Summary
Plant RNA virus‐based guide RNA (gRNA) delivery has substantial advantages compared to that of the conventional constitutive promoter‐driven expression due to the rapid and robust amplification of gRNAs during virus replication and movement. To date, virus‐induced genome editing tools have not been developed for wheat and maize. In this study, we engineered a barley stripe mosaic virus (BSMV)‐based gRNA delivery system for clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9‐mediated targeted mutagenesis in wheat and maize. BSMV‐based delivery of single gRNAs for targeted mutagenesis was first validated in Nicotiana benthamiana. To extend this work, we transformed wheat and maize with the Cas9 nuclease gene and selected the wheat TaGASR7 and maize ZmTMS5 genes as targets to assess the feasibility and efficiency of BSMV‐mediated mutagenesis. Positive targeted mutagenesis of the TaGASR7 and ZmTMS5 genes was achieved for wheat and maize with efficiencies of up to 78% and 48%. Our results provide a useful tool for fast and efficient delivery of gRNAs into economically important crops.
Keywords: Barley stripe mosaic virus, CRISPR mutagenesis, delivery, gRNA, maize, Nicotiana benthamiana, targeted mutagenesis, wheat
Introduction
Clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 systems that defend against invasive pathogens are prevalent in both bacteria and Archaea (Bhaya et al., 2011). The most widely used CRISPR/Cas9 system for molecular genetics was derived from the adaptive immunity system of Streptococcus pyogenes and consists of the endonuclease Cas9 and partly paired short RNA, crRNA and trans‐crRNA sequences that can be engineered into a single guide RNA (gRNA) (Jinek et al., 2012). The gRNA retains the Cas9 interaction structure and the ability to recognize target genes. In plants, gRNAs direct the Cas9 protein to cut double‐stranded target DNAs in cells to create double‐strand breaks that can be repaired by the error‐prone non‐homology end joining pathway and/or the homology‐directed repair pathway to modify or mutate the target genes. The great convenience and versatility provided by the CRISPR/Cas9 system permits broad applications for plant genomic research and plant improvement of economically important crops like wheat (Zhang et al., 2016), maize (Li et al., 2017) and rice (Sun et al., 2016).
Recent studies indicate that viruses can be developed to deliver gRNAs for targeted mutagenesis in plants (Ali et al., 2015, 2018; Cody et al., 2017; Gil‐Humanes et al., 2017; Jiang et al., 2019; Yin et al., 2015). Compared to the traditional gRNA delivery methods via Agrobacterium transformation, plant virus‐mediated gRNA delivery systems have several advantages: (1) the gRNAs can accumulate to high levels owing to viral replication and systemic spread in plants and may contribute to a higher genome editing efficiency, (2) multiple functional gRNAs can be expressed from a single viral genome, which provides the potential for multi‐targeted genome editing, (3) phenotypic alterations may appear in infected plants in a relatively short period of time after gene targeting by virus‐induced genome editing (VIGE), and (4) transformation and regeneration of agriculturally important crops such as wheat is laborious and time‐consuming, but VIGE may shorten this period and simplify operation and editing processes of a target gene in the specific tissues. Several plant RNA viruses including tobacco rattle virus (TRV) (Ali et al., 2015), pea early‐browning virus (PEBV) (Ali et al., 2018), tobacco mosaic virus (TMV) (Cody et al., 2017) and beet necrotic yellow vein virus (BNYVV) (Jiang et al., 2019) have been reported to enable targeted genome editing in the model plants Nicotiana benthamiana and Arabidopsis thaliana or both. The DNA virus cabbage leaf curl virus (CaLCuV) has also been engineered for plant genome editing in tobacco (Yin et al., 2015). Moreover, geminivirus viral replicons derived from bean yellow dwarf virus (BeYDV) and wheat dwarf virus (WDV) have been engineered for gene targeting in potato (Butler et al., 2016), wheat (Gil‐Humanes et al., 2017), rice (Wang et al., 2017) and tomato (Dahan‐Meir et al., 2018). However, the absence of the viral movement elements in these DNA replicons obviates the spread of these replicons and requires Agrobacterium‐mediated transformation or bombardment of the replicons into plant tissues. In contrast, agroinfiltration or mechanical inoculation can be employed for VIGE. To our knowledge, there are currently no VIGE tools for economically important crops like wheat and maize.
Barley stripe mosaic virus (BSMV) is a single‐stranded RNA virus that has a tripartite RNA genome composed of RNAs α, β and γ (Fig. 1A). In our previous work, we developed BSMV as a high‐throughput virus‐induced gene silencing (VIGS) vector for knock‐down of target genes in monocots and dicots (Yuan et al., 2011). Here, we describe a novel BSMV‐based gRNA delivery system (ge‐BSMV) for targeted mutagenesis of the model dicot N. benthamiana and the monocot crops wheat and maize.
Figure 1.
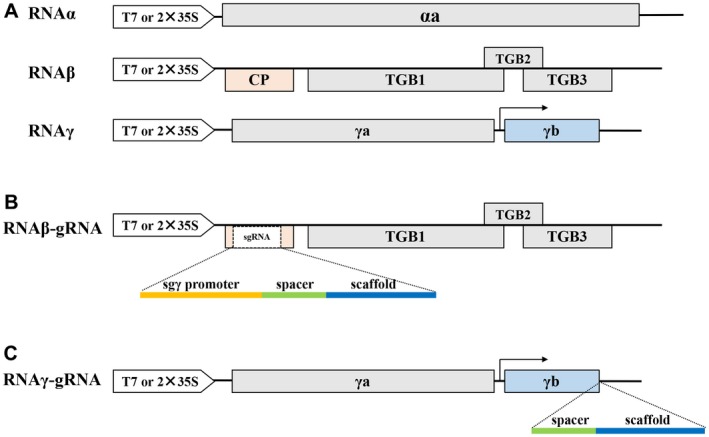
Schematic representation of BSMV‐derived constructs used in this study. (A) BSMV genome organization. The αa protein encoded by RNAα and the γa protein encoded by RNAγ form the replicase of BSMV. RNAβ encodes the coat protein (CP) and triple gene block (TGB) movement proteins. γb, which is translated from subgenomic RNAγ (sgRNAγ), is the pathogenicity determinant and a viral suppressor of RNA silencing (VSR). The arrow indicates the transcription start site of sgRNAγ. (B) ge‐BSMVβ genome organization. Part of the CP sequence (nt 74‐393) is replaced with a 192 nt sgRNAγ promoter (yellow), a gRNA consisting of a 5'‐end spacer sequence (green) and the conserved scaffold (blue). (C) ge‐BSMVγ genome organization. A gRNA is inserted downstream of the γb stop codon.
Results
Design of BSMV‐based gRNA delivery systems
To evaluate whether BSMV could be developed as a gRNA delivery tool and used for targeted mutagenesis in plants, we first used the N. benthamiana to analyse the feasibility of BSMV‐mediated genome editing. Two different systems engineered by modification of BSMV (ND18 strain) RNAs were designed for expression of gRNAs. The first system, engineered by modification of RNAβ, was designated ge‐BSMVβ and consisted of RNAα, RNAγ and RNAβ‐gRNA (Fig. 1B). A second system, designated ge‐BSMVγ, was based on an RNAγ modification and consisted of RNAα, RNAβ and RNAγ‐gRNA (Fig. 1C). To construct the ge‐BSMVβ system, the 192 nt subgenomic RNAγ (sgRNAγ) promoter that spans nt 1864‐2055 of RNAγ was cloned to isolate the sgRNAγ promoter required for transcription of BSMV sgRNAγ (Johnson et al., 2003). Then, the 192 nt sgRNAγ promoter fragment was fused to a 103 nt gRNA ‘spacer scaffold sequence’ to generate a BSMV sgRNAγ promoter‐driven gRNA. The ‘spacer scaffold sequence’ consists of a variable 20 nt spacer designed to pair with the target sequence and the 76 nt gRNA scaffold sequence followed by seven extra downstream T residues. Finally, nt 74 to 393 within the BSMV coat protein open reading frame (ORF) was deleted and replaced with the sgRNAγ promoter‐driven gRNA to generate RNAβ‐gRNA (Fig. 1B). To generate the ge‐BSMVγ system, the ‘spacer scaffold sequence’ was inserted downstream of the γb ORF stop codon to generate the RNAγ‐gRNA (Fig. 1C and Supplemental File S1).
ge‐BSMV‐mediated targeted mutagenesis in N. benthamiana
To investigate the feasibility of ge‐BSMVβ and ge‐BSMVγ systems for gene editing, we first tested the two systems in N. benthamiana by selecting the NbPDS gene for targeted editing (Supplemental Table S1). For easy manipulation of the plasmid, full‐length BSMV RNAα, RNAβ and RNAγ were cloned separately into the pCB301‐2X35S‐MCS‐HDVRZNOS plasmid (Yao et al., 2011) to generate pCB301‐BSMVα, pCB301‐BSMVβ and pCB301‐BSMVγ. DNA fragments corresponding to the NbPDS‐specific gRNA were then cloned into the RNAβ‐gRNA and RNAγ‐gRNA, respectively, as shown in Fig. 1B,C to generate the pCB301‐BSMVβ‐gNbPDS and pCB301‐BSMVγ‐gNbPDS derivatives (see Supplemental File S1). The ge‐BSMV systems consisting of pCB301‐BSMVα/pCB301‐BSMVβ‐gNbPDS/pCB301‐BSMVγ or pCB301‐BSMVα/pCB301‐BSMVβ/pCB301‐BSMVγ‐gNbPDS were designated ge‐BSMVβ‐gNbPDS and ge‐BSMVγ‐gNbPDS. Agrobacterium cultures containing different ge‐BSMV components were mixed with Agrobacterium harbouring the Cas9 expression vector pHSE401 (Xing et al., 2014) and then co‐infiltrated into wild‐type N. benthamiana leaves. The expression of Cas9 in the infiltrated leaves was confirmed by western blot analysis at 5 days post‐infiltration (dpi) (Supplemental Fig. S1). About 10 days after infiltration, symptoms characterized by downward curved and chlorosis in the leaves were observed in the upper uninoculated leaves of both ge‐BSMVβ‐gNbPDS‐ and ge‐BSMVγ‐gNbPDS‐infiltrated N. benthamiana plants (Fig. 2A). Western blot analysis confirmed the presence of TGB1 protein in the systemic leaves (Fig. 2B, upper panel). Furthermore, total RNAs extracted from the systemically infected upper leaves and the region encompassing the inserted gRNA fragment were analysed by RT‐PCR and the results revealed that the size of the RT‐PCR product derived from ge‐BSMVγ‐gNbPDS‐infected leaves was identical to that from the plasmid control and larger than that from the wild‐type BSMV‐infected leaves (Fig. 2B, bottom panel). These results indicate that the ge‐BSMV systems retain the ability to infect N. benthamiana systemically and the heterologous gRNA fragment was maintained during systemic infection.
Figure 2.
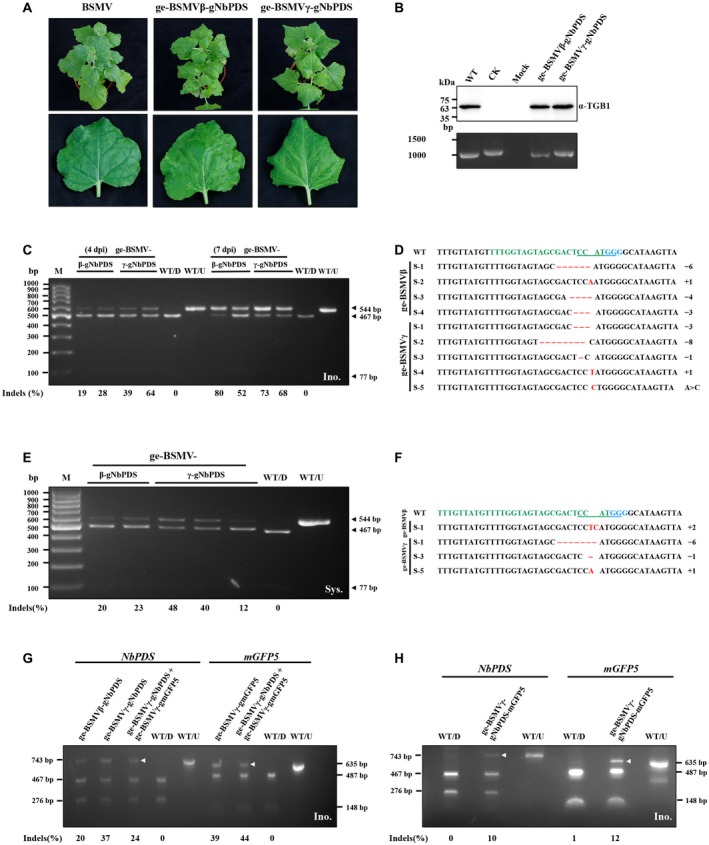
BSMV‐mediated targeted mutagenesis in Nicotiana benthamiana. (A) Symptom observations of BSMV and ge‐BSMV‐infiltrated N. benthamiana. (B) Western blot and RT‐PCR analysis of ge‐BSMVβ‐gNbPDS and ge‐BSMVγ‐gNbPDS infections in systemically infected N. benthamiana leaves. Sample designations are shown above the panels. Antibodies used for the western blot are indicated on the right of the panel. The molecular weight size markers in kDa or the size of DNA fragments in bp are positioned at the left of each panel. WT, BSMV‐infected N. benthamiana leaves; CK, pCB301‐BSMVγ‐gNbPDS plasmid as a positive control for evaluating the size of the RT‐PCR products whereas the protein loading buffer alone was used in the corresponding western blot analysis; Mock, empty agrobacterium‐infiltrated N. benthamiana plants. (C) PCR/RE analysis of ge‐BSMVβ and ge‐BSMVγ‐mediated genome editing of NbPDS in infiltrated N. benthamiana leaves at 4 and 7 days post‐infiltration (dpi). The sizes of DNA fragments in bp are indicated at the right of each panel. Arrowheads show the sizes of the target fragments. Mutation frequencies [indels (%)] were calculated by measuring band intensities with ImageJ software (v. 1.51k) and are shown below the corresponding lanes. WT/U and WT/D, DNA fragments derived from wild‐type BSMV infected N. benthamiana plants prior to or after NcoI digestion; Ino., infiltrated leaves of N. benthamiana; M, 100 bp DNA ladder. (D) Sanger sequencing analysis of the NbPDS target sites in the DNA samples from ge‐BSMVβ and ge‐BSMVγ‐infiltrated leaves. The protospacer is shown in green, the PAM (NGG) motif is in blue, underlined nucleotides indicate the NcoI recognition site, indels are shown in red, with dots indicating deleted nucleotides and red letters indicating inserted nucleotides, the numbers on the right show how many nucleotides were deleted (−) or inserted (+) in the NbPDS target site by BSMV‐mediated genome editing. (E) and (F) PCR/restriction enzyme and Sanger sequencing analysis of ge‐BSMVβ and ge‐BSMVγ‐mediated targeted genome editing of NbPDS in systemically infected leaves of N. benthamiana at 4 dpi. Sys., systemically infected leaves. (G) PCR/RE analysis of ge‐BSMVγ‐mediated multiplexed genome editing in N. benthamiana leaves co‐infiltrated with the ge‐BSMVγ‐gNbPDS, ge‐BSMVγ‐gmGFP5 and the Cas9 expression cassette. Two RNAγ‐gRNA‐derived vectors, pCB301‐BSMVγ‐gNbPDS and pCB301‐BSMVγ‐gmGFP5, which contain different gRNAs, were co‐infiltrated with the pCB301‐BSMVα, pCB301‐BSMVβ and Cas9 expression cassettes into N. benthamiana line 16C plants. Genomic DNA was extracted from the inoculated leaves at 4 dpi, and DNA fragments flanking the NbPDS target or the mGFP5 target were amplified and digested with NcoI or NdeI. Arrowheads indicate the NcoI‐ or NdeI‐resistant bands. The sizes of DNA fragments in bp are indicated at the left (for NbPDS fragments) and right (for mGFP5 fragments) of each panel. WT/D and WT/U, DNA fragments amplified from wild‐type BSMV‐infected N. benthamiana plants were digested (D) or not digested (U) with NcoI or NdeI; Ino., inoculated leaves. (H) PCR/RE analysis of ge‐BSMVγ‐mediated multiplexed genome editing in N. benthamiana leaves co‐infiltrated with the ge‐BSMVγ‐gNbPDS‐mGFP5 and Cas9 expression cassettes. Arrowheads indicate the NcoI and NdeI‐resistant bands. The sizes of the DNA fragments in bp are indicated at the left for the NbPDS fragments and right for the mGFP5 fragments in each panel.
Total RNA from the leaves shown in Fig. 2A were extracted and RT‐qPCR was conducted to analyse the corresponding mRNA levels in these leaves. The results showed that the NbPDS mRNA levels in either ge‐BSMVβ‐gNbPDS‐ or ge‐BSMVγ‐gNbPDS‐infiltrated N. benthamiana plants were similar to those of the wild‐type BSMV‐infiltrated plants, indicating that the symptoms that appeared in N. benthamiana were not due to the VIGS (Supplemental Fig. S2).
At 4 and 7 dpi, genome DNAs from the agroinfiltrated leaves were extracted, and a 544 bp DNA fragment flanking the NbPDS target was PCR amplified using the genomic DNA as the template. Because the NbPDS target contains a NcoI restriction site, editing of the NbPDS could be validated by NcoI‐digestion of the amplified PCR fragment. Gel electrophoresis of the digested products showed that a NcoI‐resistant band was present in the samples from both the ge‐BSMVβ‐gNbPDS and the ge‐BSMVγ‐gNbPDS‐infiltrated leaves (Fig. 2C), suggesting that insertions and deletions (indels) occurred within the NbPDS target gene. Sanger sequencing further revealed different indels in the NbPDS gene (Fig. 2D). To test whether the ge‐BSMV systems allowed targeted gene editing in the systemically infected leaves of the wild‐type N. benthamiana plants infiltrated with ge‐BSMVβ‐gNbPDS or ge‐BSMVγ‐gNbPDS, an Agrobacterium culture harbouring the Cas9 expression vector was infiltrated into the systemically infected leaves at 14 dpi. After an additional 4 days, the NbPDS target was analysed by a PCR/restriction enzyme (PCR/RE) assay followed by Sanger sequencing as described above. These results showed that indels also occurred at the target NbPDS site (Fig. 2E,F), indicating that both ge‐BSMVβ and ge‐BSMVγ are suitable for targeted mutagenesis in both local and systemically infected N. benthamiana leaf tissue in the presence of Cas9. The PCR/RE assay also suggested that ge‐BSMVγ had higher editing efficiency than that of ge‐BSMVβ in the systemically infected leaves (Fig. 2E), therefore the ge‐BSMVγ system was used for subsequent experiments.
We also attempted to deliver multiple gRNAs simultaneously using the ge‐BSMV system. First, Agrobacterium cultures containing pCB301‐RNAγ‐gNbPDS or pCB301‐RNAγ‐gmGFP5 were mixed and co‐infiltrated with pCB301‐BSMVα, pCB301‐BSMVβ and pHSE401 into the leaves of GFP‐transgenic N. benthamiana line 16C (Ruiz et al., 1998). PCR/RE assays showed that both the NbPDS and mGFP5 genes could be targeted simultaneously in the infiltrated leaves (Fig. 2G). Furthermore, we also tried to engineer two gRNAs in tandem with no space between the gRNAs to generate the ge‐BSMVγ‐gNbPDS‐mGFP5, and the results showed that both NbPDS and mGFP5 could be modified simultaneously in the infiltrated leaves as revealed by the presence of restriction enzyme‐resistant bands in gel images (Fig. 2H). These results indicate that ge‐BSMV‐based VIGE is able to mediate multiplex targeting in N. benthamiana inoculated leaves.
ge‐BSMV‐mediated targeted mutagenesis in wheat
As wheat is one of the natural hosts of BSMV, we sought to apply the ge‐BSMVγ system to wheat to provide additional toolkits for targeted mutagenesis in wheat. The TaGASR7 gene, which is involved in the control of grain length and weight (Zhang et al., 2016), was selected as the target (Supplemental Table S1) and the vector pCB301‐BSMVγ‐gTaGASR7 was constructed (see Supplemental File S1). The Cas9 expression cassette was first transformed into common wheat (Triticum aestivum 'Zhengmai 7698') and positive lines were confirmed by genomic PCR amplification (Supplemental Fig. S3A). Western blots were also performed to confirm the expression of the Cas9 protein in the transgenic wheat (Supplemental Fig. S3B). Nicotiana benthamiana was used as an intermediate host to recover BSMV for wheat infections as described previously (Yuan et al., 2011). For this purpose, N. benthamiana leaves were agroinfiltrated with ge‐BSMVγ‐gTaGASR7, the infiltrated leaves were harvested at 3 dpi and ground in phosphate buffer, and the leaf sap was rubbed on the leaves of Cas9‐transgenic Zhengmai7698 wheat (Fig. 3A). By 14 dpi, typical BSMV symptoms, including chlorotic spots and stripes, appeared on the upper uninoculated leaves (Fig. 3B). Western blot analyses confirmed the presence of the BSMV CP (coat protein) in the leaves (Fig. 3C, upper panel) and RT‐PCR showed that the heterologous gRNA fragment was stable during systemic movement of ge‐BSMVγ‐gTaGASR7 in wheat (Fig. 3C, bottom panel).
Figure 3.
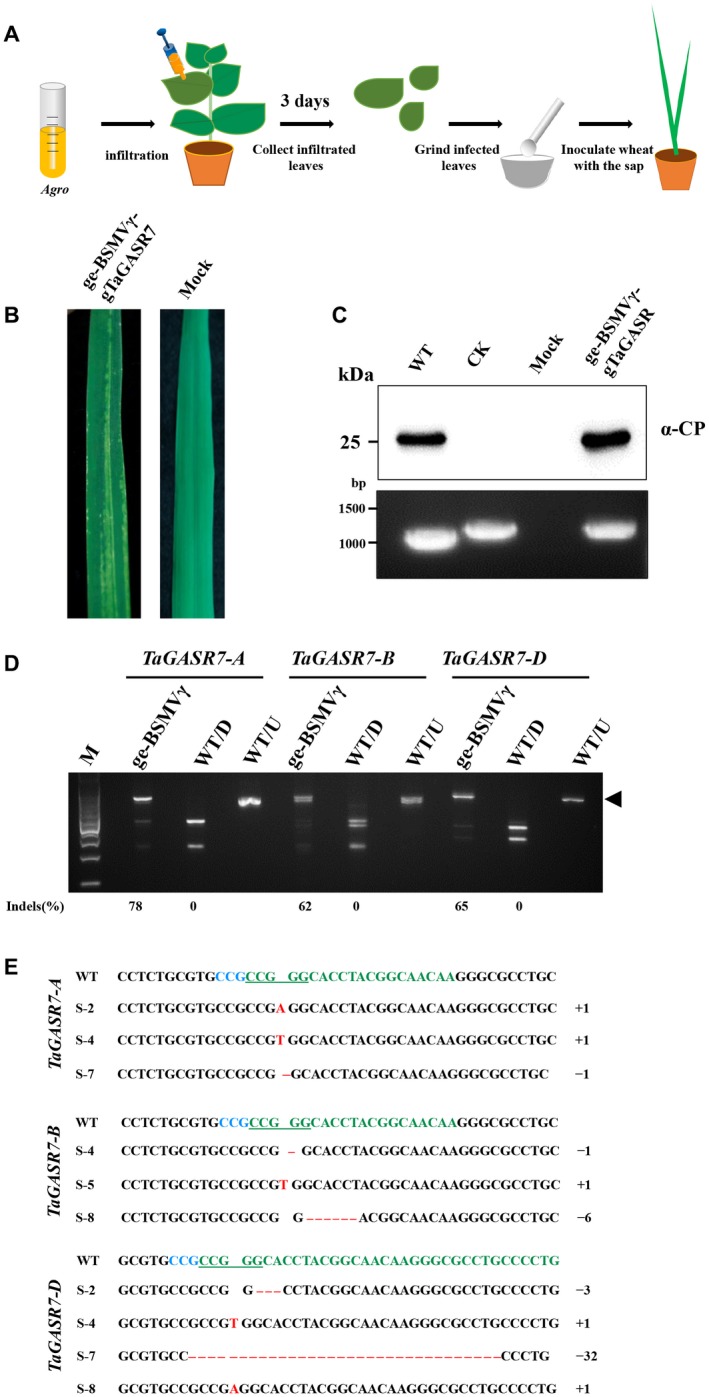
BSMV‐mediated targeted mutagenesis of TaGASR7 in wheat. (A) Workflow of ge‐BSMV‐mediated editing of target genes in wheat. Agrobacterium cultures harbouring the ge‐BSMV plasmids were infiltrated into Nicotiana benthamiana leaves, infiltrated leaves were ground at 3 days post‐infiltration (dpi) and the sap was rub‐inoculated onto leaves of Cas9‐transgenic wheat seedlings at the two‐leaf stage. (B) Symptoms of systemically infected wheat leaves at 14 dpi with ge‐BSMVγ‐gTaGASR7. Wheat seedlings rub‐inoculated with sap from empty agrobacterium‐infiltrated N. benthamiana leaves served as a control (Mock). (C) Western blot and RT‐PCR analyses of ge‐BSMVγ‐gTaGASR7 systemic infection of wheat. Sample designations are indicated above the panels. Antibodies used for western blots are indicated at the right of the panel. Molecular weight size markers in kDa or the size of DNA fragments in bp are indicated at the left side of each panel. WT, BSMV‐infected wheat leaves; CK, pCB301‐BSMVγ‐gTaGASR7 plasmid used as a positive control for evaluating the size of the RT‐PCR products. The protein loading buffer was used in the western blot controls. (D) PCR/restriction enzyme analysis of ge‐BSMVγ‐mediated genome editing of TaGASR7 in wheat. Systemically infected leaves showing typical symptoms of BSMV were collected and genomic DNA was extracted. DNA fragments flanking the wheat A, B and D genome targets were amplified and subjected to BcnI digestion. The arrowhead indicates the BcnI‐resistant bands. Mutation frequencies [indels (%)] were calculated by measuring band intensities with ImageJ software (v. 1.51k) and are shown below the corresponding lanes. WT/D and WT/U, BcnI digested (D) or undigested (U) DNA fragments amplified from wild‐type BSMV‐infected wheat. (E) Sanger sequencing of TaGASR7 target sites in DNA samples extracted from ge‐BSMVγ‐gTaGASR7 infected leaves at 20 dpi. DNA fragments derived from different wheat genomes are indicated on the left. The protospacer is shown in green, the PAM (NGG) motif is in blue, underlined nucleotides indicate the BcnI recognition site, indels are shown in red with dots indicating deleted nucleotides and red letters showing inserted nucleotides. The numbers on the right indicate how many nucleotides are deleted (−) or inserted (+) by BSMV‐mediated genome editing in the TaGASR7 target site.
About 3 weeks post‐inoculation, genomic DNA was extracted from leaves with symptoms and a DNA fragment flanking the target was amplified from the A, B and D wheat genomes. PCR/RE assays revealed indels in all of the A, B, D genome targets with mutation efficiencies of up to 78% (Fig. 3D). Sanger sequencing further confirmed that diverse TaGASR7 indels were present in the A, B, and D genomes (Fig. 3E and Supplemental Fig. S4). In addition, TaGASR7 mRNA accumulation levels in the leaves shown in Fig. 3B were analysed by RT‐qPCR. These results revealed similar TaGASR7 mRNA levels in both the ge‐BSMVγ‐gTaGASR7‐ and mock‐inoculated wheat, excluding the possibility that these symptoms resulted from VIGS (Supplemental Fig. S5). Altogether, these results demonstrate that the ge‐BSMV system can direct efficient targeted mutagenesis in common wheat.
ge‐BSMV‐mediated targeted mutagenesis in maize
We attempted to apply the ge‐BSMV system to maize, another agriculturally important crop. The Cas9 expression cassette was transformed into maize (Zea mays subsp. mays 'B73‐329') and positive lines were confirmed by genomic PCR (Supplemental Fig. S6A). Western blots were also performed to confirm the expression of Cas9 protein in the transgenic maize (Supplemental Fig. S6B). The ZmTMS5 (thermosensitive genic male‐sterile 5) gene involved in pollen fertility (Li et al., 2017) was selected as the target (Supplemental Table S1), Cas9‐transgenic maize leaves were rub‐inoculated with a mixture of the in vitro transcripts of RNAα, RNAβ and RNAγ‐gZmTMS5 (see Supplemental File S1) derived from BSMV Xinjiang (BSMVXJ), a maize‐infecting strain (Hu et al., 2015). Large‐scale chlorosis and stripes appeared on the newly emerged leaves (Fig. 4B) and western blot analysis revealed the presence BSMV CP in the upper uninoculated leaves (Fig. 4C, upper panel). RT‐PCR analysis further confirmed the maintenance of heterologous gRNA fragments during systemic infection of ge‐BSMVγ‐gZmTMS5 in maize (Fig. 4C, bottom panel).
Figure 4.
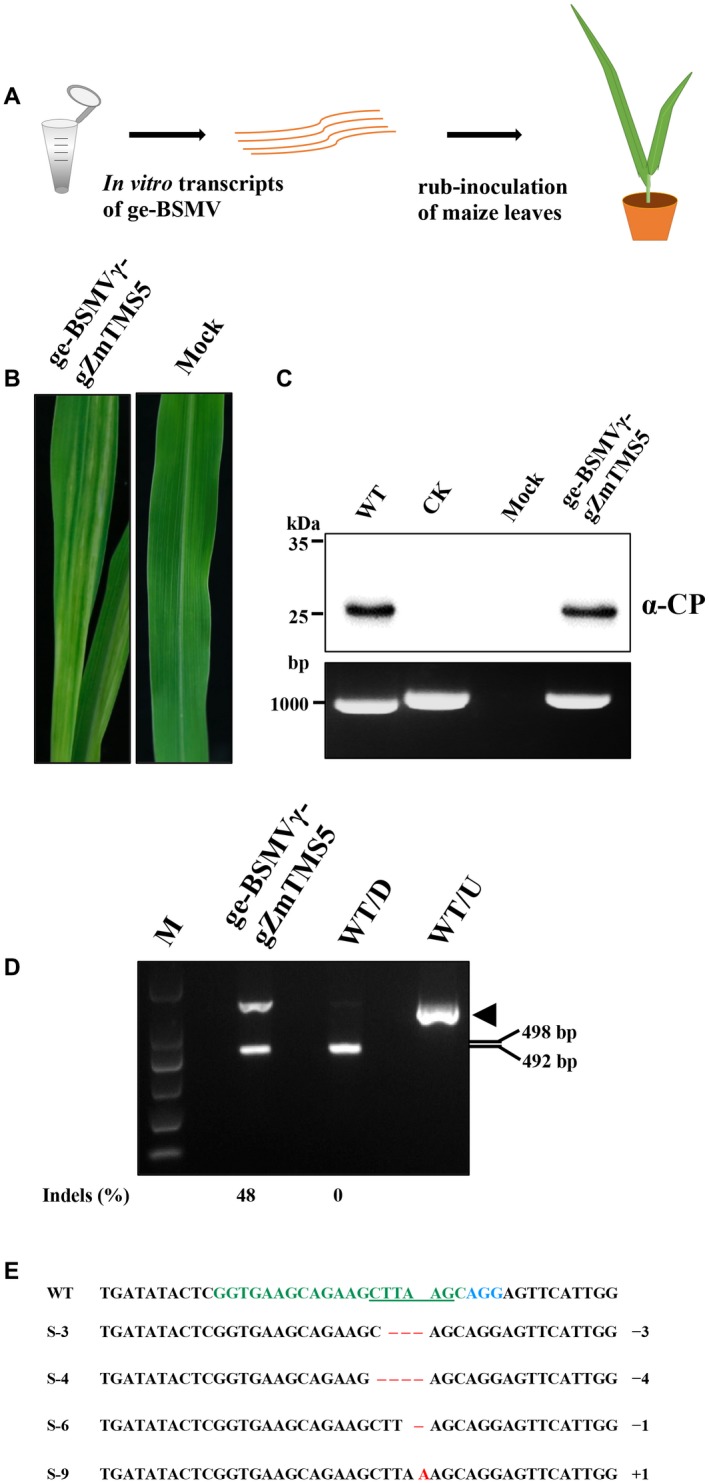
BSMV‐mediated targeted mutagenesis of ZmTMS5 in maize. (A) Workflow of ge‐BSMV‐mediated targeted gene editing in maize. In vitro‐transcripts of pT7‐αXJ, pT7‐βXJ and pT7‐γ‐gZmTMS5 were rub‐inoculated onto Cas9‐transgenic maize seedlings at the two‐leaf stage. (B) Symptoms of maize infected with ge‐BSMVγ‐gZmTMS5. Controls were FES buffer‐inoculated maize (Mock). (C) Western blot and RT‐PCR analysis of ge‐BSMVγ‐gZmTMS5 infection in the systemically infected maize leaves. Sample names are indicated above the panels. Antibodies used for western blots are indicated at the right of the panel. The molecular weight size markers in kDa or the sizes of DNA fragments in bp are indicated at the left side of each panel. WT, wild‐type BSMV‐infected maize leaves; CK, the pT7‐γ‐gZmTMS5 plasmid was used as a positive control for evaluating the size of the RT‐PCR products. The protein loading buffer alone was used as a control in western blot analyses. (D) PCR/restriction enzyme analysis of ge‐BSMVγ‐mediated genome editing of ZmTMS5 in systemically infected maize leaves at 24 days post‐inoculation (dpi). DNA fragments flanking the target site were amplified and subjected to AflII digestion. The arrowhead indicates the AflII‐resistant band (Note: due to size similarities, two of the digested DNA fragments appeared as one band in the gel). Mutation frequencies [indels (%)] were calculated by measuring band intensities with ImageJ software (v. 1.51k) and are shown below the corresponding lanes. WT/D and WT/U, DNA fragments amplified from wild‐type BSMV‐infected maize plants were digested (D) or not digested (U) with AflII. (E) Sanger sequencing analysis of the ZmTMS5 target site in DNA of leaf samples infected with ge‐BSMVγ‐gZmTMS5 at 24 dpi. The protospacer is shown in green, the PAM (NGG) motif is in blue, underlined nucleotides indicate the AflII recognition site, indels are shown in red with the dots indicating deleted nucleotides and red letters specifying inserted nucleotides. The numbers on the right indicate how many nucleotides were deleted (−) or inserted (+) in the ZmTMS5 target site by BSMV‐mediated genome editing.
About 3 weeks post‐inoculation, genomic DNA was extracted from systemically infected leaves exhibiting typical symptoms. A 994 bp DNA fragment flanking the ZmTMS5 target site was then PCR amplified by using the extracted DNA as the template and a PCR/RE assay was performed to evaluate the mutation rates within the ZmTMS5 target site. The results showed that an AflII‐resistant band was present only in the samples from ge‐BSMVγ‐gZmTMS5‐inoculated maize, but not from wild‐type BSMV infected maize (Fig. 4D), suggesting that indels occurred within the targeted gene ZmTMS5. Moreover, quantitative analyses of the AflII‐digested products indicated mutation efficiencies of up to 48% (Fig. 4D). Sanger sequencing further revealed diverse indels within the ZmTMS5 target site (Fig. 4E and Supplemental Fig. S7). These results indicate that ge‐BSMV‐based VIGE results in efficient targeted mutagenesis in maize.
Discussion
Currently, four RNA viruses have been developed as VIGE vectors. Two tobraviruses, TRV and PEBV, can be used to edit N. benthamiana and Arabidopsis genes of interest (Ali et al., 2015, 2018). TMV was also reported to be able to mediate target gene editing in the local leaves by partially substituting the CP ORF with a gRNA (Cody et al., 2017). Very recently, BNYVV has also been used for gRNA delivery in N. benthamiana (Jiang et al., 2019). BSMV‐based VIGE described above provides an alternative tool for targeted gene editing in the model plant N. benthamiana. Moreover, the broad host range of BSMV, especially its ability to infect many economically important monocots, enables its potential use as a VIGE tool in several crop plants that TRV, PEBV and TMV are unable to infect.
WDV replicons have been used for genome editing in wheat (Gil‐Humanes et al., 2017), but compared to WDV replicons, BSMV‐based VIGE does not require integration of the viral element into the wheat genome, which avoids raising additional regulatory and ethical issues. Although in vitro transcripts of CRISPR/Cas9 RNA (Zhang et al., 2016) and CRISPR/Cas9 RNP (Liang et al., 2017) were used to generate transgene‐free genome editing in wheat, the ratio of positive offspring with targeted mutations is relatively low due to rapid degradation of introduced gRNA by cellular nucleases (Woo et al., 2015). In contrast, the ge‐BSMV‐based VIGE tools used in our study showed high efficiencies of targeted gene editing in the leaves of wheat, as illustrated by the large number of gRNA‐containing fragments accompanying BSMV replication, and the protection of gRNA‐containing fragments by viral proteins such as γb silencing suppressor (Bragg et al., 2004; Yelina et al., 2002; Zhang et al., 2017).
To test the inheritance of BSMV‐mediated gene editing in the N. benthamiana leaves, we regenerated plants from leaf tissues containing the edited NbPDS gene, and several regenerated albino plants were obtained (Supplemental Fig. S8), suggesting that the edited gene can be passed to subsequent generations. Intriguingly, it has been reported that maize seedlings can be regenerated from leaf segments (Lowe et al., 2016; Yu et al., 2012). Therefore, our BSMV‐based gene editing system may provide an alternative approach to obtain genetically modified maize, which can bypass immature embryo transformation and regeneration processes. Furthermore, BSMV is capable of entering the developing ovules and embryo of barley as well as sperm and vegetative cells of barley pollen (Carroll, 1969, 1974; Carroll and Mayhew, 1976) to result in seed transmissibility of BSMV (Carroll, 1972). Unfortunately, to the best of our knowledge, seed transmission of BSMV in wheat has not been evaluated. Therefore, studies of appropriate cultivars in which the BSMVND isolate or other BSMV strains can be seed‐transmitted with high efficiency might increase the feasibility of using seed‐borne approaches for VIGE genome editing in wheat.
In addition to wheat and maize, some BSMV strains are able to infect barley (Hordeum vulgare) (Jackson et al., 2009), Brachypodium distachyon (Demircan and Akkaya, 2010; Pacak et al., 2010), oat (Pacak et al., 2010), millet (Setaria italic) (unpublished results), sorghum (Sorghum bicolor) (unpublished results) and several other cereals. Our proof‐of‐concept study of BSMV‐based VIGE in N. benthamiana, wheat and maize provides the basis for expanded use in other economically important monocot plants.
Experimental Procedures
Vector construction
To construct the pCB301‐BSMVα, pCB301‐BSMVβ and pCB301‐BSMVγ, plasmids BSMV RNAα, RNAβ and RNAγ were amplified from pT7‐αND, pT7‐βND and pT7‐γND (Petty et al., 1989), respectively, then cloned into StuI‐ and BamHI‐digested pCB301‐2X35S‐MCS‐HDVRZNOS (Yao et al., 2011) using a Seamless Assembly Cloning Kit (Clone Smarter Technologies Inc., Houston, USA) according to the manufacturer’s instructions. To construct the pCB301‐BSMVβ‐gNbPDS plasmid, nt 74–393 of the BSMV CP ORF were deleted by reverse PCR using pCB301‐BSMVβ as the template, the sequence encoding the gRNA scaffold was synthesized by GENEWIZ Inc. (GENEWIZ, Inc., South Plainfield, NJ, USA), and the gRNA scaffold sequence along with seven extra downstream T residues was then amplified and cloned into the pCB301‐BSMVβΔ74‐393 using the Seamless Assembly Cloning Kit (Clone Smarter Technologies Inc., Houston, USA). The resulting plasmid was linearized by a second round of reverse PCR, and a 192 bp sgRNAγ promoter corresponding to nt 1864–2055 of BSMV RNAγ was amplified from pCB301‐BSMVγ and cloned into the linearized vector using the same strategy as described above. The 20 bp spacer targeting the NbPDS gene sequence was inserted between the sgRNAγ promoter sequence and the sequence encoding the gRNA scaffold by a third round of reverse PCR to generate the pCB301‐BSMVβ‐gNbPDS derivative. To construct the pCB301‐BSMVγ‐gNbPDS, reverse PCR was performed to introduce NcoI, MluI, SpeI and ApaI restriction sites into the downstream γb ORF sequence to generate the plasmid pCB301‐BSMVγ‐MCS (see Supplemental File S1), and the gRNA sequence was amplified from pCB301‐BSMVβ‐gNbPDS and cloned into pCB301‐BSMVγ‐MCS via the NcoI and SpeI restriction enzyme sites.
For easy manipulation of the plasmids, a pair of reversely arranged SapI restriction enzyme sites was inserted downstream of the γb ORF by reverse PCR to generate the plasmid pCB301‐BSMVγ‐SapI (see Supplemental File S1). SmR gene was amplified from pHSE401 (Xing et al., 2014) and inserted between the two SapI sites using the Seamless Assembly Cloning Kit (Clone Smarter Technologies Inc., Houston, USA), resulting in the pCB301‐BSMVγ‐SmR. To construct the pCB301‐BSMVγ‐gTaGASR7, pCB301‐BSMVγ‐SmR was digested by SapI, ligated with annealed oligos with a 5′‐CTA and a 3′‐CAA overhangs. Oligos annealing was conducted according to previously described methods (Shan et al., 2014) with minor modifications. Briefly, a pair of 100 μM oligos were 5′ phosphorylated using T4 polynucleotide kinase (Thermo Fisher Scientific, Inc., Pittsburgh, PA, USA) and annealed in the thermal cycler using the following steps: denaturation at 95 °C for 5 min and gradually chilled to 25 °C with 1 °C drops per minute. A final step was performed by maintaining the samples at 16 °C for 10 min.
To construct the RNAγ‐gZmTMS5 vector for in vitro transcription, a pCB301‐BSMVγ‐gZmTMS5 plasmid targeting the ZmTMS5 gene was constructed as described above using oligos ZmTMS5‐T2‐F/R. The gRNA sequence was amplified from the pCB301‐BSMVγ‐gZmTMS5 and cloned into pT7‐γXJ (Hu et al., 2015) downstream of the γb stop codon to generate the pT7‐γ‐gZmTMS5 using the Seamless Assembly Cloning Kit (Clone Smarter Technologies Inc., Houston, USA).
To construct the RNAγ‐gNbPDS‐mGFP5, four primers, Fb1, F1, R2 and Rb2, were used to amplify a DNA fragment containing the two different gRNA sequences using pCB301‐BSMVγ‐SmR as the template. The concentration of primers F2 and R2 used for PCR was 1/20 of the Fb1 and Rb2 concentrations. The PCR products were then cloned into SapI‐digested pCB301‐BSMVγ‐SmR by using the seamless cloning procedure described above.
All of the primers used in this study were listed in Supplemental Table S2 and DNA sequencing was conducted to confirm the correctness of these plasmids. The sequence information of the BSMV derivatives used in this study can be found in Supplemental File S1.
Plant growth conditions
Wheat, maize and N. benthamiana plants were grown in a greenhouse under a 14/10 h light/dark photoperiod at 23–25 °C as previously described (Yuan et al., 2011). Wheat and maize seedlings were grown to the two‐leaf stage prior to rub‐inoculation, and inoculated plants were maintained in the same climate chamber as described above (Hu et al., 2015).
Transformation of the Cas9 expression cassette into wheat and maize
To construct Cas9‐transgenic wheat, the pCXUN‐Cas9 plasmid was transformed into calli of common wheat (T. aestivum 'Zhengmai 7698') by particle bombardment following the protocol described previously (Sun et al., 2019).
To create Cas9‐transgenic maize, the Cas9 expression cassette was cloned into the pCAMBIA3301 binary vector followed by transformation into A. tumefaciens strain EHA105. Agrobacterium tumefaciens‐mediated transformation of maize was carried out as described previously (Zhu et al., 2016).
Agroinfiltration and inoculation of plants with in vitro synthesized RNAs
The pCB301‐BSMVα, pCB301‐BSMVβ, pCB301‐BSMVγ, pCB301‐BSMVβ‐gRNA and pCB301‐BSMVγ‐gRNA plasmids were transformed into A. tumefaciens EHA105 strains for agroinfiltration of N. benthamiana leaves. Equal volumes of Agrobacterium strains harbouring individual plasmids were mixed to a final OD600 of 0.3 and infiltrated into leaves of 3‐ to 4‐week‐old N. benthamiana plants as described previously (Yuan et al., 2011). Agrobacterium harbouring the Cas9 expression construct was infiltrated at a final OD600 of 0.5.
For maize, the BSMV Xinjiang strain (BSMVXJ) was used for inoculation. In vitro RNA transcripts were mechanically inoculated onto two‐leaf stage maize seedlings as described previously (Hu et al., 2015; Lee et al., 2012). Briefly, in vitro transcripts from pT7‐αXJ, pT7‐βXJ and pT7‐γXJ (or pT7‐γ‐gZmTMS5) were mixed at a molar ratio of 1:1:1. These transcripts were then mixed with equal volumes of FES buffer (0.06 M potassium phosphate, 0.1 M glycine, 1% bentonite, 1% sodium pyrophosphate decahydrate, 1% celite, pH 8.5) followed by rub‐inoculation of maize leaves.
PCR/RE assay and Sanger sequencing
Nicotiana benthamiana, wheat and maize genomic DNA was extracted using the cetyltrimethylammonium bromide (CTAB) method as previously described (Doyle and Doyle, 1987). PCR/RE assay was performed according to previously described methods with minor modifications (Shan et al., 2014). Briefly, four to eight pieces of leaves from different inoculated plants were harvested for genomic DNA extraction, and a DNA fragment flanking the target site was amplified using the extracted genomic DNA as a template. The PCR products (200–300 ng) were digested with restriction enzymes corresponding to the target site followed by gel electrophoresis. For Sanger sequencing, 200–300 ng of PCR products were digested with corresponding restriction enzymes, the digested products were TOPO‐cloned into the easy vector using pClone007 Blunt Simple Vector Kit (Cat No. TSV‐007BS, Tsingke, Beijing, China), and 6–10 positive clones were sequenced by GENEWIZ Inc (GENEWIZ, Inc., South Plainfield, NJ, USA).
RT‐qPCR and statistical analysis
RT‐qPCR was performed as described previously with minor modifications (Liu et al., 2012; Zhang et al., 2013) Briefly, total RNAs were extracted and quantified by a NanoDrop ND‐1000 (Thermo Fisher Scientific, Inc., Pittsburgh, PA, USA). For RT‐qPCR, cDNA was synthesized from 2 μg of DNase‐treated total RNA using an oligo‐dT primer and M‐MLV reverse transcriptase (Promega, Madison, WI, USA). The gene fragments were amplified using 2 × SsoFast EvaGreen Supermix (Bio‐Rad, Laboratories, Inc., Hercules, CA, USA) with corresponding primers listed in Supplemental Table S2. The expression levels were normalized by reference gene PP2A for NbPDS or EF1α for TaGASR7. Values indicate mean ± SD. For NbPDS, the quantified data in each panel were subjected to statistical analysis using SPSS software (v. 22.0, IBM) and the data were compared using one‐way analysis of variance (ANOVA). Significant differences in NbPDS mRNA accumulation were determined by Duncan’s multiple range test. For TaGASR7, significant difference was analysed using Student’s t‐test. Values are mean ± SD (ns, not significant, n = 3).
Conflict of Interests
The authors declare no conflict of interests.
Supporting information
Fig. S1 Western blot analysis of transiently expressed Cas9 in N. benthamiana.
Fig. S2 RT‐qPCR analysis of NbPDS mRNA level in ge‐BSMV infected N. benthamiana leaves.
Fig. S3 Genomic PCR (A) and western blot analysis (B) of Cas9‐transgenic wheat. For western blot analysis, the Cas9‐specific monoclonal antibody (Cat No.#14697, Cell Signaling Technology, Massachusetts, USA) was used at a 1:1000 dilution.
Fig. S4 Representatives of Sanger sequencing chromatograms of indels from systemically infected wheat leaves. Black lines under the sequence indicates the target site. ‐RC, reverse complement sequence.
Fig. S5 RT‐qPCR analysis of TaGASR7 mRNA levels in ge‐BSMV infected wheat leaves. Data are represented as means ± SD (Student’s t‐test; ns, not significant; n = 3).
Fig. S6 Genomic PCR (A) and western blot analysis (B) of the Cas9‐transgenic maize. For western blot analysis, Cas9 monoclonal antibody (Cat No.#14697, Cell Signaling Technology) was used with 1:1000 dilution.
Fig. S7 Representative Sanger sequencing chromatograms showing indels from systemically infected maize leaves. Black lines under the sequence indicates the target site. ‐RC, reverse complement sequence.
Fig. S8 Regeneration of Nicotiana benthamiana leaf segments containing the edited NbPDS gene. ge‐BMVSγ‐gNbPDS‐infected systemically infected leaves of N. benthamiana were harvested at 30 days post‐inoculation (dpi), surface sterilized with 2.0–2.5% sodium hypochlorite, rinsed three times with sterile water, cut into c.2 cm² pieces and placed on medium plates (MS medium, 30.0 g/L sucrose, 1.0 mg/L zeatin, 2.0 mg/L kinetin, 1.0 mg/L indole‐3‐acetic acid, 350.0 mg/L carbenicillin, 4.0 g/L phytagel, pH 5.8) for differentiation under controlled conditions (23 °C, 16 h photoperiod). After 4–6 weeks, shoots with bleaching phenotypes were excised, transferred to rooting medium plates (1/3× MS medium, 7.0 g/L glucose, 3.0 g/L sucrose, 100.0 mg/L carbenicillin, 4.0 g/L phytagel, pH 5.8) and incubated at 23 °C. Representative N. benthamiana shoots regenerated from the leaf segments were photographed. Boxed regions in the left panels were magnified to show the regenerated albino plants.
Table S1 Cas9 targets selected in this study.
Table S2 Primers used in this study.
File S1 Sequences of RNAβ‐gNbPDS, RNAγ‐gNbPDS, RNAγ‐gTaGASR7, RNAγ‐gZmTMS5, RNAγ‐gNbPDS‐mGFP5, RNAγ‐SmR and pCB301‐BSMVγ‐MCS. Sequences in green indicate the sgRNAγ promoter, whereas sequences in red, blue and purple represent the gRNA spacer, the scaffold and the SmR gene, respectively. The underlined sequences indicate the SapI recognition sites or the multiple cloning sites.
Acknowledgements
We would like to thank Drs Xianbing Wang, Jialin Yu, Chenggui Han and Ying Wang at China Agricultural University for comments and constructive criticism. We thank Dr Andrew O. Jackson, University of California‐Berkeley, for editing the manuscript, Dr Yule Liu, Tsinghua University, for providing Cas9‐transgenic N. benthamiana seeds and Dr Qijun Chen, China Agricultural University, for the pHSE401 plasmid. We also thank Dr Daowen Wang, Chinese Academy of Sciences, Dr Huanbin Zhou, Chinese Academy of Agricultural Sciences, and Dr Yi Li, Peking University, for helpful suggestions. This work is supported by the National Key R & D Program of China (2016YFD0100502), the Transgenic Research Program of China (2016ZX08010‐001) and the Project for Extramural Scientists of SKLAB (2019SKLAB1‐14).
References
- Ali, Z. , Abul‐faraj, A. , Li, L. , Ghosh, N. , Piatek, M. , Mahjoub, A. , Aouida, M. , Piatek, A. , Baltes, N.J. , Voytas, D.F. , Dinesh‐Kumar, S. and Mahfouz, M.M. (2015) Efficient virus‐mediated genome editing in plants using the CRISPR/Cas9 system. Mol. Plant, 8, 1288–1291. [DOI] [PubMed] [Google Scholar]
- Ali, Z. , Eid, A. , Ali, S. and Mahfouz, M.M. (2018) Pea early‐browning virus‐mediated genome editing via the CRISPR/Cas9 system in Nicotiana benthamiana and Arabidopsis . Virus Res. 244, 333–337. [DOI] [PubMed] [Google Scholar]
- Bhaya, D. , Davison, M. and Barrangou, R. (2011) CRISPR‐Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 45, 273–297. [DOI] [PubMed] [Google Scholar]
- Bragg, J.N. , Lawrence, D.M. and Jackson, A.O. (2004) The N‐terminal 85 amino acids of the Barley stripe mosaic virus γb pathogenesis protein contain three zinc‐binding motifs. J. Virol. 78, 7379–7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, N.M. , Baltes, N.J. , Voytas, D.F. and Douches, D.S. (2016) Geminivirus‐mediated genome editing in potato (Solanum tuberosum L.) using sequence‐specific nucleases. Front. Plant Sci. 7, 1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, T.W. (1969) Electron microscopic evidence for the presence of Barley stripe mosaic virus in cells of barley embryos. Virology, 37, 649–657. [DOI] [PubMed] [Google Scholar]
- Carroll, T.W. (1972) Seed transmissibility of two strains of Barley stripe mosaic virus . Virology, 48, 323–336. [DOI] [PubMed] [Google Scholar]
- Carroll, T.W. (1974) Barley stripe mosaic virus in sperm and vegetative cells of barley pollen. Virology, 60, 21–28. [DOI] [PubMed] [Google Scholar]
- Carroll, T.W. and Mayhew, D.E. (1976) Occurrence of virions in developing ovules and embryo sacs of barley in relation to the seed transmissibility of Barley stripe mosaic virus . Can. J. Bot. 54, 2497–2512. [Google Scholar]
- Cody, W.B. , Scholthof, H.B. and Mirkov, T.E. (2017) Multiplexed gene editing and protein overexpression using a Tobacco mosaic virus viral vector. Plant Physiol. 175, 23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan‐Meir, T. , Filler‐Hayut, S. , Melamed‐Bessudo, C. , Bocobza, S. , Czosnek, H. , Aharoni, A. and Levy, A.A. (2018) Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant J. 95, 5–16. [DOI] [PubMed] [Google Scholar]
- Demircan, T. and Akkaya, M. (2010) Virus induced gene silencing in Brachypodium distachyon, a model organism for cereals. Plant Cell, Tissue Organ Cult. 100, 91–96. [Google Scholar]
- Doyle, J. and Doyle, J. (1987) Genomic plant DNA preparation from fresh tissue‐CTAB method. Phytochem. Bull. 19, 11–15. [Google Scholar]
- Gil‐Humanes, J. , Wang, Y. , Liang, Z. , Shan, Q. , Ozuna, C.V. , Sanchez‐Leon, S. , Baltes, N.J. , Starker, C. , Barro, F. , Gao, C. and Voytas, D.F. (2017) High‐efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. 89, 1251–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y. , Li, Z. , Yuan, C. , Jin, X. , Yan, L. , Zhao, X. , Zhang, Y. , Jackson, A.O. , Wang, X. , Han, C. , Yu, J. and Li, D. (2015) Phosphorylation of TGB1 by protein kinase CK2 promotes Barley stripe mosaic virus movement in monocots and dicots. J. Exp. Bot. 66, 4733–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, A.O. , Lim, H.S. , Bragg, J. , Ganesan, U. and Lee, M.Y. (2009) Hordeivirus replication, movement, and pathogenesis. Annu. Rev. Phytopathol. 47, 385–422. [DOI] [PubMed] [Google Scholar]
- Jiang, N. , Zhang, C. , Liu, J.Y. , Guo, Z.H. , Zhang, Z.Y. , Han, C.G. and Wang, Y. (2019) Development of Beet necrotic yellow vein virus‐based vectors for multiple‐gene expression and guide RNA delivery in plant genome editing. Plant Biotechnol. J. 17, 1302–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek, M. , Chylinski, K. , Fonfara, I. , Hauer, M. , Doudna, J.A. and Charpentier, E. (2012) A programmable dual‐RNA‐guided DNA endonuclease in adaptive bacterial immunity. Science, 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, J.A. , Bragg, J.N. , Lawrence, D.M. and Jackson, A.O. (2003) Sequence elements controlling expression of Barley stripe mosaic virus subgenomic RNAs in vivo. Virology, 313, 66–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M.Y. , Yan, L. , Gorter, F.A. , Kim, B.Y. , Cui, Y. , Hu, Y. , Yuan, C. , Grindheim, J. , Ganesan, U. , Liu, Z. , Han, C. , Yu, J. , Li, D. and Jackson, A.O. (2012) Brachypodium distachyon line Bd3‐1 resistance is elicited by the Barley stripe mosaic virus triple gene block 1 movement protein. J. Gen. Virol. 93, 2729–2739. [DOI] [PubMed] [Google Scholar]
- Li, J. , Zhang, H. , Si, X. , Tian, Y. , Chen, K. , Liu, J. , Chen, H. and Gao, C. (2017) Generation of thermosensitive male‐sterile maize by targeted knockout of the ZmTMS5 gene. J. Genet. Genom. 44, 465–468. [DOI] [PubMed] [Google Scholar]
- Liang, Z. , Chen, K. , Li, T. , Zhang, Y. , Wang, Y. , Zhao, Q. , Liu, J. , Zhang, H. , Liu, C. , Ran, Y. and Gao, C. (2017) Efficient DNA‐free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun. 8, 14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, D. , Shi, L. , Han, C. , Yu, J. , Li, D. and Zhang, Y. (2012) Validation of reference genes for gene expression studies in virus‐infected Nicotiana benthamiana using quantitative real‐time PCR. PLoS One, 7, e46451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, K. , Wu, E. , Wang, N. , Hoerster, G. , Hastings, C. , Cho, M.J. , Scelonge, C. , Lenderts, B. , Chamberlin, M. , Cushatt, J. , Wang, L. , Ryan, L. , Khan, T. , Chow‐Yiu, J. , Hua, W. , Yu, M. , Banh, J. , Bao, Z. , Brink, K. , Igo, E. , Rudrappa, B. , Shamseer, P.M. , Bruce, W. , Newman, L. , Shen, B. , Zheng, P. , Bidney, D. , Falco, C. , Register, J. , Zhao, Z.Y. , Xu, D. , Jones, T. and Gordon‐Kamm, W. (2016) Morphogenic regulators Baby Boom and Wuschel improve monocot transformation. Plant Cell, 28, 1998–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacak, A. , Geisler, K. , Jorgensen, B. , Barciszewska‐Pacak, M. , Nilsson, L. , Nielsen, T. , Johansen, E. , Gronlund, M. , Jakobsen, I. and Albrechtsen, M. (2010) Investigations of Barley stripe mosaic virus as a gene silencing vector in barley roots and in Brachypodium distachyon and oat. Plant Methods, 6, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petty, I.T.D. , Hunter, B.G. , Wei, N. and Jackson, A.O. (1989) Infectious Barley stripe mosaic virus RNA transcribed in vitro from full‐length genomic cDNA clones. Virology, 171, 342–349. [DOI] [PubMed] [Google Scholar]
- Ruiz, M.T. , Voinnet, O. and Baulcombe, D.C. (1998) Initiation and maintenance of virus‐induced gene silencing. Plant Cell, 10, 937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, Q. , Wang, Y. , Li, J. and Gao, C. (2014) Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 9, 2395–2410. [DOI] [PubMed] [Google Scholar]
- Sun, Y. , Zhang, X. , Wu, C. , He, Y. , Ma, Y. , Hou, H. , Guo, X. , Du, W. , Zhao, Y. and Xia, L. (2016) Engineering herbicide‐resistant rice plants through CRISPR/Cas9‐mediated homologous recombination of acetolactate synthase. Mol. Plant, 9, 628–631. [DOI] [PubMed] [Google Scholar]
- Sun, Y. , Sparks, C. , Jones, H. , Riley, M. , Francis, F. , Du, W. and Xia, L. (2019) Silencing an essential gene involved in infestation and digestion in grain aphid through plant‐mediated RNA interference generates aphid‐resistant wheat plants. Plant Biotechnol. J. 17, 852–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M. , Lu, Y. , Botella, J.R. , Mao, Y. , Hua, K. and Zhu, J.K. (2017) Gene targeting by homology‐directed repair in rice using a geminivirus‐based CRISPR/Cas9 System. Mol. Plant, 10, 1007–1010. [DOI] [PubMed] [Google Scholar]
- Woo, J.W. , Kim, J. , Kwon, S.I. , Corvalan, C. , Cho, S.W. , Kim, H. , Kim, S.G. , Kim, S.T. , Choe, S. and Kim, J.S. (2015) DNA‐free genome editing in plants with preassembled CRISPR‐Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164. [DOI] [PubMed] [Google Scholar]
- Xing, H.L. , Dong, L. , Wang, Z.P. , Zhang, H.Y. , Han, C.Y. , Liu, B. , Wang, X.C. and Chen, Q.J. (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, M. , Zhang, T. , Tian, Z. , Wang, Y. and Tao, X. (2011) Construction of Agrobacterium‐mediated Cucumber mosaic virus infectious cDNA clones and 2b deletion viral vector. Sci. Agric. Sin. 44, 3060–3068. [Google Scholar]
- Yelina, N.E. , Savenkov, E.I. , Solovyev, A.G. , Morozov, S.Y. and Valkonen, J.P.T. (2002) Long‐distance movement, virulence, and RNA silencing suppression controlled by a single protein in Hordei‐ and Potyviruses: complementary functions between virus families. J. Virol. 76, 12981–12991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, K. , Han, T. , Liu, G. , Chen, T. , Wang, Y. , Yu, A.Y. and Liu, Y. (2015) A geminivirus‐based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Sci. Rep. 5, 14926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H. , Wang, W. , Wang, Y. and Hou, B. (2012) High frequency wheat regeneration from leaf tissue explants of regenerated plantlets. Adv. Biosci. Biotechnol. 3, 46–50. [Google Scholar]
- Yuan, C. , Li, C. , Yan, L. , Jackson, A.O. , Liu, Z. , Han, C. , Yu, J. and Li, D. (2011) A high throughput Barley stripe mosaic virus vector for virus induced gene silencing in monocots and dicots. PLoS One, 6, e26468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, K. , Niu, S. , Di, D. , Shi, L. , Liu, D. , Cao, X. , Miao, H. , Wang, X. , Han, C. , Yu, J. , Li, D. and Zhang, Y. (2013) Selection of reference genes for gene expression studies in virus‐infected monocots using quantitative real‐time PCR. J. Biotechnol. 168, 7–14. [DOI] [PubMed] [Google Scholar]
- Zhang, Y. , Liang, Z. , Zong, Y. , Wang, Y. , Liu, J. , Chen, K. , Qiu, J.L. and Gao, C. (2016) Efficient and transgene‐free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7, 12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, K. , Zhang, Y. , Yang, M. , Liu, S. , Li, Z. , Wang, X. , Han, C. , Yu, J. and Li, D. (2017) The Barley stripe mosaic virus γb protein promotes chloroplast‐targeted replication by enhancing unwinding of RNA duplexes. PLoS Pathog. 13, e1006319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, J. , Song, N. , Sun, S. , Yang, W. , Zhao, H. , Song, W. and Lai, J. (2016) Efficiency and inheritance of targeted mutagenesis in maize using CRISPR‐Cas9. J. Genet. Genom. 43, 25–36. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Western blot analysis of transiently expressed Cas9 in N. benthamiana.
Fig. S2 RT‐qPCR analysis of NbPDS mRNA level in ge‐BSMV infected N. benthamiana leaves.
Fig. S3 Genomic PCR (A) and western blot analysis (B) of Cas9‐transgenic wheat. For western blot analysis, the Cas9‐specific monoclonal antibody (Cat No.#14697, Cell Signaling Technology, Massachusetts, USA) was used at a 1:1000 dilution.
Fig. S4 Representatives of Sanger sequencing chromatograms of indels from systemically infected wheat leaves. Black lines under the sequence indicates the target site. ‐RC, reverse complement sequence.
Fig. S5 RT‐qPCR analysis of TaGASR7 mRNA levels in ge‐BSMV infected wheat leaves. Data are represented as means ± SD (Student’s t‐test; ns, not significant; n = 3).
Fig. S6 Genomic PCR (A) and western blot analysis (B) of the Cas9‐transgenic maize. For western blot analysis, Cas9 monoclonal antibody (Cat No.#14697, Cell Signaling Technology) was used with 1:1000 dilution.
Fig. S7 Representative Sanger sequencing chromatograms showing indels from systemically infected maize leaves. Black lines under the sequence indicates the target site. ‐RC, reverse complement sequence.
Fig. S8 Regeneration of Nicotiana benthamiana leaf segments containing the edited NbPDS gene. ge‐BMVSγ‐gNbPDS‐infected systemically infected leaves of N. benthamiana were harvested at 30 days post‐inoculation (dpi), surface sterilized with 2.0–2.5% sodium hypochlorite, rinsed three times with sterile water, cut into c.2 cm² pieces and placed on medium plates (MS medium, 30.0 g/L sucrose, 1.0 mg/L zeatin, 2.0 mg/L kinetin, 1.0 mg/L indole‐3‐acetic acid, 350.0 mg/L carbenicillin, 4.0 g/L phytagel, pH 5.8) for differentiation under controlled conditions (23 °C, 16 h photoperiod). After 4–6 weeks, shoots with bleaching phenotypes were excised, transferred to rooting medium plates (1/3× MS medium, 7.0 g/L glucose, 3.0 g/L sucrose, 100.0 mg/L carbenicillin, 4.0 g/L phytagel, pH 5.8) and incubated at 23 °C. Representative N. benthamiana shoots regenerated from the leaf segments were photographed. Boxed regions in the left panels were magnified to show the regenerated albino plants.
Table S1 Cas9 targets selected in this study.
Table S2 Primers used in this study.
File S1 Sequences of RNAβ‐gNbPDS, RNAγ‐gNbPDS, RNAγ‐gTaGASR7, RNAγ‐gZmTMS5, RNAγ‐gNbPDS‐mGFP5, RNAγ‐SmR and pCB301‐BSMVγ‐MCS. Sequences in green indicate the sgRNAγ promoter, whereas sequences in red, blue and purple represent the gRNA spacer, the scaffold and the SmR gene, respectively. The underlined sequences indicate the SapI recognition sites or the multiple cloning sites.


