Abstract
Coxiella burnetii is a Gram-negative bacterium which causes Q fever, a complex and life-threatening infection with both acute and chronic presentations. C. burnetii invades a variety of host cell types and replicates within a unique vacuole derived from the host cell lysosome. In order to understand how C. burnetii survives within this intracellular niche, we have investigated the carbon metabolism of both intracellular and axenically cultivated bacteria. Both bacterial populations were shown to assimilate exogenous [13C]glucose or [13C]glutamate, with concomitant labeling of intermediates in glycolysis and gluconeogenesis, and in the TCA cycle. Significantly, the two populations displayed metabolic pathway profiles reflective of the nutrient availabilities within their propagated environments. Disruption of the C. burnetii glucose transporter, CBU0265, by transposon mutagenesis led to a significant decrease in [13C]glucose utilization but did not abolish glucose usage, suggesting that C. burnetii express additional hexose transporters which may be able to compensate for the loss of CBU0265. This was supported by intracellular infection of human cells and in vivo studies in the insect model showing loss of CBU0265 had no impact on intracellular replication or virulence. Using this mutagenesis and [13C]glucose labeling approach, we identified a second glucose transporter, CBU0347, the disruption of which also showed significant decreases in 13C-label incorporation but did not impact intracellular replication or virulence. Together, these analyses indicate that C. burnetii may use multiple carbon sources in vivo and exhibits greater metabolic flexibility than expected.
Keywords: carbon metabolism, Coxiella burnetii, glucose transport, gram-negative bacteria, intracellular bacterial pathogen
Introduction
To survive and cause disease, intracellular pathogens must obtain energy and essential nutrients from the host cell. Therefore, identifying the metabolic pathways utilized by these pathogens inside cells is key to understanding their pathogenesis [1,2]. The intracellular bacterium, Coxiella burnetii, is the causative agent of Q fever, which in humans causes a number of syndromes ranging from acute life-threatening infection to debilitating chronic disease [3]. Upon inhalation of contaminated aerosols, C. burnetii are typically phagocytosed by alveolar macrophages [4]. The C. burnetii-containing phagosome passively undergoes endocytic maturation and eventually assumes properties of a mature phagolysosome. Acidification of the vacuole leads to the conversion of the pathogen from the environmentally stable and non-replicative small cell variant (SCV) to the metabolically active and replicative large cell variant (LCV) [5,6].
Lysosomal maturation and acidification also triggers C. burnetii to activate the Dot/Icm type IV secretion system, which translocates ∼150 effector proteins into the host cell [7,8]. These effector proteins allow the bacteria to establish a specialized vacuolar compartment, termed the Coxiella-containing vacuole (CCV), in which the bacteria replicate to large numbers [9]. This unique replicative niche is highly fusogenic, with the CCV able to expand and occupy much of the host cell cytoplasm through fusion with endocytic and autophagic vesicles. This process results in the mature CCV resembling an autolysosome [10–12]. Unlike most intracellular pathogens, C. burnetii are not destroyed by the hydrolytic conditions in this niche but require the acidified environment to replicate.
Cultivation conditions for growing C. burnetii outside host cells have recently been developed. A crucial step was the development of an acidified citrate cysteine medium (ACCM) which supports axenic replication of C. burnetii in low oxygen conditions. The constituents of this media were established by studying the metabolic requirements of C. burnetii [13,14]. ACCM, and its derivatives, contains an abundance of amino acids, which are required to satisfy the auxotrophic requirements of this bacterium. Interestingly, recent metabolomic analysis of the lysosome of mammalian cells indicates that this niche may also contain amino acids at levels which are equivalent to or higher than in the cytoplasm [15]. Several lines of evidence suggest that C. burnetii may also utilize amino acids as potential carbon sources. In particular, axenically cultivated bacteria are able to proliferate in defined ACCM lacking glucose (ACCM-D) [16], while genetic disruption of the gene encoding the gluconeogenic enzyme phosphoenolpyruvate carboxykinase (PEPCK) causes a partial attenuation in virulence in animal cells [17]. Intracellular bacteria may therefore be able to switch to using amino acids, both in vitro and in vivo. However, as transcripts for glycolytic and gluconeogenic enzymes are both up-regulated during intracellular replication, it remains unclear whether intracellular bacteria also utilize or co-utilize sugars scavenged from this compartment [18].
A recent study has examined the use of a number of carbon sources, including [13C]glucose, [13C]serine and [13C]glycerol, in AX cultivated C. burnetii [1]. This study confirmed that C. burnetii is able to generate ATP and anabolic precursors via glycolysis. It also indicated that C. burnetii is auxotrophic for histidine, isoleucine, leucine, lysine, phenylalanine, proline and valine, confirming that these amino acids are scavenged from the environment [1]. However, it remains unclear how and to what extent these bacteria catabolize non-essential amino acids. In this study, we have investigated the extent to which AX and intracellularly cultivated (IC) C. burnetii use [13C]glucose and [13C]glutamate. Our results show that both AX and IC C. burnetii are able to utilize glucose and glutamate, which are primarily catabolized via glycolysis and the TCA cycle, respectively. Strikingly, we show that operation of the TCA cycle in each of these stages differ, with use of a continuous, oxidative cycle in AX and discontinuous, partial TCA cycle and increased gluconeogenesis in IC bacteria. We also provide evidence that C. burnetii express two hexose transporters, CBU0265 and CBU0347. Disruption of both transporters individually resulted in partial inhibition of glucose utilization. However, disruption of CBU0265 or CBU0347 had no impact on C. burnetii virulence in the human cell and insect models of infection, indicating that these transporters have redundant functions in vivo or perhaps intracellular bacteria are able to switch to using amino acids as their primary carbon source. These findings suggest that intracellular C. burnetii exhibit an unanticipated flexibility in the use of different carbon sources within the intracellular vacuolar compartment.
Materials and methods
Bacterial strains and tissue culture cells
Escherichia coli DH5α (F− ø80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rk−,mk+) phoA) (Clontech) was grown in Luria–Bertani medium supplemented with 50 µg/ml kanamycin (Sigma–Aldrich) or with 100 µg/ml ampicillin (A. G. Scientific) when required.
C. burnetii Phase II Nine Mile Strain RSA439, referred to as wild type (WT) throughout, 0265::Tn and 0347::Tn mutants, were routinely grown in ACCM-2 as previously described [14]. Media was supplemented with 350 µg/ml kanamycin or 3 µg/ml chloramphenicol (Boehringer Mannheim) when required.
THP-1 human monocytic cells (ATCC) were cultured in RPMI 1640 with GlutaMAX (Gibco) supplemented with 10% fetal calf serum (FCS, Gibco). HeLa human carcinoma cells (CCL2, ATCC) were cultured in Dulbecco's Modified Eagle's Media with GlutaMAX (DMEM, Gibco) supplemented with 10% FCS, or 5% FCS during infection. Both cell lines were maintained at 37°C with 5% CO2.
Cloning and plasmids
gDNA of WT C. burnetii was isolated using the Zymo gDNA extraction kit as per manufacturer's protocol. Oligonucleotides used to amplify ompA, cbu0265 and cbu0347 were obtained from Sigma–Aldrich. ompA primer sequence is as previously described [19]. cbu0265 was amplified from isolated WT C. burnetii gDNA using forward primer 5′-AAAGGATCCATGAAGTTTTCTTTTCC-3′ and reverse primer 5′-AAGCGGCCGCCTAAAAGAAATGAGAATGA-3′. cbu0347 was amplified as above with forward primer 5′-AAGGATCCATGAATTCAACCGACCAA-3′ and reverse primer 5′-AAGCGGCCGCTTATTTTCCTAAATAACG-3′. Restriction endonucleases were obtained from NEB and used according to the manufacturer's protocol. Plasmid DNA was isolated using the QIAprep spin miniprep kit (Qiagen). pJB-Kan:3xFLAG-0265 was transformed into 0265::Tn, and pJB-Kan:3xFLAG-0347 was transformed into 0347::Tn C. burnetii as described previously [10].
Quantification of C. burnetii genome equivalents using RT-PCR
Genome equivalents (GE) of C. burnetii was quantified by qPCR, using primers specific for ompA as previously described [19]. The qPCR was performed using a MX3005P qPCR system (Agilent Technologies). Data output was processed using the MxPro qPCR software and analyzed using Microsoft Excel.
C. burnetii infections of tissue culture cell lines
For labeling experiments, THP-1 cells seeded at 3.5 × 105 cells/ml into T175 flasks (Corning) in 50 ml RPMI + 10% FCS were treated and differentiated with 10 nM phorbol 12-myristate 13-acetate for 3 days. Cells were then infected with WT C. burnetii, which had been quantified using qPCR as described above, at a multiplicity of infection (MOI) of 100. At both day 3 and day 6 post-infection, bacteria were harvested for labeling experiments as described below. Infections for growth curves in HeLa CCL2 and THP-1 cells were performed as previously described [20].
Metabolite labeling, quenching and extraction
For labeling studies, IC C. burnetii were prepared by scraping infected THP-1 cells in warm 1× phosphate-buffered saline (PBS), then lysing to release intracellular bacteria by using a Dounce homogenizer (Sigma–Aldrich). Host cell debris was pelleted by centrifugation (164×g, 10 min, room temperature (RT)) and the supernatant, containing released bacteria, were collected and centrifuged (3220×g, 15 min. RT) to pellet bacteria. AX bacteria were harvested by centrifugation (3220×g, 15 min, RT). Isolated AX and IC bacteria were labeled for 10 min at 37°C 5% CO2 2.5% O2 in 5 ml fresh ACCM-2 supplemented with either 11.11 mM [13C-U] glucose or ACCM-D [16] supplemented with 3 mM [13C-U]glutamate. Cultures were then rapidly quenched to 0°C in a dry ice-ethanol bath to halt metabolic activity.
Bacterial pellets were extracted in chloroform:methanol:water (CHCl3:CH3OH:H2O, 1:3:1 v/v) containing 1 nM scyllo-inositol as internal standard. After initial addition of 300 µl methanol to 99 µl water with 1 µl of internal standard (CH3OH:H2O, 3:1 v/v), bacterial cells were sheared by repeat exposure to liquid nitrogen for 30 s, then dry ice-ethanol bath for 30 s, for a total of 10 times each. 100 µl chloroform was then added to bring the solution to the aforementioned 1:3:1 ratio. Following vortex mixing, samples were incubated on ice for 10 min. Samples were then centrifuged (17 100×g, 15 min, −2°C) to remove cell debris. The supernatant was adjusted to CHCl3:CH3OH:H2O (1:3:3 v/v) by the addition of 200 µl water, before vortex mixing and centrifuging (17 100×g, 15 min, −2°C) to induce phase separation. The upper aqueous phase, containing polar metabolites, was transferred to a fresh precooled 1.5 ml microcentrifuge tube and stored at −80°C until GC/MS analysis. Mock samples using uninfected THP-1 cells were also prepared alongside IC samples to account for contamination by host enzyme activity. THP-1 cells were lysed and cell debris was removed as per IC sample preparation. Supernatants which would normally contain the bacteria were harvested, labeled, quenched and extracted as per AX and IC conditions.
Metabolite derivatization and GC/MS analysis
Aqueous-phase samples were transferred into glass vial inserts and dried completely in a rotational vacuum concentrator (RVC-2-33; John Morris Scientific), with 30 µl of 100% methanol added at the final drying stage. Free aldehyde groups were derivatized in 20 µl methoxyamine chloride (30 mg/ml in pyridine; Sigma–Aldrich) with continuous mixing for 2 h at 37°C. 20 µl of N,O-bistrimethylsilyltrifluoroacetamide (BSTFA) containing 1% trimethylchlorosilane (TMCS; Thermo Scientific) was then added, and samples were incubated for 1 h at 37°C, with continuous shaking, using a Gerstel MPS2 autosampler robot. For GC/MS analysis, 1 µl of the derivatized sample was injected into an Agilent 7890A gas chromatograph (split/splitless inlet, 250°C) containing a VF-5ms column (30 m/250 µm/0.25 µm/10 m Eziguard precolumn) coupled to an Agilent 5975C mass selective detector. Helium was used as the carrier gas at a constant flow rate of 1 ml/min. The GC temperature was ramped from 35°C, at which it was initially held for 2 min, to 325°C, at 25°C/min, and then held for 5 min at 325°C. The 5975C mass selective detector was used in scan mode, and mass spectra data were collected at a rate of 9.19 scans/s over an m/z range of 50 to 600 atomic mass units.
Metabolite identification and calculation of 13C-label incorporation
Labeled metabolites were identified from chromatograms by using the Agilent MSD Productivity Chemstation for GC and GC/MS. Fragmented ion patterns were used to identify each metabolite using NIST, Fiehn, and Wiley libraries, and then retention times were matched to in house libraries to confirm IDs [2].
13C-label incorporation, where a representative unlabeled ion (M0), and mass isotopologues of up to M + 1 of the possible number of labeled carbon atoms were integrated using Agilent MassHunter software. Natural background 13C was subtracted from labeled samples as previously described [21]. The level of label incorporation for each replicate was normalized against the maximum labeling which occurred for glucose or glutamate for the specific replicate, and any labeling occurring in mock, uninfected samples were subtracted for each metabolite in bacterial samples. Replicates were averaged to obtain labeling percentage values. Label incorporation graphs were generated using Visualization and Analysis of Networks conTaining Experimental Data (VANTED) [22]. Student's t-test was performed using Microsoft Excel.
Mapping identified metabolites onto metabolic pathways
Metabolites identified in these studies were mapped onto predicted metabolic pathways using the C. burnetii RSA 493 strain from the KEGG (Kyoto Encyclopedia of Genes and Genomes) database as a guide. Any enzymes missing from the KEGG database annotations were searched for in C. burnetii RSA 439 using BLASTp [23], using Legionella pneumophila, E. coli, or other bacterial sequences and human sequences as references.
Western blot to detect expression of 3xFLAG-0265 and 3xFLAG-0347
Whole-cell lysates of WT, 0265::Tn, 0265::Tn pFLAG-0265, 0347::Tn and 0347::Tn pFLAG-0347 C. burnetii grown to day 6 post-inoculation were prepared for western blot analysis as previously described [24]. Proteins were transferred onto nitrocellulose membranes using the iBlot2 system (Life Technologies). Membranes were blocked in Tris-buffered saline (20 mM Tris 150 mM NaCl pH 8.0) with 0.1% Tween-20 (Thermo Scientific) (TBST) and 5% skim milk for 1 h at RT, before being incubated in primary antibodies for 1 h at RT. Anti-FLAG antibodies (Sigma–Aldrich) were diluted 1 : 2000 in TBST + 5% skim milk, and anti-RpoA antibodies (BioLegend) were diluted 1 : 5000 in TBST + 5% bovine serum albumin (Roche). Membranes were washed three times in TBST before anti-mouse-HRP secondary antibodies (PerkinElmer) diluted in TBST + 5% skim milk was added. Membranes were then incubated for 1 h at RT before being washed as before. Proteins were detected using the Clarity Western ECL reagent (Bio-Rad) and visualized using the Amersham Imager 600 (GE Healthcare) after exposure for 0.5 s.
Galleria mellonella in vivo infection model
G. mellonella larvae were grown in-house and kept at 30°C in the dark until use. Infections were conducted as described previously [25]. Briefly, 106 GE of WT, 0265::Tn, 0265::Tn pFLAG-0265, 0347::Tn, 0347::Tn pFLAG-0347 C. burnetii were injected in the right proleg of G. mellonella larvae. Larvae were incubated at 37°C, and survival was monitored every 24 h across eleven days. PBS controls were included alongside. Each condition consisted of 12 larvae.
Immunofluorescence microscopy
Infected HeLa CCL2 and THP-1 cells were stained at day 3 post-infection as previously described [20], using anti-LAMP-1 (Developmental Studies Hybridoma Bank) and anti-Coxiella antibodies (Roy Laboratory, Yale University).
Results
C. burnetii are able to catabolize [13C]glutamate to generate gluconeogenic intermediates
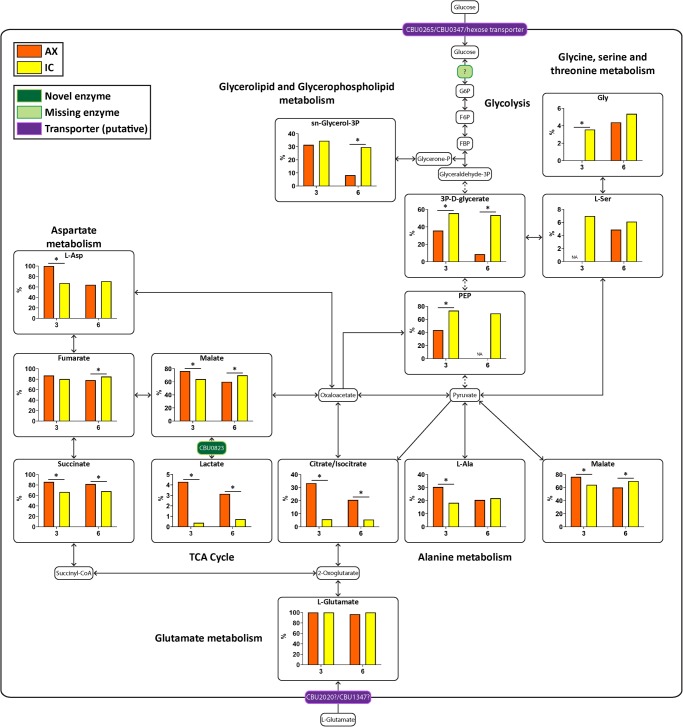
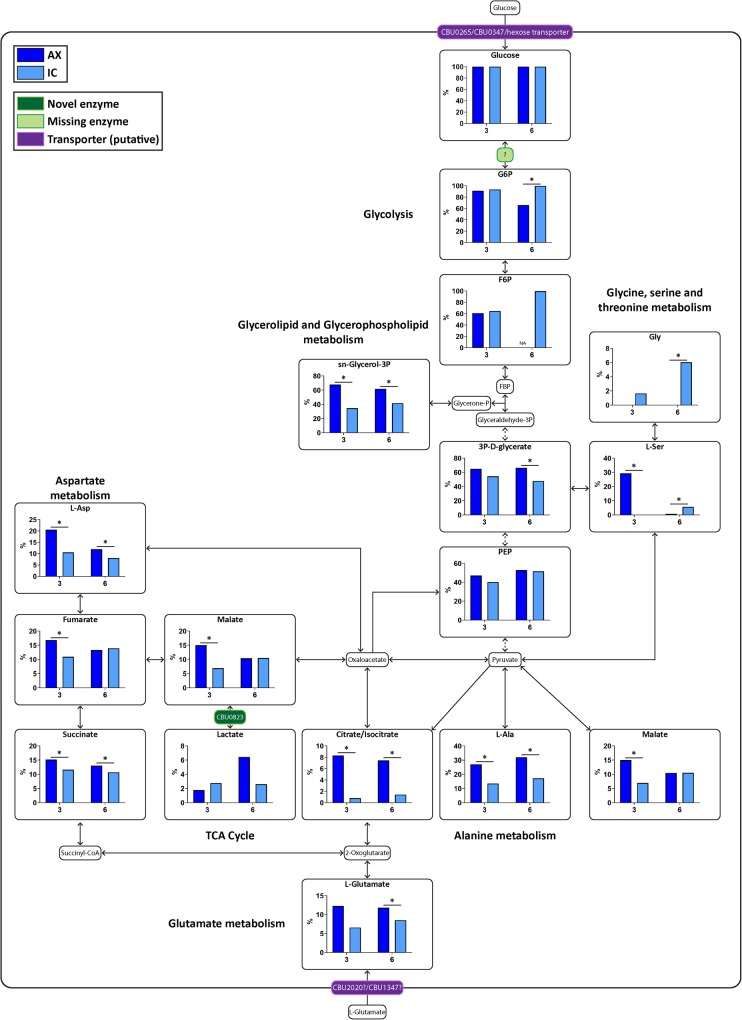
C. burnetii were cultivated AX in ACCM-2 media (axenic; AX) or extracted from macrophage-like THP-1 cells (intracellular; IC) and labeled with either [13C]glucose or [13C]glutamate at day 3 or day 6 post-inoculation/infection. Bacteria were labeled for 10 min in ACCM-2 containing either 11.11 mM [13C]glucose or ACCM-D containing 3 mM [13C]glutamate, rapidly chilled to quench metabolism and polar metabolites extracted and analyzed using gas chromatography/mass spectrometry (GC/MS). A complete metabolic pathway map of C. burnetii was generated, using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database as a guide (Supplementary Figure S1 and Supplementary Table S1). Mock or uninfected THP-1 cell samples were prepared alongside and labeled in the same manner as the IC bacteria, where any detected 13C-label incorporation in these mock samples were subtracted from the bacterial samples (Supplementary Table S2). Levels of 13C-incorporation into intermediates in C. burnetii central carbon metabolism are shown in Figures 1 and 2.
Figure 1. Both AX and IC bacteria are able to catabolize [13C]glutamate.
Metabolic pathway map of C. burnetii comparing 13C-label incorporation between AX and IC C. burnetii after 10 min incubation with [13C]glutamate. Metabolites detected on the GC/MS as having incorporated 13C-label are shown in larger boxes, with percentage labeling in bar graphs. Orange bars indicate AX samples while yellow bars indicate IC samples. Pale green boxes indicate missing enzymes based on genome annotation data in KEGG. Dotted arrows indicate pathways which have been abbreviated. * = P < 0.05. NA indicates metabolites which were unable to be detected in a given condition. Metabolite abbreviations are listed in Supplementary Table S1.
Figure 2. AX bacteria are able to utilize [13C]glucose more readily than IC.
Metabolic pathway map of C. burnetii comparing 13C-label incorporation between AX and IC C. burnetii after 10 min incubation with [13C]glucose. Metabolites detected on the GC/MS as having incorporated 13C-label are shown in larger boxes, with percentage labeling in bar graphs. Dark blue bars indicate AX samples while light blue bars indicate IC samples. Pale green boxes indicate missing enzymes based on genome annotation data in KEGG. Dotted arrows indicate pathways which have been abbreviated. * = P < 0.05. NA indicates metabolites which were unable to be detected in a given condition. Metabolite abbreviations are listed in Supplementary Table S1.
Both AX and IC bacteria utilized exogenous [13C]glutamate, which fully equilibrated with intracellular pools within 10 min of labeling. This was observed as 100% 13C-incorporation of glutamate in the raw labeling percentages from both AX and IC bacterial samples. Internalized [13C]glutamate was rapidly incorporated into TCA cycle intermediates, succinate, fumarate and malate, to similar extents in both populations indicating operation of an oxidative cycle (Figure 1). In AX bacteria, these intermediates are further catabolized to citrate and further rounds of the TCA cycle, as shown by strong labeling of intracellular pools of citrate/isocitrate (Figure 1). Some TCA cycle intermediates were also channeled into gluconeogenesis, as shown by labeling of PEP, 3P-D-glycerate and sn-glycerol-3P. Interestingly, the rate of labeling of latter intermediates decreased in stationary phase cultures, consistent with reduced demand for nucleotide precursors in this stage (Table 1 and Supplementary Figure S2). In marked contrast, IC bacteria appeared to have a highly cataplerotic TCA cycle, channeling most of the C4 dicarboxylic acids into gluconeogenesis, rather than further rounds of oxidation in the TCA cycle. High rates of 13C-incorporation into gluconeogenic intermediates was observed at both day 3 and day 6 in IC bacteria, consistent with these stages requiring ongoing nucleotide biosynthesis as they proliferate within the vacuolar compartment (Figure 1, Table 1 and Supplementary Figure S2). IC bacteria also utilized carbon backbones derived from glutamate for the synthesis of other amino acids, such as serine, glycine and l-aspartate, as well as precursors for phospholipid synthesis (sn-glycerol-3P) (Figure 1 and Supplementary Figure S2). Overall, these data suggest that while both AX and IC bacteria actively utilize glutamate, IC bacteria appear to primarily use exogenous glutamate to make anabolic precursors required for replication, while AX bacteria more efficiently catabolize this carbon source in a cyclical TCA cycle.
Table 1. Metabolites with significant day 3 vs day 6 differences within AX and IC conditions during [13C]glutamate labeling.
| Elevated at day 3 | Elevated at day 6 | |||
|---|---|---|---|---|
| Metabolite | P value | Metabolite | P value | |
| AX | 3P-d-glycerate | 2.4 × 10−3 | ||
| Malate | 3.4 × 10−7 | |||
| Fumarate | 6.4 × 10−3 | |||
| l-Ala | 1.0 × 10−2 | |||
| l-Asp | 2.9 × 10−5 | |||
| sn-Glycerol-3P | 3.2 × 10−7 | |||
| Gly | 3.4 × 10−2 | |||
| IC | Malate | 4.5 × 10−3 | ||
| Fumarate | 2.9 × 10−2 | |||
| sn-Glycerol-3P | 1.4 × 10−2 | |||
Unexpectedly, we detected low, but significant levels of 13C-enrichment in lactate for AX bacteria (Figure 1 and Supplementary Figure S2). The C. burnetii genome lacks an annotated L/D-lactate dehydrogenase, indicating the presence of an alternative mechanism for synthesizing lactate, which does not involve direct conversion from pyruvate. Lactic acid bacteria, such as Oenococcus oeni, express alternative malolactic enzymes which can convert malate to lactate [26]. Bioinformatic analysis revealed the presence of a putative malolactic enzyme encoding gene in C. burnetii. CBU0823 possesses 43% identity with the O. oeni malolactic enzyme across 63% of the predicted amino acid sequence. CBU0823 is currently annotated as a NAD-dependent malic enzyme, but like the O. oeni enzyme, may be multi-functional, converting both malate to pyruvate as well as malate to lactate (Figure 1). Lactate synthesis was barely detected in IC, possibly indicating reduced flux around the TCA cycle in this stage and/or presence of unlabeled lactate in the vacuolar compartment.
C. burnetii are able to catabolize [13C]glucose via glycolysis and the TCA cycle
Reciprocal labeling with [13C]glucose, showed that both AX and IC bacteria co-utilized sugars together with glutamate. Raw percentage labeling data showed intracellular pools of glucose 6-phosphate rapidly equilibrated with exogenous [13C]glucose, indicating high rates of uptake. Catabolism of [13C]glucose has previously been reported in AX bacteria, but not in IC bacteria [1]. It remains unclear how glucose is phosphorylated to form glucose 6-phosphate, given the absence of an annotated hexokinase or a functional phosphotransferase system (PTS) in these bacteria [27–29]. Levels of 13C-enrichment in TCA cycle intermediates was generally much lower in [13C]glucose-fed bacteria compared with [13C]glutamate-fed bacteria, indicating efficient metabolic compartmentalization of these two carbon sources. As in the [13C]glutamate labeling studies, 13C-enrichment in citrate/isocitrate in [13C]glucose labeled IC bacteria was reduced compared with AX bacteria. This suggests one or more enzymes involved in channeling glycolytically derived acetyl-CoA into the TCA cycle is down-regulated or repressed in IC bacteria. Interestingly, the levels of 13C-enrichment in C4 dicarboxylic acid intermediates in [13C]glucose-fed IC bacteria were generally higher than in citrate, suggesting that pyruvate may be converted to malate to undergo anaplerosis. 13C-enrichment was also detected in amino acids synthesized via the TCA cycle, including l-aspartate and l-glutamate, and via glycolysis, including l-alanine, l-serine and glycine.
At day 3, AX bacteria showed significantly higher incorporation of [13C]glucose (Supplementary Figure S3 and Table 2), consistent with the more rapid growth phase at this timepoint compared with the day 6 bacteria that are at stationary phase. In contrast, IC samples showed the opposite phenotype, with more 13C-label incorporation occurring at day 6 post-infection. The label was again incorporated into lactate by both AX and IC bacteria, supporting the notion that malate can also be converted to lactate (Figure 2).
Table 2. Metabolites with significant day 3 vs day 6 differences within AX and IC conditions during [13C]glucose labeling.
| Elevated at day 3 | Elevated at day 6 | |||
|---|---|---|---|---|
| Metabolite | P value | Metabolite | P value | |
| AX | Malate | 2.2 × 10−8 | ||
| Fumarate | 1.0 × 10−5 | |||
| Succinate | 4.2 × 10−2 | |||
| l-Asp | 5.4 × 10−5 | |||
| l-Ser | 4.0 × 10−5 | |||
| Lactate | 1.6 × 10−2 | |||
| IC | F6P | 2.9 × 10−2 | ||
| Malate | 8.4 × 10−3 | |||
| Fumarate | 9.0 × 10−3 | |||
| l-Ala | 4.2 × 10−4 | |||
| l-Ser | 3.9 × 10−6 | |||
| Gly | 1.8 × 10−2 | |||
CBU0265 is a glucose transporter
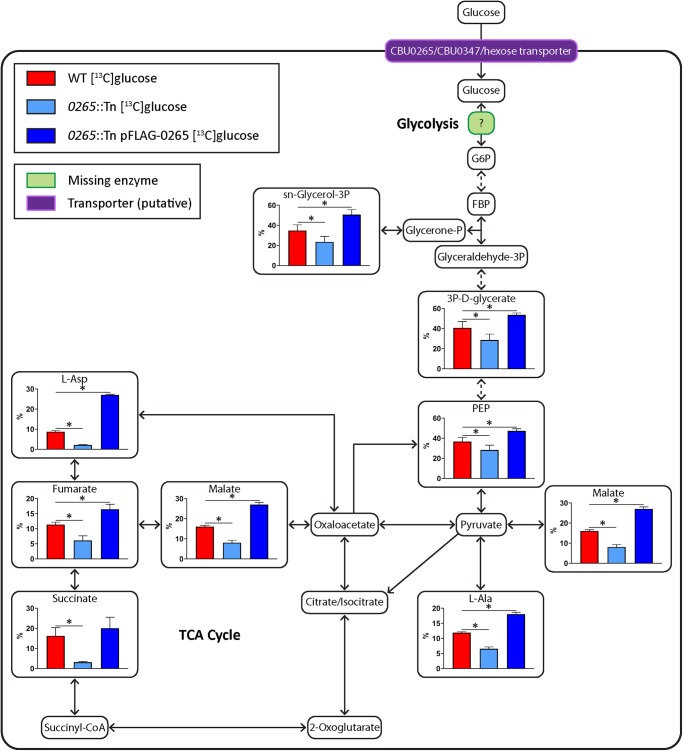
C. burnetii lacks a functional PTS capable of internalizing hexose with concomitant phosphorylation, suggesting that sugars are internalized via an active or facilitative transporter. C. burnetii CBU0265 has previously been identified as a putative proton-driven glucose/galactose transporter [1,16–18,27], and may be responsible for glucose uptake. We have previously generated a C. burnetii transposon mutant in which cbu0265 was disrupted (0265::Tn) (Supplementary Figure S4A) [10,30]. To determine whether CBU0265 is responsible for glucose uptake, and to identify if any other glucose transporters exist, [13C]glucose labeling was performed on WT C. burnetii, the 0265::Tn mutant and the 0265::Tn mutant complemented with constitutively plasmid-expressed 3xFLAG-CBU0265 (0265::Tn pFLAG-0265) at 6 days post-inoculation. Expression of 3xFLAG-0265 in the complemented strain was confirmed via western blot (Supplementary Figure S4B). While the predicted molecular mass of this protein is 47.8 kDa, H+-rich sugar transporters migrate on SDS–PAGE gels at 65–75% of their true molecular mass, and appear instead ∼35 kDa (Supplementary Figure S4B). This has been previously observed for FucP, the E. coli homolog which shares the same sugar transporter domain as CBU0265 [31,32].
All strains exhibited appreciable levels of [13C]glucose utilization and catabolism to downstream intermediates in glycolysis and the TCA cycle when axenic cultures were labeled for 10 min (Figure 3). However, 13C-enrichment in these intermediates was significantly reduced in the 0265::Tn mutant compared with WT or the complemented 0265::Tn strain (Figure 3), supporting the conclusion that CBU0265 is a glucose transporter. These data also strongly suggest that C. burnetii express additional hexose transporters to account for the residual level of [13C]glucose utilization in the 0265::Tn strain.
Figure 3. CBU0265 is required for glucose utilization in C. burnetii.
Metabolites detected on the GC/MS as having incorporated 13C-label from [13C]glucose shown in larger boxes, with percentage labeling depicted in accompanying graphs. WT in red, 0265::Tn in light blue, and 0265::Tn pFLAG-0265 C. burnetii in dark blue. Pale green boxes indicate missing enzymes based on genome annotation data in KEGG. Dotted arrows indicate pathways which have been abbreviated. Error bars indicate standard deviations. * = P < 0.05. Metabolite abbreviations are listed in Supplementary Table S1.
CBU0265 is not required for intracellular replication in human cell lines or virulence in Galleria mellonella
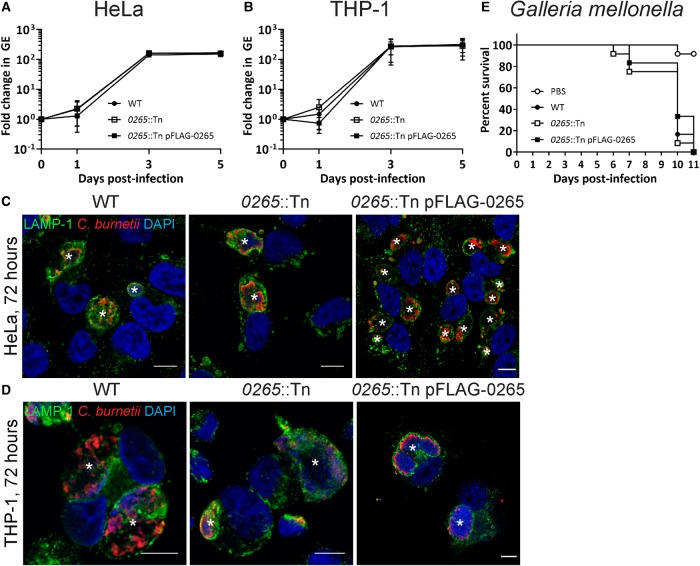
C. burnetii do not require extracellular glucose during axenic cultivation. However previous research suggests reduced glucose availability during intracellular replication can negatively impact C. burnetii growth. This was shown in a C. burnetii strain defective in gluconeogenesis, which replicated less efficiently inside Vero cells grown in glucose-restricted medium compared with WT C. burnetii [17]. To observe whether the loss of glucose uptake via CBU0265 would affect intracellular replication, growth curves were performed in HeLa and THP-1 cells using WT, 0265::Tn and complemented 0265::Tn C. burnetii (Figure 4A–D). No significant differences were observed between all three strains, suggesting that CBU0265 is not required for replication within mammalian cells.
Figure 4. CBU0265 is not required for intracellular replication or virulence.
Growth curves of WT (solid black circle), 0265::Tn (white square box) and 0265::Tn pFLAG-0265 (solid square box) C. burnetii in HeLa CCL2 cells (A) and THP-1 cells (B), n = 3. Error bars represent standard deviation. Immunofluorescence (IF) images at 3 days post-infection for HeLa CCL2 (C) and THP-1 (D) cells. Cells stained with LAMP-1 (green), C. burnetii (red) and DAPI (blue). Scale bar = 10 µm. * indicates CCV. (E) Survival of G. mellonella following infection with WT (solid black circle), 0265::Tn (white square box), 0265::Tn pFLAG-0265 (solid square box) C. burnetii at 106 GE. A PBS control (white circle) was also included. Results are shown as a representative of two replicates, each with 12 larvae per condition.
G. mellonella in vivo infection models are a relatively recent tool for studying C. burnetii virulence [12,25,33]. In this model, infection of G. mellonella larvae with Phase II C. burnetii is lethal. In order to determine whether CBU0265 is required for virulence in C. burnetii during infection, G. mellonella larvae were injected with 106 GE of WT, 0265::Tn, and 0265::Tn pFLAG-0265 C. burnetii, and survival of the larvae were monitored every 24 h for 11 days. All infected larvae died within 11 days, with similar kinetics between the three strains (Figure 4E), demonstrating that CBU0265 is not required for virulence.
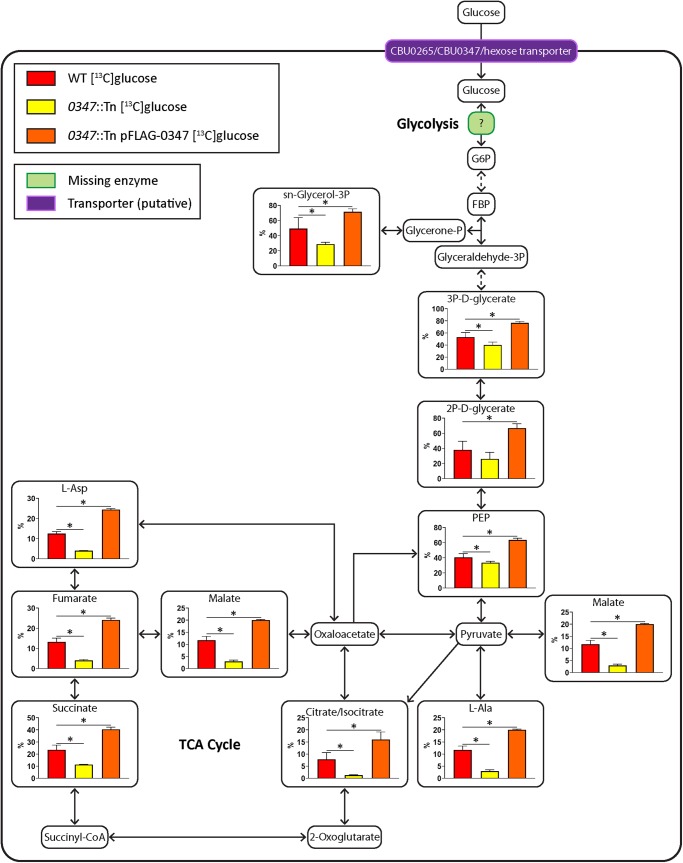
The putative xylose transporter CBU0347 is also responsible for glucose uptake in C. burnetii
The C. burnetii genome encodes a second putative sugar transporter which is currently annotated as a d-xylose-proton symporter. This protein contains a domain which is similar to the major facilitator superfamily domain present in the XylE xylose transporter of E. coli [34,35]. To determine whether CBU0347 is also responsible for glucose uptake, [13C]glucose labeling was performed using a previously generated CBU0347 transposon mutant (Supplementary Figure S4A) [10]. A complemented 0347::Tn mutant with constitutively plasmid-expressed 3xFLAG-CBU0347 (0347::Tn pFLAG-0347) was generated. Expression of 3xFLAG-0347 was confirmed via western blot (Supplementary Figure S4B). Similar to 3xFLAG-0265, this protein also migrated on SDS–PAGE at a smaller molecular mass (∼38 kDa) compared with its true molecular mass of 53.9 kDa (Supplementary Figure S4B), similar to XylE [35,36].
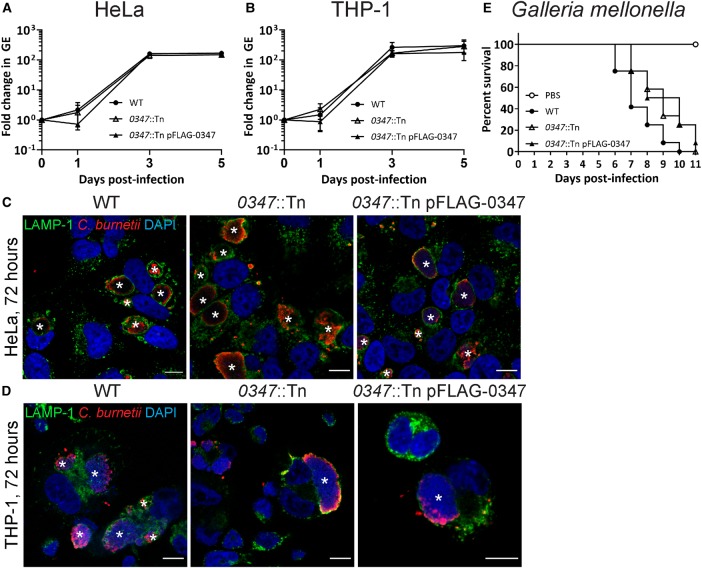
Labeling of WT C. burnetii, the 0347::Tn mutant and 0347::Tn pFLAG-0347 at day 6 post-inoculation, showed significant decreases in 13C-label incorporation for downstream pathways in the 0347::Tn mutant compared with WT (Figure 5). The complemented 0347::Tn strain showed significantly higher 13C-enrichment compared with WT, similarly to the 0265::Tn complement (Figure 5). No significant differences in replication were observed between WT, 0347::Tn and 0347::Tn pFLAG-0347 strains in both HeLa and THP-1 cell models (Figure 6A–D), which correlated with no attenuation in virulence in the G. mellonella model (Figure 6E). This demonstrates that CBU0347 is not required for replication or virulence in C. burnetii. Together, these findings confirm that both CBU0265 and CBU0347 function as hexose transporters in C. burnetii.
Figure 5. CBU0347 is also required for glucose utilization.
Metabolites detected on the GC/MS as having incorporated 13C-label from [13C]glucose shown in larger boxes, with percentage labeling depicted in accompanying graphs. WT in red, 0347::Tn in yellow, and 0347::Tn pFLAG-0347 C. burnetii in orange. Pale green boxes indicate missing enzymes based on genome annotation data in KEGG. Dotted arrows indicate pathways which have been abbreviated. Error bars indicate standard deviations. * = P < 0.05. Metabolite abbreviations are listed in Supplementary Table S1.
Figure 6. CBU0347 is not required for intracellular replication or virulence.
Growth curves of WT (solid black circle), 0347::Tn (white triangle) and 0347::Tn pFLAG-0347 (solid black triangle) C. burnetii in HeLa CCL2 cells (A) and THP-1 cells (B), n = 3. Error bars represent standard deviation. Immunofluorescence (IF) images at 3 days post-infection for HeLa CCL2 (C) and THP-1 (D) cells. Cells stained with LAMP-1 (green), C. burnetii (red) and DAPI (blue). Scale bar = 10 µm. * indicates CCV. (E) Survival of G. mellonella following infection with WT (solid black circle), 0347::Tn (white triangle) and 0347::Tn pFLAG-0347 (solid black triangle) C. burnetii at 106 GE. A PBS control (white circle) was also included. Results are shown as a representative of two replicates, each with 12 larvae per condition.
Discussion
Understanding the nutrient requirements and metabolic pathways required for C. burnetii to survive and replicate inside the host cell will assist in elucidating how the bacteria proliferate inside a normally hostile intracellular compartment. While a number of recent studies have investigated the metabolism of AX cultivated C. burnetii [1,16,37], little is known about the metabolic potential of intracellular bacteria [17]. In this study we show that both AX and IC bacteria utilize glutamate and glucose, and effectively compartmentalize the extent to which each carbon source is channeled into different arms of central carbon metabolism. We also identified significant differences in the extent to which both carbon sources are catabolized in the TCA cycle, which indicate substantial remodeling of bacterial carbon metabolism within the intracellular niche.
Current technological limitations mean in situ labeling of intracellular bacteria with 13C-labeled substrates is not possible. If labelling were to be performed during infection, it would be difficult to discern whether the incorporation of the 13C-label into detected metabolites occurred through a host or bacterial enzyme activity. Therefore, the methodology used in this study is the closest indicator to C. burnetii metabolism in vivo. While it is difficult to account for any rapid metabolic changes due to the altered environment, our data demonstrates significant differences between the IC and AX bacteria.
Recent studies suggest that the lysosomes of mammalian cells contain elevated levels of many amino acids, including glutamate, generated by proteolysis of luminal proteins and/or reverse transport from the cytosol [15]. The C. burnetii generated CCV shares many properties in common with mature lysosomes, including high fusogenicity with endosomal and autophagosome compartments, and are likely to have similar luminal nutrient levels. In this context, the utilization of glutamate by IC bacteria may represent an important adaptation to life within this niche. Based on our labeling studies, we propose that IC bacteria primarily catabolize glutamate in a discontinuous TCA cycle, involving the conversion of α-ketoglutarate to malate/oxaloacetate with production of reducing equivalents such as NADH, as well as carbon backbones for gluconeogenesis. Gluconeogenesis appears to be important in vivo, as disruption of the C. burnetii gluconeogenic enzyme PEPCK leads to attenuated virulence in animal cells [17]. Operation of a discontinuous TCA cycle may also prevent production of excess reducing equivalents and leakage of reactive oxygen species (ROS) from the respiratory chain, and thereby minimize oxidative stress in the CCV compartment. Whether TCA cycle fluxes in IC bacteria are regulated through the transcriptional regulation of key enzymes involved in citrate synthesis and/or metabolic controls remain to be determined. For instance, through increased conversion of malate/oxaloacetate to PEP and gluconeogenesis. The higher 13C-label incorporation into gluconeogenic intermediates by the IC bacteria in this study also supports preferential usage of amino acids by bacteria conditioned to the intracellular environment, suggesting that these bacteria are accustomed to generating sugars and sugar phosphates by catabolizing amino acids rather than via glycolysis. Furthermore, AX bacteria, which are cultured in ACCM-2 medium which contains a higher abundance of glucose than that present in a lysosome, more readily utilized glucose, with higher incorporation of 13C-label within the TCA cycle, particularly at day 3 when bacteria have been rapidly dividing in the late logarithmic growth phase. At day 6 however, the IC bacteria incorporated more 13C-label than day 3, particularly for [13C]glucose labeled samples. At day 6 post-infection, IC samples are likely to contain a mixture of stationary phase bacteria alongside bacteria which have newly infected cells. While it would be difficult to remove host turnover as a factor regarding these samples, further investigations are warranted to determine the cause of this apparent increase in metabolic activity in this population of bacteria.
Our labeling studies have also provided new insights into de novo amino acid biosynthesis in these bacteria. For example, the efficient labeling of intracellular pools of aspartate with both [13C]glutamate and [13C]glucose suggests that the interconversion of oxaloacetate and aspartate through transamination reactions may have an important role in regulating carbon–nitrogen metabolism. Aspartate is also the precursor for essential metabolites, such as NAD. C. burnetii express the enzyme aspartate oxidase (NadB, CBU0101), which plays an important role in de novo NAD synthesis and which may be important for intracellular replication [38].
Unexpectedly, we observed that [13C]glutamate and glucose were catabolized to lactate in AX and IC bacteria [1]. C. burnetii lacks an annotated NAD+-dependent lactate dehydrogenase (LDH), although they express a NAD+-dependent malate dehydrogenase (MDH), which in some other organisms has been postulated to also catalyze the reversible interconversion of pyruvate and lactate. However, there is no strong evidence that dual function MDHs exist in bacteria [39]. An alternative possibility is that C. burnetii express a malolactic enzyme which converts malate to lactate, as occurs in lactic acid bacteria. The bioinformatic analysis demonstrated that C. burnetii have a malolactic enzyme homolog (CBU0823). Utilization of lactate as a sole carbon source was recently demonstrated for Mycobacterium tuberculosis, an intracellular pathogen which also infects alveolar macrophages [40]. Elucidating the molecular details of this pathway in the future will assist in establishing its importance to C. burnetii pathogenesis.
Consistent with a previous study [1], we show here that AX and IC bacteria are able to utilize glucose. C. burnetii lacks an identifiable PTS or a hexokinase, raising questions as to whether they were capable of either transporting and/or phosphorylating exogenous glucose. CBU0265 has been annotated as a proton-dependent hexose transporter in C. burnetii, and in this study we provide evidence that this protein is involved in glucose transport. Specifically, disruption of CBU0265 significantly decreased catabolism of [13C]glucose, while overexpression of this protein on a multi-copy plasmid in the 0265::Tn complemented strain led to increased glucose catabolism. However the 0265::Tn mutant was still able to incorporate 13C from glucose, indicating the presence of at least one additional transporter. While CBU0347 is annotated as a putative d-xylose transporter, we show here that it is also involved in mediating the uptake of hexose sugars such as glucose, confirming the presence of at least two glucose transporters in C. burnetii. Other possible hexose transporter candidates in the C. burnetii genome include a number of ABC transporters [27]. While glucose uptake is not essential for C. burnetii growth in rich medium [16], the co-expression of redundant transporters suggests it may be more important as a nutrient source for C. burnetii than previously thought. Indeed, our studies clearly show that C. burnetii co-utilizes both hexose and amino acid carbon sources in vivo. Similarly, disruption of the gluconeogenic enzyme PEPCK only partially blocked intracellular C. burnetii growth in host cells cultured in glucose-restricted medium, indicating that the C. burnetii vacuole contains appreciable levels of glucose or other sugars.
IC C. burnetii share a number of metabolic traits in common with Leishmania species, a group of protozoan pathogens which also invade macrophages and proliferate within a similar phagolysosome compartment. Both pathogens are auxotrophic for many amino acids, purines and vitamins, suggesting that they are completely dependent on salvage of these metabolites from the host lysosome [41]. Both pathogens also utilize hexoses in vivo and exhibit an incomplete TCA cycle, minimizing the production of respiratory ROS [2]. Interestingly, intracellular Leishmania stages down-regulate use of glutamate as a carbon source compared with extracellular cultivated stages. Leishmania mutants with reduced capacity to utilize glucose adapt by increasing their capacity to uptake glutamate, although this adaptation does not restore virulence in macrophage or animal models, due to associated increase in endogenous ROS production [42]. It is possible that C. burnetii is able to tolerate higher levels of respiratory activity, or have a more efficient respiratory chain compared with Leishmania. This ability of C. burnetii to tolerate higher levels of ROS is supported by the labeling data in this current study. The pattern of 13C-incorporation suggests potential down-regulation of enzymes such as isocitrate dehydrogenase and glutamate dehydrogenase in IC bacteria, both of which when up-regulated are involved in reducing ROS build up and avoiding oxidative stress in other bacterial pathogens [43]. Further analysis into ROS tolerance in IC C. burnetii may be of interest.
This study characterized the nutrient preferences and metabolic pathways of C. burnetii cultivated both axenically and intracellularly, and investigated the importance of glucose uptake to growth of C. burnetii inside cells. Stable isotope labeling identified gaps in the existing metabolic pathway data of C. burnetii and confirmed the presence of multiple glucose transporters. Future investigation of these pathways will provide much-needed information about C. burnetii metabolism and pathogenesis.
Acknowledgements
Confocal imaging was performed at the Biological Optical Microscopy Platform, The University of Melbourne (www.microscopy.unimelb.edu.au).
Abbreviations
- ACCM
acidified citrate cysteine medium
- ACCM-D
defined ACCM lacking glucose
- AX
axenically cultivated
- BSTFA
N,O-bistrimethylsilyltrifluoroacetamide
- CCV
Coxiella-containing vacuole
- FCS
fetal calf serum
- GE
genome equivalent
- IC
intracellularly cultivated
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LCV
large cell variant
- LDH
lactate dehydrogenase
- MDH
malate dehydrogenase
- MOI
multiplicity of infection
- PBS
phosphate-buffered saline
- PEPCK
phosphoenolpyruvate carboxykinase
- PTS
phosphotransferase system
- ROS
reactive oxygen species
- RT
room temperature
- SCV
small cell variant
- TMCS
trimethylchlorosilane
- WT
wild type
Author Contribution
M.K., N.N., D.P.S., H.J.N. and F.M.S. designed the study; M.K. and N.N. performed the experimental procedures under the supervision of H.J.N. and F.M.S.; J.P.M.N. provided assistance with G. mellonella infections; M.K., N.N., D.P.S. and D.T. analyzed the GC/MS data while M.K. analyzed all other data; S.D. provided assistance with statistical analysis of GC/MS data. D.T. provided access to GC/MS. M.K. wrote the main body of the manuscript while M.J.M., F.M.S. and H.J.N. significantly contributed to drafting the manuscript. All authors have read and approved the final manuscript.
Funding
M.K. is supported by an Australian Research Training Program (RTP) Scholarship. Research in H.J.N. laboratory is supported by the Australian National Health and Medical Research Centre (NHMRC, APP1120344) and the Australian Research Council (DP180101298). M.J.M. is supported by an NHMRC Principal Research Fellowship.
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
Supplementary Material
References
- 1.Häuslein I., Cantet F., Reschke S., Chen F., Bonazzi M. and Eisenreich W. (2017) Multiple substrate usage of Coxiella burnetii to feed a bipartite metabolic network. Front. Cell. Infect. Microbiol. 7, 285 10.3389/fcimb.2017.00285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saunders E.C., Ng W.W., Kloehn J., Chambers J.M., Ng M. and McConville M.J. (2014) Induction of a stringent metabolic response in intracellular stages of Leishmania mexicana leads to increased dependence on mitochondrial metabolism. PLoS Pathog. 10, e1003888 10.1371/journal.ppat.1003888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schneeberger P.M., Wintenberger C., van der Hoek W. and Stahl J.P. (2014) Q fever in the Netherlands–2007–2010: what we learned from the largest outbreak ever. Med. Mal. Infect. 44, 339–353 10.1016/j.medmal.2014.02.006 [DOI] [PubMed] [Google Scholar]
- 4.Graham J.G., MacDonald L.J., Hussain S.K., Sharma U.M., Kurten R.C. and Voth D.E. (2013) Virulent Coxiella burnetii pathotypes productively infect primary human alveolar macrophages. Cell. Microbiol. 15, 1012–1025 10.1111/cmi.12096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coleman S.A., Fischer E.R., Howe D., Mead D.J. and Heinzen R.A. (2004) Temporal analysis of Coxiella burnetii morphological differentiation. J. Bacteriol. 186, 7344–7352 10.1128/JB.186.21.7344-7352.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hackstadt T. and Williams J.C. (1981) Biochemical stratagem for obligate parasitism of eukaryotic cells by Coxiella burnetii. Proc. Natl Acad. Sci. U.S.A. 78, 3240–3244 10.1073/pnas.78.5.3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newton H.J., McDonough J.A. and Roy C.R. (2013) Effector protein translocation by the Coxiella burnetii Dot/Icm type IV secretion system requires endocytic maturation of the pathogen-occupied vacuole. PLoS ONE 8, e54566 10.1371/journal.pone.0054566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larson C.L., Martinez E., Beare P.A., Jeffrey B., Heinzen R.A. and Bonazzi M. (2016) Right on Q: genetics begin to unravel Coxiella burnetii host cell interactions. Future Microbiol. 11, 919–939 10.2217/fmb-2016-0044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howe D., Shannon J.G., Winfree S., Dorward D.W. and Heinzen R.A. (2010) Coxiella burnetii phase I and II variants replicate with similar kinetics in degradative phagolysosome-like compartments of human macrophages. Infect. Immun. 78, 3465–3474 10.1128/IAI.00406-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Newton H.J., Kohler L.J., McDonough J.A., Temoche-Diaz M., Crabill E., Hartland E.L. et al. (2014) A screen of Coxiella burnetii mutants reveals important roles for Dot/Icm effectors and host autophagy in vacuole biogenesis. PLoS Pathog. 10, e1004286 10.1371/journal.ppat.1004286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moffatt J.H., Newton P. and Newton H.J. (2015) Coxiella burnetii: turning hostility into a home. Cell. Microbiol. 17, 621–631 10.1111/cmi.12432 [DOI] [PubMed] [Google Scholar]
- 12.Kohler L.J., Reed Sh C., Sarraf S.A., Arteaga D.D., Newton H.J. and Roy C.R. (2016) Effector protein Cig2 decreases host tolerance of infection by directing constitutive fusion of autophagosomes with the Coxiella-containing vacuole. mBio 7, e01127-16 10.1128/mBio.01127-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Omsland A., Cockrell D.C., Howe D., Fischer E.R., Virtaneva K., Sturdevant D.E. et al. (2009) Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc. Natl Acad. Sci. U.S.A. 106, 4430–4434 10.1073/pnas.0812074106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beare P.A., Gilk S.D., Larson C.L., Hill J., Stead C.M., Omsland A. et al. (2011) Dot/Icm type IVB secretion system requirements for Coxiella burnetii growth in human macrophages. mBio 2, e00175-11 10.1128/mBio.00175-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abu-Remaileh M., Wyant G.A., Kim C., Laqtom N.N., Abbasi M., Chan S.H. et al. (2017) Lysosomal metabolomics reveals V-ATPase- and mTOR-dependent regulation of amino acid efflux from lysosomes. Science 358, 807–813 10.1126/science.aan6298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandoz K.M., Beare P.A., Cockrell D.C. and Heinzen R.A. (2016) Complementation of arginine auxotrophy for genetic transformation of Coxiella burnetii by use of a defined axenic medium. Appl. Environ. Microbiol. 82, 3042–3051 10.1128/AEM.00261-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vallejo Esquerra E., Yang H., Sanchez S.E. and Omsland A. (2017) Physicochemical and nutritional requirements for axenic replication suggest physiological basis for Coxiella burnetii niche restriction. Front. Cell. Infect. Microbiol. 7, 190 10.3389/fcimb.2017.00190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuley R., Bossers-deVries R., Smith H.E., Smits M.A., Roest H.I. and Bossers A. (2015) Major differential gene regulation in Coxiella burnetii between in vivo and in vitro cultivation models. BMC Genomics 16, 953 10.1186/s12864-015-2143-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaton K., Peter O., Raoult D., Tissot J.D. and Greub G. (2013) Development of a high throughput PCR to detect Coxiella burnetii and its application in a diagnostic laboratory over a 7-year period. New Microbes New Infect. 1, 6–12 10.1002/2052-2975.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fielden L.F., Moffatt J.H., Kang Y., Baker M.J., Khoo C.A., Roy C.R. et al. (2017) A farnesylated Coxiella burnetii effector forms a multimeric complex at the mitochondrial outer membrane during infection. Infect. Immun. 85, e01046-16 10.1128/IAI.01046-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kowalski G.M., De Souza D.P., Burch M.L., Hamley S., Kloehn J., Selathurai A. et al. (2015) Application of dynamic metabolomics to examine in vivo skeletal muscle glucose metabolism in the chronically high-fat fed mouse. Biochem. Biophys. Res. Commun. 462, 27–32 10.1016/j.bbrc.2015.04.096 [DOI] [PubMed] [Google Scholar]
- 22.Rohn H., Junker A., Hartmann A., Grafahrend-Belau E., Treutler H., Klapperstück M. et al. (2012) VANTED v2: a framework for systems biology applications. BMC Syst. Biol. 6, 139 10.1186/1752-0509-6-139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W. et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Latomanski E.A. and Newton H.J. (2018) Interaction between autophagic vesicles and the Coxiella-containing vacuole requires CLTC (clathrin heavy chain). Autophagy 14, 1710–1725 10.1080/15548627.2018.1483806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Norville I.H., Hartley M.G., Martinez E., Cantet F., Bonazzi M. and Atkins T.P. (2014) Galleria mellonella as an alternative model of Coxiella burnetii infection. Microbiology 160, 1175–1181 10.1099/mic.0.077230-0 [DOI] [PubMed] [Google Scholar]
- 26.Schümann C., Michlmayr H., Del Hierro A.M., Kulbe K.D., Jiranek V., Eder R. et al. (2013) Malolactic enzyme from Oenococcus oeni: heterologous expression in Escherichia coli and biochemical characterization. Bioengineered 4, 147–152 10.4161/bioe.22988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seshadri R., Paulsen I.T., Eisen J.A., Read T.D., Nelson K.E., Nelson W.C. et al. (2003) Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc. Natl Acad. Sci. U.S.A. 100, 5455–5460 10.1073/pnas.0931379100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deutscher J., Francke C. and Postma P.W. (2006) How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70, 939–1031 10.1128/MMBR.00024-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Romero-Rodríguez A., Ruiz-Villafán B., Rocha-Mendoza D., Manzo-Ruiz M. and Sánchez S. (2015) Biochemistry and regulatory functions of bacterial glucose kinases. Arch. Biochem. Biophys 577–578, 1–10 10.1016/j.abb.2015.05.001 [DOI] [PubMed] [Google Scholar]
- 30.Crabill E., Schofield W.B., Newton H.J., Goodman A.L. and Roy C.R. (2018) Dot/Icm-translocated proteins important for biogenesis of the Coxiella burnetii-containing vacuole identified by screening of an effector mutant sublibrary. Infect. Immun. 86, e00758-17 10.1128/IAI.00758-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gunn F.J., Tate C.G. and Henderson P.J. (1994) Identification of a novel sugar-H+ symport protein, FucP, for transport of L-fucose into Escherichia coli. Mol. Microbiol. 12, 799–809 10.1111/j.1365-2958.1994.tb01066.x [DOI] [PubMed] [Google Scholar]
- 32.Spooner P.J., O'Reilly W.J., Homans S.W., Rutherford N.G., Henderson P.J. and Watts A. (1998) Weak substrate binding to transport proteins studied by NMR. Biophys. J. 75, 2794–2800 10.1016/S0006-3495(98)77722-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez E., Allombert J., Cantet F., Lakhani A., Yandrapalli N., Neyret A. et al. (2016) Coxiella burnetii effector CvpB modulates phosphoinositide metabolism for optimal vacuole development. Proc. Natl Acad. Sci. U.S.A. 113, E3260–E3269 10.1073/pnas.1522811113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis E.O. and Henderson P.J. (1987) The cloning and DNA sequence of the gene xylE for xylose-proton symport in Escherichia coli K12. J. Biol. Chem. 262, 13928–13932 PMID: [PubMed] [Google Scholar]
- 35.Maiden M.C., Davis E.O., Baldwin S.A., Moore D.C. and Henderson P.J. (1987) Mammalian and bacterial sugar transport proteins are homologous. Nature 325, 641–643 10.1038/325641a0 [DOI] [PubMed] [Google Scholar]
- 36.Henderson P.J. and Macpherson A.J. (1986) Assay, genetics, proteins, and reconstitution of proton-linked galactose, arabinose, and xylose transport systems of Escherichia coli. Methods Enzymol. 125, 387–429 10.1016/S0076-6879(86)25033-8 [DOI] [PubMed] [Google Scholar]
- 37.Omsland A. and Heinzen R.A. (2011) Life on the outside: the rescue of Coxiella burnetii from its host cell. Annu. Rev. Microbiol. 65, 111–128 10.1146/annurev-micro-090110-102927 [DOI] [PubMed] [Google Scholar]
- 38.Bitew M.A., Khoo C.A., Neha N., De Souza D.P., Tull D., Wawegama N.K. et al. (2018) De novo NAD synthesis is required for intracellular replication of Coxiella burnetii, the causative agent of the neglected zoonotic disease Q fever. J. Biol. Chem. 293, 18636–18645 10.1074/jbc.RA118.005190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masukagami Y., Tivendale K.A., Browning G.F. and Sansom F.M. (2018) Analysis of the Mycoplasma bovis lactate dehydrogenase reveals typical enzymatic activity despite the presence of an atypical catalytic site motif. Microbiology 164, 186–193 10.1099/mic.0.000600 [DOI] [PubMed] [Google Scholar]
- 40.Billig S., Schneefeld M., Huber C., Grassl G.A., Eisenreich W. and Bange F.C. (2017) Lactate oxidation facilitates growth of Mycobacterium tuberculosis in human macrophages. Sci. Rep. 7, 6484 10.1038/s41598-017-05916-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naderer T., Heng J., Saunders E.C., Kloehn J., Rupasinghe T.W., Brown T.J. et al. (2015) Intracellular survival of Leishmania major depends on uptake and degradation of extracellular matrix glycosaminoglycans by macrophages. PLoS Pathog. 11, e1005136 10.1371/journal.ppat.1005136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saunders E.C., Naderer T., Chambers J., Landfear S.M. and McConville M.J. (2018) Leishmania mexicana can utilize amino acids as major carbon sources in macrophages but not in animal models. Mol. Microbiol. 108, 143–158 10.1111/mmi.13923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh R., Mailloux R.J., Puiseux-Dao S. and Appanna V.D. (2007) Oxidative stress evokes a metabolic adaptation that favors increased NADPH synthesis and decreased NADH production in Pseudomonas fluorescens. J. Bacteriol. 189, 6665–6675 10.1128/JB.00555-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.