Abstract
Synaptic activity induces a rapid transcriptional response that is essential for the establishment of long-term neuronal plasticity. Using a differential cloning technique, we have identified a gene induced by seizure activity in the brain as RB3. RB3 is a recently cloned gene belonging to the stathmin family of phosphoproteins. Like SCG10, RB3 is brain-specific, although in situhybridization results show that the expression of RB3 is more ubiquitous than is that of SCG10. Using genomic DNA sequencing, we show that the 27 amino acid sequence unique to the RB3" transcript is encoded by an alternatively spliced exon, exon 2′. Using a peptide antibody raised against exon 2′ to detect RB3" and an anti-Flag antibody to detect an epitope-tagged version of RB3, we show that both proteins are localized to the Golgi apparatus of transfected COS7 cells. Of particular interest, RB3 mRNA, but not SCG10 mRNA, is rapidly induced in the dentate gyrus granule layer of the hippocampus after electrically induced seizure activity as well as stimuli leading to long-term potentiation (LTP). In addition, RB3 mRNA is induced in pheochromocytoma (PC12) cells treated with 250 ng/ml NGF. These results suggest that RB3 may play a role in activity-induced neuronal plasticity and neuronal differentiation.
Keywords: plasticity, RB3, SCG10, stathmin, neuronal activation, long-term potentiation
Neuronal plasticity, which is an integral component of learning and long-term memory, involves changes in gene expression and is protein synthesis dependent (Wang et al., 1997). Various rat models, such as long-term potentiation (LTP) and electrically induced seizures, can be used to examine changes at the molecular level that may be directly or indirectly responsible for these long-term changes. Via the use of differential screening techniques (Yamagata et al., 1993), a variety of known and novel genes that are induced rapidly by neuronal activation and contribute to neuronal plasticity has now been identified and characterized. These include growth factors, transcription factors, cytoskeleton-associated proteins, intracellular signaling molecules, and others, indicating the complex mechanisms underlying neuronal plasticity (Yamagata et al., 1994; Tsui et al., 1996; Brakeman et al., 1997).
Screening of a subtracted cDNA library containing genes upregulated by electrically induced seizures [maximal electroconvulsive seizures (MECS) (Cole et al., 1990)] in the adult rat brain led to the isolation of RB3, a recently identified member of the stathmin family. Stathmin (Sobel, 1991) and the related proteins SCG10, RB3, and scalo-like protein (SCLIP) constitute a highly conserved family of intracellular proteins (Maucuer et al., 1993;Okazaki et al., 1993; Ozon et al., 1997, 1998). Stathmin, a ubiquitously expressed protein that is a substrate for a variety of serine kinases, is thought to integrate intracellular signaling pathways involved in differentiation and proliferation (Sobel, 1991).
Like stathmin, SCG10 binds to tubulin, promotes tubulin disassemblyin vitro (Riederer et al., 1997), and is phosphorylated by a variety of serine and threonine kinases (Antonsson et al., 1997). However, unlike stathmin, SCG10 expression is restricted to neurons, and the protein is membrane-associated and localized to the Golgi apparatus and growth cones (Stein et al., 1988a; Antonsson et al., 1997). This has led to the proposal that SCG10 contributes to the regulation of microtubule dynamics in the growth cone of developing neurons and plays a role in neuritogenesis (Riederer et al., 1997). In support of this theory, SCG10 is induced by NGF in pheochromocytoma (PC12) cells (Stein et al., 1988b) and enhances neurite outgrowth when stably expressed in NGF-treated PC12 cells (Riederer et al., 1997). In addition, SCG10 is expressed during periods of neurite extension in the developing rat brain and in regions of neuronal plasticity in the adult rat brain (Stein et al., 1988a; Himi et al., 1994).
Little is known about the more recently cloned RB3 (the rat homolog of the Xenopus gene XB3) and SCLIP (SCG10-like protein) (Ozon et al., 1997, 1998). Like SCG10, the expression of both RB3 and SCLIP mRNA is limited to the nervous system, although their expression patterns are quite distinct. The similarity of the N-terminal sequences of both RB3 and SCLIP to that of SCG10 (Stein et al., 1988a; Di Paolo et al., 1997) suggests that both proteins may be membrane-associated proteins. In addition, sequence similarity within the so-called stathmin domain suggests that they may bind to similar target proteins (e.g., tubulin) in a phosphorylation-dependent manner. Three different transcripts of the RB3 gene—RB3, RB3′, and RB3"—have been identified. These are thought to result from alternative splicing within the RB3 gene.
Here we identify one of the genes upregulated by seizure activity as RB3. We show that the different transcripts RB3 and RB3" are indeed produced by the alternative splicing of a novel exon, exon 2′. Similar to SCG10 mRNA, RB3 mRNA is expressed in the developing and adult brains and induced in differentiating PC12 cells, and the protein is localized to the Golgi apparatus in transfected COS7 cells. Most importantly, RB3 can be distinguished from SCG10 because RB3 mRNA is induced rapidly in the dentate gyrus after electrically induced seizure activity and stimuli resulting in LTP, which is considered to be a model system for some aspects of long-term memory formation.
MATERIALS AND METHODS
Cloning and sequencing of RB3. The preparation and screening of the subtracted cDNA library used in this study have been described previously (Yamagata et al., 1993). This library was prepared from the hippocampi of rats treated with cycloheximide (20 mg/ml) 4 hr after multiple MECS (see below). The library was screened with32P-labeled cDNA derived from RNA isolated from the hippocampus of control or MECS-stimulated rats treated with cycloheximide (Yamagata et al., 1993).
A 1019 bp fragment containing a putative polyA tail, derived from this subtracted cDNA library, was used to screen 140,000 pfu of a lambda ZAP (Stratagene, La Jolla, CA) cDNA library derived from rat LTP hippocampal RNA, using standard filter hybridization techniques. This library has been used previously to clone full-length genes successfully (Lyford et al., 1995; Tsui et al., 1996; Brakeman et al., 1997). Positive clones were sequenced using the automated DNA sequencer at Perkin-Elmer/Applied Biosystems (Foster City, CA). The sequences were analyzed using the Sequencher program (Gene Codes Corporation, Ann Arbor, MI). Similarity to genes and expressed sequence tags (ESTs) in the GenBank database was analyzed using the BLAST algorithm (Altschul et al., 1990).
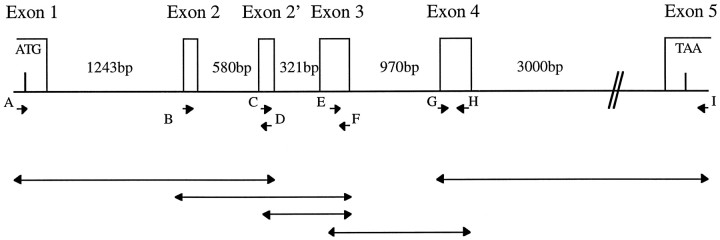
PCR amplification of genomic DNA. PCR amplification of genomic rat liver DNA (Clontech, Cambridge, UK) was performed in 0.2 mm dNTPs, 50 ng/μl template DNA, each primer at 0.4 μm, KlenTaq polymerase (Clontech), 40 mmTricine-KOH, 15 mm KOAc, 3.5 mmMg(OAc)2, and 75 μg/ml bovine serum albumin, using a DNA Engine PCR machine (MJ Research, Watertown, MA). The cycling conditions varied, depending on the primers used. Deprotected/dephosphorylated primers, obtained from Life Technologies (Gaithersburg, MD), were as follows: A, 5′-GATAAACACTCCTTGTGACTTTTGG-3′; B, 5′-CGCAGCCTATAAGGAGAAGATGAAG-3′; C, 5′-GGTGTGGGAGACAGTGTAGGAGGAA-3′; D, 5′-CTTCTTTCTCTCCAGTCAGCACTGC-3′; E, 5′-GCCACCTTCCTTTGACGGGGTGCC-3′; F, 5′-AACTCAGGCACCCCGTCAAAGGAAG-3′; G, 5′-GCCCATCTGGCTGCCATGTTGGAGC-3′; and H, 5′-CCGCTCCAACATGGCAGCCAGATGG-3′ 5′-TCACAGGTCAATCTTTTACATGTAC-3′. The positions of these primers are shown in Figure 1. For sequencing, PCR fragments were subcloned into the TA vector (using the TA cloning kit from Invitrogen, San Diego, CA) and then sequenced using the universal M13 forward and reverse primers.
Fig. 1.
The intron and exon structure of RB3 genomic DNA, determined by PCR amplification. Rat genomic DNA was amplified using the primers shown (arrows labeled A–H). The resulting PCR products are shown as unlabeled double-headed arrows below the gene structure. The sizes of the introns are indicated. The start (ATG) and stop (TAA) codons are shown in exons 1 and 5, respectively. Exon 2′ is alternatively spliced, being present in RB3" and absent in RB3.
Animals. A single MECS stimulation was induced in adult male Sprague Dawley rats using a constant-current signal generator (ECT unit; Ugo, Basil, Switzerland) as described previously (Cole et al., 1990). For RNA extraction, brains were collected 4 hr after MECS. Forin situ hybridization studies, brains were collected at 30 min and 1, 2, 4, and 8 hr after MECS.
For LTP studies, stimulating and recording electrodes were implanted bilaterally in the perforant path and hilus of the dentate gyrus of Fischer-344 rats, as described previously (Yamagata et al., 1994). After a 2 week recovery time, the rats received a high-frequency stimulation in one hemisphere and low-frequency stimulation in the another hemisphere. The electrical stimuli consisted of 200 msec diphasic, constant-current pulses given at a stimulus intensity of 500 μA. The low-frequency stimulation, which does not induce LTP, was delivered at 0.1 Hz, whereas the LTP-inducing high-frequency stimulation consisted of 50 repetitions of eight pulses delivered at 400 Hz. Brains were collected at 1 and 3 hr after stimulation, to be used for in situ hybridization studies.
For ontogeny studies, control Wistar rats (Charles River Laboratories, Wilmington, MA) of various prenatal and postnatal ages were used.
RNA extraction. Extracted brains and cells collected for RNA extraction were snap-frozen on dry ice. Poly(A+) RNA was then extracted using Invitrogen’s Micro Fast-track kit, according to the manufacturer’s instructions, quantified by UV spectrophotometry (Hewlett Packard, Palo Alto, CA), and used for Northern blot analysis and reverse transcription (RT)-PCR.
Northern blot analysis. Poly(A+) RNA samples (1 μg) were separated on a 1% denaturing gel and transferred to Optitran nitrocellulose (Schleicher & Scheull, Keene, NH). Premade Northern blots containing poly(A+) RNA from multiple rat tissues and human brain regions were obtained from Clontech. The Northern blots were prehybridized for at least 2 hr at 42°C in 50% formamide, 5× Denhardt’s solution, 6× SSC, 0.1% sodium pyrophosphate, 0.1% SDS, and 0.1 mg/ml sonicated and denatured salmon sperm DNA and then were hybridized overnight at 42°C to [32P]dCTP-labeled cDNA probes (∼5 × 106 cpm/ml) in prehybridization solution. Washes were of high stringency, including two 30 min washes in 0.1× SSC/0.1% SDS at 55°C. The blots were exposed to Biomax film (Eastman Kodak, Rochester, NY) for 1–4 d, depending on the intensity of the signal. To detect RB3 we used a 3′ cDNA probe corresponding to bases 877–1280, which should detect all three RB3 gene transcripts–RB3, RB3′, and RB3".
RT-PCR analysis. Poly(A+) RNA derived from various sources was reverse transcribed using 400 units of Superscript RNaseH-Reverse Transcriptase (Life Technologies), 0.5 μg of oligo-dT primer (Pharmacia, Piscataway, NJ), each dNTP at 0.5 mm, 50 mm Tris-HCl, pH 8.3, 75 mmKCl, and 3 mm MgCl2 for 90 min at 42°C. First-strand cDNA was then amplified using PCR primers B and F (see above) to examine alternative splicing of exon 2′. To visualize the RB3 transcripts, we transferred the PCR products to Optitran nitrocellulose using standard Southern blot techniques. The blot was then probed with a [32P]dCTP-labeled cDNA fragment corresponding to bases 877–1280 of the cDNA sequence in the following conditions: 0.2% SDS, 10 mm EDTA, 5× Denhardt’s solution, and 5× SSC at 65°C overnight.
In situ hybridization. To detect RB3 we used two cDNA fragments as probes. The first corresponded to bases 877–1280 and therefore hybridizes to both RB3 and RB3" transcripts (see below), whereas the second consisted of the entire coding region of RB3". To detect SCG10, we used a cDNA fragment corresponding to amino acids 1–70. All cDNAs were subcloned into pBluescript (Stratagene), and the constructs were linearized with the appropriate endonucleases.35S-UTP–labeled sense and antisense RNA probes were generated using the Megascript T3 and T7 kits (Ambion, Austin, TX) according to the manufacturer’s instructions.
Tissues were fresh-frozen in isopentane on dry ice and mounted into molds with freezing agent. Frozen coronal sections (8 μm) were cut from LTP, MECS, and control developing and adult brains on a Leica (Nussloch, Germany) CM3050 cryostat and thaw-mounted onto Fisher brand Superfrost microscope slides (Fisher Scientific, Houston, TX). In addition, sagittal sections were cut from E16 whole embryos. For MECS time course studies, half brains or isolated hippocampi from different time points were mounted in the same block to allow identical hybridization conditions for each section. The sections were post-fixed with 4% paraformaldehyde for 30 min at 4°C, acetylated with 0.0025% acetic anhydride in 0.1 mtriethanolamine, pH 8.0, for 10 min, dehydrated in an ascending ethanol series, and air-dried ready for hybridization. Hybridization took place overnight at 55°C, in a humidified chamber, in 70 μl of hybridization buffer consisting of 50% deionized formamide, 200 mm NaCl, 0.2% Ficoll, 0.2% polyvinylpyrrolidone, 0.2% bovine serum albumin, 10 mm Tris, pH 8, 1 mmEDTA, and 107 cpm/ml radiolabeled probe. Negative controls consisted of hybridizing adjacent sections with radiolabeled sense strand and pretreatment of others with RNase before hybridization with the antisense strand. Positive signal was detected using a Cyclone phosphorimager (Packard, Meridian, CT).
Generation of a RB3 peptide antibody. An antibody, raised against the peptide RRKGQSRKGSADWR (which is encoded by exon 2′ and is thus present only in RB3"), was generated by Genosys (The Woodlands, Texas). The peptide was conjugated to keyhole limpet hemocyanin and injected into New Zealand White rabbits. The highest titer of antibody (corresponding to the third bleed) was then used for immunoprecipitation and immunocytochemistry. A similarly produced antibody raised against the C-terminal sequence EEVRKNKELKEEASR failed to detect either RB3 or RB3" (data not shown).
Cell culture. Cell culture media were purchased from Life Technologies. COS7 and PC12 cells were obtained from American Type Culture Collection (Rockville, MD) and maintained at 37°C in humidified conditions with 5% CO2. PC12 cells were grown in DMEM (high glucose) with 10% fetal bovine serum and 5% horse serum, and COS7 cells were grown in DMEM F-12 with 10% fetal bovine serum. For NGF treatment studies, PC12 cells were treated once with 250 ng/ml NGF 2.5S (Boehringer Mannheim, Indianapolis, IL) in 1% horse serum and maintained in this medium until collected for RNA extraction (at 2, 4, 8, 24, 72, and 120 hr after treatment).
Transfection of COS7 cells. For expression of RB3 in mammalian cells, cDNAs corresponding to the coding sequences of both RB3 and RB3" were generated from brain RNA using RT-PCR. A Flag tag (encoding the peptide N-Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys-C; Eastman Kodak) was attached to the C terminal of RB3 using PCR to allow detection of the protein. The cDNA fragments were then subcloned into the EcoRI/HindIII sites of a vector containing a cytomegalovirus promoter and enhancer (Gorman et al., 1989). RB3" and RB3Flag cDNAs (10 μg) were transiently transfected into COS7 cells (grown to 50–80% confluence) using lipofectamine (Life Technologies) according to the manufacturer’s instructions.
Metabolic labeling and immunoprecipitation. Metabolic labeling was performed in COS7 cells (∼48 hr after transfection) or in PC12 cells with or without NGF treatment (250 ng/ml). Cells were starved of cysteine and methionine for 60 min before labeling. [35S]Methionine and [35S]cysteine (0.1 mCi of each) were added and allowed to incorporate for 4 hr. Cell lysates (1 ml) were collected, precleared with protein A–Sepharose (Pharmacia), and immunoprecipitated with either the rabbit peptide antibody (to detect RB3") or a Sepharose-conjugated monoclonal anti-Flag M2 antibody (Eastman Kodak) (to detect RB3Flag) at 4°C overnight. The RB3" protein–antibody complex was incubated with protein A–Sepharose for 60 min at 4°C. The protein A pellets of both samples were then washed extensively, and the protein–antibody complexes were released by incubation at 95°C for 5 min and electophoresed on a 14% Tris-glycine gel (Novex, San Diego, CA). The gel was dried, and the bands were visualized using the phosphorimager.
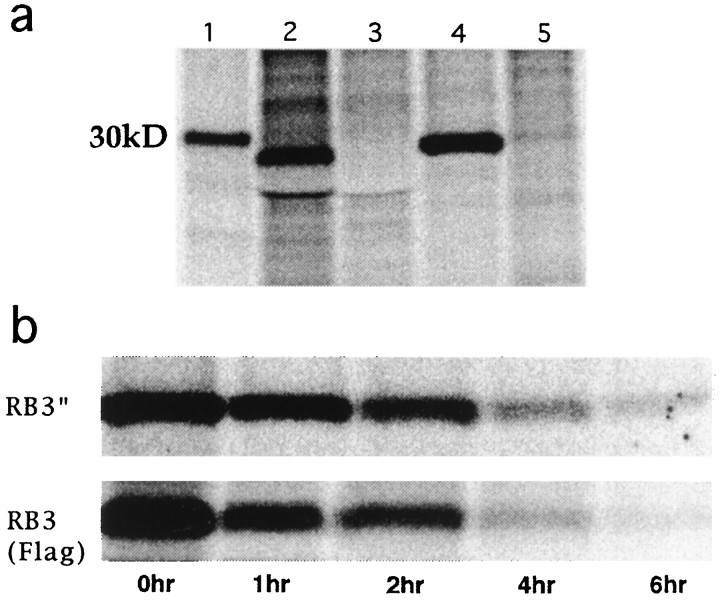
Pulse-chase experiments were performed using COS7 cells transiently transfected with RB3" or RB3Flag. Twenty-four hours after transfection the cells were starved of methionine for 30 min. The cells were then incubated for 10 min with 100 μCi/ml [35S]methionine, washed extensively with cold PBS, and incubated in DMEM with 1 mg/ml nonradioactive methionine until they were lysed at the following times: 0, 1, 2, 4, and 6 hr after labeling. Immunoprecipitation was then performed on the lysates, as described above. The intensities of the bands were quantified using the IP Lab Spectrum program (Signal Analytics, Vienna, VA), and the half-lives were calculated.
Immunocytochemistry. Cells were grown on two-well chamber slides (Nunc, Naperville, IL) for immunocytochemistry. COS7 cells were transfected with RB3" or RB3Flag 24 hr before immunocytochemistry. Immunocytochemistry was performed on PC12 cells 1, 3, and 5 d after NGF treatment. The cells were washed with PBS and fixed in methanol for 5 min at −20°C. The cells were then permeabilized in PBS containing 1% Triton X-100 and 150 mm sucrose for 15 min. After the blocking of nonspecific binding with 10% BSA in PBS for 60 min, the cells were incubated with primary antibody at the appropriate dilution in PBS, 1% BSA, and 0.1% Triton X-100 for 60 min at room temperature. After thorough washing, the cells were incubated in fluorescently labeled secondary antibody diluted at 1:500 for 30 min, mounted in Vectashield containing 4,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA), and viewed with a fluorescent microscope (Nikon, Melville, NY). Photomicrographs were taken using a 40× oil objective. To detect transfected RB3", we used the peptide antibody described above at 1:500 dilution. To detect RB3Flag, we used a rabbit polyclonal anti-Flag M2 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) at 1:500 dilution. A monoclonal antibody to 58K (Sigma, St. Louis, MO) was used at 1:50 dilution as a marker for the Golgi apparatus. Secondary antibodies were a Texas Red-conjugated goat anti-mouse antibody (Cappel, Durham, NC) and a FITC-conjugated sheep anti-rabbit antibody (Jackson ImmunoResearch, West Grove, PA). Both were used at a dilution of 1:500. As a negative control for the RB3" peptide antibody, preimmune serum was used in place of the primary antibody.
RESULTS
cDNA sequence
A partial novel cDNA clone (1019 bp) identified by its differential nature was isolated from a subtracted cDNA library. Using this to screen the LTP cDNA library using filter hybridization led to the isolation of seven positive clones. The three longest clones (∼1.3 kb) were isolated and sequenced, producing a 1280 bp consensus cDNA sequence. During the preparation of this paper, the sequences for RB3, RB3′, and RB3" were deposited in the GenBank database and published (Ozon et al., 1997, 1998). Thus we have adopted the terminology used by Ozon et al. (1997) to avoid confusion. When compared with the GenBank database, our gene was found to be almost identical to the recently cloned RB3" (accession number AF02630). The only difference is in the region immediately before the stop codon, with the deposited sequence having an additional 13 bases. However, we noticed a discrepancy between the sequence deposited in GenBank and the one reported by Ozon et al. (1997). We found that our cDNA sequence (and also the genomic sequence, see below) is identical to the published sequence (Fig. 2). We conclude that an error has been introduced into the deposited GenBank sequence and that the published sequences are correct.
Fig. 2.
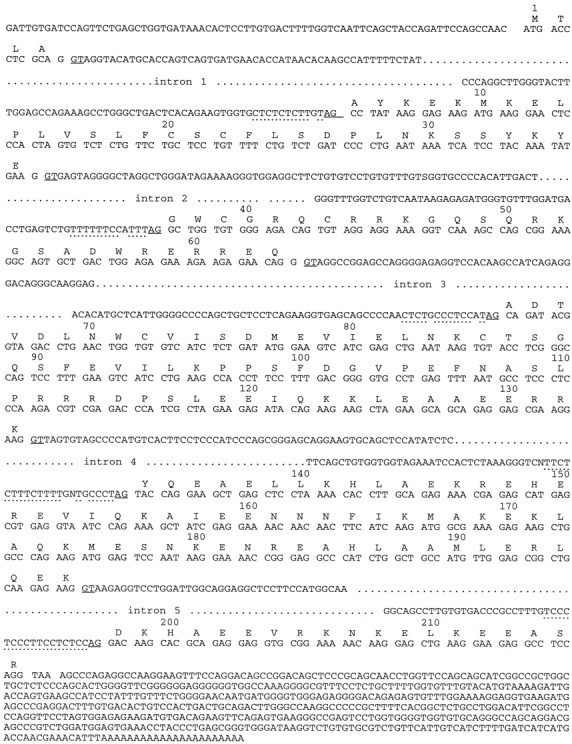
Genomic DNA sequence of RB3 showing the exons and the exon–intron junction sites. The deduced amino acid sequence of RB3 is shown above the corresponding nucleotide sequence. Splicing donor (GT) and acceptor (AG) sequences are underlined, and pyrimidine-rich regions preceding splicing acceptor sites areunderlined with dots.
Intron and exon structure
To study the gene structure of RB3, we used PCR primers (25mers) to amplify fragments generated from a rat liver genomic library. Overlapping fragments amplified using primer pairs A and D, B and F, C and F, E and H, and G and I (see Materials and Methods; Fig. 1) were sequenced, and the intron and exon boundaries were identified by consensus donor and acceptor sequences and alignment with the cDNA sequence (Fig. 2). The location of the primers and the fragments generated are indicated in Figure 1. We identified six exons separated by five introns, ranging in size from 320 bp to ∼3 kb (Fig. 1). The introns were fully sequenced with the exception of the long intron 5, although only the intron–exon junction sites are shown in Figure 2. The size of the genomic DNA is ∼7 kb, corresponding to the size of the stathmin genomic sequence (6 kb) but quite different from SCG10 (∼35 kb) (Okazaki et al., 1993). However, because we have not amplified the extreme 5′ and 3′ ends of the RB3 gene, the entire length of RB3 cannot be determined accurately.
Comparison of intron and exon boundaries of RB3, SCG10, and stathmin genes revealed a very similar primary structure, in that the exon boundaries between amino acids are equivalent. Exon 3 of RB3 contains an additional 24 bp sequence at its 5′ end that is absent in the equivalent exon of stathmin and SCG10 (Okazaki et al., 1993). Interestingly, the RB3 gene contains an additional 81 bp exon, exon 2′ (see Figs. 1, 2), that encodes the novel 27 amino acids found in RB3" but not in stathmin, SCG10, SCLIP, or the Xenopushomolog XB3 (Maucuer et al., 1993; Okazaki et al., 1993).
After the publication of the RB3 family sequences by Ozon et al. (1997), we re-examined our genomic sequence and identified exon 4′, which is ∼680 bp downstream of exon 4 (data not shown). Expression of this exon leads to the production of RB3′, a truncated transcript of RB3 (Ozon et al., 1997).
Alternative splicing of exon 2′
To study potential alternative splicing of exon 2′, we examined mRNA transcripts derived from adult rat brain using RT-PCR with primers B and F, which are situated in exons 2 and 3, respectively (see Fig.1). Two bands, of 308 and 227 bp, were amplified. Sequencing of these PCR products showed that they corresponded to two RB3 gene transcripts, with one transcript containing exon 2′ and the other lacking exon 2′, corresponding to RB3" and RB3, respectively. We then examined the expression of transcripts RB3" and RB3 in various paradigms (see below for full description): in the whole brain during development [from embryonic day 12 (E12), E16, E18, and postnatal day 1 (P1) and in the adult], in isolated hippocampi from MECS-stimulated and unstimulated animals, and in PC12 cells with and without treatment with 250 ng/μl NGF. These results demonstrated that in tissues and cells in which RB3 gene transcripts were detectable, both forms were present in approximately the same ratio (see Fig. 7). Thus, the regulation of exon 2′ alternative splicing seems to be independent of the developmental or neuronal activation state of the brain.
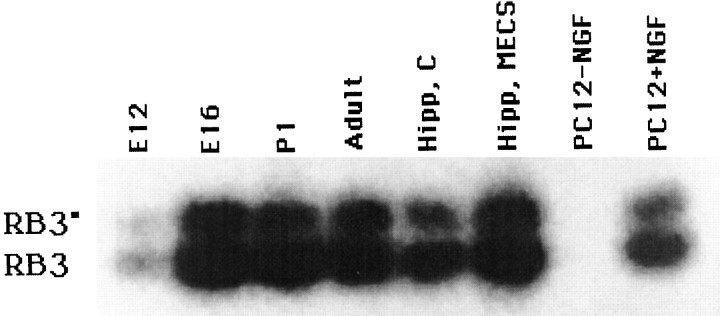
Fig. 7.
Alternative splicing of RB3. RNA was extracted from the developing brain at E12, E16, and P1, from the adult brain, from the hippocampus of control (Hipp,C) and MECS-stimulated (Hipp,MECS) brains, and from PC12 cells with (PC12 + NGF) and without (PC12 − NGF) NGF treatment. RB3" and RB3 transcripts were amplified using RT-PCR with primers B and F situated on either side of exon 2′ (see Fig. 3).
It must be noted that as these experiments were performed before the publication of the paper by Ozon et al. (1997), the primers we used were designed to detect the presence or absence of exon 2′ only and did not distinguish differences in the splicing of exon 4′. Thus the band we identified as RB3 may in fact be a combination of RB3 and RB3′. RB3′ encodes a truncated protein because of the alternative splicing of exon 4′ (Ozon et al., 1997) but does not differ from RB3 in the region being examined.
Expression patterns of RB3 mRNA
Northern blot analysis showed a 1.4 kb band present in the rat brain but not in the rat kidney, skeletal muscle, liver, spleen, lung, or heart (Fig. 3a). A very faint band could also be detected in testes with exposure times of over 3 weeks (data not shown). Northern blot analysis of RNA from multiple regions of the human brain showed that RB3 was present at similar levels in the substantia nigra, the hippocampus, the corpus callosum, the caudate nucleus, and the amygdala, with lower levels present in the thalamus and subthalamic nuclei (Fig. 3b).
Fig. 3.
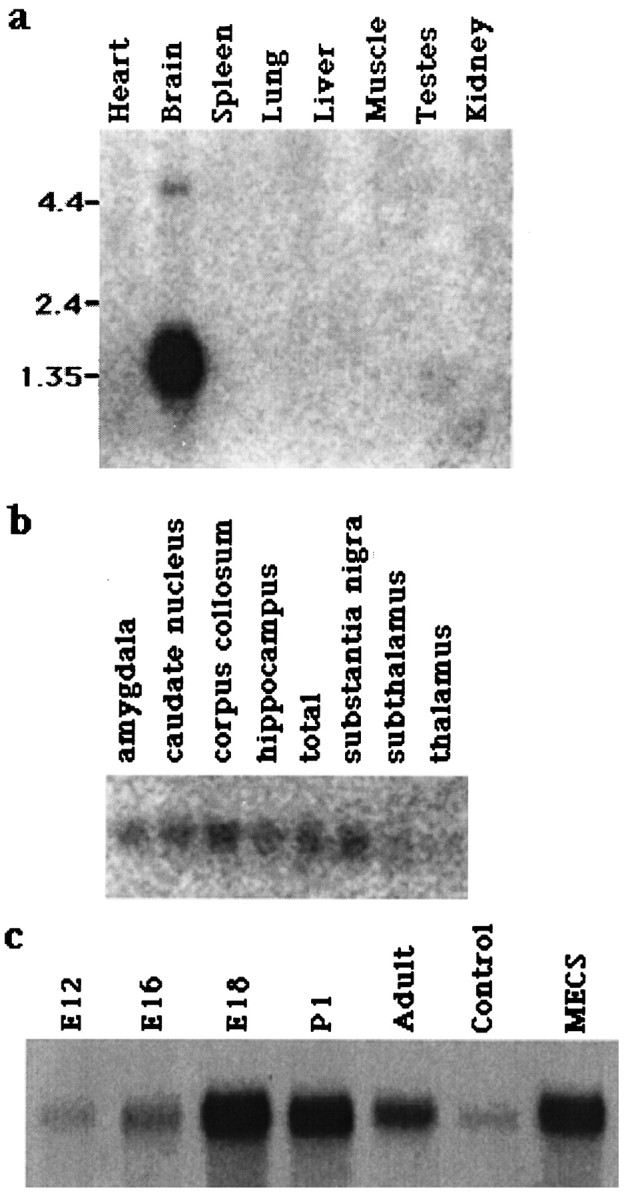
Northern analysis of RB3 mRNA expression in the rat. a, A rat multiple-tissue Northern blot showing the expression of RB3 mRNA in the heart, brain, spleen, lung, liver, skeletal muscle, kidney, and testis. b, Northern blot showing the expression of RB3 mRNA in different regions of the human brain: amygdala, caudate nucleus, corpus callosum, hippocampus, total brain, substantia nigra, subthalamic nucleus, and thalamus.c, Northern blot showing the expression of RB3 mRNA in the rat brain at E12, E16, E18, and P1, in the adult brain, and in the hippocampus of control and MECS-stimulated brains.
Northern blot analysis of RNA isolated at different developmental stages demonstrated that RB3 mRNA was detected in the rat brain at E12, E16, E18, and P1 and in the adult, although the levels at E12 were significantly lower (Fig. 3c). This result was confirmed by an independent experiment using RT-PCR (see Fig. 7). The highest levels of RB3 mRNA were detected in the E18 and P1 brains.
In situ hybridization confirmed that RB3 mRNA was expressed at low levels throughout the adult brain, with little regional specificity (Fig. 4). Both probes, one consisting of 3′ sequences and the other consisting of the entire coding sequence, gave the same results. In the hippocampus, RB3 mRNA was expressed predominantly in the dentate gyrus granule layer and the CA1–CA4 pyramidal layer, although some cells outside these layers were also labeled. RB3 mRNA appeared to be expressed at lower levels than SCG10 mRNA. As shown previously (Himi et al., 1994), the expression of SCG10 mRNA showed regional specificity, with high levels present in a number of regions, including the cortex, amygdala, hippocampus, thalamus, and hypothalamus. Although both genes were expressed in the hippocampus, the expression patterns differed. RB3 mRNA was expressed at low levels uniformly throughout the CA1–CA4 pyramidal layer and at slightly higher levels in the dentate granule layer; SCG10 mRNA was expressed strongly in CA3/CA4 pyramidal cells and less strongly in the CA1 region and dentate gyrus. Indeed, the distribution of RB3 mRNA resembled that of stathmin, which is expressed throughout the brain (Himi et al., 1994), although some differences exist. For example, the expression of stathmin in the dentate gyrus is predominantly in cells of the innermost layers (Himi et al., 1994), unlike RB3 that is more evenly distributed throughout the entire granule layer.
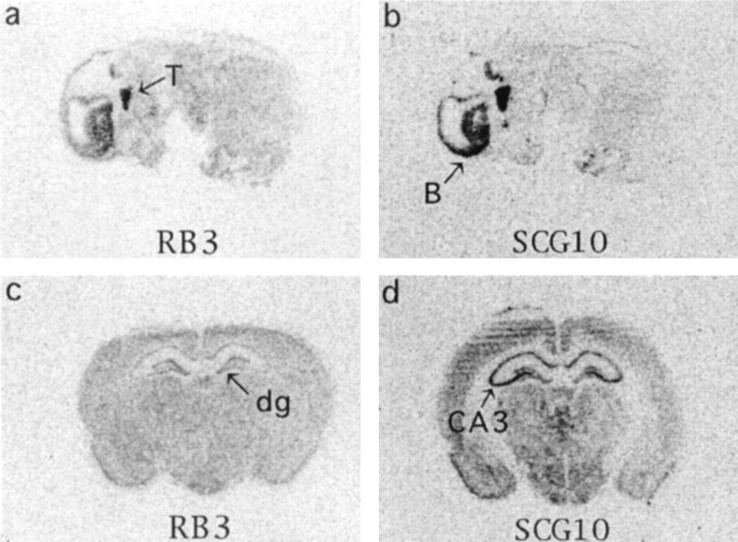
Fig. 4.
In situ hybridization showing the expression of basal RB3 and SCG10 mRNA in the rat. Expression of RB3 (a, c) and SCG10 (b,d) mRNA in sagittal sections of the whole E16 embryo (a, b) and in coronal sections of the adult brain (c, d). In aand b, the trigeminal nucleus (T) and brain (B) are indicated. In c and d, the dentate gyrus (dg) and CA3 regions of the hippocampus are marked.
In situ hybridization of coronal sections from rat brains collected at E16, E18, and P1 demonstrated that RB3 mRNA was expressed throughout the brain during development. Sagittal sections of whole E16 embryos showed that in addition to being expressed in the brain, RB3 mRNA was also expressed in the trigeminal nucleus (Fig. 4), similar to SCG10 mRNA (Himi et al., 1994). A faint signal in white matter (e.g., the corpus callosum) suggests that unlike SCG10 mRNA (Himi et al., 1994), RB3 mRNA is expressed not only in neurons but also at low levels in non-neuronal cells.
Induction of RB3 mRNA by neuronal activity
We investigated the expression of RB3 mRNA in two in vivo models of neuronal activation, MECS and LTP, by in situ hybridization and Northern blot analysis. For comparison,in situ hybridization was also used to analyze the expression of SCG10 mRNA in adjacent sections. Northern blot analysis showed a significant upregulation of RB3 mRNA in the hippocampus at 4 hr after MECS (Fig. 3c). In situ hybridization results confirmed the Northern blot results and further localized the induction of RB3 mRNA to the granule layer of the dentate gyrus. An examination of the expression in hippocampi isolated from brains at various times after MECS revealed that RB3 mRNA induction could be detected between 30 min and 1 hr, peaked at 4 hr, and was still elevated at 8 hr (Fig. 5a). Hybridization of the probe to MECS-stimulated half-brains showed that the induction was restricted to the dentate gyrus, with expression in other areas of the hippocampus and brain remaining unchanged (Fig.5c). Importantly, induction in the dentate gyrus after MECS stimulation was unique to RB3, with SCG10 mRNA remaining at basal levels (Fig. 5b,d).
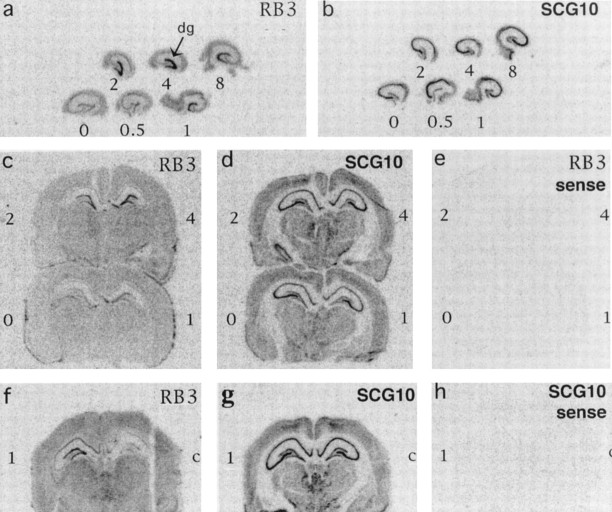
Fig. 5.
In situ hybridization showing the induction of RB3 mRNA (a, c,f) but not SCG10 mRNA (b,d, g) in the rat brain after MECS and LTP. a, b, Hippocampi isolated from brains collected at 0, 0.5, 1, 2, 4, and 8 hr after MECS stimulation. The dentate gyrus (dg) is indicated.c–e, Half-brains isolated at 0, 1, 2, and 4 hr after MECS stimulation. e, Control, probed with a RB3 sense probe. f–h, Brains collected 1 hr after unilateral LTP stimulus (high frequency) to the left hippocampus (1). The right hippocampus (control, c) received low-frequency stimulus that did not induce LTP.h, Control, probed with SCG10 sense probe.
RB3 mRNA was also induced after electrical stimulation leading to LTP, as shown by in situ hybridization (Fig. 5). Induction was seen in brains collected at 1 hr and 3 hr after LTP and was again restricted to the dentate gyrus, with other areas remaining unchanged. In contrast, the expression of SCG10 mRNA was not altered by LTP (Fig.5).
Induction of RB3 mRNA by NGF in PC12 cells
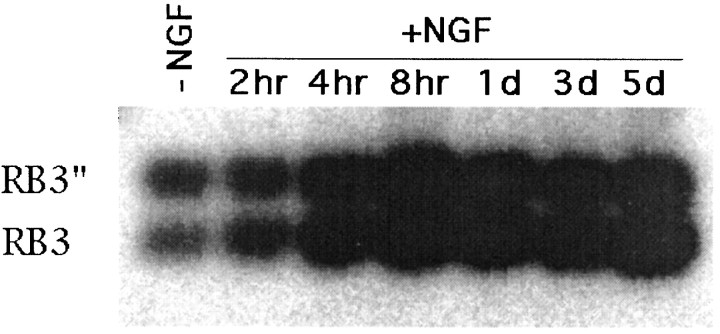
To examine whether RB3 was involved in neuronal differentiation of PC12 cells [as shown previously for SCG10 (Stein et al., 1988b)], we studied, using Northern blot analysis and RT-PCR, the levels of RB3 mRNA in PC12 cells before and after NGF (250 ng/ml) treatment. The levels of RB3 mRNA in both untreated and treated PC12 cells were so low that we could not detect it using Northern blot analysis (data not shown). This differs from the expression of SCG10 mRNA in differentiating PC12 cells, which Stein et al. (1988b) found to be easily detectable by Northern blot analysis. However, RT-PCR demonstrated that very low levels of RB3 and RB3" transcripts were present in nondifferentiated PC12 cells and that NGF treatment led to an increase in the levels of both transcripts (Figs.6,7). This increase was first detected at 4 hr after NGF treatment, before any detectable neurite outgrowth. Although we did not attempt to quantify the degree of induction, it was clear that RB3 mRNA remained elevated until 5 d, at which point the experiment was terminated. Immunocytochemistry and immunoprecipitation using an RB3"-specific peptide antibody failed to detect RB3" protein in PC12 cells (data not shown), suggesting that the protein was present at very low levels in concordance with mRNA levels.
Fig. 6.
Induction of RB3 mRNA in PC12 cells treated with NGF. RT-PCR was performed on RNA extracted from nontreated PC12 cells (−NGF) and from PC12 cells collected 2, 4, and 8 hr and 1, 3, and 5 d after the addition of 250 ng/ml NGF (+NGF). PCR primers (B and F) were situated on either side of exon 2′, allowing amplification of both RB3" and RB3 transcripts. The resulting PCR products were transferred to nitrocellulose and probed with a RB3 cDNA fragment common to both RB3" and RB3.
Expression of RB3 protein in COS7 cells
We attempted to express RB3 and RB3" in PC12 cells to test the hypothesis that they could effect neurite outgrowth and to compare their distribution with that of SCG10. Unfortunately, we failed to obtain stably or transiently transfected PC12 cells (data not shown). We speculate that this may be caused by a lethal effect of overexpressing a protein normally present in very low abundance. As an alternative, we then expressed RB3" and RB3 in COS7 cells. Although these cells are non-neuronal and less physiologically relevant, they can still provide useful information regarding protein localization and provide a model system with which to compare SCG10 with RB3. Constructs containing the cDNA encoding both transcripts in the mammalian expression vector pRK5 were generated and transiently transfected into COS7 cells (see Materials and Methods). The peptide antibody designed against the novel exon 2′ sequence was able to detect RB3". However, a peptide antibody raised against the C terminal failed to detect either form. Thus, to be able to detect RB3, we attached a Flag-tag epitope to the C terminal and used monoclonal anti-Flag antibodies for immunoprecipitation and immunocytochemistry. Metabolic labeling with [35S]methionine and immunoprecipitation using the exon 2′-specific and anti-Flag antibodies showed that both the RB3" and RB3Flag proteins were expressed localized to the intracellular fraction of transiently transfected COS7 cells. Single bands at ∼29 and 28 kDa were detected in RB3"- and RB3Flag-transfected cells, respectively; whereas no signal was detected in control untransfected cells (Fig.8a). Because the Flag tag itself is ∼1 kDa in size, we speculate that the size of the untagged RB3 protein is ∼27 kDa. Using pulse-chase followed by immunoprecipitation, we calculated the half-lives of RB3" and RB3Flag in transfected COS7 cells to be 55 and 45 min, respectively (Fig.8b).
Fig. 8.
Expression of RB3" and RB3Flag proteins in transiently transfected COS7 cells. a, Immunoprecipitation of metabolically labeled RB3" and RB3Flag 24 hr after transfection into COS7 cells. Lane 1, 30 kDa marker protein. Lanes 2, 3, RB3"-transfected (lane 2) and control (lane 3) COS7 cells immunoprecipitated with a polyclonal RB3"-specific peptide antibody. Lanes 4,5, RB3Flag-transfected (lane 4) and control (lane 5) COS7 cells immunoprecipitated with a monoclonal anti-Flag antibody. b, Pulse-chase analysis of RB3" (top) and RB3Flag (bottom). Transfected cells were pulsed-labeled with [35S]methionine for 10 min and chased with excess unlabeled methionine. The cells were harvested at 0, 1, 2, 4, and 6 hr and were immunoprecipitated using the above antibodies.
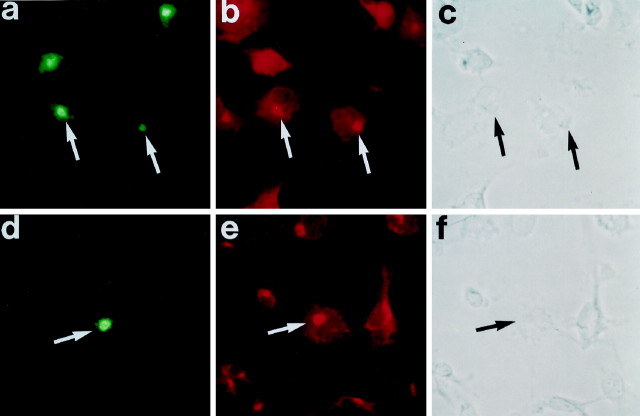
The localization of RB3" and RB3Flag was examined in transfected COS7 cells using immunocytochemistry. Both forms of the protein were localized to the perinuclear region of the cell in a punctate manner. Using double immunocytochemistry with the Golgi-specific protein 58k, we demonstrated that both RB3" and RB3Flag were localized to the area of the Golgi complex (Fig. 9). Additionally, in cells containing high concentrations of the expressed proteins, we detected both RB3" and RB3Flag immunoreactivity in cell processes (data not shown). No signal was detected in control untransformed COS7 cells.
Fig. 9.
Localization of RB3" and RB3Flag immunoreactivity to Golgi apparatus (indicated by arrows) in transfected COS7 cells. a–c, COS7 cells transfected with RB3" were double-immunostained with antibodies against RB3" (a) and 58K (b).d–f, COS7 cells transfected with RB3Flag were double-immunostained with antibodies against RB3Flag (anti-Flag;d) and 58K (e). Secondary antibodies conjugated to FITC (a, d) and Texas Red (b, f) were used to visualize the primary antibodies. c and fare phase-contrast photomicrographs of the cells in aand d, respectively.
DISCUSSION
Screening of a differential cDNA library containing genes upregulated by electrically induced seizure activity led to the identification and cloning of RB3, a newly identified member of the stathmin family. Here we report further characterization of this differentially expressed gene. Of particular interest is its induction after LTP and MECS in the brain and after NGF-induced neuronal differentiation of PC12 cells.
Analysis of the rat genomic sequence revealed that the primary structure of RB3 has been conserved evolutionarily, with intron and exon boundaries present at positions equivalent to those seen in rat stathmin and SCG10 (Okazaki et al., 1993). However, 81 bp exon 2′ is unique to the RB3 gene and encodes the 27 amino acid stretch found only in the RB3 gene. Analysis of mRNA by RT-PCR demonstrated that this exon was alternatively spliced, producing the transcripts known as RB3" (containing exon 2′) and RB3 (lacking exon 2′). Both transcripts were expressed in the rat brain. In Xenopus only the form lacking the 81 bp sequence has been reported (Maucuer et al., 1993), although the identification of ESTs in GenBank corresponding to both RB3" and RB3, from various human and mouse libraries, suggests that alternative splicing of exon 2′ in the RB3 gene is not restricted to the rat.
The biological relevance of the alternative splicing of exon 2′ remains to be determined. There was no detectable difference in transcriptional regulation of the two forms during brain development, after activity-dependent induction in the brain or in differentiating PC12 cells, suggesting that both are active in similar paradigms. However, the presence of two additional serines, with consensus sites for cGMP-dependent protein kinase and PKC (Pearson and Kemp, 1991) in RB3", suggests there may be important differences in the substrate specificity of this form. The novel 27 amino acid sequence could also potentially affect the cellular localization or half-life of RB3", thereby altering its function.
Transient expression of cDNA corresponding to RB3" and the Flag-tagged form of RB3 (RB3Flag) in COS7 cells produced proteins of ∼29 and 28 kDa, respectively. These were somewhat larger than the predicted sizes of 24 and 22 kDa (including the 1 kDa Flag tag of RB3Flag) and may be explained by the presence of various post-translational modifications, such as palmitoylation or phosphorylation, that are known to occur in SCG10 (Antonsson et al., 1997; Di Paolo et al., 1997). Pulse-chase analysis showed that RB3" and RB3Flag have half-lives of ∼55 and 45 min, respectively. It is not clear whether the relatively small difference in the half-life is physiologically significant. It is possible that it is caused by the presence of the epitope tag on RB3Flag. Indeed, because these proteins are restricted to the brain, their half-lives and the extent to which they are post-translationally modified may differ in vivo.
Like SCG10, RB3 mRNA expression is restricted to neural tissue. Northern blot analysis showed that the expression of RB3 mRNA is developmentally regulated, with the highest levels present at E18 and P1. This is similar to the expression of SCG10 and stathmin, whose RNA levels peak during embryonic development and are decreased in the adult rat brain (Anderson and Axel, 1985). However, as demonstrated byin situ hybridization, the distribution of RB3 mRNA differed from that of SCG10 in that RB3 was expressed at lower levels and more uniformly throughout the adult rat brain. Thus, although the actions of SCG10 may be restricted to distinct neuronal populations, RB3 may have a more widespread role in the adult brain.
We demonstrated that RB3 mRNA was induced between 30 min and 1 hr in dentate granule neurons in the rat brain after both MECS and LTP and remained elevated for at least 8 and 3 hr, respectively. This suggests that RB3 may play a role in the synaptic plasticity resulting from the activation of specific neuronal pathways evoked by these paradigms. RB3 mRNA was also upregulated in undifferentiated PC12 cells after 4 hr of NGF treatment and remained elevated for at least 5 d, suggesting that it is also involved in the cellular mechanisms underlying phenotypic differentiation. In a similar experiment, Ozon et al. (1997)reported that RB3 was not upregulated after NGF treatment, as demonstrated by Northern blot analysis. The differences between our results and theirs are likely to be attributable to the fact that RB3 mRNA could only be detected in PC12 cells by the more sensitive RT-PCR analysis. It has been suggested previously that SCG10 plays a role in neuronal plasticity in the adult rat brain (Mori, 1993). However, although SCG10 mRNA is upregulated in the hippocampus after entorhinal cortex lesions (Zarow and Finch, 1995), our results show that the rapid upregulation after neuronal activation in the form of MECS or LTP was specific to RB3. Thus, transcriptional regulation differs significantly between the two genes. On the other hand, RB3 and SCG10 mRNAs are induced similarly after NGF treatment of PC12 cells, although SCG10 is present at much higher levels (Stein et al., 1988b). A balance between the level and distribution of RB3 and those of related family members may have functional importance.
The function of RB3 induced in activated or differentiating neurons is not known. Because of the strong sequence similarity of RB3 with stathmin and SCG10, it is tempting to speculate that RB3 may act by contributing to the control of microtubule dynamics. Although experimental evidence is lacking, the strong sequence similarity of RB3 with the coiled-coil protein-interacting domain of stathmin and SCG10 suggests that RB3 may bind to tubulin in a similar manner, thereby promoting the disassembly of microtubules. In addition, the presence of an N terminal similar to that of SCG10 suggests that RB3 may be similarly localized, i.e., in the Golgi apparatus (Antonsson et al., 1997) and growth cones (Stein et al., 1988a). Changes in microtubule dynamics in neurons are a vital component of structural alterations that occur during both neuronal differentiation and synaptic plasticity (Ratushnyak et al., 1997). One would thus expect such alterations to occur in the brain after LTP and MECS and in differentiating PC12 cells. If RB3 does indeed play a microtubule-destabilizing role in these neurons, it could, along with microtubule-associated proteins that exhibit microtubule-stabilizing properties, be a potentially important component of the active growth cone, in which microtubules exist in a highly dynamic state (Tanaka et al., 1995).
To test the hypothesis that RB3 is localized intracellularly in a manner similar to that of SCG10, we used immunocytochemistry to determine the location of the endogenous protein in the more relevant neuronal system PC12 cells. Unfortunately, RB3 immunoreactivity was undetectable in NGF-treated PC12 cells. The failure to detect RB3 protein in PC12 cells is consistent with the inability to detect RNA using Northern blot analysis and suggests that the protein is present at low levels. However, we did find that like SCG10, both RB3" and RB3Flag were localized to the Golgi in transfected COS7 cells, as demonstrated by their colocalization with a Golgi marker. Although the expression and localization of RB3 and RB3" in COS cells cannot necessarily be extrapolated to neurons, the similarity between RB3 and SCG10 expression patterns suggests that the intracellular localization of the two is similar. Our hypothesis that RB3 may enhance neurite outgrowth when overexpressed in differentiating PC12 cells, as reported for SCG10 (Riederer et al., 1997), could not be tested, because initial attempts to produce stably transfected PC12 cells failed. Answering these questions may require inducible expression of RB3 in future transfection studies to prevent potentially lethal effects that may result from the stable expression of RB3. Localization of RB3 to the Golgi suggests that RB3 may also function in the regulation of protein transport in these neurons undergoing activity-induced plasticity. Of course, the exact localization of RB3 within neurons remains to be determined, as does its ability to influence microtubule stability.
In conclusion, we have shown that RB3, a member of the stathmin family, is induced by neuronal activity in vivo and in differentiating PC12 cells in vitro. This induction is specific to RB3 and is not seen with the closely related SCG10, suggesting different roles for these two genes. Induction of RB3 mRNA after seizure-induced neuronal activation and LTP suggests that it may be an important component of the complex activity-induced mechanisms that result in neuronal plasticity in the brain.
Footnotes
P.F.W. was supported by NIH Grants KO2, MHO1152, and AGO9219. We thank Shujun Luo for excellent technical assistance and Carol Barnes for supplying LTP tissue.
Correspondence should be addressed to Dr. Erica Beilharz, Lynx Therapeutics, 3832 Bay Center Place, Hayward, CA 94545.
REFERENCES
- 1.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Anderson DJ, Axel R. Molecular probes for the development and plasticity of neural crest derivatives. Cell. 1985;42:649–662. doi: 10.1016/0092-8674(85)90122-9. [DOI] [PubMed] [Google Scholar]
- 3.Antonsson B, Lutjens R, Di Paolo G, Kassel D, Allet B, Bernard A, Catsicas S, Grenningloh G. Purification, characterization, and in vitro phosphorylation of the neuron-specific membrane-associated protein SCG10. Protein Expr Purif. 1997;9:363–371. doi: 10.1006/prep.1996.0710. [DOI] [PubMed] [Google Scholar]
- 4.Brakeman PR, Lanahan AA, O’Brien R, Roche K, Barnes CA, Huganir RL, Worley PF. Homer: a protein that selectively binds metabotropic glutamate receptors. Nature. 1997;386:284–288. doi: 10.1038/386284a0. [DOI] [PubMed] [Google Scholar]
- 5.Cole AJ, Abu-Shakra S, Saffen DW, Baraban JM, Worley PF. Rapid rise in transcription factor mRNAs in rat brain after electroshock-induced seizures. J Neurochem. 1990;55:1920–1927. doi: 10.1111/j.1471-4159.1990.tb05777.x. [DOI] [PubMed] [Google Scholar]
- 6.Di Paolo G, Lutjens R, Pellier V, Stimpson SA, Beuchat MH, Catsicas S, Grenningloh G. Targeting of SCG10 to the area of the Golgi complex is mediated by its NH2-terminal region. J Biol Chem. 1997;272:5175–5182. doi: 10.1074/jbc.272.8.5175. [DOI] [PubMed] [Google Scholar]
- 7.Gorman CM, Gies D, McCray G, Huang M. The human cytomegalovirus major immediate early promoter can be trans-activated by adenovirus early proteins. Virology. 1989;171:377–385. doi: 10.1016/0042-6822(89)90605-3. [DOI] [PubMed] [Google Scholar]
- 8.Himi T, Okazaki T, Wang H, McNeill TH, Mori N. Differential localization of SCG10 and p19/stathmin messenger RNAs in adult rat brain indicates distinct roles for these growth-associated proteins. Neuroscience. 1994;60:907–926. doi: 10.1016/0306-4522(94)90271-2. [DOI] [PubMed] [Google Scholar]
- 9.Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- 10.Maucuer A, Moreau J, Mechali M, Sobel A. Stathmin gene family: phylogenetic conservation and developmental regulation in Xenopus. J Biol Chem. 1993;268:16420–16429. [PubMed] [Google Scholar]
- 11.Mori N. Toward understanding of the molecular basis of loss of neuronal plasticity in ageing. Age Ageing. 1993;22:S5–S18. [PubMed] [Google Scholar]
- 12.Okazaki T, Yoshida BN, Avraham KB, Wang H, Wuenschell CW, Jenkins NA, Copeland NG, Anderson DJ, Mori N (1993) Molecular diversity of the SCG10/stathmin gene family in the mouse. Genomics [Erratum (1994) 21:298] 18:360–373. [DOI] [PubMed]
- 13.Ozon S, Maucuer A, Sobel A. The stathmin family – molecular and biological characterization of novel mammalian proteins expressed in the nervous system. Eur J Biochem. 1997;248:794–806. doi: 10.1111/j.1432-1033.1997.t01-2-00794.x. [DOI] [PubMed] [Google Scholar]
- 14.Ozon S, Byk T, Sobel A. SCLIP: a novel SCG10-like protein of the stathmin family expressed in the nervous system. J Neurochem. 1998;70:2386–2396. doi: 10.1046/j.1471-4159.1998.70062386.x. [DOI] [PubMed] [Google Scholar]
- 15.Pearson RB, Kemp BE. Protein kinase phosphorylation site sequences and consensus specificity motifs: tabulations. Methods Enzymol. 1991;200:62–81. doi: 10.1016/0076-6879(91)00127-i. [DOI] [PubMed] [Google Scholar]
- 16.Ratushnyak AS, Zapara TA, Zharkikh AA, Ratushnyak OA. Effects of changes in dynamic equilibrium in microtubule and microfilament systems on the plastic responses of neurons. Neurosci Behav Physiol. 1997;27:353–359. doi: 10.1007/BF02462935. [DOI] [PubMed] [Google Scholar]
- 17.Riederer BM, Pellier V, Antonsson B, Di Paolo G, Stimpson SA, Lutjens R, Catsicas S, Grenningloh G. Regulation of microtubule dynamics by the neuronal growth-associated protein SCG10. Proc Natl Acad Sci USA. 1997;94:741–745. doi: 10.1073/pnas.94.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sobel A. Stathmin: a relay phosphoprotein for multiple signal transduction? Trends Biochem Sci. 1991;16:301–305. doi: 10.1016/0968-0004(91)90123-d. [DOI] [PubMed] [Google Scholar]
- 19.Stein R, Mori N, Matthews K, Lo LC, Anderson DJ. The NGF-inducible SCG10 mRNA encodes a novel membrane-bound protein present in growth cones and abundant in developing neurons. Neuron. 1988a;1:463–476. doi: 10.1016/0896-6273(88)90177-8. [DOI] [PubMed] [Google Scholar]
- 20.Stein R, Orit S, Anderson DJ. The induction of a neural-specific gene, SCG10, by nerve growth factor in PC12 cells is transcriptional, protein synthesis dependent, and glucocorticoid inhibitable. Dev Biol. 1988b;127:316–325. doi: 10.1016/0012-1606(88)90318-1. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka E, Ho T, Kirschner MW. The role of microtubule dynamics in growth cone motility and axonal growth. J Cell Biol. 1995;128:139–155. doi: 10.1083/jcb.128.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsui CC, Copeland NG, Gilbert DJ, Jenkins NA, Barnes C, Worley PF. Narp, a novel member of the pentraxin family, promotes neurite outgrowth and is dynamically regulated by neuronal activity. J Neurosci. 1996;16:2463–2478. doi: 10.1523/JNEUROSCI.16-08-02463.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang JH, Ko GY, Kelly PT. Cellular and molecular bases of memory: synaptic and neuronal plasticity. J Clin Neurophysiol. 1997;14:264–293. doi: 10.1097/00004691-199707000-00002. [DOI] [PubMed] [Google Scholar]
- 24.Yamagata K, Andreasson KI, Kaufmann WE, Barnes CA, Worley PF. Expression of a mitogen-inducible cyclooxygenase in brain neurons: regulation by synaptic activity and glucocorticoids. Neuron. 1993;11:371–386. doi: 10.1016/0896-6273(93)90192-t. [DOI] [PubMed] [Google Scholar]
- 25.Yamagata K, Sanders LK, Kaufmann WE, Yee W, Barnes CA, Nathans D, Worley PF. rheb, a growth factor- and synaptic activity-regulated gene, encodes a novel Ras-related protein. J Biol Chem. 1994;269:16333–16339. [PubMed] [Google Scholar]
- 26.Zarow C, Finch CE. Limited responses of neuronal mRNAs to unilateral lesions of the rat entorhinal cortex. Neurosci Lett. 1995;185:87–90. doi: 10.1016/0304-3940(94)11231-7. [DOI] [PubMed] [Google Scholar]