SUMMARY
Mammalian SWI/SNF (mSWI/SNF) ATP-dependent chromatin remodeling complexes are multi-subunit molecular machines that play vital roles in regulating genomic architecture and are frequently disrupted in human cancer and developmental disorders. To date, the modular organization and pathways of assembly of these chromatin regulators remain unknown, presenting a major barrier to structural and functional determination. Here, we elucidate the architecture and assembly pathway across three classes of mSWI/SNF complexes—canonical BRGI/BRM-associated factor (BAF), polybromo-associated BAF (PBAF), and newly defined ncBAF complexes—and define the requirement of each subunit for complex formation and stability. Using affinity purification of endogenous complexes from mammalian and Drosophila cells coupled with cross-linking mass spectrometry (CX-MS) and mutagenesis, we uncover three distinct and evolutionarily conserved modules, their organization, and the temporal incorporation of these modules into each complete mSWI/SNF complex class. Finally, we map human disease-associated mutations within subunits and modules, defining specific topological regions that are affected upon subunit perturbation.
In Brief
Mapping assembly pathways for mSWI/ SNF remodeling complexes delineates three distinct organizational modules and contextualizes human disease mutations.
Graphical Abstract
INTRODUCTION
ATP-dependent chromatin remodeling complexes are multimeric molecular assemblies that regulate chromatin architecture (Kadoch and Crabtree, 2015; Masliah-Planchon et al., 2015; Wu et al., 2009). These complexes are grouped into four major families, including switching (SWI)/sucrose fermentation (sucrose non-fermenting [SNF]), IN080 (Conaway and Conaway, 2009), ISWI (imitation SWI; Bartholomew, 2014), and CHD/M-2 (chromodomain helicase DNA-binding) groups (Murawska and Brehm, 2011), all of which contain Snf2-like ATPase subunits but differ substantially via the incorporation of distinct subunits and in their targeting and activity on chromatin (Clapier et al., 2017).
SWI/SNF complexes were originally discovered and characterized in yeast, later in Drosophila (Dingwall et al., 1995; Stern et al., 1984), and most recently in mammals (Ho et al., 2009; Kadoch et al., 2013). Mammalian SWI/SNF (mSWI/SNF) complexes are ~1- to 1.5-MDa entities combinatorially assembled from the products of 29 genes, including multiple paralogs, generating extensive diversity in composition. All complexes contain an ATPase subunit, either SMARCA4 (BRG1) or SMARCA2 (BRM), that catalyzes the hydrolysis of ATP. The roles of most other accessory subunits in complex assembly and stability as well as targeting and function remain unknown.
Over the past several years, mSWI/SNF complexes have emerged as a major focus of attention because of the striking mutational frequencies in the genes encoding their subunits across a range of human diseases, from cancer to neurologic disorders. Indeed, recent exome sequencing studies have revealed that over 20% of human cancers bear mutations in the genes encoding mSWI/SNF subunits (Kadoch et al., 2013). In addition, heterozygous point mutations in mSWI/SNF genes have been implicated as causative events in intellectual disability and autism spectrum disorders (Bogershausen and Wollnik, 2018; López and Wood, 2015)
A major barrier to our understanding of the functions, tissue-specific roles, and effect of mutations on mSWI/SNF complex mechanisms lies in the lack of information regarding subunit organization, assembly, and 3D structure. Several factors pose major challenges to such studies. Individually expressed subunits are often unstable or incorrectly folded without their appropriate binding partners, and minimal complexes pieced together via in vitro co-expression may not represent endogenous, physiologically relevant complexes in cells. Large quantities of purified endogenous complexes with minimal heterogeneity are required for downstream analyses, and selection of appropriate purification strategies cannot be informed without understanding modular architecture and assembly order. For these reasons and others, to date only low-resolution maps have been achieved using cryoelectron microscopy (cryo-EM) approaches (Dechassa et al., 2008; Leschziner et al., 2007), and X-ray crystallographic analyses have been successfully performed on only a few isolated domains (Yan et al., 2017), including the recently reported yeast Snf2 ATPase domain (Liu et al., 2017; Xia et al., 2016).
To establish a comprehensive structural framework for mSWI/ SNF complexes, we used a multifaceted series of approaches involving complex and subcomplex purification, mass spectrometry (MS), crosslinking mass spectrometry (CX-MS), systematic genetic manipulation of subunits and subunit paralog families, evolutionary analyses, and human disease genetics. These studies reveal that mSWI/SNF complexes exist in three non-redundant final form assemblies: BRGI/BRM-associated factor complexes (BAFs), polybromo-associated BAF complexes (PBAFs), and non-canonical BAFs (ncBAFs), for which we establish the assembly requirements and modular organization. We define the full spectrum of endogenous combinatorial possibilities and the effect of individual subunit deletions and mutations, including recurrent, previously uncharacterized missense and nonsense mutations, on complex architecture. These studies provide important insights into mSWI/SNF complex organization, structure, and function and the biochemical consequences of a wide range of human disease-associated mutations.
RESULTS
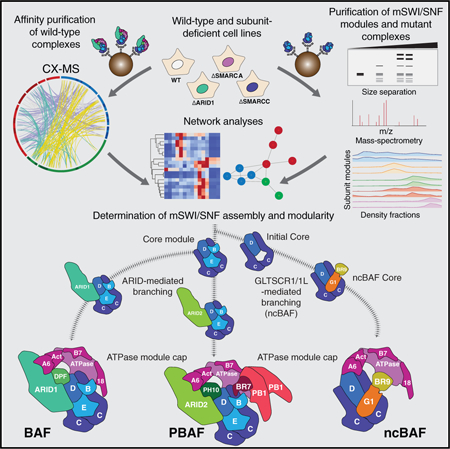
Affinity Purification of Endogenous mSWI/SNF Reveals Distinct Complex Types and Their Intermediates
To begin to probe the modular organization and assembly order of mSWI/SNF family complexes, we subjected HEK293T cell nuclear extracts to density sedimentation analyses using 10%–30% glycerol gradients, reasoning that such an approach could reveal the presence of distinct final-form SWI/SNF complexes as well as assembly pathway intermediates (Figure 1A). We identified a range of migration patterns, with subunits such as SMARCD1 and SMARCC1 exhibiting marked spreading across the gradient and complex-defining subunits migrating in a restricted set of fractions, such as DPF2 and ARID1A (fraction of [Fx] 13–14), marking canonical BAF (cB or BAF) complexes and ARID2, BRD7, and PBRM1 in higher-mass fractions, Fx 16–17, marking PBAF complexes. In addition, BRD9 and GLTSCR1/1L subunits, corresponding to a newly identified class of mSWI/SNF complexes we have termed ncBAF (Alpsoy and Dykhuizen, 2018; Ho et al., 2009; Hohmann et al., 2016; Kadoch et al., 2013; Sarnowska et al., 2016), exhibited distinct lower-molecular-weight migration patterns (Fx 9–10).
Figure 1. Distinct mSWI/SNF Complexes and Their Intermediates Revealed through Affinity Purification.
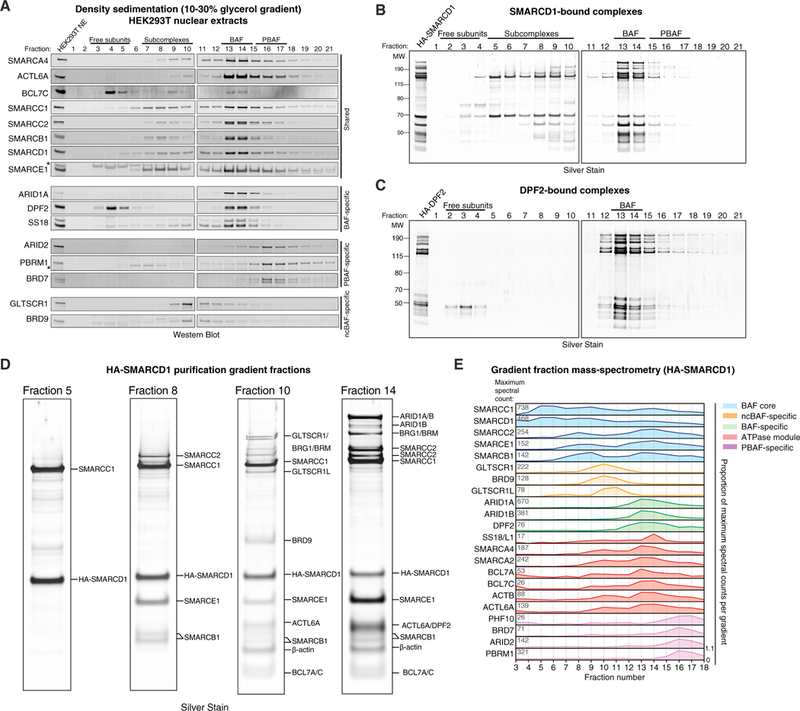
(A) Density sedimentation and immunoblot performed on HEK293T nuclear extracts. *, non-specific bands.
(B) Silver stain performed on density sedimentation of HA-SMARCD1 mSWI/SNF complexes purified from HEK293T cells.
(C) Silver stain performed on density sedimentation of HA-DPF2 BAF complexes purified from HEK293T cells.
(D) Silver staining of the indicated HA-SMARCD1 gradient fractions from (B). Identified proteins are labeled.
(E) Mass spectrometry analysis performed on selected fractions (fractions 3–18) collected from the HA-SMARCD1 density gradient in (B). Peptide proportion (0to1) representsthe fraction ofthe maximum number of peptides captured for each subunit overthe full gradient. Total spectral counts in fraction with highest peptide abundance for each subunit are indicated on the left. Colors distinguish mSWI/SNF complexes and modules.
See also Figure S1.
Using these results, we developed a robust purification strategy to capture endogenous mammalian complexes at each of these extremes with over 95% purity (Figure S1A; Table S1). SMARCD1-based purifications were used to capture all forms of mSWI/SNF complexes (because SMARCD1 is present across the full gradient), and hemagglutinin (HA)-DPF2 was used to purify fully assembled BAF complexes, which do not contain PBAF or ncBAF complex components (Figures S1B and S1C). Remarkably, density sedimentation and silver staining of purified complexes revealed that SMARCDI-captured complexes spread across the gradient, whereas DPF2 complexes marked only complete BAF complexes with no detectable intermediates (Figures 1B–1D; Figures S1D and S1E; Table S2), highlighting the utility of this approach to detect specific complexes and intermediate modules. Analysis of spectral counts from mass spectrometry performed across SMARCD1 gradient fractions confirmed the silver stain results and further identified components with lower abundance, such as ncBAF and PBAF subunits (Figure 1E; Figure S1F; Table S2). Taken together, these data suggest a stepwise, modular assembly pathway for mSWI/SNF family complexes, resulting in three distinct final complex forms, each with their own combinatorial diversity.
Cross-Linking Mass Spectrometry of Canonical BAF Complexes Globally Defines Modular Architecture
We next performed bis(sulfosuccinimidyl)suberate (BS3)-based cross-linking mass spectrometry using DPF2 and SS18 as baits to identify BAF subunit architecture and linkages. We generated high-density subunit crosslinking maps based on 1,560 identified spectra for inter-protein crosslinks and 2,373 identified spectra for intra-protein crosslinks with coverage across all BAF complex subunits, with the exception of SS18 (because of limited lysine residues) (Figures 2A and S2A; Table S3; STAR Methods). To comprehensively define regions of crosslinking between BAF complex subunits, we divided each subunit family (collapsed; i.e., SMARCD = SMARCD1, SMARCD2, or SMARCD3) into regions based on existing domain annotation, conservation, and newly defined domains stemming from this cross-linking mass spectrometry work (Figures 2A and S2B). The median distance between crosslinked residues within domains of known structure was 12.2 Å, close to the expected 11.4–30 Å distance for the BS3 crosslinking agent (Figure S2C; Table S4). In addition, the C-alpha distances between crosslinked residues mapped onto the structure of the Snf2 helicase were within expected distances for the nucleosome-bound and free conformations (Liu et al., 2017; Xia et al., 2016; Figure S2D).
Figure 2. Cross-Linking Mass Spectrometry of SWI/SNF Complexes Reveals Conserved Connectivity of Interacting Modules.
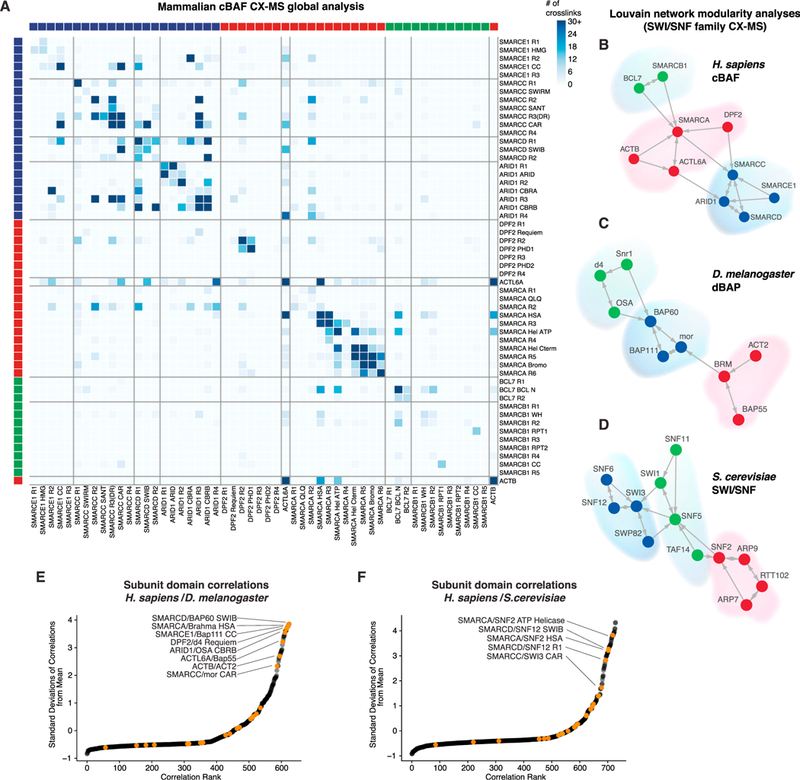
(A) Matrix heatmap of the total crosslinks identified in combined HA-SS18 and HA-DPF2 BAF complex cross-linking mass spectrometry datasets. Individual subunits are divided into domains and ordered according to modules in (B). See also Figures S2B, S2J, and S2K.
(B-D) Louvain modularity analysis performed on (B) mammalian cBAF complex cross-linking mass spectrometry datasets, (C) D. melanogaster D4 and BAP60 cross-linking mass spectrometry datasets, and (D) S. cerevisiae cross-linking mass spectrometry datasets (from Sen et al., 2017).
(E) Correlations between mammalian and Drosophila BAF or BAP subunit domain and region interactions from cross-linking mass spectrometry datasets. See also Figures S2B and S2J.
(F) Correlations between mammalian and yeast SWI/SNF subunit domain and region interactions from cross-linking mass spectrometry datasets. See also Figures S2B and S2K.
See also Figure S2.
To elucidate potential crosslinking preferences between subunits, we performed Louvain two-nearest-neighbor analysis, where nodes are subunits (or paralog families) and edges are drawn between the top two crosslinking partners for each subunit based on the number of BAF crosslinks (Blondel et al., 2008). This clustering revealed three distinct network modules: a catalytic module containing the SMARCA ATPase subunit, β-actin, and ACTL6A, an associated module containing SMARCB1 and BCL7, and a module containing SMARCC, SMARCD, SMARCE1, and ARID1 (Figure 2B), recapitulating our inferred assembly of components. In addition, correlation an analyses of total inter-subunit crosslinks for each subunit revealed similar results (Figure S2E).
Arthropods represent a parallel evolutionary branch to meta-zoans that retain at least two classes of SWI/SNF complexes, namely BAP (BAF in mammals) and PBAP (PBAF in mammals). Hence, we isolated BAP complexes from D. melanogaster S2 cells using insect orthologs of DPF2 (D4) and SMARCD1 (BAP60) as baits and performed cross-linking mass spectrometry (Figures S2F and S2G). Similar to mammalian complexes, the ATPase module clustered with BAP55 (ACTL6A ortholog) and ACT2 (β-actin ortholog), and moira (mor) (SMARCC ortholog) formed a tight network with BAP60, BAP111 (SMARCE1 ortholog), and Osa (ARID1 ortholog), whereas Snr1 (SMARCB1 ortholog) and D4 separated as a distinct module (Figures 2C and S2H; Table S5). These cross-linking mass spectrometry results suggest conserved modularity for at least two complex modules: the BAF ATPase module and the “core module” that forms around SMARCC or mor subunits. Finally, using a recently published S. cerevisiae SWI/SNF cross-linking mass spectrometry dataset (Sen et al., 2017), we found similar clustering of the majority of both core and ATPase subunits, with the SNF2-centered ATPase module containing ARP7, ARP9 (potential orthologs of ACTL6A), and RTT102. SWI3 (SMARCC ortholog) and SNF12 (SMARCD ortholog), along with yeast-specific SNF6 and SWP82, form the core module, and SWI1 (ARID1 ortholog) and SNF5 (SMARCB1 ortholog) subunits cluster and bridge the core and ATPase modules (Figures 2D and S2I–S2L). Using correlation analyses of crosslinks within individual subunit regions and domains across mammalian, fly, and yeast complexes, we found that the most highly conserved interactions were between regions of the BAF core, OSA or ARID1, and ATPase modules (Figures 2E, 2F, S2M, and S2N). Taken together, SWI/SNF complexes retain specific modular organization across evolutionarily distant branches of life, indicating functional conservation of subunit architecture.
Characterization of the BAF Core Module Components and Their Assembly
Complex purifications (Figures 1B and 1D) coupled with these cross-linking mass spectrometry analyses suggested the presence of an early subcomplex containing SMARCD and SMARCC followed by SMARCE1 and SMARCB1 subunits (Figure 3A). Indeed, SMARCC1 purifications showed enrichment of the same subcomplex module (Figure 3B; Table S2). Similar results were obtained from SMARCB1, SMARCE1, and SMARCD2 purifications (Figures S3A–S3I; Table S2) using both mass spectrometry and fluorometric approaches and demonstrated SMARCB1 association with the BAF core module of cBAF and PBAF (Figures S3A–S3G). Of note, ncBAF-specific BRD9 and GLTSCR1 and GLTSCR1L components were completely absent in these three purifications, further suggesting that these subunits mark complexes of unique composition and lack several ubiquitously expressed, highly conserved subunits.
Figure 3. Identification and Characterization of the BAF Core Module: SMARCC, SMARCD, SMARCB1, and SMARCE1 Subunits.
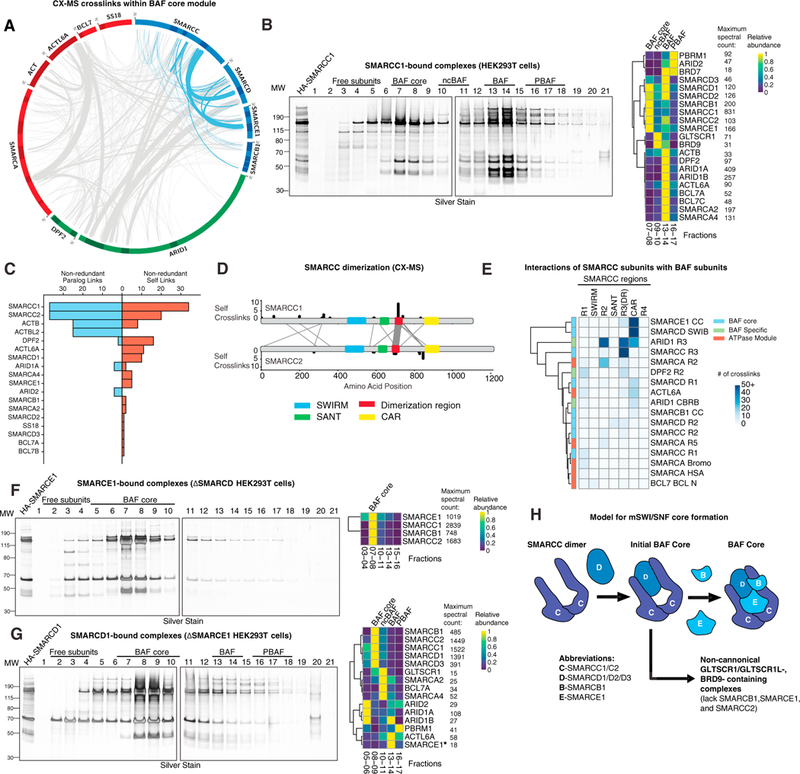
(A) Circle-plot analysis of the mammalian BAF complex cross-linking mass spectrometry dataset, with the BAF core module highlighted in blue.
(B) Silver stain performed on density sedimentation of HA-SMARCC1 complexes purified from HEK293T cells (left) and clustered heatmap of mass spec-called peptides and spectral counts on selected fractions (right).
(C) Distribution of inter-paralog crosslinks and self-crosslinks in the BAF cross-linking mass spectrometry dataset.
(D) SMARCC self-crosslinks and SMARCC1 and SMARCC2 inter-paralog crosslinks from the BAF cross-linking mass spectrometry dataset. Line width is proportional to the number of crosslinks.
(E) Heatmap depicting SMARCC crosslinks with BAF subunits from the BAF cross-linking mass spectrometry dataset.
(F) Silver stain performed on density sedimentation of HA-SMARCE1 complexes purified from ∆SMARCD HEK293T cells (left) and clustered heatmap of mass spec-called peptides and spectral counts on selected fractions (right).
(G) Silver stain performed on density sedimentation of HA-SMARCD1 complexes purified from ∆SMARCE1 HEK293T cells (left) and clustered heatmap of mass spec-called peptides and spectral counts on selected fractions (right). *, minimal SMARCE1 peptide abundance was detected despite no observed band (see also Table S2).
(H) Schematic representation of initial steps of BAF core assembly. Subunit abbreviations are indicated.
See also Figure S3.
SMARCC subunits have been shown to form homo- and heterodimers (as C1/C1, C1/C2, orC2/C2), with C1/C1 homodimers found in embryonic stem cells (ESCs) and C1/C2 heterodimers in most differentiated cell types (Ho et al., 2009; Wang et al., 1996). Cross-linking mass spectrometry analysis showed either hetero-dimerization (by crosslinking between paralog subunits) or homodimerization (by crosslinked residues mapping to the same position of the identical peptide sequence, here termed “self-crosslinks”) (Figure 3C). Self-crosslinks were abundant in SMARCC subunits and p-actin, which is known to polymerize (DPF2 also exhibited some crosslinking because of high free subunit concentrations). Immunodepletion of SMARCC1 and SMARCC2 further revealed preferential homodimerization of this subunit family in this cell context (Figure S3J). Using colloidal blue stain and fluorometric analysis of DPF2-purified complexes to approximate relative subunit stoichiometry, we found that most components of the complex are present in nearly 1:1 stoichiometry, with the exception of SMARCC1, which displayed 1:1.6, reflecting its known dimerization (Figure S3K). SMARCC2 displayed nearly 1:1 stoichiometry, most likely because of its lower expression in these cells in comparison with SMARCC1. Despite preferential homodimer formation, we identified substantial SMARCC1 and SMARCC2 crosslinks and found a region C-terminal to the SANT domain (amino acids [aa] 679–747) that contained the majority of self crosslinks with the subunit paralog, which we named the dimerization region (DR), whereas no crosslinks were identified within established domains (Figure 3D; Figure S3L). The SMARCC coiled-coil region also contained a high number of crosslinks to the SWIB domain of the SMARCD core subunit (Figure 2A). Given that we repeatedly observed a SMARCC and SMARCD heteromer without any other BAF core module components in early gradient fractions, we concluded that this trimer is the first mSWI/SNF assembly intermediate, which we termed the initial BAF core.
To determine the order of assembly for the BAF core module of SMARCC, SMARCD, SMARCB1, and SMARCE1 subunits, we systematically deleted each component using CRISPR-Cas9, removing all paralogs of each subunit family (i.e., SMARCC1 and SMARCC2, SMARCD1/2/3, SMARCE1 [one gene] and SMARCB1 [one gene]) because of structural redundancy. Importantly, removal of both SMARCC subunits resulted in near-complete degradation of all mSWI/SNF complex components (Figure S4M), supporting the role for the SMARCC dimer as a platform for mSWI/SNF formation. Indeed, SMARCC crosslinks reveal additional binding regions aside from the DR: a conserved region (core assembly region [CAR]) that interacts with core subunits SMARCE1 and SMARCD, and the R2 and CAR regions that crosslink to ARID1 subunits (Figures 2A, 3E, and S2B). Loss of SMARCD inhibited BAF complex assembly and resulted in complete disruption of ARID and ATPase subunit binding; nonetheless, we were still able to observe SMARCD- deficient BAF core formation in fractions 7–8 using SMARCE1 and SMARCB1 as baits for purification and in co-immunoprecipitation (coIP) experiments (Figures 3F and S3M–S3O; Table S2). These data suggest that all three BAF core subunits bind the SMARCC dimer platform using distinct, independent interfaces.
Loss of SMARCE1 resulted in partial complex destabilization as subunit abundance was drastically shifted toward BAF core intermediates in Fx 8–9 (Figures 3G and S3Q; Table S2). Complexes were destabilized relative to wild-type (WT) BAF, and ARID subunits were observed in Fx 5–6, suggesting that they are unable to stably bind complexes in the absence of SMARCE1. In contrast to stringent gradient sedimentation, coIP showed that SMARCE1 loss minimally affected BAF complex formation, implicating a possible role in inter-module stability (Figure S3M). Finally, SMARCB1 deletion resulted in a minimal effect on BAF complex formation, confirming our previous observations (Nakayama et al., 2017; Figure S3P; Table S2). However, we observed a shift in the migration of PBAF components to Fx 12–14 (in contrast to Fx 16–17 in WT cells), suggesting that SMARCB1 is important for normal PBAF stoichiometry or PBAF-specific subunit binding. Of note, in both DSMARCE1 and DSMARCB1 settings, ncBAF complex components were still readily detectable and unaffected (Fx 10–11), consistent with our finding that these complexes lack SMARCE1 and SMARCB1 (Figures 3G and S3P). Taken together, these data suggest that mSWI/SNF complex assembly is triggered by formation of the initial BAF core (SMARCC and SMARCD) formed around the SMARCC dimer. This initial subcomplex then acts as a platform for independent docking of SMARCE1 and SMARCB1 subunits to form the BAF core module, which is required for assembly toward fully formed cBAF and PBAF complexes (Figure 3H).
ARID Subunits Interact with the BAF Core Module to Facilitate Binding of the ATPase Subcomplex
Cross-linking mass spectrometry analyses indicated that BAF core components (SMARCD, SMARCC, SMARCB1, and SMARCE1) strongly crosslinked with ARID subunits, ARID1A and ARID1B. The C-terminal region of ARID1A and ARID1B exhibited a large number of crosslinks to the BAF core, particularly to SMARCC and SMARCD (Figure 4A). ARID1 proteins contain several distinct, conserved regions, including the N terminus, the ARID domain, and three potential domains in the C terminus that we termed core binding region (CBR) A and B and region 4 (R4) (Figures 2A, S2B, and S4A). CBRs and R4 crosslink to the BAF core and ATPase subunits, respectively (Figure 4B). For example, CBR A displays preferential binding to SMARCD R1 and SMARCE1 R2, ARID1 R3 exhibits crosslinks to several SMARCC regions, and CBR B crosslinks to SMARCC CAR and SMARCD R1 and R2. ARIDI R4 crosslinks to the ATPase component SMARCA and ACTL6A components (Figures 4B and 4C). These results were similar in both yeast and Drosophila, indicating conservation of ARID and SWI1 binding modality (Figure S4B).
Figure 4. ARID Subunits Dictate Specific Branches of BAF and PBAF Complex Assembly.
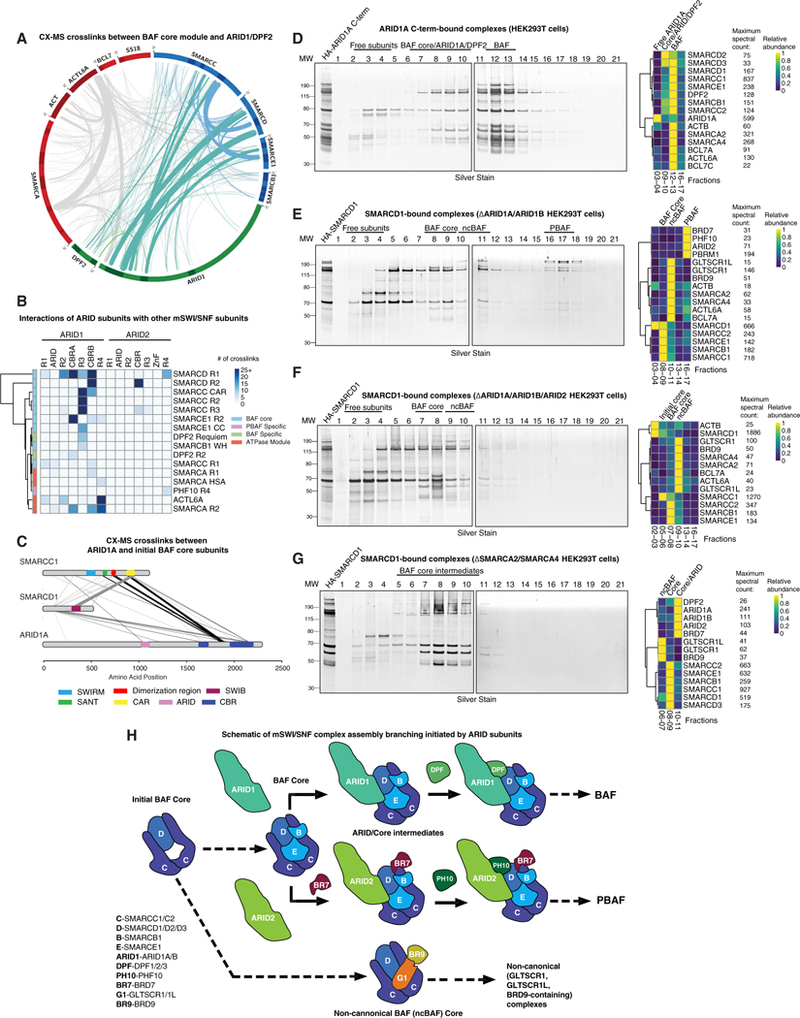
(A) Circle-plot analysis of the mammalian cross-linking mass spectrometry dataset, with BAF core subunit crosslinks in blue and ARID module subunits in teal.
(B) Clustered heatmap of cross-linking mass spectrometry data, highlighting crosslinks between ARID subunits and other complex components.
(C) ARID1A, SMARCC1, and SMARCD1 crosslinks from the BAF cross-linking mass spectrometry dataset. Line width is proportional to the number of crosslinks.
(D) Gradient and mass spectrometry heatmap of native HA-ARID1A C terminus-bound BAF complexes purified from HEK293T cells.
(E-G) Native HA-SMARCD1 purification and gradient mass spectrometry in (E) ARID1Aand ARID1B-deficient, (F) ARID1A, ARID1B, and ARID2-deficient, and (G) SMARCA4/2-deficient HEK293T cells.
(H) mSWI/SNF assembly branch points are initiated by ARID subunits. Subunit abbreviations are indicated.
See also Figure S4.
The ARID domain of ARID1 subunits displayed limited crosslinking, suggesting its involvement in complex recruitment to DNA rather than its role in assembly of the complex. Guided by these results, we cloned and expressed a C-terminal ARID1A fragment containing CBR A, CBR B, and R4 (aa 1,611–2,285), which we predicted to stably bind and facilitate the assembly of complete BAF complexes. We found that the HA-ARID1A C terminus is sufficient to interact with and capture fully formed BAF complexes (Figure 4D; Table S2). Mass spectrometry analysis of lower-molecular-weight gradient fractions revealed intermediates containing the BAF core module, ARID1A C-terminal region, and DPF2 (Figure 4D). In addition, the ARID1AC terminus was sufficient to enable incorporation of DPF2 into both ARID1 or BAF complex core intermediates as well as full BAF complexes, suggesting that the DPF2 subunit requires both modules for its binding.
To test this, we performed DPF2 affinity purifications in BAF core module subunit deletion mutant cell lines (DSMARCB1 and DSMARCE1 lines). Importantly, we observed a complete loss of BAF complex capture (and, hence, DPF2 binding) in these settings as well as in ARID1Aand ARID1B double knockout (KO) 293T cells or MIA-Pa-Ca-2 cells (deficient in ARID1A and ARID1B) (Figures S4C–S4G). DPF2 crosslinks to all modules of the BAF complex, suggesting a large interaction interface and consistent with its binding preference for fully formed cBAFcomplexes (Figure S4H). However, removal of the ATPase subunits SMARCA2 and SMARCA4 did not disrupt DPF2 assembly (Figures S4I and S4J), suggesting that the ATPase module is the last to be incorporated into mSWI/SNF complexes. These data corroborate results from DPF2 purifications (Figure 1C), explaining why DPF2 exists only as part of fully formed BAF complexes or as a free subunit and never as part of any assembly intermediates.
To define the requirement for ARID1 subunits in BAF complex assembly, we analyzed SMARCD1-bound complexes in DARID1 (DARID1A and ARID1B) KO cells (Figure 4E; Table S2). We observed normal BAF core formation in Fx 8–9, ncBAF in Fx 10–11, and PBAF in Fx 16–18. However, there were no detectible cBAF complexes in the expected Fx 13–14. These data indicate that ARID proteins interact with fully assembled BAF core modules, which then enable binding of the ATPase module through interaction of the ARID R4 region with ACTL6A and SMARCA subunits. In addition, we found ncBAF forms completely independently of the presence of ARID1 subunits, suggesting an ARID-independent ATPase recruitment mechanism. Finally, we purified SMARCD1-bound complexes in cells lacking all three mSWI/SNF family ARID proteins (DARID1A, DARID1B, and DARID2). Despite intact assembly of the BAF core module, upon losing ARID2 in addition to ARID1A and ARID1B, assembly of both BAF and PBAF complexes was completely inhibited (Figure 4F; Table S2).
These results demonstrate that ARID proteins nucleate complex-specific branching into BAF and PBAF complexes (ARID1A and ARID1B for BAF and ARID2 for PBAF). To detect ARID-containing intermediate complexes, we performed SMARCD1 purifications from HEK293T cells lacking both ATPases (DSMARCA2 and DSMARCA4), followed by native complex gradient separation and mass spectrometry (Figure 4G; Table S2). As expected, we detected complexes of smaller size, similar to DPF2-purified BAF complexes from SW13 ATPase-deficient cells (Figures S4I and S4J) which resolved partially formed ncBAF complexes (consisting of the initial core [SMARCC and SMARCD1] and BRD9 and GLTSCR but lacking the ATPase and its associated components in Fx 6–7), which we termed the ncBAF core module; BAF core module components SMARCB1 and SMARCE1, which do not bind ncBAF (Fx 8–9); and a mixture of BAF and PBAF intermediates containing the core module, ARID1 or ARID2, and the PBAF-specific subunit BRD7 (Fx 10–11) (indicating that BRD7 is the next PBAF-specific member to assemble on to the core and ARID modules) (Figure 4G). Global coIP and immunoblot confirmed findings across a range of mutant cell lines (Figure S4K).
The ATPase Module Finalizes Assembly of All Three mSWI/SNF Family Complexes
SMARCA2 and SMARCA4 ATPases crosslink extensively with components previously identified to engage with the ATPase, such as β-actin and ACTL6A (Zhao et al., 1998), as well as BCL7A, BCL7B, or BCL7C and SS18 (Figures 5A and S5A). Substantial crosslinks were detected between ACTL6A and β-actin and the SMARCA2 and SMARCA4 HSA domain and between β-actin and ACTL6A (Figure 5B). We found similar interaction preferences for the actin-like proteins and the HSA and catalytic domains across species (Figure S5B). In further support of our model in which ARID1 bridges the BAF core and ATPase modules, we detected a large number of crosslinks between ACTL6A and the ARID1 C-terminal R4 as well as between SMARCA2 and SMARCA4andARID1 CBRA and B(Figure5B). Inaddition, R2 of SMARCA crosslinks with both ARID1 subunits as well as other BAF core components, including SMARCC R2 and SMARCD R1. The N termini of both SS18 and BCL7 crosslink to the N-terminal R1 and HSA domains of SMARCA subunits, respectively.
Figure 5. The mSWI/SNF ATPases Recruit Accessory Subunits and Finalize BAF, PBAF, and ncBAF Complex Assembly.
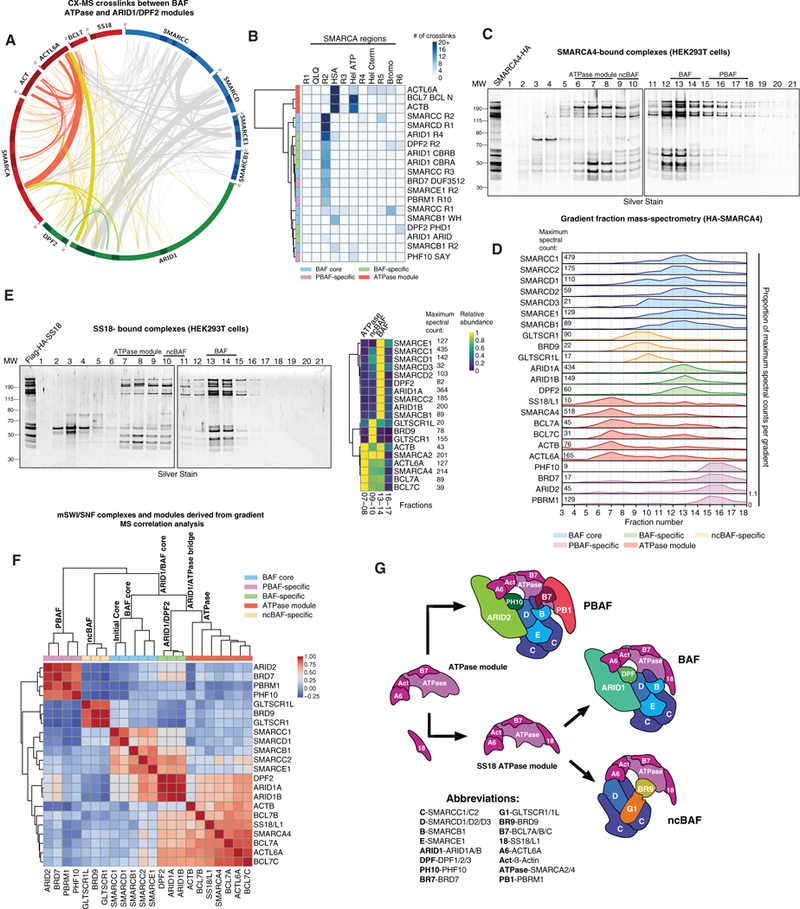
(A) Circle-plot analysis of the mammalian cross-linking mass spectrometry dataset with ATPase module subunits crosslinks in red and ATPase/ARID module crosslinks in yellow.
(B) Clustered heatmap of the cross-linking mass spectrometry analysis of the mammalian BAF complex, highlighting the occurrence of crosslinks between SMARCA and other complex components.
(C) Silver stain performed on density sedimentation of HA-SMARCA4-bound complexes purified from HEK293T cells.
(D) Gradient mass spectrometry of selected fractions collected from the HA-SMARCA4 density gradient. Total spectral counts in fraction with highest peptide abundance for each subunit are indicated on the left.
(E) Silver stain performed on density sedimentation analysis of FLAG-HA-SS18-bound BAF complexes purified from HEK293T cells (left) and clustered heatmap of mass spec-called peptides and spectral counts on selected fractions (right).
(F) Clustered correlation heatmap performed on HA-SMARCD1, HA-SMARCB1, and HA-SMARCA4 density gradient mass spectrometry results from HEK293T cells. Experimentally determined complexes and subcomplexes are indicated.
(G) Schematic of the assembly and incorporation of the BAF ATPase module. Subunit abbreviations are indicated.
See also Figure S5.
To reveal whether ATPases and their associated subunits form a separate module, we purified SMARCA4-bound complexes. Indeed, we were able to clearly separate the ATPase module in Fx 6–9 from ATPase module-containing fully formed BAF complexes (Figures 5C and 5D; Figure S5C). In addition to cBAF complexes, SMARCA4 purification captured components of ncBAF and PBAF in expected Fx 9–10 and 15–16, respectively.
In further validation of the ATPase as a distinct module, we performed purifications using satellite ATPase module subunits. SS18-bound complexes separated on gradients in a manner similar to SMARCA4-bound complexes and captured ncBAF complexes (Fx 10–11) (Figures 5E and S5D–S5F; Table S2) but not PBAF subunits because SS18 does not assemble into PBAF complexes (Nakayama et al., 2017), suggesting a mutually exclusive competition between SS18 and PBAF-specific subunits such as PBRM1. BCL7 purifications resolved all three mSWI/SNF complexes in expected fractions (Figure S5G), demonstrating that BCL7 proteins are pan-mSWI/SNF ATPase module components.
Louvain modularity analysis performed on mass spectrometry datasets from SMARCD1, SMARCB1, and SMARCA4 purifications and gradients showed clear separation of core BAF, ATPase, and ARID modules as well as separation between PBAF and ncBAF as branches connected to the main group of subunits through ARID2 and SMARCD1, respectively (Figures 5F and S5H). CoIP and immunoblot of endogenous complexes from SMARCA2 and SMARCA4 double KO HEK293T cells indicated intact assembly of the BAF core and ARID and DPF2 modules but a marked and specific loss of ATPase module stability and interaction (Figure S5I). SS18 and SS18L1 double KO cells displayed no assembly defects apart from a general increase in PBAF complex abundance, corroborating our competition model above.
Because of the lack of intermediate ATPase subcomplexes, we concluded that each of the components of this module binds independently to the large SMARCA platform, which is then incorporated as a unit into pre-assembled BAF, PBAF, and ncBAF subcomplexes. We define a split in assembly of the ATPase modules that differs between BAF, ncBAF, and PBAF because SS18-containing complexes contained only BAF and ncBAF components but were devoid of PBAF components. These data suggest that the final step of mSWI/ SNF complex assembly is controlled by both specific components of the core BAF modules as well as the elements of the ATPase subcomplex components, SS18 and PBRM1 (Figure 5G).
Assembly of PBAF and ncBAF Complexes and the Global Mammalian SWI/SNF Assembly Pathway
To define the assembly and inter-subunit linkages of PBAF complexes, we performed cross-linking mass spectrometry on BRD7- and PHF10-bound complexes, confirming that PBAF complexes contain the same common BAF core module as BAF complexes (Figure S6A; Table S5). We detected PBAF intermediates containing the BAF core module, ARID2, BRD7, and PHF10 (Figure 4G). PBAF assembly is initiated by ARID2 as loss of ARID2 completely disrupts PBAF complex assembly (Figure S4K). To dissect the last steps of PBAF assembly, we purified ARID2-bound complexes using a mini version of ARID2 predicted by cross-linking mass spectrometry to bind PBAF (mARID2, aa 1–626 fused to C-terminal aa 1,592–1,835). mARID2 displayed increased expression levels compared with full-length ARID2, sufficient to purify protein complexes (Figure 6A; Table S2). We observed fully formed PBAF complexes in expected Fx 15–17 and partial assemblies in Fx 12–13, with PBRM1 being the only subunit absent in PBAF subcomplex fractions, suggesting that it requires full-length ARID2, other PBAF-specific subunits, and the ATPase module for its incorporation. Finally, PBRM1-bound PBAF complexes migrated in expected Fx 15–17. Mass spectrometry analysis did not identify any PBRM1-containing intermediate complexes apart from its free form in Fx 5–6 (Figure 6B), suggesting that PBRM1 is one of the last subunits to be added to the PBAF complex via interaction of its C terminus with both SMARCC and ATPase module subunits, as determined by cross-linking mass spectrometry (Figure S6B; Table S6).
Figure 6. Assembly of Alternative mSWI/SNF Complexes, PBAF and ncBAF, and the Full Assembly Pathway.
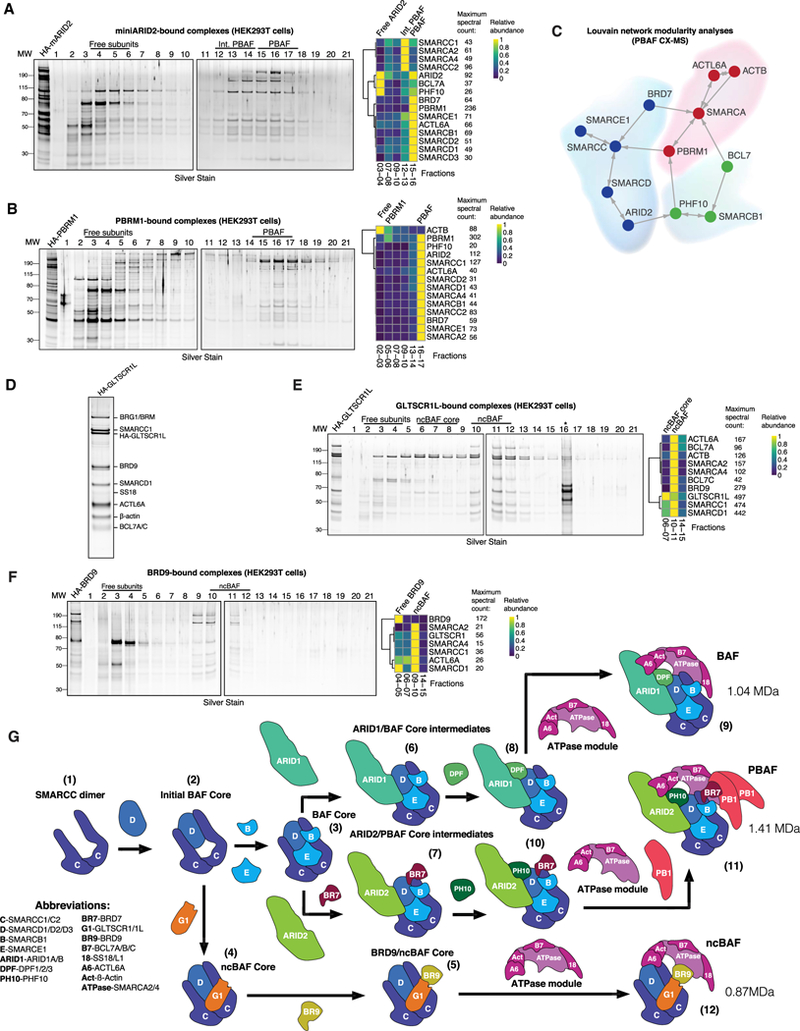
(A) Silver stain performed on density sedimentation of HA-mARID2 PBAF complexes purified from HEK293T cells (left) and clustered heatmap of mass spec-called peptides and spectral counts on selected fractions (right).
(B) Silver stain performed on density sedimentation of HA-PBRM1 PBAF complexes purified from HEK293T cells (left) and clustered heatmap of mass spec-called peptides and spectral counts on selected fractions (right).
(C) Louvain network analysis of PBAF subunit (PHF10 and BRD7) cross-linking mass spectrometry datasets.
(D) HA-GLTSCR1L-bound ncBAF complexes were purified from HEK293T, PAGE-separated, and silver stained. Individual identified proteins are indicated.
(E) Silver stain performed on density sedimentation of HA-GLTSCR1L-bound ncBAF complexes purified from HEK293T cells (left) and clustered heatmap of mass spec-called peptides and spectral counts on selected fractions (right). *, non-specific contaminants in fraction 16.
(F) Silver stain performed on density sedimentation of HA-BRD9 ncBAF complexes purified from HEK293T cells (left). Clustered heatmap of mass spec-called peptides and spectral counts on selected fractions (right).
(G) Schematic of the full mSWI/SNF complex assembly pathway. Subunit abbreviations are indicated. Numbers indicate the steps in assembly (see text).
See also Figure S6.
ATPase and BAF core modules were similar to those of cBAF complexes, whereas, interestingly, PBAF-specific subunits such as BRD7 and PBRM1 associated with both the BAF core and ATPase modules (Figures 6C and S6B). Purification of two other PBAF specific subunits, BRD7 and PHF10, yielded only full complexes without intermediates (Figures S6C and S6D). CoIP of PBAF component KO cell lines proved to be more informative regarding the order of integration of these subunits (Figure S6E). As expected, loss of ARID2 resulted in loss of stability of BRD7, PBRM1, and PHF10, confirming the early role of ARID2 in PBAF assembly. BRD7 deletion minimally affected ARID2 stability but strongly affected both PHF10 and PBRM1 interactions. Finally, PBRM1 deletion had no effect, implicating this subunit as the last to assemble into PBAF complexes. In addition, using coIP analysis, we determined that PBRM1 is present in more than one copy per PBAF complex, suggesting its multimerization (Figures S6F and S6G).
To finalize the composition and assembly of ncBAF complexes, we purified GLTSCR1L- and BRD9-containing complexes. As expected, we identified complexes containing initial core SMARCC1 and SMARCD1 subunits, ATPase module components, and BRD9; however, no other core subunits (SMARCC2, SMARCD2, SMARCD3, SMARCE1, or SMARCB1) were identified (Figures 6D and 6E). GLTSCR1L purification resolved full ncBAF complexes in Fx 10–11 and subcomplexes in fractions 6–7 (Figure 6E), highlighting the ncBAF core of SMARCC1, SMARCD1, and GLTSCR1L, the same components identified in the SMARCD1 purification from ΔATPase cells (Figure 4G). BRD9 purification captured the full ncBAF complex in fractions 9–11 but failed to resolve subcomplexes, suggesting that BRD9 functions similarly to BRD7 by forming partial assemblies that result in immediate incorporation of the ATPase module ((Figures 4G, 6F, and S6C). Loss of BRD9 had no effect on SMARCD1, whereas BRD9 and GLTSCR1 stability were substantially affected in SMARCD1 KO cells, substantiating the early assembly order and the critical role of SMARCD1 in the nucleation of all three mSWI/SNF family complexes (Figure S6I).
Based on this study, we summarize the mammalian SWI/SNF assembly pathway (Figure 6H). The main steps of complex assembly and branching are as follows: dimerization of SMARCC subunits (1); formation of the BAF initial core of SMARCC and SMARCD subunits (2); incorporation of SMARCE1 and SMARCB1 components, forming the BAF core module (3) or, alternatively, incorporation of GLTSCR1 and GLTSCR1L (4); formation of the ncBAF core module, which binds BRD9 (5); cBAF core complexes interact with ARID1 or ARID2 subunits and branch into cBAF complexes (containing ARID1) (6) and PBAF complexes (containing ARID2) (7), respectively; ARID1 or BAF complex core intermediates bind DPF2 (8) and incorporate the SS18-containing ATPase module (9), finalizing cBAF assembly. In parallel, the PBAF complex intermediate, ARID2/BAF core, incorporates BRD7 and PHF10 (10), and subsequently recruits the SS18-negative ATPase module, which finalizes its formation by binding PBRM1 (11). The alternative BRD9 and ncBAF core finalizes its formation with the integration of an SS18-containing ATPase module to form ncBAF complexes (12). The existence of multiple subunit paralogs across these three distinct mSWI/ SNF complexes results in further diversification, for which we calculated the full set of possible combinations (Figure S6J).
Disease-Associated Mutations Affect mSWI/SNF Binding Interfaces and Subunit Stability
The genes encoding mSWI/SNF complex subunits are widely mutated in human disease, most notably in cancer and intellectual disability syndromes (Bogershausen and Wollnik, 2018; Ka- doch and Crabtree, 2015; Kadoch et al., 2013; Sokpor et al., 2017). Because the large majority of mSWI/SNF subunit mutations in cancer (Figure 7A) result in protein loss, we analyzed complexes purified from KO cell lines by mass spectrometry to assess the global effect of each subunit loss on the relative abundance of other subunits in the complex (Figure 7B). Subunits that assemble at the earliest stages of BAF assembly are the most critical for complex assembly, with their deletion resulting in profound effects on complex integrity. This dataset excludes SMARCC-deleted cells because this resulted in near-complete degradation of all mSWI/SNF subunits, further underscoring the important role of this initial subunit dimer as the structural foundation of all mSWI/SNF complexes (Figure S3M). Notably, we found that loss of SMARCB1, a well-known tumor suppressor (Versteege et al., 1998), has minor effects on complex stability relative to other subunits (Figures 7B, S3M, and S3P), indicating instead a critical regulatory role exerted by the SMARCBI-containing core module on the ATPase and its associated components. Defining the proportion of crosslinked sites between subunits lost upon gene truncating mutations showed that the subunits most affected by truncating mutations in cancer are PBRM1 and ARID1A, which interact with complexes primarily via C-terminal binding regions (Figure 7C).
Figure 7. Disruption of mSWI/SNF Complex Assembly in Human Disease.
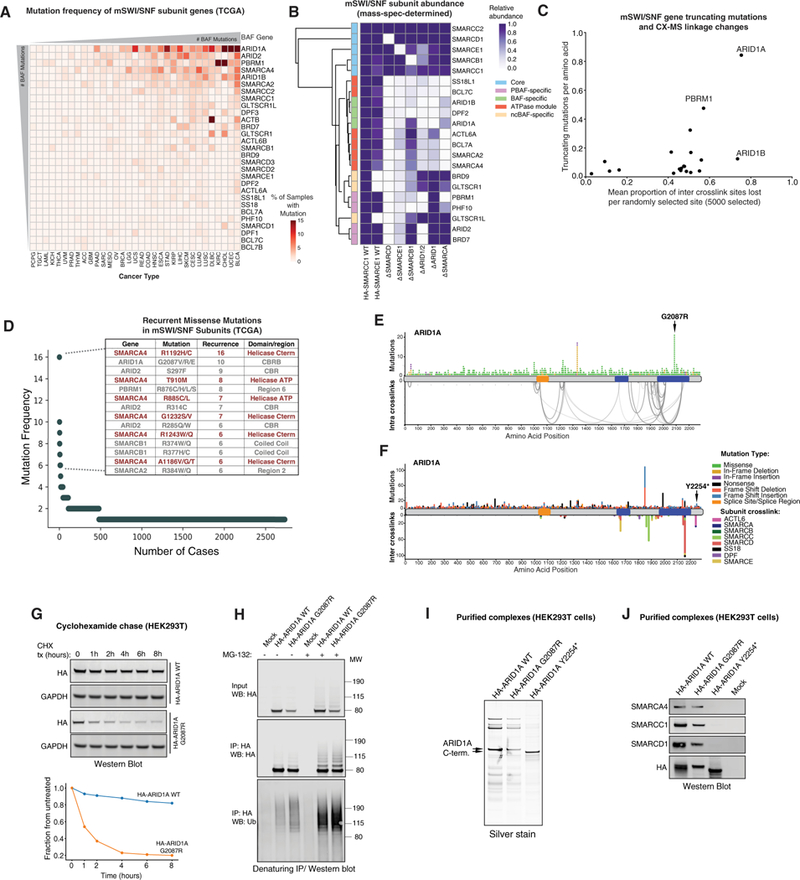
(A) Frequency of mSWI/SNF gene mutations across human cancers (The Cancer Genome Atlas [TCGA]). Hypermutated samples were excluded from analysis.
(B) Mass spectrometry analysis of mSWI/SNF complex subunit relative abundance in complexes purified from the indicated cell types (WT and subunit KO cells), normalized toWTSMARCC1 purifications. DSMARCDcomplexeswere purified using SMARCE1; ∆SMARCE1, ∆SMARCB1, ∆ARID1/2, ∆ARID1, and ∆SMARCA complexes were purified using HA-SMARCD1.
(C) Correlation analysis reflectingtheeffect oftruncating mutationson mSWI/SNF subunit linkages. Subunits mostfrequentlytruncated exhibit higherproportions of inter-crosslinked sites lost.
(D) Top-ranked cancer-associated missense mutations (TCGA). Mutations predicted to disrupt catalytic activity are shown in red.
(E) Non-truncating mutations in ARID1A across human cancers mapped over intra-crosslinks. The hotspot mutation in the highly crosslinked C-terminal CBRB region of the protein is indicated.
(F) Truncating mutations in ARIDIAacross human cancers mapped over crosslinks to other BAF subunits. The position ofthetruncating mutation Y2254* used in this study is indicated by the arrow.
(G) Top: Representative cycloheximide chase experiment assessing the half-life of ARID1A WT and G2087R mutant C-terminal region variants. Bottom: quantification of western blot (WB) above, normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH).
(H) MG-132 treatment (8 hr) of HEK293T cells expressing ARID1A WT and G2087R C-terminal regions. IP performed under denaturing conditions; immunoblot performed using indicated antibodies.
(I) Silver stain performed on ARID1A WT, G2087R, and Y2254* BAF complexes purified from HEK293T cells.
(J) Immunoblot of ARID1A WT, G2087R-, and Y2254*-bound BAF complexes purified from HEK293T cells.
See also Figure S7.
In addition to their roles in cancer, mSWI/SNF subunit muations have been linked to several developmental and neurologic conditions, including intellectual disability and autism spectrum disorders, with additional mutations continuing to emerge in other rare but well-defined disease settings (Sokpor et al., 2017). For example, heterozygous ARID1B mutations are common in Coffin-Siris syndrome (Figure S7A), and mutations of ACTL6A were identified in autism and shown to disrupt its interaction with SMARCA4 (Marom et al., 2017). Intriguingly, our analyses revealed that these map to ACTL6A and SMARCA cross-links (Figure S7B). Finally, SMARCD2 mutations were reported to drive neutrophil-specific granule deficiency (SGD) (Priam et al., 2017; Witzel et al., 2017). These mutations result in truncation before the C-terminal region, which we found removes the region containing a significant number of crosslinks to ARID1 CBR B and SMARCC, likely explaining the loss of BAF complex binding (Figure S7C). Intriguingly, the C-terminal region of theSMARCD1 paralog contains fewer crosslinks to these subunits and also failed to rescue SGD phenotypes in in vivo models of SGD (Priam et al., 2017; Witzel et al., 2017), suggesting a structural basis for paralog- and tissue-specific function of BAF subunits.
ARID1A, critical for BAF complex specification and assembly of the ATPase module, is the most frequently mutated mSWI/ SNF subunit in human cancers (Figure 7A; Davoli et al., 2013; Wu et al., 2014). ARID1A is particularly vulnerable to truncating mutations because these will result in deletion of the C-terminal binding region. However, until now, the effect of recurrent missense mutations and small deletions within the CBR regions of ARID1A remained unknown (Figure 7D). The most common single missense mutations in mSWI/SNF subunits (second only to mutations in the SMARCA4 helicase) result in substitution of glycine 2,087 of ARID1A to valine, arginine, or glutamic acid. This region corresponds to the CBRB-interacting region of the protein we identified (Figure 7E). Additional recurrent missense mutations include Y2254*, resulting in a small, 31-aa deletion in R4 of the ARID1A C terminus, involved in anchoring of the ATPase module to the BAF core module (Figure 7F). We found that the C-terminal ARID1A region containing the G2087R mutation did not result in loss of interaction with BAF complexes (Figure S7D) but that its expression was substantially lower in comparison with WT ARID1A C-terminal protein because of decreased protein stability, as revealed by cycloheximide chase experiments (Figure 7G). Further, we observed an increased poly-ubiquitin signal in the G2087R mutant compared with the WT ARID1A C-terminal protein, which further increased upon treatment with MG132, suggesting proteasome-mediated degradation (Figure 7H). In contrast, ARID1A Y2254* resulted in complete loss of interaction between ARID1A and BAF complexes (Figures 7I and 7J), indicating that any truncating mutations in preceding residues would similarly disrupt binding. Taken together, these studies evaluated different routes toward ARID1A disruption, each of which result in inhibited assembly of fully formed complexes. Loss of ARID1A is not compensated by increased expression of ARID1B, which also displays lower expression than ARID1A in most tumor samples (Figures S7E–S7G).
DISCUSSION
This study presents a comprehensive architectural framework for the mSWI/SNF chromatin remodeler complex family, including the assembly pathways and inter- and intra-module linkages across three distinct complexes. Integrating multiple complex purifications with size fractionation, mutagenesis, and cross-linking mass spectrometry, we defined intra-complex modular architecture, stoichiometry, and evolutionary relationships and explored the effects of disease-associated mutations on complex architecture and assembly.
One particularly unexpected result is that the initial core for all three mSWI/SNF family complexes is a heterotrimer consisting of two SMARCC subunits (as a dimer) and one SMARCD subunit. Although previous in vitro subunit co-purifications had suggested a “minimal BAF complex” consisting of SMARCA4, SMARCC1, SMARCC2, and SMARCB1 (Phelan et al., 1999), we found that neither complex assembly pathways nor cross-linking mass spectrometry profiles of full BAF or PBAF complexes implicated this tetramer as a physiologic core in mammalian cells. Indeed, these results may begin to explain the challenges that have been faced in obtaining high-resolution structural information on this complex and in using such minimal complexes for small molecule screening efforts. Importantly, the mSWI/SNF initial core is required for global complex stability and the interaction of the majority of subunits in all three mSWI/SNF complexes (Figure 3). Notably, the newly identified ncBAF complex assembles exclusively around a SMARCC1 and SMARCD1 initial core and lacks SMARCE1 and SMARCB1 subunits, indicating fundamental differences and/or compensation in biochemical activity.
Interestingly, network modularity analyses of cross-linking mass spectrometry data place SMARCB1 in the ATPase module, whereas biochemical purification of SMARCB1 demonstrates its presence in the BAF core module. This suggests that SMARCB1 may be involved in functionally linking the core and ATPase modules, potentially modulating ATPase or remodeling activity. Indeed, SNF5 regulates the chromatin remodeling activity of the yeast complex (Sen et al., 2017). Although the SNF5 and SMARCB1 subunits are largely dispensable for complex integrity in both yeast and human settings, respectively, we observed that these orthologs exhibit different module associations in distantly related eukaryotes, suggesting that SMARCB1 may play a dynamic role in regulating SWI/SNF complex activity.
ARID subunits (ARID1A, ARID1B, and ARID2) are among the most frequently mutated subunits in human disease. Importantly, we found that ARID subunits are the major determinants of assembly pathway branching toward BAF or PBAF complexes. ARID subunits bind the BAF core module through the CBR regions on the C terminus and N terminus of ARID1 and ARID2, respectively, likely leading to the formation of a large interaction interface and forging a structurally essential bridge between the core and ATPase modules. SMARCD subunits in particular play a major role in ARID subunit binding as their loss substantially affects ARID and subsequent ATPase module assembly. The critical role for ARID subunits is further illustrated by their interaction with the ATPase module subunits SMARCA and ACTL6A. Finally, the absence of any ARID subunits in the newly identified ncBAF complex suggests an alternative, ARID-independent mode of binding the ATPase module (likely mediated by GLTSCR1 and GLTSCR1L subunits).
Assembly of multiprotein complexes often occurs in a directed modular manner with defined and evolutionarily conserved subcomplexes (Marsh and Teichmann, 2015) Indeed, our analysis of cross-linking mass spectrometry-identified linkages within SWI/SNF complexes of two other eukaryotic species reveals evolutionary conservation of the complex modularity we identified in mammalian cells. The conserved structural properties of these complexes suggest separation and divergence of complex functions. Although the ATPase domain has been implicated in nucleosome sliding (Smith and Peterson, 2005), the role of the other modules and subunits in both ATPase activity and nucleosome remodeling remains poorly understood. Extensive DNA binding surfaces, particularly on the BAF core module, may play critical roles in directing forces from the ATPase-nucleosome-DNA interaction. Further studies will be required to define the role of each subunit and domain in complex targeting and in the modulation of ATPase-driven nucleosome remodeling.
Finally, our findings suggest that BAF inter- and intra-modular interactions are altered by mutations found in many human cancers and other diseases and that these mutations disrupt the normal complex assembly pathway or subunit protein stability. A prime example of this lies in the extensively mutated ARID1A subunit, including both nonsense mutations and missense mutations that are disproportionately skewed to the C-terminal domain that we found is required for BAF complex binding.
Taken together, these studies present new opportunities for structural and functional characterization of this family of mammalian chromatin remodeling complexes that exhibits outsized roles in human disease. Understanding the architecture and modular organization of mSWI/SNF complexes greatly potentiates the ability to assign density to subunits and modules using cryo-EM in efforts to achieve 3D structure, to link structure to binding and biochemical activity on chromatin, and to develop physiologically meaningful small-molecule screening strategies, collectively serving as a critical foundation in the quest to define mechanisms of mSWI/SNF-mediated chromatin remodeling in normal and disease states.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Cigall Kadoch (Cigall_kadoch@dfci.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mammalian cell culture
HEK293T (female), MIA-Pa-Ca-2 (male) and SW13 (female) cell lines were cultured in standard DMEM (GIBCO) media supplemented with 10% FBS (GIBCO), 1mM HEPES pH 7.5 (GIBCO), and Pen/Step (GIBCO) at 28°C and 5% CO2. HEK293T cells used in this study were routinely fingerprinted and tested for mycoplasma. Wild-type gene sequences and gene expression for mSWI/SNF complex subunit genes were confirmed using RNA-seq prior to experimentation.
D. melanogaster cell culture
Drosophila S2 cells were cultured in SFX-Insect media at 28°C with constant shaking at 112 rpm. To generate stable cell lines, cells were plated in 6-well plates at 2×106 and transfected with 2 μg of expression construct using Effectene Transfection Reagent (Quiagen) in accordance with manufacturer’s recommendation. Cells were selected using 250 mg/ml of hygromycin or 10 μg/ml of puromycin for 10 days and expanded to 1 l culture for complex purification. 1-l cultures were induced with 500 μM copper sulfite for 72 hours and collected by centrifugation at 4000 g for 5 minutes. Cell pellets were processed identically to mammalian cells (see Protein Purification).
METHOD DETAILS
Expression constructs and lentiviral infection
All constructs were PCR-amplified from cDNA using Phusion High-Fidelity DNA Polymerase with GC buffer (NEB) or with Q5 High-Fidelity Polymerase (NEB). Purified PCR products were cloned into a modified pTight vector from Clonetech (EF1-alpha promoter) containing blasticidin resistance using In-Fusion (Clontech) at the NotI cloning site. Recombination products were transformed in to One-Shot Stbl3 chemically competent E. coli (Invitrogen). For the HA-ARID1A C-term construct corresponding to aa1611–2285, the cloning region was selected based on conservation analysis and cross-linking mass spectrometry data. HA-ARID1A C-term was cloned into a modified pTight vector from Clonetech (EF1-alpha promoter) containing blasticidin resistance. For mini ARID2 (mARID2), the cloning region was selected based on cross-linking mass spectrometry data corresponding to N-terminal aa1–626 fused to C-terminal aa1592–1835. The N-terminal (aa1–626) and C-terminal (aa1592–1835) fragments were PCR amplified separately, with the primers designed at the 3′ end of the aa1–626 and the 5′ end of aa1592–1835 containing 27 base pairs of complementarity. N-terminal and C-terminal regions of ARID2 were amplified independently, gel purified as above, fused together in a second PCR reaction, and cloned into a modified pTight vector (EF1-alpha promoter) containing blasticidin resistance. SS18 was cloned into pENTR D-Topo vector and recombined into pMSCV Flag-HA IRES Puro retroviral vector (Gift from Wade Harper laboratory (HMS) and Addgene). All constructs were sequence validated.
For lentiviral infection, cells were transduced with lentivirus at 50% confluency, incubated with lentivirusfor48 hours, and selected with blasticidin at 10 μg/ml. Cell cultures were expanded to desired amounts for mSWI/SNF complex purification.
Generation of HEK293T mSWI/SNF subunit knockout cell lines
CRISPR-Cas9 KO constructs were purchased from Santa Cruz Biotechnology (SCBT) and transfected into HEK293T cells using Lip-ofectamine 3000 reagent (Invitrogen). Cells were selected with puromycin at 2 μg/ml for 5 days. Single cell clones were isolated and subsequently screened for loss of subunit expression using immunoblot and DNA sequencing.
Protein purification
Stable cell lines were cultured in 150mm dishes and expanded according to assay requirements and bait expression levels. Complexes were purified as previously described with modifications (Mashtalir et al., 2014). Cells were scraped from plates and washed with cold PBS. Suspension was centrifuged at 3000 rpm for 5 min at 4°C and pellets were resuspended in hypotonic buffer (HB) containing 10mM Tris HCl pH 7.5, 10mM KCL, 1.5 mM MgCL2, 1mM DTT, 1mM PMSF and incubated on ice for 5min. Suspension was centrifuged at 5000 rpm for 5 min at 4°C, and pellets were resuspended in 5 volumes of fresh HB containing protease inhibitor cocktail and homogenized using glass Dounce homogenizer. Suspension was layered onto HB sucrose cushion containing 30% sucrose w/v, centrifuged at 5000 rpm for 1 hour at 4°C and cytosol-containing layer was discarded. Nuclear pellets were resuspended in high salt buffer (HSB) containing 50mM Tris HCl pH 7.5, 300mM KCL, 1mM MgCL2,1mM EDTA, 1mM, 1% NP40,1mM DTT, 1mM PMSF and protease inhibitor cocktail. Homogenate was incubated on rotator for 1H. Homogenates then were centrifuged at 20,000 rpm (30,000 × g) for 1 hour at 4°C using an SW32Ti rotor. Chromatin pellets were discarded and high salt nuclear extract was filtered through a 0.45μm filter and incubated overnight with HA magnetic resin. HA beads were washed in HSB and eluted with HSB containing 1mg/ml of HA peptide for 4 times 1.5 hour each. Eluted proteins were then subjected to density gradient centrifugation or dialysis.
Density sedimentation gradients
Eluted protein complexes or nuclear extracts were loaded on top of linear, 11ml 10%–30% glycerol gradients containing 25mM HEPES pH 7.9, 0.1mM EDTA, 12.5 mM MgCl2,100mM KCl supplemented with 1mM DTT and protease inhibitors. Tubes were loaded into SW41 rotor and centrifuged at 40000 rpm for 16 hours at 4°C. 550 μL fractions were manually collected from the top of the gradient. 100 μL of each collected fraction were concentrated using 10 μL of Strataclean beads, loaded onto SDS-PAGE gels and either stained using Silver Quest staining kit, or used for western blot analysis.
Co-Immunoprecipitation
Cells were washed with cold PBS and resuspended in EB0 hypotonic buffer containing 50mM Tris pH 7.5,0.1% NP-40,1mM EDTA, 1mM MgCl2 supplemented with protease inhibitors. Lysates were pelleted at 5,000rpm for 5min at 4°C. Supernatants were discarded and nuclei were resuspended in EB300 high salt buffer containing 50mM Tris pH 7.5, 300mM NaCl, 1% NP-40, 1mM EDTA, 1mM MgCl2 supplemented with protease inhibitors. Lysates were incubated on ice for 10 min with occasional vortexing. Lysate was pelleted at 21000 g for 10 min at 4°C. Supernatants were quantified and supplemented with 1 mM DTT. 1mg of protein was used for immunoprecipitation with 2–5ug of antibodies over night at 4°C. Protein-G Dynabeads were added for 2 hours and washed with EB300. Beads were eluted with loading LDS and loaded onto SDS-PAGE.
Immunoprecipitation under denaturing conditions
Cells were grown to 80% confluency and treated with MG132 at 20uM for 8 hours. Cells were washed with PBS and lysed in buffer containing 25mM Tris pH 7.5 and 1.5% SDS. Lysates were collected and boiled for 5 minutes. Lysates were sonicated and dissolved in EB300 buffer to dilute SDS concentration to 0.1%. Diluted extracts were incubated with HA beads overnight, washed with EB300 five times and resuspended in LDS for western blot loading.
IRDye680 and colloidal blue labeling
Strataclean concentrated fractions were resuspended in denaturing staining solution containing 1x PBS, 1% SDS, and 1uM IRDye 680RD NHS Ester, heated at 70°C for 5 min and then incubated overnight at 37°C. Reactions were quenched with 4X LDS buffer and loaded onto SDS-PAGE. Upon migrations gels were scanned on Li-Cor Odyssey CLx instrument on 700 channel. Bands were quantified and analyzed as indicated below.
For stoichiometric quantification 1ug of purified DPF2 cBAF complexes were loaded onto SDS-PAGE, stained with colloidal blue according to manufacturer’s recommendations and scanned using Li-Cor Odyssey CLx in 700 channel, bands were quantified and normalized to protein molecular weight and DPF2 signal.
Western Blotting
Western blot analysis was performed using standard approaches. Samples were separate on 4%−12% PAGE gel and transferred onto PVDF membrane. Membranes were blocked with 5% milk PBST and incubated with primary antibodies for three hours or overnight. Membranes were then washed with PBST three times and incubated with secondary flurophore-conjugated species-specific secondary antibodies (Li-Cor) for one hour. Subsequently, membranes were washed 3 times with PBST and final time with PBS and imaged using Li-Cor Odyssey CLx.
Mass-spectrometric sample preparation and experiments
Sample preparation
Equal amounts of selected fractions from glycerol gradient-separated complexes were concentrated using StrataClean beads and loaded onto SDS-PAGE gels. Samples were migrated 2cm into the gel, stained with colloidal blue stain and excised for mass spectrometry analysis. All subsequent gradient mass spectrometry steps were performed at the Taplin Biological Mass Spectrometry Facility directed by Dr. Steven Gygi (Harvard Medical School).
Excised gel bands were cut into approximately 1 mm3 pieces. Gel pieces were then subjected to a modified in-gel trypsin digestion procedure (Shevchenko et al., 1996). Gel pieces were washed and dehydrated with acetonitrile for 10 min. followed by removal of acetonitrile. Pieces were then completely dried in a speed-vac. Rehydration of the gel pieces was with 50 mM ammonium bicarbonate solution containing 12.5 μg/ml modified sequencing-grade trypsin (Promega, Madison, WI) at 4°C. After 45 min., the excess trypsin solution was removed and replaced with 50 mM ammonium bicarbonate solution to just cover the gel pieces. Samples were then placed in a 37° C room overnight. Peptides were later extracted by removing the ammonium bicarbonate solution, followed by one wash with a solution containing 50% acetonitrile and 1% formic acid. The extracts were then dried in a speed-vac (~1 hr). The samples were then stored at 4°C until analysis.
On the day of analysis the samples were reconstituted in 5 −10 μl of HPLC solvent A (2.5% acetonitrile, 0.1% formic acid). A nanoscale reverse-phase HPLC capillary column was created by packing 2.6 mm C18 spherical silica beads (Accucore, ThermoFisher) into a fused silica capillary (100 μm inner diameter × ~30 cm length) with a flame-drawn tip. After equilibrating the column each sample was loaded via a Famos auto sampler (LC Packings, San Francisco CA) onto the column. A gradient was formed and peptides were eluted with increasing concentrations of solvent B (97.5% acetonitrile, 0.1% formic acid).
As peptides eluted they were subjected to electrospray ionization and then entered into an LTQ Orbitrap Elite ion-trap mass spectrometer (ThermoFisher Scientific, Waltham, MA). Peptides were detected, isolated, and fragmented to produce a tandem mass spectrum of specific fragment ions for each peptide. Peptide sequences (and hence protein identity) were determined by matching protein databases with the acquired fragmentation pattern by the software program, Sequest (Thermo Fisher Scientific, Waltham, MA). All databases include a reversed version of all the sequences, and the data were filtered to a 1% false discovery rate based on linear discriminant analysis (Huttlin et al., 2010). All raw data from all fractions of gradient mass spectrometry across all experiments are found in Appendix.
Protein Sample preparation for cross-linking mass-spectrometry
Native protein complexes were eluted in detergent free elution buffer and dialyzed over night against amine free buffer containing 25mM HEPES pH 7.9, 1 mM EDTA, 1 mM MgCl2, 100mM KCl 10% Glycerol supplemented with 1mM DTT. Samples were concentrated using Amicon Ultra centrifugal filters with 30K cutoff and subjected to BS3-based crosslinking and mass spectrometry described below.
BS3 crosslinking and cross-linking mass spectrometry analysis
Purified protein complexes in 25mM HEPES pH 7.6,150 mM KCl, 1mM EDTA, 1mM MgCl2, 1mM DTT, 1mM PMSF and 10% Glycerol, were crosslinked by addition of BS3 (Thermo Scientific; freshly prepared as 100 mM in pure water) to 2 mM for2hrsat 25°C. The protein amounts used were HA-DPF2: 70 ug; Flag-HA-SS18: 52ug; HA-BRD7: 17ug; HA-PHF10: 15ug; BAP60-HA: 52ug; HA-D4: 60ug. The reactions were quenched by addition of 10 uL of 1M ammonium bicarbonate. For the HA-DPF2, Flag-HA-SS18 and HA-BRD7 samples, an equal volume of trifluoroethanol (TFE) was added and the samples were incubated at 60°C for 30 minutes to denature the proteins. Tris(2-carboxyethyl)phosphine hydrochloride (TCEP) was added to a final concentration of 5 mM. The samples were alkylated by addition of iodoacetamide (IAA) to 10 mM. After incubating at 37°C for 2 hr in the dark, the samples were diluted 10-fold with 0.1 M ammonium bicarbonate and digested with trypsin (Promega, Madison, WI) at a ratio of 20:1 (protein:trypsin) overnight at 37°C. For the HA-PHF10, BAP60-HA and HA-D4 samples, we followed the sample preparation protocol using SP3 beads previously described (Hughes et al., 2014): 10uL of SP3 beads (10ug/uL) and an equal volume of acetonitrile were added to the crosslinked samples and incubated at 60°C for 30 minutes with shaking. Then the beads were concentrated with a magnet and washed with 70% ethanol and 100% acetonitrile. The beads were then suspended in 100 uL 8M Urea in 1 M ammonium bicarbonate and treated with TECP/IAA for 2 hr at 37°C in the dark. Then the samples were diluted 10 times with water and digested by addition of trypsin (20:1, protein:trypsin) overnight at 37°C.
All peptide samples were desalted by passage over C18 cartridges (The Nest group, Southborough, MA), and dried by Speed-Vac. The peptides were resuspended in 50 uL Buffer A (25 mM ammonium formate, 20% acetonitrile, 0.1% formic acid, pH 2.8). 1 ug of each sample was reserved for direct mass spectrometry analysis and the remaining sample was fractionated using an in-house prepared microcapillary strong cation exchange column (200 mm × 20 cm; 5 μm, 200 Å partisphere SCX, Whatman or Proteomix SCX 3um, Sepax Technologies). We used a binary HPLC pump with split flow with microcapillary flowrate at 2–3 uL/min. Peptides were loaded onto the microcapillary column equilibrated in Buffer A and washed with Buffer A. Bound peptides were eluted with 20 μL of Buffer A containing 30%, 50%, 70%, and 100% Buffer B (800 mM ammonium formate, 20% acetonitrile, pH 2.8), followed by 50 μL elutions with Buffer B containing 5%, or 10% Buffer D (0.5 M ammonium acetate, 30% acetonitrile),or just 20 mL of Buffer D. All fractions were dried in a Speed-vac, and resuspended in 0.1% trifluoroacetic acid (TFA), 2% acetonitrile.
Peptides were analyzed by electrospray ionization microcapillary reverse phase HPLC on a Thermo Scientific Fusion with HCD fragmentation and serial mass spectrometry events that included one FTMS1 event at 30,000 resolution followed by FTMS2 events at 15,000 resolution. Other instrument settings included: MS1 scan range (m/z): 400–1500; cycle time 3 s; Charge states 4–10; Filters MIPS on, relax restriction = true; Dynamic exclusion enabled: repeat count 1, exclusion duration 30 s; Filter Intensity Threshold, signal intensity 50000; Isolation mode, quadrupole; Isolation window 2Da; HCD normalized collision energy 28%, isolation width 2 Da; AGC target 500,000, Max injection time 200ms. A 90 min gradient from 5% ACN to 40% ACN was used.
QUANTIFICATION AND STATISTICAL ANALYSIS
Cross-linking mass spectrometry database search and crosslinked peptide identification
The RAW files were converted to mzXML files by Rawconverter (He et al., 2015). For crosslinked peptide searches, we used two different crosslink database searching algorithms: pLink (Yang et al., 2012) and an in-house designed Nexus. Crosslinking data were analyzed using pLink (Yang et al., 2012) with default settings (precursor monoisotopic mass tolerance: ± 10 ppm; fragment mass tolerance: ± 20 ppm; up to 4 isotopic peaks; maxevalue 1; static modification on Cysteines; 57. 0215 Da; differential oxidation modification on Methionines; 15. 9949 Da) against a database containing only BAF or PBAF protein sequences.
For Nexus searches, the same databases were used with the following parameter settings: (a) up to three miscleavages; (b) static modification on Cysteines (+57.0215 Da); (c) differential oxidation modification on Methionines (+15.9949 Da); (d) differential modification on the peptide N-terminal Glutamic acid residues (−18.0106 Da) or N-terminal Glutamine residues (−17.0265 Da); (e) differential mono-BS3 modification on Lysine residue (+156.0806 Da). A 5% of FDR cutoff was used for both pLink and Nexus. After performing the pLink and Nexus analyses, the search results were combined and each spectrum was manually evaluated for the quality of the match to each peptide using the COMET/Lorikeet Spectrum Viewer (TPP). Crosslinked peptides are considered confidently identified if at least 4 consecutive b or y ions for each peptide are observed and the majority of the observed ions are accounted for. Search results that did not meet these criteria were removed. Intralinks involving a crosslink between identical residues were only kept if the spectral evidence strongly supported the identification; that is, the major fragment ions correspond to the intralinked peptide sequence and no/few other fragment ions were observed. The percentage of spectra deleted after manual examination was: for DPF2 (11% for interlinks, 5.1% for intralinks), SS18 (30% for interlinks, 5.6% for intralinks), BRD7 (34.9% for interlinks, 15.7% for intralinks), PHF10 (25.7% for interlinks, 9.7% for intralinks), BAP60 (10.4% for interlinks, 9.4% for intralinks), HAD4 (33.7% for interlinks, 10% for intralinks). Crosslinks that met these criteria were uploaded into ProXL for viewing and data analysis(Riffle et al., 2016). All data including the spectra, linkages and structure analyses can be visualized at: https://www.yeastrc.org/proxl_public/viewProject.do?project_id=127.
Analyses of gradient-mass spectrometric data
Total spectral counts (peptides) corresponding to each protein subunit within mSWI/SNF complexes in each gradient fraction were assembled into elution profiles and used for downstream analysis. For all panels showing mSWI/SNF complex purification elution profiles, the total peptide counts are min-max normalized separately for each subunit across fractions. Peptide counts are represented both as wave plots and heatmaps. For waveplots, SS18 and SS18L1 peptide counts were combined because individually each yielded low numbers of peptides, owing to the low number of lysines in these proteins. Z-Scores were calculated for heatmaps across rows using the seaborns ‘z_score’ option with all default settings.
To calculate Pearson correlations across elution profiles, total peptide counts across all gradient fractions for each of the baits (SMARCD1, SMARCB1 and SMARCA4) were used. The profiles for each were appended to create a n × 3 m matrix where n is the number of mSWI/SNF proteins and m is the number of gradient fractions in each experiment. The correlation across these three appended sample profiles was calculated using numpy. The total peptide counts for paralogs of the baits used were excluded (i.e., SMARCD2 and SMARCD3 in the SMARCD1 purification, SMARCA2 in the SMARCA4 purification, etc.).
In order to generate the heatmap reflecting the impact of subunit loss (Figure 7B), a normalization ratio was calculated by dividing the total number of mSWI/SNF subunit peptides captured across all fractions in each experiment by the mean peptide total across all experiments. All peptide numbers in a particular experiment were multiplied by this ratio to account for potential differences in peptide abundance between experiments. After normalization, the fraction in each experiment with the most total peptides for a given protein was taken and divided by the number of (normalized) peptides in the WT SMARCC1 pull down condition, yielding the proportion of normalized peptides in the mutant condition over the wild-type condition. This was repeated for all proteins and then clustered using scipy hierarchical clustering (from inside the seaborn clustermap package); correlation between samples was used as the distance metric for the clustering. Paralogs of the bait for the mutant samples (SMARCD2 and SMARCD3), proteins that had low numbers of peptides across samples (BCL7B and SS18), and ACTB were excluded from the heatmap.
Computational Analysis
Unless otherwise noted, all data analysis was performed using Python version 2.7. Plots were generated using matplotlib and the seaborns data visualization packages.
Structural Analysis
A complete list of SWI/SNF structures was compiled from the Protein Data Bank (Table S4). If multiple structures existed for a domain or protein, the structure with the highest resolution was selected. If a single domain had structures in multiple organisms, the structure from the organism most similar to humans was selected. For each protein that had an available structure, the canonical FASTA sequence was aligned to the sequence of the structure using EMBOSS needle 6.6.0 in order to create a map from the FASTA sequence numbers and the structure residue numbers. For each internal cross link between two residues that were both in the structure, the distance between carbon alphas was calculated and recorded in angstroms. All structures were represented using PyMOL, crosslinks were displayed on the structure using the PyMol distance function.
Network schematics of SWI/SNF complexes from crosslinking data
For each complex, a directed network was built with subunits as nodes. Protein paralogs were collapsed for simplicity and number of crosslinks per region of alignment was used as measure of binding strength. Directed edges were shown between subunits with crosslinks between them. The maximum out-degree of each subunit was fixed to be two, where edges were preserved by taking the top edges ranked by number of crosslinks. Modules were colored by membership in communities as detected by the igraph implementation of Louvain clustering (cluster_louvain), hence, colors were generated as a function of the relationship between the nodes (subunits and subunit groups) within the network. Networks were plotted with igraph in R. For yeast and human networks, any edges with fewer than 10 crosslinks mapping between the subunits were removed, for Drosophila complexes, they were not removed owing to lower relative protein capture.
Crosslinking Maps
Each protein was divided in to amino acid regions (defined in Figure S2B). Crosslinks between protein regions were counted, paralog proteins were considered equivalent. A small number of proteins (BRD9, GLTSCR1, DPF1, DPF3) were excluded from this analysis because of their very low peptide counts.
When these are clustered (Figures 3E, 4B, and 5B) the matrix from above was filtered for a protein family of interest (SMARCC, ARID1/2 and SMARCA respectively). Only domains that had a total of at least 3 external crosslinks to any domain in this family of interest were included. Any external crosslinks between proteins in the family of interest were excluded (except for the SMARCC). The rows were clustered using the seaborns clustermap function with all clustering options set to default, columns were not clustered.
Circos Plots
Circos plots were generated using Circos 0.69. Thickness setting of lines displayed for external crosslinks corresponds to the number of times that that crosslink appeared in the dataset, any link that appeared 50 or more times was set to 50. Lines and proteins are colored according to module, domains annotated are shown in Figure S3B.
For protein paralog families, the bar in the plot represents the alignment of all family members. For families of 2 proteins, global alignments were done using EMBOSS needle 6.6.0 with a gap open penalty of 10 and a gap extend penalty of 0.5. Sequences for families with 3 or more proteins were aligned using Clustal Omega with all default settings. The position of the crosslink in the protein was then mapped to the appropriate position in the alignment, and the link was drawn from there. The ‘circos’ script was executed with the ‘-noparanoid’ setting.
Gel Quantification
For each band, gel quantification signal was normalized to the area of the region read. These signal per area values were then min-max normalized across the gradient giving a relative amount of signal for each protein across the gradient. Mass spec data was handled as described above in Analyses of Gradient Mass Spectrometric Data. Correlation values were calculated between the normalized gel quantification data and the mass spec data using numpy.corrcoef() with defaults settings.
Conservation analysis
Domain binding correlation
For each comparison of organisms, a matrix of external crosslinks between domains within each organism was created, as described above in Crosslinking Maps. For humans, all paralogs were collapsed and considered as single entities. All domains that were not present in both species were removed, leaving n orthologous domains. (51 for humans to flies, 38 for humans to yeast, 38 for flies to yeast,). The n × n matrices were ordered such that they had the same order of orthologous domains on both axes. The Pearson correlation between each domain di in (1 ...n) in organism i was correlated with each domain dj (1 …n) in organism to get a full set of binding correlations between every domain. A z-score was calculated for each correlation value across this set, and they were then ranked.
Mutational analysis
Global Mutations
For every mSWI/SNF gene the number of TCGA (https://cancergenome.nih.gov/) samples with non-silent mutations was calculated in each cancer type and then divided by the total number of samples. Samples in the top 7.5% for overall number mutations were considered hypermutated and removed from the analysis.
Truncation Mutation Impact
Tumor mutation data for each protein was downloaded from CBioPortal. Cell line data was excluded. For each protein, the number of mutations (nonsense, frameshift in/dels or splice site mutations) that resulted in a truncation/amino acid was calculated.
For each protein p, 5000 random integers were selected between 1 and the length of p using numpy.random.randint. Each of these integers represents the position of a random mutation. For each of these simulated ‘mutations’, the proportion of external crosslinking sites (lysines that crosslink to another mSWI/SNF protein) that occur beyond the mutation (and thus would be lost in the random ‘truncation’) was calculated. A mean fraction of sites lost was calculated over the 5000 runs for each protein.
Expression Comparison
CCLE mutation and expression data is available for download at https://portals.broadinstitute.org/ccle/users/sign_in, TCGA mutation and expression data was downloaded from https://cancergenome.nih.gov. For both the CCLE and TCGA datasets, samples were considered ARID1A deficient if they contained a truncation mutation (nonsense, splice site, frameshift mutation) in the ARID1A gene.
DATA AND SOFTWARE AVAILABILITY
All cross-linking mass-spectrometry data including the spectra, linkages and structure analyses can be visualized at: https://www.yeastrc.org/proxl_public/viewProject.do?project_id=127
All raw files relating to cross-linking mass-spectrometry are deposited at proteome Xchange (Deutsch et al., 2017): http://www.proteomexchange.org/ under PRIDE access numbers PXD010122, PXD010123, and PXD010124, for mammalian BAF, PBAF and Drosophila BAP, respectively.
ADDITIONAL RESOURCES
The Nexus program can be directly downloaded from: https://systemsbiology.org/people/labs/ranish-lab/ [Click on Nexus link]
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse Anti-SMARCA4 (BRG1) (G-7) (WB) | Santa Cruz | Cat# sc-17796; RRID: AB_626762 |
| Rabbit Anti-SMARCA4 (BRG1) (D1Q7F) (IP) | Cell Signaling Technology | Cat# 49360S |
| Rabbit Anti-BCL7C (human) | This study | N/A |
| Mouse Anti-INI1 (BAF47) (A-5) (WB) | Santa Cruz | Cat# sc-166165; RRID: AB_2270651 |
| Rabbit Anti-SMARCC1 (H-76) (WB) | Santa Cruz | Cat# sc-10756; RRID: AB_2191997 |
| Rabbit Anti-SMARCC1 D7F8S (155–7) (IP) | Cell Signaling Technology | Cat# 11956S |
| Rabbit Anti-SS18 (D6I4Z) (IP) | Cell Signaling Technology | Cat# 21792S |
| Mouse Anti-ARID1A (BAF250A) (C-7) (WB) | Santa Cruz | Cat# sc-373784; RRID: AB_10917727 |
| Rabbit Anti-ARID1A D2A8U (IP) | Cell Signaling Technology | Cat# 12354S; RRID: AB_2637010 |
| Mouse Anti-ARID2 (BAF200) (E-3) (IB) | Santa Cruz | Cat# sc-166117; RRID: AB_2060382 |
| Rabbit Anti-DPF2 (WB) | Abcam | Cat# ab134942; RRID: AB_2728668 |
| Rabbit Anti-SMARCE1 (WP) | Bethyl | Cat# A300–810A; RRID: AB_577243 |
| Mouse Anti-SMARCD1 23 (WB) | Santa Cruz | Cat# sc-135843; RRID: AB_2192137 |
| Mouse Anti-SS18 A10 (WB) | Santa Cruz | Cat# sc-365170; RRID: AB_10709584 |
| Mouse Anti-GAPDH (G-9) (WB) | Santa Cruz | Cat# sc-365062; RRID: AB_10847862 |
| Mouse Anti-V5 tag (WB) | Thermo Fisher Scientific | Cat# R960–25; RRID: AB_2556564 |
| Rabbit Anti-V5 tag (D3H8Q) (IP) | Cell Signaling Technology | Cat# 13202S |
| Normal Rabbit IgG (IP) | Cell Signaling Technology | Cat# 2729S; RRID: AB_1031062 |
| Rabbit Anti-Actl6A (WB) | Bnvirogreethyl | Cat# A301–391A; RRID: AB_1309569 |
| Rabbit Anti-PBRM1 (WB) | Millipore | Cat# ABE70; RRID: AB_10807561 |
| Mouse Anti-BRD7 B-8 (WB) | Santa Cruz | Cat# sc-376180; RRID: AB_10989389 |
| Mouse Anti-GLTSCR1 (H-10)(IB) | Santa Cruz | Cat# sc-515086 |
| Rabbit Anti-BRD9 (WB) | Abcam | Cat# ab137245 |
| Rabbit Anti-ARID1B (WB) | Abcam | Cat# ab84461; RRID: AB_1859798 |
| Rabbit Anti-HA (C29F4) (WB) | Cell Signaling Technology | Cat# 3724S; RRID: AB_1549585 |
| Mouse Anti-Ubiquitin (P4D1) (WB) | Santa Cruz | Cat# sc-8017; RRID: AB_628423 |
| Rabbit Anti-SMARCA4 (D1Q7F) (WB) | Cell Signaling Technology | Cat# 49360S |
| IRDye 680RD Goat anti-Rabbit IgG (H + L) | Li-Cor Biosciences | Cat# 925–68071; RRID: AB_2721181 |
| IRDye 800CW Donkey anti-Mouse IgG (H + L) | Li-Cor Biosciences | Cat# 925–32212; RRID: AB_2716622 |
| Bacterial and Virus Strains | ||
| One-Shot Stbl 3 chemically competent cells | Invitrogen | Cat# C7373–03 |
| One Shot T0P10 Chemically Competent E. coli | Thermos Fisher Scientific | Cat#C4040–06 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Puromycin | Sigma-Aldrich | Cat# P8833–25MG |
| Blasticidin | Life Technologies | Cat# R210–01 |
| Dimethyl sulfoxide | Sigma-Aldrich | Cat# D2650 |
| DMEM, high glucose, no glutamine | Life Technologies | Cat# 11960–069 |
| IRDye 680RD NHS Ester | Li-Cor | Cat# 929–70050 |
| Hygromycin B (50 mg/mL) | Thermos Fisher Scientific | Cat# 10687010 |
| HA peptide synthetic | GenScript | NA |
| PBS, pH 7.4 | Life Technologies | Cat# 10010–049 |
| GlutaMAX | Life Technologies | Cat# 35050–079 |
| Trypsin-EDTA (0.25%), phenol red | Life Technologies | Cat# 25200–114 |
| Sodium Pyruvate | Life Technologies | Cat# 11360–070 |
| Penicillin-Streptomycin | Life Technologies | Cat# 15140–163 |
| Polybrene | Santa Cruz Biotechnology | Cat# sc-134220 |
| Cycloheximide | Sigma-Aldrich | Cat# C7698–5G |
| Polyethylenimine (PEI) (MW 40,000) | Polysciences | Cat# 24765 |
| Goat Anti-Mouse IgG Antibody, IRDye 680RD Conjugated | LI-COR Biosciences | Cat# 926–68070 |
| Goat Anti-Rabbit IgG Antibody, IRDye 800CW Conjugated | LI-COR Biosciences | Cat# 926–32211 |
| Dynabeads Protein G | Thermo Fisher Scientific | Cat# 10004D |
| NuPage LDS Sample Buffer (4X) | Life Technologies | Cat# NP0007 |
| Effectene Transfection Reagent | QIAGEN | Cat# 301425 |
| StrataClean Resin | Agilent Technologies | Cat# 400714 |
| Pierce Anti-HA Magnetic Beads | Thermo Fisher Scientific | Cat# 88837 |
| NuPAGE Novex 4–12% Bis-Tris Gel 1.0 mm, 12 well | Thermo Fisher Scientific | Cat# NP0322 |
| SFX-Insect media | GE Healthcare | Cat# SH30278.LS |
| QIAamp DNA Mini Kit | QIAGEN | Cat# 51304 |
| MG132 | Sigma | Cat# 1211877–36-9 |
| Lipofectamine 3000 Transfection Reagent | Thermo Fisher Scientific | Cat# 13778030 |
| Effectene Transfection Reagent | QIAGEN | Cat# 301425 |
| BS3 (bis(sulfosuccinimidyl)suberate), No-Weigh Format | Thermo Fisher Scientific | Cat# 21585 |
| Critical Commercial Assays | ||
| pENTR/D-TOPO Cloning Kit | Thermos Fisher Scientific | Cat# K240020 |
| SilverQuest Silver Staining Kit | Thermo Fisher Scientific | Cat# LC6070 |
| Invitrogen Novex Colloidal Blue Staining Kit | Thermos Fisher Scientific | Cat# LC6025 |
| Deposited Data | ||
| All raw cross-linking mass spectrometry data PRIDE access numbers: PXD010122 (BAF), PXD010123 (BAF), PXD010124 (Fly PBAP) | This paper | http://www.proteomexchange.org/ |
| Visualization of cross-linking mass spectrometry data | This paper | https://www.yeastrc.org/proxl_public/viewProject.do?project_id=127 |
| Experimental Models: Cell Lines | ||
| Drosophila S2 cells | Thermo Fisher | Cat # R690-07 |
| Human: HEK293T LentiX (Female) | Clonetech | Cat# 632180 |
| Human: SW13 (Female) | ATCC | Cat# CCL-105 |
| Human: MIA-PA-Ca-2(Male) | ATCC | Cat# CRL-1420 |
| Recombinant DNA | ||
| EF-1a-MCS-PGK-Blast-HA-SMARCD1 | This study | N/A |
| EF-1a-MCS-PGK-Blast-HA-SMARCD2 | This study | N/A |
| EF-1a-MCS-PGK-Blast-HA-SMARCC1 | This study | N/A |
| EF-1a-MCS-PGK-Blast-HA-SMARCB1 | This study | N/A |
| EF-1a-MCS-PGK-Blast-HA-SMARCE1 | This study | N/A |
| EF-1a-MCS-PGK-Blast-HA-DPF2 | This study | N/A |
| EF-1a-MCS-PGK-Blast-HA-BRD7 | This study | N/A |
| EF-1a-MCS-PGK-Blast- HA-ARID1A-C-term Y2254* | This study | N/A |
| EF-1a-MCS-PGK-Blast- HA-ARID1A-C-term G2087N | This study | N/A |
| EF-1a-MCS-PGK-Blast-HA-ARID1A-C-term | This study | N/A |
| EF-1a-MCS-PGK-Blast-BCL7A-HA | This study | N/A |
| EF-1a-MCS-PGK-Blast-HA-mARID2 | This study | N/A |
| EF-1a-MCS-PGK-Blast- HA-PBRM1 | This study | N/A |
| pMSCV-Flag-HA-IRES-Puro-SS18 | This study | N/A |
| EF-1a-MCS-PGK-Blast-HA-GLRSCR1L | This study | N/A |
| EF-1a-MCS-PGK-Blast-HA-BRD9 | This study | N/A |
| EF-1a-MCS-PGK-Blast-HA-PHF10 | This study | N/A |
| EF-1a-MCS-PGK-Blast-V5-PBRM1 | This study | N/A |
| EF-1a-MCS-PGK-Blast-V5-PBRM1 Δ BAH1 | This study | N/A |
| EF-1a-MCS-PGK-Blast-GFP-PBRM1 | This study | N/A |
| pMT-Puro HA-D4 | This study | N/A |
| pMCFHBD BAP60-Flag-HA | DGRC | N/A |
| BAF57 CRISPR/Cas9 KO Plasmid (h) | Santa Cruz Biotechnology | Cat#sc-404713 |
| Ini1 CRISPR/Cas9 KO Plasmid (h) | Santa Cruz Biotechnology | Cat#sc-401485 |
| ARID1A CRISPR/Cas9 KO Plasmid (h) | Santa Cruz Biotechnology | Cat#sc-400469 |
| ARID1B CRISPR/Cas9 KO Plasmid (h) | Santa Cruz Biotechnology | Cat#sc-402365 |
| Brm CRISPR/Cas9 KO Plasmid (h) | Santa Cruz Biotechnology | Cat#sc-401049-K0–2 |
| BAF155 CRISPR/Cas9 KO Plasmid (h) | Santa Cruz Biotechnology | Cat#sc-400838 |
| BAF170 Double Nickase Plasmid (h) | Santa Cruz Biotechnology | Cat#sc-402023-NIC |
| BAF60A CRISPR/Cas9 KO Plasmid (h) | Santa Cruz Biotechnology | Cat#sc-402641 |
| BAF60B Double Nickase Plasmid (h) | Santa Cruz Biotechnology | Cat#sc-403091-NIC |
| BAF60C CRISPR/Cas9 KO Plasmid (h) | Santa Cruz Biotechnology | Cat#sc-402707 |
| BRG-1 Double Nickase Plasmid (h) | Santa Cruz Biotechnology | Cat#sc-400168-NIC |
| BRD7 Double Nickase Plasmid (h) | Santa Cruz Biotechnology | Cat#sc-416299-NIC |
| ARID2 Double Nickase Plasmid (h) | Santa Cruz Biotechnology | Cat#sc-401863-NIC |
| PBRM1 Double Nickase Plasmid (h) | Santa Cruz Biotechnology | Cat#sc-403988-NIC |
| SS18 Double Nickase Plasmid (h) | Santa Cruz Biotechnology | Cat#sc-401575-NIC |
| CREST Double Nickase Plasmid (h) | Santa Cruz Biotechnology | Cat#sc-403134-NIC-2 |
| BRD9 Double Nickase Plasmid (h) | Santa Cruz Biotechnology | Cat#sc-404933-NIC |
| Software and Algorithms | ||
| MacPyMOL | Schrodinger | https://pymol.org/2/ |
| Seaborns | Seaborns/Michael Waskom | https://seaborn.pydata.org/index.html |
| Matplotlib | Matplotlib V2.3.3/ John Hunter and colleagues | https://matplotlib.org/ |
| Circos 0.69–6 | Circos | http://circos.ca/software/download/circos/ |
| ShareLaTeX | Overleaf/ShareLaTeX | https://v2.overleaf.com/project |
| GeneiousR9 | Geneious/Biomatters Limited | https://www.geneious.com/ |
| Nexus | Ranish Laboratory (Institute for Systems Biology) | https://systemsbiology.org/people/labs/ranish-lab/ [Select: Nexus] |
Highlights.
mSWI/SNF complexes assemble in an ordered, modular pathway
mSWI/SNF core module is a required platform for BAF, PBAF, and ncBAF formation
ARID/GLTSCR subunits define complex identity and facilitate ATPase module binding
Recurrent missense and nonsense mutations affect mSWI/ SNF complex assembly
ACKNOWLEDGMENTS
We are grateful to all members of the Kadoch laboratory for helpful discussions and advice. We thank Dr. Steven Gygi for very helpful advice and guidance pertaining to mass spectrometry experiments and all members of the Taplin Mass Spectrometry Facility (Harvard Medical School). We thank G. R. Crabtree and A. Gomez for human PBRM1 cDNA. This work was supported in part by awards from the NIH DP2 New Innovator Award 1DP2CA195762–01(to C.K.), American Cancer Society Research Scholar Award RSG-14–051-01-DMC (to C.K.), the Pew-Stewart Scholars in Cancer Research Grant (to C.K.), NIH R01 GM110064 (to J.A.R.), and P50 GM076547 (to J.A.R).
Footnotes
DECLARATION OF INTERESTS
C.K. is a scientific founder, shareholder, director, scientific advisory board member, and consultant for Foghorn Therapeutics (Cambridge, MA). The other authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and six tables and can be found with this article online at https://doi.org/10.1016/j.cell.2018.09.032.
REFERENCES
- Alpsoy A, and Dykhuizen EC (2018). Glioma tumor suppressor candidate region gene 1 (GLTSCR1) and its paralog GLTSCR1-like form SWI/SNF chromatin remodeling subcomplexes. J. Biol. Chem 293, 3892–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew B. (2014). ISWI chromatin remodeling: one primary actor or a coordinated effort? Curr. Opin. Struct. Biol 24, 150–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel VD, Guillaume J-L, Lambiotte R, and Lefebvre E. (2008). Fast unfolding of communities in large networks. J. Stat. Mech 2008, P10008. [Google Scholar]
- Bögershausen N, and Wollnik B. (2018). Mutational Landscapes and Phenotypic Spectrum of SWI/SNF-Related Intellectual Disability Disorders. Front. Mol. Neurosci 11, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Iwasa J, Cairns BR, and Peterson CL (2017). Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol 18, 407–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway RC, and Conaway JW (2009). The INO80 chromatin remodeling complex in transcription, replication and repair. Trends Biochem. Sci 34, 71–77. [DOI] [PubMed] [Google Scholar]
- Davoli T, Xu AW, Mengwasser KE, Sack LM, Yoon JC, Park PJ, and Elledge SJ (2013). Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155, 948–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechassa ML, Zhang B, Horowitz-Scherer R, Persinger J, Woodcock CL, Peterson CL, and Bartholomew B. (2008). Architecture of the SWI/ SNF-nucleosome complex. Mol. Cell. Biol 28, 6010–6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch EW, Csordas A, Sun Z, Jarnuczak A, Perez-Riverol Y, Ternent T, Campbell DS, Bernal-Llinares M, Okuda S, Kawano S, et al. (2017). The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res 45 (D1), D1100–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall AK, Beek SJ, McCallum CM, Tamkun JW, Kalpana GV, Goff SP, and Scott MP (1995). The Drosophila snr1 and brm proteins are related to yeast SWI/SNF proteins and are components of a large protein complex. Mol. Biol. Cell 6, 777–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Diedrich J, Chu YY, and Yates JR 3rd. (2015). Extracting Accurate Precursor Information for Tandem Mass Spectra by RawConverter. Anal. Chem 87, 11361–11367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, and Crabtree GR (2009). An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. USA 106, 5181–5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann AF, Martin LJ, Minder JL, Roe JS, Shi J, Steurer S, Bader G, McConnell D, Pearson M, Gerstberger T, et al. (2016). Sensitivity and engineered resistance of myeloid leukemia cells to BRD9 inhibition. Nat. Chem. Biol 12, 672–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CS, Foehr S, Garfield DA, Furlong EE, Steinmetz LM, and Krijgsveld J. (2014). Ultrasensitive proteome analysis using paramagnetic bead technology. Mol. Syst. Biol 10, 757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villén J, Haas W, Sowa ME, and Gygi SP (2010). A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C, and Crabtree GR (2015). Mammalian SWI/SNF chromatin remodeling complexes and cancer: Mechanistic insights gained from human genomics. Sci. Adv 1, e1500447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoch C, Hargreaves DC, Hodges C, Elias L, Ho L, Ranish J, and Crabtree GR (2013). Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet 45, 592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leschziner AE, Saha A, Wittmeyer J, Zhang Y, Bustamante C, Cairns BR, and Nogales E. (2007). Conformational flexibility in the chromatin remodeler RSC observed by electron microscopy and the orthogonal tilt reconstruction method. Proc. Natl. Acad. Sci. USA 104, 4913–4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Li M, Xia X, Li X, and Chen Z. (2017). Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure. Nature 544, 440–445. [DOI] [PubMed] [Google Scholar]
- López AJ, and Wood MA (2015). Role of nucleosome remodeling in neuro-developmental and intellectual disability disorders. Front. Behav. Neurosci 9, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marom R, Jain M, Burrage LC, Song IW, Graham BH, Brown CW, Stevens SJC, Stegmann APA, Gunter AT, Kaplan JD, et al. (2017). Heterozygous variants in ACTL6A, encoding a component of the BAF complex, are associated with intellectual disability. Hum. Mutat 38, 1365–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JA, and Teichmann SA (2015). Structure, dynamics, assembly, and evolution of protein complexes. Annu. Rev. Biochem 84, 551–575. [DOI] [PubMed] [Google Scholar]
- Mashtalir N, Daou S, Barbour H, Sen NN, Gagnon J, Hammond-Martel I, Dar HH, Therrien M, and Affar B. (2014). Autodeubiquitination protects the tumor suppressor BAP1 from cytoplasmic sequestration mediated by the atypical ubiquitin ligase UBE2O. Mol. Cell 54, 392–406. [DOI] [PubMed] [Google Scholar]
- Masliah-Planchon J, Bièche I, Guinebretiere JM, Bourdeaut F, and De-lattre O. (2015). SWI/SNF chromatin remodeling and human malignancies. Annu. Rev. Pathol 10, 145–171. [DOI] [PubMed] [Google Scholar]
- Murawska M, and Brehm A. (2011). CHD chromatin remodelersand the transcription cycle. Transcription 2, 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama RT, Pulice JL, Valencia AM, McBride MJ, McKenzie ZM, Gillespie MA, Ku WL, Teng M, Cui K, Williams RT, et al. (2017). SMARCB1 is required for widespread BAF complex-mediated activation of enhancers and bivalent promoters. Nat. Genet 49, 1613–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan ML, Sif S, Narlikar GJ, and Kingston RE (1999). Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3, 247–253. [DOI] [PubMed] [Google Scholar]
- Priam P, Krasteva V, Rousseau P, D’Angelo G, Gaboury L, Sauvageau G, and Lessard JA (2017). SMARCD2 subunit of SWI/SNF chromatin-remodeling complexes mediates granulopoiesis through a CEBPε dependent mechanism. Nat. Genet 49, 753–764. [DOI] [PubMed] [Google Scholar]
- Riffle M, Jaschob D, Zelter A, and Davis TN (2016). ProXL (Protein CrossLinking Database): A Platform for Analysis, Visualization, and Sharing of Protein Cross-Linking Mass Spectrometry Data. J. Proteome Res 15, 2863–2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnowska E, Gratkowska DM, Sacharowski SP, Cwiek P, Tohge T, Fernie AR, Siedlecki JA, Koncz C, and Sarnowski TJ (2016). The Role of SWI/SNF Chromatin Remodeling Complexes in Hormone Crosstalk.Trends Plant Sci 21, 594–608. [DOI] [PubMed] [Google Scholar]
- Sen P, Luo J, Hada A, Hailu SG, Dechassa ML, Persinger J, Brahma S, Paul S, Ranish J, and Bartholomew B. (2017). Loss of Snf5 Induces Formation of an Aberrant SWI/SNF Complex. Cell Rep 18, 2135–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, and Mann M. (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem 68, 850–858. [DOI] [PubMed] [Google Scholar]
- Smith CL, and Peterson CL (2005). A conserved Swi2/Snf2 ATPase motif couples ATP hydrolysis to chromatin remodeling. Mol. Cell. Biol 25, 5880–5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokpor G, Xie Y, Rosenbusch J, and Tuoc T. (2017). Chromatin Remodeling BAF (SWI/SNF) Complexes in Neural Development and Disorders. Front. Mol. Neurosci 10, 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern M, Jensen R, and Herskowitz I. (1984). Five SWI genes are required for expression of the HO gene in yeast. J. Mol. Biol 178, 853–868. [DOI] [PubMed] [Google Scholar]
- Versteege I, Sévenet N, Lange J, Rousseau-Merck MF, Ambros P, Handgretinger R, Aurias A, and Delattre O. (1998). Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394, 203–206. [DOI] [PubMed] [Google Scholar]
- Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, and Crabtree GR (1996). Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev 10, 2117–2130. [DOI] [PubMed] [Google Scholar]
- Witzel M, Petersheim D, Fan Y, Bahrami E, Racek T, Rohlfs M, Puchałka J, Mertes C, Gagneur J, Ziegenhain C, et al. (2017). Chromatin-remodeling factor SMARCD2 regulates transcriptional networks controlling differentiation of neutrophil granulocytes. Nat. Genet 49, 742–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JI, Lessard J, and Crabtree GR (2009). Understanding the words of chromatin regulation. Cell 136, 200–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RC, Wang TL, and Shih, leM. (2014). The emerging roles of ARID1A in tumor suppression. Cancer Biol. Ther 15, 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Liu X, Li T, Fang X, and Chen Z. (2016). Structure of chromatin remodeler Swi2/Snf2 in the resting state. Nat. Struct. Mol. Biol 23, 722–729. [DOI] [PubMed] [Google Scholar]
- Yan L, Xie S, Du Y, and Qian C. (2017). Structural Insights into BAF47 and BAF155 Complex Formation. J. Mol. Biol 429, 1650–1660. [DOI] [PubMed] [Google Scholar]
- Yang B, Wu YJ, Zhu M, Fan SB, Lin J, Zhang K, Li S, Chi H, Li YX, Chen HF, et al. (2012). Identification of cross-linked peptides from complex samples. Nat. Methods 9, 904–906. [DOI] [PubMed] [Google Scholar]
- Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, and Crabtree GR (1998). Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95, 625–636. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All cross-linking mass-spectrometry data including the spectra, linkages and structure analyses can be visualized at: https://www.yeastrc.org/proxl_public/viewProject.do?project_id=127
All raw files relating to cross-linking mass-spectrometry are deposited at proteome Xchange (Deutsch et al., 2017): http://www.proteomexchange.org/ under PRIDE access numbers PXD010122, PXD010123, and PXD010124, for mammalian BAF, PBAF and Drosophila BAP, respectively.


