Abstract
Dopamine β-hydroxylase (DBH) catalyzes the conversion of dopamine to noradrenaline and is selectively expressed in noradrenergic and adrenergic neurons and neuroendocrine cells. Recent data from this laboratory showed that a paired-like homeodomain (HD) protein, Phox2a, interacts with the HD-binding site residing within a composite promoter of the human DBH gene, designated domain IV, in a cell-specific manner and directly controls noradrenergic-specific DBH promoter activity. In this report, we demonstrate that three additional protein-binding sites (i.e., domains I, II, and III) between domain IV and the TATA box are critical for intact DBH promoter activity in noradrenergic cells and that they activate DBH transcription in a highly concerted manner. Transient transfection assays of mutant DBH reporter constructs indicated that domain I was active in every cell line tested, whereas domain III was preferentially active in DBH-positive cells. Remarkably, mutation of domain II was associated with inactivation of DBH promoter activity exclusively in DBH-positive cell lines, defining it as another noradrenergic-specific promoter element. The cell-specific profile of the promoter function of these sequence motifs was further supported byin vitro DNA-binding studies and Southwestern analysis. Furthermore, competition and antibody supershift assays show that transcription factors Sp1 and AP2 are the cognate nuclear factors interacting with domains I and III, respectively. Parallel evidence indicates that domain II is another Phox2a-binding site, demonstrating at least two binding sites for this factor in the upstream DBH promoter. Strikingly, four tandem copies of domain II increased the promoter activity of a minimal DBH promoter by 100- to 200-fold in DBH-positive cell lines without compromising cell specificity. Cotransfection of Phox2a-expression vector dramatically increased the activity of the multiple domain II promoter only in DBH-negative cell lines, confirming that domain II is responsive to Phox2a. Collectively, this study emphasizes a critical role of Phox2a as well as its functional synergism with other transcription factors (e.g., CREB, AP2, and Sp1) in transcriptional activation of the DBH gene.
Keywords: dopamine β-hydroxylase, homeodomain protein, Phox2a, Phox2b, cell-type specific transcription, noradrenergic neuron, cis-acting element, AP2, Sp1, synergistic activation
Dopamine β-hydroxylase (DBH) is a hallmark protein of noradrenergic neurons because noradrenaline is synthesized by this enzyme (Kirshner and Goodall, 1957; Friedman and Kaufman, 1965). The highly restricted pattern of DBH expression in the nervous system predicts that this gene is subject to neuron-specific as well as to cell type-specific control mechanisms. Transgenic mice experiments have shown that 5.8 or 4 kb of the 5′ flanking sequences of the human DBH gene can drive expression of the reporter gene in neurons of the locus coeruleus as well as other noradrenergic neurons and adrenal chromaffin cells, with some ectopic expression (Mercer et al., 1991; Morita et al., 1993; Kobayashi et al., 1994). More recently, comparison of reporter gene expression in transgenic animals generated by using DBH 5′ flanking regions of different lengths indicated that the upstream region between −1.1 and −0.6 kb is necessary for expression in adult and fetal noradrenergic neurons (Hoyle et al., 1994). In addition, this and other laboratories, using cell culture systems, demonstrated that the 5′ upstream region of the DBH gene can drive reporter gene expression in a cell-specific manner (Shaskus et al., 1992, 1995; Ishiguro et al., 1993, 1995; Lamouroux et al., 1993;Kim et al., 1994; Afar et al., 1996; Seo et al., 1996; Yang et al., 1998a,b).
Deletional and site-directed mutational analyses indicated that as little as 486 bp of the upstream sequence of the human DBH gene can direct expression of a reporter gene in a cell-specific manner (Ishiguro et al., 1993; Seo et al., 1996). In the 486 bp region of the human DBH gene, the distal part spanning −486 to −263 bp appears to have a cell-specific silencer function that contributed to suppression of the promoter activity in non-neuronal cells (Ishiguro et al., 1993,1995). A recent analysis has indicated that its cell-specific silencer function may depend on several sequence motifs including two novelcis-elements (Kim et al., 1998). Transient transfection assays identified the proximal part spanning −262 to +1 bp as sufficient and essential for the high-level DBH promoter activity in DBH-positive cells (Ishiguro et al., 1993; Seo et al., 1996). In this 262 bp proximal area, four protein-binding regions (domains I–IV) have been identified by DNase I footprinting analysis (Seo et al., 1996). A cAMP response element (CRE), 5′-TGACGTCC-3′, with a single base deviation from the consensus octamer motif, 5′-TGACGTCA-3′ (Roesler et al., 1988; Goodman, 1990), was shown to be critical for both the basal and cAMP-inducible transcription in DBH-expressing cell lines (Ishiguro et al., 1993; Lamouroux et al., 1993; Kim et al., 1994; Seo et al., 1996). This CRE is included in a composite promoter structure located at −185 to −150 bp, designated domain IV, which contains several additional cis-elements such as AP1, YY1, and two core motifs of homeodomain (HD)-binding sites. Site-directed mutagenesis of each sequence motif has revealed that the CRE is essential for basal promoter activity in every cell line, YY1 is multifunctional, and the AP1-like motif may be transcriptionally inactive (Seo et al., 1996;Yang et al., 1998a). Most interestingly, mutation of either core ATTA motif within domain IV significantly reduced the promoter activity only in DBH-positive cell lines, defining the HD-binding site within domain IV as the first noradrenergic-specific promoter element (Yang et al., 1998a).
The murine paired-like HD protein, Phox2a, is selectively expressed in noradrenergic cells and is critical for development of several noradrenergic neuron populations, including the locus coeruleus (Morin et al., 1997). Phox2a, whose forced expression robustly activates the DBH promoter activity, binds to the HD site of domain IV, strongly suggesting a mechanism for noradrenergic-specific promoter function (Yang et al., 1998a). In addition, Phox2b, containing an HD identical to that of Phox2a, has been identified and shown to be widely coexpressed with Phox2a in both the central and peripheral nervous system (Pattyn et al., 1997). Cotransfection assays showed that Phox2a and Phox2b transactivate the DBH promoter activity with a comparable efficiency (Yang et al., 1998a). There is no direct biochemical evidence that Phox2b binds to the HD-binding site of domain IV. However, the fact that Phox2a and Phox2b have identical HDs and the above cotransfection data strongly suggest that both Phox2a and Phox2b directly interact with the HD-binding site of domain IV; accordingly, we designate this site as PRS1 forPhox2a/Phox2b-response site 1 in this article.
Although domain IV contains several cis-elements critical for DBH transcription, when placed 5′ to the TATA and transcription start site, it was able to recapitulate neither intact basal level nor noradrenergic cell-specific transcription of the reporter gene (Yang et al., 1998a). In addition, domain IV by itself mediated Phox2a/Phox2b-induced transcription only modestly (approximately threefold) compared with the intact DBH promoter (10- to 15-fold). In the present study, we sought to characterize the contributions of domains I, II, and III to DBH gene expression and identify their cognate nuclear factors. We now show that all three domains are critical for maximal DBH promoter activity in noradrenergic cell lines. We identify Sp1 and AP2 as the transcription factors binding to domains I and III, respectively. Domain II is found to be exclusively active in DBH-expressing cells, defining it as another noradrenergic-specific promoter element, and we provide several lines of evidence supporting domain II as an additional Phox2a-binding site. Interestingly, four tandem copies of domain II increased the DBH minimal promoter activity by 150-fold in DBH-positive cells, which is at least twofold of that by the intact DBH promoter. The promoter activity of this multiple domain II was similar to that of the DBH minimal promoter in DBH-negative cells and increased by 200-fold after cotransfecting the Phox2a-expressing plasmid. These results demonstrate that domain II is indeed a noradrenergic-specific promoter and another Phox2a/Phox2b-response site; accordingly, we designate domain II as PRS2.
MATERIALS AND METHODS
Cell culture and transient transfection assays. Human neuroblastoma SK-N-BE(2)C and SK-N-BE(2)M17 and mouse central noradrenergic neuron-derived CATH.a cell lines were maintained as described (Ishiguro et al., 1993; Suri et al., 1993; Kim et al., 1994) and used as the DBH-positive system. The HeLa and rat C6 glioma cell lines were grown in DMEM supplemented with 10% fetal calf serum (Hyclone), streptomycin, and penicillin and used as the DBH-negative system in this study.
Transfection was performed by the calcium phosphate coprecipitation method as previously described (Ishiguro et al., 1993; Seo et al., 1996). For the SK-N-BE(2)C and SK-N-BE(2)M17 cell lines, each 60 mm dish was transfected with 2 μg of the reporter construct, 1 μg of pRSV β-galactosidase, varying amounts of the effector plasmid, and pUC19 plasmid to a total of 5 μg DNA. For the other cell lines, twice as much DNA was used in transfection. Plasmids used for transient transfection assays were prepared using Qiagen (Santa Clarita, CA) columns. To correct for differences in transfection efficiencies among different DNA precipitates, chloramphenicol acetyltransferase (CAT) activity was normalized to that of β-galactosidase. CAT and β-galactosidase activities were assayed as previously described (Ishiguro et al., 1993; Seo et al., 1996).
DNA constructions. The DBH978CAT and DBH262CAT reporter constructs contain the 978 and 262 bp upstream sequences of the human DBH gene, respectively, fused to the bacterial CAT gene (Ishiguro et al., 1993). A series of human DBH promoter-CAT reporter constructs with progressive deletions of proximal protein-binding sites were generated using pBLCAT3–1 (Yang et al., 1998a), which drives significantly lower background CAT activity compared with pBLCAT3 (Luckow and Schutz, 1987). 262′CAT construct was generated by ligating the 271 bpSphI–XbaI fragment of 262CAT plasmid with the 4.3 kb SphI–XbaI backbone of pBLCAT3–1 plasmid. 114′CAT construct was made by ligating the 726 bpHindIII–NcoI fragment withHindIII–NcoI backbone of pBLCAT3–1 plasmid. To generate 142′CAT and 62′CAT plasmids, PCR was performed using oligonucleotides 5′-GACATGCATGCGCAGGCTGAGTGCTTGGC-3′ and 5′-CATTTTAGCTTCCTTAGC-3′, and 5′-GACATGCATGCGCTGCCTGGACCCACCCC-3′ and 5′-CATTTTAGCTTCCTTAGC-3′, respectively, using DBH978CAT as the template. The 163 and 71 bp fragments were isolated after digesting the PCR products with SphI and XbaI and were subcloned to pBLCAT3–1 that had been digested with SphI andXbaI, resulting in 142′CAT and 62′CAT plasmids, respectively. 38′CAT plasmid, which is identical to the TATA-CAT plasmid, has been described previously (Yang et al., 1998a). The upstream sequences and junction regions of these deletional constructs were confirmed by sequencing analysis.
Base substitutions in domains I, II, or III were generated in the context of the 978 bp upstream sequence using the QuickChange PCR-based site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer’s procedure. The following oligonucleotides were used in the mutagenesis procedure using DBH978CAT plasmid as the template: 5′-CCTGGACCCAC TATGTTCAGGACCAG-3′ and 5′-CCTGGTCCTGAA CATAGTGGGTCCAG-3′ for domain I mutant, 5′-CCGCTAGACAA GCAGACGTACCCGTGCTG-3′ and 5′-GCAGCACGGGTA CGTCTGCTTGTCTAGCG-3′ for domain II mutant, and 5′-TGAGTGCTTGGCCTGG TTAGCAAGCTTGTGGGAGG-3′ and 5′-CCCTCCCACAAGCTTGC TAACCAGGCCAAGCACTC-3′ for domain III mutant. The first set of primers represents coding strand sequences of the promoter containing the desired mutations (underlined bases), and the second set of primers represents the corresponding noncoding strand sequences. Constructs with correct mutations were screened by restriction enzyme digestion and sequencing analysis.
A single copy of the domain II oligonucleotide (see below) was subcloned to the SphI site of 38′CAT plasmid. After restriction and sequencing analyses, the 1xII-CAT construct containing a single copy of domain II in correct orientation was selected. In addition, the same domain II oligonucleotide was ligated after Klenow reaction. A DNA fragment of 92 bp was isolated and subcloned to the same SphI site of 38′CAT plasmid. 4xII-CAT plasmid, which contains four copies of domain II (three copies in the right orientation and one in the opposite orientation; see Fig. 8), was isolated and confirmed by sequence analysis. pRC/Phox2a and pRC/Phox2b, which express full lengths of Phox2a and Phox2b protein factor, respectively, under control of the CMV promoter, have been described previously (Yang et al., 1998a) and were used as effector plasmids.
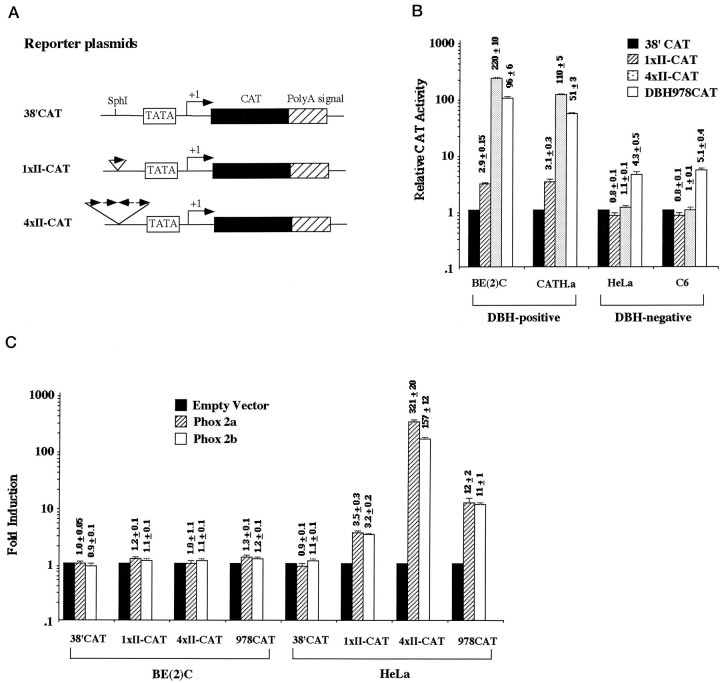
Fig. 8.
Domain II mediates transactivation of the promoter activity by Phox2a and Phox2b. A, Diagram of reporter plasmids. 38′CAT, identical to TATA-CAT (Yang et al., 1998a), is a minimal DBH-CAT reporter plasmid that contains the TATA box and the transcription start site of the human DBH gene. A single copy of domain II oligonucleotide is cloned at theSphI site upstream of the TATA box, resulting in1xII-CAT. Likewise, four tandem copies of domain II oligonucleotide are cloned at the SphI site. Sequence analysis of 4xII-CAT showed that, among the four copies of domain II, the third copy was in opposite orientation as indicated by the direction ofarrows. B, Domain II sequence motif(s) can activate the promoter activity in a noradrenergic-specific manner. Transcription activities driven by reporter constructs were compared by transient transfection assay using two DBH-positive and two DBH-negative cell lines. 4xII-CAT plasmid drives the CAT gene expression 100- to 200-fold higher than the 38′CAT plasmid, which represent at least twice activity of that by the intact DBH promoter-CAT construct (978CAT), without compromising cell specificity. The CAT activity driven by each construct is presented relative to that of 38′CAT, with mean ± SEM for six to eight determinations plotted on a logarithmic scale. This experiment was repeated once more in triplicate, using plasmid DNAs independently prepared, and resulted in similar patterns.C, Domain II can mediate transactivation by Phox2a and Phox2b. HeLa and SK-N-BE(2)C cells were transiently cotransfected with reporter plasmids and pRC/Phox2a or pRC/Phox2b with a molar ratio of 0.5. The CAT activity driven by each reporter construct itself was set to 1.0 to compare transactivation by Phox2a or Phox2b. Fold induction by Phox2a/Phox2b cotransfection is presented as mean ± SEM values from six to eight samples on a logarithmic scale.
Preparation of nuclear extracts, EMSA, and DNase I footprinting. Nuclear extracts were prepared from different cell lines according to the procedure described by Dignam et al. (1983). Sense and antisense oligonucleotides corresponding to the sequences of domains I, II, and III were synthesized (Gene Link, Inc., Thornwood, NY) with the following nucleotide sequences: 5′-GCCTGGACCCACCCCATTCA-3′ and 5′-CTGAATGGGGTGGGTCCAGG-3′ for domain I (DI), 5′-CCGCTAGACAAATGTGATTACC-3′ and 5′-GGGTAATCACATTTGTCTAGCG-3′ for domain II (DII), and 5′-TGAGTGCTTGGCCTGGGGCGCAAGCTTGTGGGAGG-3′ and 5′-CCCTCCCACAAGCTTGCGCCCCAGGCCAAGCACTC-3′ for domain III (DIII). Nucleotide sequences for mutant oligonucleotides were 5′-GCCTGGACCCAC TATGTTCA3′ and 5′-CTGAA CATAGTGGGTCCAGG-3′ for DIm, 5′-CCGCTAGACAA GCAGACGTACC-3′ and 5′-GGGTA CGTCTGCTTGTCTAGCG-3′ for DIIm, and 5′-TGAGTGCTTGGCCTGG TTAGCAAGCTTGTGGGAGG-3′ and 5′-CCCTCCCACAAGCTTGC TAACCAGGCCAAGCACTC-3′ for DIIIm. Additional mutant oligonucleotides for domain II were used and described in Figure 7A. In addition, oligonucleotides 5′-GATCGAACTGACCGCCCGCGGCCCG-3′ and 5′-ACGGGCCGCGGGCGGTCAGTTCGAT-3′ were synthesized as the consensus AP2 (Williams et al., 1988), and Sp1 oligonucleotides were previously described (Seo et al., 1996). HD oligonucleotide, which contains the HD-binding site within domain IV, was also described previously (Yang et al., 1998a). The sense and antisense oligonucleotides were annealed, gel-purified, and 32P-labeled by T4 DNA kinase and used as probes in electrophoretic mobility shift assays (EMSA). EMSA and antibody coincubation experiments were performed using 30,000–50,000 cpm of labeled probe (∼0.05–0.1 ng) and nuclear extracts (10–30 μg) in a final volume of 20 μl of 12.5% glycerol, and (in mm) 12.5 HEPES, pH 7.9, 4 Tris-HCl, pH 7.9, 60 KCl, 1 EDTA, and 1 DTT with 1 μg of poly(dI-dC) as described (Yang et al., 1998a). Competition-binding assays were performed by adding nonradioactive competitor oligonucleotides in a molar excess before adding32P-labeled oligonucleotides. For supershift assay, antibody was coincubated with the nuclear extract mix for 30 min at room temperature before adding the radiolabeled probe. Antibodies against Sp1, Sp3, Sp4, and AP2 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). A Phox2a-specific antibody (number 59) raised against a polypeptide (Y75–R88) residing immediately upstream of the HD was used in the supershift assay. Recombinant AP2 proteins were purchased from Promega (Madison, WI) and used for EMSA with32P-labeled DIII oligonucleotide and consensus AP2 sequence as probes. DNase I footprinting assay was performed using the wild-type and mutant human DBH 5′ proximal promoter fragments that had been prepared by PCR as the probe as described (Seo et al., 1996). After incubating ∼30,000 cpm of labeled probe with 150–200 μg of nuclear extracts from different cell lines, freshly diluted DNase I (1.5–2.5 units) was added to a final volume of 40 μl and incubated for 90 sec at the room temperature. The precise amount of DNase I was empirically determined for each extract to ensure an even pattern of digested bands. For the sample without nuclear extracts, much lower amounts of DNase I (approximately one-tenth) were used. The probe DNA treated with DNase I was purified, and an aliquot (∼10–20%) of each sample was analyzed on a 6% polyacrylamide/8 m urea-sequencing gel followed by autoradiography with an intensifying screen. Location of the protected area was determined by Maxam–Gilbert sequencing of labeled probes.
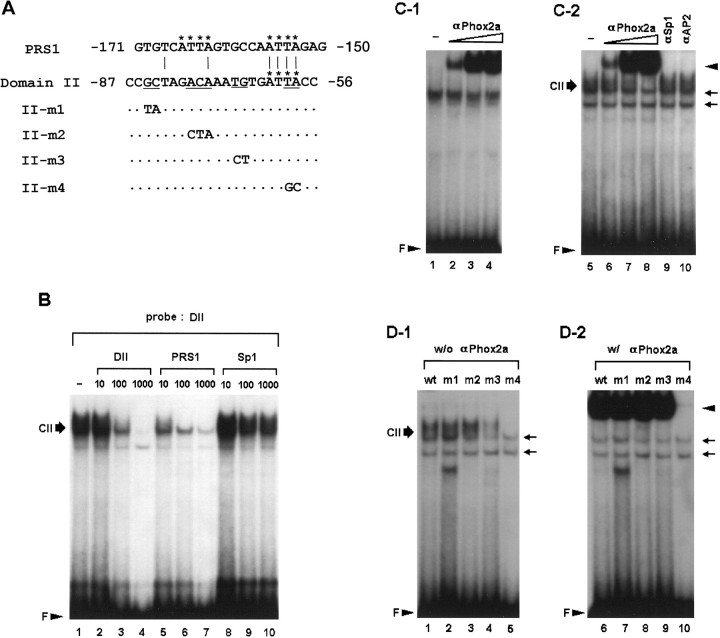
Fig. 7.
Domain II is another Phox2a-binding site.A, Nucleotide sequences of the PRS1 and wild-type and mutant of domain II oligonucleotides. The ATTA motifs are indicated byasterisks. Mutated bases are shown in the mutant oligonucleotides. Dots represent unchanged sequences.B, DNA–protein complexes formed with SK-N-BE(2)C nuclear proteins and the DII oligonucleotide were competed by molar excesses of different cold oligonucleotides as indicated above each panel. Thirty micrograms of nuclear proteins were used in each binding reaction. C, Antibody supershift assays indicate that Phox2a binds to both domain II and PRS1 sequences. Coincubation of nuclear proteins with increasing amounts of 1 μl of 10−2 (lanes 2,6), 10−1 (lanes 3, 7) and 1:3 dilution (lanes 4, 8) of Phox2a-specific antibody resulted in the generation of a supershifted band (arrowhead) in a dose-responsive manner with both PRS1 (C-1) and DII oligonucleotide (C-2). In addition, formation of domain II-specific complex, CII, was significantly diminished. In contrast, coincubation with either SP1 or AP2-specific antibody (0.1 μg each) neither generated the supershifted band nor diminished formation of CII (lanes 9, 10). Phox2a-specific antibody by itself was unable to form any complex with the radiolabeled probes (data not shown). D, Determination of nucleotide bases important for domain II-protein interaction in the absence (D-1) or presence (D-2) of Phox2a-specific antibody. m1 and m2 probes formed DNA–protein complexes as efficiently as the wild-type DII probe. m3 probe showed a limited ability to form complexes, and m4 probe shows the most severe defect for forming specific complexes (lanes 4, 5). In the presence of 1 μl of 1:3 dilution of Phox2a-specific antibody, even m3 was able to produce a robust band of supershifted complex as indicated by anarrowhead. m4 was able to generate little, if any, supershifted band, suggesting that the ATTA motif is critical for its interaction with Phox2a. Two nonspecific bands are indicated byarrows.
Southwestern blot analysis. Southwestern blotting was performed as described (Michael et al., 1988). Two sets of nuclear proteins prepared from different cell lines (100 μg each per lane) were mixed with 10 μl of 2 × sample loading buffer (4% SDS, 14% glycerol, 0.16 m Tris, pH 6.8, 0.1% brome phenol blue, 5 mm DTT) and buffer D (Dignam et al., 1983) to a final volume of 20 μl, heated to 95°C for 5 min and then separated on a denaturing SDS-10% polyacrylamide gel. The protein bands were transferred to a nitrocellulose membrane, and the nonspecific protein bands on the membrane were blocked by three washes of 45 min in 10 mm Tris, pH 7.5, 5% nonfat dry skim milk, 10% glycerol, 2.5% Nonidet P-40, 0.1 mm DTT, and 150 mm NaCl at 25°C. The membrane was then rinsed briefly in binding buffer (in mm: 10 Tris, pH 7.5, 40 NaCl, 1 EDTA, and 1 DTT, 8% glycerol, and 0.125% nonfat dry skim milk) and was incubated in 10 ml of binding buffer containing 500,000 cpm/ml end-labeled domain II oligonucleotide probe and 10 μg/ml poly(dI-dC). After incubation overnight at room temperature, the membranes were removed from the bag, washed with 10 ml of 10 mm Tris, pH 7.5, and 50 mm NaCl three times. The specific protein-domain II interactions were visualized by autoradiography. The specificity of these interactions was determined by adding a 100-fold molar excess of unlabeled domain II oligonucleotide to the second set of separate hybridization bag.
RESULTS
Transcription of the human DBH gene requires synergy of four proximal protein-binding sites
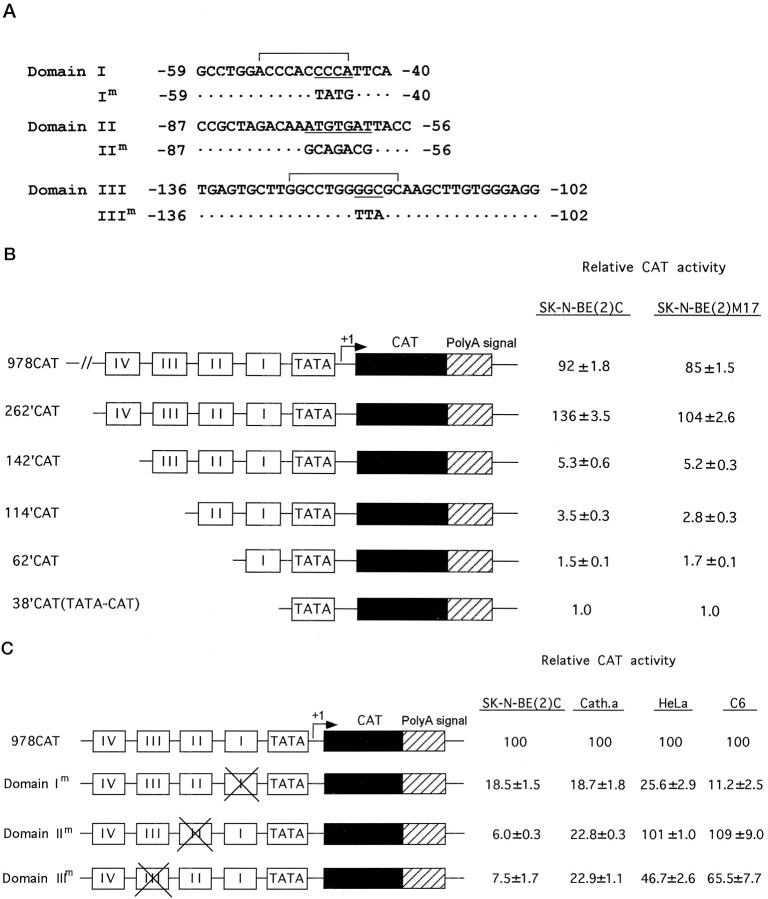
In transient transfection assays using DBH-expressing SK-N-BE(2)C and SK-N-BE(2)M17 cell lines (Fig.1B), deletion of domain IV resulted in a dramatic decrease (20- to 30-fold) of the transcription activity in both cell lines. Further deletion of domains III, II, and I resulted in a progressive decrease of the promoter activity, indicating that all three domains may act as positive regulators of the DBH promoter function in DBH-positive cells, but the proportional decrease in promoter activity was minor (30–50%) for successive deletions. Base substitutions within each motif were then introduced in the context of the intact 978 bp DBH promoter and examined for effects on promoter activity in both DBH-positive and DBH-negative cells (Fig. 1C). The CAT activity driven by the intact 978 bp DBH promoter in DBH-positive cell lines was much higher than that in DBH-negative cell lines (typically >10-fold; Ishiguro et al., 1993; Yang et al., 1998a), but was given the relative value of 100 in each cell line to compare the relative effect of individual mutation on the promoter activity. Base substitutions within domain I, II, or III diminished most (>80%) promoter activity in DBH-positive cells, suggesting that the proximal cis-elements activate DBH transcription in an interdependent manner. Mutation of domain I equally diminished most of DBH promoter activity in DBH-negative cell lines, whereas domain III mutation was severalfold less effective in the negative cell lines. Base substitutions in domain II diminished DBH promoter activity only in DBH-expressing cells, indicating that domain II is a critical noradrenergic cell-specific cis-acting element.
Fig. 1.
Interdependent activation of DBH transcription by proximal protein-binding sites in DBH-expressing cells.A, Nucleotide sequences and the locations of domains I, II, and III of the human DBH gene, as identified by DNase I footprint analysis. The Sp1-binding motif and AP2-binding motif residing in domain I and III, respectively, are indicated by brackets. In contrast, domain II did not show any significant sequence homology to known binding motifs except an ATTA motif at its 3′ side. Base substitutions within each domain that are analyzed by EMSA and transient transfection assays in this study are also indicated. B, Promoter activities of deletional DBH-CAT reporter constructs were determined by transient transfection assays in DBH-expressing SK-N-BE(2)C and SK-N-BE(2)M17 cell lines and expressed relative to that of the minimal 38′CAT construct. C, Effect of site-directed mutation of each cis-regulatory element on DBH promoter activity in the context of the upstream 978 bp sequences in DBH-expressing [SK-N-BE(2)C and CATH.a] and nonexpressing (HeLa and C6) cell lines. The normalized CAT activity driven by 978CAT in each cell line was set to 100 to compare the effect of each mutation on cell-specific promoter function of the DBH upstream sequence. The relative values are presented as mean ± SEM values from six to eight independent samples. Base substitutions in domain II diminished DBH promoter activity exclusively in DBH-expressing cells, virtually rendering the upstream DBH sequence a nonspecific promoter.
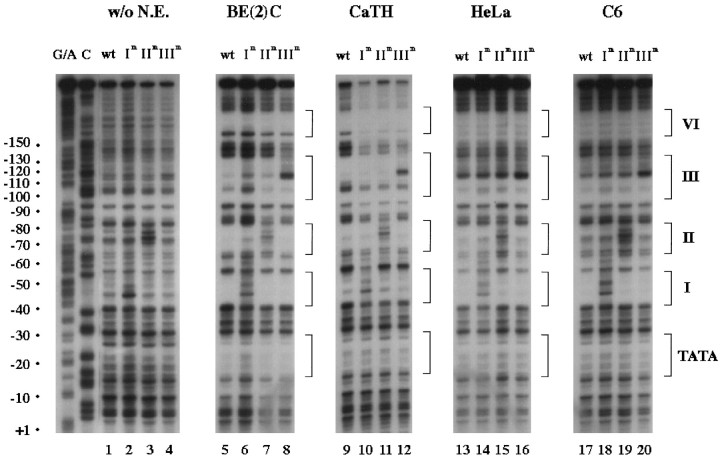
Using different nuclear extracts, we next performed DNase I footprint analyses of the wild-type and mutant promoters (Fig.2) to address (1) whether there is a positive correlation between promoter function and DNA–protein interaction and (2) whether cognate nuclear factors synergistically bind to these proximal protein-binding sites. Consistent with our previous report (Seo et al., 1996), patterns of DNA–protein interaction appeared to be significantly different between DBH-positive and DBH-negative cell lines in this study using multiple cell lines; a hypersensitive site at −161 bp appeared only with DBH-positive extracts, and footprinting at domains II and III was much more evident with DBH-positive extracts (Fig. 2, compare lanes 5and 9 with 13 and 17). Mutation of each motif specifically blocked footprinting at that site, demonstrating a direct correlation between promoter function and DNA–protein interaction at each motif. Mutation of one site did not impair DNA–protein interactions at other sites, including domain IV, suggesting that the transcription factors bind to the corresponding sites independently of each other. This conclusion was further supported by additional footprinting experiments using suboptimal amounts of nuclear extracts, which protected these domains only incompletely (data not shown).
Fig. 2.
Protein factors bind independently to domains I, II, III, and IV. Nuclear extracts from SK-N-BE(2)C, CATH.a, HeLa, and C6 cells were used for DNase I footprinting analyses of the wild-type and mutant upstream promoters of the human DBH gene. The coding strand probes were prepared using wild-type (lane 1), domain Im (lane 2), domain IIm (lane 3), and domain IIIm (lane 4) mutant constructs. Each 32P-labeled probe was digested with DNase I in the presence of each nuclear extract. The TATA box and four footprinted domains are denoted by brackets at the right side of the panel. The patterns of DNA–protein interaction at these domains appear to be conserved among DBH-expressing cell lines [SK-N-BE(2)C and CATH.a] and significantly differed from those of nonexpressing (HeLa and C6) cell lines.
Domain I interacts with the transcription factor Sp1 without cell type specificity
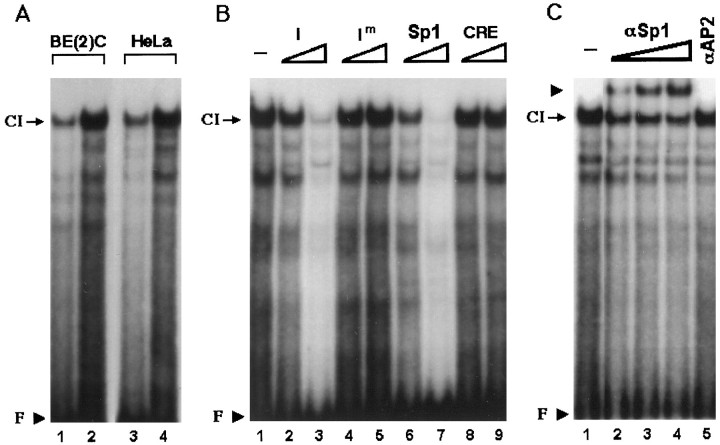
In EMSA, nuclear extracts prepared from SK-N-BE(2)C and HeLa cells formed a similar complex (CI) with an oligonucleotide representing domain I, indicating that domain I interacts with nuclear proteins in both DBH-positive and DBH-negative cells (Fig.3A). This interaction of domain I with protein factor(s) in DBH-positive and DBH-negative nuclear extracts is consistent with its general promoter function as identified in Figure 1C. In a competition assay, a 100-fold excess of cold domain I oligonucleotide almost completely abolished formation of CI, but its mutant form (Im) or the CRE oligonucleotide did not affect it at all (Fig. 3B), demonstrating that CI is a sequence-specific complex. Molar excesses of the consensus Sp1 motif competed formation of CI with a slightly higher efficiency than that of domain I oligonucleotide (Fig. 3B, compare lanes 2 with 6 and 3with 7). In addition, coincubation of SK-N-BE(2)C and HeLa nuclear extracts with specific antibody against Sp1, but not with that against AP2, diminished formation of CI and produced a new supershifted complex in a dose-dependent manner (Fig. 3C; data not shown). In addition, coincubation with different concentrations of antibodies against Sp3 and Sp4 neither diminished the complex C1 nor generated a supershifted band (data not shown).
Fig. 3.
Domain I is an Sp1-binding site and interacts with the transcription factor Sp1 without cell type-specificity.A, 20 bp oligonucleotide containing domain I sequence was radiolabeled and incubated with 10 μg (lanes 1,3) or 20 μg (lanes 2,4) of SK-N-BE(2)C or HeLa nuclear extracts. A sequence-specific complex (CI) was formed by both nuclear extracts. Unbound free probe (F) is indicated by an arrowhead. B, Complex CI was competed by cold domain I (I) or Sp1 oligonucleotide but not by the mutant form of domain I (Im) or the CRE oligonucleotide, indicating that CI is a sequence-specific complex. For competition, 10-fold (lanes 2, 4, 6,8) or 100-fold (lanes 3,5, 7, 9) molar excesses of cold oligonucleotides were added to the reaction mixture containing 20 μg of SK-N-BE(2)c nuclear extracts before the addition of the radiolabeled probe. C, Twenty micrograms of SK-BE(2)C nuclear extracts were coincubated with 0.03 (lane 2), 0.1 (lane 3), or 0.3 (lane 4) μg of Sp1-specific antibody or with 0.1 μg (lane 5) of AP2-specific antibody. Coincubation with Sp1 antibody but not with AP2 antibody diminished formation of CI and resulted in formation of a supershifted band (arrowhead) in a dose-responsive manner.
Domain II interacts with cognate protein factor(s) in a cell-specific manner
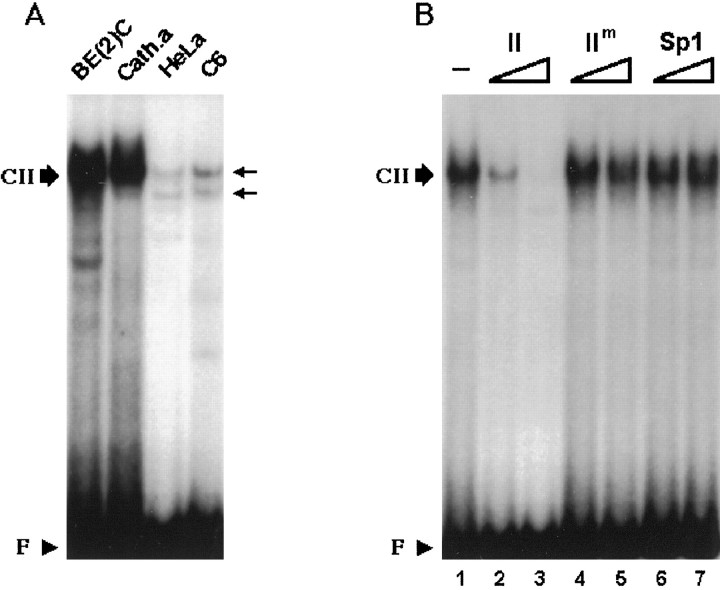
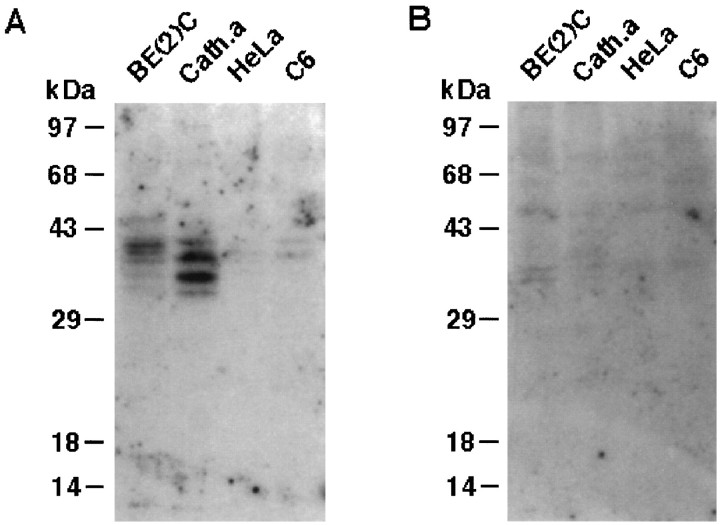
In contrast to domain I, DNase I footprint analysis suggests that domain II selectively interacts with nuclear proteins from noradrenergic cells (Fig. 2). As shown in Figure4A, nuclear extracts from DBH-positive cells robustly formed the complex CII. In competition assays (Fig. 4B), molar excesses of cold domain II oligonucleotide, but not its mutant form or unrelated Sp1 oligonucleotide, abolished formation of CII, strongly suggesting that it represents a sequence-specific complex. To further test whether the cognate protein factor(s) of domain II exist in a noradrenergic-specific manner, Southwestern analysis was performed using nuclear proteins prepared from DBH-positive and DBH-negative cells (Fig. 5). This analysis demonstrated that a few nuclear protein factors interact with domain II. Two protein bands of 39 and 40 kDa were detected in SK-N-BE(2)C cells, and those of 38 and 35 kDa were evident in CATH.a cells (Fig.5A). In contrast, nuclear proteins from DBH-negative cells did not show prominent signals. A 100-fold molar excess of cold domain II oligonucleotide abolished most of the signals (Fig. 5B), suggesting that the bands represent sequence-specific associations between domain II and cognate protein factors.
Fig. 4.
Domain II, a potentially novelcis-regulatory element, interacts with nuclear protein(s) in a cell-specific manner. A, 22 bp oligonucleotide containing the domain II sequence formed a complex (CII) with nuclear extracts (20 μg) from BE(2)C or CATH.a cells but not from HeLa or C6 cells, indicating that domain II interacts with nuclear proteins in a cell-specific manner.B, CII is a sequence-specific complex. Twenty micrograms of SK-BE(2)C nuclear extracts were incubated with the radiolabeled domain II oligonucleotide. We used 40-fold (lanes 2,4, 6) or 400-fold (lanes 3, 5, 7) molar excesses of indicated cold oligonucleotides for competition. CII was competed by domain II cold oligonucleotide but not by its mutant form (IIm) or Sp1 sequence.
Fig. 5.
Identification of noradrenergic-specific protein factors that interact with domain II by Southwestern blot analysis.A, One hundred micrograms of nuclear proteins extracted from the indicated cell line were separated on a denaturing polyacrylamide gel, transferred to two sets of nitrocellulose membranes, and probed with the P32-labeled domain II oligonucleotide probe (∼500,000 cpm/ml) in the absence (A) or presence (B) of a 100-fold molar excess of unlabeled domain II oligonucleotide. Two major proteins of ∼35–40 kDa were identified to specifically interact with domain II only in DBH-positive cells.
Domain III is a high-affinity AP2-binding site and preferentially interacts with nuclear proteins from noradrenergic cells
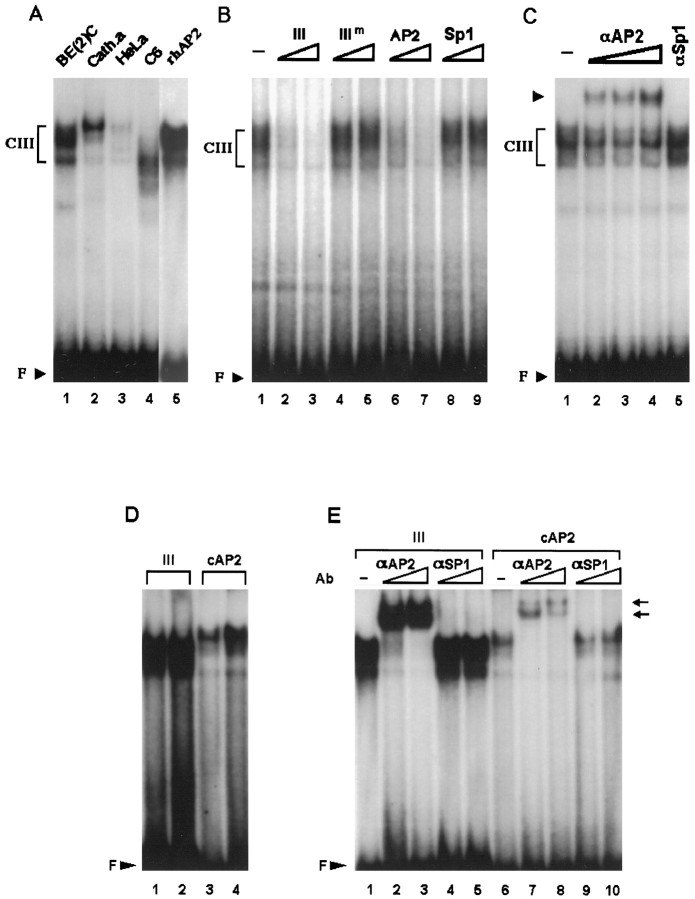
Domain III was more evidently footprinted by DBH-expressing cells (Fig. 2) and contains a sequence patch at its middle, which perfectly matches the consensus AP2 motif, 5′-GCCNNNGGC-3′ (Williams and Tjian, 1991). Figure 6Ademonstrates that nuclear extracts from SK-N-BE(2)C and CATH.a cells generated prominent complexes with an oligonucleotide that contains domain III sequence. At present, it is not clear whether these bands represent single or multiple proteins interacting with domain III, and we collectively designated these complexes as CIII. Affinity-purified human AP2 protein also generated multiple bands that migrated with mobilities similar to those formed by SK-N-BE(2)C nuclear proteins (Fig. 6A, compare lanes 1 and5). Using SK-N-BE(2)C nuclear proteins in EMSA, formation of CIII was efficiently competed by molar excesses of cold domain III or consensus AP2 oligonucleotide but not by those of mutated domain III or unrelated Sp1 sequence (Fig. 6B). These findings suggest that CIII represents sequence-specific complex(es) and that domain III is an AP2-binding site. Consistent with this, coincubation of SK-N-BE(2)C and CATH.a nuclear extracts with specific antibody against AP2, but not with that against Sp1, significantly diminished the DNA–protein complex(es) and generated a supershifted band, demonstrating that AP2 is involved in formation of CIII (Fig.6C; data not shown). Nuclear extracts from DBH-negative cells formed either much weaker bands (HeLa) or diffuse bands with faster mobilities (C6) (Fig. 6A). However, these complex(es) were shown to contain AP2 by antibody coincubation experiments (data not shown). Our competition assay indicates that cold domain III oligonucleotide competes formation of CIII more efficiently than the consensus AP2 sequence (Fig. 6B, comparelanes 2 and 6). To test whether domain III has higher affinity for AP2 protein, we incubated the affinity-purified recombinant AP2 protein with radiolabeled domain III or consensus AP2 sequence. Figure 6D indeed shows that domain III has a significantly higher affinity for AP2 than the consensus AP2-binding oligonucleotide. In addition, coincubation with AP2-specific antibody, but not with Sp1-specific antibody, completely abolished both complexes formed by recombinant AP2 protein and generated supershifted bands (Fig. 6E), demonstrating that the AP2 protein is involved in forming the complex(es) with both the domain III and the consensus AP2 sequence.
Fig. 6.
Domain III preferentially interacts with nuclear extracts from DBH-expressing cells and binds to the transcription factor AP2. A, 32P-labeled domain III oligonucleotide was incubated with 20 μg of nuclear extracts from each cell line as indicated at the top of thelanes. Three nanograms of the recombinant human AP2 protein (rhAP2) were also used in a control experiment (lane 5). B, CIII was competed by 40-fold (lanes 2, 4, 6,8) or 400-fold (lanes 3,5, 7, 9) molar excesses of domain III cold oligonucleotide or AP2 consensus sequence but not by the mutant form of domain III (IIIm) or Sp1 sequence. Twenty micrograms of SK-N-BE(2)C nuclear extracts were used for each binding reaction. C, In supershift assays, 0.03 (lane 2), 0.1 (lane 3), or 0.3 (lane 4) μg of AP2-specific antibody were used in each reaction along with 20 μg of SK-N-BE(2)C extracts. In a control experiment (lane 5), 0.1 μg of Sp1-specific antibody was used. D, Two nanograms (lanes 1, 3) or 10 ng (lanes 2,4) of recombinant AP protein is incubated with a32P-labeled domain III oligonucleotide (lanes 1, 2) or the consensus AP2 sequence (lanes 3, 4). AP2 protein bound to domain III sequence much more strongly than to the consensus sequence.E, Incubation of the recombinant AP2 protein with 0.03 (lanes 2, 7) or 0.1 (lanes 3, 8) μg of AP2-specific antibody completely diminished the specific complex and produced supershifted bands (arrows). In control experiments, 0.03 (lanes 4, 9) or 0.1 (lanes 5, 10) μg of Sp1-specific antibody did not affect the patterns of AP2-specific complexes formed with either domain III (lanes 4, 5) or the consensus AP2 sequence (lanes 9, 10).
Domain II is a Phox2a-binding site
Our findings that domain II binds to nuclear proteins in a cell-specific manner and that its mutation is associated with a severe loss of DBH promoter function only in noradrenergic cells prompted us to characterize and identify the cognate nuclear factor(s). The nucleotide sequence of domain II is A/T-rich, and our previous sequence search did not reveal significant homology to any knowncis-acting motif (Seo et al., 1996). To determine nucleotide bases important for domain II-protein factor interaction, we performed EMSA using domain II or mutant oligonucleotides containing double or triple base substitutions at different locations as probes (Fig.7A). Mutant m1 and m2 probes containing base substitutions at the 5′ side of domain II were able to form complexes with an efficiency comparable to that of the wild-type domain II oligonucleotide with nuclear proteins prepared from SK-N-BE(2)C (Fig. 7D, lanes 1-3) or CATH.a cells (data not shown). In contrast, m3 and m4 probes no longer generated signals as prominent as the wild-type sequence (Fig.7D, lanes 4-5), indicating that nucleotides residing at the 3′ side are critical for domain II-protein interactions. The m4 probe showing the most severe defect in forming the DNA–protein complex has base substitutions within the ATTA motif of the HD-binding site at the 3′ side, raising the possibility that domain II may represent another Phox2a/Phox2b-binding site in addition to the PRS1 within domain IV (Yang et al., 1998a). To test this, we further analyzed DNA–protein interaction at domain II using competition and supershift assays. Using the DII oligonucleotide as the probe, the cold PRS1 oligonucleotide was able to compete formation of DNA–protein complex(es) even more efficiently than the cold DII oligonucleotide (Fig. 7B; lanes 1-10). These competition assays thus suggest that common nuclear factor(s) interact with PRS1 and, with a lower affinity, with domain II, presumably caused by the fact that PRS1 contains two ATTA core motifs, whereas domain II has only one such motif.
To address whether domain II-protein complex(es) contain Phox2a, a supershift assay was performed using Phox2a-specific antibody. In a control experiment using radiolabeled PRS1 oligonucleotide as the probe, coincubation with Phox2a-specific antibody generated a supershifted band in a dose–response manner as described previously (Fig. 7C, lanes 1-4; Yang et al., 1998a). When the radiolabeled domain II oligonucleotide was used as the probe, coincubation of SK-N-BE(2)C nuclear extracts with Phox2a-specific antibody diminished formation of CII and generated a supershifted band in a dose-dependent manner (Fig. 7C,lanes 6-9). In contrast, coincubation with specific antibodies against Sp1 (Fig. 7C, lane 9) or AP2 (Fig. 7C, lane 10) neither diminished CII nor generated a supershifted band. Coincubation of Phox2a-specific antibody with nuclear proteins from CATH.a or PC12 cells similarly resulted in generation of a robust supershifted band (Yang et al., 1998a; data not shown). In contrast, coincubation with preimmune serum or nuclear extracts from C6 or HeLa cells did not produce any detectable signal of supershifted band using either radiolabeled PRS1 (Yang et al., 1998a) or domain II oligonucleotide (data not shown). Taken together, these data demonstrate that Phox2a is directly involved in formation of CII.
The signals of the supershifted band were significantly stronger than those of original DNA–protein complex(es) using either PRS1 or domain II probe. In the supershift assay using wild-type and mutant domain II oligonucleotides, all probes except m4 formed a robust supershifted band, strongly suggesting that the ATTA motif is the only subregion that is essential for interaction of domain II with Phox2a (Fig.7D). Similar results were obtained using lower amounts of antibodies (data not shown). Although the m3 mutant did not itself form intact amounts of DNA–protein complex(es), it was able to generate a supershifted band with a comparable signal (Fig. 7D, comparelanes 4 and 9 with 1 and6). One interpretation for this finding is that m3 has comparable affinity to Phox2a but does not bind to other binding proteins as efficiently as the wild-type domain II. Alternatively, association of Phox2a with the specific antibody may have overcome its low affinity to m3 sequence.
Domain II upregulates the DBH minimal promoter activity in a noradrenergic-specific manner and mediates Phox2a/Phox2b-induced transcriptional activation
A single copy of domain II was subcloned in the correct orientation in front of the minimal promoter region of the DBH gene containing the TATA box and transcription start site (Fig.8A,1xII-CAT), and its transcriptional activity was examined by transient transfection assays in DBH-positive and DBH-negative cell lines. As shown in Figure 8B, 1xII-CAT drives expression of the reporter gene threefold higher than that driven by 38′CAT in DBH-expressing SK-N-BE(2)C and CATH.a cells, but not in DBH-negative HeLa and C6 cells. Furthermore, cotransfection assay shows that Phox2a or Phox2b activates the reporter gene expression driven by 1xII-CAT plasmid threefold to fourfold in DBH-negative HeLa (Fig. 8C) and C6 (data not shown) cells. In DBH-expressing cell lines, in contrast, cotransfection of Phox2a or Phox2b activated the promoter activity of 1xII-CAT construct only marginally, if at all (Fig. 8C; data not shown). These data confirm that domain II is a noradrenergic-specific promoter element and mediates Phox2a-responsive transcriptional activation. However, the promoter activity of a single copy of domain II by itself represented ∼5% of the intact DBH promoter activity in the DBH-positive cell lines (Fig. 8B). To address whether multiple copies of domain II may synergistically activate the DBH minimal promoter activity in a cell-specific manner, we next subcloned four tandem copies of domain II using the same 38′CAT plasmid (Fig.8A). The resulting plasmid, 4xII-CAT plasmid, increased the DBH minimal promoter activity by 100- to 200-fold in DBH-positive cell lines. Thus, four tandem copies of domain II exhibited at least twofold of the promoter activity of the intact DBH promoter in our transient transfection assay. Strikingly, the CAT activity driven by 4xII-CAT plasmid in DBH-negative cell lines was comparable to that of 38′CAT, demonstrating a tight cell specificity. Furthermore, cotransfection with Phox2a/Phox2b-expression plasmid increased its promoter activity by 200- to 300-fold only in DBH-negative cell lines. We conclude that domain II is another Phox2a-binding site and designate it as PRS2. These results suggest that the multiple domain II promoter may be used as a valuable tool for targeted transgene expression to noradrenergic neurons.
DISCUSSION
Our present study characterizes the potential promoter function of multiple protein-binding sites residing within the 5′ proximal area of the human DBH promoter and demonstrates that their combinatorial interplay is crucial for noradrenergic-specific transcriptional activity. These protein-binding sites have been identified as protected regions in DNase I footprinting analysis using nuclear extracts prepared from DBH-expressing SK-N-BE(2)C and DBH-negative HeLa cells (Seo et al., 1996). One of these footprinted regions (domain IV), located at −185 to −150 bp upstream of the transcription start site, is a composite promoter that contains overlapping cis-acting elements, including a CRE, YY1-binding site, and two core ATTA motifs of the HD-binding site. Our previous work showed that the HD-binding site within domain IV is a noradrenergic-specific promoter element and that paired-like HD protein factors, Phox2a and Phox2b, directly transactivate DBH transcription through this sequence (Yang et al., 1998a). The studies described here further characterize the promoter function of the proximal protein-binding sites residing between domain IV and the TATA box of the human DBH gene and identify the cognate nuclear factors that interact with these sequences.
Protected region I (domain I) is located at −58 to −40 bp in the human DBH proximal promoter region and is only 10 bases from the TATA box (Figs. 1A, 2; Seo et al., 1996). The nucleotide sequences of domain I are G/C-rich and include a 9 bp fragment at the center, 5′-ACCCACCCC-3′, the opposite sequence of which shows an 8/9 match to the consensus Sp1-binding site, 5′-(G/T)(G/A)GGC(G/T)(G/A)(G/A)(G/T)−3′ (Faisst and Meyer, 1992). Consistent with the imperfect sequence match, domain I appeared to have a slightly lower affinity for the transcription factor Sp1 than for the consensus Sp1 motif. Several lines of evidence establish that domain I is an authentic Sp1-binding site: (1) formation of DNA–protein complex (CI) was specifically competed by the consensus Sp1 motif-containing oligonucleotide and domain I oligonucleotide (Fig. 3), (2) CI is shown to contain Sp1 in antibody supershift assay (Fig. 3), and (3) the recombinant Sp1 protein binds to domain I, and the complex shows an identical mobility to that of CI (data not shown). Our mutational analyses demonstrate that the Sp1-binding site in domain I is critical for the intact DBH promoter activity. Sp1 is known to be a ubiquitous transcription factor (Kadonaga et al., 1987) and controls, by interacting with its binding site(s), numerous eukaryotic genes including housekeeping, signal pathway-induced, as well as tissue-specific genes. Based on its frequent occurrence in CpG islands, one possible mechanism underlying transcriptional control of a variety of eukaryotic genes by Sp1 may be its role in maintaining methylation-free CpG islands of active genes (Macleod et al., 1994;Graff et al., 1997). Another potential structure/function link of the Sp1-binding site is that it is mostly located in proximity to the core promoter region and, thus, may facilitate the formation of the active transcriptional machinery. In support of this, Sp1 is found to directly interact with TAFII110 (Hoey et al., 1993; Gill et al., 1994).
Intriguingly, the rat phenylethanolamine N-methyltransferase gene, encoding the last step of catecholamine biosynthesis, has been shown to be regulated by Sp1 through two Sp1-binding sites at −45 and −165 bp (Ebert and Wong, 1995). In addition, our recent analysis of the tyrosine hydroxylase promoter identified a proximal Sp1-binding site that is critically required for intact promoter activity (Yang et al., 1998b). Taken together, Sp1 may represent the first known transcription factor that coregulates transcription of all three catecholamine-specific synthesizing genes.
Domain III, at −136 to −102 bp, resides immediately downstream of the composite promoter, domain IV. This region includes a nucleotide patch, 5′-GGCCTGGGGCGC-3′, which perfectly matches the consensus AP2 site (Williams and Tjian, 1991; Faisst and Meyer, 1992). In contrast to Sp1, which is ubiquitously expressed in most cells, AP2 is primarily expressed in neural crest cells and their major derivatives in a cell type-specific manner (Mitchell et al., 1991). In addition, our EMSA and DNase I footprint analyses demonstrate that domain III is more efficiently bound by nuclear proteins prepared from DBH-expressing SK-N-BE(2)C and CATH.a than by those from DBH-negative HeLa and C6 cells (Fig. 2). However, AP2 was originally purified and cloned from HeLa cells (Williams et al., 1988) and, thus, we previously proposed that a protein factor other than AP2 may bind to domain III and regulate DBH transcription (Seo et al., 1996). Nevertheless, our present studies demonstrate that AP2 interacts with the domain III sequence, because the specific DNA–protein complex was eliminated by the unlabeled consensus AP2-binding site and supershifted by AP2-specific antibody (Fig. 6). Antibody coincubation experiments showed that the DNA–protein complex(es) formed by HeLa and C6 nuclear extracts also contain, albeit to a lesser extent, AP2 (data not shown). Base substitutions within domain III strikingly reduced the DBH promoter activity in DBH-positive cell lines and reduced it modestly in DBH-negative cells (Fig. 1C). Collectively, these data indicate that HeLa and C6 cells express low levels of AP2 compared with DBH-positive cells. Consistent with these results, Greco et al. (1995)have shown that the recombinant AP2 protein binds to the corresponding motif of the rat DBH promoter and an AP2 expression vector preferentially transactivates the DBH promoter activity in DBH-negative HepG2 cells. Our EMSA and competition assays (Fig. 6) indicate that domain III has a significantly higher affinity (>5 times) for AP2 than a well characterized AP2 consensus motif residing in the human metallothioneine-IIA gene (Williams et al., 1988; Williams and Tjian, 1991). We conclude that the transcription factor AP2 is abundantly expressed in noradrenergic cells and activates transcription of the human DBH gene via interaction with this high-affinity AP2-binding site residing within domain III.
Domain II is identified as a positive noradrenergic-specificcis-element in that its mutation diminishes the DBH promoter activity exclusively in DBH-expressing cell lines. DNA binding studies using EMSA, Southwestern, and DNase I footprint analyses show that domain II interacts with nuclear factor(s) in a noradrenergic-specific manner (Figs. 4, 5, 7). We originally proposed that domain II may represent a novel cis-regulatory element because it did not show any significant homology to known protein-binding motifs (Seo et al., 1996). Domain II contains an ATTA motif at the 3′ side but beyond these four bases does not bear any sequence homology with the PRS1 residing within domain IV. However, several lines of evidence support the identification of domain II as another Phox2a/Phox2b-binding site: (1) DNA–protein complex (CII) formed by domain II oligonucleotide is specifically competed by the PRS1 sequence, (2) coincubation of nuclear extracts from DBH-positive cells with Phox2a-specific antibody produces supershifted complexes using domain II oligonucleotide as the probe, (3) domain II oligonucleotides containing mutation within the ATTA motif was unable to form complex CII, (4) the same mutant oligonucleotide does not produce the supershifted complex in the presence of the Phox2a-specific antibody, and (5) cotransfection assay shows that domain II confers Phox2a- or Phox2b-mediated transactivation in the DBH-negative cells. In a previous study, forced expression of Phox2a or Phox2b in DBH-negative cell lines activated the reporter gene expression driven by a single copy of the PRS1 approximately fourfold while it activated that driven by the intact DBH promoter 10- to 15-fold (Yang et al., 1998a). Similarly, coexpression of Phox2a or Phox2b stimulated reporter expression driven by 1xII-CAT plasmid ∼3.5-fold in DBH-negative HeLa or C6 cells (Fig. 7; data not shown). Taken together, we conclude that Phox2a/Phox2b activates DBH transcription through at least two contacts on the DBH upstream promoter.
The finding that PRS1 and domain II, both critical for noradrenergic-specific DBH promoter activity, are binding sites for Phox2a/Phox2b (Figs. 7, 8; Yang et al., 1998a) was unexpected, because these two sequence elements do not show any sequence identity beyond the single ATTA motif at the 3′ side (Fig. 7A). Because this motif may occur quite often (statistically once in every 128 bases in either orientation), it is unlikely that this motif by itself can determine noradrenergic-specific transcriptional activity of the DBH gene. Several different, but mutually nonexclusive, mechanisms may explain this rather surprising finding. First, the DBH promoter contains multiple binding sites for Phox2a/Phox2b, and their simultaneous occupation may be critical for intact transcriptional activity. In line with this, PRS1 contains two ATTA motifs, both of which are important for the intact promoter activity (Yang et al., 1998a). Thus, PRS1 may in fact represent two Phox2a/Phox2b-binding sites that may interact with Phox2a/Phox2b alternately or simultaneously. If so, the proximal DBH promoter may include three or even more PRSs, and combinatorial action of these sequences may be critical for DBH transcription. Second, as-yet-unidentified cofactor(s) may modulate DNA-binding properties, e.g., affinity and/or specificity, of Phox2a/Phox2b in vivo. This possibility is supported by our finding that, with both PRS1 and domain II, Phox2a forms DNA–protein complex(es) much more robustly when it is bound by a Phox2a-specific antibody. Many HD proteins are known to have relaxed DNA-binding specificities in vitro despite their functional specificity in vivo (Hayashi et al., 1990; McGinnis and Krumlauf, 1992). Several such cofactors, including extradenticle and αFtz-F1, have recently been identified to modify DNA-binding specificity or selectivity of several HD protein factors (Chan et al., 1994; van Dijk and Murre, 1994; Chan and Mann, 1996; Guichet et al., 1997; Yu et al., 1997).
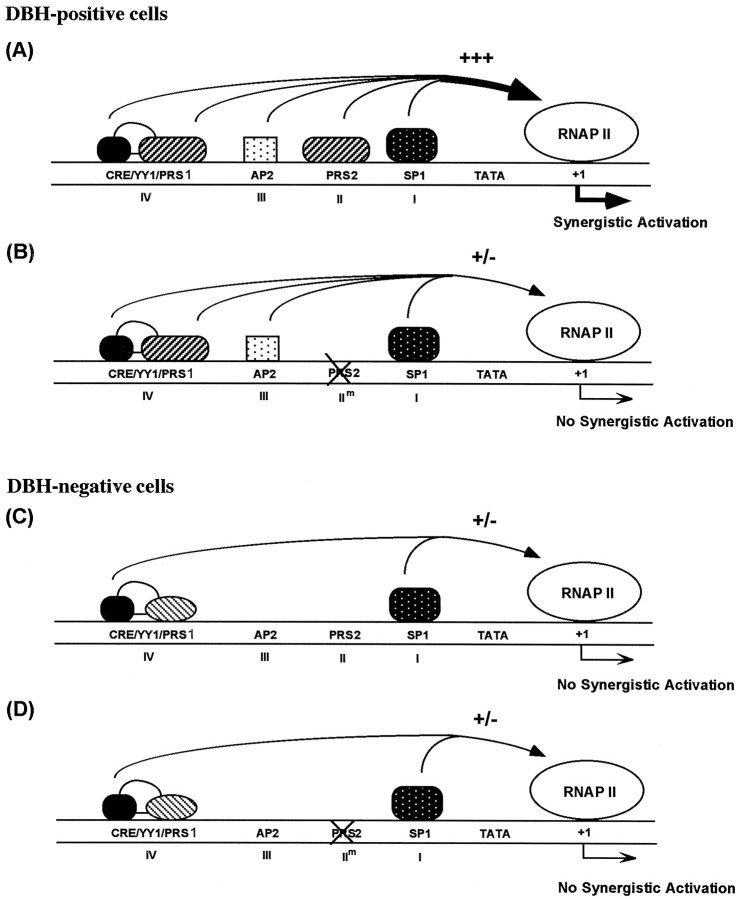
A schematic diagram of DBH promoter activation via multiple protein-binding sites is depicted in Figure9. Based on DNase I footprinting analyses using the upstream promoters containing mutations at each site, it is unlikely that synergistic binding of the cognate transcription factors to their binding sites underlies their concerted activation of DBH transcription (Fig. 2). Rather, we infer that protein–protein interactions between the nuclear factors, their putative cofactors, and/or the components of the basal RNA polymerase II machinery may be pivotal for intact transcription of the DBH gene. In this model, lack of any one component diminishes transcriptional activation as when mutation of PRS2 abolishes most of the promoter activity in noradrenergic cells (Fig. 9B). In DBH-negative cells, lack of a cell-specific nuclear factor(s), e.g., Phox2a and AP2, causes RNA polymerase II machinery to be largely inactive for the DBH gene (Fig.9C,D). In support of synergistic activation of DBH transcription by multiple cis-regulatory elements, four tandem copies of domain II increased the minimal promoter activity by 100- to 200-fold in DBH-positive cell lines, which far exceeds the additive activity of individual four copies of domain II (Fig. 8). Remarkably, the cell specificity of this multiple domain II promoter was not compromised at all, suggesting that it may be of great value for targeted expression of therapeutic genes to noradrenergic neurons. We are currently testing the promoter activity and cell-specificity with additional copies of domain II.
Fig. 9.
Proposed interaction among multiple transcription factors in the 260 bp upstream region of the human DBH gene.A, In DBH-positive cells, transcription factors interacting with domains I–IV, including cell type specific factors (e.g., Phox2a, Phox2b, and AP2) and general transcription factors (e.g., CREB and Sp1), synergistically activate the DBH transcription (+++). B, When any of these domains, domain II motif in this example, is mutated, the synergism is disrupted and robust transcription cannot occur (+/−) although DNA–protein interactions at the other sites are intact (Fig. 2). C,D, In DBH-negative cells, cell-specific Phox2a/Phox2b and AP2 are absent or present in lower amount compared with DBH-positive cells, respectively. Accordingly, synergistic activation of DBH gene transcription cannot occur whether or not domain II is intact (+/−).
Most mutant mice lacking DBH die in utero, indicating that proper expression of DBH is critical for fetal development and survival (Thomas et al., 1995). Strikingly, clinical scientists have reported that DBH is deficient in some patients and have come to recognize that DBH deficiency is a genetic disorder of cardiovascular regulation (Robertson et al., 1986; Man in’t Veld et al., 1987; for review, seeRobertson, 1995). Neonates with DBH deficiency had episodic hypothermia, hypoglycemia, and hypotension. Survivors cope relatively well until late childhood, when overwhelming orthostatic hypotension profoundly limits their activities. At present, mechanisms underlying DBH deficiency are not known. Although this disease may be associated with mutation of the coding sequence that leads to expression of nonfunctional protein, it is also possible that mutation of critical promoter element(s) or a cognate protein factor may cause this genetic disease.
In summary, our results suggest that multiple nuclear factors, including general transcription factors (Sp1 and CREB) as well as cell-specific factors (AP2, Phox2a, and Phox2b) bind to proximalcis-acting elements and synergistically activate transcription of the DBH gene in noradrenergic cells. We have previously shown that the upstream region spanning from −486 to 263 bp of the human DBH gene preferentially exhibits a silencer function in DBH-negative cells, and this region may contain several cell-specific silencer motifs (Ishiguro et al., 1993, 1995; Kim et al., 1998). Furthermore, transgenic mice experiments showed that the upstream region between −1.1 and −0.6 kb of the human DBH gene is required for expression of the reporter gene in vivo (Hoyle et al., 1994). Thus, DBH gene regulation appears to require multiple layers of regulatory mechanisms to ensure its accurate expression pattern in quite limited locations in the nervous system, and promises to be an excellent system to study neural-specific gene regulation in the normal and diseased state of the brain, as well as in response to environmental stimuli.
Footnotes
This work was supported by National Institutes of Health Grant MH48866 and the Research Contingency Fund from the University of Tennessee. We thank Drs. T. S. Nowak Jr and E. A. Park for critical reading of this manuscript.
Correspondence should be addressed to Dr. Kwang-Soo Kim, Department of Neurology, University of Tennessee, College of Medicine, 855 Monroe Avenue, Memphis, TN 38163.
REFERENCES
- 1.Afar R, Silverman R, Aguanno A, Albert VR. Positive and negative elements contribute to the cell-specific expression of the rat dopamine β-hydroxylase gene. Mol Brain Res. 1996;36:79–92. doi: 10.1016/0169-328x(95)00247-p. [DOI] [PubMed] [Google Scholar]
- 2.Chan SK, Mann RS. A structural model for a homeotic protein-extradenticle-DNA complex accounts for the choice of HOX protein in the heterodimer. Proc Natl Acad Sci USA. 1996;28:5223–5228. doi: 10.1073/pnas.93.11.5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan SK, Jaffe L, Capovilla M, Botas J, Mann RS. The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell. 1994;78:603–615. doi: 10.1016/0092-8674(94)90525-8. [DOI] [PubMed] [Google Scholar]
- 4.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ebert SN, Wong DL. Differential activation of the rat phenylethanolamine N-methyltransferase gene by Sp1 and Egr-1. J Biol Chem. 1995;270:17299–17305. doi: 10.1074/jbc.270.29.17299. [DOI] [PubMed] [Google Scholar]
- 6.Faisst S, Meyer S. Compilation of vertebrate-encoded transcription factors. Nucleic Acids Res. 1992;20:3–26. doi: 10.1093/nar/20.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedman S, Kaufman S. 3,4-Dihydroxyphenylethylamine β-hydroxylase. J Biol Chem. 1965;240:4763–4773. [PubMed] [Google Scholar]
- 8.Gill G, Pascal E, Tseng ZH, Tjian R. A glutamine-rich hydrophobic patch in transcription factor Sp-1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc Natl Acad Sci USA. 1994;91:192–196. doi: 10.1073/pnas.91.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodman RH. Regulation of neuropeptide gene expression. Annu Rev Neurosci. 1990;13:111–127. doi: 10.1146/annurev.ne.13.030190.000551. [DOI] [PubMed] [Google Scholar]
- 10.Graff JR, Herman JG, Myohanen S, Baylin SB, Vertino PM. Mapping patterns of CpG island methylation in normal and neoplastic cell implicates both upstream and downstream regions in de novo methylation. J Biol Chem. 1997;272:22322–22329. doi: 10.1074/jbc.272.35.22322. [DOI] [PubMed] [Google Scholar]
- 11.Greco D, Zellmer EJ, Zhang Z, Lewis EJ. Transcription factor AP-2 regulates expression of the dopamine β-hydroxylase gene. J Neurochem. 1995;65:510–516. doi: 10.1046/j.1471-4159.1995.65020510.x. [DOI] [PubMed] [Google Scholar]
- 12.Guichet A, Copeland JWR, Erdelyi M, Hlousek D, Zavorszky P, Ho J, Brown S, Percival-Smith A, Krause HM, Ephrussi A. The nuclear receptor homologue Ftz-Fq and the homeodomain protein Ftz are mutually dependent cofactors. Nature. 1997;385:548–552. doi: 10.1038/385548a0. [DOI] [PubMed] [Google Scholar]
- 13.Hayashi S, Scott MP. What determines the specificity of action of Drosophila homeodomain proteins? Cell. 1990;63:883–894. doi: 10.1016/0092-8674(90)90492-w. [DOI] [PubMed] [Google Scholar]
- 14.Hoey T, Weinzierl RO, Gill G, Chen JL, Dynlacht BD, Tjian R. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell. 1993;72:247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- 15.Hoyle GW, Mercer EH, Palmiter RD, Brinster RL. Cell-specific expression from the human dopamine β-hydroxylase promoter in transgenic mice is controlled via a combination of positive and negative regulatory elements. J Neurosci. 1994;14:2455–2463. doi: 10.1523/JNEUROSCI.14-05-02455.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishiguro H, Kim KT, Joh TH, Kim KS. Neuron-specific expression of the human dopamine β-hydroxylase gene requires both cAMP response element and a silencer region. J Biol Chem. 1993;268:17987–17994. [PubMed] [Google Scholar]
- 17.Ishiguro H, Kim KS, Joh TH. Identification of a negative regulatory element in the 5′-flanking region of the human dopamine β-hydroxylase gene. Mol Brain Res. 1995;34:251–261. doi: 10.1016/0169-328x(95)00170-w. [DOI] [PubMed] [Google Scholar]
- 18.Kadonaga JT, Carner KR, Masiarz FR, Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987;24:1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- 19.Kim HS, Yang C, Kim KS. The cell-specific silencer region of the human dopamine β-hydroxylase gene contains several negative regulatory elements. J Neurochem. 1998;71:41–50. doi: 10.1046/j.1471-4159.1998.71010041.x. [DOI] [PubMed] [Google Scholar]
- 20.Kim KS, Ishiguro H, Tinti C, Wagner J, Joh TH. The cAMP-dependent protein kinase regulates transcription of the dopamine β-hydroxylase gene. J Neurosci. 1994;14:7200–7207. doi: 10.1523/JNEUROSCI.14-11-07200.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirshner N, Goodall M. Separation of adrenaline, noradrenaline, and hydroxytyramine by ion exchange chromatography. J Biol Chem. 1957;226:207–212. [PubMed] [Google Scholar]
- 22.Kobayashi K, Morita S, Mizuguchi T, Sawada H, Yamada K, Nagatsu I, Jujuta K, Nagatsu T. Functional and high level expression of human dopamine β-hydroxylase in transgenic mice. J Biol Chem. 1994;269:29725–29731. [PubMed] [Google Scholar]
- 23.Lamouroux A, Houhou L, Faucon Biguet N, Serck-Hanssen G, Guibert B, Icard-Liepkalns C, Mallet J. Analysis of the human dopamine β-hydroxylase promoter: transcriptional induction by cyclic AMP. J Neurochem. 1993;60:364–367. doi: 10.1111/j.1471-4159.1993.tb05861.x. [DOI] [PubMed] [Google Scholar]
- 24.Luckow B, Schutz G. CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res. 1987;15:5490. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macleod D, Charlton J, Mullins J, Bird AP. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 26.Man in’t Veld AJ, Boomsma F, Moleman P, Schalekamp MADH. Congenital dopamine-beta-hydroxylase deficiency. Lancet. 1987;2:183–187. doi: 10.1016/s0140-6736(87)90002-x. [DOI] [PubMed] [Google Scholar]
- 27.McGinnis W, Krumlauf R. Homeobox genes and axial patterning. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 28.Mercer EH, Hoyle GW, Kapur RP, Brinster RL, Palmiter RD. The dopamine β-hydroxylase gene promoter directs expression of E. coli lacZ to sympathetic and other neurons in adult transgenic mice. Neuron. 1991;7:703–716. doi: 10.1016/0896-6273(91)90274-4. [DOI] [PubMed] [Google Scholar]
- 29.Michael N, Spector D, Mavromara-Nazos P, Kristie TM, Roizman B. The DNA-binding properties of the major regulatory protein a4 of herpes simplex viruses. Science. 1988;239:1531–1533. doi: 10.1126/science.2832940. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell PJ, Timmons PM, Hebert JM, Rigby PWJ, Tjian R. Transcription factor AP-2 is expressed in neural crest cell lineages during mouse embryogenesis. Genes Dev. 1991;5:105–119. doi: 10.1101/gad.5.1.105. [DOI] [PubMed] [Google Scholar]
- 31.Morin X, Cremer H, Hirsch MR, Kapur RP, Goridis C, Brunet JF. Defects in sensory and autonomic ganglia and absence of locus coeruleus in mice deficient for the homeobox gene Phox2a. Neuron. 1997;18:411–423. doi: 10.1016/s0896-6273(00)81242-8. [DOI] [PubMed] [Google Scholar]
- 32.Morita S, Kobayashi K, Mizuguchi T, Yamada K, Nagatsu I, Titani K, Fujita K, Hidaka H, Nagatsu T. The flanking region of the human dopamine β-hydroxylase gene promotes neuron subtype-specific gene expression in the central nervous system of transgenic mice. Mol Brain Res. 1993;17:239–244. doi: 10.1016/0169-328x(93)90007-c. [DOI] [PubMed] [Google Scholar]
- 33.Pattyn A, Morin X, Cremer H, Goridis C, Brunet JF. Expression and interactions of the two closely related homeobox genes Phox2a and Phox2b during neurogenesis. Development. 1997;124:4065–4075. doi: 10.1242/dev.124.20.4065. [DOI] [PubMed] [Google Scholar]
- 34.Robertson D. Genetics and molecular biology of hypotension. Curr Opin Nephrol Hypertens. 1995;3:13–24. doi: 10.1097/00041552-199401000-00002. [DOI] [PubMed] [Google Scholar]
- 35.Robertson D, Goldberg MR, Onrot J, Hollister AS, Wiley R, Thompson JG, Robertson RM. Isolated for impaired β-hydroxylation of dopamine. N Engl J Med. 1986;314:1494–1497. doi: 10.1056/NEJM198606053142307. [DOI] [PubMed] [Google Scholar]
- 36.Roesler WJ, Vandenbark GR, Hanson RW. Cyclic AMP and the induction of eukaryotic gene transcription. J Biol Chem. 1988;263:9063–9066. [PubMed] [Google Scholar]
- 37.Seo H, Yang C, Kim HS, Kim KS. Multiple protein factors interact with the cis-regulatory elements of the proximal promoter in a cell-specific manner and regulate transcription of the dopamine β-hydroxylase gene. J Neurosci. 1996;16:4102–4112. doi: 10.1523/JNEUROSCI.16-13-04102.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaskus J, Greco D, Asnani LP, Lewis EJ. A bifunctional genetic regulatory element of the rat dopamine β-hydroxylase gene influences cell type specificity and second messenger-mediated transcription. J Biol Chem. 1992;267:18821–18830. [PubMed] [Google Scholar]
- 39.Shaskus J, Zellmer E, Lewis EJ. A negative regulatory element in the rat dopamine β-hydroxylase gene contributes to the cell type specificity of expression. J Neurochem. 1995;64:52–60. doi: 10.1046/j.1471-4159.1995.64010052.x. [DOI] [PubMed] [Google Scholar]
- 40.Suri C, Fung BP, Tischler AS, Chikaraishi DM. Catecholaminergic cell lines from the brain and adrenal glands of tyrosine hydroxylase-SV40 T antigen transgenic mice. J Neurosci. 1993;13:1280–1291. doi: 10.1523/JNEUROSCI.13-03-01280.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas SA, Matsumoto AM, Palmiter RD. Noradrenaline is essential for mouse fetal development. Nature. 1995;374:643–646. doi: 10.1038/374643a0. [DOI] [PubMed] [Google Scholar]
- 42.van Dijk MA, Murre C. extradenticle raises the DNA binding specificity of homeotic selector gene products. Cell. 1994;78:617–624. doi: 10.1016/0092-8674(94)90526-6. [DOI] [PubMed] [Google Scholar]
- 43.Williams T, Adman A, Luscher B, Tjian R. Cloning and expression of AP-2, a cell-type-specific transcription factor that activates inducible enhancer elements. Genes Dev. 1988;2:1557–1569. doi: 10.1101/gad.2.12a.1557. [DOI] [PubMed] [Google Scholar]
- 44.Williams T, Tjian R. Analysis of the DNA-binding and activation properties of the human transcription factor AP-2. Genes Dev. 1991;5:670–682. doi: 10.1101/gad.5.4.670. [DOI] [PubMed] [Google Scholar]
- 45.Yang C, Kim HS, Seo H, Kim CH, Brunet JF, Kim KS. Paired-like homeodomain proteins, Phox2a and Phox2b, are responsible for cell-specific transcription of the dopamine β-hydroxylase gene. J Neurochem. 1998a;71:1813–1826. doi: 10.1046/j.1471-4159.1998.71051813.x. [DOI] [PubMed] [Google Scholar]
- 46.Yang C, Kim HS, Seo H, Kim KS. Identification and characterization of potential cis-regulatory elements governing transcriptional activation of the rat tyrosine hydroxylase gene. J Neurochem. 1998b;71:1358–1368. doi: 10.1046/j.1471-4159.1998.71041358.x. [DOI] [PubMed] [Google Scholar]
- 47.Yu Y, Li W, Su K, Yussa M, Han W, Perrimon M, Pick L. The nuclear hormone receptor Ftz-F1 is a cofactor for the Drosophila homeodomain protein Ftz. Nature. 1997;385:552–555. doi: 10.1038/385552a0. [DOI] [PubMed] [Google Scholar]