Abstract
Zinc inhibits NMDA receptor function through both voltage-dependent and voltage-independent mechanisms. In this report we have investigated the role that the NR1 subunit plays in voltage-independent Zn2+ inhibition. Our data show that inclusion of exon 5 into the NR1 subunit increases the IC50 for voltage-independent Zn2+inhibition from 3-fold to 10-fold when full length exon 22 is also spliced into the mature NR1 transcript and the NMDA receptor complex contains the NR2A or NR2B subunits; exon 5 has little effect on Zn2+ inhibition of receptors that contain NR2C and NR2D. Mutagenesis within exon 5 indicates that the same residues that control proton inhibition, including Lys211, also control the effects of exon 5 on Zn2+inhibition. Amino acid exchanges within the NR1 subunit but outside exon 5 (E181Q, E339Q, E342Q, N616R, N616Q, D669N, D669E, C744A, and C798A) that are known to decrease the pH sensitivity also decrease the Zn2+ sensitivity, and concentrations of spermine that relieve tonic proton inhibition also relieve Zn2+ inhibition. In summary, our results define the subunit composition of Zn2+-sensitive NMDA receptors and provide evidence for structural convergence of three allosteric regulators of receptor function: protons, polyamines, and Zn2+.
Keywords: NMDA receptors, zinc, protons, RNA splicing, polyamines, site-directed mutagenesis
The group IIB transition metal ion Zn2+ is widely and variably distributed throughout neurons in the CNS. Zn2+ is released from nerve fibers in a Ca2+-dependent manner and sequestered into neurons via uptake systems withKm values between 10 and 400 μm(for review, see Smart et al., 1994). Although the Zn2+ concentration within the narrow synaptic cleft is dynamic and difficult to quantify, estimates of the concentration of interstitial synaptically released Zn2+ range from 5 to 500 μm (Assaf and Chung, 1984; Howell et al., 1984;Charlton et al., 1985; Aniksztejn et al., 1987). Of the numerous roles that have been proposed for Zn2+ in the CNS, perhaps the best known are its actions on a host of voltage- and ligand-gated ion channels, including glutamate receptors (Smart et al., 1994; Laube et al., 1995).
Glutamate receptors can be subdivided on the basis of both structure and pharmacology into three classes: NMDA, kainate, and AMPA receptors. The potential involvement of NMDA receptors in synaptic plasticity and neuronal development (McBain and Mayer, 1994; Scheetz and Constantine-Paton, 1994; Asztely and Gustafsson, 1996) as well as neurological disorders such as epilepsy, ischemic cell death, and neurodegeneration (Choi, 1992; Bradford, 1995; Whetsell, 1996) have fueled intensive study of all aspects of this receptor, including its sensitivity to extracellular Zn2+ (Peters et al., 1987; Westbrook and Mayer, 1987; Choi and Koh, 1998). Inhibition of NMDA receptor function by extracellular Zn2+ within the predicted concentration range in the CNS raises the possibility that the modulatory effects of Zn2+ may be an important component of some of the processes and disorders that NMDA receptors are thought to participate in (Peters et al., 1987; Koh et al., 1996).
In cultured neurons, tens of micromolar Zn2+ can inhibit NMDA receptors through a voltage-dependent channel block that appears to occur within the ion channel pore and may involve residues (e.g., Asn616 in M2 region of NR1) that are known to influence other NMDA channel-blocking ions such as Mg2+ (Mayer and Vyklicky, 1989; Christine and Choi, 1990; Legendre and Westbrook, 1990; Mori et al., 1992; Sakurada et al., 1993; Kawajiri and Dingledine, 1993). Lower concentrations of Zn2+ (<5 μm) can act in a voltage-independent manner to reduce the frequency of channel opening as well as the duration of neuronal NMDA receptor-mediated bursts (Christine and Choi, 1990; Legendre and Westbrook, 1990).
Five cDNAs have been identified that encode NMDA receptor subunits (NR1, NR2A, NR2B, NR2C, and NR2D; McBain and Mayer, 1994), and zinc inhibits recombinant receptors comprised of NR1 + NR2 receptors in a dual voltage-dependent and -independent manner that appears to be controlled by the NR2 subunit (Mori et al., 1992; Kawajiri and Dingledine, 1993; Sakurada et al., 1993; Williams, 1996; Chen et al., 1997; Paoletti et al., 1997). In contrast, very little information exists about the role of alternative splicing of the NR1 subunit RNA on Zn2+ inhibition of heteromeric NMDA receptors (Paoletti et al., 1997), although the eight possible NR1 splice variants expand structural heterogeneity of the NMDA receptor class considerably. NR1 splicing occurs in a region- and developmental-specific manner (Laurie and Seeburg, 1994; Nash et al., 1997; Paupard et al., 1997), suggesting it may be used by the CNS to fine tune NMDA receptor responsiveness or influence receptor localization. We have studied the effects of Zn2+ on recombinant NMDA receptors containing different NR1 splice variants to define structural determinants of Zn2+ inhibition. In this report, we describe striking effects of the N-terminal alternative exon 5 on voltage-independent Zn2+inhibition, some of which are reminiscent of the effects of exon 5 on proton inhibition of NMDA receptors (Zheng et al., 1994; Traynelis et al., 1995). The similar effects of exon 5 on Zn2+and proton inhibition lead us to explore whether the modulatory sites for these endogenous ions share similar structural determinants.
Some of these results have appeared in preliminary form (Burgess et al., 1996; Burgess and Traynelis, 1997).
MATERIALS AND METHODS
NMDA receptor subunit cDNAs. cDNAs encoding the NR1 subunit splice variants (GenBank accession number L08228) as well as NR2A (GenBank numbers AF001423 and CD13211), NR2B (GenBank numberU11419), NR2C (GenBank numbers U08259 and M91563), and NR2D (GenBank number L31611) subunits were generously provided by Drs. J. Boulter (University of California at Los Angeles, Los Angeles, CA), S. Heinemann (Salk Institute, La Jolla, CA), S. Nakanishi (Kyoto University, Kyoto, Japan), and P. Seeburg (Max Planck Institute for Medical Research, Heidelberg, Germany). NR1(N616R) and NR1(N616Q) were generously provided by Drs. R. Dingledine (Emory University, Atlanta, GA) and K. Moriyoshi (Kyoto University, Kyoto, Japan), respectively. NR1-1a(E181Q), NR1-1a(E339Q), NR1-1a(E342Q), NR1-1b(E363Q), NR1-1a(D669N), and NR1-1a(D669E) were generously provided by K. Williams (University of Pennsylvania, Philadelphia, PA); the wild-type sequence for NR1 subunit for these mutants can be accessed by GenBank number X63255. Other NMDA receptor NR1 subunit mutations (see Fig. 7) have been previously described (Sullivan et al., 1994; Traynelis et al., 1995). All experiments were performed with NR1 subunits that contained exon 21 and exon 22 except those shown in Figures 1 and 2. NR1 splice variants that contain the two C-terminal alternate exons but lack exon 5 are referred to in the literature as NR1-1a (Hollmann et al., 1993), NR1011 (Durand et al., 1992), and NR1A (Sugihara et al., 1992). NR1-1b, NR1111, and NR1B describe the corresponding cDNA that contains exon 5. We use the nomenclature of Hollmann et al. (1993), and for simplicity, refer whenever possible to the presence or absence of NR1 exon 5.
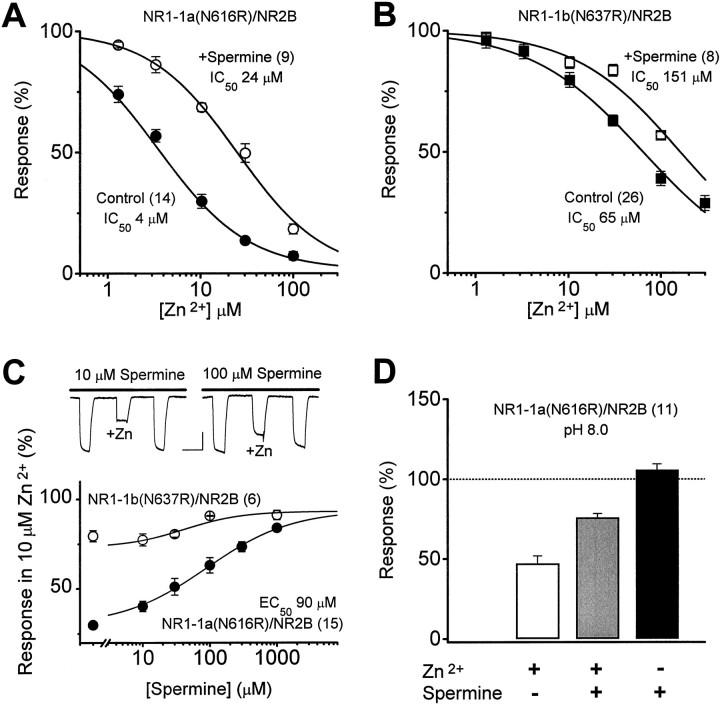
Fig. 7.
Relief of Zn2+ inhibition by spermine. A, B, To test whether spermine also relieves voltage-independent Zn2+ inhibition in an manner analogous to the effect of exon 5, Zn2+inhibition curves were constructed for exon 5-lacking NR1-1a(N616R)/NR2B receptors in the absence and presence of 300 μm spermine. This concentration of spermine induces no channel block of these mutant receptors (Fig. 5C). Spermine shifted the IC50 for Zn2+inhibition of receptor function by sixfold when exon 5 was absent (A) and by 2.3-fold for receptors that contain exon 5 (B). C, Top panel, Zn2+ inhibition of glutamate-evoked NR1-1a(N616R)/NR2B receptor-mediated currents recorded in either 10 or 100 μm spermine at −30 mV. Calibration: 150 sec, 50 nA.Bottom panel, Spermine decreases the inhibition of NR1-1a(N616R)/NR2B receptor responses observed in 10 μmZn2+ in a dose-dependent manner with an IC50 value of 90 μm; spermine can similarly relieve the small amount of inhibition of exon 5-containing NR1-b(N637R)/NR2B receptors as well (open circles).D, To test whether the actions of spermine persist in the absence of polyamine potentiation, we evaluated the effect of spermine on Zn2+ inhibition at pH 8.0 at which there is no spermine potentiation (i.e., because there is no inhibition by protons). Under these conditions, we still observe relief of Zn2+ inhibition, consistent with the idea that spermine, like exon 5, can relieve Zn2+ inhibition. Numbers in parentheses are the number of oocytes.
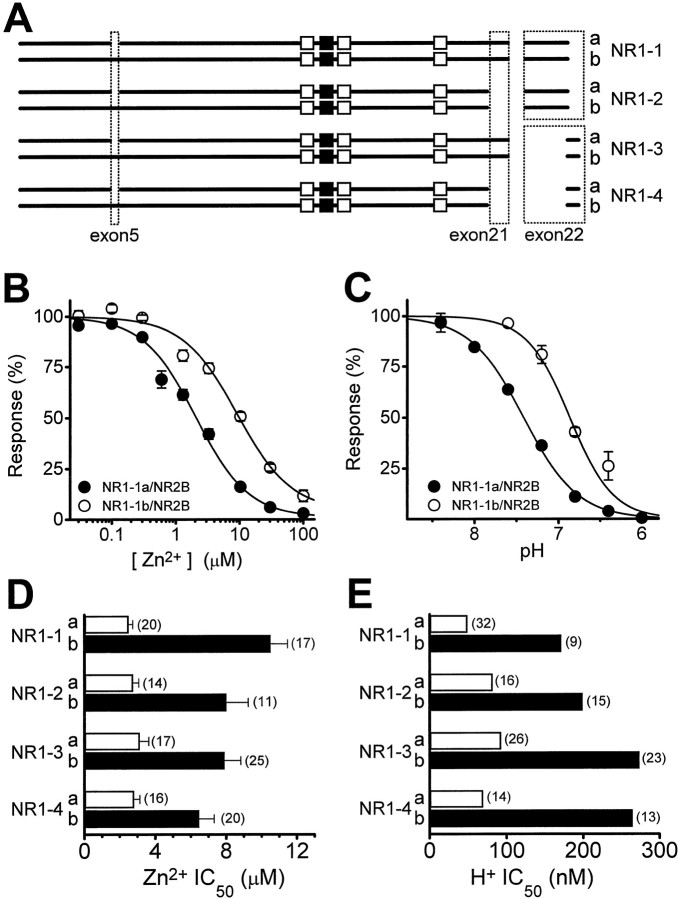
Fig. 1.
Control of Zn2+ and proton inhibition by RNA splicing of the NR1 subunit. A, Coding regions are shown schematically for eight NR1 splice variants (nomenclature according to Hollmann et al., 1993; the suffixa or b denotes absence or presence of exon 5) that arise by alternative use of exons, alternative use of acceptor splice sites, or both. The N terminal is shown to theleft and the C terminal is at the right. The position of exons 5 and 21 are shown as a broken box; two different boxes surround exon 22, each indicating the use of a different acceptor splice site. Membrane-associated regions are shown as boxes (open, transmembrane;closed, membrane-associated). NR1-3 and NR1-4 contain a new 66 base pair reading frame terminated by a previously out of frame stop codon as a result of the use of an alternative splice-acceptor site within exon 22, which deletes the original stop codon.B, Composite Zn2+ inhibition curves are shown for NR1-1 splice variants ± exon 5 coexpressed with NR2B. All error bars are SEM. Low Zn2+concentrations (0.03–0.3 μm) were achieved using tricine buffer (see Materials and Methods); all other concentrations were corrected for 0.3 μm contaminant Zn2+.C, Composite proton inhibition curves are shown for NR1-1 splice variants ± exon 5 coexpressed with NR2B reexpressed as a percent of the fitted maximum. D, Mean-fitted IC50 values for Zn2+ inhibition of wild-type NR1-a/NR2B receptors (open bars) are significantly lower than NR1-b/NR2B (filled bars;p < 0.05 for all). E, IC50 values for proton inhibition of wild-type NR1-a/NR2B receptors (open bars) are also increased by exon 5 (filled bars). The number of oocytes recorded for each experiment is shown in parentheses.
Fig. 2.
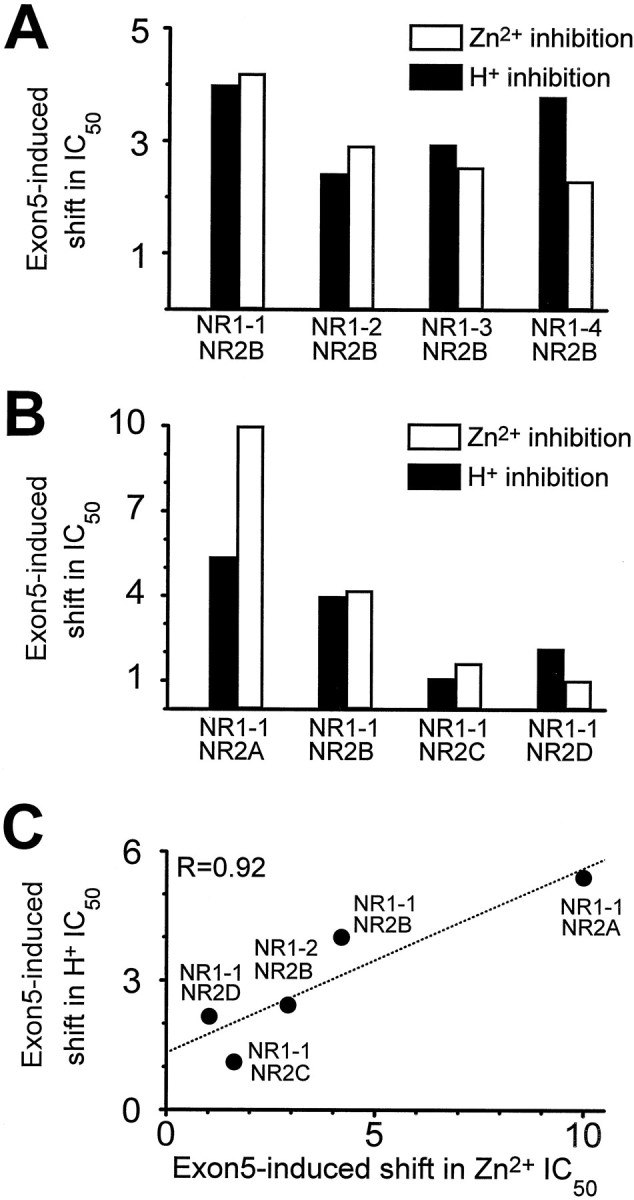
Comparison of the effects of exon 5 on proton and Zn2+ inhibition. A, Ratio of the fitted IC50 values for proton (closed bars) and Zn2+ (open bars) inhibition of NR1 splice variants coexpressed with NR2B. B, Ratio of the fitted IC50 values for proton and Zn2+ inhibition of NR1-1 splice variants coexpressed with different NR2 subunits. Data describing proton inhibition for NR2A and C are from Traynelis et al. (1995). Data for proton inhibition of NR2B are from Figure 1; IC50 values for proton inhibition of NR1-1a/NR2D and NR1-1b/NR2D were 71 (n = 5) and 154 nm (n = 10), respectively.C, Correlation between the effectiveness of exon 5 to relieve proton and Zn2+ inhibition for different NR1 splice variants coexpressed with different NR2 subunits. NR1 variants that use the alternate acceptor splice site within exon 22 are not shown because of the oocyte-dependent variability in the effects of exon 5.
Expression of NMDA receptors in Xenopus oocytes.cRNA was synthesized from linearized template cDNA according to manufacturer specifications (Ambion). Quality of synthesized cRNA was assessed by gel electrophoresis, and quantity was estimated by spectroscopy and gel electrophoresis. Stage V and VI oocytes were surgically removed from the ovaries of Xenopus laevisanesthetized with 3-amino-benzoic acid ethylester (1 gm/l). Clusters of ∼30 oocytes were incubated with 292 U/ml Worthington (Freehold, NJ) type IV collagenase or 1.3 mg/ml collagenase (Life Technologies, Gaithersburg, MD; 17018–029) for 2 hr in Ca2+-free solution comprised of (in mm) 115 NaCl, 2.5 KCl, and 10 HEPES, pH 7.5, with slow agitation to remove the follicular cell layer. Oocytes were then washed extensively in the same solution supplemented with 1.8 mm CaCl2 and maintained in Barth’s solution comprised of (in mm): 88 NaCl, 1 KCl, 24 NaHCO3, 10 HEPES, 0.82 MgSO4, 0.33 Ca(NO3)2, and 0.91 CaCl2 and supplemented with 100 μg/ml gentamycin, 40 μg/ml streptomycin, and 50 μg/ml penicillin. Oocytes were injected within 24 hr of isolation with 5 ng of NR1 subunit and 5–10 ng of NR2 subunit in a 50–100 nl volume and incubated in Barth’s solution at 18°C for 3–18 d; some oocytes were stored at 4°C after 3–5 d.
Voltage-clamp recordings from Xenopus oocytes.Two electrode voltage-clamp recordings were made 3–18 d postinjection. Oocytes were placed in a dual-track recording chamber with a single perfusion line that split to perfuse two oocytes, which increased the efficiency of data collection and allowed side-by-side comparisons to be made on oocytes injected with different subunit combinations. Dual recordings were made using Warner model OC725B two-electrode voltage clamps as recommended by the manufacturer. The bath clamps communicated across silver chloride wires placed into each side of the recording chamber, both of which were assumed to be at a reference potential of 0 mV. Oocytes were perfused with a solution comprised of (in mm) 90 NaCl, 1 KCl, 10 HEPES, and 1.0 BaCl2, pH 7.3, and held at −20 to −40 mV. ZnCl2 solutions (10 mm) were made fresh every 10 hr and added directly to the recording solution to obtain the desired nominal Zn2+ concentration for all experiments except those with NR2B- and NR2A-expressing receptors. For these receptors, we used tricine to buffer Zn2+concentrations <3 μm. We used a stability constant (KS) for Zn-tricine of 10−5m (Vieles et al., 1972; Tripathi et al., 1987; Paoletti et al., 1997) to calculate the nominal free Zn2+ concentration according to:
| Equation 1 |
in which we assume that the [Tricine] is unchanged by addition of Zn2+ to the solution (i.e.,[Tricine] is much greater than [Zn2+]TOTAL). α is defined by:
| Equation 2 |
where the KA for tricine is 10−8.15. Similar free Zn2+concentrations were obtained with the use of programs that calculate free ion concentrations (WINMAXC or BAD; Zheng et al., 1998). HEPES does not contribute to Zn2+ buffering (Benitez et al., 1991). We estimate a free Zn2+ at pH 7.4 that is ∼80% of that reported at pH 7.3 by Paoletti et al. (1997). Our contaminant Zn2+ (Paoletti et al., 1997) is ∼300 nm (Zheng et al., 1998) and arises predominantly from the NaCl. We assume that only free (or fully hydrated) Zn2+ acts at NMDA receptors, and calculated free Zn2+ concentrations at pH 8.0 using the log stability constants (KSn) for Zn(OH)n+2−n of 5.0 (n = 1), 11.1 (n = 2), 13.6 (n = 3), and 14.8 (n = 4; Martell and Smith, 1974) according to:
| Equation 3 |
Secure impalements were usually obtained in solution containing 1 mm CaCl2 before switching to the BaCl2-containing solution. Voltage and current electrodes had resistances of 2–10 MΩ and were filled with 300 mmKCl. NMDA receptor currents were evoked with 20 μmglutamate plus 7–10 μm glycine superfused for 1–2 min; washout time was 3–5 min between drug applications. For NR2A-containing receptors, all experiments were performed in 10 μm glycine, which is 10× our measured EC50value for glycine (1 μm; n = 8; data not shown). Solution exchange was computer-controlled through an eight-valve manifold. The flow rate varied minimally between various valves and throughout the experiment.
Data analysis. Only current responses to glutamate plus glycine that were >50 nA at pH 7.3 (−40 mV) and were not potentiated by 1 μm Zn2+ (Hollmann et al., 1993) were analyzed to minimize the contribution to our responses of previously described NR1 homomeric receptors, which are probably heteromeric receptors that incorporate Xenopus glutamate receptor subunits (Soloviev et al., 1996). By this criteria, ∼6% of recordings were discarded; response amplitude was typically several hundreds of nanoamperes at −20 mV. Inhibition curves were analyzed as shown in Figure 3A, Zn2+ concentrations were corrected for 0.3 μm contaminant Zn2+ for NR2B-containing receptors. For proton inhibition curves, the oocytes were prewashed with the test pH before and after agonist application. Experimental manipulations were expressed as a percent of pre-event and post-event control responses, and the data were pooled together. Zn2+ inhibition data were fitted (least squares criterion) to the equation:
| Equation 4 |
where n is the Hill slope, IC50 is the nominal concentration of Zn2+ that produces 50% inhibition, and minimum a residual current response. For all receptors, minimum was fixed to 0 if the fitting algorithm returned a value <5.6%, our estimated limit of detection for the maximal Zn2+ concentration tested (mean SEM for responses at 100 μm Zn2+ was 2.55%). For mutant receptors and NR2C- and NR2D-containing receptors,minimum was fixed to 0. Hill slopes ranged from 0.5 to 1.6; IC50 values for wild-type NR1/NR2B,C,D receptors will be slight underestimates of true IC50 for voltage-independent effects if voltage-dependent inhibition by high concentrations (30–100 μm) of Zn2+ occurs at holding potentials between −20 and −40 mV; Hill slopes will be slightly overestimated for the same reason. Throughout the text the IC50 values that are reported reflect the nominal Zn2+ concentration; effective Zn2+ activity will be approximately half the nominal concentration given the activity coefficient for Zn2+ of 0.5 (Lobo and Quaresma, 1989; Lide, 1997). Composite proton inhibition curves were constructed using pH 7.6 as a control and fitted using the same equation, except with proton activity substituted for [Zn2+]. Proton IC50values were determined from the pH-derived activities using an activity coefficient of 0.8 (Lide, 1997).
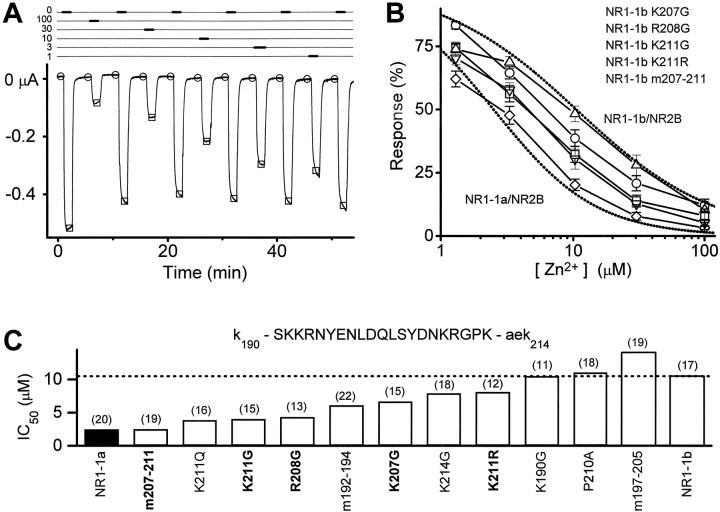
Fig. 3.
Structural determinants within exon 5 that control Zn2+ inhibition. A, Typical recording of mutant NMDA receptor NR1-1b(K211R)/NR2B current responses from which Zn2+ inhibition curves were determined. Thethick bars (top) show application of solutions nominally containing 0, 100, 30, 10, 3, or 1 μmZn2+ plus maximal glutamate and glycine. Before recording glutamate responses in the presence 30 and 100 μm Zn2+, the oocyte was prewashed with 30 and 100 μm Zn2+ to control for any shift in the preresponse leak current. Measurements of the preresponse baseline (open circles) and the peak response (open squares) were made from the same position relative to the response waveform. B, Broken linesshow the Zn2+ inhibition curves for wild-type NR1-1a/NR2B and NR1-1b/NR2B receptors. Symbols show inhibition of NR1-1b/NR2B receptors with amino acid substitutions at amino acid positions 207, 208, and 211 in the NR1-1b subunit.C, IC50 values were determined for Zn2+ inhibition of NR1-1b/NR2B receptors containing the mutations indicated (open bars). The filled bar shows the IC50 values for receptors comprised of wild-type NR1-1a/NR2B for comparison. The broken lineshows the IC50 of wild-type NR1-1b/NR2B. The amino acid sequence of exon 5 is written above (capitals letters are amino acids encoded by exon 5). Full inhibition curves for mutant receptors labeled in bold are shown in B. Numbers in parentheses show the number of oocytes used to construct each inhibition curve. m207–211 describes an NR1-1b mutant subunit that contains three amino acid substitutions (K207G, R208G, and K211G); m192–194 contains NR1-1b mutations K192G, K193G, and R194G, and m197–205 contains NR1-1b mutations E197A, D200A, and D205A.
Current–voltage (I–V) relationships were recorded in response to a 4 sec voltage ramp (−80 to +40 mV) before, during, and after drug application. The NMDA receptor I–Vcurve was determined from the difference of current responses in the presence and absence of agonist, and the reversal potential was determined from a fitted polynomial function (fifth to ninth order; least squares criterion) or interpolation.
Statistical significance for all tests was set at p < 0.05, and minimum detectable differences were calculated for a power of 0.7. Error bars in all figures are SEM. Paired and unpairedt tests and ANOVA were performed to evaluate the difference between means; for t tests we used Welch’s approximatet statistic when the underlying distributions had different variances.
Voltage-dependent channel block of wild-type and mutant NMDA receptors. Because both Zn2+ and polyamines can cause a voltage-dependent block of the NMDA receptor channel, it is important to determine whether this effect complicates any of the experiments we performed. Although we observe a weak voltage-dependent channel block of wild-type NR1-1/NR2B receptors by 10 μmZn2+ at hyperpolarized potentials (e.g., −70 mV;n = 9; p < 0.05), we can detect no voltage-dependent block by extracellular Zn2+ at our typical recording potential, as measured by the ratio of current recorded 30 mV negative to the reversal potential to that recorded 30 mV positive to the reversal potential (IVREV−30/IVREV+30), determined from current–voltage curves (controlIVREV−30/IVREV+30 = 1.0 ± 0.02, mean ± SEM; 10 μmZn2+IVREV−30/IVREV+30= 1.0 ± 0.06; p > 0.4; n = 9; minimum detectable difference, 15%). We also find no significant effects of exon 5 on channel block induced by addition of 10 μm Zn2+ (n = 9 oocytes; data not shown) or 0.5 mm Mg2+(n = 28 oocytes; data not shown), and no effects of exon 5 on the current–voltage curve recorded when all external Na+ was replaced by 61.5 mmBa2+ (n = 28 oocytes; data not shown). These data suggest the effects of exon 5 on voltage-independent Zn2+ inhibition that we describe here are unlikely to involve ion permeation or block, except perhaps at very high concentrations of Zn2+. To evaluate the ability of spermine to relieve Zn2+ inhibition (see Fig.7), we have used an NR1 subunit that contains a mutation in the M2 region of the receptor that has previously been shown to relieve voltage-dependent channel block by both Zn2+ and spermine (Mori et al., 1992; Kawajiri and Dingledine, 1993; Sakurada et al., 1993; Kashiwagi et al., 1997). To confirm that under our recording conditions these M2 mutations relieved voltage-dependent Zn2+ inhibition of NR1-1/NR2B receptors, we constructed current–voltage curves in the presence and absence of extracellular Zn2+ and spermine for wild-type and mutant receptors. We found receptors that contained an arginine residue in place of the M2 Asn616 in NR1-1a and M2 Asn637 in NR1-1b (mutant numbering differs because of inclusion of 21 amino acids encoded by exon 5) showed no detectable channel block by Zn2+ at −30 mV (pooled controlIVREV−30/IVREV+30 = 1.1 ± 0.04; 30 μm Zn2+IVREV−30/IVREV+30= 1.0 ± 0.1; p > 0.1; n = 11; minimum detectable difference, 13%) or at −70 mV (data not shown;n = 23), consistent with the findings of other laboratories (Mori et al., 1992; Kawajiri and Dingledine, 1993;Sakurada et al., 1993). Similarly, voltage-dependent channel block by 1 mm spermine (see Fig.5A,B) was also abolished by the M2 pore mutation in NR1-1a(N616R)/NR2B and NR1-1b(N637R)/NR2B receptors (pooledIVREV−30/IVREV+30 = 1.2 ± 0.03; n = 11) compared with wild-type NR1-1/NR2B receptors (IVREV−30/IVREV+30 = 0.6 ± 0.05; n = 16; p < 0.05 by unpaired t test). Similar results were found at more hyperpolarized potentials and with 100 μm spermine (n = 27; Kashiwagi et al., 1997).
Fig. 5.
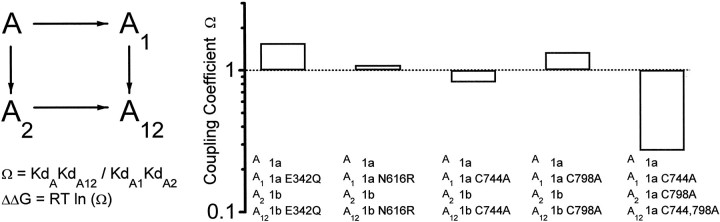
Thermodynamic cycle analysis of mutant NR1 subunits. The mutations in exon 5 containing NR1-1b subunits are labeled with the numbering of NR1-1a subunits for clarity. However, the position of these mutations should be increased by 21 residues because of insertion of exon 5. The actual numbering of the mutant pairs is (± exon 5) E342Q/E363Q, N616R/N637R, C744A/C765A, and C798A/C819A. The coupling coefficient was calculated as indicated assuming our experimental IC50 values approximate the KD values for protonation. The coupling energies (ΔΔG) in kilocalories per mole for these various mutations and splice variants are 0.23 for E342/E363Q ± exon 5, 0.05 for N616/N637R ± exon 5, −0.12 for C745/C765A ± exon 5, 0.17 for C798/C819A ± exon 5, and −0.8 for C745A and C798A. Proton IC50 values not included in Figures 3 or 4 are (in nm): 1071 NR1-1b(E363Q)/NR2B, 446 NR1-1b(N637R)/NR2B, 558 NR1-1b(C765A), 688 NR1-1b(C819A), and 135 NR1-1a(C744A, C798A)/NR2B. All inhibition curves were constructed for wild-type and mutant NR1-1a and NR1-1b subunits coexpressed with NR2B except for NR1-1b(C765A) and NR1-1b(C819A), which were expressed in Xenopus oocytes in the absence of NR2 (Sullivan et al., 1994).
RESULTS
Zn2+ sensitivity of alternatively spliced NR1 subunits
Extracellular Zn2+ is known to inhibit NMDA receptors by both voltage-independent and voltage-dependent mechanisms. Voltage-dependent inhibition appears to involve weak channel block (Mayer et al., 1989; Christine and Choi, 1990; Legendre and Westbrook, 1990) and is controlled by some of the same amino acid residues that control channel block by Mg2+ (Mori et al., 1992;Kawajiri and Dingledine, 1993; Sakurada et al., 1993; see Materials and Methods). In this study we have examined the ability of NR1 to control voltage-independent inhibition of NMDA receptors by extracellular Zn2+. To do this, we first evaluated the IC50 values for Zn2+ inhibition of responses to maximal glutamate and glycine of all NR1 splice variants expressed in combination with the NR2B subunit in Xenopusoocytes. Current responses were recorded from oocytes expressing NMDA receptor subunits that were voltage clamped at depolarized holding potentials (−20 to −40 mV) at which voltage-dependent channel block by Zn2+ is minimal (see Materials and Methods). Figure1 summarizes the effects of extracellular Zn2+ on the function of NMDA receptors comprised of different NR1 splice variants and the NR2B subunit. Inclusion of the N-terminal alternative exon 5 reduced the sensitivity to extracellular Zn2+ by more than fourfold for receptors that had spliced in exon 22 starting at the alternative acceptor site 2647 bp [numbered as in GenBank number X63255; seeHollmann et al. (1993), their Fig. 1]. By contrast, exon 5 had variable effects on the Zn2+ sensitivity of recombinant receptors that had the original stop codon deleted by use of an internal acceptor splice site at 3003 bp within exon 22 [i.e., NR1-3 and NR1-4 by the nomenclature of Hollmann et al. (1993)]. In two of four batches of oocytes, exon 5 reduced Zn2+sensitivity of NR1-3/NR2B and NR1-4/NR2B receptors by less than twofold when the alternative acceptor splice site within exon 22 was used; in the other batches of oocytes expressing NR1-3/NR2B and NR1-4/NR2B receptor, exon 5 had much larger effects. This exon 22-linked variability might be related to interactions between proteins endogenous to the oocyte and the PDZ2 site (Kornau et al., 1995) encoded in the new reading frame that results from the deletion of the stop codon (data not shown). Interestingly, the effects of exon 5 on Zn2+ inhibition were largely unaffected by the presence or absence of alternative exon 21 (Fig. 1).
Inclusion of exon 5 into the NR1 subunit has previously been shown to reduce the proton sensitivity of NR1-1/NR2 receptors (Traynelis et al., 1995). Because of the similar effects of exon 5 on proton and Zn2+ inhibition of receptors that contain the full length NR1 subunit, we tested whether C-terminal splicing of NR1 controlled the effects of exon 5 on proton inhibition of heteromeric receptors in a parallel manner to Zn2+ inhibition. Figure 1, D and E, compares the effects of NR1 RNA splicing on proton and Zn2+ inhibition of NR1/NR2B receptors and shows that exon 5 relieves proton inhibition irrespective of C-terminal splicing of NR1 subunit. Similar results for proton inhibition have previously been reported for NR1 splice variants expressed in Xenopus oocytes in the absence of NR2 subunits [Traynelis et al. (1995), their footnote 13]. Figure2A compares more directly the effects of exon 5 on proton and Zn2+inhibition of various C-terminal splice variants of the NMDA receptor.
The NR2 subunit controls the relief of voltage-independent Zn2+ inhibition of exon 5
The NR2 subunit is known to control allosteric regulation of NMDA receptors. For example, both extracellular Mg2+ and polyamines can relieve tonic proton inhibition of NMDA receptors that contain the NR2B subunit, but not other NR2 subunits (Williams et al., 1994; Zhang et al., 1994; Paoletti et al., 1995; Traynelis et al., 1995; Williams, 1995). To test whether the NR2 subunit influences the ability of exon 5 to decrease voltage-independent Zn2+ inhibition, we constructed Zn2+ inhibition curves for receptors comprised of NR1-1 ± exon 5 and each of the NR2 subunits. Table1 summarizes these data, which show that exon 5 can reduce the sensitivity to extracellular Zn2+ of receptors that contain NR1-1 and NR2A or NR2B by 4-fold to 10-fold, although the IC50 for voltage-independent Zn2+ inhibition differs by more than an order of magnitude between NR2A- and NR2B-containing receptors (Paoletti et al., 1997). By contrast, not only did we find that receptors comprised of NR1-1 plus NR2C (Williams, 1996; Chen et al., 1997) or NR2D subunits were much less sensitive to extracellular Zn2+, but exon 5 was also less effective in reducing the sensitivity of these receptors to Zn2+ (Table1). Because exon 5 exerts similar effects on both the proton and Zn2+ sensitivity of NR1 splice variants (Fig.2A), we also compared the effects of different NR2 subunits on proton and Zn2+ sensitivity of NMDA receptors. As shown in Figure 2, B and C, we found a correlation between the effects of exon 5 on proton and Zn2+ sensitivity for various heteromeric receptors. This finding, together with similar results from NR1 splice variants, raises the possibility that inhibition by Zn2+ and protons might share common structural or functional determinants.
Table 1.
Coassembly with NR2 subunits influences the effects of NR1 exon 5 on Zn2+ inhibition curves
| IC50(μm) | Ratio | n | IC50(μm) | Ratio | n | |||
|---|---|---|---|---|---|---|---|---|
| NR1-1a | NR1-1b | +exon 5/−exon 5 | NR1-1a N616R | NR1-1b N637R | +exon 5/−exon 5 | |||
| NR2A | 0.02 | 0.2 | 10 | 25 | ND | ND | ND | |
| NR2B | 2.5 | 11 | 4.2 | 37 | 4 | 50 | 12 | 25 |
| NR2C | 23 | 38 | 1.7 | 21 | ND | ND | ND | |
| NR2D | 14 | 14 | 1.0 | 24 | 27 | 83 | 3 | 30 |
Fitted IC50 values are shown for the indicated NR1-1/NR2 combinations (assuming complete inhibition for NR2B,C,D); Hill slopes ranged from 0.6 to 1.5. Mean IC50 values were similar to those determined from the composite curve. All experiments were performed with constructs that contain exon 21 and exon 22. For NR2A the inhibition curves was generated using tricine-buffered Zn2+ (see Materials and Methods). The mutation in the M2 region (N616R or N637R) enhances the effect of exon 5 on the NR1-1/NR2B and NR1-1/NR2D IC50 values, possibly by removing some Zn2+ channel block at high concentrations that probably increased overall Zn2+ inhibition. Voltage-dependent and -independent IC50 values for NR2A are different enough such that no voltage-dependent block should contaminate inhibition by nanomolar concentrations of Zn2+ (Paoletti et al., 1997). NR2C has very little voltage-dependent inhibition (Chen et al., 1997). Thus the effects of N616R mutation were not determined (ND) for these two subunit combinations. All dose–response curves were determined with five or more Zn2+ concentrations; n is the number of oocytes.
Although we can detect no channel block by 10 μmZn2+ at approximately −30 mV (see Materials and Methods), it is still possible that some of the higher Zn2+ concentrations required to inhibit exon 5-containing receptors might complicate our evaluation of voltage-independent inhibition by causing some channel block. To test whether exon 5 acts only on voltage-independent Zn2+inhibition, we examined Zn2+ inhibition of receptors that carried NR1 mutations, NR1-1a(N616R) and NR1-1b(N637R), that have been reported to relieve voltage-dependent Zn2+inhibition (Mori et al., 1992; Kawajiri and Dingledine, 1993; Sakurada et al., 1993). Evaluation of the Zn2+ effects of these mutant NR1 subunits showed that although they are somewhat less sensitive to voltage-independent inhibition of Zn2+(see Fig. 4B), exon 5 still increases the IC50 value for Zn2+ inhibition to a similar extent as in wild-type receptors (Table 1). This result supports the idea that the actions of exon 5 cannot be explained by effects on voltage-dependent block at high concentrations of Zn2+.
Fig. 4.
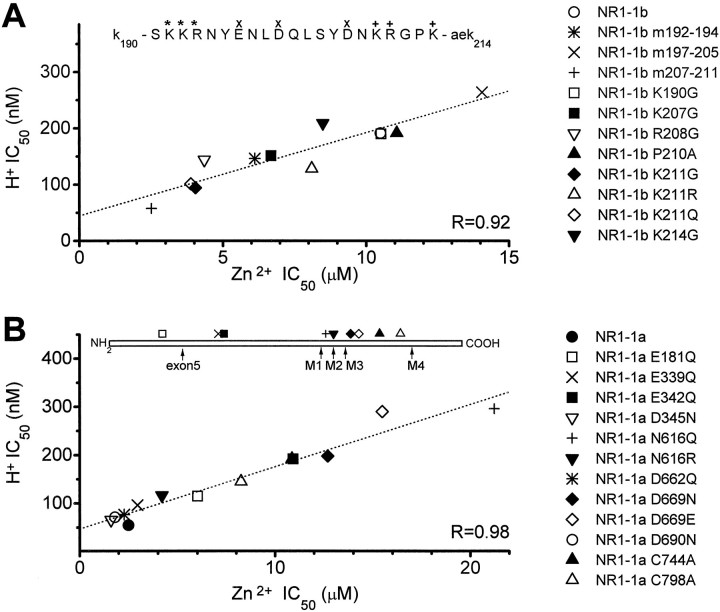
Common structural determinants of pH and Zn2+ inhibition. A, There is a strong correlation between the pH and Zn2+ sensitivity of the NMDA receptor across a variety of different NR1 exon 5 mutations. All Zn2+ inhibition curves were determined from heteromeric receptors consisting of NR1-1/NR2B subunits; IC50 values describing pH inhibition are data from NR1 subunits expressed in Xenopus oocytes in the absence of NR2 (Traynelis et al., 1995), except for wild-type NR1-1b, NR1-1b(m207–211), NR1-1b(K214G), and NR1-1b(K211G), which were determined from heteromeric receptors containing NR2B. The amino acids encoded by exon 5 are shown above, with the appropriate symbol denoting the residues that were altered in the triple mutants to G (m192–194, m207–211) or A (m197–205). B, There is a strong correlation between pH and Zn2+ sensitivity for NR1-1 mutations outside exon 5 that reduce pH sensitivity. All Zn2+ and proton IC50 values were determined from responses of heteromeric NR1-1/NR2B receptors recorded at five or more different proton or Zn2+concentrations. The position of residues with marked effects on proton and Zn2+ sensitivity are shown on a linear representation of the NR1 subunit; M1–4 denote membrane-associated regions. For all experiments, the number of oocytes recorded for each proton or Zn2+ inhibition curve ranged between 7 and 40. Broken lines show linear regressions.
Common structural determinants of proton and Zn2+ inhibition
The ability of exon 5 to reduce Zn2+ inhibition suggests that voltage-independent Zn2+ inhibition may share structural features with proton inhibition, further supporting an emerging trend of convergence among allosteric modulatory systems (see Discussion). To evaluate whether a structural argument can be made in support of the idea that Zn2+ and protons act through convergent mechanisms to inhibit NMDA receptors, we tested whether mutations in exon 5 that alter its ability to relieve proton inhibition similarly alter its ability to relieve Zn2+ inhibition. Figure3 summarizes these experiments and shows that residues including and near Lys211 control the effects of exon 5 on Zn2+ inhibition. Figure4A shows that a strong correlation exists between the ability of these various exon 5 mutations to relieve proton and Zn2+ inhibition, consistent with the idea that similar structural components within exon 5 influence both proton and Zn2+ inhibition. Does the correlation between NR1-1b mutations within exon 5 that influence Zn2+ and proton inhibition extend to other portions of the receptor? We (Burgess et al., 1996) and others (Kashiwagi et al., 1997) have observed that mutations at Asn616 in the M2 pore-forming region of the molecule can reduce the sensitivity of the receptor to protons. Figure 4B shows that this decrease in pH sensitivity is also correlated with a decrease in Zn2+ sensitivity (Table 1). In addition, we have also now extended the results of Sullivan et al. (1994) to show that two extracellular cysteine residues (Cys744 and Cys798 in NR1-1a) that control redox modulation, proton inhibition, polyamine relief of proton inhibition, and Zn2+ potentiation of NR1-1a expressed inXenopus oocytes in the absence of NR2 subunits (J. M. Sullivan and S. F. Traynelis, unpublished data) also control Zn2+ inhibition of heteromeric receptors. We also report that several acidic residues that have previously been shown to alter proton inhibition of NMDA receptor function (Williams et al., 1995; Kashiwagi et al., 1996; K. Williams, unpublished observations) cause similar changes in the Zn2+ sensitivity (Fig.4B). The tight correlation between proton and Zn2+ IC50 values across these various NMDA receptor mutants provide strong evidence that some elements of the mechanism that control proton and Zn2+ inhibition must be similar and that common structural determinants of proton and Zn2+ inhibition are spread throughout the NR1 subunit.
Interactions between exon 5 and NR1 mutations
To evaluate the interactions between exon 5 and NR1 mutations that alter proton and Zn2+ inhibition, we used thermodynamic cycles (Carter et al., 1984) to calculate the coupling energy between the effects of exon 5 and various NR1 mutations on proton inhibition. The coupling energy is the difference in free energy with respect to proton inhibition between the NR1-1 exon 5-containing mutant receptor and receptors containing wild-type NR1 + exon 5 and mutant NR1 − exon 5. For our calculations we assume that the IC50 values we measure approximate theKD for proton binding to a single site. We did not examine the coupling energy with respect to Zn2+ because its IC50 is decreased by the mutations to a point at which further reduction by exon 5 might not be fully detectable because of voltage-dependent channel block by high Zn2+ concentrations. Figure5 shows the independent effects on proton inhibition of exon 5 and NR1-1 mutations (coexpressed with NR2B) within a pore-forming domain, at a potential site of disulfide linkage, and at an acidic residue. As a control for the reliability of this approach, we examined the mutations at two conserved extracellular cysteine residues (Cys744 and Cys798 in NR1-1a) that control a variety of important features of NMDA receptor function and may combine to form a disulfide bridge (Sullivan et al., 1994). As expected, the coupling coefficient was less than that predicted for independent effects of amino acid substitutions at the two cysteine residues, consistent with the idea that the hypothesized disulfide linkage mediates the effects of these residues on the proton sensor. The predicted independence of the effects of exon 5 and mutations that alter the pH inhibition suggest that these perturbations in proton sensitivity are additive and thus occur by different mechanisms.
Polyamine relief of voltage-independent Zn2+ inhibition
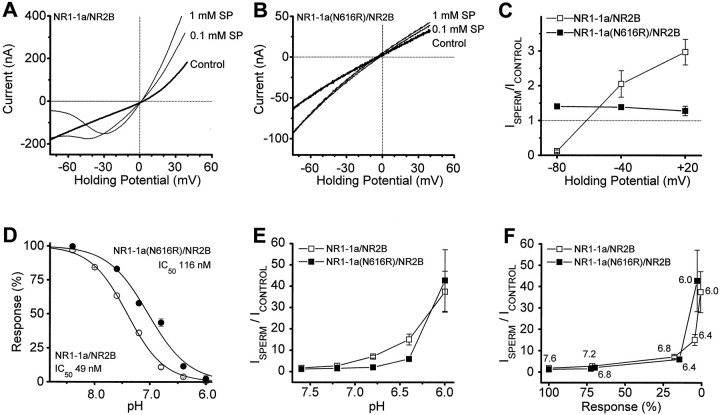
Polyamines such as spermine have been shown to relieve tonic proton inhibition in a manner analogous to the effects of exon 5, suggesting that some extracellular modulatory systems (protons and spermine) previously considered to be independent actually work through convergent pathways (Traynelis et al., 1995). To further examine the links between proton and Zn2+ inhibition, we tested whether polyamines can mimic the effects of exon 5 on Zn2+ inhibition in an manner analogous to the ability of polyamines to mimic exon 5-induced relief of proton inhibition. Because both Zn2+ (see above) and polyamines can block the NMDA receptor pore, we coexpressed NR1-1a(N616R) and NR1-1b(N637R) with NR2B to minimize complications attributable to voltage-dependent effects of Zn2+(Mori et al., 1992; Kawajiri and Dingledine, 1993; Sakurada et al., 1993) and polyamines (Kashiwagi et al., 1997; see Materials and Methods). Because polyamines can have both potentiating and inhibiting actions, we first performed experiments to ensure that these mutations, which abolished voltage-dependent polyamine block, did not abolish the ability of polyamines to relieve tonic proton inhibition and potentiate receptor function. Figure6A shows the voltage-dependent inhibition by polyamines as well as voltage-independent relief of proton block (i.e., polyamine potentiation). Figure 6, B and C, confirms that replacement of the M2 asparagine with arginine removes voltage-dependent channel block by spermine (Kashiwagi et al., 1997) and also shows that potentiation of the current response by polyamines is reduced. Such a result could occur if the M2 mutation altered the ability of spermine to interact with the receptor, or if the pore mutations we used unexpectedly decreased the proton sensitivity. To distinguish between these two possibilities, we examined the proton sensitivity of NR1-1a(N616R)/NR2B receptors. The experiments described in Figure 6D–F confirm that the M2 pore mutations decrease the proton sensitivity of the NMDA receptors and show that the level of decreased polyamine potentiation can be fully accounted for from the reduced proton sensitivity (i.e., less tonic inhibition to relieve means less potentiation by polyamines). These results suggest that the interaction between polyamines and receptors containing either NR1-1a or NR1-1a(N616R) subunits in combination with NR2B are similar. Kashiwagi et al. (1997) reported similar effects of pore mutations on pH inhibition while these experiments were in progress.
Fig. 6.
Effects of mutations in the NR1-1 M2 pore-forming region on proton inhibition and spermine relief of proton inhibition.A, Potentiation and block of NR1-1a/NR2B receptors by extracellular spermine (SP) is evident in the current–voltage relationship. B, Exchange of Asn616 in NR1 for Arg relieves voltage-dependent channel block by spermine at hyperpolarized potentials (see Materials and Methods), but also attenuates spermine potentiation of receptor responses. C, Ratio of the current response obtained in the presence of 1 mm spermine to the control response recorded at fixed holding potentials illustrates more clearly both potentiation and block of wild-type NR1-1a/NR2B receptors by spermine (open squares, n = 6–8 oocytes at each holding potential). Filled squares show relief of voltage-dependent channel block by spermine and attenuation of spermine potentiation at pH 7.3 for mutant NR1 subunits (n = 9–11 oocytes at each holding potential). Identical results were found for 0.1 mm spermine (n = 56, data not shown). D, Composite proton inhibition curves were constructed for wild-type NR1-1a/NR2B receptors (63 oocytes) and NR1-1a(N616R)/NR2B (48 oocytes) receptors by normalizing current responses recorded at different pH values to those observed at pH 7.6 and expressing the mean response at each pH as a percent of the fitted maximum (see Materials and Methods). NR1 N616R decreased the amount of tonic proton inhibition for receptors that both lack and contain exon 5 (data not shown; n = 15 oocytes). E, Spermine potentiation, defined as the current recorded in the presence of 300 μm spermine expressed as a percent of the control current response, is shown as a function of pH for NR1-exon 5/NR2B receptors (data from 45 oocytes) and NR1-1a(N616R)/NR2B receptors (n = 37). F, Plotting the degree of spermine potentiation against the degree of proton inhibition for the same data show that there is no apparent difference in the effect of spermine on wild-type and mutant receptors. Differences in spermine potentiation shown in panels C and Etherefore result from the change in pH sensitivity shown inD.
To test whether voltage-independent Zn2+ inhibition, like proton inhibition, can be relieved by both exon 5 and polyamines, we constructed Zn2+ inhibition curves in the absence and presence of 300 μm spermine for NR1-1a(N616R)/NR2B and NR1-1b(N637R)/NR2B receptors. In the absence of exon 5, spermine caused an increase in the IC50 for Zn2+inhibition from 4 to 24 μm (Fig.7A; a sixfold shift). However, for NR1 subunits that contained exon 5, spermine had much more modest effects, shifting the Zn2+ IC50 value from 65 to 151 μm (Fig. 7B; a 2.3-fold shift). These data suggest that spermine can increase the IC50value for Zn2+ in an exon 5-dependent manner. If we assume that spermine–Zn2+ binding (discussed below) only minimally influences free Zn2+ concentration, then we can evaluate the concentration–effect relationship for spermine relief of Zn2+ inhibition by examining the current response in the presence of 10 μmZn2+ with or without a variable concentration of spermine present (Fig. 7C). The result of this experiment suggests that spermine can relieve Zn2+inhibition almost completely with an EC50 value of ∼90 μm (n = 15) under these conditions.
Because spermine potentiates the NMDA receptors, any reduction by Zn2+ of polyamine potentiation would reduce current response amplitude and thus, appear as an opposing effect to polyamine relief of Zn2+ inhibition. That is, the current recorded in the presence of both spermine and Zn2+might be less than that recorded in spermine alone in part because of interference of spermine potentiation by Zn2+. This could cause an underestimation of the effects of spermine on Zn2+ inhibition. To evaluate the effects of spermine on Zn2+ inhibition without this confounding variable, we tested the effects of spermine on Zn2+inhibition at pH 8.0, a pH value at which there is almost no tonic proton inhibition, and thus, no polyamine potentiation (Fig.7D). Even in the absence of any potentiating actions of polyamines, we still see a relief of Zn2+ inhibition by polyamines to approximately the same level reported at pH 7.3. Similar results to those shown in Figure 6, C andD, were found when the above experiments were repeated with wild-type NR1-1/NR2B receptors that lack or contain exon 5 (n = 45 oocytes; data not shown).
The role of spermine–Zn(II) complexation in spermine relief of Zn2+ inhibition
Polyamines are known to chelate transition metals such as copper and zinc under certain conditions. For example, two spermine molecules can complex with Zn2+, and their interaction is stabilized at alkaline pH and by certain anions (Antonelli et al., 1984; Wojciechowska et al., 1991). Accordingly, it is important to consider whether the effects of spermine we observe on voltage-independent Zn2+ inhibition of NMDA receptor function simply reflect spermine binding of Zn2+. Although extensive data exists for Cu(II)–polyamine complexes as a function of pH, considerably less data exists for Zn(II)–polyamine complexes at different pH values. Although most Cu2+is complexed with spermine at physiological pH (Palmer and Powell, 1974; Wojciechowska et al., 1991), Zn2+ is known to form weaker complexes with polyamines (Antonelli et al., 1984;Wojciechowska et al., 1991). For example, at pH 6, <2% of the Zn2+ is complexed with spermine compared with 24% of Cu2+.
Could the relief of Zn2+ inhibition of NMDA receptor function by spermine that we observe be explained by spermine chelation of Zn2+? Two experimental observations make this unlikely. First, it is not possible to account for the different effects of spermine on receptors that lack and contain exon 5 by simply removing a fraction of the extracellular Zn2+ from the solution. Second, spermine-transition metal complexes are pH-sensitive, with metal-spermine complexes being more prevalent at alkaline pH. However, we observe similar relief by spermine of Zn2+ inhibition at pH 6.6, 7.3, and 8.0. Zn2+ at 10 μm inhibited NR1-exon 5(N616R)/NR2B responses to 48 ± 5% (pH 6.6; n = 8), 47 ± 3% (pH 7.3; n = 6), and 48 ± 4% (pH 8.0; n = 11) of control, and this level of inhibition was reduced by 300 μm spermine to 77 ± 3% (pH 6.6), 76 ± 3% (pH 7.3), and 75 ± 2% (pH 8.0) of control (p > 0.4 by ANOVA). Thus, if significant Zn2+–spermine complexation does occur at pH 7.3, our data suggest that either the complexed ion is biologically active or the spermine–water exchange rate of the complex is fast enough to allow Zn2+ to rapidly interact with a binding site on the receptor protein.
Interactions between protons and Zn2+
Does Zn2+ inhibition involve an enhancement of the proton sensitivity of the NMDA receptor such that tonic proton inhibition is enhanced in the presence of Zn2+? Such a mechanism has been proposed to underlie inhibition of NMDA receptors by phenylethanolamines such as ifenprodil and its analogs (Mott et al., 1998). We performed two experiments to evaluate the functional interaction between proton and Zn2+ inhibition. First, we measured the Zn2+ sensitivity of NMDA receptors at different pH values to assess whether alkaline pH relieved Zn2+ inhibition. We found that that NR1-1a/NR2B receptors are similarly sensitive to extracellular Zn2+ at both pH 8.0 (IC50, 2.2 μm; n = 10) and pH 6.6 (IC50, 3.0 μm; n = 10) pH values, compared with pH 7.3 (IC50, 2.5 μm; Table 1). Modest effects of pH are expected because the ionization of residues that interact with Zn2+almost certainly will be affected by this pH change. However, the lack of a strong monotonic relationship between pH and Zn2+ sensitivity is inconsistent with the idea that Zn2+ inhibits receptor function by enhancement of pH sensitivity. Second, we tested whether extracellular Zn2+ could alter proton sensitivity. Consistent with our idea that Zn2+ inhibition does not reflect an enhancement of proton inhibition, we find no significant difference in proton inhibition at pH 6.6 or 6.8 in the absence or presence of 1 μm extracellular Zn2+(n = 19; p > 0.4; minimum detectable difference 17%). Zn2+ appears to slightly enhance potentiation of current responses by pH 8.0 (122 ± 6% of control; p < 0.05). Given the limited solubility of hydroxide–Zn(II) complexes that form at alkaline pH values (see Eq. 3, Materials and Methods), we might expect an enhanced current at pH 8.0 versus pH 7.3 if a small portion of the Zn2+precipitated slowly out of solution at pH 8.0. These data support the conclusion that Zn2+ does not inhibit receptors by enhancing the pH sensitivity through a stabilization of the protonated state. Moreover, the data also show that NMDA receptor responses partially inhibited by protons are sensitive to Zn2+, and vice versa.
DISCUSSION
The two most important conclusions to emerge from this study are (1) that alternative splice variants of the NR1 subunit control voltage-independent inhibition by extracellular Zn2+and (2) that this form of Zn2+ inhibition shares several prominent structural and functional features with proton inhibition. The effects of alternative exon splicing on Zn2+ inhibition have interesting implications for the role of Zn2+ in information processing. Postsynaptic neurons that contact presynaptic terminals that release Zn2+ in a Ca2+-dependent manner might include NR1 exon 5 in the mature mRNA to either relieve tonic inhibition of postsynaptic receptor function by ambient levels of Zn2+ in the extracellular space (Zheng et al., 1998) or shift NMDA receptor responsiveness to synaptically released Zn2+. On a structural level, the three parallels that we have described between protons and Zn2+inhibition (similar sensitivity to NR1 exon 5, polyamines, and NR1 mutations) suggest these two modulatory sites either contain overlapping structural components or converge on a common downstream mechanism.
Control of Zn2+ inhibition by alternative exon splicing
NR1 exon 5 has widespread and interesting effects on NMDA receptor function (Durand et al., 1992, 1993; Sugihara et al., 1992; Hollmann et al., 1993). Exon 5 is thought to encode an extracellular surface loop that may act as a tethered ligand that can influence several extracellular allosteric modulators, including protons and polyamines, as well as potentiation by Zn2+ of NR1 subunits expressed without NR2 in Xenopus oocytes (Durand et al., 1992, 1993; Hollmann et al., 1993, Westbrook, 1994; Traynelis et al., 1995). Thus, the ability of exon 5 to influence voltage-independent inhibition by extracellular protons and Zn2+ fits well with the idea that this portion of the protein may control access of extracellular ions to a complex regulatory site on the extracellular surface of the receptor. What ramifications does this altered sensitivity have for neurobiology? Exon 5 is expressed in a region-specific manner and regulated throughout development of the nervous system (Laurie and Seeburg, 1994; Nash et al., 1997; Paupard et al., 1997; Weiss et al., 1998). Unfortunately, no data exist that allow a direct comparison of the relative abundance of NMDA receptors that lack and contain NR1 exon 5. However, it seems safe to suggest that in regions such as hippocampal CA3, in which exon 5 is expressed and Zn2+ is released, that the presence of exon 5-containing NR1 subunits should decrease the response of the neurons to both ambient levels of Zn2+ as well as synaptically released Zn2+.
Common functional and structural determinants of proton and Zn2+ inhibition
The idea that the structural determinants of proton and Zn2+ binding might overlap was first suggested on the basis of the similarity between the pKa values of amino acid residues that frequently coordinate Zn2+ and the IC50 for proton inhibition of NMDA receptors (Peters et al., 1987; Westbrook and Mayer, 1987; Traynelis and Cull-Candy, 1990,1991; Traynelis, 1998). However, it is a particularly thorny issue to address with functional studies, given the proposed mechanism of action for protons. Traynelis and Cull-Candy (1991) have argued that a protonated NMDA receptor is nonfunctional, suggesting that the proton inhibition curve reflects an increasing fraction at any moment of receptors that remain only briefly protonated. If this view proves correct, it explains the additive rather than synergistic effects of proton and Zn2+ inhibition. This is because a nonfunctional protonated receptor, regardless of whether it binds Zn2+, does not contribute to the current response. Thus the properties of residual currents at acidic pH should mirror properties of larger currents at alkaline pH because only unprotonated and thus fully functional receptors contribute to the response.
There are several possible ways to explain our data describing parallel effects of spermine, exon 5, and selected NR1 residues on proton and Zn2+ inhibition. Zn2+-binding to any protein reflects its coordination chemistry, which is flexible for Zn2+ given its pseudonoble gas configuration (d10). Zn2+ is known to form tetrahedral, trigonal-bipyramid, square pyramid, octahedral, and other complexes in free solution (Huhey, 1983), and additional geometries could be dictated by the structure and flexibility of the binding site within the protein. In other Zn2+-binding proteins, typically at least three electron pairs are provided by the protein to hybrid Zn2+ orbitals (Frausto da Silva and Williams, 1991); water or additional amino acid residues may satisfy the other 1–3 coordination sites, depending on the coordination geometry. If we assume at least three electron donors exist that define the Zn2+-binding site on the NMDA receptor (Fig.8), we then must hypothesize that the neutralization of the partial or formal negative charge on these residues caused by Zn2+ binding is sufficient to shut down receptor function. Alternatively, binding of Zn2+ might decrease the degrees of freedom of one of the electron-donating groups and thus prevent the conformational changes necessary for channel opening.
Fig. 8.
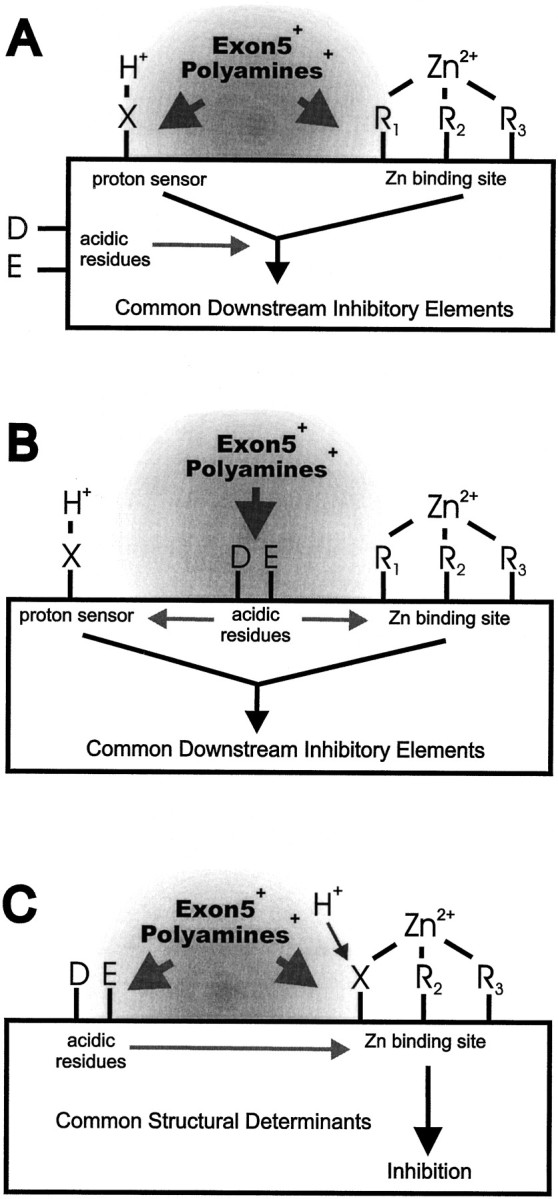
Structural models of proton and Zn2+ action. A, If the proton sensor is a single ionizable amino acid residue (X) rather than carbohydrate or lipid, it could be physically distinct from the residues that bind Zn2+ (R1–3). At least three electron pairs are usually required to coordinate Zn2+; fourth and/or fifth electron pairs could be provided by water or other amino acid residue(s) depending on coordination geometry. In this scenario, the acidic NR1 amino acid residues (D, E) studied in Figures 3 and4 could enhance the manner by which both protons or Zn2+-binding influence channel gating, rather than altering the binding of either ion. Both exon 5 and polyamines are shown influencing the local microenvironment (gray area) of each regulatory site, which could shift the pKa of the proton sensor and alter the association of Zn2+ with its binding site. B, Although the proton sensor and Zn2+-binding site might be physically distinct, certain acidic residues alternatively could enhance the electronegativity and partial negative charge of the proton sensor and electron donors to Zn2+, thereby decreasing the KD for proton and Zn2+ binding. Exon 5 and polyamines could influence the proton- and Zn2+-binding sites indirectly through association with these acidic residues, although this idea contradicts the additive effects of exon 5 and mutation E342Q in NR1. C, If a single amino acid residue constitutes the proton sensor, it might contribute to the Zn2+coordination site. This would transmit the effects of acidic residues that control the pKa of the proton sensor directly to the affinity of Zn2+ for its binding site. Exon 5 and polyamines could, by shielding the proton sensor, directly influence Zn2+ binding. Alternatively, exon 5 and polyamines might alter the pKa of the proton sensor by interacting with acidic residues that themselves influence the electronegativity of the proton sensor (but see Fig. 5).
At this juncture there are three different models that could explain our data. First, we could hypothesize that protons bind to a site separate from any of the electron donors to Zn2+(Fig. 8A), and that binding of Zn2+ or protons inhibits receptor function by controlling in a similar manner movement of the channel gate. This functional convergence could create the parallel effects we observe of nonexon 5 NR1 mutations on both proton and Zn2+effects, provided all of these mutations uncouple both Zn2+ and proton binding from the downstream mechanisms. The effects of polyamines and exon 5 that we observe on proton and Zn2+ inhibition are simplest to explain if both binding sites are close enough so that their local environments can be influenced by charged polyamines or residues near Lys211 in exon 5. Second, the proton and Zn2+-binding sites could be separate but under the control of a handful of NR1 anionic residues (Glu181, Glu342, Asp669, etc.) that increase the partial negative charge on the proton sensor and are electron donors to Zn2+. In this scenario, the positive charges on exon 5 and polyamines could interact with these residues to reduce their effects on the proton and Zn2+-binding sites (Fig.8B), although this idea is hard to reconcile with the additive effects of exon 5 and certain anionic residues (Fig. 5). Third, the proton sensor could be a part of the Zn2+-binding site. In this scenario, similar effects on protein charge distribution and conformation might follow either Zn2+ binding to its coordination site or protonation of a critical electron donor to Zn2+ (Fig.8C). Both spermine and exon 5 could reduce the effectiveness of Zn2+ and protons in this model by shielding the proton sensor to shift its pKa (Traynelis et al., 1995), while at the same time altering the ionic microenvironment of the Zn2+-binding site and/or interfering with Zn2+ access to the site. Alternatively, exon 5 and polyamines might interact with a variety of acidic residues that control the partial charge on the proton sensor (but see Fig. 5). An important part of this idea is that spermine and exon 5 must not associate so tightly with the proton sensor–electron donor so as to neutralize the partially or formally charged amino acid side chains, because such neutralization would inhibit the receptor (rather than relieve proton and Zn2+ inhibition). This last model postulates three unique molecular interactions at the same binding site: protonation, coordination of a metal ion, and shielding.
Whereas identification of the proton sensor and residues that coordinate Zn2+ would help in the evaluation of these models, efforts to find these sites using site-directed mutagenesis are likely to be fraught with interpretative difficulties. Removal of a residue that contributes, say, to Zn2+binding may cause only a shift in its IC50 rather than abolish Zn2+ binding if other nearby residues can also donate electron pairs, albeit less willingly. This reduces the search for Zn2+-binding partners to interpretation of partial shifts in the Zn2+ sensitivity. Although construction of multiple mutations might pinpoint additional candidate residues that interact with Zn2+ (Fig. 5), the number of mutants to be investigated is substantial. Similarly, removal of the proton sensor by mutagenesis is likely to reveal other ionizable residues that can be protonated at higher acid concentrations and can alter receptor function. Again, the search for a key residue may well become an evaluation of subtle shifts in pH sensitivity. For these reasons, more complicated approaches probably will be needed to advance these structural models to the next level, such as biophysical analysis of the mechanisms of pH and Zn2+ inhibition in concert with mutagenesis or perhaps biochemical cross-linking studies to identify residues that can associate with exon 5 in a polyamine-sensitive manner.
Convergence of extracellular modulatory sites on the NMDA receptor
For nearly a decade the number of allosteric regulatory sites proposed to exist on the extracellular surface of the NMDA receptors has increased, and this hyperregulation has been taken as evidence of the importance of the receptor. The advent of molecular techniques, and in particular, site-directed mutagenesis, brought the first hints that many NMDA receptor modulators might share either portions of their binding sites or common downstream elements. Sullivan et al. (1994) showed that two extracellular cysteine residues control effects of redox modulators, protons, and polyamines. We provide data in this report (Fig. 7B) further linking these two residues to voltage-independent Zn2+ inhibition. Traynelis et al. (1995) showed that polyamine potentiation reflects the relief of tonic proton inhibition, a result that has been supported now by other investigators (Williams et al., 1995; Kashiwagi et al., 1996). Furthermore, Zheng et al. (1994) showed that potentiating actions of Zn2+ on NR1 subunits expressed inXenopus oocytes had structural parallels to polyamine potentiation [Traynelis et al. (1995), see footnote 20]. Zheng et al. (1998) have recently shown that src-induced potentiation of NMDA receptors likely reflects a shift in the Zn2+sensitivity. Thus, no less than five regulatory sites now appear linked in some way, and a similar convergence has become apparent among pore-blocking ions and drugs (Mori et al., 1992; Kawajiri and Dingledine, 1993; Sakurada et al., 1993). Could this trend continue? Possibly so, because links between other regulatory compounds remain to be investigated (e.g., arachidonic acid, dynorphin, ethanol, neurosteroids, and pituitary adenylyl cyclase-activating peptide;McBain and Mayer, 1994; Chen et al., 1995; Liu and Madsen, 1997; Wu and Dun, 1997; Engblom et al., 1997; Park-Chung et al., 1997). Although it seems unlikely that all regulatory sites will end up sharing common structural elements, a prudent prediction would be that two or three classes of extracellular voltage-independent regulators will emerge. With clear links between Zn2+, pH, and polyamines, the stage has already been set for the discovery of molecules (e.g., ifenprodil; Kew et al., 1996; Mott et al., 1998) that regulate NMDA receptor function in a unique voltage-independent and noncompetitive manner through control of these modulatory sites. Such drugs at a minimum will be interesting research tools and may eventually lead to novel treatment of the multitude of neurological disorders that have been suggested to involve NMDA receptors.
Footnotes
This work was supported by the National Institute of Neurological Diseases and Stroke (S.F.T., F.Z.), by the John Merck Fund (S.F.T.), the American Epilepsy Foundation (S.F.T.), and a Kennesaw State University Faculty Development Grant (J.L.P.). We thank R. Dingledine, M. B. Gingrich, and K. Williams for critical comments on this manuscript, N. Patel for excellent technical assistance, and P. Ascher, J. Neyton, and P. Paoletti for sharing unpublished data. We are grateful to J. Boulter, R. Dingledine, S. Heinemann, M. Hollmann, K. Moriyoshi, S. Nakanish, P. Seeburg, J. Sullivan, and K. Williams for sharing cDNAs with us, K. Williams for sharing unpublished results with us, and M. Vandergrift for help with some of these experiments.
Correspondence should be addressed to Dr. Stephen F. Traynelis, Department of Pharmacology, Room 5025 Rollins Research Center, Emory University, 1510 Clifton Road, Atlanta, GA 30322-3090.
REFERENCES
- 1.Aniksztejn L, Charlton G, Ben-Ari Y. Selective release of endogenous zinc from hippocampal mossy fibers in situ. Brain Res. 1987;404:58–64. doi: 10.1016/0006-8993(87)91355-2. [DOI] [PubMed] [Google Scholar]
- 2.Antonelli ML, Balzamo S, Carunchio V, Cernia E. Complex formation of 4,9-diazadodecane-1,12-diamine with copper(II) and zinc(II) ions in aqueous solution. Thermochimica Acta. 1984;78:1–8. [Google Scholar]
- 3.Assaf SY, Chung SH. Release of endogenous Zn2+ from brain tissue during activity. Nature. 1984;308:734–738. doi: 10.1038/308734a0. [DOI] [PubMed] [Google Scholar]
- 4.Asztely F, Gustafsson B. Ionotropic glutamate receptors: their possible role in expression of hippocampal synaptic plasticity. Mol Neurobiol. 1996;12:1–11. doi: 10.1007/BF02740744. [DOI] [PubMed] [Google Scholar]
- 5.Benitez MJ, Company M, Arevalillo A, Jimenez JS. Comparative study of various hydrogen ion buffers to assay Zn2+-dependent β-lactamases. Antimicrob Agents Chemother. 1991;35:1517–1519. doi: 10.1128/aac.35.7.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford HF. Glutamate, GABA, and epilepsy. Prog Neurobiol. 1995;47:477–511. doi: 10.1016/0301-0082(95)00030-5. [DOI] [PubMed] [Google Scholar]
- 7.Burgess MF, Traynelis SF. NMDA receptor inhibition by Zn2+ is regulated by subunit composition. Soc Neurosci Abstr. 1997;23:945. [Google Scholar]
- 8.Burgess MF, Powers JL, Traynelis SF. Asparagine616 in the NR1 subunit controls spermine block, spermine potentiation, and pH sensitivity. Soc Neurosci Abstr. 1996;22:588. [Google Scholar]
- 9.Carter PJ, Winter G, Wilkinson AJ, Fersht AR. The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus). Cell. 1984;38:835–840. doi: 10.1016/0092-8674(84)90278-2. [DOI] [PubMed] [Google Scholar]
- 10.Charlton G, Rovira C, Ben-Ari Y, Leviel V. Spontaneous and evoked release of endogenous Zn2+ in the hippocampal mossy fibre zone of the rat in situ. Exp Brain Res. 1985;58:202–205. doi: 10.1007/BF00238969. [DOI] [PubMed] [Google Scholar]
- 11.Chen L, Gu Y, Huang L-YM. The opioid peptide dynorphin directly blocks NMDA receptor channels in the rat. J Physiol (Lond) 1995;482:575–581. doi: 10.1113/jphysiol.1995.sp020541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen N, Moshaver A, Raymond LA. Differential sensitivity of recombinant N-methyl-d-aspartate receptor subtypes to zinc inhibition. Mol Pharmacol. 1997;51:1015–1023. doi: 10.1124/mol.51.6.1015. [DOI] [PubMed] [Google Scholar]
- 13.Choi DW. Bench to bedside: the glutamate connection. Science. 1992;258:241–243. doi: 10.1126/science.1357748. [DOI] [PubMed] [Google Scholar]
- 14.Choi DW, Koh JY. Zinc and brain injury. Annu Rev Neurosci. 1998;21:347–375. doi: 10.1146/annurev.neuro.21.1.347. [DOI] [PubMed] [Google Scholar]
- 15.Christine CW, Choi DW. Effect of zinc on NMDA receptor-mediated channel currents in cortical neurons. J Neurosci. 1990;10:108–116. doi: 10.1523/JNEUROSCI.10-01-00108.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durand GM, Gregor P, Zheng X, Bennett MVL, Uhl G, Zukin RS. Cloning of an apparent splice variant of the rat N-methyl-d-aspartate receptor NMDAR1 with altered sensitivity to polyamines and activators of protein kinase C. Proc Natl Acad Sci USA. 1992;89:9359–9363. doi: 10.1073/pnas.89.19.9359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durand GM, Bennett MVL, Zukin RS. Splice variants of the N-methyl-d-aspartate receptor NR1 identify domains involved in regulation by polyamines and protein kinase C. Proc Natl Acad Sci USA. 1993;90:6731–6735. doi: 10.1073/pnas.90.14.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engblom AC, Courtney MJ, Kukkonen JP, Akerman KE. Ethanol specifically inhibits NMDA receptors with affinity for ifenprodil in the low micromolar range in cultured cerebellar granule cells. J Neurochem. 1997;69:2162–2168. doi: 10.1046/j.1471-4159.1997.69052162.x. [DOI] [PubMed] [Google Scholar]
- 19.Frausto da Silva JJR, Williams RJP. The biological chemistry of the elements: the inorganic chemistry of life, pp 299–318. Clarendon; Oxford: 1991. [Google Scholar]
- 20.Hollmann M, Boulter J, Maron C, Beasley L, Sullivan J, Pecht G, Heinemann SF. Zinc potentiates agonist-induced currents at certain splice variants of the NMDA receptor. Neuron. 1993;10:943–954. doi: 10.1016/0896-6273(93)90209-a. [DOI] [PubMed] [Google Scholar]
- 21.Howell GA, Welch MG, Frederickson CJ. Stimulation-induced uptake and release of zinc in hippocampal slices. Nature. 1984;308:736–738. doi: 10.1038/308736a0. [DOI] [PubMed] [Google Scholar]
- 22.Huhey JE. Inorganic chemistry. Harper and Row; New York: 1983. [Google Scholar]
- 23.Kawajiri S, Dingledine R. Multiple structural determinants of voltage-dependent magnesium block in recombinant NMDA receptors. Neuropharmacology. 1993;32:1203–1211. doi: 10.1016/0028-3908(93)90014-t. [DOI] [PubMed] [Google Scholar]
- 24.Kashiwagi K, Fukuchi J-I, Chao J, Igarashi K, Williams K. An aspartate residue in the extracellular loop of the N-methyl-d-aspartate receptor controls sensitivity to spermine and protons. Mol Pharmacol. 1996;49:1131–1141. [PubMed] [Google Scholar]
- 25.Kashiwagi K, Pahk AJ, Masuko T, Igarashi K, Williams K. Block and modulation of N-methyl-d-aspartate receptors by polyamines and protons: role of amino acid residues in the transmembrane and pore-forming regions of NR1 and NR2 subunits. Mol Pharmacol. 1997;52:701–713. doi: 10.1124/mol.52.4.701. [DOI] [PubMed] [Google Scholar]
- 26.Kew JNC, Trube G, Kemp JA. A novel mechanism of activity-dependent NMDA receptor antagonism describes the effect of ifenprodil in rat cultured cortical neurones. J Physiol (Lond) 1996;497:761–772. doi: 10.1113/jphysiol.1996.sp021807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koh JY, Suh SW, Gwag BJ, He YY, Hsu CY, Choi DW. The role of zinc in selective neuronal death after transient global cerebral ischemia. Science. 1996;272:1013–1016. doi: 10.1126/science.272.5264.1013. [DOI] [PubMed] [Google Scholar]
- 28.Kornau H-C, Schenker LT, Kennedy MB, Seeburg PH. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 29.Laube B, Kuhse J, Rundstrom N, Kirsch J, Schmieden V, Betz H. Modulation by zinc ions of native rat and recombinant human inhibitory glycine receptors. J Physiol (Lond) 1995;483:613–619. doi: 10.1113/jphysiol.1995.sp020610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laurie DJ, Seeburg PH. Regional and developmental heterogeneity in splicing of the rat brain NMDAR1 mRNA. J Neurosci. 1994;14:3180–3194. doi: 10.1523/JNEUROSCI.14-05-03180.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Legendre P, Westbrook GL. The inhibition of single N-methyl-d-aspartate-activated channels by zinc ions on cultured rat neurones. J Physiol (Lond) 1990;429:429–449. doi: 10.1113/jphysiol.1990.sp018266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lide DR. CRC handbook of chemistry and physics, 77th Ed, pp 5, 100–106. CRC; New York: 1997. [Google Scholar]
- 33.Liu GJ, Madsen BW. PACAP38 modulates activity of NMDA receptors in cultured chick cortical neurons. J Neurophysiol. 1997;78:2231–2234. doi: 10.1152/jn.1997.78.4.2231. [DOI] [PubMed] [Google Scholar]
- 34.Lobo VMM, Quaresma JL. Physical sciences data 41: handbook of electrolyte solutions, part B, pp 2160–2172. Elsevier; New York: 1989. [Google Scholar]
- 35.Martell AE, Smith RM. Critical stability constants, Vol 4. Plenum; New York: 1974. [Google Scholar]
- 36.Mayer ML, Vyklicky L. The action of zinc on synaptic transmission and neuronal excitability in cultures of mouse hippocampus. J Physiol (Lond) 1989;415:351–365. doi: 10.1113/jphysiol.1989.sp017725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mayer ML, Vyklicky L, Westbrook GL. Modulation of excitatory amino acid receptors by group IIB metal cations in cultured mouse hippocampal neurones. J Physiol (Lond) 1989;415:329–350. doi: 10.1113/jphysiol.1989.sp017724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McBain CJ, Mayer ML. N-methyl-d-aspartic acid receptor structure and function. Physiol Rev. 1994;74:723–760. doi: 10.1152/physrev.1994.74.3.723. [DOI] [PubMed] [Google Scholar]
- 39.Mori H, Masaki H, Yamakura T, Mishina M. Identification by mutagenesis of a Mg2+-block site of the NMDA receptor channel. Nature. 1992;358:673–675. doi: 10.1038/358673a0. [DOI] [PubMed] [Google Scholar]
- 40.Mott DD, Doherty JJ, Zhang S, Lyuboslavsky P, Traynelis SF, Dingledine R (1998) Phenylethanolamines inhibit NMDA receptors by potentiating proton inhibition: implications for novel neuroprotective strategies. Soc Neurosci Abstr 24, in press. [DOI] [PubMed]
- 41.Nash NR, Heilman CJ, Rees HD, Levey AI. Cloning and localization of exon 5-containing isoforms of the NMDAR1 subunit in human and rat brains. J Neurochem. 1997;69:485–493. doi: 10.1046/j.1471-4159.1997.69020485.x. [DOI] [PubMed] [Google Scholar]
- 42.Palmer BN, Powell HKJ (1974) Complex formation between 4,9-diazadodecane-1,12-diamine (spermine) and copper(II) ions and protons in aqueous solution. J Chem Soc Dalton: 2086–2089.
- 43.Paoletti P, Neyton J, Ascher P. Glycine-independent and subunit-specific potentiation of NMDA responses by extracellular Mg2+. Neuron. 1995;15:1109–1120. doi: 10.1016/0896-6273(95)90099-3. [DOI] [PubMed] [Google Scholar]
- 44.Paoletti P, Ascher P, Neyton J (1997) High-affinity zinc inhibition of NMDA NR1-NR2A receptors. J Neurosci [Erratum (1997) 17(20)] 17:5711–5725. [DOI] [PMC free article] [PubMed]
- 45.Park-Chung M, Wu F-S, Purdy RH, Malayev AA, Gibbs TT, Farb DH. Distinct sites for inverse modulation of N-methyl-d-aspartate receptors by sulfated steriods. Mol Pharmacol. 1997;52:1113–1123. doi: 10.1124/mol.52.6.1113. [DOI] [PubMed] [Google Scholar]
- 46.Paupard MC, Friedman LK, Zukin RS. Developmental regulation and cell-specific expression of N-methyl-d-aspartate receptor splice variants in rat hippocampus. Neuroscience. 1997;79:399–409. doi: 10.1016/s0306-4522(96)00677-x. [DOI] [PubMed] [Google Scholar]
- 47.Peters S, Koh J, Choi DW. Zinc selectively blocks the action of N-methyl-d-aspartate on cortical neurons. Science. 1987;236:589–593. doi: 10.1126/science.2883728. [DOI] [PubMed] [Google Scholar]
- 48.Sakurada K, Masu M, Nakanishi S. Alteration of Ca2+ permeability and sensitivity to Mg2+ and channel blockers by a single amino acid substitution in the N-methyl-d-aspartate receptor. J Biol Chem. 1993;268:410–415. [PubMed] [Google Scholar]
- 49.Scheetz AJ, Constantine-Paton M. Modulation of NMDA receptor function: implications for vertebrate neural development. FASEB J. 1994;8:745–752. doi: 10.1096/fasebj.8.10.8050674. [DOI] [PubMed] [Google Scholar]
- 50.Smart TG, Xie X, Krishek BJ. Modulation of inhibitory and excitatory amino acid receptor ion channels by zinc. Prog Neurobiol. 1994;42:393–441. doi: 10.1016/0301-0082(94)90082-5. [DOI] [PubMed] [Google Scholar]
- 51.Soloviev MM, Brierley MJ, Shao ZY, Mellor IR, Volkova TM, Kamboj R, Ishimaru H, Sudan H, Harris J, Foldes RL, Grishin EV, Usherwood PNR, Barnard EA. Functional expression of a recombinant unitary glutamate receptor from Xenopus, which contains N-methyl-d-aspartate (NMDA) and non-NMDA receptor subunits. J Biol Chem. 1996;271:32572–32579. doi: 10.1074/jbc.271.51.32572. [DOI] [PubMed] [Google Scholar]
- 52.Sugihara H, Moriyoshi K, Ishii T, Masu M, Nakanishi S. Structures and properties of seven isoforms of the NMDA receptor generated by alternative splicing. Biochem Biophys Res Commun. 1992;185:826–832. doi: 10.1016/0006-291x(92)91701-q. [DOI] [PubMed] [Google Scholar]
- 53.Sullivan JM, Traynelis SF, Chen HS, Escobar W, Heinemann SF, Lipton SA. Identification of two cysteine residues that are required for redox modulation of the NMDA subtype of glutamate receptor. Neuron. 1994;13:929–936. doi: 10.1016/0896-6273(94)90258-5. [DOI] [PubMed] [Google Scholar]
- 54.Traynelis SF. pH Modulation of ligand-gated ion channels. In: Kaila K, Ransom BR, editors. pH and brain function. Wiley-Liss; New York: 1998. pp. 407–436. [Google Scholar]
- 55.Traynelis SF, Cull-Candy SG. Proton inhibition of N-methyl-d-aspartate receptors in cerebellar neurons. Nature. 1990;345:347–350. doi: 10.1038/345347a0. [DOI] [PubMed] [Google Scholar]
- 56.Traynelis SF, Cull-Candy SG. Pharmacological properties and H+ sensitivity of excitatory amino acid receptor channels in rat cerebellar granule neurones. J Physiol (Lond) 1991;433:727–763. doi: 10.1113/jphysiol.1991.sp018453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Traynelis SF, Hartley M, Heinemann SF. Control of proton sensitivity of the NMDA receptor by RNA splicing and polyamines. Science. 1995;268:873–876. doi: 10.1126/science.7754371. [DOI] [PubMed] [Google Scholar]
- 58.Tripathi RM, Ghose R, Ghose AK. Interaction of N-[Tris(hydroxymethyl)methyl]glycine with some transition metal complexes in aqueous solution. J Indian Chem Soc. 1987;64:519–520. [Google Scholar]
- 59.Vieles P, Frezou C, Galsomias J, Bonniol A. Physicochemical study of tricine and its complexes with the transition metal ions cobalt (II), nickel (II), copper (II), and zinc (II). J Chim Phys Physicochim Biol. 1972;69:869–874. [Google Scholar]
- 60.Weiss SW, Albers DS, Iadarola MJ, Dawson TM, Dawson VL, Standaert DG. NMDAR1 glutamate receptor subunit isoforms in neostriatal, neocortical, and hippocampal nitric oxide synthase neurons. J Neurosci. 1998;18:1725–1734. doi: 10.1523/JNEUROSCI.18-05-01725.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Westbrook GL. Glutamate receptor update. Curr Opin Neurobiol. 1994;4:337–346. doi: 10.1016/0959-4388(94)90094-9. [DOI] [PubMed] [Google Scholar]
- 62.Westbrook GL, Mayer ML. Micromolar concentrations of Zn2+ antagonize NMDA and GABA responses of hippocampal neurons. Nature. 1987;328:640–643. doi: 10.1038/328640a0. [DOI] [PubMed] [Google Scholar]
- 63.Whetsell WO. Current concepts of excitotoxicity. J Neuropathol Exp Neurol. 1996;55:1–13. doi: 10.1097/00005072-199601000-00001. [DOI] [PubMed] [Google Scholar]
- 64.Williams K. Pharmacological properties of recombinant N-methyl-d-aspartate (NMDA) receptors containing the ε4 (NR2D) subunit. Neurosci Lett. 1995;184:181–184. doi: 10.1016/0304-3940(94)11201-s. [DOI] [PubMed] [Google Scholar]
- 65.Williams K. Separating dual effects of zinc at recombinant N-methyl-d-aspartate receptors. Neurosci Lett. 1996;215:9–12. doi: 10.1016/s0304-3940(96)12924-4. [DOI] [PubMed] [Google Scholar]
- 66.Williams K, Zappia AM, Prichett DB, Shen YM, Molinoff PB. Sensitivity of the N-methyl-d-aspartate receptor to polyamines is controlled by NR2 subunits. Mol Pharmacol. 1994;45:803–809. [PubMed] [Google Scholar]
- 67.Williams K, Kashiwagi K, Fukuchi J-I, Igarashi K. An acidic amino acid in the N-methyl-d-aspartate receptor that is important for spermine stimulation. Mol Pharmacol. 1995;48:1087–1098. [PubMed] [Google Scholar]
- 68.Wojciechowska A, Bolewski L, Lomozik L. A study of polyamine complex formation with H+, Cu (II), Zn (II), Pb (II), and Mg (II) in aqueous solution. Monatsh Chem. 1991;122:131–138. [Google Scholar]
- 69.Wu SY, Dun NJ. Potentiation of NMDA currents by pituitary adenylate cyclase activating polypeptide in neonatal rat sympathetic preganglionic neurons. J Neurophysiol. 1997;78:1175–1179. doi: 10.1152/jn.1997.78.2.1175. [DOI] [PubMed] [Google Scholar]
- 70.Zhang L, Zheng X, Paupard M-C, Wang AP, Santchi L, Friedman LK, Zukin RS, Bennett MVL. Spermine potentiation of recombinant N-methyl-d-aspartate receptors is affected by subunit composition. Proc Natl Acad Sci USA. 1994;91:10883–10887. doi: 10.1073/pnas.91.23.10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zheng F, Gingrich MB, Traynelis SF, Conn PJ. Tyrosine kinase modulates Zn2+ sensitivity of NMDA receptors. Nature. 1998;1:185–191. doi: 10.1038/634. [DOI] [PubMed] [Google Scholar]
- 72.Zheng X, Zhang L, Durand GM, Bennett MV, Zukin RS. Mutagenesis rescues spermine and Zn2+ potentiation of recombinant NMDA receptors. Neuron. 1994;12:811–818. doi: 10.1016/0896-6273(94)90334-4. [DOI] [PubMed] [Google Scholar]