ABSTRACT
Accumulating evidence has indicated that long noncoding RNAs (lncRNAs) play pivotal roles in the processes of cancer occurrence, progression, and treatment. FAM83A-AS1 is a novel onco-lncRNA involved in various cancers. Nevertheless, the biological function and underlying mechanism of FAM83A-AS1 in lung adenocarcinoma (LUAD) remain largely unclear. In this study, we found FAM83A-AS1 to be upregulated in LUAD tissues and closely associated with tumor size, lymph node metastasis, and TNM stage. In addition, high FAM83A-AS1 expression correlated positively with a poor prognosis. Functional investigation revealed that FAM83A-AS1 promotes LUAD cell proliferation, migration, invasion and the epithelial-mesenchymal transition (EMT) in vitro and tumor growth in vivo. Mechanistically, FAM83A-AS1 functions as an endogenous sponge of miR-150-5p by directly targeting it, removing inhibition of MMP14, a target of miR-150-5p. Furthermore, rescue assays demonstrated that FAM83A-AS1 enhances cell migration, invasion and EMT by modulating the miR-150-5p/MMP14 pathway. Collectively, we conclude that the novel FAM83A-AS1/miR-150-5p/MMP14 axis regulates LUAD progression, suggesting an innovative therapeutic strategy for this cancer.
KEYWORDS: FAM83A-AS1, miR-150-5p, MMP14, lung adenocarcinoma
1. Introduction
Lung cancer is the most common malignancy and the leading cause of global cancer mortality worldwide. Adenocarcinoma is the major histological subtype of lung cancer, accounting for approximately 60% of cases [1]. Although there has been great progress in chemotherapy, radiation and surgery, the survival time of lung cancer patients remains unsatisfactory. Indeed, fewer than 20% of patients survive more than 5 years. As high invasion and early metastasis of lung adenocarcinoma (LUAD) are the main causes contributing to poor prognoses [2], developing novel therapeutic targets is crucial for LUAD diagnosis and treatment.
Long noncoding RNAs (lncRNAs) are a class of transcripts longer than 200 nt with no protein-coding function [3]. Numerous studies have confirmed that lncRNAs are involved in the regulation of a variety of biological processes, including cell proliferation, apoptosis, metastasis and genomic stability. Dysregulated lncRNA expression has been reported to contribute to the development of diverse human malignancies, including breast cancer [4], hepatocellular carcinoma [5], colorectal cancer [6], and esophageal cancer [7]. Furthermore, an increasing number of lncRNAs have been found to participate in lung cancer regulation, yet novel lncRNAs with clinical diagnostic and basic research value in LUAD remain to be discovered. Nonetheless, accumulating evidence highlights the pivotal functions of lncRNAs as competing endogenous RNAs (ceRNAs) in the tumorigenesis and metastasis of LUAD. For instance, lncRNA DGCR5 promotes the tumor growth of LUAD by epigenetically inhibiting hsa-miR-22-3p [8], and lncRNA XIST acts as an effective sponge for let-7i, thereby increasing BAG-1 levels and promoting cisplatin resistance in LUAD cells [9]. Located at 8q24, the gene family with sequence similarity 83 member A antisense RNA 1 (FAM83A-AS1) produces a noncoding RNA that is a natural antisense transcript RNA. Based on TCGA data analysis, a recent study found that high FAM83A-AS1 expression was associated with poor prognosis and tumor metastasis in lung squamous cell carcinoma (LUSC) patients [10], though few studies have reported its role in LUAD. Thus, it is necessary to further investigate the biological function and potential mechanism of lncRNA FAM83A-AS1 in LUAD carcinogenesis.
In this study, we discovered that FAM83A-AS1 serves as an oncogenic lncRNA in LUAD and that high FAM83A-AS1 expression predicts poor survival in LUAD patients. In vitro, lncRNA gain and loss function experiments indicated that FAM83A-AS1 promotes cell proliferation, migration, invasion and the epithelial-mesenchymal transition (EMT) by sponging miR-150-5p/MMP14. In addition, FAM83A-AS1 knockdown inhibits LUAD tumor growth in vivo. Our results suggest that FAM83A-AS1 acts as a ceRNA in LUAD carcinogenesis and has potential as a therapeutic target for LUAD.
2. Methods and materials
2.1. Patient samples
Sixty pairs of LUAD samples and adjacent normal samples were collected from The First Affiliated Hospital of Xi'an Jiaotong University from January 2013 to January 2018. All patients underwent surgical excision prior to any radiation or chemotherapy, and the samples were placed directly in liquid nitrogen (−196°C) for subsequent RNA extraction. The clinical and pathological characteristics of the patients are shown in Table 1. All patients signed informed consent. The research protocol was approved by the Ethics Committee of The First Affiliated Hospital of Xi'an Jiaotong University.
Table 1.
Correlation between FAM83A-AS1expression and clinicopathologic factors. (n = 60).
| FAM83A-AS1 expression Low High |
||||
|---|---|---|---|---|
| Clinicopathological characteristics |
Numberof cases (n = 60) |
(n = 25) | (n = 35) | P |
| Age(years) | 0.108 | |||
| <55 | 36 | 12 | 24 | |
| ≥55 | 24 | 13 | 11 | |
| Gender | 0.314 | |||
| Male | 47 | 18 | 29 | |
| Female | 13 | 7 | 6 | |
| Tumor size(cm) | 0.014* | |||
| ≤ 3 | 28 | 7 | 21 | |
| >3 | 32 | 18 | 14 | |
| TNM tumor stage | 0.003** | |||
| Ⅰ+Ⅱ | 37 | 10 | 27 | |
| Ⅲ+Ⅳ | 23 | 15 | 8 | |
| Degree of differentiation | 0.526 | |||
| Well and moderate | 22 | 8 | 14 | |
| Poor | 38 | 17 | 21 | |
| Lymph node metastasis | 0.039* | |||
| Yes | 39 | 20 | 19 | |
| No | 21 | 5 | 16 | |
| Smoking history | 0.170 | |||
| Yes | 25 | 13 | 12 | |
| No | 35 | 12 | 23 | |
Note. Low/high expression was divided according to the sample mean.
The clinical data was analyzed by Peason chi-square test
TNM: Tumor Node Metastasis.
*P < 0.05, **P < 0.01.
2.2. Cell culture and transfection
Human lung adenocarcinoma cell lines (A549, H1299, HCC827 and PC9) and human lung epithelial cells (BEAS-2B) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and China Center for Type Culture Collection (CCTCC, Wuhan, China). All cells were cultured in DMEM/F12 or RPMI-1640 containing 10% fetal bovine serum (FBS) and antibiotics (1x105 U/l penicillin and 100 mg/l streptomycin). All cells were incubated at 37°C in 90% humidified air with 5% CO2. Small interfering (siRNA) and an overexpression plasmid (pcDNA3.1) for FAM83A-AS1 were designed and synthesized by GenePharm (Shanghai, China) and then transfected using Lipofectamine 3000. miR-150-5p mimic and negative control (NC) were purchased from RiboBio Co., Ltd. (Guangzhou, China). MMP14 siRNA and NC siRNA were obtained from GeneChem (Shanghai, China). Transfections were performed using a Lipofectamine 3000 kit (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer’s protocol. The MMP14-siRNA sequence was obtained from previous studies [11]. The si-FAM83A-AS1 target sequence was purchased from GenePharma (Shanghai).
2.3. RT-qPCR
TRIzol® reagent (Invitrogen) was used to isolate total RNA from tissues and cells according to the manufacturer’s protocol. Extracted RNA was reverse-transcribed into cDNAs using a reverse transcription kit (Takara, Dalian, China) under the following reaction conditions: 37°C for 15 min and 85°C for 5 sec, followed by a hold at 4°C. Quantitative real-time PCR was performed using a SYBR-Green Master Mix kit (Takara, Dalian, China) and an ABI 7000 instrument. The PCR reaction conditions were 95°C for 2 min, followed by 40 cycles at 95°C for 10 sec, annealing at 60°C for 30 sec and extension at 70°C for 5 sec. The primers are listed in Table 2. lncRNA-FAM83A-AS1 and MMP14 expression was normalized to that of GAPDH; U6 was used to normalize miR-150-5p. Relative expression of noncoding RNA and mRNA was calculated using the 2−ΔΔCT method.
Table 2.
Primer and siRNA sequences of genes.
| Gene | Primer sequence of genes |
|---|---|
| FAM83A-AS1 | F: 5′-CCC CAGAGCACTTCCTTAGC‐3′ |
| R: 5′-CAGGGCCGTCTGTGTTTACT-3′ | |
| MMP14 | F: 5′-CAACACTGCCTACGAGAGGA-3′ |
| R: 5′-GTTCTACCTTCAGCTTCTGG-3′ | |
| miR-150-5p | F: 5ʹ-TCGGCGTCTCCCAACCCTTGTAC-3’ |
| R: 5ʹ-GTCGTATCCAGTGCAGGGTCCGAGGT-3’ | |
| U6 | F: 5ʹ- TGCGGG TGCTCGCTTCGGCAGC-3’ |
| R: 5ʹ-CCAGTGCAGGGTCCGAGGT-3’ | |
| GAPDH | F: 5ʹ-TGA AGG TCG GAG TCA ACG GAT TTG GT-3’ |
| R: 5ʹ-CAT GTG GGC CAT GAG GTC CAC CAC-3’ | |
| siMMP14 | 5ʹ-GGC GGG UGA GGA ATA ACC AAG-3’ |
2.4. Fluorescence in situ hybridization (FISH)
The FAM83A-AS1 probe was purchased from RiboBio (Guangzhou, China). RNA FISH was performed using A549 and H1299 cells with a FISH kit according to the manufacturer’s instructions.
2.5. RNA immunoprecipitation (RIP)
To verify the relationship between FAM83A-AS1 and miR-150-5p, a Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Sigma-Aldrich) was utilized to perform RIP assays following the manufacturer’s protocol. Briefly, HEK293T cells were harvested and lysed in RIP lysis buffer and then incubated with RIP buffer containing magnetic beads conjugated with an anti-Argonaute2 (Ago2) antibody or normal mouse IgG as a negative control. The samples were incubated with proteinase K, and immunoprecipitated RNA was then isolated. FAM83A-AS1 and miR-150-5p in the precipitates were detected by qRT-PCR.
2.6. Western blotting
Total protein from cells was extracted using RIPA lysis buffer, and protein quantity was assessed using the BCA protein assay. The protein extracts were separated by SDS-PAGE (mini gel), transferred onto PVDF membranes, and probed using primary and then secondary antibodies. The primary antibodies were as follows: anti-MMP14 (Abcam, Cambridge, USA; 1:1000), EMT Antibody Sampler Kit (Cell Signaling Technology, Inc; 1:1000), and anti-β-actin (Santa Cruz Biotechnology, USA; 1:1500). The ECL substrate (Millipore, MA, USA) was used for signal visualization. The band images were analyzed using ImageJ software (NIH) and normalized to β-actin.
2.7. MTT and colony formation assays
Cell viability was evaluated using an MTT (3-(4,5-dimethyl-thiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay kit. Transfected cells were cultured on 96-well plates at a density of 5 × 103 cells/well. After 48 h of culture, the cells were incubated with complete medium plus MTT (5 mg/ml; Sigma-Aldrich) for 4 h. After aspirating the medium, 200 μl DMSO was added to each well to dissolve the formazan crystals produced. MTT-formazan absorbance was detected at OD490 nm using a universal microplate reader. Transfected cells were seeded in 24-well plates at 1000 cells per well. After growth for 14 ~ 21 days, the colonies were washed with PBS, fixed in methanol for 15 min, and stained with a 0.1% crystalline violet solution for 30 min. The number of colonies was counted under an inverted microscope.
2.8. Wound-healing assay
Cell migration was evaluated in vitro by wound-healing assays, as previously described. Briefly, 5 × 104 cells were plated in six-well dishes and cultured in high-glucose DMEM containing 10% FBS overnight. The cells were then cultured in high-glucose DMEM without FBS. After 24 h of serum starvation, wounds were scratched in the cell monolayers using a 200-μl sterile pipette tip. The cells were washed to remove any floating cells and incubated under serum starvation conditions. Wound closure images were photographed at different time points following wounding. Image J software was used to measure wound distances and calculate migration rates.
2.9. Transwell assay
For Transwell migration and invasion assays, 2 × 105 transfected cells were seeded into the upper chamber of Transwell filters precoated with or without Matrigel. A total of 500 μl serum-free medium was added to the upper chamber, and 200 μl of medium containing 10% FBS was added to the lower chamber. After incubation for 24 h, the migrated or invaded cells on the lower membrane surface were fixed in paraformaldehyde for 15 min and then stained with a 0.1% crystal violet solution for 15 min. Cell migration or invasion through the membrane was evaluated under an inverted microscope.
2.10. Luciferase assay
Luciferase assays were performed using Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). Briefly, a fragment of the lncRNA FAM83A-AS1 or MMP14 3′UTR sequence containing a putative target site or mutated target site for miR-150-5p was synthesized and cloned into the pGL3 reporter vector (Promega). For the luciferase reporter assay, cells were cotransfected with miR-150-5p mimic or miR-control and pGL3-FAM83A-AS1-WT/pGL3-MMP14-WT or pGL3-FAM83A-AS1-MUT/pGL3-MMP14-MUT. pRL-TK (Promega) was also cotransfected into cells for a normalization control. After transfection for 48 h, Renilla and firefly luciferase activities were detected using Dual-Luciferase Reporter Assay. The firefly luciferase activity was normalized to that of Renilla.
2.11. Xenograft mouse model
A total of eight male four-week-old BALB/c nude mice were purchased from the Kunming Institute of Zoology and housed and fed at the Animal Center of Zhengzhou University. All animal studies were carried out according to institutional guidelines and were approved by the Ethics Committee of The First Affiliated Hospital of Xi'an Jiaotong University. The mice were divided randomly into two groups (four mice for each group) and then injected subcutaneously with 5 × 106 LUAD cells transfected with control or si-FAM83A-AS1. Subsequently, tumor growth was measured every 4 days, and tumor sizes were calculated as follows: volume = (length × width2)/2. At 4 weeks after implantation, the mice were sacrificed, and the tumors formed were weighed and photographed.
2.12. Statistical analysis
All statistical analyzes were performed using SPSS 20 software (SPSS, USA) and GraphPad Prism 8.0 (San Diego, USA). The data are presented as the mean ± standard deviation (SD) of at least three separate experiments. Two groups and more than two groups were analyzed using Student’s t test and one-way ANOVA, respectively. Spearman’s correlation was used to determine the relationship between FAM83A-AS1 and MMP14 expression. P values less than 0.05 were considered statistically significant.
3. Results
3.1. FAM83A-AS1 is upregulated in LUAD tissues
TCGA data analysis results collected from GEPIA (http://gepia.cancer-pku.cn/index.html) indicated that FAM83A-AS1 expression was significantly higher in LUAD tissues than in normal lung tissues (Figure 1(a), P < 0.05). High expression of FAM83A-AS1 also correlated significantly with advanced stages according to GEPIA (Figure 1(b), P = 3.18e-0.5). To verify the above findings, we measured FAM83A-AS1 expression in 60 pairs of LUAD tissues and adjacent paracancerous tissues from our hospital. As shown in Figure 1(c,d), our results were similar to TCGA data. Clinicopathological analyzes results further indicated that increased FAM83A-AS1 expression was associated with tumor size (Table 1, P = 0.014), TNM stage (Table 1, P = 0.003) and lymph node metastasis (Table 1, P = 0.039). We used the GEPIA software program to verify the association of FAM83A-AS1 expression with the outcomes of 477 LUAD patients from TCGA and found that high FAM83A-AS1 expression was significantly associated with poor OS (n = 477, hazard ratio (HR) = 1.9, P(HR) = 5.1e-05, log-rank P = 3.9e-05) and disease-free survival (n = 477, HR = 1.6, P(HR) = 0.0025, log-rank P = 0.0024) in LUAD (Figure 1(e-f)).
Figure 1.
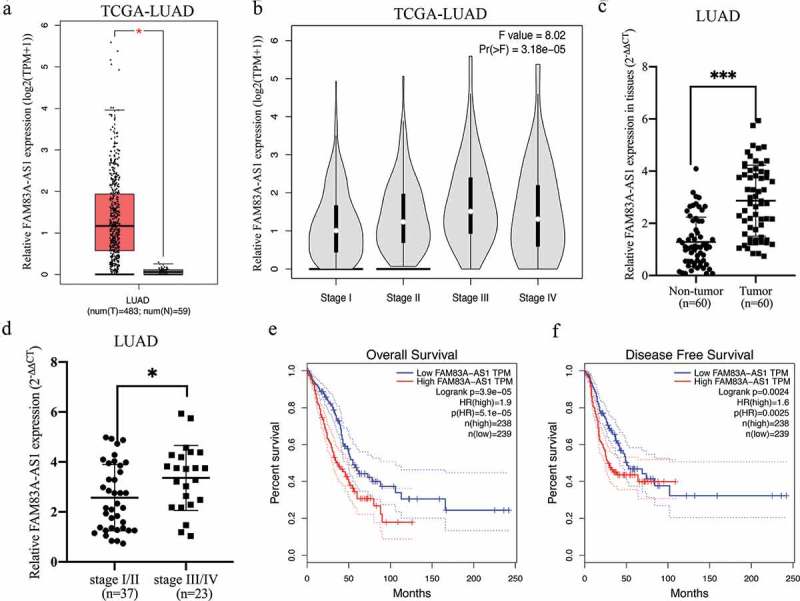
FAM83A-AS1 is highly expressed in LUAD tissues.
(a) TCGA dataset analysis of FAM83A-AS1 expression in LUAD tissues (n = 483) and normal lung tissues (n = 59). (b) TCGA dataset analysis of FAM83A-AS1 in LUAD patients at different clinical stages. (c) Quantitative RT-PCR analysis of FAM83A-AS1 expression in LUAD tissues (n = 60) and adjacent nontumor samples (n = 60). (d) FAM83A-AS1 expression correlated positively with LUAD grade. (e) Kaplan-Meier analysis of overall survival in LUAD patients (n = 477, p = 3.9e-05) according to TCGA datasets. (f) Kaplan-Meier analysis of disease-free survival in LUAD patients (n = 477, p = 0.0024) according to TCGA datasets.
3.2. FAM83A-AS1 promotes cell proliferation, migration, invasion and EMT
To further investigate the effects of FAM83A-AS1 on the biological functions of LUAD cells, we first examined FAM83A-AS1 localization in lung adenocarcinoma cell lines. Using the lncLocator Database (http://www.csbio.sjtu.edu.cn/bioinf/lncLocator/), we predicted intracellular FAM83A-AS1 to be localized to the cytosol (Table 3). The above prediction was confirmed by RNA FISH (Figure 2(a)) and subcellular fractionation (Figure 2(b)) assays in two LUAD cell lines. FAM83A-AS1 expression was also higher in A549 (P < 0.01), NCI-H1299 (P < 0.01), PC-9 (P < 0.001) and NCI-HCC827 (P < 0.001) cells than in BEAS-2B cells (Figure 3(a)). To modulate the FAM83A-AS1 level in LUAD cells, we used a specific siRNA to silence endogenous FAM83A-AS1 in HCC827 cells and a pcDNA3.0-FAM83A-AS1 recombinant plasmid to overexpress FAM83A-AS1 in H1299 cells. qRT-PCR analysis demonstrated that FAM83A-AS1 expression was significantly decreased following transfection with si-FAM83A-AS1 (Figure 3(b), P < 0.001) in HCC827 cells, whereas it was significantly increased following transfection with pcDNA3.0-FAM83A-AS1 (Figure 3(c), **P < 0.01) in H1299 cells. CCK8 and colony formation assays showed that cell proliferation was effectively inhibited in the si-FAM83A-AS1 group compared with that in the si-control group (Figure 3(d), upper row, and Figure 3(e), **P < 0.01). Conversely, forced FAM83A-AS1 expression significantly promoted cell proliferation compared to the empty vector (Figure 3(d), lower row, and Figure 3(f), **P < 0.01). Animal experiments confirmed that FAM83A-AS1 knockdown significantly inhibited tumor growth in vivo (Figure 9(a-c), **P < 0.01). As demonstrated by cell migration and invasion assays, si-FAM83A-AS1 significantly suppressed the migration and invasive capacity of HCC827 cells compared to the si-control, whereas overexpression of FAM83A-AS1 led to a marked promotion of cell motility compared with the empty vector (Figure 4(a-b)). As EMT is a major phenotypic indicator of cancer invasion and metastasis, we next examined whether changes in FAM83A-AS1 expression in LUAD cells affects key EMT markers. As expected, decreased FAM83A-AS1 expression increased that of E-cadherin while decreasing that of N-cadherin and Snail1 in HCC827 cells (Figure 4(c), upper row). Furthermore, overexpression of FAM83A-AS1 significantly decreased E-cadherin expression and increased Snail1 and N-cadherin expression in H1299 cells (Figure 4(c), lower row). These data show that FAM83A-AS1 acts as an onco-lncRNA in LUAD carcinogenesis.
Table 3.
Score of FAM83A-AS1 at different subcellular locations is obtained by lncLocator.
| Subcellular locations | Score |
|---|---|
| Cytoplasm | 0.289256702646 |
| Nucleus | 0.0209332024027 |
| Ribosome | 0.0660473379019 |
| Cytosol | 0.588540489529 |
| Exosome | 0.0352222675202 |
Note. PCC: Pearson correlation coefficient
Figure 2.
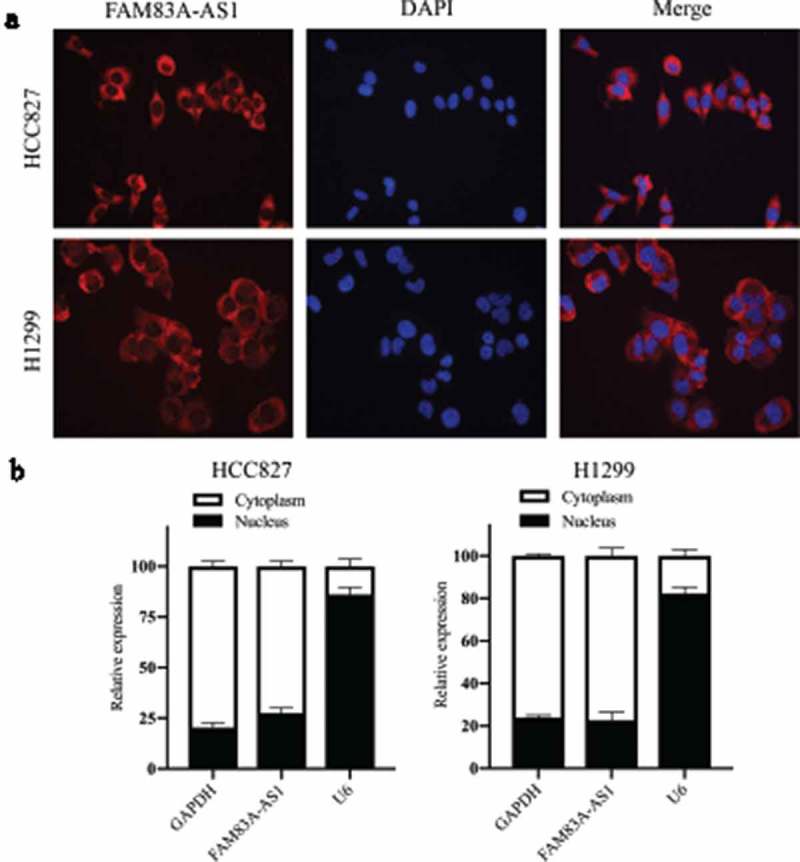
Cellular localization of FAM83A-AS1 in two LUAD cell lines.
(a) Fluorescence in situ hybridization (FISH) assays were used to show subcellular localization of FAM83A-AS1 in A549 and H1299 cells (scale bar = 50 μm). (b) A subcellular fractionation assay revealed FAM83A-AS1 to be mostly located in the cytoplasm; GAPDH and U6 acted as endogenous controls for the nucleus and cytoplasm, respectively.
Figure 3.
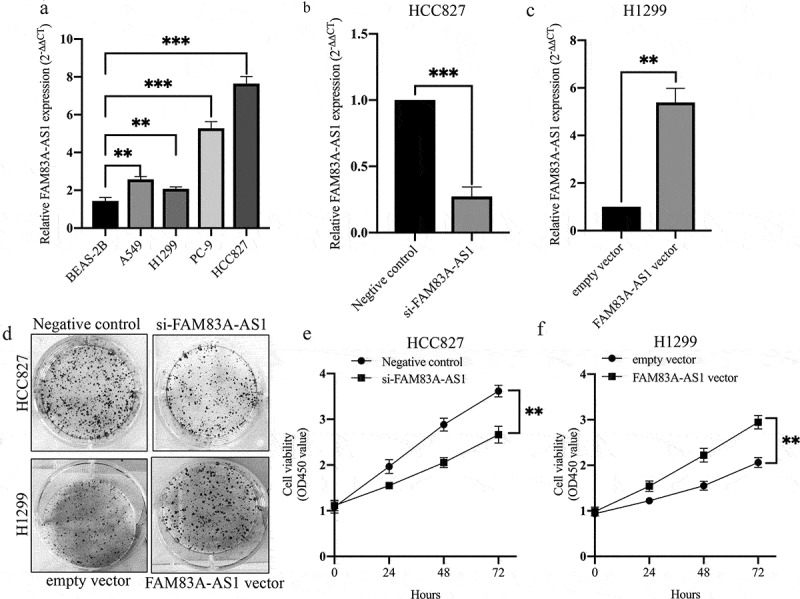
FAM83A-AS1 facilitates LUAD cell proliferation.
(a) qRT-PCR analysis of FAM83A-AS1 expression in four LUAD cell types and human bronchial epithelial cells (BEAS-2B). (b) FAM83A-AS1-knockdown HCC827 cells were generated by transfection with si-FAM83A-AS1 for 48 h. (c) FAM83A-AS1-overexpressing H1299 cells were generated by transfection with the pcDNA3.1-FAM83A-AS1 plasmid for 48 h. (d-f) Colony formation and MTT assays were used to determine the effect of FAM83A-AS1 on cell proliferation in two LUAD cell lines.
Figure 9.
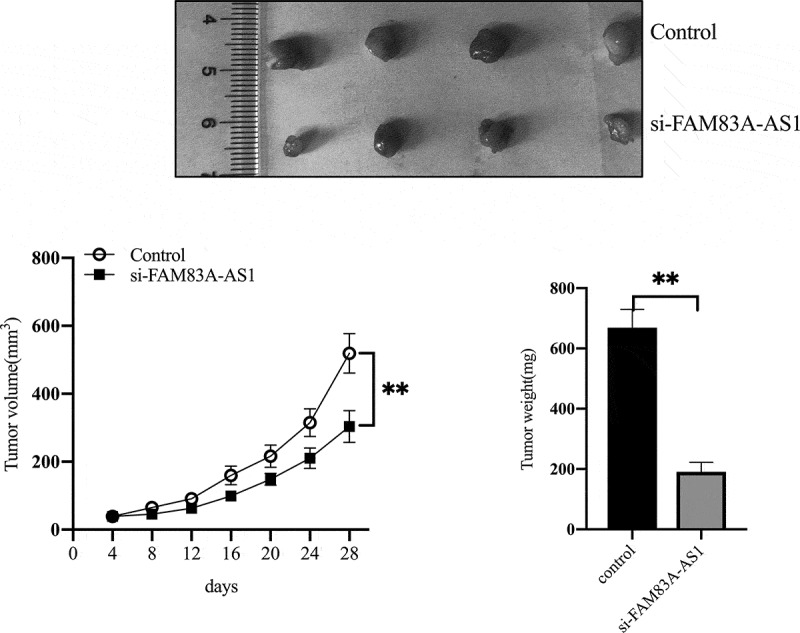
FAM83A-AS1 downregulation inhibits tumor growth in vivo.
(a) Representative images of xenograft tumors are shown. (b) Volumes of the xenograft tumors were measured and used to draw a growth curve. (c) Weights of the xenograft tumors were measured and analyzed.
Figure 4.
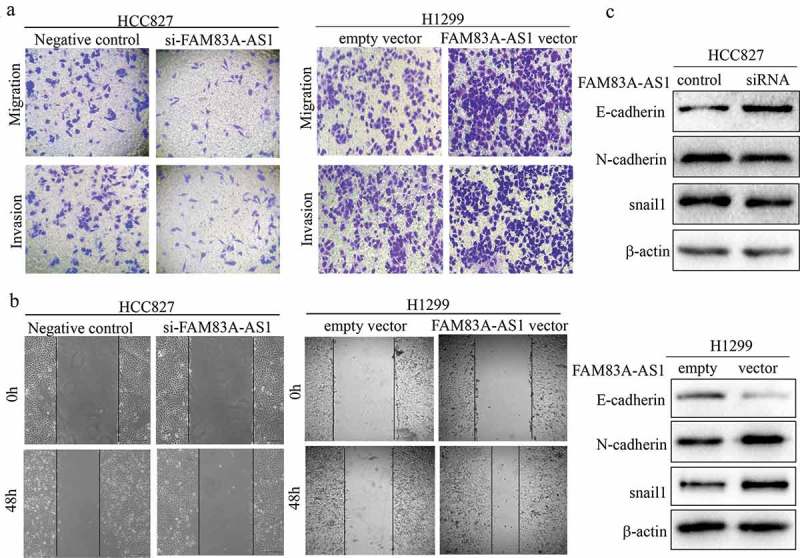
FAM83A-AS1 promotes migration, invasion and EMT in LUAD cells.
(a) Transwell assays were used to examine cell migration and invasion. (b) The effect of FAM83A-AS1 on cell migration was determined by wound-healing assay after transfection with si-FAM83A-AS1 or pcDNA3.1-FAM83A-AS1. (c) Western blot analysis was utilized to explore the effect of FAM83A-AS1 on EMT in two LUAD cell lines.
3.3. FAM83A-AS1 targets miR-150-5p to decrease its expression
It has been reported that lncRNAs can act as ceRNAs to modulate miRNA targeting of mRNA. We thus focused on the ceRNA regulatory network to search for potential target miRNAs of FAM83A-AS1. According to two bioinformatics databases (DIANA-LncBase and mirdb.org), miR-150-5p is predicted to be a target of FAM83A-AS1 (Figure 5(a)). To ascertain whether miR-150-5p is a direct target of FAM83A-AS1, we constructed two reporter plasmids carrying the full-length FAM83A-AS1 3′-UTR harboring wild-type (wt) or mutant (mut) miR-150-5p binding sites. miR-150-5p inhibited the luciferase activity of the pGL3-FAM83A-AS1-wt+miR-150-5p mimic group but not that of the pGL3-FAM83A-AS1-mut+miR-150-5p mimic group (Figure 5(b), **P < 0.01). Moreover, according to RNA-binding protein immunoprecipitation (RIP) assays, miR-150-5p recognized FAM83A-AS1 in a sequence-specific manner (Figure 5(c), **P < 0.01). Consistent with the reporter assay, FAM83A-AS1 knockdown significantly increased the level of miR-150-5p expression in HCC827 cells (Figure 5(d), **P < 0.01), though levels decreased in H1299 cells expressing the FAM83A-AS1 plasmid (Figure 5(e), **P < 0.01). These results demonstrate that miR-150-5p is a direct target of FAM83A-AS1.
Figure 5.
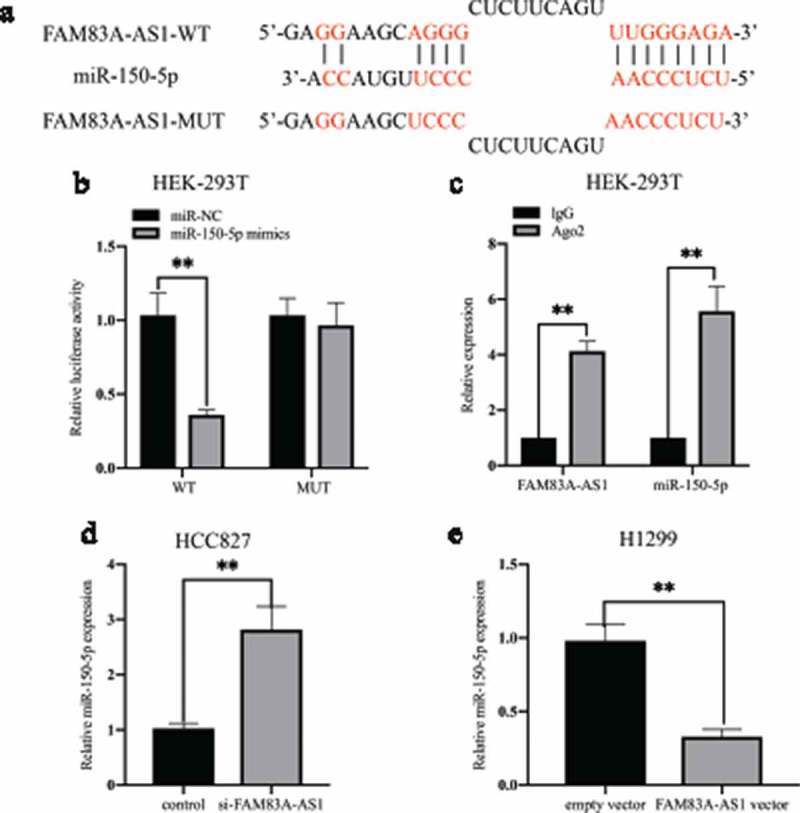
FAM83A-AS1 interacts directly with miR-150-5p.
(a) Bioinformatics predicted and mutated miR-150-5p binding sites of FAM83A-AS1. (b) Luciferase activity in HEK293T cells was detected by luciferase reporter assay after cotransfection with pGL3-FAM83A-AS1-WT or pGL3-FAM83A-AS1-MUT and miR-150-5p mimics or miR-NC. (c) The relationship among FAM83A-AS1, miR-150-5p and Ago2 was confirmed by RNA immunoprecipitation (RIP) with an Ago2 antibody or IgG antibody. Real-time PCR was used to detect changes in FAM83A-AS1 or miR-150-5p in precipitates. (d-e) qRT-PCR assays were performed to assess the effect of FAM83A-AS1 on miR-150-5p expression in transfected LAUD cells.
3.4. miR-150-5p is essential for FAM83A-AS1-promoted LUAD cell migration, invasion and EMT
Transwell and western blot assays were employed to determine whether FAM83A-AS1-dependent promotion of LUAD cell metastasis and EMT is mediated by miR-150-5p. The Transwell assay results showed that miR-150-5p mimic significantly attenuated FAM83A-AS1-induced promotion of H1299 and A549 cell migration and invasion (Figure 6(a)). Transfection with the FAM83A-AS1 plasmid decreased E-cadherin expression and increased that of N-cadherin in the two LUAD cell lines, and these effects were rescued by miR-150-5p mimic (Figure 6(b), *P < 0.05, **P < 0.01). In addition, miR-150-5p mimic significantly promoted and inhibited E-cadherin and N-cadherin protein expression, respectively, in A549 cells (Supplement Figure 3). These findings are consistent with our hypothesis and further indicate that FAM83A-AS1 directly targets miR-150-5p to promote metastasis and EMT in LUAD. These data indicate that the tumor-promoting effects of FAM83A-AS1 on metastasis and EMT in LUAD cells are mediated by miR-150-5p.
Figure 6.
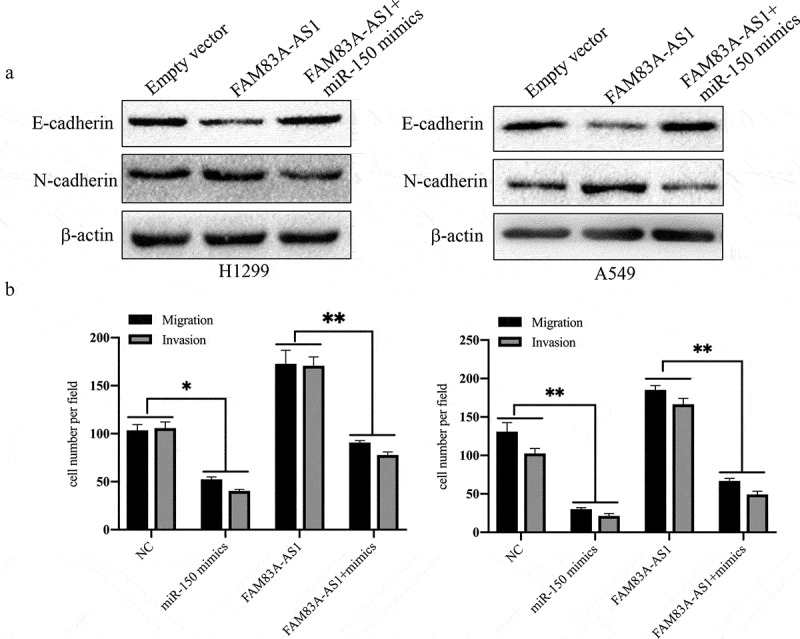
miR-150-5p reverses the effect of FAM83A-AS1 on cell motility and EMT in LUAD cells.
(a) The effect of FAM83A-AS1 overexpression on protein expression of E-cadherin and N-cadherin was reversed by miR-150-5p mimics in H1299 and A549 cells. (b) Transwell assays indicated that miR-150-5p mimics reversed the stimulating effects of FAM83A-AS1 overexpression on LUAD cell migration and invasion capacity.
3.5. MMP14 is a direct target of miR-150-5p
Next, we aimed to identify the potential target gene that participates in the FAM83A-AS1/miR-150-5p axis in LUAD. Bioinformatics tool (TargetScan and StarBase v2.0) analyzes predicted MMP14 to be a target of miR-150-5p, as the 3′-UTR of MMP14 contains an evolutionarily conserved miR-150-5p binding site (Figure 7(a)). To determine whether miR-150-5p effectively targets the 3ʹ-UTR of MMP14, we cotransfected HEK293T cell lines with a wt or mut MMP14 reporter and miR-150-5p mimic. Dual-luciferase reporter assays showed that ectopic expression of miR-150-5p mimic significantly inhibited the luciferase activity of the MMP14-3ʹ-UTR-wt reporter but not that of the MMP14-3ʹ-UTR-mut reporter (Figure 7(b)). As shown in Figure 7(c-d), qRT-PCR and western blot assays indicated that miR-150-5p mimic significantly suppressed expression of MMP14 mRNA and protein in two LUAD cell lines. Interestingly, we found that MMP14 knockdown significantly inhibited expression of N-cadherin protein and promoted that of E-cadherin in A549 cells (Supplement Figure 3). These data demonstrate that MMP14 is a target of miR-150-5p and may be involved in the regulation of EMT.
Figure 7.
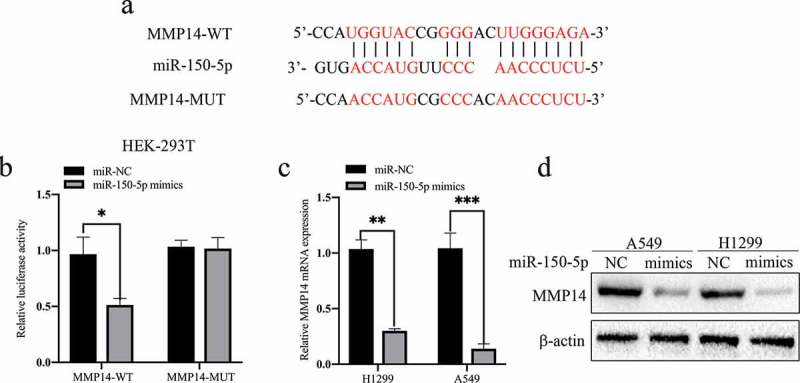
MMP14 is a direct target of miR-150-5p.
(a) The predicted binding site for miR-150-5p in the 3′-UTR of MMP14 and the mutant 3′-UTR of MMP14 (MMP14-Mut) are shown. (b) Luciferase activity in HEK293T cells was detected by luciferase reporter assays after cotransfection with pGL3-MMP14-WT or pGL3-MMP14-MUT and miR-150-5p mimic or miR-NC. (c-d) The mRNA and protein expression levels of MMP14 were determined by qRT-PCR and western blot analyzes after miR-150-5p mimic transfection.
3.6. FAM83A-AS1 affects cell migration and invasion by targeting the miR-150-5p/MMP14 pathway
To further verify whether FAM83A-AS1/miR-150-5p modulates cell migration and invasion in an MMP14-mediated manner, the protein expression levels of MMP14 in FAM83A-AS1 plasmid- and miR-150-5p mimic-transfected LUAD cell lines were detected. Transfection with the FAM83A-AS1 plasmid or miR-150-5p mimic obviously increased or decreased protein expression of MMP14, respectively (Figure 8(b)). Moreover, miR-150-5p mimic partially abrogated the increase in MMP14 protein levels induced by the FAM83A-AS1 plasmid (Figure 8(b)). We then knocked down MMP14 in LUAD cells using siRNA, which was restored by the FAM83A-AS1 plasmid (Figure 8(a)). Based on the relationship between FAM83A-AS1 and MMP14 in LUAD cells, we further analyzed the relationship between FAM83A-AS1 and MMP14 in LUAD tissues using GEPIA. Using LUAD TCGA datasets, Spearman’s correlation analysis showed that FAM83A-AS1 correlated positively with MMP14 expression in LUAD (Figure 8(c), r = 0.27, P = 2.2e-09). A similar result was observed in collected LUAD tissues (Supplement Figure 1, r = 0.4523, P = 0.0003). Transwell assays showed that MMP14 siRNA reversed the promotion of cell migration and invasion ability induced by FAM83A-AS1 overexpression (Figure 8(d-e), P < 0.01). Taken together, the data indicate that FAM83A-AS1 promotes LUAD cell migration and invasion via the miR-150-5p/MMP14 axis. Finally, to explore the underlying mechanisms of FAM83A-AS1 function in LUAD, we used GEPIA to generate a list of genes that have expression patterns similar to that of FAM83A-AS1, as ranked by the Pearson correlation coefficient (PCC). These similar genes are listed in Table S1. Gene Ontology (GO) and KEEG pathway analyzes were performed using Cytoscape. FAM83A-AS1-similar genes were revealed to be mainly involved in the regulation of cell migration, apoptotic signaling, cadherin binding and I-kappaB kinase/NF-κB signaling (Supplement Figure 2). Thus, functional enrichment analysis suggests that the effects of FAM83A-AS1 are associated with tumor growth and metastasis.
Figure 8.
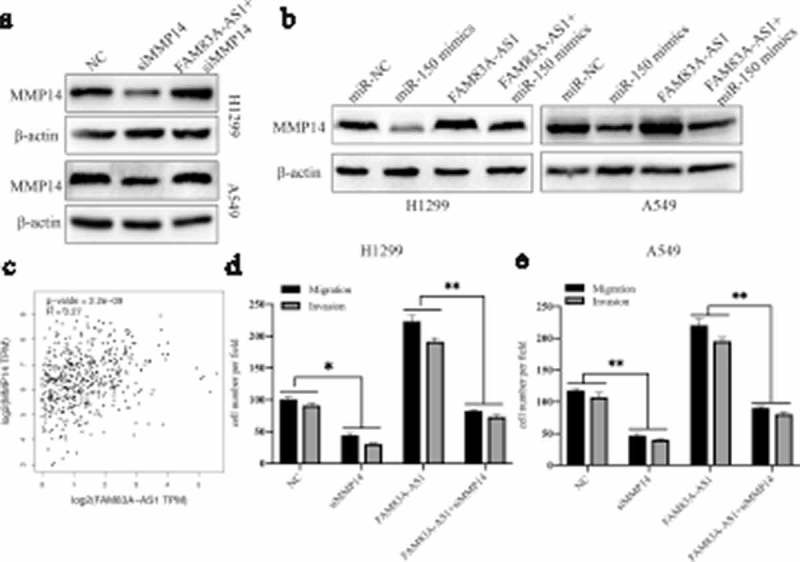
FAM83A-AS1 affects cell migration and invasion through the miR-150-5p/MMP14 axis.
(a) Protein expression of MMP14 was determined in A549 and H1299 cells by western blot assay after transfection with negative control (NC) or siMMP14 or cotransfection with the FAM83A-AS1 plasmid and siMMP14. (b) Protein expression of MMP14 was determined in two LUAD cells by western blot assay after transfection with miR-NC, miR-150-5p mimic, or the FAM83A-AS1 plasmid or cotransfection with the FAM83A-AS1 plasmid and miR-150-5p mimic. (c) Spearman’s correlation analysis indicated the relationship between FAM83A-AS1 and miR-150-5p expression in LUAD tissues based on TCGA datasets. (d-e) Transwell assays were used to estimate cell migration and invasion abilities after transfection with NC, siMMP14, or the FAM83A-AS1 plasmid or cotransfection with the FAM83A-AS1 plasmid and siMMP14 in A549 and H1299 cells.
4. Discussion
Herein, we evaluate the clinical significance and biological function of FAM83A-AS1 in LUAD. The data demonstrated that FAM83A-AS1 is upregulated in LUAD tumor tissues and that high FAM83A-AS1 expression is closely associated with advanced TNM stage, tumor size, lymph node metastasis and poor prognosis in LUAD patients. In vitro, loss- and gain-of-function assays confirmed that FAM83A-AS1 acts as an oncogenic lncRNA to promote cell proliferation, metastasis and EMT. Mechanistic experiments identified that FAM83A-AS1 sponges miR-150-5p to regulate MMP14 protein expression, cell motility and EMT in LUAD cells. These data suggest that FAM83A-AS1 affects LUAD tumorigenesis by regulating expression of the miR-150/MMP14 axis. Therefore, FAM83A-AS1 may serve as a novel biomarker and a potential therapeutic target in LUAD.
A large number of studies have demonstrated that lncRNAs play important roles in the occurrence and development of LUAD and that they might be useful as potential prognostic factors and therapeutic targets [12]. For instance, Rui L et al. (2018) indicated that lncRNA SNHG6 is upregulated in LUAD tissues and sponges components of the miR-26a-5p/E2F7 axis to promote cell motility and EMT in LAUD cells [13]. Additionally, Cong Z et al. (2019) revealed that lncRNA linc00665 can function as a ceRNA by sponging miR-98 and thereby activating AKR1B10-ERK signaling in LUAD progression [14]. However, few studies have reported the role of FAM83A-AS1 in LUAD. By retrieving and screening previous relevant literature, Braicu C et al. (2019) reported that FAM83A-AS1 is upregulated in lung cancer tissues and plays an oncogenic role [15]. Interestingly, Shi R et al. showed that FAM83A-AS1 is markedly upregulated in NSCLC and increases FAM83A protein levels, activating the downstream ERK1/2 signaling pathway [16]. In addition, Ning P et al used TCGA datasets to reveal that FAM83A-AS1 upregulation correlates significantly with poor survival in LUSC patients [10]. Based on the above studies, we speculated that FAM83A-AS1 may be a new diagnostic and therapeutic target for LUAD, and TCGA dataset and clinical sample analyzes validated this hypothesis.
Increasing evidence confirms that lncRNA-mediated ceRNA regulatory mechanisms partly explain the mutual regulation of mRNAs, miRNAs and lncRNAs. Although several studies have reported the role of lncRNA-mediated ceRNA in LUAD, whether FAM83A-AS1 can also regulate the initiation and progression of LUAD through a ceRNA mechanism remains unclear. In the present study, we propose a ceRNA model involving FAM83A-AS1, miR-150-5p, and MMP14 in LUAD. By combining bioinformatics software prediction and dual-luciferase reporter assays, we found that the effect of FAM83A-AS1 on LUAD metastasis and EMT was dependent on direct miR-150-5p targeting. Suetsugu T et al. and Li T et al. demonstrated that miR-150 inhibited invasion and migration by targeting MMP14 in LUSC [17] and hepatocellular carcinoma (HCC) [18]. Our study also confirmed the direct binding of miR-150-5p to MMP14 in LUAD cells. Using GEPIA online tool analysis, we found FAM83A-AS1 expression to significantly positively correlate with MMP14. We also showed that FAM83A-AS1 overexpression reversed miR-150-5p mimic- or siMMP14-induced MMP14 protein suppression in LUAD cells. In addition, overexpressing FAM83A-AS1 reversed the promotional effects of downregulated MMP14 on cell migration and invasion in two LUAD cell lines. In summary, these findings support our hypothesis and reveal a novel ceRNA regulatory axis (FAM83A-AS1-miR-150-MMP14) in LUAD cells.
MMP14 is a member of the matrix metalloproteinase family and is frequently upregulated in many types of cancer. MMP14 plays a pivotal role in cancer metastasis by degrading extracellular macromolecules and promoting proMMP2 and proMMP9 activation [19]. Taras et al. reported that pravastatin decreased lung metastasis in HCC in rats by decreasing MMP14 expression [20]. As a membrane-bound activator, MMP14 promotes cancer cell aggressiveness by binding to membrane proteins (receptor tyrosine kinases (RTKs) and integrins). For example, activated MMP14 accelerates proteolytic processing, which in turn induces cleavage of HB-EGF and stimulates downstream activation of MAPK and PI3K signaling pathways in NSCLC [21]. In our study, we found that overexpression of FAM83A-AS1 restored the suppression of MMP14 induced by siMMP14. Interestingly, recent research has shown that upregulated FAM83A-AS1 enhances expression of intracellular FAM83A and phosphorylation of ERK1/2 [16]. In vitro and vivo studies have also confirmed that FAM83A confers tumor cells with EGFR-TKI resistance [22–24]. Based on the above studies and our results, we speculate that the FAM83A-AS1/miR-150-5p/MMP14 regulatory axis may affect the EGFR signaling pathway. Furthermore, bioinformatics analysis demonstrated that the effects of FAM83A-AS1 are associated with I-kappaB kinase/NF-κB signaling. Therefore, we will explore whether FAM83A-AS1 affects EGFR and NF-κB signaling pathways and its effects on EGFR-TKI resistance in future studies.
Taken together, this study reveals that forced FAM83A-AS1 expression facilitates cell proliferation, metastasis and EMT in LUAD cells via miR-150-5p suppression by enhancing MMP14 expression. These findings indicate that FAM83A-AS1 is a novel oncogenic lncRNA in LUAD cells and a promising biomarker for LUAD diagnosis, prognosis and therapy. The regulatory RNA networks of FAM83A-AS1 remain to be further explored in future studies.
Acknowledgments
This study received no specific grant from any funding agency of the public, commercial or not-for-profit sector.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Siegel RL, Miller KD, Jemal A.. Cancer statistics. CA Cancer J Clin. 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- [2].Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Vikram R, Ramachandran R, Abdul KS. Functional significance of long non-coding RNAs in breast cancer. Breast Cancer. 2014;21:515–521. [DOI] [PubMed] [Google Scholar]
- [5].Yang X, Xie X, Xiao YF, et al. The emergence of long non-coding RNAs in the tumorigenesis of hepatocellular carcinoma. Cancer Lett. 2015;360:119–124. [DOI] [PubMed] [Google Scholar]
- [6].Yang Y, Junjie P, Sanjun C, et al. Long non-coding RNAs in colorectal cancer: progression and future directions. J Cancer. 2017;8:3212–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Li S, Xu Y, Sun Z, et al. Identification of a lncRNA involved functional module for esophageal cancer subtypes. Mol Biosyst. 2016;12:3312–3323. [DOI] [PubMed] [Google Scholar]
- [8].Dong HX, Wang R, Jin XY, et al. LncRNA DGCR5 promotes lung adenocarcinoma (LUAD) progression via inhibiting hsa-mir-22-3p. J Cell Physiol. 2018;233:4126–4136. [DOI] [PubMed] [Google Scholar]
- [9].Sun J, Pan LM, Chen LB, et al. LncRNA XIST promotes human lung adenocarcinoma cells to cisplatin resistance via let-7i/BAG-1 axis. Cell Cycle. 2017;16:2100–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ning P, Wu Z, Hu A, et al. Integrated genomic analyses of lung squamous cell carcinoma for identification of a possible competitive endogenous RNA network by means of TCGA datasets. PeerJ. 2018;6:e4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chang ML, Huang YH, Cheng JC, et al. Interaction between hepatic membrane type 1 matrix metalloproteinase and acireductone dioxygenase 1 regulates hepatitis C virus infection. J Viral Hepat. 2016;23:256–266. [DOI] [PubMed] [Google Scholar]
- [12].Sui J, Li YH, Zhang YQ, et al. Integrated analysis of long non-coding RNAassociated ceRNA network reveals potential lncRNA biomarkers in human lung adenocarcinoma. Int J Oncol. 2016;49:2023–2036. [DOI] [PubMed] [Google Scholar]
- [13].Liang R, Xiao G, Wang M, et al. SNHG6 functions as a competing endogenous RNA to regulate E2F7 expression by sponging miR-26a-5p in lung adenocarcinoma. Biomed Pharmacother. 2018;107:1434–1446. [DOI] [PubMed] [Google Scholar]
- [14].Cong Z, Diao Y, Xu Y, et al. Long non-coding RNA linc00665 promotes lung adenocarcinoma progression and functions as ceRNA to regulate AKR1B10-ERK signaling by sponging miR-98. Cell Death Dis. 2019;10:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Braicu C, Zimta AA, Harangus A, et al. The function of non-coding RNAs in lung cancer tumorigenesis. Cancers (Basel). 2019;11(5). pii: E605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shi R, Jiao Z, Yu A, et al. Long noncoding antisense RNA FAM83A-AS1 promotes lung cancer cell progression by increasing FAM83A. J Cell Biochem. 2019;120:10505–10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Li T, Xie J, Shen C, et al. miR-150-5p inhibits hepatoma cell migration and invasion by targeting MMP14. PLoS One. 2014;9:e115577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Li T, Xie J, Shen C, et al. Amplification of long noncoding RNA ZFAS1 promotes metastasis in hepatocellular carcinoma. Cancer Res. 2015;75:3181–3191. [DOI] [PubMed] [Google Scholar]
- [19].Pytliak M, Vargova V, Mechirova V. Matrix metalloproteinases and their role in oncogenesis: a review. Onkologie. 2012;35:49–53. [DOI] [PubMed] [Google Scholar]
- [20].Taras D, Blanc JF, Rullier A, et al. Pravastatin reduces lung metastasis of rat hepatocellular carcinoma via a coordinated decrease of MMP expression and activity. J Hepatol. 2007;46:69–76. [DOI] [PubMed] [Google Scholar]
- [21].Stawowczyk M, Wellenstein MD, Lee SB, et al. Matrix metalloproteinase 14 promotes lung cancer by cleavage of heparin-binding EGF-like growth factor. Neoplasia. 2017;19:55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Richtmann S, Wilkens D, Warth A, et al. FAM83A and FAM83B as prognostic biomarkers and potential new therapeutic targets in NSCLC. Cancers (Basel). 2019;11(5). pii: E652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Grant S. FAM83A and FAM83B: candidate oncogenes and TKI resistance mediators[J]. J Clin Invest. 2012;122(9):3048–3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lee SY, Meier R, Furuta S, et al. FAM83A confers EGFR-TKI resistance in breast cancer cells and in mice. J Clin Invest. 2012;122:3211–3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


