Abstract
Bis-indole derivatives including 1,1-bis(3’-indolyl)-1-(4-chlorophenyl)methane (DIM-C-pPhCl) and substituted quinolines such as chloroquine (CQ) and amodiaquine (AQ) are nuclear receptor 4A2 (NR4A2, Nurr1) ligands and they exhibit anti-inflammatory activities in mouse and rat models of Parkinson’s disease, respectively. However, computational modeling demonstrates that the quinoline derivatives interact with the ligand binding domain whereas the bis-indoles preferentially interact with a C-terminal cofactor binding site of NR4A2. In this study, the effects of DIM-C-pPhCl and related analogs were compared to CQ/AQ as inducers of NR4A2-responsive genes including vasoactive intestinal peptide (VIP), osteopontin (OPN), proopiomelanocortin (POMC) and neuropilin 1 (NRP1) in Panc1 and Panc28 pancreatic cancer cells. The results demonstrate that, among the bis-indole analogs, their relative potencies as inducers were structure-gene and cell context dependent. In contrast, CQ and AQ were significantly less potent than the bis-indole derivatives and, for some of the NR4A2-regulated genes, CQ and AQ were inactive as inducers. These results demonstrate that, although bis-indole and quinoline derivatives have been characterized as activators of NR4A2-dependent gene expression, these two classes of compounds exhibit different activities, indicating that they are selective NR4A2 modulators.
Keywords: NR4A2 Ligand, Bis-indole-derived, Quinoline-derived
Graphical Abstract
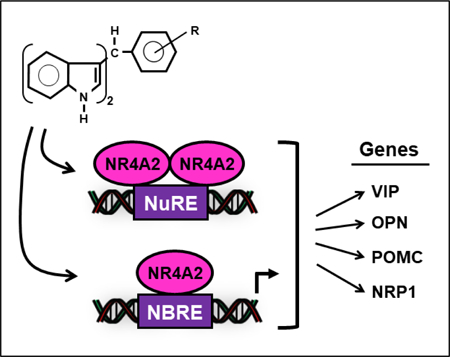
Bis-indole derived 1,1-bis(3’-indoly)-1-(p-chloro-phenyl)methane (DIM-C-pPhCl) and related analogs induce expression of nuclear receptor 4A2 (NR4A2) – dependent vasoactive intestinal peptide (VIP), osteopontin (OPN), proopiomelanocortin (POMC) and neuropilin1 (NRP1) gene expression in pancreatic cancer cells. In contrast, chloroquine (CO) and amodiaquine (AQ) which have also been characterized as NR4A2 ligands were less active or inactive as inducers of the same genes suggesting that the bis-indole analogs, CQ and AQ are selective NR4A2 modulators.
1. INTRODUCTION
Nuclear receptor 4A2 (NR4A2, Nurr1) is an orphan receptor and a member of the NR4A receptor family which also includes NR4A1 (Nur77, TR3) and NR4A3 (Nor1). NR4A receptors are immediate early genes inducible by multiple stressors and these receptors play important roles in maintaining cellular homeostasis and in disease. There is accumulating evidence for the importance of NR4A receptors in metabolic, cardiovascular, neuronal and immune functions as well as their involvement in inflammatory diseases and cancer.(Pearen & Muscat, 2010; Safe et al., 2014; Wang et al., 2014; Zhou et al., 2014) The three NR4A receptors share significant sequence similarities in their ligand binding domains (LBDs) and DNA binding domains (DBDs) while their N-terminal domains (A/B domains) containing activation function 1 (AF-1) are highly divergent.(Giguere, 1999; Wansa et al., 2002; Wansa et al., 2003) NR4A receptors were initially defined as nerve growth factor-induced clone B (NGFI-B) receptors that bind as monomers to an NGFI-B response element (NBRE) (Paulsen et al., 1995; Wang et al., 2003; Wilson et al., 1992; Wilson et al., 1991; Wilson et al., 1993) and as homo- or heterodimers to a Nur-responsive element (NurRE) which has been characterized from the proopiomelanocortin gene promoter.(Maira et al., 1999; Philips et al., 1997) Both NR4A1 and NR4A2 can also form heterodimers with the retinoid X receptor (RXR) and bind to a DR5 element.(Perlmann & Jansson, 1995; Zetterstrom et al., 1996) Among the NR4A receptors, NR4A2 plays a unique role in neuronal development and mice deficient in this receptor exhibit deficits in dopaminergic neuron development.(Zetterstrom et al., 1997) Endogenous ligands for these orphan NR4A receptors have not been identified and crystal structure analysis of the ligand binding domain indicates that the binding pocket of NR4A2 is occupied by hydrophobic amino acid side chains and unlikely to accommodate a ligand.(Wang et al., 2003) Nevertheless, several studies have characterized different structural classes of synthetic NR4A2 ligands and these include benzimidazoles, isoxazolo-pyridinone, unsaturated fatty acids, substituted quinolines and substituted bis-indole-derived compounds.(de Vera et al., 2016; Dubois et al., 2006; Hammond et al., 2018; Hintermann et al., 2007; Kim et al., 2015; Li et al., 2012)
There is evidence that NR4A2 plays a role in Parkinson’s disease and the NR4A2-active 4-aminoquinoline derivatives, chloroquine (CQ) and amodiaquine (AQ), which are antimalarial compounds, were highly neuroprotective in a rat model of Parkinson’s disease.(Kim et al., 2015) Studies in our laboratory have demonstrated that the NR4A2 ligand 1,1-bis(3’-indolyl)-1-(4-chlorophenyl)methane (DIM-C-pPhCl or C-DIM12) also exhibits neuroprotective/anti-inflammatory activities in cell culture and mouse models of Parkinson’s disease.(De Miranda et al., 2013; De Miranda et al., 2015b; Hammond et al., 2018) For example, DIM-C-pPhCl stabilized the nuclear localization of NR4A2 in dopaminergic neurons in the substantia nigra in mice lesioned with the parkinsonian neurotoxicant, 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP).(De Miranda et al., 2015b) In BV-2 microglial cells, treatment with DIM-C-pPhCl blocked NF-κB/p65 binding to its cis enhancer element in the 5’-flanking region of inducible nitric oxide synthase (Nos2) and stabilized chromatin binding of the transcriptional co-repressor proteins, NCoR2 and CoREST.(De Miranda et al., 2015a) Given that substituted bis-indole-derived compounds can interact with and modulate the activity of NR4A2 in multiple cell types, we sought to identify structural elements of these compounds mediating these specific activities. In this report, we have compared the transactivation activity of the DIM-C-pPhCl and its Bromo analog (Li et al., 2012) and several newly developed analogs with CQ and AQ and have observed different response-specific potency and efficacy, indicating that the bis-indole and substituted quinoline analogs are selective NR4A2 modulators.
2. MATERIALS AND METHODS
2.1. Cell culture
Panc1 and Panc28 pancreatic cancer cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) nutrient mixture with Ham’s F-12 (DMEM/F-12; Sigma-Aldrich, St. Louis, MO) supplemented with 5% fetal bovine serum (FBS; Sigma-Aldrich) and 10 mL/L 100× antibiotic-antimycotic solution (Gibco, Thermo Fisher Scientific, Waltham, MA). Cells were maintained at 37°C in the presence of 5% CO2.
2.2. Chemical synthesis
The bis-indole analogs were synthesized in this laboratory by condensation of indole or 1-methylindole with corresponding benzaldehyde derivatives (2:1 indole/benzaldehyde) at 80°C in a pH 5 buffer. Compounds were crystallized from benzene/hexane (≥1 time) and purities were >97% as determined by gas chromatography or gas chromatography-mass spectrometry. Indole and benzaldehyde derivatives were purchased from Sigma-Aldrich. Over 100 bis-indole analogs were synthesized and screened and six analogs were identified as NR4A2 agonists, namely 1,1-bis(3’-indolyl)-1-(4-bromophenyl)methane (4-Br), 1,1-bis(3’-indolyl)-1-(4-chlorophenyl)methane (4-Cl), 1,1-bis(3’-indolyl)-1-(4-methylthiophenyl)methane (4-SCH3), 1,1-bis(3’-indolyl)-1-(3-trifluoromethyl-4-chlorophenyl)methane (3-CF3-4-Cl), 1,1-bis(3’-indolyl)-1-(2-hydroxy-4-bromophenyl)methane (2-OH-4-Br) and 1,1-bis[3’-(1’-methylindolyl)]-1-(4-hydroxyphenyl)methane (N-Me-4-OH). Indole and 1-methyl indole were purchased from Sigma-Aldrich; the starting materials for the 6 NR4A2 ligands included the following: 4-Br (indole plus 4-bromobenzaldehyde), 4-Cl (indole plus 4-chlorobenzaldehyde), 4-SCH3 (indole plus 4-thiomethylbenzaldehyde), 3-CF3-4-Cl (indole plus 4-chloro-3-trifluoromethylbenzaldehyde), 2-OH-4-Br (4-bromo-2-hydroxybenzaldehyde) and N-Me-4-OH (1-methylindole and 4-hydroxybenzaldehyde). The structures of the new analogs, 4-SCH3, 3-CF3-4-Cl, 2-OH-4-Br and NMe-4-OH were confirmed by 1H NMR spectra on a Bruker Avance Spectrometer (400 MHz) and by LC-MS on a SHIMADZU 2010 EV using methanol as solvent. With the exception of N-Me-4-OH (94%), all compounds were > 98% pure. The substituted benzaldehydes chloroquine and amodiaquine were purchased from Sigma-Aldrich. NMR spectra for the new compounds odentified in this study as NR4A2 activators include the following; 4-SCH3; 1H NMR (400 MHz, CDCI3):ᵟ 7.86 (s, 2H), 7.40 – 7.31 (m, 5H), 7.25 (s, 1H), 7.18 – 7.14 (m, 4H), 7.02 – 6.97 (m, 2H), 6.64 (d, J = 1.6 Hz, 2H), 5.84 (s, 1H), 2.45 (s, 3H); 3-CF3-4-Cl, 1H NMR (400 MHz, CDCI3):ᵟ 7.86 (s, 2H), 7.70 (d, J = 1.4 Hz, 1H), 7.40 – 7.29 (m, 6H), 7.22 – 7.14 (m, 2H), 7.05 – 6.99 (m, 2H), 6.59 (d, J = 1.7 Hz, 2H), 5.89 (s, 1H), 2.45 (s, 3H); 2-OH-4-Br, 1H NMR (400 MHz, CDCI3):ᵟ 7.89 (s, 2H), 7.37 – 7.32 (m, 3H), 7.19 (t, J = 7.6 Hz, 2H), 7.05 – 6.95 (m, 5H), 6.69 (d, J = 1.8 Hz, 2H), 5.92 (s, 1H), 5.48 (s, 1H); N-Me-4-OH, 1H NMR (400 MHz, CDCI3):ᵟ 7.37 (d, J = 7.9 Hz, 2H), 7.27 (d, J = 8.2 Hz, 2H), 7.22 – 7.16 (m, 4H), 7.02 – 6.94 (m, 2H), 6.76 – 6.68 (m, 2H), 6.51 (s, 2H), 5.81 (s, 1H), 4.53 (s, 1H), 3.67 (s, 6H).
2.3. Plasmids
The GAL4-NR4A2 chimera was constructed by inserting full length human NR4A2 DNA (amino acids 1 – 598) into the BamHI/HindIII site of pM vector (Clontech Laboratories) and the UAS-Luc plasmid contains five tandem GAL4 response elements inserted into pGL3 luciferase reporter vector (Promega, Madison, WI).
2.4. Luciferase assay
Cells were plated on 12-well plates at 8×104 (Panc1) or 12×104 (Panc28) per well in DMEM/F12 supplemented with 2.5% charcoal-stripped FBS. After overnight attachment and growth, 400 ng UAS-Luc and 40 ng GAL4-NR4A2 were cotransfected into each well using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. After 6 hr of transfection, cells were treated with the above medium containing either solvent or compound for 18 hr. Cells were then lysed and cell extract was processed for chemiluminescence qualification. Luciferase activity values were normalized against corresponding protein concentration values determined by Bradford assay. The solvent (DMSO) used in the experiments was always below 0.15% by volume in the medium.
2.5. Real-time quantitative PCR
Cells were plated on 6-well plates at 1.8×105 (Panc1) or 2.5×105 (Panc28) per well in DMEM/F12 supplemented with 2.5% charcoal-stripped FBS. After overnight attachment and growth, cells were treated with medium containing either solvent or compound for 24 hr. Total mRNA was extracted using Qiagen’s protocol and quantitative PCR was carried out using Bio-Rad’s SYBR Green reagent and thermal cycler. Gene expression (mRNA) levels were normalized and calibrated to the expression levels of three housekeeping genes including GAPDH, 18S ribosomal RNA and TATA-binding protein (TBP). The sequences of the primers used for qPCR were: VIP sense 5’- TCA GGT TCA TTT GCT CCC TC, antisense 5’- TCT TCT CAC AGA CTT CGG CA; OPN sense 5’- TTG CAG TGA TTT GCT TTT GC, antisense 5’- GCC ACA GCA TCT GGG TAT TT; POMC sense 5’- AAG ATG CCG AGA TCG TGC TG, antisense 5’- ATG ACG TAC TTC CGG GGG TTC; NRP1 sense 5’- AAG GTT TCT CAG CAA ACT ACA GTG, antisense 5’- GGG AAG AAG CTG TGA TCT GGT C.
2.6. Statistical analysis
Statistical significance of differences in luciferase activities and gene expression levels between treatment and control was analyzed using unpaired Student’s t-test and p-values of <0.01 were considered statistically significant. Results are expressed as mean ± standard deviation (SD) for at least three independent determinations (i.e. three biologically independent experiments) for each treatment group.
3. RESULTS
Both bis-indole derivatives (4-Cl and 4-Br) and substituted quinolines such as CQ and AQ have previously been characterized as NR4A2 ligands that exhibit anti-inflammatory effects in mouse (De Miranda et al., 2013; De Miranda et al., 2015b; Hammond et al., 2018) and rat (Kim et al., 2015) models of Parkinson’s disease and this study examines their transactivation activities in pancreatic cancer cells. Both Panc1 and Panc28 cells were used since previous studies show that they express NR4A2 and they are responsive to ligand-induced NR4A2-dependent gene and reporter gene expression.(Li et al., 2012) Results in Figure 1 illustrate the effects of DIM-C-pPhCl (4-Cl), DIM-C-pPhBr (4-Br) and four additional newly synthesized analogs, 1,1-bis(3’-indolyl)-1-(4-methylthiophenyl)methane (DIM-C-pPhSCH3; 4-SCH3), 1,1-bis(3’-indolyl)-1-(3-trifluoromethyl-4-chlorophenyl)methane (DIM-C-pPhCl-3-CF3; 3-CF3-4-Cl), 1,1-bis(3’-indolyl)-1-(2-hydroxy-4-bromophenyl)methane (DIM-C-pPhBr-2-OH; 2-OH-4-Br) and 1,1-bis[3’-(1’-methylindolyl)]-1-(4-hydroxyphenyl)methane (N-Me-DIM-C-pPhOH; N-Me-4-OH) (Fig. 1A), on luciferase activity in Panc1 (Fig. 1B) and Panc28 (Fig. 1C) cells. The newly synthesized analogs were identified in a screening of over 100 structurally – related bis-indole derivatives and these compounds were selected for further study since they activated NR4A2 but had minimal effects on NR4A3 – and NR4A1 – dependent transactivation using GAL4-receptor/UAS-luc screening assays. In this assay, cells were transfected with a reporter construct containing five tandem yeast GAL4 response elements (UAS) linked to a luciferase reporter gene (UAS-Luc) and a GAL4-NR4A2 chimeric expressing construct so that exclusive NR4A2 transactivation was measured after drug treatment. The results show that the six bis-indole-derived compounds induced NR4A2-dependent reporter gene activity in both cell lines with variable potencies, however, the 3-CF3-4-Cl analog was the most active compound in both cell lines. In Panc1 cells the induction responses were similar for the 4-Br, 4-Cl, 4-SCH3 + 3-CF3-4-Cl compounds whereas in Pan28 cells the former 3 compounds exhibit lower activity. In contrast the N-Me-4-OH and 2-OH-4-Br analogs exhibited lower activity in both cell lines. Figure 2A illustrates the structures of two NR4A2-active substituted quinolines (Kim et al., 2015) and Figure 2B summarizes the effects of the quinoline compounds CQ and AQ as inducers of luciferase activity in Panc1 and Panc28 cells transfected with UAS-Luc/GAL4-NR4A2. The results show that CQ and AQ induce transactivation in the pancreatic cancer cells as previously reported in SK-N-BE(2)C neuroblastoma cells.(Kim et al., 2015) However, unlike previous studies, we noted that both CQ and AQ, but not the bis-indole-derived compounds, activated luciferase activity in Panc1 and Panc28 cells transfected only with the UAS-Luc reporter construct (Figs. 2C and 2D) and similar results for CQ/AQ were observed in these cells transfected with several luciferase reporter gene constructs containing arbitrary response/promoter elements (data not shown), suggesting unspecific activation of luciferase activity by CQ/AQ. Thus, comparing NR4A2-dependent reporter gene activity of bis-indole analogs and substituted quinolines did not determine the relative potencies of these compounds due to the NR4A2-independent effects of the quinolines on inducible luciferase activity.
Figure 1.
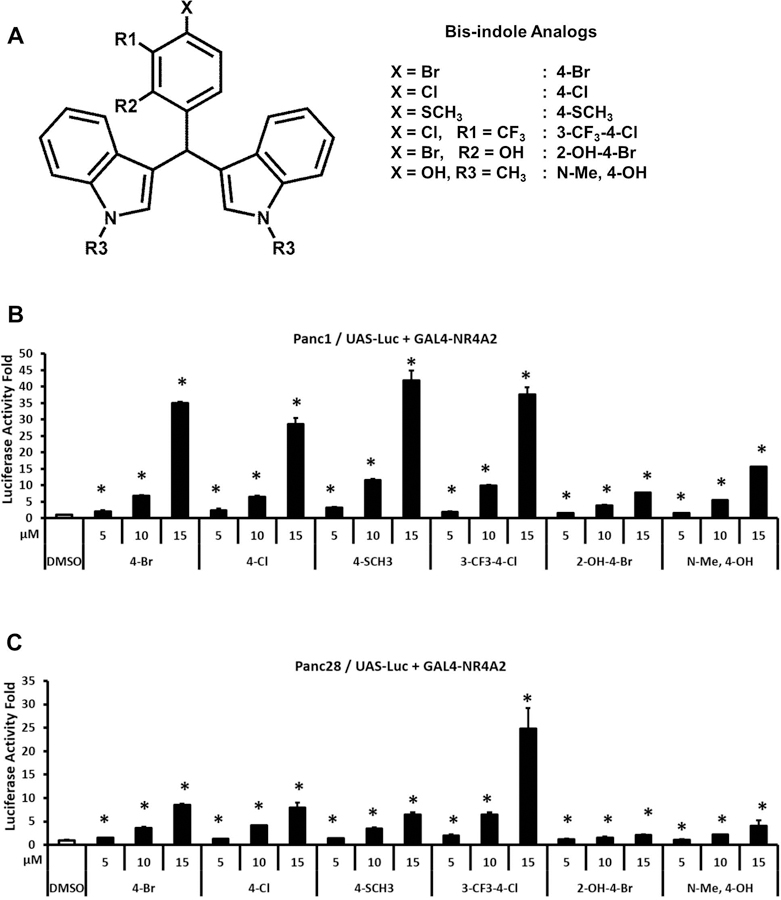
Transactivation of NR4A2 by bis-indole analogs. Six structurally related bis-indole analogs (A) were used in the transactivation assays. Panc1 (B) and Panc28 (C) cells were cotransfected with UAS-luciferase reporter constructs and GAL4-NR4A2 expression constructs and then the cells were treated with 5, 10 and 15 μM bis-indole analogs. Luciferase activity fold induction was determined as described in the Materials and Methods. Results are expressed as mean ± SD for at least three independent determinations for each treatment. *, P < 0.01, treatment vs. solvent control (DMSO).
Figure 2.
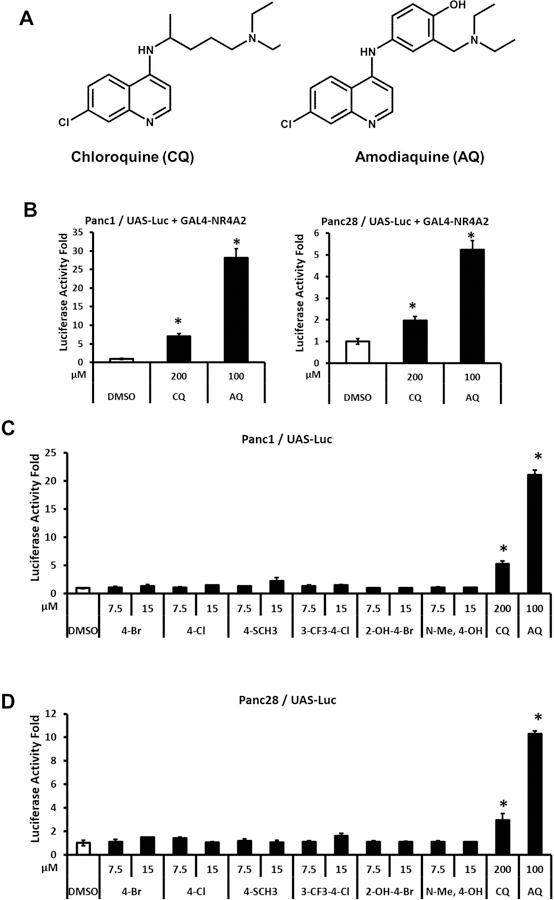
Transactivation of NR4A2 by substituted quinolines. Chloroquine and amodiaquine (A) were used in the transactivation assays. Panc1 and Panc28 cells were cotransfected with UAS-luciferase reporter constructs and GAL4-NR4A2 expression constructs followed by treatment with 200 μM CQ or 100 μM AQ (B). Panc1 (C) and Panc28 (D) cells were transfected with only UAS-luciferase plasmid and cells were treated with 7.5 and 15 μM bis-indole analogs, 200 μM CQ or 100 μM AQ. Luciferase activity fold induction was determined as described in the Materials and Methods. Results are expressed as mean ± SD for at least three independent determinations for each treatment. *, P < 0.01, treatment vs. solvent control (DMSO).
Therefore, we further investigated the effects of bis-indole and quinoline derivatives on activation of several NR4A2-regulated genes including vasoactive intestinal peptide (VIP), osteopontin (OPN), proopiomelanocortin (POMC) and neuropilin 1 (NRP1).(Hermanson et al., 2006; Lammi et al., 2004; Luo et al., 2007; Murphy & Conneely, 1997) Figure 3A shows that the bis-indole compounds significantly induce VIP in Panc1 cells with up to a 330-fold induction observed using 2-OH-4-Br (Fig. 3A). The 2-OH-4-Br and 3-CF3-4-Cl analogs were the most potent inducers of VIP in Panc1 and Panc28 cells and this was in contrast to their activation of GAL4-NR4A1 (Fig. 1) where the 2-OH-4-Br compound was a weak inducer. Although CQ and AQ induced VIP in Panc1 cells (2.3 to 4.1-fold), the magnitude of the response was significantly lower than the NR4A2-active bis-indole compounds (Fig. 3B). The differences between bis-indoles and quinolines for induction of VIP in Panc28 cells (Figs. 3C and 3D, respectively) were similar to those observed in Panc1 cells. The least active bis-indole compound, N-Me-4-OH, was a more potent inducer of VIP in Panc1 and Panc28 cells than either CQ or AQ.
Figure 3.
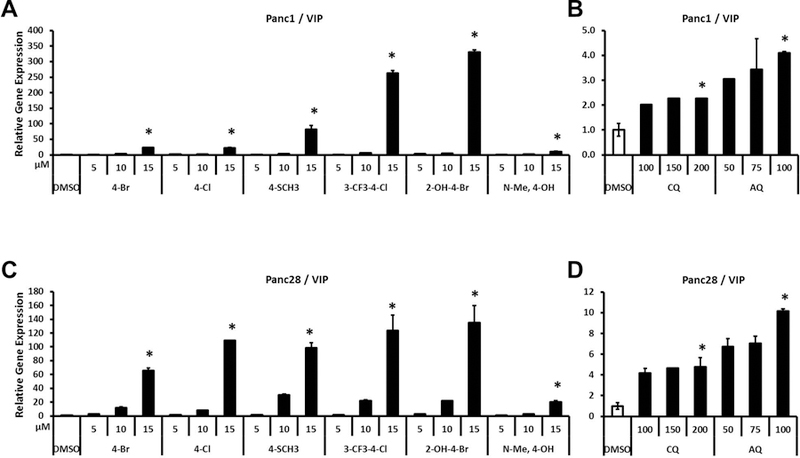
Effects of bis-indole analogs and quinoline derivatives on VIP gene expression. Panc1 (A and B) and Panc28 (C and D) cells were treated with bis-indole analogs (5, 10 and 15 μM), CQ (100, 150 and 200 μM) or AQ (50, 75 and 100 μM). Relative expression levels of VIP were determined by quantitative PCR analysis as described in Materials and Methods. Results are expressed as means ± SD for at least six independent determinations for each treatment. The asterisk (*) indicates significant gene induction (P < 0.01) of the highest concentration treatment vs. solvent control (DMSO).
OPN is also a highly inducible gene in Panc1 cells and the most potent compounds, 4-SCH3, and 3-CF3-4-Cl induced a 25–30-fold increase in OPN mRNA levels (Fig. 4A). Interestingly, the order of potency for the bis-indole compounds as inducers of OPN in Panc1 cells was different from that observed for the induction of VIP, demonstrating that the structure-dependent differences in potencies were also gene-dependent. For example, the N-Me-4-OH compound was a weak inducer of VIP (Fig. 3A) but a relatively potent inducer of OPN in Panc1 cells. In Panc1 cells, CQ and AQ induced expression of OPN (2.3 to 6.5-fold) (Fig. 4B); but the fold induction was much lower than that observed for the bis-indole compounds. In contrast, inducibility of OPN in Panc28 cells by the bis-indole NR4A2 agonists was ≤ 3-fold (by 3-CF3-4-Cl) (Fig. 4C), which was an order of magnitude lower than that observed in Panc1 cells (Fig. 4A). Both CQ and AQ did not induce OPN in Panc28 cells (Fig. 4D).
Figure 4.
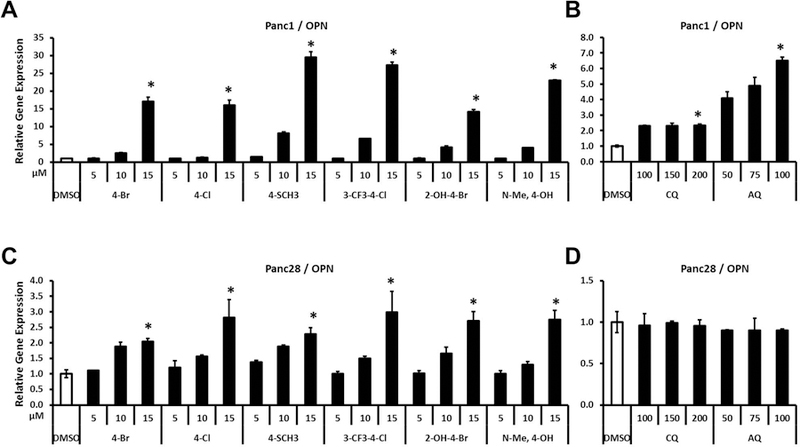
Effects of bis-indole analogs and quinoline derivatives on OPN gene expression. Panc1 (A and B) and Panc28 (C and D) cells were treated with bis-indole analogs (5, 10 and 15 μM), CQ (100, 150 and 200 μM) or AQ (50, 75 and 100 μM). Relative expression levels of OPN were determined by quantitative PCR analysis as described in Materials and Methods. Results are expressed as means ± SD for at least six independent determinations for each treatment. The asterisk (*) indicates significant gene induction (P < 0.01) of the highest concentration treatment vs. solvent control (DMSO).
POMC is induced in Panc1 (Fig. 5A) but not in Panc28 cells (data not shown) by the bis-indole analogs. The 3-CF3-4-Cl, 2-OH-4-Br and N-Me-4-OH analogs induced a robust response (>10-fold) whereas the other bis-indoles were less active and CQ/AQ did not induce POMC (Fig. 5B). Interestingly, induction of NRP1 by bis-indole analogs was highly dependent on the cell line. In Panc1 cells, bis-indoles did not induce but repressed NRP1 gene expression (Fig. 5C) and CQ/AQ did not induce or repress expression of NRP1 (Fig. 5D). In contrast, all the bis-indole compounds (Fig. 5E) but not CQ or AQ (Fig. 5F) induced NRP1 gene expression in Panc28 cells with a comparable 8 to 11-fold enhancement. The cell context-dependent antagonist (Panc1) and agonist (Panc28) activity of the bis-indole analogs as modulators of NRP1 expression was unexpected and is currently being further investigated. These results (Figs. 1–5) clearly demonstrate that the bis-indole analogs are more potent than CQ/AQ as inducers of NR4A2-dependent genes in Panc1 and Panc28 cells and the relative potencies of the individual bis-indole compound are dependent on the gene and cell context with the 3-CF3-4-Cl being among the most potent inducer for most genes.
Figure 5.
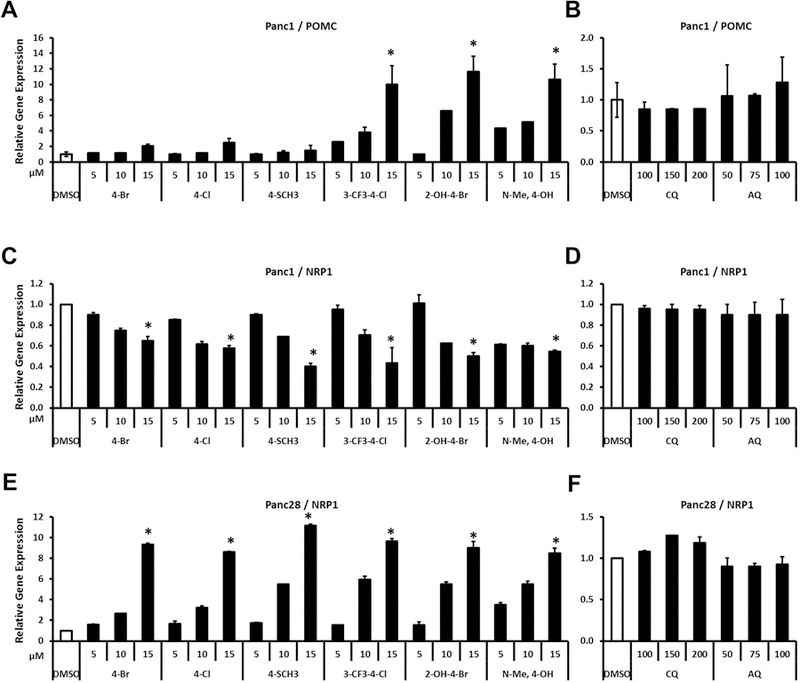
Effects of bis-indole analogs and quinoline derivatives on POMC and NRP1 gene expression. Panc1 (A, B, C and D) and Panc28 (E and F) cells were treated with bis-indole analogs (5, 10 and 15 μM), CQ (100, 150 and 200 μM) or AQ (50, 75 and 100 μM). Relative expression levels of POMC and NRP1 were determined by quantitative PCR analysis as described in Materials and Methods. Results are expressed as means ± SD for at least six independent determinations for each treatment. The asterisk (*) indicates significant gene induction/repression (P < 0.01) of the highest concentration treatment vs. solvent control (DMSO).
4. DISCUSSION
X-ray crystal structure analysis of the LBD of NR4A2 did not reveal an obvious ligand binding pocket since this space was occupied by bulky hydrophobic amino acid side chain residues.(Wang et al., 2003) However, nuclear magnetic resonance (NMR) analysis of the binding of substituted quinolines and unsaturated fatty acids such as docosahexaenoic acid (DHA) indicated that in solution both of these NR4A2 ligands exhibited similar interactions within the LBD of NR4A2.(de Vera et al., 2019; Kim et al., 2015) Moreover, mutational analysis confirmed that mutations of I403, L409, Y575 and D580 in NR4A2 resulted in loss of AQ-induced transactivation using the GAL4-NR4A2-LBD/UAS-Luc assay.(Kim et al., 2015) DIM-C-pPhCl has been extensively used as an NR4A2 ligand in transactivation and functional assays where this compound acts as an NR4A2-dependent inhibitor of inflammatory genes and pathways in cell culture and mouse models of Parkinson’s disease.(De Miranda et al., 2013; De Miranda et al., 2015b; Hammond et al., 2018; Li et al., 2012) In addition, computation-based small molecule docking studies were used to predict interactions of DIM-C-pPhCl with NR4A2.(Hammond et al., 2018) In contrast to studies with DHA and substituted quinolines, DIM-C-pPhCl was predicted to interact with the coactivator binding site with a binding energy of −73.3 kcal/mol whereas much weaker interactions of DIM-C-pPhCl with the LBD were predicted (−12.2 kcal/mol).(Hammond et al., 2018) Thus computational analysis indicates that bis-indoles and substituted quinolines interact with different domains of NR4A2 even though it has been reported that both sets of chemicals exhibit NR4A2-dependent responses in different assay systems (de Vera et al., 2019; Kim et al., 2015; Li et al., 2012).
In this study, we wanted to directly compare the NR4A2 agonist activities of bis-indoles and substituted quinolines using the same assays and cell lines and several known NR4A2-regulated genes. Initial comparisons using the GAL4-NR4A2/UAS-Luc assay demonstrated induced transactivation with both sets of compounds; however, interpretation of the results for the quinoline compounds was somewhat compromised since both CQ and AQ induced luciferase activity in pancreatic cancer cells transfected with UAS-Luc in the absence of GAL4-NR4A2. It should be pointed out that the direct effects of several heterocyclic chemotypes, including quinolines, on induction of luciferase activity have previously been reported (Thorne et al., 2012) and this is consistent with our observations using CQ and AQ. Therefore, a more direct comparison of the substituted quinolines and bis-indoles was to investigate their induction of several NR4A2-regulated genes (Hermanson et al., 2006; Lammi et al., 2004; Luo et al., 2007; Murphy & Conneely, 1997) in Panc1 and Panc28 cells and previous studies with the 4-Br analog in these cells showed that induction of VIP, OPN and NRP1 was abrogated after knockdown of NR4A2.(Li et al., 2012) The fold induction of VIP, OPN, POMC and NRP1 mRNA levels by bis-indole analogs was highly variable and both gene and cell context dependent with VIP being the most highly inducible gene. DIM-C-pPhSCH3 (4-SCH3) appeared to be a potent inducer of NR4A2-dependent gene expression; however, the rank order potencies of the other bis-indole analogs were highly variable. Moreover, we also observed that the bis-indole derivatives induced very steep dose – response curves for some genes (VIP; Panc1 and Panc28; OPN; Panc1) and these cell – and gene – specific differences in gene activation are not understood. In contrast, the effects of CQ and AQ on induction of NR4A2-dependent genes were substantially lower than that observed for the bis-indole compounds. For example, CQ and AQ had minimal effects on expression of NRP1 in Panc28 cells whereas all of the bis-indole analogs induced NRP1 and 4-SCH3 induced a >10-fold increase in expression of NRP1 mRNA (Fig. 5E).
In summary, the results indicate that among the bis-indole derivatives, their potency and efficacy as inducers of NR4A2-dependent genes is structure, cell context and gene dependent whereas CQ and AQ induce weak to minimal responses. Some of these differences in activity may be due to the NR4A2-region specific interactions of substituted quinolines with the LBD of NR4A2 versus bis-indole derivatives with the coactivator binding sites. The observed differences in these compounds as activators of NR4A2-dependent gene expression is consistent with their designation as selective NR4A2 modulators and this parallels identification of selective receptor modulators for the estrogen receptor and many other nuclear receptors (Jordan, 2007; Jordan & O’Malley, 2007; Katzenellenbogen et al., 1996; Paige et al., 1999). Thus the structure-dependent activation/inactivation of NR4A2-regulated genes in Panc1 and Panc28 cells is due to multiple factors including differential interactions of the bound receptor monomers or dimers and their interactions with cognate cis elements, differential interactions with other bound transcription factors (e.g. Sp1) and tissue/cell-specific differences in the expression of nuclear cofactors. It is also apparent from our results that the selectivity of an NR4A2 or other nuclear receptor ligand is not necessarily predictable per se and it requires further examination of specific target and non-target genes/pathways of interest.
ACKNOWLEDGEMENTS
The financial assistance of Texas AgriLife Research, the Syd Kyle Chair Endowment and the National Institutes of Health (P30-ES023512) is gratefully acknowledged.
ABBREVIATIONS:
- 2-OH-4-Br
1,1-bis(3’-indolyl)-1-(2-hydroxy-4-bromophenyl)methane
- 4-Br
1,1-bis(3’-indolyl)-1-(4-bromophenyl)methane
- 4-Cl
1,1-bis(3’-indolyl)-1-(4-chlorophenyl)methane
- 4-SCH3
1,1-bis(3’-indolyl)-1-(4-methylthiophenyl)methane
- 3-CF3-4-Cl
1,1-bis(3’-indolyl)-1-(3-trifluoromethyl-4-chlorophenyl)methane
- AF-1
activation function 1
- AQ
amodiaquine
- CQ
chloroquine
- DIM-C-pPhCl
1,1-bis(3’-indolyl)-1-(p-chlorophenyl)methane
- LBD
ligand binding domain
- N-Me-4-OH
1,1-bis[3’-(1’-methylindolyl)]-1-(4-hydroxyphenyl)methane
- NBRE
nerve growth factor binding response element
- NGF1-B
nerve growth factor-induced clone B
- NRP1
neuropilin 1
- NR4A1
nuclear receptor 4A1
- NR4A2
nuclear receptor 4A2
- NR4A3
nuclear receptor 4A3
- NuRE
Nur77-responsive element
- POMC
proopiomelanocortin
- OPN
osteopontin
- RXR
retinoic acid X receptor
- VIP
vasoactive intestinal peptide
Footnotes
Disclosure of Conflicts of Interest: There are no conflicts of interests to declare.
REFERENCES
- De Miranda BR, Miller JA, Hansen RJ, Lunghofer PJ, Safe S, Gustafson DL, … Tjalkens RB. (2013). Neuroprotective efficacy and pharmacokinetic behavior of novel anti-inflammatory para-phenyl substituted diindolylmethanes in a mouse model of Parkinson’s disease. J Pharmacol Exp Ther, 345(1), 125–138. doi: 10.1124/jpet.112.201558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Miranda BR, Popichak KA, Hammond SL, Jorgensen BA, Phillips AT, Safe S, & Tjalkens RB (2015a). The Nurr1 Activator 1,1-Bis(3’-Indolyl)-1-(p-Chlorophenyl)Methane Blocks Inflammatory Gene Expression in BV-2 Microglial Cells by Inhibiting Nuclear Factor kappaB. Mol Pharmacol, 87(6), 1021–1034. doi: 10.1124/mol.114.095398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Miranda BR, Popichak KA, Hammond SL, Miller JA, Safe S, & Tjalkens RB (2015b). Novel para-phenyl substituted diindolylmethanes protect against MPTP neurotoxicity and suppress glial activation in a mouse model of Parkinson’s disease. Toxicol Sci, 143(2), 360–373. doi: 10.1093/toxsci/kfu236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vera IM, Giri PK, Munoz-Tello P, Brust R, Fuhrmann J, Matta-Camacho E, … Kojetin DJ (2016). Identification of a Binding Site for Unsaturated Fatty Acids in the Orphan Nuclear Receptor Nurr1. ACS Chem Biol, 11(7), 1795–1799. doi: 10.1021/acschembio.6b00037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vera IMS, Munoz-Tello P, Zheng J, Dharmarajan V, Marciano DP, Matta-Camacho E, … Kojetin DJ (2019). Defining a Canonical Ligand-Binding Pocket in the Orphan Nuclear Receptor Nurr1. Structure, 27(1), 66–77.e65. doi: 10.1016/j.str.2018.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois C, Hengerer B, & Mattes H (2006). Identification of a potent agonist of the orphan nuclear receptor Nurr1. ChemMedChem, 1(9), 955–958. doi: 10.1002/cmdc.200600078 [DOI] [PubMed] [Google Scholar]
- Giguere V (1999). Orphan nuclear receptors: from gene to function. Endocr Rev, 20(5), 689–725. doi: 10.1210/edrv.20.5.0378 [DOI] [PubMed] [Google Scholar]
- Hammond SL, Popichak KA, Li X, Hunt LG, Richman EH, Damale PU, … Tjalkens RB (2018). The Nurr1 Ligand,1,1-bis(3’-Indolyl)-1-(p-Chlorophenyl)Methane, Modulates Glial Reactivity and Is Neuroprotective in MPTP-Induced Parkinsonism. J Pharmacol Exp Ther, 365(3), 636–651. doi: 10.1124/jpet.117.246389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanson E, Borgius L, Bergsland M, Joodmardi E, & Perlmann T (2006). Neuropilin1 is a direct downstream target of Nurr1 in the developing brain stem. J Neurochem, 97(5), 1403–1411. doi: 10.1111/j.1471-4159.2006.03829.x [DOI] [PubMed] [Google Scholar]
- Hintermann S, Chiesi M, von Krosigk U, Mathe D, Felber R, & Hengerer B (2007). Identification of a series of highly potent activators of the Nurr1 signaling pathway. Bioorg Med Chem Lett, 17(1), 193–196. doi: 10.1016/j.bmcl.2006.09.062 [DOI] [PubMed] [Google Scholar]
- Jordan VC (2007). SERMs: meeting the promise of multifunctional medicines. J Natl Cancer Inst, 99(5), 350–356. doi: 10.1093/jnci/djk062 [DOI] [PubMed] [Google Scholar]
- Jordan VC, & O’Malley BW (2007). Selective estrogen-receptor modulators and antihormonal resistance in breast cancer. J Clin Oncol, 25(36), 5815–5824. doi: 10.1200/jco.2007.11.3886 [DOI] [PubMed] [Google Scholar]
- Katzenellenbogen JA, O’Malley BW, & Katzenellenbogen BS (1996). Tripartite steroid hormone receptor pharmacology: interaction with multiple effector sites as a basis for the cell- and promoter-specific action of these hormones. Mol Endocrinol, 10(2), 119–131. doi: 10.1210/mend.10.2.8825552 [DOI] [PubMed] [Google Scholar]
- Kim CH, Han BS, Moon J, Kim DJ, Shin J, Rajan S, … Kim KS (2015). Nuclear receptor Nurr1 agonists enhance its dual functions and improve behavioral deficits in an animal model of Parkinson’s disease. Proc Natl Acad Sci U S A, 112(28), 8756–8761. doi: 10.1073/pnas.1509742112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammi J, Huppunen J, & Aarnisalo P (2004). Regulation of the osteopontin gene by the orphan nuclear receptor NURR1 in osteoblasts. Mol Endocrinol, 18(6), 1546–1557. doi: 10.1210/me.2003-0247 [DOI] [PubMed] [Google Scholar]
- Li X, Lee SO, & Safe S (2012). Structure-dependent activation of NR4A2 (Nurr1) by 1,1-bis(3’-indolyl)-1-(aromatic)methane analogs in pancreatic cancer cells. Biochem Pharmacol, 83(10), 1445–1455. doi: 10.1016/j.bcp.2012.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Henricksen LA, Giuliano RE, Prifti L, Callahan LM, & Federoff HJ (2007). VIP is a transcriptional target of Nurr1 in dopaminergic cells. Exp Neurol, 203(1), 221–232. doi: 10.1016/j.expneurol.2006.08.005 [DOI] [PubMed] [Google Scholar]
- Maira M, Martens C, Philips A, & Drouin J (1999). Heterodimerization between members of the Nur subfamily of orphan nuclear receptors as a novel mechanism for gene activation. Mol Cell Biol, 19(11), 7549–7557. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/10523643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy EP, & Conneely OM (1997). Neuroendocrine regulation of the hypothalamic pituitary adrenal axis by the nurr1/nur77 subfamily of nuclear receptors. Mol Endocrinol, 11(1), 39–47. doi: 10.1210/mend.11.1.9874 [DOI] [PubMed] [Google Scholar]
- Paige LA, Christensen DJ, Gron H, Norris JD, Gottlin EB, Padilla KM, … Fowlkes DM (1999). Estrogen receptor (ER) modulators each induce distinct conformational changes in ER alpha and ER beta. Proc Natl Acad Sci U S A, 96(7), 3999–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen RF, Granas K, Johnsen H, Rolseth V, & Sterri S (1995). Three related brain nuclear receptors, NGFI-B, Nurr1, and NOR-1, as transcriptional activators. J Mol Neurosci, 6(4), 249–255. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/8860236 [DOI] [PubMed] [Google Scholar]
- Pearen MA, & Muscat GE (2010). Minireview: Nuclear hormone receptor 4A signaling: implications for metabolic disease. Mol Endocrinol, 24(10), 1891–1903. doi: 10.1210/me.2010-0015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlmann T, & Jansson L (1995). A novel pathway for vitamin A signaling mediated by RXR heterodimerization with NGFI-B and NURR1. Genes Dev, 9(7), 769–782. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/7705655 [DOI] [PubMed] [Google Scholar]
- Philips A, Lesage S, Gingras R, Maira MH, Gauthier Y, Hugo P, & Drouin J (1997). Novel dimeric Nur77 signaling mechanism in endocrine and lymphoid cells. Mol Cell Biol, 17(10), 5946–5951. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/9315652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safe S, Jin UH, Hedrick E, Reeder A, & Lee SO (2014). Minireview: role of orphan nuclear receptors in cancer and potential as drug targets. Mol Endocrinol, 28(2), 157–172. doi: 10.1210/me.2013-1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne N, Shen M, Lea WA, Simeonov A, Lovell S, Auld DS, & Inglese J (2012). Firefly luciferase in chemical biology: a compendium of inhibitors, mechanistic evaluation of chemotypes, and suggested use as a reporter. Chem Biol, 19(8), 1060–1072. doi: 10.1016/j.chembiol.2012.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JR, Gan WJ, Li XM, Zhao YY, Li Y, Lu XX, … Wu H (2014). Orphan nuclear receptor Nur77 promotes colorectal cancer invasion and metastasis by regulating MMP-9 and E-cadherin. Carcinogenesis, 35(11), 2474–2484. doi: 10.1093/carcin/bgu157 [DOI] [PubMed] [Google Scholar]
- Wang Z, Benoit G, Liu J, Prasad S, Aarnisalo P, Liu X, … Perlmann T (2003). Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature, 423(6939), 555–560. doi: 10.1038/nature01645 [DOI] [PubMed] [Google Scholar]
- Wansa KD, Harris JM, & Muscat GE (2002). The activation function-1 domain of Nur77/NR4A1 mediates trans-activation, cell specificity, and coactivator recruitment. J Biol Chem, 277(36), 33001–33011. doi: 10.1074/jbc.M203572200 [DOI] [PubMed] [Google Scholar]
- Wansa KD, Harris JM, Yan G, Ordentlich P, & Muscat GE (2003). The AF-1 domain of the orphan nuclear receptor NOR-1 mediates trans-activation, coactivator recruitment, and activation by the purine anti-metabolite 6-mercaptopurine. J Biol Chem, 278(27), 24776–24790. doi: 10.1074/jbc.M300088200 [DOI] [PubMed] [Google Scholar]
- Wilson TE, Day ML, Pexton T, Padgett KA, Johnston M, & Milbrandt J (1992). In vivo mutational analysis of the NGFI-A zinc fingers. J Biol Chem, 267(6), 3718–3724. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/1740423 [PubMed] [Google Scholar]
- Wilson TE, Fahrner TJ, Johnston M, & Milbrandt J (1991). Identification of the DNA binding site for NGFI-B by genetic selection in yeast. Science, 252(5010), 1296–1300. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/1925541 [DOI] [PubMed] [Google Scholar]
- Wilson TE, Padgett KA, Johnston M, & Milbrandt J (1993). A genetic method for defining DNA-binding domains: application to the nuclear receptor NGFI-B. Proc Natl Acad Sci U S A, 90(19), 9186–9190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetterstrom RH, Solomin L, Jansson L, Hoffer BJ, Olson L, & Perlmann T (1997). Dopamine neuron agenesis in Nurr1-deficient mice. Science, 276(5310), 248–250. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/9092472 [DOI] [PubMed] [Google Scholar]
- Zetterstrom RH, Solomin L, Mitsiadis T, Olson L, & Perlmann T (1996). Retinoid X receptor heterodimerization and developmental expression distinguish the orphan nuclear receptors NGFI-B, Nurr1, and Nor1. Mol Endocrinol, 10(12), 1656–1666. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/8961274 [DOI] [PubMed] [Google Scholar]
- Zhou F, Drabsch Y, Dekker TJ, de Vinuesa AG, Li Y, Hawinkels LJ, … Dijke PT (2014). Nuclear receptor NR4A1 promotes breast cancer invasion and metastasis by activating TGF-beta signalling. Nat Commun, 5, 3388. doi: 10.1038/ncomms4388 [DOI] [PubMed] [Google Scholar]


