Abstract
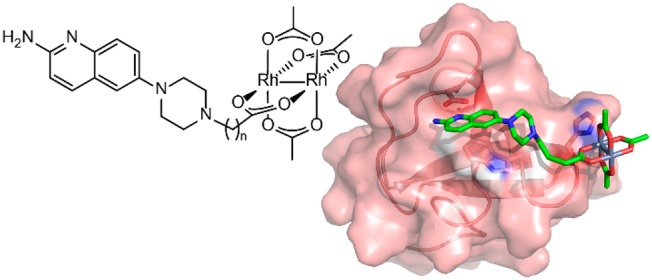
High-affinity, selective ligands are sought for a variety of biomolecules but are particularly difficult to generate in the protein–protein interaction space. Rhodium(II) conjugates provide a structure-based approach to improved affinity and specificity for targeting protein–protein interactions such as SH3 domains. In this study of small-molecule–rhodium conjugates, we report a potent ligand 4b (Kd of 27 nM) for the Lyn SH3 domain, based on an aminoquinoline fragment. The results demonstrate robust affinity gains possible from even modest small-molecule leads through cooperative inorganic–organic binding, based on specific histidine interactions. A docking study sheds light on the structural basis of binding and supports a previously proposed binding model.
Keywords: Small molecule, cooperative binding, protein−protein interaction, docking
Protein–protein interactions (PPIs) are ubiquitous in living systems and are important in a wide range of disease-relevant pathways. Significant and increasing efforts over many years have been made to “drug” PPIs, but PPI targets are especially challenging, and successes have been quite limited.1−6 Typically lacking the deep, well-defined binding pocket that characterizes traditional drug targets, PPIs often occur at relatively flat, exposed binding surfaces. The total interacting surface area can be quite large, and the hydrophobicity of these interfaces is often lower than would be expected for a deep pocket that binds small molecules. All this combines to make potent and selective inhibition difficult.
New fundamental concepts may be important to address the vexing problems presented by PPIs. The potency of small molecule ligands is inherently limited by the weak noncovalent and H-bonding interactions that contribute to the binding energy. Furthermore, ligand specificity among similar members of a protein family is a significant additional challenge.
In a new approach to selective ligands for PPIs, we introduced the concept of rhodium–small molecule conjugates as hybrid organic–inorganic inhibitors (Figure 1). Conceptually, the approach aims to improve the binding energetics of a known SH3 ligand through metal bonding to specific Lewis-basic residue(s) that flank the binding pocket. Among the benefits of this approach, metal–ligand coordination is reasonably tolerant of functional group positioning and so may be more amenable to structure-driven ligand design. We initially demonstrated a proof-of-concept for this idea using peptide–rhodium conjugates to achieve improvements in binding potency as high as 75-fold.7 Among the potential benefits of this approach, rhodium interactions with coordinating side chains (histidine, methionine)8 are energetically much stronger (∼7 kcal/mol)9 than typical nonbonding or aqueous H-bonding interactions (≤1 kcal/mol)7 and thus could deliver much larger gains in potency. Furthermore, targeting a unique peripheral Lewis-basic residue provides a new and complementary approach to binding specificity among members of a protein family. For example, a metallopeptide ligand for CALP could be designed on the basis of a unique histidine residue that is not found in the homologous protein NHERF1, which often binds natural ligands much more tightly than CALP.7,10
Figure 1.
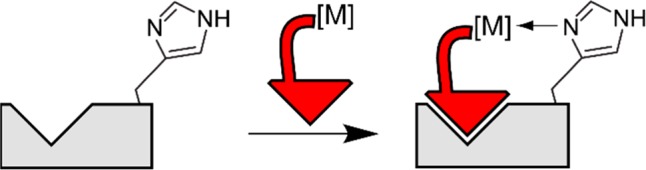
Conceptual depiction of cooperative binding of a hybrid organic–inorganic ligand to a protein/peptide binding site.
In this study, we wanted to assess whether these concepts could be extended from metal–peptide conjugates to metal–small-molecule conjugates, in part to avoid cell entry and in vivo stability concerns. Metallodrugs containing rhodium,11−14 iridium,14−17 and other metals18−22 have been shown to effectively target a variety of systems, including PPIs.23 We recently demonstrated that some small-molecule–rhodium conjugates are cell permeable and have half-lives greater than 1 h in cells.24 Additionally, metalation with dirhodium is relatively facile, allowing flexibility and creativity in conjugate design.25−27 We chose SH3 domains of the Src family kinases as a case study for this purpose. Src family proteins are a large family of multidomain proteins with important applications to human disease. Src family kinases have traditionally been targeted by ATP-competitive ligands, which bind the kinase domain with high potency but often low selectivity. The selectivity problem is particularly troublesome within the Src family due to the exceptional sequence and structure homology among its members.28 In fact, even determining each member’s precise role in various signaling processes has been difficult because of overlapping substrate scope, promiscuous inhibitors, and complex regulation pathways.29,30 Src family SH3 domains are protein-binding domains that interact with specific proline-rich sequences, recognizing natural substrates and regulating activity.28 These SH3 domains are highly conserved among the Src family (Figure 2a), and modulating the function of these domains with selective ligands has been challenging.31 However, several medically relevant SH3 domains do contain unique metal-binding residues near the SH3 binding pocket that might serve as an anchor point for a hybrid organic–inorganic ligand approach.32−34 For example, Figure 2b shows a canonical SH3 fold, with Src family kinase histidine residues shown explicitly. In theory, each unique site could be selectively targeted with proper design of a hybrid inhibitor, and peptide-based ligands have demonstrated that a rhodium conjugate approach could produce unique and specific inhibitors for multiple members of the homologous Src family of SH3 domains.32
Figure 2.
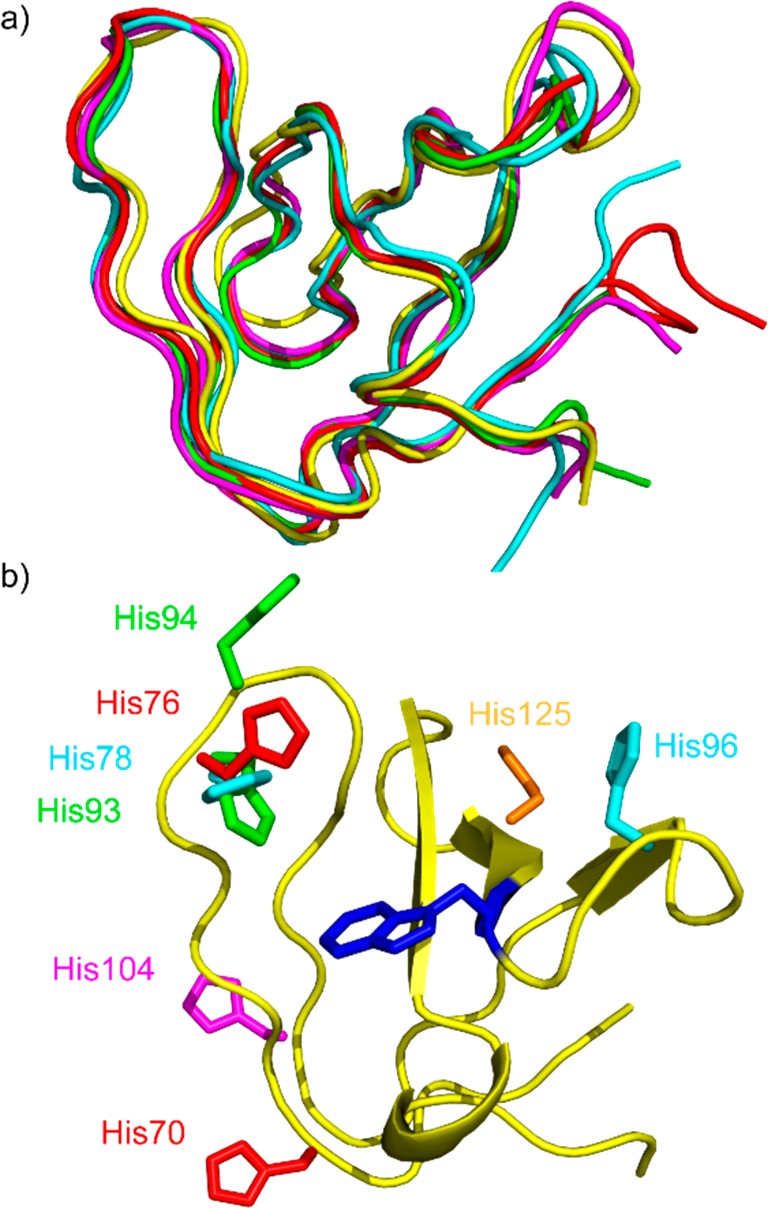
Protein sequence and 3D structure are highly conserved among Src family kinases. (a) The peptide backbones align with high similarity. (b) Locations of histidine residues vary within the Src family sequences. Backbone ribbon of Src shown in yellow. Central tryptophan (see Figure 3b) shown in blue. Histidine side chains displayed: Src (orange), Fyn (magenta), Hck (green), Lck (red), Lyn (cyan). PDB entries 4rtz, 4eik, 1bu1, 2iim, 1w1f.
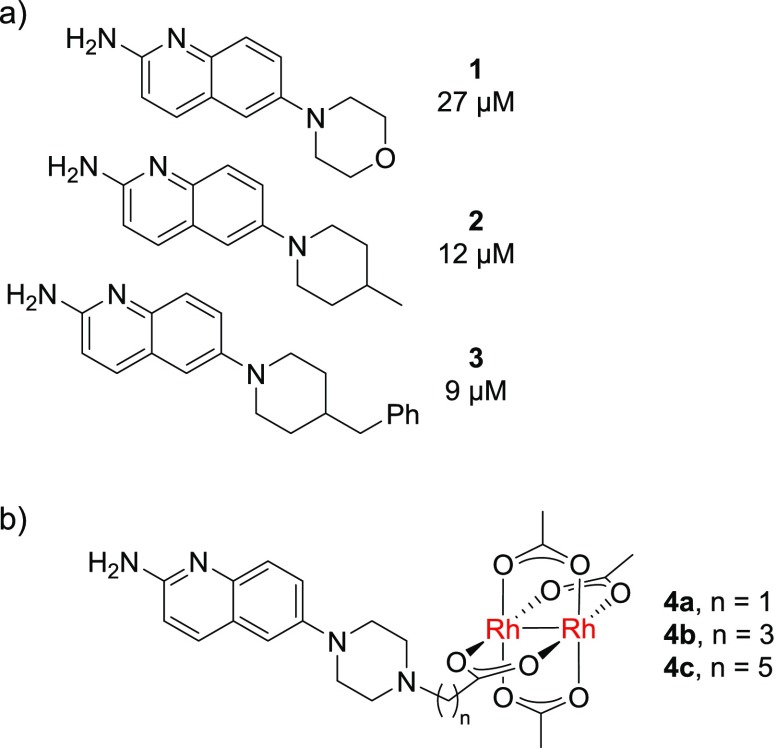
We decided to build rhodium-containing analogues of an SH3-binding scaffold to test questions of potency and selectivity. However, few examples of well-characterized small molecule ligands for Src family SH3 domains have been reported, reflecting the challenge of targeting SH3 domains.35−37 One study demonstrated that simple 2-aminoquinolines (Figure 3a) have useful (micromolar) affinity for the Tec SH3 domain.37 Although Tec is part of a separate SH3 family, it seemed reasonable to suppose that 2-aminoquinolines might interact with members of other SH3 families. Subsequent SAR studies effectively delineated permissive sites on the 2-aminoquinolines structure, providing useful information about where rhodium conjugation might be tolerated.38,39 We imagined preparing a series of rhodium-aminoquinoline conjugates, with varying linker lengths (Figure 3b) to assess the potential for cooperative binding and to understand the importance of the linker structure.
Figure 3.
(a) Small molecules featuring a functionalized 2-aminoquinoline scaffold bind Tec SH3.40 (b) Metalloconjugates consisting of a 2-aminoquinoline connected via a variable linker to a dirhodium(II) center.
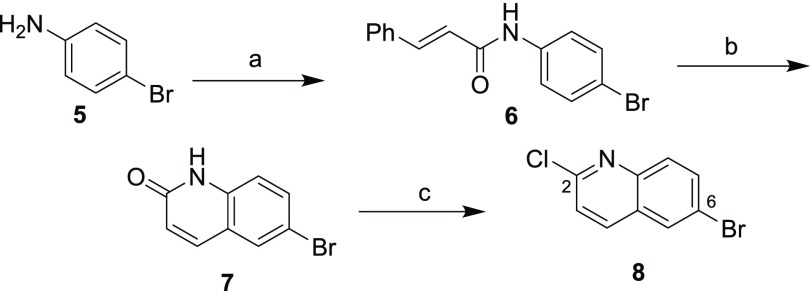
Pyke and co-workers synthesized variants of 2-aminoquinolines by coupling a bromoquinoline with various amines at the 6-position.40 As a common intermediate for the synthesis of the 2,6-diaminated quinolines desired, dihalide 8 was prepared according to Scheme 1, adapted from this previous report. 4-Bromoaniline was treated with cinnamoyl chloride to give cinnamamide 6, which undergoes an interesting cyclization with loss of benzene41 in the presence of aluminum chloride, providing quinolinone 7. Finally dehydrative chlorination in neat phosphoryl chloride42 gave 6-bromo-2-chloroquinoline (8) in 54% overall yield.
Scheme 1. Synthesis of Key Intermediate Dihalide.
Reagents and conditions: (a) cinnamoyl chloride, K2CO3, H2O/acetone, 99%; (b) (i) AlCl3, PhCl, (ii) H2O, 55%; (c) (i) POCl3, reflux, (ii) H2O, 100%.
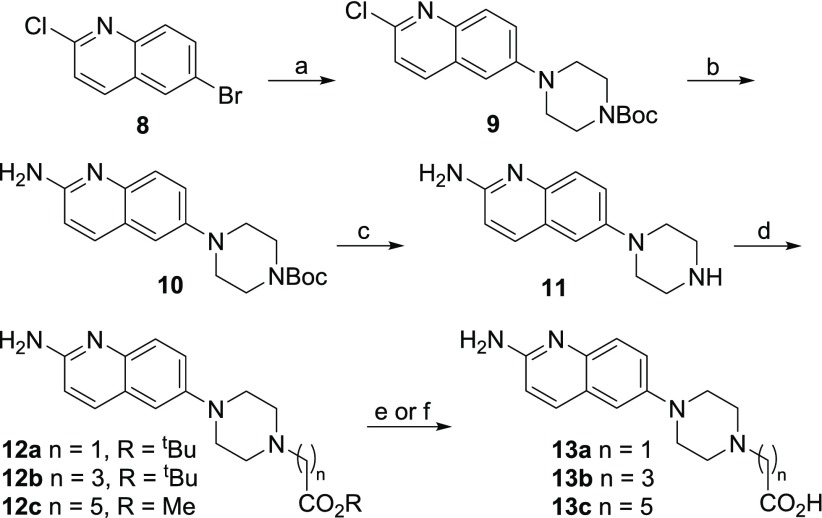
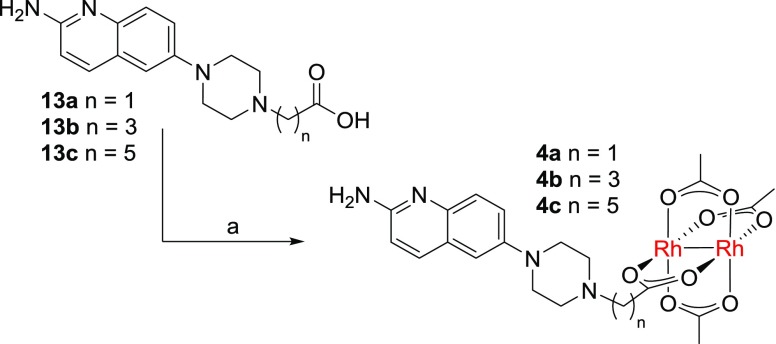
Palladium-catalyzed amination of intermediate 8 occurs first at the more reactive 6-position.40 For example, mono-N-Boc-piperazine reacts cleanly to give a 6-amino-2-chloro product 9 in 44% yield with the bulky ligand, cataCXium A.40 Palladium-catalyzed installation of the key amino group was best performed with LiHMDS as an ammonia surrogate43 to give the Boc-protected product 10. From there, a series of esters (12a–c) were formed via three-step elaboration to give carboxylic acids 13a–c (Scheme 2). Finally, the designed rhodium conjugates 4a–c were prepared by metalation under buffered aqueous conditions with the heteroleptic rhodium species, Rh2(OAc)3(tfa) (Scheme 3). We developed these metalation conditions to prepare peptide–rhodium conjugates, but they are equally useful here, as traditional methods to prepare rhodium(II) carboxylates in organic solvent were incompatible with the other functional groups present in complex 4.44,45 The rhodium conjugates 4a–c were purified by reverse-phase HPLC, isolated, and characterized by NMR and mass spectrometry.
Scheme 2. Synthesis of Aminoquinoline–Carboxylic Acids.
Reagents and conditions: (a) Boc-piperazine, Pd(OAc)2, cataCXium A, NaOtBu, toluene, 120 °C, 44%; (b) LiHMDS, Pd2(dba)3, DavePhos,43 dioxane, 100 °C, 73%; (c) TFA, CH2Cl2, 97%; (d) Br-(CH2)n-CO2R, K2CO3, DMF; (e) R = tBu:TFA/CH2Cl2, 13a: 89% over two steps; 13b: 30% over two steps; (f) R = Me:LiOH, MeOH/THF/H2O; 13c: 8% over two steps.
Scheme 3. Synthesis of Dirhodium Conjugates.
Reagents and conditions: (a) Rh2(OAc)3(tfa), MES buffer, 4a: 86%; 4b: 98%; 4c: 95%.
We examined affinity for two different Src-family SH3 domains. First, Lyn is a prototypical Src family SH3 domain with two prominent histidine residues near the binding pocket, at least one of which (His96) is completely unique within the family and is well situated to interact with the rhodium complex when bound according to a reasonable binding model (see below, Figure 5 and surrounding discussion). Separately we examined binding to Fyn. Fyn is another Src-family protein with significant structural and sequence similarity to Lyn. Indeed, most traditional peptide and small-molecule ligands exhibit overlapping activity for binding these two proteins. Fyn also has a histidine near the binding pocket (His104, ∼1.4 nm from the key Asp100 in the binding pocket) but is not well positioned to interact with rhodium in the putative bound structure. Isothermal titration calorimetry (ITC) was used to assess the binding affinity of the rhodium conjugates (Figure 4 and Table 1). The original Tec-binding lead compound 3 bound the SH3 domain of a Src-family protein (Lyn SH3 domain) with Kd = 37 μM. This affinity is within 2-fold of that reported for Tec binding, and this instance of indiscriminate binding reflects the severe challenges of developing truly selective SH3 small-molecule ligands.
Figure 5.
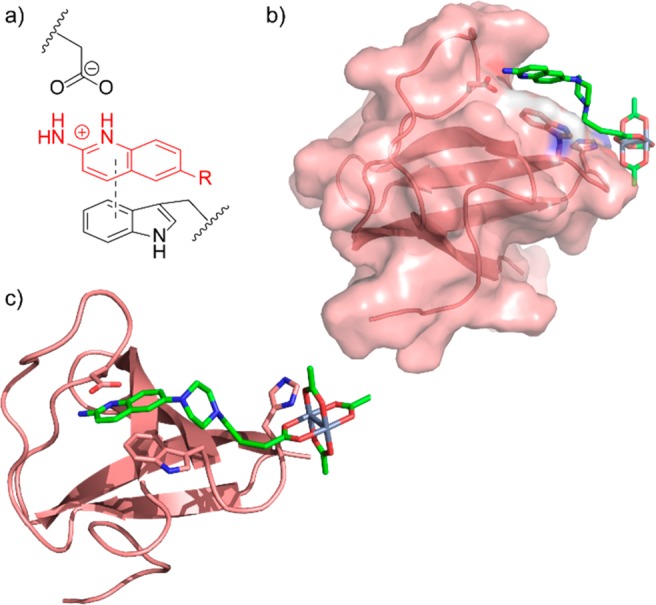
(a) The SH3 binding model proposed by Pyke et al.37 (b) Space-filling model of conjugate 4b docked to the Lyn SH3 domain, indicating the hydrophobic cleft above Trp99. (c) An alternative view of the docked model, with key binding residues shown explicitly. Docking studies were performed in GOLD (CCDC). The aminoquinoline core was then locked and a reasonable binding model built through bond rotation of the flexible alkyl linker.
Figure 4.
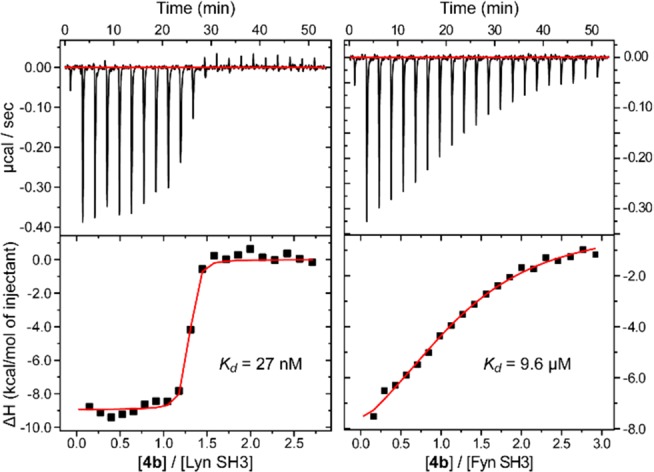
ITC curves acquired by titration of medium-length metalloligand 4b (275 μM) into Lyn SH3 and Fyn SH3 (18 μM). Raw data with baseline corrected (top) and injection peak integrals (bottom).
Table 1. Binding Affinities of Selected Compounds to Lyn SH3 and Fyn SH3.
|
Kd (μM) |
||
|---|---|---|
| Compound | Lyn SH3 | Fyn SH3 |
| 3 | 37 ± 22 | 127 ± 75 |
| 4a | 0.066 ± 0.023 | --- |
| 4b | 0.027 ± 0.012 | 9.6 ± 1.1 |
| 4c | 0.140 ± 0.032 | 12.0 ± 1.8 |
| 12c | 55 ± 17 | 7.2 ± 1.1 |
| Rh2(OAc)4 | >10 | --- |
Rhodium conjugates 4a–c were then assayed. The methyl ester of the six-carbon linker, 12c, and Rh2(OAc)4 were each tested as negative controls, and both exhibited minimal binding. Each of the rhodium–aminoquinoline conjugates exhibited strong nanomolar binding to Lyn SH3. In contrast, all three compounds showed much weaker binding to Fyn, consistent with that expected of the aminoquinoline scaffold without enhancement due to metal coordination to a nearby Lewis base. An improvement in Kd of roughly 3 orders of magnitude strongly suggests the success of the cooperativity concept. While it might be expected that increasing linker length would lead to increasing affinity due to nonspecific hydrophobic interactions, in fact we found that the intermediate linker length (n = 3, 4b) was a “sweet spot” with optimal affinity. This finding also suggests a specific interaction responsible for affinity gains.
Original reports of aminoquinoline SH3 ligands proposed an interesting binding model involving specific interactions of the cationic aminoquinoline core through π–π stacking with a conserved tryptophan residue (Lyn Trp99) and charge–charge interaction with a neighboring carboxylate side chain (Lyn Asp81, Figure 5a).38,39 We wanted to understand the extent to which rhodium conjugates fit within that binding model, and to that end we computationally docked the rhodium conjugate 4b with the Lyn SH3 domain (GOLD program, CCDC, www.ccdc.cam.ac.uk/). We did indeed find many bound states with intimate quinoline–tryptophan contacts. The most commonly occurring bound state found the aminoquinoline occupying a shallow hydrophobic cleft above the tryptophan (Figure 5b), which orients the rhodium(II) core directly toward a proximal surface-exposed histidine. Docking programs do not correctly model rhodium interactions, yet we wanted to develop a useful model for two-point binding to the SH3 domain. Manual rotation of C–C bonds of the flexible alkyl linker allowed the dirhodium core to be positioned in an ideal geometry for histidine coordination (His96) without disturbing the aminoquinoline binding. The docking studies support the previous binding model, and the near-perfect fit of the best linker length (4b) may be solid evidence of the predictive value of this binding model.
We designed, synthesized, and evaluated a series of novel rhodium–aminoquinoline conjugates as SH3-binding molecules. We are not aware of any nonpeptidic ligands with comparable affinity for SH3 domains. Introduction of the rhodium center improved the Kd for Lyn binding over 1000-fold, and varying the length of the linker between it and the organic moiety allowed some tuning of binding affinity. Docking experiments support a binding model for 2-aminoquinolines to an SH3 fold and provide a structural predictive framework for further optimization or for extending similar fragment-based ideas to other SH3 domains. This work describes an alternative approach to simultaneously achieving potency and selectivity with small-molecule ligands for SH3 domains and for PPI interactions more generally. The metalloligands reported here overcome the limitations typical of small-molecule SH3 ligands by exploiting organic–inorganic cooperativity afforded by the dirhodium core. This cooperativity is available for use in binding any protein surface with a nearby Lewis basic side chain. Future work in this program will extend these preliminary results to other members of the Src family and will examine how these or related molecules might be used to alter function in living cells.
Acknowledgments
We thank Paul Leonard for assistance with ITC measurements. We thank Jose Onuchic, Qian Wang, and Jonathan Clinger for assistance with docking and modeling. We thank Sarah Knudsen, Matthew Minus, and Farrukh Vohidov for helpful suggestions.
Glossary
Abbreviations
- CALP
CFTR-associated ligand PDZ domain
- CCDC
Cambridge Crystallographic Data Centre
- dba
dibenzylideneacetone
- ITC
isothermal titration calorimetry
- LiHMDS
lithium bis(trimethylsilyl)amide
- MES
2-(N-morpholino)ethanesulfonic acid
- NHERF1
Na+/H+ exchanger regulatory factor
- PPI
protein–protein interaction
- SAR
structure–activity relationship
- SH3
Src homology 3
Biography
Zachary Ball grew up in Columbus, Ohio before furthering his education at Harvard University (A.B.), Stanford University (Ph.D.), and the University of California–Berkeley (Post-doc). He has been a faculty member of the Department of Chemistry at Rice University since 2006, where his research interests focus on transition metals in biological and medicinal chemistry, protein chemistry, and bioconjugation methodology. Zachary is currently Professor and Director of the Institute of Biosciences and Bioengineering at Rice.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsmedchemlett.9b00309.
Synthesis of compounds, including NMR, MS, and HPLC characterization as appropriate. ITC protocol and data for compounds 3, 4a–c, and 12c. Docking experimental procedures (PDF)
This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant number 1450681 (S.C.M.). We acknowledge support from the Robert A. Welch Foundation Research Grant C-1680 and from the National Science Foundation under research grant numbers CHE-1609654 and CHE-1904865.
The authors declare no competing financial interest.
Supplementary Material
References
- Chène P. Drugs Targeting Protein–Protein Interactions. ChemMedChem 2006, 1, 400–411. 10.1002/cmdc.200600004. [DOI] [PubMed] [Google Scholar]
- Fletcher S.; Hamilton A. D. Targeting Protein–protein Interactions by Rational Design: Mimicry of Protein Surfaces. J. R. Soc., Interface 2006, 3, 215–233. 10.1098/rsif.2006.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Ruiz D.; Gohlke H. Targeting Protein-Protein Interactions with Small Molecules: Challenges and Perspectives for Computational Binding Epitope Detection and Ligand Finding. Curr. Med. Chem. 2006, 13, 2607–2625. 10.2174/092986706778201530. [DOI] [PubMed] [Google Scholar]
- Wells J. A.; McClendon C. L. Reaching for High-Hanging Fruit in Drug Discovery at Protein–Protein Interfaces. Nature 2007, 450, 1001–1009. 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- Zinzalla G.; Thurston D. E. Targeting Protein–Protein Interactions for Therapeutic Intervention: A Challenge for the Future. Future Med. Chem. 2009, 1, 65–93. 10.4155/fmc.09.12. [DOI] [PubMed] [Google Scholar]
- Ivanov A. A.; Khuri F. R.; Fu H. Targeting Protein–Protein Interactions as an Anticancer Strategy. Trends Pharmacol. Sci. 2013, 34, 393–400. 10.1016/j.tips.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu R.; Cushing P. R.; Popp B. V.; Zhao Y.; Madden D. R.; Ball Z. T. Hybrid Organic–Inorganic Inhibitors of a PDZ Interaction that Regulates the Endocytic Fate of CFTR. Angew. Chem., Int. Ed. 2012, 51, 7217–7220. 10.1002/anie.201202291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man B. Y.-W.; Chan H.-M.; Leung C.-H.; Chan D. S.-H.; Bai L.-P.; Jiang Z.-H.; Li H.-W.; Ma D.-L. Group 9 Metal-based Inhibitors of β-Amyloid (1–40) Fibrillation as Potential Therapeutic Agents for Alzheimer’s Disease. Chem. Sci. 2011, 2, 917–921. 10.1039/c0sc00636j. [DOI] [Google Scholar]
- Das K.; Simmons E. L.; Bear J. L. Thermodynamics and Kinetics of Some tetra-μ-Carboxylato-Dirhodium(II) Adduct Formation Reactions. Inorg. Chem. 1977, 16, 1268–1271. 10.1021/ic50172a002. [DOI] [Google Scholar]
- Cushing P. R.; Fellows A.; Villone D.; Boisguérin P.; Madden D. R. The Relative Binding Affinities of PDZ Partners for CFTR: A Biochemical Basis for Efficient Endocytic Recycling. Biochemistry 2008, 47, 10084–10098. 10.1021/bi8003928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu F.-M.; Lin I. W.-S.; Yan K.; Lok C.-N.; Low K.-H.; Yun-Chung Leung T.; Lam T.-L.; Che C.-M. Anticancer Dirhodium(II,II) Carboxylates as Potent Inhibitors of Ubiquitin-Proteasome System. Chem. Sci. 2012, 3, 1785–1793. 10.1039/c2sc00620k. [DOI] [Google Scholar]
- Kang T.-S.; Ko C.-N.; Zhang J.-T.; Wu C.; Wong C.-Y.; Ma D.-L.; Leung C.-H. Rhodium(III)-Based Inhibitor of the JMJD3-H3K27me3 Interaction and Modulator of the Inflammatory Response. Inorg. Chem. 2018, 57, 14023–14026. 10.1021/acs.inorgchem.8b02256. [DOI] [PubMed] [Google Scholar]
- Ma D.-L.; Wang M.; Mao Z.; Yang C.; Ng C.-T.; Leung C.-H. Rhodium Complexes as Therapeutic Agents. Dalton Trans 2016, 45, 2762–2771. 10.1039/C5DT04338G. [DOI] [PubMed] [Google Scholar]
- Graf M.; Gothe Y.; Siegmund D.; Metzler-Nolte N.; Sünkel K. Synthesis and Characterization of Cyclometallated Rhodium(III) and Iridium(III) Compounds with Antiproliferative Activities in the Nanomolar Range. Inorg. Chim. Acta 2018, 471, 265–271. 10.1016/j.ica.2017.11.003. [DOI] [Google Scholar]
- Kang T.-S.; Wang W.; Zhong H.-J.; Dong Z.-Z.; Huang Q.; Mok S. W. F.; Leung C.-H.; Wong V. K. W.; Ma D.-L. An Anti-prostate Cancer Benzofuran-conjugated Iridium(III) Complex as a Dual Inhibitor of STAT3 and NF-κB. Cancer Lett. 2017, 396, 76–84. 10.1016/j.canlet.2017.03.016. [DOI] [PubMed] [Google Scholar]
- Wu C.; Wu K.-J.; Liu J.-B.; Zhou X.-M.; Leung C.-H.; Ma D.-L. A Dual-functional Molecular Strategy for in Situ Suppressing and Visualizing of Neuraminidase in Aqueous Solution Using Iridium(III) Complexes. Chem. Commun. 2019, 55, 6353–6356. 10.1039/C9CC02189B. [DOI] [PubMed] [Google Scholar]
- Liu Z.; Sadler P. J. Organoiridium Complexes: Anticancer Agents and Catalysts. Acc. Chem. Res. 2014, 47, 1174–1185. 10.1021/ar400266c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaouen G.; Vessières A.; Top S. Ferrocifen Type Anti Cancer Drugs. Chem. Soc. Rev. 2015, 44, 8802–8817. 10.1039/C5CS00486A. [DOI] [PubMed] [Google Scholar]
- Gasser G.; Metzler-Nolte N. The Potential of Organometallic Complexes in Medicinal Chemistry. Curr. Opin. Chem. Biol. 2012, 16, 84–91. 10.1016/j.cbpa.2012.01.013. [DOI] [PubMed] [Google Scholar]
- Śmiłowicz D.; Metzler-Nolte N. Synthesis of Monofunctional Platinum(IV) Carboxylate Precursors for use in Pt(IV)–Peptide Bioconjugates. Dalton Trans 2018, 47, 15465–15476. 10.1039/C8DT03082K. [DOI] [PubMed] [Google Scholar]
- Muñoz-Osses M.; Godoy F.; Fierro A.; Gómez A.; Metzler-Nolte N. New Organometallic Imines of Rhenium(I) as Potential Ligands of GSK-3β: Synthesis, Characterization and Biological Studies. Dalton Trans 2018, 47, 1233–1242. 10.1039/C7DT04344A. [DOI] [PubMed] [Google Scholar]
- Bertrand B.; Casini A. A Golden Future in Medicinal Inorganic Chemistry: The Promise of Anticancer Gold Organometallic Compounds. Dalton Trans 2014, 43, 4209–4219. 10.1039/C3DT52524D. [DOI] [PubMed] [Google Scholar]
- Leung C.-H.; Zhong H.-J.; Yang H.; Cheng Z.; Chan D. S.-H.; Ma V. P.-Y.; Abagyan R.; Wong C.-Y.; Ma D.-L. A Metal-Based Inhibitor of Tumor Necrosis Factor-α. Angew. Chem., Int. Ed. 2012, 51, 9010–9014. 10.1002/anie.201202937. [DOI] [PubMed] [Google Scholar]
- Minus M. B.; Kang M. K.; Knudsen S. E.; Liu W.; Krueger M. J.; Smith M. L.; Redell M. S.; Ball Z. T. Assessing the Intracellular Fate of Rhodium(II) Complexes. Chem. Commun. 2016, 52, 11685–11688. 10.1039/C6CC05192H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann D. G.; Schmidt P. J.; Geigle S. N.; Chougnet A.; Woggon W.-D.; Gillingham D. G. Modular Ligands for Dirhodium Complexes Facilitate Catalyst Customization. Adv. Synth. Catal. 2015, 357, 2033–2038. 10.1002/adsc.201500050. [DOI] [Google Scholar]
- Zhao J.; Bachmann D. G.; Lenz M.; Gillingham D. G.; Ward T. R. An Artificial Metalloenzyme for Carbene Transfer Based on a Biotinylated Dirhodium Anchored Within Streptavidin. Catal. Sci. Technol. 2018, 8, 2294–2298. 10.1039/C8CY00646F. [DOI] [Google Scholar]
- Ohata J.; Ball Z. T. A Hexa-rhodium Metallopeptide Catalyst for Site-Specific Functionalization of Natural Antibodies. J. Am. Chem. Soc. 2017, 139, 12617–12622. 10.1021/jacs.7b06428. [DOI] [PubMed] [Google Scholar]
- Boggon T. J.; Eck M. J. Structure and Regulation of Src Family Kinases. Oncogene 2004, 23, 7918–7927. 10.1038/sj.onc.1208081. [DOI] [PubMed] [Google Scholar]
- Ingley E. Src family kinases: Regulation of Their Activities, Levels and Identification of New Pathways. Biochim. Biophys. Acta, Proteins Proteomics 2008, 1784, 56–65. 10.1016/j.bbapap.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Thomas S. M.; Brugge J. S. Cellular Functions Regulated by Src Family Kinases. Annu. Rev. Cell Dev. Biol. 1997, 13, 513–609. 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Bishop A.; Witucki L.; Kraybill B.; Shimizu E.; Tsien J.; Ubersax J.; Blethrow J.; Morgan D. O.; Shokat K. M. Structural Basis for Selective Inhibition of Src Family Kinases by PP1. Chem. Biol. 1999, 6, 671–678. 10.1016/S1074-5521(99)80118-5. [DOI] [PubMed] [Google Scholar]
- Vohidov F.; Knudsen S. E.; Leonard P. G.; Ohata J.; Wheadon M. J.; Popp B. V.; Ladbury J. E.; Ball Z. T. Potent and Selective Inhibition of SH3 Domains with Dirhodium Metalloinhibitors. Chem. Sci. 2015, 6, 4778–4783. 10.1039/C5SC01602A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vohidov F.; Coughlin J. M.; Ball Z. T. Rhodium(II) Metallopeptide Catalyst Design Enables Fine Control in Selective Functionalization of Natural SH3 Domains. Angew. Chem., Int. Ed. 2015, 54, 4587–4591. 10.1002/anie.201411745. [DOI] [PubMed] [Google Scholar]
- Martin S. C.; Vohidov F.; Wang H.; Knudsen S. E.; Marzec A. A.; Ball Z. T. Designing Selectivity in Dirhodium Metallopeptide Catalysts for Protein Modification. Bioconjugate Chem. 2017, 28, 659–665. 10.1021/acs.bioconjchem.6b00716. [DOI] [PubMed] [Google Scholar]
- Oneyama C.; Nakano H.; Sharma S. V. UCS15A, a Novel Small Molecule, SH3 Domain-mediated Protein–protein Interaction Blocking Drug. Oncogene 2002, 21, 2037. 10.1038/sj.onc.1205271. [DOI] [PubMed] [Google Scholar]
- Oneyama C.; Agatsuma T.; Kanda Y.; Nakano H.; Sharma S. V.; Nakano S.; Narazaki F.; Tatsuta K. Synthetic Inhibitors of Proline-Rich Ligand-Mediated Protein-Protein Interaction: Potent Analogs of UCS15A. Chem. Biol. 2003, 10, 443–451. 10.1016/S1074-5521(03)00101-7. [DOI] [PubMed] [Google Scholar]
- Inglis S. R.; Stojkoski C.; Branson K. M.; Cawthray J. F.; Fritz D.; Wiadrowski E.; Pyke S. M.; Booker G. W. Identification and Specificity Studies of Small-Molecule Ligands for SH3 Protein Domains. J. Med. Chem. 2004, 47, 5405–5417. 10.1021/jm049533z. [DOI] [PubMed] [Google Scholar]
- Inglis S.; Jones R.; Fritz D.; Stojkoski C.; Booker G.; Pyke S. Synthesis of 5-, 6- and 7-Substituted-2-Aminoquinolines as SH3 Domain Ligands. Org. Biomol. Chem. 2005, 3, 2543–2557. 10.1039/b504498g. [DOI] [PubMed] [Google Scholar]
- Inglis S. R.; Jones R. K.; Booker G. W.; Pyke S. M. Synthesis of N-Benzylated-2-Aminoquinolines as Ligands for the Tec SH3 Domain. Bioorg. Med. Chem. Lett. 2006, 16, 387–390. 10.1016/j.bmcl.2005.09.073. [DOI] [PubMed] [Google Scholar]
- Smith J. A.; Jones R. K.; Booker G. W.; Pyke S. M. Sequential and Selective Buchwald–Hartwig Amination Reactions for the Controlled Functionalization of 6-Bromo-2-chloroquinoline: Synthesis of Ligands for the Tec Src Homology 3 Domain. J. Org. Chem. 2008, 73, 8880–8892. 10.1021/jo801808r. [DOI] [PubMed] [Google Scholar]
- Johnston K. M.; Luker R. M.; Williams G. H. Friedel–Crafts Cyclisations. Part III. Synthesis of Derivatives of 2(1H)-Quinolone (Carbostyril) by Aluminium Chloride-catalysed Cycloeliminations of Cinnamanilide and Related Compounds. J. Chem. Soc., Perkin Trans. 1 1972, 1, 1648–1652. 10.1039/P19720001648. [DOI] [Google Scholar]
- Baston E.; Palusczak A.; Hartmann R. W. 6-Substituted 1H-Quinolin-2-ones and 2-Methoxy-quinolines: Synthesis and Evaluation as Inhibitors of Steroid 5α Reductases Types 1 and 2. Eur. J. Med. Chem. 2000, 35, 931–940. 10.1016/S0223-5234(00)01167-3. [DOI] [PubMed] [Google Scholar]
- Old D. W.; Wolfe J. P.; Buchwald S. L. A Highly Active Catalyst for Palladium-Catalyzed Cross-Coupling Reactions: Room-Temperature Suzuki Couplings and Amination of Unactivated Aryl Chlorides. J. Am. Chem. Soc. 1998, 120, 9722–9723. 10.1021/ja982250+. [DOI] [Google Scholar]
- Popp B. V.; Ball Z. T. Structure-selective Modification of Aromatic Side Chains with Dirhodium Metallopeptide Catalysts. J. Am. Chem. Soc. 2010, 132, 6660–6662. 10.1021/ja101456c. [DOI] [PubMed] [Google Scholar]
- Martin S. C.; Minus M. B.; Ball Z. T.. Chemical Posttranslational Modification with Designed Rhodium(II) Catalysts. In Methods in Enzymology; Pecoraro V. L., Ed.; Academic Press, 2016; Vol. 580, pp 1–19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.