Abstract
Many biological processes are influenced by the mechanical rigidity of surrounding tissues. Now, a combination of experiments and mathematical modelling has been used to describe the precise molecular and physical mechanism by which cells sense and respond to the mechanical properties of their extracellular environment through integrin-based adhesions.
Cells exert forces on other cells and their extracellular matrix (ECM) to migrate or remodel tissues and the ECM during development, wound healing and the immune response. Conversely, forces and mechanics of the environment influence transcription, cellular differentiation and inflammation. Thus, understanding how physical information is transmitted and sensed by the cell to regulate its function is critical in characterizing the molecular basis of a wide variety of physiological processes and diseases. In this issue, Roca-Cusachs and colleagues reveal important insights into the regulation of force transmission across integrin-based focal adhesions1.
Focal adhesions (FAs) are the macromolecular assemblies of proteins that constitute the primary link connecting the actin cytoskeleton to the ECM through the integrin family of receptors, and are thought to be the sites of integrin-mediated mechanosensation. Integrins are heterodimeric transmembrane receptors with a large extracellular ECM-binding domain, a single-pass transmembrane domain and a short cytoplasmic domain. As the integrin cytoplasmic domain lacks an actin binding site, other proteins are required to complete the actin–integrin–ECM linkage. These putative integrin–actin linking proteins are members of the ‘integrin adhesome’ — the more than 500 proteins associated with FAs that have been identified in proteomic studies2.
Physical forces can directly or indirectly alter the dynamics and interactions of FA proteins to change FA composition, morphology or signalling, all of which result in downstream changes in FA-dependent cellular functions including FA strengthening and nuclear trans-location of transcriptional activators that regulate gene expression3,4.
The sources and nature of forces exerted on FAs during different cellular processes have been best studied in the context of cell migration. Mesenchymal cells move by first forming a protrusion at the leading edge, which then adheres to the ECM. Cells then generate traction against the ECM adhesion to move forward5. Protrusion is driven by rapid polarized actin polymerization at free filament ends in close apposition to the leading edge plasma membrane. The force exerted by filament elongation can drive the membrane forward, but only if the pushing force is opposed by anchoring the filaments to an immobile object, such as the ECM. If the filaments are not anchored, the force of polymerization pushes them away from the leading edge, resulting in the backward motion of the filament network, known as retrograde flow6. Integrin-mediated adhesions formed at the leading edge of the cell are thought to couple actin to the ECM through FA proteins that are capable of mediating the link between integrin and actin, resulting in exertion of traction force on the ECM and net forward motion of the cell4.
A leading concept in the field to explain the mechanism of coupling between integrins and actin during cell migration, originally proposed by Mitchison and colleagues, is called the ‘molecular clutch’ hypothesis7. In this analogy, when the clutch is engaged — that is, the actin flow is coupled to the ECM through FA proteins and immobilized integrins — the force due to actin polymerization would result in slowing down of the retrograde flow, protrusion of the leading edge, and generation of rearward traction forces by which the cell can be propelled forward.
To test if the molecular clutch could function as a mechanosensitive linkage, Odde and colleagues built a simple stochastic physical model of the clutch and examined how the model dynamically responded to ECMs of different stiffness8. The model consisted of actin retrograde flow of a known velocity and force, and Hookean springs to represent the flexibility of both the ECM and the ensemble of clutch molecules that linked actin to the ECM in a FA. By simply taking into account the rates of clutch binding and unbinding, the Odde model showed that the clutch functions in two dynamic regimes, one called ‘frictional slippage’ observed with stiff substrates, and the other a ‘load-and-fail’ regime observed with flexible substrates. On stiff substrates, cellular pulling forces resulted in engaged clutches rapidly reaching their breaking strength, and the actin filament to which they were bound moving rapidly rearward before any other clutches had time to engage. This slipping between actin and the ECM resulted in a lower magnitude of traction force and a higher rate of retrograde flow. On flexible substrates, the stretching of the substrate resulted in a slower rate of loading on engaged clutches, allowing more clutches time to engage (the loading phase). This resulted in increased ECM traction forces and a decrease in retrograde flow rate. Eventually, load became high enough that failure of a single clutch resulted in failure of all clutches and loss of traction, whereupon the cycle would repeat, resulting in force fluctuations over time.
Several predictions of the Odde model have been borne out in experiments. Indeed, traction force microscopy of neuronal growth cones and migrating fibroblasts has revealed that forces at FAs fluctuate on flexible substrates, but remain low and steady on stiff substrates8,9. However, the model also predicts a biphasic increase in cellular traction forces in response to increasing substrate stiffness, whereas experiments have shown a monotonic increase in traction force as a function of increasing ECM stiffness10.
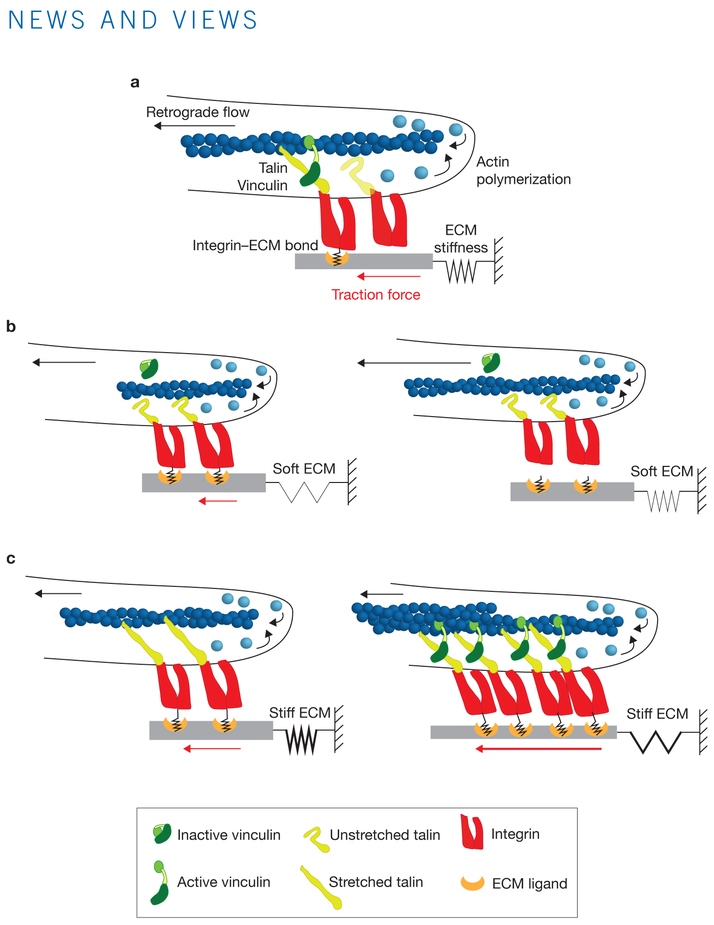
Roca-Cusachs and colleagues incorporate the molecular behaviours of integrin, talin and vinculin into a mathematical model to bring quantitative predictive understanding of the molecular mechanism of integrin-mediated force transmission and mechanosensing (Fig.1a). Talin and vinculin are critical FA proteins mediating the clutch linkage between ECM-bound integrins and actin, as knocking down either protein results in lower cellular traction forces and increased actin retrograde flow, as would be predicted from the clutch model11,12. Talin can bind directly to actin and the cytoplasmic tail of β-integrins to activate high-affinity integrin–ECM binding, and thus could mediate engagement of actin to the ECM through integrins13. Vinculin can also bind to actin and a number of other FA proteins including talin, and may strengthen the labile talin-mediated linkage between integrins and actin14. The talin sequence contains several cryptic vinculin binding sites that are only exposed following force-induced unfolding of the talin molecule15. This suggests that loading the talin-mediated linkage between integrin and actin could result in additional recruitment of vinculin, which can bind to actin and thus reinforce the clutch. However, until now, this concept has not been incorporated into a molecular clutch model.
Figure 1.
The Roca-Cusachs model of the molecular clutch. (a) An overview of the mechanosensitive molecular clutch model, as proposed in the current study1. (b) On soft substrates, the rate of load transmission is slower than the integrin–ECM bond lifetime, resulting in bond dissociation before talin can unfold or vinculin can bind. (c) On stiff substrates, load is transmitted faster than the integrin–ECM bond lifetime, resulting in talin unfolding, vinculin binding and actin-mediated reinforcement.
The authors first evaluate the role of talin in cell–ECM force transmission using cells either lacking talin or expressing a dominant-negative talin head mutant plated on substrates of different rigidity. They find that there is a rigidity threshold above which talin is needed for force transmission. This threshold correlates with the growth of vinculin-rich FAs, recruitment of integrin, the association of FAs to actin stress fibres, and the nuclear translocation of the mechanosensitive transcription factor YAP. Combining their own measurements with published data on talin unfolding time and the strength of interactions between integrins and fibronectin, the authors develop the hypothesis that the observed rigidity threshold occurs due to talin unfolding. They find that depletion of talin reveals the biphasic relationship between cellular traction and ECM stiffness predicted by the Odde model. The authors thus adapt the model, adding tension-dependent reinforcement features including the integrin-ligand catch bond, talin unfolding and vinculin-mediated clutch reinforcement.
The enhanced Roca-Cusachs clutch model shows remarkable agreement with experimental observations, particularly the ability to recapitulate a monotonic increase in traction force in response to increasing ECM stiffness. The new model predicts that the key determinant of mechanosensitivity is whether the resistive force of the ECM substrate can be transmitted to talin before the integrin–ligand bond dissociates. With low-stiffness ECMs, the loading rate on the clutch is slower than the integrin-ECM bond lifetime, and the bond fails before any force can be transmitted to talin (Fig.1b). Clutch loading gets faster as the rigidity of the substrate increases, and beyond a specific rigidity threshold, becomes faster than the integrin–ECM bond lifetime, thereby resulting in force transmission to talin. This leads to talin unfolding, binding of vinculin and adhesion reinforcement, and thus increasing traction with increasing ECM stiffness (Fig.1c). Remarkably, authors find that the predicted ECM rigidity threshold at which talin unfolds is the precise stiffness at which cells exhibit FA growth and YAP nuclear translocation.
Roca-Cusachs and colleagues further test their model by manipulating molecular functions of integrin, talin and vinculin. These include altering the amount of ECM ligand that is available for integrin binding, talin mutants that lack the ability to bind actin or integrin or that have an increase in the threshold required for force-induced unfolding, and a dominant-negative vinculin construct that blocks vinculin-mediated reinforcement. All of these conditions show the biphasic (and not the monotonic) response of force as a function of increasing mechanosensitive outputs. Thus, characterization of the biophysical and biochemical molecular behaviours of the clutch components allows the authors to gain a quantitative and predictive understanding of the physical and molecular mechanism of integrin-mediated mechanosensing.
A number of unanswered questions arise from the present study, including the mechanism of transcriptional activator trans-location, which has been previously shown to be integrin independent2. Additionally, the biphasic response of force to increasing stiffness in the absence of talin hints at a different molecule mediating the clutch in the low force regime, with a likely candidate being kindlin, another integrin activator that associates with tiny, low-force adhesions16. Just as the original molecular clutch metaphor and subsequent experiments and models resulted in a plethora of significant studies and advances, the present results set a quantitative benchmark that will lead to further studies to help us understand the fundamentals of the biophysical interaction between a cell and its environment.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Elosegui-Artola et al. Nat. Cell Biol 18, 540–548 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Winograd-Katz SE, Fässler R, Geiger B & Legate KR Nat. Rev. Mol. Cell Biol 15, 273–288 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Dupont S et al. Nature 474, 179–183 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Wolfenson H, Lavelin I & Geiger B Dev. Cell 24, 447–458 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lauffenburger D. a. & Horwitz AF Cell 84, 359–369 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Mitchison T & Cramer L Cell 84, 371–379 (1996). [DOI] [PubMed] [Google Scholar]
- 7.Mitchison T & Kirschner M Neuron 1, 761–772 (1988). [DOI] [PubMed] [Google Scholar]
- 8.Chan CE & Odde DJ Science 322, 1687–1691 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Plotnikov SV & Waterman CM Curr. Opin. Cell Biol 25, 619–626 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polte TR, Eichler GS, Wang N & Ingber DE Am. J. Physiol. Cell Physiol 286, 518–528 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Thievessen I et al. J. Cell Biol 202, 163–177 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X et al. Nat. Cell Biol 10, 1062–1068 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calderwood DA, Campbell ID & Critchley DR Nat. Rev. Mol. Cell Biol 14, 503–17 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humphries JD et al. J. Cell Biol 179, 1043–1057 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Del Rio A et al. Science 323, 638–641 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moser M, Legate KR, Zent R & Fässler R Science 324, 895–899 (2009). [DOI] [PubMed] [Google Scholar]