Supplemental Digital Content is available in the text
Keywords: Africa, cervical intraepithelial neoplasia, DNA methylation, early detection of cancer, HIV, human papillomavirus
Objective:
To determine the performance of molecular screening strategies for detection of cervical intraepithelial neoplasia grade 3 or worse (CIN3+) in comparison with cytology screening in women living with HIV.
Design:
Post-hoc analysis using data from a South African study cohort.
Methods:
Cytology and human papillomavirus (HPV)-based strategies were evaluated, including single test and FAM19A4/miR124-2 methylation triage strategies. Participants underwent cytology screening and a colposcopy-directed biopsy. Valid results on cytology, HPV status, 16/18 genotyping and histology were available for 318 women. Detection of HPV and FAM19A4/miR124-2 hypermethylation was performed on DNA from cervical scrapes. Histological diagnosis of CIN3+ was used as outcome.
Results:
Cytology provided highest specificity (91.6%), but lowest sensitivity (59.3%), whereas a single HPV test provided highest sensitivity (83.1%), but lowest specificity (66.4%). Combining cytology with methylation did not improve the performance compared with cytology alone: a slight increase in sensitivity was seen, at the cost of a decrease in specificity. Triage of high-risk HPV positive women with methylation increased specificity (76.1%) compared with a single HPV or cytology test, while maintaining acceptable sensitivity (72.9%). Similar performance was observed for HPV16/18 with methylation triage (sensitivity 79.7%, specificity 74.8%). The number of women needed to refer to detect one CIN3+ ranged from 1.5 (cytology) to 2.6 (single HPV test).
Conclusion:
Molecular screening strategies using HPV, with or without HPV16/18 genotyping, and FAM19A4/miR124-2 methylation have higher sensitivity with an acceptable loss in specificity compared with current cytology screening and are efficient for the detection of CIN3+ in South African women living with HIV.
Introduction
The implementation of organized cervical screening has caused a significant decline in cervical cancer incidence in high-income countries [1–4], yet cervical cancer remains the fourth most common cancer in women worldwide [5]. The majority of the disease burden prevails in low-income and middle-income countries (LMIC), where the absence of organized and effective cervical screening programmes contributes to a high number of preventable cervical cancer deaths. Among women living with HIV (WLHIV), who are at increased risk for cervical cancer and its precursor lesions (i.e. cervical intraepithelial neoplasia, CIN, graded 1–3) [6–8], effective cervical screening is particularly essential to lower cervical cancer morbidity and mortality.
The ideal screening method for WLHIV in LMIC remains to be defined and may vary from country-to-country, depending on locally accepted risks and available resources [9]. Cytology-based screening, requiring a clinician-collected sample, adequate training of cytopathologists, and a good-quality assurance system to compensate for the high degree of subjectivity, has proven difficult to implement effectively in developing countries. Moreover, sensitivity for cervical intraepithelial neoplasia grade 3 or worse (CIN3+) is limited and follow-up of women with abnormal cytology is challenging. Despite these drawbacks, cytology-based screening is provided on demand in some regions. Primary testing for presence of high-risk human papillomavirus (HPV) in cervical or vaginal samples is currently recommended because of its high sensitivity [10], but requires additional triage testing as specificity of the HPV test is lower than cytology, and referral of all high-risk HPV positive women would lead to unmanageable referral rates.
An alternative to HPV-based screening is a screen-and-treat approach using visual inspection with acetic acid (VIA), allowing for immediate treatment of all screen-positives. However, the clinical performance of this approach is inferior to that of primary HPV screening [11–15] and varies highly as it depends on the clinician's experience, leading to both undertreatment and overtreatment. Efforts to enhance the accuracy of VIA using artificial intelligence are ongoing, but such tools are still in research phase [16,17].
Molecular markers that are currently available and can be assessed in the original sample, but also have the potential to be used as a stand-alone test, are currently being investigated. Among these are high-risk HPV partial genotyping, methylation of viral or host cell DNA, E6 protein detection and microRNAs [18]. These tests are objective, reproducible, applicable on various sample types including self-sampled material, and have the potential for high-throughput, automated testing. The advantages could circumvent some of the main barriers to cervical screening in developing areas.
Detection of hypermethylated host cell DNA in cervical samples has been identified as a good tool for the detection of cervical cancer and CIN3 [18–22]. Significantly, methylation assays have been demonstrated to be particularly sensitive for the detection of so-called advanced CIN3 lesions, which have a cancer-like methylation profile and are thought to have a high short-term progression risk to cervical cancer [23–25]. Multiple-target genes have been identified, of which a few have been evaluated in WLHIV [26–32]. Most research has focussed on the performance of methylation analysis for triage of high-risk HPV positive women, which revealed good performance of marker panel CADM1/MAL/miR124–2[26,32]. More recently discovered markers ASCL1 and LHX8 were also identified as promising primary screening tools that can be used without prior HPV testing in WLHIV [31]. The balance between sensitivity for current CIN3 or even invasive cervical cancer (ICC) and the number of women needed to refer for colposcopy or treatment remains challenging, and further optimization of these methylation assays is ongoing.
In this study, we used data from a screening cohort of WLHIV in South Africa to further evaluate the role of methylation-based screening strategies for this population as part of a cytology-based or HPV-based strategy. The performance was also compared with currently used cytology screening and proposed HPV-based screening strategies. For this purpose we used the FAM19A4/miR124-2 methylation assay, as this test has been validated extensively [24,33–36], is commercially available (QIAsure Methylation Test; Qiagen, Hilden, Germany) and has not been evaluated in WLHIV to date. The performance of four different screening strategies using this methylation assay for the detection of histologically proven CIN3+ was evaluated. Moreover, potential screening-related harms were assessed by the number of tests and the number of referrals needed to detect one case of CIN3+ at baseline.
Methods
Study population
In accordance with the national cervical cancer prevention policy, [37] women aged 18 years and above were included in a South African screening cohort of WLHIV (n = 355) at the gynaecological outpatient clinic in Tshwane District Hospital, Pretoria, in a study conducted from March 2013 to March 2015. In this study, different cervical screening strategies for low-resource settings were evaluated. The study was approved by the Research Ethics Committee of the University of Pretoria (protocol numbers 100/2012 and 155/2014). All study participants gave written informed consent. Detailed characteristics of the study procedures and the study population have been described previously [31,32,38].
In short, WLHIV presenting for cervical screening were invited to participate in the study. On study inclusion, a routine conventional cervical cytology specimen, a liquid-based cytology (LBC) specimen and two colposcopy-directed cervical biopsies were collected by the research physician and specialist nurse. In women without visible lesions two random biopsies were collected at 0600 and 1200 h. The conventional cytology specimens were analysed and reported within routine diagnostics by a local cytopathologist according to the Bethesda 2001 classification [39]. LBC specimens were processed in The Netherlands and the presence of high-risk HPV DNA was determined using the GP5+/6+ PCR-EIA (Labo Biomedical Products B.V., Rijswijk, The Netherlands). All high-risk HPV positive samples were genotyped using a bead-based analysis of GP5+/6+ PCR products and/or the HPV-risk assay (Self-screen B.V., Amsterdam, The Netherlands). These clinically validated assays can differentiate 14 (probably) high-risk HPV types (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68) [40–43].
All LBC specimens were subjected to methylation analysis of FAM19A4 and miR124-2 using a quantitative methylation specific PCR according to the manufacturer's instructions (QIAsure Methylation Test, Qiagen). This assay runs on the Rotorgene PCR-system (Qiagen) and provides automated data analysis and interpretation.
All cervical biopsies were classified according to international guidelines as no dysplasia, CIN1, CIN2, CIN3 or ICC [44]. Women with at least high-grade squamous intraepithelial lesion (HSIL), or worse (includes atypical squamous cells-cannot exclude HSIL), or CIN2 or worse (CIN2+) on a cervical biopsy were treated according to local guidelines (large loop excision of the transformation zone, LLETZ). The worst histology diagnosis (either biopsy or LLETZ specimen) was used as study endpoint. Women with invalid test results on one or more parameters were excluded from the analysis (n = 70).
Screening strategies
We analysed and describe here the performance of screening strategies combining cytology and the FAM19A/miR124-2 methylation assay (strategies 1–3) as well as the performance of strategies combining the high-risk HPV DNA test with the methylation assay in the defined study group (strategies 4–6).
Strategies include: cytology (strategy 1), cytology with methylation triage of women with atypical squamous cells of unknown significance (ASC-US) or low-grade squamous intraepithelial lesion (LSIL) cytology (strategy 2), methylation with cytology triage (strategy 3), high-risk HPV testing (strategy 4), HPV with methylation triage (strategy 5) and HPV16/18 genotyping with methylation triage of non16/18 high-risk HPV positives (strategy 6).
Strategy 1 consists of conventional cytology screening, which is a frequently used screening method for cervical screening in South Africa and is offered only on demand (e.g. during a visit to an antenatal clinic). Only women with at least HSIL are directly referred for ablative treatment, and women with ASC-US or LSIL are advised to be rescreened using cytology. However, there is no active recall system for repeat testing of women with ASC-US/LSIL cytology and the number of women lost to follow-up is high. We therefore decided to evaluate cytology with threshold at least HSIL, which represents the current screening and referral practice most accurately. For strategy 1, women were considered screen-positive if cytology was graded at least HSIL, and all others were considered screen-negative.
Strategy 2 consists of cytology with FAM19A4/miR124-2 methylation triage of women with ASC-US or LSIL cytology. Women with ASC-US or LSIL cytology and a positive FAM19A4/miR124-2 methylation result, and women with at least HSIL cytology, irrespective of the FAM19A4/miR124-2 methylation result, were considered screen-positive. All other women (i.e. normal cytology or ASC-US/LSIL cytology and a negative FAM19A4/miR124-2 methylation result) were considered screen-negative.
Strategy 3 consists of FAM19A4/miR124-2 methylation with triage of methylation positives with cytology. For this strategy, only double positive women, that is a positive FAM19A4/miR124-2 methylation result and at least ASC-US cytology, were considered screen-positive, and all others were considered screen-negative.
Strategy 4 consists of screening with a single high-risk HPV test for which women were scored screen-positive if high-risk HPV DNA was present in the LBC sample. High-risk HPV negative women were scored screen-negative.
Strategy 5 consists of triage of high-risk HPV positive women with FAM19A4/miR124-2 methylation analysis. Women were considered screen-positive if both high-risk HPV and FAM19A4/miR124-2 methylation results were positive. All other women were considered screen-negative.
Strategy 6 consists of HPV16/18 genotyping and subsequent testing of non16/18 high-risk HPV positive samples with FAM19A4/miR124-2 methylation analysis. For this strategy, women were considered screen-positive if they were non16/18 high-risk HPV positive and had a positive FAM19A4/miR124-2 methylation result, or if they were positive for HPV16/18 (irrespective of their FAM19A4/miR124-2 methylation result). All high-risk HPV-negative women and all non16/18 high-risk HPV-positive women with a negative FAM19A4/miR124-2 methylation result were scored screen-negative.
Statistical analysis
The primary endpoint of the study was CIN3+ on histology as these are the most reproducibly classified lesions having the highest risk of cervical cancer and most in need of treatment. All analyses were also performed using the outcome CIN2+. However, classification of CIN2 is less reproducible as it constitutes very heterogeneous disease categories, including both productive CIN1-like lesions and transforming CIN3-like lesions with different clinical behaviour [19,45]. Consequently, this endpoint leads to less reproducible results. The CIN2+ data are therefore provided in the Supplementary data. The performance of each strategy was evaluated with respect to sensitivity, specificity, positive and negative predictive value and referral rate (based on the percentage test positivity). The number of tests (cytology, methylation and/or HPV test with or without integrated HPV16/18 genotyping) required to screen 1000 women and the number of women referred for colposcopy to detect one case of CIN3+ (calculated by dividing the number of screen positives by the number of true positives) was also calculated for each strategy.
Relative sensitivities and specificities (ratios of the sensitivity or specificity of one test to the sensitivity or specificity of the reference test) were calculated with 95% confidence intervals (95% CIs) and visualized using forest plots with cytology (strategy 1) as the reference for all other strategies. Relative sensitivities and specificities were also calculated for the HPV-based strategies (strategies 5 and 6) using a single high-risk HPV test (strategy 4) as a reference. McNemar testing was used to calculate P values of the relative sensitivities and specificities. A difference in sensitivity or specificity was considered significant if the 95% CI of the relative sensitivity or specificity was entirely above or below one and the corresponding P value was less than 0.05. Calculations were performed in Microsoft Excel (2016; Microsoft, Redmond, Washington, USA), SPSS (V22; IBM, Armonk, New York, USA) and STATA (V14; StataCorp, College Station, Texas, USA).
Results
In total 285 women with a median age of 40 years (interquartile range: 35–46 years) were included in the analyses, of whom 194 (68.1%) had no dysplasia or CIN1 (≤CIN1), 32 (11.3%) had CIN2, 57 (20.0%) had CIN3 and two (0.7%) had ICC.
Table 1 provides an overview of the performance characteristics of the six screening strategies evaluated. Overall, cytology-based screening strategies had highest specificities, but lower sensitivities, and HPV-based screening strategies had highest sensitivities, but lower specificities (Fig. 1). The two women with ICC were detected by all screening strategies evaluated. Similar results were obtained when CIN2+ was used as outcome, but with lower sensitivities and higher specificities for all strategies (see Table, Supplemental Digital Content 1, for CIN2+ performance characteristics, and Figure, Supplemental Digital Content 2, for relative performance).
Table 1.
Accuracy and diagnostic efficiency of screening strategies to detect cervical intraepithelial neoplasia grade 3 or worse.
| No. | Strategy | Sensitivity (95% CI) | n1/N1 | Specificity (95% CI) | n2/N2 | PPV | NPV | Referral rate | Referrals needed to detect one CIN3+ | Number of tests/1000 women screened | ||
| Cytology-based screening | ||||||||||||
| 1 | Cytology (≥HSIL) | 59.3% | (46.8–71.9) | 35/59 | 91.6% | (88.0–95.2) | 207/226 | 64.8% | 89.6% | 18.9% | 1.5 | 1000 |
| 2 | Cytology (≥HSIL) with FAM19A4/miR124-2 triage of ASC-US/LSIL | 67.8% | (55.9–79.7) | 40/59 | 85.0% | (80.3–89.6) | 192/226 | 54.1% | 91.0% | 26.0% | 1.9 | 1095 |
| 3 | FAM19A4/miR124-2 with cytology (≥ASC-US) triage | 62.7% | (50.4–75.1) | 37/59 | 87.2% | (82.2–91.5) | 197/226 | 56.1% | 90.0% | 23.2% | 1.8 | 1674 |
| HPV-based screening | ||||||||||||
| 4 | HPV | 83.1% | (73.5–92.6) | 49/59 | 66.4% | (60.2–72.5) | 150/226 | 39.2% | 93.8% | 43.9% | 2.6 | 1000 |
| 5 | HPV with FAM19A4/miR124-2 triage | 72.9% | (61.5–84.2) | 43/59 | 76.1% | (70.5–81.7) | 172/226 | 44.3% | 91.5% | 34.0% | 2.3 | 1440 |
| 6 | HPV16/18 with FAM19A4/miR124-2 triage of non16/18HPV+ | 79.7% | (69.4–89.9) | 47/59 | 74.8% | (69.1–80.4) | 169/226 | 45.2% | 93.4% | 36.5% | 2.2 | 1315 |
Cytology with threshold high-grade squamous intraepithelial lesion or worse (≥HSIL, includes atypical squamous cells – cannot exclude HSIL); cytology with threshold atypical squamous cells of unknown significance or worse (≥ASC-US). 95% CI, 95% confidence interval; ASC-US, atypical squamous cells of unknown significance; CIN3+, cervical intraepithelial neoplasia grade 3 or worse; HPV, human papillomavirus; HSIL, high-grade squamous intraepithelial lesion; LSIL, low-grade squamous intraepithelial lesion; n1, number of screen-positive disease cases; N1, total number of disease cases; n2, number of screen-negative nondisease cases; N2, total number of nondisease cases; NPV, negative predictive value; PPV, positive predictive value.
Fig. 1.
Forest plots showing the relative sensitivities and specificities for the detection of cervical intraepithelial neoplasia grade 3 or worse of different screening strategies compared with cytology (threshold ≥ high-grade squamous intraepithelial lesion).
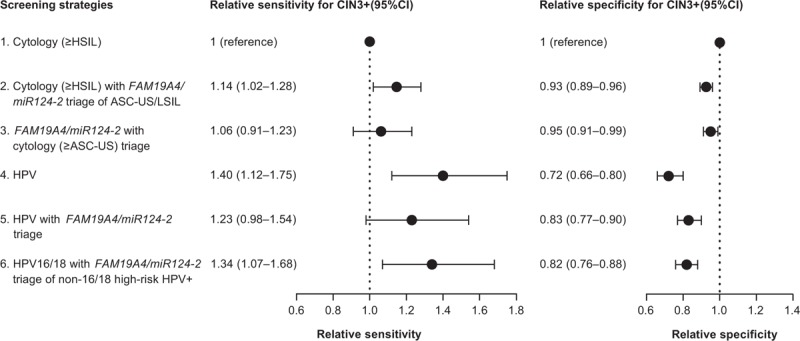
95% CI, 95% confidence interval; ASC-US, atypical squamous cells of unknown significance; HSIL, high-grade squamous intraepithelial lesion; LSIL, low-grade squamous intraepithelial lesion.
A single cytology test, referring only women with at least HSIL cytology (strategy 1), had the lowest sensitivity. This strategy detected 59.3% of CIN3+, but had the lowest referral rate (18.9%) and highest specificity (91.6%), leading to a low number of women needed to refer to detect one case of CIN3+ (1.5). Combining cytology with FAM19A4/miR124-2 methylation, either triage of women with ASC-US or LSIL with FAM19A4/miR124-2 methylation (strategy 2), or triage of FAM19A4/miR124-2 methylation positive women with cytology (strategy 3), led to a small nonsignificant increase in sensitivity compared with a single cytology test (67.8%, ratio 1.14, P value 0.06, and 62.7%, ratio 1.06, P value 0.73, respectively) and a significant decrease in specificity (85.0%, ratio 0.93, P value <0.001, and 87.2%, ratio 0.95, P value 0.04, respectively). The referral rates of these strategies were higher compared with a single cytology test (26.0 and 23.2%, respectively), as well as the number of women needed to refer to detect one case of CIN3+ (1.9 and 1.8, respectively).
The most sensitive strategy for CIN3+ detection was screening with a single high-risk HPV test, referring all high-risk HPV positive women (strategy 4). This strategy detected 83.1% of CIN3+, but also had the highest referral rate (43.9%) and lowest specificity (66.4%), leading to a relatively high number of colposcopy referrals to detect one case of CIN3+ (2.6). Compared with cytology, a single high-risk HPV test had a significantly higher sensitivity (ratio 1.40, P value 0.004), but also a significantly lower specificity (ratio 0.73, P value <0.001, Fig. 1).
Combined strategies using HPV (either high-risk HPV alone or high-risk HPV testing with 16/18 genotyping) and FAM19A4/miR124-2 methylation increased specificity compared with a single high-risk HPV test (ratios 1.15 and 1.13, respectively, P values <0.001), at the cost of a slight decrease in sensitivity (ratios 0.88 and 0.96, P values 0.03 and 0.5, respectively, Fig. 2). Both HPV with FAM19A4/miR124-2 methylation triage of high-risk HPV positives (strategy 5) and HPV16/18 genotyping with FAM19A4/miR124-2 methylation triage of non16/18 high-risk HPV positives (strategy 6) provided good sensitivities, detecting 72.9 and 79.7% of CIN3+ respectively, while retaining specificities of 76.1 and 74.8%. The referral rates of these strategies were lower compared with a single high-risk HPV test (34.0% for strategy 5 and 36.5% for strategy 6), as were the numbers of colposcopy referrals needed to detect one case of CIN3+ (2.3 and 2.2, respectively). Triaging only non16/18 high-risk HPV-positive women reduced the number of tests needed to screen 1000 women from 1440 (triage of all high-risk HPV positives) to 1315 (triage of non16/18 high-risk HPV positives). Compared with a single cytology test, strategy 6 had a significantly higher sensitivity (ratio 1.34, P value 0.017), but lower specificity (ratio 0.82, P value <0.001). Strategy 5 showed a moderate, nonsignificant increase in sensitivity (ratio 1.23, P value 0.12) and a decrease in specificity compared with cytology (ratio 0.83, P value <0.001, Fig. 1).
Fig. 2.
Forest plots showing the relative sensitivities and specificities for the detection of cervical intraepithelial neoplasia grade 3 or worse of different screening strategies compared with a single human papillomavirus test.
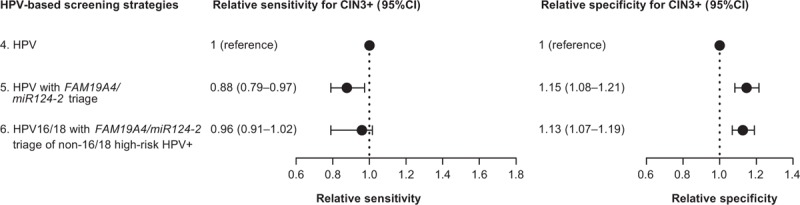
95% CI, 95% confidence interval; HPV, human papillomavirus.
The complementarity of FAM19A4/miR124-2 methylation testing in either cytology-based or HPV-based screening is shown in the supplementary data (see Table, Supplemental Digital Content 3).
Discussion
The current study provides further evidence for the potential role of methylation analysis in cervical screening of WLHIV in South Africa, indicating that FAM19A4/miR124-2 methylation analysis could be used to improve screening and facilitate full molecular screening.
Cytology screening is still common practice in South Africa, and only women with HSIL cytology or worse are referred for treatment. Loss to follow-up of women with ASC-US or LSIL is high as there is no active recall system. We evaluated whether FAM19A4/miR124-2 methylation analysis could improve this cytology-based screening by using it either as a triage test of women with ASC-US or LSIL cytology (strategy 2), thereby reducing the workload for cytopathologists and reducing the risk of loss to follow-up, or as a primary screening test with cytology triage of methylation positives (strategy 3). In this study, these strategies only provided a nonsignificant increase in sensitivity for CIN3+ (67.8% for strategy 2, 62.7% for strategy 3) compared with cytology alone (59.3%), while specificity decreased (from 91.6% for cytology alone to 85.0% and 87.2% for strategy 2 and 3, respectively). Furthermore, as the costs of these strategies will be higher due to the higher number of tests needed and because they still carry the subjectivity of cytology-based screening, these strategies are not considered to be effective.
The three HPV-based screening strategies had higher sensitivity compared with the three cytology-based screening strategies, whereas the specificity of the HPV-based strategies was lower. As shown before, a single high-risk HPV test provided the highest sensitivity for CIN3+ (83.1%), but also had the lowest specificity (66.4%). The use of a single high-risk HPV test within a screen-and-treat approach could be considered and has shown to be cost-effective [46], but will also lead to considerable overtreatment and associated adverse reproductive outcomes due to its limited specificity. Our results show that FAM19A4/miR124-2 methylation analysis, with or without HPV16/18 genotyping, could be used to improve specificity of the primary high-risk HPV test while maintaining good sensitivity. Compared with the current cytology-based screening, these strategies had a higher sensitivity for CIN3+ (59.3% for cytology versus 72.9% for HPV with methylation, and 79.7% for HPV16/18 genotyping with methylation), while the decrease in specificity remained acceptable (91.6 versus 76.1 and 74.8%, respectively).
The FAM19A4/miR124-2 methylation assay is commercially available and has been clinically validated for triage of high-risk HPV positive women in a European, high-resource setting, showing comparable performance with other currently used triage tests [sensitivity and specificity ∼70% (Bonde et al., in preparation)] [24,34]. This is the first study evaluating the assay in a low-resource setting in WLHIV, showing a good performance when combined with high-risk HPV testing and/or HPV16/18 genotyping. These full molecular screening strategies are, in contrast to cytology-based screening, objective, compatible with both cervical scrapes and self-sampled cervico-vaginal material [33], and suitable for high-throughput workflows. Although these characteristics and present findings could contribute to improving screening coverage and effectiveness, further implementation studies are required. Ongoing development of the methylation assay is expected to result in a robust and user-friendly assay, suitable for centralized testing in laboratories with HPV testing facilities and generating results within a day. Furthermore, cost-effectiveness of HPV screening with FAM19A4/miR124-2 methylation triage testing in centralised laboratories should ideally be compared with cost-effectiveness of a one day screen-and-treat strategy using a point-of-care HPV test, associated with considerable over-referral and overtreatment requiring more medically trained personnel.
Several other host cell methylation markers have been studied in WLHIV [28–31,47] and similar performance compared with HPV with FAM19A4/miR124-2 methylation triage has been shown for triage of high-risk HPV positive women with the marker panel CADM1, MAL and miR124-2 by us and others [26,32]. More recently, Kelly et al.[27] showed that methylation of EPB41L3 increased with cervical disease severity in a South African cohort of WLHIV, but the accuracy of this marker for the detection of CIN3 and cervical cancer is difficult to derive from these data, as the threshold for methylation positivity was neither determined nor validated. At this moment no commercial assays are available for these markers. Other commercially available assays, such as the GynTect assay [48], have not been evaluated in WLHIV yet.
A major strength of this study is the availability of a histology endpoint for all study participants, which minimizes the disease ascertainment bias. Yet, women were treated based only on an abnormal cervical scrape or biopsy, but not based on a positive high-risk HPV or methylation result. This may have caused a preferential effect in favour of cytology. Other limitations of this study are the small sample size and the current absence of follow-up data. Although a recent study in a Dutch screening population demonstrated that high-risk HPV positive women with a negative FAM19A4/miR124-2 methylation triage test have a lower long-term cervical cancer risk compared with high-risk HPV positive, cytology-negative women [49], longitudinal data from a low-resource setting are required to determine the long-term safety of this screening algorithm in a different population.
Conclusion
The optimal screening strategy, balancing harms and benefits of screening, depends on local circumstances such as disease prevalence and available resources. The data presented in this study show that screening strategies with HPV (with or without partial genotyping) and FAM19A4/miR124-2 methylation triage testing are feasible screening options for WLHIV in South Africa, warranting further evaluation.
Acknowledgements
We gratefully acknowledge all the women participating in the described study. Also, we are grateful for the active cooperation of the teams of the clinics of the department of Obstetrics and Gynaecology of the Steve Biko Academic Hospital and the HIV clinic at the Tshwane District Hospital. Cytological evaluations were performed at the National Health Laboratory Service, South Africa.
The current study was funded by the VU University Research Fellowship (URF) program (Amsterdam, The Netherlands), the 1st For Women Foundation (Pretoria, South Africa), and the Carl & Emily Fuchs Foundation (Pretoria, South Africa).
Conflicts of interest
R.D.M.S. and C.J.L.M.M. are minority shareholders of Self-screen B.V., a spin-off company of VUmc; Self-screen B.V. holds patents related to the work (i.e. high-risk HPV test and methylation markers for cervical screening) and has developed and manufactured the methylation assay, which is licensed to Qiagen (QIAsure Methylation Test); C.J.L.M.M. is part-time CEO of Self-screen B.V.; C.J.L.M.M. has received speakers fees from GSK, Qiagen, and SPMSD/Merck, and served occasionally on the scientific advisory board (expert meeting) of GSK, Qiagen, and SPMSD/Merck; C.J.L.M.M. has a very small number of shares of Qiagen; W.W.K., M.Z., K.L.R., E.B. and G.D. declare no conflicts of interest.
Supplementary Material
References
- 1.International Agency for Research on Cancer. Biological agents: a review of human carcinogens. IARC monographs on the evaluation of carcinogenic risks to humans, volume 100B. Lyon: International Agency for Research on Cancer; 2012. [Google Scholar]
- 2.Vaccarella S, Lortet-Tieulent J, Plummer M, Franceschi S, Bray F. Worldwide trends in cervical cancer incidence: impact of screening against changes in disease risk factors. Eur J Cancer 2013; 49:3262–3273. [DOI] [PubMed] [Google Scholar]
- 3.Arbyn M, Raifu AO, Weiderpass E, Bray F, Anttila A. Trends of cervical cancer mortality in the member states of the European Union. Eur J Cancer 2009; 45:2640–2648. [DOI] [PubMed] [Google Scholar]
- 4.Kitchener HC, Castle PE, Cox JT. Chapter 7: Achievements and limitations of cervical cytology screening. Vaccine 2006; 24 Suppl 3:S3/63–S3/70. [DOI] [PubMed] [Google Scholar]
- 5.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68:394–424. [DOI] [PubMed] [Google Scholar]
- 6.Denslow SA, Rositch AF, Firnhaber C, Ting J, Smith JS. Incidence and progression of cervical lesions in women with HIV: a systematic global review. Int J STD AIDS 2014; 25:163–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaturvedi AK, Madeleine MM, Biggar RJ, Engels EA. Risk of human papillomavirus-associated cancers among persons with AIDS. J Natl Cancer Inst 2009; 101:1120–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun XW, Kuhn L, Ellerbrock TV, Chiasson MA, Bush TJ, Wright TC., Jr Human papillomavirus infection in women infected with the human immunodeficiency virus. N Engl J Med 1997; 337:1343–1349. [DOI] [PubMed] [Google Scholar]
- 9.Meijer CJ, Berkhof J. Screening: cervical cancer – should we abandon cytology for screening?. Nat Rev Clin Oncol 2012; 9:558–559. [DOI] [PubMed] [Google Scholar]
- 10.Jeronimo J, Castle PE, Temin S, Shastri SS. Secondary prevention of cervical cancer: American Society of Clinical Oncology resource-stratified clinical practice guideline summary. J Oncol Pract 2017; 13:129–133. [DOI] [PubMed] [Google Scholar]
- 11.Viviano M, DeBeaudrap P, Tebeu PM, Fouogue JT, Vassilakos P, Petignat P. A review of screening strategies for cervical cancer in human immunodeficiency virus-positive women in sub-Saharan Africa. Int J Womens Health 2017; 9:69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fokom-Domgue J, Combescure C, Fokom-Defo V, Tebeu PM, Vassilakos P, Kengne AP, et al. Performance of alternative strategies for primary cervical cancer screening in sub-Saharan Africa: systematic review and meta-analysis of diagnostic test accuracy studies. BMJ 2015; 351:h3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sankaranarayanan R, Nessa A, Esmy PO, Dangou JM. Visual inspection methods for cervical cancer prevention. Best Pract Res Clin Obstet Gynaecol 2012; 26:221–232. [DOI] [PubMed] [Google Scholar]
- 14.Arbyn M, Sankaranarayanan R, Muwonge R, Keita N, Dolo A, Mbalawa CG, et al. Pooled analysis of the accuracy of five cervical cancer screening tests assessed in eleven studies in Africa and India. Int J Cancer 2008; 123:153–160. [DOI] [PubMed] [Google Scholar]
- 15.Sankaranarayanan R, Nene BM, Shastri SS, Jayant K, Muwonge R, Budukh AM, et al. HPV screening for cervical cancer in rural India. N Engl J Med 2009; 360:1385–1394. [DOI] [PubMed] [Google Scholar]
- 16.Thay S, Goldstein A, Goldstein LS, Govind V, Lim K, Seang C. Prospective cohort study examining cervical cancer screening methods in HIV-positive and HIV-negative Cambodian Women: a comparison of human papilloma virus testing, visualization with acetic acid and digital colposcopy. BMJ Open 2019; 9:e026887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu L, Bell D, Antani S, Xue Z, Yu K, Horning MP, et al. An observational study of deep learning and automated evaluation of cervical images for cancer screening. J Natl Cancer Inst 2019; [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luttmer R, De Strooper LM, Steenbergen RD, Berkhof J, Snijders PJ, Heideman DA, et al. Management of high-risk HPV-positive women for detection of cervical (pre)cancer. Expert Rev Mol Diagn 2016; 16:961–974. [DOI] [PubMed] [Google Scholar]
- 19.Steenbergen RD, Snijders PJ, Heideman DA, Meijer CJ. Clinical implications of (epi)genetic changes in HPV-induced cervical precancerous lesions. Nat Rev Cancer 2014; 14:395–405. [DOI] [PubMed] [Google Scholar]
- 20.Wentzensen N, Sherman ME, Schiffman M, Wang SS. Utility of methylation markers in cervical cancer early detection: appraisal of the state-of-the-science. Gynecol Oncol 2009; 112:293–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cook DA, Krajden M, Brentnall AR, Gondara L, Chan T, Law JH, et al. Evaluation of a validated methylation triage signature for human papillomavirus positive women in the HPV FOCAL cervical cancer screening trial. Int J Cancer 2019; 144:2587–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmitz M, Wunsch K, Hoyer H, Scheungraber C, Runnebaum IB, Hansel A, et al. Performance of a methylation specific real-time PCR assay as a triage test for HPV-positive women. Clin Epigenetics 2017; 9:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bierkens M, Hesselink AT, Meijer CJ, Heideman DA, Wisman GB, van der Zee AG, et al. CADM1 and MAL promoter methylation levels in hrHPV-positive cervical scrapes increase proportional to degree and duration of underlying cervical disease. Int J Cancer 2013; 133:1293–1299. [DOI] [PubMed] [Google Scholar]
- 24.De Strooper LM, Meijer CJ, Berkhof J, Hesselink AT, Snijders PJ, Steenbergen RD, et al. Methylation analysis of the FAM19A4 gene in cervical scrapes is highly efficient in detecting cervical carcinomas and advanced CIN2/3 lesions. Cancer Prev Res 2014; 7:1251–1257. [DOI] [PubMed] [Google Scholar]
- 25.Verlaat W, Snoek BC, Heideman DAM, Wilting SM, Snijders PJF, Novianti PW, et al. Identification and validation of a 3-gene methylation classifier for HPV-based cervical screening on self-samples. Clin Cancer Res 2018; 24:3456–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Vuyst H, Franceschi S, Plummer M, Mugo NR, Sakr SR, Meijer CJ, et al. Methylation levels of CADM1, MAL, and MIR124-2 in cervical scrapes for triage of HIV-infected, high-risk HPV-positive women in Kenya. J Acquir Immune Defic Syndr 2015; 70:311–318. [DOI] [PubMed] [Google Scholar]
- 27.Kelly HA, Chikandiwa A, Warman R, Segondy M, Sawadogo B, Vasiljevic N, et al. Associations of human gene EPB41L3 DNA methylation and cervical intraepithelial neoplasia in women living with HIV-1 in Africa. AIDS 2018; 32:2227–2236. [DOI] [PubMed] [Google Scholar]
- 28.Nye MD, Hoyo C, Huang Z, Vidal AC, Wang F, Overcash F, et al. Associations between methylation of paternally expressed gene 3 (PEG3), cervical intraepithelial neoplasia and invasive cervical cancer. PLoS One 2013; 8:e56325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vidal AC, Henry NM, Murphy SK, Oneko O, Nye M, Bartlett JA, et al. PEG1/MEST and IGF2 DNA methylation in CIN and in cervical cancer. Clin Transl Oncol 2014; 16:266–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wijetunga NA, Belbin TJ, Burk RD, Whitney K, Abadi M, Greally JM, et al. Novel epigenetic changes in CDKN2A are associated with progression of cervical intraepithelial neoplasia. Gynecol Oncol 2016; 142:566–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kremer WW, Van Zummeren M, Novianti PW, Richter KL, Verlaat W, Snijders PJ, et al. Detection of hypermethylated genes as markers for cervical screening in women living with HIV. J Int AIDS Soc 2018; 21:e25165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Zummeren M, Kremer WW, Van Aardt MC, Breytenbach E, Richter KL, Rozendaal L, et al. Selection of women at risk for cervical cancer in an HIV-infected South African population. AIDS 2017; 31:1945–1953. [DOI] [PubMed] [Google Scholar]
- 33.De Strooper LM, Verhoef VM, Berkhof J, Hesselink AT, de Bruin HM, van Kemenade FJ, et al. Validation of the FAM19A4/mir124-2 DNA methylation test for both lavage- and brush-based self-samples to detect cervical (pre)cancer in HPV-positive women. Gynecol Oncol 2016; 141:341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luttmer R, De Strooper LM, Berkhof J, Snijders PJ, Dijkstra MG, Uijterwaal MH, et al. Comparing the performance of FAM19A4 methylation analysis, cytology and HPV16/18 genotyping for the detection of cervical (pre)cancer in high-risk HPV-positive women of a gynecologic outpatient population (COMETH study). Int J Cancer 2016; 138:992–1002. [DOI] [PubMed] [Google Scholar]
- 35.Luttmer R, De Strooper LM, Dijkstra MG, Berkhof J, Snijders PJ, Steenbergen RD, et al. FAM19A4 methylation analysis in self-samples compared with cervical scrapes for detecting cervical (pre)cancer in HPV-positive women. Br J Cancer 2016; 115:579–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Floore A, Hesselink A, Ostrbenk A, Alcaniz E, Rothe B, Pedersen H, et al. Intra- and inter-laboratory agreement of the FAM19A4/mir124-2 methylation test: results from an international study. J Clin Lab Anal 2019; 33:e22854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.South African Government, Department of Health. Cervical Cancer Prevention and Control Policy 2017. 2017. Available at: http://www.health.gov.za/index.php/2014-08-15-12-53-24?download=1393:cervical-cancer-policy-pdf [Accessed: 15 July 2019]. [Google Scholar]
- 38.Kremer WW, van Zummeren M, Heideman DAM, Lissenberg-Witte BI, Snijders PJF, Steenbergen RDM, et al. HPV16-related cervical cancers and precancers have increased levels of host cell DNA methylation in women living with HIV. Int J Mol Sci 2018; 19:3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Solomon D, Davey D, Kurman R, Moriarty A, O’Connor D, Prey M, et al. The 2001 Bethesda System: terminology for reporting results of cervical cytology. JAMA 2002; 287:2114–2119. [DOI] [PubMed] [Google Scholar]
- 40.Polman NJ, Ostrbenk A, Xu L, Snijders PJF, Meijer C, Poljak M, et al. Evaluation of the clinical performance of the HPV-Risk Assay using the VALGENT-3 Panel. J Clin Microbiol 2017; 55:3544–3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hesselink AT, Berkhof J, van der Salm ML, van Splunter AP, Geelen TH, van Kemenade FJ, et al. Clinical validation of the HPV-risk assay, a novel real-time PCR assay for detection of high-risk human papillomavirus DNA by targeting the E7 region. J Clin Microbiol 2014; 52:890–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitt M, Bravo IG, Snijders PJ, Gissmann L, Pawlita M, Waterboer T. Bead-based multiplex genotyping of human papillomaviruses. J Clin Microbiol 2006; 44:504–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacobs MV, Snijders PJ, van den Brule AJ, Helmerhorst TJ, Meijer CJ, Walboomers JM. A general primer GP5+/GP6(+)-mediated PCR-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in cervical scrapings. J Clin Microbiol 1997; 35:791–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wright TC, Ronnett BM, Kurman RJ, Ferenczy A. Kurman RJ, Hedrick Ellenson L, Ronnett BM. Precancerous lesions of the cervix. Blaustein's pathology of the female genital tract Springer, 6th ed.New York: 2011. [Google Scholar]
- 45.Zummeren MV, Kremer WW, Leeman A, Bleeker MCG, Jenkins D, Sandt MV, et al. HPV E4 expression and DNA hypermethylation of CADM1, MAL, and miR124-2 genes in cervical cancer and precursor lesions. Mod Pathol 2018; 31:1842–1850. [DOI] [PubMed] [Google Scholar]
- 46.Campos NG, Lince-Deroche N, Chibwesha CJ, Firnhaber C, Smith JS, Michelow P, et al. Cost-effectiveness of cervical cancer screening in women living with HIV in South Africa: a mathematical modeling study. J Acquir Immune Defic Syndr 2018; 79:195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teixeira da Costa Lodi C, Michelin MA, Miranda Lima MI, Murta EFC, Braga LDC, Montes L, et al. Predicting cervical intraepithelial neoplasia recurrence in HIV-infected and –noninfected women by detecting aberrant promoter methylation in the CDH1, TIMP3, and MGMT genes. Arch Gynecol Obstet 2018; 298:971–979. [DOI] [PubMed] [Google Scholar]
- 48.Schmitz M, Eichelkraut K, Schmidt D, Zeiser I, Hilal Z, Tettenborn Z, et al. Performance of a DNA methylation marker panel using liquid-based cervical scrapes to detect cervical cancer and its precancerous stages. BMC Cancer 2018; 18:1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.De Strooper LMA, Berkhof J, Steenbergen RDM, Lissenberg-Witte BI, Snijders PJF, Meijer C, et al. Cervical cancer risk in HPV-positive women after a negative FAM19A4/mir124-2 methylation test: a post hoc analysis in the POBASCAM trial with 14 year follow-up. Int J Cancer 2018; 143:1541–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


