Abstract
A novel G-protein–coupled receptor (GRL106) resembling neuropeptide Y and tachykinin receptors was cloned from the molluscLymnaea stagnalis. Application of a peptide extract from the Lymnaea brain to Xenopus oocytes expressing GRL106 activated a calcium-dependent chloride channel. Using this response as a bioassay, we purified the ligand for GRL106,Lymnaea cardioexcitatory peptide (LyCEP), an RFamide-type decapeptide (TPHWRPQGRF-NH2) displaying significant similarity to the Achatina cardioexcitatory peptide (ACEP-1) as well as to the recently identified family of mammalian prolactin-releasing peptides. In the Lymnaeabrain, the cells that produce egg-laying hormone are the predominant site of GRL106 gene expression and appear to be innervated by LyCEP-containing fibers. Indeed, LyCEP application transiently hyperpolarizes isolated egg-laying hormone cells. In theLymnaea pericardium, LyCEP-containing fibers end blindly at the pericardial lumen, and the heart is stimulated by LyCEPin vitro. These data confirm that LyCEP is an RFamide ligand for GRL106.
Keywords: neuropeptide receptor, mollusc, HPLC, Xenopusoocyte, RFamide, orphan receptor
Neuropeptides form a structurally diverse class of signaling molecules. Despite their diversity, various families with specific sequence characteristics can be discerned. For instance, many members end in the sequence RFamide. Among these are γ1-MSH, the anti-opioids F-8-Famide (NPFF) and A-18-Famide in vertebrates, and FMRFamide and FMRFamide-related peptides in invertebrates (Yang et al., 1985; Greenberg et al., 1988). These examples represent only a limited selection from a range of naturally occurring RFamides. These peptides display a wide range of peripheral and central actions. To further our understanding of the action of RFamide peptides, it is necessary to study the intracellular signaling pathways that are triggered by these molecules, beginning with the peptide receptors involved.
In Lymnaea stagnalis, a number of RFamide neuropeptides have been characterized, including FMRFamide, FLRFamide, GDPFLRFamide, and SDPFLRFamide (Ebberink et al., 1987; Saunders et al., 1991, 1992). FMRFamide was originally identified by its cardioexcitatory actions (Price and Greenberg, 1977), but this peptide also modulates neuronal activities (e.g., Cottrell et al., 1984). As such L. stagnalis represents an attractive animal model in which to study RFamide peptide signaling. In the search for an FMRFamide receptor, an FMRFamide-gated sodium channel was characterized in neurons of the snail Helix aspersa (Lingueglia et al., 1995). Despite the clear effects of FMRFamide on the snail heart, this FMRFamide-gated channel appeared to be absent from this tissue. Moreover, evidence exists that FMRFamide can also activate a G-protein–coupled receptor (GPCR) (Brezina et al., 1987; Volterra and Siegelbaum, 1988). This receptor has not been identified yet, but it would be very interesting to know its molecular details to develop molecular tools to study its role in FMRFamide signaling.
In the past, we have cloned several GPCRs from the CNS of L. stagnalis, but none of these seems to represent a receptor for an RFamide-type neuropeptide (Tensen et al., 1994a,b; Cox et al., 1997). Here, we report the characterization of a novel GPCR that is expressed in the heart and brain of L. stagnalis as well as the identification of its ligand, designated Lymnaeacardioexcitatory peptide (LyCEP). The latter appears to be an RFamide that is akin to cardioactive peptides from the evolutionary distant snails Achatina fulica and Aplysia californica. Moreover, we have found that the LyCEP receptor is expressed in a group of defined egg-laying hormone–producing neurons that are well suited for studying intracellular signaling by LyCEP at the single-cell level.
MATERIALS AND METHODS
Isolation of a cDNA clone encoding GRL106.Total RNA isolated from Lymnaea hearts was converted into cDNA by the use of oligo-dT and reverse transcriptase as described previously (Tensen et al., 1994a). Two degenerate primers were synthesized on the basis of conserved amino acid sequences within transmembrane regions III and VI of G-protein–coupled neuropeptide receptors (Probst, 1992). The sense primer was 5′-CCGGATCCG(CT)(GC)AT(CT)(GA)(GC)(GC)IT(GT)GAC(CA)G(GC)TA-3′; the antisense primer was 5′-ACGAATTCGG(GC)(CA)ICCA(GA)CAGAI(GC)(GA)(CT)(GA)AA-3′ (BamHI and EcoRI restriction sites in italics). These primers were used in a PCR with heart cDNA using the conditions described earlier (Tensen et al., 1994a), and PCR products of the expected size (400–800 bp) were isolated, cloned, and sequenced (Sambrook et al., 1989). The nucleotide sequence of a PCR product putatively encoding part of a novel GPCR was used to design specific oligonucleotides. The latter were used to isolate the corresponding full-length clone from a Lymnaea CNS cDNA library in λZAP II, using a PCR-based screening strategy (Bloem and Yu, 1990; Gibbons et al., 1991). The cDNA insert was excised in vivo as a pBluescript SK(−) phagemid (designated pBS GRL106) and was sequenced from both strands using the chain termination method (Sanger et al., 1977).
Isolation of a cDNA clone encoding preproLyCEP. A degenerate primer was synthesized, based on the amino acid sequence of LyCEP. This primer was 5′-CCAAGCTTAC(GATC)CC(GATC)CA(TC)TGG(AC)G(GATC)CC(GATC)CA(AG)GG-3′. The oligonucleotide was provided with a HindIII restriction site (in italics) at its 5′ end and was used as a primer in a PCR, with cDNA isolated from a λZAP II cDNA library of the LymnaeaCNS as template. As reverse primer, an oligonucleotide was used complementary to a pBluescript sequence. cDNA was amplified by the use of these two primers for 40 cycles: 94°C for 20 sec, 58°C for 20 sec, and 72°C for 1 min. Amplified cDNA was cloned in M13 after digestion with HindIII and EcoRI, sequenced, and used to screen a cDNA library. We screened 20,000 clones of an amplified library of the CNS of L. stagnalis in λZAP II, using charged Nylon filters (Boehringer Mannheim, Mannheim, Germany) at a density of 1000 pfu/20 cm2 filter. Clones were purified by rescreening at a lower plaque density. Hybridization was performed in 6× SSC (1× SSC, 0.15 m NaCl and 0.015m sodium citrate) for 16 hr at 65°C. Membranes were washed in 1× SSC for 45 min at 65°C and autoradiographed.
Oocyte expression of GRL106 and electrophysiological recordings. The full open reading frame of GRL106 was amplified by 25 cycles of PCR using 100 ng of pBS GRL106 as a template, two specific oligonucleotide primers, and Ultma DNA polymerase under conditions recommended by the manufacturer (Perkin-Elmer, Norwalk, CT). The sense primer was 5′-GTTAAGCTTCCACCATGGCGATGGCGAACAGCGA-3′; the antisense primer was 5′-CATTCTAGATATCAAAGATACATATCGTTTGACAC-3′ (HindIII and XbaI restriction sites in italics). The resultant cDNA was cloned as aHindIII–XbaI fragment into the oocyte expression vector pGEMHE (Liman et al., 1992). The nucleotide sequence of the resultant construct was verified to check for any PCR-generated errors, and the plasmid was used as a template in a PCR with primers flanking the T7 and Sp6 RNA polymerase promoters. The PCR product was gel-purified and used as a template for the synthesis of capped sense cRNA with T7 RNA polymerase in the presence of 2 mmm7G(5′)ppp(5′)G according to the manufacturer’s instructions (New England Biolabs, Beverly, MA). The cRNA was diluted with double-distilled water to 10 ng/μl, and 50 nl was injected into manually defolliculated Xenopus laevisoocytes. After 2–3 d, whole-cell current measurements were performed as described previously (Meyerhof et al., 1988).
Crude peptide extract from Lymnaea brains.Samples of Lymnaea CNS were dissected, collected on dry ice, and stored at −60°C until use. The nervous systems were rapidly thawed at room temperature, sonicated in 1 m acetic acid on ice, and centrifuged at 10,000 × g for 5 min at 4°C, and the resulting supernatant was applied to a C18 solid phase extraction column (Supelclean from Supelco). Bound material was eluted with 60% acetonitrile and 7 mm trifluoroacetic acid (TFA), lyophilized, and dissolved in double-distilled water.
Purification of LyCEP. The crude peptide mixture was size-fractionated by high-performance gel permeation chromatography (HPGPC) on Protein-Pak columns I-125 and I-300 connected in series (Waters Associates, Milford, MA) with 7 mm TFA in 30% acetonitrile as a running buffer and 1 ml fractions collected at a flow rate of 1 ml/min. Fractions were lyophilized, redissolved in double-distilled water, and tested in the Xenopus oocyte assay. The bioactive fraction was then subjected to reversed-phase HPLC (rpHPLC) on a Nucleosil C18 column (Hichrom; 250 × 4.6 mm) by the application of linear gradients of acetonitrile in 7.5 mm TFA (0% acetonitrile for 10 min; then from 0 to 60% acetonitrile over 60 min). Fractions of 300 μl were collected at a flow rate of 300 μl/min. The active fraction was lyophilized, taken up in double-distilled water, and rechromatographed on a similar column (Nucleosil C18; 250 × 2.1 mm) by the application of linear gradients of acetonitrile in 0.05% HCl (from 0 to 10% acetonitrile in 10 min, then from 10 to 25% acetonitrile over 60 min, and finally to 60% acetonitrile in 10 min). At this stage, the bioactive fraction appeared to contain a single protein peak as assessed spectroscopically. It was lyophilized, and the solute was redissolved in double-distilled water.
Amino acid sequence determination, peptide synthesis, and mass determination. Amino acid sequences were determined with an automated sequencer (model 477; Applied Biosystems, Foster City, CA) and an on-line rpHPLC system (model 120A; Applied Biosystems) to detect phenylthiohydantoin amino acids using sequencing programs recommended by the manufacturer.
LyCEP was chemically synthesized on an Applied Biosystems 432A peptide synthesizer according to the manufacturer’s instructions and was HPLC-purified. The precise molecular masses of natural and synthetic LyCEP were obtained using a Quattro-BQ triple quadrupole mass spectrometer (Fisons). The mass spectrometer was equipped with an electron spray atmospheric pressure ionization source. The peptide was dissolved in 20 μl of 50% acetonitrile/1% formic acid, and the resultant solution was injected via a 10 μl loop into the electron spray source. The flow rate was 4 μl/min; the mobile phase was 50% acetonitrile/1% formic acid. Data were collected over a suitable mass range, and several 10 sec continuum scans were accumulated.
Immunohistochemistry and in situ hybridization.Dissected CNS of L. stagnalis was frozen in Freon cooled by liquid nitrogen, lyophilized in a tissue freeze dryer, and then fixed in paraformaldehyde vapor (80°C for 1 hr) and embedded in paraffin. Sections of 7 μm were mounted on chromalum- and gelatin-coated slides and dried overnight at 37°C. For some experiments alternate sections were mounted on different slides to be able to compare the various antibody stainings as well as the immunohistochemistry and in situ hybridization results.
For immunohistochemical detection of LyCEP, a polyclonalAchatina cardioexcitatory peptide (ACEP-1) antiserum (a gift from Dr. M. Kobayashi, Hiroshima, Japan) was used. This antiserum was first tested for its ability to recognize synthetic LyCEP and for the absence of cross-reactivity with nonrelated RFamide peptides in dot blot assays. Sections were incubated overnight with anti-ACEP-1 at a 1:500 dilution in PBS plus 0.5% Triton X-100 (PBST) with 0.2% gelatin, washed with PBST, and incubated with alkaline phosphatase-conjugated goat anti-rabbit IgG (1:100 dilution; DAKO, Glostrup, Denmark). After three final rinses in PBST, phosphatase activity was detected with 0.03% nitroblue tetrazolium (NBT; Boehringer Mannheim), bromochloroindolyl phosphate (BCIP; Boehringer Mannheim), and diaminobenzidine tetrahydrochloride (Sigma, St. Louis, MO) in PBST containing 0.015% H2O2. Egg-laying inducing hormone (ELH) was detected using a monoclonal anti-ELH antibody (1:3000 dilution). In this case a peroxidase-conjugated rabbit anti-mouse secondary antibody (1:100 dilution; DAKO) was used; peroxidase activity was detected with 0.03% diaminobenzidine tetrahydrochloride (Sigma) in PBST containing 0.015% H2O2.
For double immunohistochemical stainings, sections were incubated simultaneously with the ACEP-1 and ELH antibodies (1:500 and 1:3000, respectively). Next, the ACEP-1 antibody was detected as detailed above. After thorough rinsing in PBST plus 0.2% gelatin, sections were preincubated with 10% normal rabbit serum, and subsequently the ELH antibody was detected as described.
For in situ hybridization, pBS106 was used as a template to generate a PCR product with primers flanking the T7 and T3 RNA polymerase promoters. The linear fragment was gel-purified and used as a template for the synthesis of antisense cRNA in the presence of digoxigenin-labeled UTP according to the manufacturer’s instructions (Boehringer Mannheim). In a similar manner, sense cRNA was synthesized as a negative control. Sections of the CNS of L. stagnaliswere pretreated with 0.2% pepsin in 0.2 m HCl for 20 min at 37°C, rinsed with double-distilled water, and subsequently hybridized using 80 μl of hybridization buffer (per slide) containing 2–5 ng of cRNA per μl. Hybridization conditions, treatments after hybridization, as well as the detection of hybrids with an alkaline phosphatase-conjugated sheep anti-digoxigenin antibody were as described (Tensen et al., 1994a).
Effects of LyCEP on the Lymnaea heart. The auricle of the heart was dissected and attached to a displacement transducer in a 1 ml chamber filled with snail Ringer’s solution (Geraerts et al., 1984). LyCEP was added under continuous superfusion with Ringer’s solution at a flow rate of 1 ml/min, and contractions were recorded on paper. Responses of the auricle were quantified 2.5 min after addition of the peptide for a period of 2 min as described previously (Geraerts et al., 1984)
Effects of LyCEP on caudodorsal cells. Caudodorsal cells (CDCs; ELH–producing cells) were isolated from the CNS under aseptic conditions [for a detailed procedure, see Ridgway et al. (1991)]. The nervous systems were incubated for 20–25 min in Leibovitz L-15 defined medium (DM; special order; Life Technologies, Gaithersburg, MD) with 0.67 mg/ml trypsin (type III; Sigma) at room temperature (20–22°C). After enzyme treatment, the brains were rinsed for 10–15 min in DM containing 0.67 mg/ml soybean trypsin inhibitor (type I-S, Sigma). Isolated cells were plated on poly-l-lysine (Sigma)-coated 35 mm diameter Petri dishes (Falcon) filled with DM or brain-conditioned DM (CM) (see Ridgway et al., 1991). Intracellular recordings of the isolated CDCs were made 24–30 hr after plating. The cells were impaled with glass microelectrodes (TW150 F-6; WPI; resistance, 50–60 MΩ) filled with 0.5 m potassium acetate and 0.01 m potassium chloride. The recording chamber was continuously superfused with DM or standard saline. LyCEP (5–10 μm) or a control solution was applied close to the somata. The concentrations used were based on the concentration of FMRFamide that resulted in a saturated response in isolated CDCs (see Brussaard et al., 1988). Fast green (10 μm; Sigma) was also added to monitor application of the drugs and control solutions visually.
RESULTS
Molecular cloning of a novel G-protein–coupled neuropeptide receptor from L. stagnalis
On the basis of two amino acid sequence motifs that are highly conserved among many G-protein–coupled neuropeptide receptors, degenerate oligonucleotide primers were designed (see Materials and Methods) and used in a PCR with cDNA prepared from Lymnaeaheart RNA. As revealed by nucleotide sequence analysis, one of the PCR products obtained encoded part of a novel putative GPCR. Based on this nucleotide sequence, an oligonucleotide was designed to screen cDNA libraries using a PCR-based strategy (Bloem and Yu, 1990; Gibbons et al., 1991). From a CNS library, a 1.5 kb cDNA designated GRL106 was isolated and sequenced (the nucleotide sequence for GRL106 has been deposited into GenBank and is available under accession numberAF037444).
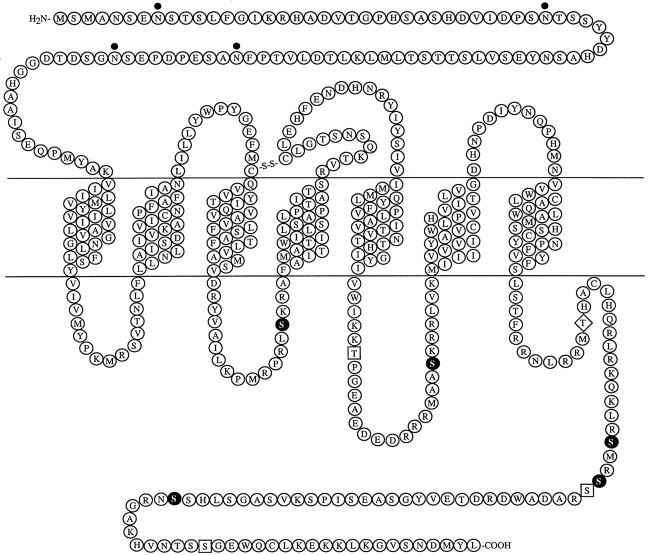
In the GRL106 cDNA sequence, an A/T-rich untranslated region of 66 nucleotides precedes a 1395 nucleotide open reading frame that contains an initiation codon obeying the Kozak consensus (Kozak, 1989). Hydrophobicity analysis of the predicted protein product of 465 amino acids suggests the presence of seven membrane-spanning segments (Fig.1), which is typical of GPCRs. Another feature of GPCRs is a putative disulfide bridge between extracellular loops I and II. Residues Cys171 and Cys247 may form such a bond in GRL106. Furthermore, as in all GPCRs, there are consensus sites for N-linked glycosylation within the predicted extracellular N terminal of GRL106, as well as sites for phosphorylation of serine and threonine residues within putative intracellular loops II and III and in the C terminal. In addition, the receptor protein harbors a typical B-B-X-X-B motif (in which B is a basic residue and X is any nonbasic residue) at the C-terminal part of the third intracellular loop. Such a conserved motif is found in many G-protein–coupled receptors and has been implicated in G-protein activation (Okamoto and Nishimoto, 1992). Database searches indicate that GRL106 shares the highest amino acid sequence similarity (∼40%) with invertebrate G-protein–coupled neuropeptide receptors such as the Drosophila NPY receptor (Li et al., 1992), the Drosophila and Stomoxys calcitranstachykinin(-like) receptors (Li et al., 1991) (accession numberU52347), and vertebrate tachykinin receptors (Shigemoto et al., 1990;Takeda et al., 1991). Similarity is highest in the transmembrane domains, especially in domain VII (50% identity between all receptors).
Fig. 1.
Deduced amino acid sequence and protein model of GRL106. Amino acids are given in single letter code. Potential N-linked glycosylation sites (dots) and putative protein kinase C (filled circles), casein kinase 2 (squares), and cAMP-dependent protein kinase phosphorylation (diamond) sites are indicated. The nucleotide sequence for GRL106 has been deposited into GenBank and is available under accession number AF037444.
Identification of a neuropeptide ligand for GRL106 inLymnaea brain extracts
Various GPCRs, including the vertebrate NPY and tachykinin receptors (Shigemoto et al., 1990; Li et al., 1991), trigger intracellular calcium mobilization after ligand binding. Because of the resemblance of GRL106 to the latter receptors, we hypothesized that it would be possible to screen for a GRL106 ligand by measuring the intracellular calcium mobilization. To do so, we made use ofXenopus oocytes, a well-established model system for expression and functional analysis of GPCRs, in which receptor-generated calcium signals can be detected easily via voltage-clamp analysis of an endogenous calcium-dependent chloride channel (Meyerhof et al., 1988). This system has been used for the expression and analysis of numerous G-protein–coupled receptors cloned from vertebrates [e.g., tachykinin receptors (Shigemoto et al., 1990)] and invertebrates [e.g., conopressin receptors (van Kesteren et al., 1995, 1996)]. Indeed, when oocytes were injected with GRL106-encoding cRNA and were subsequently challenged with a crude peptide extract from the Lymnaea brain, a dose-dependent inward chloride current was observed (Fig.2A). This response was not seen in oocytes injected with cRNA lacking GRL106-encoding sequences (data not shown).
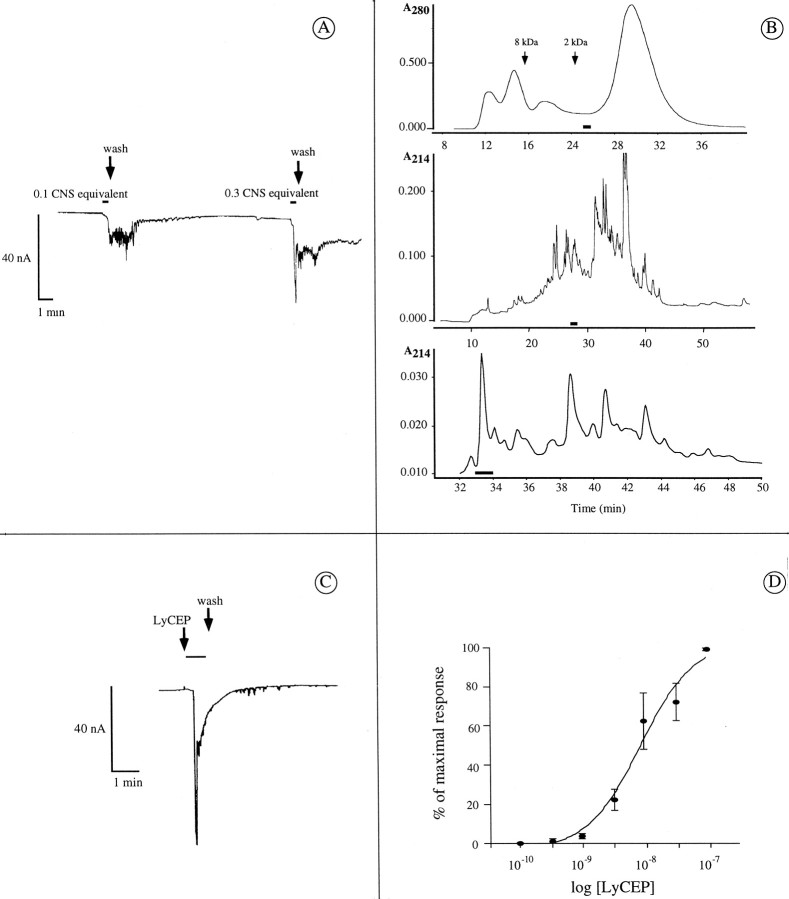
Fig. 2.
Functional expression of GRL106 inXenopus oocytes and purification of the GRL106 ligand from Lymnaea brain extracts. A, Whole-cell current of a Xenopus oocyte expressing GRL106 and responding to a Lymnaea brain peptide extract. GRL106-encoding cRNA was injected into Xenopus oocytes, and after 48 hr whole-cell currents were measured while challenging the oocytes with a crude peptide extract from the LymnaeaCNS. Different amounts of the extract were applied for 25 sec (indicated by horizontal bars) and then washed out. One CNS equivalent is equal to an amount of extract that originates from one CNS. B, Purification of the bioactive peptide(s) from the brain extract. Top, HPGPC fractionation on Protein-Pak columns I-125 and I-300 connected in series (elution times of size markers are indicated).Middle, rpHPLC fractionation of the combined bioactive fractions of the HPGPC column on a Nucleosil C18 column eluted with 7.5 mm TFA and 0–60% acetonitrile. Bottom, rpHPLC fractionation of the combined bioactive fractions of the first rpHPLC column on a Nucleosil C18 column eluted with 0.05% HCl and 0–25% acetonitrile. Horizontal bars indicate bioactive fractions. C, Whole-cell current of aXenopus oocyte expressing GRL106 and responding to 3 nm synthetic LyCEP. D, Dose–response curve of the effect of synthetic LyCEP on whole-cell currents inXenopus oocytes expressing GRL106. LyCEP was applied in different concentrations to oocytes, and the mean membrane current of four experiments was plotted versus the logarithm of the concentration. Error bars denote SEM.
To purify the GRL106-activating molecule(s), we size-fractionated the peptide extract (Fig. 2B, top) and tested fractions for their ability to trigger a chloride current in GRL106-expressing oocytes. Fractions that were active were subjected to further HPLC fractionation and testing until a single active fraction remained (Fig. 2B, middle,bottom). As revealed by mass spectrometry, the final preparation contained two peptides with molecular masses of 1281 and 1296 Da. However, amino acid sequence analysis of the two peptides yielded a single sequence of which the C-terminal residue could not be determined (TPHWRPQGRX). The difference of 146 Da between the molecular mass calculated for the nine amino acid sequence determined via sequencing and the mass of the 1281 Da peptide suggests that the 10th C-terminal residue is phenylalanine-NH2. Thus, the GRL106-activating peptide in extracts from the Lymnaea brain indeed appeared likely to be an amidated neuropeptide, TPHWRPQGRF-NH2, that belongs to the RFamide family. The identical N-terminal sequence and the difference in the molecular masses of the two peptides could be explained by the presence of an hydroxyproline in the 1296 Da peptide, as was found for another member of the RFamide family, the head peptide isolated from Aedes aegypti (Matsumoto et al., 1989) and bradykinin forms (Sasaguri et al., 1988). However, close inspection of the sequencing data did not show any indication of hydroxyproline, so the nature of the observed mass difference remains as yet unexplained.
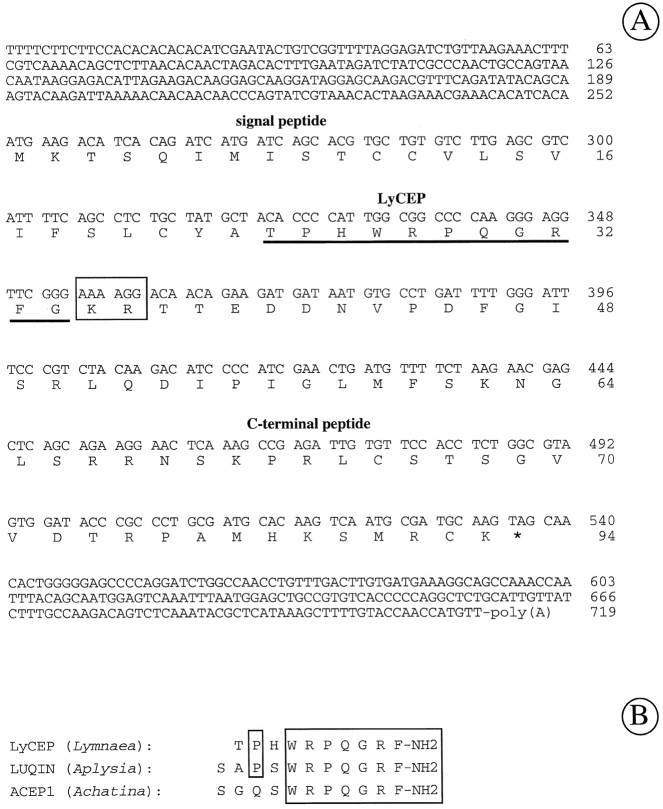
To prove further the prediction that the isolated peptide ends in RFamide, we have cloned the cDNA encoding the LyCEP precursor using the PCR technique (see Materials and Methods). This cDNA contains an open reading frame of 285 nucleotides encoding a 94 amino acid LyCEP precursor protein (Fig. 3A). The predicted sequence for the mature LyCEP is fully consistent with our protein sequencing data and continues after the Arg at position 9 with a Phe residue followed by the sequence Gly-Lys-Arg, the common signal for processing and subsequent amidation (Bradbury and Smyth, 1991).
Fig. 3.
Nucleotide sequence of preproLyCEP cDNA, the derived amino acid sequence, and comparison of LyCEP with related peptides. A, Nucleotide sequence and conceptual translation of the LyCEP precursor cDNA. The putative proteolytic processing site (Lys-Arg) is boxed, and the LyCEP domain is underlined. Nucleotide numberis indicated at the right of each line. The predicted amino acid sequence of preproLyCEP isnumbered (on the right) with the first methionine designated position 1. The stop codon is indicated by anasterisk. The nucleotide sequence of the preproLyCEP cDNA has been deposited into GenBank under accession numberAF047683. B, Alignment of LyCEP with ACEP-1 fromAchatina fulica (Fujimoto et al., 1990) and with LUQIN from Aplysia californica (Aloyz and DesGroseillers, 1995). Amino acids identical in LyCEP and either of the other peptides are boxed.
To verify whether LyCEP is the endogenous ligand for the GRL106 receptor, we made a synthetic form of the purified peptide and used this form to challenge Xenopus oocytes expressing the receptor. The synthetic peptide elicited a response similar to that of the purified peptide (Fig. 2C) and activated the chloride current with an EC50 of 8 ± 0.4 nm(n = 4; Fig. 2D). In contrast, GRL106-expressing oocytes did not respond to FMRFamide, SDPFLRFamide, or GDPFLRFamide in concentrations up to 10 μm. Also, oocytes expressing other receptors such as the conopressin receptors LSCPR1 and LSCPR2 (van Kesteren et al., 1995, 1996) did not respond to the peptide.
Because the GRL106 ligand displays a striking similarity to ACEP-1 of the African giant snail Achatina fulica(Fujimoto et al., 1990) (Fig. 3B), the Lymnaeapeptide was designated LyCEP (Ly for Lymnaea). LyCEP and ACEP-1 are also similar to the neuropeptide LUQIN, which has been recently isolated from Aplysia californica (Aloyz and DesGroseillers, 1995) (Fig. 3B).
ELH–producing neurons in the Lymnaea CNS express GRL106 and are innervated by LyCEP-containing fibers
Being interested in the function of neuropeptides in interneuronal communication, we sought to identify neurons within theLymnaea CNS that express GRL106 and thus are the target of the LyCEP peptide. In situ hybridization of the snail brain with GRL106 cRNA as a probe revealed that the cognate mRNA is expressed predominantly in the egg-laying hormone–producing CDCs, some 150 peptidergic neurons arranged in two clusters in the right and left cerebral ganglia of the CNS (Fig.4A) (Geraerts et al., 1988). CDCs were unequivocally identified in alternate sections of the cerebral ganglia by immunostaining with an antibody recognizing the ELH produced in these cells (Fig. 4B). GRL106-specific mRNA was also found in a limited number of neurons within the cerebral, pleural, parietal, and visceral ganglia of the CNS, whereas sense probes were negative in all cases (data not shown).
Fig. 4.
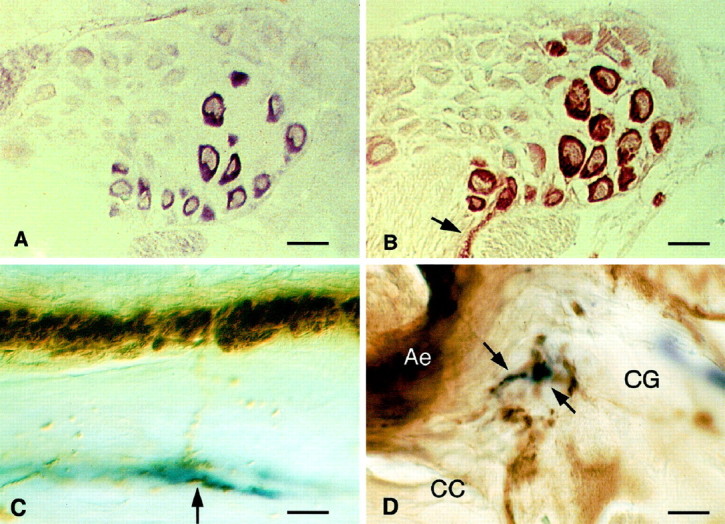
Localization of GRL106 and LyCEP in theLymnaea CNS. A, GRL106 expression revealed by in situ hybridization of a section of theLymnaea CNS with a GRL106-specific probe. Within the cerebral ganglia, one of which is shown here, cells located at the position of CDC neurons hybridize with the GRL106 probe.B, Identification of CDCs by immunohistochemical staining of a section consecutive to the one in A with an antibody against ELH. The cells hybridizing with the GRL106 probe inA are clearly identified as CDCs by their strong reaction with the ELH antibody; in the commissure, immunopositive axon bundles and endings of the CDCs can be seen (arrow). Scale bars: A, B, 80 μm.C, Immunohistochemical double staining of theLymnaea CNS with an anti-ACEP-1 antibody (blue reaction product) and an anti-ELH antibody (brown reaction product). Shown is part of the cerebral commissure, where ELH-positive fibers seem to run to and make contact with fibers that react with the ACEP-1 antibody (arrow). Scale bar, 10 μm. D, In the neuropil of the cerebral ganglion (CG), axons that are immunoreactive to the ACEP-1 antibody and closely opposing axons containing ELH (arrows), which may indicate a site of contact between the LyCEP-expressing neurons and the CDCs. CC, Cerebral commissure containing the axon endings of the CDCs (Ae). Scale bar, 5 μm.
To assess whether the CDCs are indeed contacted by nerve endings containing LyCEP, we performed an immunohistochemical double staining of the Lymnaea CNS using the ACEP-1 and ELH antibodies. With the ACEP-1 antibody, prominent staining was found in neurons within the pedal ganglia. Immunoreactive axons were observed throughout the CNS, including the commissure of the cerebral ganglia, where extensive ELH immunoreactivity also occurs (Fig. 4C). In fact, axon ramifications immunopositive for the ACEP-1 antibody were observed in close apposition to ELH-immunoreactive fibers (Fig.4C,D). These findings are in keeping with the notion that LyCEP is an endogenous ligand for GRL106.
LyCEP inhibits the activity of CDC neurons
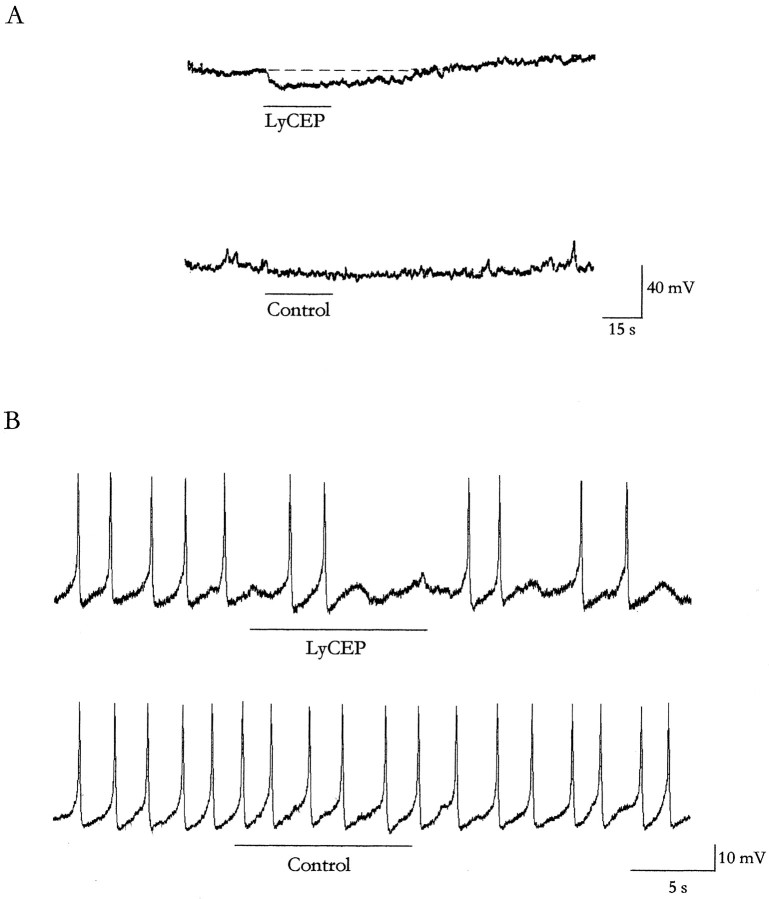
To obtain evidence that LyCEP has an effect on CDCs, we applied the synthetic peptide to CDC neurons isolated from theLymnaea CNS and made intracellular recordings of the membrane potential and activity. After local application of 10 μm LyCEP, 80% of the isolated CDCs showed a hyperpolarization within 5 sec after the application (Fig.5A, top;n = 16). The effect lasted several tens of seconds and outlasted the presence of the peptide. On average, the membrane potential returned to its pretreatment value within 45 ± 9 sec (mean ± SEM; n = 13). The amplitude of the response decreased when the neuron was hyperpolarized (data not shown). The application of vehicle did not affect the membrane potential of isolated CDCs (Fig. 5A, bottom; n= 4). Suprathreshold stimulation (0.5–0.8 nA; 100 msec; 3 Hz) for 20–30 sec induced spontaneous regular spiking activity in 17% of the cells (3 out of 18). Application of LyCEP (5 μm) resulted in an immediate disruption of the beating activity in all cells (Fig.5B, top). The neurons resumed their regular spiking activity 30–40 sec after application. The application of vehicle never changed the spiking activity of the cells (Fig.5B, bottom; n = 3). Thus, it seems that LyCEP has a direct inhibitory effect on the activity of CDC neurons.
Fig. 5.
Effect of LyCEP on isolated CDC neurons.A, Effect of LyCEP on membrane potential. A representative intracellular recording of an isolated CDC at the resting membrane potential (−60 mV) to which 10 μm LyCEP (top) or vehicle (bottom) was applied under continuous superfusion. The horizontal lineindicates the moment and duration of application. B, Effect of LyCEP on spiking activity. A representative intracellular recording of an isolated CDC with spontaneous beating activity to which 5 μm LyCEP (top) or vehicle (bottom) was applied under continuous superfusion. Thehorizontal line indicates the moment and duration of application.
Is LyCEP a cardioactive neuropeptide?
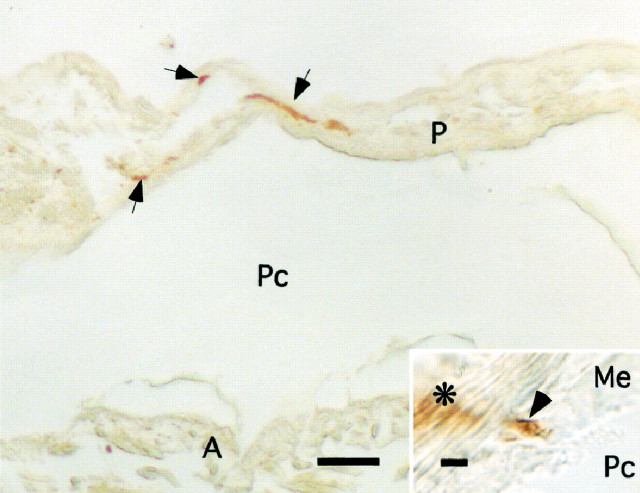
Because of the close similarity of LyCEP to ACEP-1 as well as the fact that we originally amplified the GRL106-encoding DNA fragment from cDNA synthesized from mRNA isolated from the heart ofLymnaea, it seems likely that LyCEP could stimulate the snail heart. In fact, immunohistochemical staining of theLymnaea heart with an antiserum raised against ACEP-1 (recognizing LyCEP but not FMRFamide; data not shown) revealed immunopositive nerve fibers entering the heart (Fig.6). Ramifications occur in the pericardium, ending blindly. This would suggest release of LyCEP into the pericardial lumen.
Fig. 6.
LyCEP immunoreactivity in the pericard. A strongly immunoreactive axon bundle (arrows) runs through the pericard (P). Inset, Occasionally small branches arise from this tract (asterisk, out of focus in this micrograph), penetrate the layer of mesothelial cells (Me; arrowhead), and project toward the pericardial cavity (Pc), which may suggest that the immunoreactive material is released into this cavity. A, Atrium. Scale bar: 100 μm; inset, 5 μm.
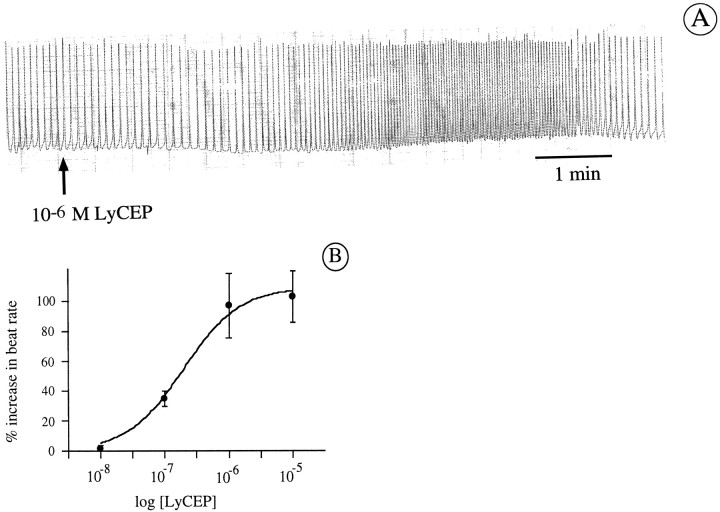
To assess whether LyCEP indeed has an effect on Lymnaeaheart activity, we applied the synthetic peptide to isolated auricle preparations. Auricles challenged with 1 μm LyCEP responded with an increase in spontaneous beat rate (Fig.7A), an effect that is dose-dependent having an EC50 of ∼200 nm(Fig. 7B). Thus, LyCEP seems to be a cardioactive peptide in much the same way as the ACEP-1 peptide of Achatina(Fujimoto et al., 1990). LyCEP is ∼25-fold less active in increasing the beat rate of the isolated auricle compared with its activity for GRL106 expressed in Xenopus oocytes. This difference can be explained by differences in receptor densities or in receptor versus G-protein stoichiometry, parameters that are known to influence EC50 values (Kenakin, 1997). Heterologous expression inXenopus oocytes is likely to give rise to high numbers of receptor protein, whereas the receptor density on theLymnaean heart might be low.
Fig. 7.
Effect of LyCEP on an isolatedLymnaea auricle. A, Contractions of an auricle that was dissected from the heart and placed in a displacement chamber. At the indicated time (arrow), 1 μm LyCEP was applied directly onto the auricle under continuous superfusion. B, Dose–response curve of the increase in beat rate induced by synthetic LyCEP on the isolated auricle. Each data point is the mean of at least seven determinations. Error bars denote SEM.
DISCUSSION
The RFamide neuropeptides, carrying the sequence Arg-Phe-amide at their C terminals, form a large family of signaling molecules comprising both vertebrate (e.g., γ1-MSH) and invertebrate (e.g., FMRFamide) members (Greenberg et al., 1988). Despite the importance of having a detailed knowledge of cellular signal transduction by RFamides, molecular data on the receptors that transduce RFamide-mediated signals are lacking. Only recently was an ionotropic FMRFamide receptor characterized (Lingueglia et al., 1995). Although previous evidence suggested the existence of metabotropic RFamide receptors (Brezina et al., 1987; Volterra and Siegelbaum, 1988), to date no such receptor has been characterized at the molecular level. Here, we present the characterization of a novel GPCR from the CNS of the mollusc Lymnaea stagnalis and the isolation and identification of its ligand, an RFamide neuropeptide designated LyCEP. We initially performed PCR on cDNA synthesized from RNA isolated from the heart of Lymnaea, because we reasoned that an FMRFamide receptor, or a receptor for a related peptide, might be identified among the GPCRs expressed in this tissue because of the clear effects of FMRFamide and its congeners on the snail heart (Buckett et al., 1990a,b). In addition, we expected, compared with the CNS, a less complex mixture of transcripts encoding neuropeptide receptors. Indeed, with a combination of oligonucleotide primers shown previously to be successful for the amplification of cDNAs encoding GPCRs in the CNS of L. stagnalis, partial cDNA sequences encoding several potential neuropeptidergic GPCRs from the Lymnaea heart could be cloned. Next, we used a Lymnaea brain cDNA library to isolate the corresponding full-length clone of one of these cDNAs that was designated GRL106.
GRL106: a novel G-protein–coupled receptor in Lymnaeaheart and brain
Primary structure comparisons suggest that the putative protein product predicted from the GRL106 cDNA is a GPCR. This is based on the presence of seven hydrophobic segments that could serve as transmembrane domains, conserved amino acid residues within these domains, as well as the Cys171–Cys247 combination that may form a stabilizing disulfide bridge. Like all GPCRs, GRL106 contains N-linked glycosylation sites within the N terminal and serine and threonine phosphorylation sites within putative intracellular segments. Moreover, GRL106 seems to be most closely related to GPCRs of the neuropeptide receptor type, such as the Drosophila NPY receptor and vertebrate tachykinin receptors (Shigemoto et al., 1990;Li et al., 1992). Amino acid sequence similarity is highest in the seven transmembrane domains, as is seen for all GPCRs. However, unequivocal proof that GRL106 is a GPCR comes from expression inXenopus oocytes in which we show the protein to activate a G-protein–coupled pathway after addition of LyCEP (see below).
LyCEP: a brain-derived RFamide ligand for GRL106
We set out to identify the putative neuropeptide ligand of GRL106 using a Xenopus oocyte expression system. Because the NPY and tachykinin receptors are known to trigger the liberation of calcium from intracellular stores, we surmised that GRL106 might do so as well. Therefore, we took advantage of the possibility to assay calcium mobilization in Xenopus oocytes by voltage-clamp analysis of a calcium-dependent chloride channel (Meyerhof et al., 1988).
A significant inward chloride current was observed after the addition of a crude peptide extract from the Lymnaea brain to GRL106-expressing oocytes (Fig. 2A). Thus, at least in the heterologous expression system, GRL106 indeed seems to be coupled to calcium signaling, most probably by an endogenous G-protein that triggers phospholipase C-mediated phosphoinositide hydrolysis (Meyerhof et al., 1988). Combining HPLC fractionation of the peptide extract with the oocyte response as a bioassay, we were able to purify an RFamide decapeptide that was responsible for the activation of GRL106. The peptide activating GRL106 was designated LyCEP because of its high similarity to the cardioexcitatory ACEP-1 peptide fromAchatina fulica (Fujimoto et al., 1990), and these peptides, together with LUQIN from Aplysia (Aloyz and DesGroseillers, 1995), seem to constitute a distinct subfamily of RFamides. Interestingly, the C-terminal part of LyCEP (RPQGRF amide) is nearly identical to the C-terminal part of the recently identified family of mammalian prolactin-releasing peptides (RPVGRF amide) (Hinuma et al., 1998).
Different lines of evidence suggest LyCEP to be the natural ligand for GRL106. First, synthetic LyCEP exhibits an EC50 of 8 nm in the oocyte assay. Other calcium-mobilizing GPCRs challenged with their cognate ligand in this system exhibit EC50 values that are somewhat higher than the value observed for the LyCEP–GRL106 combination (Kimura et al., 1992;Mahlmann et al., 1994; van Kesteren et al., 1995). This is a strong indication that LyCEP is, in fact, the GRL106 ligand. Second, not only have we found that LyCEP-containing nerve fibers are present at sites of GRL106 expression in the brain (Fig. 4) and the heart (Fig. 6) ofLymnaea, but we have also established a modulatory action of LyCEP on the activity of innervated brain and targeted heart cells (Figs. 5, 7, respectively; see below).
LyCEP modulates a distinct set of identified GRL106-expressing neurons within the Lymnaea brain
GRL106 was found to be predominantly expressed in the so-called caudodorsal cells (CDCs; Fig. 4A). These neurons express a small family of genes encoding related yet distinct prohormones from which various neuropeptides can be derived that regulate different aspects of egg laying and egg-laying behavior, among which is the ovulation-inducing egg-laying hormone (Geraerts et al., 1988).
As deduced from immunohistochemical staining of the LymnaeaCNS with the anti-ACEP-1 antibody, the CDCs seem to be contacted by LyCEP-containing nerve endings (Fig.4C,D). Furthermore, isolated CDCs respond to the LyCEP peptide by a hyperpolarization and with disruption of the regular spiking activity (Fig. 5). Thus, it seems likely that the LyCEP-containing fibers are involved in inhibiting the snail’s egg-laying system. Conversely, the neurons synthesizing LUQIN inAplysia are inhibited by peptides from the bag cells, theAplysia equivalents of the CDCs in Lymnaea(Rothman et al., 1983). If the LyCEP-producing neurons were to be inhibited by the CDCs in a similar manner, this suggests that the activities of the two systems are mutually exclusive.
LyCEP is a cardioactive neuropeptide
Because its primary structure is so closely related to that of the ACEP-1 peptide of Achatina (Fig. 3B), one would expect LyCEP to exhibit cardioactive properties. Indeed, this seems to be the case. First, in the pericardium surrounding theLymnaea heart, immunoreactive axon endings end blindly at the pericardial lumen (Fig. 6). This suggests that the immunoreactive material is released into the pericardial cavity. The anti-ACEP-1 antibody clearly discriminates LyCEP from other RFamides, such as FMRFamide, as was shown by immunoblots (Fujiwarasakata and Kobayashi, 1994; C. P. Tensen, unpublished data). Therefore, we conclude that the staining by the anti-ACEP-1 antibody reveals fibers that contain LyCEP. Similarly, the Aplysia heart has been found to be innervated by the LUQ neurons producing LUQIN (Skelton et al., 1992;Aloyz and DesGroseillers, 1995). Second, the frequency of beating is increased when isolated auricle preparations are challenged with synthetic LyCEP (Fig. 7). This effect is similar to that of ACEP-1 onAchatina ventricles (Fujimoto et al., 1990). Thus, LyCEP is both present and active at the Lymnaea heart. Because LyCEP-containing fibers are seen to end blindly at the pericardial lumen in immunohistochemical sections, the LyCEP peptide will not function as a neurotransmitter. Rather, it seems that LyCEP fulfills a modulatory role.
The ACEP-1 peptide was found to stimulate other muscles than the heart, such as the penis retractor and buccal muscles (Fujimoto et al., 1990). In addition, the LUQIN peptide of Aplysia is synthesized by neurons that not only innervate the heart but also the kidney, where they seem to be involved in stimulating circular muscles that close the renal pore (Koester and Alevizos, 1989). Whether LyCEP can also modulate other Lymnaea muscles apart from the heart has to await further investigations.
From the idea that GRL106 is not the FMRFamide receptor and from the observations that FMRFamide and related RFamides other than LyCEP do have (differential) effects on snail heart activity and neurons (Cottrell and Davies, 1987; Payza, 1987), we expect that many more receptors for RFamide peptides remain to be identified inLymnaea.
Footnotes
This work was supported by a European Molecular Biology short-term fellowship awarded to C.P.T. and by the Human Frontier Science Program Organization to D.R. We gratefully acknowledge Drs. E. R. Liman and M. Kobayashi for generous gifts of the pGEMHE plasmid and the anti-ACEP-1 antibody, respectively. We also thank Günther Ellinghausen and Hans-Hinrich Hönck for their contributions to the oocyte experiments.
Correspondence should be addressed to Dr. Harm van Heerikhuizen, Department of Biochemistry and Molecular Biology, De Boelelaan 1083, 1081 HV Amsterdam, The Netherlands.
Dr. Tensen’s present address: Amsterdam Leiden Institute for Immunology, Department of Dermatology, Academisch Ziekenhuis, De Boelelaan 1117, 1081 HV Amsterdam, The Netherlands.
Dr. Meyerhof’s present address: Department of Molecular Genetics, German Institute of Human Nutrition, University of Potsdam, Arthur Scheunert Allee 114-116, D-14558, Potsdam-Rehbrücke, Germany.
Dr. Hermann’s present address: Department of Medical Physiology, The University of Calgary, Faculty of Medicine, 3330 Hospital Drive, Calgary, Canada T2N 4N1.
Dr. Vreugdenhil’s present address: Department of Medical Pharmacology, Sylvius Laboratories, P.O. Box 9503, 2300 RA Leiden, The Netherlands.
REFERENCES
- 1.Aloyz RS, DesGroseillers L. Processing of the L5–67 precursor peptide and characterization of LUQIN in the LUQ neurons of Aplysia californica. Peptides. 1995;16:331–338. doi: 10.1016/0196-9781(94)00140-5. [DOI] [PubMed] [Google Scholar]
- 2.Bloem LJ, Yu L. A time-saving method for screening cDNA or genomic libraries. Nucleic Acids Res. 1990;18:2830. doi: 10.1093/nar/18.9.2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradbury AF, Smyth DG. Peptide amidation. Trends Biochem Sci. 1991;16:112–115. doi: 10.1016/0968-0004(91)90044-v. [DOI] [PubMed] [Google Scholar]
- 4.Brezina V, Eckert R, Erxleben C. Modulation of potassium conductances by an endogenous neuropeptide in neurones of Aplysia californica. J Physiol (Lond) 1987;382:267–290. doi: 10.1113/jphysiol.1987.sp016367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brussaard AB, Kits KS, Ter Maat A, Van Minnen J, Moed PJ. Dual inhibitory action of FMRFamide on neurosecretory cells controlling egg laying behavior in the pond snail. Brain Res. 1988;447:35–51. doi: 10.1016/0006-8993(88)90963-8. [DOI] [PubMed] [Google Scholar]
- 6.Buckett KJ, Dockray GJ, Osborne NN, Benjamin PR. Pharmacology of the myogenic heart of the pond snail Lymnaea stagnalis. J Neurophysiol. 1990a;63:1413–1425. doi: 10.1152/jn.1990.63.6.1413. [DOI] [PubMed] [Google Scholar]
- 7.Buckett KJ, Peters M, Dockray J, van Minnen J, Benjamin PR. Regulation of heartbeat in Lymnaea by motoneurons containing FMRFamide-like peptides. J Neurophysiol. 1990b;63:1426–1435. doi: 10.1152/jn.1990.63.6.1426. [DOI] [PubMed] [Google Scholar]
- 8.Cottrell GA, Davies NW. Multiple receptor sites for a molluscan peptide (FMRFamide) and related peptides of Helix. J Physiol (Lond) 1987;382:51–68. doi: 10.1113/jphysiol.1987.sp016355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cottrell GA, Davies NW, Green KA. Multiple actions of a molluscan cardioexcitatory neuropeptide and related peptides on identified Helix neurones. J Physiol (Lond) 1984;356:315–333. doi: 10.1113/jphysiol.1984.sp015467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox KJ, Tensen CP, van der Schors RC, Li KW, van Heerikhuizen H, Vreugdenhit E, Geraerts WP, Burke JF. Cloning, characterization, and expression of a G-protein-coupled receptor from Lymnaea stagnalis and identification of a leucokinin-like peptide, PSFHSWS amide, as its endogenous ligand. J Neurosci. 1997;17:1197–1205. doi: 10.1523/JNEUROSCI.17-04-01197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebberink RHM, Price DA, van Loenhout H, Doble KD, Riehm JP, Geraerts WPM, Greenberg MJ. The brain of Lymnaea contains a family of FMRFamide-like peptides. Peptides. 1987;8:515–522. doi: 10.1016/0196-9781(87)90018-0. [DOI] [PubMed] [Google Scholar]
- 12.Fujimoto K, Ohta N, Yoshida M, Kubota I, Muneoka Y, Kobayashi M. A novel cardio-excitatory peptide isolated from the atria of the African giant snail, Achatina fulica. Biochem Biophys Res Commun. 1990;167:777–783. doi: 10.1016/0006-291x(90)92093-f. [DOI] [PubMed] [Google Scholar]
- 13.Fujiwarasakata M, Kobayashi M. Localization of FMRFamide- and ACEP1-like immunoreactivities in the nervous system of Achatina fulica. Cell Tissue Res. 1994;278:451–460. doi: 10.1007/BF00331363. [DOI] [PubMed] [Google Scholar]
- 14.Geraerts WPM, de With ND, Vreugdenhil E, van Hartingsveldt W, Hogenes TM. Studies on the physiological role of a partially purified small cardioactive neuropeptide of Lymnaea stagnalis. J Comp Physiol [B] 1984;154:29–34. [Google Scholar]
- 15.Geraerts WPM, Ter Maat A, Vreugdenhil E. The peptidergic neuroendocrine control of egg-laying behavior in Aplysia and Lymnaea. In: Laufer H, Downer RGH, editors. Invertebrate endocrinology, Vol 2. Liss; New York: 1988. pp. 141–231. [Google Scholar]
- 16.Gibbons IR, Asai DJ, Ching NS, Dolecki GJ, Mocz G, Phillipson CA, Ren H, Tang W-JY, Gibbons BH. A PCR procedure to determine the sequence of large polypeptides by rapid walking through a cDNA library. Proc Natl Acad Sci USA. 1991;88:8563–8567. doi: 10.1073/pnas.88.19.8563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenberg MJ, Payza K, Nachman RJ, Holman GM, Price DA. Relationships between the FMRFamide-related peptides and other peptide families. Peptides. 1988;9[Suppl 1]:125–135. doi: 10.1016/0196-9781(88)90236-7. [DOI] [PubMed] [Google Scholar]
- 18.Hinuma S, Habata Y, Fujii R, Kawamata Y, Hosoya M, Fukusumi S, Kitada C, Masuo Y, Asano T, Matsumoto H, Sekiguchi M, Kurokawa T, Nishimura O, Onda H, Fujino M. A prolactin-releasing peptide in the brain. Nature. 1998;393:272–276. doi: 10.1038/30515. [DOI] [PubMed] [Google Scholar]
- 19.Kenakin T. Differences between natural and recombinant G protein-coupled receptors with varying receptor/G protein stoichiometry. Trends Pharmacol Sci. 1997;18:456–464. doi: 10.1016/s0165-6147(97)01136-x. [DOI] [PubMed] [Google Scholar]
- 20.Kimura T, Tanizawa O, Mori K, Brownstein MJ, Okayama H. Structure and expression of a human oxytocin receptor. Nature. 1992;356:526–529. doi: 10.1038/356526a0. [DOI] [PubMed] [Google Scholar]
- 21.Koester J, Alevizos A. Innervation of the kidney of Aplysia by L10, the LUQ cells, and an identified peripheral motoneuron. J Neurosci. 1989;9:4078–4088. doi: 10.1523/JNEUROSCI.09-11-04078.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kozak M. The scanning model for translation: an update. J Cell Biol. 1989;108:229–241. doi: 10.1083/jcb.108.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li XJ, Wolfgang W, Wu YN, North RA, Forte M. Cloning, heterologous expression and developmental regulation of a Drosophila receptor for tachykinin-like peptides. EMBO J. 1991;10:3221–3229. doi: 10.1002/j.1460-2075.1991.tb04885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li XJ, Wu YN, North A, Forte M. Cloning, functional expression, and developmental regulation of a neuropeptide Y receptor from Drosophila melanogaster. J Biol Chem. 1992;267:9–12. [PubMed] [Google Scholar]
- 25.Liman ER, Tytgat J, Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- 26.Lingueglia E, Champigny G, Lazdunski M, Barbry P. Cloning of the amiloride-sensitive FMRFamide peptide-gated sodium channel. Nature. 1995;378:730–733. doi: 10.1038/378730a0. [DOI] [PubMed] [Google Scholar]
- 27.Mahlmann S, Meyerhof W, Hausmann H, Heierhorst J, Schönrock C, Zwiers H, Lederis K, Richter D. Structure, function, and phylogeny of [Arg8]vasotocin receptors from teleost fish and toad. Proc Natl Acad Sci USA. 1994;91:1342–1345. doi: 10.1073/pnas.91.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsumoto S, Brown MR, Crim JW, Vigna SR, Lea AO. Isolation and primary structure of neuropeptides from the mosquito, Aedes aegypti, immunoreactive to FMRF amide antiserum. Insect Biochem. 1989;17:277–283. [Google Scholar]
- 29.Meyerhof W, Morley S, Schwarz J, Richter D. Receptors for neuropeptides are induced by exogenous poly(A)+ RNA in oocytes from Xenopus laevis. Proc Natl Acad Sci USA. 1988;85:714–717. doi: 10.1073/pnas.85.3.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okamoto T, Nishimoto I. Detection of G protein-activator regions in M4 subtype muscarinic, cholinergic, and alpha 2-adrenergic receptors based upon characteristics in primary structure. J Biol Chem. 1992;267:8342–8346. [PubMed] [Google Scholar]
- 31.Payza K. FMRFamide receptors in Helix aspersa. Peptides. 1987;8:1065–1074. doi: 10.1016/0196-9781(87)90138-0. [DOI] [PubMed] [Google Scholar]
- 32.Price DA, Greenberg MJ. Structure of a molluscan cardioexcitatory neuropeptide. Science. 1977;197:670–671. doi: 10.1126/science.877582. [DOI] [PubMed] [Google Scholar]
- 33.Probst WC, Lenore AS, Schuster DI, Brosius J, Sealfon SC. Sequence alignment of the G-protein-coupled receptor superfamily. DNA Cell Biol. 1992;11:1–20. doi: 10.1089/dna.1992.11.1. [DOI] [PubMed] [Google Scholar]
- 34.Ridgway RL, Syed NI, Lukowiak K, Bulloch AGM. Nerve growth factor (NGF) induces sprouting of specific neurons of the snail, Lymnaea stagnalis. J Neurobiol. 1991;22:377–390. doi: 10.1002/neu.480220406. [DOI] [PubMed] [Google Scholar]
- 35.Rothman B, Mayeri E, Brown RO, Yan PM, Shively JE. Primary structure and neuronal effects of α-bag cell peptide, a second candidate neurotransmitter encoded by a single gene in bag cell neurons of Aplysia. Proc Natl Acad Sci USA. 1983;80:5753–5757. doi: 10.1073/pnas.80.18.5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory; New York: 1989. [Google Scholar]
- 37.Sanger F, Nicklen S, Coulson AB. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sasaguri M, Ikeda M, Ideishi M, Arakawa K. Identification of [hydroxyproline 3]-bradykinin released from human plasma and plasma protein Cohn’s Fraction IV-4 by trypsin. Biochem Biophys Res Commun. 1988;157:210–217. doi: 10.1016/s0006-291x(88)80034-2. [DOI] [PubMed] [Google Scholar]
- 39.Saunders SE, Bright K, Kellett E, Benjamin PR, Burke JF. Neuropeptides Gly-Asp-Pro-Phe-Leu-Arg-Phe-NH2 (GDPFLRFamide) and Ser-Asp-Pro-Phe-Leu-Arg-Phe-NH2 (SDPFLRFamide) are encoded by an exon 3′ to Phe-Met-Arg-Phe-NH2 (FMRFamide) in the snail Lymnaea stagnalis. J Neurosci. 1991;11:740–745. doi: 10.1523/JNEUROSCI.11-03-00740.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saunders SE, Kellett E, Bright K, Benjamin PR, Burke JF. Cell-specific alternative splicing of an FMRFamide gene transcript in the brain. J Neurosci. 1992;12:1033–1039. doi: 10.1523/JNEUROSCI.12-03-01033.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shigemoto R, Yokota Y, Tsuchida K, Nakanishi S. Cloning and expression of a rat neuromedin K receptor cDNA. J Biol Chem. 1990;265:623–628. [PubMed] [Google Scholar]
- 42.Skelton M, Alevizos A, Koester J. Control of the cardiovascular system of Aplysia by identified neurons. Experientia. 1992;48:809–817. doi: 10.1007/BF02118413. [DOI] [PubMed] [Google Scholar]
- 43.Takeda Y, Chou KB, Takeda J, Sachais BS, Krause JE. Molecular cloning, structural characterization and functional expression of the human substance P receptor. Biochem Biophys Res Commun. 1991;179:1232–1240. doi: 10.1016/0006-291x(91)91704-g. [DOI] [PubMed] [Google Scholar]
- 44.Tensen CP, van Kesteren ER, Planta RJ, Cox K, Burke JF, van Heerikhuizen H, Vreugdenhil E. A G protein-coupled receptor with low density lipoprotein-binding motifs suggests a role for lipoproteins in G-linked signal transduction. Proc Natl Acad Sci USA. 1994a;91:4816–4820. doi: 10.1073/pnas.91.11.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tensen CP, Cox KJA, Burke JF, van der Schors RC, Meyerhof W, Richter D, Planta RJ, Vreugdenhil E, van Heerikhuizen H. A novel successful strategy towards the identification of endogenous ligands for cloned “orphan neuropeptide receptors.”. Soc Neurosci Abstr. 1994b;20:28. [Google Scholar]
- 46.van Kesteren RE, Tensen CP, Smit AB, van Minnen J, van Soest PF, Kits KS, Meyerhof W, Richter D, van Heerikhuizen H, Vreugdenhil E, Geraerts WPM. A novel G protein-coupled receptor mediating both vasopressin- and oxytocin-like functions of Lys-conopressin in Lymnaea stagnalis. Neuron. 1995;15:897–908. doi: 10.1016/0896-6273(95)90180-9. [DOI] [PubMed] [Google Scholar]
- 47.van Kesteren RE, Tensen CP, Smit AB, van Minnen J, Kolakowski LF, Jr, Meyerhof W, Richter D, van Heerikhuizen H, Vreugdenhil E, WPM G. Co-evolution of ligand-receptor pairs in the vasopressin/oxytocin superfamily of bioactive peptides. J Biol Chem. 1996;271:3619–3626. doi: 10.1074/jbc.271.7.3619. [DOI] [PubMed] [Google Scholar]
- 48.Volterra A, Siegelbaum SA. Role of two different guanine nucleotide-binding proteins in the antagonistic modulation of the S-type K+ channel by cAMP and arachidonic acid metabolites in Aplysia sensory neurons. Proc Natl Acad Sci USA. 1988;85:7810–7814. doi: 10.1073/pnas.85.20.7810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang H-YT, Fratta W, Majane EA, Costa E. Isolation, sequencing, synthesis, and pharmacological characterization of two brain neuropeptides that modulate the action of morphine. Proc Natl Acad Sci USA. 1985;82:7757–7761. doi: 10.1073/pnas.82.22.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]