Abstract
We have cloned cDNAs for two closely related connexins (Cx), Cx35 and Cx34.7, from a perch retinal cDNA library. Sequencing of PCR products from genomic DNA revealed that both connexins have an intron 71 bp after the translation initiation site; in Cx35, the intron is 900 bp in length, whereas in Cx34.7 it is ∼20 kb. Southern blots of genomic DNA suggest that the two connexins represent independent single copy genes. In Northern blots, Cx35 and Cx34.7 transcripts were detected in retina and brain; Cx34.7 also showed a weak signal in smooth muscle (gut) RNA. Antibodies against Cx35 labeled a 30 kDa band on a Western blot of retinal membranes, and in histological sections, the pattern of antibody recognition was consistent with labeling of bipolar cells and unidentified processes in the inner plexiform and nerve fiber layers. When expressed in Xenopus oocytes, Cx35 and Cx34.7 formed homotypic gap junctions, but the junctional conductance between paired oocytes expressing Cx35 was 10-fold greater than that recorded for gap junctional channels formed by Cx34.7. The homotypic gap-junctional channels were closed in a voltage-dependent manner but with relatively weak voltage sensitivity. Heterotypic gap junctions formed by Cx35 and Cx34.7 displayed junctional conductances similar to those of Cx34.7 homotypic pairs and showed a slightly asymmetric current–voltage relationship; the side expressing Cx35 exhibited a higher sensitivity to transjunctional potentials. An analysis of the sequence and gene structure of the connexin family revealed that perch Cx35 and Cx34.7, skate Cx35, and mouse Cx36 constitute a novel γ subgroup.
Keywords: retinal connexins, cloning, γ connexin subgroup, oocytes, neurons, gap junctions, channels, intercellular communication
In almost every vertebrate tissue, direct electrical and chemical communication between groups of contiguous cells are mediated by gap junctions: membrane-spanning aqueous pores that permit free intercellular diffusion of ions, metabolites, and other small molecules (≤1 kDa). The membrane specializations forming these channels consist of hexameric assemblies (connexons) of connexin (Cx) proteins in each of the adjoining cells, and molecular biological studies have shown that all of the connexins cloned and sequenced thus far belong to a superfamily of gap-junctional genes (Beyer, 1993; Bennett et al., 1994; Bruzzone et al., 1996). Despite the overall degree of homology, the proteins for which they code often exhibit regions of unique sequence that probably adapt channel properties to special tissue requirements.
In the vertebrate retina, electrical coupling is widespread, and virtually every cell type forms an electrical “synapse” with its neighbors. Thus, gap junctions constitute an important component of the retinal circuitry and acting in concert with a wide variety of chemical neurotransmitters, expand the range of intercellular interactions. However, the physiological and morphological properties of gap junctions in different retinal cell populations are diverse (Vaney, 1994; Cook and Becker, 1995; Zahs and Newman, 1997), and there is a great deal of indirect evidence suggesting that connexins of different types are present in retinal neurons and glia (Dacheux and Raviola, 1982; Vaney, 1991; Qian et al., 1993; Mills and Massey, 1995).
To gain a better understanding of the molecular basis of the unique properties of retinal gap junctions, it is necessary to identify the connexin proteins that form the gap junctional channels. We had previously cloned a gap junction protein having a molecular mass of 35 kDa, referred to as Cx35, which was preferentially expressed in the skate retina and found to be evolutionarily divergent from other connexins (O’Brien et al., 1996). In the present study, we have extended our findings to the retina of a teleost fish, the white perch. Screening for connexins in this species has yielded two connexins closely related to Cx35 that are expressed primarily in retina and brain. We have examined the ability of the two connexins to form homotypic and heterotypic gap-junctional channels in a paired oocyte expression system. In addition, phylogenetic analysis of the sequences of the retinal connexins and consideration of their gene structure suggests that they may form a unique subgroup of the connexin family.
MATERIALS AND METHODS
Library screening and clone analysis. A white perch (Morone americana) retinal cDNA library (Qian et al., 1997) prepared in the λZapII vector (Stratagene, La Jolla, CA) was generously provided by Dr. Haohua Qian (University of Illinois, Chicago, IL). Approximately 500,000 plaque-forming units were applied to an Escherichia coli strain (XL1Blue MRF′) and plated in 150 mm Petri dishes. Replicas were lifted onto nylon filters (Micron Separations, Westborough, MA) and screened at low stringency with a skate Cx35 cDNA clone (O’Brien et al., 1996). The probe, containing only the coding sequence, was labeled with 32P by nick translation, using a Boehringer Mannheim (Indianapolis, IN) nick translation kit according to the manufacturer’s protocol. The hybridization solution contained 40% formamide, 0.45 mNaCl, 30 mm NaH2PO4, pH 7.4, 3 mm EDTA, 0.5% nonfat dry milk, 1% SDS, and 0.5 mg/ml salmon sperm DNA; hybridizations were performed at 37°C. The hybridized filters were washed at 42°C sequentially for 20 min in each of the following solutions: (1) 0.5% SDS and 2× SSC (1× SSC = 150 mm NaCl and 15 mm sodium citrate, pH 7.0); (2) 0.5% SDS and 0.5× SSC; and (3) 0.5% SDS and 0.1× SSC.
Fuji RX film was exposed to the washed filters for 1–2 d with intensifying screens (DuPont Cronex). Top agar sectors containing positive plaques were cut out and rescreened under the same conditions until individual plaques were isolated. In vivo phagemid excision and rescue were performed as described in the Stratagene protocols for λZapII libraries.
The pBluescript clones were categorized by restriction analysis and sequenced with vector-specific and gene-specific primers. The complete sequence was obtained on both strands in the coding regions and throughout most of the noncoding regions. Sequencing was performed with a cycle-sequencing protocol using a dye-terminator cycle sequencing kit (Applied Biosystems, Foster City, CA). The reaction products were run and analyzed on an Applied Biosystems 377 automated DNA sequencer at the University of Illinois College of Medicine core facility. DNA sequence analysis was performed with PCGene software (Oxford Molecular Group, Campbell, CA) and Sequence Navigator software (Applied Biosystems).
Intron cloning. Hybrid bass (Morone americana andM. saxatilis) were obtained from AquaFutures (Turners Falls, MA), and genomic DNA was isolated from brain tissue by established procedures (Sambrook et al., 1989). Introns were amplified by PCR from genomic DNA using an Expand Long Template PCR kit (Boehringer Mannheim). Specifically, primers for PCR were designed to amplify sequences homologous to the region surrounding the skate Cx35 intron (O’Brien et al., 1996). Thus, to amplify the region containing the perch Cx35 intron, the primers used were 5′-CGCTTTGGAGACTGAGAACAACGAG-3′ and 5′-TCTTATGTGTGAAATGGGAAATGCC-3′. The PCR product was cloned into a pGem T vector (Promega, Madison, WI) and sequenced from both ends with vector-specific primers.
For perch Cx34.7, the primers used to amplify the intron from genomic DNA were 5′-GGAGAATGGACCATCCTAGAGCGC-3′ and 5′-ATAAAGCAGAGGCTGGGCGTGC-3′. Because of its size, the PCR product could not be cloned directly into a T vector; it was therefore digested with PstI, and fragments containing the ends were subcloned into pBluescript II KS (Stratagene). The end fragments were identified by Southern hybridization of the digested intron PCR product with probes made from a 5′ end BamHI fragment or a 3′ endBamHI fragment of the Cx34.7 cDNA. The probes were labeled with digoxygenin by random priming using a Boehringer Mannheim High Prime labeling kit. Hybridization and washings were as described below for Southern blots, and the labeled fragments were detected by probing with an alkaline phosphatase-linked anti-digoxygenin Fab fragment (Boehringer Mannheim), using 5-bromo-4-chloro-3-indolyl phosphate and nitro blue tetrazolium as the color development substrates.
Southern blot analysis. Aliquots (12 μg) of genomic DNA from hybrid bass were digested for 18 hr with 40–50 units of the following restriction enzymes: XbaI, NcoI,PstI, HindIII, EcoRI, andBamHI (New England Biolabs, Beverly, MA, or Life Technologies, Gaithersburg, MD). The digested DNA samples were ethanol precipitated, dissolved in water, and 9 μg of each was resolved on a 0.8% agarose gel in 1× Tris–acetate–EDTA buffer. DNA in the gel was denatured for 45 min with 0.5N NaOH, 1.5m NaCl, neutralized with 1 m Tris, pH 7.6, and 1.5 m NaCl, and transferred to a Nytran Plus (Schleicher & Schuell, Keene, NH) membrane by capillary diffusion in 10× SSC.
The blot was hybridized to probes containing only the DNA sequence of the intracellular loops of Cx35 or Cx34.7. The probes were prepared by PCR amplification of the loops from cDNA clones. The primers used for Cx35 were 5′-AAGGAGCGGCGGTACTCAAC-3′ and 5′-AACCTGGAGATGCCCTCTTG-3′, and the primers used for Cx34.7 were 5′-AAGACCGGCGTTACTCCTTTC-3′ and 5′-AAGCGGGAGATCCCTTCTTG-3′. The PCR products were labeled with 32P by random primer extension using a Boehringer Mannheim random primed DNA labeling kit. Hybridization was performed overnight at 42°C in a solution containing 50% formamide, 225 mm NaCl, 15 mmNaH2PO4, pH 7.4, 1.5 mmEDTA, 0.5% nonfat dry milk, 2% SDS, and 0.5 mg/ml salmon sperm DNA. After hybridization, the blot was washed at 55°C for 20 min each in solutions containing 0.5% SDS, 2× SSC, 0.5% SDS, 0.5× SSC, and 0.5% SDS, 0.1× SSC. Kodak X-Omat AR film was exposed to the blot probed with the Cx35 probe for 6 d with two intensifying screens (DuPont Cronex). The blot was then stripped, probed with the Cx34.7 probe, and used to expose Kodak X-Omat AR film for 3 d with two intensifying screens.
Northern blot analysis. Total RNA was isolated from frozen tissues collected from hybrid bass using the Trizol reagent (Life Technologies). RNAs were isolated from lens, brain, retina, heart, smooth muscle (gut), spleen, liver, kidney, and skeletal muscle. Thirty-five micrograms of each RNA was run on a 1% agarose formaldehyde gel according to Sambrook et al. (1989). The RNA was transferred to a Nytran Plus filter (Schleicher & Schuell) by capillary diffusion in 20× SSC. The blot was probed with 32P-labeled intracellular loop probes made as described above. The hybridization and washing conditions were the same as described for the Southern blot, except that hybridizations were performed at 45°C and washes at 57°C. After washing, Kodak X-Omat AR film was exposed to the blots for 5 d with two intensifying screens.
Preparation of anti-Cx35 antibodies. Antisera against a fusion protein containing the entire intracellular loop of perch Cx35 were raised in rabbits; this region contains extensive amino acid sequences that distinguish it from other connexins. The fusion protein was created by cloning a 235 bp fragment of the Cx35 cDNA coding for the intracellular loop into the pMal c2 vector (New England Biolabs).BamHI and XbaI sites, introduced by PCR into the 5′ and 3′ ends, respectively, of the Cx35 fragment were used to clone the fragment into the vector. The resulting construct coded for amino acids 101–177 of Cx35 fused to the C terminus of E. colimaltose binding protein. The fusion protein was grown in E. coli strain TB1 under isopropyl thiogalactoside induction and was purified on an amylose column (New England Biolabs) according to the manufacturer’s instructions. Injections of the purified fusion protein into rabbits and subsequent bleeds were performed at Chemicon (Temecula, CA).
The crude antisera were affinity purified by chromatography over an immobilized His-tagged Cx35 intracellular loop peptide. The peptide was produced by cloning a 241 bp fragment coding for the Cx35 intracellular loop into the NdeI and BamHI sites of the vector pET15b (Novagen, Madison, WI). Restriction sites and a termination codon were introduced into the Cx35 sequence as described above. The fusion protein consisted of the same 76 amino acid stretch of the Cx35 intracellular loop used for the MBP fusion, with the addition of an amino terminal 6× His tag. The protein was produced in E. coli strain BL21(DE3) and purified by chromatography on a Zinc-nitrilotriacetic acid column (Pharmacia, Piscataway, NJ). The purified His-tagged Cx35 intracellular loop peptide was coupled to cyanogen bromide-activated Sepharose 4B (Pharmacia) according to the manufacturer’s protocol. Crude antisera diluted in PBS were passed over the Cx35 I-loop column, and the column was then washed sequentially with (1) PBS, (2) 50 mm Tris-Cl, pH 8.5, and 0.5 m NaCl, and (3) 40 mm sodium citrate, pH 5.5, and 20 mm NaCl. Antibodies were eluted with 50 mm sodium citrate, pH 3.2, and 20 mm NaCl and neutralized with 1 m Tris, pH 8.7.
Immunofluorescent localization of Cx35. Hybrid bass eyecups were fixed overnight in Davidson’s fixative (Moore et al., 1954). The eyecups were washed, cryoprotected in 30% sucrose in PBS, and embedded in OCT (Miles, Elkhart, IN). Sections (12 μm) were cut with a cryostat and preincubated with 2% nonfat dry milk in PBS for 2 hr. They were probed overnight at 4°C with the affinity-purified anti Cx35 polyclonal antibodies at a dilution of 1:10 in PBS containing 0.5m NaCl. FITC-conjugated goat anti-rabbit secondary antibody (Boehringer Mannheim) was applied for 1 hr at a dilution of 1:50 in PBS. After washes in PBS, slides were mounted with Vectashield (Vector Laboratories, Burlingame, CA) and viewed with a Zeiss Axiophot microscope.
Western blotting. Samples of hybrid bass retina, brain, heart, spleen, and liver were homogenized in 10 volumes (w/v) of 0.1m NaCl, 20 mm HEPES, pH 7.2, 2 mmEDTA, and 0.5 mm PMSF. Membrane fractions were prepared by centrifugation for 15 min at 10,000 × g, and the resulting pellets were resuspended in the same medium. For brain and retina samples, the supernatant fraction of the 10,000 ×g spin was used for analysis. Protein content was assayed using the BCA reagent (Pierce, Rockford, IL). Protein samples were dissolved in Laemmli sample buffer and resolved on 13% polyacrylamide gels. Samples were transferred to polyvinylidene difluoride membranes (BioRad, Hercules, CA) in a BioRad Trans-blot apparatus. The membrane was blocked with 2% nonfat dry milk in Tris-buffered saline (TBS) (0.5 m NaCl, 3 mm KCl, 25 mm Tris, pH 7.5, and 0.05% Tween 20) and probed with affinity-purified anti-Cx35 antibodies at 1:500 dilution in TBS containing 0.5 m NaCl. Peroxidase-conjugated anti-rabbit secondary antibody (Pierce) was applied at 1:15,000 dilution in TBS containing 0.375 m NaCl, and labeled bands were detected by chemiluminescence (SuperSignal; Pierce).
In vitro transcription. Cx34.7 (a 1871 bp fragment) and Cx35 (a 1320 bp fragment) were excised by EcoRI/XbaI (Boehringer Mannheim) digestion of pBluescript clones, blunted with the Klenow fragment of DNA polymerase I (Pharmacia-Biotech, Orsay, France), and subcloned into the BglII site of the expression vector pSP64T (Krieg and Melton, 1984). Constructs were linearized withXbaI, and capped cRNAs were produced in vitrowith SP6 RNA polymerase, using the mMessage mMachine Kit (Ambion, Austin, TX) according to the manufacturer’s instructions. The purity and yield of transcribed cRNAs were determined by visualizing their integrity on agarose gels stained with ethidium bromide and by absorbance measurements at 260 and 280 nm.
Preparation of Xenopus oocytes. Ovarian lobes were surgically removed under cold anesthesia from female Xenopus laevis, purchased from the colony of the Institut für Entwicklungsbiologie (Hamburg, Germany). Oocytes (stage V-VI) were defolliculated after collagenase treatment and processed for the paired oocyte expression assay as described previously (Swenson et al., 1989;Bruzzone et al., 1994). For physiological analysis, manually defolliculated oocytes were injected with an antisense oligonucleotide corresponding to a portion of the coding sequence of XenopusCx38 (3 ng/oocyte: 5′-CTGACTGCTCGTCTGTCCACACAG-3′) to eliminate the possible contribution of endogenous intercellular channels to the measured conductance (Barrio et al., 1991; Bruzzone et al., 1993). After an overnight incubation at 18°C, each antisense-treated oocyte was then injected with 40 nl of either water or the appropriate dilutions of the various cRNAs. Microinjected oocytes were immersed for a few minutes in hypertonic solution to strip the vitelline envelope (Methfessel et al., 1986), transferred to Petri dishes containing modified Barth’s medium, and manually paired with the vegetal poles apposed.
Electrophysiological measurements of junctional currents.Intercellular communication was quantitated by double voltage-clamp recordings (Spray et al., 1981) obtained 24–48 hr after pairing. Electrodes had resistances of 0.5–2 MΩ and were filled with 3m KCl, 10 mm EGTA, and 10 mm HEPES, pH 7.4. Voltage clamping of oocyte pairs was performed using two GeneClamp 500 amplifiers (Axon Instruments, Foster City, CA) controlled by a compatible personal computer (Kenitec) via a Digidata 1200 interface (Axon Instruments). pCLAMP 6.0 software (Axon Instruments) was used to program stimulus and data collection paradigms. Current outputs were filtered at 10 Hz, and the sampling interval was 7.5 msec. For simple measurements of junctional conductance, both cells of a pair were initially clamped at −40 mV to ensure zero transjunctional potential, and alternating pulses of ±10–20 mV were imposed to one cell. Current delivered to the cell clamped at −40 mV during the voltage pulse was equal in magnitude to the junctional current and was divided by the voltage to yield the conductance. Families of junctional currents were generated by applying transjunctional potentials of increasing amplitude and opposite polarity to one cell, in 10 mV steps, while clamping the second cell at a constant voltage (−40 mV). To ensure adequate control of voltage across the transjunctional membrane and to avoid the risk of overestimating the actual junctional conductance at steady-state (Wilders and Jongsma, 1992), oocyte pairs exhibiting conductance <5 μS were selected for analysis of voltage dependence.
Connexin gene family analysis. Relationships between members of the connexin gene family were studied by sequence comparisons at the amino acid and DNA levels. The sequences compared included representatives of all vertebrate connexin genes available in the GenBank database. The sequences were selected to minimize repetition of known gene homologs within the mammals but to include all information from vertebrates of other classes. Twenty-eight sequences were compared that included the following: mouse connexins 26, 30.3, 31, 31.1, 32, 36, 37, 40, 43, 45, and 50; rat connexins 33 and 46; bovine connexin 44, a homolog of rat Cx46 (Gupta et al., 1994); chicken connexins 42, 45, 45.6, and 56; Xenopus connexins 30, 38, 41, and 43; connexins 32.2 and 32.7 from Atlantic croaker (Micropogonius undulatus), 43.4 from zebrafish (Danio rerio), 35 and 34.7 from white perch (M. americana); and connexin 35 from little skate (Raja erinacea).
All of the amino acid sequences were aligned using the Clustal algorithm contained in the Omiga software package (Oxford Molecular Group). From the multiple alignment, poorly conserved portions of the intracellular loop and carboxyl terminal tail were identified and eliminated from the sequences. The remaining sequences coded for the amino terminus through the second transmembrane domain corresponding to residues 1–100 of skate Cx35 and the third through fourth transmembrane domains corresponding to residues 173–255 of skate Cx35. The nucleotide sequences of the remaining regions were then aligned with Clustal W (Thompson et al., 1994), and gap positions were adjusted to match codons, if necessary. Once fully aligned in this way, the third base pair of each codon was stripped, leaving 370 characters in each sequence of the aligned data set (including gaps).
The nucleotide sequence data were analyzed using the Phylip 3.57c software package (Felsenstein, 1995). One thousand bootstrapped replicates of the aligned data set were made and analyzed directly by parsimony or were analyzed further by a distance matrix method. In the latter case, genetic distances were calculated by the Kimura two-parameter method, and tree structure was calculated by the Fitch–Margoliash method. Consensus trees were calculated for both types of analyses. Finally, genetic distances were calculated as described above for the intact data set (without bootstrapping), and the distance matrix was used to calculate branch lengths for a tree with the topology of the consensus trees from the two bootstrapped analyses.
RESULTS
Cloning of two retinal connexins
A low-stringency screen of a perch retinal cDNA library with a skate Cx35 probe yielded 12 positive plaques out of the ∼500,000 pfu that were screened. Nine of these were ultimately isolated and fell into two categories based on restriction digestion analysis. Seven clones contained a single internal BamHI site, whereas two clones had a similar BamHI site and also contained an internal PstI site. DNA sequence analysis demonstrated that all clones contained full coding sequences of connexins. The majority of clones coded for a 304 aa protein with a predicted molecular mass of 35,096 Da, whereas the two clones containing an internalPstI site coded for a 306 aa protein with a molecular mass of 34,713 Da. In keeping with the traditional nomenclature for connexins, these will be referred to as perch Cx35 and Cx34.7, respectively. Figure 1Ashows the alignment of the putative amino acid sequences of the two perch connexins with that of skate Cx35 and mouse Cx36 (Condorelli et al., 1998). The predicted transmembrane domains are outlined in Figure1A, and several other features are indicated. The four connexins have a high degree of homology throughout most of their sequences (Fig. 1A, asterisks). The amino termini, transmembrane domains, and first extracellular loops are nearly identical in the four connexins. However, small differences are evident in the second extracellular loop and C terminus, and substantial differences are apparent in the intracellular loop. There are also differences in the nucleotide sequences of the untranslated regions (data not shown). Sequences are available under Genbank accession numbers AF059183 and AF059184.
Fig. 1.
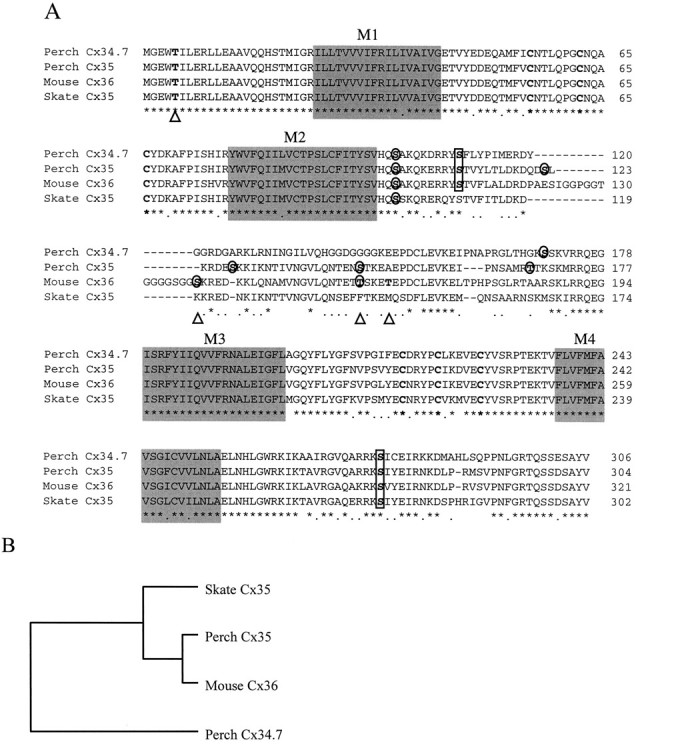
Amino acid sequences of retinal connexins.A, Alignment of perch Cx34.7 and Cx35, mouse Cx36, and skate Cx35 illustrates the extensive homology of this novel group of connexins. Completely conserved amino acids are indicated withasterisks, and conservative replacements are indicated by periods. The four predicted transmembrane domains (M1–M4) are shown as shaded boxes, and the conserved extracellular loop cysteine residues are boldfaced. Consensus sequences for PKC phosphorylation sites areoutlined with circles, and consensus PKA/G phosphorylation sites are outlined withrectangles. Putative casein kinase II sites are indicated by triangles below the alignment and include some of the same sites predicted for other kinases. B, The dendrogram of the multiple amino acid sequence alignment shown inA was calculated using the unweighted pair group method. Similarity scores used for the analysis use the Dayhoff MDM-78 similarity matrix. Although there is a close relationship among all of the connexins, perch Cx34.7 is removed from the tight group formed by perch Cx35, mouse Cx36, and skate Cx35.
Consensus sequences consistent with protein kinase phosphorylation sites were found at several locations in both connexins. A predicted protein kinase C site in the beginning of the intracellular loop and a predicted protein kinase A or G site in the C terminus are conserved in each of the connexins (Fig. 1), and additional predicted phosphorylation sites are present in the intracellular loops of the perch and mouse connexins. Perch Cx35 and Cx34.7 and mouse Cx36 share a predicted PKA/G site early in the intracellular loop (the skate sequence differs from the consensus in only one amino acid at this site), whereas four additional PKC sites were found in perch Cx35, two in mouse Cx36, and one site in perch Cx34.7. The last 10 amino acids of the C terminus of Cx35 and Cx34.7 are largely conserved and contain a sequence element related to a conserved motif present in the α group connexins (Bennett et al., 1994).
The high degree of amino acid sequence homology between the connexins illustrated in Figure 1A is shown quantitatively in Table 1. For example, perch Cx35, skate Cx35, and mouse Cx36 show a minimum sequence identity of 84% and similarities (i.e., identity plus conservative amino acid replacements) that exceed 91%. It is noteworthy that when the intracellular loop is excluded these values increase significantly, e.g., the identity between perch Cx35 and mouse Cx36 increases from 85.9 to 94.2%. There is less correspondence between perch Cx34.7 and the other connexins, ranging from 74.2 to 78.6% amino acid identity. The dendrogram of the multiple amino acid sequence alignment (Fig. 1B) illustrates the close relationship between perch Cx35, skate Cx35, and mouse Cx36 and their more distant relationship to perch Cx34.7.
Table 1.
Percent amino acid identity and similarity of Cx35 group members
| Skate Cx35 | Perch Cx35 | Mouse Cx36 | |
|---|---|---|---|
| Perch Cx35 | 87.1 (92.1) | ||
| Mouse Cx36 | 84.1 (91.1) | 85.9 (93.4) | |
| Perch Cx34.7 | 74.2 (83.4) | 78.6 (86.5) | 77.1 (85.6) |
Amino acid identities calculated from pairwise alignments of the complete sequences of each connexin are listed, with similarities shown in parentheses. Similarity scores include both identical residues and conservative substitutions, as scored using the Dayhoff MDM-78 similarity matrix.
Genomic organization of Cx35 and Cx34.7
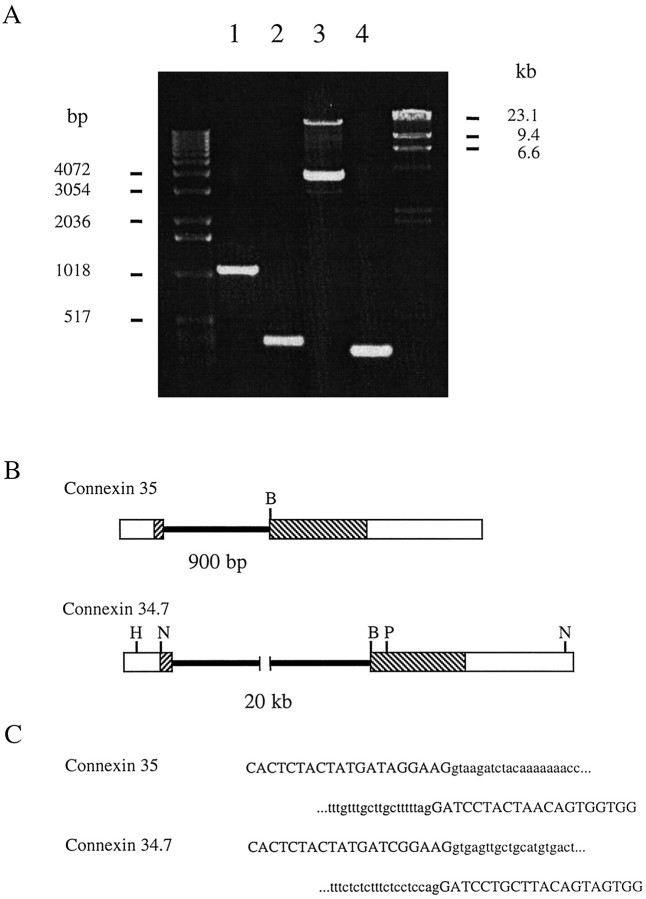
The genes of most known connexins lack introns in their coding regions and contain a single intron in the 5′ untranslated region (Beyer, 1993; Bruzzone et al., 1996). Skate Cx35 was an exception to this pattern, lacking the 5′ intron but containing a single intron within the coding sequence (O’Brien et al., 1996). PCR amplification and cloning of the introns in perch Cx35 and Cx34.7 revealed that the same pattern holds as well for these connexins. Amplification of the Cx35 intron from genomic DNA (Fig.2A, lane 1) resulted in a PCR product ∼900 bp larger than the control (lane 2). Likewise, amplification of the Cx34.7 intron (Fig.2A, lane 3) yielded a PCR product ∼20 kb larger than control (lane 4). An additional 4 kb product was consistently amplified with Cx34.7 primers; this product was sequenced and proved to be unrelated to any connexin. Sequencing of the intron PCR products revealed that both introns were located 71 bp after the translation initiation sites (Fig. 2B). This location is identical to that observed earlier in the skate Cx35 gene and corresponds to the junction between the amino terminal domain and the first predicted transmembrane domain. The intron splice junctions conform to the typical consensus sequence AGgt … agGA (cf. Padgett et al., 1986) (Fig. 2C).
Fig. 2.
Structure of the Cx35 and Cx34.7 genes.A, PCR amplification of Cx35 and Cx34.7 introns from hybrid bass genomic DNA. Introns were amplified by long PCR, as described in Materials and Methods. Amplification of Cx35 fragments from 100 ng of genomic DNA is shown in lane 1, and control amplification from ∼1 pg of cDNA clone pcx7 is shown inlane 2. The genomic product is ∼900 bp larger than the control. Amplification of Cx34.7 fragments from 100 ng of genomic DNA is shown in lane 3, with control amplification from ∼1 pg of cDNA clone pcx1 in lane 4; a 20 kb product was obtained in the genomic amplification. The ∼4 kb product inlane 3 was cloned, sequenced, and found to be spurious.B, Gene maps of the known sequences of perch Cx35 and Cx34.7 genes based on sequence analysis of the intron PCR products. Regions comprising the cDNA sequences are enclosed bywide rectangles, with the hatched portion representing the coding sequence. Introns, indicated by thenarrow bars, occur at the same location within the coding region as was described previously for skate Cx35. Known restriction sites within the cDNA sequences are coded as follows:BamHI (B); HindIII (H);NcoI (N); PstI (P).C, DNA sequences at the intron splice junctions for Cx35 and Cx34.7. Intron sequences are in lowercaseletter, and the cDNA sequences are in uppercaseletters. Both sets of intron splice junctions conform to the consensus sequence AGgt … agGA.
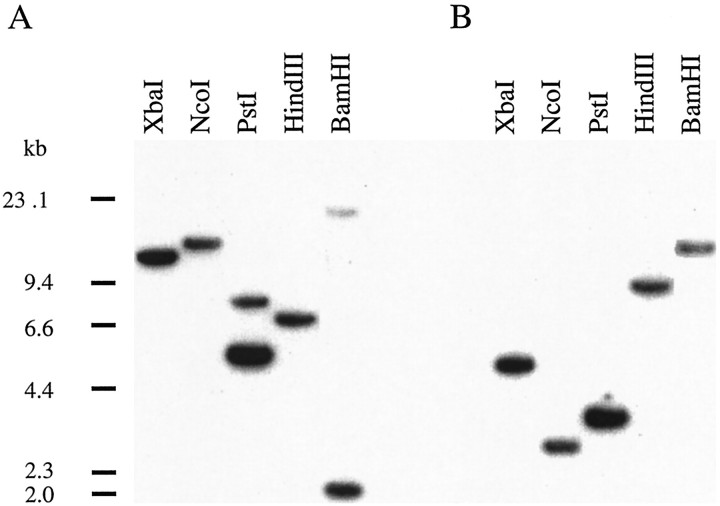
A Southern blot analysis of hybrid bass genomic DNA is shown in Figure3. For both connexins, the probes were derived from the intracellular loop and did not contain introns or restriction sites for the enzymes that were used (Fig.2B). The fragments labeled by the two probes are different, and single bands were labeled in most of the digested DNA samples. In the case of Cx35 in Fig. 3A, two bands were labeled in the PstI lane and in the BamHI lane, probably caused by differences in the restriction sites of the two parental species of the hybrid bass.
Fig. 3.
Southern blot analysis of hybrid bass genomic DNA. Each lane contains 8 μg of genomic DNA digested withXbaI, NcoI, PstI,HindIII, or BamHI and resolved on an agarose gel. Molecular weights of HindIII digested λ phage DNA are shown as markers. A, Blot probed with the perch Cx35 intracellular loop fragment. A single band was labeled inlanes containing DNA digested with XbaI,NcoI, or HindIII, whereas two bands were labeled in lanes containing DNA digested withPstI or BamHI. B, Blot probed with the perch Cx34.7 intracellular loop fragment. A single band was labeled in each lane. The labeling of different bands in A and B indicate that different genes are involved, and suggest that both are single-copy genes. See Results for further details.
Tissue expression
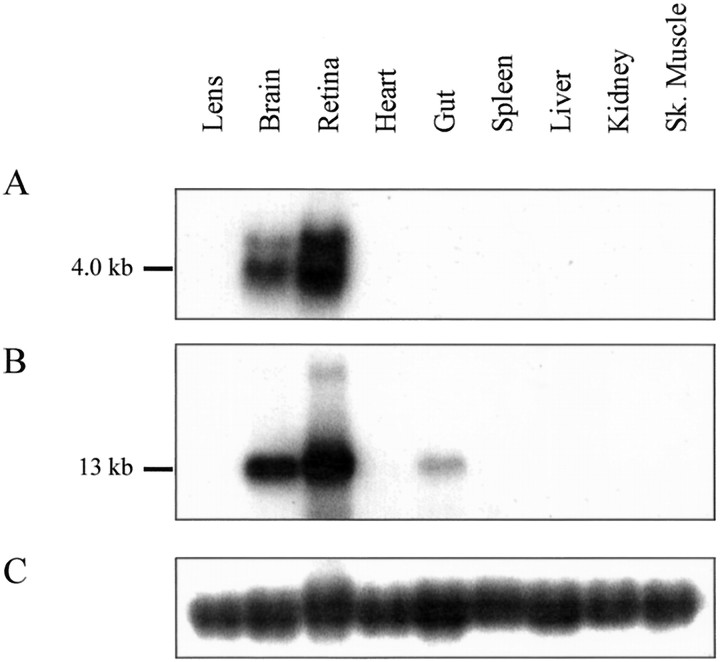
The expression of perch Cx35 and Cx34.7 was examined by Northern blot analysis of total RNA samples from nine tissues and probed with the intracellular loop probes described above. These probes contain the most divergent portions of the coding sequence and have little homology to each other. Figure4A shows that the Cx35 probe labeled a strong 4.0 kb and a weak 4.8 kb transcript in both retina and brain. No detectable labeling was seen in RNA samples from lens, heart, gut, spleen, liver, kidney, or skeletal muscle. On the other hand, the Cx34.7 probe labeled a 13 kb transcript that was detected in the retina and brain and in very low abundance in gut RNA (Fig. 4B). No other tissues showed detectable labeling, and there was no evidence of cross-hybridization with the transcripts labeled by the Cx35 probe. An apparently larger RNA species was also labeled weakly in retina RNA, but it seems likely that the smeared signal resulted from incomplete denaturation of the retina RNA sample or perhaps the presence of genomic DNA in the sample.
Fig. 4.
Northern blot analysis of connexin distribution in hybrid bass tissues. The blot contains 35 μg of total RNA from each of the tissues indicated. A, Hybridization with the Cx35 intracellular loop probe labeled a 4.0 kb transcript in retina and brain RNA and weakly labeled a 4.8 kb transcript in both tissues. Other tissues lacked detectable transcripts. B, The Cx34.7 intracellular loop probe labeled a 13 kb transcript in retina and brain RNA, with very weak labeling of a transcript of the same size in smooth muscle (gut) RNA. C, Labeling of the 28 S rRNA shows approximately equal loading of alllanes.
Although the 13 kb transcript of Cx34.7 is larger than expected to code for a 306 aa protein, large messages are not without precedent among the connexins. For example, lens connexins from mouse (Cx50) and chicken (Cx56 and Cx45.6) have transcripts that range from 8.0 to 9.4 kb (White et al., 1992; Rup et al., 1993; Jiang et al., 1994). The only feature shared by these connexins is their restricted tissue distribution, but the significance of the large message size is not known. However, untranslated portions of the transcript may contain regulatory elements that can be functional at the transcriptional or translational levels.
Localization of Cx35 in the retina
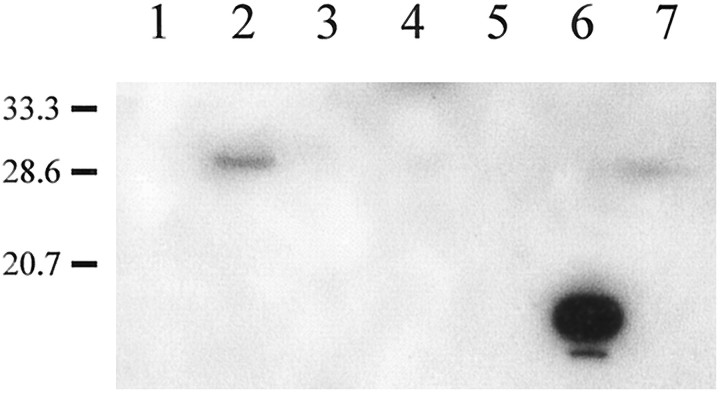
Affinity-purified antibodies raised against the intracellular loop of perch Cx35 labeled a single band in Western blots of hybrid bass retinal homogenates membrane fractions (Fig.5), with weak labeling of other protein bands. The apparent Mr of the recognized band was 30 kDa, which is consistent with the aberrant migration of connexin proteins observed in SDS gels (cf. Green et al., 1988; Beyer, 1993). The 30 kDa band was not observed in other tissues, although weakly labeled background bands were present. A small amount of the 30 kDa band was present in the 10,000 × g pellet of the retinal homogenate (Fig. 5, lane 7), but the majority was detected in the supernatant (lane 2), suggesting the presence of Cx35 in a light membrane fraction.
Fig. 5.
Western blot analysis of hybrid bass tissue homogenates probed with affinity-purified anti-Cx35 antisera.Lane 6 contains ∼1 μg of a crude lysate of the bacterial strain expressing the 6× His-tagged Cx35 intracellular loop peptide. Lanes 1 and 2 contain 10 μg of supernatant fractions of brain and retina, respectively. Ten micrograms of membrane pellets of the remaining tissues were loaded as follows: 3, heart; 4, spleen; 5, liver; 7, retina. Molecular weight markers are shown on the left. A 30 kDa band was strongly labeled in the retina supernatant fraction and was barely detectable in the 10,000 × g pellet.
Cx35 antibodies labeled several structures in cryostat sections of hybrid bass retina. Punctate labeling was observed in the distal and proximal regions of the inner nuclear layer, corresponding to the loci of bipolar cell dendrites and axon terminals, respectively (Fig.6A,B); slender processes extending between these sites were often labeled (Fig. 6C). In addition, punctate labeling of fine processes was observed consistently in regions corresponding to the inner plexiform, ganglion cell, and nerve fiber layers, but no definitive pattern emerged. Identification of the cell types associated with these processes will require further experiments with EM immunochemistry and the use of double-labeling with cell-specific markers. Control sections in which the primary antibody was omitted showed no labeling, although the autofluorescence of receptor inner and outer segments and the outer limiting membrane was clearly evident (Fig. 6D).
Fig. 6.
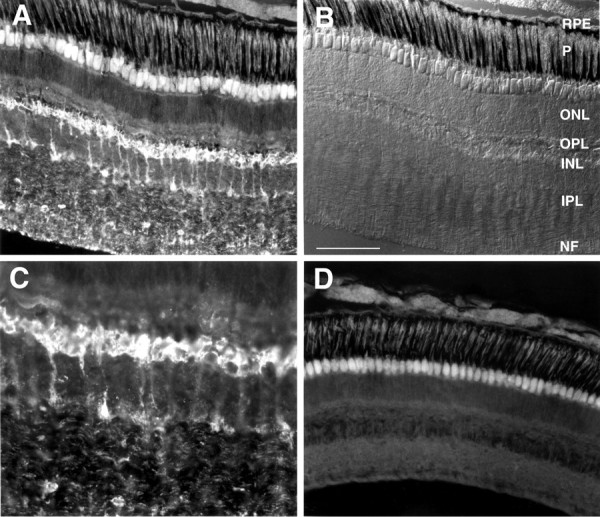
Immunofluorescent labeling of hybrid bass retina with affinity-purified anti-Cx35 antisera. A, Low-power micrograph of a transverse retinal section shows labeling of neurons in the inner nuclear, inner plexiform, and nerve fiber layers. No labeling was observed distal to the outer plexiform layer, nor were signals detected in the optic nerve (data not shown). B, Nomarski image of the field shown in A reveals the structure of the hybrid bass retina. RPE, Retinal pigment epithelium; P, photoreceptor layer;ONL, outer nuclear layer; OPL, outer plexiform (synaptic) layer; INL, inner nuclear layer;IPL, inner plexiform layer; NF, ganglion cell and nerve fiber layer. C, Higher-power view of the inner nuclear and inner plexiform layers from the section shown inA. Strongly stained cells in the inner nuclear layer appear to be bipolar cells. In addition, punctate labeling is present throughout much of the inner plexiform and ganglion cell layers; the cellular origin of this labeling is unclear. D, Control section lacking primary antibody shows the autofluorescence of the photoreceptor inner segments that was seen in A. Scale bar (in B): A, B,D, 100 μm; C, 50 μm.
Functional expression of perch connexins
Intercellular channels are defined as homotypic when both connexons forming the gap junction contain the same connexin or as heterotypic when each connexon is composed of a different connexin. The ability of perch connexins to form functional channels in both configurations was tested using the paired Xenopus oocyte expression system (Dahl, 1992). To eliminate any possible contribution of the endogenous Xenopus Cx38 to our recordings, we used antisense oligonucleotides to deplete oocytes of the endogenously expressed connexin. As reported previously (Barrio et al., 1991;Bruzzone et al., 1993), water-injected cells showed no detectable coupling under these conditions (Fig. 7, Table 2). Because several connexins from other species readily interact with Xenopus Cx38 (Hennemann et al., 1992a,b; Bruzzone et al., 1993), we constructed Cx34.7/water and Cx35/water pairs to test whether perch connexins could also recruitXenopus Cx38 if antisense treatment was omitted for water-injected oocytes. Neither Cx34.7 nor Cx35 formed channels with endogenous Xenopus Cx38, i.e., the levels of conductance never exceeded background values (Table 2).
Fig. 7.
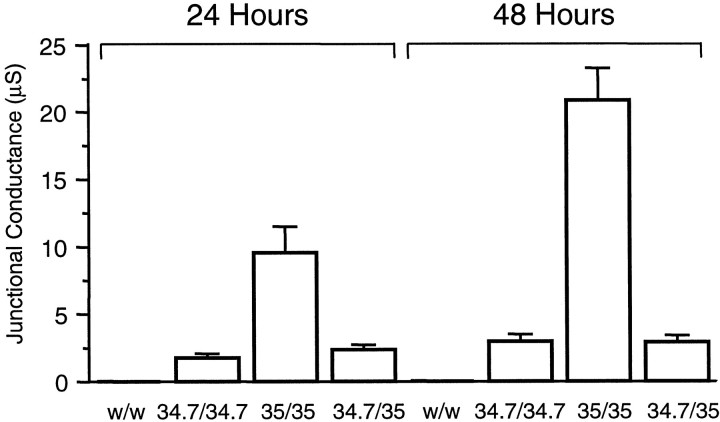
Expression of perch connexins induces the formation of both homotypic and heterotypic intercellular channels. Oocytes pretreated with an oligonucleotide antisense to a sequence within the coding region of Xenopus Cx38 were injected with either connexin cRNAs or water (w) and paired for 24–48 hr before measuring junctional conductance by dual voltage clamp. Cells received similar amounts of Cx34.7 (34.7) and Cx35 (35) cRNAs. Junctional conductances increased approximately twofold between 24 and 48 hr for homotypic pairs, but the heterotypic pairing showed little change. Values are mean ± SEM of 8–11 oocyte pairs.
Table 2.
Perch connexins fail to form heterotypic channels with endogenous Xenopus Cx38
| Oocyte injection (cell 1/cell 2) | Gj(μS) | # of pairs |
|---|---|---|
| antisense/antisense | 0.02 ± 0.01 | 8 |
| Cx34.7/water | 0.02 ± 0.01 | 8 |
| Cx35/water | 0.05 ± 0.02 | 8 |
| Cx34.7/Cx34.7 | 2.72 ± 0.35 | 5 |
| Cx35/Cx35 | 2.81 ± 1.10 | 3 |
Oocytes were either injected with an oligonucleotide antisense to a region within the coding sequence of Xenopus Cx38 (see Materials and Methods) or mock-treated (water), before receiving the specified cRNAs. The development of junctional conductance (Gj) was analyzed using a dual voltage-clamp procedure, and measurements of steady-state Gjwere performed 24–48 hr after pairing. Homotypic pairs expressing Cx34.7 and Cx35 were used as positive controls of cRNA injections and oocyte viability. Results are shown as mean ± SEM of the specified number of pairs.
In contrast, injection of cRNAs for Cx34.7 and Cx35 efficiently assembled both homotypic and heterotypic channels (Fig. 7) and induced conductance levels of the same order of magnitude as those developed by pairs injected with similar amounts of cRNAs encoding mammalian connexins (Dahl et al., 1992; Nicholson et al., 1993; White et al., 1995; Barrio et al., 1997). Although homotypically paired oocytes injected with either Cx34.7 or Cx35 received approximately the same amount of cRNA (6 and 6.4 ng/cell, respectively), Cx35 consistently induced higher values of junctional conductance, and this difference persisted even when measurements were taken 48 hr after pairing. The observation that Cx34.7 and Cx35 formed functional heterotypic pairs indicated that these two perch connexins are compatible partners. However, in the heterotypic pairs, the functional ability of Cx34.7 proved to be a limiting factor, i.e., the gap-junctional conductance developed by heterotypic pairs (Cx34.7/Cx35) was of the same magnitude as that measured for Cx34.7 homotypic pairs.
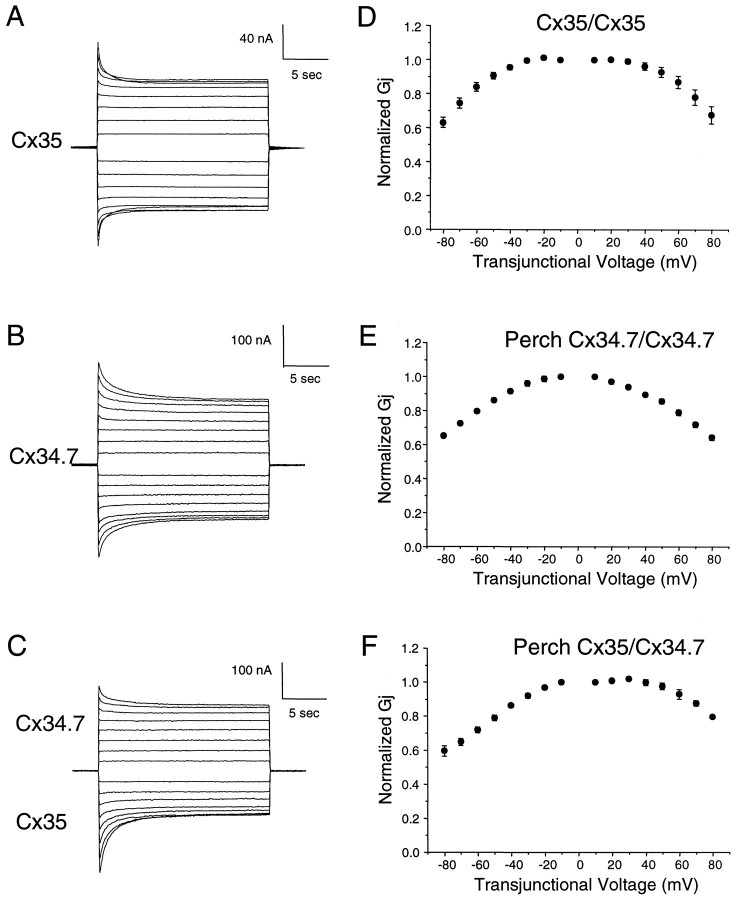
The analysis of junctional currents revealed that homotypic intercellular channels made of Cx34.7 or Cx35 were closed in a voltage-dependent manner (Fig. 8). Voltage steps of opposite polarity were imposed in 10 mV increments from a holding potential of −40 mV and lasted 22.5 sec to allow currents to reach equilibrium values. Junctional currents of Cx34.7 channels decreased relatively slowly with time after reaching a threshold transjunctional potential of ±40 mV (Fig.8A). Similarly, Cx35 exhibited voltage dependence, with currents decaying for transjunctional potentials of either polarity ≥50 mV (Fig. 8B). The rate of this slow decay increased with increasing transjunctional voltage. In the heterotypic configuration, channel closure was slightly asymmetric and exhibited a different threshold for positive and negative potentials (Fig. 8C). Thus, currents showed a higher voltage threshold for relative positivity of the cell expressing Cx34.7, whereas a decreased threshold was observed for relative positivity of the cell expressing Cx35.
Fig. 8.
Homotypic and heterotypic channels composed of perch connexins are gated by transjunctional voltage.A–C, Time-dependent decay of junctional currents developed by pairs of antisense-treated oocytes (see Materials and Methods) injected with cRNAs coding for perch connexins. Both cells were initially clamped at −40 mV to ensure zero transjunctional potential. While one cell was held at a constant potential, depolarizing or hyperpolarizing voltage steps were applied sequentially in 10 mV increments to the other cell, and the resulting junctional currents were recorded. The currents reflect voltage-induced closure for transjunctional potentials greater than ±40 mV in the case of Cx35 (A) and ±30 mV in the case of Cx34.7 (B). In the heterotypic configuration (C), channel closure was asymmetric and exhibited a different threshold for positive and negative potentials. Thus, currents showed a higher voltage threshold for relative positivity of the cell expressing Cx34.7, whereas a reduced voltage threshold was observed for relative positivity of the cell expressing Cx35. These characteristics were quantitated by plotting steady-state conductance, normalized as described in Materials and Methods, as a function of transjunctional voltage (D–F). Results represent the mean ± SEM of five to six oocyte pairs. In many cases, the error bars are contained within the plot symbols.
Relationships of retinal connexins to the connexin gene family
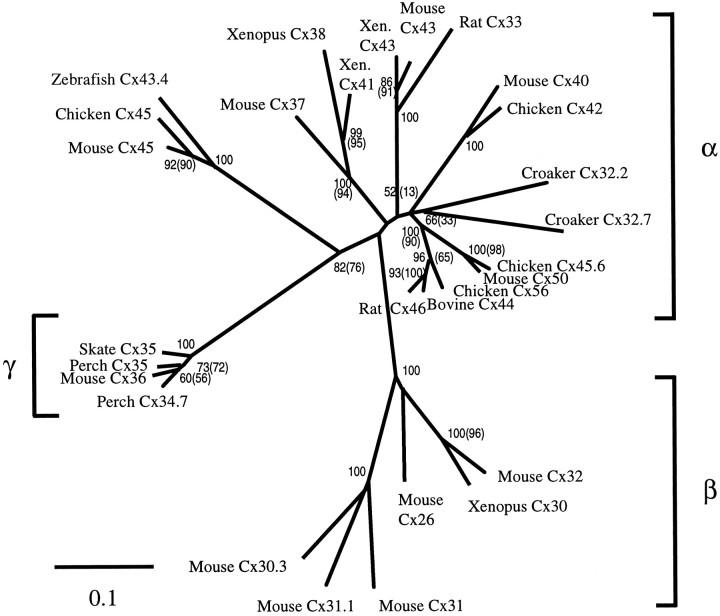
A phylogenetic analysis of the connexin gene family is shown in Figure 9. Bootstrap analyses using a distance matrix method and parsimony were used to estimate the degree of support for each node in the tree. In Figure 9, numbersat the nodes indicate the percentage of 1000 bootstrap replicates in which that node occurred in the distance analysis, andnumbers in parentheses indicate bootstrap values for the parsimony analysis. Estimates of branch lengths, using the distance algorithm (see Materials and Methods), indicate that the two perch connexins form a tight cluster with skate Cx35 and the newly cloned mouse Cx36 (Condorelli et al., 1998). These connexins diverge widely from the α group of connexins; the sequence divergence from the Cx35 cluster to the nearest node in the α group is 18.9%, which is the longest single branch in the tree. In contrast, the distance between the α and β groups is 14.6%. The divergence of the Cx35 cluster occurs near the base of the α group. Only the Cx45 homologs group with the Cx35 cluster, and the degree of support for that grouping, based on the bootstrap analyses, is moderate. Cx45 and its homologs, furthermore, are the most divergent elements of the α group, with a distance of 15.3% from the nearest node. The large genetic distance between the Cx35 cluster and the α group of connexins, together with the acquisition of an intron in the coding region, suggest a unique line of descent for this subgroup.
Fig. 9.
Relationships of retinal connexins to the connexin gene family. The tree represents a distance matrix analysis of the nucleotide sequences of the conserved portions of the coding regions of each connexin (see Materials and Methods). A sequence divergence of 10% is indicated by the scale bar. Bootstrap analyses were performed using both distance matrix and parsimony methods to estimate the support for each node in the tree. Both analyses gave the same topology, and bootstrap values (percent of 1000 bootstrap replicates containing the node) are indicated at the nodes. The bootstrap value for the distance method is written first, followed inparentheses by the value for the parsimony analysis, if different. Bootstrap values <50% were not included. The α and β subgroups of connexin genes are labeled with brackets on the right. The novel γ subgroup defined by this study is bracketed on the left.
DISCUSSION
The identity of retinal gap junctions
We have identified two connexins, Cx35 and Cx34.7, that are expressed in the perch retina. The connexins were cloned from a retinal cDNA library and transcripts of both were detected in greatest abundance in RNA from retina (Fig. 4). Unlike skate Cx35, which was found only in retina by Northern blot analysis (O’Brien et al., 1996), perch Cx35 and perch Cx34.7 were found in the brain, as well. Mouse Cx36, a homolog of Cx35, has also been found in both retina and brain (Condorelli et al., 1998).
Immunocytochemical labeling in hybrid bass retina showed Cx35 to be present in a subset of retinal cells. Punctate labeling of processes in the distal portion of the inner nuclear layer and within the inner plexiform layer suggests that the antigen is present at the dendrites and axon terminals of bipolar cells. This pattern of labeling is consistent with the observations of gap junctions at the dendrites and synaptic terminals of bipolar cells visualized by electron microscopy (Raviola, 1976; Marc et al., 1988; Cuenca et al., 1993) and tracer coupling experiments (Saito and Kujiraoka, 1988; Vaney, 1991, 1994;Umino et al., 1994; Poznanski and Umino, 1997). On the other hand, the labeling in the region of the ganglion cell and inner plexiform layers is difficult to correlate with structural features. Few gap junctions are seen in this area, although there is evidence from tracer studies that ganglion cell and amacrine cell processes located in this region may be coupled by gap junctions (Vaney, 1991, 1994; Mills and Massey, 1995). In addition to neurons, astrocytes are extensively coupled in vertebrate retinas, and coupling between astrocytes and Müller cells has been shown to occur in several vertebrate species (Robinson et al., 1993; Zahs and Newman, 1997). Although further investigation is needed to identify the cellular origins of the labeled processes, it is noteworthy that the labeling pattern we observed in the inner retina with Cx35 is consistent with the observations of Condorelli et al. (1998), who detected mouse Cx36 transcripts in the ganglion cell layer and inner border of the inner nuclear layer by in situhybridization.
Other connexin proteins have also been reported to occur in the vertebrate retina. Cx43, cloned recently from the giantDanio (teleost) retina by RT-PCR and cDNA library screening (Wagner et al., 1997), has been seen with immunohistochemical methods in astrocytes, Müller cells, and retinal pigment epithelial cells of various vertebrate species (Finch and Paul, 1989; Jones et al., 1992; Janssen-Bienhold et al., 1996; Giblin and Christensen, 1997;), as well as in a type of amacrine cell in zebrafish retina (Janssen-Bienhold et al., 1996). In addition, Cx32 immunoreactivity has been described in various neuronal cell types in mammalian retinas (Finch and Paul, 1989; Jones et al., 1992), a further indication that multiple connexins are expressed in retinal neurons. This molecular diversity may account for the selective pharmacology and unique gating behavior of gap junction channels in the retina (McMahon, 1994; Vaney, 1994; Cook and Becker, 1995; Bruzzone and Ressot, 1997).
Functional properties of Cx35 and Cx34.7
We have provided evidence that both perch connexins are functionally competent to assemble intercellular channels. However, the reasons for the lower values of junctional conductance measured in oocyte pairs expressing Cx34.7 either homotypically or heterotypically are not clear. It is possible that the unitary conductance of homotypic Cx34.7 channels is approximately an order of magnitude smaller than that of Cx35 and that heterotypic interactions drastically reduce the unitary conductance of Cx35 below that of homotypic channels. Alternatively, a reduced propensity to functionally interact with the partners tested in this study may severely limit the magnitude of the macroscopic junctional conductance observed in channels containing Cx34.7 (cf. Swenson et al., 1989; Bruzzone et al., 1993)
Cx34.7 and Cx35 exhibit voltage-dependent closure that distinguishes their behavior from that of previously characterized connexins (cf.Nicholson et al., 1993; White and Bruzzone, 1996). For example, zebra fish Cx43.4 and Atlantic croaker Cx32.2 form channels that are more sensitive to voltage and shut off more completely in response to transjunctional potentials of increasing amplitude, whereas Atlantic croaker Cx32.7 is functionally incompetent in the paired oocyte expression system (Bruzzone et al., 1995; Barrio et al., 1997). A distinct feature of perch connexins is the high level of conductance observed at the largest (±80 mV) voltage steps (Fig. 8), a property attributable to their weak voltage dependence. Indeed, perch connexins are among the least voltage-sensitive members of the connexin family; only Cx26 channels show a comparable half-maximal decrease in junctional conductance with voltage steps >80 mV (Barrio et al., 1991). It seems likely that the reduced voltage sensitivity of Cx34.7 and Cx35 allows intercellular channels to remain open, independent of voltage shifts that may occur as a result of changes in their normal cellular environment.
The observation that Cx34.7 and Cx35 are compatible partners implies that, if expressed by adjacent cells, they could form functional heterotypic channels in vivo. Analysis of other heterotypic channels has demonstrated that novel gating properties often result from interactions between different connexins (Barrio et al., 1991;Hennemann et al., 1992b; Bruzzone et al., 1994; White et al., 1994,1995; Chen and DeHaan, 1996). However, the interaction of perch connexins produces currents that, by and large, conserve the electrical characteristics exhibited by the respective homotypic channels; Cx35 appears to be slightly more sensitive to voltage when paired heterotypically, compared with the homotypic Cx35/Cx35 pairs. The opposite is true for Cx34.7, which begins to close at greater transjunctional potentials when paired with Cx35. If, as suggested by the phylogenetic analysis, this group of connexins can be regarded as separate from the other members of the family, it is likely that the divergence of primary sequence will be translated into distinct forms of channel regulation. Further studies will determine how the activity of Cx34.7 and Cx35 channels is controlled by more physiological stimuli, such as phosphorylation and changes in cytosolic ions.
The connexin 35 subfamily of gap junctions
The data presented in Figures 1 and 3 provide strong evidence that the two perch connexins, Cx34.7 and Cx35, cloned in this study represent distinct genes. Although the dendrogram of the multiple amino acid sequence alignment (Fig. 1B) shows that these connexins are related to each other, to skate Cx35, and to the recently cloned mouse Cx36 (Condorelli et al., 1998), Cx34.7 is a more distant relative and contains a substantially different intracellular loop sequence (Fig. 1A). Support for the notion that Cx35 and Cx34.7 are different genes is provided by the observation that different fragments of genomic DNA were labeled in the Southern blots probed with unique sequences from a single exon of each connexin (Fig.3) and that there are large sequence differences in both untranslated regions of the mRNAs. However, we cannot exclude the possibility that Cx35 and Cx34.7 represent alternative splice products. Further investigation involving genomic cloning will be required to resolve this issue. On the other hand, Cx35 from perch and skate, and mouse Cx36 may represent the same gene product in the three species. Their amino acid sequences are >84% identical (Table 1), there is a high degree of sequence homology even within the intracellular loop, and the dendrogram groups these connexins closely.
The gene structure of members of the Cx35 group suggests that they are different from other connexins. Each of these connexins contains an intron at a conserved location within the coding region of the gene, unlike any member of the α or β groups of connexins. The Cx35 group also appears to have lost the intron located in the 5′ untranslated region of other connexin genes (cf. Miller et al., 1988). However, we cannot exclude the presence of introns elsewhere in the gene, because the cDNA sequences examined encompass a relatively small portion of the full transcript of either gene.
Phylogenetic analysis of the connexin family (Fig. 9) indicates that the Cx35 group is divergent from all other connexins. This was noted earlier for the skate Cx35 (O’Brien et al., 1996), but the significance could not be evaluated at that time because the skate is the most primitive vertebrate from which any connexin had been cloned. The current analysis, which includes a mammalian and two teleost members of the Cx35 group, shows that they are tightly clustered and remain disparate from other connexins. Three other teleost connexins, zebrafish Cx43.4 and Atlantic croaker Cx32.2 and Cx32.7, fell into the α group in this analysis. Cx32.2 and Cx32.7 lie well within the α group, whereas Cx43.4 clusters with mammalian and avian Cx45. This is a good indication that the phylogenetic positions of the species studied are not the cause of the divergence of the Cx35 group. Indeed, the analysis reveals a deep division within the connexin gene family, with the branch leading to the Cx35 group being the longest in the tree. These results, together with the gene structure data, provide evidence that the Cx35 group has followed a unique line of descent. From this perspective, it seems appropriate to identify this group of connexins as the γ group. In accordance with this nomenclature, Cx35 would be referred to as γ1 and Cx34.7 as γ2.
At present, it is not clear whether the evolutionary divergence of the connexin subgroups correlates with any distinct pattern of gap-junctional properties. Moreover, the notion that there may be functional incompatibility between the different groups of connexins has not been supported by experimental data. For example, Cx40, an α connexin, fails to form channels with most other α connexins, whereas another member of the α group, Cx46, forms heterotypic gap junctions with a wide variety of α and β connexins (White et al., 1995). Given the divergence between the connexin subgroups and the reported presence of α and β connexins in retinal neurons, it will be interesting to determine the degree of functional compatibility of the γ connexins with α and β connexins and the properties of the heterotypic gap junctions they form.
Footnotes
This research was supported by National Eye Institute Grants EY-06516, EY-02430, EY-11376, and EY-01792, an unrestricted award from Research to Prevent Blindness, Inc., and the Association Française Contre les Myopathies. We thank Scott Lindell of AquaFuture (Turners Falls, MA) for providing the hybrid bass used in this study, Danielle Gomès for help with subcloning, and Jane Zakevicius for expert assistance in the histochemical studies.
Correspondence should be addressed to Dr. Harris Ripps, Department of Ophthalmology and Visual Sciences, University of Illinois College of Medicine 1855 West Taylor Street, Chicago, IL 60612.
REFERENCES
- 1.Barrio LC, Suchyna T, Bargiello T, Xu LX, Roginski RS, Bennett MVL, Nicholson BJ. Gap junctions formed by connexins 26 and 32 alone and in combination are differently affected by applied voltage. Proc Natl Acad Sci USA. 1991;88:8410–8414. doi: 10.1073/pnas.88.19.8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrio LC, Capel J, Jarillo JA, Castro C, Revilla A. Species-specific voltage-gating properties of connexin-45 junctions expressed in Xenopus oocytes. Biophys J. 1997;73:757–769. doi: 10.1016/S0006-3495(97)78108-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennett MVL, Zheng X, Sogin ML. The connexins and their family tree. In: Fambrough DM, editor. Molecular evolution of physiological processes. Rockefeller UP; New York: 1994. [Google Scholar]
- 4.Beyer EC. Gap junctions. Int Rev Cytol. 1993;137C:1–37. [PubMed] [Google Scholar]
- 5.Bruzzone R, Ressot C. Connexins, gap junctions and cell-cell signalling in the nervous system. Eur J Neurosci. 1997;9:1–6. doi: 10.1111/j.1460-9568.1997.tb01346.x. [DOI] [PubMed] [Google Scholar]
- 6.Bruzzone R, Haefliger J-A, Gimlich RL, Paul DL. Connexin40, a component of gap junctions in vascular endothelium, is restricted in its ability to interact with other connexins. Mol Biol Cell. 1993;4:7–20. doi: 10.1091/mbc.4.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruzzone R, White TW, Paul DL. Expression of chimeric connexins reveals new properties of the formation and gating behavior of gap junction channels. J Cell Sci. 1994;17:955–967. doi: 10.1242/jcs.107.4.955. [DOI] [PubMed] [Google Scholar]
- 8.Bruzzone R, White TW, Yoshizaki G, Patiño R, Paul DL. Intercellular channels in teleosts: functional characterization of two connexin genes from Atlantic croaker. FEBS Lett. 1995;358:301–304. doi: 10.1016/0014-5793(94)01457-c. [DOI] [PubMed] [Google Scholar]
- 9.Bruzzone R, White TW, Paul DL. Connections with connexins: the molecular basis of direct intercellular signaling. Eur J Biochem. 1996;238:1–27. doi: 10.1111/j.1432-1033.1996.0001q.x. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y-H, DeHaan RL. Asymmetric voltage dependence of embryonic cardiac gap junction channels. Am J Physiol. 1996;270:C276–C285. doi: 10.1152/ajpcell.1996.270.1.C276. [DOI] [PubMed] [Google Scholar]
- 11.Condorelli DF, Parenti R, Spinella F, Salinaro AT, Belluardo N, Cardile V, Cicirata F. Cloning of a new gap junction gene (Cx36) highly expressed in mammalian brain neurons. Eur J Neurosci. 1998;10:1202–1208. doi: 10.1046/j.1460-9568.1998.00163.x. [DOI] [PubMed] [Google Scholar]
- 12.Cook JE, Becker DL. Gap junctions in the vertebrate retina. Microsc Res Tech. 1995;31:408–419. doi: 10.1002/jemt.1070310510. [DOI] [PubMed] [Google Scholar]
- 13.Cuenca N, Fernández E, García M, DeJuan J. Dendrites of rod dominant ON-bipolar cells are coupled by gap junctions in carp retina. Neurosci Lett. 1993;162:34–38. doi: 10.1016/0304-3940(93)90553-w. [DOI] [PubMed] [Google Scholar]
- 14.Dacheux RF, Raviola E. Horizontal cells in the retina of the rabbit. J Neurosci. 1982;2:1486–1493. doi: 10.1523/JNEUROSCI.02-10-01486.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dahl G. The Xenopus oocyte cell–cell channel assay for functional analysis of gap junction proteins. In: Stevenson BR, Gallin WJ, Paul DL, editors. Cell–cell interactions: a practical approach. Oxford UP; New York: 1992. pp. 143–165. [Google Scholar]
- 16.Dahl G, Werner R, Levine E, Rabadan-Diehl C. Mutational analysis of gap junction formation. Biophys J. 1992;62:172–182. doi: 10.1016/S0006-3495(92)81803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsenstein J (1995) PHYLIP (Phylogeny Inference Package) version 3.57c. Distributed by the author. Department of Genetics, University of Washington, Seattle.
- 18.Finch EA, Paul DL. Expression and localization of different gap junction proteins in retina and other regions of the CNS. J Neurosci [Abstr] 1989;15:127. [Google Scholar]
- 19.Giblin LJ, Christensen BN. Connexin43 immunoreactivity in the catfish retina. Brain Res. 1997;755:146–150. doi: 10.1016/s0006-8993(97)00174-1. [DOI] [PubMed] [Google Scholar]
- 20.Green CR, Harfst E, Gourdie RG, Severs NJ. Analysis of the rat liver gap junction protein: clarification of the anomalies in its molecular size. Proc R Soc Lond B Biol Sci. 1988;233:165–174. doi: 10.1098/rspb.1988.0016. [DOI] [PubMed] [Google Scholar]
- 21.Gupta VK, Berthoud VM, Atal N, Jarillo JA, Barrio LC, Beyer EC. Bovine connexin44, a lens gap junction protein: molecular cloning, immunologic characterization, and functional expression. Invest Ophthalmol Vis Sci. 1994;35:3747–3758. [PubMed] [Google Scholar]
- 22.Hennemann H, Dahl E, White JB, Schwarz H-J, Lalley PA, Chang S, Nicholson BJ, Willecke K. Two gap junction genes, connexin 31.1 and 30.3, are closely linked on mouse chromosome 4 and preferentially expressed in skin. J Biol Chem. 1992a;267:17225–17233. [PubMed] [Google Scholar]
- 23.Hennemann H, Suchyna T, Lichtenberg-Fraté H, Jungbluth S, Dahl E, Schwarz J, Nicholson BJ, Willecke K. Molecular cloning and functional expression of mouse connexin40, a second gap junction gene preferentially expressed in lung. J Cell Biol. 1992b;117:1299–1310. doi: 10.1083/jcb.117.6.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janssen-Bienhold U, Dermietzel R, Weiler R. Distribution of connexin43 immunoreactivity in the retinas of different vertebrates. J Neurosci [Abstr] 1996;22:882. [PubMed] [Google Scholar]
- 25.Jiang JX, White TW, Goodenough DA, Paul DL. Molecular cloning and functional characterization of chick lens fiber connexin 45.6. Mol Biol Cell. 1994;5:363–373. doi: 10.1091/mbc.5.3.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones CR, Becker DL, Cook JE. Gap junctions in the rat retina: immunoreactivity with antisera raised to oligopeptides of connexins Cx32 & Cx43. Neurosci Lett [Suppl] 1992;42:S34. [Google Scholar]
- 27.Krieg PA, Melton DA. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 1984;12:7057–7070. doi: 10.1093/nar/12.18.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marc RE, Liu W-L S, Muller JF. Gap junctions in the inner plexiform layer of the goldfish retina. Vision Res. 1988;28:9–24. [PubMed] [Google Scholar]
- 29.McMahon DG. Modulation of electrical synaptic transmission in zebrafish retinal horizontal cells. J Neurosci. 1994;14:1722–1734. doi: 10.1523/JNEUROSCI.14-03-01722.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Methfessel C, Witzemann V, Takahashi T, Mishina M, Numa S, Sakmann B. Patch clamp measurements on Xenopus laevis oocytes: currents through endogenous channels and implanted acetylcholine receptor and sodium channels. Pflügers Arch. 1986;407:577–588. doi: 10.1007/BF00582635. [DOI] [PubMed] [Google Scholar]
- 31.Miller T, Dahl G, Werner R. Structure of a gap junction gene:rat connexin-32. Biosci Rep. 1988;8:455–464. doi: 10.1007/BF01121644. [DOI] [PubMed] [Google Scholar]
- 32.Mills SL, Massey SC. Differential properties of two gap junctional pathways made by AII amacrine cells. Nature. 1995;377:734–738. doi: 10.1038/377734a0. [DOI] [PubMed] [Google Scholar]
- 33.Moore KL, Graham MA, Barr ML. Nuclear morphology, according to sex, in human tissues. Acta Anat (Basel) 1954;21:197–208. doi: 10.1159/000140928. [DOI] [PubMed] [Google Scholar]
- 34.Nicholson BJ, Suchyna T, Xu LX, Hammernick P, Cao FL, Fourtner C, Barrio L, Bennett MVL. Divergent properties of different connexins expressed in Xenopus oocytes. Prog Cell Res. 1993;3:3–13. [Google Scholar]
- 35.O’Brien J, Al-Ubaidi MR, Ripps H. Connexin 35: a gap-junctional protein expressed preferentially in the skate retina. Mol Biol Cell. 1996;7:233–243. doi: 10.1091/mbc.7.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Padgett RA, Grabowski PJ, Konarska MM, Seiler S, Sharp PA. Splicing of messenger RNA precursors. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- 37.Poznanski RR, Umino O. Syncytial integration by a network of coupled bipolar cells in the retina. Prog Neurobiol. 1997;53:273–291. doi: 10.1016/s0301-0082(97)00037-3. [DOI] [PubMed] [Google Scholar]
- 38.Qian H, Malchow RP, Ripps H. Gap-junctional properties of electrically coupled skate horizontal cells in culture. Vis Neurosci. 1993;10:287–295. doi: 10.1017/s0952523800003680. [DOI] [PubMed] [Google Scholar]
- 39.Qian H, Hyatt G, Schanzer A, Hazra R, Hackam AS, Cutting GR, Dowling JE. A comparison of GABAC and ρ subunit receptors from the white perch retina. Vis Neurosci. 1997;14:843–851. doi: 10.1017/s0952523800011585. [DOI] [PubMed] [Google Scholar]
- 40.Raviola E. Intercellular junctions in the outer plexiform layer of the retina. Invest Ophthalmol. 1976;15:881–895. [Google Scholar]
- 41.Robinson SR, Hampson ECGM, Munro MN, Vaney DI. Unidirectional coupling of gap junctions between neuroglia. Science. 1993;262:1072–1074. doi: 10.1126/science.8093125. [DOI] [PubMed] [Google Scholar]
- 42.Rup DM, Veenstra RD, Wang H-Z, Brink PR, Beyer EC. Chick connexin-56, a novel lens gap junction protein: molecular cloning and functional expression. J Biol Chem. 1993;268:706–712. [PubMed] [Google Scholar]
- 43.Saito T, Kujiraoka T. Characteristics of bipolar–bipolar coupling in the carp retina. J Gen Physiol. 1988;91:275–287. doi: 10.1085/jgp.91.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual, Ed 2. Cold Spring Harbor Laboratory; Plainview, NY: 1989. [Google Scholar]
- 45.Spray DC, Harris AL, Bennett MVL. Equilibrium properties of a voltage-dependent junctional conductance. J Gen Physiol. 1981;77:77–93. doi: 10.1085/jgp.77.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swenson KI, Jordan JR, Beyer EC, Paul DL. Formation of gap junctions by expression of connexins in Xenopus oocyte pairs. Cell. 1989;57:145–155. doi: 10.1016/0092-8674(89)90180-3. [DOI] [PubMed] [Google Scholar]
- 47.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Umino O, Maehara M, Hidaka S, Kita S, Hashimoto Y. The network properties of bipolar–bipolar cell coupling in the retina of teleost fishes. Vis Neurosci. 1994;11:533–548. doi: 10.1017/s0952523800002443. [DOI] [PubMed] [Google Scholar]
- 49.Vaney DI. Many diverse types of retinal neurons show tracer coupling when injected with biocytin or neurobiotin. Neurosci Lett. 1991;125:197–190. doi: 10.1016/0304-3940(91)90024-n. [DOI] [PubMed] [Google Scholar]
- 50.Vaney DI. Patterns of neuronal coupling in the retina. Prog Ret Eye Res. 1994;13:301–355. [Google Scholar]
- 51.Wagner TLE, McMahon DG, Beyer EC. Functional expression and characterization of a gap junction channel gene from the retina of the giant danio. Soc Neurosci Abstr. 1997;23:2359. [Google Scholar]
- 52.White TW, Bruzzone R. Multiple connexin proteins in single intercellular channels: connexin compatibility and functional consequences. J Bioenerg Biomembr. 1996;28:339–350. doi: 10.1007/BF02110110. [DOI] [PubMed] [Google Scholar]
- 53.White TW, Bruzzone R, Goodenough DA, Paul D. Mouse Cx50, a functional member of the connexin family of gap junction proteins, is the lens fiber protein MP70. Mol Biol Cell. 1992;3:711–720. doi: 10.1091/mbc.3.7.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.White TW, Bruzzone R, Wolfram S, Paul DL, Goodenough DA. Selective interactions among the multiple connexin proteins expressed in the vertebrate lens: the second extracellular domain is a determinant of compatibility between connexins. J Cell Biol. 1994;125:879–892. doi: 10.1083/jcb.125.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.White TW, Paul DL, Goodenough DA, Bruzzone R. Functional analysis of selective interactions among rodent connexins. Mol Biol Cell. 1995;6:459–470. doi: 10.1091/mbc.6.4.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilders R, Jongsma HJ. Limitations of the dual voltage clamp method in assaying conductance and kinetics of gap junction channels. Biophys J. 1992;63:942–953. doi: 10.1016/S0006-3495(92)81664-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zahs KR, Newman EA. Asymmetric gap junctional coupling between glial cells in the rat retina. Glia. 1997;20:10–22. [PubMed] [Google Scholar]