Abstract
AMPA receptors are hetero-oligomers composed of subsets of four distinct subunits, termed GluR1, GluR2, GluR3, and GluR4. Using quantitative reverse transcription-PCR analysis, we have found that light-induced degeneration of rat retina dramatically suppresses developmental progression of the flip-to-flop alternative splicing switch of retinal GluR1 mRNA. When animals were raised under standard conditions of a 12 hr light/dark cycle (LD 12:12), the flop-to-flip ratio in GluR1 and GluR2 dramatically increased between postnatal day 10 (P10) and P28, and the ratios continued to increase gradually up to P84. When animals were raised in complete darkness, this increase was delayed in GluR1 between P21 and P42. In addition, the increase of the flop-to-flip ratio in GluR1 was strongly suppressed after P21 under conditions of continuous illumination from P2. This is significant because P21 is just after the eye opening and is the timing of the onset of light-induced retinal degeneration. This suppression of the increase of the flop-to-flip ratio was specific to GluR1 and was not observed in GluR2–4. Immunocytochemistry and immunoblot analysis suggested no changes in either the distribution or expression of GluR1 protein in the light-damaged retina measured at P84. When rats were raised under continuous illumination from P2 to P21 followed by LD 12:12 from P22 to P84, retinal degeneration did not progress after P22. In such animals the flop-to-flip ratio, once decreased to ∼50% of the control (LD 12:12) at P21, was restored to the control level at P84. These findings demonstrate that developmental progression of the flip-to-flop exon switch in retinal GluR1 is affected by lighting conditions, and that light-induced retinal degeneration contributes to the mechanism of suppression of this splicing switch.
Keywords: flip and flop, AMPA, retina, quantitative reverse transcription-PCR, light/dark condition, retinal degeneration
Glutamate is a major excitatory neurotransmitter that plays an important role in neuronal plasticity and neurotoxicity in the CNS. In the vertebrate retina, synapses connecting photoreceptors (cones and rods) to bipolar and horizontal cells and those connecting bipolar cells to ganglion and amacrine cells are thought to use Glu as a neurotransmitter (Ayoub and Copenhagen, 1990; Tachibana et al., 1993). Photoreceptors respond to light through graded electrical potentials, and depolarization triggers release of Glu from the photoreceptor terminals in the outer plexiform layer (OPL) (Ehinger and Dowling, 1987). Both ionotropic and metabotropic Glu receptors are believed to be involved in retinal glutamatergic synaptic transmission (Hamassaki-Britto et al., 1993;Masu et al., 1995). In the ionotropic receptors, AMPA-type Glu receptors are thought to mediate fast excitatory postsynaptic currents (Collingridge and Lester, 1989; Monaghan et al., 1989).
AMPA receptors are hetero-oligomeric complexes composed of various combinations of four subunits termed GluR1, GluR2, GluR3, and GluR4. Each subunit exists in “flip” and “flop” isoforms that are generated by alternative mRNA splicing (Sommer et al., 1990). The two isoforms differ by seven residues in a 38 amino acid cassette that forms part of the large extracellular loop of the receptor molecules (Hollmann et al., 1994; Stern-Bach et al., 1994; Bennett and Dingledine, 1995). The splice variant composition of AMPA receptors affects the kinetics of the onset of and recovery from desensitization. Receptors containing flip subunits exhibit significantly slower desensitization kinetics and a greater steady-state component in their response to Glu relative to those containing flop subunits (Sommer et al., 1990; Seeburg, 1993; Hollmann and Heinemann, 1994; Lambolez et al., 1996). Recently, Otis et al. (1996) reported direct evidence that AMPA receptors desensitize after glutamatergic neurotransmission. Furthermore, flip-to-flop alternative splicing is developmentally regulated in a cell-specific manner (Monyer et al., 1991); prenatal AMPA receptors are composed mostly of the flip form, with the flop form appearing postnatally in the rat brain (Standley et al., 1995). These findings suggest that the flip-flop region plays a critical role in AMPA receptor function in some synapses as demonstrated by Trussell and Fischbach (1989).
In retina, all four subunits of AMPA receptors have been localized byin situ hybridization (Hamassaki-Britto et al., 1993;Duvoisin et al., 1995) and immunocytochemistry (Hughes et al., 1992;Peng et al., 1995). However, the existence of flip and flop subunit isoforms has not yet been determined. In the present study, we monitored the postnatal progression of flip-to-flop alternative mRNA splicing in retinal AMPA receptor subunits as a function of rat development under conditions of either continuous illumination or darkness. For this purpose, we found that isoform-specific in situ hybridization proved insufficient because of low sensitivity. Therefore, reverse transcription-PCR (RT-PCR) was used with total RNA isolated from the retina of rats reared under different light/dark conditions. Our results show that light-induced retinal degeneration dramatically suppresses developmental progression of flip-to-flop alternative mRNA splicing in the retinal GluR1 subunit but not in GluR2–4.
MATERIALS AND METHODS
Experimental animals. All experiments were performed on Wistar rats in accordance with the Association for Research in Vision and Ophthalmology statement for the Use of Animals in Ophthalmic and Vision Research. The animals were housed in a room with controlled temperature and a fixed lighting schedule of a 12 hr light/dark cycle (LD 12:12, control condition) unless otherwise described. Food and tap water were given ad libitum.
The rats were divided into five groups: LD 12:12 between postnatal day 2 (P2) and P84; 24 hr constant darkness between P2 and P84; 24 hr constant illumination between P2 and P21 followed by LD 12:12 to P84; 24 hr constant illumination between P2 and P42 followed by LD 12:12 to P84; and 24 hr constant illumination between P2 and P84. Light intensity inside the cages ranged from 100 to 200 lux under LD 12:12, whereas 1200–2000 lux was used for 24 hr constant illumination to effect light-induced retinal degeneration (Noell, 1980; Harada et al., 1996).
Amplification of GluR1–4 cDNAs. Total RNA was extracted from rat retina with the acid guanidinium thiocyanate–phenol–chloroform method (Chomczynski and Sacchi, 1987) and subsequently used as a template for RT-PCR. Three micrograms of total RNA were reverse-transcribed into first-strand cDNA using the Superscript preamplification system (BRL, Bethesda, MD) and oligo-dT primers. Total RNAs were extracted from two or three animals per developmental stage, and RT-PCR was performed in triplicate for every total RNA sample.
Oligonucleotide primers used for PCR amplification.The oligonucleotide primers used for PCR amplification were as follows: 5′-AGAGGGACGAGACCAGACAACCAG-3′ (GluR1-forward, 1713–1736); 5′-GCTGGTCTTGTCCTTGGAGTCACC-3′ (GluR1 flop-reverse, 2374–2397); 5′-GCTGGTCTTGTCCTTACTTCCGGA-3′ (GluR1 flip-reverse, 2374–2397); 5′-AGATGGAAGAGAAACACAAAGTAG-3′ (GluR2-forward, 2049–2072); 5′-ACTGGTCTTTTCCTTGGAATCACC-3′ (GluR2 flop-reverse, 2710–2733); 5′-ACTGGTCTTTTCCTTACTTCCCGA-3′ (GluR2 flip-reverse, 2710–2733); 5′-ACCTCGTGACCCACAAAGCCCTCC-3′ (GluR3-forward, 1749–1772); 5′-ACTGGTCTTGTCCTTGGAGTCACC-3′ (GluR3 flop-reverse, 2410–2433); 5′-ACTGGTCTTGTCCTTACTCCCGGA-3′ (GluR3 flip-reverse, 2410–2433); 5′-GGATGGGAAGGAAGGACCCAGTGA-3′ (GluR4-forward, 1895–1918); 5′-ACTCGTCTTGTCCTTGGAGTCACC-3′ (GluR4 flop-reverse, 2559–2582); 5′-ACTCGTCTTGTCCTTGCTTCCCGA-3′ (GluR4 flip-reverse, 2559–2582); 5′-ACCACAGTCCATGCCATCAC-3′ [glyceraldehyde 3-phosphate dehydrogenase (G3PDH)-forward, 586–605]; and 5′-TCCACCACCCTGTTGCTGTA-3′ (G3PDH-reverse, 1018–1037).
These oligonucleotide primers encompassed the regions of maximum nucleotide sequence dissimilarity between the flip and flop exons, as well as between the four receptor sequences. The last two primers were used to amplify a fragment of the gene encoding G3PDH, and this fragment served as an internal standard. Complete nucleotide sequences encoding the flip-containing polypeptides are available in the EMBL/GenBank database under accession numbers M38060 (GluR1), M38061(GluR2), M38062 (GluR3), and M38063 (GluR4), and the corresponding flop versions under M36418 (GluR1), M36419 (GluR2), M36420 (GluR3), andM36421 (GluR4).
Quantitative RT-PCR analysis. Each cDNA product from the reverse transcription procedure was used as the template for PCR amplification in a reaction mixture containing PCR buffer (in mm: 10 Tris-HCl, pH 8.3, 50 KCl, and 1.5 MgCl2), 0.2 mm dNTPs, a 0.2 mm concentration of each set of oligonucleotide primers, and 2.5 U of Taq DNA polymerase in 100 μl of final volume. PCR was performed after initial denaturation at 94°C for 3 min. Each cycle consisted of a heat denaturation step at 94°C for 15 sec, annealing of primers at 60°C for 2 min, and polymerization at 72°C for 2 min. Negative controls for PCR were performed using templates derived from reverse transcription reactions lacking either reverse transcriptase or total RNA. After the first 12 cycles, the primers for G3PDH mRNA were added to the reaction mixture, and additional 20 cycles of amplification were performed.
To construct a standard curve, 3.75–60 ng of total RNA was reverse-transcribed, and the resulting cDNA was subjected to 32 cycles of PCR. Ten microliters of each reaction mixture were removed after each cycle during cycles 23–32 and electrophoresed on a 2% Tris borate–EDTA agarose gel. The gel was stained with ethidium bromide to detect the bands of amplified fragments, which were quantitated using a CCD image sensor (Densitograph AE 6920, Atto, Japan) (Nakayama et al., 1992). The results were plotted on a semilogarithmic scale against the PCR cycle number or on a logarithmic scale against the amount of template RNA used in the reverse transcription reaction.
To determine relative expression levels, the intensity of the band from GluR was normalized to the intensity of the band from G3PDH at every reaction. This normalized value was used to calculate the flop-to-flip ratio.
Histology and morphometric studies. Rats were anesthetized with diethylether and perfused transcardially with saline, followed by 4% paraformaldehyde in 0.1 m phosphate buffer containing 0.5% picric acid at room temperature. For the dark-reared animals, anesthesia was administered under complete darkness, and the rat’s face was covered with a black bag that was impervious to light. Eyes were removed and post-fixed overnight in the same fixative and then embedded in paraffin. The posterior portion of the eye was sectioned sagittally at 7 μm thickness, mounted, and stained with hematoxylin and eosin.
For immunocytochemical staining, the sections were incubated in PBS containing 10% normal goat serum for 30 min at room temperature. The sections were then incubated overnight with rabbit anti-GluR1 affinity-purified polyclonal antibody (2 mg/ml; Chemicon, Temecula, CA) at room temperature. This antibody recognizes rat GluR1 protein and exhibits no cross-reactivity with GluR2–4, as determined by the supplier. The sections were then incubated at room temperature with secondary antibodies for 1 hr and with avidin–biotin–peroxidase complex for 30 min using a Histofine SAB-PO(R) kit (Nichirei Corp., Tokyo, Japan). The immunoreaction was visualized after addition of 3,3′-diaminobenzidine.
For the estimation of the thickness of the retinal layers, measurements were performed with a calibrated reticule at 80× magnification (Imaging System KS100; Kontron Elektronik).
Immunoblot. Membrane extracts were prepared by homogenizing rat retinas in 8–10 volumes of ice-cold 50 mm Tris-HCl, pH 7.5, containing (in mm): 250 sucrose, 25 KCl, and 4 MgCl2. After centrifugation for 10 min at 1000 ×g, the protein concentration was determined by Lowry’s method (BCA protein assay reagent; Pierce, Rockford, IL). Twenty micrograms of the protein samples were electrophoresed on 10% SDS-PAGE and subsequently transferred to a nitrocellulose membrane. The membrane was incubated with an anti-GluR1, affinity-purified polyclonal antibody (1 mg/ml) (Chemicon, Temecula, CA) and visualized using a chemiluminescence Western blotting kit (Boehringer Mannheim, Mannheim, Germany).
Statistics. Data are presented as mean ± SEM except as noted. When statistical analysis was performed, Student’st test was used to estimate the significance of the results. Statistical significance was accepted at p < 0.05.
RESULTS
Identification of alternative splicing variants
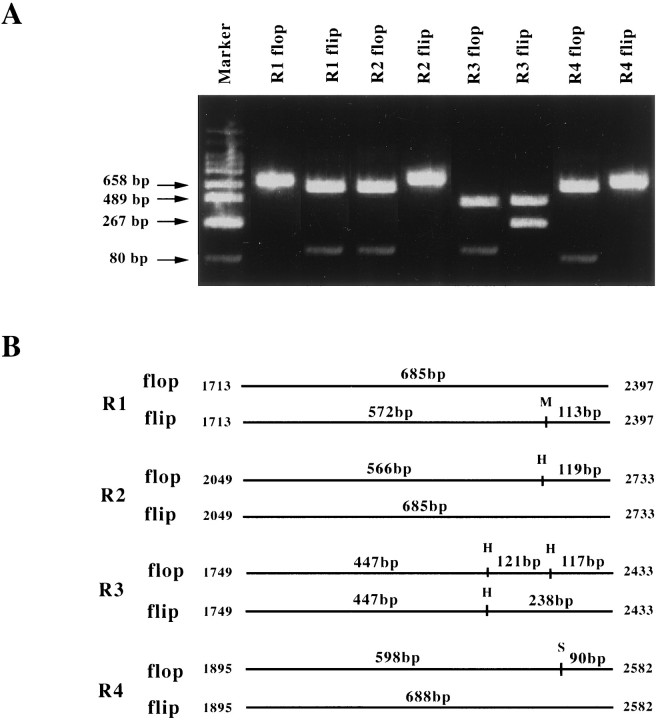
To reveal the existence of alternatively spliced variants of AMPA receptor subunits in retina, we performed RT-PCR using total RNA isolated from P21 rat retina and primers specific for individual GluR1–4 flip and flop isoforms. PCR products were separated on an agarose gel and visualized using ethidium bromide (Fig.1). In each reaction, a band migrating slightly above the 658 bp DNA marker was evident, consistent with the size expected for the GluR variants. Digestion of the products with restriction enzymes produced fragments of the following sizes: 572 and 113 bp for GluR1-flip cut by MaeI, whereas the GluR1-flop was uncut; 566 and 119 bp for GluR2-flop cut by HincII, whereas the GluR2-flip was uncut; 447, 121, and 117 bp for GluR3-flop, and 447 and 238 bp for GluR3-flip cut by HincII; and 598 and 90 bp for GluR4-flop cut by StuI, whereas the GluR4-flip was uncut. The size of each fragment was as expected with respect to the enzyme restriction sites. From these findings, we concluded that both flip and flop isoforms of all four GluR subunit mRNAs are expressed in P21 rat retina.
Fig. 1.
Amplification by PCR of DNA fragments specific for flop and flip sequences. A, Total RNA isolated from rat retinas was reverse-transcribed, and the DNA products were used as templates of PCR. Eight sets of primers specific for GluR1–4 (flip and flop) were used. The PCR products were digested with restriction endonucleases and then electrophoresed on a 2% agarose gel. The digested fragments were visualized after staining with ethidium bromide. B, Schematic representation of the specific GluR-flip and -flop fragments amplified as in A. The restriction endonuclease cleavage patterns were used to confirm specific amplification. M, MaeI;H, HincII; S,StuI.
Standard curves
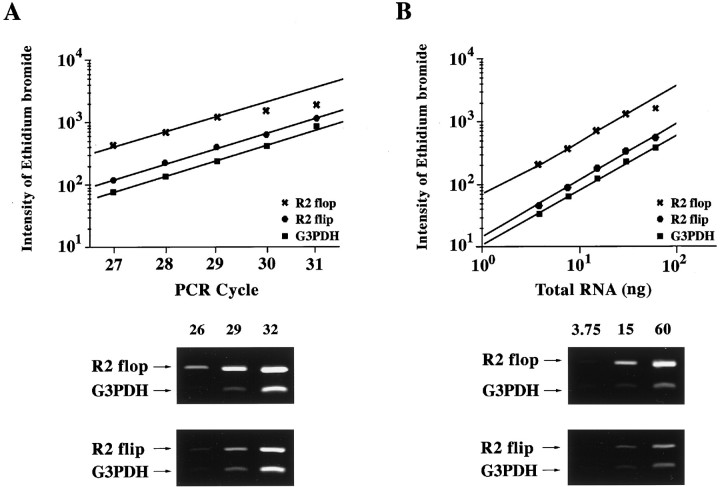
For quantitative comparison, we determined the linear range of PCR product accumulation as a function of both the number of cycles and the amount of total RNA template in the reverse transcription reaction. Standard curves were constructed for all GluR-flip and -flop isoforms and for the internal control, G3PDH. For GluR2 and G3PDH, amplification curves were linear between 25 and 29 cycles and when 3.75–30 ng of total RNA was used in the reverse transcription reactions (Fig.2). A plateau was observed when >29 cycles were performed or when >30 ng of total RNA was used (data not shown). The amplification curves for other subunits were similar to that of the GluR2-flip and -flop. On the basis of these results, subsequent RT-PCR analyses were performed for 28 cycles using 15 ng of total RNA in the reverse transcription reactions.
Fig. 2.
Standard curves for quantitative RT-PCR of GluR2-flip, GluR2-flop, and G3PDH. A, Product formation as a function of the number of PCR cycles. The cDNA template for each reaction was reverse-transcribed from 15 ng of total RNA isolated from rat retina. B, Product formation as a function of total rat retinal RNA used to produce the cDNA template for PCR. In each case, 28 cycles of PCR were performed. The intensity of the bands from GluR2-flip (685 bp), GluR2-flop (685 bp), and G3PDH (452 bp) detected by ethidium bromide staining (bottom) were quantitated using a CCD image sensor.
AMPA receptor mRNA splicing in rat retina under LD 12:12
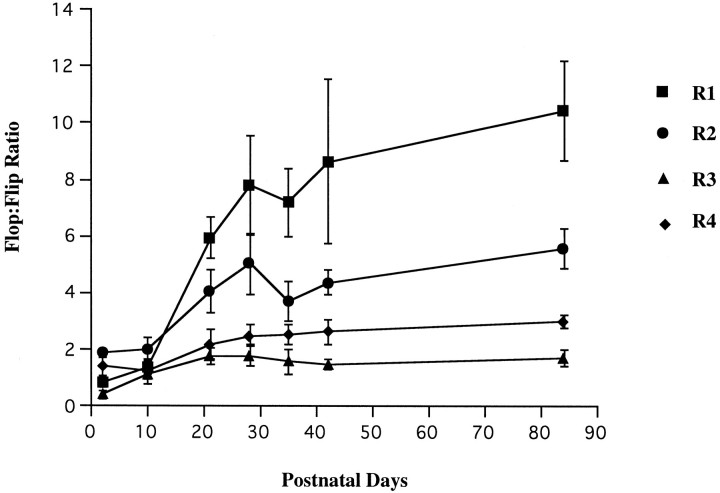
The flop-to-flip ratio of each AMPA receptor subunit was calculated between P2 and P84 (Fig. 3). The initial flop-to-flip ratio was <2.0 and remained relatively static in all four GluR subtypes during the first 10 postnatal days. However, in GluR1 and GluR2 the ratio increased dramatically between P10 and P28, to ∼6- and 2.5-fold, respectively. Between P28 and P84 the GluR2 ratio plateaued, whereas that of GluR1 increased to ∼10 at P84. The ratios for GluR3 and GluR4 did not change significantly from P10 to P84.
Fig. 3.
GluR flop-to-flip ratios in the developing rat retina under LD 12:12. The ratio is calculated by RT-PCR analysis as described in Materials and Methods. Note the dramatic increase of the flop-to-flip ratio in GluR1 and GluR2 between P10 and P28. Each data point indicates the mean ± SEM of the values obtained from three or four independent experiments.
AMPA receptor mRNA splicing in dark- and light-reared animals
To determine the effect of continuous illumination or darkness on AMPA receptor mRNA splicing, experiments similar to those shown in Figure 3 were performed using rats raised under three different lighting conditions: LD 12:12, continuous dark from P2, and continuous light from P2. The results are summarized in Figure4. The continuous dark condition did not significantly alter developmental changes of the flop-to-flip ratio in GluR2, GluR3, and GluR4 relative to the LD 12:12 condition. However, there were significant changes in the ratio for GluR1. As shown in Figure 4A, the flop-to-flip ratios at P28 and P35 were significantly lower in dark-reared animals than in the LD 12:12 animals (p = 0.014 at P28; p = 0.017 at P35). However, the flop-to-flip ratios in these two lighting conditions were not significantly different at P42 and P84. These results suggest that complete darkness delays the switch to the flop isoform in GluR1 mRNA splicing although the effects are restricted between P21 and P42.
Fig. 4.
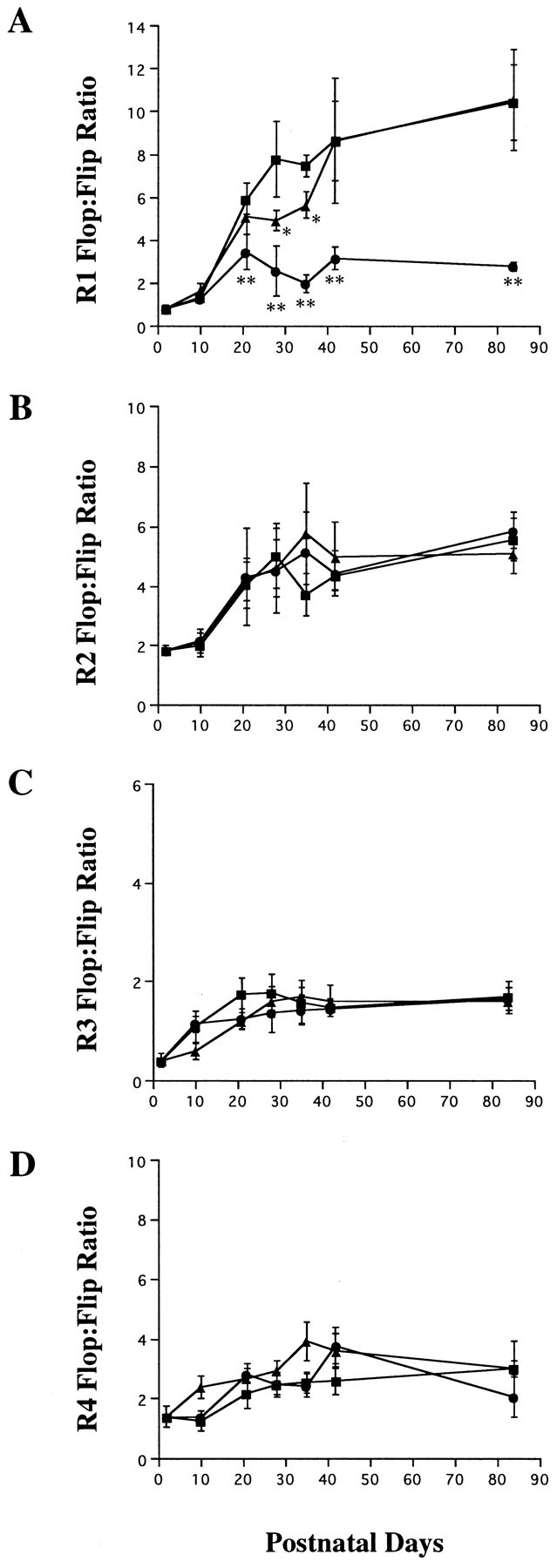
GluR flop-to-flip ratio in the developing rat retina under a 12 hr light/dark cycle (▪), constant dark conditions (▴), and constant illumination (•). Note the significantly low GluR1 flop-to-flip ratio under the light-reared conditions compared with that under a 12 hr light/dark cycle (A). Each data point represents the mean ± SEM of the values obtained from three or four independent experiments. The LD 12:12 data from Figure 3 are shown for comparison in each subunit. *p < 0.05; **p < 0.01.
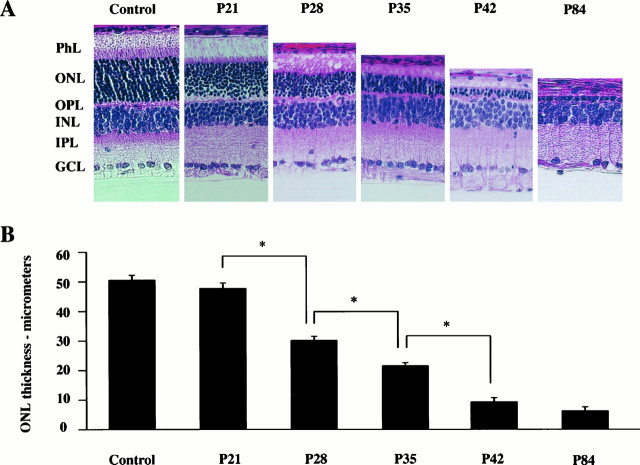
To determine the effect of constant illumination on the flop-to-flip ratio, RT-PCR analysis was performed using total RNA extracted from the retinas of rats raised under 24 hr continuous illumination. It is well documented that continuous illumination induces irreparable damage to retinal rod photoreceptors, quantified both as a reduction in the amplitude of the electroretinogram and as a decrease in photoreceptor density and thickness of the outer nuclear layer (ONL) (Noell, 1980). In the present study, continuous illumination began at P2, and the morphology of the retina was not changed at P21 when compared with that of the LD 12:12 retina (Fig. 5). This is probably attributable to the fact that eye opening occurs after P15 in the case of Wistar rats. The thickness of the ONL decreased rapidly between P21 and P42, and light-damage to the retina was near complete at P42 (Fig. 5). As shown in Figure 4, constant illumination suppressed the normal progression of alternative splicing only in GluR1. The flop-to-flip ratio of GluR1 at P21 was significantly lower in light-reared animals than in LD 12:12 animals (p< 0.001). In addition, the flop-to-flip ratio in GluR1 decreased between P21 and P35 as light-induced retinal degeneration progressed and was only ∼25% of that of LD 12:12 at P84 (Figs.4A, 6).
Fig. 5.
The damaging effect of constant light on rat retina. A, Light micrograph of rat retinal sections taken from P21, P28, P35, P42, and P84 rats raised under constant illumination (1200–2000 lux). The control section was taken from a P21 rat raised under LD 12:12 conditions. Note the progressive thinning of the ONL and PhL between P21 and P42. Almost all of the ONL and PhL are absent at P84. B, Progressive thinning of theONL. Each data point indicates the mean ± SEM of the values obtained from three independent experiments.PhL, Photoreceptor layer; ONL, outer nuclear layer; OPL, outer plexiform layer;INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer. *p < 0.05.
Fig. 6.
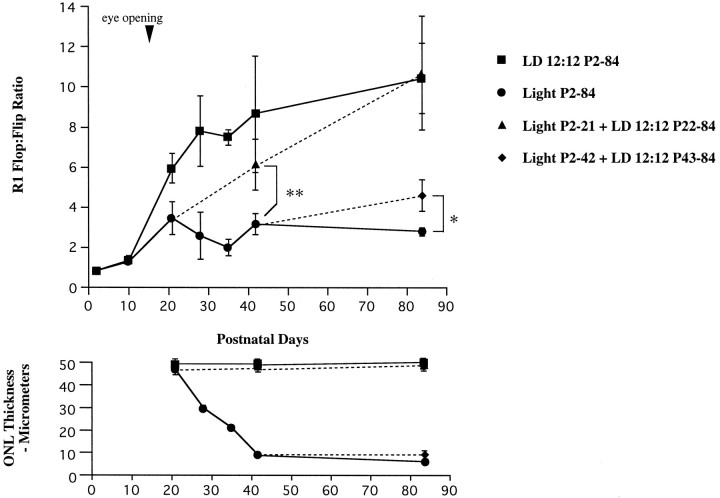
GluR1 flop-to-flip ratio in rat retina under LD 12:12 or constant illumination (top). Note the recovery of flop-to-flip ratio under LD 12:12 (dotted lines) when removed from constant illumination. The bottom shows theONL thickness in each group. When light-reared animals were removed to LD 12:12 at P22, retinal degeneration did not progress after P21. Each data point indicates the mean ± SEM of the values obtained from three or four independent experiments. *p < 0.05; **p < 0.01.
To determine whether the low flop-to-flip ratio in GluR1 resulted from the loss of photoreceptors, we divided the rats into two subgroups: light-reared until P21 or P42, followed by LD 12:12 to P84 (Fig. 6). The flop-to-flip ratio in the first group, which exhibited normal ONL thickness, fell to ∼50% of the control (LD 12:12) value but was completely restored by P84. In the second group in which the retina was damaged, the recovery was only marginal.
Retinal degeneration does not induce downregulation of GluR1 expression
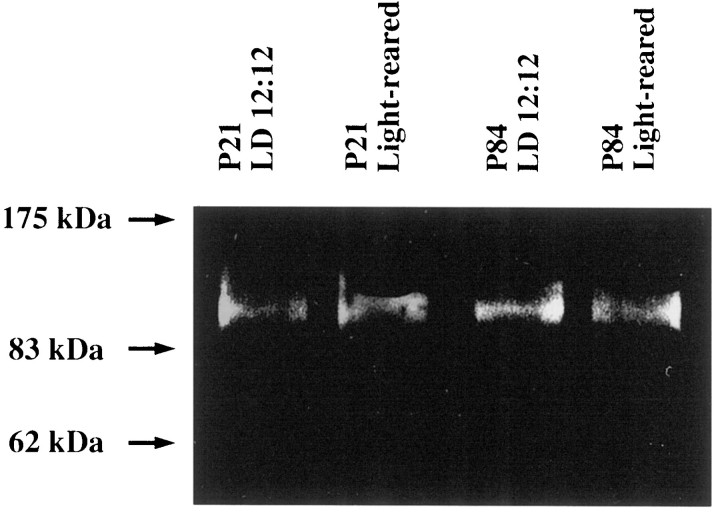
After retinal degeneration, glutamatergic input from photoreceptors is greatly reduced, but no significant change in the expression levels of the genes encoding GluR1–4 is apparent in therd mouse retina (Duvoisin et al., 1995). To determine the expression level of GluR1 protein in our retinal degeneration model, we performed immunoblots using extracts of P21 and P84 rat retina and an affinity-purified polyclonal antibody to GluR1. GluR1 protein (102–108 kDa) was detected in both the control and light-damaged retinas (Fig.7). The intensities of the bands corresponding to GluR1 were approximately equal, demonstrating that no apparent downregulation of GluR1 expression occurred between P21 and P84. In addition, the immunostaining of GluR1 in the light-damaged retina from a P84 rat was unchanged compared with that of normal retina from a P84 rat (data not shown). These results are consistent with the previous findings that inner layers of retina (bipolar, horizontal, amacrine, Müller, and ganglion cells) are not altered by light-induced retinal degeneration (Noell, 1980; Harada et al., 1996).
Fig. 7.
GluR1 in rat retina demonstrated by immunoblotting. Total tissue homogenates (P21 or P84) from rat retina (20 mg/lane) were electrophoresed by SDS-PAGE and transferred to nitrocellulose membrane. Immunoblotting was performed using antibodies specific for GluR1. A single immunoreactive band appeared at ∼100 kDa. The density of the band was almost identical in two different lighting conditions at both P21 and P84 as quantified by CCD imaging.
DISCUSSION
Subunit-specific development of AMPA receptor flip-to-flop mRNA splicing
We used a quantitative RT-PCR technique to examine the development of AMPA receptor alternative mRNA splicing in rat retina. A similar quantitative RT-PCR method was previously used to estimate the ratio of flop-to-flip mRNAs in rat cortical neurons and human brain (Lambolez et al., 1996; Eastwood et al., 1997). Here, we have demonstrated the dynamic molecular changes that occur in AMPA receptors in rat retina during postnatal development. It is reported that not only flip-to-flop alternative splicing but also subunit compositions and R/G site editing are important factors that determine AMPA receptor kinetics (Lomeli et al., 1994; Mosbacher et al., 1994; Geiger et al., 1995; Angulo et al., 1997). Therefore, further investigations, including single-cell PCR to determine subunit composition or relative abundance of R/G site-edited versions, are necessary to understand developmental changes of AMPA receptor function in retina. Our results described in the present study, developmental changes of flip-to-flop alternative splicing, seem to provide fundamental information for such understandings. Under control conditions of equal dark and light (LD 12:12), the fold increase in the flop-to-flip ratio was larger for GluR1 and GluR2 mRNAs than for GluR3 and GluR4 mRNAs (Fig. 3). In the rat retina, amacrine and ganglion cells express GluR1 predominantly, bipolar cells express GluR2 predominantly, horizontal cells express GluR3 predominantly, and glial Müller cells express GluR4 predominantly (Hughes et al., 1992; Peng et al., 1995). After the onset of vision, vertical synaptic connections consisting of photoreceptors, bipolar cells, and ganglion cells become the principal pathway by which visual information is transmitted from the retina to the visual cortex through the optic nerve. In GluR1- and GluR2-positive cells, the alteration of AMPA receptor kinetics by flip-to-flop alternative splicing may have physiological significance in relation to the development of the visual system.
GluR1 mRNA splicing in light/dark conditions
Flip-to-flop alternative splicing is thought to be regulated in a cell-specific manner under developmental control (Monyer et al., 1991; Standley et al., 1995). In addition, we have determined that environmental light/dark conditions affect the regulation of GluR1 alternative splicing in rat retina. Although it has been reported that environmental light/dark conditions affect the expression level of mRNAs encoding proteins involved in phototransduction, such as Fos (Yoshida et al., 1993, 1996; Harada et al., 1996), transducin (Brann and Cohen, 1987), and opsin (Bowes et al., 1988), we are not aware of data showing the relationship between light/dark conditions and the process of alternative mRNA splicing.
The suppression of the developmentally regulated increase of the flop-to-flip ratio in GluR1 was observed under conditions of either complete light or dark, and this suppression preceded any morphological signs of photoreceptor cell death (Figs. 4A, 6). This suggests that GluR1 alternative splicing is affected differentially by cyclic (LD 12:12) or constant light conditions. Amacrine and ganglion cells, which predominantly contain GluR1, can be divided into three subtypes: ON cells that are rapidly depolarized when the retina is illuminated; OFF cells that are depolarized when the light is turned off; and ON–OFF cells that are depolarized transiently both at the onset and offset of illumination (Miller, 1989; Schiller, 1992). Because specific light conditions are needed to stimulate each type of cell effectively, constant light conditions may have failed to stimulate each type of GluR1-containing cell. The heterogeneity of GluR1-containing retinal cells may underlie the process of GluR1-specific alternative splicing.
GluR1 in a retinal degeneration model
We found that light-induced degeneration of rat retina dramatically suppressed developmental progression of the flip-to-flop switch in retinal GluR1. The suppressive effect was specific to GluR1 and was not observed in the other three GluR subunits. Immunoblot analysis demonstrated that GluR1 protein expression was not affected by retinal degeneration (Fig. 7). When rats were removed from constant light to LD 12:12 before the onset of photoreceptor cell death, the flop-to-flip ratio at P84 was normal, although the low ratio did not recover in light-damaged retina (Fig. 6). Therefore, the suppression of the increase of the GluR1 flop-to-flip ratio correlates with retinal degeneration (Fig. 6). This may reflect a decrease of glutamatergic input from photoreceptors to GluR1-containing retinal cells during degeneration. These findings suggest that retinal degeneration has secondary effects on the kinetics of AMPA receptors containing GluR1, and that these effects may modulate synaptic transmission in light-damaged retina.
GluR1 and the other three subunits
Both light and dark conditions, as well as retinal degeneration, induced changes in GluR1-specific flip-to-flop alternative splicing in rat retina. The alteration of subunit-specific splicing has been observed in the hippocampus after seizure activity in GluR1–3 and after transient global ischemia in GluR2 (Pollard et al., 1993;Kamphuis et al., 1994). Because GluR2-containing receptors are Ca2+-impermeable (Gasic and Hollmann, 1992; Brusa et al., 1995), Ca2+ permeability may be selectively altered in epileptic and ischemic hippocampi in a GluR2-dependent manner. However, in the case of GluR1, specific pharmacological and/or physiological functions remain unknown.
Regulation of the function of GluR subunits is specified not only by flip-to-flop alternative splicing but also by mRNA editing in the R/G site (Lomeli et al., 1994). The R/G site is located at the N-terminal side of the flip-flop segments in mature proteins. Although this editing occurs in GluR2–4 mRNAs, it does not occur in that of GluR1 (Lomeli et al., 1994). The mRNA editing is thought to be catalyzed by adenosine deaminases that recognize specific double-stranded RNAs consisting of complementary intron and exon sequences in GluR2–4 mRNAs (Higuchi et al., 1993). The exon-complementary intron sequences are absent from the GluR1 intron 13 that is just upstream of the flop exon (exon 14). Although it is unknown whether the mechanisms regulating the R/G site editing are related to those of the flip-to-flop alternative splicing, the extent of R/G site editing is reported to be different according to the alternating splice forms of a subunit (Lomeli et al., 1994). For example, in GluR4, the flop version is almost fully edited at P7, whereas the flip version is edited only 55% in the adult rat brain. Given that the exon-complementary intron sequences are thought to be important for adenosine deaminase activity (Higuchi et al., 1993), the uniqueness of GluR1 intron 13 may be responsible for the observed specific effect of light/dark conditions, as well as retinal degeneration, on GluR1 mRNA splicing.
In conclusion, our data suggest that flip-to-flop alternative splicing in GluR1 is a mechanism of transcriptional regulation under conditions of light and dark and during retinal degeneration. Further investigations of events leading to the modification of AMPA receptor mRNAs may lead to a better understanding of the mechanisms of both retinal development and degeneration and may further our understanding of retinitis pigmentosa and macular degeneration.
Footnotes
This study was supported in part by grants from the Ministry of Health and Welfare, the Ministry of Education, Science, Sports, and Culture, the Science and Technology Agency of Japan, and the Japan Foundation for Neuroscience and Mental Health. We are very grateful to Dr. K. Yoshida for critical comments and Drs. O. Nakabayashi, H. Onodera, and J. Imaki for technical assistance.
Correspondence should be addressed to Dr. Takayuki Harada, Department of Degenerative Neurological Diseases, National Institute of Neuroscience, National Center of Neurology and Psychiatry, 4-1-1 Ogawahigashi, Kodaira, Tokyo 187-8502, Japan.
REFERENCES
- 1.Angulo MC, Lambolez B, Audinat E, Hestrin S, Rossier J. Subunit composition, kinetic, and permeation properties of AMPA receptors in single neocortical nonpyramidal cells. J Neurosci. 1997;17:6685–6696. doi: 10.1523/JNEUROSCI.17-17-06685.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayoub GS, Copenhagen DR. Applications of a fluorometric method to measure glutamate release from single retinal photoreceptors. J Neurosci Methods. 1990;37:7–14. doi: 10.1016/0165-0270(91)90016-s. [DOI] [PubMed] [Google Scholar]
- 3.Bennett JA, Dingledine R. Topology profile for a glutamate receptor: three transmembrane domains and a channel-lining re-entrant membrane loop. Neuron. 1995;14:373–384. doi: 10.1016/0896-6273(95)90293-7. [DOI] [PubMed] [Google Scholar]
- 4.Bowes C, Venn TV, Farber DB. Opsin, G-protein and 48 kDa protein in normal and rd mouse retinas: developmental expression of mRNAs and proteins and light/dark cycling of mRNAs. Exp Eye Res. 1988;47:369–390. doi: 10.1016/0014-4835(88)90049-8. [DOI] [PubMed] [Google Scholar]
- 5.Brann MR, Cohen LV. Diurnal expression of transducin mRNA and translocation of transducin in rods of rat retina. Science. 1987;235:585–587. doi: 10.1126/science.3101175. [DOI] [PubMed] [Google Scholar]
- 6.Brusa R, Zimmermann F, Koh DS, Felder D, Gass P, Seeburg PH, Sprengel R. Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science. 1995;270:1677–1680. doi: 10.1126/science.270.5242.1677. [DOI] [PubMed] [Google Scholar]
- 7.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 8.Collingridge GL, Lester RA. Excitatory amino acid receptors in the vertebrate central nervous system. Pharmacol Rev. 1989;41:143–210. [PubMed] [Google Scholar]
- 9.Duvoisin RM, Zhang C, Hamassaki-Britto DE, Britto LRG. Changes in expression of glutamate receptor subunits following photoreceptor degeneration in the rd mouse retina. Neurosci Lett. 1995;183:83–86. doi: 10.1016/0304-3940(94)11120-8. [DOI] [PubMed] [Google Scholar]
- 10.Eastwood SL, Burnet PWJ, Harrison PJ. GluR2 glutamate receptor subunit flip and flop isoforms are decreased in the hippocampal formation in schizophrenia: a reverse transcriptase-polymerase chain reaction (RT-PCR) study. Mol Brain Res. 1997;44:92–98. doi: 10.1016/s0169-328x(96)00195-7. [DOI] [PubMed] [Google Scholar]
- 11.Ehinger B, Dowling JE. Retinal neurocircuitry and transmission. In: Bjorklund A, Hokfelt T, Swanson LW, editors. Handbook of chemical neuroanatomy, Vol 5. Elsevier; Amsterdam: 1987. pp. 389–446. [Google Scholar]
- 12.Gasic GP, Hollmann M. Molecular neurobiology of glutamate receptors. Annu Rev Physiol. 1992;54:507–536. doi: 10.1146/annurev.ph.54.030192.002451. [DOI] [PubMed] [Google Scholar]
- 13.Geiger JR, Melcher T, Koh DS, Sakmann B, Seeburg PH, Jonas P, Monyer H. Relative abundance of subunit mRNAs determines gating and Ca2+ permeability of AMPA receptors in principal neurons and interneurons in rat CNS. Neuron. 1995;15:193–204. doi: 10.1016/0896-6273(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 14.Hamassaki-Britto DE, Hermans-Borgmeyer I, Heinemann S, Hughes TE. Expression of glutamate receptor genes in the mammalian retina: the localization of GluR1 through GluR7 mRNAs. J Neurosci. 1993;13:1888–1898. doi: 10.1523/JNEUROSCI.13-05-01888.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harada T, Imaki J, Ohki K, Ono K, Ohashi T, Matsuda H, Yoshida K. Cone-associated c-fos gene expression in the light-damaged rat retina. Invest Ophthalmol Vis Sci. 1996;37:1250–1255. [PubMed] [Google Scholar]
- 16.Higuchi M, Single FN, Kohler M, Sommer B, Sprengel R, Seeburg PH. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 17.Hollmann M, Heinemann S. Cloned glutamate receptors. Annu Rev Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- 18.Hollmann M, Maron C, Heinemann S. N-glycosylation site tagging suggests a three transmembrane domain topology for the glutamate receptor GluR1. Neuron. 1994;13:1331–1343. doi: 10.1016/0896-6273(94)90419-7. [DOI] [PubMed] [Google Scholar]
- 19.Hughes TE, Hermans-Borgmeyer I, Heinemann S. Differential expression of glutamate receptor genes (GluR1–5) in the rat retina. Vis Neurosci. 1992;8:49–55. doi: 10.1017/s0952523800006489. [DOI] [PubMed] [Google Scholar]
- 20.Kamphuis W, De Rijk TC, Talamini LM, Lopes da Silva FH. Rat hippocampal kindling induces changes in the glutamate receptor mRNA expression patterns in dentate granule neurons. Eur J Neurosci. 1994;6:1119–1127. doi: 10.1111/j.1460-9568.1994.tb00609.x. [DOI] [PubMed] [Google Scholar]
- 21.Lambolez B, Ropert N, Perrais D, Rossier J, Hestrin S. Correlation between kinetics and RNA splicing of α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptors in neocortical neurons. Proc Natl Acad Sci USA. 1996;93:1797–1802. doi: 10.1073/pnas.93.5.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger JRP, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg PH. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 23.Masu M, Iwakabe H, Tagawa Y, Miyoshi T, Yamashita M, Fukuda Y, Sasaki H, Hiroi K, Nakamura Y, Shigemoto R, Takada M, Nakamura K, Nakao K, Katsuki M, Nakanishi S. Specific deficit of the ON response in visual transmission by targeted disruption of the mGluR6 gene. Cell. 1995;80:757–765. doi: 10.1016/0092-8674(95)90354-2. [DOI] [PubMed] [Google Scholar]
- 24.Miller RF. The physiology and morphology of the vertebrate retina. In: Ryan SJ, editor. Retina, Vol 1. Mosby; St. Louis: 1989. pp. 83–106. [Google Scholar]
- 25.Monaghan DT, Bridges RJ, Cotman CW. The excitatory amino acid receptors: their classes, pharmacology, and distinct properties in the function of the central nervous system. Annu Rev Pharmacol Toxicol. 1989;29:365–402. doi: 10.1146/annurev.pa.29.040189.002053. [DOI] [PubMed] [Google Scholar]
- 26.Monyer H, Seeburg PH, Wisden W. Glutamate-operated channels: developmentally early and mature forms arise by alternative splicing. Neuron. 1991;6:799–810. doi: 10.1016/0896-6273(91)90176-z. [DOI] [PubMed] [Google Scholar]
- 27.Mosbacher J, Schoepfer R, Monyer H, Burnashev N, Seeburg PH, Ruppersberg JP. A molecular determinant for submillisecond desensitization in glutamate receptors. Science. 1994;266:1059–1062. doi: 10.1126/science.7973663. [DOI] [PubMed] [Google Scholar]
- 28.Nakayama H, Yokoi H, Fujita J. Quantification of mRNA by non-radioactive RT-PCR and CCD imaging system. Nucleic Acids Res. 1992;20:4939. doi: 10.1093/nar/20.18.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noell WK. Possible mechanism of photoreceptor damage by light in mammalian eyes. Vision Res. 1980;20:1163–1171. doi: 10.1016/0042-6989(80)90055-3. [DOI] [PubMed] [Google Scholar]
- 30.Otis T, Zhang S, Trussell LO. Direct measurement of AMPA receptor desensitization induced by glutamatergic synaptic transmission. J Neurosci. 1996;16:7496–7504. doi: 10.1523/JNEUROSCI.16-23-07496.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng YW, Blackstone CD, Huganir RL, Yau KW. Distribution of glutamate receptor subtypes in the vertebrate retina. Neuroscience. 1995;66:483–497. doi: 10.1016/0306-4522(94)00569-q. [DOI] [PubMed] [Google Scholar]
- 32.Pollard H, Heron A, Moreau J, Ben-Ari Y, Khrestchatisky M. Alterations of the GluR-B AMPA receptor subunit flip/flop expression in kainate-induced epilepsy and ischemia. Neuroscience. 1993;57:545–554. doi: 10.1016/0306-4522(93)90004-y. [DOI] [PubMed] [Google Scholar]
- 33.Schiller PH. The on and off channels of the visual system. Trends Neurosci. 1992;15:86–92. doi: 10.1016/0166-2236(92)90017-3. [DOI] [PubMed] [Google Scholar]
- 34.Seeburg PH. The molecular biology of mammalian glutamate receptor channels. Trends Neurosci. 1993;16:359–365. doi: 10.1016/0166-2236(93)90093-2. [DOI] [PubMed] [Google Scholar]
- 35.Sommer B, Keinanen K, Verdoorn TA, Wisden W, Burnashev N, Herb A, Kohler M, Takagi T, Sakmann B, Seeburg PH. Flip and flop: a cell-specific functional switch in glutamate-operated channels of the CNS. Science. 1990;249:1580–1585. doi: 10.1126/science.1699275. [DOI] [PubMed] [Google Scholar]
- 36.Standley S, Tocco G, Tourigny MF, Massicotte G, Thompson RF, Baudry M. Developmental changes in alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate receptor properties and expression in the rat hippocampal formation. Neuroscience. 1995;67:881–892. doi: 10.1016/0306-4522(95)00075-t. [DOI] [PubMed] [Google Scholar]
- 37.Stern-Bach Y, Bettler B, Hartley M, Sheppard PO, O’Hara PJ, Heinemann SF. Agonist-selectivity of glutamate receptors is specified by two domains structurally related to bacterial amino acid binding proteins. Neuron. 1994;13:1345–1357. doi: 10.1016/0896-6273(94)90420-0. [DOI] [PubMed] [Google Scholar]
- 38.Tachibana M, Okada T, Arimura T, Kobayashi K, Piccolino M. Dihydropyridine sensitive calcium current mediates neurotransmitter release from bipolar cells of the goldfish retina. J Neurosci. 1993;13:2898–2909. doi: 10.1523/JNEUROSCI.13-07-02898.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trussell LO, Fischbach GD. Glutamate receptor desensitization and its role in synaptic transmission. Neuron. 1989;3:209–218. doi: 10.1016/0896-6273(89)90034-2. [DOI] [PubMed] [Google Scholar]
- 40.Yoshida K, Kawamura K, Imaki J. Differential expression of c-fos mRNA in rat retinal cells: regulation by light/dark cycle. Neuron. 1993;10:1049–1054. doi: 10.1016/0896-6273(93)90053-t. [DOI] [PubMed] [Google Scholar]
- 41.Yoshida K, Imaki J, Fujisawa H, Ohki K, Harada T, Matsuda H, Hagiwara M. Differential distribution of CaM kinases and induction of c-fos expression by flashing and sustained light in rat retinal cells. Invest Ophthalmol Vis Sci. 1996;37:174–179. [PubMed] [Google Scholar]