Abstract
Migrating neuronal growth cones exert traction forces that are generated by ATP-driven F-actin/myosin interactions. Sustained generation of these forces may require an energy supply mediated by the guanidino kinases, creatine kinase and arginine kinase. We cloned and sequenced grasshopper arginine kinase and examined its expression during embryogenesis and its subcellular localization in vivo and in vitro. During the first half of embryogenesis, arginine kinase is expressed selectively in a small percentage of ectodermal cells (dorsal closure cells), in a small percentage of mesodermal cells (muscle pioneers), and throughout the developing CNS. Most of these cell types are motile, including nascent neurons, muscle pioneers, dorsal closure cells, and many CNS glia. Neuroblasts also strongly express arginine kinase; they are nonmotile but are undergoing repeated rounds of (ATP-dependent) mitosis. Arginine kinase is colocalized with F-actin in a narrow band along the leading edges of lamellipodia of migrating glia. In neurons undergoing axonogenesis, arginine kinase is concentrated in growth cones and extends to the tips of filopodia. The amount of arginine kinase varies widely between growth cones, even between different growth cones of the same neuron. Energy for growth cone migration appears to be mobilized by (1) selective expression of arginine kinase by neurons, (2) localization of arginine kinase within growth cones, and (3) concentration of arginine kinase within specific growth cones, depending on the traction forces being generated. Mobilization of guanidino kinases may participate in the selective growth of specific growth cones.
Keywords: arginine kinase, creatine kinase, growth cone, cell motility, axonogenesis, neurogenesis
Cells and extending cell processes translocate by applying traction forces to the substrate (Mitchison and Cramer, 1996). These forces have been measured in the range of 100–500 μdyn for growth cones (Lamoureux et al., 1989; Heidemann et al., 1990). Tension generation within motile cells and growth cones appears to involve forces applied to actin filaments by myosin-ATPase (Verkhovsky, 1995; Kelly et al., 1996; Lin et al., 1996). Forces generated during cell migration and process outgrowth make special energy demands on the cell. We investigated whether the energy requirements of actin-based motility of growth cones are met by specialized enzyme/substrate systems.
Creatine kinases (CK) and arginine kinase (argK) are ATP/guanidino phosphotransferases that provide ATP by catalyzing the conversion of ADP and phosphorylcreatine or phosphorylarginine to ATP and creatine or arginine (Eppenberger et al., 1967; Blethen and Kaplan, 1968; Morrison, 1973; Bessman, 1985; Wallimann, 1994; Wyss et al., 1995). Both phosphotransferases belong to a family that is so highly conserved that functional hybrid enzymes can be formed (Reddy and Watts, 1994). Vertebrates have four CK genes that encode tissue-specific isozymes (Wallimann, 1994; Muhlebach et al., 1996). Although echinoderms and annelids can express both CK and argK (Thoai and Robin, 1964; Moreland et al., 1967), only argK has been reported in arthropods (Wallimann and Eppenberger, 1973). ArgK is represented by a single gene (James and Collier, 1988; Munneke and Collier, 1988), and the sequence or partial sequence is available from Drosophila (L. B. Hecht, L. M. Scott, and G. E. Collier, GenBank U26939 information, 1995), Limulus (Strong and Ellington, 1995), lobster (Homarus; Dumas and Camonis, 1993), shrimp (Panuliris), and abalone (Suzuki and Furukohri, 1994).
CK and argK are abundant in muscle, where they maintain ATP homeostasis during muscle contraction (Wallimann et al., 1977; Newsholme et al., 1978; Lang et al., 1980; van Deursen et al., 1993). CK also is heavily expressed in vertebrate brain (Hemmer and Wallimann, 1993; Friedman and Roberts, 1994; Kaldis et al., 1996a), where it may provide energy for ion pumps, synaptic transmission, and photo transduction and in other energy-demanding tissues, including kidney and Torpedoelectroplaque (Wallimann and Hemmer, 1994). Motility of sperm flagellae, which is microtubule-based, is mediated by high levels of CK or argK (Tombes and Shapiro, 1985; Tombes et al., 1988; Strong and Ellington, 1993). During cell division, microtubule motors driving spindle elongation in anaphase require ATP, which appears to be provided by CK (Koons et al., 1982; Cande, 1983). Although the role of phosphotransferases in actin-based cell motility has not been determined, CK is colocalized with F-actin stress fibers in PtK1 cells (Koons et al., 1982). In intestinal epithelial cells it also appears to support the ATP-dependent actin/myosin-based contractions of the circumferential ring underlying microvilli (Gordon and Keller, 1992).
CK can be positioned subcellularly (Manos and Bryan, 1993), and different isozymes can be confined to specific cell regions (Kaldis et al., 1996b). Subcellular localization mechanisms include membrane anchoring (Quest et al., 1992), tethering to nonsoluble proteins (Wallimann et al., 1977; Lang et al., 1980), and physically restricted mobility of soluble enzyme (Wegmann et al., 1992). CK is a phosphoprotein for which the activity can be regulated by phosphorylation (Mahadevan et al., 1984; Quest et al., 1990; Hemmer et al., 1995). Consequently, CK, and probably argK, can be controlled by at least three mechanisms: selective expression, subcellular positioning, and phosphorylation.
To investigate the role and control of argK in growth cone motility, we used in situ hybridization to determine the expression pattern during neurogenesis and axonogenesis in vivo, and we used immunocytochemistry to characterize protein localization in vivo and in dissociated cells in vitro.
MATERIALS AND METHODS
cDNA and RNA analysis. Grasshopper (Schistocerca americana) argK cDNA was identified via a kinase screen, using a 40% grasshopper nerve cord cDNA library that was kindly provided by Dr. Michael Bastiani (University of Utah, Salt Lake City, UT). ArgK cDNA was subcloned into plasmids according to the manufacturer’s directions (Stratagene, La Jolla, CA) and sequenced with Sequenase reagents (United States Biochemical, Cleveland, OH). Sequence data were analyzed with LaserGene software (DNASTAR, Madison, WI). The BLAST programs (Altschul et al., 1990) were used for homology searches of the GenBank and Swissport databases.
Total RNA was isolated from different stages of embryos, using the method of Chomczynski and Sacchi (1987). Per lane, 20 μg of RNA was electrophoresed through a 6% formaldehyde/1% agarose gel and transferred to a GeneScreen Plus filter (DuPont NEN, Boston, MA). The filter was hybridized to 32P-labeled argK cDNA, using Church hybridization conditions (Church and Gilbert, 1984), and was exposed to Kodak XAR-5 film (Rochester, NY) with an intensifying film.
For whole-mount in situ hybridization, 35% stage embryos were processed according to Zachow and Bentley (1996). Both sense and antisense in situ hybridization probes were made from a full-length cDNA (2.4 kb). From the full-length transcript a pool of shorter probes in the range of 300 bases was prepared by chemical cleavage.
Protein analysis. Different stages of S. americana were dissected and homogenized in sample buffer (50 mm Tris-HCl, pH 6.8, 0.02% bromophenol blue, 1% SDS, 1% β-mercaptoethanol, and 8% glycerol). Protein samples were separated by 10% SDS-PAGE and transferred to nitrocellulose. The filter was stained first with Ponceau S (Sigma, St. Louis, MO) to detect molecular weight markers. The filter was blocked with PBT/BS (150 mmNaCl, 200 mm Na2HPO4, 10% bovine serum, and 0.05% Triton X-100) for 30 min, incubated with a 1:100 dilution of anti-argK antibody (kindly provided by Dr. W. Ross Ellington, Florida State University, Tallahassee, FL) in PBT/BS for 1 hr, washed three times for 10 min each with PBT/BS, and incubated with a 1:1000 dilution of goat anti-rabbit IgG conjugated with horseradish peroxidase (HRP; Jackson ImmunoResearch Laboratories, West Grove, PA) in PBT/BS for 1 hr. The filter was washed with PBT three times, equilibrated with 50 mm Tris-HCl, pH 7.5, and developed with fresh DAB solution (0.05% 3,3′ diaminobenzidine tetrahydrochloride, 0.015% H2O2, and 50 mm Tris-HCl, pH 7.5). We affinity-purified anti-argK polyclonal antibody according to Harlow and Lane (1988).
Enzyme analysis. Manipulations were based on Manchenko (1994), with modifications. Different stages of S. americanawere dissected and homogenized with 20 μl of grinding solution (10% sucrose, 1% Triton X-100, 1 mm Tris-citrate, pH 7.1, and 0.1% bromophenol blue) on ice to preserve enzymatic activity. After centrifugation for 3 min at 16,000 rpm at 4°C, equal aliquots of the supernatant were loaded and electrophoresed at 4°C on a 5% acrylamide nondenaturing gel in 1,1,2,2-tetrabromoethane (TBE) buffer, pH 8.9, until bromophenol blue reached the bottom of the gel. After electrophoresis, the gel was cut in half. One-half of the gel was incubated in a staining solution (10 mg of phospho-l-arginine, 900 mg of glucose, 900 mg of fructose, 5 mm MgCl2, 50 mg of ADP, 20 mg of NADP+, 140 U of hexokinase, 40 U of glucose-6-dehydrogenase, 20 mg of nitroblue tetrazolium, and 0.1m TBE, pH 7.4; all chemicals are from Sigma, St. Louis, MO) that contained substrates for argK enzymatic activity. Because adenylate kinase also uses the same substrates (glucose, fructose, and cofactors) as argK, the other one-half of the gel was assayed solely for adenylate kinase (everything in the staining solution minus phospho-l-arginine, the substrate for argK). The gels were incubated in staining solution for 30 min at 37°C and developed by adding 4 mg of Phenazine methosulfate. The reaction was stopped by rinsing the gels with several changes of H2O. Under such electrophoresis and staining conditions, adenylate kinase is inactive (data not shown). Only argK enzymatic activity appeared on the gel (see Fig. 2C). The gels were fixed with gel fixative (glacial acetic acid/methanol/H2O 1:5:9) for 10 min, rinsed with H2O, air-dried overnight between two sheets of Gelwrap (BioDesign, Carmel, NY), and scanned.
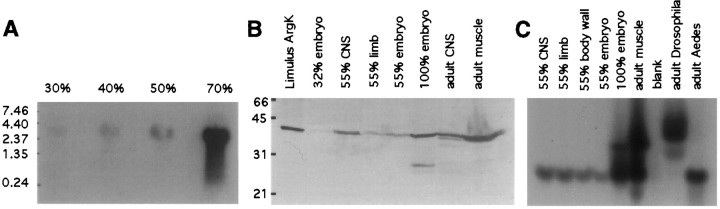
Fig. 2.
A, Northern analysis of argK mRNA in different stages of grasshopper embryogenesis detects a single transcript of ∼2.4 kb. There is a gradual increase in the amount of transcript in 30, 40, and 50% stage embryos. In 70% stage embryos, argK is transcribed at a high level, probably because of muscle maturation. In each lane, 20 μg of total RNA was loaded. The blot was probed with 32P-labeled grasshopper argK cDNA. RNA size standards are in kilobases. B, Immunoblotting withLimulus anti-argK polyclonal antibody shows grasshopper argK is ∼40 KDa, and there is only one form (the faster migrating band in 100% embryo is likely a degradation product). Equal amounts of total protein were loaded in each lane. There is an increase of argK expression from 32 to 55% and 100% stage total embryo. Within 55% stage embryos, with the same amount of total protein loaded, there is more argK in the CNS than in the limb. In the adult, more argK is detected in the muscle than in CNS ganglia. Protein size standards are in kilodaltons. C, Enzymatic activity of grasshopper argK is present in different tissues and in different stages of development. Equal amounts of total protein were loaded in each lane. Grasshopper argK migrated to the same distance as that ofAedes (mosquito) but faster thanDrosophila argK. Within grasshopper, an abrupt increase in enzyme activity was observed after the 55% stage, concomitant with the maturation of muscle.
Embryo immunocytochemistry. All manipulations were performed according to Zachow and Bentley, 1996. Embryos were dissected in grasshopper saline [containing (in mm) 140 NaCl, 5 KCl, 4 CaCl2·2 H2O, 2 MgSO4·7 H2O, 2 TES, and 60 sucrose, pH 7.2] and fixed for 45 min in 3.7% formaldehyde in PEM (100 mm PIPES, 2 mm EGTA, and 1 mm MgSO4, pH 6.9). After three rinses with PBS (150 mm NaCl and 20 mm Na2HPO4, pH 7.2) and permeabilization with PBT (PBS with 0.5% BSA and 0.5% Triton X-100) for 5 min, the embryos were incubated in a 1:50 dilution of affinity-purified anti-argK antibody diluted in PBT overnight at 4°C. After three PBT washes for 10 min each, the embryos were incubated in a 1:1000 dilution of goat anti-rabbit IgG conjugated with HRP (Jackson ImmunoResearch) for 4 hr at room temperature. After three PBT washes for 10 min each, the embryos were developed with fresh DAB solution (with Ni enhancement), cleared in glycerol, mounted in Hanker–Yates medium, and examined on a Nikon DIC microscope. For epifluorescence, the embryos were labeled with a 1:1000 dilution of donkey anti-rabbit IgG conjugated with Cy3 (Jackson ImmunoResearch), mounted in Vectashield (Vector Labs, Burlingame, CA), and examined on a Nikon epifluorescence microscope (Tokyo, Japan) or a Bio-Rad 1024 confocal microscope (Richmond, CA).
Cell culture and immunocytochemistry. For cell culture, eggs with 50% embryos were sterilized in 0.2% benzethonium chloride for 3 min. Three thoracic ganglia per embryo were dissected, rinsed in sterile Leibovitz’s L-15 medium (Life Technologies, Grand Island, NY), and incubated in 200 μl of enzyme solution (2 mg/ml collagenase dispase, 0.5% ficin, and 2 mg/ml papain, in L-15) for 1 hr at 31°C. After five changes of L-15 (200 μl each), a final volume of 30 μl of L-15 was added to the ganglia. The ganglia were triturated 50 times at a 20 μl setting with a P20 Pipetman that was fitted with a BSA-coated gel-loading tip “Gel Saver II” (USA/Scientific Plastics, Ocala, FL). Dissociated cells were plated in the center of Falcon 3001 dishes (Becton Dickinson Labware, Lincoln Park, NJ) that were prefilled with 2 ml of L-15 (one ganglion per plate for optimum cell density). The dishes sat for 20 min to allow the cells to settle to the bottom. Then the dishes were wrapped with Parafilm and incubated at 31°C for 22–44 hr.
For substrate coating, five independent drops (1 μl per drop) of substrate solution (1.5 mg/ml FITC-conjugated goat anti-HRP antibody and 30 mg/ml FITC-conjugated dextran in L-15) were plated on Falcon 3001 dishes and allowed to air dry. L-15 (2 ml) was added to the dishes, and the dissociated cells were plated.
For double-labeling for argK and F-actin (see Fig. 6C,D), we followed the fixation protocol of Welnhofer et al. (1997). Rhodamine phalloidin (1 μg/ml; Molecular Probes, Eugene, OR) was used to label F-actin. Anti-argK antibody was used at a 1:200 dilution. A 1:500 dilution of FITC-conjugated anti-rabbit IgG antibody was used.
Fig. 6.
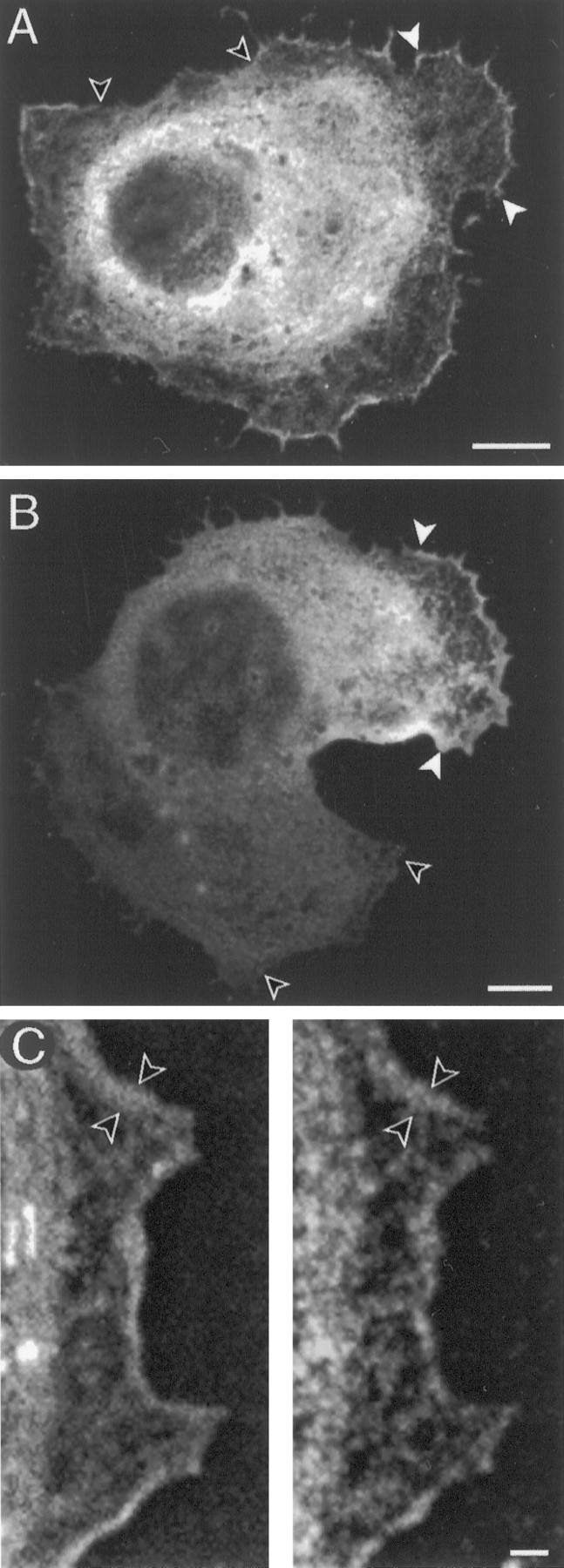
ArgK localization in lamellae of CNS glial cells in culture. A, In a cell with nonmotile morphology, argK is concentrated in protrusive lamellae (between white arrows) and is reduced or absent in retracted regions (between unfilled arrows) of the cell perimeter.B, ArgK is concentrated at the leading edge (between white arrows) and is absent from the trailing edge (between unfilled arrows) of an elongated (motile) cell. C, F-actin (phalloidin label, left) and argK (antibody label, right) are colocalized in a narrow band (between arrows) along a glial cell lamella. Scale bars: A, B, 5 μm;C, 1 μm.
For double-labeling for argK and neuron membrane (see Figs. 7, 8,9B,C, 10), plated cells were fixed in 3.7% formaldehyde in PEM for 20 min, washed in PBS three times for 5 min each, and blocked in PBS/0.2% BSA for 30 min. To label the membrane, we incubated cells with a 1:500 dilution of goat anti-HRP antibody (Jackson ImmunoResearch) in PBS for 1 hr, incubated them with a 1:500 dilution of either Cy5- or FITC-conjugated donkey anti-goat IgG antibody for 1 hr, and washed them in PBS three times for 5 min each. Next, they were permeabilized with PBS/0.02% saponin for 5 min, washed in PBS, incubated with affinity-purified rabbit anti-argK polyclonal antibody (see below for concentration) in PBS, washed with PBS three times for 5 min each, incubated with a 1:500 dilution of Cy3-conjugated donkey anti-rabbit IgG antibody for 1 hr, rinsed in PBS, cleared in glycerol, and mounted with Vectashield. Three concentrations of anti-argK antibody were used. For highest sensitivity to examine argK in filopodia (see Fig. 7), we used a 1:10 dilution; for medium sensitivity to reveal differences in argK concentration among filopodia (see Fig.10), we used 1:100; for low sensitivity to reveal differences in argK concentration between growth cones (see Fig. 8), we used a 1:200 dilution. The cells were imaged with a Bio-Rad 1024 Confocal microscope and printed with Adobe Photoshop (Adobe Systems, Mountain View, CA).
Fig. 7.
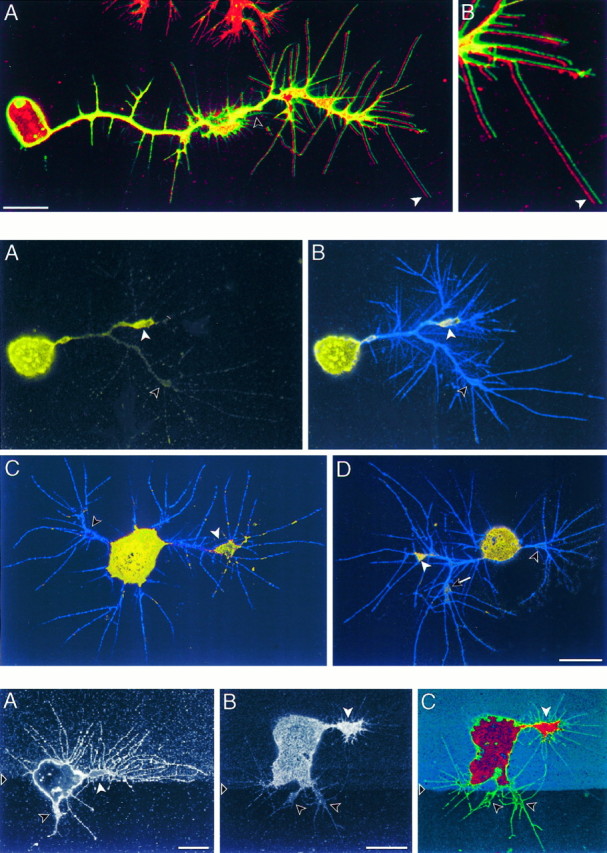
Top. ArgK extends to the tips of filopodia at the leading edges of neuronal growth cones.A, A CNS neuron in culture is identified by anti-HRP antibodies (green), which label a set of membrane proteins, and also is labeled with anti-argK antibody (red). The two images are offset horizontally by a few pixels to show that the red label (white arrow) extends to the tips of filopodia. Note the absence of red label in the membrane tracks (unfilled arrow) of retracted filopodia and the absence of green label in the neuron soma and in processes of a glial cell (top of panel). B, Enlargement of the leading filopodia from A. Scale bar, 10 μm.
Fig. 8.
Middle. ArgK is concentrated in neuronal growth cones and can be distributed differentially among growth cones of the same neuron. Three CNS neurons (A/B, C, D) are double-labeled for argK (yellow) and membrane proteins (anti-HRP, blue). ArgK is shown separately in A, and the two labels are shown combined in B–D. The neurons are bipolar (C, D) or have bifurcating axons (A/B, D). Each neuron has one growth cone (white arrowhead) that labels brightly relative to the axon and other growth cones (unfilled arrowhead). The neuron in D has a third growth cone (arrow) that labels at an intermediate level. A low antibody concentration (1:200; see Materials and Methods) was used to reveal differences in argK concentration. Scale bar, 10 μm (all panels).
Fig. 9.
Bottom. ArgK can be more concentrated in a growth cone extending on a preferred substrate.A, A neuron (labeled with anti-HRP) at the border between tissue culture plastic (below triangle) and plastic coated with anti-HRP antibodies (mixed with FITC-dextran) extends a longer growth cone with more filopodia (white arrow) on the coated substrate and a shorter growth cone with few filopodia (black arrow) on the uncoated substrate. More filopodia also extend directly from the cell body onto the coated substrate than onto the uncoated substrate. B/C, A neuron double-labeled with anti-argK antibodies (B;red channel in C) and anti-HRP antibodies (green channel in C) at the border between uncoated substrate (below triangles) and substrate coated with anti-HRP antibodies (FITC-dextran, blue channel in C). The neuron extends a large growth cone (white arrows) with many filopodia onto the coated substrate and two small growth cones (black arrows) with few filopodia onto the uncoated substrate. ArgK (B) is found predominantly in the large growth cone that appears to be extending more actively on the coated substrate. Scale bars, 10 μm.
Fig. 10.
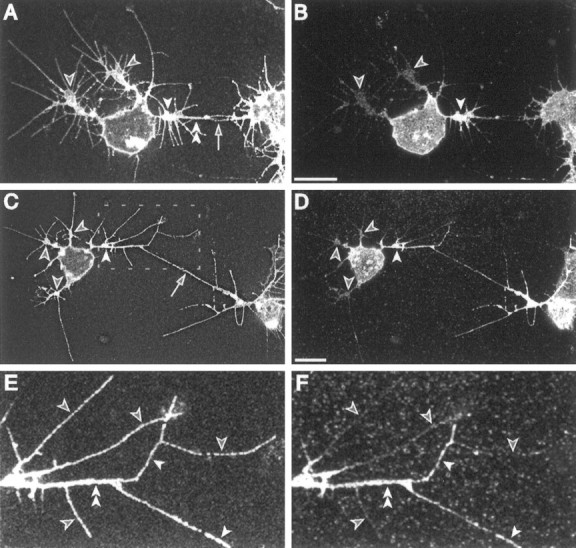
ArgK can be more concentrated in a growth cone engaged in an attractive interaction with another neuron and can be restricted to a subset of filopodia. A,B, In a mixed CNS culture, neurons often extend growth cones along each other’s processes. Here, a multipolar neuron (left) has a thickened branch (double arrowheads) and filopodia (arrow) in contact with another neuron (right). The branch is extended from one growth cone (white arrowhead), whereas two other growth cones (black arrowheads) migrate in other directions. Viewed with a membrane label (A, anti-HRP antibodies), the three growth cones are similar in size, distance from the cell body, and number of filopodia. Antibody labeling (B) shows a higher concentration of argK in the growth cone (white arrowhead) extending toward the other cell. C, D, A second similar neuron again has a higher level of argK (D) in the growth cone (C, D, white arrowheads) making contact (C, arrow) with another neuron than in three other growth cones (C,D, black arrowheads). E,F, An enlargement (box inC) of the growth cone contacting the other neuron. Although all filopodia appear similar with membrane label (E, anti-HRP antibodies), the level of argK (F) is high in some filopodia (white arrowheads) and much lower in others (black arrowheads). The filopodia with a relatively high level of argK are mediating, or are close to, the contact with the other cell. A thickened branch (E, F, double arrowheads) extending from the growth cone along the line of contact to the other cell also is heavily labeled for argK. Scale bars, 10 μm.
We evaluated the relative concentration of argK in different growth cones from the same cell as follows: CNS cells were dissociated, plated, cultured 22 or 44 hr, and double-labeled with anti-argK and either phalloidin or anti-HRP antibody. In each of four dishes (two at 22 hr; two at 44 hr), the first 30 multipolar neurons were scored that had two to four cylindrical axons at least one cell diameter in length, that were tipped by a growth cone, and in which the axons did not differ in length by more than a factor of two. Cells were rated as meeting acceptance criteria before argK labeling was viewed. Cells were scored as positive if argK labeling in one growth cone appeared at least twice as bright as that in another growth cone of the same cell. Brightness was estimated by eye, after making unsaturated confocal images of several sample cells and measuring growth cone brightness with National Institutes of Health Image morphometrics. By these criteria, 51 of 117 scored neurons (44%) had different concentrations of argK in their growth cones.
RESULTS
Cloning and sequence analysis of grasshopper argK
Grasshopper argK was cloned by screening a grasshopper nerve cord cDNA library (kindly provided by M. Bastiani) for kinase consensus sequences. From M. Bastiani’s 40% nerve cord cDNA library, we isolated 20 putative clones. One cDNA showed sequence similarity to argK. We cloned and sequenced overlapping cDNAs to obtain the full transcript of this cDNA. The transcript was 2030 bp in length. The clone extending farthest 5′ contained 84 untranslated bp 5′ of the ATG codon. It was followed by an open reading frame of 1071 bp, ending at termination codon TAA. The coding region was followed by a 875 bp 3′-untranslated region that included a putative polyadenylation signal (AATAAA) 11 bp upstream of a poly(A+) tail. The cDNA deduced protein sequence was 356 amino acids long (Fig.1A) with a calculated molecular weight of 40,018 Da. Sequence similarity identifies this transcript as grasshopper argK (GenBank accession number U77580). It contains the signature sequence pattern of ATP-guanidino kinase: “C-P -(S/T)-N-(I/L)-G-T” (Bairoch, 1991), with the reactive cysteine at residue 271.
Fig. 1.

A, Alignment of argK sequences of grasshopper (Schistocerca americana; this study), fly (Drosophila melanogaster; L. B. Hecht, L. M. Scott, G. E. Collier, GenBank U26939), horseshoe crab (Limulus polyphemus; Strong and Ellington, 1995), lobster (Homarus vulgaris; Dumas and Camonis, 1993), abalone (Nordotis madaka; Suzuki and Furukohri, 1994), and CK amino acid sequences of human MiCK (Haas et al., 1989), BCK (Mariman et al., 1989), and MCK (Perryman et al., 1986). B, Phylogenetic tree of argK and CK, using the CLUSTAL method from LaserGene software (Higgens and Sharp, 1988). Thenumbers represent the percentage of divergence. Grasshopper and fly are 16% (8 + 8) dissimilar, i.e., 84% homologous.
Sequence comparison with other argKs and CK
Available argK sequences and three representative forms of CK sequences were aligned by using the CLUSTAL method of LaserGene software (Higgens and Sharp, 1988). Grasshopper argK displays extensive homology to both argKs and CK (black boxes in Fig.1A). It shares 82% positional identity toDrosophila argK. A phylogenetic tree was constructed from the eight ATP-guanidino phosphotransferase sequences shown in Figure1A. The phylogenetic tree indicates that there are major clusters corresponding to CK and argK (Fig.1B). Abalone argK does not belong to either cluster. Within the argK cluster, grasshopper is more similar toDrosophila than to lobster or horseshoe crab. The data indicate that grasshopper argK belongs to the evolutionarily conserved family of ATP-guanidino phosphotransferases.
RNA, protein, and enzyme analysis
Northern blot analysis of grasshopper argK revealed a single ∼2.4 kb transcript expressed from at least the 30–100% stages of embryogenesis (Fig.2A). This occurred in two distinct phases, an early phase of relatively low expression (30, 40, and 50% in Fig. 2A; in whole embryo assays) and a second phase of upregulation that appeared to be associated with muscle maturation (70% in Fig. 2A).
A polyclonal antibody generated against Limulus argK (Strong and Ellington, 1993) enabled us to do a Western blot analysis of expression throughout grasshopper embryogenesis (Fig.2B). This antibody recognizes grasshopper argK, which is shown to be a single tight band of similar molecular weight (40 kDa) to Limulus argK (41,000 ± 210 Da) (Strong and Ellington, 1993). This result indicates that there is only one form of grasshopper argK, and the active form is a monomer. In early embryogenesis (32% in Fig. 2B), grasshopper argK is expressed at low levels. By 55%, argK is expressed at higher levels; the CNS has more argK than the limb (55% in Fig.2B). ArgK is expressed throughout embryonic development and is abundant in adult CNS and muscle (Fig.2B).
We assayed grasshopper embryo lysates in native gels for enzymatic activity of argK (Fig. 2C). An argK-specific substrate/product reaction revealed active enzyme at all embryonic stages in which protein was present, as well as in adult CNS and muscle. The level of activity was similar in grasshopper,Drosophila, and Aedes (mosquito). On the basis of motility of the enzyme in the gel, we did not see evidence for isozymes or other alleles. This result suggests that there may not be separate cytoplasmic and mitochondrial forms of argK in grasshopper.
ArgK expression in muscle pioneer cells
Muscle pioneers comprise a specialized subclass of mesodermal cells that are founder cells for embryonic and larval muscles (Ho et al., 1983; Ball et al., 1985). They differentiate at the sites of origin of muscles, extend processes to the insertion sites of muscles, and form a scaffold on which myoblasts later accrue.
Muscle pioneer cells have distinctively high levels of argK expression. In specific muscle pioneers, such as the retractor unguis pioneers (Fig. 3A–C,E), expression begins early in the differentiation of the cell. Antibody labeling is detectable as soon as the cell begins to elongate and is heavy before process extension begins (Fig. 3A). When process extension commences, in situ hybridization shows argK mRNA throughout the cytoplasm (Fig. 3B). Strong expression is maintained while the muscle pioneer growth cones migrate to the point of muscle insertion (Fig. 3C) and also while the cells shorten (Fig. 3E), concomitant with the epithelial invagination that will form the retractor unguis apodeme.
Fig. 3.
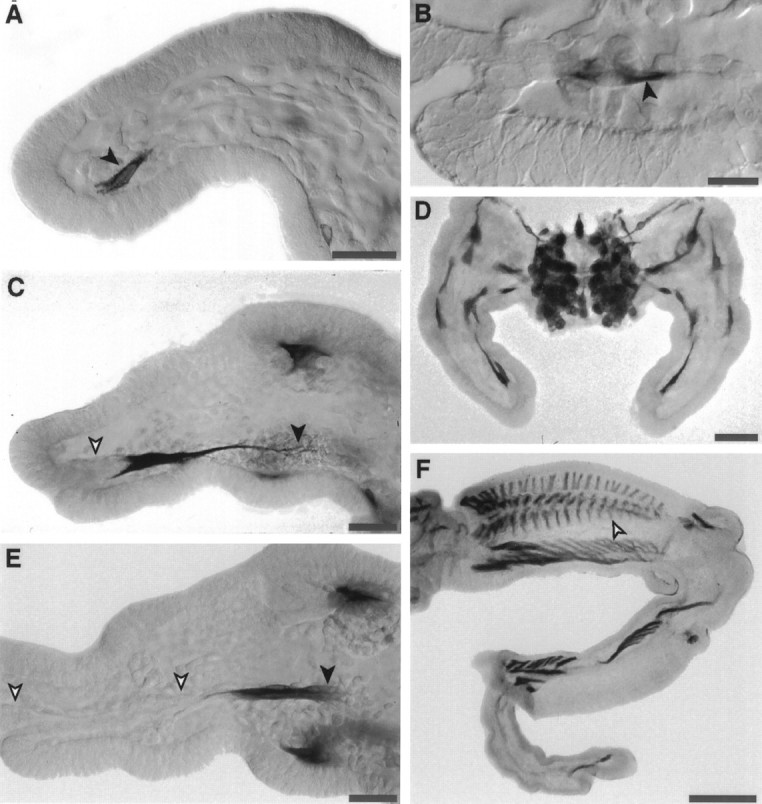
ArgK is strongly expressed in muscle pioneer cells and their extending processes. A, C,E, The two retractor unguis muscle pioneer cells express argK protein (antibody label) at the onset of their differentiation (A, black arrow), while they are extending their processes to the site of retractor unguis muscle insertion in the femur (C, black arrow), and as they shorten (E, black arrow) with the invagination of the tube of epithelial cells that will form the muscle apodeme (C, E, white arrows). B, In situ hybridization reveals argK mRNA throughout the cytoplasm of the retractor unguis muscle pioneers early in their differentiation. D, In mesothoracic limb buds at the 33% stage, argK is expressed in all limb muscle pioneer cells (and in the CNS); it is not expressed in afferent neurons, epithelial cells, or mesodermal cells (other than muscle pioneers). F, At the 55% stage, argK is still expressed in muscle pioneer cells (white arrow) of the extensor tibia (and other) muscles, but not in extensor tibia myoblasts or in afferent neurons or ectoderm. Scale bars: A, C, E, 50 μm; B, 25 μm; D, 100 μm;F, 200 μm.
All muscle pioneers appear to express high levels of argK (Fig.3D). They continue to express through the stage of myoblast adhesion (Fig. 3F), which precedes the onset of strong expression in contractile muscles. Notably, the muscle pioneers are the only mesodermal or ectodermal cells in limb bud that do express argK, at level that we could detect with antibodies or with in situ hybridization, during the first half of embryogenesis. This highly selective expression pattern suggests that argK plays a specific role in muscle pioneers.
ArgK expression in neuroblasts
During generation of the grasshopper CNS, stem cells called neuroblasts differentiate from the ventral neuroectoderm (Bate, 1981;Doe and Goodman, 1985). They undergo repeated rounds of mitosis to generate ganglion mother cells, which divide once to produce neurons. Each body segment has a specific set of individually identifiable neuroblasts.
Neuroblasts strongly express argK. In fact, the most striking feature of argK immunocytochemistry is the intense strip of neuroblast labeling along the ventral midline of the embryo (Fig.4A). In situhybridization (Fig. 4B) and antibody labeling (Fig.4C) indicate that all neuroblasts express argK to some degree. At any particular stage of embryogenesis, there is considerable variation in the amount of argK protein present in different neuroblasts. The pattern of these variations between specific neuroblasts is repeated in adjacent segments (Fig. 4C). Similar patterns are seen in different embryos at the same stage, at least for the most distinctive neuroblasts.
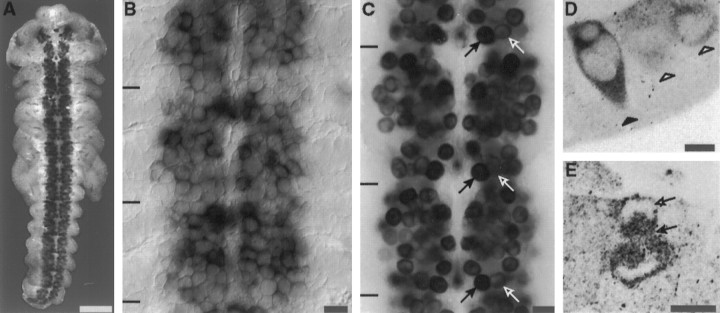
Fig. 4.
ArgK expression in CNS neuroblasts in situ. A, ArgK protein is abundant throughout the length of the neuroblast plate; labeling is not detected in the epithelium (antibody label; 33% stage). B, In situ hybridization confirms the presence of argK mRNA in neuroblasts and not in the adjacent epithelium (black bars, border between neuromeres). C, There is a stage-specific pattern of intensity of argK expression in specific neuroblasts. This pattern is iterated segmentally. For example, at the 33% stage, neuroblast 7-1 (black arrows) expresses more intensely than neuroblast 7-2 (white arrows).D, ArgK is strongly expressed early in the differentiation of neuroblasts. In this cross section, a neuroblast (black arrow) expresses argK before it has rounded up at the inner surface of the neuroblast plate (ventral surface at thebottom of the section; white arrows, two neuroblasts rounded up at the inner surface). E, In dividing neuroblasts argK is present in the spindle (black arrow) and excluded from the chromosomes (white arrow). Scale bars: A, 250 μm;B, C, 25 μm; D,E, 10 μm.
As neuroblasts differentiate, they delaminate from the ventral surface of the embryo and enter a subepidermal layer. The distinctive expression of argK begins early in the differentiation of neuroblasts and can be seen in cells that have not yet fully delaminated and still have a process that is retracting from the ventral surface (Fig.4D). Neuroblasts continue to express argK throughout the cell cycle. In mitosis, argK is excluded from the region occupied by the chromosomes but is present in the spindle and elsewhere in the cytoplasm (Fig. 4E). Ganglion mother cells, the initial progeny of neuroblasts, also express argK (Fig.4D, above neuroblast).
ArgK expression in CNS neurons in situ
Embryonic CNS neurons strongly express argK. During the period of process outgrowth, heavy labeling by argK antibodies is seen in the developing ganglionic cortex, neuropil, commissures, and connectives (Fig. 5A). Most cell bodies and most axons are labeled. In contrast, in situhybridization labeling is much more intense in the neuropil region, commissures, and connectives and is considerably lighter in the surrounding ganglionic cortex containing the neuronal cell bodies (Fig.5B). No labeling was seen with the pool of sense probes (data not shown). This indicates that argK mRNA is present throughout the cells processes and at higher concentrations than in the perinuclear region.
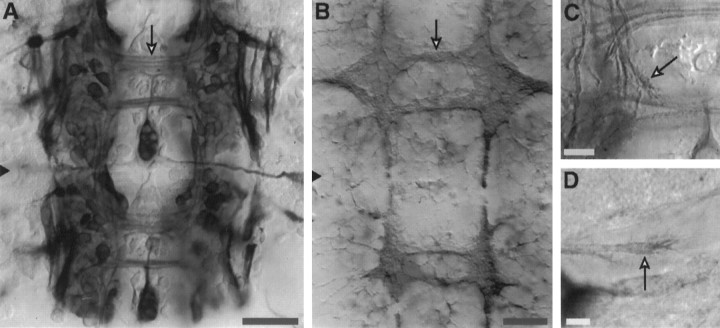
Fig. 5.
ArgK expression in CNS neurons in situ. A, A 50% stage embryo labeled with anti-argK antibody showing two neuromeres (black triangle, border): neuron cell bodies and nascent axons (arrow) are strongly labeled. B,In situ hybridization with the pool of antisense probes (see Materials and Methods) at the 50% stage shows mRNA localization in thoracic (upper) and abdominal (lower) ganglionic neuropil and in commissures and connectives (arrow, anterior commissure). No labeling was seen with the pool of sense probes. ArgK protein is labeled in situ in a CNS growth cone (C, arrow) turning from the connective to join the posterior commissure) and in a motor neuron growth cone entering the hindlimb (D, arrow). Scale bars: A, B, 50 μm; C, D, 10 μm.
Growth cones in the CNS (Fig. 5C) and motor neuron growth cones in the periphery (Fig. 5D) are labeled with argK antibody, and labeling also can be seen in filopodia. In some cases, labeling appears stronger in the growth cone than in the adjacent axon (Fig. 5D). This disposition corresponds well with labelingin vitro (see below).
As with the muscle pioneer expression, it is striking how strong the CNS argK labeling is when compared with the absence of argK in surrounding non-CNS cells (Fig. 4A). It is particularly notable that argK is not expressed in the cell bodies, axons, or growth cones of the peripheral sensory neurons in vivo or in vitro (data not shown).
ArgK expression in CNS glial cells in vitro
In vitro cultures of dissociated CNS embryonic ganglia comprise neurons, which are positive for anti-HRP antibody labeling, and glia, which are negative for anti-HRP. Glial cells are abundant in these cultures. They typically have large nuclei, flattened cytoplasm, and approximately ovoid shape, with relatively short, broad protrusions. Many are motile and, when migrating, are elongate with microspikes on the leading edge and retraction fibers on the trailing edge.
In fixed cultures labeled with anti-argK antibodies, most CNS glial cells are seen to strongly express argK (Fig.6). Thus the intense CNS labeling seenin vivo likely comprises both neural and glial expression. The brightest labeling is perinuclear (Fig. 6A), but the central cytoplasm in general is well labeled. Labeling intensity is low in fringing lamellae, except at the edge.
Because these motile cells translocate by lamellipodia, we examined argK localization at the leading edges of cells with motile and nonmotile morphology. Nonmotile appearing cells were more rounded, with protrusive lamellae at various points around the cell circumference (Fig. 6A). ArgK labeled brightly in leading edges and microspikes of these protrusions (Fig. 6A). By contrast, argK concentration was relatively low in retracted areas of the cell perimeter (Fig. 6A). Thus a higher concentration of argK in the perimeter region was associated with protrusive morphology.
In elongated glia with motile morphology (Fig. 6B), argK was absent from the trailing edge and was highly concentrated in a narrow band that spanned the leading edge. It also was present in microspikes along the leading edge. Thus there was a clear association of argK concentration with the traction force-generating region of the cell.
In a variety of motile fibroblastic cells, F-actin is concentrated in a narrow band at the edge of the leading lamellipodium. In CNS glial protrusions double-labeled for F-actin and argK, there was a close spatial correspondence between F-actin and argK localization (Fig.6C). Both were concentrated in a band at the leading edge, and at different locations along the band there was a tight correlation between the width of the F-actin and argK label.
ArgK localization during CNS neuron axonogenesisin vitro
CNS neurons dissociated from 50% stage ganglia initiate axonogenesis in vitro and extend lengthy processes. In growing neurons, argK protein is present in the cell body, along the whole length of the axon, and in the growth cone (Fig.7A). This corresponds well with observations in situ (Fig. 5A,C).
Grasshopper growth cones in vivo and in vitrohave numerous filopodia that extend 20–30 μm. In neurons double-labeled with antibodies that bind the extracellular domain of membrane proteins (anti-HRP; Snow et al., 1987) and with anti-argK antibodies, argK protein is seen to occur along the length of filopodia and to extend all the way to the tip (Fig. 7B).
Whereas labeling with a higher concentration of anti-argK antibody revealed the presence of argK protein throughout the cell, labeling with a lower concentration could expose intracellular differences in argK protein concentration (Fig. 8). In many neurons, labeling was relatively uniform along the length of the axon, and axonal labeling was dimmer than labeling in growth cones (Fig. 8A–D). z-Axis sampling showed that the axons and growth cones were of similar thickness (cell bodies were much thicker) so that argK often was more concentrated in the growth cone than in the adjacent axon. In other cases labeling in the growth cone was similar in intensity to that in the axon (Fig. 8). These growth cones may have been quiescent or retracting.
Many dissociated CNS neurons are multipolar or, if monopolar, have bifurcated axons. Thus a single cell can have multiple growth cones. An unexpected result was that, in neurons with multiple growth cones, there often were substantial differences in argK labeling intensity in different growth cones (Fig. 8). This pattern was common. In a scored set (see Materials and Methods) of 117 neurons with multiple growth cones, 44% had differentially labeled growth cones. The growth cones with brighter labeling were not distinguished morphologically by being larger in area or by having longer axons or by having more numerous or longer filopodia (Fig. 8). These results show that neurons are able to concentrate argK in growth cones locally and, more surprisingly, can concentrate it selectively in different growth cones.
The level of concentration of argK in growth cones may be associated with the rate of growth cone migration and/or the amount of traction force being generated. We evaluated this possibility by examining multiple growth cones from the same neuron on different substrates. These neurons extend processes on Falcon 3001 plastic dishes (Figs. 7,8). However, they prefer a substrate coated with anti-HRP antibodies (which bind the extracellular domains of a variety of membrane proteins; Snow et al., 1987). When given a choice, they extend axons as well as more numerous and longer filopodia on the preferred substrate (Fig. 9A). In dishes with domains of each of these substrates, neurons were found that satisfied the criteria of straddling the boundary and extending multiple growth cones, at least one of which was on each substrate. We observed several such cases in which processes extended on the preferred substrate had brighter argK labeling (Fig. 9B,C) and no cases in which growth cones on the less-preferred substrate had brighter label.
Another situation in which different growth cones of the same neuron can interact with different substrates in vitro is encounters with the surfaces of other cells. In cultures of grasshopper neurons, neuronal surfaces often are a favorable substrate for growth cones (Condic et al., 1989). We scanned cultures of dissociated ganglia for instances in which one growth cone of a multipolar neuron was undergoing a positive interaction with the surface of another neuron. Positive interactions are indicated by (1) filopodial contact, (2) by filopodia that are straight between their growth cone and the point of contact, suggesting that they are under tension, and (3) by thickening or dilation of growth cone branches leading to the contact region (O’Connor et al., 1990). Of nine cells observed with this configuration, six did not have differential labeling of argK in their growth cones and may have been quiescent. The other three cells did have differential labeling, and in each of these cases the growth cone with the high concentration of argK was the growth cone in contact with the other cell (Fig. 10). The cell shown in Figure 10, A and B, has three robust growth cones. Each is approximately the same distance from the cell body, has approximately the same surface area, and has approximately the same number of filopodia. One growth cone has made contact with an adjacent cell, and the contacting filopodium has dilated substantially (Fig. 10B), suggesting that the cell is beginning to extend toward the contact zone. Imaging for argK antibody labeling shows just detectable argK label in the two growth cones extending on plastic and bright labeling in the growth cone making the contact. A similar result in seen in Figure 10, C and D, where the growth cone making contact is one of four growth cones and is the only one with a high level of argK label.
Because argK can be concentrated differentially in different growth cones, it is possible that it also could be concentrated differentially among filopodia, even those extending from the same growth cone. In growth cones migrating on a uniform substrate, such differences were not evident (Fig. 7B). However, in growth cones contacting other cells, different filopodia could have substantially different levels of argK labeling. Several lengthy filopodia (Fig.10E) extend from the growth cone in Figure10D contacting another cell. In some of these, argK is barely detectable (Fig. 10F), whereas others label brightly. The brightly labeled filopodia either are in contact with the other cell or extend from the growth cone branch from which the contacting filopodium is protruded.
DISCUSSION
We investigated expression and distribution of the guanidino phosphotransferase, argK, during grasshopper embryogenesis. We have two main findings. First, the presence, concentration, and localization of argK are highly controlled. ArgK is expressed only in a small percentage of cells of the embryo. In those cells it is localized in certain subcellular domains, and its concentration can vary greatly between different domains of the same type. Second, a new function with which argK appears to be strongly associated is actin-based motility. It is colocalized with F-actin in leading lamellae of motile glia. In neurons it is concentrated in growth cones and filopodia, is differentially expressed in different growth cones of the same neuron, and appears to be more concentrated in more active growth cones.
Grasshopper argK is a 356 amino acid protein with 82% positional identity to Drosophila argK (Fig. 1). Its calculated molecular weight of 40,018 Da corresponds well with the band position observed on Western blots (Fig. 2B). Northern analysis reveals a single transcript of 2.4 kb (Fig.2A). With the single enzymatically active band in native gels (Fig. 2C), this suggests that, like other arthropods (James and Collier, 1988; Munneke and Collier, 1988), grasshoppers have a single argK gene and isoform.
Northern and Western analyses indicate that grasshopper argK is expressed in two phases during embryogenesis. During the first half of embryogenesis, it is expressed at low but steadily increasing levels, first detectable by our methods at the ∼30% stage. After the onset of striated muscle maturation in the second half of embryogenesis, it is much more heavily expressed. Antibody and in situhybridization labeling of embryos show that during the first half of embryogenesis it is expressed in a select group of cells comprising muscle pioneers, dorsal closure cells, neuroblasts, glia, and neurons.
ArgK expression is an early and sustained marker of neuroblast differentiation (Fig. 4). A possible reason for this selective expression is the energy demands of microtubule dynamics during mitosis. Neuroblasts undergo many rounds of cell division throughout much of embryogenesis (Bate, 1981; Doe and Goodman, 1985). Creatine kinase and/or argK are required for the microtubule-based motility of sperm (Tombes and Shapiro, 1985; Tombes et al., 1988; Strong and Ellington, 1993). During cell division mitotic spindle lengthening in anaphase also is driven by microtubule motors, and creatine kinase supports the special ATP demands of this process (Koons et al., 1982;Cande, 1983). ArgK might play a similar role in neuroblast division.
Except for neuroblasts and their daughter cells (ganglion mother cells), the other cell types that strongly express argK have in common a special cell motility or tension generation function. Dorsal closure cells (data not shown) migrate across the dorsal surface of the embryo to complete the formation of the body wall. As in other closure or “wound-healing” type cells, they are likely to be under tension. Muscle pioneers generate a growth cone that can migrate a considerable distance through the embryo to the site of muscle insertion (Fig.5B). For some muscles this step is followed by invagination of a long tube of ectoderm (Fig. 5E) to form the apodeme on which individual muscle fibers later insert. If this morphogenetic movement requires tension generated by the muscle pioneer cell, then muscle pioneers would have two special actin-based ATP requirements: growth cone migration and intracellular tension generation. These requirements could underlie the high level of argK expressed by this cell class.
Mammalian CNS glia strongly express creatine kinase (Manos and Edmond, 1992; Molloy et al., 1992), and grasshopper CNS glia also have a high level of guanidino phosphotransferase (argK) expression (Fig. 6). In glial cells in vitro, argK is abundant in the perinuclear and central cytoplasm, is generally sparse in lamellar regions, but is highly concentrated in a narrow band along the perimeter of lamellae and in microspikes. On the same cell it is prominent in protrusive lamellae (Fig. 6A) and in leading lamellae of cells with motile morphology (Fig.6B) and is much reduced in retracted lamellae and in trailing edges of motile cells. Leading or protrusive lamellae have a narrow perimeter band of F-actin of varying width (Fig. 6C). In these perimeter bands argK is highly concentrated, and its location is tightly correlated with the location of F-actin. These relationships suggest that argK is providing ATP for the actin/nonmuscle myosin system that generates traction force in these motile cells.
In migrating neuronal growth cones, translocation also is driven by actin/nonmuscle myosin force generation in leading lamellae and in filopodia (Lin et al., 1996). ArgK is present throughout neurons undergoing axonogenesis in vivo (Fig. 5) and in vitro (Fig. 7). It extends to the tips of most filopodia (Fig.7B). Its presence in filopodia, which mostly comprise tightly packed actin filaments (Bridgman and Dailey, 1989) and which lack mitochondria and microtubules, suggests a role in ATP generation for F-actin force production. This supposition is supported by the relatively high concentration of argK specifically in filopodia that are in contact with other cells and which appear to be applying tension to those cells (Fig. 10F).
Labeling with lower levels of antibody indicates that argK is not distributed uniformly in growing neurons and often is highly concentrated in growth cones (Figs. 8-10). Growth cones provide the traction forces required for axon elongation (Lamoureux et al., 1989;Heidemann et al., 1990), and these forces appear to be generated at least in part by actin/myosin (Lin et al., 1996). The high level of argK found in growth cones may supply ATP for this process. Three results support this association of argK with growth cone migration. First, on a uniform substrate, different growth cones of the same neuron often have very different concentrations of argK (Fig. 8). These differences might reflect the rate of migration of the growth cone and the amount of force it is generating. Second, when growth conesin vitro contact the surface of another neuron, they often react positively by enlarging filopodia and growth cone branches along the line of contact and eventually by extending over the contacted cell. In multipolar neurons that appeared to be fixed during this response (Fig. 10), the growth cone engaged in the response sometimes had a much higher level of argK (and never had a lower level). Third, neurons observed with growth cones on two substrates that varied in promoting outgrowth had higher levels of argK in the growth cone on the more favored substrate (Fig. 9). These observations suggest that more active growth cones have higher levels of argK. The high level of argK could support multiple growth associated functions, but one of these is likely to be actin/myosin force generation.
An unexpected feature of the analysis of argK in early embryogenesis is the degree to which its expression is restricted and the precision with which its intracellular localization is controlled. Given its association with cell motility in muscle pioneers, CNS glia, and CNS neurons, a surprising omission in the expression pattern is afferent neurons. One possible explanation would be a second argK gene. This seems unlikely, because only single bands are seen in Northerns, Westerns, and native gels (Fig. 2) and because only a single (highly homologous) gene is present in Drosophila (James and Collier, 1988; Munneke and Collier, 1988). A more plausible alternative is that a different phosphate source, such as adenylate kinase (Savabi, 1994; Dzeja et al., 1996), is involved in ATP generation in afferent neurons.
The concentration of argK, as revealed by antibody labeling, varies in different subcellular regions of glia and neurons. An interesting issue is how this is regulated. CK can be diffusible, or it can be tethered to membrane or to nonsoluble proteins (Wallimann et al., 1977; Lang et al., 1980; Quest et al., 1992). In neuronal growth cones and in leading lamellae of glia, the lack of barriers blocking diffusion out of the actin domain and the absence of separated plasma membrane domains favor argK tethering to actin, myosin, or an actin-associated protein. Another option is regulated local synthesis of argK. CK mRNAs can be localized differentially (Wilson et al., 1995), and in the grasshopper CNS in situ, argK mRNA also is localized differentially in axons and in neuropil (Fig. 5B). Whichever control options are in effect, the striking differences in concentration of argK among growth cones and even among filopodia raise the issue of whether regulation of this enzyme is one target of signaling cascades that determine the direction of neuron outgrowth.
Footnotes
Support was provided by National Science Foundation Grant IBN 94-10068, National Institutes of Health Grant NS 09074, and the W. M. Keck Foundation. We are grateful to W. Ross Ellington for the gift of anti-arginine kinase antibody, to Michael Bastiani for grasshopper cDNA libraries, and to Leonard Munstermann for advice and assistance with the enzyme analysis.
Correspondence should be addressed to Dr. Yu-mei E. Wang, Department of Molecular and Cell Biology, 142 Life Sciences Addition, University of California, Berkeley, CA 94720-3200.
REFERENCES
- 1.Altschul SF, Warren G, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Bairoch A. PROCITE: a dictionary of sites and patterns in proteins. Nucleic Acids Res. 1991;16:2241–2245. doi: 10.1093/nar/19.suppl.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball EE, Ho RK, Goodman CS. Muscle development in the grasshopper embryo. I. Muscles, nerves, and apodemes in the metathoracic leg. Dev Biol. 1985;111:383–398. doi: 10.1016/0012-1606(85)90492-0. [DOI] [PubMed] [Google Scholar]
- 4.Bate CM. Embryogenesis of an insect nervous system. I. A map of the thoracic and abdominal neuroblasts in Locusta migratoria. J Embryol Exp Morphol. 1981;35:107–123. [PubMed] [Google Scholar]
- 5.Bessman SP. The creatine–creatine phosphate energy shuttle. Annu Rev Biochem. 1985;54:831–862. doi: 10.1146/annurev.bi.54.070185.004151. [DOI] [PubMed] [Google Scholar]
- 6.Blethen SL, Kaplan NO. Characteristics of arthropod arginine kinases. Biochemistry. 1968;7:2123–2135. doi: 10.1021/bi00846a015. [DOI] [PubMed] [Google Scholar]
- 7.Bridgman PC, Dailey ME. The organization of myosin and actin in rapid frozen nerve growth cones. J Cell Biol. 1989;108:95–110. doi: 10.1083/jcb.108.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cande WZ. Creatine kinase role in anaphase chromosome movement. Nature. 1983;304:557–558. doi: 10.1038/304557a0. [DOI] [PubMed] [Google Scholar]
- 9.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 10.Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Condic ML, Lefcort F, Bentley D. Selective recognition between embryonic afferent neurons of grasshopper appendages in vitro. Dev Biol. 1989;135:221–230. doi: 10.1016/0012-1606(89)90174-7. [DOI] [PubMed] [Google Scholar]
- 12.Doe CQ, Goodman CS. Early events in insect neurogenesis. I. Development and segmental differences in the pattern of neuronal precursor cells. Dev Biol. 1985;111:193–205. doi: 10.1016/0012-1606(85)90445-2. [DOI] [PubMed] [Google Scholar]
- 13.Dumas C, Camonis J. Cloning and sequence analysis of the cDNA for arginine kinase of lobster muscle. J Biol Chem. 1993;268:21599–21605. [PubMed] [Google Scholar]
- 14.Dzeja PP, Zeleznikar RJ, Goldberg ND. Suppression of creatine kinase-catalyzed phosphotransfer results in increased phosphoryl transfer by adenylate kinase in intact skeletal muscle. J Biol Chem. 1996;271:12847–12851. doi: 10.1074/jbc.271.22.12847. [DOI] [PubMed] [Google Scholar]
- 15.Eppenberger ME, Eppenberger HM, Kaplan NO. Evolution of creatine kinase. Nature. 1967;214:239–241. doi: 10.1038/214239a0. [DOI] [PubMed] [Google Scholar]
- 16.Friedman DL, Roberts R. Compartmentation of brain-type creatine kinase and ubiquitous mitochondrial creatine kinase in neurons: evidence for a creatine phosphate energy shuttle in adult rat brain. J Comp Neurol. 1994;343:500–511. doi: 10.1002/cne.903430311. [DOI] [PubMed] [Google Scholar]
- 17.Gordon PV, Keller TC. Functional coupling to brush border creatine kinase imparts a selective energetic advantage to contractile ring myosin in intestinal epithelial cells. Cell Motil Cytoskeleton. 1992;21:38–44. doi: 10.1002/cm.970210105. [DOI] [PubMed] [Google Scholar]
- 18.Haas RC, Korenfeld C, Zhang Z, Perryman B, Roman D, Strauss AW. Isolation and characterization of the gene and the cDNA encoding human mitochondrial creatine kinase gene. J Biol Chem. 1989;266:18058–18065. [PubMed] [Google Scholar]
- 19.Harlow E, Lane D. Antibodies: a laboratory manual, p 498. Cold Spring Harbor Laboratories; Cold Spring Harbor, NY: 1988. Purifying antibodies from immunoblots. [Google Scholar]
- 20.Heidemann SR, Lamoureux P, Buxbaum RE. Growth cone behavior and production of traction force. J Cell Biol. 1990;111:1949–1957. doi: 10.1083/jcb.111.5.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hemmer W, Wallimann T. Functional aspects of creatine kinase in brain. Dev Neurosci. 1993;15:249–260. doi: 10.1159/000111342. [DOI] [PubMed] [Google Scholar]
- 22.Hemmer W, Furter-Graves EM, Frank G, Wallimann T, Furter R. Autophosphorylation of creatine kinase: characterization and identification of a specifically phosphorylated peptide. Biochim Biophys Acta. 1995;1251:81–90. doi: 10.1016/0167-4838(95)00083-7. [DOI] [PubMed] [Google Scholar]
- 23.Higgens DG, Sharp PM. CLUSTAL: a package for performing multiple sequence analysis on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 24.Ho RK, Ball EE, Goodman CS. Muscle pioneers: large mesodermal cells that erect a scaffold for developing muscles and motoneurones in grasshopper embryos. Nature. 1983;301:66–69. doi: 10.1038/301066a0. [DOI] [PubMed] [Google Scholar]
- 25.James JM, Collier GE. Distribution and genetic basis of arginine kinase in wild type and flightless mutants of Drosophila melanogaster. J Exp Zool. 1988;248:185–191. [Google Scholar]
- 26.Kaldis P, Hemmer W, Zanolla E, Holtzman D, Wallimann T. “Hot spots” of creatine kinase localization in brain: cerebellum, hippocampus, and choroid plexus. Dev Neurosci. 1996a;18:542–554. doi: 10.1159/000111452. [DOI] [PubMed] [Google Scholar]
- 27.Kaldis P, Stolz M, Wyss M, Zanolla E, Rothen-Rutishauser B, Vorherr T, Wallimann T. Identification of two distinctly localized mitochondrial creatine kinase isoenzymes in spermatozoa. J Cell Sci. 1996b;109:2079–2088. doi: 10.1242/jcs.109.8.2079. [DOI] [PubMed] [Google Scholar]
- 28.Kelly C, Sellers J, Gard D, Bui D, Adelstein R, Baines I. Xenopus nonmuscle myosin heavy chain isoforms have different subcellular localizations and enzymatic activities. J Cell Biol. 1996;134:675–688. doi: 10.1083/jcb.134.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koons SJ, Eckert BS, Zobel CR. Immunofluorescence and inhibitor studies on creatine kinase and mitosis. Exp Cell Res. 1982;140:401–409. doi: 10.1016/0014-4827(82)90130-6. [DOI] [PubMed] [Google Scholar]
- 30.Lamoureux P, Buxbaum RE, Heidemann SR. Direct evidence that growth cones pull. Nature. 1989;13:159–162. doi: 10.1038/340159a0. [DOI] [PubMed] [Google Scholar]
- 31.Lang AB, Wyss C, Eppenberger HM. Localization of arginine kinase in muscle fibres of Drosophila melanogaster. J Muscle Res Cell Motil. 1980;1:147–161. doi: 10.1007/BF00711796. [DOI] [PubMed] [Google Scholar]
- 32.Lin C, Espreafico E, Mooseker M, Forscher P. Myosin drives retrograde F-actin flow in neuronal growth cones. Neuron. 1996;16:769–782. doi: 10.1016/s0896-6273(00)80097-5. [DOI] [PubMed] [Google Scholar]
- 33.Mahadevan LC, Whatley SA, Leung TK, Lim L. The brain isoform of a key ATP-regulating enzyme, creatine kinase, is a phosphoprotein. Biochem J. 1984;222:139–144. doi: 10.1042/bj2220139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manchenko GP. Handbook of detection of enzymes on electrophoretic gels. CRC; Boca Raton, FL: 1994. [Google Scholar]
- 35.Manos P, Bryan GK. Cellular and subcellular compartmentation of creatine kinase in brain. Dev Neurosci. 1993;15:271–279. doi: 10.1159/000111344. [DOI] [PubMed] [Google Scholar]
- 36.Manos P, Edmond J. Immunofluorescent analysis of creatine kinase in cultured astrocytes by conventional and confocal microscopy: a nuclear localization. J Comp Neurol. 1992;326:273–282. doi: 10.1002/cne.903260209. [DOI] [PubMed] [Google Scholar]
- 37.Mariman ECM, Schepens JTG, Wieringa B. Complete nucleotide sequence of the human creatine kinase B gene. Nucleic Acids Res. 1989;17:6385–6390. doi: 10.1093/nar/17.15.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitchison T, Cramer L. Actin-based cell motility and cell locomotion. Cell. 1996;84:371–380. doi: 10.1016/s0092-8674(00)81281-7. [DOI] [PubMed] [Google Scholar]
- 39.Molloy GR, Wilson CD, Benfield P, de Vellis J, Kumar S. Rat brain creatine kinase messenger RNA levels are high in primary cultures of brain astrocytes and oligodendrocytes and low in neurons. J Neurochem. 1992;59:1925–1932. doi: 10.1111/j.1471-4159.1992.tb11028.x. [DOI] [PubMed] [Google Scholar]
- 40.Moreland B, Watts DC, Virden R. Phosphagen kinases and evolution in the Echinodermata. Nature. 1967;214:458–462. doi: 10.1038/214458a0. [DOI] [PubMed] [Google Scholar]
- 41.Morrison JF. Arginine and other invertebrate guanidino kinases. In: Boyer PD, editor. The enzymes, Vol 8. Academic; New York: 1973. pp. 457–486. [Google Scholar]
- 42.Muhlebach SM, Wirz T, Brandle U, Perriard J-C. Evolution of the creatine kinases. J Biol Chem. 1996;271:11920–11929. doi: 10.1074/jbc.271.20.11920. [DOI] [PubMed] [Google Scholar]
- 43.Munneke LR, Collier GE. Cytoplasmic and mitochondrial arginine kinases in Drosophila: evidence for a single gene. Biochem Genet. 1988;26:131–141. doi: 10.1007/BF00555494. [DOI] [PubMed] [Google Scholar]
- 44.Newsholme EA, Beis I, Leech AR, Zammit VA. The role of creatine kinase and arginine kinase in muscle. Biochem J. 1978;172:533–537. doi: 10.1042/bj1720533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Connor TP, Duerr JS, Bentley D. Pioneer growth cone steering decisions mediated by single filopodial contacts in situ. J Neurosci. 1990;10:3935–3946. doi: 10.1523/JNEUROSCI.10-12-03935.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perryman MB, Kerner SA, Bohlmeyer TJ, Roberts R. Isolation and sequence analysis of a full-length cDNA for human M creatine kinase. Biochem Biophys Res Commun. 1986;140:981–989. doi: 10.1016/0006-291x(86)90732-1. [DOI] [PubMed] [Google Scholar]
- 47.Quest AF, Soldati T, Hemmer W, Perriard JC, Eppenberger HM, Wallimann T. Phosphorylation of chicken brain-type creatine kinase affects a physiologically important kinetic parameter and gives rise to protein microheterogeneity in vivo. FEBS Lett. 1990;269:457–464. doi: 10.1016/0014-5793(90)81215-a. [DOI] [PubMed] [Google Scholar]
- 48.Quest AF, Chadwick JK, Wothe DD, McIlhinney RA, Shapiro BM. Myristoylation of flagellar creatine kinase in the sperm phosphocreatine shuttle is linked to its membrane association properties. J Biol Chem. 1992;267:15080–15085. [PubMed] [Google Scholar]
- 49.Reddy SRR, Watts DC. Hybridization of matrix-bound MM-creatine kinase with BB-creatine kinase and arginine kinase. Comp Biochem Physiol [B] 1994;108:73–78. doi: 10.1016/0305-0491(94)90167-8. [DOI] [PubMed] [Google Scholar]
- 50.Savabi F. Interaction of creatine kinase and adenylate kinase systems in muscle cells. Mol Cell Biochem. 1994;133–134:145–152. doi: 10.1007/BF01267953. [DOI] [PubMed] [Google Scholar]
- 51.Snow P, Patel N, Harrelson A, Goodman C. Neural-specific carbohydrate moiety shared by many surface glycoproteins in Drosophila and grasshopper embryos. J Neurosci. 1987;7:4137–4144. doi: 10.1523/JNEUROSCI.07-12-04137.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Strong SJ, Ellington WR. Horseshoe crab sperm contain a unique isoform of arginine kinase that is present in midpiece and flagellum. J Exp Zool. 1993;267:563–571. [Google Scholar]
- 53.Strong SJ, Ellington WR. Isolation and sequence analysis of the gene for arginine kinase from the chelicerate arthropod, Limulus polyphemus: insights into catalytically important residues. Biochim Biophys Acta. 1995;1246:197–200. doi: 10.1016/0167-4838(94)00218-6. [DOI] [PubMed] [Google Scholar]
- 54.Suzuki T, Furukohri T. Evolution of phosphagen kinase: primary structure of glycocyamine kinase and arginine kinase from invertebrates. J Mol Biol. 1994;237:353–357. doi: 10.1006/jmbi.1994.1237. [DOI] [PubMed] [Google Scholar]
- 55.Thoai NV, Robin Y. Distribution of phosphagens in errant and sedentary polychaeta. In: Munday K, editor. Studies in comparative biochemistry. Pergamon; London: 1964. pp. 152–161. [Google Scholar]
- 56.Tombes RM, Shapiro BM. Metabolite channeling: a phosphorylcreatine shuttle to mediate high energy phosphate transport between sperm mitochondrion and tail. Cell. 1985;41:325–334. doi: 10.1016/0092-8674(85)90085-6. [DOI] [PubMed] [Google Scholar]
- 57.Tombes RM, Farr A, Shapiro BM. Sea urchin sperm creatine kinase: the flagellar isozyme is a microtubule-associated protein. Exp Cell Res. 1988;178:307–317. doi: 10.1016/0014-4827(88)90401-6. [DOI] [PubMed] [Google Scholar]
- 58.van Deursen J, Heerschap A, Oerlemans F, Ruitenbeek W, Jap P, ter Laak H, Wieringa B. Skeletal muscles of mice deficient in muscle creatine kinase lack burst activity. Cell. 1993;74:621–631. doi: 10.1016/0092-8674(93)90510-w. [DOI] [PubMed] [Google Scholar]
- 59.Verkhovsky A, Svitkina T, Borisy G. Myosin II filament assemblies in the active lamella of fibroblasts: their morphologies and role in the formation of actin filament bundles. J Cell Biol. 1995;131:989–1002. doi: 10.1083/jcb.131.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wallimann T. Bioenergetics. Dissecting the role of creatine kinase. Curr Biol. 1994;4:42–46. doi: 10.1016/s0960-9822(00)00008-7. [DOI] [PubMed] [Google Scholar]
- 61.Wallimann T, Eppenberger HM. Properties of arginine kinase from Drosophila melanogaster. Eur J Biochem. 1973;38:180–184. doi: 10.1111/j.1432-1033.1973.tb03048.x. [DOI] [PubMed] [Google Scholar]
- 62.Wallimann T, Hemmer W. Creatine kinase in non-muscle tissues and cells. Mol Cell Biochem. 1994;133–134:193–220. doi: 10.1007/BF01267955. [DOI] [PubMed] [Google Scholar]
- 63.Wallimann T, Turner DC, Eppenberger HM. Localization of creatine kinase isoenzymes in myofibrils. I. Chicken skeletal muscle. J Cell Biol. 1977;75:297–317. doi: 10.1083/jcb.75.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wegmann G, Zanolla E, Eppenberger HM, Wallimann T. In situ compartmentation of creatine kinase in intact sarcomeric muscle: the acto-myosin overlap zone as a molecular sieve. J Muscle Res Cell Motil. 1992;13:420–435. doi: 10.1007/BF01738037. [DOI] [PubMed] [Google Scholar]
- 65.Welnhofer EA, Zhao L, Cohan CS. Actin dynamics and organization during growth cone morphogenesis in Helisoma neurons. Cell Motil Cytoskeleton. 1997;37:54–71. doi: 10.1002/(SICI)1097-0169(1997)37:1<54::AID-CM6>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 66.Wilson IA, Brindle KM, Fulton AM. Differential localization of the mRNA of the M and B isoforms of creatine kinase in myoblasts. Biochem J. 1995;308:599–605. doi: 10.1042/bj3080599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wyss M, Maughan D, Wallimann T. Re-evaluation of the structure and physiological function of guanidino kinases in fruitfly (Drosophila), sea urchin (Psammechinus milaris), and man. Biochem J. 1995;309:255–261. doi: 10.1042/bj3090255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zachow KZ, Bentley D. Blackjack, a novel protein associated with microtubules in embryonic neurons. J Cell Sci. 1996;109:1497–1507. doi: 10.1242/jcs.109.6.1497. [DOI] [PubMed] [Google Scholar]