Abstract
Understanding the diversity and evolution of eukaryotic microorganisms remains one of the major challenges of modern biology. In recent years, we have advanced in the discovery and phylogenetic placement of new eukaryotic species and lineages, which in turn completely transformed our view on the eukaryotic tree of life. But we remain ignorant of the life cycles, physiology and cellular states of most of these microbial eukaryotes, as well as of their interactions with other organisms. Here, we discuss how high-throughput genome-wide gene expression analysis of eukaryotic single cells can shed light on protist biology. First, we review different single-cell transcriptomics methodologies with particular focus on microbial eukaryote applications. Then, we discuss single-cell gene expression analysis of protists in culture and what can be learnt from these approaches. Finally, we envision the application of single-cell transcriptomics to protist communities to interrogate not only community components, but also the gene expression signatures of distinct cellular and physiological states, as well as the transcriptional dynamics of interspecific interactions. Overall, we argue that single-cell transcriptomics can significantly contribute to our understanding of the biology of microbial eukaryotes.
This article is part of a discussion meeting issue ‘Single cell ecology’.
Keywords: scRNA-seq, protists, environmental sampling, gene expression analysis, unicellular eukaryotes
1. Introduction
Microbial eukaryotes, also known as protists, display an astonishing variety of life cycles, cell morphology and structures, metabolism, and ecological strategies. In fact, phylogenetic analyses show that protists represent the vast majority of eukaryotic clades and lineages [1,2], and environmental sampling continues to reveal hidden eukaryotic diversity that remains uncharacterized [3]. Microbial eukaryotes are invisible to the naked eye, but they are present almost everywhere—from human guts and the soils we stand on to acidic hot springs and Arctic ice shelves—and they play major roles in these diverse environments. For example, about half of carbon dioxide fixed globally is attributed to photosynthetic microbial eukaryotes [4]. Moreover, many protists are characterized by processes and features rarely or never found in multicellular organisms: silicification, mixotrophy, apicoplasts, extensively fragmented genomes, and much more. However, our understanding of the cell biology, physiology and evolution of most microbial eukaryotes remains extremely limited owing to both historical reasons (biased research focus on animal, plant, and fungal model species and a few parasitic protists) and technical constraints (associated with unculturability, small cell sizes, low abundance, etc.) [5,6].
Community sequencing and single-cell genomics technologies are contributing to mitigate this bias. Metabarcoding studies profile the diversity of ensemble communities, usually by performing targeted sequencing of selected markers such as the 18S ribosomal gene [7]. Metagenomic and metatranscriptomic analyses further improve our knowledge on the functions and physiology of protist communities in the environment [8–10]. By contrast, single-cell genomics approaches target individual, selected cells [11]. A single cell generally contains only one or two copies of each DNA molecule and, for this reason, single-cell genomic methods require massive amplification prior to sequencing. This amplification very often results in biased and partial coverage of the genome sequence [12,13]. An alternative to single-cell DNA sequencing that is gaining popularity is the sequencing of transcriptomes from single cells [14,15]. However, the obvious limitation of single-cell transcriptomics is that we can only sample genes and only those that are expressed above certain thresholds defined by the technical sensitivity.
So far, single-cell genomics/transcriptomics in microbial eukaryotes have been employed to characterize taxonomic diversity. These studies shed light on uncultured species and improved our understanding of the eukaryotic tree of life [16,17]. However, rapid advances in single-cell transcriptomics should allow us to go beyond phylogenetic charting of protist diversity and to begin quantitatively analysing the life cycles, cell states, physiology and ecological interactions of unicellular eukaryotes from the perspective of gene expression.
2. Single-cell transcriptomics: a brief state-of-the-art and specific challenges for unicellular eukaryotes
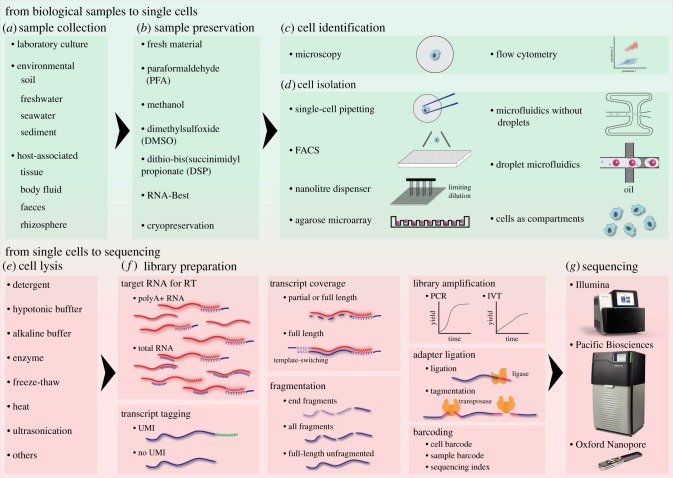
The last few years have seen tremendous development of single-cell RNA sequencing (scRNA-seq) technologies and a rapid increase in the number of studies using this method. Most of these studies sampled vertebrate cells, but the same procedures have also been successfully applied to other eukaryotes [18–22]. The growing diversity of scRNA-seq protocols and analytical tools can seem overwhelming. To help ecologists and protistologists who are interested in using this technique, here we divide scRNA-seq into four basic steps and summarize common options for each step (figure 1): sampling, cell isolation, cell lysis, and library preparation. Additionally, we discuss specific challenges relevant to applying scRNA-seq to microbial eukaryotes. We recommend excellent specific reviews for detailed discussions on the molecular biology [23,24], performance and estimated costs [25,26], and data analysis [27,28] aspects of scRNA-seq.
Figure 1.
Overview of the main steps and alternative strategies in single-cell RNA sequencing methods. (a,b) Biological samples taken from the laboratory or the field are either processed immediately or preserved for later use. (c,d) A variety of single-cell isolation platforms provide different throughput and possibilities to characterize individual cells. (e,f) Each single cell is usually lysed in its own compartment, and the targeted RNA is converted to cDNA, tagged with a specific cell barcode to identify the cellular origin of each RNA molecule. Various library preparation protocols offer different options for transcript quantification, coverage, and fragmentation, signal amplification, adapter ligation, and barcoding strategies, which depend on the sequencing technology (g) as well as the study design. RT, reverse transcription; UMI, unique molecular identifier. (Online version in colour.)
(a). Sampling and preservation
Sampling strategy is a critical component in capturing the native transcriptomes of the cells of interest (figure 1a). Ideally, fresh cells should be isolated and immediately lysed in an RNA-stabilizing buffer. However, this is usually not the case, even in a laboratory setting with model organisms. The time needed for harvesting samples, dissociating them into single cells, and isolating cells of interest may affect the RNA composition of the cells. This can be particularly dramatic in the case of unicellular eukaryotes, as changes in cell states/life stages have been shown to be extremely fast in many species [29,30].
Although most single-cell studies are performed on fresh cells, several chemicals have been employed to try to preserve native cell states in animal samples, including methanol [31], dimethylsulfoxide [32], crosslinkers that require later reverse cross-linking such as paraformaldehyde [33] and dithio-bis(succinimidyl propionate) [34], and proprietary reagents such as RNA-Best [35] (figure 1b). After preservation, cells can be stored either at 4°C [34,35] or cryopreserved at −80°C [31–33]. It is important to note that, with the exception of methanol fixation, these different preservation methods have only been tested for specific tissues/cell types and its applicability to other samples is still unclear and may require optimization.
There are two major advantages to sample preservation in addition to reducing artefacts in RNA pools. First, by fixing cells in their native states, sampling can be decoupled in time and space from cell isolation, allowing more flexible study design and scheduling that are less dependent on equipment availability. Thus, it not only enables studies involving intensive sampling in the laboratory, but also sampling in the field where there might be no access to facilities for cell isolation and downstream processing. The second advantage is that fixation increases cell permeability, which can be a critical factor for some protist species (see below).
(b). Isolation of single cells
The second key step in all single-cell techniques is cell isolation. The different isolation strategies for scRNA-seq differ in: (i) processing time, (ii) cost per cell, (iii) throughput (i.e. number of cells processed in a single experiment); (iv) doublet rates (i.e. fraction of two or more cells encapsulated together), and (v) the possibility to obtain phenotypic information to further identify and select/filter cells (figure 1c,d). Single-cell pipetting with microcapillary tubes, manually or using automated devices, is the simplest yet probably the most widely used approach to isolating single eukaryotic cells for diversity and ecological studies [14–16,36]. Albeit slow and tedious, accurate observation of cells under the microscope is sometimes essential to find the target cells in complex communities where only morphological information is available for cell identification. For high-throughput separation of single cells, fluorescence-activated cell sorting (FACS) systems have been the most widely used instruments, both in model systems and for studies concerning microbial ecology and diversity in the environment [37]. In scRNA-seq applications, FACS is used to select specific populations of interest and collect single-cells in 96- or 384-well plates [38]. FACS can simultaneously record multiple parameters for each sorted cell and these metadata can be used together with single-cell gene expression. FACS-enabled scRNA-seq offers additional advantages such as strict filtering of cell doublets and non-cellular particles (which can be very abundant in environmental samples). Moreover, in the case of complex communities in the field, FACS can be used to further target specific cell populations either using intrinsic cell properties (e.g. presence of pigments) or in combination with staining (e.g. against specific organelles).
A different technology commonly used to isolate single cells is microfluidics [39]. With lab-on-a-chip devices, cell isolation, lysis, and other reactions can be integrated into a single chip. The earliest available commercial platform designed specifically for single-cell analyses was the Fluidigm C1 system [40]. Although it is not possible to select cells, the C1 system allows imaging of each capture site for the presence of single cells and their viability [41]. Fluidigm C1 has been successfully used to isolate single cells of unicellular algae for RNA sequencing or quantitative polymerase chain reaction (qPCR) [14,42]. However, the restrictive cell size ranges of these chips [40] can be a limiting factor for cells out of the size range [14] or for a mixture of cells of very different sizes. Other limitations of the C1 system are the low throughput (tens to hundreds of cells) and high cost. To overcome this limitation, high-throughput droplet microfluidics-based protocols have been developed, such as Indrop [43], Drop-seq [44] and the commercial 10× Geonomics Chromium platform. In these methods, ideally a single cell is co-encapsulated with a hydrogel or bead (carrying primers with a barcode to identify each cell) in a nanolitre-volume aqueous droplet surrounded by oil. This strategy allows efficient capture of thousands of cells in just a few hours, but it also has specific drawbacks. The first limitation is the presence of cell doublets, given that the encapsulation is a random Poisson process. This effect can be controlled by diluting the samples and leaving most of the droplets empty, but in practice the doublet rates are still up to 25–30% in many microfluidics platforms [45]. Another limitation is the impossibility to select or discard specific cells and non-cellular particles. Finally, it is difficult to distinguish cells from empty droplets, which complicates downstream data analyses [46].
Another cell isolation strategy for high-throughput scRNA-seq is based on ultra-small wells, such as nanolitre wells in the ICELL8 method [47] and picolitre wells in the agarose microwells methods [48]. Single cells are trapped in wells (each with a unique cell barcode attached or receiving a barcoded bead) using a limiting dilution strategy: most wells are left empty in order to maximize the fraction of wells with a single cell under Poisson distribution. Both ICELL8 chips and agarose plates can be manually inspected under a microscope. The ICELL8 system can be further coupled to an imaging and analysis system for automatic identification of empty, doublet, or multiplet wells, allowing processing of only single cell-containing wells. The same imaging can also be used to evaluate cell viability [47].
Finally, while it is essential to distinguish RNA contents of individual cells for scRNA-seq, it does not necessarily mean that cells need to be in separated physical compartments like the aforementioned wells, droplets or microwells. Two recently published methods, sci-RNA-seq [49] and SPLiT-seq [50] take a radically different approach and use the cells themselves as compartments to keep RNA isolated. The key to this strategy is the usage of two or more rounds of random molecular barcoding (using barcoded oligonucleotides during reverse transcription, ligation and/or PCR amplification reactions). Different groups of intact (but permeabilized) cells are first barcoded, then pooled together, and finally randomly separated in groups again and receiving new barcodes. This can be repeated multiple times and potentially separate millions of single cells [50,51].
(c). Cell lysis
Following cell isolation, a critical step is to lyse each single cell to efficiently release RNA (figure 1e). Little attention is devoted to this key step by most scRNA-seq methods because animal cells are easily lysed when placed into a detergent-containing hypotonic buffer. But the situation with protists is very different, as many have diverse, often poorly characterized, cellular structures that might make cell lysis extremely difficult (cell walls, cysts, spores, tests, exoskeletons, sheaths, thecae, etc. [52]). Therefore, simple detergent or hypotonic buffers might not work for cells with these protective outer structures or contractile vacuoles capable of osmoregulation [53]. If the components of these structures are known and specific degrading enzymes are available, they can be added to the lysis buffer, as in the use of zymolase for yeast scRNA-seq [20]. An alkaline solution may also be effective for cell lysis, and is commonly used to lyse bacterial cells for single-cell genome sequencing [54]. There are also physical approaches to lyse cells, including freeze–thaw cycles or heat as employed by microbial single-cell genome sequencing protocols [55]. If all of these are unsuitable for the cells of interest, acoustic, electrical, optical (laser), mechanical (nanoknifes), or other physical methods may be considered as summarized by Brown & Audet [56]. However, these physical disruptions are only compatible with multi-well plate-based methods such as MARS-seq [38] and ICELL8 [47], but not with microfluidic isolation platforms.
(d). Library preparation and sequencing
Library preparation for scRNA-seq converts the RNA molecules in each single cell into a uniquely barcoded DNA library that can be pooled with other single-cell libraries and sequenced together. Beyond this general principle, scRNA-seq protocols are diverse in terms of (i) which type of RNAs are captured (only mRNAs versus all RNAs), (ii) how transcripts will be quantified (with or without transcript barcodes), (iii) which part of the transcripts to sequence (5′, 3′ or full-length), (iv) library amplification, and (v) barcoding strategies (figure 1f). Below we go through the basic parts of scRNA-seq library construction, and illustrate the different options with example protocols.
(i) Targeting RNAs. The first step in all scRNA-seq library preparation protocols is the conversion of RNAs into complementary DNAs (cDNAs) by reverse transcription (RT). Barcodes are usually introduced at this step to uniquely identify the transcriptome of each cell. Because the reverse transcriptase requires the 3′-end of an oligonucleotide primer to start synthesis, the transcripts we want to analyse determines the choice of primers. The vast majority of scRNA-seq studies target mRNA molecules and, to this end, primers with oligo-dT sequences (complementary to the poly-A tail of typical eukaryotic mRNA) are used. Note that oligo-dT enriches mRNA in the library, but it does not exclude other RNA from the library, especially rRNA [22], which comprise up to over 90% of a cellular transcriptome. On the other hand, there are protocols that target RNAs other than polyadenylated mRNA. For example, in host–pathogen systems, specific primers can be added to capture pathogen transcripts, as in virus-inclusive scRNA-seq analyses [57,58]. There are also protocols like scDual-seq [59] that target all types of RNAs by priming the RT with random hexamers (oligonucleotides with six random nucleotides at their 3′ end). An issue with random hexamer priming is that rRNA will represent the largest fraction of sequenced molecules. To avoid this, RamDA-seq uses not-so-random primers: random six-nucleotide sequences that avoid matching any rRNA sequence [60].
(ii) Transcript tagging. Aside from barcodes for cells, it is increasingly common to add barcodes to transcripts [61]. These unique molecular identifiers (UMIs) are random sequences (usually 8 to 20 nt in length) included in the RT primer and incorporated into the cDNA during RT, such that each transcript from a gene will receive a different barcode. Single-cell cDNAs are massively amplified (by PCR and/or in vitro transcription, see below) before sequencing and UMIs are used to avoid amplification biases and accurately quantify gene expression. In contrast to read-based expression metrics, UMI counts do not require normalization, are less affected by amplification bias, and provide absolute count with a defined zero that can be compared between cells [26,62]. The main drawback of using UMI is that they must be attached to either 3′ or 5′ end of the cDNA. With Illumina short-read sequencing, this means that only one end of the transcript can be sequenced together with the UMI.
(iii) Transcript coverage. An important difference between scRNA-seq protocols is the sequencing of full transcripts or just the 5′ or 3′ ends. In full-transcript protocols such as Smart-seq2, which uses template-switching oligonucleotides for full-length RT [63], all the fragments from cDNA will be sequenced and can provide information on the internal sequence and structure of RNA molecules, alternative splicing, and relative abundances of isoforms. Full-length sequencing methods show higher sensitivity [25], but they do not have strand-specific information and, importantly, are incompatible with the use of UMIs. By contrast, in partial sequencing protocols only one end of the transcript (generally the 3′, sometimes the 5′) will be sequenced. With the advent of long-read sequencing technologies like Oxford Nanopore and PacBio (figure 1g), methods to sequence full unfragmented transcripts with UMIs are under development [64,65]. When a reference genome/transcriptome is available, partial sequencing methods are generally preferable as they allow accurate gene expression estimation using UMIs. But when sampling, for example, protists from a complex environment, full-length transcript sequencing is advantageous and could be potentially used to reconstruct and quantify unannotated transcriptomes, as discussed in §4.
(iv) Library amplification. Almost all library preparation protocols involve the amplification of signal and the addition of adapters for specific sequencing platforms. For scRNA-seq, which usually deals with hundreds to tens of thousands of cells, amplification is crucial, but can also be a source of biases. Compared with the more commonly used PCR, in which the amplification is nonlinear and depends on the sequence composition [26], in vitro transcription (IVT) with the T7 promoter (as adopted in some scRNA-seq protocols [38,43,66]) provides linear amplification. However, IVT has the disadvantages of another round of RT and 3′-end coverage biases [26]. Two options are available for adding sequencing adapters. In addition to the traditional way of ligating one adapter to one end of a fragment, tagmentation, a portmanteau of tagging and fragmentation, relies on a specially engineered transposase Tn5 to fragment DNA and add adapters at the same time, thus greatly streamlining scRNA-seq protocol [63].
(v) Barcoding. As aforementioned, barcoding is key to scRNA-seq for distinguishing between individual cells. It can work at different levels (cellular barcodes, sample or pool barcodes, and sequencing indices) and in different steps (RT priming, RT template-switching, ligation, extension through PCR, etc.). The barcoding strategy, together with the cell isolation method and sequencing throughput, determines the number of single-cell transcriptomes that can be profiled at once. The simplest way for barcoding cells is to use a single set of sequencing indices, as in Smart-seq2 [63]. Combinatorial indexing strategies allow scaling up of the number of cells that can be processed together. For example, in MARS-seq [38], 384 cells are barcoded during RT and then different pools of 384 cells are distinguished with additional barcodes introduced by ligation.
In summary, there is no single scRNA-seq strategy that would be the best for all studies, especially given the diversity of unicellular eukaryotes and ecological questions to be addressed. Variables like the number of cells to be sampled, the cell size, the types and abundances of transcripts, the availability of a reference genome/transcriptome or the need to discriminate specific cell populations will define the most suitable strategy in each scenario.
3. Single-cell transcriptomics of cultured protists
In recent years, an increasing number of protist species from diverse lineages have been studied under laboratory culture conditions, providing novel insights into their cell biology and life cycles. The ability to regularly grow and differentiate these organisms in culture provided an opportunity for diverse functional genomics studies. For example, cell stage-specific transcriptomes for protists with complex life cycles allowed the dissection of gene expression phenotypes associated with cell growth (e.g. in the diatom Thalassiosira pseudonana [67]) and with temporal cell types in organisms like the ichthyosporean Creolimax fragrantissima [68], the choanoflagellate Salpingoeca rosetta, the amoebozoans Dictyostelium discoideum and Dictyostelium purpureum [69], the apicomplexan Plasmodium falciparum [70,71] or the filasterean Capsaspora owczarzaki [30]. Furthermore, genome-wide chromatin profiling experiments in some of these species defined the regulatory genome dynamics underlying temporal cell differentiation [72–74].
However, bulk genome-wide profiling methods require precise synchronization of large cell populations. This is not possible for many protist species and, even for those where some level of staging can be achieved, we might be completely missing additional heterogeneity and rare cell states. scRNA-seq bypasses these limitations and offers an unbiased tool to characterize the transcriptional programmes underlying distinct cell states, as well as the temporal transcriptional dynamics associated with growth and cell differentiation. The power of scRNA-seq to dissect protistan molecular phenotypes is illustrated by two hallmark single-cell studies in Plasmodium species. Reid et al. [22] sampled full transcripts in 500 P. falciparum and Plasmodium berghei single cells using high-coverage Smart-seq2, which sequences full-transcript fragments without UMIs. The authors found sharp, non-continuous transcriptional transitions between life cycle stages, in contrast with the smooth transitions previously reported by bulk transcriptome analyses and probably resulting from averaging of imperfectly synchronized cell populations (see also [71]). In another study, Poran et al. [75] opted for a different approach, sampling many more cells (18 000 single cells at different time-points), but at a relatively lower coverage using Drop-seq (average of approx. 500 genes per cell, compared with 1900 in Reid et al.), which uses UMIs and samples the 3′-end of transcripts. This dense sampling allowed the authors to define cell differentiation transcriptional signatures in P. falciparum at high resolution and to model the gene regulatory network of sexual commitment, which involves serial activation of three AP-2 transcription factors. More recently, UMI-based scRNA-seq has been applied to characterize transcriptional heterogeneity and stress responses in Saccharomyces cerevisiae [20] and Schizosaccharomyces pombe [21].
scRNA-seq also creates the potential to systematically study microbial interactions at high resolution, including symbioses, predation and infections. In contrast to single-cell genome sequencing, scRNA-seq allows us to go beyond the identification of the interacting organisms and to characterize the gene expression signatures and dynamics of interactions. Pioneering work in this direction comes from studies of human/mouse pathogens [76], including influenza [77], dengue and Zika viruses [57], and Salmonella [78]. For example, transcriptomic profiles of single cells helped establish the causality between pathogen variability and heterogeneous host cell immune responses [78]. As discussed above, most scRNA-seq methods target polyadenylated RNAs (i.e. mRNAs and a few long non-coding RNAs). To study a known pathogen without polyadenylated RNAs, one potential strategy is the use of targeted oligonucleotides (e.g. against Zika and dengue viruses [57]), but this requires a priori knowledge of the genome and only detects copies of the RNA genome or a specific transcript. Another strategy that allows both host and pathogen whole-transcriptome profiling is the use of random primers to capture both polyA and non-polyA types of RNAs. This strategy has been successfully applied to bacterial infections [59].
Finally, scRNA-seq technologies should enable the characterization of multicellular structures found in many protists, such as colonies, cell aggregates, or fruiting bodies [79]. Progress in this direction will shed light on exciting questions, such as the gene regulation involved in colony development, the existence of cell differentiation within them, the molecular phenotypes of potential cell types, and much more. Unfortunately, a severe limitation of current scRNA-seq methods is the low efficiency of cell recovery (fraction of profiled cells to the initial input). This precludes the analysis of single small pluricellular structures. Instead, current studies on such structures, for example early-stage animal embryos, rely on the accurate synchronization and pooling of multiple specimens [80,81]. This strategy might not be adequate for protists where synchronization is not possible or when inter-colony/aggregate variability is expected.
4. Towards single-cell transcriptomics of microbial eukaryote communities
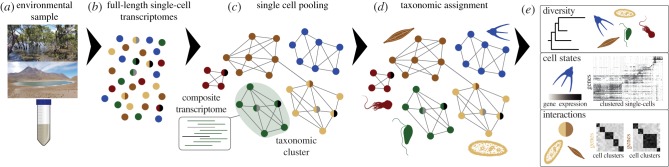
A natural extension of scRNA-seq analysis of protists in culture is the direct single-cell characterization of environmental communities. Briefly, cells would be isolated from the environmental sample (figure 2a), ensuring that only single cells (but also any interactors such as prey, pathogens, parasites or symbionts within them) are isolated in each compartment. Then the transcriptome of each single cell (plus any potential interactors) would be sequenced (figure 2b). Single cells would be aggregated into low-granularity clusters, representing taxonomic groups (figure 2c) and then the composite transcriptome of these clusters would be used to taxonomically identify the clusters (figure 2d), for example using rRNA sequences (see below). Finally, downstream analysis within and between clusters of cells would allow the systematic characterization of (figure 2e): (i) the components/diversity of the community, similar to what we would obtain from metabarcoding or single-cell genomics; (ii) physiological states among each of these components, analogous to what we would obtain from a metatranscriptomic analysis but with single-cell resolution; (iii) life cycle stages or temporal cell types for each of the members of the community; and (iv) microbial interactions and their associated transcriptional phenotypes. Many of the predictions of these analyses could be validated by using multiplexed fluorescence in situ hybridization [82,83], targeting markers for different temporal cell states/cell types in a particular species or against markers for multiple species (to validate interactions and community composition).
Figure 2.
Main steps in a potential application of scRNA-seq for functional and ecological studies on natural communities of microbial eukaryotes. (a) Isolation of single cells from environmental samples. (b) Sequencing of single-cell full-length transcripts. (c) Grouping of single cells into taxonomic clusters based on transcriptome similarity. (d) Cell clusters taxonomic assignment based on database searches with the cluster composite transcriptome. (e) Downstream analysis, including community components, diversity of physiological states or cell types of a particular organism, or patterns of interactions (symbiosis, pathogenicity, predation, etc.) between two or more species. (Online version in colour.)
However, considering the methodological state-of-the-art discussed in §2, we foresee important technical and analytical challenges associated with realizing single-cell ecology experiments of this nature.
(i) Sampling bias. An essential aspect when sampling a heterogeneous community will be the use of a methodology agnostic to distinct cell properties. For example, differences in cell lysis efficiency can be a major source of bias. In this sense, plate-based methods could be preferable to droplet-based methods, as harsher lysis conditions can be applied (freezing cycles, sonication, etc.). Another source of biases is cell size, but this is mainly an issue with Fluidigm C1 arrays. Finally, if protist-pathogen interactions are of interest, the most commonly used methods that target only polyA-transcripts may not be the best option. Instead, random hexamers should be used to capture all kinds of RNAs. This might have the (usually undesired) side effect of profiling rRNAs. However, in this specific application, this could be favourable and aid in the taxonomic assignment of single cells, given the existence of well-curated taxonomic databases based on 18S ribosomal sequences [3]. In practice, most current polyA-based scRNA-seq methods profile rRNA, either because some rRNAs can be polyadenylated and/or because rRNAs are generally very highly expressed. Finally, the strategy described here aims to be essentially unbiased, so in communities unevenly dominated by a few species, only deep single-cell sampling would uncover rarer community components.
(ii) Targeting eukaryotic cells and identifying cellular interactions. In complex environmental samples it would be necessary to discriminate eukaryotic cells from non-cellular particles and non-eukaryotes. Cell selection through FACS can solve this issue by targeting only cells of interest. FACS additionally allows the strict selection of only single cells, which may contain within them endosymbionts, prey, parasites and/or pathogens. This is another major advantage when studying cell–cell interactions, as it is possible to distinguish between true intracellular interactions and random aggregates. The latter could be removed during cell dissociation/homogenization before sampling, and FACS would provide a second level of exclusion of doublet or multiplet event. In contrast to FACS, droplet microfluidic applications with higher doublet rates may result in some false positive interactions owing to random co-encapsulation events.
-
(iii) Lack of reference genomes. Another problematic aspect of the single-cell analysis of a multi-species ensemble would be the absence of reference genomes/transcriptomes to map sequencing reads, at least for the majority of sampled cells. To address this problem, full-length scRNA-seq methods such as Smart-seq2 would be advantageous, with the downside of losing strand-specific information, the lower cell throughput of this method, and the impossibility to use UMIs to accurately quantify gene expression. Future developments in PacBio- or Oxford Nanopore-based scRNA-seq methods [64,65] may provide a solution for obtaining tagged full-length transcripts.
Even with full-length transcript sequencing, the transcriptomes recovered for each cell are expected to be extremely partial. One solution is to use a pooling strategy: de novo transcriptomes could be assembled by pooling similar cells into taxonomic clusters, generating a composite reference transcriptome [13] (figure 2c). This pooling strategy relies on the repeated sampling of similar cells from a population (in this case, from the same species) and it is commonly used for the identification of cell types in animal scRNA-seq data (e.g. with MetaCell analysis [84]). The only limitation of this resampling strategy is imposed by throughput: with more single cells profiled (ideally thousands), rarer species/cell types can be detected.
(iv) Phylogenetic classification and interspecific interactions. Composite transcriptomes, even if far from complete, would easily enable classification of these single-cell taxonomic clusters, using standard phylogenetic procedures and public molecular databases. These same searches should allow us to identify composites containing two or more species and to zoom in and characterize both the identity and the frequency of these interactions, as well as the transcriptional signatures of different cells states in the interacting species (figure 2).
(v) Single-cell morphotyping. Extremely valuable additional information would be to image cells before transcriptome profiling and in a way that transcriptomes can be unambiguously associated with specific images. A similar approach has been applied to low-throughput single-cell genomic sequencing of marine protists, providing complementary information on the sampled organisms [16,36]. In a recent study in fission yeast, over 2000 single-cells were manually isolated in 96 well-plates, imaged, and then transcriptomically profiled [21]. While this could in principle be adapted to environmental samples, the method is extremely time-consuming and the scalability very limited. A more promising future direction is the development of high-throughput microscopy/imaging cytometry coupled to cell sorting, for which proof of concept is emerging [85].
Overall, single-cell transcriptomics has transformed the study of animal systems, from tissue and cell type characterization to cell state dynamics and development. We anticipate its growing use for studies on the regulation and heterogeneity of gene expression in microbial eukaryotes, with pioneering studies in Plasmodium cell differentiation and yeast growth. Furthermore, we argue for its potential value for studies on natural communities, not only for profiling taxonomic diversity, but also for characterizing life stages, cell physiological states, and ecological interactions in microbial eukaryotes.
Acknowledgements
We wish to thank Assaf Vardi for discussions and suggestions, Bennett Lambert for carefully reading our manuscript, and three anonymous reviewers for precious advice and comments.
Data accessibility
This article does not contain any additional data.
Authors' contributions
C.K. and A.S.-P. wrote this article.
Competing interests
We declare we have no competing interests.
Funding
C.K. is supported by the Institute of Plant and Microbial Biology, Academia Sinica. A.S-P. is supported by CRG Severo Ochoa.
References
- 1.Burki F. 2014. The eukaryotic tree of life from a global phylogenomic perspective. Cold Spring Harb. Perspect. Biol. 6, a016147 ( 10.1101/cshperspect.a016147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adl SM, et al. 2012. The revised classification of eukaryotes. J. Eukaryot. Microbiol. 59, 429–514. ( 10.1111/j.1550-7408.2012.00644.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.del Campo J, et al. 2018. EukRef: phylogenetic curation of ribosomal RNA to enhance understanding of eukaryotic diversity and distribution. PLoS Biol. 16, e2005849 ( 10.1371/journal.pbio.2005849) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. 1998. Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281, 237–240. ( 10.1126/science.281.5374.237) [DOI] [PubMed] [Google Scholar]
- 5.del Campo J, Sieracki ME, Molestina R, Keeling P, Massana R, Ruiz-Trillo I. 2014. The others: our biased perspective of eukaryotic genomes. Trends Ecol. Evol. 29, 252–259. ( 10.1016/j.tree.2014.03.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keeling PJ, et al. 2014. The marine microbial eukaryote transcriptome sequencing project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 12, e1001889 ( 10.1371/journal.pbio.1001889) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Vargas C, et al. 2015. Eukaryotic plankton diversity in the sunlit ocean. Science 348, 1261605 ( 10.1126/science.1261605) [DOI] [PubMed] [Google Scholar]
- 8.Caron DA, et al. 2017. Probing the evolution, ecology and physiology of marine protists using transcriptomics. Nat. Rev. Microbiol. 15, 6–20. ( 10.1038/nrmicro.2016.160) [DOI] [PubMed] [Google Scholar]
- 9.Alexander H, Rouco M, Haley ST, Wilson ST, Karl DM, Dyhrman ST. 2015. Functional group-specific traits drive phytoplankton dynamics in the oligotrophic ocean. Proc. Natl Acad. Sci. USA 112, E5972–E5979. ( 10.1073/pnas.1518165112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marchetti A, Schruth DM, Durkin CA, Parker MS, Kodner RB, Berthiaume CT, Morales R, Allen AE, Armbrust EV. 2012. Comparative metatranscriptomics identifies molecular bases for the physiological responses of phytoplankton to varying iron availability. Proc. Natl Acad. Sci. USA 109, E317–E325. ( 10.1073/pnas.1118408109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoon HS, Price DC, Stepanauskas R, Rajah VD, Sieracki ME, Wilson WH, Yang EC, Duffy S, Bhattacharya D. 2011. Single-cell genomics reveals organismal interactions in uncultivated marine protists. Science 332, 714–717. ( 10.1126/science.1203163) [DOI] [PubMed] [Google Scholar]
- 12.López-Escardó D, Grau-Bové X, Guillaumet-Adkins A, Gut M, Sieracki ME, Ruiz-Trillo I. 2017. Evaluation of single-cell genomics to address evolutionary questions using three SAGs of the choanoflagellate Monosiga brevicollis. Sci. Rep. 7, 11025 ( 10.1038/s41598-017-11466-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mangot J-F, et al. 2017. Accessing the genomic information of unculturable oceanic picoeukaryotes by combining multiple single cells. Sci. Rep. 7, 41498 ( 10.1038/srep41498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z, Hu SK, Campbell V, Tatters AO, Heidelberg KB, Caron DA. 2017. Single-cell transcriptomics of small microbial eukaryotes: limitations and potential. ISME J. 11, 1282–1285. ( 10.1038/ismej.2016.190) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolisko M, Boscaro V, Burki F, Lynn DH, Keeling PJ. 2014. Single-cell transcriptomics for microbial eukaryotes. Curr. Biol. 24, R1081–R1082. ( 10.1016/j.cub.2014.10.026) [DOI] [PubMed] [Google Scholar]
- 16.Strassert JFH, et al. 2018. Single cell genomics of uncultured marine alveolates shows paraphyly of basal dinoflagellates. ISME J. 12, 304–308. ( 10.1038/ismej.2017.167) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang S, et al. 2017. Between a pod and a hard test: the deep evolution of amoebae. Mol. Biol. Evol. 34, 2258–2270. ( 10.1093/molbev/msx162) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poran A, Nötzel C, Aly O, Mencia-Trinchant N, Harris CT, Guzman ML, Hassane DC, Elemento O, Kafsack BFC. 2017. Single-cell RNA sequencing reveals a signature of sexual commitment in malaria parasites. Nature 551, 95–99. ( 10.1038/nature24280) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sebé-Pedrós A, et al. 2018. Early metazoan cell type diversity and the evolution of multicellular gene regulation. Nat. Ecol. Evol. 2, 1176–1188. ( 10.1038/s41559-018-0575-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nadal-Ribelles M, Islam S, Wei W, Latorre P, Nguyen M, de Nadal E, Posas F, Steinmetz LM.. 2019. Sensitive high-throughput single-cell RNA-seq reveals within-clonal transcript correlations in yeast populations. Nat. Microbiol. 4, 683–692. ( 10.1038/s41564-018-0346-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saint M, Bertaux F, Tang W, Sun X-M, Game L, Köferle A, Bähler J, Shahrezaei V, Marguerat S. 2019. Single-cell imaging and RNA sequencing reveal patterns of gene expression heterogeneity during fission yeast growth and adaptation. Nat. Microbiol. 4, 480–491. ( 10.1038/s41564-018-0330-4) [DOI] [PubMed] [Google Scholar]
- 22.Reid AJ, Talman AM, Bennett HM, Gomes AR, Sanders MJ, Illingworth CJRR, Billker O, Berriman M, Lawniczak MKN. 2018. Single-cell RNA-seq reveals hidden transcriptional variation in malaria parasites. Elife 7, 1–29. ( 10.7554/eLife.33105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lafzi A, Moutinho C, Picelli S, Heyn H. 2018. Tutorial: guidelines for the experimental design of single-cell RNA sequencing studies. Nat. Protoc. 13, 2742–2757. ( 10.1038/s41596-018-0073-y) [DOI] [PubMed] [Google Scholar]
- 24.Picelli S. 2017. Single-cell RNA-sequencing: the future of genome biology is now. RNA Biol. 14, 637–650. ( 10.1080/15476286.2016.1201618) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ziegenhain C, et al. 2017. Comparative analysis of single-cell RNA sequencing methods. Mol. Cell 65, 631–643. ( 10.1016/j.molcel.2017.01.023) [DOI] [PubMed] [Google Scholar]
- 26.Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA. 2015. The technology and biology of single-cell RNA sequencing. Mol. Cell 58, 610–620. ( 10.1016/j.molcel.2015.04.005) [DOI] [PubMed] [Google Scholar]
- 27.Tanay A, Regev A. 2017. Scaling single-cell genomics from phenomenology to mechanism. Nature 541, 331–338. ( 10.1038/nature21350) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stegle O, Teichmann SA, Marioni JC. 2015. Computational and analytical challenges in single-cell transcriptomics. Nat. Rev. Genet. 16, 133–145. ( 10.1038/nrg3833) [DOI] [PubMed] [Google Scholar]
- 29.Preston TM, King CA. 2003. Locomotion and phenotypic transformation of the amoeboflagellate Naegleria gruberi at the water-air interface. J. Eukaryot. Microbiol. 50, 245–251. ( 10.1111/j.1550-7408.2003.tb00128.x) [DOI] [PubMed] [Google Scholar]
- 30.Sebé-Pedrós A, Irimia M, Del Campo J, Parra-Acero H, Russ C, Nusbaum C, Blencowe BJ, Ruiz-Trillo I. 2013. Regulated aggregative multicellularity in a close unicellular relative of Metazoa. Elife 2, e01287 ( 10.7554/eLife.01287) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alles J, et al. 2017. Cell fixation and preservation for droplet-based single-cell transcriptomics. BMC Biol. 15, 1–14. ( 10.1186/s12915-017-0383-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guillaumet-Adkins A, et al. 2017. Single-cell transcriptome conservation in cryopreserved cells and tissues. Genome Biol. 18, 45 ( 10.1186/s13059-017-1171-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomsen ER, et al. 2016. Fixed single-cell transcriptomic characterization of human radial glial diversity. Nat. Methods 13, 87–93. ( 10.1038/nmeth.3629) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Attar M, et al. 2018. A practical solution for preserving single cells for RNA sequencing. Sci. Rep. 8, 2151 ( 10.1038/s41598-018-20372-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruf-Zamojski F, et al. 2018. Single-cell stabilization method identifies gonadotrope transcriptional dynamics and pituitary cell type heterogeneity. Nucleic Acids Res. 46, 11 370–11 380. ( 10.1093/nar/gky991) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gawryluk RMR, del Campo J, Okamoto N, Strassert JFH, Lukeš J, Richards TA, Worden AZ, Santoro AE, Keeling PJ. 2016. Morphological identification and single-cell genomics of marine diplonemids. Curr. Biol. 26, 3053–3059. ( 10.1016/j.cub.2016.09.013) [DOI] [PubMed] [Google Scholar]
- 37.Davey HM, Kell DB. 1996. Flow cytometry and cell sorting of heterogeneous microbial populations: the importance of single-cell analyses. Microbiol. Rev. 60, 641–696. ( 10.1016/j.mimet.2004.10.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jaitin DA, et al. 2014. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343, 776–779. ( 10.1126/science.1247651) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shields CW IV, Reyes CD, López GP. 2015. Microfluidic cell sorting: a review of the advances in the separation of cells from debulking to rare cell isolation. Lab Chip 15, 1230–1249. ( 10.1039/C4LC01246A) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valihrach L, Androvic P, Kubista M. 2018. Platforms for single-cell collection and analysis. Int. J. Mol. Sci. 19, 22–24. ( 10.3390/ijms19030807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xin Y, et al. 2016. Use of the Fluidigm C1 platform for RNA sequencing of single mouse pancreatic islet cells. Proc. Natl Acad. Sci. USA 113, 3293–3298. ( 10.1073/pnas.1602306113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenwasser S, Sheyn U, Frada MJ, Pilzer D, Rotkopf R, Vardi A. 2019. Unmasking cellular response of a bloom-forming alga to viral infection by resolving expression profiles at a single-cell level. PLoS Pathog. 15, e1007708 ( 10.1371/journal.ppat.1007708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Klein AM, et al. 2015. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201. ( 10.1016/j.cell.2015.04.044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macosko EZ, et al. 2015. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214. ( 10.1016/j.cell.2015.05.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang YJ, Schug J, Lin J, Wang Z, Kossenkov A, Consortium TH, Kaestner KH.2019. Comparative analysis of commercially available single-cell RNA sequencing platforms for their performance in complex human tissues. bioRxiv . ( ) [DOI]
- 46.Lun ATL, Riesenfeld S, Andrews T, Dao TP, Gomes T, Marioni JC. 2019. EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 20, 63 ( 10.1186/s13059-019-1662-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goldstein LD, et al. 2017. Massively parallel nanowell-based single-cell gene expression profiling. BMC Genomics 18, 1–10. ( 10.1186/s12864-017-3893-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Han X, et al. 2018. Mapping the mouse cell atlas by Microwell-seq. Cell 172, 1091–1107. ( 10.1016/j.cell.2018.02.001) [DOI] [PubMed] [Google Scholar]
- 49.Cao J, et al. 2017. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667. ( 10.1126/SCIENCE.AAM8940) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosenberg AB, et al. 2018. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 360, 176–182. ( 10.1126/science.aam8999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cao J, et al. 2019. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502. ( 10.1038/s41586-019-0969-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corliss JO. 2002. Biodiversity and biocomplexity of the protists and an overview of their significant roles in maintenance of our biosphere. Acta Protozool. 41, 199–219. [Google Scholar]
- 53.Allen RD, Naitoh Y. 2002. Osmoregulation and contractile vacuoles of protozoa. Int. Rev. Cytol. 215, 351–394. ( 10.1016/S0074-7696(02)15015-7) [DOI] [PubMed] [Google Scholar]
- 54.Raghunathan A, Ferguson HR, Bornarth CJ, Song W, Driscoll M, Lasken RS. 2005. Genomic DNA amplification from a single bacterium. Appl. Environ. Microbiol. 71, 3342–3347. ( 10.1128/AEM.71.6.3342-3347.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stepanauskas R. 2012. Single cell genomics: an individual look at microbes. Curr. Opin. Microbiol. 15, 613–620. ( 10.1016/j.mib.2012.09.001) [DOI] [PubMed] [Google Scholar]
- 56.Brown RB, Audet J. 2008. Current techniques for single-cell lysis. J. R. Soc. Interface 5, S131–S138. ( 10.1098/rsif.2008.0009.focus) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zanini F, Pu SY, Bekerman E, Einav S, Quake SR. 2017. Single-cell transcriptional dynamics of flavivirus infection. Elife 7, e32942 ( 10.1101/203331) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zanini F, et al. 2018. Virus-inclusive single-cell RNA sequencing reveals the molecular signature of progression to severe dengue. Proc. Natl Acad. Sci. USA 115, E12 363–E12 369. ( 10.1073/PNAS.1813819115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Avital G, Avraham R, Fan A, Hashimshony T, Hung DT, Yanai I. 2017. scDual-Seq: mapping the gene regulatory program of Salmonella infection by host and pathogen single-cell RNA-sequencing. Genome Biol. 18, 200 ( 10.1186/s13059-017-1340-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hayashi T, Ozaki H, Sasagawa Y, Umeda M, Danno H, Nikaido I. 2018. Single-cell full-length total RNA sequencing uncovers dynamics of recursive splicing and enhancer RNAs. Nat. Commun. 9, 619 ( 10.1038/s41467-018-02866-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M, Lönnerberg P, Linnarsson S.. 2014. Quantitative single-cell RNA-seq with unique molecular identifiers. Nat. Methods 11, 163–166. ( 10.1038/nmeth.2772) [DOI] [PubMed] [Google Scholar]
- 62.Conesa A, et al. 2016. A survey of best practices for RNA-seq data analysis. Genome Biol. 17, 13 ( 10.1186/s13059-016-0881-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Picelli S, Faridani OR, Björklund ÅK, Winberg G, Sagasser S, Sandberg R. 2014. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181. ( 10.1038/nprot.2014.006) [DOI] [PubMed] [Google Scholar]
- 64.Karlsson K, Linnarsson S. 2017. Single-cell mRNA isoform diversity in the mouse brain. BMC Genomics 18, 126 ( 10.1186/s12864-017-3528-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Volden R, Palmer T, Byrne A, Cole C, Schmitz RJ, Green RE, Vollmers C. 2018. Improving nanopore read accuracy with the R2C2 method enables the sequencing of highly multiplexed full-length single-cell cDNA. Proc. Natl Acad. Sci. USA 115, 9726–9731. ( 10.1073/pnas.1806447115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hashimshony T, et al. 2016. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 17, 77 ( 10.1186/s13059-016-0938-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ashworth J, Coesel S, Lee A, Armbrust EV, Orellana MV, Baliga NS. 2013. Genome-wide diel growth state transitions in the diatom Thalassiosira pseudonana. Proc. Natl Acad. Sci. USA 110, 7518–7523. ( 10.1073/pnas.1300962110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Mendoza A, Suga H, Permanyer J, Irimia M, Ruiz-Trillo I.. 2015. Complex transcriptional regulation and independent evolution of fungal-like traits in a relative of animals. Elife 4, e08904 ( 10.1007/s13398-014-0173-7.2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Parikh A, et al. 2010. Conserved developmental transcriptomes in evolutionarily divergent species. Genome Biol. 11, R35 ( 10.1186/gb-2010-11-3-r35) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hall N. 2005. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science 307, 82–86. ( 10.1126/science.1103717) [DOI] [PubMed] [Google Scholar]
- 71.Howick VM, et al. 2019. The Malaria Cell Atlas: a comprehensive reference of single parasite transcriptomes across the complete Plasmodium life cycle. bioRxiv . ( ) [DOI]
- 72.Ay F, Bunnik EM, Varoquaux N, Bol SM, Prudhomme J, Vert J-P, Noble WS, Le Roch KG.. 2014. Three-dimensional modeling of the P. falciparum genome during the erythrocytic cycle reveals a strong connection between genome architecture and gene expression. Genome Res. 24, 974–988. ( 10.1101/gr.169417.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Toenhake CG, Fraschka SA-K, Vijayabaskar MS, Westhead DR, van Heeringen SJ, Bártfai R.. 2018. Chromatin accessibility-based characterization of the gene regulatory network underlying Plasmodium falciparum blood-stage development. Cell Host Microbe 23, 557–569. ( 10.1016/j.chom.2018.03.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sebé-Pedrós A, Ballaré C, Parra-Acero H, Chiva C, Tena JJ, Sabidó E, Gómez-Skarmeta JL, Di Croce L, Ruiz-Trillo I.. 2016. The dynamic regulatory genome of Capsaspora and the origin of animal multicellularity. Cell 165, 1224–1237. ( 10.1016/j.cell.2016.03.034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nötzel C, Poran A, Kafsack BFC. 2018. Single-cell transcriptome profiling of protozoan and metazoan parasites. Trends Parasitol. 34, 731–734. ( 10.1016/j.pt.2018.04.009) [DOI] [PubMed] [Google Scholar]
- 76.Chattopadhyay PK, Roederer M, Bolton DL. 2018. A deadly dance: the choreography of host–pathogen interactions, as revealed by single-cell technologies. Nat. Commun. 9, 1–11. ( 10.1038/s41467-018-06214-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Russell AB, Trapnell C, Bloom JD. 2018. Extreme heterogeneity of influenza virus infection in single cells. Elife 7, 1–26. ( 10.7554/eLife.32303) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Avraham R, et al. 2015. Pathogen cell-to-cell variability drives heterogeneity in host immune responses. Cell 162, 1–13. ( 10.1016/j.cell.2015.08.027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sebé-Pedrós A, Degnan BM, Ruiz-Trillo I. 2017. The origin of Metazoa: a unicellular perspective. Nat. Rev. Genet. 18, 498–512. ( 10.1038/nrg.2017.21) [DOI] [PubMed] [Google Scholar]
- 80.Karaiskos N, Wahle P, Alles J, Boltengagen A, Ayoub S, Kipar C, Kocks C, Rajewsky N, Zinzen RP. 2017. The Drosophila embryo at single cell transcriptome resolution. Science 3235, 117382 ( 10.1101/117382) [DOI] [PubMed] [Google Scholar]
- 81.Wagner DE, Weinreb C, Collins ZM, Briggs JA, Megason SG, Klein AM. 2018. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987. ( 10.1126/science.aar4362) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lukumbuzya M, Schmid M, Pjevac P, Daims H. 2019. A multicolor fluorescence in situ hybridization approach using an extended set of fluorophores to visualize microorganisms. Front. Microbiol. 10, 1–13. ( 10.3389/fmicb.2019.01383) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wagner M, Haider S. 2012. New trends in fluorescence in situ hybridization for identification and functional analyses of microbes. Curr. Opin. Biotechnol. 23, 96–102. ( 10.1016/j.copbio.2011.10.010) [DOI] [PubMed] [Google Scholar]
- 84.Baran Y, Sebé-Pedrós A, Lubling Y, Giladi A, Chomsky E, Meir Z, Hoichman M, Lifshitz A, Tanay A.2018. MetaCell: analysis of single cell RNA-seq data using k-NN graph partitions. bioRxiv ( ) [DOI] [PMC free article] [PubMed]
- 85.Nitta N, et al. 2018. Intelligent image-activated cell sorting. Cell 175, 266–276. ( 10.1016/j.cell.2018.08.028) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article does not contain any additional data.