Abstract
We describe a novel synaptic vesicle protein called SVOP that is distantly related to the synaptic vesicle proteins SV2A, SV2B, and SV2C (20–22% sequence identity). Both SVOP and SV2 contain 12 transmembrane regions. However, SV2 is highly glycosylated, whereas SVOP is not. Databank searches revealed that closely related homologs of SVOP are present in Caenorhabditis elegans andDrosophila (48% sequence identity), suggesting that SVOP is evolutionarily ancient. In contrast, no invertebrate orthologs of SV2 were detected. The sequences of SVOP and SV2 exhibit homology with transport proteins, in particular with mammalian organic cation and anion transporters. SVOP and SV2 are more distantly related to eukaryotic and bacterial phosphate, sugar, and organic acid transporters. SVOP is expressed at detectable levels only in brain and endocrine cells where it is primarily localized to synaptic vesicles and microvesicles. SVOP is present in all brain regions, with particularly high levels in large pyramidal neurons of the cerebral cortex. Immunocytochemical staining of adjacent rat brain sections for SVOP and SV2 demonstrated that SVOP and SV2 are probably coexpressed in most neurons. Although the functions of SV2 and SVOP remain obscure, the evolutionary conservation of SVOP, its hydrophobic nature, and its homology to transporters strongly support a role in the uptake of a novel, as yet unidentified component of synaptic vesicles. Thus synaptic vesicles contain two classes of abundant proteins with 12 transmembrane regions that are related to transporters, nonglycosylated SVOP and highly glycosylated SV2, suggesting that the transport functions of synaptic vesicles may be more complex than currently envisioned.
Keywords: synaptic vesicle protein, SV2, transport protein, synaptic-like microvesicles, chromaffin granules, synapse structure
The synaptic vesicle protein SV2 was identified by a monoclonal antibody that was raised against synaptic vesicles isolated from the electric organ ofDiscopyge ommata (Buckley and Kelly, 1985). SV2 is a transmembrane glycoprotein that constitutes a conserved and abundant component of synaptic vesicles in all vertebrate species tested. In addition, SV2 is found on endocrine secretory granules. SV2 is composed of a protein backbone of ∼80 kDa that is highly glycosylated. Glycosylation of SV2 differs between synaptic vesicles and endocrine secretory granules. SV2 on secretory granules is more extensively modified than is synaptic vesicle SV2 (Buckley and Kelly, 1985). In synaptic vesicles from fish electric organs, SV2 was proposed to be a proteoglycan containing keratan sulfate (Scranton et al., 1993), although this has not yet been confirmed for mammalian SV2. Independent of the nature of the glycosylation of SV2, it represents the most glycosylated component of synaptic vesicles in vertebrates.
By the use of the monoclonal SV2 antibody, cDNA clones encoding SV2 were isolated from rat, bovine, and elasmobranch brains (Bajjalieh et al., 1992; Feany et al., 1992; Gingrich et al., 1992; Bindra et al., 1993). The sequence of SV2 revealed that it contains 12 transmembrane regions and exhibits significant homology to transport proteins. SV2 was observed to be similar to bacterial sugar transporters that use a proton gradient as a driving force. This led to the hypothesis that SV2 functions to transport an unknown substrate into synaptic and secretory vesicles (Bajjalieh et al., 1992; Feany et al., 1992; Gingrich et al., 1992). Two additional isoforms of SV2 were identified more recently, and the three forms of SV2 were named SV2A, SV2B, and SV2C (Bajjalieh et al., 1993; R. Janz and T. C. Südhof, unpublished observations). Studies of the distribution of SV2A and SV2B in brain byin situ hybridization and immunocytochemistry revealed that SV2A is ubiquitously present in most synapses (Bajjalieh et al., 1994). SV2B has a more restricted pattern of expression that partially overlaps with that of SV2A. The distribution of SV2C is unknown. The expression of SV2A or SV2B does not correlate with known properties of neurons, such as neurotransmitter type, indicating that different SV2 isoforms are not directly associated with distinct functional characteristics of neurons.
All three SV2 isoforms (SV2A, SV2B, and SV2C) react with the monoclonal SV2 antibody, are localized to synaptic vesicles, and are similarly homologous to transport proteins (Bajjalieh et al., 1993; Janz and Südhof, unpublished observations). This suggests that SV2A, SV2B, and SV2C are functionally similar. Although the abundant presence of SV2 on synaptic vesicles indicated an important function, no specific role for SV2 has been discovered. Initial proposals that SV2 proteins serve as neurotransmitter transporters (Feany et al., 1992) were made unlikely by the ubiquitous expression of SV2 proteins that is indicative of a function common to all synaptic vesicles (Bajjalieh et al., 1994). Furthermore, no SV2 genes were identified in invertebrates. The lack of SV2-related sequences in invertebrates was particularly puzzling in view of the abundance and conservation of SV2 in vertebrates. Because no specific transport activity for SV2 could be identified in spite of a large effort, it is possible that SV2 isoforms were evolutionarily derived from transporters but adopted a new function as structural vesicle components. This hypothesis was supported by the extensive glycosylation of SV2, suggesting that they may represent matrix proteins for synaptic vesicles with a structural function.
Here we describe the cloning and characterization of a novel synaptic vesicle protein that is similar to SV2. Because of its distant relation to SV2, we named this protein SVOP ( SVtw o-related protein) SVOP is also a component of synaptic vesicles, probably in colocalization with SV2, but is not glycosylated. SVOP may represent an evolutionary precursor of SV2 because homologous genes are found in the invertebrates Caenorhabditis elegans and Drosophila melanogaster. However, the lack of extensive glycosylation eliminates a matrix function for SVOP. Our data indicate that vertebrate synaptic vesicles contain at least two classes of proteins related to transporters, only one of which appears to be evolutionarily conserved.
MATERIALS AND METHODS
Cloning of SVOP and construction of expression vectors. A rat brain λZAP cDNA library (Stratagene, La Jolla, CA) was screened with a 690 bp PstI/BamHI fragment from mouse EST clone IMAGE #312052. Three overlapping clones were isolated and sequenced. Their assembled sequence contained an open reading frame of 548 amino acids (see Fig. 1). We combined inserts from two cDNA clones to generate a eukaryotic expression vector for full-length SVOP (pCMV-SVOP) containing a 2.05 kbEcoRI/HindIII insert in pCMV5 (Andersson et al., 1989). The bacterial expression vectors pGEX-SVOP and pMAL-SVOP encoding amino acids 2–85 of SVOP fused to glutathioneS-transferase (GST) or maltose-binding protein (MBP) were constructed in pMAL-c2 (New England Biolabs, Beverly, MA) and pGEX-KG (Guan and Dixon, 1991). pMal-Cgyr was described previously (Janz and Südhof, 1998). Expression of bacterial fusion proteins was performed as described (Li et al., 1995; Janz and Südhof, 1998). The expression vectors pCMV-myc-SV2A and -SV2B were generated by cloning the coding sequence of the rat SV2A and SV2B cDNAs (gifts of Drs. Bajjalieh and Scheller, Stanford University) (Bajjalieh et al., 1992, 1993) into pCMV5-myc. The rat SV2C clone was similarly cloned into pCMV5 to generate pCMV-SV2C (Janz and Südhof, unpublished observations).
Fig. 1.

Alignment of SVOP and SV2 sequences. The amino acid sequences of rat (R), C. elegans (C), andDrosophila (D)SVOPs, rat SV2A, SV2B, andSV2C, and an organic cation transporter (OCT1) from rat are aligned with each other. Gaps are indicated by hyphens. Residues conserved in >50% of all sequences are shown on a red background, residues conserved only in SVOPs are on a green background, and residues conserved in SV2 isoforms are on a bluebackground. The 12 transmembrane domains are marked by numbered lines above the sequences. Asparagine residues in N-linked glycosylation consensus sequences are shown in black. Sequences are named on theleft and numbered on the right. TheC. elegans and Drosophila SVOP sequences were assembled from genomic databanks. The N-terminal exon of theDrosophila SVOP sequence is absent because it could not be identified in the genomic sequence. The SV2A and SV2B sequences are from Bajjalieh et al. (1993), and the SV2C sequence is from Janz and Südhof (unpublished observations).
Sequence analyses. These analyses were performed exclusively at the amino acid level. Analyses were initially made using Dnastar, with manual optimization of sequence alignments. Databank searches were executed using BLAST (Altschul et al., 1990) with the default settings of National Center for Biotechnology Information (for GenBank), the Sanger Center (for the C. elegans database), and the Berkeley Drosophila Genome Project (for theDrosophila database) using the Netscape 3.0 browser. Profiles of the SVOP, SV2, and sugar transporter families were constructed using the pftools package (Bucher et al., 1996). The profiles were used for searching current DNA and protein databases as described (Brose et al., 1995; Bucher et al., 1996). Phylogenetic trees were calculated from profile-derived multiple alignments by the CLUSTALW V1.74 program (Thompson et al., 1994) using the neighbor-joining algorithm (Saitou and Nei, 1987). Before tree construction, nonconserved regions and regions containing insertions or deletions were removed from the alignment. Tree reliability was tested by bootstrap analysis as described (Brose et al., 1995; Bucher et al., 1996).
Immunological procedures. Rabbits were immunized with GST-SVOP fusion protein. The resulting antiserum was affinity-purified on the MBP-SVOP fusion protein coupled to cyanogen bromide-activated Sepharose (Pharmacia, Piscataway, NJ). The purified SVOP polyclonal antibody and ascites from the SV2 monoclonal cell line (Buckley and Kelly, 1985) (gift of Dr. R. Jahn, Goettingen) were used in a dilution of 1:2000 for immunoblots and 1:200 for immunohistochemistry. Secondary peroxidase-coupled goat anti-rabbit or anti-mouse antibodies were from Cappel (West Chester, PA) and were used in a dilution of 1:10,000. For the blocking experiments, MBP-SVOP and MBP-Cgyr were added to the primary antibody at a concentration of 5 μg/ml during the immunocytochemical staining reaction. Immunohistochemistry of adult rat brain was performed as described in Rosahl et al. (1995).
Protein analysis. Samples were mixed with 2× SDS-PAGE sample buffer, incubated at 37°C for 5 min but not boiled, and separated on 8 or 10% SDS-PAGE gels (Laemmli, 1970). The proteins were electrotransferred onto nitrocellulose filters according to the method of Towbin et al. (1979). Blots were blocked for 1 hr in TBST (0.1% Tween 20, 150 mm NaCl, and 10 mm Tris-HCl, pH 7.0) containing 5% nonfat dry milk and 5% goat serum, incubated for 1 hr with the primary antibody in the same buffer, and washed five times for 5 min each with TBST. Blots were then reacted for 1 hr with peroxidase-coupled secondary antibodies in TBST with 5% dry milk and 5% goat serum, washed five times for 5 min each with TBST, and processed for enhanced chemiluminescence using the Amersham kit (Arlington Heights, IL). For the analysis of N-glycosylation, 0.2 mg of total brain protein was denatured in 0.5% SDS and 1% β-mercaptoethanol in 0.1 ml at 90°C for 10 min and cooled to room temperature. Triton X-100 (1% final concentration) and 50 mm Na2PO4, pH 7.5, were added, and the sample was incubated for 10 min at 37°C with or without 2500 units of PNGase F (New England Biolabs). Samples (20 μg/lane) were then analyzed by SDS-PAGE and immunoblotting as described above.
Subcellular fractionations. Rat brain fractionations were performed essentially as described by Huttner et al. (1983). Two rat brains were homogenized in 30 ml of homogenization buffer (0.32m sucrose, 10 mm HEPES-NaOH, pH 7.4, 0.1 mm phenylmethylsulfonyl fluoride, 1 mg/l pepstatin, 10 mg/l leupeptin, and 10 mg/l aprotinin) using a glass teflon homogenizer (10 strokes; 900 rpm). The homogenate was centrifuged at 750 rpm in an HB4 rotor. The pellet was saved (P1), and the supernatant was centrifuged again at 7600 rpm in an HB4 rotor. The pellet was resuspended in 40 ml of homogenization buffer and recentrifuged at 7600 rpm in an HB4 rotor to yield the synaptosomal pellet (P2). The supernatants of the last two spins were pooled (S2). The synaptosomes in P2 were resuspended in 5 ml of homogenization buffer, lysed hypo-osmotically by dilution with 45 ml of 5 mm HEPES-NaOH, pH 7.4, containing protease inhibitors (PMSF, leupeptin, pepstatin, and aprotinin), and homogenized with a glass teflon homogenizer (10 strokes; 900 rpm) followed by shaking at 4°C for 15 min. The lysed synaptosomes were centrifuged for 20 min at 10,000 rpm in an HB4 rotor to obtain the LP1 pellet, and the supernatant of this spin was centrifuged again for 1 hr at 60,000 rpm in a TL 100.4 rotor to obtain the LP2 pellet and the LS2 supernatant. Synaptic vesicles purified by controlled pore-glass chromatography (Jahn et al., 1986) were a gift of S. Butz. Chromaffin granules and adrenal microsomes were prepared by homogenizing bovine adrenal medullae in 5 vol of 0.3m sucrose and 10 mm HEPES-NaOH, pH 7.4, followed by a 10 min centrifugation at 800 × g to remove debris. Chromaffin granules were pelleted from the resulting supernatant by centrifugation (20 min at 26,000 × g) and further purified over a 1.6 m sucrose step gradient (Smith and Winkler, 1967). The microsomes in the supernatant of the 26,000 × g centrifugation were pelleted by a 1 hr centrifugation at 100,000 × g. The protein concentrations of all fractions were determined using the Bio-Rad (Hercules, CA) Coomassie protein assay. For purification of synaptic vesicles by organelle immunoprecipitation, beads coated with antibodies to synaptotagmin and synaptobrevin or control beads containing only glycine were used to specifically isolate synaptic vesicles from brain homogenates as described (Baumert et al., 1990; Burger et al., 1991). Equivalent amounts of all fractions were analyzed by immunoblotting as described below.
Cell culture. COS cells were cultured in DMEM with 10% FCS and were transfected using DEAE-dextran with chloroquine and a 2 min glycerol shock as described by Gorman (1985), with 6.6 μg of DNA for 900,000 cells in a 10 cm dish. Seventy-two hours after transfection, cells were washed once with PBS, harvested in SDS sample buffer with a rubber policeman, and sheared by 10 passages through a 25 gauge needle before analysis by SDS-PAGE.
Northern blots. A 690 bp PstI/BamHI fragment from the mouse SVOP cDNA clone IMAGE #312052 with 160 bp of 5′-untranslated sequence and with a 530 bp coding region was labeled with [α-32P]dCTP and used as a probe on multitissue RNA blots from Clontech (Cambridge, UK). Blots were washed once for 10 min with 2× SSC at room temperature and twice for 30 min at 55°C in 0.1× SSC and 0.1% SDS. Blots were exposed to film at −70°C with a screen.
RESULTS
Identification of SVOP
Homologs of most synaptic vesicle proteins are present in invertebrates and in vertebrates in which they perform similar functions (see for example McIntire et al., 1997; Nonet et al., 1997). To investigate whether isoforms of the vertebrate vesicle protein SV2 are expressed in invertebrates, we searched the C. elegansand Drosophila genome and EST databases. We identified sequences in the C. elegans and the Drosophilagenomes with low but significant homology to vertebrate SV2 proteins. In C. elegans, this sequence corresponds to a gene of unknown function called YOU1 (GenBank accession #586797). YOU1 encodes a protein of 536 amino acids (Fig.1). The Drosophila sequence was assembled from the sequence of a P1 clone (#DS00543; subclone 1-f8) by linking putative exons. However, the first exon of theDrosophila gene could not be identified in the genomic sequences, possibly because of a low degree of sequence conservation.
The C. elegans and Drosophila sequences are highly homologous to each other (45.8% amino acid sequence identity) but only distantly related to SV2A, SV2B, and SV2C (20–22% amino acid sequence identity; Fig. 1). Analysis of the C. elegans andDrosophila sequences predicts that they include 12 transmembrane domains similar to that of SV2, with the highest conservation observed in the transmembrane regions. Thus we have identified a conserved gene in C. elegans andDrosophila that is related to vertebrate SV2. We found no sequences in the invertebrate databanks with a higher degree of homology to SV2s than C. elegans you1 and itsDrosophila counterpart. The databanks searched include theC. elegans genome sequence that currently contains >75% of the total C. elegans genome, indicating that C. elegans may have no other SV2 homologs. A partial sequence resembling SV2 from mosquito has been reported in GenBank (accession #AF049228). However, the incompleteness of this sequence and its low degree of homology to SV2 or to YOU1 make it unclear whether the corresponding protein corresponds to an invertebrate SV2 or to another related protein.
Do C. elegans YOU1 and its Drosophila counterpart represent an invertebrate SV2 ortholog or a separate, independently conserved protein? To address this, we searched for YOU1-related sequences in the human and mouse EST databases. A mouse EST sequence (IMAGE clone #312052) was found that exhibits a higher degree of homology to C. elegans YOU1 than to vertebrate SV2s. This suggests that the mouse EST sequence may be part of a vertebrate ortholog of YOU1 and that YOU1 and SV2 are independently conserved in vertebrate evolution. To clarify this question, we isolated and sequenced full-length rat brain cDNA clones corresponding to the mouse EST (Fig. 1). The protein encoded by the cDNAs is composed of 548 amino acids with a high degree of homology to YOU1 (48.3% amino acid sequence identity) and its Drosophila counterpart (48.5% identity) but with only a low degree of homology to SV2 (20–22% identity). We named the new protein SVOP for SVtwo-related protein because it is distantly related to SV2 and shares with SV2 the presence of 12 transmembrane regions and homology to bacterial transporters. However, SVOP is not an isoform of SV2 because its sequence is quite dissimilar from SV2, it lacks some of the structural features of SV2 (see below), and it is separately conserved in evolution.
Homologies of SVOP
The sequences of rat, C. elegans, andDrosophila SVOP are aligned with each other and with the sequences of rat SV2A, SV2B, and SV2C in Figure 1. The sequence of the most closely related protein, the organic cation transporter OCT1 (see below), is also included in the alignment. The alignment reveals that these proteins share a similar overall structure with 12 putative transmembrane domains. Their sequence homology extends over the entire proteins except for the N and C terminals. SVOP and SV2 are most homologous in their transmembrane regions and cytoplasmic loops connecting the transmembrane regions. They are most dissimilar in the intravesicular loops in addition to the cytoplasmic N and C terminals. Most striking is the large intravesicular sequence of SV2 that is located between the seventh and eighth transmembrane regions. This sequence is highly conserved in all isoforms of SV2 but absent from SVOP (Fig. 1).
In agreement with previous findings (Bajjalieh et al., 1992, 1993;Feany et al., 1992; Gingrich et al., 1993), databank searches revealed that SV2 is significantly related to a variety of transport proteins (Fig. 2). These include bacterial, yeast, and vertebrate transporters for sugars, phosphates, organic acids, and organic cations. Does this also apply to SVOP, and are there differences between SV2 and SVOPs in their homology patterns? This was evaluated by a systematic databank analysis using the profile method (Bucher et al., 1996). Generalized profiles are position-specific scoring tables that are typically derived from multiple alignments and used for sequence database searches and alignments. In addition to the pure sequence information, profiles also contain alignment-based information, such as the degree of conservation or the occurrence of deletion and/or insertion gaps. The resulting benefits, compared with single sequence-based methods, are a higher sensitivity in database searches and an improved alignment accuracy (Bucher et al., 1996).
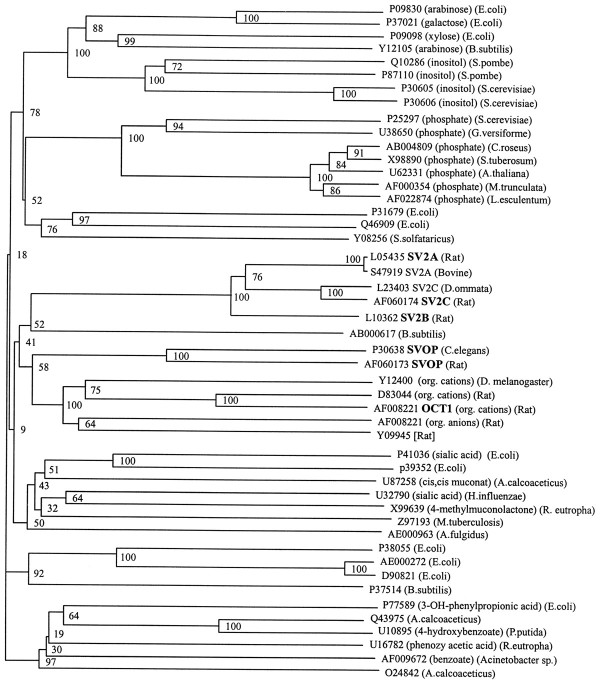
Fig. 2.
Phylogenetic tree of the SVOP/SV2 gene family: relation to transport proteins. Genes are identified on theright by their GenBank accession numbers. The substrates for known transporters and the species of origin for each gene are listed in parentheses on the right. Genes were grouped into classes based on their nearest-neighbor relations. Note that SV2, SVOP, the organic cation and anion transporters, and a single bacterial sequence, the YceI gene from B. subtilis, form a single subgroup of related sequences. Bifurcation points were confirmed by bootstrapping; thenumbers of replications per 100 runs are shownnext to the bifurcation points. Names of sequences used in Figure 1 are bolded.
Profiles of the SV2 and SVOP subfamilies were constructed from corresponding multiple alignments and used for establishing sequence relationships between the two families and the sugar transporter superfamily. Subsequently, profiles of the sugar transporter superfamily were used to align all sequences analyzed in Figure2. From the resulting superfamily alignment, a phylogenetic tree was constructed using the neighbor-joining algorithm (Saitou and Nei, 1987). Tree reliability was tested by bootstrap analysis that calculates multiple trees using different subsets of the available data. The robustness of a predicted branch point is evaluated by counting how many of the subtrees exhibit the same particular branching pattern, and the results are depicted in a dendrogram (Fig. 2).
As expected, SVOP and SV2 form two separate but related families. The closest relative of SVOP is a family of organic anion and cation transporters that includes OCTs, ROAT, and ORCT (Koepsell, 1998). For SV2, the most homologous sequence is aBacillus subtilis gene of unknown function named YceI (Fig.1). SVOP, SV2, organic cation and anion transporters, and YceI belong to a single subgroup of homologous proteins as indicated by a bootstrapping value of 41 (Fig. 2). They are more distantly related to a large number of transporters, including a group of sugar and phosphate transporters shown at the top of the dendrogram (bootstrapping value 18), and to a variety of transporters for complex organic acids, shown in the bottom half of the dendrogram (Fig. 2).
This analysis places SV2 and SVOP into the context of transport proteins that are mostly localized to the plasma membrane. Although this result strongly suggests that SV2 and SVOP function as transporters, it is difficult to predict the nature of the transported molecule. The proteins that are related to SVOP and SV2 transport both anions and cations and transport inorganic as well as organic compounds. The majority of the proteins to which SVOP and SV2 are homologous are involved in transporting organic anions and cations, suggesting that SVOP and SV2 also transport a charged molecule. A possible function for SVOP and SV2 in transporting a positively charged molecule is supported by the presence of conserved negatively charged amino acids in the first transmembrane region (Fig. 1).
Tissue distribution of SVOP mRNA
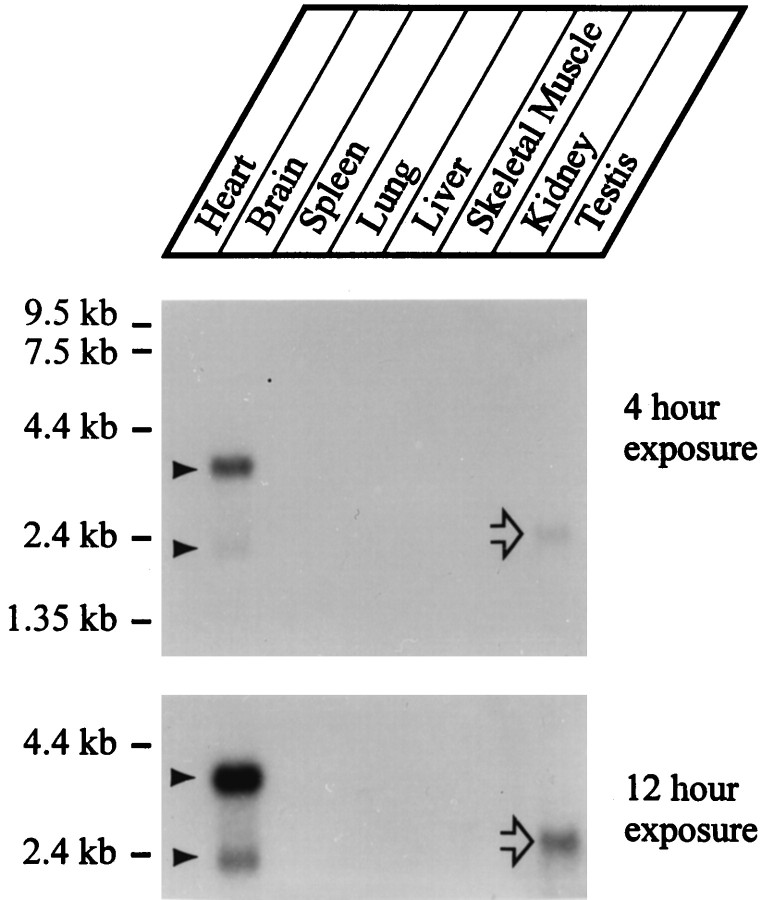
We hybridized a rat multitissue RNA blot with SVOP probes under high stringency using a conserved region of the mouse EST clone (Fig.3). The autoradiograms revealed that among the tissues tested, SVOP mRNAs were only detectable in brain. An abundant mRNA of ∼4 kb and a less abundant mRNA of ∼2 kb were observed. Even long exposures did not reveal additional cross-hybridizing mRNAs except in testis where we detected a signal at ∼2.4 kb (Fig. 3, open arrow). This signal probably represents an artifact because we have observed similar testis-specific signals with other unrelated probes. Immunoblots with a SVOP-specific antibody (see below) confirmed the tissue distribution deduced from the RNA blot (data not shown).
Fig. 3.
Tissue distribution of SVOP expression. A rat multitissue RNA blot was hybridized with a cDNA probe for SVOP at high stringency and exposed for 4 hr (top) or 12 hr (bottom). Two mRNA species are observed only in brain even after long exposures (filled arrowheads). The single cross-hybridizing mRNA observed in testis (open arrow) is probably an artifact because it does not correspond in size to SVOP brain mRNAs and because a similar band is also observed with other unrelated probes in testis.
Properties of the SVOP protein
To study the SVOP protein, we generated a polyclonal antibody against a GST-fusion protein containing the N-terminal 85 residues of SVOP. The antibody was affinity-purified on immobilized MBP-SVOP fusion protein and tested by immunoblotting using brain homogenates and transfected COS cells. For this purpose we transfected COS cells with a control expression vector and with expression vectors encoding SVOP, SV2A, SV2B, and SV2C.
In brain and SVOP-transfected COS cells, we observed a single protein of ∼60 kDa that reacted with the SVOP antibody (Fig.4). This band was not present in COS cells transfected with control DNA or with SV2 expression vectors, suggesting that the 60 kDa protein corresponds to SVOP (Fig. 4). The apparent size of SVOP in transfected COS cells agrees well with its predicted molecular weight and is identical to that observed in brain. In addition to the 60 kDa band, the SVOP antibody recognized an endogenous COS cell protein of ∼90 kDa (Fig. 4, open arrow). This band was absent from brain tissues in which the SVOP antibody was monospecific. When we analyzed transfected COS cells and brain homogenates with the SV2 monoclonal antibody, we detected a ladder of immunoreactive bands. These bands were observed only in SV2A-, SV2B-, and SV2C-transfected COS cells and not in control or SVOP-transfected COS cells. In brain, the SV2 monoclonal antibody reacted with a fuzzy band that was less heterogeneous than that in COS cells (Fig. 4).
Fig. 4.
Expression of SVOP in transfected COS cells and brain: specificity of antibodies. COS cells transfected with SV2A (lane 1), SV2B (lane 2), SV2C (lane 3), SVOP (lane 4), and control expression vectors (lane 5) and rat brain homogenate (lane 6) were analyzed by SDS-PAGE and immunoblotting using a polyclonal antibody against SVOP (top) or the monoclonal antibody against SV2 (bottom). Positions of specific bands are indicated byarrowheads on the right. The cross-reacting unrelated band observed with the SVOP antibody in COS cells but not in brain is marked by an open arrow.Numbers on the left indicate positions of molecular weight markers. Note the multiple heterogeneous bands for SV2A, SV2B, and SV2C in transfected COS cells that are caused by incomplete glycosylation (Janz and Südhof, unpublished observations).
The heterogeneous ladder of bands detected with the SV2 monoclonal antibody in the transfected COS cells is probably caused by incomplete glycosylation of SV2 in transfected COS cells. Similar to other synaptic proteins, SV2 is more extensively glycosylated in COS and endocrine cells than in synaptic vesicles (Buckley and Kelly, 1985;Johnston et al., 1989). In contrast, the sharpness of the SVOP band and its exact correspondence to the predicted molecular weight of SVOP suggested that SVOP is not glycosylated. To test this hypothesis, we used PNGase F, a glycohydrolase that specifically cleaves N-linked sugars. Proteins in brain homogenates were unfolded with SDS to make the glycosylation sites accessible. SDS was then quenched with Triton X-100, and the samples were incubated with PNGase F or control buffer. Immunoblotting revealed that this treatment decreased the apparent size of SV2 by 15–20 kDa (Fig. 5). The apparent size of SVOP, however, was unchanged, confirming that SV2 is N-glycosylated but SVOP is not. Similar experiments with transfected COS cells gave the same results (data not shown).
Fig. 5.
SV2 but not SVOP is N-glycosylated. Rat brain proteins (20 μg/lane) were incubated with (left lanesfor SVOP and SV2) or without (right lanes for SVOP and SV2) PNGase F. Protein extracts were analyzed by SDS-PAGE and immunoblotting for SVOP and SV2 as indicated. Molecular weight standards are indicated on the right.
Time course of SVOP expression in development
In mammals, most synapses are formed postnatally in the first 3 weeks of life. This leads to a corresponding increase in the levels of synaptic vesicle proteins as a function of development. To compare the developmental profiles of SVOP and SV2, we immunoblotted equivalent amounts of proteins from rat brain at different developmental ages (embryonic day 19; postnatal days 1, 4, 11, and 18; and adult). The results revealed that SVOP and SV2 exhibit remarkably different patterns of developmental expression. SVOP was present during late embryonic development and displayed a continuous postnatal increase (Fig. 6). In contrast, SV2 was undetectable before birth under the low-sensitivity conditions used but experienced a major increase in levels after birth. We observed that the relative amount of SV2 more than doubled between P18 and adult brain, whereas SVOP levels decreased slightly during the same time period.
Fig. 6.
Developmental time course of SVOP and SV2 expression in brain. Equivalent amounts of rat brain protein from embryonic day 19 (E19), postnatal days 1, 4, 11, and 18 (P1, P4, P11, andP18), and adult animals were analyzed by immunoblotting with the polyclonal SVOP (top) and the monoclonal SV2 (bottom) antibody. Numbers on theleft indicate positions of molecular weight markers.
Subcellular localization of SVOP
The brain-specific expression of SVOP and its similarity to SV2 suggest that it may be a synaptic vesicle protein. To test this, we isolated different subcellular fractions from rat brain (Huttner et al., 1983). Crude synaptosomes (P2) were lysed hypo-osmotically and fractionated into a low-speed pellet (LP1) comprising primarily synaptic plasma membranes, myelin, and mitochondria; a high-speed pellet (LP2) enriched in synaptic vesicles; and a supernatant (LS2) containing soluble synaptosomal proteins. Finally, we also examined highly purified synaptic vesicles obtained by controlled pore-glass chromatography. We analyzed equivalent amounts of protein from each fraction by SDS-PAGE and immunoblotting with SVOP and SV2 antibodies (Fig. 7). The results show that both proteins are highly enriched in synaptic vesicles. The degree of enrichment is very similar for SVOP and SV2, indicating that both are similarly concentrated on small synaptic vesicles (Fig. 7).
Fig. 7.
Analysis of the localization of SVOP by subcellular fractionation. The following rat brain fractions were analyzed: brain homogenate (Total); low-speed pellet (P1); crude synaptosomes (P2); supernatant of synaptosomal fraction (S2); low-speed pellet of lysed synaptosomes containing synaptic plasma membranes, myelin, and mitochondria (LP1); high-speed pellet from lysed synaptosomes enriched in synaptic vesicles (LP2); synaptosomal cytosol (LS2); and synaptic vesicles purified from LP2 by controlled pore-glass chromatography (SV). Equivalent amounts of each fraction were immunoblotted with antibodies to SVOP (top) and SV2 (middle). The SV2 blot was reprobed with antibodies to NMDA-receptor and munc18–1 (bottom) to show that these proteins de-enrich during synaptic vesicle purification. Numbers on theright indicate positions of molecular weight markers.
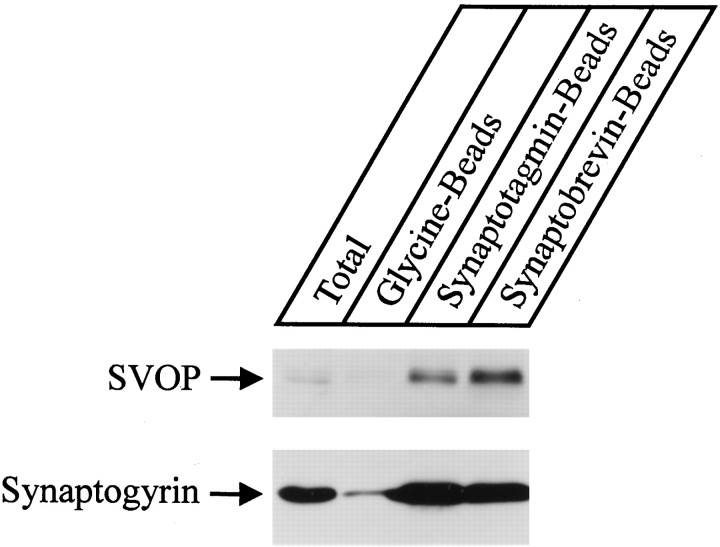
To confirm that SVOP is enriched on synaptic vesicles, we performed immunoprecipitations of synaptic vesicles from brain homogenates with beads coated with glycine (control) or with antibodies to synaptotagmin and synaptobrevin (Fig. 8). The starting material and the pellets were analyzed by immunoblotting for SVOP and for synaptogyrin as a synaptic vesicle marker (Baumert et al., 1990). Although the signal for synaptogyrin on the blots is much stronger than the SVOP signal (possibly because of the superior quality of the synaptogyrin antibodies), SVOP clearly copurified with synaptogyrin and was highly enriched in the immunoprecipitated vesicles (Fig. 8).
Fig. 8.
Copurification of SVOP with synaptogyrin on immunoprecipitated synaptic vesicles. Synaptic vesicles were captured on immunobeads from brain homogenates (Total) with beads coated with glycine only (Glycine-Beads; control) or with antibodies to synaptotagmin (Synaptotagmin-Beads) or synaptobrevin (Synaptobrevin-Beads). Bead fractions were immunoblotted for SVOP (top) or synaptogyrin (bottom).
Immunohistochemical localization of SVOP
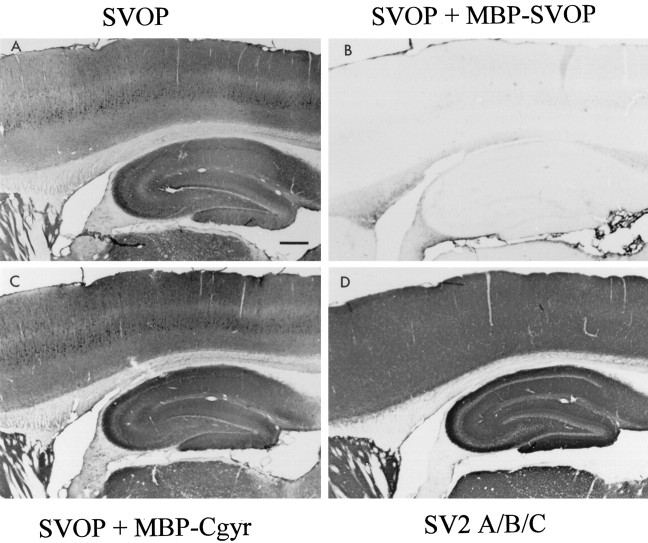
The subcellular fractionation shows that SVOP and SV2 are both present on synaptic vesicles but does not disclose their relative distributions in brain. To determine which brain areas express SVOP, we performed immunocytochemical experiments using the sensitive peroxidase–antiperoxidase method with signal amplification by heavy metal enhancement. Analysis of sagittal sections of the cerebellum with affinity-purified antibodies to SVOP demonstrated reactivity in all synaptic layers, with dense labeling of the molecular layer and less intense staining in the granule cell layer (Fig.9A). To test the specificity of the staining pattern, we added MBP-SVOP fusion protein as a blocking agent to the SVOP antibody during immunocytochemistry. The MBP-SVOP fusion protein used for blocking contains the same SVOP sequences as the GST-SVOP fusion protein used for immunization. SVOP staining was completely blocked by MBP-SVOP (Fig. 9B) but was unaffected by an unrelated MBP-fusion protein applied at the same concentration (Fig. 9C). When we probed an adjacent section of the cerebellum for SV2, we obtained a pattern of immunoreactivity indistinguishable from that of SVOP (Fig. 9D). Together these data establish the specificity of the SVOP immunocytochemistry and demonstrate that SVOP and SV2 are present throughout the cerebellum with similar localizations.
Fig. 9.

Immunocytochemical localization of SVOP in rat cerebellum. Sagittal sections were stained with the following antibodies using peroxidase detection with heavy metal enhancement: polyclonal SVOP antibody without additions (A), SVOP antibody incubated with MBP-SVOP fusion protein as a specific blocking agent (B), SVOP antibody incubated with MBP-cellugyrin fusion protein as a nonspecific blocking agent (C), or SV2 monoclonal antibody (D). Note the exact correspondence in staining patterns between SVOP and SV2. Scale bar, 0.5 mm.
We next analyzed the distribution of SVOP in the hippocampus and cerebral cortex. Again we observed immunolabeling of all areas containing synapses in a specific reaction that was blocked by MBP-SVOP fusion protein but not by an unrelated fusion protein (Fig.10A–C). In the hippocampus, staining for SVOP was very similar to that for SV2 (Fig.10D). In the cerebral cortex, however, SVOP was present in a more complex pattern that differed from that of SV2. Although SVOP was found in all cortical layers, it exhibited a heterogeneous distribution with relatively high levels in layers 3 and 5 and low levels in layers 4 and 5 (Fig. 10A). The higher magnification pictures (Fig.11A,C) revealed strong staining for SVOP in large pyramidal cells in layers 3 and 5. In contrast, these pyramidal cells were not labeled by SV2 antibodies (Fig. 11B,D). These data show that the overall staining patterns for SVOP and SV2 are very similar and suggest a colocalization to synapses. However, SVOP and SV2 do not completely overlap, indicating that SVOP is also localized in nonsynaptic compartments.
Fig. 10.
Immunocytochemical localization of SVOP in hippocampus and cortex. Rat brain sections were stained by immunoperoxidase labeling with polyclonal antibody to SVOP (A), SVOP antibody blocked with MBP-SVOP fusion protein (B), SVOP-antibody blocked with MBP-cellugyrin fusion protein (C), or SV2 monoclonal antibody (D). Scale bar, 0.5 mm.
Fig. 11.

Comparison of the distributions of SVOP and SV2 in rat brain cortex. Sections were stained with polyclonal SVOP antibody (A, C) or monoclonal SV2 antibody (B, D). Cortical layers are identified in A and B. Cand D show a higher magnification of layer 5 fromA and B. The large pyramidal cells stained with the SVOP antibody but not the SV2 antibody are marked witharrows (C, D). Scale bars: A, B, 0.25 mm;C, D, 25 μm.
SVOP in adrenal chromaffin granules and microvesicles
Extensive studies on endocrine cells demonstrated that they express homologs of synaptic vesicle proteins on two separate compartments: relatively large secretory granules and small synaptic-like microvesicles (Thomas-Reetz and De Camilli, 1994). The granules are exocytosed in response to stimulation, whereas the role of the synaptic-like microvesicles is unknown. Interestingly, synaptic vesicle protein isoforms are differentially targeted to these two compartments. For example, chromaffin cells express two isoforms of rab3, rab3A and rab3C. Rab3A is preferentially localized on synaptic-like microvesicles, whereas rab3C is concentrated on chromaffin granules (Fischer von Mollard et al., 1990; Li et al., 1994). SV2 is found on chromaffin granules, indicating that it plays a role in both synaptic exocytosis and endocrine exocytosis (Buckley and Kelly, 1985). Synaptophysin, in contrast, is enriched on synaptic-like microvesicles (Lowe et al., 1988; Thomas-Reetz and De Camilli, 1994).
To evaluate the distribution of SVOP in chromaffin cells, we analyzed chromaffin granules and microsomes (that include synaptic-like microvesicles) purified from bovine adrenal medulla. The relative amount of SVOP in these fractions and in bovine brain was then compared with the levels of other synaptic proteins (Fig.12). SVOP was highly enriched in microsomes and relatively excluded from chromaffin granules in contrast to SV2 that exhibited the opposite pattern. The distribution of SVOP resembled that of rab3A and synaptogyrin, two unrelated synaptic vesicle proteins that are enriched on synaptic-like microvesicles (Baumert et al., 1990; Fischer von Mollard et al., 1990). The synaptic vesicle proteins synaptotagmin I and rab3C, conversely, exhibited patterns resembling those of SV2 (Fig. 12). We detected no synapsins in the chromaffin cell fractions at our level of detection, thereby confirming that the signal observed is not attributable to nerve terminal contamination. These data show that SV2 and SVOP have distinct localizations in endocrine cells in spite of their apparent colocalization to synaptic vesicles in neurons. They also suggest that SVOP is not a general component of all secretory vesicles.
Fig. 12.

Relative distributions of SVOP and SV2 in adrenal chromaffin granules and microsomes containing synaptic-like microvesicles. Equivalent amounts of protein from bovine brain (lane 1), bovine chromaffin granules (lane 2), and bovine adrenal medulla microsomes (lane 3) were analyzed by immunoblotting with antibodies to the indicated proteins. Numbers on the leftindicate positions of molecular weight markers.
DISCUSSION
Synaptic vesicles are relatively simple and uniform organelles that are abundant in neurons. Because of their small size, synaptic vesicles contain comparatively few proteins. A large amount of effort from many laboratories has resulted in a molecular description of synaptic vesicles that seems to be nearly complete (for review, seeSüdhof, 1995). As a result of these studies, synaptic vesicles are thought to be composed of a limited number of protein families, most of which contain several isoforms that are differentially distributed. Quantitative evaluations of the amounts of the different vesicle proteins suggest that the majority of the synaptic vesicle proteins has been discovered (Jahn and Südhof, 1992). In view of this it is a surprise that synaptic vesicles contain an additional general component, SVOP, that was not recognized previously.
SVOP is a nonglycosylated synaptic vesicle protein expressed in all brain areas. It contains 12 transmembrane regions with few cytoplasmic sequences and has a highly hydrophobic character. SVOP is evolutionarily conserved, with 48% amino acid sequence identity between vertebrate and invertebrate forms. It is distantly related to the different isoforms of SV2 (SV2A, SV2B, and SV2C), with which it shares 20–22% sequence identity. SVOP and SV2 have the same transmembrane topology, and both localize to synaptic vesicles. SVOP differs from SV2, however, because it contains fewer cytoplasmic and intravesicular sequences and lacks the large intravesicular glycosylated loop between the seventh and eighth transmembrane regions. Although SVOP and SV2 are both highly enriched in synaptic vesicles and exhibit almost identical staining patterns in brain, they also display differences in distribution. In brain, SVOP appears to be at least partially present outside of synapses, for example, in the cell bodies of cortical pyramidal neurons. Conversely, in chromaffin cells SV2 is enriched on secretory granules and localized at lower levels on synaptic-like microvesicles, whereas SVOP is not abundant in the granules but enriched in microvesicles.
One of the most interesting results from the molecular anatomy of synaptic vesicles is that most proteins are conserved between vertebrates and invertebrates. Synaptic vesicle proteins are often represented by multiple isoforms in vertebrates and a single isoform in invertebrates. This suggests that in evolution, the architecture of synaptic vesicles was primarily conserved but the number of genes was amplified. In vertebrates, the generation of multiple genes encoding vesicle protein isoforms may have occurred to allow for fine tuning of expression patterns and/or functions. Because SV2 is such an abundant and ubiquitous component of synaptic vesicles and secretory granules, it was puzzling that no SV2 isoforms could be identified in invertebrates. Although it is currently unclear whether invertebrates express SV2, it is certain that SVOP is an evolutionarily conserved component of synaptic vesicles and that SVOP and SV2 are evolutionarily related. It is possible that SV2 is not present in invertebrates and represents an evolutionary offshoot of SVOP and related proteins. This hypothesis implies that SVOP performs a very basic function in all metazoa, whereas SV2 represents a specialized vertebrate version. Completion of the sequence of C. elegans will allow testing of this hypothesis.
What could be the physiological function of SVOP? Systematic sequence analyses showed that SVOP and SV2 are related to transporters for organic anions and cations, phosphates, and sugars. This suggests that SVOP and SV2 represent a vesicular transporter for a molecule that is probably charged. The presence of conserved charged residues in the transmembrane regions of SVOP and SV2 supports this suggestion. Because these residues differ between SVOP and SV2, SVOP and SV2 presumably transport different substrates. However, extensive efforts have failed to identify a transport activity for SV2 (Gingrich et al., 1992; M. Caron, personal communication; R. Jahn, personal communication; R. Scheller, personal communication). Preliminary studies for SVOP have also been unsuccessful (Janz and Südhof, unpublished observations). Thus it is unclear whether SVOP and SV2, in spite of their sequence homologies, are in fact transporters. A least three transport activities of synaptic vesicles have not yet been associated with known proteins: glutamate transport, ATP transport, and chloride fluxes. The ubiquitous distributions of SVOP and SV2 argue against functions as glutamate or ATP transporters because these molecules are thought to be present only in subsets of synaptic vesicles. Furthermore, SVOP and SV2 exhibit no sequence homology to neurotransmitter transporters that have been cloned previously. Although the possibility that SVOP and/or SV2 represent chloride channels cannot be excluded, their lack of homology to known chloride channels, including the nonsynaptic endomembrane chloride transporter (Landry et al., 1993), make such a role unlikely. It is certainly possible that SVOP or SV2 mediate the flux of unknown cofactors into the vesicles or regulate other transporters. Thus, if either SVOP or SV2 function as transporters, they probably perform transport activities that are not anticipated by our current understanding of the biology of synaptic vesicles.
An alternative hypothesis would be that SVOP and SV2 were evolutionarily derived from transporters but now perform a structural role. This hypothesis is more probable for SV2 than for SVOP because SV2 is highly glycosylated and contains significant amounts of extramembranous sequences. It is possible that the sugar modification of SV2 serves as a stabilizing gel in the intravesicular space. This would also offer a potential explanation for the apparent late evolutionary emergence of SV2. For example, in vertebrates, synaptic vesicles have to travel much longer distances on the average than in invertebrates before they reach their destination, the synaptic nerve terminals. It may be necessary to supply synaptic vesicles in vertebrates with a longer lifetime than in invertebrates to maintain efficient functioning in nerve terminals that are as far as a meter away from the cell body. Although this argument applies to SV2, it fails for SVOP that is conserved in nematodes, insects, and mammals. Genetic approaches or identification of new transport activities in synaptic vesicles may be necessary to address the interesting question of the relative functions of SVOP and SV2.
Footnotes
This work was supported by fellowships from the Deutsche Forschungsgemeinschaft and the Max-Planck-Gesellschaft to R.J. and by National Institutes of Health Grant MH52804. We thank Drs. S. Butz, R. Jahn, J. Albanesi, S. Bajjalieh, R. Scheller, M. Missler, and M. Caron for experimental reagents, advice, and helpful discussions.
Correspondence should be addressed to Dr. Thomas C. Südhof, Howard Hughes Medical Institute, Room Y5.322, 5323 Harry Hines Boulevard, Dallas TX 75235.
REFERENCES
- 1.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Andersson S, Davis DN, Dahlbeck H, Jörnvall H, Russell DW. Cloning, structure and expression of the mitochondrial cytochrome P-450 sterol 26-hydroxylase, a bile acid biosynthetic enzyme. J Biol Chem. 1989;264:8222–8229. [PubMed] [Google Scholar]
- 3.Bajjalieh SM, Peterson K, Shinghal R, Scheller RH. SV2, a brain synaptic vesicle protein homologous to bacterial transporters. Science. 1992;257:1271–1273. doi: 10.1126/science.1519064. [DOI] [PubMed] [Google Scholar]
- 4.Bajjalieh SM, Peterson K, Linial M, Scheller RH. Brain contains two forms of synaptic vesicle protein 2. Proc Natl Acad Sci USA. 1993;90:2150–2154. doi: 10.1073/pnas.90.6.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bajjalieh SM, Franz GD, Weimann JM, McConnell SK, Scheller RH. Differential expression of synaptic vesicle protein 2 (SV2) isoforms. J Neurosci. 1994;14:5223–5235. doi: 10.1523/JNEUROSCI.14-09-05223.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumert MK, Takei K, Hartiger J, Burger PM, Fisher von Mollard G, Maycox PR, DeCamilli P, Jahn R. P29: a novel tyrosine-phosphorylated membrane protein present in small clear vesicles of neurons and endocrine cells. J Cell Biol. 1990;110:1285–1294. doi: 10.1083/jcb.110.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bindra PS, Knowles R, Buckley KM. Conservation of the amino acid sequence of SV2, a transmembrane transporter in synaptic vesicles and endocrine cells. Gene. 1993;137:299–302. doi: 10.1016/0378-1119(93)90024-w. [DOI] [PubMed] [Google Scholar]
- 8.Brose N, Hoffman K, Hata Y, Südhof TC. Mammalian homologues of C. elegans unc-13 gene define novel family of C2-domain proteins. J Biol Chem. 1995;270:25273–25280. doi: 10.1074/jbc.270.42.25273. [DOI] [PubMed] [Google Scholar]
- 9.Bucher P, Karplus K, Moeri N, Hofmann K. A flexible motif search technique based on generalized profiles. Comput Chem. 1996;20:3–23. doi: 10.1016/s0097-8485(96)80003-9. [DOI] [PubMed] [Google Scholar]
- 10.Buckley K, Kelly RB. Identification of a transmembrane glycoprotein specific for secretory vesicles of neural and endocrine cells. J Cell Biol. 1985;100:1284–1294. doi: 10.1083/jcb.100.4.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burger PM, Hell J, Mehl E, Krasel C, Lottspeich F, Jahn R. GABA and glycine in synaptic vesicles: storage and transport characteristics. Neuron. 1991;7:287–293. doi: 10.1016/0896-6273(91)90267-4. [DOI] [PubMed] [Google Scholar]
- 12.Feany MB, Lee S, Edwards RH, Buckley KM. The synaptic vesicle protein SV2 is a novel transmembrane transporter. Cell. 1992;70:861–867. doi: 10.1016/0092-8674(92)90319-8. [DOI] [PubMed] [Google Scholar]
- 13.Fischer von Mollard G, Mignery GA, Baumert M, Perin MS, Burger PM, Jahn R, Südhof TC. Rab3 is a small GTP-binding protein exclusively localized to synaptic vesicles. Proc Natl Acad Sci USA. 1990;87:1988–1992. doi: 10.1073/pnas.87.5.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gingrich JA, Andersen PH, Tiberi M, Mestikawy SE, Jorgensen PN, Fremeau RT, Jr, Caron M. Identification, characterization, and molecular cloning of a novel transporter-like protein localized to the central nervous system. FEBS Lett. 1992;132:115–122. doi: 10.1016/0014-5793(92)80917-6. [DOI] [PubMed] [Google Scholar]
- 15.Gorman C. DNA cloning, Vol II (Glover DM, ed), pp 143–190. IRL; Oxford: 1985. [Google Scholar]
- 16.Guan KL, Dixon JD. Eukaryotic proteins expressed in E. coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 17.Huttner WB, Schiebler W, Greengard P, De Camilli P. Synapsin I (protein I), a nerve terminal-specific phosphoprotein. III. Its association with synaptic vesicles studied in a highly purified synaptic vesicle preparation. J Cell Biol. 1983;96:1374–1388. doi: 10.1083/jcb.96.5.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jahn R, Südhof TC. Synaptic vesicle traffic: rush hour in the nerve terminal. J Neurochem. 1992;61:12–21. doi: 10.1111/j.1471-4159.1993.tb03533.x. [DOI] [PubMed] [Google Scholar]
- 19.Jahn R, Schiebler W, Oiumet C, Greengard P. A 38,000 dalton membrane protein (p38) present in synaptic vesicles. Proc Natl Acad Sci USA. 1986;82:4137–4141. doi: 10.1073/pnas.82.12.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janz R, Südhof TC. Cellugyrin, a novel ubiquitous form of synaptogyrin that is phosphorylated by pp60c-src. J Biol Chem. 1998;273:2851–2857. doi: 10.1074/jbc.273.5.2851. [DOI] [PubMed] [Google Scholar]
- 21.Johnston PA, Cameron PL, Stukenbrok H, Jahn R, De Camilli P, Südhof TC. Synaptophysin is targeted to similar microvesicles in CHO- and PC12-cells. EMBO J. 1989;8:2863–2872. doi: 10.1002/j.1460-2075.1989.tb08434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koepsell H. Organic cation transporters in intestine, kidney, liver, and brain. Annu Rev Physiol. 1998;60:243–266. doi: 10.1146/annurev.physiol.60.1.243. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli UK. Cleavage of structural proteins during the head assembly of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Landry D, Sullivan S, Nicolaides M, Redhead C, Edelman A, Field M, al-Awqati Q, Edwards J. Molecular cloning and characterization of p64, a chloride channel protein from kidney microsomes. J Biol Chem. 1993;268:14948–14955. [PubMed] [Google Scholar]
- 25.Li C, Takei K, Geppert M, Daniell L, Stenius K, Chapman ER, Jahn R, De Camilli P, Südhof TC. Synaptic targeting of rabphilin-3A, a synaptic vesicle Ca2+/phospholipid-binding protein, depends on rab3A/3C. Neuron. 1994;13:885–898. doi: 10.1016/0896-6273(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 26.Li C, Ullrich B, Zhang ZZ, Anderson RGW, Südhof TC. Ca2+-dependent and Ca2+-independent activities of neural and nonneural synaptotagmins. Nature. 1995;375:594–599. doi: 10.1038/375594a0. [DOI] [PubMed] [Google Scholar]
- 27.Lowe AW, Madeddu L, Kelly RB. Endocrine secretory granules and neuronal synaptic vesicles have three integral membrane proteins in common. J Cell Biol. 1988;106:51–59. doi: 10.1083/jcb.106.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McIntire SL, Reimer RJ, Schuske K, Edwards RH, Jorgensen EM. Identification and characterization of the vesicular GABA transporter. Nature. 1997;389:870–876. doi: 10.1038/39908. [DOI] [PubMed] [Google Scholar]
- 29.Nonet ML, Staunton JE, Kilgard MP, Fergestad T, Hartwieg E, Horvitz HR, Jorgensen EM, Meyer BJ. Caenorhabditis elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. J Neurosci. 1997;17:8061–8073. doi: 10.1523/JNEUROSCI.17-21-08061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosahl TW, Spillane D, Missler M, Herz J, Selig D, Wolff JR, Hammer RE, Malenka RC, Südhof TC. Essential functions of synapsins I and II in synaptic vesicle regulation. Nature. 1995;375:488–493. doi: 10.1038/375488a0. [DOI] [PubMed] [Google Scholar]
- 31.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 32.Scranton TW, Iwata M, Carlson SS. The SV2 protein of synaptic vesicles is a keratan sulfate proteoglycan. J Neurochem. 1993;61:29–44. doi: 10.1111/j.1471-4159.1993.tb03535.x. [DOI] [PubMed] [Google Scholar]
- 33.Smith AD, Winkler H. A simple method for the isolation of adrenal chromaffin granules on a large scale. Biochem J. 1967;103:480–482. doi: 10.1042/bj1030480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Südhof TC. The synaptic vesicle cycle: a cascade of protein–protein interactions. Nature. 1995;375:645–653. doi: 10.1038/375645a0. [DOI] [PubMed] [Google Scholar]
- 35.Thomas-Reetz AC, De Camilli P. A role for synaptic vesicles in non-neuronal cells: clues from pancreatic beta cells and from chromaffin cells. FASEB J. 1994;8:209–216. doi: 10.1096/fasebj.8.2.7907072. [DOI] [PubMed] [Google Scholar]
- 36.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignments through sequence weighting, positions-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose filters: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]