Abstract
Background
Osimertinib yields significant tumor responses and durations of progression‐free survival (PFS) in patients with acquired T790M mutations. However, the evidence supporting liquid biopsy‐guided treatment is still limited. This study examined the real‐world benefits of osimertinib in patients with tissue or plasma T790M mutations.
Methods
From January 2016 to June 2018, a total of 183 non‐small‐cell lung cancer patients were enrolled. The presence of the T790M mutation was assessed by either tissue or plasma. The PFS, overall survival, and tumor response rates of the patients were calculated and compared with those of previous clinical trials.
Results
T790M mutations were detected in 51.5% of the patients, including 64 of 140 (45.7%) who underwent liquid biopsies and 23 of 29 (79.3%) who underwent tumor biopsies. After excluding those in clinical trials, 46 patients received osimertinib, including 33 with positive plasma and 13 with positive tissue results for T790M mutations. The median PFS was 11.3 months (interquartile range: 5.2‐NR) in all the T790M‐positive patients and 10.1 months (interquartile range: 5.9‐NR) in the plasma T790M‐positive patients. The overall survival, meanwhile, was not reached, whereas the one‐year survival rate was 66.1% in all the patients and 61.4% in those who were plasma T790M‐positive. The objective response rate and disease control rate were 37.8% and 91.9% in all the patients and 34.6% and 92.3% in the plasma T790M‐positive group, respectively. Using a Cox proportional hazards regression, we determined that male gender was a poor prognostic factor for PFS.
Conclusions
In this retrospective real‐world analysis, it was determined that both tissue and plasma T790M mutations can be used to guide treatment with osimertinib. Similar disease control rates and survival durations were observed in comparison to those of phase 3 clinical trials.
Keywords: circulating tumor DNA, NSCLC, osimertinib, progression‐free survival, T790M mutation
This study examined the real‐world benefits of osimertinib in patients with tissue or plasma T790M mutations. The progression‐free survival, overall survival, and tumor response rates of 46 non‐small‐cell lung cancer patients were calculated and compared with those of previous clinical trials. It was determined that both tissue and plasma T790M mutations can be used to guide treatment with osimertinib.

1. BACKGROUND
Epidermal growth factor receptor (EGFR) mutations are key genetic drivers of non‐small‐cell lung cancer (NSCLC) and are present in about 10% of the Caucasian population and 40%‐50% of the Asian population.1 The response rates to first‐generation EGFR tyrosine kinase inhibitors (TKIs) range between 56% and 74%, and the median progression‐free survival (PFS) durations range from 9 to 13 months.2 However, most patients experience disease progression, and about 50%‐70% of them develop newly acquired resistance EGFR p.Thr790Met (T790M) point mutations.3, 4, 5, 6, 7 These acquired mutations enhance the binding affinity of adenosine triphosphate on the EGFR kinase domain and, in turn, decrease the efficacy of first‐ and second‐generation EGFR‐TKIs. Osimertinib, a third‐generation inhibitor, was designed to and has been proven to selectively target T790M.8, 9, 10 Previous clinical trials have shown the promising efficacy of this third‐generation EGFR‐TKI in patients who experience disease progression after treatment with first‐ and second‐generation EGFR‐TKIs.9, 10, 11 A phase 3 clinical trial (AURA 3) also reported enhanced PFS associated with osimertinib in comparison to standard chemotherapy in NSCLC patients with an acquired T790M mutation.12
In order to use osimertinib clinically in a patient, that patient must be shown to have the T790M mutation in his or her tissue. However, tumor rebiopsy continues to present challenges, especially in patients with poor performance status and tumor inaccessibility. For these reasons, researchers have developed a number of liquid biopsy platforms that can be utilized as complements to or in lieu of tissue biopsy.13, 14, 15 With the advancement of genotyping assays—namely, droplet digital polymerase chain reactions (ddPCRs) and beads, emulsions, amplification, and magnetics (BEAMing) digital polymerase chain reaction (dPCR)—the mutation status of a patient can be detected using cell‐free plasma DNA.5, 16, 17, 18, 19 However, studies on the efficacy of osimertinib using plasma testing remain scarce and limited in terms of patient numbers.20 In this study, therefore, we evaluated the feasibility of performing T790M identification through circulating tumor DNA (ctDNA) in blood samples among EGFR‐mutant NSCLC patients exhibiting disease progression after treatment with first‐ or second‐generation EGFR‐TKIs. We also retrospectively evaluated the efficacy of osimertinib in plasma and tissue T790M‐positive patients.
2. METHODS
2.1. Patients
From January 2016 to June 2018, patients with advanced EGFR‐mutated NSCLC who experienced radiological or clinical progression after treatment with one or more first‐ or second‐generation EGFR‐TKIs were recruited; that is, no maximum limit was set for the number of previous EGFR inhibitors or systemic therapies a patient had received. All of the patients were positive for EGFR mutation at their initial diagnoses. Upon disease progression, genotyping detection based on tissue, liquid, or both was performed based on the clinical judgment of the treating physician. The detection of the EGFR T790M mutation in cell‐free plasma DNA was conducted in the Department of Pathology of National Cheng Kung University Hospital. When a positive result for the T790M mutation was found in a tissue or liquid biopsy, 80 mg per day of osimertinib was prescribed through the compassionate‐use programs of Astrazeneca until the occurrence of disease progression or the appearance of unacceptable adverse effects. The study protocol was approved by the institutional ethics committee of National Cheng Kung University Hospital (IRB number: A‐ER‐106‐205).
2.2. Outcomes
All the patients received a chest computed tomography scan before the initiation of the osimertinib treatment and every 12 weeks thereafter to evaluate their tumor responses. Brain imaging and bone scans were performed if there were related symptoms. The primary endpoint was PFS. The secondary endpoints included the disease control rate, overall response rate, overall survival (OS), and the percentage of T790M mutation‐positive patients identified using liquid biopsy. PFS was calculated from the date of osimertinib initiation until the date of radiological progression according to the Response Evaluation Criteria in Solid Tumors (RECIST) v1.121 or death, with censoring at the date of the last follow‐up in the event that the patient had not progressed. The overall response rate was calculated as the percentage of patients who exhibited a partial response or complete response in the first image study after the initiation of the osimertinib treatment, while the disease control rate was defined as the percentage of patients who exhibited a partial response, complete response, or stable disease. Furthermore, the duration of OS was defined as the period from the initiation date of osimertinib treatment until the date of death.
2.3. Tumor mutation analysis
To perform the EGFR mutation analysis, tumor tissues were collected from the primary lung cancer or metastatic lesions, with tissue samples having a tumor content of more than 80% as determined using microscopy with hematoxylin and eosin staining being selected for the analysis. DNA was extracted using a QIAamp DNA FFPE tissue kit (Qiagen) eluted in ATE (QIAmp Tissue Elution) buffer in combination with the QIAcube automated extractor (Qiagen) according to the manufacturer's instructions. The EGFR PCR Kit (EGFR RUO Kit) and the therascreen EGFR RGQ PCR Kit (EGFR IVD Kit, Qiagen) were used to identify mutated EGFR DNA. These kits utilized a combination of Scorpions real‐time PCR technology and amplification‐refractory mutation system (ARMS) technology for the detection of the mutations with real‐time quantitative PCR.22
2.4. Cell‐free DNA extraction
Cell‐free DNA was isolated from 1 mL of plasma with the QIAamp Circulating Nucleic Acid kit (Qiagen) according to the manufacturer's instructions. Briefly, samples were centrifuged at 1000 g for 2 minutes, and the resulting supernatant was then transferred to a clean tube before the DNA extraction. Subsequently, lysate buffer (ACL) (800 μL) and carrier RNA (5 μL) were added, and the supernatant was incubated at 60°C. After supplementing the mixture with binding buffer (ACB), the mixture was transferred to ice for 5 minutes, treated with QIAvacuum, and washed with buffer. The eluted cell‐free DNA was stored at −80°C until analysis.
2.5. In‐house T790M detection method
The test was performed in duplicate on samples containing 21 µL of Super Therm Gold Master Mix (Bionovas Biotechnology), 2 µL of T790M primers (10 µmol/L), 2 µL of control primers (2.5 µmol/L), and 2 µL of working DNA (15 ng/µL) at a final volume of 27 µL. The following conditions were utilized in performing the PCR assay: 5 minutes at 95°C, followed by 35 cycles of 30 seconds at 95°C, 30 seconds at 63°C, 30 seconds at 72°C, and 20 seconds at 73°C. The human AF4 gene primers (exon 3 and exon 11; GenBank accession no. Z83687) were amplified products of 100 bp used to indicate the quality of the PCR reaction.23 The expected PCR product sizes of T790M and the control alleles were 150 bp and 100 bp, respectively.
Analysis of the PCR products was conducted using high‐resolution capillary electrophoresis with the QIAxcel® DNA high‐resolution gel cartridge (Qiagen, Hilden, Germany) on the QIAxcel system (Qiagen) according to the manufacturer's instructions. The sizes of the PCR products were calculated, and the genotypes were determined using Qiagen BioCalculator® software. The DNA size markers (Qiagen) ranged from 50 to 800 bp. The alignment marker (Qiagen) consisting of 15 bp and 1000 bp fragments corresponded to a 15‐second sample injection time at 5 kV and a 420‐second separation time at 5 kV.
2.6. Statistical analysis
The clinical characteristics of the patients, including, sex, age, the presence or absence of brain metastasis, tissue genotyping at initial diagnosis, and preceding EGFR‐TKIs, were recorded. The Kaplan–Meier method was used to estimate the PFS and OS. A Cox proportional hazards regression was also performed to evaluate the determinants of PFS and OS. Age, sex, tumor size, nodal stage, EGFR mutation subtypes, and sites of disease progression were chosen as the prognostic factors, and the statistical analysis was carried out with SAS version 9.4 (SAS Institute). All the reported P‐values were two‐sided.
3. RESULTS
3.1. Patient characteristics and T790M mutation rate
Of the total of 248 patients with advanced EGFR‐mutated NSCLC recruited from January 2016 to June 2018, 183 underwent examinations for T790M. Among these patients, 140 and 29 patients received liquid and tissue biopsies, respectively, while 14 of the patients received both types of biopsies. Figure 1 shows the flow chart for enrolling the subjects. T790M mutations were detected in 51.5% of the patients who received only one type of biopsy, including 64 of the 140 (45.7%) who underwent liquid biopsies and 23 of the 29 (79.3%) who underwent tumor biopsies. Among the patients who had received only one EGFR‐TKI therapy before receiving osimertinib, the rates for positive T790M results were 62% (47/76), 44% (14/32), and 45% (20/44) in patients receiving gefitinib, erlotinib, and afatinib, respectively. There were no significant differences in these rates of positive results between the patients who had received the different EGFR‐TKIs (P = .11). After excluding patients in clinical trials, a total of 46 T790M‐positive patients were enrolled in the study.
Figure 1.

Flow chart for patient enrollment. (EGFR, epidermal growth factor receptor; NSCLC, non‐small‐cell lung cancer; TKI, tyrosine kinase inhibitor)
The baseline characteristics of the T790M‐positive patients are summarized in Table 1. All the patients had adenocarcinoma histology and were at the advanced stage. The mean age of the patients was 61.8 (range 52.9‐75.2) years, and there were 13 male patients (28.3%) and 33 female patients (71.7%). Fourteen (30.4%) patients had brain metastases. The EGFR genotyping at the initial diagnosis showed Del 19 mutations in 19 (41.3%), L858R mutations in 22 (47.8%), and rare mutations in five (10.9%) of these NSCLC patients. All the patients were pretreated with EGFR‐TKIs: 25 (54.3%), 14 (30.4%), and 7 (15.2%) received gefitinib, erlotinib, and afatinib, respectively.
Table 1.
Baseline characteristics of T790M‐mutated patients
| Characteristic |
No. of patients (%) N = 46 |
|---|---|
| Age | |
|
Median (range), (y) |
62 (53‐74) |
| <60 y | 21 (45.8%) |
| >60 y | 25 (54.2%) |
| Sex | |
| Female | 33 (71.7%) |
| Male | 13 (28.3%) |
| Brain metastases | 14 (30.4%) |
| EGFR mutation | |
| Exon 19 deletion | 19 (41.3%) |
| L858R | 22 (47.8%) |
| Others | 5 (10.9%) |
| First‐line EGFR‐TKIs | |
| Gefitinib | 25 (54.3%) |
| Erlotinib | 7 (15.2%) |
| Afatinib | 14 (30.4%) |
Abbreviations: EGFR, epidermal growth factor receptor; TKI, tyrosine kinase inhibitor.
3.2. Response rate of osimertinib
Of the 46 patients with tissue‐ or plasma‐positive T790M NSCLC, 37 had measurable target lesions that were evaluated for responses. Among the 9 patients without measurable target lesions, six had pleural effusion only, and three deteriorated rapidly. These nine patients were also enrolled in the study. Of the 37 other patients, one had a complete response, 13 had partial responses, 20 had stable diseases, and four experienced disease progression. The overall response rate was 37.8%, and the disease control rate was 91.9% (Figure 2). In the 26 patients with plasma‐positive T790M and measurable lesions, one had a complete response, eight had partial responses, 15 had stable diseases, and two experienced disease progression. The disease control rate was 92.3%, while the objective response rate was calculated to be 34.6%.
Figure 2.
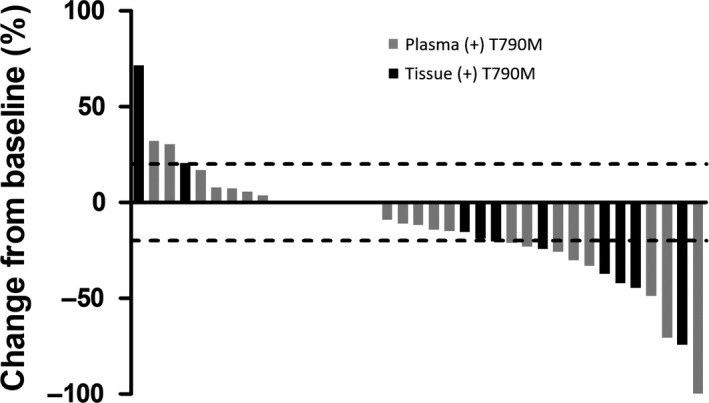
Tumor responses with osimertinib in plasma (gray bar) and tissue (black bar) T790M‐positive patients
3.3. Progression‐free survival and overall survival
After the median follow‐up period of 17.1 months, the median PFS of all 46 patients was 11.3 months (interquartile range, 5.2 to NR); however, the median OS was not reached (Figure 3A,B). Using a Cox proportional hazards regression, we determined that male gender was a poor prognostic factor for PFS (Table 2). In the subgroup of patients with positive plasma T790M, the median PFS was 10.1 months (interquartile range, 5.9 to NR), but the median OS also was not reached (Figure 3C,D). The median PFS durations of the patients receiving osimertinib immediately after a first‐line EGFR‐TKI and later than a third‐line treatment were 11.0 months (interquartile range, 2.5 to NR) and 13.9 months (interquartile range, 6.7 to NR), respectively (Figure S1A), while the median OS in both groups was not reached (Figure S1B). There were no significant between‐group differences for the PFS and OS groups (log rank P = .781 and .335, respectively).
Figure 3.

A, PFS in all patients treated with osimertinib. (B), OS in all patients treated with osimertinib. (C), PFS in patients treated with osimertinib according to positive plasma T790M. (D), OS in patients treated with osimertinib according to positive plasma T790M. PFS, progression‐free survival; OS, overall survival
Table 2.
Cox proportional hazards regression results for progression‐free survival and overall survival
| Progression‐free survival | Overall survival | ||||
|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | ||
| Age | >65 versus <65 | 2.23 (0.83‐6.01) | .114 | 3.15 (0.71‐14.03) | .132 |
| Sex | Male versus female | 2.73 (1.03‐7.24) | .043 | 4.27 (0.96‐18.95) | .056 |
| Tumor size | >3 cm versus <3 cm | 1.67 (0.65‐4.27) | .288 | 1.24 (0.30‐5.07) | .766 |
| Nodal involvement | N2 versus N1 | 0.54 (0.12‐2.52) | .431 | 0.25 (0.02‐4.08) | .333 |
| N3 versus N1 | 2.92 (0.99‐8.62) | .052 | 3.85 (0.69‐21.35) | .123 | |
| EGFR mutation | Del 19 versus others | 0.51 (0.19‐1.38) | .186 | 0.70 (0.17‐2.80) | .609 |
| Sequential | Second‐line versus ≥third‐line cohorts | 0.64 (0.25‐1.66) | .360 | 0.86 (0.22‐3.32) | .828 |
| Disease progression | Intracranial versus extracranial | 0.57 (0.17‐1.92) | .362 | 0.36 (0.07‐1.98) | .242 |
Abbreviation: EGFR, epidermal growth factor receptor.
4. DISCUSSION
In comparison to liquid biopsies, tissue biopsies have many limitations. Patients may suffer from complications after rebiopsy, such as pneumothorax, which is reported to occur in 17%‐27% of patients.24 Also, about 25% of cases provide too few tumor cells or none at all, such that an EGFR mutation assay cannot be conducted.25 A single biopsy specimen may miss cancer heterogeneity, but plasma ctDNA can potentially capture it.26, 27 Many technologies have been developed to improve the sensitivity of tests used to detect genetic mutations in ctDNA. We developed an in‐house method based on an ARMS and compared our results to those of four well‐established technologies, namely, the Bio‐Rad ddPCR system, PANAMutype, the therascreen EGFR Plasma RGQ PCR Kit, and the Cobas EGFR Mutation kit. The clinical performance was analyzed in a clinical cohort of 46 EGFR‐mutated NSCLC patients with acquired EGFR‐TKI resistance. It was found that the detection rate of our platform was equivalent to those of the other four platforms (manuscripts in submission). We employed a straightforward ARMS strategy that combined a higher melting temperature with high‐resolution capillary electrophoresis to enhance the specificity and sensitivity of T790M detection in ctDNA. In two recent prospective studies using a ctDNA‐guided osimertinib treatment, the positivity of plasma T790M was enhanced by 60% using tagged amplicon sequencing and by 71% using dPCR.20, 28 The detection rate of T790M mutations in liquid biopsies from EGFR‐mutant patients progressing after treatment with an EGFR‐TKI in the current study was 45.7%, which was only slightly lower than the rates for the two studies referenced above, while the detection rates for other retrospective studies have ranged from 50% to 70%.5, 20, 28, 29, 30 Genotyping using a plasma sample is minimally invasive and has thus become an attractive alternative method for T790M detection.
The AURA 3 phase III trial revealed that osimertinib has efficacy higher than that of platinum plus pemetrexed in T790M‐mutated advanced NSCLC patients who have experienced disease progression while receiving first‐line EGFR‐TKI therapy, and osimertinib has become a standard treatment for these patients.12 Before treatment with osimertinib can be commenced, it is required that the presence of a T790M mutation in tumor cells be established by either tissue or plasma genotyping.12, 20, 28 Recently, a review conducted jointly by American Society of Clinical Oncology (ASCO) and the College of American Pathologists proposed that the evidence for the clinical validity and utility of the majority of ctDNA assays for advanced cancer is insufficient. However, a growing range of new NGS platforms have been developed in recent years.31, 32, 33 Moreover, the International Association for the Study of Lung Cancer recently issued a consensus statement holding that liquid biopsies are reliable and should be considered as surrogates for a tissue biopsy in the event that a tissue biopsy cannot be performed, especially with respect to the detection of T790M mutations.34 Nonetheless, there is still no definite conclusion regarding the use of liquid biopsies as surrogates for tissue biopsies because some studies have reported that the level of validity needed for widespread use in routine clinical diagnostics has not yet been achieved for liquid biopsies.35
There have been a limited number of trials in which the outcomes of treatment for a targeted therapy that was selected solely based on the result of a ctDNA assay were prospectively tested. Oxnard et al conducted a retrospective analysis comparing osimertinib efficacy guided by either plasma or tissue genotyping for advanced EGFR‐mutant NSCLC.5 In that study, the reported ORR and median PFS results for the patients with T790M‐positive plasma (ORR, 63%; PFS, 9.7 months) and the patients with T790M‐positive tumor (ORR, 62%; PFS, 9.7 months) samples were similar. The first prospective study investigating the efficacy of osimertinib guided by plasma genotyping was conducted by Remon et al Among 16 patients with positive plasma T790M and evaluable lesions, the ORR was 62.5% and the rate of disease control was 100%. The 1‐year PFS, meanwhile, was as high as 52%.20 However, the number of patients was relatively low, and patients with T790M‐positive tumors were not enrolled. Despite the fact that plasma genotyping has many advantages, its low sensitivity and high false‐negative rate are major concerns.36 In Oxnard's study,5 the ORR of patients with T790M‐negative plasma was found to be 46%, and the PFS was 8.2 months. The tumor genotyping was successful in distinguishing between a subset of patients who were negative for T790M and had poor outcomes (ORR, 25%; PFS, 2.8 months) and another subset of patients who were positive for T790M and had better outcomes (ORR, 69%; PFS, 16.5 months). Based on these findings, the paradigm for the use of plasma genotyping for T790M proposed by several experts emphasizes that if the plasma genotyping is negative for T790M, then tissue genotyping should be performed.5, 34, 37, 38 Following this algorithm, Buder et al conducted a clinical trial to evaluate osimertinib response. In that study, the objective response rate was found to be 70% while the disease control rate was 81% in patients with positive plasma T790M as determined using ddPCR or by tissue analyses if they were plasma T790‐negative. However, in real‐world settings, the detection of T790M based on the analysis of cell‐free plasma DNA or supported by tissue biopsy as determined by the treating physician may be an alternative choice for guiding treatment with osimertinib.
Of the various types of studies, randomized controlled trials (RCTs) provide the strongest possible form of evidence, and their results are usually the bases for the decision‐making guidelines used in clinical practice. However, RCTs do have some drawbacks, including the underrepresentation of vulnerable patient groups, a lack of long‐term safety data, and limited generalizability. For example, a variety of patient groups who do not meet the strict criteria for inclusion in RCTs are seen in clinical practice. As such, real‐world data from observational studies or databases can be helpful for answering a wide spectrum of clinical questions and addressing the drawbacks of clinical trials. The present study constitutes the first real‐world study suggesting that T790M detection via cell‐free plasma DNA analysis or supported by the results of a tissue biopsy can be used to guide treatment with osimertinib. In the AURA 3 study trial,12 patients with T790M‐positive tumors had a disease control rate of 93% and a median PFS of 9.6 months. In the current study, we confirmed that treatment guided by plasma or tissue T790M mutations could result in a similar median PFS (11.3 months) and disease control rate (91.9%). In patients positive for T790M in plasma, the disease control rate was 92.3%, and the median PFS was 10.1 months. Though the OS data from the Aura 3 study are not available, the updated results from a pooled analysis of two Phase II studies (namely, the AURA extension and AURA2 studies) indicated that there was no significant difference between the second‐line and ≥third‐line cohorts, who received 80 mg of osimertinib once daily, in terms of median OS (95% CI, 25.8 versus 24.0). The 12‐month and 24‐month survival rates were 80% and 56%, respectively.39 In this study, we found that the 1‐year OS rate was 66.1%, which was slightly lower than that found in a previous report. We further conducted a Cox proportional hazards regression to identify the prognostic factors that would indicate when patients could benefit from receiving osimertinib in a real‐world setting. Similar to the results for the study referenced above, our results for this study indicated that the sequential use of chemotherapy and osimertinib did not affect survival (Table 2 and Figure S1). Also, the observation that intracranial progress after the failure of previous treatment did not affect the OS implied good penetration of osimertinib into the CNS. However, male patients tended to have poor prognoses. Although male gender was previously found to be a poor prognostic factor in patients receiving first‐line EGFR‐TKI,40 the significance of gender in patients receiving osimertinib has not been previously reported. In the AURA 3 study, male patients tended to have a higher hazard ratio of PFS after receiving osimertinib compared to chemotherapy as compared to female patients (0.43 versus 0.34), but that difference was not statistically significant.
This study had several limitations that should be acknowledged. First, the present study was a retrospective study of patients seen at a single center, and any patients taking part in clinical trials were excluded. However, the patients who joined the clinical trials also had positive results for tissue T790M, and if they had been included in the analyses, the response rate would not have been so low. In contrast to a review article,41 our real‐world data did not show a substantial difference from the results of clinical trials.5, 20, 28 Second, the sensitivity and specificity of the liquid biopsies could not be examined since most of the patients did not concomitantly receive both tissue and liquid biopsies. Given this lack of tissue biopsies for comparison, differences in the PFS and OS in the patients could not be assessed, which potentially biased the results. A prospective study comparing the survival rates of two such groups of patients would be worthwhile in the future. Finally, it is not clear whether the results of this study can be extrapolated to various other ctDNA platforms. A larger cohort study comparing more methods, including dPCR, BEAMing PCR, and next‐generation sequencing, would also be warranted in the future to guide the selection of patients who would benefit from osimertinib use.
5. CONCLUSIONS
We verified the role of plasma genotyping and tissue genotyping in guiding osimertinib treatment in the real world. Moreover, the PFS, objective response rate, and disease control rates found in the present study were similar to those found in previous trials. Overall, the study results indicate that both plasma genotyping and tissue genotyping for T790M detection can be used to predict the response to osimertinib in a real‐world setting.
AUTHOR CONTRIBUTIONS
Po‐Lan Su, Szu‐Chun Yang and Yi‐Lin Chen contributed equally to this work. PLS, SCY, YLW, CLH, and CCL conceived and designed the experiments. PLS, SCY, YLW, CCL, WYC, YLT, WWL, and WCS contributed materials. PLS, SCY, YLW, and CCL analyzed and interpreted the patient data. YLC and CLH performed the histological and genetic examinations. CYL analyzed the radiologic data. PLS, SCY, YLC, and CCL were major contributors to the writing of the manuscript. All the authors read and approved the final manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study protocol was approved by the institutional ethics committee of National Cheng Kung University Hospital (IRB number: A‐ER‐106‐205). Consent to participate was sought from all participants.
COMPETING INTERESTS
The authors declare that they have no competing interests.
Supporting information
ACKNOWLEDGMENTS
We are grateful to Wen‐Ping Su, Shang‐Yin Wu, Yu‐Ming Yeh, and Yi‐Ting Yen, for their valuable assistance in recruiting subjects. This study was based in part on data sourced from the Cancer Data Bank of National Cheng Kung University Hospital.
Su P‐L, Yang S‐C, Chen Y‐L, et al. Real‐world outcomes of NSCLC patients receiving tissue or circulating tumor DNA‐guided osimertinib treatment. Cancer Med. 2019;8:5939–5947. 10.1002/cam4.2485
Funding information
This study received support from the Center of Applied Nanomedicine at National Cheng Kung University and from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project of the Ministry of Education (MOE), Taiwan, as well as via grant MOHW106‐TDU‐B‐211‐144‐004 from the Ministry of Health and Welfare, Taiwan; grants MOST104‐2314‐B‐006‐046‐MY3 and MOST105‐2314‐B‐076‐MY2 from the Ministry of Science and Technology, Taiwan; and grant NCKUH‐10503002 from National Cheng Kung University Hospital, Tainan, Taiwan.
Contributor Information
Chung‐Liang Ho, Email: clh9@mail.ncku.edu.tw.
Chien‐Chung Lin, Email: joshcclin@gmail.com.
DATA AVAILABILITY STATEMENT
The datasets for this manuscript are not publicly available due to legal restrictions imposed by the government of Taiwan in relation to the “Personal Information Protection Act.” Requests for access to the datasets should be directed to Chien‐Chung Lin, MD, PhD.
REFERENCES
- 1. Bulbul A, Husain H. First‐line treatment in EGFR mutant non‐small cell lung cancer: Is there a best option? Front Oncol. 2018;8:94 10.3389/fonc.2018.00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lee CK, Davies L, Wu YL, et al. Gefitinib or erlotinib vs chemotherapy for EGFR mutation‐positive lung cancer: individual patient data meta‐analysis of overall survival. J Natl Cancer Inst. 2017;109(6):djw279 10.1093/jnci/djw279 [DOI] [PubMed] [Google Scholar]
- 3. Kuiper JL, Heideman D, Thunnissen E, et al. Incidence of T790M mutation in (sequential) rebiopsies in EGFR‐mutated NSCLC‐patients. Lung Cancer. 2014;85:19‐24. 10.1016/j.lungcan.2014.03.016 [DOI] [PubMed] [Google Scholar]
- 4. Li W, Ren S, Li J, et al. T790M mutation is associated with better efficacy of treatment beyond progression with EGFR‐TKI in advanced NSCLC patients. Lung Cancer. 2014;84:295‐300. 10.1016/j.lungcan.2014.03.011 [DOI] [PubMed] [Google Scholar]
- 5. Oxnard GR, Thress KS, Alden RS, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non‐small‐cell lung cancer. J Clin Oncol. 2016;34:3375‐3382. 10.1200/JCO.2016.66.7162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ricciuti B, Baglivo S, Paglialunga L, et al. Osimertinib in patients with advanced epidermal growth factor receptor T790M mutation‐positive non‐small cell lung cancer: rationale, evidence and place in therapy. Ther Adv Med Oncol. 2017;9:387‐404. 10.1177/1758834017702820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR‐TKI therapy in 155 patients with EGFR‐mutant lung cancers. Clin Cancer Res. 2013;19:2240‐2247. 10.1158/1078-0432.CCR-12-2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sequist LV, Soria JC, Goldman JW, et al. Rociletinib in EGFR‐mutated non‐small‐cell lung cancer. N Engl J Med. 2015;372:1700‐1709. 10.1056/NEJMoa1413654 [DOI] [PubMed] [Google Scholar]
- 9. Jänne PA, Yang JH, Kim DW, et al. AZD9291 in EGFR inhibitor‐resistant non‐small‐cell lung cancer. N Engl J Med. 2015;372:1689‐1699. 10.1056/NEJMoa1411817 [DOI] [PubMed] [Google Scholar]
- 10. Cross D, Ashton SE, Ghiorghiu S, et al. AZD9291, an irreversible EGFR TKI, overcomes T790M‐mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov. 2014;4:1046‐1061. 10.1158/2159-8290.CD-14-0337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goss G, Tsai CM, Shepherd FA, et al. Osimertinib for pretreated EGFR Thr790Met‐positive advanced non‐small‐cell lung cancer (AURA2): a multicentre, open‐label, single‐arm, phase 2 study. Lancet Oncol. 2016;17:1643‐1652. 10.1016/S1470-2045(16)30508-3 [DOI] [PubMed] [Google Scholar]
- 12. Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or platinum‐pemetrexed in EGFR T790M‐positive lung cancer. N Engl J Med. 2017;376:629‐640. 10.1056/NEJMoa1612674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Buder A, Tomuta C, Filipits M. The potential of liquid biopsies. Curr Opin Oncol. 2016;28:130‐134. 10.1097/CCO.0000000000000267 [DOI] [PubMed] [Google Scholar]
- 14. Jiang T, Ren S, Zhou C. Role of circulating‐tumor DNA analysis in non‐small cell lung cancer. Lung Cancer. 2015;90:128‐134. 10.1016/j.lungcan.2015.09.013 [DOI] [PubMed] [Google Scholar]
- 15. Santarpia M, Karachaliou N, González‐Cao M, Altavilla G, Giovannetti E, Rosell R. Feasibility of cell‐free circulating tumor DNA testing for lung cancer. Biomark Med. 2016;10:417‐430. 10.2217/bmm.16.6 [DOI] [PubMed] [Google Scholar]
- 16. Oxnard GR, Paweletz CP, Kuang Y, et al. Noninvasive detection of response and resistance in EGFR‐mutant lung cancer using quantitative next‐generation genotyping of cell‐free plasma DNA. Clin Cancer Res. 2014;20:1698‐1705. 10.1158/1078-0432.CCR-13-2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sacher AG, Komatsubara KM, Oxnard GR. Application of plasma genotyping technologies in non‐small cell lung cancer: a practical review. J Thorac Oncol. 2017;12:1344‐1356. 10.1016/j.jtho.2017.05.022 [DOI] [PubMed] [Google Scholar]
- 18. Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol. 2016;2:1014‐1022. 10.1001/jamaoncol.2016.0173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang L, Guo Q, Yu W, et al. Quantification of plasma EGFR mutations in patients with lung cancers: comparison of the performance of ARMS‐Plus and droplet digital PCR. Lung Cancer. 2017;114:31‐37. 10.1016/j.lungcan.2017.10.007 [DOI] [PubMed] [Google Scholar]
- 20. Remon J, Caramella C, Jovelet C, et al. Osimertinib benefit in EGFR‐mutant NSCLC patients with T790M‐mutation detected by circulating tumour DNA. Ann Oncol. 2017;28:784‐790. 10.1093/annonc/mdx017 [DOI] [PubMed] [Google Scholar]
- 21. Tirkes T, Hollar MA, Tann M, Kohli MD, Akisik F, Sandrasegaran K. Response criteria in oncologic imaging: review of traditional and new criteria. Radiographics. 2013;33:1323‐1341. 10.1148/rg.335125214 [DOI] [PubMed] [Google Scholar]
- 22. Chen YL, Lu CC, Yang SC, et al. Verification of wild‐type EGFR status in non‐small cell lung carcinomas using a mutant‐enriched PCR on selected cases. J Mol Diagn. 2014;16:486‐494. 10.1016/j.jmoldx.2014.05.007 [DOI] [PubMed] [Google Scholar]
- 23. van Dongen J, Langerak AW, Brüggemann M, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T‐cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED‐2 concerted action BMH4‐CT98‐3936. Leukemia. 2003;17:2257‐2317. 10.1038/sj.leu.2403202 [DOI] [PubMed] [Google Scholar]
- 24. Yoon HJ, Lee HY, Lee KS, et al. Repeat biopsy for mutational analysis of non‐small cell lung cancers resistant to previous chemotherapy: adequacy and complications. Radiology. 2012;265:939‐948. 10.1148/radiol.12112613 [DOI] [PubMed] [Google Scholar]
- 25. Chouaid C, Dujon C, Do P, et al. Feasibility and clinical impact of re‐biopsy in advanced non small‐cell lung cancer: a prospective multicenter study in a real‐world setting (GFPC study 12–01). Lung Cancer. 2014;86:170‐173. 10.1016/j.lungcan.2014.08.016 [DOI] [PubMed] [Google Scholar]
- 26. Piotrowska Z, Niederst MJ, Karlovich CA, et al. Heterogeneity underlies the emergence of EGFRT790 wild‐type clones following treatment of T790M‐positive cancers with a third‐generation EGFR inhibitor. Cancer Discov. 2015;5:713‐722. 10.1158/2159-8290.CD-15-0399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Piotrowska Z, Drapkin B, Engelman JA, Nagy RJ, Lanman RB, Sequist LV. Plasma T790M result alters treatment options in a previously T790 wild‐type EGFR‐mutant lung cancer. J Thorac Oncol. 2016;11:e95‐e97. 10.1016/j.jtho.2016.03.020 [DOI] [PubMed] [Google Scholar]
- 28. Buder A, Hochmair MJ, Schwab S, et al. Cell‐free plasma DNA‐guided treatment with osimertinib in patients with advanced EGFR‐mutated NSCLC. J Thorac Oncol. 2018;13:821‐830. 10.1016/j.jtho.2018.02.014 [DOI] [PubMed] [Google Scholar]
- 29. Sundaresan TK, Sequist LV, Heymach JV, et al. Detection of T790M, the acquired resistance EGFR mutation, by tumor biopsy versus noninvasive blood‐based analyses. Clin Cancer Res. 2016;22:1103‐1110. 10.1158/1078-0432.CCR-15-1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reckamp KL, Melnikova VO, Karlovich C, et al. A highly sensitive and quantitative test platform for detection of NSCLC EGFR mutations in urine and plasma. J Thorac Oncol. 2016;11:1690‐1700. 10.1016/j.jtho.2016.05.035 [DOI] [PubMed] [Google Scholar]
- 31. Malapelle U, Mayo de‐Las‐Casas C, Rocco D, et al. Development of a gene panel for next‐generation sequencing of clinically relevant mutations in cell‐free DNA from cancer patients. Br J Cancer. 2017;116:802–810. 10.1038/bjc.2017.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Newman AM, Lovejoy AF, Klass DM, et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat Biotechnol. 2016;34:547‐555. 10.1038/nbt.3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20:548‐554. 10.1038/nm.3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rolfo C, Mack PC, Scagliotti GV, et al. IASLC statement paper: liquid biopsy for advanced non‐small cell lung cancer (NSCLC). J Thorac Oncol. 2018;13:1248‐1268. 10.1016/j.jtho.2018.05.030 [DOI] [PubMed] [Google Scholar]
- 35. Arneth B. Update on the types and usage of liquid biopsies in the clinical setting: a systematic review. BMC Cancer. 2018;18:527 10.1186/s12885-018-4433-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: a cross‐platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer. 2015;90:509‐515. 10.1016/j.lungcan.2015.10.004 [DOI] [PubMed] [Google Scholar]
- 37. Merker JD, Oxnard GR, Compton C, et al. Circulating tumor DNA analysis in patients with cancer: American society of clinical oncology and college of American pathologists joint review. J Clin Oncol. 2018;36:1631‐1641. 10.1200/JCO.2017.76.8671 [DOI] [PubMed] [Google Scholar]
- 38. Tan D, Yom SS, Tsao MS, et al. The international association for the study of lung cancer consensus statement on optimizing management of EGFR mutation‐positive non‐small cell lung cancer: status in 2016. J Thorac Oncol. 2016l;11:946‐963. 10.1016/j.jtho.2016.05.008 [DOI] [PubMed] [Google Scholar]
- 39. Mitsudomi T, Ahn M‐J, Bazhenova L, et al. 1348P overall survival (OS) in patients (pts) with EGFR T790M‐positive advanced non‐small cell lung cancer (NSCLC) treated with osimertinib: results from two phase II studies. Ann Oncol. 2017;28(suppl_5):mdx380.050 10.1093/annonc/mdx380.050 [DOI] [Google Scholar]
- 40. Choi YW, Jeon SY, Jeong GS, et al. EGFR exon 19 deletion is associated with favorable overall survival after first‐line gefitinib therapy in advanced non‐small cell lung cancer patients. Am J Clin Oncol. 2018;41:385‐390. 10.1097/COC.0000000000000282 [DOI] [PubMed] [Google Scholar]
- 41. Singal AG, Higgins PD, Waljee AK. A primer on effectiveness and efficacy trials. Clin Transl Gastroenterol. 2014;5:e45 10.1038/ctg.2013.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets for this manuscript are not publicly available due to legal restrictions imposed by the government of Taiwan in relation to the “Personal Information Protection Act.” Requests for access to the datasets should be directed to Chien‐Chung Lin, MD, PhD.


