Abstract
Motivation
The analysis of biological samples in untargeted metabolomic studies using LC-MS yields tens of thousands of ion signals. Annotating these features is of the utmost importance for answering questions as fundamental as, e.g. how many metabolites are there in a given sample.
Results
Here, we introduce CliqueMS, a new algorithm for annotating in-source LC-MS1 data. CliqueMS is based on the similarity between coelution profiles and therefore, as opposed to most methods, allows for the annotation of a single spectrum. Furthermore, CliqueMS improves upon the state of the art in several dimensions: (i) it uses a more discriminatory feature similarity metric; (ii) it treats the similarities between features in a transparent way by means of a simple generative model; (iii) it uses a well-grounded maximum likelihood inference approach to group features; (iv) it uses empirical adduct frequencies to identify the parental mass and (v) it deals more flexibly with the identification of the parental mass by proposing and ranking alternative annotations. We validate our approach with simple mixtures of standards and with real complex biological samples. CliqueMS reduces the thousands of features typically obtained in complex samples to hundreds of metabolites, and it is able to correctly annotate more metabolites and adducts from a single spectrum than available tools.
Availability and implementation
https://CRAN.R-project.org/package=cliqueMS and https://github.com/osenan/cliqueMS.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
The analysis of biological samples in untargeted metabolomic studies using liquid chromatography coupled to electrospray mass spectrometry results in tens of thousands of ion signals or features. It is now well accepted that this large number of features is an overestimation of the real number of different compounds in the sample, mainly because single metabolites can be detected as multiple ions of different mass in either positive or negative ionization mode. This redundancy of features is mostly due to in-source phenomena including cation adduction, multimerization and in-source fragmentation, plus contaminants. However, few studies have attempted to estimate which percentage of unique metabolites, out of the total number of detected features, are being profiled in an untargeted metabolomic experiment. These studies, in addition, have reported very disparate numbers (Brown et al., 2009, 2011; Jankevics et al., 2012; Mahieu and Patti, 2017), ranging from as low as only 3% to more than 50% unique endogenous metabolites. This scenario reflects that the annotation of features, understood as their feature relationships in MS1 mode, is a challenging task and represents a serious obstacle for the real high-throughput analysis of metabolomics data. While computational solutions for the structural identification of metabolites from MS2 data (Aguilar-Mogas et al., 2017; Allen et al., 2014; Dührkop et al., 2015; Heinonen et al., 2012; Ridder et al., 2014; Ruttkies et al., 2016; Tsugawa et al., 2016) have recently demonstrated substantial progress (Nishioka et al., 2014; Schymanski et al., 2017; Schymanski and Neumann, 2013), automated tools aimed at the complete exploitation of LC-MS1 data through the successful annotation of metabolite features have not reached the same maturity level.
The two main grouping principles for detecting and annotating features related to a metabolite are chromatographic peak-shape similarity (i.e. coeluting features) and peak-abundance correlation, or a combination thereof. Pairwise intensity correlation analysis across multiple samples is the basis of computational tools such as AStream (Alonso et al., 2011), MSClust (Tikunov et al., 2012), RAMClust (Broeckling et al., 2014), MS-FLO (DeFelice et al., 2017), compMS2Miner (Edmands et al., 2017), xMSannotator (Uppal et al., 2017) or findMAIN (Jaeger et al., 2017) among other similar approaches (Lee et al., 2013; Zeng et al., 2014). On the other hand, peak-shape similarity is used by CAMERA (Kuhl et al., 2012) and MZmine2 (Pluskal et al., 2010). MetAssign (Daly et al., 2014) or xMSannotator (Uppal et al., 2017) has also included a probabilistic score to measure the confidence in particular assignments based on statistical clustering. More recently knowledge-driven annotation tools have also been proposed by de la Fuente and collaborators (Gil de la Fuente et al., 2018).
To aid in the automatization of LC-MS1 data processing we have developed CliqueMS, a computational tool that annotates redundant LC-MS1 features using the similarity between coelution profiles and a calculated natural frequency of adduct formation observed in real complex biological samples and pure compounds. As a result, in contrast to the majority of existing tools, CliqueMS can produce accurate annotations for a single LC-MS1 spectrum.
To do so, CliqueMS implements a novel mathematical approach to obtain the most plausible groupings of features according to a similarity network. Next, CliqueMS annotates features and ranks annotations using an estimated frequency of dominant adducts and in-source fragments in complex biological samples and from all available compounds in the National Institute of Standards and Technology 14 MS/MS library (Fig. 1 and Supplementary Material). CliqueMS correctly identifies and annotates a larger number of adducts, leading to more correct parental ion neutral masses than existing available approaches such as CAMERA (Kuhl et al., 2012), xMSannotator (Uppal et al., 2017) and MS-FLO (DeFelice et al., 2017).
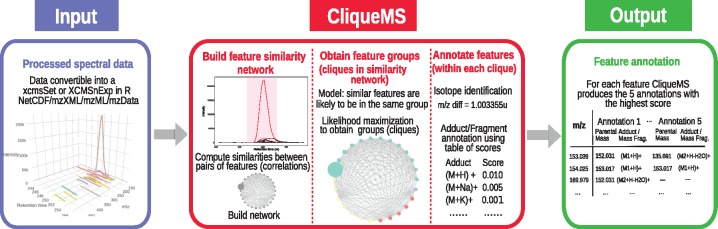
Fig. 1.
Schematic representation of CliqueMS. CliqueMS identifies the features belonging to the same metabolite. CliqueMS uses as input LC-MS1 data in any format that can be converted into either an ‘xcmsSet’ or an ‘XCMSnExp’ object in R such as mzML, mzXML, mzData and NetCDF. First, CliqueMS determines peak-shape (i.e. coeluting) similarities between all pairs of features in the LC-MS1 spectrum. Then CliqueMS finds groups of features based on the network of similarities. The assumption is that the more similar a pair of features, the more likely they are to belong to the same group. Following a maximum likelihood procedure, CliqueMS finds the best division into fully connected groups of features (or cliques). Then, for each clique, CliqueMS proceeds to annotate each feature by establishing the parental ion neutral mass. Annotations are scores based on a table of empirically observed frequencies for each adduct. The final output is, for each feature, the five annotations with the highest score specifying the adducts/in-source fragment and its corresponding parental mass. See Supplementary Figure S1 for a detailed description of the installation process, input and output formats as well as the parameters and modules within CliqueMS
2 Materials and methods
2.1 Description of CliqueMS
Formally, CliqueMS addresses the following problem. Our spectral data are comprised of a set of features characterized by an m/z value and intensity vector [note that the list of features is obtained by running the peak peaking algorithm available in XCMS (see Supplementary Material S2)]. For each feature i, we obtain the intensity vector discretizing the feature into K equal bins so that , where is the measured intensity at retention time tk, where and (in our analysis, depends on the mass detector operational parameters and the spectral data processing program). Given this data CliqueMS aims at providing a set of plausible annotations for complex samples based on the assumptions that: (i) features of the same metabolite corresponding to in-source phenomena, including adducts (e.g. Na, K) and fragments (e.g. loss of water), display similar chromatographic elution profiles and (ii) in-source phenomena (such as adducts or fragments) occur with a probability equal to the frequency with which they are observed in experiments.
To achieve this goal, we have identified three main steps for annotating features (Fig. 1): (i) the construction of a similarity network, where each node represents a feature and edges are weighted according to the similarity between features; (ii) the identification of the most plausible division of the similarity network into cliques (fully connected groups of features) and (iii) the annotation of the features corresponding to the same parental mass within each clique.
Step 1: construction of a similarity network between features
In order to provide meaningful annotations, first we need to group features so that features corresponding to the same metabolite are grouped together. CliqueMS is based on the expectation that all features resulting from in-source transformations of the same metabolite have a similar chromatographic retention pattern. A critical step is thus to select an appropriate measure of similarity between features that reflects our expectations and allows the construction of a similarity network to obtain reliable groups of features.
A possible choice of similarity function is the Pearson correlation between intensity vectors as considered in CAMERA (Kuhl et al., 2012). However, the caveat of the Pearson correlation coefficient is that it is only suited to detect similarity of features that are linearly growing/decreasing and therefore it is a priori not an optimal option when features are non-linear such as the spectral data we consider. To overcome this caveat, we propose to use the cosine similarity, a simple measure that assesses the alignment between intensity vectors:
| (1) |
where . Note that the sum runs over all time bins, and therefore we are not restricting the comparison between features to a specific retention time window; we are considering the full window of retention times.
To compare the ability of Pearson and cosine similarities to discriminate between features corresponding to the same metabolite from coeluting features corresponding to different metabolites, we performed a validation experiment in which we manually simulate the coelution of features (see Supplementary Fig. S2). The results from this experiment show that the cosine similarity has a superior discriminatory power than the Pearson correlation, therefore justifying our choice for similarity metric.
We then construct a weighted undirected similarity network in which each node corresponds to a feature and the weight of each edge between nodes (i, j) corresponds to . Note that this is not a fully connected network, because features that have non-overlapping intensity vectors are not connected (cij = 0).
Step 2: principled identification of groups of features (cliques) in the similarity network
Our next step is to identify groups of features that are similar. Because CliqueMS assumes that all features of the same metabolite must have cij > 0, we aim at identifying cliques of features in the network, that is groups of features that are fully connected so that for any pair of features within a clique.
Formally, the task of finding these groups is equivalent to a label assignment problem in which we want to assign a group label to each feature σi with the constraint that cij > 0 for any pair of nodes with the same label. Note that this problem is different from the problem of community detection in complex networks in which nodes with the same group labels do not have to be necessarily connected (Guimerà and Amaral, 2005). For this reason, we cannot use community detection algorithms for this purpose. Instead, we follow a probabilistic approach to identify the most plausible label assignments (groupings) of features.
To that end, we propose a generative model for node label assignments by noting that the cosine similarity between two features is a good proxy for how likely two features are to be adducts of the same metabolite. A plausible assumption is thus that the probability of two features (i, j) having the same group label (i.e. belonging to the same clique) given a certain similarity cij between intensity vectors is precisely a function of that similarity:
| (2) |
Conversely, the probability that two nodes (i, k) have different labels given their similarity cik is .
To specify the precise dependency of on cij, we note that needs to fulfill the following conditions: (i) it has to be equal to zero if cij = 0 (i.e. two nodes whose intensity vectors do not overlap cannot belong to the same clique) and (ii) it has to be equal to one if cij = 1 (i.e. features with proportional intensity vectors, eg. similar peak shapes, have to belong to the same clique). Because in our sample , any power of the cosine similarity will satisfy these two conditions. Hence, we assume that
| (3) |
where the proportionality is due to a necessary but irrelevant normalization constant and .
Under these assumptions, we can express the probability of an assignment of group labels conditioned on the observed network of similarities as
| (4) |
where is the Kronecker delta function. This conditional probability is the likelihood of the model.
Assuming that we have no prior information about how labels are assigned to nodes, the most plausible group label assignment is the one that maximizes Equation (4) or, equivalently, the log-likelihood . To obtain this label assignment, we use the following algorithm:
Start from a configuration in which each node has a different label.
Propose a new label assignment.
Accept the new label assignment if increases.
Return to step (ii) and iterate until no more changes are accepted.
In step (ii), in order to propose a new label assignment we use a combined strategy that alternates between: (i) merging existing cliques and (ii) moving nodes from one clique to another clique. In our implementation, we alternate these two possible configuration changes with a ratio of 10:1. To merge existing cliques, we follow the heuristic approach in Blondel et al. (2008) which is computationally fast. Specifically, we compute the mean-similarity between nodes within each pair of cliques. We then propose to merge the pair of cliques with the largest mean-similarity. To move a node (i.e. to change the label of that node to that of a different clique), we select the label assignment that produces the largest increase in . As a last step, when cannot be increased by merging any pair of cliques in the network, we use the Kernighan–Lin algorithm (Kernighan and Lin, 1970) to propose single-node moves between cliques. The algorithm stops when we cannot further increase the log-likelihood with single-node switches. We set a lower bound (by default ) for the relative change in necessary to consider that a change in node label assignments results in a significant increase in the log-likelihood.
In order to estimate the best value for the parameter α in Equation (3), we measure the accuracy of our algorithm at assigning group label to features that have similar retention time patterns.
Specifically, starting from the spectral data for the mixture of 9 standards as in the validation of Step 1 (see Section 2.2 and Supplementary Fig. S2), we simulated differences in the coelution of metabolites by manually displacing all the features of the same metabolite along the retention time axis. We then simulate the coelution of two, three and four compounds for different time shifts, and evaluate the accuracy of our algorithm at correctly labeling features using the adjusted mutual information (AMI) (Vinh et al., 2010). The AMI measures the accuracy of the labeling by comparing the ‘true’ and the proposed assignment while taking into account the number of features associated to each metabolite. The AMI value is scaled, so that for a random assignment of features to groups.
In Supplementary Figure S3, we show the accuracy of our algorithm for different values of and 2. For reference, we also show the results obtained with the feature grouping algorithm in CAMERA, which is also network based [see Kuhl et al. (2012) for details]. We find that for any choice of α, our algorithm outperforms the feature grouping algorithm in CAMERA. This is because the feature grouping algorithm in CAMERA tends to produce a large number of groups of features and therefore the AMI is close to zero independently of the time shift. We also find that for our algorithm, larger values of α result into too many cliques when the coelution is not as accentuated, slightly decreasing the algorithm’s accuracy. Therefore, we use a value of α = 2 as a default in CliqueMS and in what follows.
Step 3: Annotation of adducts by isotope and parental mass identification
After obtaining the maximum likelihood configuration of label assignments to nodes , we use the differences in values for all the features within a clique to identify isotopes, and putative adducts and in-source fragments associated to the parental neutral mass of the metabolite.
Consider we have a clique γ comprising Γ features . The first step is to identify features corresponding to isotopic variants of the same metabolite, as they can be determined by the exact mass difference between features and their relative intensities. Whenever the mass difference between two features corresponds to 1.003355 (Da), ϵI being the relative error of the isotope search, the two features are candidates for being isotopes. If their intensity ratios also correspond to the relative abundance of such isotopes, then these two features are considered to be two isotopic variants of the same metabolite Note that we take into account other differences in m/z between isotopes with z > 1 (see details in the Supplementary Material).
For the remaining features , NI being the number of isotopes in the clique, our goal is to associate each one of the features to an adduct or fragment, and therefore to establish the mass of the corresponding neutral compound. In order to do that, CliqueMS considers a list of possible adducts and fragments and their associated mass difference taken from the NIST database (National Institute of Standards and Technology, 2014) spectra with positive and negative ionization (see Supplementary Tables S1 and S2).
First, we determine the possible annotations for each feature that are compatible with the observed mass differences. In what follows, for simplicity we refer to all possible annotations of features as adducts, but bear in mind that annotations can also correspond to metabolite in-source fragments considered in the previously mentioned list of mass differences. Specifically, for feature , we obtain all the possible parental masses Mk that are compatible with feature i being adduct Ak (), i.e. those that fulfill
| (5) |
Note that in order to get mi, we consider zi to be the charge of adduct Ak: In our analysis, we set ppm, but this parameter can be tuned by the user. For the remaining features , we establish that is compatible with being adduct Al with parental neutral mass Mk if:
| (6) |
Following this procedure for all the features , we obtain for clique γ all possible parental masses that are compatible with at least two features being annotated. For each such parental mass Mk, we construct an adduct vector in which each component corresponds to the adduct annotation of feature i compatible with parental mass Mk. If there is no compatible annotation for feature i then .
The second step is to assess the plausibility of each one of these annotations. In order to do this, we note two facts. First, we note that in manual annotation the observation of some adducts such as [M + H]+ or [M+Na]+ is more frequent than that of other adducts such as [M + H-OH]+ or [2M+Na]+. As a result, the former couple of adducts are more commonly sought for in manual annotations than the latter couple of adducts. To formalize this intuition and quantify the plausibility of a specific annotation, CliqueMS uses observed frequencies of adducts and fragments from available LC-MS1 spectra for pure compounds available in the NIST database and biological in-house samples (see Supplementary Tables S1 and S2). Specifically, for each Mk the log-plausibility sk of annotation is then:
| (7) |
where is the frequency of observation of adduct x and, . In our analysis, we set , so that the frequency of a non-annotated feature is lower than that of the least common adduct or fragment in our database. Note that since available LC-MS1 spectra are likely to increase in the future, these parameters can be changed by the user as needed.
Second, we note that, in the clique identification procedure, features corresponding to different metabolites that coelute sometimes are assigned to the same clique. Taking this into consideration, CliqueMS allows for the annotation of adducts corresponding to more than one parental neutral mass in the same clique. Therefore, given the set of parental masses and their associated annotations we can in principle obtain complex annotations with multiple compatible parental masses, so that and with Mk not necessarily equal to . These annotations are also subject to the constraint that we have at least two annotated features for each parental mass. Nonetheless, because we expect the number of metabolites in coelution to be low, we assume the plausibility of annotations with a large number of parental masses NM to be low. To formalize this idea, the log-plausibility of such complex annotations sc is then:
| (8) |
where we have introduced an exponential penalty if the number of parental masses is larger than one and a = 10 in our analysis. While this may seem a rather large penalty, we note that the most common adducts have and rarest adducts have . Therefore, because our priority is to annotate large amounts of adducts or fragments (including rare ones) associated to the same parental mass rather than annotating the same features with two different parental masses and more common adducts, we need to introduce exponentially large penalties. On the other hand, the penalty has to be low enough to enable the use of more than one parental mass when no other annotations are possible. Using a value of a = 10 strikes the balance between both undesirable situations.
Unfortunately, the number of possible annotations grows very fast with the number of features in a clique, so that even for moderately small cliques (30 features) it is unfeasible to produce and score all annotations exhaustively. Because of this CliqueMS focuses on producing only a few annotations with the largest plausibilities. To that end, we follow a greedy procedure to produce complex annotations. Specifically, we limit the list of parental masses to include: (i) those masses that have the largest overall plausibilities sk and (ii) consider the top scoring masses for annotating each feature . In our analysis, we use Mks with the 15 top overall sks and the most plausible Mk for each feature; these parameter choices show a good compromise between speed of the calculations for large cliques and the retrieval of the most plausible annotations obtained from exhaustive annotation searches.
Finally, we rank annotations according to their plausibility and produce for each clique the five most plausible annotations. In this way, unlike other methods which produce a unique annotation, CliqueMS allows researchers to compare alternative annotations. Note that annotation scores depend on the size of the clique/group of features, therefore annotation scores for different groups of features are not comparable.
2.2 Spectral data acquisition
Mixture of standards: LC-MS1 spectrum of a mixture of the following standards in solution: (-)riboflavin, 1, 2-distearoyl-sn-glycero-3-phosphocholine, biotin, cholic acid, deoxycholic acid, L-methionine sulfoxide, thymine and uracil (see Supplementary Material for details on preparation and acquisition of LC-MS1 spectra). The mzXML file is available at Zenodo with id 1480659, doi: 10.5281/zenodo.1480659.
Complex sample 1: IRS2 KO—LC-MS1 spectra for retina samples from Irs-2-deficient mice [see Hennige et al. (2003), Withers et al. (1998) and Supplementary Materials for preparation, metabolite extraction and LC-MS1 spectra acquisition]. The mzXML file is available at Zenodo with id 1480659, doi: 10.5281/zenodo.1480659.
Complex sample 2: MTBLS103—LC-MS1 spectra of a subset of serum samples of young females within the control group in the study by Samino et al. (2015), which are available at https://www.ebi.ac.uk/metabolights/MTBLS103. We consider a subset of samples of both positive ionization C18 (18 samples) and HILIC (13 samples) found in MBTLS103 dataset; all samples belong to the control group.
3 Results and discussion
To validate the accuracy of CliqueMS we perform three kinds of experiments. First, we look at the accuracy at annotating a relatively simple sample corresponding to mixture of standards for which we have a manual MS1 annotation and the identity of the eluting standards was confirmed via MS2 fragmentation patterns. Second, we use CliqueMS to annotate a complex sample for which we also have manual MS1 annotations for metabolites confirmed via MS2 fragmentation (for the retina samples we provide the manual annotation of metabolites whose concentrations were significantly different from that of wild type animals). We look at the accuracy of CliqueMS at correctly annotating the identified compounds in the sample for LC-MS1 spectra obtained in positive and negative ionization modes separately. In these two cases, because we have a single LC-MS1 spectrum, we compare the accuracy of CliqueMS to annotate metabolites with that of CAMERA (Kuhl et al., 2012), which is the only available tool that can annotate single LC-MS1 spectra. As a general result, we find that Clique MS groups feature in a smaller number of groups (cliques) than CAMERA (see Table 1); this makes CliqueMS able to annotate a larger number of adducts than CAMERA.
Table 1.
Summary of the full set of annotations for each sample
| Sample | Tool | Features | Number of cliques/ groups/clusters | Annotated unique parental masses | Annotated features (%) |
|---|---|---|---|---|---|
| Standards | CliqueMS | 275 | 69 | 49 (48) | 64 (55) |
| CAMERA | 164 | 25 | 32 | ||
| Retina IRS2 KO(+ ionization) | CliqueMS | 8489 | 606 | 1231 (1512) | 70 (57) |
| CAMERA | 2836 | 1303 | 43 | ||
| Retina IRS2 KO (− ionization) | CliqueMS | 3893 | 349 | 334 (494) | 44 (36) |
| CAMERA | 1083 | 552 | 32 | ||
| MTBLS103 HILIC | CliqueMS | 16 160a | 387a | 3186 (3703)a | 84 (68)a |
| CAMERA | 13 048 | 488 | 2947 | 62 | |
| xMSannotator | 230 | 5314 | 46 | ||
| MS-FLO | NA | 2875 | 57 | ||
| MTBLS103 C18 | CliqueMS | 24 620a | 927a | 3980 (4769)a | 74 (61)a |
| CAMERA | 19 871 | 1332 | 13131 | 58 | |
| xMSannotator | 540 | 6226 | 48 | ||
| MS-FLO | NA | 3283 | 41 |
Note: For single spectrum datasets, we show the total number of features in the LC-MS1 spectrum. For MTBLS103 datasets, we report the number of features after sample alignment with XCMS. For single spectrum datasets, we report results for CliqueMS and CAMERA. For multiple spectrum datasets, we report the results for CliqueMS, CAMERA, xMSannotator and MS-FLO. We report the total number of groups identified by the algorithm, the number of unique parental neutral masses identified and the percentage of features each algorithm associated to a parental neutral mass. For single spectrum datasets, we consider the five annotations with the highest scores produces by CliqueMS and report: (i) the average number of unique parental masses over annotations, and, in parenthesis, the number of unique parental masses in the annotation with the best score; (ii) the percentage of features with at least one annotation within the five annotations with best scores, and, in parenthesis, the percentage of features annotated within the best ranked annotation.
For MTBLS103 datasets, we run CliqueMS for each individual sample. For each dataset, we report the the average number of features and the average number of cliques obtained across samples. We also report: (i) the average number of unique parental neutral masses and, in parenthesis, the average number of unique parental masses in the top annotation and (ii) the average of the percentage of features with at least one annotation within the five top annotations and only considering the top annotation (in parenthesis).
Third, because other available methods need more than one spectrum to produce annotations, we also consider another two datasets with 13 and 18 LC-MS1 spectra from Samino et al. (2015). We compare the performance of CliqueMS with that of CAMERA, xMSannotator (Uppal et al., 2017) and MS-FLO (DeFelice et al., 2017) (all other tools mentioned in the abstract were not in working condition at the time of our analysis). We find that CliqueMs is able to consistently provide better, more complete annotations than the other methods for the identified metabolites in the samples.
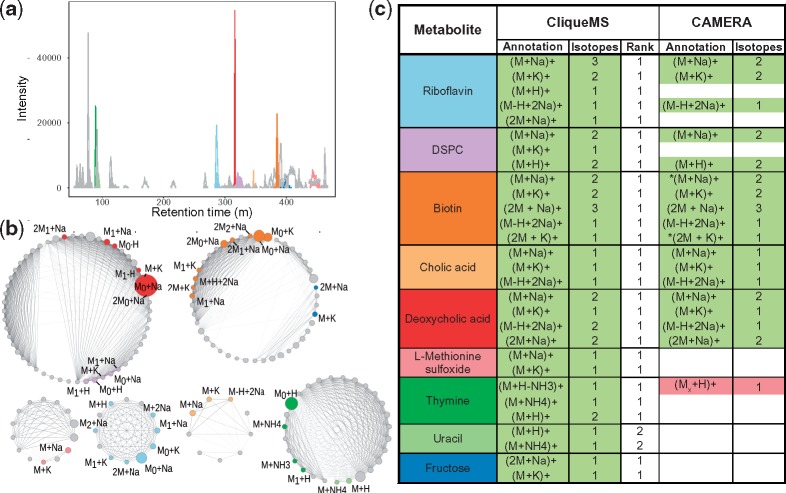
Mixture of standards: Table 1 and Figure 2 show that overall CliqueMS produces better annotations than CAMERA. CliqueMS is able to correctly identify more manually annotated metabolites, and correctly annotate more features associated to these metabolites by both correctly identifying adducts/in-source fragments and their isotopes. The reason for this superior performance is that CliqueMS identifies a smaller number of feature groups so that features associated to the same metabolite are in the same group (Fig. 2a–b). By contrast, CAMERA generates a larger number of groups which results in assigning features corresponding to the same metabolite to different groups, usually annotating them as different metabolites or as non-annotated (Fig. 2c).
Fig. 2.
Feature annotation for a mixture of standards. (a) Extracted ion chromatogram. The nine ionized metabolites were annotated with CliqueMS. We show features that are adducts of each metabolite in different colors (shades of grey), as annotated by CliqueMS in (c). (b) Cliques identified by CliqueMS in the same experiment, after computing cosine correlation and maximizing clique likelihood. The intensity of the link is proportional to the correlation, and the area of each node is proportional to feature intensity. The colors are the same as in (a). For each feature, we show the annotation given by CliqueMs as shown in (c). We denote isotopes by adding a subindex to M, so that M0 corresponds to the monoisotopic mass and M1 to the first isotope. (c) Feature annotation by CliqueMS and CAMERA. For each metabolite, we show the different adducts annotated and the total number of isotopic variants of that particular adduct. Correctly annotated features are shown in green; incorrectly annotated features are shown in red (darker shade of grey), with indicating that the associated parental neutral mass was incorrect; non-annotated features are shown in white. For CliqueMS, we also show the ranking of the feature annotation that matches manual annotation. For CAMERA the * indicates those features for which the algorithm returned two possible annotations. DSPC stands for 1, 2-distearoyl-sn-glycero-3-phosphocholine. See Supplementary Material for CliqueMS annotations
Overall CliqueMS is able to correctly annotate all nine metabolites within the two most plausible annotations for each clique (since for each metabolite, CliqueMS provides the correct annotation for at least one adduct/in-source fragment and its corresponding isotopes within the two highest ranked annotations). The total number of annotated features corresponding to the standard compounds is 42 (of which 29 correspond to adducts/in-source fragments and 13 to isotopes). Instead CAMERA annotates correctly 5 metabolites and a total of 27 features. Note that even if we only considered the highest ranked annotation provided by CliqueMS, the number of correctly annotated metabolites (8) would be higher than for CAMERA (5).
Note that overall CliqueMS identifies a number of unique parental neutral masses that is substantially larger than 9 (48 if we consider the best ranked annotation—see Table 1). The main reason for this is that during the process to obtain the LC-MS1 data, metabolites can break down into smaller fragments that can also become ionized. Because the fragments that one might expect are different for each metabolite in the annotation step, CliqueMS is not considering these effects in the annotation step, therefore these fragments are assigned different parental neutral masses. Despite this issue, the difference in the percentage of features for which a parental neutral mass is reported—64% (or 55% if we consider exclusively the annotation with the largest score) versus 32%—is substantial and is a direct effect of the aforementioned high quality feature grouping which leads to a more accurate adduct annotation.
Single spectrum from complex samples: To evaluate the capacity of CliqueMS to identify adducts in complex LC-MS1 data from a single spectrum, we analyze real retina samples from a mouse model in which the gene irs2 has been knocked out (see Supplementary Material). We analyze spectral data with both positive and negative ionizations. The positive ionization spectrum contained 8489 features reduced to 606 cliques by CliqueMS, whereas the negative ionization spectrum comprised 3893 features reduced to 349 cliques. Instead, as for the previously studied sample, CAMERA identifies a much larger number of groups: 2836 for positive ionization spectra, and 1083 for the negative ionization spectra.
CliqueMS groups the features into a smaller number of groups than CAMERA does. However, in contrast to the results for the mixture of standards, each clique is not necessarily associated to a single metabolite. In fact, because metabolite coelution is so frequent in samples with a large number of features, CliqueMS can group features corresponding to different metabolites within the same clique (see Supplementary Fig. S3).
In Tables 2 and 3 and in Supplementary Figure S4, we show that, overall, CliqueMS provides a better annotation than CAMERA; specifically, CliqueMS is able to annotate a larger number of the identified (via MS/MS) metabolites than CAMERA. Furthermore, CliqueMS is able to correctly annotate a larger number of features, including adducts, in-source fragments and isotopes.
Table 2.
Summary of the performance of different algorithms for complex samples
| Sample | Identified and annotated metabolites | Tool | Annotated metabolites |
Adducts/mass fragments | Annotated features | |
|---|---|---|---|---|---|---|
| Multiple adducts | Single adduct | |||||
| Retina IRS2 KO (+ ionization) | 20 | CliqueMS | 15 | — | 50 | 95 |
| CAMERA | 8 | — | 25 | 45 | ||
| Retina IRS2 KO (− ionization) | 18 | CliqueMS | 6 | — | 16 | 35 |
| CAMERA | 5 | — | 14 | 33 | ||
| MTBLS103 HILIC | 6 (78)a | CliqueMS | 5/6/56b | — | 18/26/213b | 44/72/318b |
| 6 | CAMERA | 3 | — | 13 | 21 | |
| xMSannotator | 1 | 4 | 10 | 10 | ||
| MS-FLO | 1 | — | 2 | 3 | ||
| MTBLS103 C18 | 9 (162)a | CliqueMS | 6/8/104b | — | 17/29/304b | 46/66/524b |
| 9 | CAMERA | 3 | — | 11 | 20 | |
| xMSannotator | 3 | 6 | 13 | 13 | ||
| MS-FLO | 0 | — | 0 | 0 | ||
Note: For single spectrum samples (Retina IRS2 KO in positive and negative ionization mode), we report results for CAMERA and CliqueMS. For the datasets in MTBLS103 (Samino et al., 2015), we report results for the chromatographic column operating in two different conditions: RP-C18 and HILIC. For the MTBLS103 datasets, we show results for CliqueMS, CAMERA, xMSAnnotator and MS-FLO. The multiple adduct and single adduct columns indicate the number of correctly annotated metabolites through the identification of at least two adducts with the same parental neutral mass, and the number of metabolites annotated through the annotation of a single adduct [annotated single adducts are assigned to (M + H)+ by xMSannotator].
CliqueMS analyzes individual samples, therefore in parenthesis we show the total number of annotated metabolites in all samples.
Because CliqueMS produces an individual annotation for each sample (13 for HILIC and 18 for RP-C18), we report three results : r1 shows the number of unique metabolites/adducts/features that are correctly annotated in of the samples; r2 shows the number of unique metabolites/adducts/features which are correctly annotated in at least one sample and r3 shows the aggregate numbers over samples.
Table 3.
Feature annotation for complex samples
| Metabolite | CliqueMS |
CAMERA |
|||
|---|---|---|---|---|---|
| Annotation | Iso- topes | Rank | Annotation | Iso- topes | |
| Uracil | (M+H)+ | 2 | 1 | (M+H)+ | 2 |
| (M+H-H2O)+ | 1 | 1 | (M+H-H2O)+ | 1 | |
| (M+H-NH3)+ | 1 | 1 | (M+H-NH3)+ | 1 | |
| Taurine | (M+H)+ | 2 | 2 | (M+H)+ | 2 |
| (M+Na)+ | 2 | 3 | (M+Na)+ | 2 | |
| (M+H-H2O)+ | 1 | 2 | (M+H-H2O)+ | 1 | |
| (2M+H)+ | 1 | 2 | (2M+H)+ | 1 | |
| (M2+Na)+ | 3 | 1 | (M-H+2Na)+ | 3 | |
| Adenine | (M+H)+ | 2 | 1 | (M2+NH4)+ | 2 |
| (M+H-NH3)+ | 2 | 1 | (M2+H)+ | 2 | |
| L-glutamic acid | (M+H)+ | 3 | 1 | (M2+NH4)+ | 3 |
| (M+H-H2O)+ | 2 | 1 | — | — | |
| (M+Na-H2O)+ | 1 | 3 | (M2+H-H2O)+ | 1 | |
| (M+Na)+ | 3 | 3 | (M2+H)+ | 3 | |
| (M-H+2Na)+ | 3 | 3 | (M2+Na)+ | 3 | |
| (M-2H+3Na)+ | 2 | 3 | (M2-H+2Na)+ | 3 | |
| Guanine | (M+H-H2O)+ | 1 | 1 | (M+H-H2O)+ | 1 |
| (M+H-NH3)+ | 2 | 1 | (M+H-NH3)+ | 2 | |
| (M+H)+ | 2 | 1 | (M+H)+ | 3 | |
| Xanthine | (M+Na)+ | 1 | 1 | — | — |
| (M+H-NH3)+ | 1 | 1 | — | — | |
| (M+H)+ | 2 | 1 | — | — | |
| L-2-aminoadipic acid | (M+H-H2O)+ | 1 | 2 | *(M+H-H2O)+ | 1 |
| (M+H)+ | 1 | 2 | *(M+H)+ | 1 | |
| L-ascorbic acid | (M+Na)+ | 1 | 1 | (M+Na)+ | 1 |
| (M+H)+ | 1 | 1 | (M+H)+ | 1 | |
| PC | (M+K)+ | 1 | 1 | (M2+K-H2O)+ | 1 |
| (M+Na)+ | 2 | 1 | (M2+Na-H2O)+ | 2 | |
| (M+H)+ | 3 | 1 | — | — | |
| Inosine | (M+K)+ | 2 | 1 | (M+K)+ | 1 |
| (2M+H)+ | 2 | 1 | (2M+H)+ | 3 | |
| (2M+Na)+ | 3 | 1 | (2M+Na)+ | 3 | |
| (M+H)+ | 3 | 1 | (M+H)+ | 3 | |
| (M+Na)+ | 2 | 1 | (M+Na)+ | 2 | |
| Guanosine | (2M+H)+ | 2 | 1 | (2M+H)+ | 2 |
| (M+Na)+ | 1 | 1 | (M+Na)+ | 1 | |
| (M+H)+ | 4 | 1 | (M+H)+ | 4 | |
| Glutathione | (M+Na)+ | 1 | 1 | (M+Na)+ | 1 |
| (M+H)+ | 2 | 1 | (M+H)+ | 3 | |
| (M+H-H2O)+ | 3 | 1 | (2M2+H)+ | 3 | |
| Oxigluthatione | (M+Na)+ | 2 | 1 | (2M2+Na)+ | 2 |
| (M+K)+ | 3 | 1 | (2M2+K)+ | 3 | |
| (M+H)+ | 3 | 1 | (2M2+H)+ | 4 | |
| NAD | (M+Na)+ | 1 | 1 | (2M2+Na)+ | 1 |
| (M+2H)2+) | 3 | 1 | (M2+H)+ | 1 | |
| (M+H)+ | 4 | 1 | (2M2+H)+ | 4 | |
Note: Detail of the adducts and in-source fragments annotated by CliqueMS and CAMERA for the retina samples of IRS2 deficient mice (+ ionization). For each molecule, we show the different adducts and in-source fragments annotated; in parenthesis we show the total number of isotopic variants of that particular adduct/in-source fragment. Correctly annotated features are shown in green (light grey); incorrectly annotated features are shown in red (darker grey), with indicating that the associated parental mass was incorrect; non-annotated features are shown in white. For CliqueMS, we also show the ranking of the feature annotation that matches manual annotation. For CAMERA the* indicates those features for which the algorithm returned two possible annotations (see Supplementary Material for the complete results obtained for this sample using CliqueMS and for the complete list of manually annotated metabolites).
The differences in number of metabolites and features annotated are specially remarkable for the positive ionization mode spectrum, in which the number of adducts is larger mainly due to the influence of mobile phase additives and organic solvents (Kruve and Kaupmees, 2017), and therefore more features can coelute. In this case CliqueMS is able to assign a parental neutral mass to 70% of the features overall (and 57% if we only consider the top-ranked annotation), whereas CAMERA only assigns a parental mass to 43% of the features. In the negative ionization mode, the number adducts is much smaller and therefore the differences between both algorithms are not as stark.
Multiple spectra from complex samples: In contrast to CAMERA, xMSannotator and MS-FLO, CliqueMS only produces annotations for each individual spectrum. Our results show that there is in fact an advantage to analyze individual spectra, since overall the performance of CliqueMS is consistently better than that of the other methods.
CliqueMS is able to correctly annotate more metabolites than the other methods (see Table 2 and Supplementary Material). The only exception is xMSannotator, which annotates correctly more metabolites for the C18 dataset because it annotates single features as (M + H)+ by default without having another adduct for the same parental neutral mass (DeFelice et al., 2017). Remarkably, CliqueMS is consistently able to correctly annotate substantially more adducts and identify more isotopic variants than the other methods. As an illustration (see Table 2), CliqueMS correctly annotates 17 adducts in the majority of samples of the HILIC dataset (29 if we consider all correct unique annotations across samples), whereas CAMERA, xMSAnnotator and MS-FLO identify 13, 10 and 2 different adducts/in-source fragments, respectively.
4 Conclusions
Annotating features in LC-MS1 metabolomic experiments is of the utmost importance. Without reliable annotation, however, questions as fundamental as, e.g. how many metabolites are there in a given sample or what is the best adduct for MS/MS experiments cannot be properly addressed. Here, we have shown that CliqueMS provides high quality annotations for biological samples from LC-MS1 single spectra. With simple and synthetic datasets we have provided evidence that explains the performance of CliqueMS: (i) it uses a highly discriminatory feature similarity metric; (ii) it treats the similarities between peaks in a transparent way by means of a simple generative model; (iii) it uses a well-grounded maximum likelihood inference approach to group features; (iv) it uses empirical adduct frequencies to identify the parental neutral mass and (v) it deals flexibly with the identification of the parental neutral mass by proposing and ranking alternative annotations. With real complex biological samples, we have demonstrated that annotating single spectra produces correct annotations for a larger number of features and metabolites than currently available tools for annotating both single and aligned spectra.
Funding
We acknowledge the support of Generalitat de Catalunya [program FI-DGR 2014 to O.S.]; the Ministry of Economy and Competitiveness of Spain [grant numbers FIS2013-47532-C3-1-P, FIS2016-78904-C3-1-P to R.G. and M.S.-P., BFU2014-57466-P to O.Y. and BES-2012-052585 (SAF2011-30578) to M.N.]; and the Ministerio de Ciencia e Innovación [grant SAF2011-28331 to D.B. and L.N.]. O.Y., D.B. and L.N. also acknowledge the support of the Spanish Biomedical Research Centre in Diabetes and Associated Metabolic Disorders (CIBERDEM), an initiative of Instituto de Investigacion Carlos III (ISCIII).
Conflict of Interest: none declared.
Supplementary Material
References
- Aguilar-Mogas A. et al. (2017) iMet: a Network-Based Computational Tool To Assist in the Annotation of Metabolites from Tandem Mass Spectra. Anal. Chem., 89, 3474–3482. [DOI] [PubMed] [Google Scholar]
- Allen F. et al. (2014) CFM-ID: a web server for annotation, spectrum prediction and metabolite identification from tandem mass spectra. Nucleic Acids Res., 42, W94–W99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A. et al. (2011) AStream: an R package for annotating LC/MS metabolomic data. Bioinformatics, 27, 1339–1340. [DOI] [PubMed] [Google Scholar]
- Blondel V.D. et al. (2008) Fast unfolding of communities in large networks. J. Stat. Mech. Theor. Exp., 2008, P10008. [Google Scholar]
- Broeckling C.D. et al. (2014) RAMClust: a Novel Feature Clustering Method Enables Spectral-Matching-Based Annotation for Metabolomics Data. Anal. Chem., 86, 6812–6817. [DOI] [PubMed] [Google Scholar]
- Brown M. et al. (2009) Mass spectrometry tools and metabolite-specific databases for molecular identification in metabolomics. Analyst, 134, 1322.. [DOI] [PubMed] [Google Scholar]
- Brown M. et al. (2011) Automated workflows for accurate mass-based putative metabolite identification in LC/MS-derived metabolomic datasets. Bioinformatics, 27, 1108.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly R. et al. (2014) MetAssign: probabilistic annotation of metabolites from LC-MS data using a Bayesian clustering approach. Bioinformatics, 30, 2764–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFelice B.C. et al. (2017) Mass Spectral Feature List Optimizer (MS-FLO): a tool to minimize false positive peak reports in untargeted liquid chromatography-mass spectroscopy (LC-MS) data processing. Anal. Chem., 89, 3250–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dührkop K. et al. (2015) Searching molecular structure databases with tandem mass spectra using CSI: FingerID. Proc. Natl. Acad. Sci. USA, 112, 12580–12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmands W.M.B. et al. (2017) compMS2Miner: an Automatable Metabolite Identification, Visualization, and Data-Sharing R Package for High-Resolution LC-MS Data Sets. Anal. Chem., 89, 3919–3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil de la Fuente A. et al. (2018) Knowledge-based metabolite annotation tool: CEU Mass Mediator. J. Pharm. Biomed. Anal., 154, 138–149. [DOI] [PubMed] [Google Scholar]
- Guimerà R., Amaral L.A.N. (2005) Functional cartography of complex metabolic networks. Nature, 433, 895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinonen M. et al. (2012) Metabolite identification and molecular fingerprint prediction through machine learning. Bioinformatics, 28, 2333–2341. [DOI] [PubMed] [Google Scholar]
- Hennige A.M. et al. (2003) Upregulation of insulin receptor substrate-2 in pancreatic beta cells prevents diabetes. J. Clin. Invest., 112, 1521–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger C. et al. (2017) Compound annotation in liquid chromatography/high-resolution mass spectrometry based metabolomics: robust adduct ion determination as a prerequisite to structure prediction in electrospray ionization mass spectra. Rapid. Commun. Mass Spectrom., 31, 1261–1266. [DOI] [PubMed] [Google Scholar]
- Jankevics A. et al. (2012) Separating the wheat from the chaff: a prioritisation pipeline for the analysis of metabolomics datasets. Metabolomics, 8, 29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernighan B., Lin S. (1970) An efficient heuristic procedure for partitioning graphs. At&T Tech. J., 49, 291–307. [Google Scholar]
- Kruve A., Kaupmees K. (2017) Adduct formation in ESI/MS by mobile phase additives. J. Am. Soc. Mass Spectrom., 28, 887–894. [DOI] [PubMed] [Google Scholar]
- Kuhl C. et al. (2012) CAMERA: an integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal. Chem., 84, 283–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.S. et al. (2013) Precursor mass prediction by clustering ionization products in LC-MS-based metabolomics. Metabolomics, 9, 1301–1310. [Google Scholar]
- Mahieu N.G., Patti G.J. (2017) Systems-Level Annotation of a Metabolomics Data Set Reduces 25000 Features to Fewer than 1000 Unique Metabolites. Anal. Chem., 89, 10397–10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institute of Standards and Technology (2014) NIST/EPA/NIH Mass Spectral Library v2014. US Secretary of Commerce, Gaithersburg, MD, USA. [Google Scholar]
- Nishioka T. et al. (2014) Winners of CASMI2013: automated Tools and Challenge Data. Mass Spectrom., 3, S0039.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluskal T. et al. (2010) MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics, 11, 395.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridder L. et al. (2014) In silico prediction and automatic LC-MSn annotation of green tea metabolites in urine. Anal. Chem., 86, 4767–4774. [DOI] [PubMed] [Google Scholar]
- Ruttkies C. et al. (2016) MetFrag relaunched: incorporating strategies beyond in silico fragmentation. J. Cheminform., 8, 3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samino S. et al. (2015) Metabolomics reveals impaired maturation of HDL particles in adolescents with hyperinsulinaemic androgen excess. Sci. Rep., 5, 11496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymanski E., Neumann S. (2013) The Critical Assessment of Small Molecule Identification (CASMI): challenges and Solutions. Metabolites, 3, 517–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schymanski E.L. et al. (2017) Critical Assessment of Small Molecule Identification 2016: automated methods. J. Cheminformatics, 9, 22.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikunov Y.M. et al. (2012) MSClust: a tool for unsupervised mass spectra extraction of chromatography-mass spectrometry ion-wise aligned data. Metabolomics, 8, 714–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugawa H. et al. (2016) Hydrogen rearrangement rules: computational MS/MS fragmentation and structure elucidation using MS-FINDER software. Anal. Chem., 88, 7946–7958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppal K. et al. (2017) xMSannotator: an R package for network-based annotation of high-resolution metabolomics data. Anal. Chem., 89, 1063–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinh N.X. et al. (2010) Information theoretic measures for clusterings comparison: variants, properties, normalization and correction for chance. J. Mach. Learn. Res., 11, 2837–2854. [Google Scholar]
- Withers D. et al. (1998) Disruption of IRS-2 causes type 2 diabetes in mice. Nature, 391, 900–904. [DOI] [PubMed] [Google Scholar]
- Zeng Z. et al. (2014) Ion fusion of high-resolution LC-MS-based metabolomics data to discover more reliable biomarkers. Anal. Chem., 86, 3793–3800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.