Abstract
Basic helix-loop-helix (bHLH) genes have emerged as important regulators of neuronal determination and differentiation in vertebrates. Three putative neuronal differentiation factors [NEX for neuronal helix-loop-helix protein-1 (mammalian atonal homolog-2), neuroD (β-2), and NDRF for neuroD-related factor (neuroD2)] are highly homologous to each other in the bHLH region and comprise a new bHLH subfamily. To study the role of NEX, the first bHLH protein identified in this group, we have disrupted the NEX gene by homologous recombination. NEX-deficient mice have no obvious developmental defect, and CNS neurons appear fully differentiated. To investigate further whether the absence of NEX is compensated for by neuroD and NDRF, we compared the spatiotemporal expression of all three genes. We demonstrate, by in situ hybridization, that the transcription patterns of NEX, neuroD, and NDRF genes are highly overlapping in the developing CNS of normal rats between embryonic day 12 and adult stages but are not strictly identical. The most prominent transcription of each gene marks the dorsal neuroepithelium of the telencephalon in early development and is sustained in the adult neocortex, hippocampus, and cerebellum. In general, neuroD provides the earliest marker of neuronal differentiation in any given region compared with NDRF or NEX. Whereas a few CNS regions are specific for neuroD, no region was detected in which solely NEX or NDRF is expressed. This suggests that the function of the mutant NEX gene in neuronal differentiation is compensated for by neuroD and NDRF and that, in analogy with myogenic bHLH proteins, neuronal differentiation factors are at least in part equivalent in function.
Keywords: neuronal differentiation factors, forebrain development, basic helix-loop-helix proteins, neuroD, NDRF, NEX-1, homologous recombination
Transcription factors of the basic helix-loop-helix (bHLH) superfamily of proteins play a central role in the determination and differentiation of cells in various lineages, from invertebrates to mammals. The function of bHLH proteins in mammalian myogenesis (Weintraub, 1993) and Drosophila neural development is remarkably similar (for review, see Jan and Jan, 1993). Functional similarities in regulating neurogenesis have now emerged between Drosophila bHLH proteins, encoded by genes in the achaete-scute and enhancer-of-split complex, and their mammalian homologs Mash-1 and Hes-1 (Guillemot et al., 1993; Ishibashi et al., 1994, 1995). More recently, a novel family of vertebrate bHLH proteins has been identified that is distantly related to theDrosophila atonal gene product. In the developing peripheral nervous system of Drosophila, atonal acts as a proneural gene for chordotonal organ formation (Jarman et al., 1993). The mammalian atonal-related genes are expressed in neuronal cells and/or neuronal precursor cells (Bartholomae and Nave, 1994; Akazawa et al., 1995; Lee et al., 1995; Shimizu et al., 1995; Gradwohl et al., 1996;Kume et al., 1996; Ma et al., 1996; Sommer et al., 1996; Yasunami et al., 1996; Takebayashi et al., 1997).
Three of these atonal-related genes are predominantly expressed at later stages of development and are thus more likely to be involved in neuronal differentiation (and adult neuronal function) than in early determination events. The neuronal helix-loop-helix protein-1 (NEX) gene was originally cloned from the adult brain with highly degenerate PCR primers directed against the most-conserved residues of unrelated bHLH proteins (Bartholomae and Nave, 1994). This gene was independently cloned as mammalian atonal homolog-2 (MATH-2) (Shimizu et al., 1995). A second neuronal bHLH gene, neuroD, has been cloned by virtue of a functional interaction with the daughterless gene product in a yeast two-hybrid screen (Lee et al., 1995). Importantly, when its mRNA was injected into Xenopus embryos, ectopic neuroD had the ability to induce ectopic neurogenic conversion of cells in the ectoderm. NeuroD is not strictly specific to the neuronal lineage, however, and has been identified independently as a transcription factor of the insulin gene (Β-2) in pancreatic β cells (Naya et al., 1995). A third atonal-related family member, neuroD-related factor (NDRF/KW8/neuroD2), was identified by structural homology to NEX and neuroD (Kume et al., 1996; McCormick et al., 1996; Yasunami et al., 1996). As shown for neuroD, ectopic overexpression of NDRF/neuroD2 inXenopus causes neurogenic conversion (McCormick et al., 1996). However, the late expression of the entire bHLH subfamilyin vivo makes these genes more likely to regulate neuronal differentiation (for review, see Lee, 1997).
If the NEX, neuroD, and NDRF subfamily were required for neuronal differentiation, corresponding mouse mutants might yield an informative differentiation defect, unless these genes have overlapping expression domains and are functionally equivalent. Here we report that transgenic mice with a targeted disruption of the NEX gene are fully viable. To substantiate further the possibility that neuroD and NDRF compensate for NEX function, we have mapped by in situ hybridization the spatiotemporal expression pattern of all three genes. In the developing CNS, analyzed between embryonic day 12 and adult ages (in the rat), we reveal side-by-side a highly overlapping pattern of expression but also distinct differences. Along with the structural homology, our genetic and morphological data suggest that neuronal differentiation factors are in part functionally equivalent.
MATERIALS AND METHODS
Gene targeting. A λ mouse 129Sv genomic library (Stratagene, La Jolla, CA) was screened with a full-length NEX cDNA, and one clone containing the two-exon gene (Bartholomae and Nave, 1994), including 4.5 kb of 5′- and 5 kb of 3′-flanking sequences, was isolated. A gene-targeting vector for positive–negative selection (Mansour et al., 1988) was constructed by subcloning the neomycin resistance (neoR) gene (from pMCneo; Stratagene) in sense orientation into the XhoI–HindIII linearized vector pKS (Stratagene). A 1.0 kb ApaI fragment (“short arm”), comprising most of intron 1 and the first 154 bp of the NEX open reading frame, was cloned from the 5′ end to the neomycin resistance gene. For negative selection, the HSV-tk gene (from pMC1TK; Stratagene) was inserted into the ClaI–NdeI linearized construct at the 3′ end of the polylinker. Finally, a 5.2 kbSpeI fragment (“long arm”), containing part of exon 2 and downstream sequences, was inserted between neo and HSV-tk (Fig.1A). The targeting construct lacked most of the NEX coding region (amino acids 53–337, including the bHLH domain), but transcription of the mutant gene could be monitored at the RNA level using a neoR specific probe.
Fig. 1.
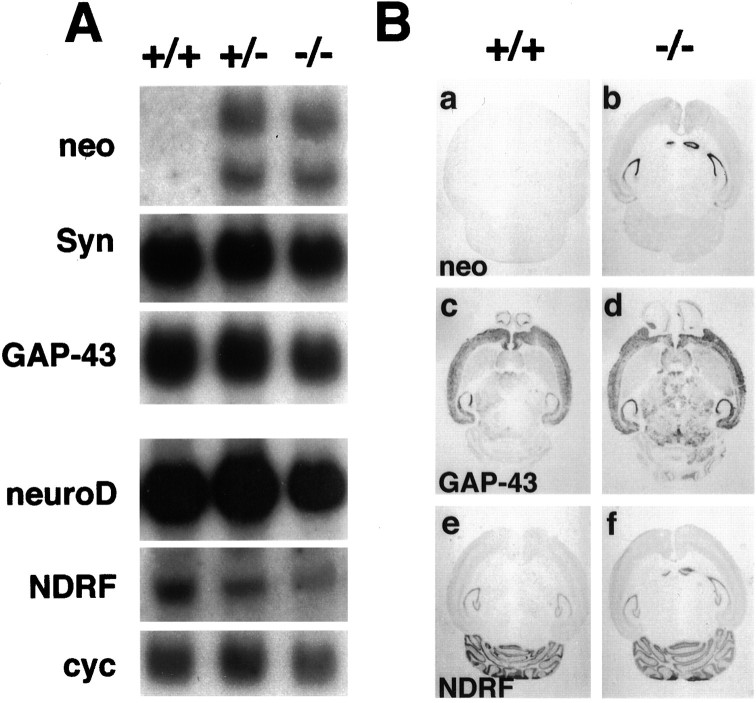
Targeted inactivation of the NEX gene in mice.A, Structure of the wild-type NEX gene (top), the targeting construct (middle), and the inactivated gene (bottom). The NEX locus comprises two exons (boxes). The entire open reading frame is located on exon 2 (filled black box). After homologous recombination, most of the coding region (residues 53–337, including the bHLH domain) is replaced by the neomycin resistance gene (neoR, stippled box) transcribed in sense orientation relative to NEX. Note that after recombination a (nonfunctional) neoRcontaining RNA is transcribed from the NEX promoter (see Fig.3A). Positions of the 5′ probe (for Southern blot analysis), primers 1 and 2 (for ES cell screening), and primers 3 and 4 (for routine genotyping) are indicated. Restriction sites areE (EcoRI), H(HindIII), and S (SpeI).HSV-tk, Herpes simplex virus thymidine kinase gene (stippled box) used for negative selection.B, Southern blot analysis of genomic DNA from wild-type (+/+) and from heterozygous (+/−) and homozygous (−/−) NEX mutant mice. Correct genomic targeting introduces a HindIII site (in neoR) and increases the genomic region between primers 3 and 4 by 200 bp. When hybridized to the 5′ probe,HindIII fragments are 7.5 kb in the wild-type and 6.5 kb in the mutant allele, as indicated in A. When stripped and rehybridized to a neoR-specific probe, a single band is obtained with DNA from heterozygous (+/−) and homozygous (−/−) mutant mice but not with DNA from wild-type (+/+) mice (data not shown). C, Northern blot analysis of total brain RNA from postnatal day 5. Transcription of the NEX gene in wild-type (+/+) mice yields differentially polyadenylated mRNAs (top) of 2.6 and 2.2 kb in length, with reduced abundance in brains of heterozygous (+/-) mice. Because the deleted part of the full-length NEX cDNA was used as a hybridization probe, no NEX mRNA was detectable in brains of homozygous (−/−) mutant mice. A cyclophilin probe hybridizing to a 1.2 kb mRNA (bottom) was included to verify equal loading of RNA.
R1 mouse embryonic stem (ES) cells (kindly provided by A. Nagy, Toronto) were cultured on mitomycin-treated primary mouse embryonic fibroblasts (feeders). For electroporation (240 V, 500 μF; Gene Pulser; BioRad, Munich, Germany), ∼107 ES cells were suspended in 0.8 ml of PBS containing 50 μg of the linearized (SacII) targeting construct. Transfected cells were cultured on gelatinized dishes (Falcon) in the presence of 103 U/ml leukemia inhibitory factor (GIBCO), and selection with 300 μg/ml G418 (GIBCO) and 2 μmganciclovir (Syntex) was started after 48 and 72 hr, respectively. Between 8 and 10 d after electroporation, double-resistant colonies were picked and trypsinized. Approximately one third of each colony was plated onto feeder cells. The remaining cells of eight clones each were pooled, suspended in 50 μl of water, heat-lysed at 95°C for 10 min, and digested with 10 μg of proteinase K at 55°C for 30 min.
To identify homologous recombinants, we used aliquots of the pooled lysates to PCR amplify a 1.5 kb genomic fragment. Two forward primers at position 1 (NEX2s, NEX1s) corresponded to a genomic sequence immediately 5′ to the short arm (NEX2s, 5′-CTGTCCGATATTAGCTAGAGGG-3′; NEX1s, 5′-AGACTTCCGTGGCTCTTAGAAC-3′). Reverse primers at position 2 (Neo3as, Neo6as) were derived from the neo gene (Neo3as, 5′-GCCTCTCCACCCAAGCGGCCGGAGAACCTGCGTGC-3; Neo6as, 5′-GCAATCCATCTTGTTCAATGGC-3; see Fig. 1A for positions of primers). In a 50 μl reaction, “nested” PCRs were performed with denaturation at 94°C (30 sec), annealing at 56°C (45 sec), and extension at 72°C (60 sec) for 19 cycles (using NEX2s and Neo3as) followed by 35 cycles with primers NEX1s and Neo6as. Amplification products were visualized on ethidium bromide-stained gels and verified by Southern blot analysis. Correct targeting was also confirmed by Southern blotting of ES cell DNA (10 μg), hybridized to a 5′ genomic probe (see location in Fig. 1A) or to a restriction fragment of the neo gene. Microinjection of selected ES cells into C57Bl/6J blastocysts was performed by standard procedures. Highly chimeric males were crossed to C57Bl/6J females. Heterozygous offspring were interbred to yield homozygous NEX−/− mutants.
For routine analysis, offspring genotype was determined by PCR with primers flanking the inserted neo fragment. Amplifications with primers NEX250s at position 3 (5′-CAAGTTGTCCTTCGAGGAAAGAGC-3′) and NEX1350as at position 4 (5′-GATACAGACAAGAGGGAAGGG-3′) yielded a single 1.1 kb product for the wild-type gene and a 1.3 kb fragment for the mutant allele.
RNA analysis. Total RNA was prepared from frozen rat brains by the method of Chomczynski and Sacchi (1987) or from mouse brains by column purification (RNeasy; Quiagen, Hilden, Germany). For Northern blot analysis, RNA was size-separated on 1% denaturing agarose gels (5 μg/lane) and blotted onto nylon membranes (Nytran; Schleicher & Schuell, Dassel, Germany). Blots were probed with random-primed ([32P]dCTP) DNA probes encompassing the coding region of rat or mouse NEX (Bartholomae and Nave, 1994), neuroD (Lee et al., 1995; Kawakami et al., 1996), NDRF (Kume et al., 1996; Yasunami et al., 1996), rat GAP-43 (Basi et al., 1987; kindly provided by L. Schrama, Utrecht), rat synaptophysin (Leube et al., 1987), and rat cyclophilin (Danielson et al., 1988). A neo-specific probe was derived from pMCneo (Stratagene). Overnight hybridizations were performed at 42°C in the presence of 50% formamide, 5× Denhardt’s solution, 3× SSC, 120 mm phosphate, 1% SDS, 200 ng/ml salmon sperm DNA, and 106 cpm/ml radiolabeled probe. The final washing stringency was 0.1% SSC and 0.1% SDS at 50°C. Blots were autoradiographically exposed to Kodak X-AR5 film with intensifying screens (24–72 hr), and hybridization signals were quantified with a phosphoimager (Fuji).
In situ hybridization. In mouse and rat (Sprague Dawley), the day of a vaginal plug was defined as embryonic day 0. Embryos and brains were dissected without fixation, frozen on dry ice, and stored at −70°C. For in situ hybridization, sections of 15 μm thickness were cut on a cryostat (AS620M; Shandon) at −15°C, thaw-mounted onto poly-l-lysine-coated glass slides, dried at room temperature (RT), fixed in 4% PBS-buffered paraformaldehyde, pH 7.5, for 5 min, and washed in PBS. Slides were dehydrated in 70 and 95% ethanol and stored in 95% ethanol at 4°C (for hybridization with radiolabeled oligonucleotides), or slides were dried at RT and stored at −70°C (for hybridization with digoxigenin-labeled cRNAs). Preparation of oligonucleotide probes, hybridization, and development of tissue sections were essentially as described previously (Bartholomae and Nave, 1994).
For in vitro transcription of cRNA probes, species-specific templates were generated by PCR amplification of genomic DNA and by subcloning of the protein coding regions into vectors pKS (Stratagene) or pGEM-T (Invitrogen, San Diego, CA). Sense and antisense RNA probes were transcribed in vitro using T7, T3, or SP6 RNA polymerase according to the manufacturer’s instructions (Boehringer Mannheim, Mannheim, Germany). cRNA probes were subjected to partial alkaline hydrolysis to reduce the probe size to 200–300 nucleotide fragments. Probes were diluted 1:100 in hybridization buffer and stored at −20°C. Prehybridization, hybridization, and immunological detection of digoxigenin-labeled cRNAs were performed as described bySchaeren-Wiemers and Gerfin-Moser (1993). Briefly, sections were thawed at RT, rinsed in 0.1 m triethanolamine at pH 8.0 (Sigma, Deisenhofen, Germany), acetylated by immersion into 0.1 mtriethanolamine containing freshly added 0.25% (v/v) acetic anhydride (Sigma), and washed three times in PBS. Prehybridization of sections (at RT for 3 hr) was performed with 0.5 ml of hybridization buffer containing 50% formamide, 5× SSC, 2% blocking agent (Boehringer Mannheim), 0.02% SDS, and 0.1% sarcosyl. Sections were hybridized for 16 hr at 65°C, using 150 μl of hybridization buffer containing ∼200 ng of cRNA probe. After a brief wash in 2× SSC at RT, slides were incubated at 37°C for 30 min with RNase A (50 μg/ml) in TNE (10 mm Tris, pH 8.0, 500 mm NaCl, and 1 mm EDTA). The final wash was in 0.2× SSC at 68°C for 10 min and at RT for 5 min. For immunological detection of digoxigenin, slides were pre-equilibrated in buffer 1 (0.1 m maleic acid and 0.15 m NaCl, pH 7.5), blocked for 1 hr in buffer 2 (buffer 1 containing 1× blocking agent), and overlayed with alkaline phosphatase-conjugated anti-DIG antibody (Boehringer Mannheim) that was diluted 1:2000 in buffer 2. Slides were washed twice for 30 min each in buffer 1 and once for 5 min in buffer 3 (100 mm Tris, pH 9.5, 100 mm NaCl, and 50 mmMgCl2). For detection of hybridization signals, slides were placed into a color substrate solution composed of 45 μl of 4-nitroblue tetrazolium chloride (NBT; 75 mg/ml in 70% dimethylformamide) and 35 μl of 5-bromo-4-chloro-3-indolylphosphate (BCIP; 50 mg/ml in dimethylformamide) per 10 ml of buffer 3. Reactions at RT (1–24 hr) were stopped by immersion into TE (10 mmTris, pH 8.0, and 1 mm EDTA). Sections were counterstained with Hoechst dye 33258 (Calbiochem, Lucerne, Switzerland), washed in PBS, and mounted in Kaiser’s gelatin (Merck, Darmstadt, Germany). For overviews, sections were photographed (Kodak Ektachrome film), and images were digitalized with a slide scanner (Nikon LS1000) and combined for composites (Macintosh with Adobe Photoshop). For pictures for the microscopy studies, sections were visualized using a Leitz Axiophot, and digitalized images were imported for further processing. Neuroanatomical localizations were verified using the maps of Paxinos and Watson (1986) and Swanson (1992).
Histology and immunohistochemistry. For immunohistochemistry, animals were killed by transcardiac perfusion under deep anesthesia (pentobarbital). After perfusion with 0.9% NaCl, brains were fixed in situ with 4% (w/v) buffered paraformaldehyde, pH 7.4. Brains were removed and post-fixed overnight in the same fixative before paraffin embedding or vibratome sectioning. Histological examination of CNS structures was performed on routine hematoxilin–eosin (H&E)- and Nissl-stained paraffin sections (3 μm). Coronal serial paraffin sections were obtained from the brains of adult NEX−/− mice, NEX+/− heterozygotes, and wild-type littermates (n = 3 each). Immunohistological analysis was performed on serial coronal free-floating vibratome sections (50 μm) from adult NEX−/− animals and wild-type littermates (n = 3), incubated with a panel of primary antibodies against neural marker proteins. The following antisera (rabbit) were kindly provided by R. Bravo (Bristol-Myers Squibb, Princeton, NJ): anti c-Fos (diluted 1:40,000), anti c-Jun (1:10,000), and anti Krox-24 (1:20,000). Additionally, we used the following monoclonal mouse antibodies against calcium-binding proteins: anti-parvalbumin (1:1000; Sigma), anti-calbindin-D28k (1:4000; Sigma), and anti-calretinin (1:1000; SWANT). A mouse monoclonal antibody against tyrosine hydroxylase (Boehringer Mannheim) was used at a dilution of 1:500; a rat monoclonal anti-somatostatin antibody (Paesel and Lorrei, Frankfurt) was used at a dilution of 1:1000. Polyclonal antisera against glial fibrillary acidic protein and myelin basic protein (both from Dako, Hamburg, Germany) were used at a dilution of 1:400. Vibratome sections were incubated at 4°C in buffer A (10% normal swine serum in PBS, pH 7.4, with 0.2% Triton X-100) for 30 min, followed by incubation with the primary antibody diluted in buffer A for 48 hr. Immunoreactivity was visualized by the avidin–biotin complex method (Vectastain; Vector Laboratories, Burlingame, CA) described previously (Gass et al., 1992). Sections were developed in 0.02% diaminobenzidine with 0.02% hydrogen peroxide, dehydrated in an ascending alcohol series, and embedded in eutelan (Eukitt; Kindler GmbH, Freiburg, Germany). For pictures for the microscopy studies, sections were processed as described previously.
RESULTS
Generation of mice lacking NEX
To inactivate NEX function in transgenic mice, we designed a targeting vector that replaced most of the coding region (amino acids 53–337, including the bHLH domain) with a neomycin resistance (neoR) cassette, as shown in Figure1A. After selection of mouse ES cells with G418 and ganciclovir, one clone with a correct targeting event (identified in 360 double-resistant clones) yielded highly chimeric mice (F0). Heterozygous F1 offspring (NEX+/−) had no apparent abnormalities and were interbred to yield homozygous (NEX−/−) mice. F2 animals were genotyped by either PCR (data not shown) or Southern blotting (Fig.1B). Among >150 offspring tested, wild-type and heterozygous and homozygous mutant genotypes were represented at the expected Mendelian frequencies.
No obvious behavioral differences were observed between newborn or adult NEX−/− mice and wild-type littermates. Male and female mutants were fertile, and female mice were able to conceive and nurse their pups. No behavioral or motor abnormalities were observed when mice were tested in a standard open field setting or challenged in a specialized locomotor test (“rotarod test”) at 2, 4, and 10 months of age. In these experiments, we used age-matched C57Bl/6J and 129SvJ inbred mice representing the parental strains as additional controls (data not shown). Although subtle behavioral differences cannot be excluded, this suggests that brain development is not dramatically impaired in the absence of NEX.
Neuronal differentiation in the absence of NEX
The analysis of brain sections stained with Nissl or H&E revealed no obvious histological difference between NEX−/− mice and wild-type controls (Fig.2A,B). The absence of gliosis was corroborated by immunostainings for GFAP (data not shown). Immunostaining of the neuronally expressed Krox-24 gene yielded a regular, constitutive expression pattern in NEX−/− mice (Fig. 2C,D), and expression of the immediate early genes c-fos and c-jun was also unaltered (data not shown). Immunostaining with antibodies against calbindin-D28k (Fig.2E,F), parvalbumin (Fig.2G,H), calretinin, and somatostatin (data not shown) revealed the presence of distinct subpopulations of interneurons throughout the forebrain that were differentiated normally in the absence of NEX. Calbindin-D28k immunoreactivity also demonstrated the regular presence of hippocampal granule cell dendrites and mossy fibers, as well as dendrites of CA1 pyramidal neurons (Fig.2E,F). Similarly, calretinin marked dendrites of dentate gyrus granule cells that were differentiated normally both in mutant and wild-type brain. An antibody against tyrosine hydroxylase revealed normal numbers of neuronal cell bodies and normal density of fibers in the substantia nigra and the ventral tegmental area (data not shown).
Fig. 2.

Hippocampal formation of adult NEX mutant mice.A, B, Nissl staining demonstrates a regular morphology of the hippocampal fields in 30-d-old homozygous mutants (−/−) compared with wild-type controls (+/+).C, D, By immunohistochemical analysis, the transcription factor Krox-24 marks the hippocampal CA1 field and is unaltered. E, F, The regular development of granule cell dendrites and mossy fibers is shown by immunostaining of calbindin-D28k that also labels dendrites and perikarya of CA1 pyramidal neurons. G, H, The presence of normal numbers of interneurons is visualized by immunostaining for parvalbumin.
Expression of neuronal bHLH genes in NEX−/− mice
To detect possible effects of the NEX null mutation on the steady-state levels of neuroD and NDRF mRNAs, we performed a comparative Northern blot analysis at three ages. As shown for postnatal day 10 (P10), neither neuroD nor NDRF mRNA levels were obviously altered in NEX−/− mice (Fig.3A). Similar results were obtained at P5 and with adult brains (data not shown), suggesting that sustained neuroD and NDRF expression does not require NEX and that neuroD and NDRF are not upregulated in the absence of NEX. At these ages, transcripts encoding the distantly related, “early” atonal homologs (MATH-1, MATH-3, and Neurogenin-1) were no longer detectable (data not shown). We also observed no differences between wild-type and NEX−/− mice when analyzing for GAP-43 and synaptophysin mRNAs as markers of neuronal differentiation (Fig. 3A), and byin situ hybridization, the spatial distribution of these mRNAs was unaltered (Fig. 3B; data not shown).
Fig. 3.
Transcription of neuronal genes in the absence of NEX. A, Northern blot analysis of total brain RNA isolated at postnatal day 10 from wild-type (+/+) and from heterozygous (+/−) and homozygous (−/−) NEX mutant mice. Homozygous mutants show no major difference in the steady-state level of two marker genes for neuronal differentiation, synaptophysin (Syn) andGAP-43. With a neomycin-specific probe (neo), two mRNAs of 2.8 and 2.4 kb were detected in heterozygous and homozygous brains. Transcription of these mRNAs is initiated at the endogenous NEX promoter (compare with Fig.1A). NeuroD andNDRF mRNAs show little change in abundance. Cyclophilin (cyc) was used as an internal standard of RNA loading (5 μg/lane). B, In situ hybridization of horizontal brain sections from wild-type (+/+) and from homozygous (−/−) NEX mutant mice obtained at postnatal day 5 (c,d) and postnatal day 10 (a,b, e, f). Sections were hybridized with a neomycin-specific probe (neo;a, b), revealing NEX promoter activity, and with probes for GAP-43 (c,d) and NDRF (e,f). Note that the neomycin-specific probe shows the same overall expression pattern as a NEX-specific probe (see Fig.7A to compare), revealing the apparently normal differentiation of neurons that lack NEX. Distribution ofGAP-43 and NDRF mRNAs appears unaltered in NEX−/− mice.
We predicted from the gene-targeting strategy that the mutant NEX allele has no protein-coding capacity but that RNA transcripts derived from this gene contain neo-related sequences (Fig.1A). This was confirmed by Northern blot analysis of brain RNA isolated at P5 from wild-type, NEX+/−, and NEX−/− mice. With a NEX cDNA probe, no mRNA was detected in NEX−/− mice, and heterozygous brains showed ∼50% of the wild-type NEX mRNA level (Fig. 1C). In contrast, a neoR-specific probe revealed a novel transcript in heterozygous and homozygous mutants (Fig. 3A), demonstrating that the promoter of the targeted gene is active. The same results were obtained with RNA from older animals (data not shown). When a neomycin-specific probe was used forin situ hybridization, the NEX gene showed a normal pattern of expression in the absence of the NEX gene product (Fig.3B).
Overlapping expression domains of NEX, neuroD, and NDRF
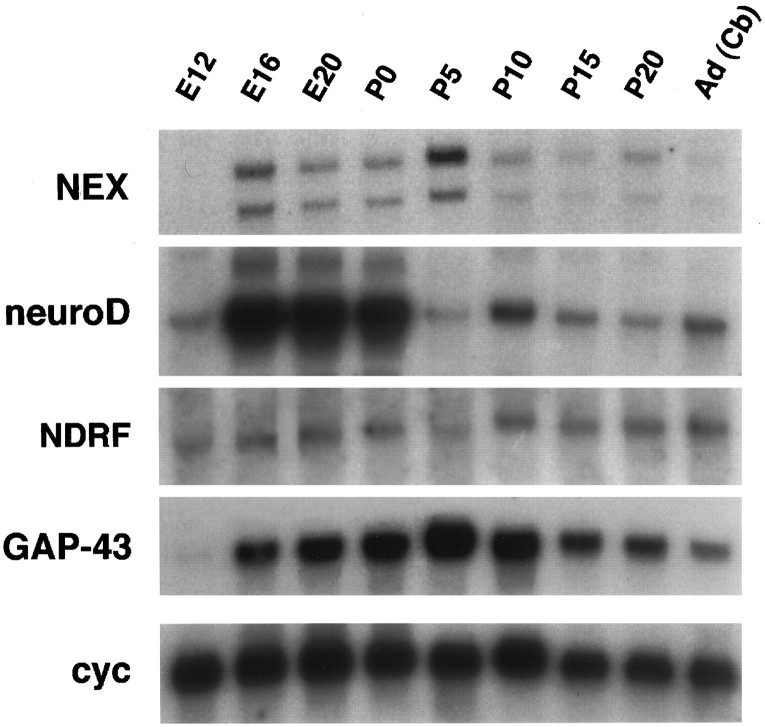
The apparent lack of a developmental defect in NEX-deficient mice suggested that the NEX-related bHLH proteins neuroD and NDRF may compensate in the mutants, provided their genes are temporally and spatially coexpressed. By Northern blot analysis, using RNA isolated from normal embryonic, postnatal, and adult rats, we noticed similarities but also striking differences (Fig.4). Both neuroD and NDRF mRNAs were detectable before NEX mRNA at embryonic day 12 (E12). Throughout late embryonic development (E16–P0), neuroD was clearly the most abundant family member. In contrast, NEX mRNA levels peaked in the first week after birth, when neuroD transcripts were reduced to a rather low abundance. Whereas both neuroD and NEX transcripts were temporally regulated, NDRF expression appeared relatively unchanged between embryonic and adult stages. Transcription of all three genes was sustained in the adult. Their overall temporal expression resembles the profile of GAP-43, a known marker of neuronal differentiation.
Fig. 4.
Temporal expression of NEX,neuroD, and NDRF correlates with neuronal differentiation. For Northern blot analysis, total RNA was prepared from normal rat brains of different ages (top) and hybridized to cDNA probes for NEX,neuroD, NDRF, GAP-43, and cyclophilin (cyc) (left). The steady-state level of neuroD mRNA is highest during late embryonic development, whereas NEX peaks during the first postnatal week. NDRF expression shows little variation between early and late stages. Note thatneuroD and NDRF mRNAs are detectable 1–2 d earlier than is NEX mRNA. GAP-43 has been used as a marker for neuronal differentiation, and cyclophilin is an internal control of RNA loading. Specific activities and exposure times of blots were the same for each probe. E, Embryonic day; P, postnatal day; Ad, adult; Cb, cerebellum.
Neuronal bHLH proteins in late embryonic development
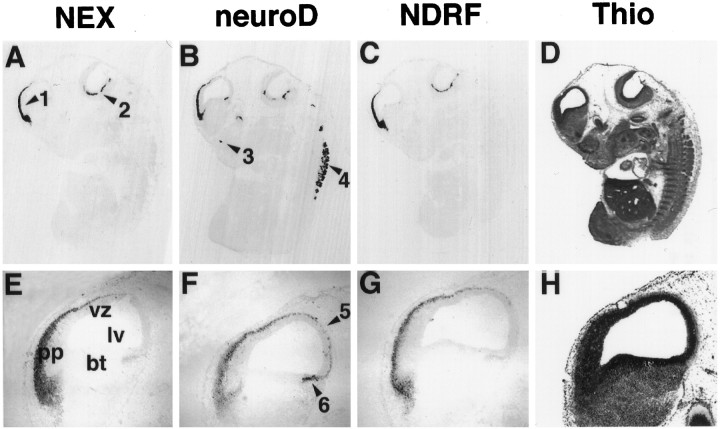
To determine the spatial colocalization of NEX, neuroD, and NDRF, we performed a comparative in situ hybridization analysis. The earliest time point of this analysis was E12, just before the induction of NEX in the rat CNS. At this stage, only neuroD was detectable by in situ hybridization in the dorsal part of the neural tube and the dorsal telencephalon, in cranial ganglia, and in thalamus and hypothalamus (data not shown). Prominent expression of all three genes was visible at E14, with staining of the cortical neuroepithelium in the telencephalon (Fig.5A–C). Strong expression of neuroD was also detectable in dorsal root ganglia (DRG) (Fig. 5B) that express NDRF at very low level (not visible on the overview shown). At this age, several expression domains seemed to be unique for neuroD, such as the olfactory neuroepithelium (Fig.5B), thalamus, the ventral hypothalamus, cranial ganglia (trigeminal, vestibular, glossopharyngeal, and vagal), and the superior cervical ganglia (data not shown). Within the cortical neuroepithelium, expression of all three genes was restricted to the preplate and was absent from the ventricular zone (Fig. 5E–G). For each gene, there were sharp rostral and caudal boundaries within the telencephalon, as a result of which expression was excluded from the basal telencephalon (Fig. 5E–G). The caudal boundary was located between the intermediate part (no expression) and the posterior part of the basal telencephalic plate (expression) that includes the neuroepithelium of the amygdala. In the posterior neuroepithelium of the hippocampus and the amygdala, steady-state levels of neuroD mRNA appeared higher than did those of NEX or NDRF mRNAs (Fig. 5E–G). Only very low expression levels of all three genes were observed in the mes-, met-, and myelencephalon. In the spinal cord, NEX expression was strong compared with that for neuroD and NDRF (data not shown). Specifically, NEX and NDRF transcripts were present in a broad area of the dorsal spinal cord, and expression of either gene was absent from the ventricular zone.
Fig. 5.
Colocalization of neuronal bHLH differentiation factors by in situ hybridization (embryonic day 14). Immediately adjacent parasagittal sections of rat embryos were hybridized with riboprobes specific for NEX(A, E), neuroD(B, F), and NDRF(C, G). For better orientation, one adjacent section was stained with thionin (Thio;D, H).A–C, Overviews reveal highly overlapping expression patterns of NEX, neuroD, andNDRF in the cortical neuroepithelium of the telencephalon (arrow 1 in A). Lining the 4th ventricle (arrow 2), the spinal trigeminal nucleus and the principal sensory trigeminal nucleus are also stained with all three probes. Note the absence of expression of either bHLH gene outside the nervous system (the pancreatic expression ofneuroD/β2 is not visible in the plane of this section). Some regions are only expressing the neuroDgene, including the olfactory neuroepithelium (arrow 3in B) and the dorsal root ganglia (arrow 4). E–G, Higher magnification of the dorsal telencephalon reveals colocalization of all three transcripts in the cortical preplate (pp inE) and the absence of the transcripts from the ventricular zone (vz) lining the lateral ventricle (lv). Note the rostral and caudal expression boundaries and the absence of either transcript from the basal telencephalon (bt). At E14, a more prominent expression ofneuroD (compared with NEX andNDRF expression) is seen in the neuroepithelium of the hippocampus (arrow 5 in F) and amygdala (arrow 6). No hybridization signal was obtained with sense-orientated probes (data not shown).
At E18, the strongest expression of all three bHLH genes was seen in the telencephalon (Fig.6A–C), again with no signal in cells of the ventricular zone. In the olfactory bulb (Fig. 6E–G), neuroD showed strong expression in the olfactory neuroepithelium of turbinate I–IV but was absent from cells lining the nasal cavity (Fig. 6F). NDRF expression in the olfactory neuroepithelium was very low. Some other CNS regions seemed to express predominantly or exclusively neuroD. These included the medial and lateral habenular nucleus (Fig.6I,K,L), the pineal gland (Fig. 6K), nuclei of the medial hypothalamus, nuclei in the dorsal medulla (hypoglossal and tegmental), and the nucleus of the solitary tract (data not shown). All three genes were expressed in mammillary bodies (data not shown). In the developing cerebellum (Fig. 6N–P), a strong expression was noted for neuroD in the external granular layer (EGL), where NDRF was very low and NEX was virtually absent. There was also neuroD expression in the inferior colliculus, where both NEX and NDRF appeared absent (Fig. 6N–P). In the pituitary, neuroD was more strongly expressed than was NDRF, and NEX was undetectable (data not shown).
Fig. 6.

Colocalization of neuronal bHLH differentiation factors by in situ hybridization (embryonic day 18). Immediately adjacent midsagittal sections of rat embryos were hybridized with riboprobes specific for NEX(A, E, I,N), neuroD (B,F, K, O), andNDRF (C, G,L, P). For better orientation, one adjacent section was stained with thionin (Thio;D, H, M,Q). A–C, Overviews show prominent expression of all three genes in the neuroepithelium of the dorsal telencephalon (arrowheads).E–G, Higher magnification of the anterior telencephalon, olfactory bulb (ob inF), and olfactory neuroepithelium (on) reveals coexpression in the cortical plate and the intermediate cortical layer. In the olfactory bulb, NEX,neuroD, and NDRF seem to be restricted to the mitral cell layer. The olfactory neuroepithelium shows strongneuroD expression (F) but no NEX and very little NDRF expression.I–L, Similarly, neuroD(but only little NEX or NDRF) mRNA is found in the medial and lateral habenular nucleus (hnin K) and in the pineal gland (pg). N–P, In the mesencephalon, neuroD expression marks the inferior colliculus (ic in O) and ceases at the boundary to the central gray matter. In the developing cerebellum (cb in O), external granular cells prominently express neuroD but very little if anyNDRF and NEX. cxp, Cortical plate.
Neuronal bHLH proteins in postnatal development
Total NEX expression peaked in abundance around P5 when NEX mRNA was detected throughout the cortex, hippocampus (CA1–CA3), subiculum, and entorhinal cortex and in the pontine nucleus. NEX expression was clearly absent from granular cells of the dentate gyrus, whereas neuroD and NDRF were detected in both CA1–CA3 regions of the hippocampus and the dentate gyrus (with neuroD expression being higher in the dentate gyrus). In the developing cerebellum, NEX transcripts were present only in migrating and postmigratory granular cells (IGL), and no expression was detectable in the EGL (Fig.7K). NeuroD and NDRF (Fig. 7L,M), on the other hand, were expressed in both EGL and IGL cells. We were unable to detect neuroD and NDRF transcripts in Purkinje cells at any stage tested (see Fig. 8D; data not shown). In the olfactory bulb, strong neuroD expression was restricted to the glomerular layer, whereas only weak labeling for neuroD and NDRF transcripts was present in the mitral layer. During postnatal and adult stages, no NEX expression was detectable in the olfactory bulb (data not shown).
Fig. 7.

Overlapping expression of NEX,neuroD, and NDRF in the postnatal brain. Horizontal adjacent sections, obtained from normal 10-d-old (A–I) and 6-d-old (K–M) rat brains, were hybridized with digoxigenin-labeled probes for NEX(A, D, G,K), neuroD (B,E, H, L), andNDRF (C, F,I, M) mRNAs. In K, a 35S-labeled NEX-specific oligonucleotide probe was used. A–C, The overviews demonstrate the overall similarity of the postnatal expression domains but also clear differences in expression levels.D–F, In the hilar region (h in D) of the hippocampus, polymorphic cells show prominent NEX expression that is not matched by neuroD or NDRF at this age.G–I, Also the subiculum and the entorhinal cortex (ehc in G) show strong labeling for NEX, the absence of neuroD, and only minor expression of NDRF.K–M, Earlier (at P6), the cerebellum shows a high level expression of neuroD, most prominently in the external granular layer (egl inL) and less so in the internal granular layer (igl). In contrast, NDRFtranscripts are more uniformly distributed, and NEXtranscripts are absent from the external granular layer but appear in migratory (postmitotic) granule cells and the internal granular layer (autoradiographic dark-field signal in K; bright-field microscopy in all others). dg, Dentate gyrus.
Fig. 8.
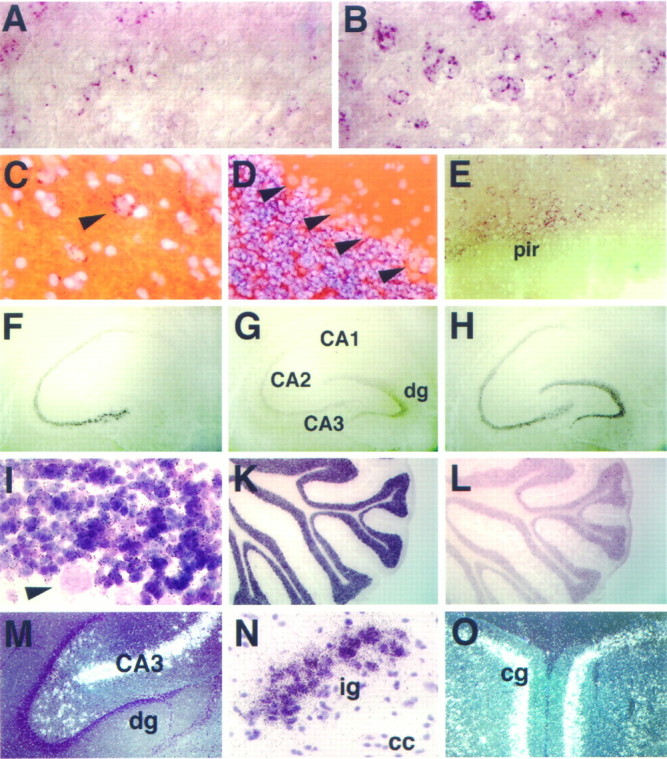
Sustained expression of NEX, neuroD, and NDRF in subsets of mature neurons. Brain sections from young adult rats (postnatal day 35) were hybridized to probes specific for NEX (left and bottom; A,C, F, I,M–O), neuroD (middle;D, G, K), and NDRF (right; B, E,H, L). Expression of NEX (A) and NDRF (B) is sustained in some but not all neurons of the adult neocortex, most strongly in layers II–V. Signals appear as perinuclear red granules (B) of large-sized cells in layer V of the neocortex that are most likely pyramidal neurons. By comparison, neuroD transcripts are of low abundance in the adult neocortex (data not shown). NEX is also sustained in single neurons of the hilar region (arrowhead in C), but the number varies with the plane of the section (overview inM). NDRF is detected in the piriform cortex (pir in E) and in the amygdala, colocalizing with NEX (data not shown). In the adult hippocampus, NEX expression (F) is strongest in the CA fields but absent from the dentate gyrus, where neuroD (G) and NDRF (H) mRNAs can be readily visualized. The strongest neuroD signals (K) were found in the adult cerebellum, confined to the granular cell layer (D), and absent from Purkinje cells (arrowheads inD). Likewise, NEX transcription (autoradiographic grains in I) is maintained in granule cells and not detectable in the large Purkinje cells (arrowhead inI). In comparison with neuroD (K), both cerebellar NDRF (L) and NEX transcripts are less abundant. Additional regions of strong NEX expression in the adult brain include the induseum griseum (ig in N) and the cingulate gyrus (cg in O). (I, M–O are autoradiographs of an35S-labeled NEX oligonucleotide probe, withM and O photographed in dark field and all others by bright-field microscopy). Sections were coronal (A, B, E,N, O), sagittal (C,D), and horizontal (F–M), with A–B,F–H, and K–L immediately adjacent.cc, Corpus callosum; dg, dentate gyrus.
At P10, the “adult” patterns of gene expression could be recognized, with reduced neuroD expression in the cortex and with NEX mRNA most prominent in the cingulate gyrus (Fig.7A–C). In the hippocampus, we noted strong labeling of NEX-transcribing neurons in the polymorphic layer of the dentate gyrus (Paxinos, 1995) that were rarely labeled with a neuroD or NDRF probe (Fig. 7D–F; see also Fig.8C,M). NEX expression was also strong in the subiculum and parasubiculum and in the entorhinal cortex where neuroD and NDRF were low by comparison (Fig.7G–I). A closer inspection of these areas in the CNS of P10 NEX-deficient mice revealed no obvious morphological differences, at least after routine histological staining (data not shown). In the rat, all three bHLH genes showed similar expression in the induseum griseum, tenia tecta, and the piriform cortex (data not shown).
Neuronal differentiation factors in the adult CNS
The expression of NEX and NDRF in the adult rat brain has been noted (Bartholomae and Nave, 1994; Shimizu et al., 1995; Yasunami et al., 1996). NEX was strongest in the CA1–CA3 region of the hippocampus (but absent from the dentate gyrus) and in the subiculum and parasubiculum, entorhinal cortex, cingulate cortex, induseum griseum, and tenia tecta (Fig.8C,F,M–O; data not shown). Lower levels of NEX were detectable in the neocortex (Fig. 8A), piriform cortex, anterior olfactory nucleus, and the amygdala (data not shown). All of these adult brain regions could be stained also with a probe for neuroD and NDRF, although neuroD mRNA hybridization signals of the neocortex, olfactory bulb, and hippocampus were reduced compared with the signals of earlier postnatal stages (Fig. 8G; data not shown). A similar pattern of expression persisted for NDRF in the adult brain, marking neurons of the neocortex, cingulate gyrus, piriform cortex, hippocampus, induseum griseum, medial habenular nucleus, and cerebellum (Fig.8B,E,H,L; data not shown). We have not been able to detect the expression of any of these genes in the adult spinal cord (data not shown).
Taken together, NEX, neuroD, and NDRF are clearly colocalized throughout the developing and adult CNS. Expression domains that have not been recognized before include the induseum griseum, tenia tecta, habenular nucleus, pontine nucleus, and the anterior olfactory nucleus. Our data suggest that the functions of neuronal bHLH proteins in differentiation must be region specific. Their functions in subsets of mature neurons remain to be defined.
DISCUSSION
In an attempt to identify genes that regulate the terminal differentiation of neurons and glia in the mammalian CNS, we have cloned previously the gene for a CNS-specific bHLH protein, termed NEX (for neuronal helix-loop-helix protein), which is distantly related to a bHLH protein encoded by the Drosophila proneural atonal gene (Bartholomae and Nave, 1994; Shimizu et al., 1995). To analyze the function of mammalian NEX in CNS development, we have generated NEX-deficient mice. The molecular and histological analysis has revealed that the mutant mice are viable and that the major expression domains of NEX, in particular the hippocampus, the neocortex, and the cerebellum, develop and differentiate without obvious morphological defects. NEX-deficient mice are fertile and appear on first inspection behaviorally unaffected.
While these gene-targeting experiments were in progress, two additional bHLH genes were discovered that encode proteins closely related to NEX, termed neuroD and NDRF, and that induce ectopic neuronal differentiation when overexpressed in microinjected frog oocytes (Lee et al., 1995; McCormick et al., 1996). Molecular cloning demonstrated the existence of an entire bHLH subfamily of putative neuronal differentiation factors, a finding reminiscent of the myoD subfamily of bHLH proteins required for muscle differentiation (Weintraub, 1993). Preliminary expression data, compiled from the initial description of these proteins, suggested that NEX, neuroD, and NDRF are associated with later stages of neurogenesis, i.e., after neuronal determination. This late expression profile of neuronal bHLH proteins that is documented in detail in the present study is in marked contrast to the transient and early expression profile of previously described bHLH genes in the mammalian nervous system, such as MASH-1 (Lo et al., 1991;Guillemot and Joyner, 1993), NSCL-1 and -2 (Begley et al., 1992; Gobel et al., 1992), HES-1 and -5 (Akazawa et al., 1992; Sasai et al., 1992), and neurogenins (Gradwohl et al., 1996; Ma et al., 1996, 1997; Sommer et al., 1996).
The emergence of a family of NEX-related genes suggested to us that the three bHLH proteins may serve overlapping functions in neuronal differentiation and adult neuronal function, provided that they are coexpressed. If so, a high degree of functional redundancy could compensate for the loss of any one bHLH gene in a mouse with a targeted mutation. The latter was suggested by the results of the NEX gene-targeting experiment summarized here. To investigate this working hypothesis further, we have performed a comparative in situhybridization analysis. Using adjacent rat brain sections, we have established the normal spatiotemporal expression of NEX, neuroD, and NDRF, beginning with the induction of NEX at embryonic day 12. Normal Sprague Dawley rats were chosen to ease correlations with anatomical landmarks in rodent brain development (Paxinos and Watson, 1986;Swanson, 1992). These expression data were confirmed for several developmental stages in the mouse (data not shown).
CNS development of NEX-deficient mice
The principal observation, and unexpected finding at first, was the fact that homozygous (NEX-deficient) mice were indistinguishable from littermates in terms of behavioral abnormalities. In agreement, the histological analysis with neuronal differentiation markers failed to reveal morphological differences of the NEX expression domains or secondary alterations in gene expression. Although necessarily not exhaustive, these studies suggested that neuronal differentiation and function of CNS neurons are primarily unimpaired. At the molecular level, we detected no compensatory upregulation of neuroD and/or NDRF in the absence of NEX.
The promoter of the mouse NEX gene has multiple bHLH protein-binding sites (E boxes), and at least one of these is recognized by NEX/E12 heterodimers, suggesting a positive autoregulation (Bartholomae and Nave, 1994; Bartholomae et al., 1995). However, in the CNS of adult NEX mutant mice, the steady-state level of RNA transcripts driven by the NEX promoter seemed not to be reduced, when monitored with a neo-specific probe. Thus, NEX may be tied into a complex auto- and cross-regulatory network, as shown previously for myoD and related myogenic factors (Thayer et al., 1989; Zingg et al., 1994; Cheng et al., 1995; Naidu et al., 1995). A hierarchical auto- and cross-regulatory network of neuronal bHLH genes is also suggested by the induction of XneuroD after ectopic Xenopusneurogenin-related protein 1A expression (Ma et al., 1996) and by transfection experiments in which neuroD and NDRF were able to stimulate the NDRF promoter (McCormick et al., 1996).
Differential expression of neuronal bHLH genes
Transcription of the neuroD gene during early neurogenesis shows temporal but little spatial overlap with neurogenin (Lee et al., 1995;Ma et al., 1996; Sommer et al., 1996). We have compared the expression of neuroD, NDRF, and NEX at later stages and are able to show a related but clearly distinct temporal expression pattern of the three genes. The relative abundance of its mRNA suggests that neuroD is predominantly associated with early aspects of neuronal differentiation, and its “transient” expression in CNS neurons has been noted before (Lee et al., 1995). In our analysis, neuroD and NDRF precede the induction of NEX by 1–2 d. None of the three genes was expressed in cells of the ventricular zone, supporting their predicted role in the differentiation of postmitotic, and presumably committed, neurons. NEX mRNA levels peaked in early postnatal development at the time of most active neurite outgrowth and synaptogenesis in the rat cortex (Aghajanian and Bloom, 1967), indicating that NEX is the “latest” family member, possibly associated, more than neuroD, with terminal aspects of neuronal differentiation.
Several neuronal subpopulations appeared to express neuroD exclusively, as shown for cells of the olfactory neuroepithelium and cranial ganglia. In other regions, NEX was absent, but neuroD and NDRF were detectable, as shown for granular cells of the dentate gyrus and the external granular layer of the cerebellum. The latter indicates that NEX is restricted to postmitotic neurons. We did not find regions in which the NEX gene was exclusively transcribed, one possible exception being the hilar region of the hippocampus in which relatively high levels of NEX mRNA were found in many large-sized neurons that were neuroD and NDRF negative. Interestingly, there was no increase of neuroD and NDRF signals in these neurons when NEX mutant mice were analyzed.
We were also unable to detect neuroD, NDRF, or NEX transcripts in cerebellar Purkinje cells at any age (see Fig.8D,I; data not shown), in contrast to a previous report on NDRF expression (Yasunami et al., 1996). We also confirmed our original finding (Bartholomae and Nave, 1994) that NEX is absent from neurons of the dentate gyrus (Figs. 7D,8M) in both mouse and rat, which contradicts the observation by Shimizu et al. (1995) of NEX/MATH-2 mRNA in these cells. This raises the possibility that additional bHLH protein family members exist (in Purkinje cells) and that cross-hybridization of probes (when used singly) must be considered a potential caveat of these studies.
Expression of bHLH proteins in mature neurons
Expression of neuroD, NDRF, and NEX is sustained in specific subpopulations of fully differentiated neurons in the adult brain. The principal expression domains are overlapping and include the pyramidal neurons of the CA1–CA3 region of the hippocampus, granule cells of the dentate gyrus (except for NEX), and granule cells of the cerebellum. In the hippocampus, virtually all principal neurons stain positively for either gene transcript, indicating that the neuronal bHLH proteins are coexpressed at the single cell level. In the cerebral cortex, only a subset of cells (predominantly pyramidal neurons as judged by shape and location) can be labeled. Different from the early postnatal stages, the level of neuroD mRNA is relatively low in the adult neocortex and difficult to detect by in situ hybridization. A fine analysis of bHLH protein colocalization in the adult neocortex (at the single cell level) is now in progress, a study that will also address the colocalization with subtype-specific neuronal markers.
Analyzed together, the available data strongly suggest that the genes for neuroD, NDRF, and NEX are controlled differently in CNS development but have related developmental profiles and highly overlapping expression domains. The combined spatiotemporal pattern of all three genes suggests that in nearly all regions of the CNS at least two neuronal bHLH proteins are jointly expressed. This pattern provides a molecular basis for the functional compensation of bHLH proteins in a mutant mouse, provided that the structural homology of any two family members is sufficiently high. We note that there is functional compensation of myogenic bHLH proteins (Rudnicki et al., 1993; Braun and Arnold, 1995; Wang et al., 1996) that are structurally not as highly related to each other when compared with the neuroD subfamily (∼83 vs ∼95% within the bHLH domain). However, this compensation may operate at the level of different populations of myogenic precursors (Braun and Arnold, 1996). The most likely explanation for the lack of a recognizable developmental defect in NEX-deficient mice is also the functional compensation of neuronal differentiation factors. Because expression of all three genes primarily coincides with the generation of postmitotic neurons, the postulated compensatory mechanism is more likely to operate at the level of individual cells in which these genes are coexpressed.
It is intriguing that the expression of neuronal bHLH proteins is sustained in the adult rodent brain (but not in spinal cord), marking mature cortical neurons that have been associated with neuronal plasticity and higher brain functions. This suggests that neuronal bHLH proteins control target genes that are currently unknown but most likely common to developmental processes and adult neuronal functions. As shown here for NEX-deficient mice, the consequences associated with the loss of any one gene may be subtle. A detailed behavioral analysis may yield results that indicate a function of these genes in the adult brain. These experiments await the establishment of each mutation on a strictly defined genetic background.
While this manuscript was in review, Tsai and coworkers reported the targeted disruption of the β2/neuroD gene (Naya et al., 1997), expressed outside the nervous system in pancreatic β cells. In agreement with our suggestion of a partial functional compensation in this gene family, neuroD mutants lack an obvious neuronal phenotype but die early postnatally with a pancreatic failure.
Footnotes
This work was supported by Grant SFB229 from the Deutsche Forschungsgemeinschaft to K.-A.N. We thank Drs. J. Lee for providing a mouse neuroD cDNA, L. Schrama for supplying a rat GAP-43 cDNA, R. Bravo for providing various antibodies, and A. Nagy for making available ES cells. We also thank H. Krischke for excellent technical assistance, L. Dimou and H. Werner for helpful discussions, and J. Rami for secretarial help.
Correspondence should be addressed to Dr. Klaus-Armin Nave, Zentrum für Molekulare Biologie, Universität Heidelberg, Im Neuenheimer Feld 282, D-69120 Heidelberg, Germany.
REFERENCES
- 1.Aghajanian GK, Bloom FE. The formation of synaptic junctions in developing rat brain: a quantitative electron microscopic study. Brain Res. 1967;6:716–727. doi: 10.1016/0006-8993(67)90128-x. [DOI] [PubMed] [Google Scholar]
- 2.Akazawa C, Sasai Y, Nakanishi S, Kageyama R. Molecular characterization of a rat negative regulator with a basic helix-loop-helix structure predominantly expressed in the developing nervous system. J Biol Chem. 1992;267:21879–21885. [PubMed] [Google Scholar]
- 3.Akazawa C, Ishibashi M, Shimizu C, Nakanishi S, Kageyama R. A mammalian helix-loop-helix factor structurally related to the product of Drosophila proneural gene atonal is a positive transcriptional regulator expressed in the developing nervous system. J Biol Chem. 1995;270:8730–8738. doi: 10.1074/jbc.270.15.8730. [DOI] [PubMed] [Google Scholar]
- 4.Bartholomae A, Nave KA. NEX-1: a novel brain-specific helix-loop-helix protein with autoregulation and sustained expression in mature cortical neurons. Mech Dev. 1994;48:217–228. doi: 10.1016/0925-4773(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 5.Bartholomae A, Schwab MH, Schrama L, Gass P, Nave KA. The brain-specific helix-loop-helix protein NEX-1: a neuronal activator of GAP-43? Soc Neurosci Abstr. 1995;21:1290. [Google Scholar]
- 6.Basi GS, Jacobson RD, Virag I, Schilling J, Skene JH. Primary structure and transcriptional regulation of GAP-43, a protein associated with nerve growth. Cell. 1987;49:785–791. doi: 10.1016/0092-8674(87)90616-7. [DOI] [PubMed] [Google Scholar]
- 7.Begley CG, Lipkowitz S, Gobel V, Mahon KA, Bertness V, Green AR, Gough NM, Kirsch IR. Molecular characterization of NSCL, a gene encoding a helix-loop-helix protein expressed in the developing nervous system. Proc Natl Acad Sci USA. 1992;89:38–42. doi: 10.1073/pnas.89.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun T, Arnold HH. Inactivation of Myf-6 and Myf-5 genes in mice leads to alterations in skeletal muscle development. EMBO J. 1995;14:1176–1186. doi: 10.1002/j.1460-2075.1995.tb07101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun T, Arnold HH. Myf-5 and myoD genes are activated in distinct mesenchymal stem cells and determine different skeletal muscle cell lineages. EMBO J. 1996;15:310–318. [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng TC, Tseng BS, Merlie JP, Klein WH, Olson EN. Activation of the myogenin promoter during mouse embryogenesis in the absence of positive autoregulation. Proc Natl Acad Sci USA. 1995;92:561–565. doi: 10.1073/pnas.92.2.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 12.Danielson PE, Forss Petter S, Brow MA, Calavetta L, Douglass J, Milner RJ, Sutcliffe JG. p1B15: a cDNA clone of the rat mRNA encoding cyclophilin. DNA. 1988;7:261–267. doi: 10.1089/dna.1988.7.261. [DOI] [PubMed] [Google Scholar]
- 13.Gass P, Herdegen T, Bravo R, Kiessling M. Induction of immediate early gene encoded proteins in the rat hippocampus after bicuculline-induced seizures: differential expression of KROX-24, FOS and JUN proteins. Neuroscience. 1992;48:315–324. doi: 10.1016/0306-4522(92)90493-l. [DOI] [PubMed] [Google Scholar]
- 14.Gobel V, Lipkowitz S, Kozak CA, Kirsch IR. NSCL-2: a basic domain helix-loop-helix gene expressed in early neurogenesis. Cell Growth Differ. 1992;3:143–148. [PubMed] [Google Scholar]
- 15.Gradwohl G, Fode C, Guillemot F. Restricted expression of a novel murine atonal-related bHLH protein in undifferentiated neural precursors. Dev Biol. 1996;180:227–241. doi: 10.1006/dbio.1996.0297. [DOI] [PubMed] [Google Scholar]
- 16.Guillemot F, Joyner AL. Dynamic expression of the murine achaete-scute homologue Mash-1 in the developing nervous system. Mech Dev. 1993;42:171–185. doi: 10.1016/0925-4773(93)90006-j. [DOI] [PubMed] [Google Scholar]
- 17.Guillemot F, Lo LC, Johnson JE, Auerbach A, Anderson DJ, Joyner AL. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75:463–476. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- 18.Ishibashi M, Moriyoshi K, Sasai Y, Shiota K, Nakanishi S, Kageyama R. Persistent expression of helix-loop-helix factor HES-1 prevents mammalian neural differentiation in the central nervous system. EMBO J. 1994;13:1799–1805. doi: 10.1002/j.1460-2075.1994.tb06448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishibashi M, Ang SL, Shiota K, Nakanishi S, Kageyama R, Guillemot F. Targeted disruption of mammalian hairy and enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 1995;9:3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- 20.Jan YN, Jan LY. HLH proteins, fly neurogenesis, and vertebrate myogenesis. Cell. 1993;75:827–830. doi: 10.1016/0092-8674(93)90525-u. [DOI] [PubMed] [Google Scholar]
- 21.Jarman AP, Grau Y, Jan LY, Jan YN. Atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell. 1993;73:1307–1321. doi: 10.1016/0092-8674(93)90358-w. [DOI] [PubMed] [Google Scholar]
- 22.Kawakami H, Maruyama H, Yasunami M, Ohkubo H, Hara H, Saida T, Nakanishi S, Nakamura S. Cloning and expression of a rat brain basic helix-loop-helix factor. Biochem Biophys Res Commun. 1996;221:199–204. doi: 10.1006/bbrc.1996.0569. [DOI] [PubMed] [Google Scholar]
- 23.Kume H, Maruyama K, Tomita T, Iwatsubo T, Saido TC, Obata K. Molecular cloning of a novel basic helix-loop-helix protein from the rat brain. Biochem Biophys Res Commun. 1996;219:526–530. doi: 10.1006/bbrc.1996.0267. [DOI] [PubMed] [Google Scholar]
- 24.Lee JE. Basic helix-loop-helix genes in neural development. Curr Opin Neurobiol. 1997;7:13–20. doi: 10.1016/s0959-4388(97)80115-8. [DOI] [PubMed] [Google Scholar]
- 25.Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 26.Leube RE, Kaiser P, Seiter A, Zimbelmann R, Franke WW, Rehm H, Knaus P, Prior P, Betz H, Reinke H, Beyreuther K, Wiedenmann B. Synaptophysin: molecular organization and mRNA expression as determined from cloned cDNA. EMBO J. 1987;6:3261–3268. doi: 10.1002/j.1460-2075.1987.tb02644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lo LC, Johnson JE, Wuenschell CW, Saito T, Anderson DJ. Mammalian achaete-scute homolog 1 is transiently expressed by spatially restricted subsets of early neuroepithelial and neural crest cells. Genes Dev. 1991;5:1524–1537. doi: 10.1101/gad.5.9.1524. [DOI] [PubMed] [Google Scholar]
- 28.Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- 29.Ma Q, Sommer L, Cserjesi P, Anderson DJ. Mash1 and neurogenin1 expression patterns define complementary domains of neuroepithelium in the developing CNS and are correlated with regions expressing notch ligands. J Neurosci. 1997;17:3644–3652. doi: 10.1523/JNEUROSCI.17-10-03644.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mansour SL, Thomas KR, Capecchi MR. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- 31.McCormick MB, Tamimi RM, Snider L, Asakura A, Bergstrom D, Tapscott SJ. NeuroD2 and neuroD3: distinct expression patterns and transcriptional activation potentials within the neuroD gene family. Mol Cell Biol. 1996;16:5792–5800. doi: 10.1128/mcb.16.10.5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naidu PS, Ludolph DC, To RQ, Hinterberger TJ, Konieczny SF. Myogenin and MEF2 function synergistically to activate the MRF4 promoter during myogenesis. Mol Cell Biol. 1995;15:2707–2718. doi: 10.1128/mcb.15.5.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naya FJ, Stellrecht CM, Tsai MJ. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 1995;9:1009–1019. doi: 10.1101/gad.9.8.1009. [DOI] [PubMed] [Google Scholar]
- 34.Naya FJ, Huang HP, Qiu Y, Mutoh H, DeMayo FJ, Leiter AB, Tsai MJ. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/neuroD-deficient mice. Genes Dev. 1997;11:2323–2334. doi: 10.1101/gad.11.18.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paxinos G. The rat nervous system. Academic; New York: 1995. [Google Scholar]
- 36.Paxinos G, Watson C. The rat brain in stereotaxic coordinates. Academic; New York: 1986. [DOI] [PubMed] [Google Scholar]
- 37.Rudnicki MA, Schnegelsberg PN, Stead RH, Braun T, Arnold HH, Jaenisch R. MyoD or Myf-5 is required for the formation of skeletal muscle. Cell. 1993;75:1351–1359. doi: 10.1016/0092-8674(93)90621-v. [DOI] [PubMed] [Google Scholar]
- 38.Sasai Y, Kageyama R, Tagawa Y, Shigemoto R, Nakanishi S. Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and enhancer of split. Genes Dev. 1992;6:2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- 39.Schaeren-Wiemers N, Gerfin-Moser A. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- 40.Shimizu C, Akazawa C, Nakanishi S, Kageyama R. MATH-2, a mammalian helix-loop-helix factor structurally related to the product of Drosophila proneural gene atonal, is specifically expressed in the nervous system. Eur J Biochem. 1995;229:239–248. doi: 10.1111/j.1432-1033.1995.tb20461.x. [DOI] [PubMed] [Google Scholar]
- 41.Sommer L, Ma Q, Anderson DJ. Neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogeneity in the developing CNS and PNS. Mol Cell Neurosci. 1996;8:221–241. doi: 10.1006/mcne.1996.0060. [DOI] [PubMed] [Google Scholar]
- 42.Swanson LW. Brain maps: structure of the rat brain. Elsevier; Amsterdam: 1992. [Google Scholar]
- 43.Takebayashi K, Takahashi S, Yokota C, Tsuda H, Nakanishi S, Asashima M, Kageyama R. Conversion of ectoderm into a neural fate by ATH-3, a vertebrate basic helix-loop-helix gene homologous to Drosophila proneural gene atonal. EMBO J. 1997;16:384–395. doi: 10.1093/emboj/16.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thayer MJ, Tapscott SJ, Davis RL, Wright WE, Lassar AB, Weintraub H. Positive autoregulation of the myogenic determination gene MyoD1. Cell. 1989;58:241–248. doi: 10.1016/0092-8674(89)90838-6. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Schnegelsberg PN, Dausman J, Jaenisch R. Functional redundancy of the muscle-specific transcription factors Myf5 and myogenin. Nature. 1996;379:823–825. doi: 10.1038/379823a0. [DOI] [PubMed] [Google Scholar]
- 46.Weintraub H. The MyoD family and myogenesis: redundancy, networks, and thresholds. Cell. 1993;75:1241–1244. doi: 10.1016/0092-8674(93)90610-3. [DOI] [PubMed] [Google Scholar]
- 47.Yasunami M, Suzuki K, Maruyama H, Kawakami H, Nagai Y, Hagiwara M, Ohkubo H. Molecular cloning and characterization of a cDNA encoding a novel basic helix-loop-helix protein structurally related to Neuro-D/BHF1. Biochem Biophys Res Commun. 1996;220:754–758. doi: 10.1006/bbrc.1996.0476. [DOI] [PubMed] [Google Scholar]
- 48.Zingg JM, Pedraza Alva G, Jost JP. MyoD1 promoter autoregulation is mediated by two proximal E-boxes. Nucleic Acids Res. 1994;22:2234–2241. doi: 10.1093/nar/22.12.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]