Abstract
Cerebellar granule neurons isolated from postnatal day 7 (P7) rats and grown in normal K+ medium begin to degenerate at approximately 4 d in vitro (DIV) and die. To search for genes upregulated in the process of neuronal cell death, differential hybridization was performed with subtracted cDNA probes and a cDNA library from 5 DIV. One of the genes isolated was microglial response factor-1 (mrf-1), which encoded a sequence of 177 amino acids with a single EF-hand calcium-binding motif. By Northern blots, the transcript was upregulated in cerebellar culture at 4 DIV, peaked at 6 DIV, and decreased at 7 DIV. Upregulation was also found when the apoptosis of granule cells was induced by replacing high K+ medium with normal K+ medium. However, when non-neuronal cells were thoroughly eliminated with aphidicolin, an antimitotic agent, the upregulation at 4–7 DIV did not occur. By immunocytochemistry, MRF-1 was detected at 5 DIV in OX-42-positive cells (microglia), and it exhibited an increase in response to granule cell death. MRF-1 levels in microglia purified from cerebral cortex also upregulated in the presence of 5 DIV granule cells. In the developing cerebellum in vivo, levels ofmrf-1 mRNA transiently increased in the early postnatal stages, reaching a peak at P7 when cerebellar neurons and astrocytes undergo extensive apoptosis. In adult brain sections, MRF-1 was detected in the perikarya and processes of ramified/resting microglia, and peripheral motor nerve dissection prominently increased the expression in activated microglia surrounding injured central motoneurons. Therefore, mrf-1 appears to be one of the microglial genes that respond to neuronal cell death and degeneration.
Keywords: microglia, neuron, degeneration, apoptosis, programmed cell death, gene expression
Microglia/brain macrophages are involved in brain function under both normal and pathological conditions (Dickson et al., 1993; McGeer et al., 1993). In the adult brain, resident ramified/resting microglia are activated to become rod-shaped or ameboid types in response to injury, infection, and inflammation of the nervous system. Activated microglia proliferate, engulfing degenerating elements (Giulian et al., 1989; Stoll et al., 1989), while secreting cytotoxic agents that induce neuronal death and demyelination (Thery et al., 1991; Giulian et al., 1994).
In the development of the mammalian nervous system, supernumerary neurons typically die around the time of their functional contact with targets via apoptosis (Oppenheim, 1991; Johnson and Deckwerth, 1993;Henderson, 1996). This is referred to as programmed cell death (PCD) in neurons. Apoptosis is characterized morphologically (Kerr and Harmon, 1991; Jacobson et al., 1997) by cell shrinkage and chromatin condensation, followed by cytoplasmic condensation. The degenerated cytoplasmic organelles become compartmentalized into a membrane-bound apoptotic body. Microglia are capable of recognizing these degenerated neurons and clearing them out through their phagocytic activity. The genes involved in these activities, however, have not been adequately characterized.
PCD in the development of Caenorhabditis elegans is subject to strict genetic regulation (Ellis et al., 1991): seven genes, including ced-7, are defined as genes that relate to the engulfment of dead cells. Luciani and Chimini (1996) have reported that the ATP binding cassette 1 transporter (ABC1) is expressed in macrophages localized in areas that contain cells undergoing developmental PCD in mice. Moreover, they have shown that ABC1 is required for the engulfment of dead cells and may possibly be a mammalian homolog to ced-7. These results suggest that engulfment is also genetically controlled in PCD, occurring as a natural part of mammalian development.
It is known that cultured cerebella granule neurons are a good model system for studying neuronal apoptosis. If granule neurons dissociated from postnatal day 7–8 (P7–8) rats are incubated under a high K+ (25–30 mm) concentration, which induces depolarization, the neurons grow to their differentiated state at approximately 10 d in vitro (DIV). When the K+ concentration of the culture medium is shifted to normal (5.4 mm), the cultured granule neurons undergo apoptotic cell death (D’Mello et al., 1993; Miller and Johnson, 1996;Armstrong et al., 1997). In addition, granule neurons grown in vitro under normal K+ conditions mimic thein vivo situation (Gallo et al., 1987; Balazs et al., 1988;Koike, 1991): granule neurons undergo the initial differentiation steps and then begin to degenerate at approximately 4 DIV and die progressively via an apoptotic cascade by 8 DIV. This neuronal death is an active process requiring RNA and protein synthesis (Suzuki and Koike, 1997). We have searched for genes that are upregulated in the process of cell death by differential hybridization and have found a new microglial gene, microglial response factor-1 (mrf-1). An extensive Northern blot analysis and immunocytochemical and immunohistochemical analyses using anti-MRF-1 antibody were performed, with the results indicating that the MRF-1 protein is localized in the microglia and upregulated in response to neuronal cell death or degeneration.
These results have been published previously in abstract form (Tanaka et al., 1997).
MATERIALS AND METHODS
Cell culture. A cerebellar cell culture was prepared from the cerebella of P7 rats (Sprague Dawley), as described previously (Suzuki and Koike, 1997). In brief, dissected cerebella were minced, treated with Dispase (250 U/ml; Godo Shusei Co., Ltd., Tokyo, Japan) at 37°C for 30 min, and then triturated in a Ca2+-free Krebs’-Ringer’s bicarbonate buffer. The dissociated cells were collected by centrifugation and resuspended in Eagle’s MEM (Life Technologies, Grand Island, NY) containing 10% heat-inactivated fetal calf serum (FCS; J.R.H. Biosciences, Lenexa, KS), 50 U/ml penicillin, and 50 μg/ml streptomycin (Sigma, St. Louis, MO). The cells were plated on poly-l-lysine (Sigma)-coated 60 mm dishes (1 × 107 cells/dish), 35 mm dishes (0.3 × 107 cells/dish), or 13.5 mm plastic sheets (5 × 105 cells/sheet) (Celldesk LF1; Sumitomo Bakelite Inc., Tokyo, Japan) for RNA isolation, sandwich culture, or immunocytochemistry, respectively. The plated cells were cultured at 36°C in a humidified atmosphere of 5% CO2/95% air, and the medium was changed only once at 3 DIV. To minimize proliferation of non-neuronal cells, neurons were treated with 50 μm fluorodeoxyuridine (FudR) for 1 d at 2 DIV, and then maintained in a 10% FCS MEM containing 10 μm FudR (standard culture) (Suzuki and Koike, 1997). Alternatively, to thoroughly eliminate the contamination of non-neuronal cells, cerebellar cells were incubated in the presence of 10 μm aphidicolin from 2 DIV (Miller and Johnson, 1996). Unless noted otherwise, cerebellar cells were grown in the standard culture. For a long-time culture (∼10 d) of granule neurons, a high concentration of potassium (at final 30 mm) was added to the culture medium at 2 DIV. The contamination of Vimentin-positive (or GFAP-positive) cells in the 7 DIV culture maintained with a high potassium medium for 5 d was 8.9 ± 0.8 (2.0 ± 0.2)% or 3.3 ± 0.2 (0.6 ± 0.1)% for the standard culture or the culture in the presence of aphidicolin, respectively. When ∼60% of the granule neurons died in 7 DIV standard culture with a normal potassium medium, the rate of Vimentin-positive cells among surviving granule neurons was ∼20%.
Microglia were isolated and purified according to the method ofSuzumura et al. (1984), with some modifications. The cerebral cortices were dissected from neonatal rat pups. Special care was taken to remove all meninges and blood vessels during dissection to minimize contamination by blood monocytes and macrophages. The dissected cortices were dissociated with 250 U/ml Dispase for 60 min at 37°C and then triturated. The dissociated cells were collected by centrifugation and resuspended in DMEM nutrient mixture F-12 Ham (Sigma) containing 10% heat-inactivated FCS and penicillin/streptomycin. The cells were plated on a flask and cultured until confluency (∼8–10 d). Microglia were collected by shaking (60 rpm for 1.5 min) and centrifugation, and then they were replated on 35 mm dishes or plastic sheets (φ 13.5 mm, Celldesk LF1). For sandwich culture, both purified cortical microglia and cerebellar cells were separately prepared on plastic sheets and dishes, respectively. The plastic sheets on which microglia were cultured were turned upside down and laid over 3 or 5 DIV cerebellar cells in plastic dishes. They were then co-incubated for 12 hr with the two cell populations in contact with each other. Conditioned medium was obtained from the culturing medium of 5 DIV cerebral cells. After filtration (0.2 μm pore size), the medium was added directly to purified microglial culture. For an immunocytochemical assay, the replated purified microglia were cultured for >2 d before use in the experiments. Identification of the microglia was performed by an immunocytochemical method using the marker OX-42 (mouse monoclonal antibody; BMA, Tavistock Square, London, UK) (Robinson et al., 1986). The microglia constituted >95% of all cells on the culture plate after replating.
Extraction and purification of RNA. Cells were washed with a Ca2+-free, Mg2+-free PBS, pH 7.2, and solubilized with 4 m guanidinium thiocyanate, pH 7.0, containing 25 mm sodium citrate, 0.5% sarkosyl, and 0.1m 2-mercaptoethanol (Chomczynski and Sacchi, 1987). The total RNA fraction was extracted from one or two 60 mm dishes (1 × 107 cells per dish at plating) for cerebellar cell culture or 1 × 105 cells for microglia culture, and precipitated by isopropanol. After centrifugation for 15 min, the pellet was washed with 70% ethanol and dried with flowing air. Poly(A+) RNA was purified by oligo(dT)-cellulose affinity column chromatography (Pharmacia Biotech, Piscataway, NJ).
Differential hybridization and isolation of a partial cDNA fragment of the mrf-1 clone.Poly(A+) RNA was isolated from both 3 DIV and 5 DIV cells of the original cerebellar cell culture, in which granule neurons were surviving and degenerating, respectively (Suzuki and Koike, 1997). To construct each of the cDNA libraries, the cDNAs were synthesized using a TimeSaver cDNA Synthesis Kit (Pharmacia), andEcoRI/NotI adaptor-ligated double-strand cDNAs were ligated into a λgt11 vector followed by in vitro packaging using Gigapack II Gold (Stratagene, La Jolla, CA). Differential hybridization was performed using a subtracted cDNA probe to screen a cDNA library from 5 DIV mRNA, as described (Sambrook et al., 1989). In brief, we isolated mRNA from 5 DIV cells (degenerating cells) and generated 32P-labeled cDNA from it by reverse transcription with random hexamer as a primer. After hydrolysis of RNA with alkali, the labeled cDNA was hybridized with ∼50-fold excess of mRNA isolated from 3 DIV cells (healthy cells), and then unhybridized labeled cDNA was purified as a probe with a hydroxyapatite column. These steps for the construction of subtracted cDNA probe were repeated twice. Approximately 5000 plaques of the cDNA library of 5 DIV cells were screened with the subtracted probe. Clones that expressed messages only in the culture containing degenerating-granule neurons were subcloned into pUC18 plasmid vectors with the SureClone Ligation Kit. The nucleotide sequence of the 3′-partial cDNA clone ofmrf-1 (421 bp) was determined using a DNA sequencing system with dye primer cycle sequencing (model 373A; Applied Biosystems, Foster City, CA).
Cloning of mrf-1 cDNA. Based on a 3′-partialmrf-1 cDNA fragment, the mrf-1 cDNA clones were isolated by the rapid amplification of cDNA ends (RACE) (Frohman, 1993) using the Marathon cDNA Amplification Kit (Clontech, Palo Alto, CA). The RACE cDNA library was constructed from the total RNA of the 5 DIV cerebellar cell culture. The RACE reaction products were subjected to a 6% PAGE, and specific products were excised, which was followed by subcloning to pUC18. A nucleotide sequence analysis was performed as described above. Sequence searches of the nucleotides and the predicted amino acids of the GenBank database were performed using fasta software (DDBJ, Mishima, Sizuoka, Japan), and motif searches were generated using GENETYX-MAC software (Ver. 8; Software Development Co. Ltd., Shibuya, Tokyo, Japan).
Northern blot. Equal amounts of total RNA (10–20 μg) or poly(A+) RNA (3 μg) were loaded per lane onto formaldehyde denaturing gel (1% agarose) and separated by electrophoresis. The RNAs were transferred to a nylon membrane (Hybond-N+; Amersham, Arlington Heights, IL). Hybridization was performed as described previously (Suzuki and Koike, 1997). The cDNA probes were labeled with 32P-dCTP to a specific activity of 1–1.5 × 106 dpm/ng by the random priming method using the Ready-To-Go DNA Labeling Kit (Pharmacia). Themrf-1 or G3PDH probe was prepared from a cloned-3′ partial cDNA fragment (421 bp, as described above), or a PCR product subcloned into pUC18 (983 bp) (Suzuki and Koike, 1997), respectively. The washed membranes were visualized, and the amount of radioactivity of specific transcripts was measured using a Bio-imaging Analyzer (BAS2000; Fuji Photo Film Corp., Tokyo, Japan).
In situ hybridization. Under deep pentobarbital anesthesia, adult rat brains were freshly obtained 5 d after axotomy of the right hypoglossal nerve. Fresh frozen sections through the hypoglossal nuclei were prepared in the coronal plane by cryostat (20 μm in thickness). As specific probes for mrf-1 mRNA, two nonoverlapping antisense oligonucleotides were synthesized with sequences complementary to the mrf-1 nucleotide residues at 85–129 and 439–483. Oligonucleotides were radiolabeled with35S-dATP using terminal deoxynucleotidyl transferase (Life Technologies). Fixation, prehybridization, and hybridization of sections were performed as described previously (Watanabe et al., 1993). Briefly, hybridization occurred overnight at 42°C in hybridization buffer containing 50% formamide, 50 mmTris-HCl, pH 8.0, 1× Denhardt’s solution, 0.6 m NaCl, 0.025% SDS, 200 μg/ml tRNA, 1 mm EDTA, 10% dextran sulfate, 0.1 m dithiothreitol, and 10,000 dpm/μl radiolabeled probe. After washing with 0.1× SSC containing 0.1% sarkosyl at 55°C, sections were dipped in nuclear track emulsion (NTB-2, Kodak) and exposed for 2 months. After development, sections were photographed by dark-field and bright-field microscopy.
Protein expression studies and Western blot. Full-length recombinant MRF-1 was expressed as glutathione S-transferase (GST) fusion proteins, using a pGEX-4T-2 plasmid vector (Pharmacia) andEscherichia coli BL21. The fusion proteins were purified with glutathione Sepharose 4B, according to the manufacturer’s instruction (Pharmacia). After thrombin digestion, the MRF-1 polypeptide was separated from GST by reversed-phase HPLC. The purified polypeptides (100–300 μg) were emulsified with Freund’s complete adjuvant (Difco, Detroit, MI), and were injected subcutaneously into female New Zealand White rabbits at intervals of 2 weeks. From antisera sampled 2 weeks after the sixth injection, immunoglobulins were separated using Protein G-Sepharose (Pharmacia). Antibodies specific to the MRF-1 were then affinity-purified using fusion proteins coupled to CNBr-activated Sepharose 4B (Pharmacia). Tissues or cultured cells were homogenized in a 1× SDS sample buffer. Protein extracts (50 μg) were separated on 15% SDS-polyacrylamide gels and then transferred to a polyvinylidene difluoride membrane (PVDF) (Mirripore, Bedford, MA). Immunostaining was performed with an enhanced chemoluminescence detection kit (ECL; Amersham) according to the manufacturer’s directions, with a 1 μg/ml concentration of the primary antiserum.
Immunocytochemical analysis. Cultured cells on plastic sheets were fixed with 4% paraformaldehyde (PFA)/0.12 mNa+-phosphate buffer, pH 7.2. After being washed free of the fixative, the cells were preincubated with PBS containing 10% horse serum for 1 hr. The cells were then incubated with anti-MRF-1 antibody (0.5 μg/ml) or OX-42 (mouse IgG, 1:300 dilution) for 1 hr at room temperature. Detection of the primary antiserum was performed using biotinylated IgG according to the procedure provided by the manufacturer (Histofine kit; Nichirei, Tokyo, Japan). Staining was made visible by horseradish peroxidase-conjugated streptavidin and aminoethercarbazol (AEC) reaction product (Zymed, South San Francisco, CA). Finally, the cells were exposed to counterstaining with Mayer’s hematoxylin solution. The numbers of AEC-stained cells were counted at three to four randomly selected, nonoverlapping areas (φ 2.5 mm circle, 5.0 mm2). The number of OX-42-positive cells per area was ∼200 for the cerebellar cell culture at 7 DIV. When both anti-MRF-1 antibody and OX-42 were used for the same sample, the former antibodies were detected with horseradish peroxidase-conjugated anti-rabbit IgG (1:200 dilution) and visualized with AEC reaction product. The latter primary antibodies were detected with biotinylated anti-mouse IgG (Nichirei) and then visualized with both alkaline phosphatase-conjugated streptavidin (Zymed) and the newfuchsine substrate kit (Nichirei).
Immunohistochemical analysis. Under deep pentobarbital anesthesia, adult rats were perfused transcardially with 4% PFA in a 0.1 m sodium phosphate buffer, pH 7.2. Fixed brains were sectioned by microslicer (50 μm in thickness) or embedded in paraffin wax to prepare paraffin sections (5 μm). After they were blocked with 3% normal goat serum, sections were incubated overnight at room temperature with anti-MRF-1 antibody at 0.5 μg/ml for immunofluorescence or at 0.1 μg/ml for immunoperoxidase. For immunofluorescence, sections were then incubated with FITC-labeled anti-rabbit IgG for 2 hr (1:100, Jackson ImmunoResearch, West Grove, PA) and photographed with a confocal laser scanning microscope (MRC 1024, Bio-Rad, Hercules, CA). For immunoperoxidase, sections were processed by the ABC method, using a Histofine kit, and photographed with a bright-field microscope (AX-80, Olympus, Japan).
Phagocytotic activity. The cells were incubated in a medium containing 10 μg/ml fluorescein-conjugated particles [Zymosan A (Saccharomyces cerevisiae) BioParticle; Molecular Probes, Eugene, OR] for 12 hr, washed with PBS, and then fixed with 4% PFA for immunocytochemical analysis.
RESULTS
Isolation and characterization of mrf-1 cDNA clone
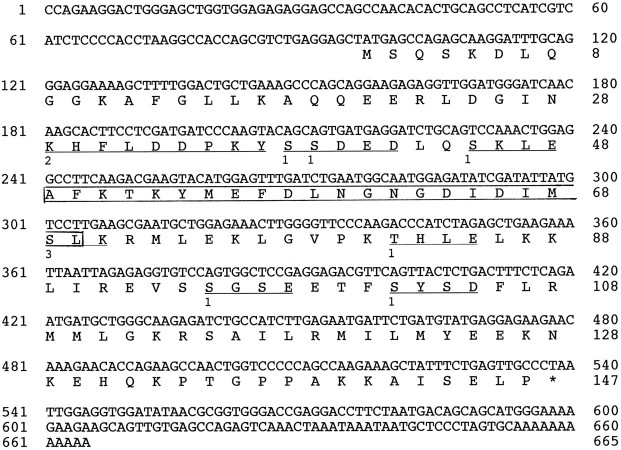
In cerebellar cell culture with normal (5.4 mm) K+ medium, granule neurons begin to degenerate around 4 DIV, and most of the neurons die by 8 DIV (Gallo et al., 1987;Balazs et al., 1988; Koike, 1991; Suzuki and Koike, 1997). Thisin vitro cell death shares major features with that of developing sympathetic and sensory neurons undergoing apoptosis after NGF deprivation: it requires protein and RNA synthesis (Suzuki and Koike, 1997); chronic depolarization with a high concentration of K+ prevents neuronal death via Ca2+ influx through L-type Ca2+channels (Gallo et al., 1987); and BDNF prevents neuronal death (Suzuki and Koike, 1997). To investigate the specific genes that are regulated during neuronal cell death, mRNAs from both 3 and 5 DIV [∼70% survival of granule neurons (Suzuki and Koike, 1997)] cells of the cerebellar cell culture were isolated and analyzed by differential hybridization. We isolated five upregulated genes that were specifically induced in 5 DIV cells. The 3′-partial mrf-1 cDNA fragment, one of the five genes, was subcloned and sequenced. This subcloned fragment has 421 bp and contains a poly(A+) tail. On the basis of information from this partial sequence, we designed specific primer sets and performed a RACE reaction to isolate full-length clones of mrf-1. The full-length cDNA (Fig. 1) was 665 bp long and contained a 116 bp 5′-untranslated region, an entire open-reading frame of 441 bp encoding 117 amino acid residues, and a 108 bp 3′-untranslated region including a polyadenylation signal 20 bp upstream from the poly(A+) tail.
Fig. 1.
Nucleotide and deduced amino acid sequences ofmrf-1 cDNA. Both nucleotide numbering and amino acid numbering (on only the right side) are indicated. Theboxed area indicates the region of the putative EF-hand calcium-binding motif. With regard to other putative motifs ofmrf-1 (number): 1, casein kinase II phosphorylation site (6); 2, tyrosine kinase phosphorylation site (1); 3, protein kinase C phosphorylation site (1).
Database searches revealed a homology with three different genes. Figure 2 shows the homology of the alignment of the MRF-1 amino acid sequence and others: allograft inflammatory factor-1 [AIF-1; Utans et al. (1995)], ionized calcium binding adapter molecule-1 (Iba-1; Imai et al., 1996), and balloon angioplasty responsive transcript-1 [BART-1; Autieri et al. (1996)] have a 100, 94.1, or 54.1% similarity to the MRF-1 sequence, respectively. There is a difference in the nucleotide sequences betweenmrf-1 and AIF-1 cDNA; mrf-1 mRNA is 46 bp longer than AIF-1 in the 5′ upstream region. Moreover, MRF-1 has one EF-hand-like motif that is characteristic of an evolutionary family of calcium-binding proteins (Strynadka and James, 1989), and this domain is completely conserved within all four transcripts mentioned above. These transcripts may consist of a protein subfamily having a single EF-hand motif, because these region are also homologous with calmodulin (Heinzmann and Hunziker, 1991).
Fig. 2.

Comparison of the amino acid sequences among MRF-1, AIF-1, Iba-1, and BART-1. Amino acid numbering is indicated on both sides. Amino acids that are fully conserved are indicated byshadowed boxes. Dashes denote gaps. The EF-hand-like motif is indicated by an open box.
Upregulation of mrf-1 during PCD of cultured granule neurons
To study the expression pattern of an mrf-1 gene during the PCD of granule neurons in cerebellar cell culture, we examined the relative transcript levels as a function of incubation time by Northern blot analysis. The mrf-1 mRNA was detected at ∼0.7 kilobase pair (kbp) in Northern blots, which is consistent with the expected length of the full-length mrf-1 cDNA. As shown in Figure 3A, in the cerebellar culture containing a normal concentration of K+, the transcript level was upregulated at 4 DIV, at which point the granule neurons began to degenerate, and had a peak at 6 DIV, followed by a decrease at 7 DIV, at which time many granule neurons died and were left on the surface of the dishes. It is known that granule neurons maintained in a medium that contains a high concentration of K+ will undergo apoptosis when the high K+ medium is replaced with a normal K+ medium (D’Mello et al., 1993; Yan et al., 1994;Miller and Johnson, 1996). Upregulation of mrf-1 mRNA was also detected in this “low” K+-induced apoptosis of granule neurons (Fig. 3B). These results indicate that the expression of the mrf-1 gene is upregulated in response to the neuronal death of granule neurons.
Fig. 3.
Northern blot showing the upregulation ofmrf-1 mRNA in cerebellar cell culture during the PCD of granule neurons. Sister cerebellar cells were incubated in normal K+ (5.4 mm) medium for 3–7 d (A) or in high K+ (30 mm) medium for 10 d following a switch from the high K+ medium to a serum-free, normal K+ medium (B). At each day or hour indicated at the top of the autoradiograms, the cells were subjected to RNA isolation according to the guanidinium thiocyanate method. Total RNA (20 μg) was run on a 1% agarose gel and transferred to a nylon membrane.32P-labeled mrf-1 or G3PDHcDNA was used as a hybridization probe. Both mrf-1 (0.7 kbp) and G3PDH (1.3 kbp) mRNAs were detected on the same membrane. The positions of the rRNA are indicated on theleft. The amount of separated rRNA in each lane was almost equal when the transferred membrane was stained with methylene blue.
Immunocytochemical localization of the MRF-1 antigen in the cerebellar cell culture
To assess which cell type in the cerebellar cell culture synthesizes the MRF-1, a recombinant MRF-1 protein was prepared and immunized to a rabbit. The purified antiserum was analyzed by Western blot. A single band of ∼17 kDa was detected with anti-MRF-1 antibody in extracts from cerebellar cell cultures, an infant whole brain, and an adult rat spleen (Fig. 4). Spleen tissues have many monocytes or macrophages. This Western blot shows that the MRF-1 level increased in response to granule cell death. The anti-MRF-1 antibody was used for the immunostaining of cultured cerebellar cells. When cells of a 5 DIV cerebellar cell culture were immunostained with this rabbit anti-MRF-1 antibody, partial cytoplasmic areas of non-neuronal cells, which are morphologically characteristic of ameboid microglia, were detected. The cells were then immunostained with both anti-MRF-1 antibody and OX-42 (Robinson et al., 1986), as a marker for microglia (Fig.5A); MRF-1 positive cells were overlapped with a population of OX-42 positive cells. MRF-1 was not detected in all microglia, as shown in Figure 5Da, indicating that the mrf-1 mRNA was expressed in a subpopulation of microglia but not granule neurons. To examine whether the PCD of granule neurons influences the immunoreactivity of MRF-1 in microglia, sets of sister cerebellar cells were fixed at 3, 5, 7, and 9 DIV and immunostained with anti-MRF-1 antibody or OX-42. The photographs of the 3 and 5 DIV cells treated with anti-MRF-1 antibody are shown in Figure 5B. The number of MRF-1-positive cells increased and peaked on the 7 DIV cell; the cell number relative to that of the 3 DIV cell was ∼4, 6, and 4 on the 5, 7, and 9 DIV cells, respectively. Because the number of OX-42-positive cells increased slightly during the incubation from 3 to 7 DIV (∼1.4-fold) (Table1), the ratio of MRF-1-positive cells to the OX-42-positive cells became 0.2, 0.5, 0.8, and 0.5 on the 3, 5, 7, and 9 DIV cells, respectively. The increase in number of MRF-1-positive cells from 3 to 7 DIV is summarized in Table 1. As shown in Figure5B, the immunoreactivity of MRF-1-positive cells also increased dramatically from the 3 DIV to the 5 DIV cells. The upregulation of MRF-1 in response to neuronal death was further confirmed by the following sandwich culture (Fig. 5C). Both purified cortical microglia and cerebellar cells (3 or 5 DIV) were prepared separately and then co-incubated for 12 hr with the two cell populations in contact with each other. After fixation, the cortical microglia were immunostained with anti-MRF-1 antibody. Upregulation of MRF-1 was observed when the sandwich culture of cortical microglia was performed with 5 DIV but not 3 DIV cerebellar granule neurons (Fig. 5Cb,c). This indicates that degenerated granule neurons stimulate the synthesis of MRF-1 in microglia, although morphological alterations do not occur under these conditions. Moreover, induction of MRF-1 did not occur when cortical microglia were incubated with a conditioned medium of 5 DIV cerebellar cells for 12 hr (Fig. 5Cd). Similar data were obtained from the culture cells of purified cerebellar microglia (data not shown). These data indicate that mrf-1 transcripts in microglia are upregulated in response to the PCD of granule neurons and that this induction may require cell-to-cell contact between the microglia and the degenerated granule neurons or secreted signals that lose their activity because of the extreme diffusion.
Fig. 4.
Western blot showing immunoreactivity of the anti-MRF-1 antibody. Granule cells were cultured until 5 DIV in the presence of a high concentration (30 mm;HK+) or normal (5.4 mm;Normal) K+. Proteins (50 μg) extracted from the cultured cells or the indicated tissues were run on a 15% acrylamide gel and then transferred to a PVDF membrane. Chemoluminescent detection (see Materials and Methods) shows a band of ∼17 kDa in each extract. The positions of molecular markers (kD) are indicated on theleft.
Fig. 5.
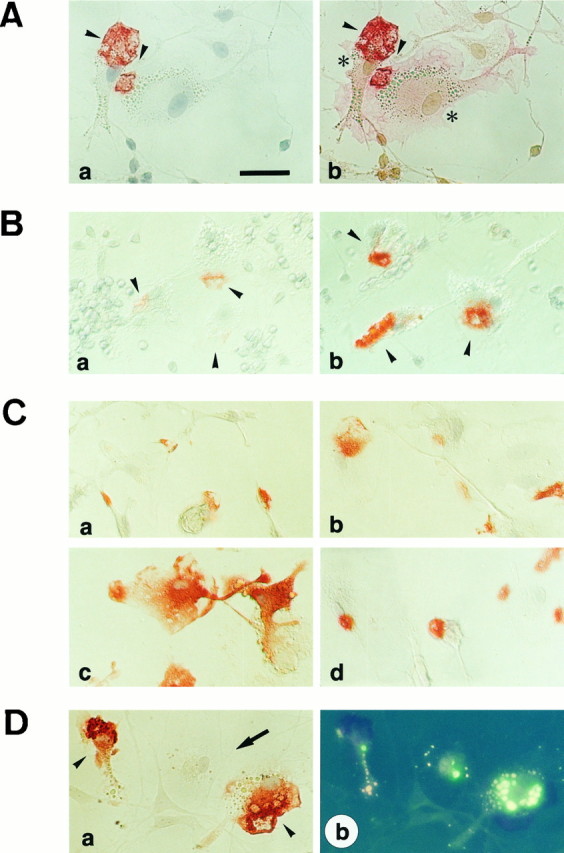
Immunocytochemical analysis of MRF-1 synthesis in cultured cells. A, Immunocytochemical staining showing that the synthesized MRF-1 was localized in the microglia but not in the granule neurons in the cerebellar cell culture. Cerebellar cells cultured in normal K+ medium were fixed at 5 DIV and treated with anti-MRF-1 antibody. Staining of MRF-1 was made visible by the AEC reaction product (dark red). The cells were exposed to counterstaining with Mayer’s hematoxylin solution and photographed (Aa). The cells were then treated with OX-42, a microglial marker. Staining of OX-42 was made visible by the newfuchsine reaction product (pink) (Ab). Arrowheads show MRF-1-positive cytoplasmic areas. Asterisks show OX-42-positive cells/microglia. Scale bar, 50 μm. B, Immunocytochemical staining showing the upregulation of the MRF-1 on microglia during the PCD of granule neurons in cerebella cell culture. Cerebellar cells cultured in normal K+ medium were fixed at 3 DIV (Ba) or 5 DIV (Bb) and treated with anti-MRF-1 antibody. Cells were stained as described above. Arrowheads show the MRF-positive microglia.C, Immunocytochemical staining showing upregulation of MRF-1 on microglia during the PCD of granule neurons in cortical microglia culture. Purified-cortical microglia cells were sandwich-cultured with 3 DIV (Cb) or 5 DIV (Cc) cerebellar cells (see Results) or incubated in a conditioned medium of 5 DIV cerebellar cells (Cd) for 12 hr. The cells were stained as described above. No morphological changes in the microglia were observed after any of the treatments. Untreated microglia (control) are shown in Ca. D, Immunostaining showing that upregulation of MRF-1 on microglia is not associated with microglial phagocytic activity. Cultured cerebellar cells (5 DIV) were exposed to fluorescein-conjugated beads, Zymosan A, for 12 hr, and then fixed and immunostained with anti-MRF-1 antibody. The cells were stained as described above, photographed with differential interference contrast [contrast micrograph (Da)] and under UV light [fluorescent photographs (Db)]. Arrowheads or anarrow show MRF-1-positive or -negative microglia, respectively. All three microglia were fluorescence positive (Db).
Table 1.
Increase in the number of MRF-1-positive microglia in response to death of granule neurons
| Number of positive cells per 5.0 mm2(n = 4) | Ratio (DIV 7/DIV 3) | ||
|---|---|---|---|
| DIV 3 | DIV 7 | ||
| MRF-1 | 34 ± 5 | 190 ± 26 | 5.59 |
| OX-42 | 165 ± 36 | 237 ± 21 | 1.44 |
| Ratio (MRF-1/OX-42) | 0.21 | 0.80 | 3.89 |
Microglial upregulation of mrf-1 mRNA under conditions inhibiting the proliferation of microglia
Microglia proliferated to a slight extent under our standard culture where FudR was supplemented as an antimitotic agent; contamination of OX-42-positive microglia was ∼5% of the number of granule neurons surviving at 7 DIV. To exclude the possibility that this mrf-1 upregulation was simply caused by the proliferation of microglia, we examined the mrf-1 mRNA level in the presence of a DNA polymerase inhibitor aphidicolin (Miller and Johnson, 1996). When cerebellar cells were cultured for 6 d under high K+ condition in the presence of aphidicolin (10 μm), contamination of Vimentin-positive non-neuronal cells was reduced to 3.3 ± 0.2%. Aphidicolin completely inhibited the incorporation of bromodeoxyuridine to microglia, and the contamination of OX-42-positive microglia in the presence of aphidicolin was reduced to one-fifth (37.3 ± 2.1 cells/5 mm2) of the standard culture at 7 DIV in normal K+ medium. This number of OX-42-positive cells was <1% of the surviving granule neurons. Northern blot shows that themrf-1 mRNA was undetectable at 4 DIV cerebellar culture, when granule neurons normally begin to degenerate (Fig.6), and had not upregulated at 5–7 DIV in the presence of aphidicolin. However, when purified microglia were added to the 3 DIV cerebellar culture and incubated for 18 hr in the presence of aphidicolin, mrf-1 mRNA was upregulated (Fig.6). These results indicate that the microglial upregulation ofmrf-1 in response to neuronal cell death is independent of the proliferation of microglia.
Fig. 6.
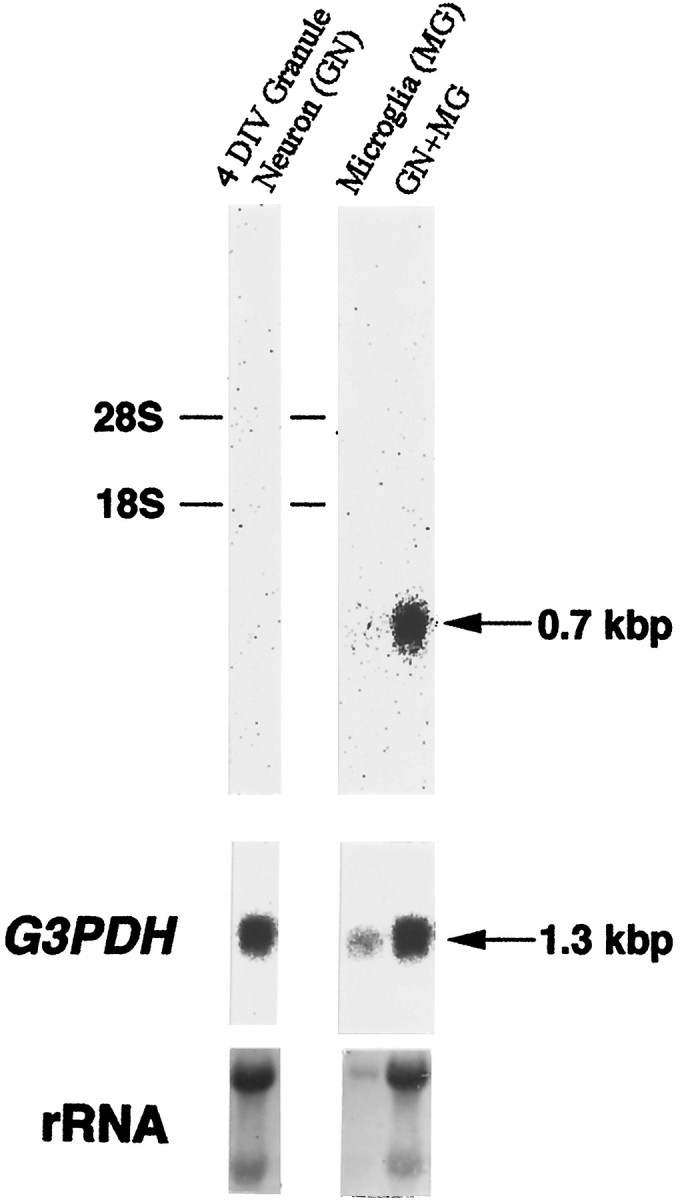
Autoradiogram showing the upregulation ofmrf-1 mRNA under the inhibitory condition of cell proliferation. Both granule cells (35 mm dishes) incubated in the presence of 10 μm aphidicolin, and cortical cells were prepared separately. Purified microglia obtained from the cortical culture (1 × 105 cells) were replated or added into the 3 DIV granule culture and incubated for an additional 18 hr in the presence of aphidicolin. All RNA was extracted from 4 DIV granule cells (GN), 18 hr incubated purified microglia (MG), or the mixed cells (GN+MG), and separated in an agarose gel. Hybridization was performed as shown in the legend of Figure 3. The rRNA positions are indicated on theleft. rRNA was visualized by stain with methylene blue.
The relationship between MRF-1 induction and the phagocytic activity of microglia
To determine whether the phagocytic activity of microglia is associated with MRF-1 induction, we exposed cultured cerebellar cells (5 DIV) to fluorescein-conjugated beads, Zymosan A, for 12 hr. After fixation, the cells were immunostained with anti-MRF-1 antibody. As shown in Figure 5D, not all microglia cells labeled with beads were MRF-1-positive cells. Moreover, when purified cortical microglia were exposed to the fluorescein-conjugated beads, the level of MRF-1 protein was unaltered in the microglia engulfing the beads (data not shown). These data indicate that the induction of MRF-1 may not be associated with the phagocytic activity of microglia.
Transient increase in the mrf-1 message in the postnatal development of the cerebellum
To explore the expression level of mrf-1 in vivo, its mRNA levels were examined by Northern blot as a function of the postnatal development of the cerebellum. As shown in Figure7, the mrf-1 mRNA was detected at all postnatal days examined (P0–28), although its level of expression varied dramatically. The expression level reached a peak at P7 and was then downregulated at later postnatal days. We then prepared a section of P7 rat cerebella and immunostained it with anti-MRF-1 antibody (Fig. 8). Most of the MRF-1-positive signals were detected in the white matter (WM) and/or internal granular layer (IGL). These MRF-1-positive signals did not overlap with GFAP-positive cells and were present in the interior of the calbindin-positive Purkinje cell layer (data not shown). Because the anti-MRF-1 antibody specifically recognizes microglia in the adult rat brain (see below) as well as in in vitro culture, it is thought that the distribution of MRF-1-positive signals in a cerebellar section suggests the presence of MRF-1-positive microglia. The immunohistochemical data with anti-MRF-1 antibody in P7 cerebellum is consistent with the finding that in the rat cerebellum, programmed cell death occurs most prominently at approximately P7 in these regions (Krueger et al., 1995). These results indicate a temporally increased expression of mrf-1, suggesting the possibility that MRF-1 plays an important role in cerebellar development.
Fig. 7.
Autoradiogram showing the expression ofmrf-1 mRNA in postnatal development of the cerebellum. Cerebella were dissected from rat pups on the indicated postnatal days and homogenized in guanidinium thiocyanate solution. Poly (A+) RNA (3 μg) was run on a 1% agarose gel and transferred to a nylon membrane. Hybridization was performed as shown in the legend of Figure 3. The presumed rRNA positions are indicated on the left.
Fig. 8.
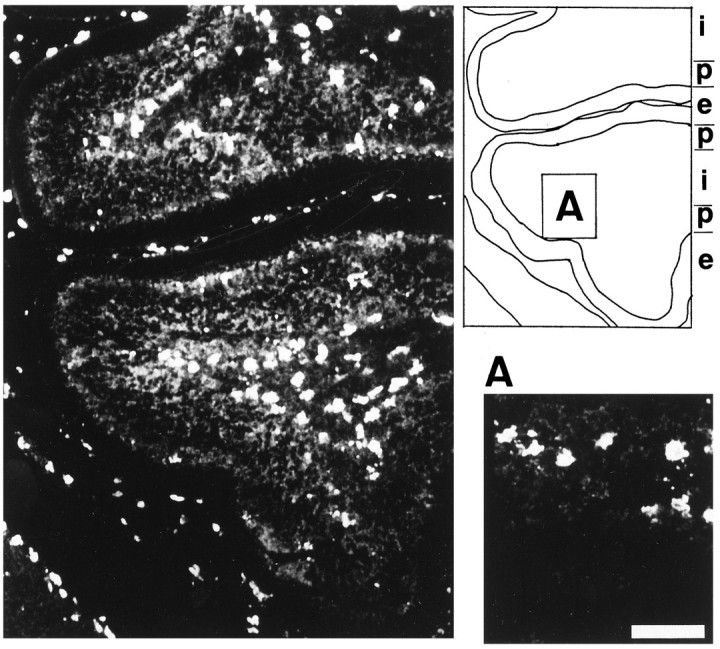
Confocal microscopic image showing the distribution of MRF-1-immunoreactive antigen in P7 rat cerebellum. A section of the P7 rat cerebellum was immunostained with anti-MRF-1 antibody and photographed. Scale bar (shown in A): 50 μm. i, Internal granule layer and/or white matter;p, Purkinje cell layer; e, external granule layer.
Localization of the MRF-1 antigen in ramified microglia and its upregulation after axotomy
In adult brain sections, the distribution of cells expressing MRF-1 and their morphology were examined by immunohistochemistry (Fig.9A,B). Immunostained cells were found in various brain regions. In the cerebellum, they were evenly distributed in the molecular layer, granular layer, and medullary zone (Fig. 9A). Their cell bodies were spindle-shaped and contained an elongated nucleus (Fig. 9B). From the cell body, a few thin processes extended in different directions and were further branched in the periphery. No significant staining was detected in any of the neuronal elements in the cerebellum and other brain regions. From the characteristic morphology, MRF-1 was judged to be localized in ramified or resting microglia in the adult normal rat brain.
Fig. 9.
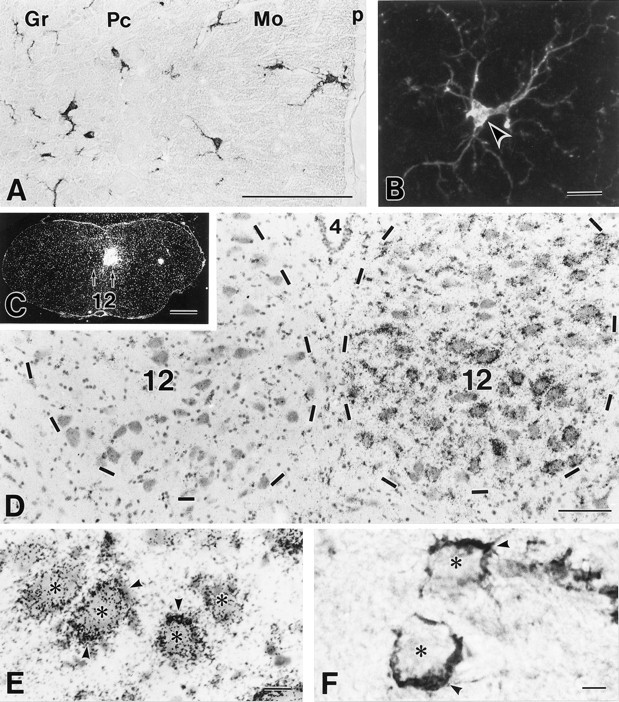
MRF-1 in ramified/resting microglia in the cerebellum (A, B) and its prominent upregulation in activated microglia of the hypoglossal nucleus 5 d after right hypoglossal nerve dissection (C–F). A, B, Immunoperoxidase (A) and confocal immunofluorescence (B) images showing the distribution and morphology of MRF-1-positive, ramified/resting microglia in the adult cerebellum. Arrowhead indicates the cell body of immunostained microglia. Gr, Granular layer; Pc, Purkinje cell layer; Mo, molecular layer; P, pia matter. C–E,In situ hybridization showing elevatedmrf-1 mRNA in ipsilateral, axotomized hypoglossal nucleus (right). Strong signals for themrf-1 mRNA are found in small cells (arrowheads) that surround cell bodies of injured hypoglossal neurons (asterisks). 4, Fourth ventricle; 12, hypoglossal nucleus.F, Immunohistochemistry showing intensified MRF-1 immunoreactivity in activated microglia (arrowheads) surrounding cell bodies of axotomized hypoglossal neurons (asterisks). Scale bars: A, D, 100 μm; B, E, F, 10 μm; C, 1 mm.
Changes after neuronal injury were experimentally examined by dissecting hypoglossal motor nerves at the hyoid bone. Five days after the axotomy, the ipsilateral hypoglossal nucleus displayed a prominent increase in mrf-1 mRNA (Fig. 9C,D). The signals were detected in small cells around hypoglossal motoneurons having large cell bodies (Fig. 9E). By immunohistochemistry, strong immunostainings were found in cell bodies and processes of activated microglia, which surrounded the injured neuronal cell bodies (Fig.9F). In the contralateral hypoglossal nucleus, however, no significant changes were found in the levels ofmrf-1 mRNA and MRF-1 immunoreactivity (Fig.9C,D). Moreover, cells with low mrf-1 mRNA and immunoreactivity in the contralateral nucleus were distributed evenly within the nucleus but not surrounded by neuronal cell bodies. These histological findings indicate that mrf-1 is expressed in ramified/resting microglia in vivo and that its expression is prominently upregulated in microglia activated by neuronal injury.
DISCUSSION
We searched for genes that are upregulated in the process of neuronal death of granule cells in cerebellar cell culture. In this study, we cloned one of these genes, mrf-1, which is a gene upregulated in response to granule cell death in microglia in vitro. In the developing cerebellum in vivo,mrf-1 mRNA was transiently increased in the early postnatal stages when cerebellar neurons and astrocytes undergo active degeneration. In adult brain sections, MRF-1 was detected in ramified/resting microglia, and upregulated in activated microglia surrounding injured central motoneurons by peripheral nerve dissection.
A homology search of the full-length mrf-1 sequence revealed the presence of three cognate genes that are expressed in macrophages or microglia: AIF-1 (Utans et al., 1995), Iba-1 (Imai et al., 1996), or BART-1 (Autieri et al., 1996). The AIF-1 protein has the same amino acid sequence as MRF-1, although the 5′ noncoding region of AIF-1 mRNA is shorter than that of mrf-1 (Utans et al., 1995). Southern blots using AIF-1 cDNA as a probe have revealed a single band with a high intensity in rat genomic DNA (Utans et al., 1995). It is thus likely that the difference between mrf-1 and AIF-1 mRNAs may reflect an alternative start site derived from a single gene. MRF-1 is different from the Iba-1 protein, which is also synthesized in microglia, in the upstream region containing the 5′ noncoding region and some N-terminal amino acid sequences (Imai et al., 1996). This indicates that, although the differences between these genes are small, they may be subject to differential regulation at transcriptional and/or translational levels. It is thought that these four genes, including BART-1, code for one EF-hand-like motif. Two or more EF-hand motifs are typical for the family of intracellular calcium-binding proteins that includes calmodulin and troponin C (for review, seeHeinzmann and Hunziker, 1991). They may be members of a calcium-binding protein family that has a single EF-hand motif. Nevertheless,mrf-1 was the first of four genes to show that it is a microglial gene responding to neuronal cell death.
The upregulated expression of mrf-1 in cerebellar cell culture was correlated with the degeneration of granule neurons. The increased expression of mrf-1 mRNA peaked in the 6 DIV cerebellar cells when the cells were cultured under conditions that allowed a limited proliferation of non-neuronal cells. An immunocytochemical analysis of the MRF-1 staining pattern revealed that the protein was synthesized in the microglia and was upregulated in response to neuronal degeneration. Compared with the mrf-1 mRNA level, which peaked at 6 DIV, the immunoreactivity of the MRF-1 peaked at 7 DIV under normal K+ conditions. The 1 d delay in MRF-1 synthesis may reflect the difference between the transcription and translation of mrf-1. When purified microglia were co-incubated with 5 DIV cerebellar cells under a sandwich culture in which the two cell populations were in contact with each other, MRF-1 in the purified microglia was upregulated. In contrast, when purified microglia were incubated with culturing medium obtained from a 5 DIV cerebellar cell culture, that is, conditioned medium of 5 DIV cerebellar cells, there was no effect on MRF-1 synthesis in the purified microglia. These data suggest that the upregulation of mrf-1 may require either cell-to-cell contact between microglia and degenerated neurons or secreted signals that are released from degenerated neurons or second stimulated non-neuronal cells (e.g., astrocyte) and lose their activity because of extreme diffusion.
What kind of role does MRF-1 play in neuronal apoptosis? It has been reported that microglia are rapidly activated and proliferate around damaged neurons in response to cerebral ischemia and peripheral neuronal injury (Svensson et al., 1993; Gehrmann et al., 1995). It is generally accepted that activated microglia engage in phagocytotic activity to eliminate neuronal debris. Also, activated microglia upregulate the expression of several surface molecules [e.g., major histocompatibility complex (MHC) class I and II antigens], cytokines (e.g., interleukin-1, transforming growth factor β1), and cytotoxic substrate (e.g., reactive oxygen species, nitric oxygen). Some of these may contribute to repair of tissue or promotion of neuronal cell death. It is thought that cultured microglia is not in a resting state. In our culture system, mrf-1 mRNA was upregulated in microglia under the inhibitory conditions for cell proliferation. This indicates that MRF-1 may not be associated with the proliferational activity of microglia. Moreover, when fluorescein-conjugated particles were phagocytized to cultured microglia, not all of the fluorescein-labeled microglia were MRF-1 positive. These data indicate that MRF-1 may especially be synthesized in microglia that are capable of recognizing signs of neuronal damage, and that the induction of MRF-1 is not directly related to phagocytotic activity in microglia. Phagocytosis is a complex and still poorly understood phenomenon (Savill et al., 1993). The first phase of phagocytosis involves the specific recognition of the eliciting particle via the engulfment of one or multiple surface receptors and is followed by ingestion per se. It has been reported that six engulfment genes controlling at least two distinct and parallel processes have been identified by the genetic analysis of PCD-deficient mutants inC. elegans. Recently, it has been shown that ABC1 is required for the engulfment of corpses generated by apoptotic cell death in mouse macrophage, and ABC1 might be a mammalian homolog toced-7, one of the engulfment genes of C. elegans(Luciani and Chimini, 1996). It is possible that MRF-1 plays a role in the pathway that allows the recognition or incorporation of neuronal debris. This is further substantiated by the fact that MRF-1 is similar to AIF-1, which has a possible functional role in macrophage activation and function (Utans et al., 1995). Taking into account the similarities between the two proteins, it is likely that these proteins are involved in a converged signal transduction cascade elicited by the recognition of tissues or cells. Utans et al. (1995) showed that the upregulation of AIF-1 occurs when cardiac allografts, but not syngrafts, are transplanted. This suggests that AIF-1 responds to the cells or debris of another species. In contrast, MRF-1 responds to the degenerated cells or debris derived from the same species. Another possible role for MRF-1 may be related to microglial cytotoxicity or the trophic effect for damaged neurons. We have found that the immunoreactivity of MRF-1 is localized to a limited cytoplasmic region of microglia in our cultures. The profile generated by the Kyte-Doolittle algorithm and others predicted a hydrophilic polypeptide having a single EF-hand motif without significant hydrophobic stretches. It is thus possible that MRF-1 functions as a signal-transducing molecule regulated by the level of intracellular Ca2+. It remains to be determined, however, whether Ca2+ indeed binds to the EF-hand motif of MRF-1 and regulates the expression of themrf-1 gene.
We also found that mrf-1 is expressed in resting microglia on adult rat brain to a lesser extent compared with the level in ameboid microglia. All the markers currently available to identify microglia including MHC class II, complement type 3 receptor, etc., are surface antigens located on the cell surface of microglia (for review, see Streit, 1995). It is likely that the antibodies against MRF-1 offer a unique tool to identify the cytoplasm of microglia both in vivo and in vitro, although the exact function of MRF-1 remains to be resolved. Expression of mrf-1 mRNA in a hypoglossal nucleus was prominently upregulated in microglia that surrounded injured neuronal cell bodies activated by dissecting hypoglossal motor nerves. It is thought that hypoglossal motor nerves degenerate, but do not die, after axotomy (for review, see Sevensson et al., 1993). Activated microglia around axotomized motoneurons, at least, will not have a neurotoxic effect. When non-neuronal cells were thoroughly eliminated from cerebellar cell cultures in the presence of aphidicolin (see the Materials and Methods), granule cell death apparently proceeded with a time course similar to that in the standard culture. Moreover, we found that the supplement of cultured microglia to aphidicolin-treated cell cultures appeared to neither promote nor suppress the cell death of granule neurons under our culture conditions (data not shown). Thus, it is likely that the upregulation ofmrf-1 per se is not causally connected with granule cell death but rather is probably secondary to this event.
We were able to show that mrf-1 mRNA increases temporally in the developing cerebellum of rats. In particular, the mrf-1 mRNA level peaks at P7. In the postnatal development of the cerebellum, differentiating granule cells migrate from the external granular layer (EGL) to the IGL and form synaptic contacts (Altman, 1972). Granule neurons that do not have enough synaptic connections may be eliminated at this stage by an apoptotic mechanism. Analysis of pyknotic cells in the developing postnatal rat cerebellum have revealed that the majority of pyknotic cells are in the developing WM, where their number peaks at ∼P7, whereas those in the IGL peak at P10 (Krueger et al., 1995). Moreover, it appears that 50–70% of the pyknotic cells in the WM and IGL are astrocytes. The period of mrf-1 activation in the cerebellum corresponded to the time of apoptotic death of the astrocytes in the WM and/or IGL. It is possible that mrf-1 may respond to the death of astrocytes as well. The majority of neuronal cell deaths for the granule neurons in the rat cerebellum occur in the EGL at ∼2–4 weeks postnatal. However, the number of pyknotic cells in the EGL is less than that of WM and/or IGL around P7 (Krueger et al., 1995). For this reason, it is not clear that there is an upregulation of mrf-1 expression levels in response to the neuronal death in the EGL. The immunoreactivity of MRF-1 was also detected in the ramified/resting microglia in the adult rat brain. This expression of mrf-1 in the adult brain indicates that MRF-1 may have functions other than just the role it plays in cell death.
In summary, we have provided here evidence that a microglial gene,mrf-1, is upregulated in response to neuronal cell death and degeneration both in vitro and in vivo. The specific physiological function of MRF-1 is unknown, although it may possibly play a role both in developmental PCD and in recovery from brain injuries.
Footnotes
This work was supported in part by a special Grant-in-Aid for the Promotion of Education and Research in Hokkaido University, Grants-in-Aid for the Ministry of Education, Science, Culture, and Sports, and the Program for the Promotion of Fundamental Studies in Health Sciences.
Correspondence should be addressed to Shuuitsu Tanaka, Molecular Neurobiology Laboratory, Graduate Program in Biological Sciences, Hokkaido University, Faculty of Science, Kita 10 Nishi 8, Kita-ku, Sapporo 060–0810, Japan.
Mr. Suzuki’s present address: Pharmaceutical Research Institute, Kyowa Hakko Kogyo Co., Ltd., Sizuoka 411–8731, Japan.
REFERENCES
- 1.Altman J. Postnatal development of the cerebellar cortex in the rat. I. The external germinal layer and the transitional molecular layer. J Comp Neurol. 1972;145:353–397. doi: 10.1002/cne.901450305. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong RC, Aja TJ, Hoang KD, Bai X, Alnemri ES, Litwack G, Karanewsky DS, Fritz LC, Tomaselli KJ. Activation of the CED3/ICE-related protease CPP32 in cerebellar granule neurons undergoing apoptosis but not necrosis. J Neurosci. 1997;17:553–562. doi: 10.1523/JNEUROSCI.17-02-00553.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Autieri MV, Prystowsky MB, Ohlstein EH. Isolation and characterization of BART-1: a novel balloon angioplasty responsive transcript in rat carotid arteries. DNA Cell Biol. 1996;15:297–304. doi: 10.1089/dna.1996.15.297. [DOI] [PubMed] [Google Scholar]
- 4.Balazs R, Jorgensen OS, Hack N. N-methyl-d-aspartate promotes the survival of cerebellar granule cells in culture. Neuroscience. 1988;27:437–451. doi: 10.1016/0306-4522(88)90279-5. [DOI] [PubMed] [Google Scholar]
- 5.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 6.Dickson DW, Sunhee CL, Mattiace LA, Yen SHC, Brosnan A. Microglia and cytokines in neurological disease, with special reference to AIDS and Alzheimer’s disease. Glia. 1993;7:75–83. doi: 10.1002/glia.440070113. [DOI] [PubMed] [Google Scholar]
- 7.D’Mello SR, Galli C, Ciotti T, Calissano P. Induction of apoptosis in cerebellar granule neurons by low potassium: inhibition of death by insulin-like growth factor I and cAMP. Proc Natl Acad Sci USA. 1993;90:10989–10993. doi: 10.1073/pnas.90.23.10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellis RE, Yuan J, Horvitz HR. Mechanisms and functions of cell death. Annu Rev Cell Biol. 1991;7:663–698. doi: 10.1146/annurev.cb.07.110191.003311. [DOI] [PubMed] [Google Scholar]
- 9.Frohman MA. Rapid amplification of complementary DNA ends for generation of full-length complementary cDNAs: thermal RACE. Methods Enzymol. 1993;218:340–358. doi: 10.1016/0076-6879(93)18026-9. [DOI] [PubMed] [Google Scholar]
- 10.Gallo V, Kingsbury A, Balazs R, Jorgensen OS. The role of depolarization in the survival and differentiation of cerebellar granule cells in culture. J Neurosci. 1987;7:2203–2213. doi: 10.1523/JNEUROSCI.07-07-02203.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gehrmann J, Banati RB, Wiessner C, Hossmann KA, Kreutzberg GW. Reactive microglia in cerebral ischaemia: an early mediator of tissue damage? Neuropathol Appl Neurobiol. 1995;21:277–289. doi: 10.1111/j.1365-2990.1995.tb01062.x. [DOI] [PubMed] [Google Scholar]
- 12.Giulian D, Chen J, Ingeman JE, George JK, Noponen M. The role of mononuclear phagocytes in wound healing after traumatic injury to adult mammalian brain. J Neurosci. 1989;9:4416–4429. doi: 10.1523/JNEUROSCI.09-12-04416.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giulian D, Li J, Leara B, Keenen C. Phagocytic microglia release cytokines and cytotoxins that regulate the survival of astrocytes and neurons in culture. Neurochem Int. 1994;25:227–233. doi: 10.1016/0197-0186(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 14.Heinzmann CW, Hunziker W. Intracellular calcium-binding proteins: more sites than insight. Trends Biochem Sci. 1991;16:98–103. doi: 10.1016/0968-0004(91)90041-s. [DOI] [PubMed] [Google Scholar]
- 15.Henderson CE. Programmed cell death in the developing nervous system. Neuron. 1996;17:579–585. doi: 10.1016/s0896-6273(00)80191-9. [DOI] [PubMed] [Google Scholar]
- 16.Imai Y, Ibata I, Ito D, Ohsawa K, Kohsaka S. A novel gene iba1 in the major histocompatibility complex class III region encoding an EF hand protein expressed in a monocytic lineage. Biochem Biophys Res Commun. 1996;224:855–862. doi: 10.1006/bbrc.1996.1112. [DOI] [PubMed] [Google Scholar]
- 17.Jacobson MD, Weil M, Raff M. Programmed cell death in animal development. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 18.Johnson EM, Jr, Deckwerth TL. Molecular mechanisms of developmental neuronal death. Annu Rev Neurosci. 1993;16:31–46. doi: 10.1146/annurev.ne.16.030193.000335. [DOI] [PubMed] [Google Scholar]
- 19.Kerr JFR, Harmon BV. Definition and incidence of apoptosis: an historical perspective. In: Tomei LD, Cope FO, editors. Apoptosis: the molecular basis of cell death. Cold Spring Harbor Lab; New York: 1991. pp. 5–29. [Google Scholar]
- 20.Koike T. Neuronal survival of cerebellar granule cells in vitro is regulated by levels of intracellular calcium. Soc Neurosci Abstr. 1991;17:1499. [Google Scholar]
- 21.Krueger BK, Burne JF, Raff MC. Evidence for large-scale astrocyte death in the developing cerebellum. J Neurosci. 1995;15:3366–3374. doi: 10.1523/JNEUROSCI.15-05-03366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luciani M-F, Chimini G. The ATP binding cassette transporter ABC1, is required for the engulfment of corpses generated by apoptotic cell death. EMBO J. 1996;15:226–235. [PMC free article] [PubMed] [Google Scholar]
- 23.McGeer PL, Kawamata T, Walker DG, Akiyama H, Tooyama I, McGree EG. Microglia in degenerative neurological disease. Glia. 1993;7:84–92. doi: 10.1002/glia.440070114. [DOI] [PubMed] [Google Scholar]
- 24.Miller TM, Johnson EM., Jr Metabolic and genetic analyses of apoptosis in potassium/serum-deprived rat cerebellar granule cells. J Neurosci. 1996;16:7487–7495. doi: 10.1523/JNEUROSCI.16-23-07487.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oppenheim RW. Cell death during development of the nervous system. Annu Rev Neurosci. 1991;14:453–501. doi: 10.1146/annurev.ne.14.030191.002321. [DOI] [PubMed] [Google Scholar]
- 26.Robinson AP, White TM, Mason DW. Macrophage heterogeneity in the rat as delineated by two monoclonal antibodies MCR OX-41 and MRC OX-42, the latter recognizing complement receptor type 3. Immunology. 1986;57:239–247. [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual, Ed 2. Cold Spring Harbor Lab; New York: 1989. [Google Scholar]
- 28.Savill J, Fadok V, Henson P, Hastell C. Phagocyte recognition of cells undergoing apoptosis. Immunol Today. 1993;14:131–135. doi: 10.1016/0167-5699(93)90215-7. [DOI] [PubMed] [Google Scholar]
- 29.Stoll G, Trapp BD, Griffin JW. Macrophage function during Wallerian degeneration of the rat optic nerve: clearance of degenerating myelin and Ia expression. J Neurosci. 1989;9:2327–2335. doi: 10.1523/JNEUROSCI.09-07-02327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Streit WJ. Microglial cells. In: Kettenmann H, Ransom BR, editors. Neuroglia. Oxford UP; New York: 1995. pp. 85–96. [Google Scholar]
- 31.Strynadka NCJ, James MNG. Crystal structures of the helix-loop-helix calcium-binding proteins. Annu Rev Biochem. 1989;58:951–998. doi: 10.1146/annurev.bi.58.070189.004511. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki K, Koike T. Brain-derived neurotrophic factor suppresses programmed death of cerebellar granule cells through a post-translational mechanism. Mol Chem Neuropathol. 1997;30:101–124. doi: 10.1007/BF02815153. [DOI] [PubMed] [Google Scholar]
- 33.Suzumura A, Bhat S, Eccleston PA, Lisak RP, Silberberg DH. The isolation and long-term culture of oligodendrocytes from newborn mouse brain. Brain Res. 1984;324:379–383. doi: 10.1016/0006-8993(84)90054-4. [DOI] [PubMed] [Google Scholar]
- 34.Svensson M, Eriksson P, Persson JKE, Molander C, Arvidsson J, Aldskogius H. The response of central glia to peripheral nerve injury. Brain Res Bull. 1993;30:499–506. doi: 10.1016/0361-9230(93)90284-i. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka S, Suzuki K, Watanabe M, Tone S, Koike T. Up-regulation of a new microglial gene (mrf-1) in response to programmed neuronal cell death. Soc Neurosci Abstr. 1997;23:1988. doi: 10.1523/JNEUROSCI.18-16-06358.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thery C, Chamak B, Mallat M. Cytotoxic effect of brain macrophages on developing neurons. Eur J Neurosci. 1991;3:1155–1164. doi: 10.1111/j.1460-9568.1991.tb00050.x. [DOI] [PubMed] [Google Scholar]
- 37.Utans U, Arceci RJ, Yamashita Y, Russell ME. Cloning and characterization of allograft inflammatory factor-1: a novel macrophage factor identified in rat cardiac allografts with chronic rejection. J Clin Invest. 1995;95:2954–2962. doi: 10.1172/JCI118003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watanabe M, Inoue Y, Sakimura K, Mishina M. Distinct distributions of five NMDA receptor channel subunit mRNAs in the forebrain. J Comp Neurol. 1993;338:377–390. doi: 10.1002/cne.903380305. [DOI] [PubMed] [Google Scholar]
- 39.Yan G-M, Ni B, Weller M, Wood KA, Paul SM. Depolarization or glutamate receptor activation blocks apoptotic cell death of cultured cerebellar granule neurons. Brain Res. 1994;656:43–51. doi: 10.1016/0006-8993(94)91364-1. [DOI] [PubMed] [Google Scholar]