ), FCER2 (Fc fragment of IgE receptor II) (cg12261095; ), B3GALT5 (beta-1,3-galactosyltransferase 5) (cg26390598; ), and LCN8 (lipocalin 8) (cg14145338; and cg13764516; ). After adjustment for age and sex, six of the nine associations between expression and methylation persisted (). Among the HEALS participants, genotyping information was available for 389 participants, and we examined whether the top arsenic-associated CpGs identified in the meta-analysis () were associated with genetic variants (within 1 megabase window) (Table S6). Twenty-five out of the 41 arsenic-associated CpGs had one or more cis-mQTL pairs (), suggesting that these CpGs and the genetic variants that influence their methylation status could be further explored in gene environment studies.
We attempted replication of the arsenic-associated CpGs identified in this meta-analysis in a group of Andean women from Argentina, who were exposed to arsenic via drinking water, () with DNA methylation measured on the array, as previously described by Ameer et al. (2017). Among the arsenic-associated CpGs from the meta-analysis (), only 16 CpGs were associated with urinary arsenic () among the Andean women and 13 (81.3%) with consistent direction of association (Excel Table S4). While these results suggest some consistency among the arsenic-associated CpGs identified in our study, there is heterogeneity with respect to study population, sample size, study design, and analysis approach.
Enrichment of Arsenic-Associated CpGs (from Meta-Analysis) in Genomic Features and Gene Sets
Similar to our discovery analysis in HEALS using the EPIC array, we examined enrichment of genomic features among the urinary arsenic–associated CpGs identified in the meta-analysis. Arsenic-associated CpGs identified in the meta-analysis were enriched in shores and non-CpG islands and depleted in islands compared with CpGs not associated with arsenic () (Figure 5A). Arsenic-associated CpGs were depleted in promoters () and enriched in DHS regions () (Figure 5B). Notably, arsenic-associated CpGs were 2.6-fold more likely to be located within enhancers compared with CpGs not associated with arsenic (). When we stratified by the direction of association, the genomic enrichment in DHS regions and enhancers and depletion in promoters persisted among negatively arsenic-associated CpGs (see Figure S8).
GSEA was applied to the urinary arsenic–associated CpGs identified in the meta-analysis among the associated CpGs with FDR of 0.05 (153 unique genes) and among the top 500 arsenic-associated CpGs (338 unique genes). Among both sets of arsenic-associated CpGs, we observed enrichment of genes annotating to the , allograft rejection, reactive oxygen species pathway, inflammatory response, and signaling via hallmarks () (Table 3). We observed enrichment of 13 KEGG pathways among the arsenic-associated CpGs with FDR of 0.05 (Table S4). Among the top 500 arsenic-associated CpGs, genes in this set annotated to KEGG pathways related to cell adhesion molecules (CAMs) (nine genes; ), mitogen-activated protein kinase signaling pathway (14 genes; ), estrogen signaling pathway (eight genes; ), hematopoietic cell lineage (four genes; ), cysteine and methionine metabolism (three genes; ), and signaling pathway (five genes; ).
Discussion
This EWAS of arsenic exposure provides evidence that arsenic exposure is associated with DNA methylation levels at specific CpG sites in the leukocytes of Bangladeshi adults. In our discovery analysis, we identified 34 novel CpGs associated with arsenic exposure assessed in urine in HEALS prior to any arsenic mitigation efforts. Sixteen of these novel arsenic-associated CpGs were also present on the array, and ten replicated in an independent cohort (BEST). For the 34 novel arsenic-associated CpGs observed in HEALS, results for arsenic exposure measured in drinking water were highly consistent with results based on urinary arsenic. Our meta-analysis of HEALS and BEST identified 221 CpGs associated with arsenic exposure assessed in urine in Bangladeshi adults. Arsenic-associated CpGs were more likely to be hypomethylated and were enriched in CpG shores, DHS regions, and enhancers.
Our results are relevant to understanding how arsenic impacts the epigenome and how alteration of the epigenome may be a mechanism involved in arsenic toxicity. Among our top CpGs from both the discovery analysis and meta-analysis, we observed that higher arsenic exposure tends to be associated with hypomethylation, a phenomenon not observed in prior studies (Ameer et al. 2017; Argos et al. 2015; Liu et al. 2014; Seow et al. 2014), potentially due to the increased coverage of the EPIC array of intergenic and non-promoter regulatory genomic regions compared with the array. While the prior adult EWAS of arsenic exposure identified more CpGs associated with hypermethylation than hypomethylation (Argos et al. 2015; Liu et al. 2014), arsenic exposure was associated with decreased global methylation, assessed using the incorporation assay in Bangladeshi adults (Niedzwiecki et al. 2013). In addition, methylation is more variable in CpG shores and may be more susceptible to changes due to environmental exposures such as arsenic (Jones 2012). While the location of arsenic-associated CpG methylation in relation to CpG islands needs to be further studied, arsenic-associated CpGs were enriched in CpG shores in the cord blood from low-exposed infants (Koestler et al. 2013) and in non-CpG island regions in the umbilical artery and placenta from Bangladeshi infants (Cardenas et al. 2015a). A meta-analysis of maternal smoking and genome-wide methylation in cord blood observed similar enrichment in CpG shores and within enhancers and DHS regions and depletion in CpG islands and promoter regions (Joubert et al. 2016).
While the mechanisms for alteration of DNA methylation in blood by arsenic remains to elucidated, two proposed mechanisms involve the influence of arsenic: a) on the expression or function of DNA methyltransferases, potentially resulting in less downstream site-specific methylation; or b) on consumption of methyl groups during arsenic metabolism, resulting in depletion of methyl groups available to DNA methyltransferase, affecting DNA methylation synthesis (Eckstein et al. 2017). In addition, it is possible that specific genes are expressed in response to arsenic exposure, and the induction of these genes may change the local epigenetic state, including DNA methylation. Arsenic-associated methylation in DHS regions and enhancers warrants further exploration in both experimental and genome-wide methylation studies.
In our discovery and replication analysis, two of the top CpGs were located upstream of the ABR gene, encoding the active BCR-related protein. ABR is a regulator of the RHO family of small GTPases, signaling proteins involved in cytosketal dynamics, and a paralog of the BCR (BCR activator of RhoGEF and GTPase) gene. ABR has a GTPase-activating protein domain and may be important in cellular signaling and immune processes (Cho et al. 2007; Chuang et al. 1995; Cunnick et al. 2009). In addition, ABR may have a pivotal role in human embryonic stem cell mitosis, suggesting that ABR may be important for maintaining the genomic integrity and renewal of stem cells (Ohgushi et al. 2017). A study of newborns () in the Biomarkers of Exposure to Arsenic (BEAR) pregnancy cohort from Mexico identified two CpGs also negatively associated with maternal water arsenic in the ABR gene body (Rojas et al. 2015). Arsenic increased ABR expression in an in vitro study of arsenic exposure in human epidermal keratinocytes (Perez et al. 2008). No prior methylation or experimental evidence has been identified for the other discovered urinary arsenic–associated CpGs.
Among the arsenic-associated CpGs from our meta-analysis, we observed some consistency with prior EWAS and gene expression studies of arsenic exposure. In the BEAR pregnancy cohort of Mexican infants, arsenic exposure was associated with methylation at CpGs within the following genes that also annotated to CpGs identified in our meta-analysis: TBC1D24 (TBC1 domain family member 24), ERC2, and PRRC2A (proline rich coiled-coil 2A) (Rojas et al. 2015). TBC1D24 is a protein that is hypothesized to be involved in vesicle transport and oxidative stress response. The PRRC2A gene is within the vicinity of the and encoding regions and may be involved in inflammatory response. In a cohort of Bangladeshi infants, methylation in the first exon of ERC2 was differentially methylated with maternal water arsenic (Kile et al. 2014). Maternal arsenic exposure increased EML2 protein levels in the cord blood of infants from the BEAR cohort (Bailey et al. 2014). In vitro arsenic exposure increased SQSTM1 and UNKL expression and decreased SPSB1 transcript stability in human fibroblasts (Qiu et al. 2015) and increased MCC (mutated in colorectal cancer) expression in human epidermal keratinocytes (Perez et al. 2008). The SQSTM1 encodes a protein that is involved in selective autophagy and cell senescence and is within the inflammatory response pathway. In Excel Table S5, we identified arsenic-associated CpGs from the prior literature and reported the replication results from HEALS and our meta-analysis.
There was no overlap between our identified arsenic-associated CpGs and metal-associated CpGs reported in prior studies of in utero exposure to mercury (Cardenas et al. 2015b), lead (Sen et al. 2015), and cadmium (Kippler et al. 2013; Mohanty et al. 2015; Sanders et al. 2014). A meta-analysis of in utero cadmium exposure identified a differentially methylated CpG (cg16768966) within GAS7, encoding growth arrest protein 7, and arsenic was also associated with methylation at this same gene but not the same CpG (Everson et al. 2018). Overall, this lack of overlap between arsenic-associated CpGs and CpGs associated with other metals suggests that arsenic may have a toxicant specific effect on the epigenome.
GSEA enabled us to identify pathways potentially affected by arsenic-associated DNA methylation alterations in whole blood. Our arsenic-associated CpGs annotated to genes in hallmark gene sets related to reactive oxygen species, cancer and aging, and inflammatory response pathways, specifically signaling via . Arsenic primarily undergoes biotransformation via methylation by AS3MT but may also involve glutathione conjugation and other antioxidant and xenobiotic metabolizing enzymes (Jomova et al. 2011). Arsenic exposure also induces expression of numerous proteins involved in response, signaling, and inflammation in exposed infants and children (Bailey et al. 2014; Fry et al. 2007; Smeester et al. 2017) and adults (Dutta et al. 2015). The KEGG pathway for CAMs also has been identified as enriched in a study of Bangladeshi infants (Kile et al. 2014) and low-exposed U.S. adults (Liu et al. 2014). CAMs are universally important in hemostasis, immune response, and development. Among a subset of BEST participants, baseline arsenic exposure was associated with increased circulating CAMs at baseline and an increase in CAMs between baseline and 6-month follow-up (Chen et al. 2007). In a study of Bangladeshi infants, arsenic-associated CpGs were also enriched in KEGG pathways, including hematopoietic cell lineage, calcium signaling pathway, Notch signaling, and mTOR signaling (Kile et al. 2014). While there is heterogeneity in GSEA approaches, the results from our GSEA and prior EWAS suggest some associations between arsenic exposure and DNA methylation may be consistent across different stages of development and exposure levels. Within the GWAS Catalog (https://www.ebi.ac.uk/gwas/), several variants within or near genes that annotated to the arsenic-associated CpGs were associated with a broad range of health traits with potential connections to arsenic exposure. These health traits included many types of cancer, lymphocyte and red blood cell phenotypes and composition, pulmonary function, and blood pressure (Table S7), suggesting that genetic variation and potentially epigenetic variation in these genes may contribute to the risk of arsenic-associated disease and health traits.
A major strength of our study was the large sample size, with individuals in both the discovery and replication datasets, resulting in 796 arsenic-exposed adults for meta-analysis. Utilizing methylation data from BEST, we were able to conduct replication analyses, enabling us to validate discovered associations between urinary arsenic and CpG methylation. Our meta-analysis increased the power to detect putative arsenic-associated CpGs that can be examined in future studies. The EPIC array measures methylation at almost twice the number of CpGs compared with the array used in prior studies and improves coverage within intergenic regions, enhancers, and distal regulatory elements (Pidsley et al. 2016). In HEALS, water and urinary arsenic were assessed at baseline prior to interventions to remediate exposure; thus, these measures are likely to represent historical exposure status, allowing us to potentially identify CpGs associated with long-term exposure, as opposed to more acute responses to exposure. The exposure range in both HEALS and BEST participants was wide, enabling us to evaluate a broad spectrum of environmental arsenic exposure and its effect on genome-wide methylation.
Several limitations must be considered when interpreting the results of our study. While urinary arsenic is the most common biomarker of recent arsenic exposure, sampling and interindividual factors, such as age, sex, and metabolism, can contribute to variation in urinary arsenic measurements (Marchiset-Ferlay et al. 2012). Unmeasured confounding, from unknown biologic and technical variation, and measurement error can also potentially bias our results. DNA methylation was measured in whole blood, and the associations we observed in blood may not be present in other tissues. The EPIC array measures methylation at of CpGs in the genome (Stevens et al. 2013) and is biased toward gene regions, and DMR analysis preferentially identifies DMRs in gene regions since intergenic CpGs are more sparsely distributed on the array (Peters et al. 2015). We currently cannot replicate results for CpGs specific to the EPIC array. The association between gene expression and methylation (measured on EPIC) cannot be assessed in HEALS since RNA is not available. Genetic background can influence DNA methylation, and the arsenic–CpG associations observed in this work may not be present in other arsenic-exposed populations due to genetic differences (Bell et al. 2011; McRae et al. 2014). Even among our significant arsenic-associated CpGs, the effect sizes of the associations between arsenic and DNA methylation at CpGs are small, and it is unknown whether these small effects have functional consequences.
Future studies using the EPIC array (or more comprehensive measurement technologies) will be necessary to confirm the novel CpGs we identified and the enrichment of arsenic-associated CpGs among CpG shores and enhancers. Other factors, such as individual genetic susceptibility to arsenic exposure and timing and duration of arsenic exposure, need to be explored in studies of arsenic-associated methylation. While the EPIC array enables us to study the effect of arsenic on genic regions, we still do not know how arsenic affects global methylation, and bisulfite genomic sequencing approaches are needed to understand the effect of arsenic on the global epigenome, especially in intergenic regions. The relationship between arsenic-related variation in DNA methylation and local histone and chromatin features require further study, and these features likely have important implications for regulation of arsenic response genes as well as arsenic toxicity and disease risk. Finally, arsenic-associated CpGs identified here and elsewhere should be explored as potential biomarkers of arsenic exposure, susceptibility, and toxicity. Because some of the arsenic-associated CpGs identified in our analysis annotate to important inflammatory, oxidative response, and other cell regulation pathways, alterations in DNA methylation may be important biologic responses and/or markers of arsenic exposure that may better inform our understanding of and predict arsenic-associated health effects and disease.
Conclusions
Among Bangladeshi adults chronically exposed to arsenic via drinking contaminated water from tube wells, we identified novel and reproducible arsenic-associated DNA methylation alterations in blood. Our meta-analysis that combines our results with a prior study identified additional CpGs showing a putative association with arsenic exposure. Arsenic-associated CpGs annotated to genes involved in signaling via , CAMs, inflammatory processes, and important signaling pathways in cancer and aging. The implications of these arsenic-associated CpGs for exposure and risk assessment and potential toxicity prevention should be further investigated.
Supplementary Material
Acknowledgments
We acknowledge fellowship support for Dr. Demanelis provided by the National Institute on Aging (NIA) Specialized Demography and Economics of Aging Training Program (2T32AG000243) at the University of Chicago. Research support for this project is provided by active and past National Institutes of Health (NIH) grants (R01ES020506, R35ES028379, P42ES010349, R01CA102484, R01CA107431, P30CA014599, P30ES027792). We would like to acknowledge the HEALS and BEST study participants and research staff for their contributions to these cohorts and Dr. M. Kibriya for his contributions to generating the DNA methylation data utilized in this analysis.
Footnotes
Supplemental Material is available online (https://doi.org/10.1289/EHP3849).
The authors declare they have no actual or potential competing financial interests.
Note to readers with disabilities: EHP strives to ensure that all journal content is accessible to all readers. However, some figures and Supplemental Material published in EHP articles may not conform to 508 standards due to the complexity of the information being presented. If you need assistance accessing journal content, please contact ehponline@niehs.nih.gov. Our staff will work with you to assess and meet your accessibility needs within 3 working days.
References
- Ahsan H, Chen Y, Parvez F, Argos M, Hussain AI, Momotaj H, et al. 2006a. Health effects of arsenic longitudinal study (HEALS): description of a multidisciplinary epidemiologic investigation. J Expo Sci Environ Epidemiol 16(2):191–205, PMID: 16160703, 10.1038/sj.jea.7500449. [DOI] [PubMed] [Google Scholar]
- Ahsan H, Chen Y, Parvez F, Zablotska L, Argos M, Hussain I, et al. 2006b. Arsenic exposure from drinking water and risk of premalignant skin lesions in Bangladesh: baseline results from the health effects of arsenic longitudinal study. Am J Epidemiol 163(12):1138–1148, PMID: 16624965, 10.1093/aje/kwj154. [DOI] [PubMed] [Google Scholar]
- Ahsan H, Perrin M, Rahman A, Parvez F, Stute M, Zheng Y, et al. 2000. Associations between drinking water and urinary arsenic levels and skin lesions in Bangladesh. J Occup Environ Med 42(12):1195–1201, PMID: 11125683, 10.1097/00043764-200012000-00016. [DOI] [PubMed] [Google Scholar]
- Ameer SS, Engstrom K, Hossain MB, Concha G, Vahter M, Broberg K. 2017. Arsenic exposure from drinking water is associated with decreased gene expression and increased DNA methylation in peripheral blood. Toxicol Appl Pharmacol 321:57–66, PMID: 28242323, 10.1016/j.taap.2017.02.019. [DOI] [PubMed] [Google Scholar]
- Anawar HM, Akai J, Mostofa KM, Safiullah S, Tareq SM. 2002. Arsenic poisoning in groundwater: health risk and geochemical sources in Bangladesh. Environ Int 27(7):597–604, PMID: 11871394, 10.1016/S0160-4120(01)00116-7. [DOI] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, et al. 2014. An atlas of active enhancers across human cell types and tissues. Nature 507(7493):455–461, PMID: 24670763, 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argos M. 2015. Arsenic exposure and epigenetic alterations: recent findings based on the Illumina 450k DNA methylation array. Curr Environ Health Rep 2(2):137–144, PMID: 26231363, 10.1007/s40572-015-0052-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argos M, Chen L, Jasmine F, Tong L, Pierce BL, Roy S, et al. 2015. Gene-specific differential DNA methylation and chronic arsenic exposure in an epigenome-wide association study of adults in Bangladesh. Environ Health Perspect 123(1):64–71, PMID: 25325195, 10.1289/ehp.1307884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argos M, Kalra T, Rathouz PJ, Chen Y, Pierce B, Parvez F, et al. 2010. Arsenic exposure from drinking water, and all-cause and chronic-disease mortalities in Bangladesh (HEALS): a prospective cohort study. Lancet 376(9737):252–258, PMID: 20646756, 10.1016/S0140-6736(10)60481-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argos M, Rahman M, Parvez F, Dignam J, Islam T, Quasem I, et al. 2013. Baseline comorbidities in a skin cancer prevention trial in Bangladesh. Eur J Clin Invest 43(6):579–588, PMID: 23590571, 10.1111/eci.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. 2007. GenABEL: an R library for genome-wide association analysis. Bioinformatics 23(10):1294–1296, PMID: 17384015, 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- Bailey KA, Fry RC. 2014. Arsenic-associated changes to the epigenome: what are the functional consequences? Curr Environ Health Rep 1:22–34, PMID: 24860721, 10.1007/s40572-013-0002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey KA, Laine J, Rager JE, Sebastian E, Olshan A, Smeester L, et al. 2014. Prenatal arsenic exposure and shifts in the newborn proteome: interindividual differences in tumor necrosis factor (TNF)-responsive signaling. Toxicol Sci 139(2):328–337, PMID: 24675094, 10.1093/toxsci/kfu053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, et al. 2011. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol 12(1):R10, PMID: 21251332, 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buja A, Eyuboglu N. 1992. Remarks on parallel analysis. Multivariate Behav Res 27(4):509–540, PMID: 26811132, 10.1207/s15327906mbr2704_2. [DOI] [PubMed] [Google Scholar]
- Cardenas A, Houseman EA, Baccarelli AA, Quamruzzaman Q, Rahman M, Mostofa G, et al. 2015a. In utero arsenic exposure and epigenome-wide associations in placenta, umbilical artery, and human umbilical vein endothelial cells. Epigenetics 10(11):1054–1063, PMID: 26646901, 10.1080/15592294.2015.1105424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas A, Koestler DC, Houseman EA, Jackson BP, Kile ML, Karagas MR, et al. 2015b. Differential DNA methylation in umbilical cord blood of infants exposed to mercury and arsenic in utero. Epigenetics 10(6):508–515, PMID: 25923418, 10.1080/15592294.2015.1046026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Santella RM, Kibriya MG, Wang Q, Kappil M, Verret WJ, et al. 2007. Association between arsenic exposure from drinking water and plasma levels of soluble cell adhesion molecules. Environ Health Perspect 115(10):1415–1420, PMID: 17938729, 10.1289/ehp.10277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Zheng Y, Mortlock R, Van Geen A. 2004. Rapid multi-element analysis of groundwater by high-resolution inductively coupled plasma mass spectrometry. Anal Bioanal Chem 379(3):512–518, PMID: 15098084, 10.1007/s00216-004-2618-x. [DOI] [PubMed] [Google Scholar]
- Cho YJ, Cunnick JM, Yi SJ, Kaartinen V, Groffen J, Heisterkamp N. 2007. Abr and Bcr, two homologous Rac GTPase-activating proteins, control multiple cellular functions of murine macrophages. Mol Cell Biol 27(3):899–911, PMID: 17116687, 10.1128/MCB.00756-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang TH, Xu X, Kaartinen V, Heisterkamp N, Groffen J, Bokoch GM. 1995. Abr and Bcr are multifunctional regulators of the Rho GTP-binding protein family. Proc Natl Acad Sci U S A 92(22):10282–10286, PMID: 7479768, 10.1073/pnas.92.22.10282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortessis VK, Thomas DC, Levine AJ, Breton CV, Mack TM, Siegmund KD, et al. 2012. Environmental epigenetics: prospects for studying epigenetic mediation of exposure-response relationships. Hum Genet 131(10):1565–1589, PMID: 22740325, 10.1007/s00439-012-1189-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnick JM, Schmidhuber S, Chen G, Yu M, Yi SJ, Cho YJ, et al. 2009. Bcr and Abr cooperate in negatively regulating acute inflammatory responses. Mol Cell Biol 29(21):5742–5750, PMID: 19703997, 10.1128/MCB.00357-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta K, Prasad P, Sinha D. 2015. Chronic low level arsenic exposure evokes inflammatory responses and DNA damage. Int J Hyg Environ Health 218(6):564–574, PMID: 26118750, 10.1016/j.ijheh.2015.06.003. [DOI] [PubMed] [Google Scholar]
- Eckstein M, Eleazer R, Rea M, Fondufe-Mittendorf Y. 2017. Epigenomic reprogramming in inorganic arsenic-mediated gene expression patterns during carcinogenesis. Rev Environ Health 32(1–2):93–103, PMID: 27701139, 10.1515/reveh-2016-0025. [DOI] [PubMed] [Google Scholar]
- Everson TM, Punshon T, Jackson BP, Hao K, Lambertini L, Chen J, et al. 2018. Cadmium-associated differential methylation throughout the placental genome: epigenome-wide association study of two U.S. birth cohorts. Environ Health Perspect 126(1):017010, PMID: 29373860, 10.1289/EHP2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan SV, Johnston RB, Zheng Y. 2012. Arsenic in tube well water in Bangladesh: health and economic impacts and implications for arsenic mitigation. Bull World Health Organ 90(11):839–846, PMID: 23226896, 10.2471/BLT.11.101253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry RC, Navasumrit P, Valiathan C, Svensson JP, Hogan BJ, Luo M, et al. 2007. Activation of inflammation/NF-kappaB signaling in infants born to arsenic-exposed mothers. PLoS Genet 3(11):e207, PMID: 18039032, 10.1371/journal.pgen.0030207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamboa-Loira B, Cebrian ME, Franco-Marina F, Lopez-Carrillo L. 2017. Arsenic metabolism and cancer risk: a meta-analysis. Environ Res 156:551–558, PMID: 28433864, 10.1016/j.envres.2017.04.016. [DOI] [PubMed] [Google Scholar]
- Gao J, Roy S, Tong L, Argos M, Jasmine F, Rahaman R, et al. 2015. Arsenic exposure, telomere length, and expression of telomere-related genes among Bangladeshi individuals. Environ Res 136:462–469, PMID: 25460668, 10.1016/j.envres.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geeleher P, Hartnett L, Egan LJ, Golden A, Raja Ali RA, Seoighe C. 2013. Gene-set analysis is severely biased when applied to genome-wide methylation data. Bioinformatics 29(15):1851–1857, PMID: 23732277, 10.1093/bioinformatics/btt311. [DOI] [PubMed] [Google Scholar]
- Green BB, Karagas MR, Punshon T, Jackson BP, Robbins DJ, Houseman EA, et al. 2016. Epigenome-wide assessment of DNA methylation in the placenta and arsenic exposure in the New Hampshire Birth Cohort Study (USA). Environ Health Perspect 124(8):1253–1260, PMID: 26771251, 10.1289/ehp.1510437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Y, Benjamini Y. 1990. More powerful procedures for multiple significance testing. Stat Med 9(7):811–818, PMID: 2218183, 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. 2012. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13:86, PMID: 22568884, 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. 2007. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8(1):118–127, PMID: 16632515, 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Jomova K, Jenisova Z, Feszterova M, Baros S, Liska J, Hudecova D, et al. 2011. Arsenic: toxicity, oxidative stress and human disease. J Appl Toxicol 31:95–107, PMID: 21321970, 10.1002/jat.1649. [DOI] [PubMed] [Google Scholar]
- Jones PA. 2012. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13(7):484–492, PMID: 22641018, 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Joubert BR, Felix JF, Yousefi P, Bakulski KM, Just AC, Breton C, et al. 2016. DNA methylation in newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am J Hum Genet 98(4):680–696, PMID: 27040690, 10.1016/j.ajhg.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karagas MR, Gossai A, Pierce B, Ahsan H. 2015. Drinking water arsenic contamination, skin lesions, and malignancies: a systematic review of the global evidence. Curr Environ Health Rep 2(1):52–68, PMID: 26231242, 10.1007/s40572-014-0040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal A, Zhang H, Karmaus WJJ, Ray M, Torres MA, Smith AK, et al. 2017. Comparison of different cell type correction methods for genome-scale epigenetics studies. BMC Bioinformatics 18(1):216, PMID: 28410574, 10.1186/s12859-017-1611-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile ML, Houseman EA, Baccarelli AA, Quamruzzaman Q, Rahman M, Mostofa G, et al. 2014. Effect of prenatal arsenic exposure on DNA methylation and leukocyte subpopulations in cord blood. Epigenetics 9(5):774–782, PMID: 24525453, 10.4161/epi.28153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kippler M, Engstrom K, Mlakar SJ, Bottai M, Ahmed S, Hossain MB, et al. 2013. Sex-specific effects of early life cadmium exposure on DNA methylation and implications for birth weight. Epigenetics 8(5):494–503, PMID: 23644563, 10.4161/epi.24401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koestler DC, Avissar-Whiting M, Houseman EA, Karagas MR, Marsit CJ. 2013. Differential DNA methylation in umbilical cord blood of infants exposed to low levels of arsenic in utero. Environ Health Perspect 121(8):971–977, PMID: 23757598, 10.1289/ehp.1205925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamm SH, Ferdosi H, Dissen EK, Li J, Ahn J. 2015. A systematic review and meta-regression analysis of lung cancer risk and inorganic arsenic in drinking water. Int J Environ Res Public Health 12(12):15498–15515, PMID: 26690190, 10.3390/ijerph121214990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. 2012. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28(6):882–883, PMID: 22257669, 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Storey JD. 2007. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet 3(9):1724–1735, PMID: 17907809, 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. 2010. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 34(8):816–834, PMID: 21058334, 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. 2015. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst 1(6):417–425, PMID: 26771021, 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zheng Y, Zhang W, Zhang X, Lioyd-Jones DM, Baccarelli AA, et al. 2014. Blood methylomics in response to arsenic exposure in a low-exposed US population. J Expo Sci Environ Epidemiol 24(2):145–149, PMID: 24368509, 10.1038/jes.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchiset-Ferlay N, Savanovitch C, Sauvant-Rochat MP. 2012. What is the best biomarker to assess arsenic exposure via drinking water? Environ Int 39(1):150–171, PMID: 22208756, 10.1016/j.envint.2011.07.015. [DOI] [PubMed] [Google Scholar]
- Martin TC, Yet I, Tsai PC, Bell JT. 2015. coMET: visualisation of regional epigenome-wide association scan results and DNA co-methylation patterns. BMC Bioinformatics 16:131, PMID: 25928765, 10.1186/s12859-015-0568-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin EM, Fry RC. 2018. Environmental influences on the epigenome: exposure-associated DNA methylation in human populations. Annu Rev Public Health 39:309–333, PMID: 29328878, 10.1146/annurev-publhealth-040617-014629. [DOI] [PubMed] [Google Scholar]
- McGregor K, Bernatsky S, Colmegna I, Hudson M, Pastinen T, Labbe A, et al. 2016. An evaluation of methods correcting for cell-type heterogeneity in DNA methylation studies. Genome Biol 17:84, PMID: 27142380, 10.1186/s13059-016-0935-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRae AF, Powell JE, Henders AK, Bowdler L, Hemani G, Shah S, et al. 2014. Contribution of genetic variation to transgenerational inheritance of DNA methylation. Genome Biol 15(5):R73, PMID: 24887635, 10.1186/gb-2014-15-5-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty AF, Farin FM, Bammler TK, MacDonald JW, Afsharinejad Z, Burbacher TM, et al. 2015. Infant sex-specific placental cadmium and DNA methylation associations. Environ Res 138:74–81, PMID: 25701811, 10.1016/j.envres.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon KA, Oberoi S, Barchowsky A, Chen Y, Guallar E, Nachman KE, et al. 2017. A dose-response meta-analysis of chronic arsenic exposure and incident cardiovascular disease. Int J Epidemiol 46(6):1924–1939, PMID: 29040626, 10.1093/ije/dyx202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naujokas MF, Anderson B, Ahsan H, Aposhian HV, Graziano JH, Thompson C, et al. 2013. The broad scope of health effects from chronic arsenic exposure: update on a worldwide public health problem. Environ Health Perspect 121(3):295–302, PMID: 23458756, 10.1289/ehp.1205875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navas-Acien A, Sharrett AR, Silbergeld EK, Schwartz BS, Nachman KE, Burke TA, et al. 2005. Arsenic exposure and cardiovascular disease: a systematic review of the epidemiologic evidence. Am J Epidemiol 162(11):1037–1049, PMID: 16269585, 10.1093/aje/kwi330. [DOI] [PubMed] [Google Scholar]
- Niedzwiecki MM, Hall MN, Liu X, Oka J, Harper KN, Slavkovich V, et al. 2013. A dose-response study of arsenic exposure and global methylation of peripheral blood mononuclear cell DNA in Bangladeshi adults. Environ Health Perspect 121(11-12):1306–1312, PMID: 24013868, 10.1289/ehp.1206421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon DE, Mussmann GV, Eckdahl SJ, Moyer TP. 1991. Total arsenic in urine: palladium-persulfate vs nickel as a matrix modifier for graphite furnace atomic absorption spectrophotometry. Clin Chem 37(9):1575–1579, PMID: 1893592. [PubMed] [Google Scholar]
- Ohgushi M, Minaguchi M, Eiraku M, Sasai Y. 2017. A RHO small GTPase regulator ABR secures mitotic fidelity in human embryonic stem cells. Stem Cell Reports 9(1):58–66, PMID: 28579391, 10.1016/j.stemcr.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez DS, Handa RJ, Yang RS, Campain JA. 2008. Gene expression changes associated with altered growth and differentiation in benzo[a]pyrene or arsenic exposed normal human epidermal keratinocytes. J Appl Toxicol 28(4):491–508, PMID: 17879257, 10.1002/jat.1301. [DOI] [PubMed] [Google Scholar]
- Peters TJ, Buckley MJ, Statham AL, Pidsley R, Samaras K, V Lord R, et al. 2015. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin 8:6, PMID: 25972926, 10.1186/1756-8935-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipson B, Maksimovic J, Oshlack A. 2016. missMethyl: an r package for analyzing data from Illumina's HumanMethylation450 platform. Bioinformatics 32(2):286–288, PMID: 26424855, 10.1093/bioinformatics/btv560. [DOI] [PubMed] [Google Scholar]
- Pidsley R, Zotenko E, Peters TJ, Lawrence MG, Risbridger GP, Molloy P, et al. 2016. Critical evaluation of the Illumina MethylationEPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol 17(1):208, PMID: 27717381, 10.1186/s13059-016-1066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce BL, Kibriya MG, Tong L, Jasmine F, Argos M, Roy S, et al. 2012. Genome-wide association study identifies chromosome 10q24.32 variants associated with arsenic metabolism and toxicity phenotypes in Bangladesh. PLoS Genet 8(2):e1002522, PMID: 22383894, 10.1371/journal.pgen.1002522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu LQ, Abey S, Harris S, Shah R, Gerrish KE, Blackshear PJ. 2015. Global analysis of posttranscriptional gene expression in response to sodium arsenite. Environ Health Perspect 123(4):324–330, PMID: 25493608, 10.1289/ehp.1408626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, McHale CM, Skibola CF, Smith AH, Smith MT, Zhang L. 2011. An emerging role for epigenetic dysregulation in arsenic toxicity and carcinogenesis. Environ Health Perspect 119(1):11–19, PMID: 20682481, 10.1289/ehp.1002114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7):e47, PMID: 25605792, 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas D, Rager JE, Smeester L, Bailey KA, Drobná Z, Rubio-Andrade M, et al. 2015. Prenatal arsenic exposure and the epigenome: identifying sites of 5-methylcytosine alterations that predict functional changes in gene expression in newborn cord blood and subsequent birth outcomes. Toxicol Sci 143(1):97–106, PMID: 25304211, 10.1093/toxsci/kfu210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez TR, Powers M, Perzanowski M, George CM, Graziano JH, Navas-Acien A. 2018. A meta-analysis of arsenic exposure and lung function: Is there evidence of restrictive or obstructive lung disease? Curr Environ Health Rep 5(2):244–254, PMID: 29637476, 10.1007/s40572-018-0192-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders AP, Smeester L, Rojas D, DeBussycher T, Wu MC, Wright FA, et al. 2014. Cadmium exposure and the epigenome: exposure-associated patterns of DNA methylation in leukocytes from mother-baby pairs. Epigenetics 9(2):212–221, PMID: 24169490, 10.4161/epi.26798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A, Heredia N, Senut MC, Hess M, Land S, Qu W, et al. 2015. Early life lead exposure causes gender-specific changes in the DNA methylation profile of DNA extracted from dried blood spots. Epigenomics 7(3):379–393, PMID: 26077427, 10.2217/epi.15.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seow WJ, Kile ML, Baccarelli AA, Pan WC, Byun HM, Mostofa G, et al. 2014. Epigenome-wide dna methylation changes with development of arsenic-induced skin lesions in Bangladesh: a case-control follow-up study. Environ Mol Mutagen 55(6):449–456, PMID: 24677489, 10.1002/em.21860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalin AA. 2012. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 28(10):1353–1358, PMID: 22492648, 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeester L, Bommarito PA, Martin EM, Recio-Vega R, Gonzalez-Cortes T, Olivas-Calderon E, et al. 2017. Chronic early childhood exposure to arsenic is associated with a TNF-mediated proteomic signaling response. Environ Toxicol Pharmacol 52:183–187, PMID: 28433805, 10.1016/j.etap.2017.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens M, Cheng JB, Li D, Xie M, Hong C, Maire CL, et al. 2013. Estimating absolute methylation levels at single-CpG resolution from methylation enrichment and restriction enzyme sequencing methods. Genome Res 23(9):1541–1553, PMID: 23804401, 10.1101/gr.152231.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, et al. 2013. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 29(2):189–196, PMID: 23175756, 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, et al. 2012. The accessible chromatin landscape of the human genome. Nature 489(7414):75–82, PMID: 22955617, 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, Tibshirani R, et al. 2001. Missing value estimation methods for DNA microarrays. Bioinformatics 17(6):520–525, PMID: 11395428, 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- Van Geen A, Ahsan H, Horneman AH, Dhar RK, Zheng Y, Hussain I, et al. 2002. Promotion of well-switching to mitigate the current arsenic crisis in Bangladesh. Bull World Health Organ 80(9):732–737, PMID: 12378292, 10.1590/S0042-96862002000900010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Iterson M, van Zwet EW, BIOS Consortium, Heijmans BT. 2017. Controlling bias and inflation in epigenome- and transcriptome-wide association studies using the empirical null distribution. Genome Biol 18(1):19, PMID: 28129774, 10.1186/s13059-016-1131-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. 2010. Metal: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26(17):2190–2191, PMID: 20616382, 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.