Macrophages are professional phagocytes that are essential for host defense and tissue homeostasis. Proper membrane trafficking and degradative functions of the endolysosomal system are known to be critical for the function of these cells. We have found that PIKfyve, the kinase that synthesizes the endosomal phosphoinositide phosphatidylinositol-3,5-bisphosphate, is an essential regulator of lysosomal biogenesis and degradative functions in macrophages.
Keywords: PIKfyve; phosphatidylinositol-3,5-bisphosphate; macrophage; lysosome homeostasis; lysosomes; macrophages
ABSTRACT
Macrophages are professional phagocytes that are essential for host defense and tissue homeostasis. Proper membrane trafficking and degradative functions of the endolysosomal system are known to be critical for the function of these cells. We have found that PIKfyve, the kinase that synthesizes the endosomal phosphoinositide phosphatidylinositol-3,5-bisphosphate, is an essential regulator of lysosomal biogenesis and degradative functions in macrophages. Genetically engineered mice lacking PIKfyve in their myeloid cells (PIKfyvefl/fl LysM-Cre) develop diffuse tissue infiltration of foamy macrophages, hepatosplenomegaly, and systemic inflammation. PIKfyve loss in macrophages causes enlarged endolysosomal compartments and impairs the lysosomal degradative function. Moreover, PIKfyve deficiency increases the cellular levels of lysosomal proteins. Although PIKfyve deficiency reduced the activation of mTORC1 pathway and was associated with increased cleavage of TFEB proteins, this does not translate into transcriptional activation of lysosomal genes, suggesting that PIKfyve modulates the abundance of lysosomal proteins by affecting the degradation of these proteins. Our study shows that PIKfyve modulation of lysosomal degradative activity and protein expression is essential to maintain lysosomal homeostasis in macrophages.
INTRODUCTION
Lysosomes are acidic organelles that are essential for the degradation of macromolecules delivered by endocytosis, phagocytosis, and autophagy (1). Lysosomal degradation requires the hydrolytic enzymes and lysosomal membrane proteins that are continuously synthesized in the endoplasmic reticulum (ER), trafficked to the trans-Golgi network (TGN), and then sorted to the endolysosomal system (2). During this synthetic process, some lysosomal enzymes undergo a series of modifications that include cleavage of signal peptides in the ER, glycosylation by the addition of mannose-6-phosphate in the Golgi apparatus, and proteolytic processing of inactive zymogens into enzymatically active mature enzymes. Thus, proper functioning and homeostasis of lysosomes critically depend on intracellular trafficking.
Intracellular trafficking events are modulated by several signaling molecules, which include members of the phosphoinositide family (3–5). In particular, phosphatidylinositol-3,5-bisphosphate [PI(3,5)P2] is critical for the trafficking along the endolysosomal system (6–8) and has been implicated in lysosome biogenesis and autophagy (9–12). Phosphatidylinositol-3,5-bisphosphate is synthesized on the membranes of late endosomes and lysosomes (13, 14) by the lipid kinase PIKfyve (phosphoinositide kinase, FYVE finger containing) (15–17). The physiological functions of PIKfyve have been recently elucidated in genetically engineered mice. PIKfyve-null mice are embryonic lethal, which indicates a critical role for PIKfyve during development (18, 19). A hypomorphic PIKfyve mouse model is viable, but it dies early due to defects in multiple organs and tissues, including neural tissues, heart, lung, kidney, thymus, and the hematopoietic system (20). Subsequently, conditional PIKfyve knockout mice were developed using the Cre-Lox system and demonstrated the essential roles of PIKfyve in specific tissues (19, 21–26). In particular, we previously showed that mice with platelet-specific deletion of PIKfyve have impaired lysosomal homeostasis and develop aberrant inflammatory and prothrombotic responses (22). Similarly, intestine-specific PIKfyve knockout mice develop defective polarization of epithelial cells, which leads to a severe inflammatory bowel disease (19). However, recently generated myeloid-cell-specific PIKfyve knockout mice did not show any abnormalities in the resident macrophages in spleen, liver, or bone marrow; the knockout affected only some populations of alveolar macrophages in the lung (25). We were surprised by the latter observation given the known importance of lysosome function within professional phagocytic cells and the published effect of PIKfyve inhibitors on innate immunity (27, 28). Therefore, we chose to revisit the role of PIKfyve in myeloid cells using a myeloid-cell-specific PIKfyve knockout mouse that was generated using our previously reported PIKfyve flox mice (22).
We found that PIKfyve deficiency in myeloid cells leads to proliferation of granulocytes and monocytes associated with elevation of inflammatory cytokines and that PIKfyve is a critical regulator of the morphology, degradative activity, and protein turnover of the endolysosomal system in macrophages. In summary, our study demonstrates that PIKfyve is essential for maintaining lysosomal homeostasis and function in macrophages.
RESULTS
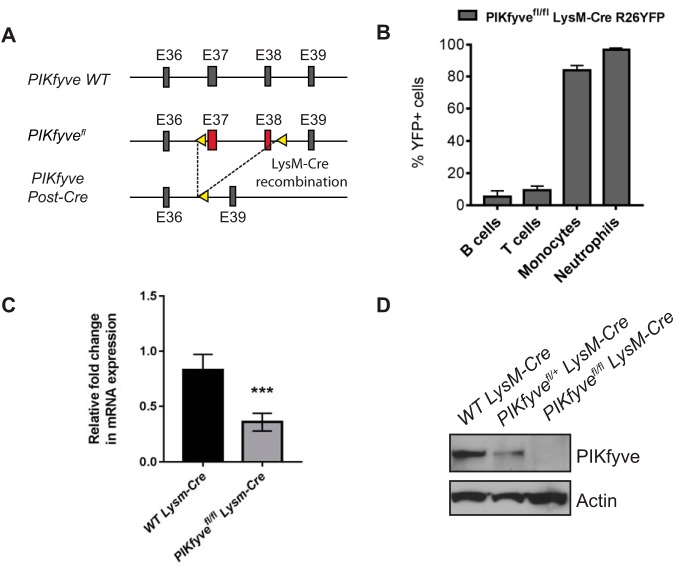
Generation and validation of mice lacking PIKfyve in myeloid lineage.
Previously reported myeloid-cell-specific PIKfyve knockout mice were generated using mice with loxP sites flanking exon 5 of the PIKfyve gene (25). Since this targeting approach can sometimes produce truncated proteins from cryptic start sites and lead to hypomorphic phenotypes, we used mice with loxP sites flanking exons 37 and 38 (corresponding to the kinase domain) of the PIKfyve gene (PIKfyvefl) (22) to breed with mice expressing the recombinase Cre under the control of the LysM promoter and generate PIKfyvefl/fl LysM-Cre mice (Fig. 1A). To validate the tissue specificity of LysM-Cre, PIKfyvefl/fl LysM-Cre mice were bred with Cre-dependent yellow fluorescent protein (YFP) reporter mice to obtain PIKfyvefl/fl LysM-Cre Rosa26YFP mice. The peripheral blood of PIKfyvefl/fl LysM-Cre Rosa26YFP mice was analyzed for YFP expression by flow cytometry, which demonstrated that the LysM-Cre promoter induced Cre expression in about 80% of the monocytes, in about 90% of the neutrophils, and in only about 1% to 5% of the circulating B cells and T cells (Fig. 1B). Primary tissue macrophages were isolated from the spleen or bone marrow using F4/80 antibody by immunomagnetic separation and were analyzed for the expression of PIKfyve. PIKfyve mRNA expression was significantly reduced in the macrophages of PIKfyvefl/fl LysM-Cre mice compared to wild-type (WT) Lysm-Cre mice as determined by quantitative reverse transcription-PCR (qRT-PCR) analysis (Fig. 1C). Furthermore, PIKfyve protein expression in macrophages was partially reduced in PIKfyvefl/+ LysM-Cre mice compared to that in WT LysM-Cre mice and completely undetectable in PIKfyvefl/fl LysM-Cre mice (Fig. 1D). Given that our gene targeting removed exons which encoded essential components of the PIKfyve kinase domain but still allowed expression of a truncated mRNA, this finding suggests that the resulting truncated PIKfyve mRNA is likely unstable and undergoes degradation, leading to the complete loss of PIKfyve protein in their macrophages.
FIG 1.
Generation and validation of mice lacking PIKfyve in myeloid cells. (A) Schematic depicting the genetic targeting of PIKfyve. PIKfyve-floxed alleles (PIKfyvefl) were generated by targeting exons 37 and 38 with loxP sites (arrowheads). PIKfyvefl mice were crossed with LysM-Cre mice to generate a myeloid-cell-specific homologous recombination of PIKfyvefl. (B) Flow cytometry analysis of percentage of YFP-expressing cells in the peripheral blood of PIKfyvefl/fl LysM-CreR26 YFP mice (n = 4 mice). (C) qRT-PCR analysis of PIKfyve gene expression relative to GAPDH in F4/80+ spleen macrophages (n = 3 mice). (D) Immunoblotting analysis of PIKfyve protein expression in the F4/80+ spleen macrophages. Statistical analysis was performed using unpaired two-tailed Student’s t test. All error bars indicate SEM. ***, P < 0.001.
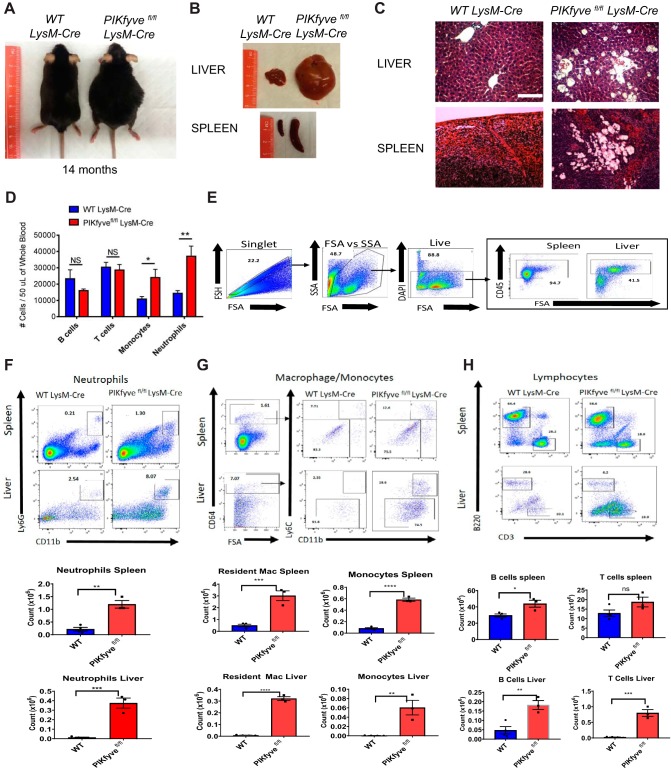
PIKfyvefl/fl LysM-Cre mice develop hepatosplenomegaly due to tissue influx of inflammatory cells.
Previously reported myeloid-cell-specific PIKfyve knockout mice did not develop any gross abnormalities (25). Although our PIKfyvefl/fl LysM-Cre mice were born at the expected Mendelian frequency and displayed no discernible morphological abnormalities at birth, they developed progressive abdominal distention as they matured (Fig. 2A). Necropsy at different ages showed that PIKfyvefl/fl LysM-Cre mice developed enlargement of their livers and spleens compared to those of their WT LysM-Cre littermates (Fig. 2B). Histological analysis of these organs revealed tissue accumulation of highly vacuolated macrophages (Fig. 2C). Immunophenotyping analysis of circulating leukocytes from PIKfyvefl/fl LysM-Cre mice showed significantly increased numbers of neutrophils and monocytes (Fig. 2D).
FIG 2.
PIKfyvefl/fl LysM-Cre mice display increased infiltration of inflammatory cells in various tissues. (A) General appearance of mice at about 14 months of age. Note the characteristic abdominal distention in the PIKfyvefl/fl LysM-Cre mouse. (B) Representative images of the liver and spleen of mice at 14 months of age, illustrating the marked hepatosplenomegaly in the PIKfyvefl/fl LysM-Cre mouse. (C) Representative images of tissue sections of the liver and spleen stained with hematoxylin and eosin. Note the tissue accumulation of engorged cells with translucent cytoplasmic vacuoles in the PIKfyvefl/fl LysM-Cre mouse. Scale bar: 100 μM. (D) Flow cytometry analysis of the numbers of B lymphocytes, T lymphocytes, monocytes, and neutrophils in the peripheral blood of mice at age of 4 to 20 weeks of age (n = 7 for WT LysM-Cre and n = 11 for PIKfyvefl/fl LysM-Cre). (E) Flow cytometry analysis of isolated leukocytes from the spleens and livers of WT LysM-Cre and PIKfyvefl/fl LysM-Cre mice. Left to right, general flow gating scheme (singlets, forward scatter [FSA]/side scatter [SSA], live) for all tissues and CD45 gating for spleen and liver off all live cells. (F) Flow cytometry gating for neutrophils (CD11b+ Ly6G+) for the spleen and liver along with cell number per tissue. (G) Flow cytometry gating for monocytes/macrophages first with CD64+, followed by monocytes (CD11b+ Ly6C+) and resident macrophages (CD11b+ Ly6C−) for the spleen and liver. (H) Flow cytometry gating for B lymphocytes (B220) and T lymphocytes (CD3). Statistical analysis was performed using unpaired two-tailed Student’s t test (NS, not significant [P > 0.05]; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). All error bars indicate SEM.
Given the elevated inflammatory myeloid cells in the peripheral blood of PIKfyvefl/fl LysM-Cre mice, we hypothesized that their recruitment to the liver and spleen was contributing to the aberrant increase of their liver and spleen. To test this hypothesis, we isolated single-cell suspensions from the livers and spleens of WT LysM-Cre and PIKfyvefl/fl LysM-Cre mice. Cells were then stained with fluorescent antibodies against various immune cell markers and analyzed by flow cytometry. Upon gating for singlets, size, and viability, we identified lineage-positive leukocytes in the liver and spleen by gating for CD45+ cells (Fig. 2E). We identified neutrophil infiltration from the tissues by gating for CD45+ CD11b+ Ly6G+ populations. Our results showed a significant increase of neutrophil infiltration in PIKfyvefl/fl LysM-Cre mice in the spleen and liver compared to that in WT LysM-Cre mice (Fig. 2F). To examine macrophage/monocyte populations, we first gated for cells positive for macrophage-specific Fc receptor CD64 expression and then examined CD11b+ and Ly6C+ populations within the CD64+gate. In the spleen, we noticed that the resident macrophage populations, defined by CD11bintermediate cells, Ly6Cnegative cells, and autofluorescence (29, 30), were increased in PIKfyvefl/fl LysM-Cre mice compared with WT LysM-Cre mice (Fig. 2G). The inflammatory/recruited monocytes, defined by CD11bhigh Ly6Chigh populations, were upregulated significantly in PIKfyvefl/fl LysM-Cre mice as well (Fig. 2G). We noticed a similar trend in the liver, where resident macrophages (i.e., Kupffer cells), defined by CD64+ CD11bintermediate Ly6Cnegative cells (31), along with the inflammatory monocyte populations defined by CD64+ CD11b+ Ly6C+ cells (32), were both significantly upregulated in PIKfyvefl/fl LysM-Cre mice (Fig. 2G). Lastly, we analyzed the lymphocyte populations by examining B220 and CD3+ populations. In the spleen, T cell numbers were similar, whereas B cell numbers were slightly increased in PIKfyvefl/fl LysM-Cre mice (Fig. 2H). In the liver, both B cell and T cell populations were increased in PIKfyvefl/fl LysM-Cre mice (Fig. 2H). These findings demonstrate that the enlargement of the spleens and livers in PIKfyvefl/fl LysM-Cre mice is due to significant influx of inflammatory cells, of both myeloid and lymphoid lineages.
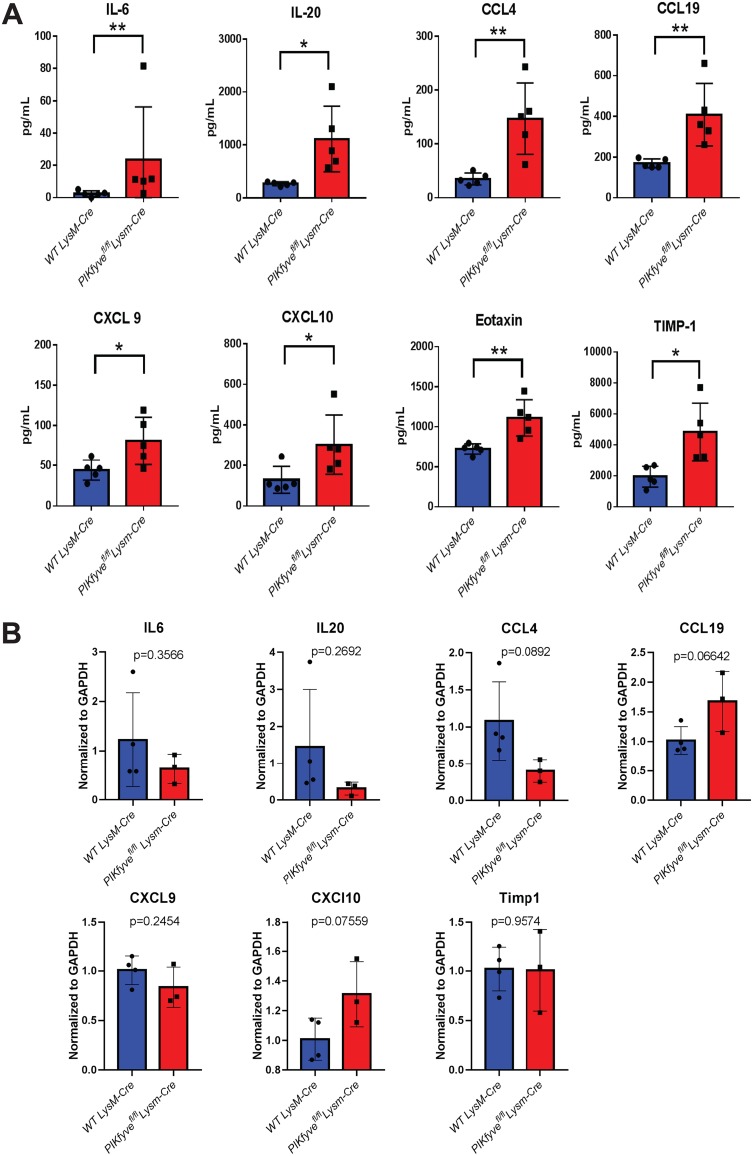
PIKfyve ablation in myeloid cells promotes systemic inflammation.
We next analyzed plasma samples for cytokine profiling by multiplex assay. We found elevated levels of several proinflammatory and chemotactic cytokines, including interleukin 6 (IL-6), IL-20, CCL-4, CCL-19, CXCL-9, CXCL-10, eotaxin, and TIMP-1, in PIKfyvefl/fl LysM-Cre mice compared to WT LysM-Cre littermates (Fig. 3A). To further explore the cellular source of inflammation, we analyzed spleen macrophages for mRNA expression of proinflammatory genes. Interestingly, the inflammatory genes were not elevated in PIKfyve-null macrophages compared with WT macrophages (Fig. 3B). These findings suggest that other inflammatory myeloid cells such as granulocytes might be responsible for the proinflammatory cytokine expression and release. Together, these findings demonstrate that PIKfyve deficiency in myeloid cells results in systemic inflammation.
FIG 3.
PIKfyvefl/fl LysM-Cre mice develop systemic inflammation. (A) Cytokine analysis by multiplex array of plasma samples of mice at 8 to 16 weeks of age (n = 5 per group). (B) Gene expression analysis of inflammatory cytokines by qRT-PCR (n = 3 per group). Statistical analysis was performed using unpaired two-tailed Student’s t test (*, P < 0.05; **, P < 0.01). All error bars indicate SEM.
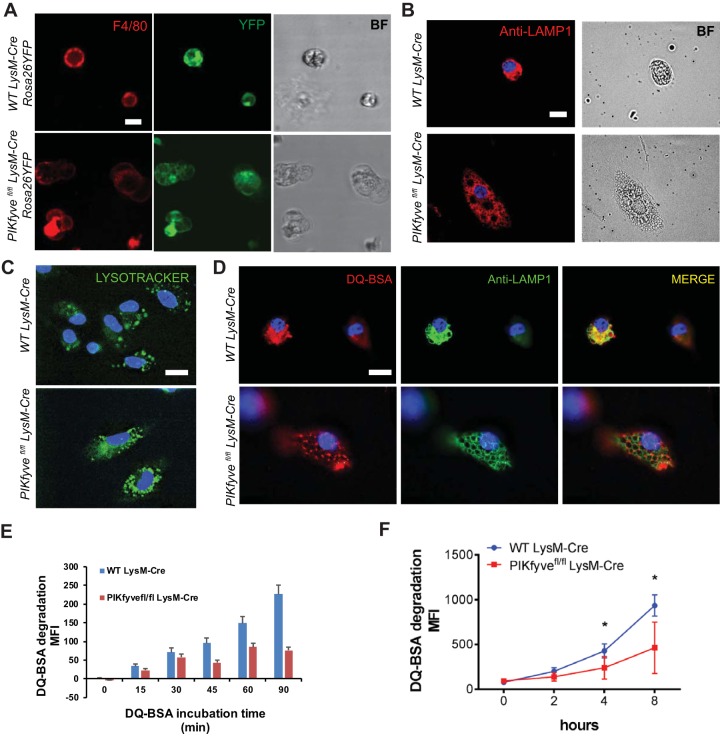
PIKfyve regulates lysosomal structure and proteolytic function in macrophages.
To investigate the effect of PIKfyve deficiency in macrophages, we first examined the morphology of macrophages isolated from the bone marrow or spleens of PIKfyvefl/fl LysM-Cre Rosa26YFP mice. We confirmed that macrophages isolated using F4/80 antibody were YFP+, indicating LysM-Cre expression in F4/80+ macrophages (Fig. 4A). As expected, PIKfyve-null macrophages displayed cytoplasmic vacuolation (Fig. 4A), which was similar to the vacuolation that has been previously reported for other PIKfyve-null cells (19, 20, 22). The enlarged cytoplasmic vacuoles in PIKfyve-null macrophages expressed LAMP1, which is a marker of late endosomes or lysosomes (Fig. 4B). Together, these data confirm that PIKfyve is necessary to maintain the endolysosomal morphology.
FIG 4.
PIKfyve modulates lysosomal morphology and degradative function in macrophages. (A) Live-cell fluorescence microscopy images of spleen macrophages isolated and stained with F4/80 antibody. The intrinsic YFP expression is driven by LysM-Cre. Note the presence of multiple cytoplasmic vacuoles of various sizes in the macrophages of a PIKfyvefl/fl LysM-Cre Rosa26YFP mouse. BF, bright-field. (B) Immunofluorescence images of bone marrow-derived macrophages stained with anti-LAMP1 antibody. (C) Images of bone marrow-derived macrophages incubated with LysoTracker for 20 min. Note the accumulation of LysoTracker in the acidic endolysosomes. (D) Immunofluorescence images of bone marrow-derived macrophages incubated with DQ BSA for 60 min and costained with anti-LAMP1 antibody. (E) Quantification of DQ BSA degradation over 90 min visualized by live microscopy. MFI, mean fluorescence intensity. (F) Proteolytic DQ BSA degradation over 8 h in the F4/80+ spleen macrophages as measured by increasing fluorescence of quenched dye on a spectrophotometer. Statistical analysis was performed using unpaired two-tailed Student’s t test (*, P < 0.05). All error bars indicate SEM. Scale bars: 10 μm.
As a low pH is necessary for normal proteolytic function of lysosomal enzymes, we further analyzed whether the absence of lysosomal proteolysis within PIKfyve-null macrophages was due to a necessary role for PIKfyve in lysosomal acidification. This process was analyzed using LysoTracker, a fluorescent dye that accumulates in acidic compartments such as lysosomes. We found that LysoTracker accumulated in the enlarged cytoplasmic vacuoles in the macrophages of PIKfyvefl/fl LysM-Cre mice (Fig. 4C). This finding is consistent with a previous report by Ho et al. demonstrating that PIKfyve is not necessary to maintain the pH of lysosomes (33). Thus, our result confirms that PIKfyve is not required for the acidification of endolysosomal compartments.
We next investigated whether PIKfyve was essential for the degradative function of macrophage lysosomes. Lysosomal proteolytic degradation was determined using self-quenched DQ bovine serum albumin (BSA), which is a protease substrate that is taken up by endocytosis and emits fluorescence upon proteolytic degradation within acidic compartments such as late endosomes and lysosomes. Proteolysis of DQ BSA was detected by two independent techniques. First, to determine the effect of PIKfyve on lysosomal proteolytic degradation in live cells, we analyzed the cellular localization of DQ BSA degradation. As anticipated, WT macrophages displayed a robust ability to catabolize DQ BSA within the LAMP1-demarcated compartments. In contrast, PIKfyve-null macrophages showed undetectable proteolytic degradation of DQ BSA within their enlarged LAMP1-positive late endosomes and lysosomes (Fig. 4D and E). For the second method, we analyzed the proteolytic degradation of DQ BSA in the lysates of macrophages via spectrophotometry. PIKfyve-null macrophages had significantly impaired ability to proteolytically degrade DQ BSA (Fig. 4F).
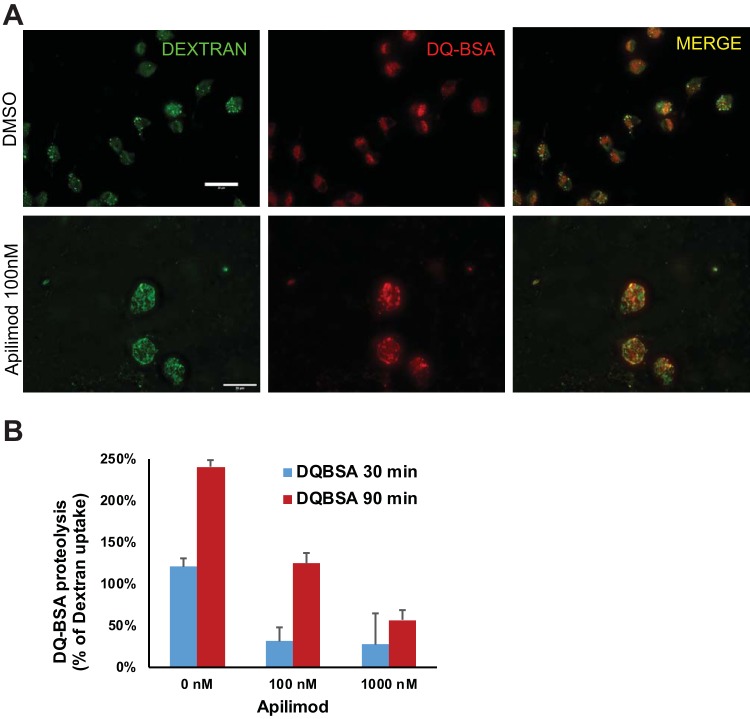
We also examined the effect of acute PIKfyve inhibition on lysosomal proteolytic degradation. Bone marrow-derived macrophages (BMDM) were treated for 2 h with apilimod at 100 nM or 1 μM. Cells were then analyzed for endocytosis of a fluid-phase marker dextran and for proteolysis of DQ BSA over 30 or 90 min. Compared with dimethyl sulfoxide (DMSO)-treated macrophages, apilimod-treated macrophages had a reduced DQ BSA degradation when normalized to dextran uptake (Fig. 5). Together, these results demonstrate the critical role of PIKfyve in the ability of macrophages to degrade proteins within their lysosomes.
FIG 5.
Acute inhibition of PIKfyve by apilimod alters lysosomal morphology and reduces lysosomal proteolysis in macrophages. (A) Fluorescence microscopy images of bone marrow-derived macrophages treated with apilimod (100 nM) for 2 h and incubated with dextran (green) and DQ BSA (red) over 90 min. (B) Measurement of lysosomal proteolysis normalized to fluid-phase dextran uptake in panel A (n = 3). Statistical analysis was performed using unpaired two-tailed Student’s t test. All error bars indicate SEM. Scale bars: 20 μm.
PIKfyve modulates lysosomal protein abundance independently of transcription.
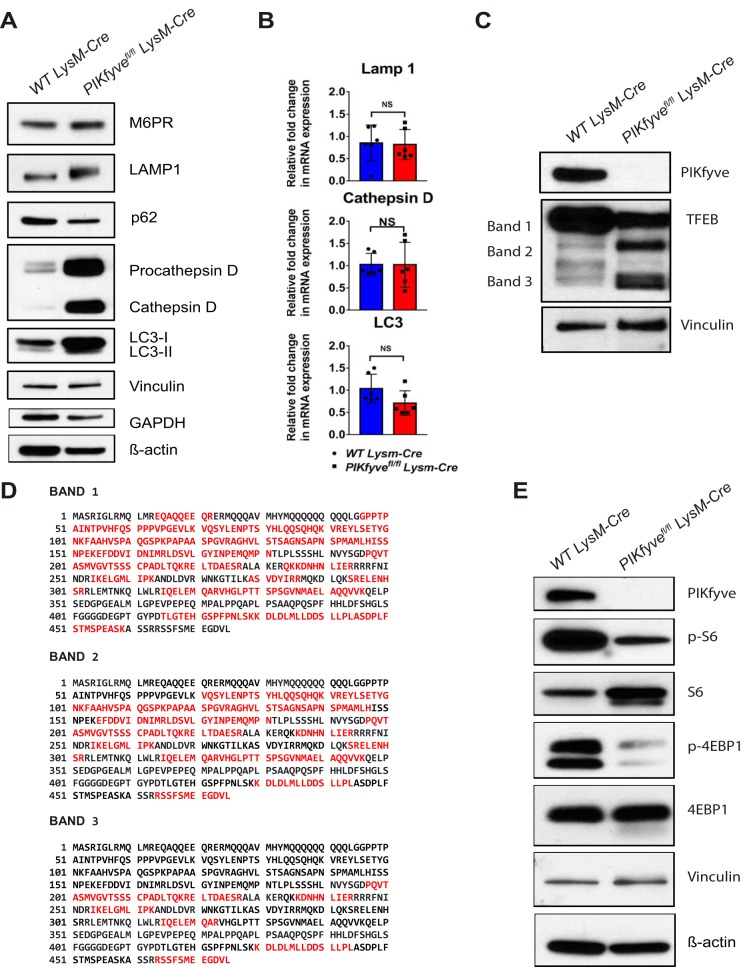
Given the importance of PIKfyve in lysosomal morphology and function, we next investigated the role of PIKfyve in lysosomal biogenesis. First, we examined the abundance of lysosomal and autophagy-related proteins in the F4/80+ macrophages from WT LysM-Cre mice and PIKfyvefl/fl LysM-Cre mice by immunoblotting analysis. Compared to WT macrophages, PIKfyve-null macrophages showed increased levels of lysosomal proteins such as LAMP1, procathepsin D, and cathepsin D (Fig. 6A). In addition, PIKfyve-null macrophages had significant reduction in the ratio of LC3-II to LC3-I (Fig. 6A), which suggests a defective basal autophagy. Lysosomal function and biogenesis are closely regulated by the transcription factor TFEB, which is often referred to as the “master regulator” of the lysosomal gene network (34). Based on this premise, we hypothesized that the elevated lysosomal proteins in PIKfyve-null macrophages is secondary to activation of TFEB promoting transcriptional upregulation of lysosomal genes. In contrast to our prediction, the quantity of expressed mRNA for LAMP1 or cathepsin D was not significantly higher in the PIKfyve-null macrophages than in the WT macrophages by qRT-PCR analysis (Fig. 6B). Intriguingly, we found that PIKfyve-null macrophages had reduced levels of full-length TFEB of about 60 kDa (band 1 in Fig. 6C) but increased amounts of the shorter variants of the TFEB protein that were between 40 and 50 kDa (bands 2 and 3 in Fig. 6C). Further analysis of the different forms of TFEB by mass spectrometry confirmed that these variants were N-terminally truncated variants of TFEB (Fig. 6D). Although the presence and significance of TFEB truncation variants have not been previously reported, we propose that these truncated variants are likely inactive forms of TFEB, since their presence was not associated with upregulated lysosomal gene expression. Together, these findings show that PIKfyve ablation leads to TFEB degradation, which might be an uncovered mechanism of downregulating TFEB activity.
FIG 6.
PIKfyve modulates lysosomal protein expression independently of transcription. (A) Immunoblot analysis of F4/80+ spleen macrophages for lysosomal and autophagy-related proteins: M6PR, LAMP1, p62, procathepsin D, cathepsin D, LC3-I, and LC3-II. Vinculin was used as a loading control. (B) qRT-PCR analysis of cathepsin D, LAMP1, and LC3 relative to GAPDH in F4/80+ spleen macrophages (n = 3 mice). (C) Immunoblot analysis of F4/80+ spleen macrophages probed with TFEB antibody. TFEB protein bands 1, 2, and 3 were cut for mass spectrometry analysis. (D) Proteomic analysis of TFEB protein bands 1, 2, and 3 in panel C. In red are the tryptic peptides identified in each band by mass spectrometry analysis. The sequence with UniProt number Q3UKG7 was used for the amino acid sequence of murine TFEB. (E) Immunoblot analysis of the F4/80+ spleen macrophages for PIKfyve, phospho-S6, S6, phospho-4EBP1, and 4EBP1. Probing for vinculin was used as the loading control. Statistical analysis was performed using unpaired two-tailed Student’s t test (NS, P > 0.05). All error bars indicate SEM.
Previous studies showed that the activity of TFEB is regulated primarily by mTORC1-mediated phosphorylation (35–37). Thus, we examined the effect of PIKfyve ablation in mTORC1 activation. This was done by analyzing the effect of the PIKfyve-null mutation on the mTORC1 substrates phospho-S6 and phospho-4EBP1. Interestingly, PIKfyve-null macrophages had decreased levels of phospho-S6 and phospho-4EBP1 compared to WT macrophages as determined by immunoblotting (Fig. 6E), which indicated reduced activity of mTORC1. Since this reduced activation of mTORC1 did not result in activation of TFEB and subsequent upregulation of lysosomal gene expression, our findings suggest that PIKfyve deficiency modulates the degradation of TFEB downstream of mTORC1 activation. Furthermore, our findings suggest that PIKfyve deficiency leads to the accumulation of lysosomal proteins, likely from reduced degradation of lysosomal proteins and not from TFEB-mediated transcriptional activation of lysosomal genes.
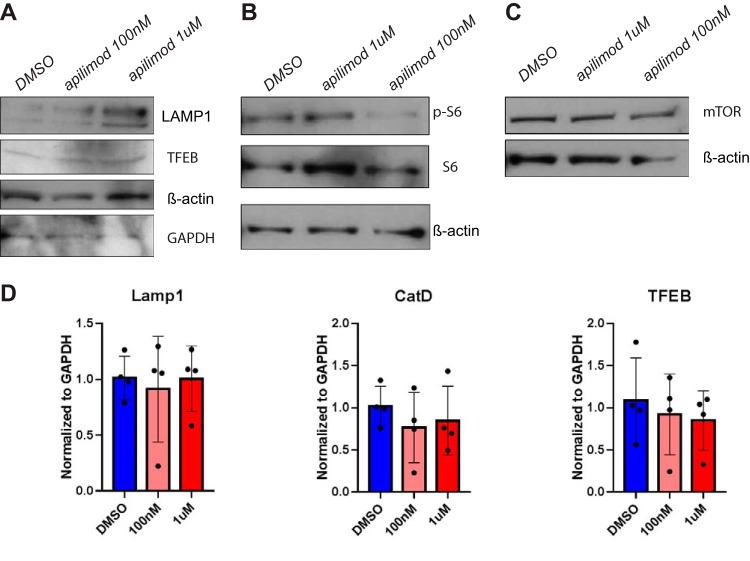
We also examined the effects of acute PIKfyve inhibition by apilimod on lysosomal biogenesis. Spleen macrophages were isolated from WT mice using F4/80 antibody by immunomagnetic separation, treated for 2 h with DMSO or with apilimod at 100 nM or 1 μM, and analyzed for the expression lysosomal genes by qRT-PCR as well as protein levels by immunoblot analyses. We found that the level of a lysosomal protein, i.e., LAMP1, was increased in apilimod-treated macrophages (Fig. 7A). However, mTORC1 activity and mTORC1 expression were not significantly changed in apilimod-treated macrophages compared with DMSO-treated macrophages (Fig. 7B and C). In addition, the mRNA levels of lysosomal proteins were comparable between DMSO-treated and apilimod-treated macrophages (Fig. 7D). These findings demonstrate that acute PIKfyve inhibition modifies the levels of lysosomal proteins independently of gene expression changes. The changes observed after brief PIKfyve inhibition by apilimod are similar to the changes seen with long-term disruption of PIKfyve in PIKfyve-null macrophages. Together, our data show that both acute and chronic inhibitions of PIKfyve do not alter the gene expression of lysosomal proteins but rather induce intracellular accumulation of lysosomal proteins, likely by reducing protein turnover.
FIG 7.
Acute inhibition of PIKfyve by apilimod increases lysosomal protein abundance independently of transcription in macrophages. (A to C) Immunoblot analysis of F4/80+ spleen macrophages treated for 2 h with DMSO, apilimod at 100 nM and 1 μM for LAMP1 and TFEB (A), phosph-S6 and S6 (B), and mTOR (C). β-Actin and GAPDH were used as loading controls. (D) qRT-PCR analysis of LAMP1, cathepsin D, and TFEB relative to GAPDH in F4/80+ spleen macrophages treated for 2 h with DMSO or apilimod at 100 nM or 1 μM. Statistical analysis was performed using unpaired two-tailed Student’s t test. All error bars indicate SEM.
DISCUSSION
In this study, using mice genetically engineered to lack PIKfyve in their myeloid cells, we found that PIKfyve is an essential regulator of the lysosomal morphology, degradative activity, and protein abundance in macrophages. Our findings are consistent with the phenotypes described in previous reports of using apilimod to inihibit PIKfyve (27, 28) as well as studies that analyzed genetic loss of function mutations in embryonic stem (ES) cells (18, 20). However, these findings differ from the phenotypes reported in a previous study of myeloid-cell-specific PIKfyve knockout mice, which showed that PIKfyve was dispensable for most tissue-resident macrophages except for alveolar macrophages in the lung (25). There are several explanations for why we (and others) find PIKfyve so essential for macrophage biology while other investigators did not. (i) Our mice were generated in a pure C57BL/6 background, whereas Kawasaki et al. (25) generated their mice by injection of ES cells from JM8/A3 strain into C57BL/6 background mice. (ii) While Kawasaki et al. targeted exon 5 of the PIKfyve gene, we targeted exons 37 and 38 of the PIKfyve gene. It is possible that our targeting approach, which removes critical components of the kinase domain, results in a truncated form that functions as a dominant negative gene. However, the absence of a phenotype in the heterozygous PIKfyvefl/+ LysM-Cre mice argues against this possibility. (iii) It is possible that the targeting strategy utilized by Kawasaki et al., which removes the start codon, still permits expression from a cryptic start site that generates a small amount of truncated protein that is catalytically active and produces a hypomorph. Such a residual minor amount of PIKfyve protein might be difficult to detect but still could be sufficient to generate a small amount of phosphatidylinositol-3,5-bisphosphate that attenuates the macrophage phenotype. (iv) Differences in the mouse housing environments used by Kawasaki et al. and our group may result in phenotypic plasticity.
With the exception of the above-referenced publication, studies using genetic or pharmacological ablation of PIKfyve in mammals, or of its orthologue fab1p in Saccharomyces cerevisiae, consistently demonstrate the development of enlarged endolysosomal compartments (38–41). Interestingly, fab1-null yeasts have impaired acidification of their enlarged vacuoles (38), whereas our PIKfyve-impaired macrophages have preserved lysosomal acidification. Consistent with prior studies using PIKfyve inhibitors (27, 33), we found that mammalian PIKfyve is dispensable for the regulation of lysosomal pH, since the enlarged late endosomes and lysosomes maintained the acidification in the PIKfyve-null macrophages. Additionally, our data showing the absence of DQ BSA cleavage within the LAMP1-demarcated compartments demonstrate that despite normal acidification, the enlarged late endosomes and lysosomes in PIKfyve-null macrophages have impaired proteolytic activity. Curiously, proteolysis still occurred in some compartments that were not demarcated by LAMP1. This suggests that DQ BSA is internalized and proteolyzed, perhaps in earlier endosomal compartments, but it cannot be degraded in the downstream compartments, such as in the late endosomes and lysosomes.
The defective degradation within the lysosomes could be explained by the following. First, proteins that are targeted for degradation could require PIKfyve in order for them to be transported to the late endosomes or lysosomes. Second, PIKfyve could be necessary for the trafficking of critical degradative proteases to late endosomes or lysosomes. Lastly, PIKfyve could be required for an essential step in the posttranslational processing of lysosomal degradative proteases that is required for their enzymatic activity. Consistent with several observations in models of PI(3,5)P2 deficiency (20, 22, 42), PIKfyve-null macrophages have disproportionally increased levels of procathepsin D, which is an unprocessed form of cathepsin D. These findings suggest that PIKfyve is critical for the processing and maturation of lysosomal proteins.
Our study shows that defective lysosomal degradative function is associated with increased levels of lysosomal proteins in PIKfyve-deficient macrophages. Because TFEB activity and lysosomal gene expression are expected to increase as a compensatory mechanism of lysosomal dysfunction, we initially hypothesized that loss of PIKfyve would activate TFEB and consequently increase transcriptional expression of lysosomal genes. In contrast to this hypothesis, PIKfyve-null macrophages expressed truncated forms of TFEB and showed unchanged levels of lysosomal gene expression. Although the significance of TFEB truncation is unknown, given the association of TFEB truncation with the absence of changes in the gene expression of lysosomal proteins, we speculate that truncation of TFEB might be a mechanism of downregulating TFEB activity. Because PIKfyve deficiency also results in excessive accumulation of lysosomal proteins despite unchanged gene expression, we hypothesize that this is likely due to reduced protein turnover and defective autophagy, which may paradoxically trigger truncation of TFEB downregulating TFEB activity. Together, our findings suggest that PIKfyve deficiency does not directly modulate lysosomal gene expression but instead is important for the turnover of lysosomal proteins. However, the exact mechanism of PIKfyve modulating lysosomal protein turnover still remains to be elucidated.
To further delineate the direct versus indirect effects mediated by PIKfyve inhibition, we compared the effects of acute PIKfyve inhibition on lysosomal function and biogenesis with those of chronic PIKfyve deficiency. Consistent with our findings in PIKfyve-null macrophages, we observed that acute inhibition of PIKfyve using apilimod resulted in reduced lysosomal proteolysis and increased lysosomal protein levels without altering TFEB expression or lysosomal gene expression. Consistent with previous studies (43–45), we observed that acute inhibition of PIKfyve with apilimod did not significantly modulate mTORC1 expression or activity. However, PIKfyve-null macrophages showed reduced mTORC1 activity, suggesting that this is an indirect effect of prolonged PIKfyve deficiency.
We and others show that abnormalities in lysosomal biogenesis and functions are linked to aberrant inflammatory responses in PIKfyve-deficient mice (19, 22, 25). These findings are not surprising since abnormal lysosomal functions and lysosomal storage disorders have long been known to associate with local and systemic inflammation due to activation of several inflammatory pathways (46, 47). Although the exact mechanisms are unknown, lyososomal dysfunction and defective autophagy have been shown to mediate several inflammatory changes (48). Thus, it is conceivable that PIKfyve deficiency induces aberrant inflammatory responses through their defective lysosomal function and autophagy.
In conclusion, our study shows that PIKfyve is essential to maintain lysosomal homeostasis in macrophages by regulating the morphology, trafficking, degradative function, and protein expression in the lysosome.
MATERIALS AND METHODS
Mice.
Mice expressing PIKfyve floxed alleles (PIKfyvefl) with exons 37 (AATTTTCTGATGCTAATGCCAAGTTTTACTGTCGGCTGTACTACGCGGGAGAGTTCCACAAGATGCGTGAAGTGATTCTGGGCAGCAGTGAGGAGGAATTCATCCGTTCCCTTTCTCACTCATCTCCCTGGCAGGCCCGGGGAGGCAAGTCAGGAGCTGCTTTCTATGCCACCGAAG) and 38 (ATGATAGATTCATTCTGAAGCAAATGCCTCGTTTGGAAGTCCAGTCTTTCC TTGACTTTGCACCACACTACTTCAATTATATCACAAATGCTGTTCAACA) of the PIKfyve gene flanked by loxP sites were generated as previously described (22). To generate myeloid-cell-specific PIKfyve-deficient mice, PIKfyvefl mice were crossed to mice expressing Cre recombinase under the control of the endogenous lysozyme 2 promoter (LysM-Cre) (B6.129P2-Lyz2tm1(cre)Ifo/J; stock number 004781; Jackson Laboratory). The resulting PIKfyvefl/fl LysM-Cre mice were crossed to a Cre reporter strain that expresses enhanced YFP (EYFP) upon cre-mediated recombination (B6.Cg-Gt(ROSA)26Sortm3(CAG-EYFP)Hze/J; stock number 007903; Jackson Laboratory). For all studies, both female and male mice were used. All mice were maintained on standard chow and tap water under pathogen-free conditions. All animal procedures were approved by and performed in accordance with the Institutional Animal Care and Use Committee at the University of Pennsylvania.
PCR genotyping.
Genomic DNA was isolated from mouse tail biopsy specimens for PCR genotyping. Genotyping for PIKfyvefl was performed as previously described (22). Briefly, loxP integration in intron 36 was identified with 5′-CCATTGCCTGGCTTAGAACAGAG-3′ and 5′-GAACTCTCCCGCGTAGTACAGC-3′ primers. LysM-Cre was identified with mutant primer 5′-CCC AGA AAT GCC AGA TTA CG-3′, common primer 5′-CTT GGG CTG CCA GAA TTT CTC-3′, and WT primer 5′-TTA CAG TCG GCC AGG CTG AC-3′. The Rosa26/EYFP transgene was identified with WT forward primer 5′-AAG GGA GCT GCA GTG GAG TA-3′, WT reverse primer 5′- CCG AAA ATC TGT GGG AAG TC-3′, mutant forward primer 5′-ACA TGG TCC TGC TGG AGT TC-3′, and mutant reverse primer 5′-GGC ATT AAA GCA GCG TAT CC-3′.
Histochemistry.
Tissues were harvested, fixed overnight in 10% formalin, paraffin embedded, and sectioned. Paraffin sections were deparaffinized and stained with hematoxylin and eosin.
Whole-blood white blood cell (WBC) analysis.
Whole blood (50 μl) was obtained by retro-orbital bleed, and red blood cells (RBCs) were lysed in ACK lysing buffer (Lonza). The remaining cells were stained with LIVE DEAD aqua (Life Technologies) and incubated with cell culture supernatants from anti-CD16/32 (clone 24G2)-expressing cells to block nonspecific binding to the Fc receptor. Cells were subsequently stained with fluorophore-labeled monoclonal antibodies (MAb) to the following antigens: CD3 (clone 145-2C11), CD19 (clone 6D5), CD115 (clone AFS98), CCR2 (clone 475301), Ly6C (clone HK1.4), and Ly6G (clone 1A8), which were obtained from BD Pharmingen, Biolegend, or R&D Systems. Samples were analyzed on a MacsQuant flow cytometer (Miltenyi), and flow cytometric analysis was performed using FlowJo software (Tree Star).
Immunomagnetic isolation of macrophages from the spleen or bone marrow.
Macrophages from tissues were magnetically enriched using protocols provided by Stemcell Technologies. Bone marrow was flushed from mouse femurs, aspirated with a syringe, and filtered with a cell strainer to derive a single-cell suspension. Spleens were dissected from the abdominal cavities of mice and filtered through a 50-μm nylon strainer to make single-cell suspensions of splenocytes. Red blood cells were lysed with ACK buffer. Next, the cells were blocked with FcR block (Miltenyi Biotec; 130-092-575), incubated with anti-F4/80 antibodies conjugated with fluorescein isothiocyanate (FITC) or phycoerythrin (PE) (Miltenyi Biotec; 130-102-988 and 130-102-943), and incubated in an EasySep FITC or PE positive-selection kit which labels FITC or PE epitopes with magnetic beads (Stemcell Technologies; 18557 and 18555). Each step was followed by a wash and spin. The prepared cells (in 15-ml centrifuge tubes) were placed in EasySep magnets (Stemcell Technologies; 18001) which retained labeled cells during washes. After several washes to remove unlabeled cells, the tubes were removed from the magnets and the isolated cells were collected.
Flow cytometry analysis of immune cells in the liver and spleen.
Spleens from WT and PIKfyvefl/fl LysM-Cre mice were ground and filtered through 70-μm nylon mesh, and RBCs were lysed in ACK lysing buffer (Lonza). Mouse livers were first minced with scissors and then digested in Hanks’ balanced salt solution (HBSS) containing 200 U/ml of collagenase I (Sigma/Aldrich) for 45 min at 37°C. The liver digests were then filtered through 70-μm nylon mesh and treated with ACK buffer for RBC lysis. Single-cell suspensions from spleen and liver were resuspended in HBSS plus 3% fetal bovine serum (FBS) plus Fc blocking antibodies for 30 min on ice prior to staining. For immune cell staining, anti-CD45 (biotin plus allophycocyanin [APC] S/A), anti-Ly6G (FITC), anti-CD11b (PE-Cy7), anti-CD64 (PE), anti-Ly6C (Alexa Fluor 488), anti-CD3 (FITC), and anti-B220 (PE) were used (Biolegend), with 4′,6-diamidino-2-phenylindole (DAPI) used as a viability dye. Cells were stained for 2 to 3 h on ice and analyzed using LSR-Fortessa (BD Biosciences). Fluorescence-activated cell sorting (FACS) plotting, gating, and quantification were performed using FlowJo (Tree Star). The specific gating for each population is as follows: neutrophil (CD45+ CD11b+ Ly6G+), monocytes (CD45+ CD64+ Ly6C+ CD11b+), macrophages (resident) (CD45+ CD64+ Ly6C− and CD11bintermediate), B cells (CD45+ B220+), and T cells (CD45+ CD3+). Inflammatory cells were enumerated by multiplying percent of event within specific gates of total processed events by total cells within the tissues.
Macrophage culture.
To generate bone marrow-derived macrophages (BMDM), bone marrow cells were extracted from femurs and tibias of mice at 8 to 12 weeks of age and cultured in Dulbecco modified Eagle medium (DMEM)–F-12 (Thermo Fisher). The cells were supplemented with 10% FBS, 1% penicillin and streptomycin, and recombinant mouse macrophage colony-stimulating factor (M-CSF; Calbiochem) at a final concentration of 10 ng/ml for 7 days. Supernatant cells were discarded, and BMDM were harvested from dishes by adding Accutase (Sigma-Aldrich; A6964) and washing with DMEM.
Apilimod treatment of cells.
Macrophages were treated with apilimod (Axon Mechem; number 1369) at the desired concentrations and times. Cells were then used for coendocytosis of dextran and DQ BSA, immunoblotting, and qRT-PCR.
Real-time quantitative PCR.
Total RNA was isolated from macrophages using an Illustra RNA spin minikit (GE Healthcare). cDNA was made with a high-capacity cDNA kit (Thermo Fisher Scientific). A total of 100 to 400 ng of cDNA was used for qPCR. Primers and probes were acquired from Thermo Fisher Scientific for the genes for GAPDH (Mm99999915_g1), PIKfyve (Mm00440793_m1), LC3 (Mm00458724_m1), cathepsin D (Mm00515586_m1), and LAMP1 (Mm00495262_m1). The primers used to analyze proinflammatory genes are listed in Table 1.
TABLE 1.
Primers used in qPCR to analyze proinflammatory genes
| Primer | Sequence |
|---|---|
| IL6 F | CTTCCATCCAGTTGCCTTCTTG |
| IL6 R | AATTAAGCCTCCGACTTGTGAAG |
| IL20 F | GGGAGACATCGATAGCCCTG |
| IL20 R | TCCAAAGACACACTATCCCGA |
| CCL4 F | CCTTGCTCCTCACGTTCAGA |
| CCL4 R | CATCTCCATGGGAGACACGC |
| CCL19 F | ATGTGAATCACTCTGGCCCAGGAA |
| CCL19 R | AAGCGGCTTTATTGGAAGCTCTGC |
| CXCL9 F | TAGGGACCACAGACTATTCCCC |
| CXCL9 R | GCCTGGTGTGTAACAGCCTT |
| CXCL10 F | CTTCTGAAAGGTGACCAGCC |
| CXCL10 R | GTCGCACCTCCACATAGCTT |
| TIMP1 F | GCAACTCGGACCTGGTCATAA |
| TIMP1 R | CGCTGGTATAAGGTGGTCTCG |
Samples were run on a Step One Plus qPCR instrument (Applied Biosystems) and analyzed using the threshold cycle (ΔΔCT) method to calculate fold change. All samples were first normalized to the GAPDH gene and then compared to WT controls.
Immunoblotting.
Tissues or cells were harvested and homogenized in radioimmunoprecipitation assay (RIPA) buffer that was supplemented with a protease inhibitor cocktail (Sigma-Aldrich) and a phosphatase inhibitor cocktail (Thermo Fisher Scientific). The protein concentrations were measured by bicinchoninic acid (BCA) protein assay (Thermo Fisher Scientific). The protein samples were analyzed by Novex NuPage SDS-PAGE gradient gels under reducing conditions (Invitrogen) and then transferred onto a polyvinylidene difluoride membrane (Invitrogen). The membrane was blotted with the indicated primary antibodies against PIKfyve (Sigma-Aldrich number P0054; 1:400), M6PR (Abcam number ab32815; 1:1,000), LAMP1 (Developmental Studies Hybridoma Bank clone 1D4B; 1:2,000), p62 (Cell Signaling Technology; 1:1,000), cathepsin D (Calbiochem; number IM16; 1:1,000), LC3 (Cell Signaling Technology; 1:1,000), TFEB (Bethyl; A303-673A; 1:2,000), phospho-S6 (Cell Signaling Technology; number 4858; 1:1,000), S6 (Cell Signaling Technology; number 2217; 1:1,000), phospho-4EBP1 (Cell Signaling Technology; number 2855; 1:1,000), 4EBP1 (Cell Signaling Technology; number 9644; 1:1,000), mTOR (Cell Signaling Technology; number 2983; 1:1,000), β-actin (Cell Signaling Technology; number 4970; 1:2,000), GAPDH (Cell Signaling Technology; number 8884; 1:10,000), and vinculin (Santa Cruz Biotechnology; number sc-7649; 1:1,000). The following horseradish peroxidase-conjugated secondary antibodies were used: anti-rabbit antibodies (GE Healthcare and Cell Signaling Technology; 1:3,000 and 1:1,000, respectively), anti-rat antibody (Santa Cruz Biotechnology; 1:5,000), anti-goat antibody (Santa Cruz Biotechnology; 1:3,000), and anti-mouse antibody (Santa Cruz Biotechnology; 1:3,000). Membranes were visualized with enhanced chemiluminescence substrate (GE Healthcare Life Sciences).
Mouse cytokine multiplex assay.
Plasma samples were obtained from mice and analyzed by Eve Technologies (Calgary, Canada) using the mouse cytokine array/chemokine array 44-plex according to the manufacturer’s instructions. The following biomarkers were analyzed: eotaxin, erythropoietin, 6Ckine, fractalkine, granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), interferon β1 (IFN-β1), IFN-γ, IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-9, IL-10, IL-11, IL-12 (p40), IL-12 (p70), IL-13, IL-15, IL-16, IL-17, IL-20, IP-10 (CXCL10), KC, LIF, LIX, MCP-1, MCP-5, M-CSF, MDC, MIG (CXCL9), MIP-1α (CCL3), MIP-1β (CCL4), MIP-2 (CXCL2), MIP-3α (CCL20), MIP-3B (CCL19), RANTES, TARC, TIMP-1, tumor necrosis factor alpha (TNF-α), and vascular endothelial growth factor (VEGF).
Immunofluorescence microscopy.
BMDM were grown on coverslips and fixed with cold methanol (MeOH)-acetone, permeabilized with PBT (phosphate-buffered saline [PBS] and Triton X-100 [0.1%]), and blocked with starting block buffer T20 (Thermo Fisher Scientific). Slides were probed with primary antibody LAMP1 (Development Study Hybridoma Bank) overnight at 4°C. Slides were imaged with Leica DM6000, and images were deconvolved with Leica LAS Autoquant software.
Live-cell imaging with LysoTracker, dextran, and DQ BSA.
Macrophages were grown overnight on glass chamber slides (Ibidi; number 80827) coated with 0.1% gelatin. For all live-stain solutions and washes, a 10% FBS–DMEM (without phenol red; Invitrogen) staining buffer was used. Macrophages were gently washed with staining buffer and labeled with one of the live stains LysoTracker green, dextran green (Thermo Fisher; P35368), and DQ red BSA (Thermo Fisher; D12051). To visualize lysosomes, LysoTracker green (100 μM) was applied to each well for 15 min. To measure proteolytic activity, DQ red BSA (10 μg/ml) was added to each well for different periods. For each stain, the wells were washed once with 10% trypan blue in staining buffer followed by a wash with staining buffer and then fixed with 4% paraformaldehyde. Images were acquired with Metamorph software on a spinning-disk confocal microscope (Nikon Eclipse Ti-U) at the University of Pennsylvania Molecular Pathology & Imaging Core Service. Images were acquired on a Leica DM6000 wide-field microscope with a Photometric HQ2 high-resolution monochrome charge-coupled-device (CCD) camera using LAS AF software. The images were taken at 13 levels at 0.5-μm steps. The images were deconvolved using the AutoQuant deconvolution package at 10 iterations. The equipment was maintained by the Cell and Developmental Biology core facility at the University of Pennsylvania. The image stacks were flattened by Z projection (max intensity), and mean fluorescence was quantified with FIJI software.
Proteolysis assay.
BMDM were seeded into a 96-well plate and incubated overnight at 37°C. Cells were then incubated with PBS or DQ red BSA (Thermo Fisher Scientific) at a final concentration of 10 μg/ml and incubated at 37°C for 0, 2, 4, and 8 h. Fluorescence of DQ red BSA was measured on a Molecular Devices spectrophotometer microplate reader at an excitation wavelength of 584 nm and an emission wavelength of 612 nm.
Immunoprecipitation of TFEB.
Isolated macrophages from the mouse spleen were lysed in RIPA (or in 1% NP-40 in PBS) lysis buffer containing a protease inhibitor cocktail (Roche) and sodium orthovanadate (2 mM). The lysates were precleared with unconjugated protein A-agarose beads (Invitrogen). The precleared lysates were subjected to immunoprecipitation using anti-TFEB antibody (Bethyl Laboratories; A303-673A) prebound to protein A-agarose beads. The immunoprecipitated sample was eluted by addition of Novex NuPAGE LDS sample buffer (Invitrogen) and Novex NuPAGE sample reducing agent (Invitrogen) and heating at 95°C for 10 min. The sample was spun down at 500 × g for 5 min, and the supernatant was run on 4% to 12% Novex NuPAGE gels (Invitrogen). The gels were Coomassie blue stained, and the lanes corresponding to TFEB bands were excised for digestion and mass spectrometry analysis.
In-gel digestion.
Each sample was excised from the gel and cut into 1-mm3 cubes (49). They were destained with 50% methanol–1.25% acetic acid, reduced with 5 mM dithiothreitol (Thermo Fisher Scientific), and alkylated with 40 mM iodoacetamide (Sigma-Aldrich). Gel pieces were then washed with 20 mM ammonium bicarbonate (Sigma-Aldrich) and dehydrated with acetonitrile (Thermo Fisher Scientific). Trypsin (Promega; 5 ng/ml in 20 mM ammonium bicarbonate) was added to the gel pieces, and proteolysis was allowed to proceed overnight at 37°C. Peptides were extracted with 0.3% trifluoroacetic acid (J. T. Baker) followed by 50% acetonitrile. Extracts were combined and the volume was reduced by vacuum centrifugation.
Mass spectrometry analysis.
Tryptic digests were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) on a QExactive HF mass spectrometer (Thermo Fisher Scientific) coupled with an Ultimate 3000. Peptides were separated by reverse-phase high-performance liquid chromatography (RP-HPLC) on a nanocapillary column (75 μm [inside diameter] by 25-cm 2-μm PepMap Acclaim column). Mobile phase A consisted of 0.1% formic acid (Thermo Fisher Scientific) and mobile phase B of 0.1% formic acid–acetonitrile. Peptides were eluted into the mass spectrometer at 300 nl/min, with each RP-LC run comprising a 90-min gradient from 10 to 25% mobile phase B in 65 min to 25 to 40% mobile phase B in 25 min. The mass spectrometer was set to repetitively scan m/z from 300 to 1,400 (R = 240,000), followed by data-dependent MS/MS scans on the 20 most abundant ions, minimum AGC 1e4, dynamic exclusion with a repeat count of 1, repeat duration of 30 s (R = 15,000), FTMS full scan AGC target value of 3e6, and MSn AGC of 1e5. MSn injection time was 160 ms; microscans were set at 1. Rejection of unassigned and 1+, 6 to 8 charge states was set.
Data processing for proteomics analysis.
Raw MS files were processed using MaxQuant, version 1.5.7.4, for identification of proteins (50). The peptide MS/MS spectra were searched against the UniProtKB/Swiss-Prot Mouse Reference Proteome database (proteome name, Mus musculus C57BL/6J, reference proteome; proteins, 49,838; proteome identifier [ID], UP000000589; strain, C57BL/6J; taxonomy, 10090 [Mus musculus]; last modified 9 July 2016; Genome assembly, GCA_000001635.6). Precursor ion tolerance was 4.5 ppm, with semitryptic specificity, and MS2 fragment ion tolerance was set to 20 ppm. Oxidation of methionine, acetylation of the protein N terminus, and conversion of glutamine to pyroglutamic acid were used as variable modifications, and carbamidomethylation of cysteine was set as a fixed modification. The minimal length required for a peptide was 7 amino acids. A target-decoy approach was used to control false-discovery rate (FDR). A maximum FDR of 1% at both the peptide and the protein levels was allowed. The MaxQuant match-between-runs (0.7 min) feature was enabled.
Statistical analysis.
Statistical analysis was performed using GraphPad Prism. Data were expressed as means ± standard errors of the means (SEM). Two-tailed Student’s t test was used for comparisons of two groups. A P value of less than 0.05 was considered statistically significant.
ACKNOWLEDGMENTS
This study was supported by the National Institutes of Health (NIH; PO1 HL120846, PO1 HL40387, and K08 HL119597) and the American Society of Hematology Scholar Award (S.H.M.).
S.H.M. designed, conducted, and analyzed experiments, cosupervised the project, and wrote the manuscript. A.S., L.W., and Y.C. conducted experiments and wrote the manuscript. J.G., H.J., F.G., and C.T. conducted some experiments. L.Z. analyzed the data. L.A.S. conducted some experiments. S.H.S. designed and analyzed some experiments. E.M.B. analyzed the data. C.S.A. analyzed the data, cosupervised the project, and wrote the manuscript. All the authors reviewed the manuscript.
We declare that we have no competing interests.
REFERENCES
- 1.Saftig P, Klumperman J. 2009. Lysosome biogenesis and lysosomal membrane proteins: trafficking meets function. Nat Rev Mol Cell Biol 10:623–635. doi: 10.1038/nrm2745. [DOI] [PubMed] [Google Scholar]
- 2.Luzio JP, Hackmann Y, Dieckmann NM, Griffiths GM. 2014. The biogenesis of lysosomes and lysosome-related organelles. Cold Spring Harb Perspect Biol 6:a016840. doi: 10.1101/cshperspect.a016840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Paolo G, De Camilli P. 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature 443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 4.Dove SK, Dong K, Kobayashi T, Williams FK, Michell RH. 2009. Phosphatidylinositol 3,5-bisphosphate and Fab1p/PIKfyve underPPIn endo-lysosome function. Biochem J 419:1–13. doi: 10.1042/BJ20081950. [DOI] [PubMed] [Google Scholar]
- 5.De Craene JO, Bertazzi DL, Bar S, Friant S. 2017. Phosphoinositides, major actors in membrane trafficking and lipid signaling pathways. Int J Mol Sci 18:E634. doi: 10.3390/ijms18030634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rutherford AC, Traer C, Wassmer T, Pattni K, Bujny MV, Carlton JG, Stenmark H, Cullen PJ. 2006. The mammalian phosphatidylinositol 3-phosphate 5-kinase (PIKfyve) regulates endosome-to-TGN retrograde transport. J Cell Sci 119:3944–3957. doi: 10.1242/jcs.03153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Lartigue J, Polson H, Feldman M, Shokat K, Tooze SA, Urbe S, Clague MJ. 2009. PIKfyve regulation of endosome-linked pathways. Traffic 10:883–893. doi: 10.1111/j.1600-0854.2009.00915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kerr MC, Wang JT, Castro NA, Hamilton NA, Town L, Brown DL, Meunier FA, Brown NF, Stow JL, Teasdale RD. 2010. Inhibition of the PtdIns(5) kinase PIKfyve disrupts intracellular replication of Salmonella. EMBO J 29:1331–1347. doi: 10.1038/emboj.2010.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferguson CJ, Lenk GM, Meisler MH. 2009. Defective autophagy in neurons and astrocytes from mice deficient in PI(3,5)P2. Hum Mol Genet 18:4868–4878. doi: 10.1093/hmg/ddp460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin S, Harper CB, May LM, Coulson EJ, Meunier FA, Osborne SL. 2013. Inhibition of PIKfyve by YM-201636 dysregulates autophagy and leads to apoptosis-independent neuronal cell death. PLoS One 8:e60152. doi: 10.1371/journal.pone.0060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sano O, Kazetani K, Funata M, Fukuda Y, Matsui J, Iwata H. 2016. Vacuolin-1 inhibits autophagy by impairing lysosomal maturation via PIKfyve inhibition. FEBS Lett 590:1576–1585. doi: 10.1002/1873-3468.12195. [DOI] [PubMed] [Google Scholar]
- 12.Sharma G, Guardia CM, Roy A, Vassilev A, Saric A, Griner LN, Marugan J, Ferrer M, Bonifacino JS, DePamphilis ML. 15 March 2019. A family of PIKFYVE inhibitors with therapeutic potential against autophagy-dependent cancer cells disrupt multiple events in lysosome homeostasis. Autophagy doi: 10.1080/15548627.2019.1586257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, Wang X, Zhang X, Zhao M, Tsang WL, Zhang Y, Yau RG, Weisman LS, Xu H. 2013. Genetically encoded fluorescent probe to visualize intracellular phosphatidylinositol 3,5-bisphosphate localization and dynamics. Proc Natl Acad Sci U S A 110:21165–21170. doi: 10.1073/pnas.1311864110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shisheva A, Rusin B, Ikonomov OC, DeMarco C, Sbrissa D. 2001. Localization and insulin-regulated relocation of phosphoinositide 5-kinase PIKfyve in 3T3-L1 adipocytes. J Biol Chem 276:11859–11869. doi: 10.1074/jbc.M008437200. [DOI] [PubMed] [Google Scholar]
- 15.Shisheva A, Sbrissa D, Ikonomov O. 1999. Cloning, characterization, and expression of a novel Zn2+-binding FYVE finger-containing phosphoinositide kinase in insulin-sensitive cells. Mol Cell Biol 19:623–634. doi: 10.1128/MCB.19.1.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shisheva A. 2012. PIKfyve and its lipid products in health and in sickness. Curr Top Microbiol Immunol 362:127–162. doi: 10.1007/978-94-007-5025-8_7. [DOI] [PubMed] [Google Scholar]
- 17.Hasegawa J, Strunk BS, Weisman LS. 2017. PI5P and PI(3,5)P2: minor, but essential phosphoinositides. Cell Struct Funct 42:49–60. doi: 10.1247/csf.17003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikonomov OC, Sbrissa D, Delvecchio K, Xie Y, Jin JP, Rappolee D, Shisheva A. 2011. The phosphoinositide kinase PIKfyve is vital in early embryonic development: preimplantation lethality of PIKfyve−/− embryos but normality of PIKfyve+/− mice. J Biol Chem 286:13404–13413. doi: 10.1074/jbc.M111.222364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takasuga S, Horie Y, Sasaki J, Sun-Wada GH, Kawamura N, Iizuka R, Mizuno K, Eguchi S, Kofuji S, Kimura H, Yamazaki M, Horie C, Odanaga E, Sato Y, Chida S, Kontani K, Harada A, Katada T, Suzuki A, Wada Y, Ohnishi H, Sasaki T. 2013. Critical roles of type III phosphatidylinositol phosphate kinase in murine embryonic visceral endoderm and adult intestine. Proc Natl Acad Sci U S A 110:1726–1731. doi: 10.1073/pnas.1213212110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zolov SN, Bridges D, Zhang Y, Lee WW, Riehle E, Verma R, Lenk GM, Converso-Baran K, Weide T, Albin RL, Saltiel AR, Meisler MH, Russell MW, Weisman LS. 2012. In vivo, Pikfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P. Proc Natl Acad Sci U S A 109:17472–17477. doi: 10.1073/pnas.1203106109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikonomov OC, Sbrissa D, Delvecchio K, Feng HZ, Cartee GD, Jin JP, Shisheva A. 2013. Muscle-specific Pikfyve gene disruption causes glucose intolerance, insulin resistance, adiposity, and hyperinsulinemia but not muscle fiber-type switching. Am J Physiol Endocrinol Metab 305:E119–E1131. doi: 10.1152/ajpendo.00030.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Min SH, Suzuki A, Stalker TJ, Zhao L, Wang Y, McKennan C, Riese MJ, Guzman JF, Zhang S, Lian L, Joshi R, Meng R, Seeholzer SH, Choi JK, Koretzky G, Marks MS, Abrams CS. 2014. Loss of PIKfyve in platelets causes a lysosomal disease leading to inflammation and thrombosis in mice. Nat Commun 5:4691. doi: 10.1038/ncomms5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mironova YA, Lenk GM, Lin JP, Lee SJ, Twiss JL, Vaccari I, Bolino A, Havton LA, Min SH, Abrams CS, Shrager P, Meisler MH, Giger RJ. 2016. PI(3,5)P2 biosynthesis regulates oligodendrocyte differentiation by intrinsic and extrinsic mechanisms. Elife 5:e13023. doi: 10.7554/eLife.13023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Venkatareddy M, Verma R, Kalinowski A, Patel SR, Shisheva A, Garg P. 2016. Distinct requirements for vacuolar protein sorting 34 downstream effector phosphatidylinositol 3-phosphate 5-kinase in podocytes versus proximal tubular cells. J Am Soc Nephrol 27:2702–2719. doi: 10.1681/ASN.2015050555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawasaki T, Ito K, Miyata H, Akira S, Kawai T. 2017. Deletion of PIKfyve alters alveolar macrophage populations and exacerbates allergic inflammation in mice. EMBO J 36:1707–1718. doi: 10.15252/embj.201695528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liggins MC, Flesher JL, Jahid S, Vasudeva P, Eby V, Takasuga S, Sasaki J, Sasaki T, Boissy RE, Ganesan AK. 2018. PIKfyve regulates melanosome biogenesis. PLoS Genet 14:e1007290. doi: 10.1371/journal.pgen.1007290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim GH, Dayam RM, Prashar A, Terebiznik M, Botelho RJ. 2014. PIKfyve inhibition interferes with phagosome and endosome maturation in macrophages. Traffic 15:1143–1163. doi: 10.1111/tra.12199. [DOI] [PubMed] [Google Scholar]
- 28.Dayam RM, Sun CX, Choy CH, Mancuso G, Glogauer M, Botelho RJ. 2017. The lipid kinase PIKfyve coordinates the neutrophil immune response through the activation of the Rac GTPase. J Immunol 199:2096–2105. doi: 10.4049/jimmunol.1601466. [DOI] [PubMed] [Google Scholar]
- 29.Gupta P, Lai SM, Sheng J, Tetlak P, Balachander A, Claser C, Renia L, Karjalainen K, Ruedl C. 2016. Tissue-resident CD169(+) macrophages form a crucial front line against Plasmodium infection. Cell Rep 16:1749–1761. doi: 10.1016/j.celrep.2016.07.010. [DOI] [PubMed] [Google Scholar]
- 30.Nagelkerke SQ, Bruggeman CW, den Haan JMM, Mul EPJ, van den Berg TK, van Bruggen R, Kuijpers TW. 2018. Red pulp macrophages in the human spleen are a distinct cell population with a unique expression of Fc-gamma receptors. Blood Adv 2:941–953. doi: 10.1182/bloodadvances.2017015008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lynch RW, Hawley CA, Pellicoro A, Bain CC, Iredale JP, Jenkins SJ. 2018. An efficient method to isolate Kupffer cells eliminating endothelial cell contamination and selective bias. J Leukoc Biol 104:579–586. doi: 10.1002/JLB.1TA0517-169R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rolot M, A MD, Javaux J, Lallemand F, Machiels B, Martinive P, Gillet L, Dewals BG. 2019. Recruitment of hepatic macrophages from monocytes is independent of IL-4Ralpha but is associated with ablation of resident macrophages in schistosomiasis. Eur J Immunol doi: 10.1002/eji.201847796. [DOI] [PubMed] [Google Scholar]
- 33.Ho CY, Choy CH, Wattson CA, Johnson DE, Botelho RJ. 2015. The Fab1/PIKfyve phosphoinositide phosphate kinase is not necessary to maintain the pH of lysosomes and of the yeast vacuole. J Biol Chem 290:9919–9928. doi: 10.1074/jbc.M114.613984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, Banfi S, Parenti G, Cattaneo E, Ballabio A. 2009. A gene network regulating lysosomal biogenesis and function. Science 325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 35.Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. 2012. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal 5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martina JA, Chen Y, Gucek M, Puertollano R. 2012. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8:903–914. doi: 10.4161/auto.19653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, Facchinetti V, Sabatini DM, Ballabio A. 2012. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 31:1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gary JD, Wurmser AE, Bonangelino CJ, Weisman LS, Emr SD. 1998. Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J Cell Biol 143:65–79. doi: 10.1083/jcb.143.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cooke FT, Dove SK, McEwen RK, Painter G, Holmes AB, Hall MN, Michell RH, Parker PJ. 1998. The stress-activated phosphatidylinositol 3-phosphate 5-kinase Fab1p is essential for vacuole function in S. cerevisiae. Curr Biol 8:1219–1222. doi: 10.1016/S0960-9822(07)00513-1. [DOI] [PubMed] [Google Scholar]
- 40.Yamamoto A, DeWald DB, Boronenkov IV, Anderson RA, Emr SD, Koshland D. 1995. Novel PI(4)P 5-kinase homologue, Fab1p, essential for normal vacuole function and morphology in yeast. Mol Biol Cell 6:525–539. doi: 10.1091/mbc.6.5.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ikonomov OC, Sbrissa D, Shisheva A. 2001. Mammalian cell morphology and endocytic membrane homeostasis require enzymatically active phosphoinositide 5-kinase PIKfyve. J Biol Chem 276:26141–26147. doi: 10.1074/jbc.M101722200. [DOI] [PubMed] [Google Scholar]
- 42.Gayle S, Landrette S, Beeharry N, Conrad C, Hernandez M, Beckett P, Ferguson SM, Mandelkern T, Zheng M, Xu T, Rothberg J, Lichenstein H. 2017. Identification of apilimod as a first-in-class PIKfyve kinase inhibitor for treatment of B-cell non-Hodgkin lymphoma. Blood 129:1768. doi: 10.1182/blood-2016-09-736892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang W, Gao Q, Yang M, Zhang X, Yu L, Lawas M, Li X, Bryant-Genevier M, Southall NT, Marugan J, Ferrer M, Xu H. 2015. Up-regulation of lysosomal TRPML1 channels is essential for lysosomal adaptation to nutrient starvation. Proc Natl Acad Sci U S A 112:E1373–81. doi: 10.1073/pnas.1419669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krishna S, Palm W, Lee Y, Yang W, Bandyopadhyay U, Xu H, Florey O, Thompson CB, Overholtzer M. 2016. PIKfyve regulates vacuole maturation and nutrient recovery following engulfment. Dev Cell 38:536–547. doi: 10.1016/j.devcel.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choy CH, Saffi G, Gray MA, Wallace C, Dayam RM, Ou ZA, Lenk G, Puertollano R, Watkins SC, Botelho RJ. 2018. Lysosome enlargement during inhibition of the lipid kinase PIKfyve proceeds through lysosome coalescence. J Cell Sci 131:jcs213587. doi: 10.1242/jcs.213587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fiorenza MT, Moro E, Erickson RP. 2018. The pathogenesis of lysosomal storage disorders: beyond the engorgement of lysosomes to abnormal development and neuroinflammation. Hum Mol Genet 27:R119–R129. doi: 10.1093/hmg/ddy155. [DOI] [PubMed] [Google Scholar]
- 47.Rigante D, Cipolla C, Basile U, Gulli F, Savastano MC. 2017. Overview of immune abnormalities in lysosomal storage disorders. Immunol Lett 188:79–85. doi: 10.1016/j.imlet.2017.07.004. [DOI] [PubMed] [Google Scholar]
- 48.Deretic V, Klionsky DJ. 2018. Autophagy and inflammation: a special review issue. Autophagy 14:179–180. doi: 10.1080/15548627.2017.1412229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shevchenko A, Wilm M, Vorm O, Mann M. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal Chem 68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 50.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]