Abstract
Cells interact intimately with complex microdomains in their extracellular matrix (ECM) and maintain a delicate balance of mechanical forces through mechanosensitive cellular components. Tissue injury results in acute degradation of the ECM and disruption of cell-ECM contacts, manifesting in loss of cytoskeletal tension, leading to pathological cell transformation and the onset of disease. Recently, microscale hydrogel constructs have been developed to provide cells with microdomains to form focal adhesion binding sites, which enable restoration of cytoskeletal tension. These synthetic anchors can recapitulate the complex 3D architecture of the native ECM to provide microtopographical cues. The mechanical deformation of proteins at the cell surface can activate signaling cascades to modulate downstream gene-level transcription, making this a unique materials-based approach for reprogramming cell behavior. An overview of the mechanisms underlying these mechanosensitive interactions in fibroblasts, stem and other cell types is provided to review their effects on cellular reprogramming. Recent investigations on the fabrication, functionalization and implementation of these materials and microtopographical features for drug testing and therapeutic applications are discussed.
Keywords: Microtopography, Microfabrication, Mechanotransduction, Extracellular matrix, Hydrogel, Drug delivery, Focal adhesion, Cytoskeleton
1. Introduction
Cells exist in a sensitive biochemical and mechanical equilibrium presented by their surrounding extracellular matrix (ECM) and other tissue components. Disruption of this equilibrium due to acute injury or progressive pathology manifests in the development of chronic disease (Jaalouk and Lammerding 2009; Lindsey et al. 2003; Midwood et al. 2004; Prabhu and Frangogiannis 2016; Raposo and Schwartz 2014). Classical therapeutic approaches to regenerate function to the affected area involve the administration of pharmacological agents or biologics to modulate cell behavior or to recruit cells to the region. However, it is becoming increasingly recognized that treatment with soluble factors is insufficient for comprehensive tissue repair. The native tissue environment is a complex milieu of signaling factors and biophysical microdomains with specific spatiotemporal distribution. Therefore, failure to recapitulate these myriad factors has significantly limited the efficacy of current tissue regeneration strategies.
An emerging paradigm in tissue engineering focuses on the generation of appropriate mechanical environments for cell attachment, growth and differentiation. Stem cells, for example, can be instructed to differentiate into specific lineages based on the stiffness of their substrate alone (Engler et al. 2006; Huebsch et al. 2010). These mechanical factors are so crucial that without an appropriate substrate stiffness, introduction of osteogenic factors is unable to instruct these cells towards an osteogenic lineage, highlighting the complex interplay between growth factor signaling and mechanical stimuli and its crucial role in dictating cell behavior. Researchers have focused on engineering matrices that recapitulate the topography and bulk mechanical properties of the native ECM to promote proper tissue growth. The development of these materials has been supported by advancements in fabrication technologies, such as photolithography, microfluidics and 3D printing, which have been extensively reviewed (Annabi et al. 2010; Khademhosseini et al. 2006b; Khademhosseini and Langer 2007; Leijten et al. 2017; Ovsianikov et al. 2012; Slaughter et al. 2009; Yanagawa et al. 2016; Zorlutuna et al. 2012). Unfortunately, complex matrices with tailored microchannels and other topographies cannot be injected in vivo and are limited to in vitro studies. Injectable materials, on the other hand, are easily administered but typically comprise of bulk hydrogels that are homogeneous and isotropic in nature, failing to recapitulate the complex architecture of the ECM. Due to these challenges, the use of mechanically instructive materials for therapeutic applications remain quite limited.
Recently, discrete topographical features have been used to reprogram cell phenotype. These materials can be fabricated with a wide variety of stiffnesses, geometries and porosities and are fully formed and characterized before use. Introduction of these materials into the extracellular matrix provides cells with anchor points that enable restoration of intracellular tension and subsequent modulation of signaling pathways via complex mechanisms (Ayala et al. 2010; Curtis et al. 2010, 2013; Le et al. 2018; Norman et al. 2008; Pinney et al. 2014a). Further, these materials are a versatile technology that can be modified and tuned in a variety of ways, such as drug loading, surface modification or physical alignment to meet the demands of varying disease states. Here, we review the mechanisms by which cells interact with discrete topographical cues and highlight novel developments in this technology for tissue engineering applications.
2. Cellular machinery to sense the physical microenvironment
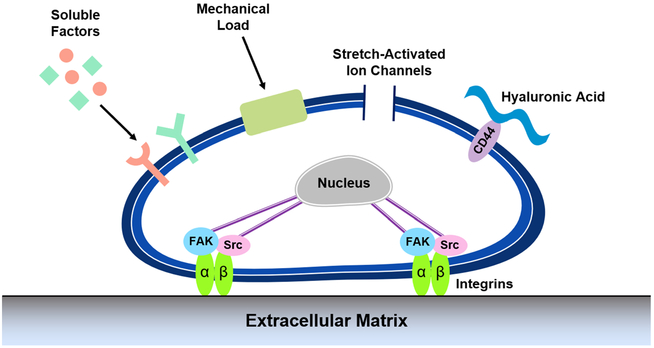
Cells intimately interact with their local microenvironment and are exposed to a variety of externally applied stresses. As such, they are equipped with an abundance of cellular machinery that allow them to detect and respond to these stimuli (Fig. 1). Cells interpret such mechanical forces by mechanotransduction, the process by which extracellular physical stimuli deform molecular structure permitting post translational modification of proteins in the cell, and leading to gene expression changes in the nucleus. Taken together, cellular behavior is regulated. These processes of mechanosensing, mechanotransduction and response occur at multiple locations in the cell, including the plasma membrane, the cytoskeleton and the nucleus.
Fig. 1. Cellular mechanisms for detecting external mechanical stimuli.
Cells are equipped with a wide range of cellular machinery to interact with the extracellular environment, including focal adhesions, ion channels and cell surface receptors. Once adhered to the extracellular matrix, activation of focal adhesion kinases and other signaling pathways, or direct propagation of force to the nucleus can initiate signaling cascades to modulate gene expression and cellular phenotype
The cell membrane serves as the most direct link between the cell and the surrounding environment. Mechanosensitive ion channels, for example, are transmembrane proteins that can be physically stretched by external forces, causing an immediate influx of ions that distorts previously established ionic equilibrium (Martinac 2004). These ion currents are capable of initiating signaling cascades and modulating the expression of key transcriptional factors that reprogram cell behavior. In fibroblasts, stretch activation of calcium ion channels has been linked to short-term increases in the production of alpha smooth muscle actin (αSMA), which promotes the differentiation of fibroblasts into a contractile myofibroblast phenotype (Asazuma-Nakamura et al. 2009).
Cells primarily interact with the surrounding ECM through transmembrane receptors on the cell surface. While receptors for a variety of ECM components exist, such as CD44 and syndecans, the most studied receptors are the family of integrins, which are able to bind to fibronectin, laminins and collagens present in the ECM (Geiger et al. 2009; Katsumi et al. 2004; Martino et al. 2018; Orr et al. 2006; Puklin-Faucher and Sheetz 2009). Activation of integrins enables cells to form intimate attachments to the surrounding matrix for cell adhesion and migration. Further maturation of these attachments is characterized by the recruitment of key adapter proteins, such as talin, paxillin and vinculin, that make up a focal adhesion complex (FAC). This assembly of proteins couples the extracellular environment to the intracellular cytoskeleton, enabling cells to experience externally applied forces (Parsons et al. 2010; Ziegler et al. 2008). Further, these complexes act as a hub for activation of downstream biochemical signaling pathways. This is typically initiated by phosphorylation of signaling molecules at the integrins’ cytosolic domains by protein tyrosine kinases such as focal adhesion kinase (FAK) and Src which induce signaling cascades involving ERK1/2 and MAPK, culminating in altered cell migration, proliferation and differentiation (Westhoff et al. 2004).
Lipid signaling at the cell membrane has also been shown to rapidly affect focal adhesion assembly, facilitating conversion of mechanical cues to signaling events. Phosphatidylinositol 4,5-biphosphate (PIP2) signaling has been shown to modulate organization of the actin cytoskeleton and lamellar architecture (Mkrtschjan et al. 2018a). In the context of focal adhesions and the actin cytoskeleton, PIP2 plays key roles in the recruitment of adaptor proteins, such as vinculin, talin and paxillin, to the focal adhesions enabling propagation of forces to the cytoskeleton (Zhang et al. 2012). While focal adhesions are an integral component of mechanosensing discussed in this review, a more detailed analysis of the intricacies of focal adhesion formation and maturation are found elsewhere (Geiger et al. 2009; Katsumi et al. 2004; Puklin-Faucher and Sheetz 2009; Wozniak et al. 2004).
Focal adhesions are the anchoring complex to which the cytoskeleton attaches. The actin cytoskeleton regulates cell shape, function, and overall cellular mechanics and is able to orchestrate responses to mechanical perturbations. Cells exist in a state of cytoskeletal pre-stress that balances the intracellular tension with the physical properties of the ECM. Upon encountering external forces that perturb this pre-stress, activation of contractility dependent mechanisms enacted by myosin-II motors act to crosslink, organize and affect sliding between actin microfilaments to restore cytoskeletal tension (Ingber et al. 2014; Ingber 1997, 2006). These responses are dependent on activation of small Rho GTPases, such as RhoA, and subsequent phosphorylation of rho-associated protein kinase (ROCK). The actin cytoskeleton also facilitates propagation of extracellular stimuli from the outside of the cell to the nuclear membrane with its own skeleton comprised of lamins formed into filaments that provide structural support for the nucleus (Wang et al. 2009). Chromosomes are anchored to the lamina at the periphery of the nucleus and are susceptible to physical perturbations that are propagated from the cytoskeleton. Force propagation can mechanically distort the cell nucleus, changing overall nuclear shape and reorganizing residing chromosomes to ultimately affect gene transcription (McNamara et al. 2012).
Cardiomyocytes (CMs) are extremely responsive to mechanical cues in order to accommodate rapid changes in blood volume and subsequent altered contractile demand, as well as long-term adaptation to static load. Cardiomyocytes have evolved to respond to changes in tensions through a variety of internal sensors, ranging from components of the focal adhesion complex such as vinculin and talin to proteins further downstream within the sarcomere, including titin and Z-disc components (Hoshijima 2006; Pandey et al. 2018). The near immediate response to functional demand is reliant primarily on length-dependent activation, widely considered the predominant effector in the Frank-Starling Law of the heart, via increased myofilament Ca2+ sensitivity (de Tombe et al. 2010). Long-term alteration of myocyte contractility is often a product of changes to the cytoskeleton and structural remodeling of the myocyte. In particular, cyclic mechanical stretch and increasing stiffness of the underlying substrata have been used as a mechanical, hypertrophic stimuli, resulting in increased cell size and sarcomere assembly dynamics (Li et al. 2014; Lin et al. 2013; Mkrtschjan et al. 2018b). Upon presentation of a stimulus, this signal is transduced to the sarcomere where the actin capping protein, CapZ, is phosphorylated (e.g. PKCε), and its association with the barbed end of actin is decreased. The resultant effect is increased assembly rate of sarcomeric actin and overall cellular and tissue hypertrophy. Moreover, due to the individual cardiomyocyte’s limited capacity to undergo cell division, myofibril addition is the primary way the heart muscle mass can increase.
In sum, cells are equipped with a wide array of mechanisms to sense, interact and respond to their local physical environments. These biophysical events are capable of reprogramming cell behavior and as noted above, mechanical cues have gained much attention as an adjustable tool for tissue engineering applications. In this review, we will focus on the influence of topography in modulating cellular phenotype.
3. Microtopographical regulation of cellular phenotype
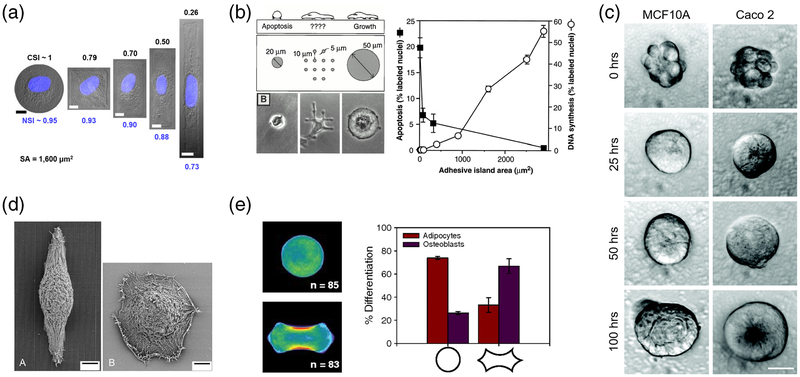
Cells are capable of detecting environmental geometries through a complex mechanism known as contact guidance (Al-Haque et al. 2012; Rørth 2011). These intimate interactions with local topographical features provide instructive cues for directional cell spreading, migration and in some cases, differentiation (Cortese et al. 2013; Jiang et al. 2011; Kilian et al. 2010; Kim et al. 2009; Teixeira 2003). Researchers have shown that simply culturing cells on scaffolds of confined micron-scale geometries can promote the reprogramming of epigenetic state for generation of induced pluripotent stem cells (iPSCs), direct stem cells towards specific lineages, or even guide multi-cell clusters to polarize and form lumenized microtissues (Fig. 2) (Cerchiari et al. 2015; Downing et al. 2013; Kilian et al. 2010). Despite significant interest in utilizing topography for tissue engineering applications, the underlying mechanisms behind cell-microtopography interactions remain incompletely understood.
Fig. 2. Topographic cues are potent modulators of cell proliferation, migration and differentiation.
a Endothelial cell and nuclear shape can be controlled by culturing cells on islands of varying shape and size (scale bars = 10 μm) (Versaevel et al. 2012). b Control of endothelial cell size can induce apoptosis or cell growth (Chen et al. 1997). c Geometric confinement of multi-cell clusters within microwells can induce cell polarization into fully lumenized spheroids (scale bar = 20 μm) (Cerchiari et al. 2015). d Corneal epithelial cells stretch and align with underlying microchannels compared to cells cultured on flat, which remain rounded (Teixeira 2003). e Changes in stem cell shape modulates myosin II activity and cell contractility to guide differentiation of stem cells into adipocytes or osteoblasts (Kilian et al. 2010). Figures reproduced with permission
Seminal work from the Desai group described the interaction of cardiomyocytes with topographical features utilizing microtextured silicone membranes (Deutsch et al. 2000). Myocytes were found to have terminated with blunted protrusions ending on a micropost, frequently bridging between two microposts. This was in contrast to untextured membranes that have randomly oriented myocytes with tapered protrusions. The textured membranes had 4-fold increases in cellular attachment with cells adopting more in vivo-like morphologies despite equivalent adhesion motifs present on the surface, suggesting increased interaction via alternative mechanisms. The incorporation of microposts likely presents the cell with 3D cues not present on flat surfaces used in culture dishes and, by mechanisms to be discussed, is able to recapitulate a complex 3D mechanical environment to generate more in vivo-like phenotypes.
Fibroblasts were similarly grown in the presence of microposts and exhibited high preference for micropost attachments as well (Boateng et al. 2003). After contacting a micropost, fibroblast migration velocity was significantly decreased and the attachments matured with extensive accumulation of FAK around the microposts. These interactions manifested in global reduction of cell proliferation, although these effects are believed to occur on the single-cell level. If this is the case, the observed effects on migration and proliferation would not be affected by micropost size, but rather by the ability to establish contact and exert force on the microposts. Indeed, when presented with large microposts that provided additional contact area, there were no differences in fibroblast proliferation (Thakar et al. 2008). Higher micropost density increased the antiproliferative effects in culture, supporting the hypothesis that single-cell level transcriptional changes were occurring. This was confirmed by BrdU staining, where cells in direct contact with the microposts had reduced BrdU incorporation, whereas non-contacting cells were similar to cells on non-patterned substrates. These results were consistent with C2C12 mouse skeletal myoblasts and mouse embryonic stem cells as well (Fig. 3a) (Biehl et al. 2009; Thakar et al. 2008).
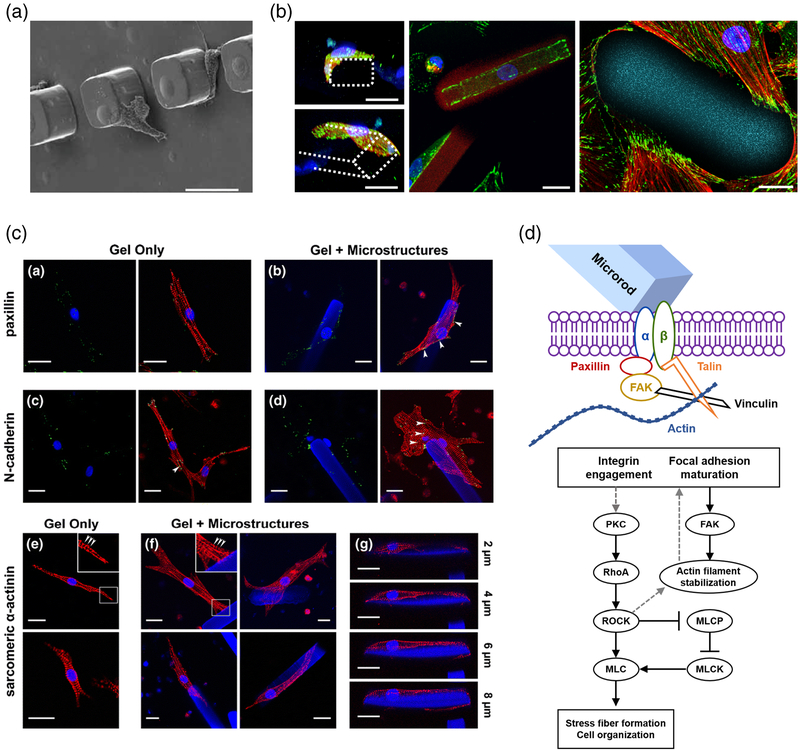
Fig. 3. Cellular interactions with discrete topographical cues.
a Fibroblasts form intimate interactions with microscale posts by stretching to the top of microscale posts or pushing up against the base (scale bars = 25 μm) (Thakar et al. 2008). b Primary cardiac fibroblasts wrap around microrods and spread across the entire structure, accumulating focal adhesions on the edges of the rods (Green = paxillin, red = actin, blue = nuclei, cyan = microrod, scale bars = 20 μm) (Le et al. 2018). c Primary ventricular myocytes form strong associations with microrods in 3D that result in accumulation of focal adhesions at the cell-microrod interface. Clusters of cells are also able to interact with the microrods as well (red = α-actinin, green = paxillin, N-cadherin, blue = nuclei, scale bars = 20 μm) (Curtis et al. 2010). d Possible mechanism of microrod-affected mechanics. The schematic depicts the signaling pathways involved upon cell binding to microrods. Addition of these microrods introduces local heterogeneity, which guides the cells towards integrin engagement and focal adhesion formation. The maintenance of PKC and RhoA/ROCK signaling mediate the microrod-related changes in contractile behavior. Detailed pathways and actin stress fiber assembly vary with cell type. Figures reproduced with permission
Such mechanisms also carried over to fully three-dimensional environments in which cells experience forces from all directions, leading to significantly different experience of extracellular forces and the adoption of more physiological cell morphologies (Cukierman et al. 2001; Geiger 2001). This is particularly relevant in damaged tissues where the softened microenvironment and disruption of cell-cell and cell-ECM connections removes native instructional cues for development (Lindsey et al. 2003; Midwood et al. 2004; Prabhu and Frangogiannis 2016). Discrete micron-scale hydrogels, termed microrods, were incorporated into these soft 3D matrices to act as anchors for cell attachment and recapitulate native ECM mechanics (Fig. 3b–c) (Ayala et al. 2010; Collins et al. 2010a; Curtis et al. 2010, 2013; Pinney et al. 2014a). The structural guidance provided by these materials can instruct specific alignment and maturation of neurons, cardiomyocytes and fibro-blasts in three dimensions (Ayala et al. 2010; Curtis et al. 2010; Le et al. 2018; Norman et al. 2008; Pinney et al. 2014a; Rose et al. 2017, 2018).
Myocytes cultured with these microrods formed intimate attachment to the microstructures and were able to pull and displace the microrods within the soft 3D matrix (Curtis et al. 2010, 2013). As a whole, myocytes generated greater magnitude of displacement in the presence of microrods compared to controls. The long axis length, perimeter and area of myocytes grown with microrods was much larger than those grown without. This interaction also significantly decreases shortening time and increased protein synthesis. Similar to previous studies, focal adhesions were accumulated at the myocyte-microstructure contact regions. Interestingly, these interactions resulted in a 2-fold higher rate of spontaneous contraction. Together, these results suggest that addition of microstructures, which introduce heterogeneity in stiffness and topography when culturing neonatal rat ventricular myocytes (NRVMs) in 3D, is sufficient to alter the hypertrophy, spontaneous contraction and gene expression. Neural cells are also responsive to these topographical cues in 3D, growing parallel to the discrete features (Rose et al. 2017). At low concentrations of discrete microstructures, dorsal root ganglions (DRGs) orient and grow unidirectionally along the structures despite minimal geometric constraint. Addition of adhesive motifs to the surface of these microstructures facilitated the interactions with DRGs, which were able to better align with those instructive features (Rose et al. 2018). These results suggest a mechanism that is dependent on adhesion strength.
A contractility-based mechanism was thus proposed to explain these changes. While challenge with ROCK and myosin light-chain kinase (MLCK) inhibitors did not reverse the global effect of microposts on cell proliferation, closer examination revealed that inhibitor treatment increased the percentage of proliferating cells that were in contact with microstructures (Thakar et al. 2008). Additionally, inhibitor treatment reduced the number of adherent cells, likely due to reduced ability to pull on the substrates and form fully matured focal adhesions (Boateng et al. 2003). Patel et al utilized time lapse imaging to examine the interaction of single cells with microtopographical cues (Patel et al. 2010). It was observed that cells cultured on micropost-textured scaffolds had greater mean tether lengths compared to cells cultured on flat substrates. After coming into contact with these microstructural anomalies, cell de-adhesion from the microposts was also slowed. Both results suggest that micropost interactions alter the balance between cell contractility and adhesion strength. While ROCK inhibition abolished the differences between micropost versus flat culture, the differences persisted during treatment with MLCK inhibitor, with micropost-textured scaffolds having similar tether lengths to flat scaffolds without inhibitor treatment. This suggests that adhesion to microposts make up for the reduced contractility induced by MLCK treatment, but is not sufficient to affect upstream ROCK activity (Fig. 3d). The dependence on contractility and exertion of active forces was confirmed by comparing the interaction of fibroblasts with immobilized and free microspheres on a flat 2D substrate (Boateng et al. 2003). Free microspheres were easily moved around by the fibroblasts and had no impact on fibroblast proliferation, while immobilized microspheres elicited effects similar to microposts. The generation of active forces by the cell must be responsible for changes in these behavioral changes. While the mechanics of the cell were unaffected by these interactions, significant upregulation of myosin contractility pathway elements, including Rho GTPase and myosin II heavy chain suggest local changes in cellular mechanobiology at the adhesion sites. These results were corroborated with later studies investigating the effect of microenvironment topography on cardiomyocyte subdomain contractility (Broughton and Russell 2015).
While many of these responses are thought to be focused at the cell surface, several reports have suggested modulation of gene expression by altering nuclear shape (Downing et al. 2013; McNamara et al. 2012; Thomas et al. 2002). Fibroblast nuclei were observed to move closer to microtopographical features and become distorted after binding. The nuclear distortion is believed to be the result of reactive forces that are exerted on the cell membrane and transmitted to the rest of the cell when the cell pushes against a microstructure. This deformation has been shown to affect cell proliferation and gene expression (Thomas et al. 2002; Versaevel et al. 2012). These responses are believed to be caused by reorganization of the nuclear lamina and repositioning of chromosomes. Depending on new position, the chromosomes can become more heterochromatic and have reduced expression of genes (Finlan et al. 2008; McNamara et al. 2012). Previous studies using fibroblasts cultured in microchannels have demonstrated that nuclear shape, chromosome localization, and histone modifications are affected by topography and can be used to facilitate the restoration of epigenetic state for cell reprogramming (Downing et al. 2013). These mechanical effects are strong enough to replace the use of histone deacetylase and histone demethylase inhibitors for generation of iPSCs.
These observations suggest that interaction with topography can modulate cell gene expression via both direct chromosomal modifications and by transcriptional regulation of genes initiated at focal adhesions. Despite the complex mechanisms at play, it is clear that topography is an effective tool for modulating cell behavior. The ability to reprogram cells to a more physiological phenotype would enable us to harness intrinsic healing mechanisms and promote healthy repair of tissues.
4. Using microstructural cues for therapeutic applications
Modifying the physical and biochemical properties of extra-cellular matrix of diseased tissues at the micron scale is an emerging strategy for enhancing our currently limited capabilities in regenerating complex tissues. It is widely recognized that the success of cellular replacement therapies has largely been hampered by poor engraftment with the local microenvironment. For example, after acute myocardial infarction (MI), widespread cell death and the release of intracellular components initiate a complex cascade of signaling events that lead to pathological matrix remodeling and disruption of the physicochemical and biochemical homeostasis of the ECM (Frangogiannis 2015; Li et al. 2018; Segura et al. 2014; Talman and Ruskoaho 2016). While these perturbations are often resolved through intrinsic wound healing mechanisms in some tissues, myocardial tissue damage is characterized by continued aberrant tissue remodeling and the formation of a stiff myocardial scar that permanently disrupts cell-cell and cell-ECM interactions. Further, the lack of adequate nutrient supply in this avascular scar challenges the survival of transplanted stem or differentiated cell types seeking to replace the lost tissue.
Injectable hydrogel materials have been the focus of bio-engineering approaches to create suitable extracellular environments that promote cell engraftment (Dimatteo et al. 2018; Tibbitt and Anseth 2009; Zhu et al. 2017). While it is possible to engineer the biophysical and biochemical makeup of these materials to support cell attachment, growth and differentiation, these materials tend to be physically and chemically homogeneous and anisotropic in nature. This is unrepresentative of the native ECM which consists of chemical gradients and biophysical microdomains that facilitate directional cell migration and physiological behavior. There are, indeed, engineering techniques available to introduce spatial heterogeneity into these hydrogel materials, but these approaches tend to involve foaming agents, porogens, or other cytotoxic materials and are not suitable for coadministration with live cells (Annabi et al. 2010).
To address this, discrete microengineered materials have been used to introduce mechanical anomalies to target tissues, promoting cellular attachment and modulating the behavior of both native and transplanted cell types. Xin et al reported the assembly of poly(ethylene glycol) microgels that can be co-injected with cells and later be annealed into a porous 3D scaffold, termed granular hydrogel, in vivo (Xin et al. 2018). These hydrogels were designed to have highly interconnected microporous structure and have Young’s moduli that span the range of physiological stiffnesses. Human mesenchymal stem cells (hMSCs) cultured with these microgels spread and surrounded the structures, growing within and throughout the pores to form a 3D cellular network. This was in contrast to bulk gels which had limited cell spreading. Further, it was observed that YAP nuclear intensity was two-fold higher for hMSCs cultured in the stiff (35 kPa) hydrogels compared to the softer hydrogels. Normally, hMSCs are observed to have decrease YAP nuclear staining on stiff hydrogels but in this 3D environment, the effects appear to be reversed. This can be attributed to differences in cytoskeletal tension in 3D matrices compared to 2D scaffolds. In two dimensional environments, YAP and TAZ expression levels are decreased on softer substrates where there is low cellular tension and reduced stress fiber formation (Dupont et al. 2011). Soft 3D environments, however, lead to reactive stiffening of fibroblasts and mechanically stressed cellular responses, such as increased production of ECM proteins. Rigid microstructures within this soft environment can act as anchors to facilitate appropriate generation of cellular tension and reduces production of YAP/TAZ compared to conditions without domains for cell attachment. This highlights the importance of mechanical microdomains within a 3D scaffold to act as anchors for cell attachment and alleviate aberrant cellular tension to decrease stress-induced mechanotransductive pathways.
While the above study illustrates the benefits of providing microdomains of increased stiffness to promote cell engraftment, others have utilized microstructural cues to directly modulate cellular phenotype and prevent pathological conditions. The application of microstructures to provide instructive signals and modulate cell behavior is highlighted in early studies by Norman et al. (2008). Anisotropic microscale hydrogels, termed microrods, were fabricated via photolithography and used to recapitulate the complex 3D patterning and regional architecture of native tissues (Fig. 4a). These microrods measured 15 μm × 15 μm × 100 μm to approximate the size of the cardiac muscle cell. This anisotropy has also been demonstrated to inhibit phagocytosis by macrophages, a desired trait for therapeutic applications (Champion et al. 2007; Champion and Mitragotri 2006, 2009). It was demonstrated that incorporation of these microrods into a 3D construct provided microdomains of high stiffness facilitated fibroblast attachment. Upon attaching to these microrods, the fibroblasts lost their dendritic morphology that was common to fibroblasts in 3D matrices and took on a morphology that was similar to 2D substrates, with stress fibers forming between the focal adhesions on the microrods. This intimate interaction stunted proliferation of fibroblasts and suggested potential applications for fibrosis.
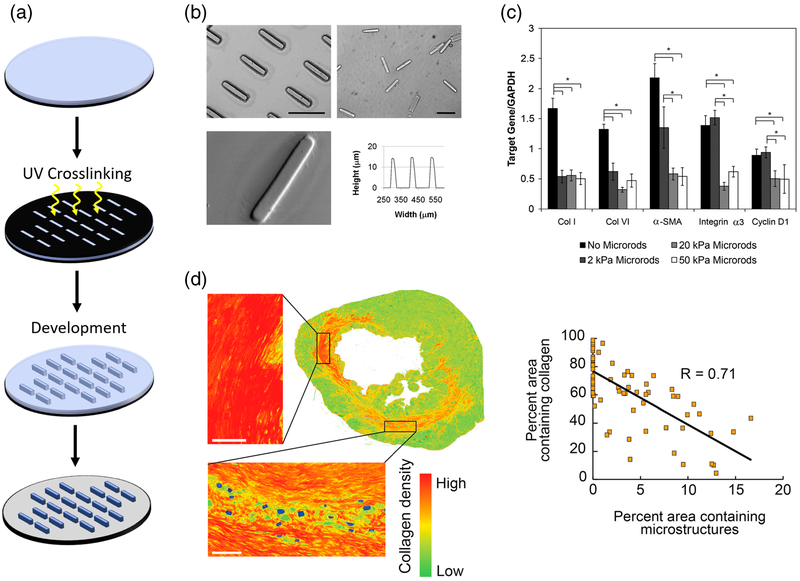
Fig. 4. Discrete microstructural cues attenuate chronic cardiac fibrosis.
a Anisotropic micron-scale hydrogels, termed “microrods” can be reproducibly fabricated by UV photolithography (Le et al. 2018). b Microrod dimensions can be controlled and are uniform across the wafer (scale bars = 100 μm) (Ayala et al. 2010). c PEGDMA-based microrods of different stiffness have varied response on extracellular matrix synthesis and gene expression in 3D gels (Ayala et al. 2010). d Collagen production is reduced proximal to microstructures in infarcted rat hearts, with increasing numbers of microstructures reducing total collagen area (scale bar = 100 μm) (Pinney et al. 2014a). Figures reproduced with permission
Ayala et al extended this work, utilizing free poly(ethylene glycol) dimethacrylate (PEGDMA) based microrods in 3D culture, to attenuate fibroblast proliferation and inhibit expression of key myofibroblast markers implicated in cardiac fibrosis (Fig. 4b–c) (Ayala et al. 2010). Similar to the microrods generated by Norman et al, these PEGDMA microrods are reproducibly fabricated on silicon wafers via a scalable photolithography approach and can be easily removed from the wafer and stored until use (Norman et al. 2008). When added to fibroblast cell culture in a 3D Matrigel scaffold, fibroblasts interacted directly with the microrods, stretching along individual microrods as well as across multiple structures. Similarly, this interaction led to reduced proliferation of fibro-blast cells. Key genes associated with the fibroblast to myofibroblast transformation were examined in the presence of microrods of varying Young’s moduli. Cells cultured with microrods that were at least 20 kPa had significantly reduced expression of collagen I, collagen VI and αSMA, genes known to be upregulated in fibrosis (Fig. 4c). Collagen I is the primary form of fibrillar collagen that makes up the bulk of scar tissue and is responsible for pathological stiffening of tissues. Expression of αSMA in fibroblast is a key marker for the transformation into the contractile myofibroblast phenotype and has been shown to be influenced by the mechanical microenvironment (Hinz 2007, 2010; Santiago et al. 2010). These effects were also found to be dependent on contractility, as treatment of the cells with MLCK or ROCK inhibitors (ML7 and Y27632, respectively) abolished the observed effects. This is in agreement with previous studies illustrating that contractile force generation is required for the cells to interact with microtopographical cues (Boateng et al. 2003; Curtis et al. 2010; Patel et al. 2010).
The potential for these PEGDMA microrods to be used as a therapeutic strategy in vivo was investigated using a rat model of myocardial infarction (Pinney et al. 2014a). As previously mentioned, the acute tissue damage arising from a MI culminates in the formation of a dense avascular scar that obstructs tissue regeneration, both intrinsic healing mechanisms and exogenous interventions (Frangogiannis 2015; Li et al. 2018; Segura et al. 2014; Talman and Ruskoaho 2016). The ability to attenuate this runaway fibrotic response would promote the survival of native and transplanted cell populations, making way for healthy tissue repair. Pinney et al probed the effect of microstructure administration on matrix remodeling by fibroblasts, namely matrix metalloproteinase 2 (MMP-2) production and transforming growth factor β (TGFβ) signaling (Pinney et al. 2014a). MMP-2 is upregulated within the first few days after MI and is responsible for degradation of the ECM to make way for macrophages to clear out cellular debris (Chen et al. 2005). This ECM degradation releases latent TGFβ that triggers myofibroblast transformation and overproduction of ECM components and pathological tissue repair (Frangogiannis 2012, 2014; Prabhu and Frangogiannis 2016).
When injected into the infarct zone of the myocardium via transthoracic injection, rats treated with PEGDMA microrods had significantly reduced deterioration of cardiac function compared to saline controls (Pinney et al. 2014a). The observed functional improvements appeared to be dose dependent, with increased numbers of delivered microstructures resulting in better cardiac output. These functional improvements were accompanied by reduced collagen deposition in the periphery of the microstructures, increased vascularization of the left ventricle and reduced TGFβ production (Fig. 4d). These outcomes are believed to be the result of introducing artificial anchors for cells to bind to in the softened, post-infarct matrix. This attachment alleviates mechanical stress-induced upregulation of TGFβ release and collagen deposition and promotes deposition of softer ECM components that make up the healthy uninjured microenvironment. Such events would allow for native tissue repair processes to occur and be amenable to cell replacement therapies.
It is important to note that the effects of these anisotropic microstructural cues vary from cell type to cell type. While proliferation appears to be stunted by fibroblastmicrostructure interactions, the opposite was observed for hMSCs (Collins et al. 2010a). Similar to studies by Xin et al, hMSCs spread out with fingerlike projections to attach to the PEGDMA microrods (Xin et al. 2018). The hMSCs adopted a more flattened and polarized morphology in the presence of microrods. This change in morphology was accompanied by increased proliferation, leading to larger cluster sizes in 3D culture. Additionally, hMSCs had reduced penchant for osteogenic differentiation when cultured with these microrods, as depicted by reduced bone morphogenic protein 6 (BMP-6) and Col1A1 gene expression. It is unclear whether the topographical interactions or paracrine signaling due to larger clusters are responsible for this change. The many possible mechanisms highlight that topographical cues can affect cells in ways that have yet to be elucidated.
5. Synergistic effects of biochemical signaling and microtopography
The discussion of topographical signaling thus far has focused on the use of bio-inert micromaterials, enabling isolation of biophysical effects alone. While a powerful and instructive mechanism, it is crucial to recognize the complex interplay of biophysical cues and biochemical factors, both soluble and immobilized, that are present in the native ECM contribute to cell function (Adams and Watt 1993; Frantz et al. 2010; Hynes 2009). Among the major components of the ECM is hyaluronic acid (HA), a widely distributed glycosaminoglycan that plays a wide array of roles in inflammation and wound healing (Chen and Abatangelo 1999; Gao et al. 2010; Litwiniuk et al. 2016; Petrey and de la Motte 2014). Many cell types express cell surface receptors, such as CD44 and receptor for hyaluronan-mediated motility (RHAMM), that can directly bind with the HA backbone and undergo changes in cell proliferation and migration (Chen and Abatangelo 1999; Jiang et al. 2007; Litwiniuk et al. 2016; Petrey and de la Motte 2014; Toole 2004; Turley et al. 2002). As such, HA has been used for tissue engineering applications (Burdick and Prestwich 2011; Ifkovits et al. 2010; Tous et al. 2011; Yoon et al. 2009). Sideris et al utilized microfluidics devices to fabricate injectable HA microparticles that can be annealed into a microporous 3D scaffold in the presence of cells with limited cytotoxicity (Sideris et al. 2016). These scaffolds supported the attachment and growth of human dermal fibroblasts, which eventually adopted a spread morphology in only 2 days. HA microparticles with guest-host motifs on the particle surface were developed to generate shear-thinning granular hydrogels that can be injected into tissues and immediately set (Mealy et al. 2018). MMP degradable motifs were incorporated into these hydrogel materials and demonstrated to degrade in 2 weeks when injected into the infarcted myocardium, due to upregulated expression of MMP relative to healthy controls.
Le et al demonstrated the fabrication of discrete HA-based microstructures for direct reprogramming of fibroblasts (Le et al. 2018). Studies have demonstrated the interplay of CD44 signaling with integrin-mediated mechanotransduction, suggesting improved mechanosensitivity on HA substrates compared to bioinert substrates (Chopra et al. 2012, 2014). Indeed, the interactions of fibroblasts with HA microrods appeared to be more intimate than bio-inert PEGDMA microrods, resulting in more dramatic downregulation of fibrosis markers, col1A2 and αSMA, as well as MMP-2 and TGFβ signaling components. These phenotypic alterations in vitro were translated into functional outcomes in a rat model of MI. Rats treated with HA microrods were able to attenuate the loss in cardiac function and preserve myocardial wall structure compared to saline controls. Soluble HA was also administered as a material control and was able to demonstrate modest improvements as well. These effects are likely attributed to the anti-inflammatory effects of HA which can alleviate the extreme cellular responses after acute tissue injury. The combination of material and topographical cues is hypothesized to synergistically improve these outcomes. It should be noted that the number/volume of HA microrods administered was much less than PEGDMA microrods, which were previously shown to have a dose dependent effect. Together, these data suggest that the synergistic effects of biochemical signaling of HA with microtopographical cues are more potent than either effect alone.
An additional advantage of using hyaluronic acid is the inherent biodegradability of the material. HA is degraded by endogenous enzymatic mechanisms, releasing small oligosaccharides that have been implicated in promoting cell migration and growth (Cyphert et al. 2015; Gao et al. 2010; Jiang et al. 2007; Litwiniuk et al. 2016; Noble 2002). A key limitation of bio-inert materials is their longevity in tissues after injection. Complete repair of tissue necessitates the clearance of exogenous materials to make way for cell migration and growth to repopulate the damaged tissue. Both HA microrods and granular HA hydrogels were shown to degrade after a few weeks in vitro and in vivo (Le et al. 2018; Mealy et al. 2018).
Aside from microgel backbone, there is great promise in functionalizing the surfaces of these materials to present binding motifs that are capable of eliciting specific cell responses. Anisotropic PEG based microgels functionalized with RGD motifs encouraged cell attachment via integrin binding, increasing total cell contact area (Rose et al. 2018). Additionally, cells in contact with RGD-PEG microgels had strong stress fiber signals and more elongated cell nuclei, suggesting significant cytoskeletal forces. These forces are likely due to the microdomains of high local stiffness relative to the surrounding matrix. This was supported by studies indicating increased shuttling of YAP into the nucleus in the presence of microgels, with modified microgels having higher YAP localization to the nucleus. When aligned in one uniform direction, PEGDMA microgels that were modified with RGD were able to promote neuron alignment to a greater degree than unmodified PEGDMA microgels. The difference between modified and unmodified microgels was muted when cultured with chicken derived dorsal root ganglions. These cells are able to produce their own fibronectin and generate binding sites on the microstructures. This mechanism is consistent with observations that fibronectin can readily adsorb to hydrogels and facilitate binding of cells with otherwise non-adhesive materials (Chopra et al. 2014; Khademhosseini et al. 2006a).
This complex interplay between surface receptors and topography is not limited to traditionally mechanosensitive cell types. CD8+ T cells have also been shown to have increased activation and proliferation when presented with ellipsoidal, rather than spherical, antigen-presenting microparticles with equivalent antigen dose, co-localization and density (Sunshine et al. 2014). These effects are hypothesized to be a result of contact angle with the microparticles – when a T cell interacts with an isotropic microparticle, the membrane reorganizes and orients to maximize contact with the long axis of the micro-particle. This results in increased frequency and size of the T cell-microparticle contact compared to spherical particles. When injected into a B16 melanoma tumor model, the ellipsoidal antigen-presenting microparticles elicited reduced tumor size and improved survival as compared to spherical particles and T cells alone.
6. Discrete microstructures as drug delivery devices
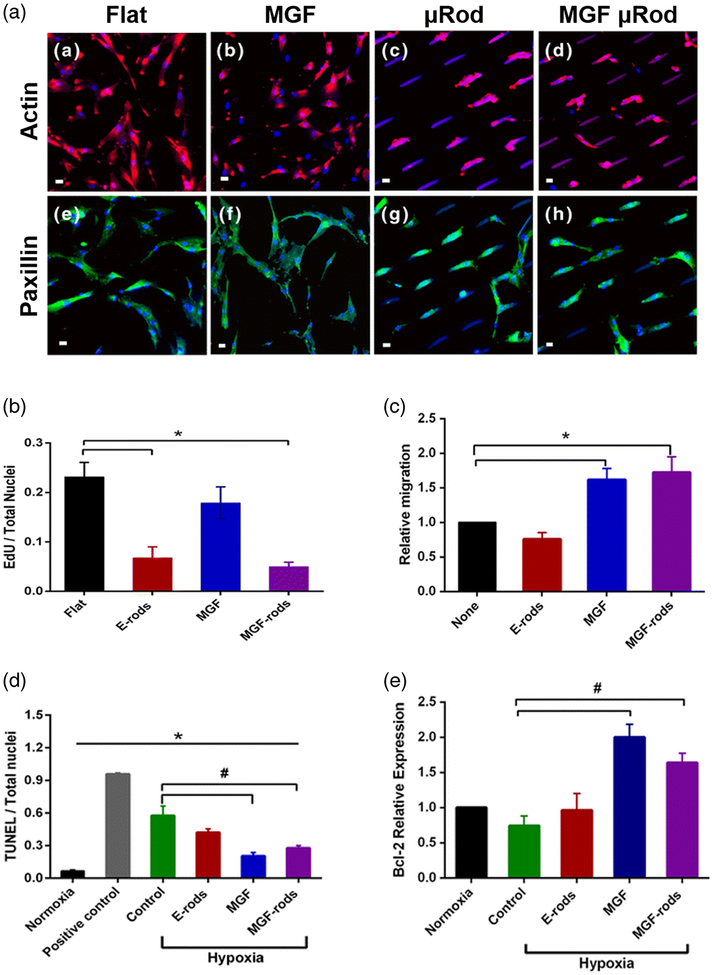
Proteins and small molecule drugs can be incorporated into the microparticle bodies for simultaneous delivery of therapeutics and microtopographical cues. These materials are thus able to modulate both the biomechanical and biochemical microenvironment to encourage healthy tissue repair. Doroudian et al described loading of PEGDMA microrods with mechano-growth factor (MGF) peptide to reduce cell apoptosis after myocardial infarction (Fig. 5) (Doroudian et al. 2014). Native MGF has been shown to block the apoptosis of injured myocytes as well as attract stem cell populations (Ates et al. 2007; Carpenter et al. 2008; Collins et al. 2010b; Mills et al. 2007; Musaro et al. 2004). MGF-peptide was eluted from PEGDMA microrods over the course of 2 weeks and maintained their biological activity, as indicated by reduced TUNEL staining and increased Bcl-2 expression (Fig. 5c–e) (Doroudian et al. 2014). These biochemical effects were maintained and independent from the mechanical effects exerted by topography, which remodeled hMSC morphology and stunted hMSC growth (Fig. 5a, b). Interestingly, this was in contrast to previous studies with hMSCs and PEGDMA microrods, likely due to differences in cytoskeletal tension that is generated in 2D compared to 3D culture (Collins et al. 2010a). Direct intracardiac injection of these MGF-peptide loaded microrods into a rat model of MI resulted in improved cardiac function and long-term survival as compared to saline injections and empty microrod injections (Peña et al. 2015).
Fig. 5. MGF-peptide loaded microrods exert biophysical effects of topography and biochemical effects of peptide administration on hMSCs and myocytes (Doroudian et al. 2014).
a Microrods remodel hMSC morphology, the actin cytoskeleton and focal adhesions while MGF administration has no effect (red = actin, green = paxillin, blue = nuclei and microrods, scale bar = 20 μm). b Microrods blunt the proliferation of hMSCs through mechanical signals and c increase migration via eluted MGF, which remains bioactive throughout microrod fabrication. Eluted MGF protected NRVM from apoptosis induced by hypoxia, as assessed by d TUNEL and e Bcl-2 expression. Figures reproduced with permission
Loading of a drug into microrods may also be useful for wound healing applications. PEGDMA microrods were loaded with high concentrations of neomycin, a small molecule drug capable promoting cell migration in vitro through modulation of lipid signaling, and delivery was maintained for over 12 h. When cultured with primary rat primary fibroblasts, the drug released to the media significantly increased the migration velocity of cells into the gap in a wound closure assay (Mkrtschjan et al. 2018a). Hence, development of these neomycin microrod devices for a therapeutic application might improve wound healing.
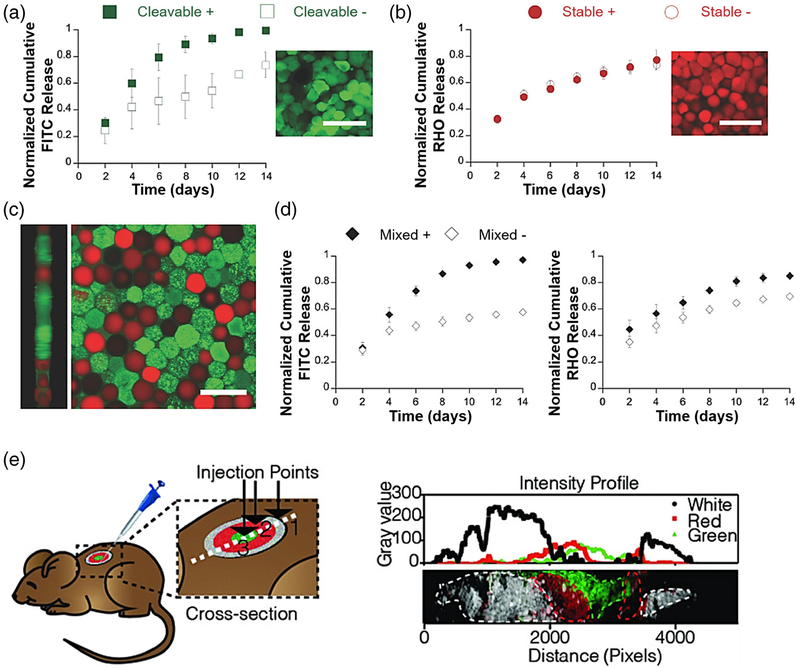
As discrete hydrogel materials, microstructures can also be independently fabricated and loaded with drug. It is possible to combine multiple types of drug-loaded microstructures that exhibit different release profiles to specifically address a variety of disease states (Fig. 6a–d) (Mealy et al. 2018). Further, the spatial distribution of multiple microstructure formulations in tissues can be controlled in order to recapitulate the biochemical gradients observed in native tissues (Fig. 6e) (Darling et al. 2018; Mealy et al. 2018). These capabilities are otherwise impossible using bulk hydrogel injections which are homogenous and are difficult to tune for multidrug release. In sum, discrete hydrogel materials are a versatile technology that can be used for therapeutic applications where tight spatiotemporal control of biochemical gradients is necessary for healthy tissue repair.
Fig. 6. Drug elution and spatial distribution of injected microparticles can be tuned for therapeutic applications in vivo.
a, b FITC-Albumin and rhodamine-dextran release from hydrogel microparticles can be tuned by inclusion of cleavable linkers (Mealy et al. 2018). c, d Microparticles can be combined and simultaneously elute two drugs with differential release kinetics. e Spatial distribution of three types of microparticles can be controlled during injection (Darling et al. 2018). Figures reproduced with permission
7. Remote control of injectable topographical cues
The random orientation of discrete microstructural cues after injection makes these systems unpredictable directors of cell behavior. Several groups have attempted to address this by incorporating magnetic nanoparticles into the microstructure scaffold. Utilizing anisotropic materials, application of an external magnetic field allows for controlled alignment of the microstructures. This is particularly beneficial for tissues in which proper alignment is crucial for organ function, such as in neural or cardiac spaces. Pinney et al demonstrated the successful alignment of PEGDMA based microstructures that were loaded with PEGDMA-coated superparamagnetic iron oxide nanoparticles (SPIONs) (Pinney et al. 2014b). Covalent attachment of SPIONs to the microstructure matrix allowed for stronger responses to an external magnetic field. Through soft lithography, Rose et al were able to fabricate anisotropic PEG-based microstructures that could also be aligned via an external magnetic field (Rose et al. 2018; Rose et al. 2017). These materials oriented the growth of fibroblasts and dorsal root ganglions via contact guidance. Additionally, these interactions induced the shuttling of YAP into the nucleus. Further conjugation with adhesive ligands did not result in improved neurite growth and alignment, confirming mechanosensory mechanisms as the dominant effector.
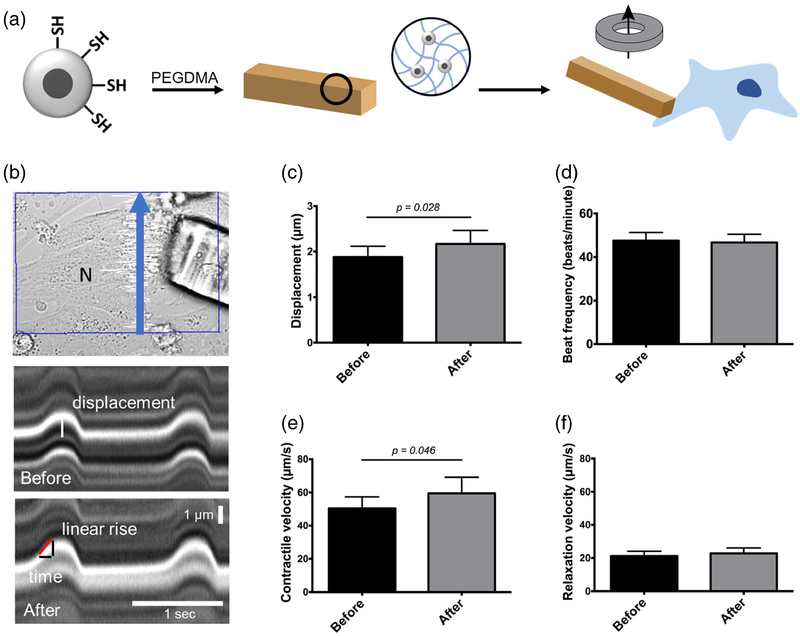
The ability to remotely control these topographical cues also have important implications for tissue engineering. Work in our lab has utilized SPION-loaded microrods as force actuators to study the contractile behavior of living cardiomyocytes following acute loading. This application of load was compatible with live cell imaging techniques. Briefly, SPIONs were synthesized via precipitation of iron salts and surface modified to add an additional silica-thiol coating for improved particle stability in solution. The addition of thiol functional groups on the silica shell surface provides reactive handles that enabled covalently crosslinking of SPIONs to a PEGDMA polymer matrix via thiolene-click chemistry (Fig. 7a). Magnetic microstructures, named micromagnets, were then fabricated using previously described photolithographic techniques.
Fig. 7. Magnetic nanoparticle-loaded microstructures enable remote manipulation of cardiomyocytes to study myocyte mechanics.
a Incorporation of silica thiol-capped superparamagnetic iron oxide nanoparticles (SPIONs) into microstructures generates micromagnets that enable remote manipulation of cells upon focal-adhesion attachment. b Line scan kymographs before and after addition of magnet to provide load. Contractile velocities are obtained by measuring displacement and time of linear portion of contraction or relaxation and taking the slope. c Contractile displacement of myocytes increases with load, p < 0.05. d Beat frequency is unchanged following load from micromagnets. e Contractile velocity increases with load, p < 0.05. f Relaxation velocity is unchanged following load from SPION micromagnets
Once the cells were attached, line scans were taken to measure beating before and after loading cardiomyocytes (CM) through the micromagnets (Fig. 7b). Kymographs showed an increase in contractile displacement, as well as permitting measurements for the contractile rising and relaxation falling phases. Changes in contractile metrics due to micromagnet loading was studied in 30 cells. As determined by an increase in contractile displacement, success rate for eliciting a response through micromagnet loading was approximately 66.6% (20 out of 30 cells), though all experiments were included in the analysis. Contractile displacement was significantly different in cells following loading, increasing from 1.88 μm to 2.17 μm (p < 0.05) (Fig. 7c). Additionally, a change in contractile velocity was observed between the two conditions, increasing from 50.47 μm/s to 59.48 μm/s following loading (p < 0.05) (Fig. 7e).
CMs interacted directly with the micromagnets such that simple placement of an external neodymium magnet allowed accurate measurement of transverse loading on CM contractility using phase microscopy. The focal adhesions formed between the microrods and the CMs were strong enough to load and displace CMs when the external magnet was introduced. The displacement resulted in significant changes to the rate of rise in tension of the CMs. With this experimental arrangement, the microrods provide load mainly in a transverse direction across the myocyte. Changing the position of the external magnet to the side of the culture dish would pull the SPION microrods along the longitudinal axis of the CM that could allow for sarcomere length dependent studies at the single cell level, potentially recapitulating the Frank-Starling law of the heart (Katz 2002; de Tombe and ter Keurs 2016).
To date, the ability to study the effect of mechanical load on living single cells has been limited by the need for high resolution spatial and temporal optical imaging in a realistic myocyte configuration. Attempts have been made using magnetically actuated micropost surfaces to provide forces along the underlying surface of a cell, but these do not mimic the type of three-dimensional strain experienced by cells in vivo (Bidan et al. 2018; Sniadecki et al. 2008). Microgroove-aligned CMs were cyclically strained using the Flexcell device, but these cells could not be imaged while mechanically deformed (Motlagh et al. 2003; Senyo et al. 2007). Signals and remodeling were also studied after static strain or after bouts of exercise, but these were not live cell recordings (Li and Russell 2013; Lin et al. 2013; Mansour et al. 2004; Yu and Russell 2005). More recently, advances were made using a system that combines PDMS microgrooves with an optically ready uniaxial stretching device to capture sarcomerogenesis of a uniaxially stretched cell, a phenomenon that had previously been observed without the benefit of live imaging (Yang et al. 2016; Yu and Russell 2005).
A relatively new approach is to study cardiac mechanics through the use of engineered heart tissues. This approach can seed primary rodent myocytes or human induced pluripotent stem cell-derived myocytes onto protein-based scaffolds or decellularized tissues between two pillars (Schwan et al. 2016). This method is advantageous, as cells derived from patient with specific mutations can be studied in a tissue-like preparation. A similar method has been used recently while adjusting the afterload experienced by the spontaneously contracting construct in order to mature the tissue, highlighting the importance of the mechanical conditions used in cell and tissue culture (Leonard et al. 2018). The application of tissue engineering approaches is advancing rapidly both for replacement organs and for drug testing (Ronaldson-Bouchard and Vunjak-Novakovic 2018; Zhang et al. 2018).
Strain deforms the proteins at the cell membrane, which is known to trigger a variety of signaling pathways such as PKA and PLC and activates electrophysiological channels leading to the contractile changes observed (Pfeiffer et al. 2014). Stretch-activated channels include the L-type calcium channel, which results in an increased intracellular calcium. Troponin C (TnC) binds the calcium allowing the myosin ATPase to produce the force with the actin in the thin filament (Puglisi et al. 2014). The angiotensin II receptor type 1 is also activated by mechanical stimulation and could potentially play a role in altered contractile behavior (Yasuda et al. 2008). The incorporation of magnetic nanoparticles into micron-scale hydrogels creates a novel and unique device that can be used for remote manipulation of single cells for advanced cell and molecular physiological studies. Its use could be extended for studies of mechanotransduction and sarcomere assembly.
8. Conclusions and perspectives
The extracellular matrix provides tissues with mechanical support and bioactive signals for the maintenance of cell survival and function. Tissue injury results in disruption of the established biochemical and mechanical equilibrium in the ECM, requiring restoration of these properties to their original states for healthy tissue repair. Micron-scale hydrogels with tunable geometry and mechanical properties provide cells with anchor points that serve as replacements for binding sites present in the native ECM. While there are still unknown intricacies to the underlying mechanisms of these cell-microtopography interactions, there is a clear dependence on intracellular architecture and tension to elicit the reported effects described in this review. These mechanisms are consistent between multiple cell types and carry over from 2D to 3D environments. This understanding enhances our ability to control the behavior of both transplanted and endogenous cell types for improved survival and function. Recent work has highlighted the versatility of this technology – discrete topographical features can be loaded with drug or surface modified to target biochemical pathways in addition to their biophysical effects.
The significant interplay between mechanical and biochemical cues has been demonstrated but the details of these interactions are still largely elusive. More in-depth study of these synergies would be made possible by simultaneous physical manipulation of cells in vitro via micromagnets and treatment with growth factors or signaling ligands. Further, the optimal spatial and temporal distribution of these mechanical and biochemical effects has yet to be determined. When are topographical cues necessary for treating pathological conditions and what time-frame of growth factor release is necessary? The use of degradable materials for these approaches provides additional knobs to tune for fabrication of patient-and disease-specific microstructures. Research in the past two decades using bioengineered microtopography clearly shows that how cells “hang on tight” in three dimensions is a major driver of cell structure and function. However, more work needs to be done to bring this ever-growing knowledge of basic discovery into the practical realm for translation into useful clinical application.
Acknowledgements
This work was supported by the National Institutes of Health (P01-HL062426 and R01-HL137209).
Footnotes
Conflict of interest The authors declare that they have no conflict of interest.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Adams JC, Watt FM, Development 117, 1183 (1993) [DOI] [PubMed] [Google Scholar]
- Al-Haque S, Miklas JW, Feric N, Chiu LLY, Chen WLK, Simmons CA, Radisic M, Macromol. Biosci 12, 1342 (2012) [DOI] [PubMed] [Google Scholar]
- Annabi N, Nichol JW, Zhong X, Ji C, Koshy S, Khademhosseini A, Dehghani F, Tissue Eng. Part B Rev. 16, 371 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asazuma-Nakamura Y, Dai P, Harada Y, Jiang Y, Hamaoka K, Takamatsu T, Exp. Cell Res. 315, 1190 (2009) [DOI] [PubMed] [Google Scholar]
- Ates K, Yang SY, Orrell RW, Sinanan ACM, Simons P, Solomon A, Beech S, Goldspink G, Lewis MP, FEBS Lett. 581, 2727 (2007) [DOI] [PubMed] [Google Scholar]
- Ayala P, Lopez JI, Desai TA, Tissue Eng. Part A 16, 2519 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidan CM, Fratzl M, Coullomb A, Moreau P, Lombard AH, Wang I, Balland M, Boudou T, Dempsey NM, Devillers T, Dupont A, Sci. Rep 8, 1464 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biehl JK, Yamanaka S, Desai TA, Boheler KR, Russell B, Dev. Dyn 238, 1964 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boateng SY, Hartman TJ, Ahluwalia N, Vidula H, Desai TA, Russell B, Am. J. Physiol. Physiol 285, C171 (2003) [DOI] [PubMed] [Google Scholar]
- Broughton KM, Russell B, Biomech. Model. Mechanobiol 14, 589 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdick JA, Prestwich GD, Adv. Mater 23, H41 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter V, Matthews K, Devlin G, Stuart S, Jensen J, Conaglen J, Jeanplong F, Goldspink P, Yang S-Y, Goldspink G, Bass J, McMahon C, Hear. Lung Circ. 17, 33 (2008) [DOI] [PubMed] [Google Scholar]
- Cerchiari A, Garbe JC, Todhunter ME, Jee NY, Pinney JR, LaBarge MA, Desai TA, Gartner ZJ, Tissue Eng. Part C Methods 21, 541 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion JA, Mitragotri S, Proc. Natl. Acad. Sci 103, 4930 (2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion JA, Mitragotri S, Pharm. Res 26, 244 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion JA, Katare YK, Mitragotri S, J. Control. Release 121, 3 (2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WYJ, Abatangelo G, Wound Repair Regen. 7, 79 (1999) [DOI] [PubMed] [Google Scholar]
- Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE, Science 276, 1425 (1997) [DOI] [PubMed] [Google Scholar]
- Chen J, Tung C-H, Allport JR, Chen S, Weissleder R, Huang PL, Circulation 111, 1800 (2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra A, Lin V, McCollough A, Atzet S, Prestwich GD, Wechsler AS, Murray ME, Oake SA, Yasha Kresh J, Janmey PA, J. Biomech 45, 824 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra A, Murray ME, Byfield FJ, Mendez MG, Halleluyan R, Restle DJ, Raz-Ben Aroush D, Galie PA, Pogoda K, Bucki R, Marcinkiewicz C, Prestwich GD, Zarembinski TI, Chen CS, Puré E, Kresh JY, Janmey PA, Biomaterials 35, 71 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JM, Ayala P, Desai TA, Russell B, Small 6, 355 (2010a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JM, Goldspink PH, Russell B, J. Mol. Cell. Cardiol 49, 1042 (2010b) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese B, Riehle MO, D’Amone S, Gigli G, J. Colloid Interface Sci. 394, 582 (2013) [DOI] [PubMed] [Google Scholar]
- Cukierman E, Pankov R, Stevens DR, Yamada KM, Science 294(80), 1708 (2001) [DOI] [PubMed] [Google Scholar]
- Curtis MW, Sharma S, Desai TA, Russell B, Biomed. Microdevices 12, 1073 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MW, Budyn E, Desai TA, Samarel AM, Russell B, Biomech. Model. Mechanobiol 12, 95 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyphert JM, Trempus CS, Garantziotis S, Int. J. Cell Biol. 2015, 1 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darling NJ, Sideris E, Hamada N, Carmichael ST, Segura T, Adv. Sci 5, 1801046 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Tombe PP, ter Keurs HEDJ, J. Mol. Cell. Cardiol 91, 148 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Tombe PP, Mateja RD, Tachampa K, Mou YA, Farman GP, Irving TC, J. Mol. Cell. Cardiol 48, 851 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch J, Motlagh D, Russell B, Desai TA, J. Biomed. Mater. Res 53, 267 (2000) [DOI] [PubMed] [Google Scholar]
- Dimatteo R, Darling NJ, Segura T, Adv. Drug Deliv. Rev 127, 167 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroudian G, Pinney J, Ayala P, Los T, Desai TA, Russell B, Biomed. Microdevices 16, 705 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downing TL, Soto J, Morez C, Houssin T, Fritz A, Yuan F, Chu J, Patel S, Schaffer DV, Li S, Nat. Mater 12, 1154 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S, Nature 474, 179 (2011) [DOI] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE, Cell 126, 677 (2006) [DOI] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA, PLoS Genet. 4, e1000039 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangogiannis NG, Circ. Res 110, 159 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangogiannis NG, Nat. Rev. Cardiol 11, 255 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangogiannis NG, In Compr. Physiol (John Wiley & Sons, Inc., Hoboken, 2015), pp. 1841–1875 [DOI] [PubMed] [Google Scholar]
- Frantz C, Stewart KM, Weaver VM, J. Cell Sci. 123, 4195 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Liu Y, He Y, Yang C, Wang Y, Shi X, Wei G, Matrix Biol. 29, 107 (2010) [DOI] [PubMed] [Google Scholar]
- Geiger B, Science 294(80), 1661 (2001) [DOI] [PubMed] [Google Scholar]
- Geiger B, Spatz JP, Bershadsky AD, Nat. Rev. Mol. Cell Biol. 10, 21 (2009) [DOI] [PubMed] [Google Scholar]
- Hinz B, J. Invest. Dermatol 127, 526 (2007) [DOI] [PubMed] [Google Scholar]
- Hinz B, J. Biomech 43, 146 (2010) [DOI] [PubMed] [Google Scholar]
- Hoshijima M, Am. J. Physiol. Circ. Physiol 290, H1313 (2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebsch N, Arany PR, Mao AS, Shvartsman D, Ali OA, Bencherif SA, Rivera-Feliciano J, Mooney DJ, Nat. Mater 9, 518 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO, Science 326(80), 1216 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ifkovits JL, Tous E, Minakawa M, Morita M, Robb JD, Koomalsingh KJ, Gorman JH, Gorman RC, a Burdick J, Proc. Natl. Acad. Sci 107, 11507 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingber DE, Annu. Rev. Physiol 59, 575 (1997) [DOI] [PubMed] [Google Scholar]
- Ingber DE, FASEB J. 20, 811 (2006) [DOI] [PubMed] [Google Scholar]
- Ingber DE, Wang N, Stamenović D, Rep. Prog. Phys 77, 046603 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaalouk DE, Lammerding J, Nat. Rev. Mol. Cell Biol. 10, 63 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Liang J, Noble PW, Annu. Rev. Cell Dev. Biol 23, 435 (2007) [DOI] [PubMed] [Google Scholar]
- Jiang Y, Lu S, Zeng Y, Tissue Eng J. Regen. Med 5, 402 (2011) [DOI] [PubMed] [Google Scholar]
- Katsumi A, Orr AW, Tzima E, Schwartz MA, J. Biol. Chem 279, 12001 (2004) [DOI] [PubMed] [Google Scholar]
- Katz AM, Circulation 106, 2986 (2002) [DOI] [PubMed] [Google Scholar]
- Khademhosseini A, Langer R, Biomaterials 28, 5087 (2007) [DOI] [PubMed] [Google Scholar]
- Khademhosseini A, Eng G, Yeh J, Fukuda J, Blumling J, Langer R, Burdick JA, J. Biomed. Mater. Res. Part A 79A, 522 (2006a) [DOI] [PubMed] [Google Scholar]
- Khademhosseini A, Langer R, Borenstein J, Vacanti JP, Proc. Natl. Acad. Sci 103, 2480 (2006b) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian KA, Bugarija B, Lahn BT, Mrksich M, Proc. Natl. Acad. Sci 107, 4872 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-H, Han K, Gupta K, Kwon KW, Suh K-Y, Levchenko A, Biomaterials 30, 5433 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le LV, Mohindra P, Fang Q, Sievers RE, Mkrtschjan MA, Solis C, Safranek CW, Russell B, Lee RJ, Desai TA, Biomaterials 169, 11 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leijten J, Seo J, Yue K, Trujillo-de Santiago G, Tamayol A, Ruiz-Esparza GU, Shin SR, Sharifi R, Noshadi I, Álvarez MM, Zhang YS, Khademhosseini A, Mater. Sci. Eng. R. Rep 119, 1 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard A, Bertero A, Powers JD, Beussman KM, Bhandari S, Regnier M, Murry CE, Sniadecki NJ, J. Mol. Cell. Cardiol 118, 147 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Russell B, Am. J. Physiol. Heart Circ. Physiol 305, H1614 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Tanhehco EJ, Russell B, Am. J. Physiol. Circ. Physiol 307, H1618 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhao Q, Kong W, Matrix Biol. 490, 68–69 (2018) [DOI] [PubMed] [Google Scholar]
- Lin Y-H, Li J, Swanson ER, Russell B, J. Appl. Physiol 114, 1603 (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey ML, Mann DL, Entman ML, Spinale FG, Ann. Med 35, 316 (2003) [DOI] [PubMed] [Google Scholar]
- Litwiniuk M, Krejner A, Speyrer MS, Gauto AR, Grzela T, Wounds 28, 78 (2016) [PubMed] [Google Scholar]
- Mansour H, de Tombe PP, Samarel AM, Russell B, Circ. Res 94, 642 (2004) [DOI] [PubMed] [Google Scholar]
- Martinac B, J. Cell Sci. 117, 2449 (2004) [DOI] [PubMed] [Google Scholar]
- Martino F, Perestrelo AR, Vinarský V, Pagliari S, Forte G, Front. Physiol 9, 1 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara LE, Burchmore R, Riehle MO, Herzyk P, Biggs MJP, Wilkinson CDW, Curtis ASG, Dalby MJ, Biomaterials 33, 2835 (2012) [DOI] [PubMed] [Google Scholar]
- Mealy JE, Chung JJ, Jeong H-H, Issadore D, Lee D, Atluri P, Burdick JA, Adv. Mater 30, 1705912 (2018) [DOI] [PubMed] [Google Scholar]
- Midwood KS, Williams LV, Schwarzbauer JE, Int. J. Biochem. Cell Biol. 36, 1031 (2004) [DOI] [PubMed] [Google Scholar]
- Mills P, Lafrenière J-F, Benabdallah BF, El Fahime EM, Tremblay J-P, Exp. Cell Res. 313, 527 (2007) [DOI] [PubMed] [Google Scholar]
- Mkrtschjan MA, Gaikwad SB, Kappenman KJ, Solís C, Dommaraju S, Le LV, Desai TA, Russell B, J. Cell. Physiol 233, 3672 (2018a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mkrtschjan MA, Solís C, Wondmagegn AY, Majithia J, Russell B, Cytoskeleton 75, 363 (2018b) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motlagh D, Hartman TJ, Desai TA, Russell B, J. Biomed. Mater. Res 67A, 148 (2003) [DOI] [PubMed] [Google Scholar]
- Musaro A, Giacinti C, Borsellino G, Dobrowolny G, Pelosi L, Cairns L, Ottolenghi S, Cossu G, Bernardi G, Battistini L, Molinaro M, Rosenthal N, Proc. Natl. Acad. Sci 101, 1206 (2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble PW, Matrix Biol. 21, 25 (2002) [DOI] [PubMed] [Google Scholar]
- Norman JJ, Collins JM, Sharma S, Russell B, Desai TA, Tissue Eng. Part A 14, 379 (2008) [DOI] [PubMed] [Google Scholar]
- Orr AW, Helmke BP, Blackman BR, Schwartz MA, Dev. Cell 10, 11 (2006) [DOI] [PubMed] [Google Scholar]
- Ovsianikov A, Mironov V, Stampfl J, Liska R, Expert Rev. Med. Devices 9, 613 (2012) [DOI] [PubMed] [Google Scholar]
- Pandey P, Hawkes W, Hu J, Megone WV, Gautrot J, Anilkumar N, Zhang M, Hirvonen L, Cox S, Ehler E, Hone J, Sheetz M, Iskratsch T, Dev. Cell 44, 326 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JT, Horwitz AR, Schwartz MA, Nat. Rev. Mol. Cell Biol. 11, 633 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel AA, Thakar RG, Chown M, Ayala P, Desai TA, Kumar S, Biomed. Microdevices 12, 287 (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña JR, Pinney JR, Ayala P, Desai TA, Goldspink PH, Biomaterials 46, 26 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrey AC, de la Motte CA, Front. Immunol 5, 1 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer ER, Tangney JR, Omens JH, McCulloch AD, J. Biomech. Eng 136, 021007 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinney JR, Du KT, Ayala P, Fang Q, Sievers RE, Chew P, Delrosario L, Lee RJ, Desai TA, Biomaterials 35, 8820 (2014a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinney JR, Melkus G, Cerchiari A, Hawkins J, Desai TA, ACS Appl. Mater. Interfaces 6, 14477 (2014b) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu SD, Frangogiannis NG, Circ. Res 119, 91 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puglisi JL, Goldspink PH, Gomes AV, Utter MS, Bers DM, Solaro RJ, Arch. Biochem. Biophys 50, 552–553 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puklin-Faucher E, Sheetz MP, J. Cell Sci. 122, 179 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo C, Schwartz M, Glia 62, 1895 (2014) [DOI] [PubMed] [Google Scholar]
- Ronaldson-Bouchard K, Vunjak-Novakovic G, Cell Stem Cell (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rørth P, Dev. Cell 20, 9 (2011) [DOI] [PubMed] [Google Scholar]
- Rose JC, Cámara-Torres M, Rahimi K, Köhler J, Möller M, De Laporte L, Nano Lett. 17, 3782 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JC, Gehlen DB, Haraszti T, Köhler J, Licht CJ, De Laporte L, Biomaterials 163, 128 (2018) [DOI] [PubMed] [Google Scholar]
- Santiago J-J, Dangerfield AL, Rattan SG, Bathe KL, Cunnington RH, Raizman JE, Bedosky KM, Freed DH, Kardami E, Dixon IMC, Dev. Dyn 239, 1573 (2010) [DOI] [PubMed] [Google Scholar]
- Schwan J, Kwaczala AT, Ryan TJ, Bartulos O, Ren Y, Sewanan LR, Morris AH, Jacoby DL, Qyang Y, Campbell SG, Sci. Rep 6, 32068 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura AM, Frazier OH, Buja LM, Heart Fail. Rev 19, 173 (2014) [DOI] [PubMed] [Google Scholar]
- Senyo SE, Koshman YE, Russell B, FEBS Lett. 581, 4241 (2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sideris E, Griffin DR, Ding Y, Li S, Weaver WM, Di Carlo D, Hsiai T, Segura T, ACS Biomater. Sci. Eng 2, 2034 (2016) [DOI] [PubMed] [Google Scholar]
- Slaughter BV, Khurshid SS, Fisher OZ, Khademhosseini A, Peppas NA, Adv. Mater 21, 3307 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniadecki NJ, Lamb CM, Liu Y, Chen CS, Reich DH, Rev. Sci. Instrum 79, 044302 (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunshine JC, Perica K, Schneck JP, Green JJ, Biomaterials 35, 269 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talman V, Ruskoaho H, Cell Tissue Res. 365, 563 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira AI, J. Cell Sci. 116, 1881 (2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakar RG, Chown MG, Patel A, Peng L, Kumar S, Desai TA, Small 4, 1416 (2008) [DOI] [PubMed] [Google Scholar]
- Thomas CH, Collier JH, Sfeir CS, Healy KE, Proc. Natl. Acad. Sci 99, 1972 (2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbitt MW, Anseth KS, Biotechnol. Bioeng 103, 655 (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toole BP, Nat. Rev. Cancer 4, 528 (2004) [DOI] [PubMed] [Google Scholar]
- Tous E, Ifkovits JL, Koomalsingh KJ, Shuto T, Soeda T, Kondo N, Gorman JH, Gorman RC, a Burdick J, Biomacromolecules 12, 4127 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turley EA, Noble PW, Bourguignon LYW, J. Biol. Chem 277, 4589 (2002) [DOI] [PubMed] [Google Scholar]
- Versaevel M, Grevesse T, Gabriele S, Nat. Commun 3, 671 (2012) [DOI] [PubMed] [Google Scholar]
- Wang N, Tytell JD, Ingber DE, Nat. Rev. Mol. Cell Biol. 10, 75 (2009) [DOI] [PubMed] [Google Scholar]
- Westhoff MA, Serrels B, Fincham VJ, Frame MC, Carragher NO, Mol. Cell. Biol 24, 8113 (2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak MA, Modzelewska K, Kwong L, Keely PJ, Biochim. Biophys. Acta, Mol. Cell Res. 1692, 103 (2004) [DOI] [PubMed] [Google Scholar]
- Xin S, Wyman OM, Alge DL, Adv. Healthc. Mater 7, e1800160 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa F, Sugiura S, Kanamori T, Regen. Ther 3, 45 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Schmidt LP, Wang Z, Yang X, Shao Y, Borg TK, Markwald R, Runyan R, Gao BZ, Sci. Rep 6, 20674 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda N, Akazawa H, Qin Y, Zou Y, Komuro I, Naunyn Schmiedeberg’s Arch. Pharmacol 377, 393 (2008) [DOI] [PubMed] [Google Scholar]
- Yoon SJ, Fang YH, Lim CH, Kim BS, Son HS, Park Y, Sun K, J. Biomed. Mater. Res. Part B Appl. Biomater 91B, 163 (2009) [DOI] [PubMed] [Google Scholar]
- Yu J-G, Russell B, J. Histochem. Cytochem 53, 839 (2005) [DOI] [PubMed] [Google Scholar]
- Zhang L, Mao YS, Janmey PA, and Yin HL, In Subcell. Biochem pp. 177–215 (2012) [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhu W, Radisic M, Vunjak-Novakovic G, Circ. Res (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Matsumura Y, Wagner WR, Biomaterials 129, 37 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler WH, Gingras AR, Critchley DR, Emsley J, Biochem. Soc. Trans 36, 235 (2008) [DOI] [PubMed] [Google Scholar]
- Zorlutuna P, Annabi N, Camci-Unal G, Nikkhah M, Cha JM, Nichol JW, Manbachi A, Bae H, Chen S, Khademhosseini A, Adv. Mater 24, 1782 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]