Abstract
Background:
The sperm DNA methylation landscape is unique and critical for offspring health. If gamete-derived DNA methylation escapes reprograming in early embryos, epigenetic defects in sperm may be transmitted to the next generation. Current techniques to assess sperm DNA methylation show bias toward CpG-dense regions and do not target areas of dynamic methylation, those predicted to be environmentally sensitive and tunable regulatory elements.
Objectives:
Our goal was to assess variation in human sperm DNA methylation and design a targeted capture panel to interrogate the human sperm methylome.
Methods:
To characterize variation in sperm DNA methylation, we performed whole genome bisulfite sequencing (WGBS) on an equimolar pool of sperm DNA from a wide cross section of 30 men varying in age, fertility status, methylenetetrahydrofolate reductase (MTHFR) genotype, and exposures. With our targeted capture panel, in individual samples, we examined the effect of MTHFR genotype ( 677CC, 677TT), as well as high-dose folic acid supplementation (, per genotype, before and after supplementation).
Results:
Through WGBS we discovered nearly 1 million CpGs possessing intermediate methylation levels (20–80%), termed dynamic sperm CpGs. These dynamic CpGs, along with 2 million commonly assessed CpGs, were used to customize a capture panel for targeted interrogation of the human sperm methylome and test its ability to detect effects of altered folate metabolism. As compared with MTHFR 677CC men, those with the 677TT genotype (50% decreased MTHFR activity) had both hyper- and hypomethylation in their sperm. High-dose folic acid supplement treatment exacerbated hypomethylation in MTHFR 677TT men compared with 677CC. In both cases, of altered methylation was found in dynamic sperm CpGs, uniquely measured by our assay.
Discussion:
Our sperm panel allowed the discovery of differential methylation following conditions affecting folate metabolism in novel dynamic sperm CpGs. Improved ability to examine variation in sperm DNA methylation can facilitate comprehensive studies of environment–epigenome interactions. https://doi.org/10.1289/EHP4812
Introduction
A significant decrease in sperm counts over the last 50 y has been reported in men from Western countries, and environmental exposures to developing male germ cells have been suggested as one of the potential causes (Barouki et al. 2018; Levine et al. 2017). Human and animal studies have demonstrated that various paternal exposures, including environmental, diet, drug, and psychological stress, can have consequences for the next generation (Carone et al. 2010; Lumey et al. 2007; Watkins and Sinclair 2014). Besides mutations and effects directly on genomic sequence, such exposures can reach measureable differences in DNA methylation, histone posttranslational modifications and small noncoding RNA expression (reviewed by Nilsson et al. 2018). DNA methylation undergoes well-characterized patterns of erasure and reestablishment during male germ cell development, is unique in sperm as compared with somatic tissue, and is a strong candidate for an epigenetic mark that can be altered, with the resulting epimutations potentially transmitted to the next generation (Ly et al. 2015; Ziller et al. 2013). To accurately determine how different paternal exposures impact the sperm DNA methylome, there is a need to develop more comprehensive and cost-effective approaches to assess the presence and transmissibility of altered DNA methylation in sperm and its impact on the health of future generations.
DNA methylation can affect sites across the human genome (Edwards et al. 2017), mainly occurring in a 5′-cytosine-phosphate-guanine-3′ (CpG) dinucleotide context. In male germ cells of the fetal testis, DNA methylation is erased in primordial germ cells and then reestablished, including at imprinted genes, in mitotically quiescent prospermatogonia between weeks 11 and 16 of gestation (Gkountela et al. 2015; Tang et al. 2015). It is also at this time that a mother’s gestational exposure might impact the germ cell epigenome of her male fetus (Wu et al. 2017a). At puberty, with the resumption of postnatal spermatogenesis, the majority of DNA methylation patterns acquired in prenatal germ cells need to be maintained in dividing spermatogonia, but continuous remodeling also occurs in meiotic spermatocytes and postmeiotic spermatids (Gaysinskaya et al. 2018; Ly et al. 2015). Thus, spermatogenesis, taking about 3 months in men, is an ongoing process and, as such, represents a window from puberty onward when the male germ cell epigenome could be susceptible to environmental insults.
Whole genome bisulfite sequencing (WGBS) provides comprehensive coverage of the epigenome. However, WGBS is challenging to adapt to large studies and, to date, only one human sperm WGBS data set of genome-wide CpG coverage has been published (Molaro et al. 2011). The Illumina® Infinium HumanMethylation450 BeadChip () arrays are the most commonly used approach to assess the methylation of human sperm. With this approach, various factors including age (Jenkins et al. 2014), smoking (Alkhaled et al. 2018; Jenkins et al. 2017; Laqqan et al. 2017), phthalates (Wu et al. 2017b), and infertility (Aston et al. 2015; Jenkins et al. 2016) have been reported to alter the sperm DNA methylome. However, the arrays provide limited coverage of the epigenome and focus on genic and CpG-rich regions. Reduced representation bisulfite sequencing (RRBS), which targets primarily classical CpG islands and other high-GC content sequences, and low coverage unbiased WGBS, were used to examine sperm of men exposed to high-dose folic acid (Aarabi et al. 2015) or dioxin (Pilsner et al. 2018), respectively. These results suggest that intergenic regions and regions of intermediate methylation (20–80%), where distal regulatory regions reside, may be particularly susceptible to paternal exposures. Although the newer Illumina® Infinium MethylationEPIC BeadChip () arrays allow assessment of some intergenic and enhancer CpG methylation sites, they are not specifically designed to interrogate the specialized sperm epigenome.
Targeted capture sequencing panels offer an alternative to WGBS, allowing the customization needed to target the sperm epigenome with enrichment of sequences of interest in both genic and intergenic regions. To this end, we recently implemented methylC-capture sequencing (MCC-Seq) for targeted assessment of DNA methylation in a tissue-specific manner (Allum et al. 2015). This approach has been particularly useful for the analysis of intermediate levels (20–80%) of methylation, or dynamic sites, that are postulated to be susceptible to environmental exposures (Ziller et al. 2016). Using this approach, we showed that DNA methylation variation linked to disease traits is enriched within intergenic and enhancer-associated regions, with such regions characterized by intermediate levels of methylation (Allum et al. 2015). With sperm possessing unique epigenetic patterns, differing dramatically from those of somatic cells, ascertaining and targeting the susceptible/variable regions of the sperm DNA methylome, in addition to genic regions, would aid in more efficiently assessing the effect of environmental exposures.
The goal of the current study was to identify regions of variable and/or dynamic DNA methylation in human sperm, to use this information to design a customized human sperm methylation capture panel for DNA methylation profiling, and then to test it in sperm samples of men exposed to low or high levels of methyl donors through either perturbations in folate metabolism or high doses of folic acid supplements, respectively. To accomplish this, we first generated a human sperm WGBS data set that differed from the published one (Molaro et al. 2011) by examining a sperm DNA sample pooled from a wide cross section of 30 men in order to represent common epigenetic diversity. Using the data, we then designed a targeted human sperm-specific MCC-Seq panel. We validated the approach by assessing the impact of the common 5,10-methylenetetrahydrofolate reductase (MTHFR) polymorphism and response of high-dose folic acid supplementation on DNA methylation of human sperm in different cohorts of men. This customized panel can be used to accurately assess sperm DNA methylation profiles at single CpGs, with enriched coverage targeting putative environmentally susceptible sequences in human sperm. Improvements in our ability to examine sperm DNA methylation following different environmental impacts (i.e., toxicants, exposures, stressors) may optimize assessment of the risks associated with alterations to the germline epigenome and the subsequent health of future generations.
Methods
Sample Collection
In order to capture/introduce variability in sperm DNA methylation, a group of 39 men, representing diverse subtypes, varying by fertility status, age, smoking status, MTHFR genotype, and folic acid use, were selected. Participants were recruited from three Canadian cities. From Toronto, 24 healthy normospermic male participants were recruited from the Canadian Reproductive Assisted Technology (CReATe) fertility clinic and provided a single semen sample. These men were considered fertile given that they had normal sperm parameters (Cooper et al. 2010) and that the couple presented to the clinic due to known female factor infertility; indeed, 67% (16/24) of the participants had achieved a previous pregnancy. The Toronto samples were chosen to introduce diversity through differing MTHFR genotypes, smoking status, and, at least in part, those who had previously fathered children. Twelve men from the Montreal area were selected from the McGill University Reproductive Centre or the OVO Clinic. Although the Montreal participants were normospermic, they were considered idiopathic infertile because their partners had no known causes for female infertility. The 12 individuals from Montreal, after consulting with their andrologists, received high-dose folic acid supplementation (), and sperm samples were collected prior to and within 1 week following 6 months of supplementation. These samples were chosen to include differing MTHFR genotypes, use of folic acid supplements, and idiopathic infertility. Finally, three additional participants of unknown fertility status were recruited from the Ottawa Fertility Clinic; these men were included due to their advanced age. In all cohorts, semen samples were collected by masturbation following a recommended 3-day minimum of abstinence. Following semen liquefaction (20–30 min at room temperature), an aliquot was taken for sperm counts and the rest of the sample was immediately frozen at . Informed consent was obtained from all participants. The study was approved by all respective research and ethics boards.
DNA Isolation and MTHFR Genotyping
Sperm were lysed in a buffer containing a final concentration of Tris, ethylenediaminetetraacetic acid, dithiothreitol, proteinase K, and 0.1% sarkosyl detergent and were incubated overnight at 37°C. DNA was then extracted using the QIAamp® DNA Mini kit (Qiagen) according to the manufacturer’s protocols. The common single nucleotide polymorphism, MTHFR , was genotyped from all sperm DNA samples using polymerase chain reaction (PCR)–restriction fragment length polymorphism, as originally described by Frosst et al. (1995) and detailed by Sener et al. (2014). Briefly, following DNA amplification, a PCR product of in size is produced. Presence of the MTHFR polymorphism creates a Hinf I restriction cut site; when cleaved, this results in fragments of 175 and , that can be visualized through gel electrophoresis.
Bisulfite Pyrosequencing
We screened sperm DNA samples for possible somatic cell contamination through bisulfite pyrosequencing of the imprinting control regions (ICRs) for H19 imprinted maternally expressed transcript (H19) and mesoderm-specific transcript (MEST), a paternally and maternally methylated imprinted gene, respectively, on all subjects. As well, this technique was used to validate differentially methylated cytosines (DMCs) found to be altered following analysis of human sperm capture sequencing data (see below). Here, primers were designed to overlap the areas where differential methylation was observed (i.e., intron 4 of sterile alpha motif domain containing 11 (SAMD11) and an intergenic region). For the validation, pyrosequencing results from five participants from each the MTHFR 677CC and 677TT groups were compared with their associated human sperm capture panel results. The average methylation between genotypes, as well as individual patient’s methylation data, was compared with the average/same patient’s capture sequencing data.
For all pyrosequencing assays, of sperm DNA underwent bisulfite conversion with the EpiTect® bisulfite kit (Qiagen) according to the manufacturer’s protocol. Bisulfite PCR was conducted using primers (see Excel Table S1) and pyrosequencing was performed as previously described (Dejeux et al. 2009). Briefly, regions of interest were PCR amplified with one of the primers being biotinylated. Capture of the biotinylated strand was performed with streptavidin-coated sepharose beads and washed using the PyroMark® Q24 Vacuum Workstation (Qiagen). A sequencing primer was annealed to the isolated captured template strand and the pyrosequencing reaction was conducted using the PyroMark® Q24 kit (Qiagen) as per the manufacturer’s protocol.
For somatic cell contamination, sperm DNA samples were considered not to be contaminated if methylation across all analyzed CpGs in both imprinted genes did not deviate from the expected high levels of methylation for H19 () and low levels for MEST () (Kläver et al. 2013). All samples in the current study met the criteria for lack of somatic cell contamination, and thus no sperm DNA samples were excluded (Table 1).
Table 1.
Demographics of participants for the pooled human sperm WGBS (WGBS-Pool) library.
| Characteristics | , , and/or range |
|---|---|
| Age | |
| Mean | |
| Range | 27–61 y |
| Imprinted gene methylation | |
| H19 (mean, range) | , |
| MEST (mean, range) | , |
| Fertility statusa | |
| Fertile | 21 |
| Infertile | 6 |
| Unknown | 3 |
| Sperm counts (million/mL) | |
| Mean | ()b |
| Range | 3.8–497.7 |
| Percentage DFI (%) | |
| Mean | ()b |
| Range | 6.05–30.9 |
| Smoking status (n) | |
| Smokers | 6 |
| Nonsmokers | 23 |
| Unknown | 1 |
| MTHFR genotype (n) | |
| MTHFR CC | 13 |
| MTHFR CT | 2 |
| MTHFR TT | 15 |
| High-dose folic acid use [ (n)] | |
| Yes | 6 |
| No | 24 |
Note: DFI, DNA fragmentation index; MTHFR, methylenetetrahydrofolate reductase; SD, standard deviation; WGBS, whole genome bisulfite sequencing.
A fertile status was determined if participants were normospermic and presented to the clinic due to known female factor infertility (see “Methods” section).
Sperm counts and %DFI were not measured in all individuals due to limited amount of sample available upon collection.
WGBS and Targeted Capture Sequencing
WGBS and targeted bisulfite sequencing were performed as previously described (Allum et al. 2015; Cheung et al. 2017). To examine the variability in human sperm DNA methylation, a subset of the men (total 30 participants; Table 1) was chosen in order to produce a single WGBS library pool (WGBS-Pool). A subset of the total of 39 men was chosen in order to ensure sufficient depth of sequencing from all participants in the single WGBS library. More specifically, samples (total ) were chosen to reflect differing MTHFR genotypes and smoking status from the Toronto cohort (); MTHFR 677TT genotype, idiopathic infertility and folic acid supplementation use from the Montreal cohort (); and advanced aged from the Ottawa cohort (). Equal amounts of sperm DNA from these 30 participants were combined in order to make the pooled sperm DNA sample used. The WGBS-Pool library was constructed using the KAPA® High Throughput Library Preparation kit (Roche/KAPA® Biosystems). Briefly, of the sperm DNA was spiked with 0.1% (w/w) unmethylated and pUC19 DNA (Promega). DNA was sonicated (S220 Focused-ultrasonicator, Covaris) and fragment sizes of were controlled on a Bioanalyzer DNA 1000 LabChip® (Agilent). Following fragmentation, DNA-end repair of double-stranded DNA breaks, 3′-end adenylation, adaptor ligation, and clean-up steps were conducted according to KAPA® Biosystems’ protocols. The sample was then bisulfite converted using the EpiTect® Fast DNA bisulfite kit (Qiagen) following the manufacturer’s protocol. The resulting bisulfite DNA was quantified with OliGreen® (Life Technology) and amplified with 9–12 PCR cycles using the KAPA® HiFi HotStart DNA Polymerase kit (Roche/KAPA® Biosystems) according to suggested protocols. The final WGBS library was purified using Agencout® AMPure® Beads (Beckman Coulter), validated on Bioanalyzer High Sensitivity DNA LabChip® kits (Agilent) and quantified by PicoGreen® (ThermoFisher).
Targeted bisulfite sequencing was performed on the same 30-participant pooled sperm DNA sample used for WGBS (Capture-Pool) to compare the technique as well as on 45 individual samples (Table 2). These samples were chosen in order to examine the effect of MTHFR genotype alone from the Toronto cohort (MTHFR 677CC , 677TT ), and to examine the effect of folic acid supplementation and MTHFR genotype on a cohort of idiopathic infertile men from Montreal ( per genotype, before and after supplementation; i.e., 24 total samples). Following WGBS library preparations for all individual samples (as described above), the MCC-Seq protocol developed and optimized by Roche NimbleGen® was applied. Briefly, the SeqCap® Epi Enrichment System protocol (Roche NimbleGen®) was used to capture the regions of interest. Equal amounts of multiplexed libraries ( of each, 12 samples per capture) were combined to obtain of total input library, which was hybridized to the capture panel at 47°C for 72 h. Washing, recovery, and PCR amplification of the captured libraries, as well as final purification were conducted as recommended by the manufacturer. Bioanalyzer High Sensitivity DNA LabChip® kits (Agilent) were used to determine quality, concentration, and size distribution of the final captured libraries.
Table 2.
Demographics of samples assessed with the human sperm capture panel.
| Characteristic | Cohort | ||
|---|---|---|---|
| Toronto | Montreal, pre-folic acid | Montreal, post-folic acid | |
| , , or range | , , or range | , , or range | |
| Total number of participants () | 21 | 12 | 12 |
| Age | |||
| Range | 28–61 y | 26–53 y | 26–53 y |
| Fertility statusa | Fertile | Infertile | Infertile |
| Sperm counts (million/mL) | |||
| Range | 38.0–140.0 | 27.5–536.5 | 46.9–674.8 |
| Percentage DFI (%) | |||
| Range | 6.1–30.9 | 7.7–39.42 | 7.64–27.6 |
| Smoking status () | |||
| Smokers | 6 | 0 | 0 |
| Nonsmokers | 15 | 12 | 12 |
| MTHFR genotype () | |||
| MTHFR CC | 13 | 6 | 6 |
| MTHFR TT | 8 | 6 | 6 |
| High-dose folic acid use [ ()] | |||
| Yes | 0 | 0 | 12 |
| No | 21 | 12 | 0 |
Note: DFI, DNA fragmentation index; MTHFR, methylenetetrahydrofolate reductase; SD, standard deviation.
A fertile status was determined if participants were normospermic and presented to the clinic due to known female factor infertility (see “Methods” section).
The single WGBS-pool library was sequenced over four lanes using the Illumina® HiSeq2000 system, whereas the capture libraries were sequenced over eight lanes on the Illumina® HiSeq4000 system. All sequencing used paired-end sequencing.
Sequencing Data Processing
WGBS HiSeq reads were aligned to the bisulfite-converted reference genome hg19/GRCh37 using BWA (version 0.6.1) (Li and Durbin 2009). Low-quality sequences (Phred score ) were trimmed from the 3′ end of paired reads. Following alignment, read-pairs not mapped at the expected distance based on the library insert size, as well as reads a) that were clonal/duplicates, b) with low mapping quality, c) mapping to both forward and reverse strands, and d) with mismatches were removed, as previously described (Johnson et al. 2012). Individual CpG methylation calling was extracted using SAMtools (version 0.1.18) in mpileup mode.
Targeted MCC-Seq HiSeq reads were aligned using an in-house GenAP_pipe pipeline (https://bitbucket.org/mugqic/genpipes). Specifically, the MCC-Seq paired-end raw reads were first trimmed for quality (), length (), and Illumina® adapters using Trimmomatic (version 0.36) (Bolger et al. 2014). The trimmed reads were then aligned, per sequencing lane, to the pre-indexed reference genome hg19/GRCh37 using Bismark (version 0.18.2) (Krueger and Andrews 2011) with Bowtie 2 (version 2.3.1) (Langmead and Salzberg 2012) in pair-end mode and default parameters. Lane BAM files were merged and then de-duplicated using Picard (Broad Institute, version 2.9.0). Methylation calls were obtained using Bismark. BisSNP (version 0.82.2) (Liu et al. 2012) was run on the de-duplicated BAM files to call variants. For both WGBS and MCC-Seq data, CpGs that were found to be overlapping with SNPs (dbSNP 137), the Data Analysis Center (DAC) Blacklisted Regions or Duke Excluded Regions [both generated by the Encyclopedia of DNA Elements (ENCODE) project] were removed. CpG sites with less then coverage were also discarded and genomic locations were annotated with HOMER, using default parameters. Sequencing data from WGBS and from MCC-Seq have been submitted to the European Genome-phenome Archive under the accession number EGAS00001003617.
Comparison with Other Human Sperm WGBS Data
Our WGBS-Pool data were compared with publically available data on human sperm from [Molaro et al. 2011 (GSE3040; WGBS-Prev)] where sperm DNA methylation data from two anonymous donors were pooled after sequencing. The processed data were downloaded (reference genome hg18) and were converted to the reference genome hg19 using the University of California, Santa Cruz (UCSC) Genome Browser tool Batch Coordinate Conversion (liftOver). Similar to our data, only sites with coverage were used for analysis. For the comparison of common sites between the two data sets (WGBS-Pool vs. WGBS-Prev), the intersectBed feature of bedtools (version 2.27.0) was used to select overlapping sites.
Statistical Analyses
Generalized linear regression models (GLMs) were built using the methylation proportion inferred from the combination of methylated reads and unmethylated reads as a binomially distributed response variable to look for associations between DNA methylation and a) MTHFR genotype (e.g., MTHFR 677CC vs. MTHFR 677TT) or b) high-dose folic acid supplementation (e.g., before vs. after). We used the R function glm (R Development Core Team, version 3.2.1) and the binomial family to fit the model, and calculated p-values for variables of interest. We corrected the obtained p-values by generating false discovery proportion q-values using the R package q-values (Chung and Storey 2015). We selected significant DMCs as q or nominal p , with a minimum of methylation level difference of . Specifically, for the comparison of MTHFR genotype (Toronto cohort; 677CC: , 677TT: ), results were significant for q and a minimum methylation difference of 10%. For the effect of folic acid supplementation in the Montreal cohort ( per genotype, per time point), due to the relatively smaller sample size, significant results were reported for nominal p and a minimum methylation difference of 10%.
Graphs were made and statistical analyses performed using GraphPad Prism (version 6.01), and statistical significance was set at for the analyses described below. Absolute values were compared by Fisher’s exact test, when comparing smoking and alcohol consumption distributions among participants. t-Tests were performed to compare the age (unpaired), serum/red blood cell (RBC) folate levels (paired), and overall sperm DNA methylation levels (paired) between MTHFR genotypes or pre/post-folic acid supplementation. Unpaired t-test and analysis of variance (ANOVA) was used in examining DNA methylation variation. Enrichment of different publically available data sets was compared between the total number of sites analyzed through our human sperm capture panel and the different folate metabolism–related DMCs using test with Yates’ correction.
Results
Deep WGBS on a Pooled Sperm DNA Sample
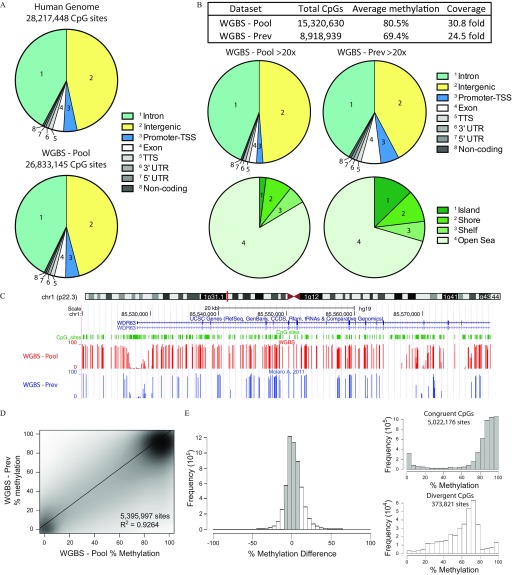
We assembled an equimolar pool of sperm DNA from a subset of the participants recruited (30 men) varying in age, fertility status, MTHFR genotype, and exposures (folic acid supplements, smoking) (Table 1). By having such a wide cross section of men, we wished to assess the variation in human sperm DNA methylation caused by having a complex pool of sperm DNA. The men came from three Canadian cities: a) Toronto, a fertile cohort; b) Montreal, an idiopathic infertility cohort; and c) Ottawa, an aged cohort (see Excel Table S2). Imprinted gene methylation was assessed by bisulfite pyrosequencing to assess sample purity and rule out samples with somatic cell contamination (i.e., abnormal imprinted gene methylation affected equally across all imprinted gene loci); all samples were accepted because they each showed the expected methylation for sperm with methylation at the paternally methylated H19 locus and methylation at the maternally methylated MEST locus (Table 1). A single WGBS library from the 30 men (WGBS-Pool) was prepared and sequenced to a depth of 1,672,735,160 raw reads, yielding an average CpG coverage of , which corresponded to an average genome-wide methylation of 74.1% (see Excel Table S3). Over 95% of CpG dinucleotides had sequence coverage (Figure 1A).
Figure 1.
Whole genome bisulfite sequencing (WGBS) of human sperm DNA. (A) Examination of the genomic distribution of CpG sites found within the human genome (top) and from WGBS (bottom) of a pooled DNA sample (WGBS-Pool). (B) Analysis of highly covered () sites from WGBS-Pool and that of WGBS-Prev examining their genomic distribution and their location within CpG islands, shores and shelves. (C) Genome browser view of DNA methylation tracks from WGBS-Pool and WGBS-Prev data sets. (D) Scatter plot of common CpGs sequenced at between WGBS data sets. (E) Difference in DNA methylation of common CpG sites (WGBS-Pool minus WGBS-Prev, left). DNA methylation of sites congruent ( methylation difference, shaded bars) and divergent ( methylation difference, open bars) sites, right top and bottom, respectively. Human genome total CpG sites were derived from the human genome sequence and downloaded through the WashU EpiGenome Browser (https://egg.wustl.edu/d/hg19/CpGsites.gz). Note: chr, chromosome; CpG, 5′-cytosine-phosphate-guanine-3′; TSS, transcriptional start site; TTS, transcriptional termination site; UTR, untranslated region; WGBS-pool, WGBS library pool; CCDS, consensus coding sequence; WGBS-Prev, publically available data on human sperm from Molaro et al. (2011).
Comparing our WGBS-Pool with a previous WGBS data set looking at human sperm DNA methylation from two individual participants (WGBS-Prev; Molaro et al. 2011), we covered 1.7-times more CpG sites at coverage (Figure 1B,C). The average methylation of the highly covered sites was found to be 80.5% and 69.4% for our WGBS-Pool and WGBS-Prev, respectively (Figure 1B, table). This difference in methylation may be explained by the fact that a greater proportion of highly covered sites from the WGBS-Prev was found within promoter–transcriptional start site (TSS) regions as well as within CpG islands (Figure 1B, bottom); these features generally show hypomethylation and would therefore reduce the overall DNA methylation. Although the WGBS-Pool data sequenced more CpG sites at high coverage, comparing the methylation of common sites with the published data demonstrated similar DNA methylation levels (Figure 1C,D); a total of 5,395,997 common CpGs were sequenced and showed a strong correlation (). When examining the difference in methylation of the common/overlapping sites between the two data sets, a large majority of the CpGs (5,022,176 sites; 93.1%) demonstrated congruent/similar ( difference) methylation (Figure 1E, shaded bars) and displayed mainly low () or high () levels of methylation. Conversely, divergent CpGs, where a methylation difference between data sets was observed (373,821 sites; open bars), demonstrated mainly intermediate levels of methylation, between 20% and 80%.
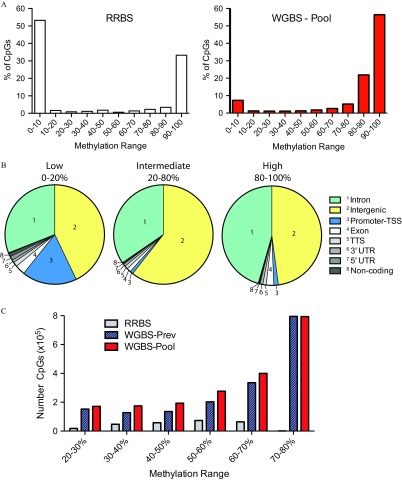
Because our WGBS-Pool was generated from DNA from 30 individuals, creating a complex pool, we hypothesized that sites of intermediate methylation would contain sites of dynamic methylation due to variability in sperm methylation states (intra- and inter-individual). Interestingly, in a recent study using blood and adipose tissue, it was demonstrated that environmental differentially methylated regions (DMRs) possess intermediate methylation, outside of the hypomethylated promoter areas (Busche et al. 2015). Techniques, such as RRBS, interrogate high-CpG density sequences, which normally show relatively stable hypomethylation (Figure 2A, left panel; data from Aarabi et al. (2015). In line with this, we found from our WGBS-Pool data (Figure 2A, right) that a significant proportion of CpGs possessing 0–20% methylation mapped within promoter-TSS regions (defined as to from TSS), which tend to harness low variation (Figure 2B, Low). More specifically, 74.6% of all CpGs found within promoter-TSS regions within the genome possessed methylation. Our data also demonstrated that the large majority of sequenced CpGs possessed methylation between 80% and 100% (Figure 2A, right panel). Intermediate-level methylation (i.e., 20–80%) was observed in the WGBS-Pool for 2.01 million CpGs, as compared with 0.26 and 1.75 million sites from RRBS and WGBS-Prev, respectively (Figure 2C); these intermediate sites were concentrated to intergenic CpGs as well as sites in intronic regions, and both areas contained distal sequences that can regulate gene activity (Figure 2B, Intermediate).
Figure 2.
Distribution of sperm DNA methylation from our WGBS-Pooled sample. (A) Percentage of CpGs with specific DNA methylation ranges sequenced from RRBS (left) the WGBS-Pool (, right). (B) Genomic distribution of CpG sites with low (0–20%), intermediate (20–80%), and high (80–100%) DNA methylation from WGBS-Pool data. (C) Number of CpG sites demonstrating intermediate levels of DNA methylation interrogated by RRBS, WGBS-Prev, and WGBS-Pool. WGBS-Prev data is from Molaro et al. 2011. RRBS data is from Aarabi et al. 2015. Note: CpG, 5′-cytosine-phosphate-guanine-3′; RRBS, reduced representation bisulfite sequencing; TSS, transcriptional start site; TTS, transcriptional termination site; UTR, untranslated region; WGBS, whole genome bisulfite sequencing; WGBS-pool, WGBS library pool; WGBS-Prev, publically available data on human sperm from Molaro et al. (2011).
Sperm Methylation Capture Design for MCC-Sequencing
We designed a capture panel targeting regions of intermediate methylation in human sperm as a cost-efficient alternative to shotgun WGBS. Specifically, in order to target regions of intermediate levels of methylation, sliding windows of bins, containing a minimum of two consecutive CpGs, were constructed using our WGBS-Pool data. The methylation levels of each bin were calculated, sorted and binned regions with a minimum absolute difference from the 50% methylation level were retained (250,000 bins above and below); overlapping bins were further merged. In addition, to allow comparisons with human sperm DNA methylation profiles generated by us and others using Illumina® arrays (Aston et al. 2015; Chan et al. 2017; Jenkins et al. 2014, 2017; Krausz et al. 2012), the complete set of CpGs from the array () was added. Upstream and downstream flanking regions were added to each individual probe location and were then added to those determined through our WGBS data, with overlapping regions merged.
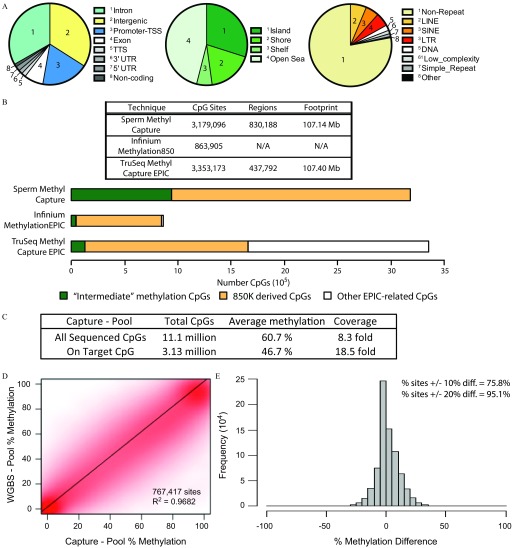
Our design for the human sperm methyl capture yielded of sequence targetable by Roche NimbleGen® for synthesis of a custom SeqCap® Epi probe panel, containing 830,188 regions capturing 3,179,096 CpG sites. These targeted sites were found mainly in a) intergenic (34%), intronic (33%), and promoter-TSS (19%); b) CpG island; and c) nonrepetitive regions and were dispersed throughout the genome (Figure 3A; see also Figure S1A). Of the CpG sites on the human sperm capture panel, 937,141 sites represented those derived from our WGBS-Pool data possessing regions of intermediate methylation (i.e. intermediate methylation captured CpGs); the remainder represented -derived sites (Figure 3B).
Figure 3.
Creation and validation of the human sperm capture panel. (A) Sites captured by the human sperm capture panel analyzing their distribution by genomic regions (left); within CpG island, shores, and shelves (middle); and within repetitive elements (right). (B) Summary of different techniques and their features (top, table). Comparison of the number of sites analyzed that are intermediate methylation captured CpGs (labeled “Intermediate” methylation CpGs), 850K dereived CpGs and other EPIC-related CpGs from different assays. (C) Capture sequencing information of the 30-participant pooled sperm DNA sample (Capture-Pool). (D) Scatter plot analysis of DNA methylation of common sites found by sequencing the pooled DNA samples using the targeted human sperm capture panel (Capture-Pool) and by WGBS (WGBS-Pool). (E) Difference in methylation (Capture-Pool minus WGBS-Pool) of the common CpG sites. Note: CpG, 5′-cytosine-phosphate-guanine-3′; diff, difference; LINE, long interspersed nuclear elements; LTR, long terminal repeats; SINE, short interspersed nuclear elements; TSS, transcriptional start site; TTS, transcriptional termination site; UTR, untranslated region; WGBS-pool, WGBS library pool.
Comparing the human sperm methyl capture with other techniques available, we noted that the commercially available TruSeq® Methyl Capture EPIC (EPIC capture) from Illumina® targets a similar DNA sequence footprint and total number of CpG sites, although fewer regions (Figure 3B). The largest differences are observed for intermediate methylation captured CpG sites captured, and although these constitute approximately one-third of our human sperm capture panel, they represent a small proportion of the sites analyzed with the array or the EPIC capture (5.2% and 3.8% of sites, respectively) (Figure 3B, bottom and Figure S1B). Notably, the intermediate methylation captured CpGs sites are found mainly in intergenic and outside of CpG-dense areas of the genome, whereas CpG sites covered by the other techniques include a greater proportion of promoter-TSS regions and CpG islands (see Figure S1C). EPIC-related CpGs not sequenced by our human sperm methyl capture panel, were mainly found in repetitive elements and immediately flanking CpG-dense areas in the EPIC design (see Figure S2). Finally, as compared with microarray technologies, the capture sequencing allowed for the measurement of genetic in parallel with epigenetic variation (see below and the “Methods” section).
Variation in DNA Methylation in Intermediate Methylation Captured CpG Sites
We hypothesized that the intermediate methylation captured CpGs sites on our human sperm capture panel, derived from our WGBS-Pool data, would represent sites with dynamic methylation. In other words, these sites would demonstrate higher variability in methylation compared with other sites sequenced. To test this, the panel was used to capture and sequence 45 individual human sperm samples (21 individuals from Toronto and 12 individuals with two time points from Montreal, discussed further below). Examining only sites targeted and sequenced with coverage in at least 30 of the 45 participants, we calculated the standard deviation of each CpG. As shown in Figure S3, the intermediate methylation captured CpG sites () had significantly higher average variation () compared with -derived sites found on our capture panel (). Because the numbers of sites differed between the intermediate methylation captured and -derived CpGs, we randomly chose subsets of the -derived CpGs in order to obtain similar numbers (permutated five times); the results were similar, with intermediate methylation captured CpGs demonstrating significantly greater variation. With these results, hereafter, we denote intermediate methylation captured CpGs as dynamic sperm CpG sites.
Validation with Targeted Sequencing of Pooled Sperm DNA
In order to validate the use of the human sperm capture panel, the same pooled sperm DNA used for the WGBS-Pool was sequenced following targeted capture (Capture-Pool). We observed a minimum of coverage at a total of 11.1 million CpGs (average ) (Figure 3C). A large proportion of these CpGs sequenced were found in regions not intentionally targeted by our panel due to nonspecific/off-target capture of other genomic sequences; these sites are covered at low depth and demonstrated an average coverage of (see Figure S4A). By contrast, we saw enrichment of our targeted sites where 3.13 million CpGs ( of targets) were sequenced, demonstrating an intermediate methylation (46.7%) at an elevated average coverage of (Figure 3C; see also Figure S4B).
Comparing common sites sequenced from the WGBS-Pool and Capture-Pool data, 767,417 CpG sites were sequenced at a minimum coverage using both techniques and showed a correlation of (Figure 3D). Examining the difference in methylation detected between the two sequencing methods, greater than 75% of common sites demonstrated methylation difference between the techniques; at a level of 20% difference in methylation, 95% of common sites were included (Figure 3E). In addition, the sequencing coverage affected the correlation particularly at dynamic sperm CpG sites (see Figure S4C). When examining sites specifically targeted by our human sperm capture panel, from our deep sequencing of the WGBS-Pool, we obtained approximately coverage for these sites from over 1.67 billion reads (see Excel Table S3). In comparison with our Capture-Pool targeted sequencing, with only a little over 46.5 million reads, we obtained a similar coverage of targeted sites (). Thus, targeted CpGs were equally covered at only 3.4% raw data depth of the shotgun WGBS, improving cost-efficiency for population studies.
MCC-Sequencing in Sperm from Individual Men
We next utilized the human sperm methyl capture panel to assess the effect of two different, yet related, perturbations of folate metabolism: a) samples from the Toronto fertile cohort, to examine the effect of MTHFR genotype; and b) participants from the Montreal infertile cohort, to examine the interaction of MTHFR genotype and high-dose folic acid supplementation (before vs. after; Table 2). Specifically, a different subset of sperm from 21 men from the CReATE fertility clinic in Toronto was used. Because they presented at the clinic due to known female factor infertility, they were considered to be fertile. As well, 12 healthy normospermic participants were used from the Montreal cohort, where semen samples were collected before and after a folic acid supplementation (i.e., a total of 24 samples); this subset of participants was previously analyzed using RRBS (Aarabi et al. 2015). These men presented with idiopathic infertility given that female factor infertility was excluded. Although a significant difference in sperm counts was observed between the Toronto and Montreal cohorts (Table 2), all men analyzed with the human sperm capture (45 men in total) were considered normospermic because they met the criteria according to WHO guidelines (; Cooper et al. 2010). This difference in sperm counts may be due to methodology or counting biases used at the different sites.
Principal component analysis (PCA) was performed for all the individual libraries based on genotyping data extracted from the methylome sequencing (see “Methods” section; see also Figure S4D). Given that of the variance can be explained by the first two principal components ( and ), the genetic ancestry of the participants is comparable and would not be considered a confounder. Similar to the Capture-Pool library preparation, on average approximately CpGs were sequenced at a minimum coverage, whereas targeted regions showed an enrichment of highly covered CpGs (average CpGs at -fold coverage; see Figure S4E and Excel Table S3).
Imprinted Gene DNA Methylation Patterns from Targeted Sequencing
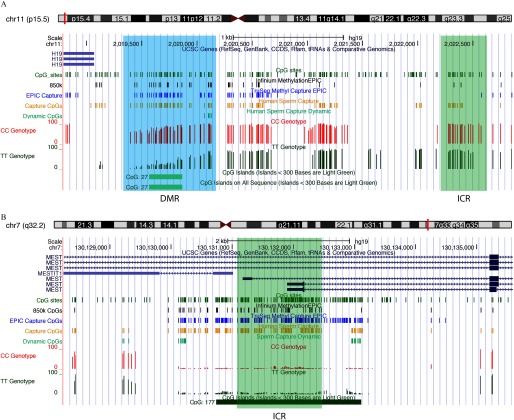
We examined in greater detail the sperm DNA methylation patterns at several imprinted genes because abnormal sperm DNA methylation patterns at imprinted loci have previously been observed in men suffering from infertility (Kobayashi et al. 2009; Li et al. 2013; Poplinski et al. 2010). Figure 4 depicts regions of two imprinted genes, H19 and MEST, and shows the methylation of highly covered CpG sites from an individual CC and TT patient (CC and TT genotype tracks, respectively). On inspection of the imprinted loci and their ICRs, we observed that the array interrogated only a subset of the CpG sites found within the regions. This can be seen particularly at the paternally methylated H19 ICR (Figure 4A), where only two -array probes are located ( track), compared with the human sperm capture, where 17 sites are targeted (Capture CpG track). As with many maternally methylated ICRs, the MEST ICR is found within a CpG island and is well covered with the different assays (Figure 4B).
Figure 4.
Examination of imprinted gene methylation from human sperm capture sequencing data. UCSC Genome Browser view of the imprinting control centers (ICRs, green regions) of (A) the paternally methylated gene H19, and (B) the maternally methylated gene MEST. Custom tracks indicate CpG sites (green), CpGs analyzed by Infinium MethylationEPIC array (black), TruSeq® Methyl Capture EPIC (blue), our human sperm Capture (gold), and Dynamic CpG sites (light green). Tracks for representative capture sequencing data (all sequenced sites with coverage are shown) for a CC individual (red) and a TT individual (dark green) are shown. As compared with the gold tracks, which show only on-target CpGs (of a total of 3.13 million; see Figure 3C), more CpG sites are marked in the CC and TT tracks as sites sequenced at coverage are shown (of a total of 11.1 million as shown in Figure 3C). Note: chr, chromosome; CpG, 5′-cytosine-phosphate-guanine-3′; DMR, differentially methylated region; ICR, imprinting control region; UCSC, University of California, Santa Cruz; CDDS, consensus coding sequence.
Earlier bisulfite pyrosequencing of these imprinted loci examined only a few sites within the ICR of two imprinted genes (5 and 10 CpG sites for H19 and MEST, respectively). The high-density data (17 and 126 sites for H19 and MEST, respectively) obtained using the human sperm capture panel on the 45 individual samples demonstrated no significant MTHFR genotype- or folic acid-dependent variation in these and other ICRs (see Figure S5). Although no differences were found in ICR methylation, a DMR could be observed just upstream of the H19 promoter when comparing MTHFR genotypes of two individual subjects (Figure 4A, shaded area).
Impact of Lifelong MTHFR Deficiency on Sperm DNA Methylation
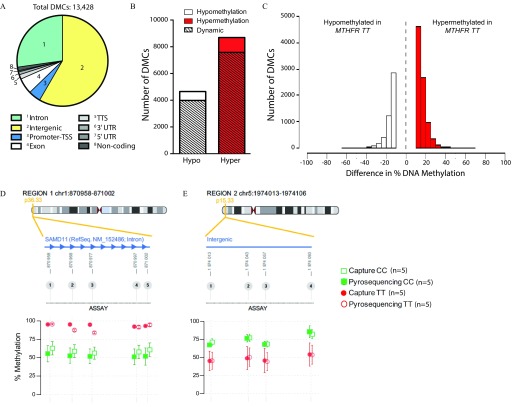
The first phenotypic correlations we carried out using the human sperm capture panel interrogated the effect of a common human functional polymorphism in MTHFR on sperm DNA methylation given that we have previously demonstrated that functional Mthfr variation in mice is a global sperm methylation modifier (Aarabi et al. 2018). In mice, MTHFR is expressed at higher levels in the testis than any other tissue (Chen et al. 2001). Because MTHFR 677TT individuals have a genetic deficiency that results in a thermolabile form of the enzyme with of residual activity (Kang et al. 1991), they have the equivalent of a lifelong MTHFR deficiency; such deficiency could impact DNA methylation patterning in the fetal or postnatal testis. Here, sperm from 13 MTHFR 677CC and 8 677TT fertile men from Toronto were analyzed (mean genome coverage of and , respectively). Both groups were similarly distributed based on age ( and ), smoking (4 of 13 and 2 of 8 participants) and alcohol consumption (3 of 13 and 2 of 8 participants, for 677CC and TT, respectively). Figure S6A shows Q-Q and Manhattan plots of association p-values for tested sites. From these, a total of 13,428 DMCs were found to be significantly altered due to MTHFR genotype, at a false discovery rate of and a minimum of 10% difference in methylation, and were found mainly in intergenic and intronic regions of the genome (Figure 5A). A greater number of sites were found to have increased methylation in MTHFR 677TT compared with MTHFR 677CC men (8,756 hyper- vs. 4,672 hypomethylated DMCs; Figure 5B,C). Interestingly, a vast majority (86.7%) of the DMCs discovered were those shown to be dynamic sperm CpG sites, uniquely targeted in our assay. DMCs were commonly annotated to the same gene/region within the genome and predominantly found clustered together (see Excel Table S4). Examples include several intergenic regions, the promoter and first exon/intron of the noncoding RNA LOC100130872, and within the introns of SAMD11, transcription elongation factor B polypeptide 3C-like (TCEB3CL), and disks large-associated protein (DLGAP2). The differential methylation of several DMCs was validated using bisulfite pyrosequencing: An intron of SAMD11 (5 CpGs; Figure 5D) and an intergenic region (4 CpGs; Figure 5E) demonstrated excellent concordance in DNA methylation between bisulfite pyrosequencing and human sperm capture panel results. Inter-individual variability between participants within each group was observed and also validated (see Figure S7 and Excel Table S5).
Figure 5.
Effect of MTHFR genotype, in Toronto cohort men, on sperm DNA methylation. Total number of DMCs between human MTHFR 677CC () and 677TT () genotypes with their (A) genomic region distribution, (B) demonstrating mainly hypermethylation and changes in dynamic sperm CpG sites, and (C) differences in methylation between genotypes. Pyrosequencing® validation of several DMCs found within (D) an intron of SAMD11 and (E) an intergenic region. . Note: chr, chromosome; CpG, 5′-cytosine-phosphate-guanine-3′; DMCs, differentially methylated cytosines; hyper, hypermethylated; hypo, hypomethylated; MTHFR, methylenetetrahydrofolate reductase; SAMD11, sterile alpha motif domain containing 11; TSS, transcriptional start site; TTS, transcriptional termination site; UTR, untranslated region.
The MTHFR 677TT genotype-associated differences in methylation are reminiscent of those we reported in mice heterozygous for a targeted mutation in Mthfr, a model for MTHFR deficiency similar to that found in MTHFR 677TT men (Aarabi et al. 2018). Our previous sperm DNA methylation data generated with RRBS were reanalyzed to generate individual CpG data. Similar to the present results, the majority of the DMCs were found in intergenic and intronic regions (see Figure S6B) and revealed hypermethylation in mice compared with their wild-type littermates (Figure S6C,D); 8,549 DMCs were found to have an increased level of methylation in mice, whereas 407 sites showed decreased levels.
Impact of Short-Term Folic Acid Exposure on Sperm DNA Methylation
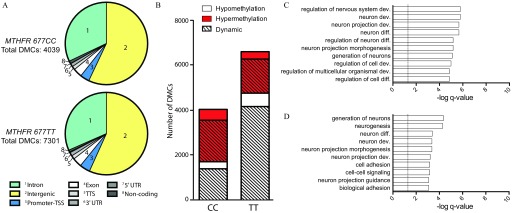
We selected a subset of men ( MTHFR 677CC and MTHFR 677TT) who had also been examined earlier by us using RRBS (Aarabi et al. 2015), and we used the human sperm methyl capture panel to analyze the effect of 6 months of treatment with high-dose folic acid supplements () on sperm DNA methylation. Six months of folic acid supplement treatment covers two rounds of spermatogenesis and can be considered a short-term perturbation of folate metabolism when compared with the long-term (lifelong) effect of MTHFR genotype described above. All men in this cohort were nonsmokers, and were equally distributed based on age ( and for 677CC and TT, respectively); alcohol consumption was not recorded for this cohort. Serum and RBC folate levels were previously measured (Aarabi et al. 2015), and reanalysis of the subset of men used in our present study demonstrated similar results, where no differences between MTHFR genotype were observed; however, elevated serum (see Figure S8A) and RBC (see Figure S8B) folate levels were observed following supplementation. We compared group effects of supplementation on DNA methylation in both MTHFR genotypes separately (see Figure S9A,B). Because of the small number of participants with each genotype, sites were nominated to be differentially methylated with and a minimum of 10% difference in methylation. Similar to our previous results, the overall sperm DNA methylation from each MTHFR genotype was not affected following high-dose folic acid supplementation (677CC: and ; 677TT: and , baseline and following supplementation, respectively). However, supplementation resulted in 4,039 and 7,301 DMCs in the MTHFR 677CC and 677TT groups, respectively, and were distributed similarly in terms of genomic regions (Figure 6A; see also Excel Tables S6 and S7). The 677CC genotype was associated with a slight tendency for increased DNA methylation (2,343 hyper- and 1,696 hypomethylated DMCs; Figure 6B). Similar to our reported RRBS findings, men with the MTHFR 677TT genotype showed a significantly higher proportion of hypomethylated sites in sperm (, test with Yates’ correction); 4,765 sites demonstrated loss, whereas 2,535 sites showed increases in methylation. Similar to genotype effects seen in the Toronto samples, approximately 80% of DMCs for both genotypes were discovered to be dynamic sperm sites. In addition, paralleling MTHFR genotype effects, altered methylation was found close to or within SAMD11 and DLGAP2 following folic acid supplementation in both 677CC and 677TT subjects
Figure 6.
Effect of folic acid supplementation on the human sperm DNA methylome. (A) Genomic distribution of DMCs found in MTHFR 677CC (top, ) and 677TT (bottom, ) following 6-month folic acid supplementation. (B) DMCs breakdown by hypo- and hypermethylation as well as dynamic sperm CpG sites. Gene Ontology (GO) analysis for enrichment of biological processes in DMCs found within genic regions from subjects of (C) 677CC and (D) 677TT genotypes. Note: CpG, 5′-cytosine-phosphate-guanine-3′; dev, development; diff, differentiation; DMCs, differentially methylated cytosines; MTHFR, methylenetetrahydrofolate reductase; TSS, transcriptional start site; TTS, transcriptional termination site; UTR, untranslated region.
To ensure low sampling bias (comparing our earlier larger RRBS study to the current proof-of-principle capture study), we restricted the comparison of RRBS and human sperm capture panel results to the same 12 MTHFR 677CC and 677TT individuals included in both studies. We observed convergence of measured effects by the independent methods: 677TT men being more affected than 677CC men and also having a greater loss of sperm DNA methylation following supplementation (see Figure S9C). Gene ontology (GO) analysis, using DMCs found within genes, identified an enrichment of biological processes related to nervous system development and neuron differentiation for MTHFR 677CC and 677TT genotypes in both the human sperm capture panel data (Figures 6C,D, respectively) and the reanalysis of RRBS data (see Figure S9D,E, respectively).
Response to Long- vs. Short-Term Perturbations in Folate Metabolism
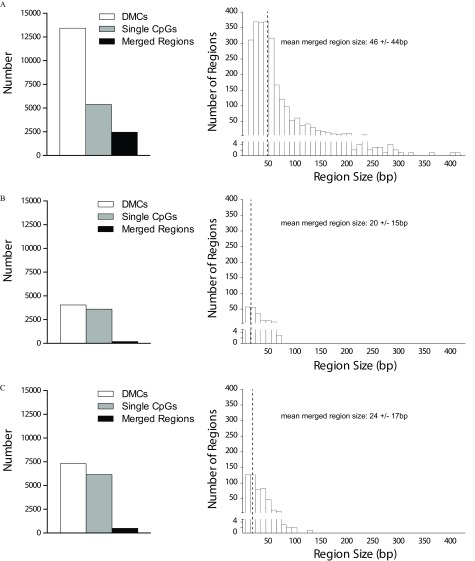
As previously mentioned, many sites demonstrating differential methylation were annotated within the same gene/region. We therefore merged neighboring DMCs found within close proximity (within ) to determine whether there were regions of differential methylation. When examining the effect of the MTHFR genotype, 40% of DMCs (5,384 sites) remained as isolated CpG sites (Figure 7A, left). Combining DMCs within resulted in 2,450 merged regions, with the largest found to be (Figure 7A, right). In contrast, the use of folic acid supplementation for both MTHFR 677CC and 677TT groups showed that many of the altered DMCs remained as individual CpG sites (92% and 84%, respectively; Figure 7B,C, respectively); only 188 (maximum ) and 495 (maximum ) merged regions were discovered, respectively. The largest 10 merged regions from each comparison are listed in Excel Table S8. Thus, folic acid supplementation resulted in fewer and smaller merged regions.
Figure 7.
Regions of differential methylation following folate metabolism perturbations. (A) Comparisons of MTHFR 677CC vs. 677TT genotypes and effect of folic acid supplementation in (B) 677CC or (C) 677TT participants were examined: left panels—total DMCs, isolated single CpG sites, and merged regions obtained after neighboring DMCs found within were merged; right panels—size distribution of merged regions. Note: CpG, 5′-cytosine-phosphate-guanine-3′; DMCs, differentially methylated cytosines; MTHFR, methylenetetrahydrofolate reductase.
The stark contrast between the effects of MTHFR genotype effect vs. high-dose folic acid supplementation can be observed in Figure 8. MTHFR 677TT subjects showed higher levels of methylation compared with MTHFR 677CC subjects within the seventh intron of DLGAP2 (Figure 8A). Here, six regions (ranging from a single CpG site to in size) were found encompassing 25 DMCs showing 15–20% hypermethylation due to the MTHFR polymorphism. Interestingly, all the altered sites were found to be dynamic sperm CpGs. Examining the effect of folic acid supplementation in MTHFR 677TT subjects, the largest DMR ( in size) was found within the first intron of epidermal growth factor receptor (EGFR) and contained four dynamic sperm CpG sites demonstrating decreased methylation (12–20% loss) after supplementation.
Figure 8.
Observed differential methylation due to MTHFR genotype and following folic acid supplementation. Examples showing regions of differential methylation seen between (A) men with MTHFR 677CC compared with 677TT genotype or (B) following high-dose folic acid supplementation in 677TT subjects. Note: chr, chromosome; MTHFR, methylenetetrahydrofolate reductase; UCSC, University of California, Santa Cruz; CCDS, consensus coding sequence.
Functional Correlates of Folate Metabolism–Related Differential DNA Methylation
Finally, it is possible that the folate metabolism–associated altered sperm DNA methylation relates to functional or sensitive areas in the genome. We therefore determined whether the DMCs or the merged regions we identified overlapped with putative functional regions from published studies. Although some overlap was found with histone modifications, evolutionarily constrained elements, and conserved sperm DNA methylation patterns, no enrichment was seen above background of the human sperm capture panel (see Figure S10A–F).
We next examined whether sites or regions would be similarly affected following different perturbations or exposures. Within our present results, few DMCs/regions overlapped when comparing effects of MTHFR genotype vs. those of folic acid supplementation. Interestingly, few altered DMCs were in common between MTHFR 677CC and 677TT subjects following folic acid supplementation (see Figure S10H). WGBS was recently used to assess the effect of serum dioxin concentrations on the sperm DNA methylome. Again, few regions intersected with the DMCs found in our current study (see Figure S10G). Although there appears to be no specific susceptible regions in human sperm DNA methylation, exposure-specific signatures may be present.
Discussion
We applied WGBS on a population pooled sperm DNA sample to develop a targeted capture panel for the analysis of variable human sperm DNA methylation. This human sperm methyl capture panel not only targets the commonly assessed gene promoter/CpG island regions but also captures novel dynamic sperm DNA methylation sites at innocuous putative distal regulatory elements. These dynamic sperm sites represented the majority of CpGs differing in methylation levels between individuals with altered one-carbon metabolism (MTHFR deficiency) and those given high-dose folic acid supplements.
Several targeted capture panels have been designed including those that examine the DNA methylome across different cells and tissues (Ziller et al. 2016) and those that are more tissue-specific, for instance, an adipose tissue–targeted panel (Allum et al. 2015). The sperm epigenome, with its low retention of histones ( in humans) (Hammoud et al. 2009, 2014) and specialized DNA methylome (Ziller et al. 2013), differs greatly from that of other cell types, indicating that development of a customized sperm panel is warranted. The approach we chose to use, in order to discover variable regions in the sperm DNA methylome, utilized a pool of sperm DNA from a diverse group of men. Although we were able to cover nearly twice as many CpG sites at coverage, our new human sperm WGBS data set compared well with the published WGBS data set from two normospermic men (Molaro et al. 2011), especially for sites with methylation levels or . In contrast, CpG sites where DNA methylation differed by between the two WGBS data sets were mostly found to possess intermediate levels of methylation (20–80%). In addition, with our WGBS-Pool data set we were able to identify about 260,000 more novel sites of intermediate methylation, likely due to inter-individual variation in the pooled discovery cohort, compared with RRBS (Aarabi et al. 2015) or other WGBS data on single/two pooled samples (Molaro et al. 2011). Our results underscore the importance of assessing population as well as tissue lineage-dictated differences in epigenetic landscapes.
Our customized sperm capture design incorporated the regions of intermediate methylation identified by WGBS, which were found to be more variable than other CpG sites targeted and were denoted as dynamic sperm CpGs. These sites represented roughly a third of the targeted CpGs; in contrast, of dynamic sperm CpGs are detected by the array or EPIC Capture. Using the WGBS-pool sample to test our capture panel allowed us to confirm that the panel we designed was able to accurately, and at high coverage, capture close to 100% () of the targeted CpGs. Moving on to individual samples, the sperm capture results recapitulated DNA methylation levels of genes, such as imprinted genes, that have been well studied in sperm.
Folate metabolism is important for the synthesis of nucleic acid precursors and amino acids and the production of S-adenosylmethionine (SAM), the universal methyl donor (Bailey et al. 2010). It is therefore not surprising that polymorphisms in MTHFR, a crucial enzyme within this pathway, would lead to altered DNA methylation. With our capture panel, participants homozygous for the polymorphism demonstrated altered sperm DNA methylation, with a greater tendency for hypermethylation. This is in line with our previous animal study demonstrating increased sperm DNA methylation in mice haplo-insufficient for the Mthfr gene and considered a model for MTHFR 677TT individuals (Aarabi et al. 2018). A large proportion of the observed changes were found to be dynamic sperm sites and would not have been detected with other currently available techniques. Furthermore, many of the sites altered due to genotype were increases in sperm DNA methylation. Altered methyl pools and SAM due the MTHFR 677TT variant may affect other epigenetic marks, such as H3K4 methylation. Particularly, H3K4me3 is anticorrelated with DNA methylation (Ooi et al. 2007); therefore, decreases in this histone modification may have resulted in increased levels of DNA methylation at specific sites.
The human sperm methyl capture panel was also used to reevaluate the effect of high-dose folic acid supplementation on a cohort of infertile participants (Aarabi et al. 2015). Use of folic acid resulted in altered DNA methylation patterns in sperm, dependent on the participants’ MTHFR genotype. A greater tendency for hypermethylation was observed in MTHFR 677CC subjects, whereas significant hypomethylation was seen in those homozygous for the T allele. Results here, along with similar results from our previous human and animal model studies, provide further support for our proposal that high circulating and testicular folate levels down-regulate MTHFR, decreasing methyl group availability and leading to loss of DNA methylation (Aarabi et al. 2018). Here, we again discovered that the vast majority of sites with altered methylation, after short-term use of high-dose folic acid supplements, were dynamic sperm CpGs.
Along with infertility, the MTHFR polymorphism has been associated with cancer, vascular, neurological, and psychiatric diseases (Liew and Gupta 2015). MTHFR 677TT subjects were found to have significantly increased methylation at several CpG sites/regions within SAMD11 and DLGAP2. SAMD11 has been found to be a strong candidate gene for autism spectrum disorders through a whole exome sequencing study (Chapman et al. 2015). Similarly, in an animal model of post-traumatic stress disorder caused by an environmental stress, specific hypermethylation within Dlgap2 and decreased expression were associated with the effects of a traumatic environmental stressor (Chertkow-Deutsher et al. 2010). High-dose folic acid supplementation also resulted in altered methylation around these two genes, although only at a few CpG sites. Interestingly however, GO analysis from both MTHFR 677CC and 677TT individuals showed enrichment in biological processes involving neurogenesis following supplementation.
In addition to enrichment for GO analysis, we examined other functional aspects of the altered sites found in our study. It has been reported, so far, that histone retention in sperm is found predominantly at promoters, which are hypomethylated (Brykczynska et al. 2010; Hammoud et al. 2009, 2014); thus, it would not be expected to show great enrichment in our data. Along similar lines, a large proportion of constrained elements, often indicating regions of functional importance, are found mainly in known exons, as well as 5′ and 3′ untranslated regions (UTRs) (Davydov et al. 2010), regions showing little alterations in sperm DNA methylation following perturbations. Finally, it has been reported that an evolutionary expansion of hypomethylated regions in the genome was found (Qu et al. 2018); the majority of DMCs we discovered were within dynamic sperm sites, possessing intermediate levels of methylation; therefore, no enrichment was observed as well.
A differential response was observed between our different perturbations to folate metabolism. We discovered that men homozygous for the MTHFR polymorphism demonstrated a larger number of DMCs when compared with MTHFR 677CC individuals; altered sites detected were found within close proximity, resulting in regions of differential methylation. An altered sperm DNA methylome due to MTHFR genotype is the result of a persistent/lifelong perturbation given that the polymorphism was present since fertilization. The highest enzyme activity of MTHFR in mice was observed in the testes (Chen et al. 2001), and studies from our lab have demonstrated that protein expression was detected in pre- and postnatal mouse germ cells (Garner et al. 2013). Specifically, in mice, the expression of MTHFR was detected at the highest levels during the major time of DNA methylation acquisition (embryonic day 15–18; Garner et al. 2013). Therefore, the observed results may be due to a lifetime altered methyl pool availability and SAM, particularly during in utero development, causing disruptions during male germ cell DNA methylation pattern establishment over larger regions in the genome.
Although genotypic differences of MTHFR examined lifelong perturbations, the effects seen in our supplementation study reflect short-term/acute perturbations. Subjects within this group were given high-dose folic acid supplementation for a 6-month period. This timeframe only covers approximately two rounds of spermatogenesis and sperm maturation. With this limited window of exposure, we did not expect to see as many dramatic and consistent changes in sperm DNA methylation. Indeed, fewer altered DMCs were observed following folic acid supplementation and few neighboring sites were combined to create regions of differential methylation. Studying longer exposures to folic acid supplements and/or examining whether any alterations in the sperm methylome persist following the cessation of supplements, would allow us to determine the reversibility of any changes and whether perturbations affect stem cells, resulting in permanent alterations. Nonetheless, this modest exposure altered the sperm DNA methylome in a manner that could be detected, thereby demonstrating the sensitivity of both the sperm methylome and the human sperm capture panel.
As mentioned previously, a vast majority of the sites altered in methylation were found to be the dynamic sperm CpGs and would not have been detected with many of the commercially available techniques. Whether these dynamic sites are more likely to be affected following other types of exposures in adult men requires further study. Indeed, studies that have examined the effect of different environmental stressors and/or factors—such as smoking (Jenkins et al. 2017), cannabis use (Murphy et al. 2018), obesity (Donkin et al. 2016), phthalates (Wu et al. 2017b), BPA (Tian et al. 2018), childhood abuse (Roberts et al. 2018), and even exercise (Denham et al. 2015)—have demonstrated alterations in human sperm DNA methylation. All but one of these studies (Tian et al. 2018) used different versions of Illumina®’s methylation array or RRBS. The use of the human sperm capture panel would allow the targeting of many of these same regions, with the added benefit of analyzing sites of dynamic methylation, at high coverage and not requiring full and costly assessment of the entire sperm DNA methylome. Examining how the risk of different environmental exposures affects the sperm DNA methylation is of great importance given that these germ cells can influence the health of future generations.
In addition to strengths of our study, there are several limitations. The greatest limitation is the small sample size of patients. Although in observational studies, large cohort sizes are expected, our motivation was to test the applicability of our human capture panel to experimentally detect previously studied alterations due to perturbations in one-carbon metabolism and the folic acid pathway. Although patient numbers in each of our experimental groups were low, we were able to obtain results similar to previous studies in animal models (MTHFR genotype effect) and from larger patient numbers (folic acid effects), demonstrating the suitability of our human sperm capture panel to detect alterations in sperm DNA methylation. Some patient characteristics that could affect the sperm DNA methylome, such as BMI, were not known for our participants, something that is worth following up in future studies using our sperm capture panel. Age may also affect the sperm DNA methylome and represents another factor that should be examined in more detail with our approach in a larger cohort of men. Another limitation of our study is the inability of bisulfite sequencing to distinguish between 5-methylcytosine () and 5-hydroxymethylcytosine (). can be actively demethylated through ten-eleven translocation mediated oxidation to and has been found to be important in the epigenetic reprograming of germ cells in mouse and humans (Hackett et al. 2013; Tang et al. 2015; Yamaguchi et al. 2013). Although is found at several orders of magnitude less than in sperm, recent studies relying on immunofluorescence (Efimova et al. 2017), enzyme-linked immunosorbent assay (ELISA) (Jenkins et al. 2013), and immunoprecipitation followed by sequencing (Zheng et al. 2017) have found altered amounts of in sperm due to semen quality, age, and exposures to bisphenol A, respectively. It would be interesting to determine the impact of different exposures on both and levels through oxidative bisulfite sequencing (Booth et al. 2013), which can be amenable to capture sequencing.
Our pooled approach for developing customized tools for epigenome variability, in a tissue-targeted manner, addresses specific variation in the human sperm epigenome. With our human sperm methylation capture panel, we discovered differential DNA methylation following conditions affecting folate metabolism, most of which was found to be in novel dynamic sperm CpG sites. Our customized panel allows for accurate assessment of sperm DNA methylation profiles at single CpGs with an unprecedented coverage, targets putative environmentally sensitive sequences in human sperm and improves our ability to examine environmental impacts on DNA methylation in human sperm.
Supplementary Material
Acknowledgments
We thank the team at the McGill University and Génome Québec Innovation Centre for performing the sequencing of the WGBS- and MCC-Seq library preparations. This research was enabled in part by support provided by Calcul Québec and Compute Canada.
This work was supported by grants from the Canadian Institutes of Health Research (CIHR) to J.M.T. (FDN-148425, EPT-142875), T.P. and G.B. (EP1-120608, CEE-151618, EPT-142875), S.K. (358654), B.R. (TE1-138298), and J.L.B. (TE1-138294).
Footnotes
Supplemental Material is available online (https://doi.org/10.1289/EHP4812).
These authors contributed equally to this work.
Current address: Xiaojian Shao, Digital Technologies Research Centre, National Research Council Canada, Ottawa, Ontario, Canada.
The authors declare they have no actual or potential competing financial interests
Note to readers with disabilities: EHP strives to ensure that all journal content is accessible to all readers. However, some figures and Supplemental Material published in EHP articles may not conform to 508 standards due to the complexity of the information being presented. If you need assistance accessing journal content, please contact ehponline@niehs.nih.gov. Our staff will work with you to assess and meet your accessibility needs within 3 working days.
References
- Aarabi M, Christensen KE, Chan D, Leclerc D, Landry M, Ly L, et al. 2018. Testicular MTHFR deficiency may explain sperm DNA hypomethylation associated with high dose folic acid supplementation. Hum Mol Genet 27(7):1123–1135, PMID: 29360980, 10.1093/hmg/ddy021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aarabi M, San Gabriel MC, Chan D, Behan NA, Caron M, Pastinen T, et al. 2015. High-dose folic acid supplementation alters the human sperm methylome and is influenced by the MTHFR C677T polymorphism. Hum Mol Genet 24(22):6301–6313, PMID: 26307085, 10.1093/hmg/ddv338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhaled Y, Laqqan M, Tierling S, Lo Porto C, Amor H, Hammadeh ME. 2018. Impact of cigarette-smoking on sperm DNA methylation and its effect on sperm parameters. Andrologia 50(4):e12950, PMID: 29315717, 10.1111/and.12950. [DOI] [PubMed] [Google Scholar]
- Allum F, Shao X, Guénard F, Simon MM, Busche S, Caron M, et al. 2015. Characterization of functional methylomes by next-generation capture sequencing identifies novel disease-associated variants. Nat Commun 6:7211, PMID: 26021296, 10.1038/ncomms8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aston KI, Uren PJ, Jenkins TG, Horsager A, Cairns BR, Smith AD, et al. 2015. Aberrant sperm DNA methylation predicts male fertility status and embryo quality. Fertil Steril 104(6):1388–1397, PMID: 26361204, 10.1016/j.fertnstert.2015.08.019. [DOI] [PubMed] [Google Scholar]
- Bailey RL, Dodd KW, Gahche JJ, Dwyer JT, McDowell MA, Yetley EA, et al. 2010. Total folate and folic acid intake from foods and dietary supplements in the United States: 2003–2006. Am J Clin Nutr 91(1):231–237, PMID: 19923379, 10.3945/ajcn.2009.28427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouki R, Melén E, Herceg Z, Beckers J, Chen J, Karagas M, et al. 2018. Epigenetics as a mechanism linking developmental exposures to long-term toxicity. Environ Int 114:77–86, PMID: 29499450, 10.1016/j.envint.2018.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120, PMID: 24695404, 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth MJ, Ost TW, Beraldi D, Bell NM, Branco MR, Reik W, et al. 2013. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nat Protoc 8(10):1841–1851, PMID: 24008380, 10.1038/nprot.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, et al. 2010. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol 17(6):679–687, PMID: 20473313, 10.1038/nsmb.1821. [DOI] [PubMed] [Google Scholar]
- Busche S, Shao X, Caron M, Kwan T, Allum F, Cheung WA, et al. 2015. Population whole-genome bisulfite sequencing across two tissues highlights the environment as the principal source of human methylome variation. Genome Biol 16:290, PMID: 26699896, 10.1186/s13059-015-0856-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R, et al. 2010. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell 143(7):1084–1096, PMID: 21183072, 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan D, McGraw S, Klein K, Wallock LM, Konermann C, Plass C, et al. 2017. Stability of the human sperm DNA methylome to folic acid fortification and short-term supplementation. Hum Reprod 32(2):272–283, PMID: 27994001, 10.1093/humrep/dew308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman NH, Nato AQ Jr, Bernier R, Ankenman K, Sohi H, Munson J, et al. 2015. Whole exome sequencing in extended families with autism spectrum disorder implicates four candidate genes. Hum Genet 134(10):1055–1068, PMID: 26204995, 10.1007/s00439-015-1585-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Karaplis AC, Ackerman SL, Pogribny IP, Melnyk S, Lussier-Cacan S, et al. 2001. Mice deficient in methylenetetrahydrofolate reductase exhibit hyperhomocysteinemia and decreased methylation capacity, with neuropathology and aortic lipid deposition. Hum Mol Genet 10(5):433–443, PMID: 11181567, 10.1093/hmg/10.5.433. [DOI] [PubMed] [Google Scholar]
- Chertkow-Deutsher Y, Cohen H, Klein E, Ben-Shachar D. 2010. DNA methylation in vulnerability to post-traumatic stress in rats: evidence for the role of the post-synaptic density protein Dlgap2. Int J Neuropsychopharmacol 13(3):347–359, PMID: 19793403, 10.1017/S146114570999071X. [DOI] [PubMed] [Google Scholar]
- Cheung WA, Shao X, Morin A, Siroux V, Kwan T, Ge B, et al. 2017. Functional variation in allelic methylomes underscores a strong genetic contribution and reveals novel epigenetic alterations in the human epigenome. Genome Biol 18(1):50, PMID: 28283040, 10.1186/s13059-017-1173-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung NC, Storey JD. 2015. Statistical significance of variables driving systematic variation in high-dimensional data. Bioinformatics 31(4):545–554, PMID: 25336500, 10.1093/bioinformatics/btu674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TG, Noonan E, von Eckardstein S, Auger J, Baker HW, Behre HM, et al. 2010. World Health Organization reference values for human semen characteristics. Hum Reprod Update 16(3):231–245, PMID: 19934213, 10.1093/humupd/dmp048. [DOI] [PubMed] [Google Scholar]
- Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. 2010. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput Biol 6(12):e1001025, PMID: 21152010, 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejeux E, abdalaoui HE, Gut IG, Tost J. 2009. Identification and quantification of differentially methylated loci by the Pyrosequencing™ technology. Methods Mol Biol 507:189–205, PMID: 18987816, 10.1007/978-1-59745-522-0_15. [DOI] [PubMed] [Google Scholar]
- Denham J, O’Brien BJ, Harvey JT, Charchar FJ. 2015. Genome-wide sperm DNA methylation changes after 3 months of exercise training in humans. Epigenomics 7(5):717–731, PMID: 25864559, 10.2217/epi.15.29. [DOI] [PubMed] [Google Scholar]
- Donkin I, Versteyhe S, Ingerslev LR, Qian K, Mechta M, Nordkap L, et al. 2016. Obesity and bariatric surgery drive epigenetic variation of spermatozoa in humans. Cell Metab 23(2):369–378, PMID: 26669700, 10.1016/j.cmet.2015.11.004. [DOI] [PubMed] [Google Scholar]
- Edwards JR, Yarychkivska O, Boulard M, Bestor TH. 2017. DNA methylation and DNA methyltransferases. Epigenetics Chromatin 10:23, PMID: 28503201, 10.1186/s13072-017-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimova OA, Pendina AA, Tikhonov AV, Parfenyev SE, Mekina ID, Komarova EM, et al. 2017. Genome-wide 5-hydroxymethylcytosine patterns in human spermatogenesis are associated with semen quality. Oncotarget 8(51):88294–88307, PMID: 29179435, 10.18632/oncotarget.18331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, et al. 1995. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet 10(1):111–113, PMID: 7647779, 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- Garner JL, Niles KM, McGraw S, Yeh JR, Cushnie DW, Hermo L, et al. 2013. Stability of DNA methylation patterns in mouse spermatogonia under conditions of MTHFR deficiency and methionine supplementation. Biol Reprod 89(5):125, PMID: 24048573, 10.1095/biolreprod.113.109066. [DOI] [PubMed] [Google Scholar]
- Gaysinskaya V, Miller BF, De Luca C, van der Heijden GW, Hansen KD, Bortvin A. 2018. Transient reduction of DNA methylation at the onset of meiosis in male mice. Epigenetics Chromatin 11(1):15, PMID: 29618374, 10.1186/s13072-018-0186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkountela S, Zhang KX, Shafiq TA, Liao WW, Hargan-Calvopiña J, Chen PY, et al. 2015. DNA demethylation dynamics in the human prenatal germline. Cell 161(6):1425–1436, PMID: 26004067, 10.1016/j.cell.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett JA, Sengupta R, Zylicz JJ, Murakami K, Lee C, Down TA, et al. 2013. Germline DNA demethylation dynamics and imprint erasure through 5-hydroxymethylcytosine. Science 339(6118):448–452, PMID: 23223451, 10.1126/science.1229277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Low DH, Yi C, Carrell DT, Guccione E, Cairns BR. 2014. Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell 15(2):239–253, PMID: 24835570, 10.1016/j.stem.2014.04.006. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. 2009. Distinctive chromatin in human sperm packages genes for embryo development. Nature 460(7254):473–478, PMID: 19525931, 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TG, Aston KI, Cairns BR, Carrell DT. 2013. Paternal aging and associated intraindividual alterations of global sperm 5-methylcytosine and 5-hydroxymethylcytosine levels. Fertil Steril 100(4):945–951, PMID: 23809503, 10.1016/j.fertnstert.2013.05.039. [DOI] [PubMed] [Google Scholar]
- Jenkins TG, Aston KI, Meyer TD, Hotaling JM, Shamsi MB, Johnstone EB, et al. 2016. Decreased fecundity and sperm DNA methylation patterns. Fertil Steril 105(1):51–57, PMID: 26453269, 10.1016/j.fertnstert.2015.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TG, Aston KI, Pflueger C, Cairns BR, Carrell DT. 2014. Age-associated sperm DNA methylation alterations: possible implications in offspring disease susceptibility. PLoS Genet 10(7):e1004458, PMID: 25010591, 10.1371/journal.pgen.1004458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins TG, James ER, Alonso DF, Hoidal JR, Murphy PJ, Hotaling JM, et al. 2017. Cigarette smoking significantly alters sperm DNA methylation patterns. Andrology 5(6):1089–1099, PMID: 28950428, 10.1111/andr.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MD, Mueller M, Game L, Aitman TJ. 2012. Single nucleotide analysis of cytosine methylation by whole-genome shotgun bisulfite sequencing. Curr Protoc Mol Biol 99:21.23.1–21.23.28, PMID: 22870857, 10.1002/0471142727.mb2123s99. [DOI] [PubMed] [Google Scholar]
- Kang SS, Wong PW, Susmano A, Sora J, Norusis M, Ruggie N. 1991. Thermolabile methylenetetrahydrofolate reductase: an inherited risk factor for coronary artery disease. Am J Hum Genet 48(3):536–545, PMID: 1998339. [PMC free article] [PubMed] [Google Scholar]
- Kläver R, Tüttelmann F, Bleiziffer A, Haaf T, Kliesch S, Gromoll J. 2013. DNA methylation in spermatozoa as a prospective marker in andrology. Andrology 1(5):731–740, PMID: 23970452, 10.1111/j.2047-2927.2013.00118.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Hiura H, John RM, Sato A, Otsu E, Kobayashi N, et al. 2009. DNA methylation errors at imprinted loci after assisted conception originate in the parental sperm. Eur J Hum Genet 17(12):1582–1591, PMID: 19471309, 10.1038/ejhg.2009.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krausz C, Sandoval J, Sayols S, Chianese C, Giachini C, Heyn H, et al. 2012. Novel insights into DNA methylation features in spermatozoa: stability and peculiarities. PLoS One 7(10):e44479, PMID: 23071498, 10.1371/journal.pone.0044479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. 2011. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27(11):1571–1572, PMID: 21493656, 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9(4):357–359, PMID: 22388286, 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laqqan M, Tierling S, Alkhaled Y, Porto CL, Solomayer EF, Hammadeh ME. 2017. Aberrant DNA methylation patterns of human spermatozoa in current smoker males. Reprod Toxicol 71:126–133, PMID: 28576685, 10.1016/j.reprotox.2017.05.010. [DOI] [PubMed] [Google Scholar]
- Levine H, Jørgensen N, Martino-Andrade A, Mendiola J, Weksler-Derri D, Mindlis I, et al. 2017. Temporal trends in sperm count: a systematic review and meta-regression analysis. Hum Reprod Update 23(6):646–659, PMID: 28981654, 10.1093/humupd/dmx022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Li JB, Xiao XF, Ma YF, Wang J, Liang XX, et al. 2013. Altered DNA methylation patterns of the H19 differentially methylated region and the DAZL gene promoter are associated with defective human sperm. PLoS One 8(8):e71215, PMID: 24015185, 10.1371/journal.pone.0071215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760, PMID: 19451168, 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew SC, Gupta ED. 2015. Methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism: epidemiology, metabolism and the associated diseases. Eur J Med Genet 58(1):1–10, PMID: 25449138, 10.1016/j.ejmg.2014.10.004. [DOI] [PubMed] [Google Scholar]
- Liu Y, Siegmund KD, Laird PW, Berman BP. 2012. Bis-SNP: combined DNA methylation and SNP calling for Bisulfite-seq data. Genome Biol 13(7):R61, PMID: 22784381, 10.1186/gb-2012-13-7-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumey LH, Stein AD, Kahn HS, van der Pal-de Bruin KM, Blauw GJ, Zybert PA, et al. 2007. Cohort profile: the Dutch Hunger Winter families study. Int J Epidemiol 36(6):1196–1204, PMID: 17591638, 10.1093/ije/dym126. [DOI] [PubMed] [Google Scholar]
- Ly L, Chan D, Trasler JM. 2015. Developmental windows of susceptibility for epigenetic inheritance through the male germline. Semin Cell Dev Biol 43:96–105, PMID: 26265561, 10.1016/j.semcdb.2015.07.006. [DOI] [PubMed] [Google Scholar]
- Molaro A, Hodges E, Fang F, Song Q, McCombie WR, Hannon GJ, et al. 2011. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell 146(6):1029–1041, PMID: 21925323, 10.1016/j.cell.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SK, Itchon-Ramos N, Visco Z, Huang Z, Grenier C, Schrott R, et al. 2018. Cannabinoid exposure and altered DNA methylation in rat and human sperm. Epigenetics 13(12):1208–1221, PMID: 30521419, 10.1080/15592294.2018.1554521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson EE, Sadler-Riggleman I, Skinner MK. 2018. Environmentally induced epigenetic transgenerational inheritance of disease. Environ Epigenet 4(2):dvy016, PMID: 30038800, 10.1093/eep/dvy016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, et al. 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448(7154):714–717, PMID: 17687327, 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilsner JR, Shershebnev A, Medvedeva YA, Suvorov A, Wu H, Goltsov A, et al. 2018. Peripubertal serum dioxin concentrations and subsequent sperm methylome profiles of young Russian adults. Reprod Toxicol 78:40–49, PMID: 29550351, 10.1016/j.reprotox.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poplinski A, Tüttelmann F, Kanber D, Horsthemke B, Gromoll J. 2010. Idiopathic male infertility is strongly associated with aberrant methylation of MEST and IGF2/H19 ICR1. Int J Androl 33(4):642–649, PMID: 19878521, 10.1111/j.1365-2605.2009.01000.x. [DOI] [PubMed] [Google Scholar]
- Qu J, Hodges E, Molaro A, Gagneux P, Dean MD, Hannon GJ, et al. 2018. Evolutionary expansion of DNA hypomethylation in the mammalian germline genome. Genome Res 28(2):145–158, PMID: 29259021, 10.1101/gr.225896.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AL, Gladish N, Gatev E, Jones MJ, Chen Y, MacIsaac JL, et al. 2018. Exposure to childhood abuse is associated with human sperm DNA methylation. Transl Psychiatry 8(1):194, PMID: 30279435, 10.1038/s41398-018-0252-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sener EF, Oztop DB, Ozkul Y. 2014. MTHFR gene C677T polymorphism in autism spectrum disorders. Genet Res Int 2014:698574, PMID: 25431675, 10.1155/2014/698574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WW, Dietmann S, Irie N, Leitch HG, Floros VI, Bradshaw CR, et al. 2015. A unique gene regulatory network resets the human germline epigenome for development. Cell 161(6):1453–1467, PMID: 26046444, 10.1016/j.cell.2015.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Zhou X, Miao M, Li DK, Wang Z, Li R, et al. 2018. Association of bisphenol A exposure with LINE-1 hydroxymethylation in human semen. Int J Environ Res Public Health 15(8):E1770, PMID: 30126118, 10.3390/ijerph15081770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins AJ, Sinclair KD. 2014. Paternal low protein diet affects adult offspring cardiovascular and metabolic function in mice. Am J Physiol Heart Circ Physiol 306(10):H1444–H1452, PMID: 24658019, 10.1152/ajpheart.00981.2013. [DOI] [PubMed] [Google Scholar]
- Wu H, Ashcraft L, Whitcomb BW, Rahil T, Tougias E, Sites CK, et al. 2017a. Parental contributions to early embryo development: influences of urinary phthalate and phthalate alternatives among couples undergoing IVF treatment. Hum Reprod 32(1):65–75, PMID: 27927842, 10.1093/humrep/dew301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Estill MS, Shershebnev A, Suvorov A, Krawetz SA, Whitcomb BW, et al. 2017b. Preconception urinary phthalate concentrations and sperm DNA methylation profiles among men undergoing IVF treatment: a cross-sectional study. Hum Reprod 32(11):2159–2169, PMID: 29024969, 10.1093/humrep/dex283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Hong K, Liu R, Inoue A, Shen L, Zhang K, et al. 2013. Dynamics of 5-methylcytosine and 5-hydroxymethylcytosine during germ cell reprogramming. Cell Res 23(3):329–339, PMID: 23399596, 10.1038/cr.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Zhou X, Li DK, Yang F, Pan H, Li T, et al. 2017. Genome-wide alteration in DNA hydroxymethylation in the sperm from bisphenol A-exposed men. PLoS One 12(6):e0178535, PMID: 28582417, 10.1371/journal.pone.0178535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Gu H, Müller F, Donaghey J, Tsai LT, Kohlbacher O, et al. 2013. Charting a dynamic DNA methylation landscape of the human genome. Nature 500(7463):477–481, PMID: 23925113, 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziller MJ, Stamenova EK, Gu H, Gnirke A, Meissner A. 2016. Targeted bisulfite sequencing of the dynamic DNA methylome. Epigenetics Chromatin 9:55, PMID: 27980681, 10.1186/s13072-016-0105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.