Abstract
Objective:
Assess clinician perceptions of a machine learning-based early warning system to predict severe sepsis and septic shock (EWS 2.0)
Design:
Prospective observational study
Setting:
Tertiary teaching hospital in Philadelphia, PA
Patients:
Non-ICU admissions November-December 2016
Interventions:
During a six-week study period conducted five months after EWS 2.0 alert implementation, nurses and providers were surveyed twice about their perceptions of the alert’s helpfulness and impact on care, first within 6 hours of the alert, and again 48 hours post-alert.
Measurements and Main Results:
For the 362 alerts triggered, 180 nurses (50% response rate) and 107 providers (30% response rate) completed the First Survey. Of these, 43 nurses (24% response rate) and 44 providers (41% response rate) completed a Second Survey. Few (24% nurses, 13% providers) identified new clinical findings after responding to the alert. Perceptions of the presence of sepsis at the time of alert were discrepant between nurses (13%) and providers (40%). The majority of clinicians reported no change in perception of the patient’s risk for sepsis (55% nurses, 62% providers). A third of nurses (30%) but few providers (9%) reported the alert changed management. Almost half of nurses (42%) but less than a fifth of providers (16%) found the alert helpful at 6 hours.
Conclusions:
In general, clinical perceptions of EWS 2.0 were poor. Nurses and providers differed in their perceptions of sepsis and alert benefits. These findings highlight the challenges of achieving acceptance of predictive and machine learning-based sepsis alerts.
Keywords: severe sepsis, septic shock, electronic medical record, predictive medicine, machine learning, early warning system
INTRODUCTION
Sepsis is a leading cause of mortality among hospitalized patients (1). Mortality from hospital-acquired sepsis is two to three times higher than sepsis present on admission (2, 3). Delayed recognition delays life-saving interventions, increasing the risk of progression to shock, organ failure, and death (4). Many hospitals have developed electronic health record (EHR)-based sepsis surveillance and alert systems to improve early detection and intervention (5).
The first surveillance tools targeted detection of the systemic inflammatory response syndrome (SIRS). With good diagnostic accuracy, detection systems prompted increased frequency of and improved time to diagnostic testing, and escalation of care (6-12). Our group previously developed an automated SIRS-based sepsis detection tool (EWS 1.0) that resulted in a non-significant trend toward reduced mortality (9).
Our group and others have more recently developed predictive tools to identify high-risk patients before clinical criteria are apparent (13-21). Using machine learning (ML) algorithms, large patient datasets can be mined for novel clinical patterns and characteristics predictive of clinical decompensation (18, 22, 23). ML algorithms to predict septic shock in ICU patients have demonstrated good predictive accuracy in retrospective validation (22, 24, 25), though few have reported prospective implementation outcomes. One small non-academic hospital reported improved sepsis-related mortality (26), and a small randomized trial demonstrated decreased length of stay and improved mortality in ICU patients (23).
To our knowledge, our group is the first to evaluate large-scale prospective implementation of a machine learning-based sepsis prediction tool (EWS 2.0) in non-ICU patients. Algorithm validation suggested excellent predictive characteristics for severe sepsis and septic shock, with a positive likelihood ratio of 13 (27). We linked EWS 2.0 to predictive alerts deployed to clinical care teams on non-ICU inpatient services and performed a prospective pre-implementation and post-implementation analysis of its impact on clinical care processes and patient outcomes (27).
In addition to good algorithm performance, stakeholder acceptance of clinical decision support systems (CDSSs) is crucial for sepsis care improvement. We previously reported that a minority of providers perceived our earlier sepsis detection system, EWS 1.0, to be helpful despite observed changes in management resulting in increases in early sepsis care and documentation (28). We postulated that acceptance was limited by alert fatigue. Provider acceptance of ML algorithm prediction tools may be further limited by their complexity and lack of transparency (28). This study describes clinician perceptions of our predictive machine learning-based EWS 2.0 deployed prospectively across our healthcare system.
MATERIALS AND METHODS
Study Design, Setting, and Subjects
This was a prospective observational study. The EWS 2.0 alert was deployed throughout our multi-hospital academic healthcare system, the same study site as EWS 1.0. This study was conducted in our flagship 782-bed academic hospital given the higher volume of alerts at this location and on-site availability of the research team. Study subjects were bedside clinicians caring for patients who triggered the alert, including registered nurses (nurses) and physicians or advanced practitioners (providers). This project was reviewed and determined to qualify as Quality Improvement by the University of Pennsylvania’s Institutional Review Board.
Early Warning System Protocol
To create EWS 2.0, we trained an ML algorithm to predict severe sepsis or septic shock. The algorithm was developed using a random forest classifier trained on EHR data from adult patients discharged from July 2011 to June 2014 from any of our three urban acute care hospitals (n=162,212). Positive cases (n=943) were defined as having an ICD9 code of 995.92 (Severe Sepsis) or 785.52 (Septic Shock) associated with their encounter, with a positive blood culture and either a lactate > 2.2 mmol/L or systolic blood pressure < 90 mm Hg. The earliest of these events was labeled as “time zero” of sepsis onset. Only non-ICU patients were included in the derivation population.
The algorithm’s sensitivity threshold was set to generate approximately 10 alerts across the hospital system per day, with the goal of generating a feasible alert response workload and minimizing false positives that would exacerbate alert fatigue and erode alert confidence. This target daily alert rate was determined a priori and informed by our experience with EWS 1.0, which, based on a threshold determined to capture the patients most likely to decompensate, generated about 6 alerts per day (9). We retrospectively validated the algorithm on hospitalized patients from October to December 2015 (n=10,448, screen positive=314). The positive likelihood ratio for “severe sepsis or septic shock” was 13, with positive and negative predictive values of 29% and 97%, respectively.
Clinicians throughout our hospital system were educated about EWS 2.0 via informational emails before alert deployment. On June 16, 2016, EWS 2.0 was activated. When EWS 2.0 was triggered, an EHR-based alert was sent to the patients’ nurse, and a text message alert was sent to the patient’s provider as well as to a rapid response coordinator who ensured clinical teams received the alert and completed an immediate bedside assessment. Alerts stated that EWS 2.0 had fired for a given patient, and included relevant recent laboratory data along with 48 hours of vital sign trends.
Survey Deployment and Administration
We created two web-based questionnaires to assess clinician perceptions of EWS 2.0 (Supplemental Digital Content 1-2). The surveys were adapted from a previous questionnaire used to evaluate perceptions of EWS 1.0 (28) and refined through an iterative process with feedback from an interdisciplinary team of critical care and general medicine attendings, medical residents, and nurses. The final questionnaires included categorical and Likert-scale survey items with options for open-ended response. Questions were designed to assess clinicians’ perceptions regarding: 1) the patient’s condition; 2) new information discovered at the time of alert; 3) whether and how the alert changed management; and 4) whether and how the alert was useful and/or improved patient care.
Surveys were administered over six consecutive weeks (11/07/2016–12/19/2016) five months after the EWS 2.0 alert was deployed across the healthcare system. For every alert triggered, a rapid response coordinator directed the covering nurse and provider to the first 16-item questionnaire (First Survey) to be completed confidentially and independently within 6 hours of the alert.
Clinicians who completed the First Survey were emailed a link to the 11-item Second Survey 48 hours after the initial alert. The Second Survey included a subset of questions from the First Survey, with a focus on clinicians’ perceptions of patients’ clinical state and the alert’s utility and impact on care after 48 hours of clinical evolution. Up to two reminders were sent by email or text to non-responders 12–24 hours after the initial Second Survey request. Completion of surveys was strictly voluntary.
Data Analysis
Study data were collected and managed using Research Electronic Data Capture (REDCap), a secure, web-based application designed to support data capture for research studies (30). To facilitate interpretation of Likert-scale survey responses, grades 1 and 2 were grouped and considered as negative, grade 3 was considered neutral, and grades 4 and 5 were grouped and considered positive. Categorical questions included options for open-ended responses; these were reviewed for themes and some were re-coded to the appropriate corresponding categorical response groups. Results were calculated as percentages of total responses within each group (provider and nurse) and comparisons were made between clinician types using the chi square test and two-tailed Fisher’s exact test, as appropriate. P values <0.05 were considered significant and are reported here.
RESULTS
Survey Response
During the six-week study period, 362 EWS 2.0 alerts were triggered, resulting in a median of 8 alerts per day (IQR 7–10, range 4–15). For the 724 potential First Survey responses (one each for a nurse and provider per alert), 287 First Surveys were completed by 252 individual clinicians (overall response rate 40%). Nurses completed 180 First Surveys (50% response rate) and providers completed 107 First Surveys (30% response rate). Of these, 43 nurses who completed a First Survey completed a Second Survey (24% response rate) and 44 providers who completed a First Survey completed a Second Survey (41% response rate), with an overall Second Survey response rate of 30%. Of these 77 respondents, 49 (64%, 33 providers, 16 nurses) reported sufficient continuity with the alerted patient to accurately complete the Second Survey.
Findings and Management at the Time of Alert
The alert and subsequent patient assessment infrequently provided new information (Table 1). Few clinicians (13% providers, 24% nurses) reported new clinical findings at the time of alert trigger (p=0.03 for provider vs. nurses). Perceptions of the presence of sepsis at the time of patient evaluation after alert were discrepant between providers (40%) and nurses (13%) (p<0.001). In addition, following the alert most clinicians remained unchanged in their impression that the patient would develop critical illness (62% providers, 55% nurses). At 48 hours, fewer clinicians in both groups believed the patient was septic (26% providers, 6% nurses), when compared to their impressions within the first 6 hours post-alert.
Table 1.
Clinical Impressions After Early Warning System 2.0 Alert
| Clinical Assessment, n (%) | Provider | Nurse | P-value |
|---|---|---|---|
| The alert resulted in new clinical findingsa | 14 (13) | 43 (24) | 0.03 |
| Vital sign change | 10 (71) | 39 (91) | |
| New symptoms | 6 (43) | 3 (7) | |
| Physical exam finding | 2 (14) | 2 (5) | |
| Lab finding | 0 (0) | 7 (16) | |
| Before the alert triggered, I thought the patient had sepsis | <0.001 | ||
| Yes | 40 (38) | 21 (12) | |
| Maybe | 29 (27) | 49 (28) | |
| No | 37 (35) | 106 (60) | |
| Within 6 hours after the alert triggered, I thought the patient had sepsis | <0.001 | ||
| Yes | 42 (40) | 23 (13) | |
| Maybe | 28 (26) | 54 (31) | |
| No | 36 (34) | 99 (56) | |
| By 48 hours after the alert triggered, I thought the patient had sepsisb | 0.06 | ||
| Yes | 8 (26) | 1 (6) | |
| Maybe | 4 (13) | 0 (0) | |
| No | 19 (61) | 15 (94) | |
| The alert affected my expectation that the patient would develop critical illness | 0.30 | ||
| Unchanged expectation | 66 (62) | 99 (55) | |
| Increased expectation | 13 (12) | 24 (13) | |
| The patient is newly critically ill | 1 (1) | 4 (2) | |
| The patient remains critically ill | 24 (2) | 47 (26) | |
| The patient is progressing in their critical illness | 3 (3) | 5 (3) |
Results are reported as number of responses for each item divided by the total number of respondents.
Clinicians could select more than one clinical finding, so percentages may add up to greater than 100% for this question.
For sepsis assessment at 48 hours, Provider n=44, nurse n=43. For all other questions, Provider n=107, nurse n=180.
Sepsis was thought to be the primary driver of alert trigger in about one third of cases (40% providers, 21% nurses, p<0.001), followed by dehydration (14% providers, 14% nurses) (Table 2). One tenth of providers (11%) and one fifth of nurses (21%) did not know why the alert triggered, as they discovered no clinical change. While providers’ impressions of sepsis driving alert trigger remained consistent over time (40% within 6 hours of alert, 39% at 48 hours after alert), nurses were less likely to attribute alert firing to sepsis at 48 hours (21% within 6 hours of alert, 0% at 48 hours after alert, p<0.05).
Table 2.
Perceived Reason for Alert Trigger
| Etiology of Alert Trigger, n (%) | First Surveya Provider (n=107) |
Nurse (n=180) | Second Surveyb Provider (n=44) |
Nurse (n=43) |
|---|---|---|---|---|
| The alert was primarily triggered by | ||||
| Sepsis | 42 (40) | 37 (21) | 13 (39) | 0 (0) |
| Dehydration | 15 (14) | 24 (14) | 6 (18) | 3 (18) |
| Cancer | 10 (9) | 13 (7) | 5 (15) | 1 (6) |
| Infection, not septic | 4 (4) | 11 (6) | 1 (3) | 0 (0) |
| Bleeding | 3 (3) | 5 (3) | 1 (3) | 1 (6) |
| Arrhythmia | 3 (3) | 8 (5) | 0 (0) | 1 (6) |
| Pulmonary problem | 3 (3) | 2 (1) | 0 (0) | 0 (0) |
| Post-operative state | 3 (3) | 8 (5) | 1 (3) | 3 (18) |
| End-stage organ failurec | 2 (2) | 7 (4) | 2 (6) | 0 (0) |
| Drug effect | 1 (1) | 3 (2) | 0 (0) | 1 (6) |
| Pain/anxiety | 1 (1) | 4 (2) | 0 (0) | 0 (0) |
| Cardiogenic shock | 0 (0) | 0 (0) | 2 (6) | 0 (0) |
| Pulmonary embolus | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Otherd | 5 (5) | 9 (5) | 0 (0) | 1 (6) |
| There was no clinical change, I don’t know why the alert triggered | 12 (11) | 37 (21) | 2 (6) | 6 (35) |
| There was a clinical change, but I don’t know why the alert triggered | 2 (2) | 7 (4) | 0 (0) | 0 (0) |
Because each percentage value has been rounded to the nearest whole number, total percentages do not equal 100%.
Completed within 6 hours of alert.
Completed 48 hours after alert.
Includes cirrhosis, end stage renal disease, dialysis, organ transplant rejection, and ventricular assist device.
Includes deconditioning, cardiac arrest, transfusion reaction, electrolyte imbalance, vasovagal, and single reading of transient hypotension.
Few providers (9%) but a third of nurses (30%) reported that the alert changed management (Table 3, p<0.001). Clinicians most commonly reported increased frequency of bedside rounding, followed by increased frequency of vital sign checks, and ordering of new diagnostic tests.
Table 3.
Effect of Early Warning System 2.0 on Patient Care
| Change In Management, n (%) | Provider (n=107) | Nurse (n=178) | P-value |
|---|---|---|---|
| Any change in management | 10 (9) | 54 (30) | <0.001 |
| Bedside rounding frequency was increased | 6 (60) | 32 (59) | |
| Vital sign check frequency was increased | 1 (10) | 19 (35) | |
| New diagnostic tests were ordered | 0 (0) | 25 (46) | |
| New therapeutic interventions were started | 0 (0) | 5 (9) | |
| New diagnoses were considered | 0 (0) | 14 (26) | |
| Goal of care discussions were initiated | 1 (10) | 1 (2) | |
| A rapid response was called | 0 (0) | 3 (6) | |
| The patient was transferred to the ICU | 5 (50) | 6 (11) |
Results are reported as number of responses for each item divided by the total number of respondents. Because clinicians could select more than one response, percentages may add up to greater than 100%.
Overall Impressions
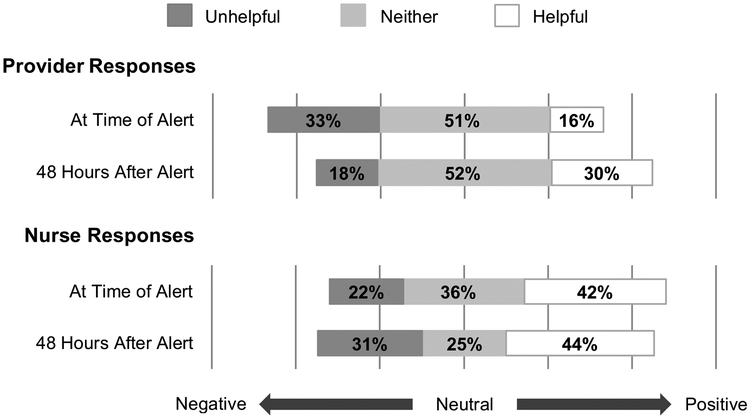
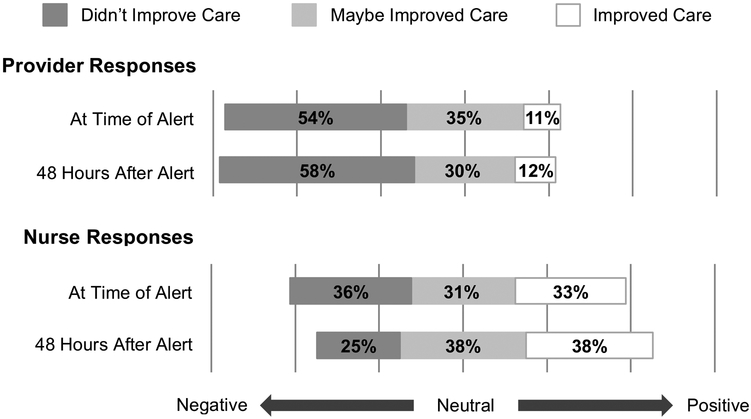
Overall impressions of EWS 2.0’s utility to clinical teams and impact on patient care were mixed (Figures 1 and 2). Almost half of nurses (42%) but less than a fifth of providers (16%) found the alert helpful at 6 hours (p<0.001). Though the proportion of providers finding the alert helpful nearly doubled by 48 hours (30%), this was not statistically significant. The proportion of nurses finding the alert helpful or unhelpful remained stable over time (helpful: 42% at 6 hours, 44% at 48 hours; unhelpful: 22% at 6 hours, 31% at 48 hours). Nurses were more likely than providers to describe the alert as improving care, at both 6 hours (11% providers, 33% nurses, p<0.001) and 48 hours (12% providers, 38% nurses, p=0.05).
Figure 1: Clinician Perceived Utility of Alert.
Unhelpful: Combined percentage of those responding, “Very unhelpful” and “Unhelpful” on a Likert Scale.
Neither: Percentage of those responding “Neither helpful nor unhelpful” on a Likert Scale.
Helpful: Combined percentage of those responding “Very helpful” and “Helpful” on a Likert Scale.
At Time of Alert: First Survey responses, submitted within 6 hours following the alert.
48 Hours After Alert: Second Survey responses, submitted at least 48 hours after alert.
Figure 2: Clinician Perceived Impact on Patient Care.
Improved Care: Combined percentage of those responding, “Definitely improved care” and “Probably improved care” on a Likert Scale.
Maybe Improved Care: Percentage of those responding “Maybe improved care” on a Likert Scale.
Didn’t Improve Care: Combined percentage of those responding, “Definitely did not improve care” and “Probably did not improve care” on a Likert Scale.
At Time of Alert: First Survey responses, submitted within 6 hours following the alert.
48 Hours After Alert: Second Survey responses, submitted at least 48 hours after alert.
Of the 26 clinicians reporting helpful features of the alert, 73% cited improved team communication and 46% cited more frequent monitoring; fewer cited the prompting of diagnostic testing (23%) or interventions (2%). Of the 19 clinicians reporting unhelpful features, 37% cited triggering for known clinical abnormalities, 21% cited patients’ clinical stability, 16% each believed the alert was a poor use of resources or that it fired too late, and 11% reported irrelevant clinical abnormalities. When asked for suggestions for improvements, clinicians most frequently requested transparency regarding factors leading to alert trigger (44% of 48 suggestions).
DISCUSSION
Perceptions
Nurses and providers frequently differed in their perceptions of alerted patients and EWS 2.0 in general. Nurses were less likely to think patients were septic; by 48 hours, none of the surveyed nurses attributed the alert to sepsis. Given that nurses are often the most proximal caregiver and may be the first to encounter signs of sepsis, this finding of differing sepsis assessments may reveal a crucial opportunity for improved sepsis awareness, and highlights the potential importance of objective automated monitoring systems. Despite infrequent concerns for sepsis, nurses were more likely to report perceived changes in management and favorable overall impressions of the alert compared to providers, with nearly one third reporting changed management, half finding the alert helpful, and one third reporting improved care. Discrepancy in nurse and provider perceptions of EWS 2.0’s impact on care suggests it conferred differential benefits and prognostic value to each group. Reported improved interdisciplinary communication may be particularly important given discrepant clinician impressions of sepsis risk in these patients.
EWS 2.0 was less favorably received than EWS 1.0. As previously reported, clinicians reported that EWS 1.0 changed management in about half of cases (56% nurses, 44% providers). Clinicians reported less frequent management changes with EWS 2.0 (30% nurses, 9% providers). Moreover, while nurses’ impressions of the two systems were similar, providers more frequently reported that EWS 1.0 was helpful (40% nurses, 33% providers) and improved care (35% nurses, 24% providers), and less frequently reported that EWS 2.0 was helpful (42% nurses and 16% providers at 6 hours, 42% nurses and 32% providers at 48 hours) or improved care (33% nurses and 11% providers). The poorer perceptions of EWS 2.0 may reflect poor clinician acceptance of predictive alert systems more generally compared to alerts designed to detect clinical deterioration.
Challenges
While others have reported on the development and small-scale implementation of predictive alerts informed by ML algorithms, this is the first study to report on clinician perceptions of such tools. These results reveal potential barriers to positive clinical reception of EWS 2.0 including: 1) relative clinical stability of patients at the time of alert, 2) confidence in clinician judgment, 3) lack of transparency of the machine learning algorithm, and 4) uncertain response to alerts on high-risk patients who are not yet decompensating. These may be generalizable to other predictive alert systems, particularly those informed by ML algorithms.
Patients’ clinical stability at the time of alert may have contributed to poor confidence in the alerts’ clinical accuracy and relevance. As a predictive tool, EWS 2.0 triggered at a median of 5–6 hours, and in some cases several days before the onset of severe sepsis and septic shock. We suspect that many clinicians perceived EWS 2.0 as a traditional detection tool and dismissed its firing as erroneous or unhelpful when they discovered no evidence of clinical deterioration. The expectation of an immediate bedside evaluation likely contributed to this false perception that the alert was monitoring for decompensation requiring an immediate response. While implementation campaigns may mitigate such misperceptions, optimal lead-time of predictive alerts remains unclear.
Poor acceptance of EWS 2.0 may reflect little perceived added value to clinicians’ judgment given clinician confidence in their clinical reasoning and prognostication. Though EWS 2.0 demonstrated positive predictive values comparable to other widely accepted screening tools (31, 32), its ability to identify at risk patients may not exceed that of clinicians. In fact, clinicians reported already suspecting sepsis in almost half of patients who triggered the alert. While objective risk assessment through predictive alerts may help standardize otherwise subjective clinical impressions, clinicians may not find the alert helpful if they believe they already know which patients to monitor closely. The utility of such predictive alerts may thus be limited by a relatively small target population: high risk patients not yet viewed as high risk by clinicians. Further studies are needed to identify the subset of patients most likely to benefit from predictive alerts.
Clinicians may find it difficult to trust alerts developed using complex algorithms. ML algorithms in particular have been described as “black box models” because the variables informing their prediction are often not explicit or easily available to the user (29). Because ML algorithms can incorporate hundreds of variables, the factors that contribute to a prediction may be too unwieldy to distill into a meaningful narrative for clinicians. Furthermore, because the machine learning process identifies important variables that may not have previously been associated with particular outcomes, predictions based on these variables may be less clinically intuitive. Though challenging, transparency in machine learning algorithm design and alert trigger may help to justify risk assessments from the clinicians’ perspective.
Lack of established action items to implement after an alert may have also contributed to the perceived lack of alert value. It is unclear what, if any, management changes clinicians should implement for high-risk patients before clinical onset of severe sepsis or septic shock. Though there are several interventions one might expect to improve sepsis outcomes if implemented prior to clinically apparent disease, including increased monitoring, there is a paucity of data regarding their efficacy and cost-effectiveness. Further research is needed to avoid increasing unnecessary cost, inappropriate testing, and poor antibiotic stewardship.
Limitations
While response rates for the First Survey were comparable to that expected for web-based clinician surveys (33, 34), we cannot exclude non-responder bias. Low response rates and limited continuity reported by clinicians at 48 hours may limit the interpretation of the Second Survey results. However, it is unclear in which direction non-responder bias would influence our results, as both clinicians who are satisfied and dissatisfied might respond more frequently.
Next Steps
To be most useful, systems to predict severe sepsis and septic shock will require an iterative development process informed by clinician perceptions. Thorough implementation campaigns are important to familiarize clinicians with the role and utility of predictive systems that are distinct from detection systems. Whether alerts are the optimal modality for communicating risk predictions remains in question. Rather than triggers for rapid response deployment, sepsis prediction systems may be most useful as longitudinal risk stratification tools to inform objective risk assessments during team handoffs and diagnostic decision-making. More research is needed to determine the most useful lead-time and the most cost-effective and high impact interventions to deploy when patients are predicted to be high risk but do not yet have disease. Future predictive systems may be strengthened by screening for real-time sepsis-related orders from the EHR to more specifically target at-risk patients who would otherwise go undetected. In order to be trusted and adopted, machine learning predictive systems will need to be both accurate and interpretable (29). Interpretability will require transparency, ideally through interactive explanations and data visualization to translate the complex logic behind “black box models”.
CONCLUSIONS
Clinician perceptions of EWS 2.0 were mixed and, in general, poor. Despite excellent predictive characteristics, the EWS 2.0 alert infrequently provided new clinical information or changed management. Alerted patients’ relative clinical stability may have contributed to alert skepticism and uncertainty in response. Clinicians may find it difficult to trust complex predictive algorithms over their own clinical intuition if not provided with explanations to facilitate alert interpretation.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Neil O. Fishman, MD, William C. Hanson III, MD, Mark E. Mikkelsen MD, and Joanne Resnic MBA, BSN, RN for their contributions to the design, testing, and implementation of the clinical decision support intervention examined in this manuscript. These data were presented as a poster at the 2017 Society of Hospital Medicine Annual Meeting, Las Vegas, NV.
Financial Support:
Dr. Umscheid’s contribution to this project was supported in part by the National Center for Research Resources (grant no. UL1RR024134), which is now at the National Center for Advancing Translational Sciences (grant no. UL1TR000003). The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Dr. Schweickert has received funding from the American College of Physicians and Arjo. Dr. Umscheid’s institution has received funding from the National Institutes of Health, Food and Drug Administration, and Agency for Healthcare Research and Quality Evidence-based Practice Center contracts, and he has received funding from the Patient-Centered Outcomes Research Institute Advisory Panel and Northwell Health (grand rounds honoraria). The remaining authors have disclosed that they do not have any potential conflicts of interest.
Footnotes
Institution: Hospital of the University of Pennsylvania
REFERENCES
- 1.Rhee C, Dantes R, Epstein L, et al. : Incidence and Trends of Sepsis in US Hospitals Using Clinical vs Claims Data, 2009–2014. JAMA 2017; 318(13):1241–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones SL, Ashton CM, Kiehne LB, et al. : Outcomes and resource use of sepsis-associated stays by presence on admission, severity, and hospital type. Medical Care 2016; 54(3):303–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levy MM, Rhodes A, Phillips GS, et al. : Surviving Sepsis Campaign: association between performance metrics and outcomes in a 7.5-year study. Intensive Care Med 2014; 40(11):1623–1633 [DOI] [PubMed] [Google Scholar]
- 4.Kumar A, Roberts D, Wood KE, et al. : Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 2006; 34(6):1589–1596 [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharjee P, Edelson DP, Churpek MM: Identifying patients with sepsis on the hospital wards. Chest 2017; 151(4):898–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buck KM: Developing an early sepsis alert program. J Nurs Care Qual 2014; 29(2):124–132 [DOI] [PubMed] [Google Scholar]
- 7.Palleschi MT, Sirianni S, O’Connor N, et al. : An Interprofessional Process to Improve Early Identification and Treatment for Sepsis. J Healthc Qual 2014; 36:23–31 [DOI] [PubMed] [Google Scholar]
- 8.Brandt BN, Gartner AB, Moncure M, et al. : Identifying severe sepsis via electronic surveillance. Am J Med Qual 2015; 30(6):559–565 [DOI] [PubMed] [Google Scholar]
- 9.Umscheid CA, Betesh J, VanZandbergen C, et al. : Development, implementation, and impact of an automated early warning and response system for sepsis. J Hosp Med 2015; 10(1):26–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amland RC, Hahn-Cover KE.. Clinical decision support for early recognition of sepsis. Am J Med Qual 2016; 31(2):103–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McRee L, Thanavaro JL, Moore K, et al. : The impact of an electronic medical record surveillance program on outcomes for patients with sepsis. Heart Lung 2014; 43(6):546–549 [DOI] [PubMed] [Google Scholar]
- 12.Kurczewski L, Sweet M, Halbritter K: Reduction in time to first action as a result of electronic alerts for early sepsis recognition. Crit Care Nurs Q 2015; 38(2):182–187 [DOI] [PubMed] [Google Scholar]
- 13.Hackmann G, Chen M, Chipara O, et al. : Toward a two-tier clinical warning system for hospitalized patients. AMIA Annu Symp Proc 2011; 2011:511–519 [PMC free article] [PubMed] [Google Scholar]
- 14.Bailey TC, Chen Y, Mao Y, et al. : A trial of a real-time alert for clinical deterioration in patients hospitalized on general medical wards. J Hosp Med 2013; 8(5):236–242 [DOI] [PubMed] [Google Scholar]
- 15.Kollef MH, Chen Y, Heard K, et al. : A randomized trial of real-time automated clinical deterioration alerts sent to a rapid response team. J Hosp Med 2014; 9(7):424–4299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Escobar GJ, LaGuardia JC, Turk BJ, et al. : Early detection of impending physiologic deterioration among patients who are not in intensive care: development of predictive models using data from an automated electronic medical record. J Hosp Med 2012; 7(5):388–395 [DOI] [PubMed] [Google Scholar]
- 17.Churpek MM, Yuen TC, Park SY, et al. : Using electronic health record data to develop and validate a prediction model for adverse outcomes in the wards. Crit Care Med 2014; 42(4):841–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Churpek MM, Yuen TC, Winslow C, et al. : Multicenter comparison of machine learning methods and conventional regression for predicting clinical deterioration on the wards. Crit Care Med 2016; 44(2):368–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Escobar GJ, Ragins A, Scheirer P, et al. : Nonelective rehospitalizations and postdischarge mortality: predictive models suitable for use in real time. Medical Care 2015; 53(11):916–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thiel SW, Rosini JM, Shannon W, et al. : Early prediction of septic shock in hospitalized patients. J Hosp Med 2010; 5(1):19–25 [DOI] [PubMed] [Google Scholar]
- 21.Sawyer AM, Deal EN, Labelle AJ, et al. : Implementation of a real-time computerized sepsis alert in nonintensive care unit patients. Crit Care Med 2011; 39(3):469–473 [DOI] [PubMed] [Google Scholar]
- 22.Henry KE, Hager DN, Pronovost PJ, et al. : A targeted real-time early warning score (TREWScore) for septic shock. Sci Transl Med 2015; 7(299):299ra122. [DOI] [PubMed] [Google Scholar]
- 23.Shimabukuro DW, Barton CW, Feldman MD, et al. : Effect of a machine learning-based severe sepsis prediction algorithm on patient survival and hospital length of stay: a randomised clinical trial. BMJ Open Respir Res 2017; 4(1):e000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calvert JS, Price DA, Chettipally UK, et al. : A computational approach to early sepsis detection. Comput Biol Med 2016; 74:69–73 [DOI] [PubMed] [Google Scholar]
- 25.Desautels T, Calvert JS, Hoffman JL, et al. : Prediction of sepsis in the intensive care unit with minimal electronic health record data: a machine learning approach. JMIR Med Inform 2016; 4(3):e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCoy A, Das R: Reducing patient mortality, length of stay and readmissions through machine learning-based sepsis prediction in the emergency department, intensive care unit and hospital floor units. BMJ Open Qual 2017; 6:e000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Giannini HM, Ginestra JC, Chivers C, et al. : A machine learning algorithm to predict severe sepsis and septic shock: Development, implementation and impact on clinical practice. Critical Care Medicine. 2019. (In press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guidi JL, Clark K, Upton MT, et al. : Clinician Perception of the Effectiveness of an Automated Early Warning and Response System for Sepsis in an Academic Medical Center. Ann Am Thoracic Soc 2015; 12(10):1514–1519 [DOI] [PubMed] [Google Scholar]
- 29.Cabitza F, Rasoini R, Gensini GF.:Unintended consequences of machine learning in medicine. JAMA 2017; 318(6):517–518 [DOI] [PubMed] [Google Scholar]
- 30.Harris PA, Taylor R, Thielke R, et al. : Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009; 42(2):377–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Humphrey LL, Deffeback M, Pappas M, et al. : Screening for Lung Cancer with Low-Dose Computed Tomography: A Systematic Review to Update the U.S. Preventive Services Task Force Recommendation. Ann Intern Med 2013;159(6):411–420 [DOI] [PubMed] [Google Scholar]
- 32.Sprague BL, Arao RF, Miglioretti DL, et al. : National Performance Benchmarks for Modern Diagnostic Digital Mammography: Update from the Breast Cancer Surveillance Consortium. Radiology 2017; 283(1): 59–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beebe TJ, Jacobson RM, Jenkins SM, et al. : Testing the Impact of Mixed-Mode Designs (Mail and Web) and Multiple Contact Attempts within Mode (Mail or Web) on Clinician Survey Response. Health Serv Res 2018; 53 Suppl 1:3070–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cunningham CT, Quan H, Hemmelgarn B, et al. : Exploring physician specialist response rates to web-based surveys. BMC Med Res Methodol 2015; 15:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.