Abstract
Microbial eukaryotes (protists) are structurally, developmentally and behaviourally more complex than their prokaryotic cousins. This complexity makes it more difficult to translate genomic and metagenomic data into accurate functional inferences about systems ranging all the way from molecular and cellular levels to global ecological networks. This problem can be traced back to the advent of the cytoskeleton and endomembrane systems at the origin of eukaryotes, which endowed them with a range of complex structures and behaviours that still largely dominate how they evolve and interact within microbial communities. But unlike the diverse metabolic properties that evolved within prokaryotes, the structural and behavioural characteristics that strongly define how protists function in the environment cannot readily be inferred from genomic data, since there is generally no simple correlation between a gene and a discrete activity or function. A deeper understanding of protists at both cellular and ecological levels, therefore, requires not only high-throughput genomics but also linking such data to direct observations of natural history and cell biology. This is challenging since these observations typically require cultivation, which is lacking for most protists. Potential remedies with current technology include developing a more phylogenetically diverse range of model systems to better represent the diversity, as well as combining high-throughput, single-cell genomics with microscopic documentation of the subject cells to link sequence with structure and behaviour.
This article is part of a discussion meeting issue ‘Single cell ecology'.
Keywords: protist, genomics, evolution, ecology, behaviour, eukaryotic
1. Introduction
Humans have long sought to explore and classify the nature and extent of Earth's biodiversity. However, measuring biodiversity is surprisingly challenging, and the scope of the problem is much larger than was recognized for most of the history of science. Perhaps ignorance was bliss, as this was a much easier problem when life consisted of what we could see and touch, and the only distinction was between animals and plants. The discovery of microbial life by Leeuwenhoek in the late seventeenth century should have shattered that cozy dichotomy, but instead the problem was kicked to the long grass by simply classifying microbial life as either microbial-plants or microbial-animals for another two centuries, including the obviously problematic fungi (for a more thorough and scholarly treatment of this history, see [1]). Modern concepts of distinct microbial kingdoms evolved throughout the twentieth century, but the abundance, diversity and importance of the microbial world consistently remained a footnote, and microbial kingdoms were depicted as somehow ‘lower' [2,3].
In many ways, this is not surprising since it is impossible to study nearly any aspect of microbial life without some level of technological assistance, so diversity will always be ‘seen' through the lens of whichever technology the observation is based on. If the method biases criteria that are unrepresentative, then even the best estimates will be misleading. Early reliance on simple morphology was tied to the technology available at the time (basic light microscopy), and this bias led to underestimates since we did not understand, and could not see, features to distinguish most microbes. The dependence on cultivation was and continues to be a major source of bias, since only a very small fraction of natural diversity can be cultured [4–7], and many methods require pure cultures. Ultimately, the first real insights into the breadth and depth of the microbial world came from the use of DNA sequence data to address two issues: the evolutionary problem of how organisms are related phylogenetically, and the ecological question of what genetic diversity exists in natural communities. Early attempts to reconstruct a global tree of life quickly showed that most phylogenetic diversity in the tree of life is microbial [8,9], and as the tree has grown in detail this impression has only grown stronger, with new microbial diversity constantly being discovered. At the same time, using molecular data to measure the genetic diversity of natural communities has revealed that the bulk of biodiversity in the environment is also consistently microbial [10–15], and again the more we look in detail, the greater this diversity grows: even microbial ‘species' with marker genes identical in sequence can be functionally diverse. Theoretically, this diversity should be quantifiable using molecular tools, but in practice, this goal remains elusive. Units of biodiversity are seldom clear-cut, might not refer to functionally relevant differences [16–20], estimates never encompass all the major lineages of life and often draw conclusions from non-overlapping subsets of taxa, all biased to underestimating whatever the unit of diversity is. For example, relatively recent and aspirationally definitive studies to estimate ‘global species diversity’ had little taxonomic overlap, used different measures of diversity and different strategies, and varied by more than five orders of magnitude [21,22].
Even the word ‘microbe’ has become biased: it should ideally refer to all microscopic life inclusively, but it is more commonly used to more narrowly refer to bacteria and archaea, excluding microbial eukaryotes (e.g. most of the studies on species concepts and numbers cited above). Eukaryotes include the plants, animals and fungi we know so well, but eukaryotic diversity is dominated by microbial lineages, collectively called protists [23]. Protists inhabit nearly all known ecosystems, and carry out diverse, unique and significant roles in global carbon and nutrient cycling. Their roles in these processes are complex and we are beginning to acknowledge that our models for microbial eukaryotic ecology are over-simplistic [24]. Similarly, many major evolutionary transitions also took place in protist lineages, including the foundations for the evolution of multicellularity that gave us the eukaryotes with which we are most familiar [25–27]. Understanding the nature and evolution of these innovations requires a comprehensive and accurate assessment of protist diversity—one that we do not have.
Despite the important questions surrounding protist diversity, the pace of advances in microbial eukaryotic research has lagged behind that of almost every other part of the tree of life, including other microbes. One result of this is that protist research has tended to follow in the methodological footsteps of bacterial research. This has not been without benefit; indeed, many technological advances made in bacterial research have been quickly adapted to protists [13–15,28–33]. However, microbial eukaryotes are in many respects fundamentally different from bacteria, so there is also the potential to sidetrack progress or even mislead how we interpret data. The easy availability of tools and strategies that were devised to answer questions about bacterial diversity can entice one to focus on problems of marginal significance, rather than developing new methods (or even new approaches using the same tools) to focus directly on more important questions.
Here, I will summarize some of the unique practical challenges that must be overcome to better fit microbial eukaryotic diversity into our understanding of global biodiversity at both evolutionary time-scales (e.g. the divergence of a new kingdom) and ecological time-scales (e.g. eat a bacterium), and also examine whether existing strategies successfully developed to understand bacterial diversity will translate across domains of life. A strongly genome-centric focus has been very successful in reconstructing major events in the deep evolution of eukaryotes, but it is not as clear whether the same can be said for ecology. This is not to say that the adopted methods of environmental tag sequencing, metagenomics, and more recently single-cell genomics and transcriptomics are uninformative. However, we cannot simply assume that these methods will yield the same answers as they do in bacterial ecology, or can be analysed in the same contexts. Because of the different ways that microbial eukaryotes interact with their environment and neighbouring microbial communities, understanding how protists function will be much more dependent on coupling genomic data with direct observations of cell structure and behaviour.
2. Microbial eukaryotes: a marriage of inconveniences
Protists are understudied because, in practical terms, they embody many of the worst characteristics of both microbes and eukaryotes. In the first instance, the challenging issues that make the microbial world as a whole difficult to grasp are just as true of protists as they are of bacteria. They are tiny, single-celled organisms that are difficult to observe to the point of abstraction: they live on the same planet as us, but in a world that is so different from ours that physical factors that most affect their activities and survival can be different from those that we find important [34]. Where known, they have huge population sizes and the structures of these populations and demarkations between ‘species’, if they exist, are poorly defined. They encompass an enormous amount of diversity in the tree of life (figure 1), and much of this is cryptic diversity, where two morphologically indistinguishable cells (to all but the most expert observers) can be separated by a great deal of divergence at molecular or functional levels. Also like bacteria, the vast majority of microbial eukaryotes are not available in culture, and such culture systems and genomic data as do exist are strongly biased to photosynthetic and parasitic species [35–37].
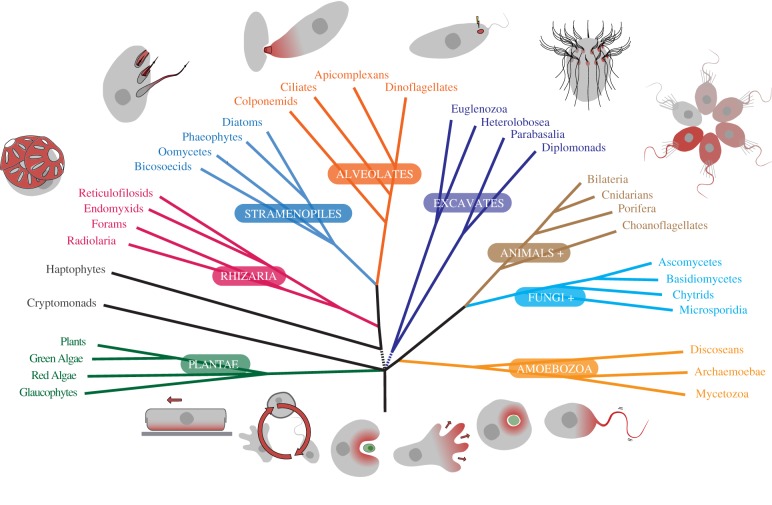
Figure 1.
Tree of eukaryotes. A schematic tree summarizing current data on the relationships between major lineages of eukaryotes with selected ancient and recent examples of structural complexity and behaviour. Examples of basic behaviours (and complex behaviours emerging from them) are indicated at the root of the tree, and are predicted to be present in the last eukaryotic common ancestor (LECA). These include (left to right) gliding motility, ontogenetic development, phagocytosis, amoeboid movement, forming endosymbiotic associations and flagellar swimming. These have all existed since the LECA or soon after, and in some cases, all modern examples are homologous (e.g. flagella), whereas others are constantly reinvented in parallel (e.g. forming endosymbiotic associations or gliding motility). Indicated around the branches of the tree are selected examples of complex structures and behaviours (often resulting from strings of basic behaviours) that emerged many times in parallel. These include (left to right) creating cell walls or armour (by aggregating material, crystalization or scaffolding organic material either inside the cell or by secretion), building and expelling extrusomes, intracellular infection, sensory organelles (light sensing eye spots is shown as an example), structural complexity though symmetrical repetition and colony formation (by reproduction or aggregation). These traits have originated many times throughout the evolution of eukaryotes. (Online version in colour.)
On the other hand, microbial eukaryotes also share many complex features with macroscopic eukaryotes, and these severely test many of the high-throughput methods that have been used to tackle the problems of large populations, extensive diversity and lack of culture in bacteria. For example, many protists have large genomes (sometimes far larger than typical animal genomes: [38–41]) with more genes, complex gene families, repeating elements, and an architecture that makes comprehensive characterization a rarity even among ‘complete genomes'. Like other eukaryotes, protist cells also have many layers of structural complexity, regulation networks, developmental programs and growth requirements absent in bacteria. Perhaps most challenging, however, is that protists also exhibit a wide range of complex activities, akin to animal behaviour. In addition to more universal behaviours like taxis or modifying metabolism according to resource availability, protists also actively hunt prey, shoot projectiles to attack and defend themselves, stun prey and tear them up, or suck out their cytoplasm, actively sort and filter particles, or ingest large items of food. Such behaviours occur via multiple, independently evolved and complex strategies involving novel and physically dynamic structures [23]. Understanding such features is critical to understanding both the evolution and ecology of microbial eukaryotes. However, their secrets will not be easily revealed through high-throughput sequencing alone, because these characteristics cannot be inferred from a genome in the same way that we can infer bacterial metabolic networks [37,42]. Instead, determining what microbial eukaryotes are really doing in natural communities requires at least both genomics and traditional cell biology approaches (and ideally also more biomechanics and physics). This, in turn, depends on well-developed model systems, or at very least culture systems, which are both few and heavily biased.
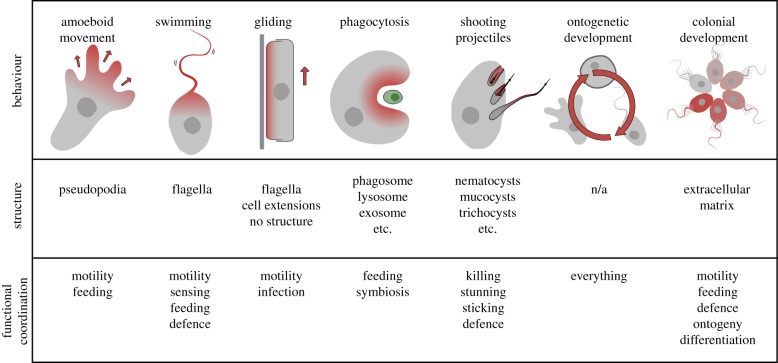
And this problem is even more complex than it first seems, because complex structures can be associated with a range of behaviours in several different ways. For example, consider the 9 + 2 microtubule-based flagella and their associated 9 + 0 basal bodies. These are easily identified, homologous structures with an obvious footprint in the genome [43,44], so the existence of the structure can be predicted from genomics relatively easily. However, these structures can be used in a variety of ways that cannot be predicted from genomics. Flagella beat to effect cell motility, but a wide variety of beat patterns and/or appendages strongly modulate this function [45]. On another level, flagella can also effect motility by a mechanism completely independent of beating: gliding motility [46,47]. And further, flagella can also possess different functions altogether, including sensing and feeding [48–51]. A single cell can use one structure for all of these strategies, or can use several different strategies and structures for one function: cells can swim with flagella, but also glide or use amoeboid movement in varying contexts, or during different life cycle stages [43,47]. This wide-ranging utility is also found in several other basic behaviours (figure 2). Moreover, even if one did meticulously work out the functional basis for one of these behaviours (e.g. a particular flagellar beat pattern), it is not clear that this could be used to infer the same behaviour from the genome of another taxon, because at this level of detail such characters likely evolved many times in parallel, and could, in this case, have non-orthologous underlying mechanisms.
Figure 2.
Examples of eukaryotic behaviours, structures and functional coordination between them. Some basic cell behaviours that are common to many or all eukaryotes are shown at the top, and under each example structures associated with that behaviour are indicated. Where more than one structure is listed, structures might all work together to effect the behaviour, or alternatively in different systems different structures are associated with the behaviour. The bottom panel provides examples of complex processes that can result from coordination of these behaviours and structures with other systems (with an emphasis on processes that are most directly affected by the behaviour). This is not a comprehensive summary, but rather a few examples selected to present a range of complexity, age and functional effects. (Online version in colour.)
On another level again, multiple relatively simple behavioural ‘building blocks' can be assembled into complex and coordinated cell functions with emergent ecological properties. For example, some microbial eukaryotes actively ‘hunt' for prey, which can also be broken down into a highly coordinated combination of behavioural traits including swimming, environmental sensing, taxis, ejecting projectiles, prey differentiation, phagocytosis and exocytosis. Without a way to break down these actions into their simpler building blocks, or even a way to infer the smaller behavioural units from the kind of data we tend to collect, it is impossible to realistically fit such complex ‘metabehaviours' into our ecological models without collecting more kinds of data.
These characteristics can be traced all the way back to the origin and early evolution of eukaryotes. Most of the main features of the complex and dynamic cytoskeleton and endomembrane systems that allow the range of behaviours in modern eukaryotes today are predicted to have been present in the last eukaryotic common ancestor, or LECA (figure 1), and were arguably the most significant changes in the origin of eukaryotes [52]. While this increased the capacity to create new cell types across eukaryotic diversity, early eukaryotes also greatly expanded the range of cell types that a single species could assume through an increase in ontogenetic complexity; once cell cycles commonly expanded to include different cell types, organisms gained flexibility to adapt to changing conditions by altering major structures or even basic body plan, on top of more universal responses like initiating stress responses or changing expression of metabolic pathways. The increasing importance of these systems on cellular function would also have precipitated a major shift in the mode of evolution of early eukaryotic microbes—away from selection on their fit within metabolic networks and towards the behavioural characteristics of the cell. These kinds of characteristics would be subject to change through different kinds of evolutionary processes. Horizontal gene transfer (HGT), for example, is important to shaping bacterial function because of the relatively high degree of modularity and interchangeability of metabolic enzymes. Changes to eukaryotic behaviour, on the other hand, are more likely to be affected by altering developmental and expression networks owing to point mutations in regions controlling the expression of regulators, or perhaps even epigenetic switches, that affect downstream expression patterns and ontogenetic programs. For example, selection to swim faster is unlikely to lead to HGT from fast swimming cells to slower ones. This is not to say that HGT does not affect microbial eukaryotes, but rather that we cannot assume that it is as significant in protist evolution as it is in bacteria.
The advent of these systems—the endomembrane and cytoskeleton—also opened up completely new ecological niches, so nascent eukaryotes would compete with prokaryotic microbes in mixed microbial communities very differently from the ways in which prokaryotes would compete with one another. Differences in modes of mobility, speed of replication and population sizes could all have seen significant changes, as would sensory functions. The most obvious and probably most important change, however, was the ability to eat bacteria. There is no evidence that predation predated eukaryotes, and the emerging ability to efficiently engulf and digest other members of a community to extract their energy and nutrients must have been a game-changing invention for early eukaryotes. It probably had a very strong impact on their early evolution, as it still does today for much of eukaryotic diversity, where it is also a major factor in how protists interact in microbial communities. However, despite the importance of this central characteristic in both evolutionary and ecological time scales, we are only just beginning to test whether it can be inferred from genomic data [53].
3. Combining genomics and morphological data from uncultivated protists
Genomic data have been so effective in bacterial systems not only because it is very informative for these organisms, but also because we have devised ways to generate and analyse it in a high-throughput fashion. Ideally, the high-throughput nature of genomic data could be combined with equally efficient ways to directly observe the structure and behaviour of protist cells. Yet, how to scale-up these kinds of observations is not obvious, especially in the absence of culture systems and other tools. Therefore, moving forward one question to ask is: what information is sacrificed by each of the strategies that are currently available?
Environmental sequence tag data (almost always SSU rRNA) was the earliest molecular strategy to quickly assess overall environmental diversity [13,14]. This continues to be a common and useful method owing to its speed and efficiency. However, it reveals only the taxonomic identity of the cells and nothing about their morphology or behaviour, with the exception of cases where it is coupled to in situ identification [54–56], or by inferring how similar sequences are to homologues from better-studied relatives and extrapolating similar behaviours and lifestyles. Metagenomic and related ‘meta-omic' methods can give a broad picture of the functions of a whole community [28,29,57] but, given the size and nature of eukaryotic genomes, likely not much about the function of individual species or populations since few genes will be physically linked in any sample. Like sequence tag methods, all non-genomic information is also lost. Another general approach that goes back to the cell as a unit of biology is to use microfluidics strategies to make some kind of measurements on individual cells. Microfluidics strategies to measure or sort specific activities are increasingly accessible technologically, but they have not yet been widely used in protists. Fluorescence-activated cell sorting (FACS) has been used to enrich for a subset of taxa from complex communities based on pre-defined criteria, allowing more targeted analyses of certain members of a community at the genomic level [30,58–65]. This is essentially a metagenomic approach, but by disentangling some of the complexity it allows for deeper sampling of whatever kind of organisms has been targeted. Related to this, individual cells can also be sorted and coupled to single-cell genomics methods such as single amplified genomes (SAGs), or single amplified transcriptomes (SATs) [30,59,62,65–69]. These methods are an important technical break from the mixed nature of ‘meta-omic' data, because they retain the individuality of the sample, so all the genes in a dataset can ideally be considered to have come from the same genome (though in practice there is often contamination). These methods still require a priori criteria for sorting, and none of these approaches directly links the genomic data with information about the physical nature of the organism from which the sequence is derived. In theory, single-cell sorting could provide such information, but unfortunately, no system currently combines microscopic imaging with cell sorting in a way that retains a link between an image and a sorted cell.
This common weakness—the absence of a link between high-throughput genomic data and structural and behavioural data from uncultivated protist cells—represents a major methodological challenge. One way forward is a massive increase in the diversity and taxonomic distribution of well-developed model systems. This would allow for much more accurate inferences about the function of various systems from genomic data, as well as the likely biological characteristics of a taxon represented by sequence data. This would, however, require a great leap forward in cell biology and genetics along the lines envisioned by evolutionary cell biology [70,71], more direct mechanical observations of cellular behaviour and how they effect function (e.g. [51]), in addition to a greater general appreciation of skills sometimes sadly considered old fashioned, like natural history and cultivation [36,72,73]. However, a compromise between throughput and information retention is possible with existing resources, by linking single-cell genomics methods to directly isolated cells. For example, a single cell can be imaged or filmed at high resolution to document the basic features of its morphology and whatever behaviours are currently on display, and then manually isolated to couple that information with genomic data from the same cell generated by SAG or SAT methods [31,68,69]. This retains the direct link between genomic data and basic information about the appearance and activities of the cell from which the sequence data are derived, but is not entirely high-throughput since the initial observations and isolation are not automated. Even simple morphological observations such as these can actually reveal a lot that would not be obvious in sequence data alone. Whether a cell is pigmented or colourless can be determined, as can whether it is motile and by what means (e.g. by flagellar swimming, amoeboid movement, or gliding). Phagocytosis and exocytosis can be observed, food can be documented and sometimes identified in a cell (and correlated to genomic data), as can other kinds of characters and interactions like size, biomass, shape, parasitism and symbiotic associations. This certainly does not constitute an exhaustive understanding of the nature of a cell from which genomic data are derived, but given that these single-cell technologies currently exist, it could represent a step in the right direction for gaining a more complete picture of the diversity of uncultured eukaryotes in microbial communities.
4. Concluding remarks: taming the technology
It is true that the future of this field will be defined in large part by what tools are readily available. However, it is critical that we carefully assess what questions need asking before simply applying available technology and allowing the presence of that technology to define these questions. Furthermore, understanding the evolution and ecology of microbial eukaryotes requires ‘old-fashioned' skills and methods that we need to support: we need more expertise in cultivation and documenting natural history, as well as improved skills to observe what a eukaryote is doing in its environment. Indeed, a number of recent papers have described new and unexpected diversity in protists, highlighting results of such methodologies, and they can be combined with genomics and advanced microscopy [74–76]. One way forward will require balancing methodological development to couple this kind of information to genomics methods with higher-throughput, while at the same time developing a more diverse set of model microbial eukaryotic systems that are cultivable and tractable. Together, such advances will allow inferences about functions in natural communities to be more accurate, and in turn increase the predictive power of environmental survey data.
Acknowledgements
Thanks to Naomi Fast, Javier del Campo and Martin Kolisko for critical reading of the manuscript and helpful discussions, and Liz Cooney for assistance with illustrations.
Data accessibility
This article has no additional data.
Competing interests
I declare I have no competing interests.
Funding
This work was supported by a grant from the Natural Sciences and Engineering Research Council of Canada (grant no. 227301).
References
- 1.Ragan MA. 1997. A third kingdom of life: history of an idea. Arch. Protistenkd. 148, 225–243. ( 10.1016/S0003-9365(97)80004-8) [DOI] [Google Scholar]
- 2.Copeland HF. 1938. The kingdoms of organisms. Quart. Rev. Biol. 13, 383–420. ( 10.1086/394568) [DOI] [Google Scholar]
- 3.Whittaker RH. 1969. New concepts of kingdoms of organisims. Science 163, 150–163. ( 10.1126/science.163.3863.150) [DOI] [PubMed] [Google Scholar]
- 4.Staley JT, Konopka A. 1985. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39, 321–346. ( 10.1146/annurev.mi.39.100185.001541) [DOI] [PubMed] [Google Scholar]
- 5.Hugenholtz P. 2002. Exploring prokaryotic diversity in the genomic era. Genome Biol. 3, REVIEWS0003 ( 10.1186/gb-2002-3-2-reviews0003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rappe MS, Giovannoni SJ. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57, 369–394. ( 10.1146/annurev.micro.57.030502.090759) [DOI] [PubMed] [Google Scholar]
- 7.Spang A, Caceres EF, Ettema TJG. 2017. Genomic exploration of the diversity, ecology, and evolution of the archaeal domain of life. Science 357, aaf3883 ( 10.1126/science.aaf3883) [DOI] [PubMed] [Google Scholar]
- 8.Woese CR, Fox GE. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl Acad. Sci. USA 74, 5088–5090. ( 10.1073/pnas.74.11.5088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woese CR. 1987. Bacterial evolution. Microbiol. Rev. 51, 221–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giovannoni SJ, Britschgi TB, Moyer CL, Field KG. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345, 60–63. ( 10.1038/345060a0) [DOI] [PubMed] [Google Scholar]
- 11.Ward DM, Weller R, Bateson MM. 1990. 16S rRNA sequences reveal uncultured inhabitants of a well-studied thermal community. FEMS Microbiol. Rev. 6, 105–115. ( 10.1111/j.1574-6968.1990.tb04088.x) [DOI] [PubMed] [Google Scholar]
- 12.Fuhrman JA, McCallum K, Davis AA. 1992. Novel major archaebacterial group from marine plankton. Nature 356, 148–149. ( 10.1038/356148a0) [DOI] [PubMed] [Google Scholar]
- 13.Lopez-Garcia P, Rodriguez-Valera F, Pedros-Alio C, Moreira D. 2001. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 409, 603–607. ( 10.1038/35054537) [DOI] [PubMed] [Google Scholar]
- 14.Moon-van der Staay SY, De Wachter R, Vaulot D. 2001. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409, 607–610. ( 10.1038/35054541) [DOI] [PubMed] [Google Scholar]
- 15.Moreira D, Lopez-Garcia P. 2002. The molecular ecology of microbial eukaryotes unveils a hidden world. Trends Microbiol. 10, 31–38. ( 10.1016/S0966-842X(01)02257-0) [DOI] [PubMed] [Google Scholar]
- 16.Rossello-Mora R, Amann R. 2001. The species concept for prokaryotes. FEMS Microbiol. Rev. 25, 39–67. ( 10.1111/j.1574-6976.2001.tb00571.x) [DOI] [PubMed] [Google Scholar]
- 17.Finlay BJ. 2004. Protist taxonomy: an ecological perspective. Phil. Trans. R Soc. B 359, 599–610. ( 10.1098/rstb.2003.1450) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Achtman M, Wagner M. 2008. Microbial diversity and the genetic nature of microbial species. Nat. Rev. Microbiol. 6, 431–440. ( 10.1038/nrmicro1872) [DOI] [PubMed] [Google Scholar]
- 19.Caron DA, Countway PD, Savai P, Gast RJ, Schnetzer A, Moorthi SD, Dennett MR, Moran DM, Jones AC. 2009. Defining DNA-based operational taxonomic units for microbial–eukaryote ecology. Appl. Environ. Microbiol. 75, 5797–5808. ( 10.1128/AEM.00298-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konstantinidis KT, Rossello-Mora R. 2015. Classifying the uncultivated microbial majority: a place for metagenomic data in the Candidatus proposal. Syst. Appl. Microbiol. 38, 223–230. ( 10.1016/j.syapm.2015.01.001) [DOI] [PubMed] [Google Scholar]
- 21.Mora C, Tittensor DP, Adl S, Simpson AG, Worm B. 2011. How many species are there on Earth and in the ocean? PLoS Biol. 9, e1001127 ( 10.1371/journal.pbio.1001127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Locey KJ, Lennon JT. 2016. Scaling laws predict global microbial diversity. Proc. Natl Acad. Sci. USA 113, 5970–5975. ( 10.1073/pnas.1521291113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee J, Leedale G, Bradbury P. 2000. An illustrated guide to the protozoa. Lawrence, KS: Allen Press Inc. [Google Scholar]
- 24.Worden AZ, Follows MJ, Giovannoni SJ, Wilken S, Zimmerman AE, Keeling PJ. 2015. Environmental science. Rethinking the marine carbon cycle: factoring in the multifarious lifestyles of microbes. Science 347, 1257594 ( 10.1126/science.1257594) [DOI] [PubMed] [Google Scholar]
- 25.Domozych DS, Domozych CE. 2014. Multicellularity in green algae: upsizing in a walled complex. Front. Plant Sci. 5, 649 ( 10.3389/fpls.2014.00649) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brunet T, King N. 2017. The origin of animal multicellularity and cell differentiation. Dev. Cell 43, 124–140. ( 10.1016/j.devcel.2017.09.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sebe-Pedros A, Degnan BM, Ruiz-Trillo I. 2017. The origin of Metazoa: a unicellular perspective. Nat. Rev. Genet. 18, 498–512. ( 10.1038/nrg.2017.21) [DOI] [PubMed] [Google Scholar]
- 28.Massana R, Karniol B, Pommier T, Bodaker I, Beja O. 2008. Metagenomic retrieval of a ribosomal DNA repeat array from an uncultured marine alveolate. Environ. Microbiol. 10, 1335–1343. ( 10.1111/j.1462-2920.2007.01549.x) [DOI] [PubMed] [Google Scholar]
- 29.Not F, del Campo J, Balague V, de Vargas C, Massana R. 2009. New insights into the diversity of marine picoeukaryotes. PLoS ONE 4, e7143 ( 10.1371/journal.pone.0007143) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoon HS, Price DC, Stepanauskas R, Rajah VD, Sieracki ME, Wilson WH, Yang EC, Duffy S, Bhattacharya D. 2011. Single-cell genomics reveals organismal interactions in uncultivated marine protists. Science 332, 714–717. ( 10.1126/science.1203163) [DOI] [PubMed] [Google Scholar]
- 31.Kolisko M, Boscaro V, Burki F, Lynn DH, Keeling PJ. 2014. Single-cell transcriptomics for microbial eukaryotes. Curr. Biol. 24, R1081–R1082. ( 10.1016/j.cub.2014.10.026) [DOI] [PubMed] [Google Scholar]
- 32.Leray M, Knowlton N. 2016. Censusing marine eukaryotic diversity in the twenty-first century. Phil. Trans. R. Soc. B 371, 20150331 ( 10.1098/rstb.2015.0331) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Z, Hu SK, Campbell V, Tatters AO, Heidelberg KB, Caron DA. 2017. Single-cell transcriptomics of small microbial eukaryotes: limitations and potential. ISME J. 11, 1282–1285. ( 10.1038/ismej.2016.190) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purcell EM. 1977. Life at low Reynolds number. Am. J. Phys. 45, 3–11. ( 10.1119/1.10903) [DOI] [Google Scholar]
- 35.Burki F, Keeling PJ. 2014. Rhizaria. Curr. Biol. 24, R103–R107. ( 10.1016/j.cub.2013.12.025) [DOI] [PubMed] [Google Scholar]
- 36.del Campo J, Sieracki ME, Molestina R, Keeling P, Massana R, Ruiz-Trillo I. 2014. The others: our biased perspective of eukaryotic genomes. Trends Ecol. Evol. 29, 252–259. ( 10.1016/j.tree.2014.03.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keeling PJ, Campo JD. 2017. Marine protists are not just big bacteria. Curr. Biol. 27, R541–R549. ( 10.1016/j.cub.2017.03.075) [DOI] [PubMed] [Google Scholar]
- 38.Lynch M, Conery JS. 2003. The origins of genome complexity. Science 302, 1401–1404. ( 10.1126/science.1089370) [DOI] [PubMed] [Google Scholar]
- 39.Kapraun DF. 2005. Nuclear DNA content estimates in multicellular green, red and brown algae: phylogenetic considerations. Ann. Bot. 95, 7–44. ( 10.1093/aob/mci002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keeling PJ, Slamovits CH. 2005. Causes and effects of nuclear genome reduction. Curr. Opin. Genet. Dev. 15, 601–608. ( 10.1016/j.gde.2005.09.003) [DOI] [PubMed] [Google Scholar]
- 41.Wong JTY. 2019. Architectural organization of dinoflagellate liquid crystalline chromosomes. Microorganisms 7, 27 ( 10.3390/microorganisms7020027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keeling PJ. 2013. Elephants in the room: protists and the importance of structure and behaviour. Environ. Microbiol. Rep. 5, 4–5. [Google Scholar]
- 43.Fritz-Laylin LK, et al. 2010. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell 140, 631–642. ( 10.1016/j.cell.2010.01.032) [DOI] [PubMed] [Google Scholar]
- 44.Dutcher SK, O'Toole ET. 2016. The basal bodies of Chlamydomonas reinhardtii. Cilia 5, 18 ( 10.1186/s13630-016-0039-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sleigh MA, Barlow DI. 1982. How are different ciliary beat patterns produced? Symp. Soc. Exp. Biol. 35, 139–157. [PubMed] [Google Scholar]
- 46.Heintzelman MB. 2006. Cellular and molecular mechanics of gliding locomotion in eukaryotes. Int. Rev. Cytol. 251, 79–129. ( 10.1016/S0074-7696(06)51003-4) [DOI] [PubMed] [Google Scholar]
- 47.Shih SM, Engel BD, Kocabas F, Bilyard T, Gennerich A, Marshall WF, Yildiz A. 2013. Intraflagellar transport drives flagellar surface motility. Elife 2, e00744 ( 10.7554/eLife.00744) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mitchell DR. 2007. The evolution of eukaryotic cilia and flagella as motile and sensory organelles. Adv. Exp. Med. Biol. 607, 130–140. ( 10.1007/978-0-387-74021-8_11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kiorboe T, Jiang H, Goncalves RJ, Nielsen LT, Wadhwa N. 2014. Flow disturbances generated by feeding and swimming zooplankton. Proc. Natl Acad. Sci. USA 111, 11 738–11 743. ( 10.1073/pnas.1405260111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nielsen LT, Kiorboe T. 2015. Feeding currents facilitate a mixotrophic way of life. ISME J. 9, 2117–2127. ( 10.1038/ismej.2015.27) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dolger J, Nielsen LT, Kiorboe T, Andersen A. 2017. Swimming and feeding of mixotrophic biflagellates. Sci. Rep. 7, 39892 ( 10.1038/srep39892) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stanier RY. 1970. Some aspects of the biology of cells and their possible evolutionary significance. Symp. Soc. Gen. Mircrobiol. 20, 1–38. [Google Scholar]
- 53.Burns JA, Pittis AA, Kim E. 2018. Gene-based predictive models of trophic modes suggest Asgard archaea are not phagocytotic. Nat. Ecol. Evol. 2, 697–704. ( 10.1038/s41559-018-0477-7) [DOI] [PubMed] [Google Scholar]
- 54.Massana R, Guillou L, Diez B, Pedros-Alio C. 2002. Unveiling the organisms behind novel eukaryotic ribosomal DNA sequences from the ocean. Appl. Environ. Microbiol. 68, 4554–4558. ( 10.1128/AEM.68.9.4554-4558.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Not F, Valentin K, Romari K, Lovejoy C, Massana R, Tobe K, Vaulot D, Medlin LK. 2007. Picobiliphytes: a marine picoplanktonic algal group with unknown affinities to other eukaryotes. Science 315, 253–255. ( 10.1126/science.1136264) [DOI] [PubMed] [Google Scholar]
- 56.Jones MD, Forn I, Gadelha C, Egan MJ, Bass D, Massana R, Richards TA. 2011. Discovery of novel intermediate forms redefines the fungal tree of life. Nature 474, 200–203. ( 10.1038/nature09984) [DOI] [PubMed] [Google Scholar]
- 57.West PT, Probst AJ, Grigoriev IV, Thomas BC, Banfield JF. 2018. Genome-reconstruction for eukaryotes from complex natural microbial communities. Genome Res. 28, 569–580. ( 10.1101/gr.228429.117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cuvelier ML, et al. 2010. Targeted metagenomics and ecology of globally important uncultured eukaryotic phytoplankton. Proc. Natl Acad. Sci. USA 107, 14 679–14 684. ( 10.1073/pnas.1001665107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bhattacharya D, Price DC, Yoon HS, Yang EC, Poulton NJ, Andersen RA, Das SP. 2012. Single cell genome analysis supports a link between phagotrophy and primary plastid endosymbiosis. Sci. Rep. 2, 356 ( 10.1038/srep00356) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vaulot D, et al. 2012. Metagenomes of the picoalga Bathycoccus from the Chile coastal upwelling. PLoS ONE 7, e39648 ( 10.1371/journal.pone.0039648) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Worden AZ, Janouskovec J, McRose D, Engman A, Welsh RM, Malfatti S, Tringe SG, Keeling PJ. 2012. Global distribution of a wild alga revealed by targeted metagenomics. Curr. Biol. 22, R675–R677. ( 10.1016/j.cub.2012.07.054) [DOI] [PubMed] [Google Scholar]
- 62.Roy RS, Price DC, Schliep A, Cai G, Korobeynikov A, Yoon HS, Yang EC, Bhattacharya D. 2014. Single cell genome analysis of an uncultured heterotrophic stramenopile. Sci. Rep. 4, 4780 ( 10.1038/srep04780) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lopez-Escardo D, Grau-Bove X, Guillaumet-Adkins A, Gut M, Sieracki ME, Ruiz-Trillo I. 2017. Evaluation of single-cell genomics to address evolutionary questions using three SAGs of the choanoflagellate Monosiga brevicollis. Sci. Rep. 7, 11025 ( 10.1038/s41598-017-11466-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mangot JF, et al. 2017. Accessing the genomic information of unculturable oceanic picoeukaryotes by combining multiple single cells. Sci. Rep. 7, 41498 ( 10.1038/srep41498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Seeleuthner Y, et al. 2018. Single-cell genomics of multiple uncultured stramenopiles reveals underestimated functional diversity across oceans. Nat. Commun. 9, 310 ( 10.1038/s41467-017-02235-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heywood JL, Sieracki ME, Bellows W, Poulton NJ, Stepanauskas R. 2011. Capturing diversity of marine heterotrophic protists: one cell at a time. ISME J. 5, 674–684. ( 10.1038/ismej.2010.155) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Martinez-Garcia M, Brazel D, Poulton NJ, Swan BK, Gomez ML, Masland D, Sieracki ME, Stepanauskas R. 2012. Unveiling in situ interactions between marine protists and bacteria through single cell sequencing. ISME J. 6, 703–707. ( 10.1038/ismej.2011.126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gawryluk RMR, Del Campo J, Okamoto N, Strassert JFH, Lukes J, Richards TA, Worden AZ, Santoro AE, Keeling PJ. 2016. Morphological identification and single-cell genomics of marine diplonemids. Curr. Biol. 26, 3053–3059. ( 10.1016/j.cub.2016.09.013) [DOI] [PubMed] [Google Scholar]
- 69.Strassert JFH, et al. 2018. Single cell genomics of uncultured marine alveolates shows paraphyly of basal dinoflagellates. ISME J. 12, 304–308. ( 10.1038/ismej.2017.167) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lynch M, Field MC, Goodson HV, Malik HS, Pereira-Leal JB, Roos DS, Turkewitz AP, Sazer S. 2014. Evolutionary cell biology: two origins, one objective. Proc. Natl Acad. Sci. USA 111, 16 990–16 994. ( 10.1073/pnas.1415861111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Titus MA, Goodson HV. 2018. Developing evolutionary cell biology. Dev. Cell 47, 395–396. ( 10.1016/j.devcel.2018.11.006) [DOI] [PubMed] [Google Scholar]
- 72.del Campo J, Not F, Forn I, Sieracki ME, Massana R. 2013. Taming the smallest predators of the oceans. ISME J. 7, 351–358. ( 10.1038/ismej.2012.85) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Heger TJ, Edgcomb VP, Kim E, Lukes J, Leander BS, Yubuki N. 2014. A resurgence in field research is essential to better understand the diversity, ecology, and evolution of microbial eukaryotes. J. Eukaryot. Microbiol. 61, 214–223. ( 10.1111/jeu.12095) [DOI] [PubMed] [Google Scholar]
- 74.Janouškovec J, Tikhonenkov DV, Burki F, Howe AT, Rohwer FL, Mylnikov AP, Keeling PJ. 2017. A new lineage of eukaryotes illuminates early mitochondrial genome reduction. Curr. Biol. 27, 3717–3724. ( 10.1016/j.cub.2017.10.051) [DOI] [PubMed] [Google Scholar]
- 75.Lax G, Eglit Y, Eme L, Bertrand EM, Roger AJ, Simpson AGB. 2018. Hemimastigophora is a novel supra-kingdom-level lineage of eukaryotes. Nature 564, 410–414. ( 10.1038/s41586-018-0708-8) [DOI] [PubMed] [Google Scholar]
- 76.Gawryluk RMR, Tikhonenkov DV, Hehenberger E, Husnik F, Mylnikov AP, Keeling PJ. 2019. Non-photosynthetic predators are sister to red algae. Nature 572, 240–243. ( 10.1038/s41586-019-1398-6) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.