Abstract
Hemorphins are atypical endogenous opioid peptides produced by the cleavage of hemoglobin beta chain. Several studies have reported the therapeutic potential of hemorphin in memory enhancement, blood regulation, and analgesia. However, the mode of interaction of hemorphin with its target remains largely elusive. The decapeptide LVV-hemorphin-7 is the most stable form of hemorphin. It binds with high affinity to mu-opioid receptors (MOR), angiotensin-converting enzyme (ACE) and insulin-regulated aminopeptidase (IRAP). In this study, computational methods were used extensively to elucidate the most likely binding pose of mammalian LVV-hemorphin-7 with the aforementioned proteins and to calculate the binding affinity. Additionally, alignment of mammalian hemorphin sequences showed that the hemorphin sequence of the camel harbors a variation – a Q > R substitution at position 8. This study also investigated the binding affinity and the interaction mechanism of camel LVV-hemorphin-7 with these proteins. To gain a better understanding of the dynamics of the molecular interactions between the selected targets and hemorphin peptides, 100 ns molecular dynamics simulations of the best-ranked poses were performed. Simulations highlighted major interactions between the peptides and key residues in the binding site of the proteins. Interestingly, camel hemorphin had a higher binding affinity and showed more interactions with all three proteins when compared to the canonical mammalian LVV-hemorphin-7. Thus, camel LVV-hemorphin-7 could be explored as a potent therapeutic agent for memory loss, hypertension, and analgesia.
Subject terms: Protein function predictions, Molecular modelling
Introduction
Initial efforts in the 1980s to recognize endogenously produced opioid peptides led to the characterization of hemoglobin-derived (Hb-derived) peptides that have opiate-like activity1. These peptides are short, 4–10 amino acids in length and are released during physiological or physiopathological hemoglobin beta-chain proteolytic degradation2. These Hb beta chain derived peptides, named hemorphins, share a central tetrapeptide core (Tyr-Pro-Trp-Thr) from the N-terminal region. Interestingly, both N- and C-terminal extensions of hemorphin have been isolated from human and bovine tissue3. Hemorphin peptides of varying lengths can be produced by the degradation of hemoglobin beta chain through numerous enzymatic digestions including lysosomal proteases, pepsin, chymotrypsin-like proteases, aspartic endoprotease cathepsin and macrophagic enzymes4–6. It has been reported that the inhibition of intraerythrocytic proteasome blocked the generation of hemorphin, supporting the involvement of proteasomes in the processing and generation of hemorphin7.
The first characterized hemoglobin derived opioid peptide was hemorphin-4 (YPWT). Hemorphin-4 was initially isolated from bovine blood treated with a mixture of gastrointestinal enzymes1. Later, a series of peptides, including hemorphin-4 to -7 and LVV-hemorphin-4, -6, and -7, were identified that contained the YPWT sequence8–10. These are naturally occurring peptides produced in the brain, spinal cord, plasma and cerebrospinal fluid5,11–14. Hemorphins have been shown to possess numerous biological activities including effects on spatial learning, inflammation, analgesia and transient hypotension15–18. Among hemorphin peptides, LVV-hemorphin-7 (LVV-H7 or LVVYPWTQRF) is the longest with 10 amino acids. Functional activity of LVV-hemorphin-7 derived from either Hb β, γ, δ and ε chains have been widely studied. Several reports have indicated that LVV-hemorphin-7 is the most abundant and has the highest hydrophobicity in mammalian central nervous system (CNS)13,19.
The discovery of stereospecific opioid binding sites in the mammalian brain initiated a series of studies to identify their natural ligands. Met- and Leu-enkephalin were the first described endogenous opioid peptides20. Later, various studies revealed the existence of new classes of bioactive peptides produced from limited proteolysis of protein precursors. Enkephalins, dynorphins, and beta-endorphin are the classical opioid peptides released by the cleavage of proenkephalin, prodynorphin and proopiomelanocortin, respectively, and are expressed in the CNS14,21,22. Mu, delta and kappa are three opioid receptors that have been investigated extensively. Partial proteolysis of proteins leads to the production of several groups of peptides. For instance, cytochrophins are released from mitochondrial cytochrome b, casomorphins from the milk protein beta-casein, and hemorphins from hemoglobin1,3. Agonistic binding to opioid receptors produces profound analgesia in inflamed tissue, highlighting the importance of investigating such molecules23. It has been shown that all hemorphin peptides, including LVV-hemorphin-7, show an affinity for opioid receptors5,12. Additionally, hemorphin-5, -6, and -7 have shown opioid-like activity in electrically stimulated myenteric plexus/longitudinal muscle preparation of the guinea pig ileum. Glucose phosphate isomerase (GPI) bioassay had also shown agonistic opioid-like effects of different hemorphins24. Further, hemorphin’s opiod receptor affinity was investigated in rat brain membrane and results showed IC50 values in the micromolar range24. Moreover, it has also been suggested that hemorphins imitate opioid peptides in terms of their analgesic activities. Hemorphins have also produced a dose-dependent antinociceptive response in mice25. Chow and colleagues reported that intrathecal doses of angiotensin IV (Ang IV) and LVV-hemorphin-7 cause anti-hyperalgesia in rats26.
The renin-angiotensin system (RAS) is one of the most thoroughly investigated enzyme-neuropeptide systems. RAS originated from the classical renin/angiotensin-converting enzyme (ACE)/angiotensin II (Ang II)/angiotensin II type I (AT1) receptor axis and has a physiological role in the regulation of cardiovascular and renal function, aldosterone biosynthesis and release, blood pressure and body salt, and fluid balance27. In recent years, a lot of progress has been made in identifying new proteins of this system as well as elucidating their roles and signal transduction mechanisms. Therefore, renin/ACE/Ang II/AT1 and AT2 axis are not the exclusive signaling pathways for the system. The prorenin/PRR/MAP kinases ERK1/2 axis, the ACE2/Ang (1–7)/Mas receptor axis and the Ang IV/AT4/IRAP (insulin-regulated aminopeptidase, IRAP) axis are three new axes that have been reported recently28,29. LVV-hemorphin-7’s role in blood pressure regulation has already been suggested by several studies. Intraperitoneal injection of LVV-hemorphin-7 was found to cause a substantial decrease in blood pressure and heart rate in hypertensive rats30. LVV-hemorphin-7 also potentiated the hypotensive effect of bradykinin in rats31. LVV-hemorphin-7 also inhibited ACE, a key component of RAS, indicating a major role in the regulation of blood pressure32.
RAS in the brain is believed to be involved in processes other than homeostatic control. It has been associated with neuronal differentiation, nerve regeneration, learning and memory33. Various physiological functions have been associated with Ang IV, including the facilitation of memory34. LVV-hemorphin-7 was identified as an endogenous high-affinity ligand of the presumed angiotensin IV receptor (AT4R)19. Further characterization of this putative receptor suggested that the protein was analogous to IRAP, a type II integral membrane protein whose catalytic activity is inhibited by LVV-hemorphin-7 and Ang IV35–37. In vivo and in vitro studies revealed that LVV-hemorphin-7 could play a potential role in learning and memory. An intracerebral administration of LVV-hemorphin-7 enhanced spatial learning in rats and diminished the effects of scopolamine-induced learning deficits in spatial learning tests and fear conditioning17,38. LVV-hemorphin-7 has also been shown to modify the behavior of rats. Additionally, reduced depression and increased locomotion, induced by LVV-hemorphin-7, depended on the activation of oxytocin receptors most likely due to changes in the oxytocinase activity of AT4R39. It was assumed that by inhibiting IRAP activity, LVV-hemorphin-7 protects the substrates of IRAP such as oxytocin and vasopressin, which are known to play a key role in memory and learning40. In vivo microdialysis revealed that LVV-hemorphin-7 enhanced spatial working memory without a significant increase in blood flow or hippocampal glucose uptake41.
In vivo and in vitro studies have demonstrated the potential therapeutic role of hemorphin in several pathological conditions. However, the precise binding behavior of hemorphin on target proteins have not been clearly elucidated yet. In this study, using in silico techniques, we provide early insight into the binding of LVV-hemorphin-7 with MOR, ACE and IRAP. Determination of the likely binding mode of peptide is a difficult task and protein-peptide docking is often used for identifying potential modes of interactions. Molecular docking generates several possible binding solutions, and the most appropriate binding pose is frequently judged by comparison of docking scores42,43. With the advent of high performance and rigorous methods, binding free energy calculations and long timescale MD simulations could be employed to study protein-peptide interactions at an atomic level44–47. In this study, protein-peptide docking and binding free energy calculations using molecular mechanics-generalized Born surface area (MM-GBSA) were used to identify the most probable binding mode of LVV-hemorphin-7 in these proteins. Additionally, the solvation effects were examined by using explicit solvent MD simulations. The hemorphin sequence is highly conserved among mammals. However, multiple sequence alignment of hemoglobin beta protein from several mammals showed that the camel hemorphin sequence harbors one variation. This study explored both variants of LVV-hemorphin-7 - camel and other mammals (referred to as non-camel here onwards) - to identify the differences in binding mode, stability and interactions.
Results
Multiple sequence alignment of hemoglobin beta protein sequences
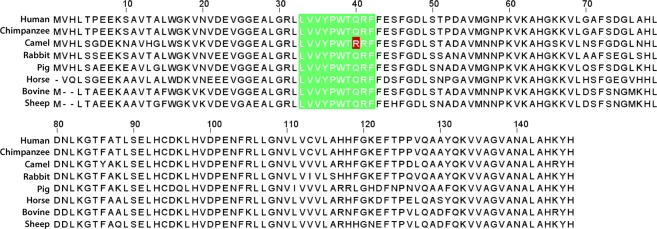
Hemoglobin beta (HBB) protein reference sequences of closely related mammals were retrieved from NCBI protein database. Multiple sequence alignment was performed using ClustalW to identify the similarities and differences in the HBB protein sequences. The decapeptide LVV-hemorphin-7 sequence is highly conserved among mammals. In human, chimpanzee, camel, pig, and rabbit it lies between amino acid positions 32 to 41 in the HBB protein sequence. In the bovine and sheep protein, it ranges from amino acids 30 to 39 and in the horse the decapeptide is located between amino acids 31 to 40. The sequence comparison clearly showed a single amino acid substitution in the camel HBB protein at position 40, where a glutamine has been changed to an arginine (Q > R). All other residues are identical (Fig. 1).
Figure 1.
Multiple sequence alignment of hemoglobin beta protein sequences from closely related mammals - Homo sapiens (human), Pan troglodytes (chimpanzee), Camelus dromedarius (camel), Oryctolagus cuniculus (rabbit), Sus scrofa (wild pig), Equus caballus (horse), Bos taurus (bovine), and Ovis aries (sheep).
Structure-based homology modeling of mu-type opioid receptor
The three-dimensional homology model of the active state of the human mu-type opioid receptor (MOR) (UniProt accession: P35372) was generated using the X-ray crystallographic structure of MOR from Mus musculus (PDB ID: 5C1M) (Fig. 2A). Structural model of the active conformation of MOR is analogous to the conserved seven-transmembrane topology of G protein-coupled receptors (GPCRs). The Ramachandran plot showed that the overall stereochemical quality of the generated model was good with almost all the residues in the favorable and allowed regions (Supplementary Fig. 1).
Figure 2.
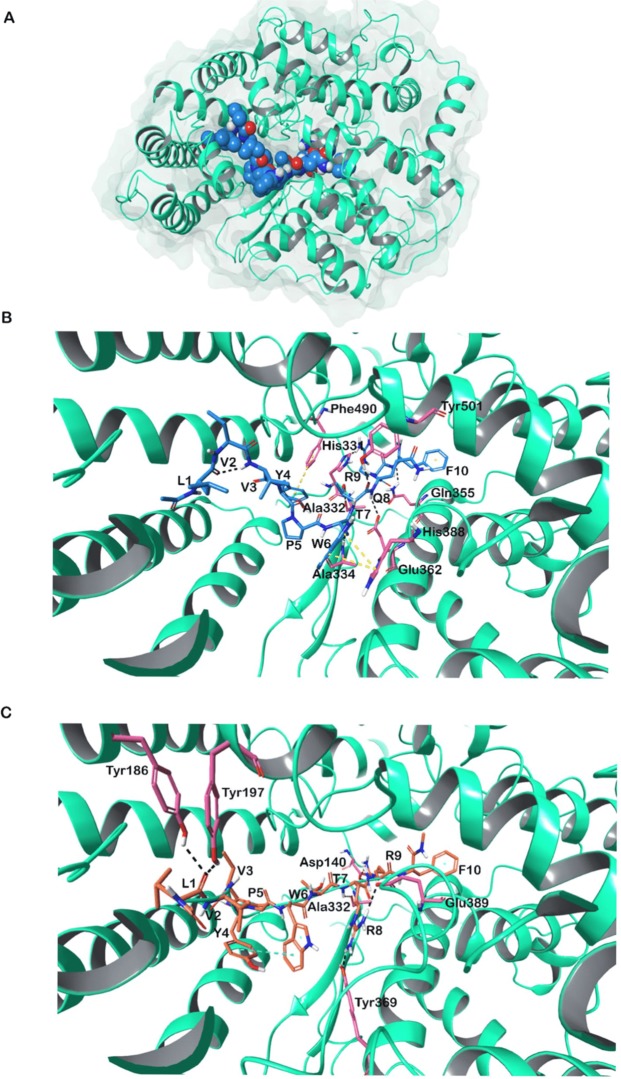
Modeled active human MOR. (A) Side view (B) Top view. (C) LVVYPWTQRF (non-camel LVV-hemorphin-7) docked in the binding pocket of the putative active conformation of human MOR. (D) LVVYPWTRRF (camel LVV-hemorphin-7) docked in the binding pocket of the putative active conformation of human MOR. Hydrogen bonds are represented by black dotted lines and π-π stacking by yellow dotted lines.
Molecular docking
By means of exhaustive docking and free energy calculations, the binding mode of both non-camel (LVVYPWTQRF) and camel (LVVYPWTRRF) LVV-hemorphin-7 peptides were identified. The binding free energies were obtained for each binding mode with the molecular mechanics generalized Born and surface area (MM-GBSA) method.
Mu-opioid receptor
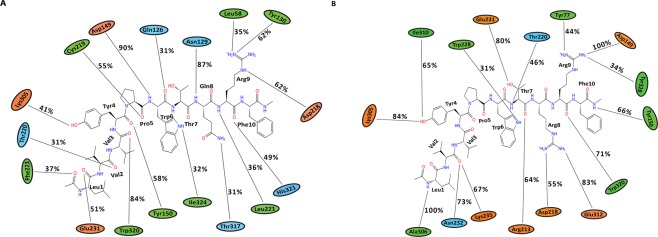
The docking of both camel and non-camel LVV-hemorphin-7 with MOR generated 10 models each. The best-docked pose of non-camel LVV-hemorphin-7, which was in the active site of MOR, produced a Glide docking score, GlideScore (GScore), of −10.88 kcal/mol and an MM-GBSA binding energy of −97.82 kcal/mol (Table 1). The best-docked conformation of non-camel LVV-hemorphin-7 revealed that the N-terminal residues of the peptide interacted with residues in transmembrane helices 5 and 6 (TM5 and TM6) lining the bottom of the binding site. The first three residues (Leu1-Val2-Val3) of the peptide produced extensive hydrophobic interactions with TM6 residues Ile298, Tyr301, Val302, Ala306, and Val308. The N-terminal leucine formed a hydrogen bond with the backbone amino group of the hydrophilic Asn232 present on TM5. Tyr4 in the non-camel LVV-hemorphin-7 interacted with Asp149 through a hydrogen bond and, along with Pro5, it produced hydrophobic interactions with Ile146 and Tyr150 present on TM3. Pro5, Trp6, and Thr7 interacted with Arg213, Ile217, Cys219, Thr220, and Leu221 on the beta sheet. Hydrophobic interactions were formed between the C-terminal residues (Thr7-Gln8-Arg9-Phe10) and Trp320, His321, Ile324, and Tyr328 on TM7. Trp6 and Gln8 interacted via π- π stacking and hydrogen bond, respectively, with His321 on TM7. Gln8 formed additional hydrogen bonds with the side chain of the Glu312 present on the third extracellular loop and with Lys305 on TM5 (Fig. 2C and Supplementary Fig. 2A).
Table 1.
Interacting residues of the best pose of non-camel (LVVYPWTQRF) and camel (LVVYPWTRRF) LVV-hemorphin-7 with MOR, ACE, and IRAP.
| Protein | Peptide | Glide docking score – GScore (kcal/mol) | MM-GBSA (kcal/mol) | Residues forming hydrogen bonds | Residues forming hydrophobic interactions | Residues forming π interactions |
|---|---|---|---|---|---|---|
| MOR | LVVYPWTQRF | −10.88 | −97.82 | Gln126, Asn129, Leu131, Asp149, Asn232, Lys305, Glu312, His321 | Leu58, Pro61, Pro65, Tyr77, Tyr130, Met132, Ile146, Tyr150, Ile217, Cys219, Leu221, Trp228, Ile298, Tyr301, Val302, Ala306, Val308, Trp320, Ile324, Tyr328 | His321 |
| MOR | LVVYPWTRRF | −10.53 | −115.08 | Asp149, Lys211, Tyr212, Glu231, Lys305, Ala306, Glu312 | Ile146, Tyr150, Cys219, Leu221, Phe223, Trp228, Leu234, Ile236, Val238, Ile298, Val302, Ile303, Leu307, Val308, Trp320, Ile324, Tyr328 | |
| ACE | LVVYPWTQRF | −10.66 | −119.32 | His331, Ala332, Ala334, Gln355, Glu362, Tyr501 | Val36, Tyr111, Leu139, Phe178, Tyr186, Tyr197, Trp198, Trp201, Trp257, Val329, Cys330, Ala335, Tyr338, Tyr369, Tyr372, Pro385, Phe435, Pro497, Val495, Tyr501, Phe505 | His388, Phe490 |
| ACE | LVVYPWTRRF | −12.31 | −151.57 | Asp140, Tyr186, Tyr197, Ala332, Tyr369, Glu389 | Leu32, Val36, Ala94, Leu98, Ala101, Tyr111, Leu115, Phe178, Tyr197, Trp201, Cys330, Ala332, Cys348, Ala334, Trp335, Phe435, Phe490, Val495, Tyr498, Tyr501, Phe505 | His388 |
| IRAP | LVVYPWTQRF | −14.86 | −148.04 | Gly428, Glu441, Lys460, Glu509, Asp510, Glu541, Glu818, Glu825, Glu895 | Tyr272, Pro296, Ala427, Ala429, Met430, Ala453, Leu457, Ile461, Tyr495, Ala514, Phe544, Tyr549, Phe550, Ala763, Leu769, Ala822, Phe826, Tyr961 | |
| IRAP | LVVYPWTRRF | −13.65 | −163.03 | Gly428, Glu441, Asp510, Glu541, Glu818, Glu825, Glu895 | Tyr272, Pro296, Ala427, Ala429, Met430, Ala453, Leu457, Ile461, Leu469, Tyr495, Ala514, Phe544, Tyr549, Phe550, Ala822, Phe826, Pro957, Tyr961 |
The best pose of camel LVVYPWTRRF had a GScore of −10.53 kcal/mol and MM-GBSA binding energy of −115.08 kcal/mol (Table 1). The N-terminal residues of camel LVV-hemorphin-7 interacted with residues present on TM5 (Glu231 and Ile236) and TM6 (Val302, Ile303, and Leu307). Differing from non-camel LVV-hemorphin-7, the residues at positions 5 and 6 (Pro5 and Trp6) formed hydrogen bonds with Tyr212 and Lys211, respectively, and produced hydrophobic interaction with the β-sheet residues. The C-terminal residues interacted with residues on TM3, TM5 and TM7. Arg9 also formed a hydrogen bond with Asp149 on TM3 and the substituted arginine at position 8 produced a hydrogen bond with the side chain of Glu312 on the third extracellular loop. Additionally, Glu231 on TM5 produced hydrogen bonds with Val3, Tyr4 and Thr7 (Fig. 2D). Hydrophobic interactions are shown in Supplementary Fig. 2B.
Angiotensin-converting enzyme
The binding mode and the affinity of the non-camel and the camel LVV-hemorphin-7 with ACE were predicted using molecular docking and binding free energy calculations. Table 1 lists the binding scores and the ACE amino acids that interacted with the peptides. The positioning of both non-camel and camel LVV-hemorphin-7 in the ACE active site was well defined, encompassing the S1 and S2 subsites. It is evident from the 2D ligand interaction diagrams that an extensive network of hydrogen bonds, hydrophobic, and electrostatic interactions were formed between ACE and hemorphin peptides (Supplementary Fig. 3).
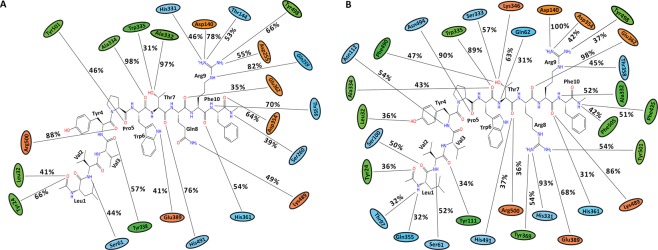
The best binding pose of non-camel LVVYPWTQRF had a GScore of −10.66 kcal/mol. The MM-GBSA binding energy calculated for this pose was −119.32 kcal/mol. The N-terminal of LVV-hemorphin-7 extended towards the lid of the active site. The C-terminal bound deep in the active site of ACE with Arg9 binding in the S2 subsite with its side chain oriented towards S1′. The sidechain of Arg9 formed a hydrogen bond with His331 in the S2 subsite and electrostatic interaction with Asp140 in S1′. Thr7 and Gln8 occupied the S1 subsite and Gln8 made hydrophobic interactions with Phe435 and Phe505. The backbone amino and carboxyl group of Thr7 formed hydrogen bonds with Glu362, and Tyr501 respectively and the side chain formed a hydrogen bond with Ala332 in the S1 subsite. The backbone carboxyl group of Trp6 formed a hydrogen bond with Ala334 and the side chain formed a π-π interaction with His388. It also produced hydrophobic interactions with Trp335, Tyr338, Tyr369, Tyr372, and Pro385. Residues at 4 and 5 (Tyr4 and Pro5) showed hydrophobic interactions with Val329, Pro385, Val495 and Pro497 and a π-π stacking was formed between Tyr4 and Phe490. The N-terminal residues oriented towards the lid and showed hydrophobic interactions with Tyr111, Phe178, Tyr186, Tyr197, Trp198, and Trp201 (Fig. 3B and Supplementary Fig. 3A).
Figure 3.
(A) Three dimensional structure of ACE. (B) LVVYPWTQRF (non-camel LVV-hemorphin-7) docked in the active site of ACE. (C) LVVYPWTRRF (camel LVV-hemorphin-7) docked in the active site of ACE. Hydrogen bonds are represented by black dotted lines and π-π stacking represented by yellow dotted lines.
Binding mode of LVVYPWTRRF in the ACE active site was different from the non-camel peptide. More specifically, the positioning of the C-terminal in S1′ and S2 pockets was different. The best docked pose had a GScore of −12.31 kcal/mol and an MM-GBSA binding energy of −151.57 kcal/mol (Table 1). Arg9 and Phe10 were stabilized in the S2 subsite, through hydrophobic interactions with Phe435, Tyr498, Tyr501, and Phe505. The side chain of Arg9 interacted with the S1′ subsite and formed a hydrogen bond with Asp140. Interestingly, the arginine at position 8 in the camel LVV-hemorphin-7 bound in the S1 subsite and produced more interactions with the active site residues. The side chain of Arg8 made hydrogen bonds with Tyr369 and Glu389 and showed hydrophobic interaction with Ala334. The backbone amino group of Thr7 interacted with Ala332 through a hydrogen bond. Residues at positions 4, 5, and 6 showed hydrophobic interactions with Leu32, Val36, Trp335, Tyr338, and Phe435. The N-terminal residues extended towards the active site lid and produced hydrophobic interactions with Ala65, Ala94, Leu98, Ala101, Tyr111, Leu115, Trp201, and Val495 (Fig. 3C and Supplementary Fig. 3B).
Insulin-regulated aminopeptidase
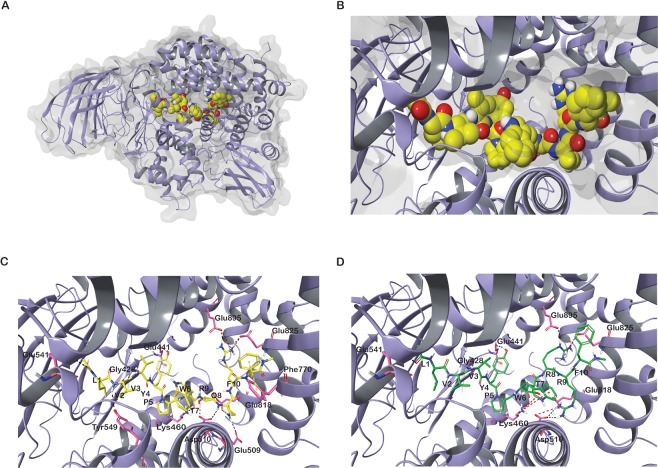
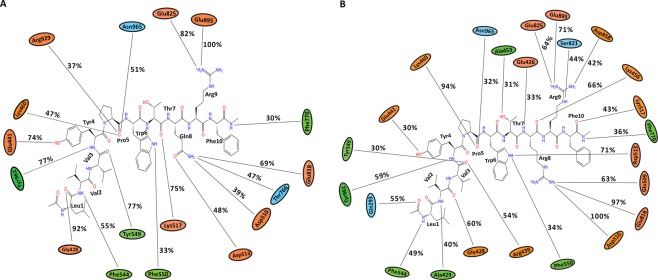
The MM-GBSA binding free energy calculations performed for the different docking conformations of LVVYPWTQRF revealed that the pose with GScore −14.86 kcal/mol exhibited the highest MM-GBSA binding free energy of −148.04 kcal/mol. The N-terminal of both peptides bound in the S1 pocket, which is adjacent to the active site. Figure 4C shows the binding mode for non-camel LVV-hemorphin-7. In the predicted binding mode, the first three residues of LVV-hemorphin-7 occupied the S1 pocket and formed hydrogen bond with Leu541, Tyr549, and Gly428. All three residues formed hydrophobic interactions with residues in the S1 pocket - Tyr272, Pro296, Phe425, Ala427, Ala429, Met430, Phe544, and Tyr961. The aromatic residues Tyr4 and Trp6 formed hydrogen bonds with Glu441 and Lys460, respectively, and showed hydrophobic interactions with Ala453, Leu457, Ile461, Tyr495, Ala514, and Phe550. The C-terminal residues formed hydrophobic interactions with Ala763, Leu769, Phe770, Ala822, and Phe826. Thr7 formed a hydrogen bond with Asp510, Gln8 formed hydrogen bonds with Glu509, and Glu818 and Arg9 displayed hydrogen bonds with the side chain of residues Glu825 and Glu895 (Fig. 4C and Supplementary Fig. 4A). The binding score and interacting residues are summarized in Table 1.
Figure 4.
(A) Three dimensional structure of IRAP. (B) Binding pocket of IRAP. (C) LVVYPWTQRF (non-camel hemorphin) docked in the binding pocket of IRAP. (D) LVVYPWTRRF (camel hemorphin) docked in the binding pocket of IRAP. Hydrogen bonds are represented by black dotted lines and cation-π interactions are represented by red dotted lines.
The camel LVV-hemorphin-7 exhibited a similar binding mode with the N-terminal binding in the S1 pocket. The first three residues of the N-terminal bound in the S1 pocket and showed hydrophobic interactions with Tyr272, Pro296, Ala427, Ala429, Met430, Phe544, Tyr549, Pro957, and Tyr961. The amino group of Gly428 of the GAMEN loop (Gly428-Ala429-Met430-Glu431-Asn432) produced a hydrogen bond with Val2 residue in the N-terminal. The residues at positions 4, 5, and 6 formed hydrophobic interactions with the active site residues Leu457, Ile461, Tyr495, Ala514, and Phe550 and Trp6 formed a hydrogen bond with Lys460. Arg8 formed two hydrogen bonds with the side chain of Glu825 and Glu895, and Arg9 formed hydrogen bonds with Asp510 and Glu818. The C-terminal residues formed hydrophobic interactions with Leu769, Ala822, and Phe826 (Fig. 4D and Supplementary Fig. 4B). The best binding pose of camel LVV-hemorphin-7 with IRAP had a GScore of −13.65 kcal/mol and binding free energy of −163.03 kcal/mol (Table 1).
Molecular dynamics simulations
To evaluate the stability and dynamics of the docked complexes, triplicate all-atom MD simulations were performed using Desmond48.
Simulations of camel and non-camel LVV-hemorphin-7 bound to MOR
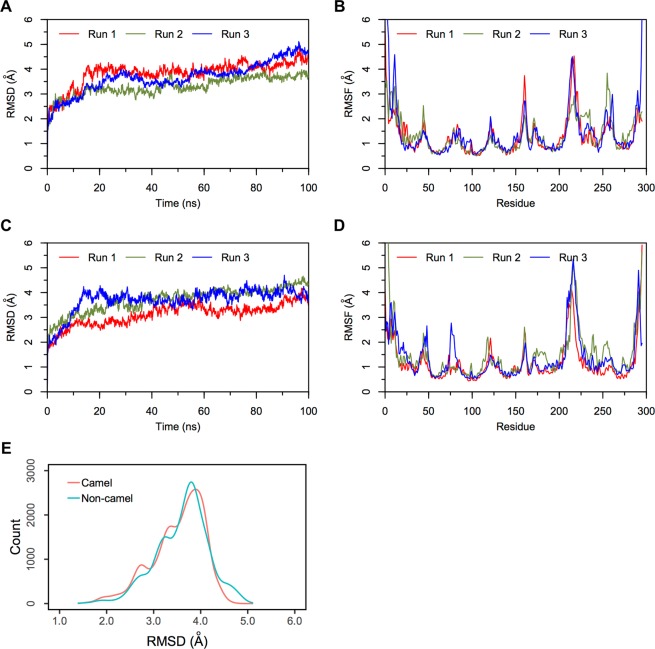
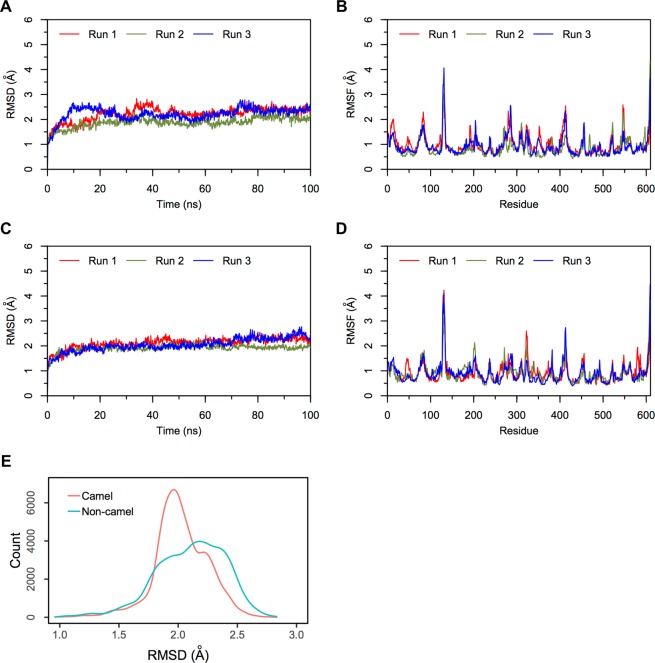
Triplicate 100 ns equilibrium MD simulations of the non-camel LVV-hemorphin-7 bound to modelled MOR structure embedded in a dipalmitoylphosphatidylcholine (DPPC) membrane remained stable with a protein Cα root mean square deviation (RMSD) under 5 Å (Fig. 5A). The per-residue root mean square fluctuation (RMSF) showed that the third intracellular (IL-3) and extracellular loop (EL-2) regions were the most flexible while the membrane-embedded regions were found to be the least flexible (Fig. 5B). The peptide interacted with residues on TM3 and TM7 in MD simulations. Tyr4 and Trp6 maintained hydrogen bond and hydrophobic interactions with Tyr150 and Asp149 respectively on TM3 and Leu1 interacted with the TM5 residue Glu231. Tyr4 interacted with Lys305 while residue Val3, Trp6, and Arg9 maintained its interactions with Trp320, Ile324 and His321 respectively on TM7 during the simulations. The average percentage of equilibrated simulation time during which the non-camel LVV-hemorphin-7 interacted with MOR residues is given in Fig. 6A. Contributions from individual simulations are provided in Supplementary Fig. 5A.
Figure 5.
RMSD and RMSF plots of triplicate 100 ns simulations of MOR. Data from the three runs are plotted with red, blue and green lines. (A) RMSD of protein Cα atoms from the MOR-LVVYPWTQRF simulations. (B) RMSF of protein Cα atoms from the MOR-LVVYPWTQRF simulations. (C) RMSD of protein Cα atoms from the MOR-LVVYPWTRRF simulations. (D) RMSF of protein Cα atoms from the MOR-LVVYPWTRRF simulations. (E) Density functions corresponding to the distribution of RMSD values from triplicate hemorphin-bound simulations.
Figure 6.
Average percentage of equilibrium simulation time during which MOR residues maintain contact with non-camel and camel LVV-hemorphin-7 from three 100 ns simulations. For equilibrium simulation data, the first 50 ns of run 1 was discarded, while the first 30 ns of runs 2 and 3 were discarded. Histograms representing the interaction from each of the 3 simulations can be found in Supplementary Fig. 5. Charged, hydrophobic and polar amino acids are represented with orange, green and blue color respectively. (A) Average percentage of time an MOR residue maintains contact with LVVYPWTQRF. (B) Average percentage of time an MOR residue maintains contact with LVVYPWTRRF.
In the camel LVV-hemorphin-7 bound simulations, the protein structure stabilized with RMSD under 4.5 Å in all three simulations (Fig. 5C). A distribution of the RMSD values obtained from the camel and non-camel hemorphin bound simulations is shown in Fig. 5E. EL-2 and IL-3 regions showed more fluctuations. More specifically, IL-3 showed more fluctuations compared to the MOR/non-camel LVV-hemorphin-7 complex (Fig. 5D). Camel LVV-hemorphin-7 showed interactions with residues from TM2, TM3, TM5, TM6, TM7 and the β-sheet. Arg9 interacted with Asp149 for the full duration of all three simulations while Phe10 interacted with Tyr150 during the simulations. LVVYPWTRRF also interacted with TM5 residues Glu231, Asn232, and Lys235. Leu1 and Tyr4 residues maintained continuous interactions with Ala306 and Lys305, respectively, on TM6 and the C-terminal residues interacted with Trp320, and Tyr328 on TM7. It also produced good interactions with Ile310 and Glu312 on the third extracellular loop. The average percentage of equilibrated simulation time during which the non-camel LVV-hemorphin-7 interacted with MOR residues is given in Fig. 6B. Contributions from individual simulations are provided in Supplementary Fig. 5B.
Simulations of camel and non-camel LVV-hemorphin-7 bound to ACE
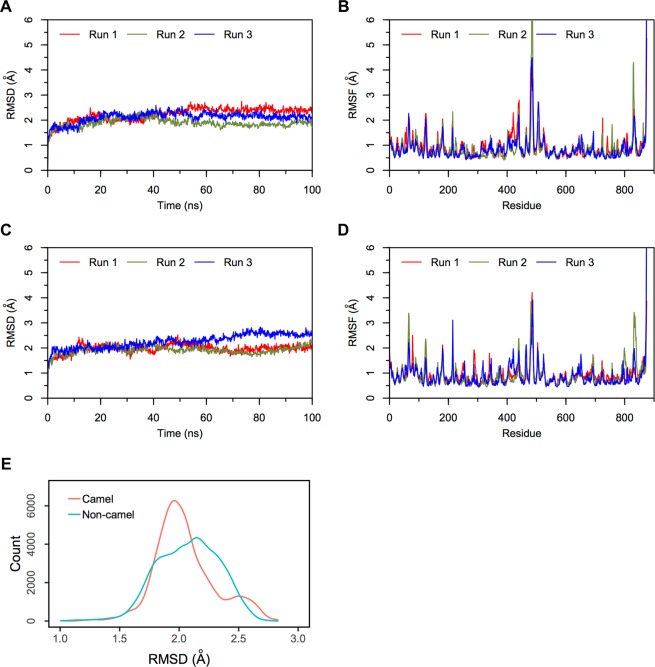
Molecular dynamics simulations of ACE with camel and non-camel LVV-hemorphin-7 showed stable interactions throughout three 100 ns simulations. In the non-camel LVV-hemorphin-7 bound simulation, the ACE structure stabilized with RMSD under 3 Å (Fig. 7A). Most of the residues in the protein produced very limited fluctuation throughout the simulations (Fig. 7B). The C-terminal residues of hemorphin were stabilized in specific subsites of the active site by forming hydrogen bonds and hydrophobic interactions. C-terminal residues Gln8, Arg9, and Phe10 maintained the interaction with Asp140, Thr144, Asp255, Gln259, His331, His361, and Lys489. MD trajectory analysis showed that Arg9 occupied the S2 subsite by interacting with Asp255 and His331, as well as Gln259. The side chain of Arg9 interacted with Asp140 in the S1′ subsite. Trp6 and Thr7 interacted with residues Ala334, Trp335, Glu389, and His491, indicating its stable presence in S1. The non-camel LVV-hemorphin-7 also interacted with Tyr24, Ser61, Ser260, Tyr338, Thr358, Arg500, and Tyr501 during the simulations. The average percentage of equilibrated simulation time during which the non-camel LVV-hemorphin-7 interacted with ACE residues is given in Fig. 8A. Contributions from individual simulations are provided in Supplementary Fig. 6A.
Figure 7.
RMSD and RMSF plots of triplicate 100 ns simulations of ACE. Data from the three runs are plotted with red, blue and green lines. (A) RMSD of protein Cα atoms from the ACE-LVVYPWTQRF simulations. (B) RMSF of protein Cα atoms from the ACE-LVVYPWTQRF simulations. (C) RMSD of protein Cα atoms from the ACE-LVVYPWTRRF simulations. (D) RMSF of protein Cα atoms from the ACE-LVVYPWTRRF simulations. (E) Density functions corresponding to the distribution of RMSD values from triplicate hemorphin-bound simulations.
Figure 8.
Average percentage of equilibrium simulation time during which ACE residues maintain contact with non-camel and camel LVV-hemorphin-7 from three 100 ns simulations. For equilibrium simulation data, the first 30 ns of each of the three simulations were discarded. Histograms representing the interaction from each of the 3 simulations can be found in Supplementary Fig. 6. Charged, hydrophobic and polar amino acids are represented with orange, green and blue color respectively. (A) Average percentage of time an ACE residue maintains contact with LVVYPWTQRF. (B) Average percentage of time an ACE residue maintains contact with LVVYPWTRRF.
In the camel LVV-hemorphin-7 bound simulations, the protein structure showed structural stability throughout the simulations and the RMSD plot indicated that the structures stabilized under 2.5 Å after the initial few nanoseconds of the simulation (Fig. 7C). Residues of the protein showed lower fluctuations (Fig. 7D). The secondary structure composition remained intact throughout the course of the simulations. A distribution of the RMSD values obtained from the camel and non-camel hemorphin bound simulations are shown in Fig. 7E. Camel LVV-hemorphin-7 remained stably bound by maintaining hydrogen bonds between the arginine residues (Arg8 and Arg9) located near the C-terminal and Asp140, His331, and Tyr369. The side chain of Arg9 interacted with Asp140 in the S1′ pocket for the full duration of all three simulations. The substituted Arg8 maintained the hydrogen bond with His331, Tyr369, and Glu389, and also maintained the hydrophobic interaction with Tyr501 in the S1 pocket. Additionally, the peptide also interacted with the HEXXH motif, specifically His361 and Glu362, during the simulations. The average percentage of equilibrated simulation time during which the non-camel LVV-hemorphin-7 interacted with ACE residues is given in Fig. 8B. Contributions from individual simulations are provided in Supplementary Fig. 6B.
Simulations of camel and non-camel LVV-hemorphin-7 bound to IRAP
The stability of the bound peptide was studied using 100 ns molecular dynamics simulation runs. In non-camel LVV-hemorphin-7 bound simulations, the IRAP structures stabilized with RMSD under 2.5 Å (Fig. 9A). There were limited fluctuations in the protein residues and the secondary structure remained intact throughout these simulations (Fig. 9B). The N-terminal residues of non-camel LVV-hemorphin-7 were stabilized in the S1 pocket with hydrogen bonds as well as hydrophobic interactions. Leu1, Val2 and Val3 maintained hydrogen bonds with Tyr549 and Gly428 during the simulations and also interacted with Phe544. Tyr4, Pro5, and Trp6 maintained their interactions with Glu441, Phe550, and Asn965. While C-terminal residues maintained their interactions with the residues Asp510, Asp513, Glu818, Gu825, and Glu895. During the simulations, new contacts were formed between the first three residues of N-terminal and Trp6 with Arg929, and Asn965. The average percentage of equilibrated simulation time during which the non-camel LVV-hemorphin-7 interacted with IRAP residues is given in Fig. 10A. Contributions from individual simulations are provided in Supplementary Fig. 7A.
Figure 9.
RMSD and RMSF plots of triplicate 100 ns simulations of IRAP. Data from the three runs are plotted with red, blue and green lines. (A) RMSD of protein Cα atoms from the IRAP-LVVYPWTQRF simulations. (B) RMSF of protein Cα atoms from the IRAP-LVVYPWTQRF simulations. (C) RMSD of protein Cα atoms from the IRAP-LVVYPWTRRF simulations. (D) RMSF of protein Cα atoms from the IRAP-LVVYPWTRRF simulations. (E) Density functions corresponding to the distribution of RMSD values from triplicate hemorphin-bound simulations.
Figure 10.
Average percentage of equilibrium simulation time during which IRAP residues maintain contact with non-camel and camel LVV-hemorphin-7 from three 100 ns simulations. For equilibrium simulation data, the first 30 ns of each of the three simulations were discarded. Histograms representing the interaction from each of the 3 simulations can be found in Supplementary Fig. 7. Charged, hydrophobic and polar amino acids are represented with orange, green and blue color respectively. (A) Average percentage of time an IRAP residue maintains contact with LVVYPWTQRF. (B) Average percentage of time an IRAP residue maintains contact with LVVYPWTRRF.
Camel LVV-hemorphin-7 simulations remained stable with an RMSD under 2.75 Å (Fig. 9C). There were limited fluctuations in IRAP residues throughout the camel peptide-bound simulations (Fig. 9D). The secondary structure elements also remained intact throughout the simulations. A distribution of the RMSD values obtained from the camel and non-camel hemorphin bound simulations are shown in Fig. 9E. The N-terminal of camel LVV-hemophin-7 was embedded in the S1 pocket of IRAP by maintaining hydrogen bonds and hydrophobic interactions. Val2 maintained a hydrogen bond with Gly428 in the GAMEN loop during these simulations and the N-terminal residues maintained the hydrophobic interactions with Ala429, Phe544, Tyr549, and Tyr961. Pro5 produced sustained interactions with Lys460, while Trp6 interacted with Asp513. The substituted Arg8 showed strong hydrogen bonds with the negatively charged residues Glu509, Asp510 and Glu818. Arg9 and Phe10 also interacted with Lys517, Phe770, Glu825 and Glu895 stabilizing it in the docked pose. The average percentage of equilibrated simulation time during which the non-camel LVV-hemorphin-7 interacted with IRAP residues is given in Fig. 10B. Contributions from individual simulations are provided in Supplementary Fig. 7B.
Discussion
Several in vivo and in vitro studies have reported the therapeutic potential of hemorphin16,17,26,32. The ability of LVV-hemorphin-7 to bind to several proteins with high affinity emphasizes the importance of understanding the binding mode and activity of this peptide at the molecular level. Computational modeling often complements experimental studies. This study employed computational approaches to investigate the binding mode of both camel and non-camel LVV-hemorphin-7 with MOR, ACE, and IRAP to understand how the peptide interacts with these unrelated proteins.
Camel and non-camel LVV-hemorphin-7 adopted different poses in the MOR. The key residues involved in the binding are consistent with previous experimental and theoretical studies49–51. Both camel and non-camel LVV-hemorphin-7 were found to interact with the acidic residue Asp149 of MOR with a hydrogen bond (Fig. 6A,B). Camel LVV-hemorphin-7 interacted for the entire duration of the three simulations while the interaction was sustained for an average of 90% of the three non-camel LVV-hemorphin-7 simulations. This finding supports various structural models and site-directed mutagenesis studies that highlighted the important role played by Asp149 (Asp147 in murine MOR) in opioid ligand recognition and the interaction with various opioid ligands49,52–55. Therefore, consideration should be given to this vital interaction for future novel design of MOR ligands. Hydrophobic interactions were formed between camel LVV-hemorphin-7 and Tyr328, which is consistent with a previous report that indicated that hemorphin interacts with Tyr326 of mouse MOR49,51,56. The interaction with Trp320 on TM7 is also consistent with previous studies51,57,58. Interestingly, camel LVV-hemorphin-7 interacted with key residues in the binding pocket for a longer duration than non-camel hemorphin (Fig. 6). LVVYPWTRRF interacted for a longer duration than LVVYPWTQRF with Thr220, Glu231, and Tyr328 in the human MOR model. The corresponding residues in the murine MOR – Thr200, Glu299, and Tyr326 – have been shown to interact with agonists49,51,56,58. RMSF values were found to be high for IL-3 residues when bound to camel LVV-hemorphin-7. Major interactions for camel LVV-hemorphin-7 on MOR were observed in TM3, TM6, and TM7 which has been suggested to contain the ionic lock responsible for GPCR activation59. As expected, the peptide-bound simulations led to greater flexibility of the IL-3 region, which is in agreement with downstream protein complex interactions and signaling pathways activated by receptor agonists60. Camel LVV-hemorphin-7 bound simulations had a lower RMSD when compared to non-camel LVV-hemorphin-7 (Fig. 5E). EL-2 showed lower fluctuations upon binding to both camel and non-camel-LVV-hemorphin-7. This may help with binding and peptide selectivity. This is consistent with various experimental studies of mutagenesis which confirmed the role of EL-2 in ligand allosteric and orthosteric activation61–63. Moreover, residues Trp320 and Tyr328 are important in controlling biased MOR signaling and are critical in effector coupling64. Interestingly, camel LVV-hemorphin-7 interacted with both residues, while the non-camel LVV-hemorphn-7 interacted with only Trp320. Thus, it is possible that camel hemorphin could produce their effect on MOR as a result of ligand bias.
Molecular modeling and simulation were used to elucidate the behavior of the binding of LVV-hemorphin-7 to ACE. Both camel and non-camel peptides were found to stably bind in the active site of ACE with the C-terminal bound in specific subsites of the active site and the N-terminal oriented towards the active site lid. Camel LVV-hemorphin-7 bound simulations had a lower RMSD when compared to non-camel LVV-hemorphin-7 (Fig. 7E). The ACE active site has three subsites – S1, S2, and S1′. S1 includes Ala332, Glu362, and Tyr501; S2 includes Gln259, His331, Lys489, His491, and Tyr498, while S1′ contains the residue Glu140 (which is mutated to Asp140 in PDB ID: 2XYD)65. In both ACE non-camel and camel LVV-hemorphin-7 complexes, the C-terminal residues from positions 7 to 10 occupied the S1, S2, and S1′ subsites (Fig. 3). Furthermore, ACE has a Zn2+ ion in its active site that coordinates with His361, His365, and Glu389. These regions are essential for the formation of ACE–inhibitor complex66. In the docked poses of non-camel LVV-hemorphin-7, the C-terminal Arg9 and Phe10 anchored the peptide in the active site through a hydrogen bond interaction with His331 in the S2 subsite. Thr7 and Gln8 bound in the S1 subsite and maintained the polar interaction with His361 and His491 (Fig. 8A). The positioning of camel hemorphin in the ACE active site was different from non-camel hemorphin. Specifically, the C-terminal was positioned differently in the S1′ and S2 pockets (Fig. 3). Arg9 and Phe10 occupied the S2 subsite with the side chain of Phe10 stabilized by interactions with S1 and S2 subsite residues Ala332, His361, and Lys489. The side chain of Arg9 formed a hydrogen bond with Asp140 present in the S1′ subsite for the full duration of all three simulations (Fig. 8B). Additionally, Arg8 showed more interactions with active site residues than Gln8 in non-camel LVV-hemorphin-7 (Figs 3 and 8). Arg8 interacted with His331 in the S2 subsite and with Tyr501 in the S1 subsite, as well as Glu362 and Glu389 potentially displaying the ability to replace the active site Zn2+ ion. Additionally, camel LVV-hemorphin-7 showed more sustained interactions than non-camel LVV-hemorphin-7 with His331, Glu362, Tyr369, and Lys489 throughout the simulation runs which are important for ACE-inhibitor complex formation. This could be the reason for the better binding free energy in the docked pose of camel hemorphin in the ACE active site. Importantly, known inhibitors of ACE interacts with similar residues – His331, Ala332, Glu362, and Tyr501– indicating that these amino acids play a major role in binding to ACE67–69. Here, camel LVV-hemorphin-7 interacted for a longer duration to these residues than non-camel LVV-hemorphin-7, which could support its role as a better ACE inhibitor. Hydrophobic interactions between the peptide and key amino acids in ACE also contributed to the stabilization of the binding pose.
Camel LVV-hemorphin-7 produced similar binding interactions with IRAP when compared to non-camel LVV-hemorphin-7. In the predicted binding pose of non-camel and camel LVV-hemorphin-7, the N-terminal residues were stabilized in the S1 pocket by maintaining hydrogen bonds with Gly428 and Tyr549 (Fig. 10). Tyr549 is highly conserved in the M1 family of metalloproteases and was shown to be important for binding and stabilization of the catalytic transition state70. Furthermore, it also interacted with Phe544 and Tyr961 in the active site. The interaction with Phe544 was previously reported as a key interaction for substrate and ligand binding71,72. IRAP is characterized by a catalytic domain that contains two conserved motifs, the HEXXH Zn2+ binding motif and the GXMEN exopeptidase motif. Mutational analysis of these motifs indicated that Gly428, Ala429, and Asn432 are important for binding of both peptide substrates and inhibitors, and also confirmed that peptide IRAP inhibitors competitively bind to its catalytic site73. In this study, the N-terminal residues of camel LVV-hemorphin-7 interacted with the GAMEN loop residues Gly428 and Ala429, while non-camel LVV-hemorphin-7 interacted with only Gly428 (Fig. 10). Specifically, Val2 of camel LVV-hemorphin-7 interacted with Gly428 and Ala429 (Fig. 10B). Val3 of both non-camel and camel LVV-hemorphin-7 interacted with Tyr549. Val3 is crucial for LVV-hemorphin-7 binding to IRAP; removal of Val3 leads to the abolition of binding to IRAP74. Tyr4 and Trp6 interacted with Glu441 and Phe550. It was reported that monosubstitutions of Tyr4 and Trp6 with alanine resulted in a 10-fold reduction in affinity74. N-terminal residues of hemorphin are crucial for IRAP inhibition forming strong interactions with critical IRAP residues. It was shown that the C-terminal deletion of LVV-hemorphin-7 (Thr7, Gln8, Arg9, and Phe10) did not significantly affect their affinity for IRAP. In fact, a modest decrease in affinity was reported with deletion of Arg9 residue in cerebellar membranes74. In the simulations, LVVYPWTQRRF residues 8 and 9 were observed to form sustained interactions with charged Lys456, Glu509, Asp510, Asp513, and Glu818 when compared to LVVYPWTQRF bound simulations (Fig. 10A,B). These interactions could result in a better affinity of camel LVV-hemorphin-7 in the IRAP catalytic pocket.
Materials and Methods
Multiple sequence alignment of hemoglobin beta protein
Hemoglobin beta protein sequences of Homo sapiens (NP_000509), Camelus dromedarius (XP_010988890), Equus caballus (XP_001504239), Bos taurus (NP_776342), Sus scrofa (NP_001138313), Ovis aries (NP_001091117), Pan troglodytes (NC_036890) and Oryctolagus cuniculus (NP_001075729) were retrieved from NCBI protein database. Multiple sequence alignment of these sequences was performed using ClustalW75.
Protein structure identification and preparation
To dock the peptides to the human MOR, a homology model was generated using Schrödinger Prime76. A Protein Data Bank (PDB) BLAST search was performed to identify homologous protein structures. The three-dimensional homology model was generated for the active human MOR (UniProt accession: P35372) using the X-ray crystallographic structure of the active MOR from Mus musculus (PDB ID: 5C1M) as the template. Loops were refined and the structure was verified using the protein refinement module of Schrödinger Prime. The three-dimensional structure of ACE (PDB ID: 2XYD) and IRAP (PDB ID: 5MJ6) were downloaded from the PDB77. The downloaded structures were prepared using the Protein Preparation Wizard of Schrödinger Suite78. Protein preparation was performed to remove unwanted water molecules, metals, and cofactors. The procedure simplified multimeric complexes, created disulfide bonds, assigned bond orders properly, adjusted ionization states, and fixed the orientation of misoriented groups. Hydrogen atoms were added to the protein structures, and standard protonation states at pH 7 were used. The preprocessed structures were then optimized and minimized to generate geometrically stable structures79.
Active site identification and grid generation
Receptor grids were generated for the prepared protein structures to assist with the docking. Receptor grids were generated with the default parameters for van der Waals scaling factor (1.00) and charge cutoff (0.25) employing the OPLS 2001 force field. A cubic search box was defined centered on the centroid of the binding site residues for each receptor.
Glide standard precision docking
Schrödinger Glide’s Standard Precision (SP) method was used to dock peptides with default parameters. SP flexible ligand docking was carried out using Schrödinger Glide version 2016-4 with penalties applied for cis amide bonds80. Peptides were docked starting from multiple random conformations, which were generated by the ConfGen algorithm81. The poses were partially optimized using the standard OPLS 2005 molecular mechanics force field (SP minimization). Ten representative peptide conformations were selected after clustering the conformers generated. Finally, all ten poses were subjected to post-docking minimization in the gridded protein field. Top poses were rescored and ranked by the GlideScore (GScore) scoring function82. The best-docked pose with lowest GScore value was recorded for each peptide.
Binding free energy calculation
The SP docking poses were subjected to molecular mechanics generalized Born surface area (MM-GBSA) to evaluate the binding free energy in an implicit solvent model. MM-GBSA binding energy was calculated using Schrödinger Prime employing the OPLS 2005 force field and the VSGB 2.0 implicit solvent model76,83. For the MM-GBSA calculations, the receptor was treated rigidly and the peptide was minimized.
Molecular dynamics simulation
In order to confirm the stability of the binding pose of the non-camel and camel LVV-hemorphin-7 in the active site of the three proteins, triplicate MD simulations were performed using Desmond employing the OPLS 2005 force field82.
Simulations of MOR
Three systems – peptide-free MOR, MOR with LVVYPWTQRF (non-camel hemorphin), and MOR with LVVYPWTRRF (camel hemorphin) – were prepared. Three runs each of the peptide bound simulations were performed with different initial velocities. The modeled structure of MOR and the complexes of MOR with non-camel and camel LVV-hemorphin-7 were embedded into a pre-equilibrated DPPC membrane in an orthorhombic box84. All systems were solvated with a water box, using SPC water model85, with a buffer distance of 10 Å. Counterions were added and sufficient number of ions were added to maintain a salt concentration of 0.15 M NaCl. The systems were subjected to steepest descent minimization with Desmond’s default protocol prior to performing MD simulations.
All three systems were first relaxed using the default relaxation protocol for membrane proteins86. The relaxation protocol consists of eight stages that included minimization with restraints on solute heavy atoms, minimization without any restraints, simulation with heating from 0 K to 300 K, H2O barrier and gradual restraining, simulation under NPT equilibration with H2O barrier with heavy atoms restrained, NPT equilibration of solvent and lipids, simulation under the NPT ensemble with protein heavy atoms restraint reduced from 10.0 to 2.0 kcal/mol, NPT equilibration with Cα atoms restrained at 2 kcal/mol, and simulation for 1.5 ns under the NPT ensemble with no restraints. After relaxation, unrestrained simulation run was performed for 100 ns for each system.
The simulations were performed under NPT ensemble using the Nose-Hoover thermostat to maintain a constant temperature of 300 K and isotropic Martyna-Tobias-Klein barostat to maintain the pressure at 1 atm87,88. The short-range Coulombic interactions were analyzed with a cut-off value of 9.0 Å using the short-range method. A time-reversible reference system propagator algorithm (RESPA) integrator was used with a time step of 2.0 fs89. The trajectories were saved at 100 ps intervals for analysis. After simulations were performed, RMSD, RMSF, and protein-ligand contacts were evaluated.
Simulation of ACE and IRAP
The systems were solvated with SPC solvent model in an orthorhombic box to minimize the volume. Simulation systems were neutralized by adding sufficient number of Na+/Cl− counterion and a salt concentration of 0.15 M NaCl was maintained. The systems were then minimized with Desmond’s default protocol before performing MD simulations. Three 100 ns simulations were performed for each system with different initial velocities. The simulations were performed under NPT ensemble using the Nose-Hoover thermostat to maintain a constant temperature of 300 K and isotropic Martyna-Tobias-Klein barostat to maintain the pressure at 1 atm87,88. The short-range coulombic interactions were analyzed with a cut-off value of 9.0 Å using the short-range method. After simulations were performed RMSD, RMSF, and protein-ligand contacts were evaluated.
Conclusion
This study provides the first structural insight into the binding of LVV-hemorphin 7 with MOR, ACE and IRAP. Interestingly, among mammals, camel LVV-hemorphin-7 is unique due to an amino acid substitution. Results showed that camel LVV-hemorphin-7 produced more sustained interactions with all three proteins – MOR, ACE and IRAP – than non-camel LVV-hemorphin-7. Substitution of glutamine with an arginine in camel hemorphin seems to produce a more stable interaction in the proteins studied. Further studies could investigate the effect of camel LVV-hemorphin-7 at the cellular and molecular level for its potential as a therapeutic agent for memory loss, hypertension, and analgesia. Finally, it would also be interesting to extend the study to other proteins involved in the regulation of RAS homeostasis.
Supplementary information
Acknowledgements
This study was funded by a UPAR grant (31S243) from United Arab Emirates University to S.S.S. and R.V. The funder had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Author Contributions
A.A., B.B., S.S.S. and R.V. designed the experiments. A.A. and B.B. performed the experiments and analyzed the data. A.A., B.B., S.S.S. and R.V. wrote and reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Amanat Ali and Bincy Baby contributed equally.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-50619-w.
References
- 1.Brantl V, et al. Novel opioid peptides derived from hemoglobin: hemorphins. Eur. J. Pharmacol. 1986;125:309–310. doi: 10.1016/0014-2999(86)90044-0. [DOI] [PubMed] [Google Scholar]
- 2.Ivanov VT, Karelin AA, Philippova MM, Nazimov IV, Pletnev VZ. Hemoglobin as a source of endogenous bioactive peptides: The concept of tissue‐specific peptide pool. Pept. Sci. 1997;43:171–188. doi: 10.1002/(SICI)1097-0282(1997)43:2<171::AID-BIP10>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 3.Nyberg F, Sanderson K, Glamsta EL. The hemorphins: a new class of opioid peptides derived from the blood protein hemoglobin. Biopolymers. 1997;43:147–156. doi: 10.1002/(SICI)1097-0282(1997)43:2<147::AID-BIP8>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 4.Fruitier, I., Garreau, I. & Piot, J.-M. Cathepsin D Is a Good Candidate for the Specific Release of a Stable Hemorphin from Hemoglobin In Vivo: VV-Hemorphin-7. Biochem. Biophys. Res. Commun. 246, 719–724 (1998). [DOI] [PubMed]
- 5.Glamsta EL, Meyerson B, Silberring J, Terenius L, Nyberg F. Isolation of a hemoglobin-derived opioid peptide from cerebrospinal fluid of patients with cerebrovascular bleedings. Biochem. Biophys. Res. Commun. 1992;184:1060–1066. doi: 10.1016/0006-291X(92)90699-L. [DOI] [PubMed] [Google Scholar]
- 6.Dagouassat N, Garreau I, Zhao Q, Sannier F, Piot J. Kinetic of in vitro generation of some hemorphins: early release of LVV-hemorphin-7, precursor of VV-hemorphin-7. Neuropeptides. 1996;30:1–5. doi: 10.1016/S0143-4179(96)90047-5. [DOI] [PubMed] [Google Scholar]
- 7.Song CZ, Wang QW, Liu H, Song CC. Inhibition of intraerythrocytic proteasome retards the generation of hemorphins. Peptides. 2012;33:170–173. doi: 10.1016/j.peptides.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 8.Dagouassat N, Garreau I, Sannier F, Zhao Q, Piot JM. Generation of VV-hemorphin-7 from globin by peritoneal macrophages. FEBS Lett. 1996;382:37–42. doi: 10.1016/0014-5793(96)00144-5. [DOI] [PubMed] [Google Scholar]
- 9.Moisan S, et al. Structural requirements and mechanism of the pressor activity of Leu-Val-Val-hemorphin-7, a fragment of hemoglobin beta-chain in rats. Peptides. 1998;19:119–131. doi: 10.1016/S0196-9781(97)00273-8. [DOI] [PubMed] [Google Scholar]
- 10.Piot JM, Zhao Q, Guillochon D, Ricart G, Thomas D. Isolation and characterization of two opioid peptides from a bovine hemoglobin peptic hydrolysate. Biochem. Biophys. Res. Commun. 1992;189:101–110. doi: 10.1016/0006-291X(92)91531-T. [DOI] [PubMed] [Google Scholar]
- 11.Erchegyi J, Kastin AJ, Zadina JE, Qiu XD. Isolation of a heptapeptide Val Val Tyr Pro Trp Thr Gln (valorphin) with some opiate activity. Int. J. Pept. Protein Res. 1992;39:477–484. doi: 10.1111/j.1399-3011.1992.tb00277.x. [DOI] [PubMed] [Google Scholar]
- 12.Glamsta EL, Morkrid L, Lantz I, Nyberg F. Concomitant increase in blood plasma levels of immunoreactive hemorphin-7 and beta-endorphin following long distance running. Regul. Pept. 1993;49:9–18. doi: 10.1016/0167-0115(93)90378-L. [DOI] [PubMed] [Google Scholar]
- 13.Karelin AA, Philippova MM, Karelina EV, Ivanov VT. Isolation of endogenous hemorphin-related hemoglobin fragments from bovine brain. Biochem. Biophys. Res. Commun. 1994;202:410–415. doi: 10.1006/bbrc.1994.1943. [DOI] [PubMed] [Google Scholar]
- 14.Nishimura K, Hazato T. Isolation and identification of an endogenous inhibitor of enkephalin-degrading enzymes from bovine spinal cord. Biochem. Biophys. Res. Commun. 1993;194:713–719. doi: 10.1006/bbrc.1993.1880. [DOI] [PubMed] [Google Scholar]
- 15.Basbaum AI, Gautron M, Jazat F, Mayes M, Guilbaud G. The spectrum of fiber loss in a model of neuropathic pain in the rat: an electron microscopic study. Pain. 1991;47:359–367. doi: 10.1016/0304-3959(91)90229-Q. [DOI] [PubMed] [Google Scholar]
- 16.Cheng BC, Tao PL, Cheng YY, Huang EY. LVV-hemorphin 7 and angiotensin IV in correlation with antinociception and anti-thermal hyperalgesia in rats. Peptides. 2012;36:9–16. doi: 10.1016/j.peptides.2012.03.019. [DOI] [PubMed] [Google Scholar]
- 17.Lee J, et al. Effect of ICV injection of AT4 receptor ligands, NLE1-angiotensin IV and LVV-hemorphin 7, on spatial learning in rats. Neuroscience. 2004;124:341–349. doi: 10.1016/j.neuroscience.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 18.Sanderson K, Nyberg F, Khalil Z. Modulation of peripheral inflammation by locally administered hemorphin-7. Inflamm. Res. 1998;47:49–55. doi: 10.1007/s000110050266. [DOI] [PubMed] [Google Scholar]
- 19.Moeller I, et al. The globin fragment LVV hemorphin 7 is an endogenous ligand for the AT4 receptor in the brain. J. Neurochem. 1997;68:2530–2537. doi: 10.1046/j.1471-4159.1997.68062530.x. [DOI] [PubMed] [Google Scholar]
- 20.Hughes J, et al. Identification of two related pentapeptides from the brain with potent opiate agonist activity. Nature. 1975;258:577–580. doi: 10.1038/258577a0. [DOI] [PubMed] [Google Scholar]
- 21.Kakidani H, et al. Cloning and sequence analysis of cDNA for porcine β-neo-endorphin/dynorphin precursor. Nature. 1982;298:245. doi: 10.1038/298245a0. [DOI] [PubMed] [Google Scholar]
- 22.Noda M, et al. Cloning and sequence analysis of cDNA for bovine adrenal preproenkephalin. Nature. 1982;295:202–206. doi: 10.1038/295202a0. [DOI] [PubMed] [Google Scholar]
- 23.Stein C, Schäfer M, Machelska H. Attacking pain at its source: new perspectives on opioids. Nat. Med. 2003;9:1003. doi: 10.1038/nm908. [DOI] [PubMed] [Google Scholar]
- 24.Yukhananov R, Glämsta E-L, Nyberg F. Interaction of hemorphins with opioid receptors in the rat vas deferens and guinea-pig ileum. Regul. Pept. 1994;53:S239–S242. doi: 10.1016/0167-0115(94)90329-8. [DOI] [Google Scholar]
- 25.Davis TP, Gillespie TJ, Porreca F. Peptide fragments derived from the beta-chain of hemoglobin (hemorphins) are centrally active in vivo. Peptides. 1989;10:747–751. doi: 10.1016/0196-9781(89)90107-1. [DOI] [PubMed] [Google Scholar]
- 26.Chow L-H, et al. A possible correlation between oxytocin-induced and angiotensin IV-induced anti-hyperalgesia at the spinal level in rats. Peptides. 2013;39:21–28. doi: 10.1016/j.peptides.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 27.Carey RM, Siragy HM. Newly recognized components of the renin-angiotensin system: potential roles in cardiovascular and renal regulation. Endocr. Rev. 2003;24:261–271. doi: 10.1210/er.2003-0001. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen GR. (pro)renin and receptor: an update. Clin. Sci. (Lond) 2011;120:169–178. doi: 10.1042/CS20100432. [DOI] [PubMed] [Google Scholar]
- 29.Santos RA, Ferreira AJ, Verano-Braga T, Bader M. Angiotensin-converting enzyme 2, angiotensin-(1–7) and Mas: new players of the renin–angiotensin system. J. Endocrinol. 2013;216:R1–R17. doi: 10.1530/JOE-12-0341. [DOI] [PubMed] [Google Scholar]
- 30.Cejka J, Zelezna B, Velek J, Zicha J, Kunes J. LVV-hemorphin-7 lowers blood pressure in spontaneously hypertensive rats: radiotelemetry study. Physiol. Res. 2004;53:603–607. [PubMed] [Google Scholar]
- 31.Ianzer D, et al. Hemorphin and hemorphin-like peptides isolated from dog pancreas and sheep brain are able to potentiate bradykinin activity in vivo. Peptides. 2006;27:2957–2966. doi: 10.1016/j.peptides.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 32.Fruitier-Arnaudin I, Cohen M, Bordenave S, Sannier F, Piot JM. Comparative effects of angiotensin IV and two hemorphins on angiotensin-converting enzyme activity. Peptides. 2002;23:1465–1470. doi: 10.1016/S0196-9781(02)00083-9. [DOI] [PubMed] [Google Scholar]
- 33.Llorens-Cortes C, Mendelsohn FA. Organisation and functional role of the brain angiotensin system. J. Renin-Angiotensin-Aldosterone Syst. 2002;3:39–48. doi: 10.3317/jraas.2002.029. [DOI] [PubMed] [Google Scholar]
- 34.Wright J, et al. Angiotensin II (3–8)(ANG IV) hippocampal binding: potential role in the facilitation of memory. Brain Res. Bull. 1993;32:497–502. doi: 10.1016/0361-9230(93)90297-O. [DOI] [PubMed] [Google Scholar]
- 35.Albiston AL, et al. Evidence that the angiotensin IV (AT4) receptor is the enzyme insulin-regulated aminopeptidase. J. Biol. Chem. 2001;276:48623–48626. doi: 10.1074/jbc.C100512200. [DOI] [PubMed] [Google Scholar]
- 36.Chai SY, et al. The angiotensin IV/AT4 receptor. Cell Mol. Life Sci. 2004;61:2728–2737. doi: 10.1007/s00018-004-4246-1. [DOI] [PubMed] [Google Scholar]
- 37.Moeller I, Albiston AL, Lew RA, Mendelsohn FA, Chai SY. A globin fragment, LVV-hemorphin-7, induces [3H]thymidine incorporation in a neuronal cell line via the AT4 receptor. J. Neurochem. 1999;73:301–308. doi: 10.1046/j.1471-4159.1999.0730301.x. [DOI] [PubMed] [Google Scholar]
- 38.Albiston AL, et al. Attenuation of scopolamine-induced learning deficits by LVV-hemorphin-7 in rats in the passive avoidance and water maze paradigms. Behav. Brain Res. 2004;154:239–243. doi: 10.1016/j.bbr.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 39.da Cruz KR, et al. The hemoglobin derived peptide LVV-hemorphin-7 evokes behavioral effects mediated by oxytocin receptors. Neuropeptides. 2017;66:59–68. doi: 10.1016/j.npep.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 40.Engelmann M, Wotjak T, Neumann C, Ludwig I, Landgraf M. R. Behavioral consequences of intracerebral vasopressin and oxytocin: focus on learning and memory. Neurosci. Biobehav. Rev. 1996;20:341–358. doi: 10.1016/0149-7634(95)00059-3. [DOI] [PubMed] [Google Scholar]
- 41.De Bundel D, et al. Angiotensin IV and LVV-haemorphin 7 enhance spatial working memory in rats: effects on hippocampal glucose levels and blood flow. Neurobiol Learn Mem. 2009;92:19–26. doi: 10.1016/j.nlm.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 42.Shan Y, et al. How does a drug molecule find its target binding site? J. Am. Chem. Soc. 2011;133:9181–9183. doi: 10.1021/ja202726y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wichapong K, et al. Structure-Based Design of Peptidic Inhibitors of the Interaction between CC Chemokine Ligand 5 (CCL5) and Human Neutrophil Peptides 1 (HNP1) J. Med. Chem. 2016;59:4289–4301. doi: 10.1021/acs.jmedchem.5b01952. [DOI] [PubMed] [Google Scholar]
- 44.Buch I, Giorgino T, De Fabritiis G. Complete reconstruction of an enzyme-inhibitor binding process by molecular dynamics simulations. Proc. Natl. Acad. Sci. 2011;108:10184–10189. doi: 10.1073/pnas.1103547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Decherchi S, Berteotti A, Bottegoni G, Rocchia W, Cavalli A. The ligand binding mechanism to purine nucleoside phosphorylase elucidated via molecular dynamics and machine learning. Nat. Commun. 2015;6:6155. doi: 10.1038/ncomms7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang J, Morin P, Wang W, Kollman PA. Use of MM-PBSA in reproducing the binding free energies to HIV-1 RT of TIBO derivatives and predicting the binding mode to HIV-1 RT of efavirenz by docking and MM-PBSA. J. Am. Chem. Soc. 2001;123:5221–5230. doi: 10.1021/ja003834q. [DOI] [PubMed] [Google Scholar]
- 47.Wichapong K, et al. Application of docking and QM/MM-GBSA rescoring to screen for novel Myt1 kinase inhibitors. J. Chem. Inf. Model. 2014;54:881–893. doi: 10.1021/ci4007326. [DOI] [PubMed] [Google Scholar]
- 48.Desmond Molecular Dynamics System, D. E. Shaw Research, New York, NY, 2016.
- 49.Cui X, Yeliseev A, Liu R. Ligand interaction, binding site and G protein activation of the mu-opioid receptor. Eur. J. Pharmacol. 2013;702:309–315. doi: 10.1016/j.ejphar.2013.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mansour A, et al. Key Residues Defining the μ‐Opioid Receptor Binding Pocket: A Site‐Directed Mutagenesis Study. Journal of neurochemistry. 1997;68:344–353. doi: 10.1046/j.1471-4159.1997.68010344.x. [DOI] [PubMed] [Google Scholar]
- 51.Serohijos AW, et al. Structural basis for μ-opioid receptor binding and activation. Structure. 2011;19:1683–1690. doi: 10.1016/j.str.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li JG, et al. ASP147 in the third transmembrane helix of the rat mu-opioid receptor forms ion-pairing with morphine and naltrexone. Life Sci. 1999;65:175–185. doi: 10.1016/S0024-3205(99)00234-9. [DOI] [PubMed] [Google Scholar]
- 53.Spivak CE, et al. Naloxone activation of μ-opioid receptors mutated at a histidine residue lining the opioid binding cavity. Mol. Pharmacol. 1997;52:983–992. doi: 10.1124/mol.52.6.983. [DOI] [PubMed] [Google Scholar]
- 54.Tang Y, et al. Molecular modeling of mu-opioid receptor and its interaction with ohmefentanyl. Zhongguo Yao Li Xue Bao. 1996;17:156–160. [PubMed] [Google Scholar]
- 55.Xu H, et al. Opioid peptide receptor studies, 11: involvement of Tyr148, Trp318 and His319 of the rat mu-opioid receptor in binding of mu-selective ligands. Synapse. 1999;32:23–28. doi: 10.1002/(SICI)1098-2396(199904)32:1<23::AID-SYN3>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 56.Gentilucci L, Tolomelli A, De Marco R, Artali R. Molecular docking of opiates and opioid peptides, a tool for the design of selective agonists and antagonists, and for the investigation of atypical ligand-receptor interactions. Curr. Med. Chem. 2012;19:1587–1601. doi: 10.2174/092986712799945030. [DOI] [PubMed] [Google Scholar]
- 57.Eguchi M. Recent advances in selective opioid receptor agonists and antagonists. Med. Res. Rev. 2004;24:182–212. doi: 10.1002/med.10059. [DOI] [PubMed] [Google Scholar]
- 58.Noori HR, Mucksch C, Urbassek HM. A structural feature of the non-peptide ligand interactions with mice mu-opioid receptors. Curr. Comput. Aided Drug Des. 2014;10:354–360. doi: 10.2174/1573409910666141031093504. [DOI] [PubMed] [Google Scholar]
- 59.Huang W, et al. Structural insights into µ-opioid receptor activation. Nature. 2015;524:315. doi: 10.1038/nature14886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waldhoer M, Bartlett SE, Whistler JL. Opioid receptors. Annu. Rev. Biochem. 2004;73:953–990. doi: 10.1146/annurev.biochem.73.011303.073940. [DOI] [PubMed] [Google Scholar]
- 61.Wheatley M, et al. Lifting the lid on GPCRs: the role of extracellular loops. Br. J. Pharmacol. 2012;165:1688–1703. doi: 10.1111/j.1476-5381.2011.01629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Venkatakrishnan AJ, et al. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 63.Kmiecik S, Jamroz M, Kolinski M. Structure prediction of the second extracellular loop in G-protein-coupled receptors. Biophys. J. 2014;106:2408–2416. doi: 10.1016/j.bpj.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hothersall J, et al. Residues W320 and Y328 within the binding site of the μ-opioid receptor influence opiate ligand bias. Neuropharmacology. 2017;118:46–58. doi: 10.1016/j.neuropharm.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 65.Pina A, Roque A. Studies on the molecular recognition between bioactive peptides and angiotensin‐converting enzyme. J. Mol. Recognit: An Interdisciplinary J. 2009;22:162–168. doi: 10.1002/jmr.905. [DOI] [PubMed] [Google Scholar]
- 66.Abdelhedi O, et al. In silico analysis and molecular docking study of angiotensin I-converting enzyme inhibitory peptides from smooth-hound viscera protein hydrolysates fractionated by ultrafiltration. Food Chem. 2018;239:453–463. doi: 10.1016/j.foodchem.2017.06.112. [DOI] [PubMed] [Google Scholar]
- 67.Jimsheena V, Gowda LR. Arachin derived peptides as selective angiotensin I-converting enzyme (ACE) inhibitors: structure–activity relationship. Peptides. 2010;31:1165–1176. doi: 10.1016/j.peptides.2010.02.022. [DOI] [PubMed] [Google Scholar]
- 68.Mayer M, Meyer B. Mapping the active site of angiotensin-converting enzyme by transferred NOE spectroscopy. J. Med. Chem. 2000;43:2093–2099. doi: 10.1021/jm990194x. [DOI] [PubMed] [Google Scholar]
- 69.Wu Q, Du J, Jia J, Kuang C. Production of ACE inhibitory peptides from sweet sorghum grain protein using alcalase: Hydrolysis kinetic, purification and molecular docking study. Food Chem. 2016;199:140–149. doi: 10.1016/j.foodchem.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 70.Wong AH, Zhou D, Rini JM. The X-ray crystal structure of human aminopeptidase N reveals a novel dimer and the basis for peptide processing. J. Biol. Chem. 2012;287:36804–36813. doi: 10.1074/jbc.M112.398842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Albiston AL, et al. Phenylalanine-544 plays a key role in substrate and inhibitor binding by providing a hydrophobic packing point at the active site of insulin-regulated aminopeptidase. Mol. Pharmacol. Mol. 2010;110:065458. doi: 10.1124/mol.110.065458. [DOI] [PubMed] [Google Scholar]
- 72.Papakyriakou A, et al. Novel selective inhibitors of aminopeptidases that generate antigenic peptides. Bioorganic Med. Chem. Lett. 2013;23:4832–4836. doi: 10.1016/j.bmcl.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 73.Ye S, Chai SY, Lew RA, Albiston AL. Insulin-regulated aminopeptidase: analysis of peptide substrate and inhibitor binding to the catalytic domain. Biol. Chem. 2007;388:399–403. doi: 10.1515/BC.2007.044. [DOI] [PubMed] [Google Scholar]
- 74.Lee J, et al. Structure-activity study of LVV-hemorphin-7: angiotensin AT4 receptor ligand and inhibitor of insulin-regulated aminopeptidase. J. Pharmacol. Exp. Ther. 2003;305:205–211. doi: 10.1124/jpet.102.045492. [DOI] [PubMed] [Google Scholar]
- 75.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schrödinger Release 2016-4: Prime, Schrödinger, LLC, New York, NY, 2016.
- 77.Sussman JL, et al. Protein Data Bank (PDB): database of three-dimensional structural information of biological macromolecules. Acta Crystallogr. D. Biol. Crystallogr. 1998;54:1078–1084. doi: 10.1107/S0907444998009378. [DOI] [PubMed] [Google Scholar]
- 78.Schrödinger Release 2016-4: Protein Preparation Wizard, Schrödinger, LLC, New York, NY, 2016.
- 79.Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 80.Friesner RA, et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 81.Watts KS, et al. ConfGen: a conformational search method for efficient generation of bioactive conformers. J. Chem. Inf. Model. 2010;50:534–546. doi: 10.1021/ci100015j. [DOI] [PubMed] [Google Scholar]
- 82.Friesner RA, et al. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 83.Li J, et al. The VSGB 2.0 model: a next generation energy model for high resolution protein structure modeling. Proteins. 2011;79:2794–2812. doi: 10.1002/prot.23106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Matyszewska D, Bilewicz R. DPPC monolayers as simple models of biological membranes for studies of interactions with perfluorinated compounds. Annales UMCS, Chemistry. 2008;63:201–210. [Google Scholar]
- 85.Mark P, Nilsson L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. 2001;105:9954–9960. doi: 10.1021/jp003020w. [DOI] [Google Scholar]
- 86.Zhang J. LBFGS Quasi-Newtonian Methods for Molecular Modeling Prion AGAAAAGA Amyloid Fibrils. Nat. Sci. 2015;04:1097–1108. [Google Scholar]
- 87.Martyna GJ, Klein ML, Tuckerman M. Nosé–Hoover chains: The canonical ensemble via continuous dynamics. J. Chem. Phys. 1992;97:2635–2643. doi: 10.1063/1.463940. [DOI] [Google Scholar]
- 88.Martyna GJ, Tobias DJ, Klein ML. Constant pressure molecular dynamics algorithms. J. Chem. Phys. 1994;101:4177–4189. doi: 10.1063/1.467468. [DOI] [Google Scholar]
- 89.Tuckerman MBBJM, Berne BJ, Martyna GJ. Reversible multiple time scale molecular dynamics. J. Phys. Chem. 1992;97:1990–2001. doi: 10.1063/1.463137. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.