Abstract
Motivation
In contrast to population-based Hi-C data, single-cell Hi-C data are zero-inflated and do not indicate the frequency of proximate DNA segments. There are a limited number of computational tools that can model the 3D structures of chromosomes based on single-cell Hi-C data.
Results
We developed single-cell lattice (SCL), a computational method to reconstruct 3D structures of chromosomes based on single-cell Hi-C data. We designed a loss function and a 2 D Gaussian function specifically for the characteristics of single-cell Hi-C data. A chromosome is represented as beads-on-a-string and stored in a 3 D cubic lattice. Metropolis–Hastings simulation and simulated annealing are used to simulate the structure and minimize the loss function. We evaluated the SCL-inferred 3 D structures (at both 500 and 50 kb resolutions) using multiple criteria and compared them with the ones generated by another modeling software program. The results indicate that the 3 D structures generated by SCL closely fit single-cell Hi-C data. We also found similar patterns of trans-chromosomal contact beads, Lamin-B1 enriched topologically associating domains (TADs), and H3K4me3 enriched TADs by mapping data from previous studies onto the SCL-inferred 3 D structures.
Availability and implementation
The C++ source code of SCL is freely available at http://dna.cs.miami.edu/SCL/.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Chromosome conformation capture (3C) and its derivative methods, such as 4C, 5C and Hi-C, make it possible to detect genome conformations ranging from a selection of loci to the whole genome. In particular, the Hi-C technique (Lieberman-Aiden et al., 2009) can detect the spatial proximity of DNA regions on a genome-wide scale and has been widely applied to many different types of cells (Darrow et al., 2016; Kim et al., 2017; Rao et al., 2014). However, all of these techniques, including Hi-C, are based on millions of cells and capture only the average conformation of a population of cells. Recently, a single-cell Hi-C technique has been developed that can capture the conformation of a single cell and reveal cell-to-cell variability (Bonev et al., 2017; Liu and Wang, 2017; Nagano et al., 2013; Ramani et al., 2017; Stevens et al., 2017).
Computational methods have been developed to reconstruct the 3D structure of chromosomes based on population-based Hi-C data. Bau et al. (2011) designed a general approach that combined 5C with the integrated modeling platform (IMP) to generate chromatin structures. Duan et al. (2010) developed a method based on 4C data to build yeast genome structures. In the work of Tanizawa et al. (2010), long-range association regions on the fission yeast genome were explored by combining next-generation sequencing and 3C. ShRec3D (Lesne et al., 2014) builds 3D chromosome structures by combining multidimensional scaling and the shortest-path distance on a graph constructed based on Hi-C contacts. Zhang et al. (2013) developed ChromeSDE, which applies semidefinite programing techniques to find the best structure fitting the observed data. Trieu and Cheng (2014) modeled the in-contact and not-in-contact relationships between bead-pairs and reconstructed chromosome 3D structures based on a specifically designed objective function. MCMC5C (Rousseau et al., 2011) models chromosomal structures using Monte Carlo sampling based on a Gaussian model. PASTIS (Varoquaux et al., 2014) and BACH (Hu et al., 2013) use metric multidimensional scaling and a Bayesian-based approach to reconstruct chromosome 3D structures, respectively. HSA (Zou et al., 2016) can jointly analyze multiple Hi-C contact maps to infer 3D chromosomal structures. Other methods for constructing 3D genome structure based on population-based Hi-C include (Adhikari et al., 2016; Oluwadare et al., 2018; Serra et al., 2015; Trieu and Cheng, 2017; Van Berkum et al., 2010).
It is a challenge to model 3D chromosomal structures based on single-cell Hi-C data. First, single-cell Hi-C captures only the existence of a contact instead of the contact frequencies obtained by population-based Hi-C. Second, only a small portion of cis-chromosomal contacts are available, which makes the contact matrix extremely sparse, i.e. containing many zeros. These properties make the previous methods designed for population-based Hi-C not ideal for using single-cell Hi-C data. Methods have been developed to model 3D chromosomal structures based on single-cell Hi-C data, such as Carstens et al. (2016), which is based on Bayesian inferential structure determination, and Nagano et al. (2013) and Stevens et al. (2017), which are based on molecular dynamics.
Here, we present a new approach to this challenging problem, in which the 3D structure of a chromosome is represented as beads-on-a-string and reconstructed inside a 3D cubic lattice. A 2D Gaussian imputation is used to estimate the propensity for the bead-pairs that do not have a single-cell Hi-C contact. A specifically designed loss function was applied to handle three cases: (i) the bead-pairs that have a single-cell Hi-C contact, (ii) the bead-pairs that do not have a single-cell Hi-C contact but are sequentially adjacent to bead-pairs that do and (iii) the bead-pairs that are far away from the bead-pairs that have single-cell Hi-C contacts. Metropolis–Hastings simulation and simulated annealing are performed to construct the 3D structure.
Carstens et al. (2016) uses Bayesian inference to build chromosome structures based on single-cell Hi-C data. However, the posterior distribution is typically of a non-standard form and presents a cluster of models. Therefore, they need to perform random sampling techniques, such as Markov Chain Monte Carlo, to select a representative model and output model parameters. In comparison, our single-cell lattice (SCL) directly simulates the 3D structure in the cubic lattice and then outputs a single 3D structure.
Stevens et al. (2017) built nuc_dynamics, a tool to infer chromosome structures based on single-cell Hi-C data using molecular dynamics. However, they do not consider the influence of a single-cell Hi-C contact on its sequentially neighboring beads. Our SCL uses a 2D Gaussian imputation directly on the 2D contact map to model the influence of each single-cell Hi-C contact on its surrounding beads.
Different single-cell Hi-C data may have different degrees of sparseness. The loss function of SCL uses three different terms to model different types of bead-pairs in terms of their sequential distance from the bead-pairs that have single-cell Hi-C contacts. Moreover, SCL allows users to freely control almost every parameter in the loss function to fit the specific single-cell Hi-C data. This approach allows SCL to handle a wide range of data, from extremely sparse single-cell Hi-C data to relatively abundant data, which will be shown later in the Section 3.
2 Materials and methods
2.1 Overview
There are two central ideas for the design of SCL: (i) the cubic lattice representation of a chromosome 3D structure and (ii) the imputation of single-cell Hi-C contact matrices using a 2D Gaussian function. The cubic lattice representation allows a bead to move only from its current cell in the lattice to its neighboring cells in each attempt of the Metropolis–Hastings simulation. Compared to using continuous 3D coordinates, this approach can greatly decrease computational costs, particularly considering that a chromosome at 50 kb resolution can easily contain thousands of beads. In very sparse single-cell Hi-C contact matrices, most of the bead-pairs have no Hi-C contacts even after imputation. We believe this property may make a continuous 3D coordinate system unnecessary. Moreover, a continuous 3D coordinate system may also significantly increase the complexity of the simulation process and the number of local minimums, making it more difficult for the simulated annealing algorithm to find the optimal conformation. The 2D Gaussian imputation is directly applied to the single-cell contact matrices. It is straightforward and relies on the intuition that if two beads are spatially proximate, their sequential adjacent beads should not be far away from each other.
2.2 Cubic lattice framework and loss function
A chromosome is represented as a continuous chain of beads, each with the same size as the resolution value stored in a 3D cubic lattice. The number of cubic cells or volume of the cubic lattice is , where is the number of beads of the target chromosome. This larger space allows enough free space to simulate the 3D structure. The side of each cell in the cubic lattice is considered to have a length of 1. A DNA bead can be placed only at the eight corners of a cubic cell in the lattice. Detailed descriptions of the simulation process will be discussed later.
We designed the following cost function specifically for zero-inflated single-cell Hi-C data:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
In Equation (1), represents the Euclidean distance between beads and , and represents the 3D coordinates of the beads. In Equations (1)–(3), is a matrix indicating the estimated propensity for beads i and j to form a contact. If there is a single-cell Hi-C contact between a pair of beads, their will be 1 based on Equation (3) ( and are then both zero, making ). For the bead-pairs that do not have a single-cell Hi-C contact, the closer they are to other bead-pairs that do have a single-cell Hi-C contact, the higher the value they will have. This behavior is modeled by the 2D Gaussian function in Equation (3). The values in the matrix cannot be larger than 1.
The first term in Equation (1), , models the bead-to-bead distances for the bead-pairs that do have single-cell Hi-C contact(s) (these bead-pairs have ) or have no Hi-C contact but with (caused by the situation that many of its closely surrounding bead-pairs have Hi-C contacts). Notice that after changing the original single-cell Hi-C data to resolutions of 500 or 50 kb, it is possible that some bead-pairs have >1 single-cell contacts. In this case, we consider the value to be 1 because single-cell Hi-C indicates only a proximate relationship instead of the probability of being in contact, as for population-based Hi-C data. The shape of the first term in Equation (1) is illustrated by the red line in Figure 1 (when the target distance ). The function of the first term does not have an upper bound. In other words, a deviation between the current distance in the 3D structure and the target distance will result in a relatively large value of the cost function, making this term the most influential and stringent in Equation (1), so that the target distances of these bead-pairs are achieved as much as possible. All of the bead-pairs that have a Hi-C contact or will have the same target distance .
Fig. 1.
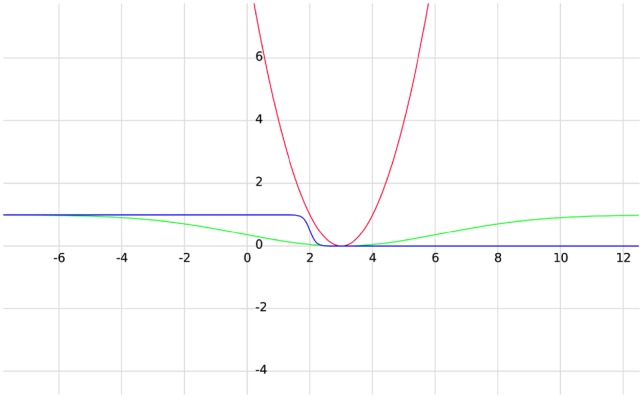
The shapes of the three mathematical terms in Equation (1). The red line represents the first term (); the green line indicates the second term () and the blue line is for the third term ()
The second term in Equation (1), , represents the bead-pairs that have no single-cell Hi-C contact but (close to the other bead-pairs that do have a Hi-C contact; the calculation of will be discussed later). The second term in Equation (1) is a Gaussian-like term that is illustrated by the green line in Figure 1. In the example shown in Figure 1, when , this term reaches its minimum value of 0. The value of controls the steepness of the curve. In contrast to the first term, this function has an upper bound of 1 and guides the optimization algorithm to achieve a target distance of . However, the cost or penalty of not equaling has an upper bound of 1. The term is designed in this way because there are a large number of bead-pairs that have no Hi-C contact but are close to other bead-pairs that have Hi-C contacts. Having an upper bound for these bead-pairs ensures that the sum of their loss values is not overwhelming.
The third term in Equation (1), , is a sigmoid function designed to represent the bead-pairs that have no single-cell Hi-C contact and (far away from the bead-pairs that do have Hi-C contact). It is illustrated by the blue line in Figure 1 (when target distance and ). Notice that this function has a value very close to zero when equals a target distance of 3, remains close to zero for , and quickly jumps to a value close to 1 when . The smoothness of the function is controlled by . This term is designed to ensure that the vast majority of the bead-pairs, having no Hi-C contact and far away from the bead-pairs that do have Hi-C contact (indicated by ), will have a distance larger than . It does not specify how much larger than is optimal but as long as the distance is larger than , the loss value will be small, making this term the least stringent one in Equation (1). It was designed in this way because (i) there are a large number of zero single-cell Hi-C contacts, and a less stringent term will not let these bead-pairs dominate the structure but will allow the bead-pairs that have Hi-C contact or are close to the bead-pairs with Hi-C contacts to mostly determine the structure; (ii) it was possible that a zero Hi-C contact might be caused by experimental imperfection or limitation, and therefore, two beads having zero Hi-C contact might actually be in contact, i.e. false negative cases; the design of the third term in Equation (1) can prevent the sum loss value gathered from these false negative cases from being overwhelming but still allow them to influence the structure.
For the same reason, the Lennard Jones potential is not used as the third term in Equation (1). The Lennard Jones potential results in sharply increasing repulsion when two particles/beads are closer than their equilibrium distance (Supplementary Fig. S1). This property essentially forbids the distance to be smaller than the equilibrium distance, which would be set to the target distance in this case. However, there is a large chance that the large number of no-contact bead-pairs are actually in contact but failed to be captured by single-cell Hi-C experiments. Moreover, the Lennard Jones potential clearly leads to a higher computational cost because of its high order of magnitude mathematical formula.
Equation (2) shows how to calculate the target distance for the bead-pairs with zero single-cell Hi-C contact. Motivated by polymer physics and previous 3D modeling methods (Varoquaux et al., 2014) using population-based Hi-C contacts, the 3D distance and the number of Hi-C contacts follow the relationship , where is the number of population-based Hi-C contacts, and is a constant scale parameter usually set to 1. The values of the bead-pairs that have a single-cell Hi-C contact are set to 1, and their target distances are set to . For the bead-pairs with no single-cell Hi-C contact, we model the target distances to be proportional (the power of −1/3) to with respect to their values.
Equation (3) is a 2D Gaussian function to smooth the zero-inflated single-cell Hi-C contact profile. The heuristic behind it is that if beads and have a single-cell Hi-C contact, i.e. are spatially proximate in the 3D space, their adjacent bead-pairs, namely, beads and , and , and and , should also be close to each other (even if they do not have a Hi-C contact), with a degree that can be controlled by . A larger will make the Gaussian-like curve flatter so that a single-cell Hi-C contact can influence more bead-pairs surrounding it. The value in Equation (3) is a cutoff value preventing this type of influence from extending beyond a certain distance.
Overall, for the bead-pairs with single-cell Hi-C contact(s) or with no Hi-C contacts but with , the optimization algorithm uses the first term in Equation (1) to guide their distance toward . For the bead-pairs with no single-cell Hi-C contact but with in the range of (), the second term in Equation (1) is used to guide their distance toward as defined in Equation (2). For the rest of the bead-pairs with no single-cell Hi-C contact and with smaller than a threshold , the third term in Equation (1) is used to make their distance larger than as calculated in Equation (4). The parameters and in Equation (1) are used to control the weights of the last two cases.
Any sequentially adjacent beads must have a distance no greater than , which is reinforced as a constraint indicated in Equation (5).
2.3 Initialization, Metropolis–Hastings simulation and simulated annealing
DNA beads are randomly added into the 3D cubic lattice to initialize a 3D structure. During this process, a newly added bead must be in the range of with excluded (Binder, 1995) with all currently existing beads in the 3D cubic lattice.
During the simulated annealing process, SCL uses a cooling schedule defined in (Kirkpatrick et al., 1983), in which we set the starting temperature . The decrement of temperature is defined as , where c is the number of times the temperature has been decremented, and is the current temperature.
We allow enough tries at each temperature to let the system stabilize at that temperature. At each temperature, if on average there are 10 accepted moves per DNA bead or the number of tries exceeds 100 times the number of beads (Kirkpatrick et al., 1983), the algorithm decreases the temperature and then keeps running with the new temperature. In every attempt, we use the Metropolis–Hastings algorithm to randomly select a DNA bead to randomly move from its current site to one of its 9 (from the level below the current site) +9 (from the level on top of the current site) +8 (from the same level of current site) =26 neighboring corners in the 3D lattice. The move is accepted with probability if or always accepted if , where is the current temperature, and is the change in the value of the loss function defined in Equation (1): . If the desired acceptance number, i.e. on average 10 accepted moves per bead, is not achieved for three consecutive temperatures, the annealing process is stopped (Kirkpatrick et al., 1983).
To generate a structure with a resolution higher than 500 kb, SCL first generates the 3D structure at 500 kb resolution using the protocol defined above and then adds high-resolution beads between every pair of consecutive low-resolution beads. The method of adding high-resolution beads is the same as the method of initializing the 3D structure before simulations. Then, simulations are performed at temperature 0.1 so that the 3D structure will not be greatly altered but only refined. The number of tries is but can be freely changed by a parameter if needed.
2.4 Selection of the representative model
For each experiment, we independently generated 50 models, each starting with a different randomly initialized 3D structure. After that, we used a Q-score (Wang et al., 2011) to select the top structure. The Q-score of a structure is the average of pairwise comparisons (measured by TM-score (Zhang and Skolnick, 2004)) between this structure and all other structures in the pool. The TM-score (Zhang and Skolnick, 2004) is a metric originally designed to measure the structural similarity between two protein 3D structures, in which a TM-score of 0 indicates no similarity between the two input structures and 1 indicates identical structures. When comparing SCL with the existing tool nuc_dynamics (Stevens et al., 2017), we also generated 50 structures using nuc_dynamics and then used the same method to select the top structure.
2.5 Clustering of models
When comparing different models generated by SCL, the root mean square deviation (RMSD) was calculated. The Kabsch algorithm (Kabsch, 1978) was first used to superimpose two 3D models, after which the RMSD was calculated. To cluster the models, a hierarchical clustering algorithm was performed, treating the RMSD values as distances. From an ensemble of models, the root mean square fluctuation (RMSF) was calculated for each DNA bead in the model. The RMSF has been used as a measure of conformational variance. The tool bio3d (Grant et al., 2006) was used to calculate the RMSF and RMSD values.
2.6 Comparison with another single-cell Hi-C-based modeling tool
We downloaded and executed nuc_dynamics (Stevens et al., 2017) on mouse embryonic stem (ES) cells. The single-cell Hi-C contact profiles of cell 1 were downloaded from Stevens et al. (2017) in the NCC data format (description about NCC format can be found at https://github.com/tjs23/nuc_processing/blob/release_1.0/README.txt). We considered the first base position of the two paired-end reads as the in-contact position, based on which we then executed our SCL program.
2.7 Definitions of topologically associating domains, lamin-B1, H3K4me3, and trans-chromosomal contacts
The definition of 1403 topologically associating domains (TADs) detected on a population-based Hi-C map of TH1 cells was downloaded from Nagano et al. (2013). The H3K4me3 ChIP-Seq data on TH1 cells were downloaded from Nagano et al. (2013) with threshold applied. The mean mESC nuclear laminB1-DamID enrichment (Peric-Hupkes et al., 2010) for each TAD was downloaded from Nagano et al. (2013) with a threshold of >0.3 applied. The trans-chromosomal contact profiles of TH1 cells were downloaded from Nagano et al. (2013). The DNA beads that had single-cell trans-chromosomal Hi-C contacts were highlighted (for details, see Section 3).
2.8 3D-FISH
We downloaded the distances between eight probe pairs detected by 3D-FISH in the mES cells (Beagrie et al., 2017). These eight probe pairs are located on chromosomes 3 and 11. Each probe has a size of 500 kb. We calculated the Pearson’s correlation between the distances of these eight probe pairs in our inferred 3D structure and the distances detected by 3D-FISH. For the 500 kb resolution structures, we directly calculated the Pearson’s correlation. For a 50 kb resolution structure, nine 50 kb beads will be added between every two consecutive 500 kb beads. Therefore, we used every fifth 50 kb bead to calculate the distances of the probe pairs from our inferred 3D structures. The same method was used for the structures generated by nuc_dynamics.
3 Results
3.1 3D structure of the X-chromosome of a mouse TH1 cell
The single-cell Hi-C data were obtained from Nagano et al. (2013), in which single-cell Hi-C experiments were conducted on male mouse TH1 cells. There were 10 cells that had high quality, as described in Nagano et al. (2013). We used the most promising, cell 1 that was associated with 616 X-chromosome cis-contacts. These single-cell Hi-C contacts were mapped to both 500 and 50 kb resolution beads since we used the beads-on-a-string representation of the chromosome. Self-contacts (contacts within the same bead) were removed, which resulted in 438 contacts. If there were contacts between the same bead pair, we kept only one contact.
Figure 2a and b shows the 3D structures of the X-chromosome inferred by SCL at 500 and 50 kb resolutions, respectively. The rainbow coloring scheme shows the segments of the X-chromosome from the centromere (blue) to the telomere (red). The 3D structures shown here depict the top 1 representative structure selected from 50 structures each with a randomly generated initial structure. The parameters of the structures are as follows: (all structures were generated using these parameters unless specified). The value represents the total number of beads of the chromosome.
Fig. 2.
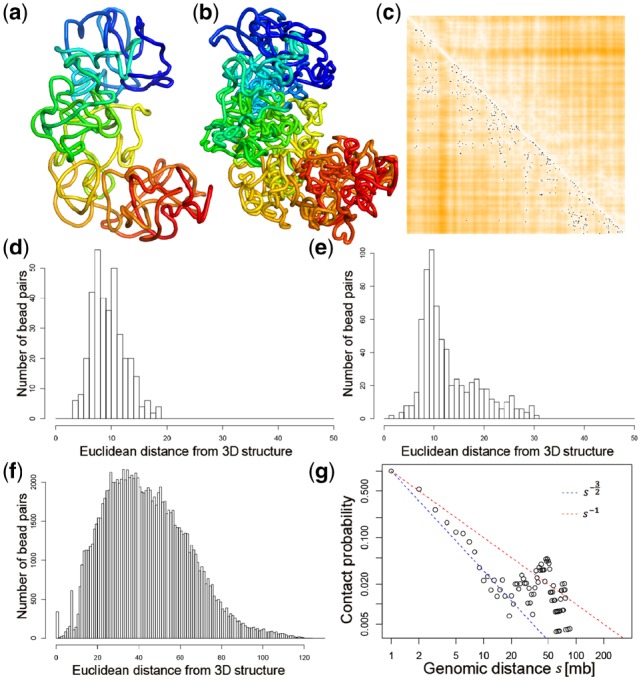
(a) The 3D structure inferred by SCL for the X-chromosome of a mouse TH1 cell (cell 1) at 500 kb resolution. (b) The 3D structure of the same chromosome at 50 kb resolution. (c) Each black dot indicates a single-cell Hi-C contact, and the heatmap indicates the Euclidean distances parsed from the inferred 3D structure (darker orange color indicates larger distance). (d)–(f) Show the distributions of bead-pairs with values of , , and . (g) The relationship between contact probability and genomic distance . The two straight lines are , which indicates a fractal globule, and , which indicates an ideal chain/equilibrium globule
We tested different values for all ten parameters, resulting in 22 different combinations of parameters. The 3D structures and their evaluations can be found in Supplementary Figures S2–S23. Default parameters were selected to generate the structures with the most reasonable evaluation results that were most consistent with the structures generated by other methods (Carstens et al., 2016; Nagano et al., 2013). Six different values of , the parameter that controls the degree of imputation, were tested (Supplementary Figs S18–S24). Notice that in addition to the default value of , the value = 5 can also generate a reasonable structure.
Figure 2c shows the Euclidean distances parsed from the inferred 3D structure superimposed with the single-cell Hi-C contacts (black dots). A darker orange color in the distance heatmap indicates a longer Euclidean distance, whereas a lighter color indicates a shorter distance. The distances parsed from the inferred structure are highly consistent with the single-cell Hi-C contacts.
The (d), (e) and (f) plots in Figure 2 show the number of bead-pairs with different Euclidean distances parsed from the inferred 3D structures. The (d), (e) and (f) plots are for the bead-pairs with values of , and , respectively, i.e. the cases modeled by the first, second and third term in Equation (1) respectively. The bead-pairs with values of 1 have a peak value smaller than the bead-pairs with values in , and the bead-pairs with values in have significantly larger Euclidean distances than the other two cases. Note that the distribution was plotted based on the original 3D coordinates in the 3D lattice (minimum coordinate 0 and maximum coordinate ). We provide a PERL script to convert the 3D coordinates into a different range, e.g. max value 20. The user can easily adjust this max value by a parameter when executing the program.
The (g) plot in Figure 2 displays the relationship between contact probability and genomic distance . The two straight lines are , which indicates a fractal globule, and , which indicates an ideal chain/equilibrium globule. The fractal globule packing of chromosomes was proposed in Duan et al. (2010) and Lieberman-Aiden et al. (2009) based on population-based Hi-C data. The segment of the X-chromosome between 5 and 20 Mb shows an equilibrium globule state, whereas the other regions are between equilibrium and fractal globule.
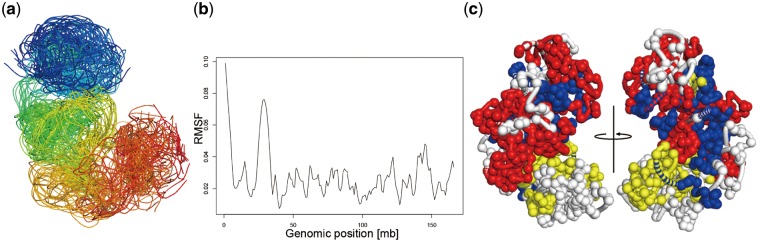
Figure 3a shows the ensemble of the X-chromosome at 500 kb resolution. We generated 50 structures each with a random initial structure and then clustered the structures based on their RMSD values. The 24 structures in a cluster of models are displayed in Figure 3a. Figure 3b shows the RMSF values for each DNA bead. The two peaks in the RMSF plot at positions of ∼1–5 Mb and 25–33 Mb positions were caused by the non-mappable regions in the single-cell Hi-C experiments. Figure 3c highlights the TADs that are enriched with lamin-B1 (yellow), H3K4me3 (blue) and trans-chromosomal contact beads (red) on a 500 kb resolution structure. Similar to Carstens et al. (2016) and Nagano et al. (2013), we also found spatial partitioning of the active (indicated by H3K4me3) and trans-contacting regions from the lamin-enriched TADs (inactive regions). The overall locations of the enrichment patterns are similar to those shown in Carstens et al. (2016).
Fig. 3.
(a) The ensemble of a cluster of 500 kb resolution structures. The beads in the Hi-C unmappable regions were omitted. The rainbow color from blue to red indicates chromosomal regions from centromere to telomere. (b) RMSF of a 1 Mb resolution structure. (c) A 50 kb resolution structure with the following features highlighted: trans-chromosomal contact beads (red), Lamin-B1 enriched topologically associating domains (yellow) and H3K4me3 enriched topologically associating domains (blue)
We executed five tools developed for population Hi-C data: 3Dmax (Oluwadare et al., 2018), ChromSDE (Zhang et al., 2013), PASTIS (Varoquaux et al., 2014), HSA (Zou et al., 2016) and 3DChrom (Liu and Wang, 2018) at 500 kb resolution (Supplementary Fig. S25). It is obvious that they failed to generate reasonable structures based on single-cell Hi-C data.
3.2 3D structures of active and inactive X-chromosome of a human GM12878 cell
We tested SCL on the haplotype-resolved single-cell Hi-C data of the X-chromosome of a GM12878 cell (cell 3) (Tan et al., 2018). Figure 4a–f shows the 3D structures generated using the single-cell Hi-C data after multiple rounds of imputations conducted by Tan et al. (2018) [GSM3271349_gm12878_03.impute3.round4.con.txt.gz in Tan et al. (2018)]. Specifically, Figure 4a and b shows the inactive X-chromosome, and (e) and (f) show the active X-chromosome. To test the performance of SCL with extremely sparse single-cell Hi-C data, we also executed SCL using the single-cell Hi-C data without imputation [GSM3271349_gm12878_03.clean.con.txt.gz in Tan et al. (2018)]. Figure 4k and o shows that the single-cell Hi-C contacts are extremely sparse. Figure 4i and j is the structures for the inactive X-chromosome, and Figure 4m and n is the structures for the active X-chromosome. The 3D structures generated with these extremely sparse single-cell Hi-C data still fit the patterns of the Hi-C contacts.
Fig. 4.
(a), (b), (e) and (f): the SCL-inferred structures generated based on the single-cell Hi-C data after an imputation performed by Tan et al. (2018). (a) and (b) The SCL-inferred 3D structures of the inactive human X-chromosome of GM12878 [cell 3 in Tan et al. (2018)] at 500 and 50 kb resolutions, respectively. (e) and (f) The 3D structures of the active X-chromosome of a GM12878 cell at 500 and 50 kb resolutions. (i), (j), (m) and (n) The structures were generated based on the single-cell Hi-C data downloaded from Tan et al. (2018) without imputation. The 3D structures were generated with parameter . (i) and (j) The structures of the inactive X-chromosome. (m) and (n) The structures of the active X-chromosome. (c), (g), (k) and (o) The single-cell Hi-C contacts superimposed on the heatmap of the Euclidean distances parsed from the SCL-inferred 3D structures. (d), (h), (l) and (p): the relationship between contact probability and genomic distance . The two straight lines are , which indicates a fractal globule, and , which indicates an ideal chain/equilibrium globule
3.3 3D structure of chromosome 11 of a mouse ES cell and comparisons with an existing tool
We applied SCL to a mouse ES cell (cell 1 in (Stevens et al., 2017)) and then compared the 3D structures generated by SCL with the ones inferred by nuc_dynamics (Stevens et al., 2017). Figure 5a and b is the 500 and 50 kb resolution 3D structures of chromosome 11 generated by SCL. Figure 5c–g shows the statistics of the SCL-inferred 3D structures: single-cell Hi-C contacts superimposed on the Euclidean distances parsed from the inferred 3D structure; the distributions of bead-pairs with values of , , and ; and the relationship between contact probability and genomic distance.
Fig. 5.
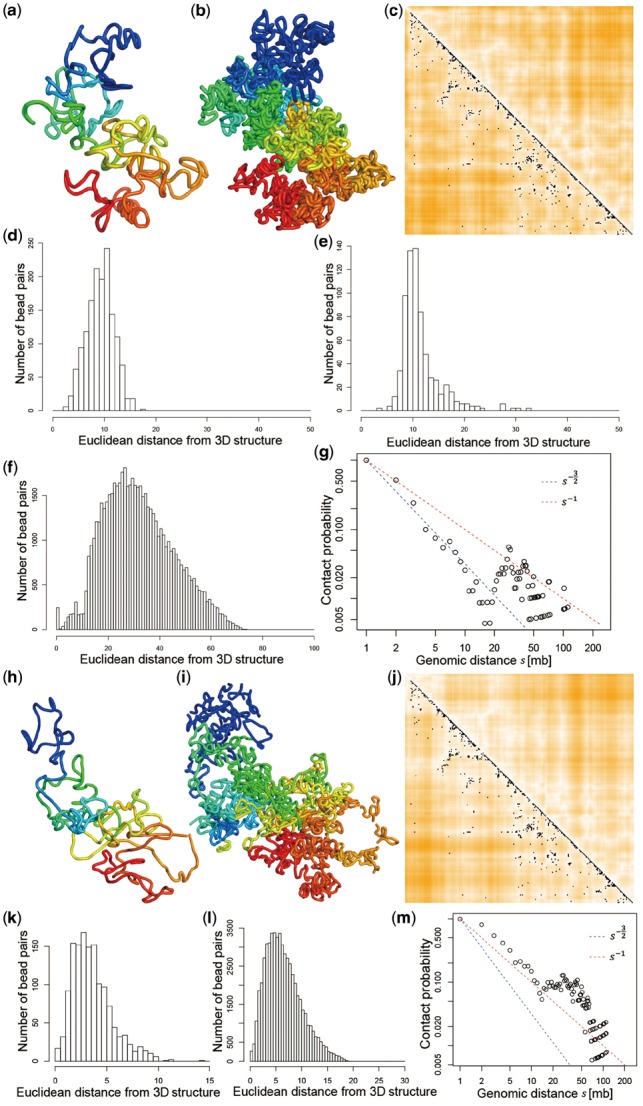
(a) The 3D structure inferred by SCL for chromosome 11 of an mES cell [cell 1 in Stevens et al. (2017)] at 500 kb resolution. (b) The 3D structure of the same chromosome at 50 kb resolution. (c) Each black dot indicates a single-cell Hi-C contact, and the heatmap indicates the Euclidean distances parsed from the inferred 3D structure (a darker orange color indicates a larger distance). (d)–(f) The distributions of bead-pairs with values of , , and . (g) The relationship between contact probability and genomic distance . The two straight lines are , which indicates a fractal globule, and , which indicates an ideal chain/equilibrium globule. (h)–(m) The 3D structure of the same chromosome generated by nuc_dynamics and statistics. (h) and (i) The 3D structures generated by nuc_dynamics. (j) Each black dot indicates a single-cell Hi-C contact, and the heatmap indicates the Euclidean distances parsed from the inferred 3D structure. (k) The distribution of distances of bead-pairs that have single-cell Hi-C contact(s). (l) The distribution of distances of bead-pairs that do not have single-cell Hi-C contacts. (m) The relationship between contact probability and genomic distance
Figure 5h and i shows the 3D structure of chromosome 11 at 500 and 50 kb resolution that were generated by nuc_dynamics. Figure 5j–m shows the statistics generated on the 3D structures generated by nuc_dynamics.
3.4 Validation with 3D-FISH
We calculated the Pearson’s correlation between the distances of eight probe pairs in the SCL-inferred (or nuc_dynamics-inferred) 3D structure and the distances detected by 3D-FISH (Beagrie et al., 2017). The eight probe pairs are located on chromosomes 3 and 11. At 500 kb resolution, the correlation based on the top one SCL-inferred structure is 0.15, whereas it is 0.08 based on the top one nuc_dynamics-inferred structure. At 50 kb resolution, the correlation based on the top one SCL-inferred structures is 0.37, whereas it is 0.21 based on the top one structure generated by nuc_dynamics.
3.5 Correlation between target distances and the distances parsed from the 3D structures
We calculated the Pearson’s correlation coefficients between the target distances and the Euclidean distances parsed from the inferred 3D structures. The correlation values were reported with different values of all of the parameters, including a wide range of values for and (Supplementary Fig. S26). It can be found that does not have an obvious influence on the correlation, but a larger will result in a higher correlation. For example, when , the correlation is 0.53; when , the correlation is 0.01. This behavior occurs because a larger will cause more bead-pairs to have higher values. In this way, more bead-pairs will be modeled by the first and second terms in Equation (1), which are relatively stringent to better enforce the fit of the distances in the 3D structure to the target distances, which eventually leads to higher correlation. However, when we selected the default parameters, we also needed to consider other evaluation criteria, as previously mentioned.
3.6 Computational time
Modeling one 500 kb resolution structure of the mouse X-chromosome of TH1 cells takes ∼35 min with an Intel Xeon CPU at 2.70 GHz. To model the structure at 50 kb resolution takes ∼4 h.
Funding
This work was supported by the National Institutes of General Medical Sciences Grant [R15GM120650 to Z.W.]; and start-up funding from the University of Miami (to Z.W.).
Conflict of Interest: None declared.
Supplementary Material
References
- Adhikari B. et al. (2016) Chromosome3D: reconstructing three-dimensional chromosomal structures from Hi-C interaction frequency data using distance geometry simulated annealing. BMC Genomics, 17, 886.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bau D. et al. (2011) The three-dimensional folding of the alpha-globin gene domain reveals formation of chromatin globules. Nat. Struct. Mol. Biol., 18, 107–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beagrie R.A. et al. (2017) Complex multi-enhancer contacts captured by genome architecture mapping. Nature, 543, 519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder K. (1995) Monte Carlo and Molecular Dynamics Simulations in Polymer Science. Oxford University Press, Oxford. [Google Scholar]
- Bonev B. et al. (2017) Multiscale 3D genome rewiring during mouse neural development. Cell, 171, 557–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstens S. et al. (2016) Inferential structure determination of chromosomes from single-cell Hi-C data. PLoS Comput. Biol., 12, e1005292.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrow E.M. et al. (2016) Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc. Natl. Acad. Sci. USA, 113, E4504–E4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z. et al. (2010) A three-dimensional model of the yeast genome. Nature, 465, 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B.J. et al. (2006) Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics, 22, 2695–2696. [DOI] [PubMed] [Google Scholar]
- Hu M. et al. (2013) Bayesian inference of spatial organizations of chromosomes. PLoS Comput. Biol., 9, e1002893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. (1978) A discussion of the solution for the best rotation to relate two sets of vectors. Acta Crystallogr. Sec. A, 34, 827–828. [Google Scholar]
- Kim S. et al. (2017) The dynamic three-dimensional organization of the diploid yeast genome. Elife, 6, e23623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick S. et al. (1983) Optimization by simulated annealing. Science, 220, 671–680. [DOI] [PubMed] [Google Scholar]
- Lesne A. et al. (2014) 3D genome reconstruction from chromosomal contacts. Nat. Methods, 11, 1141–1143. [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E. et al. (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science, 326, 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Wang Z. (2017) scHiCNorm: a software package to eliminate systematic biases in single-cell Hi-C data. Bioinformatics, 1, 2.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Wang Z. (2018) Measuring the three-dimensional structural properties of topologically associating domains In: 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM). pp. 21–28. IEEE. [Google Scholar]
- Nagano T. et al. (2013) Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature, 502, 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oluwadare O. et al. (2018) A maximum likelihood algorithm for reconstructing 3D structures of human chromosomes from chromosomal contact data. BMC Genomics, 19, 161.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peric-Hupkes D. et al. (2010) Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol. Cell, 38, 603–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani V. et al. (2017) Massively multiplex single-cell Hi-C. Nat. Methods, 14, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S.S. et al. (2014) A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell, 159, 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau M. et al. (2011) Three-dimensional modeling of chromatin structure from interaction frequency data using Markov chain Monte Carlo sampling. BMC Bioinformatics, 12, 414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra F. et al. (2015) Restraint-based three-dimensional modeling of genomes and genomic domains. FEBS Lett., 589, 2987–2995. [DOI] [PubMed] [Google Scholar]
- Stevens T.J. et al. (2017) 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature, 544, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L. et al. (2018) Three-dimensional genome structures of single diploid human cells. Science, 361, 924–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizawa H. et al. (2010) Mapping of long-range associations throughout the fission yeast genome reveals global genome organization linked to transcriptional regulation. Nucleic Acids Res., 38, 8164–8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu T., Cheng J. (2014) Large-scale reconstruction of 3D structures of human chromosomes from chromosomal contact data. Nucleic Acids Res., 42, e52–e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu T., Cheng J. (2017) 3D genome structure modeling by Lorentzian objective function. Nucleic Acids Res., 45, 1049–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Berkum N.L. et al. (2010) Hi-C: a method to study the three-dimensional architecture of genomes. J. Vis. Exp., 1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoquaux N. et al. (2014) A statistical approach for inferring the 3D structure of the genome. Bioinformatics, 30, i26–i33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. et al. (2011) APOLLO: a quality assessment service for single and multiple protein models. Bioinformatics, 27, 1715–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. et al. (2013) Inference of spatial organizations of chromosomes using semi-definite embedding approach and Hi-C data In: Research in Computational Molecular Biology. pp. 317–332. Springer. [Google Scholar]
- Zhang Z. et al. (2013) 3D chromosome modeling with semi-definite programming and Hi-C data. J. Comput. Biol., 20, 831–846. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Skolnick J. (2004) Scoring function for automated assessment of protein structure template quality. Proteins, 57, 702–710. [DOI] [PubMed] [Google Scholar]
- Zou C. et al. (2016) HSA: integrating multi-track Hi-C data for genome-scale reconstruction of 3D chromatin structure. Genome Biol., 17, 40.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.