Abstract
To analyze the human red, green, and red-green hybrid cone pigments in vivo, we studied 41 male dichromats, each of whose X chromosome carries only a single visual pigment gene (single-gene dichromats). This simplified arrangement avoids the difficulties of complex opsin gene arrays and overlapping cone spectral sensitivities present in trichromats and of multiple genes encoding identical or nearly identical cone pigments in many dichromats. It thus allows for a straightforward correlation between each observer’s spectral sensitivity measured at the cornea and the amino acid sequence of his visual pigment. For each of the 41 single-gene dichromats we determined the amino acid sequences of the X-linked cone pigment as deduced from its gene sequence. To correlate these sequences with spectral sensitivities in vivo, we determined the Rayleigh matches to different red/green ratios for 29 single-gene dichromats and measured psychophysically the spectral sensitivity of the remaining green (middle wavelength) or red (long wavelength) cones in 37 single-gene dichromats. Cone spectral sensitivity maxima obtained from subjects with identical visual pigment amino acid sequences show up to a ∼3 nm variation from subject to subject, presumably because of a combination of inexact (or no) corrections for variation in preretinal absorption, variation in photopigment optical density, optical effects within the photoreceptor, and measurement error. This variation implies that spectral sensitivities must be averaged over multiple subjects with the same genotype to obtain representative values for a given pigment. The principal results of this study are that (1) ∼54% of the single-gene protanopes (and ∼19% of all protanopes) possess any one of several 5′red-3′green hybrid genes that encode anomalous pigments and that would be predicted to produce protanomaly if present in anomalous trichromats; (2) the alanine/serine polymorphism at position 180 in the red pigment gene produces a spectral shift of ∼2.7 nm; (3) for each exon the set of amino acids normally associated with the red pigment produces spectral shifts to longer wavelengths, and the set of amino acids normally associated with the green pigment produces spectral shifts to shorter wavelengths; and (4) changes in exons 2, 3, 4, and 5 from green to red are associated with average spectral shifts to long wavelengths of ∼1 nm (range, −0.5 to 2.5 nm), ∼3.3 nm (range, −0.5 to 7 nm), ∼2.8 nm (range, −0.5 to 6 nm), and ∼24.9 nm (range, 22.2–27.6 nm).
Keywords: visual pigments, cone pigments, red-green color blindness, spectral sensitivity, human retina, protein sequences
Normal human color vision is mediated by three light-sensitive cone photopigments.a Two of these, the red pigment and the green pigment, are encoded by highly homologous genes that reside in a head-to-tail tandem array on the X chromosome (for review, see Nathans et al., 1992). The common variants of red-green color vision arise from unequal homologous exchanges within this gene array that cause a loss of one of the two types of visual pigment genes and/or the production of hybrid genes. Simple loss of one of the X-linked pigments produces dichromacy, whereas replacement of one of the X-linked pigments with a hybrid pigment results either in dichromacy or anomalous trichromacy, depending on the extent to which the absorbance spectrum of the hybrid pigment differs from that of the remaining normal pigment.
Two factors complicate the interpretation of genotype–phenotype correlations in the study of red-green color vision. First, the number of visual pigment genes within an array varies among X chromosomes. In general, each visual pigment gene array has only a single red pigment gene, whereas the number of green pigment genes varies from one to at least five (Nathans et al., 1986a; Drummond-Borg et al., 1989; Macke and Nathans, 1997; Yamaguchi et al., 1997) [although see Neitz and Neitz (1995) and Neitz et al. (1995a) for a dissenting view]. The presence of more than one green pigment gene, or in the case of deuteranomalous trichromats (subjects with an altered green cone sensitivity; abbreviated here G′R+) more than one 5′green-3′red hybrid gene and/or green pigment gene, complicates the correlation of genotype and phenotype because recent evidence indicates that only a subset of the green pigment genes is expressed (Winderickx et al., 1992b; Yamaguchi et al., 1997). There is currently no method for determining from the genotype which green or 5′green-3′red hybrid pigment genes are expressed in those individuals who carry multiple copies of these genes in their array. A second complication arises from the observation that single nucleotide polymorphisms create variant pigments in which spectral sensitivity may be shifted by as much as ∼5 nm. One of the most common polymorphic variations occurs at codon 180 in the red pigment gene where site-directed mutagenesis experiments suggest that the presence of an alanine or a serine results in a shift to shorter or longer wavelengths, respectively, of ∼4 nm (Merbs and Nathans, 1992a) or 2–7 nm (Asenjo et al., 1994) (also see below). The exact spectral shifts referable to this and other polymorphisms are of considerable interest because these sequence differences are likely to account for much of the variation in color vision between normal trichromats.
Experiments aimed at defining the cone spectral sensitivities that underlie both normal and anomalous color vision have been pursued for over a century. One approach has been to use psychophysical techniques that isolate single cone sensitivities in vivo by exploiting the selective desensitization caused by either steady (Stiles, 1939,1964; De Vries, 1948a,b; Wald, 1964) or transient (Stockman et al., 1993b) chromatic adaptation. With these techniques it is possible to isolate the red and green cones of normals throughout the spectrum (Stockman et al., 1993a,b) and the blue cones from short wavelengths to ∼540 nm (Stiles, 1953; . However, the psychophysical isolation of cones containing red-green hybrid pigments from the accompanying normal red or green cones has been difficult or impossible to achieve in X chromosome-linked anomalous trichromats, owing to the similarities between the spectral sensitivities of these pigments (see Stockman et al., Piantandida and Sperling, 1973a,b; Pokorny et al., 1973;Rushton et al., 1973b).
The simplest and most commonly used method for isolating the red or green cone sensitivities is to study red-green dichromats who lack either red or green cone photopigments (Pitt, 1935; Hecht, 1949;Willmer, 1950; Hsia and Graham, 1957; Mitchell and Rushton, 1971;Rushton et al., 1973a; Smith and Pokorny, 1975). On the assumption that their color vision is a “reduced” form of normal color vision (Maxwell, 1860; König and Dieterici, 1886), that is, that their surviving cones have the same spectral sensitivities as their counterparts in color-normal trichromats, these data have been used to define normal cone spectral sensitivities. Not all dichromats, however, conform to the reduction hypothesis, either because they have hybrid photopigments or because they have multiple photopigment genes (Nathans et al., 1986b; Deeb et al., 1992). Only red-green dichromats with a single normal photopigment gene or with multiple genes that produce identical photopigments will conform completely to the reduction hypothesis. This recently appreciated genetic complexity calls into question the conclusions of previous studies in which the genotypes of the red-green dichromats were unknown.
Over the past several decades the techniques of fundus reflectometry (Rushton, 1963, 1965), microspectrophotometry (Dartnall et al., 1983), single-cell electrophysiology (Schnapf et al., 1987), and electroretinography (Neitz et al., 1995b) also have been applied to the study of human cone pigment spectral sensitivities. The most recent approach to this problem has come from the in vitrostudy of recombinant cone pigments produced in tissue culture cells (Oprian et al., 1991; Merbs and Nathans, 1992a,b; Asenjo et al., 1994). These experiments have defined the absorption properties of the normal cone pigments, their common polymorphic variants, and each of the several hybrid pigments that underlie anomalous trichromacy. However, the reported measurements of visual pigment absorbance in solution suffer from two limitations: they are only accurate within approximately one log unit of the peak absorbance and thus encompass only a limited range of wavelengths, and they differ from measurements in the living eye owing, for example, to waveguiding (Enoch, 1961,1963) and to self-screening (Brindley, 1953) effects in the cone photoreceptor.
To analyze red, green, and red-green hybrid pigments in vivowhile avoiding the difficulties of complex opsin gene arrays and overlapping cone spectral sensitivities, we have chosen to study male dichromats each of whose X-chromosome carries only a single visual pigment gene. This simplified arrangement allows for a straightforward correlation to be made between spectral sensitivity and the individual’s visual pigment sequence, and it eliminates problems associated with dichromats who carry multiple genes that may differ subtly in spectral sensitivity. In this paper we report the spectral sensitivities of red, green, and 5′red-3′green hybrid pigments determined in the living human eye by psychophysical methods, together with the amino acid sequences of the corresponding proteins as deduced from their gene sequences. The results permit a direct correlation between single amino acid substitutions and in vivo shifts in spectral sensitivity in both normal and anomalous pigments.
MATERIALS AND METHODS
Human subjects. Males with severe color vision deficiencies were recruited in Freiburg, Tübingen, and Vienna via word of mouth and by advertising in local newspapers and cinemas. Most subjects were native Germans or Austrians; ∼15% of subjects were natives of the Balkan peninsula. Subjects were between 16 and 45 years old, with a mean age of 28. Their ages are given in Tables 3 and 4. To identify and classify the dichromats within this group, we examined the subjects by anomaloscopy as described below.
Table 3.
Best-fitting photopigment λmax values and macular and lens densities according to the full spectrum analysis
| Code | Age | λmax ±SE | Lens density ± SE (400 nm) | Macular density ± SE (peak) | rmserror | Genotype | |
|---|---|---|---|---|---|---|---|
| 1 | HS2182 | 27 | 557.79 ± 0.51 | 1.15 ± 0.04 | 0.44 ± 0.03 | 0.07 | R(A180) |
| 2 | HS2232 | 25 | 556.81 ± 0.77 | 1.48* | 0.27 ± 0.04 | 0.07 | R(A180) |
| 3 | HS2244 | 32 | R(A180) | ||||
| 4 | HS2245 | 23 | 558.13 ± 1.44 | 1.62 ± 1.02 | 0.36 ± 0.09 | 0.09 | R(A180) |
| 5 | HS2247 | 43 | 558.78 ± 1.46 | 1.14 ± 1.03 | 0.48 ± 0.09 | 0.09 | R(A180) |
| 6 | HS2248 | 18 | R(A180) | ||||
| 7 | HS2219 | 24 | 556.88 ± 0.49 | 1.31 ± 0.04 | 0.35 ± 0.02 | 0.08 | R(G2;A180) |
| 8 | HS2171 | 27 | 559.27 ± 0.34 | 1.82 ± 0.04 | 0.56 ± 0.02 | 0.06 | R(S180) |
| 9 | HS2173 | 28 | 560.61 ± 0.68 | 1.46 ± 0.05 | 0.83 ± 0.03 | 0.08 | R(S180) |
| 10 | HS2176 | 22 | 559.95 ± 0.57 | 1.51 ± 0.05 | 0.44 ± 0.03 | 0.08 | R(S180) |
| 11 | HS2177 | 20 | 559.92 ± 0.59 | 1.62 ± 0.06 | 0.26 ± 0.03 | 0.07 | R(S180) |
| 12 | HS2181 | 23 | 561.76 ± 0.41 | 1.44 ± 0.04 | 0.48 ± 0.02 | 0.07 | R(S180) |
| 13 | HS2185 | 27 | 560.58 ± 0.50 | 0.97 ± 0.04 | 0.37 ± 0.02 | 0.07 | R(S180) |
| 14 | HS2218 | 25 | 560.53 ± 0.52 | 1.13 ± 0.03 | 0.21 ± 0.02 | 0.07 | R(S180) |
| 15 | HS2220 | 42 | 560.54 ± 0.60 | 1.45 ± 0.07 | 0.37 ± 0.03 | 0.07 | R(S180) |
| 16 | HS2221 | 27 | 558.94 ± 0.65 | 0.86 ± 0.08 | 0.17 ± 0.03 | 0.07 | R(S180) |
| 17 | HS2229 | 25 | 558.01 ± 0.56 | 1.47 ± 0.05 | 0.38 ± 0.02 | 0.07 | R(S180) |
| 18 | HS2231 | 30 | 559.23 ± 0.72 | 1.29 ± 0.07 | 0.43 ± 0.03 | 0.06 | R(S180) |
| 19 | HS2243 | 23 | 559.11 ± 0.97 | 1.48* | 0.01 ± 0.05 | 0.08 | R(S180) |
| 20 | HS2249 | 22 | 561.76 ± 0.72 | 1.48* | 0.42 ± 0.05 | 0.06 | R(S180) |
| 21 | HS2299 | 32 | 561.87 ± 0.32 | 1.67 ± 0.03 | 0.50 ± 0.01 | 0.05 | R(S180) |
| 22 | HS2304 | 26 | 562.09 ± 0.31 | 1.76 ± 0.02 | 0.38 ± 0.01 | 0.05 | R(S180) |
| 23 | HS2310 | 21 | R(S180) | ||||
| 24 | HS2319 | 28 | R(S180) | ||||
| 25 | HS2322 | 41 | 561.42 ± 0.43 | 1.70 ± 0.03 | 0.58 ± 0.02 | 0.06 | R(S180) |
| 26 | HS2357 | 37 | 560.32 ± 0.44 | 1.66 ± 0.04 | 0.42 ± 0.02 | 0.07 | R(S180) |
| 27 | HS2184 | 27 | 558.84 ± 0.59 | 0.81 ± 0.05 | 0.28 ± 0.02 | 0.08 | R(S180) |
| 28 | HS2313 | 22 | 558.52 ± 0.23 | 1.66 ± 0.02 | 0.28 ± 0.01 | 0.04 | R(G2;S180) |
| 29 | HS2305 | 23 | 535.45 ± 0.51 | 1.60 ± 0.04 | 0.25 ± 0.02 | 0.08 | R4G5(A180) |
| 30 | HS2202 | 29 | 534.18 ± 0.40 | 1.25 ± 0.05 | 0.34 ± 0.02 | 0.07 | R4G5(S180) |
| 31 | HS2188 | 30 | 530.68 ± 0.51 | 1.14 ± 0.04 | 0.73 ± 0.03 | 0.07 | R3G4(S180) |
| 32 | HS2198 | 29 | 529.75 ± 0.49 | 1.39 ± 0.06 | 0.43 ± 0.03 | 0.08 | R3G4(S180) |
| 33 | HS2235 | 23 | 530.71 ± 0.58 | 1.48* | 0.08 ± 0.04 | 0.07 | R3G4(S180) |
| 34 | HS2237 | 16 | 532.14 ± 0.51 | 1.57 ± 0.06 | 0.49 ± 0.03 | 0.08 | R3G4(S180) |
| 35 | HS2307 | 23 | 534.15 ± 0.47 | 1.79 ± 0.04 | 0.44 ± 0.02 | 0.07 | R3G4(S180) |
| 36 | HS2197 | 27 | 527.16 ± 0.52 | 1.82 ± 0.05 | 0.39 ± 0.07 | 0.07 | R2G3(A180) |
| 37 | HS2201 | 39 | 528.84 ± 0.69 | 1.68 ± 0.06 | 0.01 ± 0.04 | 0.06 | R2G3(A180) |
| 38 | HS2298 | 18 | 529.61 ± 0.67 | 1.81 ± 0.12 | 0.30 ± 0.03 | 0.09 | R2G3(A180) |
| 39 | HS2196 | 35 | 525.58 ± 0.68 | 1.81 ± 0.08 | 0.14 ± 0.04 | 0.07 | R1G2(A180) |
| 40 | HS2234 | 42 | 528.80 ± 0.71 | 1.55 ± 0.06 | 0.26 ± 0.04 | 0.07 | R1G2(A180) |
| 41 | HS2241 | 19 | 528.88 ± 0.74 | 1.71 ± 0.06 | 0.32 ± 0.04 | 0.09 | R1G2(A180) |
| 42 | HS2288 | 24 | 529.73 ± 0.45 | 1.28 ± 0.03 | 0.28 ± 0.02 | 0.07 | R2G3+G(A180) |
| 43 | HS2293 | 32 | 529.59 ± 0.53 | 1.54 ± 0.03 | 0.55 ± 0.03 | 0.07 | R2G3+G(A180) |
| 44 | HS2303 | 45 | 529.69 ± 0.40 | 1.83 ± 0.05 | 0.61 ± 0.02 | 0.06 | R1G2+G(A180) |
| 45 | HS2312 | 18 | R4G5(G2)+G(A180) |
Best-fitting photopigment λmax values (in nanometers), lens densities at 400 nm, peak macular densities (at 460 nm), and root mean-squared (rms) errors for the fit of the full spectrum model (Eqs. 1, 2, and 3a) to each subject’s spectral sensitivity data. The ± SE values are plus and minus the SE of each fitted parameter. Subject ages are noted also. *In four subjects (HS2232, HS2243, HS2249, and HS2235), in whom short-wavelength measurements below 470 nm were not made, the lens density could not be estimated reliably. For them, we fixed the density at 1.48, the mean density for all subjects (see Table 5).
Table 4.
Best-fitting photopigment λmax values according to the partial spectrum (≥520 nm) analysis, and slopes of the regression lines fit to Rayleigh matches
| Code | Age | λmax ±SE | rms error | Slope ± SE (König–Helmholtz) | Genotype | |
|---|---|---|---|---|---|---|
| 1 | HS2182 | 27 | 557.70 ± 0.64 | 0.07 | 0.4191 ± 0.0135 | R(A180) |
| 2 | HS2232 | 25 | 556.99 ± 0.82 | 0.07 | 0.4128 ± 0.0129 | R(A180) |
| 3 | HS2244 | 32 | R(A180) | |||
| 4 | HS2245 | 23 | 558.28 ± 0.85 | 0.09 | 0.5672 ± 0.0374 | R(A180) |
| 5 | HS2247 | 43 | 558.50 ± 0.60 | 0.09 | 0.4378 ± 0.0300 | R(A180) |
| 6 | HS2248 | 18 | R(A180) | |||
| 7 | HS2219 | 24 | 556.17 ± 0.50 | 0.08 | 0.4171 ± 0.0119 | R(G2;A180) |
| 8 | HS2171 | 27 | 559.88 ± 0.29 | 0.06 | 0.4002 ± 0.0127 | R(S180) |
| 9 | HS2173 | 28 | 560.54 ± 0.79 | 0.08 | 0.3754 ± 0.0145 | R(S180) |
| 10 | HS2176 | 22 | 559.95 ± 0.45 | 0.08 | 0.3775 ± 0.0122 | R(S180) |
| 11 | HS2177 | 20 | 560.30 ± 0.66 | 0.07 | 0.3669 ± 0.0201 | R(S180) |
| 12 | HS2181 | 23 | 561.90 ± 0.51 | 0.07 | 0.3966 ± 0.0161 | R(S180) |
| 13 | HS2185 | 27 | 560.05 ± 0.43 | 0.07 | 0.3907 ± 0.0124 | R(S180) |
| 14 | HS2218 | 25 | 559.92 ± 0.46 | 0.07 | 0.3486 ± 0.0087 | R(S180) |
| 15 | HS2220 | 42 | 560.59 ± 0.67 | 0.07 | 0.3483 ± 0.0172 | R(S180) |
| 16 | HS2221 | 27 | 558.30 ± 1.05 | 0.07 | 0.4095 ± 0.0145 | R(S180) |
| 17 | HS2229 | 25 | 557.72 ± 0.54 | 0.07 | 0.3771 ± 0.0171 | R(S180) |
| 18 | HS2231 | 30 | 559.01 ± 0.68 | 0.06 | 0.3434 ± 0.0146 | R(S180) |
| 19 | HS2243 | 23 | 559.11 ± 1.02 | 0.08 | 0.5070 ± 0.0243 | R(S180) |
| 20 | HS2249 | 22 | 561.65 ± 0.59 | 0.06 | 0.4541 ± 0.0205 | R(S180) |
| 21 | HS2299 | 32 | 562.35 ± 0.38 | 0.05 | R(S180) | |
| 22 | HS2304 | 26 | 562.70 ± 0.38 | 0.05 | R(S180) | |
| 23 | HS2310 | 21 | R(S180) | |||
| 24 | HS2319 | 28 | R(S180) | |||
| 25 | HS2322 | 41 | 562.22 ± 0.48 | 0.06 | R(S180) | |
| 26 | HS2357 | 37 | 561.12 ± 0.45 | 0.07 | R(S180) | |
| 27 | HS2184 | 27 | 558.06 ± 0.67 | 0.08 | 0.4025 ± 0.0188 | R(S180) |
| 28 | HS2313 | 22 | 558.78 ± 0.26 | 0.04 | R(G2;S180) | |
| 29 | HS2305 | 23 | 535.36 ± 0.45 | 0.08 | R4G5(A180) | |
| 30 | HS2202 | 29 | 533.91 ± 0.27 | 0.07 | 0.6622 ± 0.0267 | R4G5(S180) |
| 31 | HS2188 | 30 | 530.47 ± 0.48 | 0.07 | 0.7101 ± 0.0256 | R3G4(S180) |
| 32 | HS2198 | 29 | 529.51 ± 0.66 | 0.08 | 0.7351 ± 0.0193 | R3G4(S180) |
| 33 | HS2235 | 23 | 530.43 ± 0.60 | 0.07 | 0.7391 ± 0.0256 | R3G4(S180) |
| 34 | HS2237 | 16 | 532.47 ± 0.58 | 0.08 | 0.6989 ± 0.0293 | R3G4(S180) |
| 35 | HS2307 | 23 | 534.26 ± 0.47 | 0.07 | R3G4(S180) | |
| 36 | HS2197 | 27 | 527.69 ± 0.49 | 0.07 | 0.7752 ± 0.0235 | R2G3(A180) |
| 37 | HS2201 | 39 | 528.56 ± 0.48 | 0.06 | 0.7655 ± 0.0330 | R2G3(A180) |
| 38 | HS2298 | 18 | 530.10 ± 0.57 | 0.09 | R2G3(A180) | |
| 39 | HS2196 | 35 | 525.50 ± 0.77 | 0.07 | 0.7110 ± 0.0136 | R1G2(A180) |
| 40 | HS2234 | 42 | 528.21 ± 0.60 | 0.07 | 0.7870 ± 0.0326 | R1G2(A180) |
| 41 | HS2241 | 19 | 528.35 ± 0.83 | 0.09 | 0.9036 ± 0.0431 | R1G2(A180) |
| 42 | HS2288 | 24 | 529.50 ± 0.46 | 0.07 | 0.8006 ± 0.0217 | R2G3+G(A180) |
| 43 | HS2293 | 32 | 529.85 ± 0.47 | 0.07 | R2G3+G(A180) | |
| 44 | HS2303 | 45 | 529.99 ± 0.36 | 0.06 | R1G2+G(A180) | |
| 45 | HS2312 | 18 | R4G5(G2)+G(A180) |
Best-fitting photopigment λmax values (in nanometers) and root mean-squared (rms) errors for the fit of the partial spectrum (≥520 nm) model (Eqs. 1, 2, and 3b) to each subject’s mean spectral sensitivity data. The ±SE values are plus and minus the SE of the λmax estimates. Slopes (König–Helmholtz) of the regression lines fit to the Rayleigh matches are given for those 29 single gene dichromats who were tested with a König–Helmholtz colorimeter. The ±SE values are plus and minus the SE of the slope estimates. The green, yellow, and red primaries for this instrument were 546, 590, and 674 nm, respectively. The König–Helmholtz data have not been corrected for preretinal absorption.
Genotyping. Venous blood samples were obtained from 94 dichromat subjects, and genomic DNA was purified by proteinase K digestion and equilibrium centrifugation in cesium chloride. Southern blots of BamHI and EcoRI-digested DNA were probed with a 350 bp EcoRI–BamHI fragment from the 5′ end of a full-length human red pigment cDNA clone (hs7; Nathans et al., 1986a) to visualize two restriction fragments from the green pigment gene (referred to as Bg and Cg), and two restriction fragments from the red pigment gene (referred to as Br and Cr; Fig.1B). Those DNA samples that showed three or more hybridizing fragments must have two or more visual pigment genes within the X chromosome. Those DNA samples with only one version of fragment B and one version of fragment C were presumed to carry a single visual pigment gene. In each Southern blot pattern of this type, visual inspection of the hybridization intensity of the B and C bands showed that they corresponded to the lowest intensity level seen among all of the subjects, consistent with the presence in these samples of only a single visual pigment gene. With the use of primers in the flanking intron regions (Nathans et al., 1993), exons 2–5 were amplified by PCR from each of the 41 DNA samples that were presumed to carry a single visual pigment gene, together with four samples predicted to carry more than one visual pigment gene. The PCR products were purified by absorption to an affinity column (Qiagen, Hilden, Germany), and both strands were sequenced by the use of thermocycle sequencing with fluorescent chain terminators (Applied Biosystems, Foster City, CA).
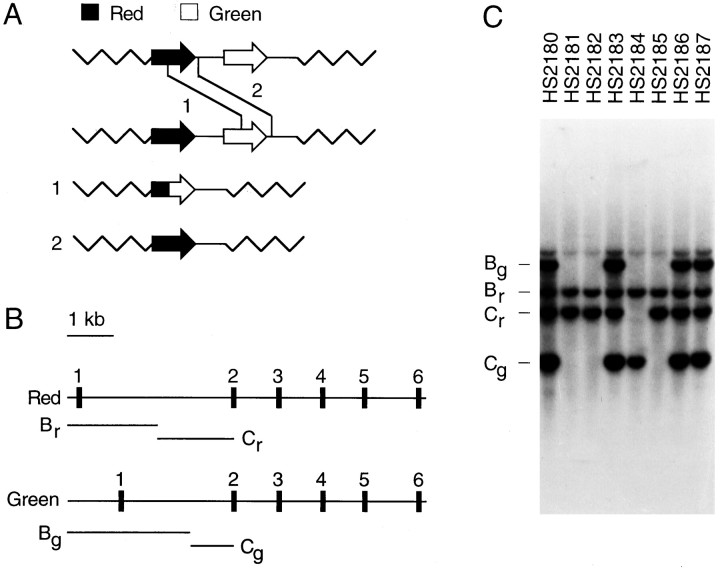
Fig. 1.
Homologous recombination between red and green pigment genes in dichromat subjects. A, Schematic diagram of unequal homologous recombination between the most 5′ gene in one visual pigment gene array (a red pigment gene) and the most 3′ gene in a second visual pigment gene array (a green pigment gene). For simplicity, only one of the usually multiple green pigment genes is shown in each of the wild-type arrays. Recombination events within the coding regions produce an array with a single 5′red-3′green hybrid gene (crossover 1), resulting in a G+R− or a G′R− phenotype; recombination events 3′ to the coding regions produce an array with a single red pigment gene (crossover 2), resulting in a G−R+ phenotype.B, Diagram of the red and green pigment genes showing the locations of the six exons and theBamHI–EcoRI restriction fragments Bg and Cg (derived from the green pigment gene) and Br and Cr (derived from the red pigment gene). The B fragments encompass 5′ flanking DNA, exon 1, and part of intron 1, and the C fragments encompass part of intron 1 and the 5′ half of exon 2 [see Nathans et al. (1986) for a detailed restriction map]. C, Representative whole genome Southern blot hybridization to identify dichromats with X chromosome visual pigment gene arrays with only a single gene. A human red pigment cDNA probe encompassing exon 1 and the 5′ half of exon 2 (5′ of theBamHI site) was hybridized to a Southern blot of human genomic DNA cut with both EcoRI andBamHI. This probe hybridizes with equal efficiency to the red and green pigment genes. In the Southern blot shown here, HS2180, HS2183, HS2186, and HS2187 carry two or more X-linked visual pigment genes as judged by the presence of both red and green versions of fragments B and C. HS2181, HS2182, HS2184, and HS2185 are presumed to carry a single visual pigment gene as judged by the presence of only one version of fragment B and one version of fragment C.
Rayleigh matches. X chromosome-linked (red-green) dichromacy traditionally is established by Rayleigh matches. The subject must be able to match fully a spectral yellow primary light to a juxtaposed mixture of spectral red and green primary lights merely by adjusting the intensity of the yellow, regardless of the red-to-green ratio. This is consistent with quantal absorptions in a single photopigment being responsible for the matches (the choice of the wavelengths and intensities of the primaries as well as the field size mainly precludes absorptions in the blue cones or rods from influencing the matches).
Subjects were tested for their Rayleigh matches on one or more of four anomaloscopes: a König–Helmholtz colorimeter and a Nagel II anomaloscope and two Nagel I anomaloscopes. Because they were recruited and tested over a span of several years and at several locations, only a subset of subjects was tested on all the instruments. The field of view (2–2.6° in diameter) and the wavelengths and intensities of the primary lights varied among the four anomaloscopes.
All the dichromat subjects listed in Tables 2-4 (n = 45) made repeated matches (three to five times) in random order for 17 different red-to-green mixture ratios solely by adjusting the intensity of the yellow primary light. Their individual matching range slopes (i.e., the slopes of regression lines fit to their yellow intensity settings for the 17 red-green mixtures) and intercepts (i.e., the yellow intensity required to match the red primary alone) then were determined by a least-squares criterion. From the extent of the matching range (i.e., whether the subjects could match both the red and green primary lights) and the slope of the regression line, the subjects were categorized as G+R− (protanopes), G−R+ (deuteranopes), or anomalous trichromats (for classification details, see Pokorny et al., 1979; Wyszecki and Stiles, 1982).
Table 2.
Amino acid sequences and summary genotypes of X-linked cone visual pigments among 45 dichromat males
| Exon 2 | Exon 3 | Exon 4 | Exon 5 | Genotype | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 65 | 111 | 116 | 153 | 171 | 174 | 178 | 180 | 230 | 233 | 236 | 249 | 274 | 275 | 277 | 279 | 285 | 298 | 309 | |||
| 1 | HS2182 | T | I | S | M | V | V | V | A | I | A | V | V | I | F | Y | V | T | A | Y | R(A180) |
| 2 | HS2232 | T | I | S | M | V | V | V | A | I | A | M | V | I | F | Y | V | T | A | Y | R(A180) |
| 3 | HS2244 | T | I | S | L | V | A | I | A | I | A | M | V | I | F | Y | V | T | A | Y | R(A180) |
| 4 | HS2245 | T | I | S | L | V | A | I | A | I | A | M | V | I | F | Y | V | T | A | Y | R(A180) |
| 5 | HS2247 | T | I | S | L | V | A | I | A | I | A | M | V | I | F | Y | V | T | A | Y | R(A180) |
| 6 | HS2248 | T | I | S | L | V | A | I | A | I | A | M | V | I | F | Y | V | T | A | Y | R(A180) |
| 7 | HS2219 | T | V | Y | M | V | A | I | A | I | A | M | V | I | F | Y | V | T | A | Y | R(G2;A180) |
| 8 | HS2171 | T | I | S | L | I | A | I | S | I | A | M | V | I | F | Y | V | T | A | Y | R(S180) |
| 9 | HS2173 | T | I | S | L | V | A | I | S | I | A | M | V | I | F | Y | V | T | A | Y | R(S180) |
| 10 | HS2176 | T | I | S | L | V | A | I | S | I | A | M | V | I | F | Y | V | T | A | Y | R(S180) |
| 11 | HS2177 | T | I | S | L | V | A | I | S | I | A | M | V | I | F | Y | V | T | A | Y | R(S180) |
| 12 | HS2181 | T | I | S | L | V | A | I | S | I | A | M | V | I | F | Y | V | T | A | Y | R(S180) |
| 13 | HS2185 | T | I | S | L | V | A | I | S | I | A | M | V | I | F | Y | V | T | A | Y | R(S180) |
| 14 | HS2218 | T | I | S | L | V | A | I | S | I | A | M | V | I | F | Y | V | T | A | Y | R(S180) |
| 15 | HS2220 | T | I | S | L | V | A | I | S | I | A | M | V | I | F | Y | V | T | A | Y | R(S180) |
| 16 | HS2221 | T | I | S | M | V | A | I | S | I | A | M | V | I | F | Y | V | T | A | Y | R(S180) |
| 17 | HS2229 | T | I | S | L | V | A | I | S | I | A | M | V | I | F | Y | V | T | A | Y | R(S180) |
| 18 | HS2231 | T | I | S | M | V | A | I | S | I | A | M | V | I | F | Y | V | T | A | Y | R(S180) |
| 19 | HS2243 | T | I | S | M | V | V | I | S | I | A | M | V | I | F | Y | V | T | A | Y | R(S180) |
| 20 | HS2249 | T | I | S | L | V | A | I | S | I | A | M | V | I | F | Y | V | T | A | Y | R(S180) |
| 21 | HS2299 | T | I | S | M | V | V | I | S | I | A | V | M | I | F | Y | V | T | A | Y | R(S180) |
| 22 | HS2304 | T | I | S | L | V | A | I | S | I | A | M | V | I | F | Y | V | T | A | Y | R(S180) |
| 23 | HS2310 | T | I | S | L | V | A | I | S | I | A | M | V | I | F | Y | V | T | A | Y | R(S180) |
| 24 | HS2319 | T | I | S | M | V | V | I | S | I | A | M | V | I | F | Y | V | T | A | Y | R(S180) |
| 25 | HS2322 | T | I | S | L | I | A | I | S | I | A | M | V | I | F | Y | V | T | A | Y | R(S180) |
| 26 | HS2357 | T | I | S | L | V | A | I | S | I | A | M | V | I | F | Y | V | T | A | Y | R(S180) |
| 27 | HS2184 | T | V | S | L | V | A | I | S | I | A | V | V | I | F | Y | V | T | A | Y | R(S180) |
| 28 | HS2313 | I | V | Y | M | V | V | I | S | I | A | M | V | I | F | Y | V | T | A | Y | R(G2;S180) |
| 29 | HS2305 | T | I | S | M | V | V | I | A | I | A | V | V | V | L | F | F | A | P | F | R4G5(A180) |
| 30 | HS2202 | T | I | S | M | V | A | I | S | I | A | M | V | V | L | F | F | A | P | F | R4G5(S180) |
| 31 | HS2188 | T | I | S | L | V | A | I | S | T | S | V | V | V | L | F | F | A | P | F | R3G4(S180) |
| 32 | HS2198 | T | I | S | L | V | A | I | S | T | S | V | V | V | L | F | F | A | P | F | R3G4(S180) |
| 33 | HS2235 | T | I | S | L | V | A | I | S | T | S | V | V | V | L | F | F | A | P | F | R3G4(S180) |
| 34 | HS2237 | T | I | S | L | V | A | I | S | T | S | V | V | V | L | F | F | A | P | F | R3G4(S180) |
| 35 | HS2307 | T | I | S | L | V | A | I | S | T | S | V | V | V | L | F | F | A | P | F | R3G4(S180) |
| 36 | HS2197 | T | I | S | M | V | A | I | A | T | S | V | V | V | L | F | F | A | P | F | R2G3(A180) |
| 37 | HS2201 | T | I | S | M | V | V | V | A | T | S | V | M | V | L | F | F | A | P | F | R2G3(A180) |
| 38 | HS2298 | T | I | S | L | V | A | I | A | T | S | V | V | V | L | F | F | A | P | F | R2G3(A180) |
| 39 | HS2196 | I | V | Y | M | V | V | V | A | T | S | V | V | V | L | F | F | A | P | F | R1G2(A180) |
| 40 | HS2234 | I | V | Y | M | V | V | V | A | T | S | V | V | V | L | F | F | A | P | F | R1G2(A180) |
| 41 | HS2241 | I | V | Y | M | V | A | I | A | T | S | V | V | V | L | F | F | A | P | F | R1G2(A180) |
| 42 | HS2288 | T | I = V | S = Y | M | V | A | I | A | T | S | V | V | V | L | F | F | A | P | F | R2G3+G(A180) |
| 43 | HS2293 | T | I = V | S = Y | M | V | A | I | A | T | S | V | V | V | L | F | F | A | P | F | R2G3+G(A180) |
| 44 | HS2303 | I | V | Y | M | V | A | I | A | T | S | V | V | V | L | F | F | A | P | F | R1G2+G(A180) |
| 45 | HS2312 | I > T | V | Y | M | V | A | I | A | I > T | S > A | V > M | V | V | L | F | F | A | P | F | R4G5(G2)+G(A180) |
| R | genomic | T | I | S | L | V | A | I | S | I | A | M | V | I | F | Y | V | T | A | Y | |
| R | cDNA | T | I | S | M | V | A | I | A | I | A | M | V | I | F | Y | V | T | A | Y | |
| G | genomic | I | V | Y | M | V | A | I | A | T | S | V | V | V | L | F | F | A | P | F | |
| G | cDNA | I | V | Y | L | V | A | I | A | T | S | V | V | V | L | F | F | A | P | F | |
X chromosome-encoded visual pigment amino acid sequences were obtained by sequencing PCR-amplified genomic DNA. Each number at the top of the table indicates the position along the polypeptide chain where one or more amino acid substitutions were observed among the 45 dichromats. HS2288, HS2293, HS2303, and HS2312 have more than one X-linked visual pigment gene; at those codon positions where the DNA sequence showed a superposition of bases, the identities of the encoded amino acids are indicated. “=” indicates an equal ratio of the encoded amino acids; “>” indicates that the DNA sequence encoding one amino acid is present at a greater mole ratio than the other, as might occur in a subject with three genes, two of which are identical but differ from the third. Red (R) and green (G) genomic and cDNA sequences listed at the bottom of the table are from Nathans et al. (1986a).
Flicker photometry: apparatus. A Maxwellian view optical system produced the flickering test stimuli and the steady adapting field. All optical channels originated from a 75 W xenon arc lamp (Osram, Berlin, Germany) run at constant current. Two channels provided the 2° in visual diameter flickering test and reference lights. Their wavelengths were shaped by grating monochromators (H-10 Vis, Jobin-Yvon, Longjumaneau, France) with 0.5 mm entrance and exit slits into a triangular profile having a full bandwidth at half-maximum (FWHM) of ∼4 nm. The wavelength of the reference light was always set to 560 nm, whereas that of the test light was varied from 400 to 700 nm in 5 nm steps. At wavelengths longer than 560 nm a glass cutoff filter (Schott OG550), which blocked short wavelengths but transmitted wavelengths higher than 550 nm, was inserted after the exit slit of both monochromators. This filter reduces the skirt of shorter wavelength stray light. The third channel provided the 18° diameter adapting field. The wavelength of the third channel was shaped by a grating monochromator (Jobin-Yvon H-10 Vis) with 2 mm entrance and exit slits into a triangular profile, peaking at 430 nm (FWHM = 17 nm). Infrared radiation was eliminated by heat-absorbing glass (Schott, Mainz, Germany) placed early in each beam.
The images of the xenon arc were <1.5 mm in diameter at the plane of the observer’s pupil. Circular field stops placed in collimated portions of each beam defined the test and adapting fields as seen by the observer. Mechanical shutters driven by a computer-controlled square-wave generator were positioned in each channel near focal points of the xenon arc. The optical waveforms so produced were monitored periodically with a Pin-10 diode (United Detector Technology, Santa Monica, CA) and oscilloscope.
Fine control over the luminance of the stimuli was achieved by variable 2.0 log unit linear (Spindler and Hoyer) or 4.0 log unit circular (Rolyn Optics) neutral density wedges positioned at image points of the xenon arc lamp and by insertion of fixed neutral density filters in parallel portions of the beams. The position of the observer’s head was maintained by a rigidly mounted dental wax impression.
Flicker photometry: calibration. The radiant fluxes of the test and adapting fields were measured at the plane of the observer’s pupil with a calibrated radiometer (Model 80X Optometer, United Detector Technology) or with a Pin-10 diode connected to a picoammeter (Model 486, Keithley, Taunton, MA). The calibration of the silicon detector was checked against a similar model (now produced by Graseby), both of which had been calibrated by the manufacturers against standards traceable to the National Bureau of Standards, USA. In addition, the UDT radiometer was checked against a photodiode that was calibrated against the German national standard (Braunschweig). The fixed and variable neutral density filters were calibrated in situ for all test and field wavelengths. Particular care was taken in calibrating the monochromator and interference filters: spectroradiometers were used to measure the center wavelength and the FWHM at each wavelength in the experiments conducted in Freiburg, Germany (Spectra Scan, Photo Research) and Tübingen, Germany (Instrument Systems).
The radiometric accuracy of the calibrations was better than 0.01 log unit from 400 to 700 nm, whereas the resolution of the individual settings was better than 0.02 log unit. The absolute wavelength accuracy was better than 0.5 nm (Freiburg) or 0.2 nm (Tuebingen), whereas the resolution of the wavelength settings was better than 0.15 nm.
Spectral sensitivity measurements. Corneal spectral sensitivities were measured by heterochromatic flicker photometry. The measurements were confined to the central 2° of the fovea. A reference light (560 nm) was alternated at a rate of 16 or 25 Hz, in opposite phase with a superimposed test light, the wavelength of which was varied in 5 nm steps over the spectrum from 400 to 680 nm. In some subjects the shorter and longer wavelength regions were measured during separate runs. To saturate the rods and to prevent the blue cones from contributing to spectral sensitivity, we superimposed the flickering stimuli on a large violet (430 nm) background with an intensity of 11.0 log quanta · sec−1 · degree−2, which is more than one log unit above rod saturation.
At the start of the spectral sensitivity experiment, the subject adjusted the intensity of the 560 nm reference flickering light until he was satisfied that the flicker was just at threshold. After five settings had been made, the mean threshold setting was calculated and the reference light was set 0.2 log unit above this value. Then the test light was added to the reference light in counterphase. The subject adjusted the intensity of the flickering test light until the flicker percept disappeared or was minimized. This procedure was repeated five times at each wavelength. After each setting the intensity of the flickering test light was randomly reset to a higher or lower intensity so that the subject had to readjust the intensity to find the best setting. The target wavelength was varied randomly in 5 nm steps from 400 to 700 nm. From two to six complete runs were performed by each subject. Thus, each data point represents between 10 and 30 threshold settings.
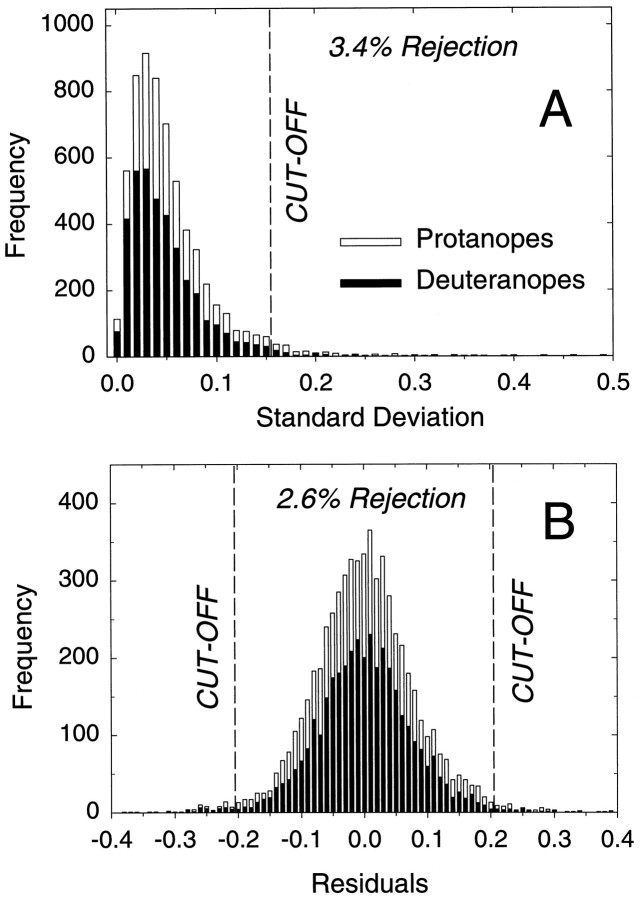
Analysis of flicker photometry data. Measurements were made in naïve subjects. Consequently, although a majority of the measurements were acceptable, a small minority were of doubtful quality. To avoid distorting the λmax (wavelength of maximal absorption) estimates, we adopted a standard method for eliminating doubtful data points from the analysis. Because each data point was averaged from five individual settings, its quality could be judged from their SD. Figure2A shows the frequency of the SDs of the five settings for the data points for all observers. To eliminate the most doubtful data, we rejected points for which the SD of the five settings was >0.15 log unit. In terms of the frequency distribution shown in Figure 2A, this cutoff is 2 SDs higher than the mean and results in a rejection rate of 3.4%.
Fig. 2.
Elimination of unreliable data from the analysis.A, Frequency of the SDs of the five individual settings (in log units) averaged to produce each of the data points for protanopes (open bars; 2410 data points) and deuteranopes (filled bars; 3820 data points); 3.4% of the data points, the five settings of which had SDs > 0.15 log unit, were eliminated from the analysis. The SD axis is truncated at 0.5 log unit. A few points had SDs higher than 0.5.B, Frequency of the log residuals between the data points and the first fit of the spectral sensitivity model [see Eqs.1, 2, and 3a for protanopes (open bars; 2290 points) and deuteranopes (filled bars; 3730 points)]; 2.6% of the data points, the residuals of which were >0.20 log unit, were eliminated from the second fit of the model.
Some data points had small SDs yet were clearly discrepant from the surrounding points and from the predictions of the models described below. They arose, we believe, because subjects occasionally did not adjust the target radiance to find the appropriate flicker null. Figure2B shows the frequency of the residuals between the data points (after the points with a high SD had been eliminated) and the predictions of the full spectrum model (see below). To avoid the discrepant points from distorting the λmax estimates (which is a problem particularly at longer wavelengths), we eliminated all data points for which the residuals were >0.20 log unit. In terms of the frequency distribution shown in Figure 2B, the cutoffs are 2.5 SDs above and below the mean, and they result in a rejection rate of 2.6% (so that the cumulative rejection rate for both steps was ∼6%).
The λmax of each subject’s red, green, or red-green hybrid spectral sensitivity was estimated by fitting a photopigment template to his flicker photometric data. The photopigment template was derived from the green and red cone spectral sensitivities of Stockman et al. (1993a). First, the Stockman et al. (1993a) green and red cone spectral sensitivities were corrected individually to the retinal level by removing the effects of macular and lens pigmentation. Next, they were corrected to photopigment optical density (or absorbance) spectra by adjusting them to infinitely dilute photopigment concentrations. The assumptions required for these corrections are given in Stockman et al. (1993a), in which the red and green cone spectra also are illustrated [Stockman et al. (1993a), their Fig. 15, lower panel]. We next derived a simple polynomial function to describe the logarithm of the red and green cone photopigment spectra after the red cone spectrum had been shifted horizontally along a log10 wavelength [log(λ)] scale to align it with the green cone spectrum. The log photopigment optical density spectrum, log[POD(x)], normalized to zero peak, is:
| Equation 1 |
where x is log(λ), a = 3593840.5764, b = −48574668.0585, c = 262378945.6275, d = −708007074.0766, e= 954435497.8303, f = −514228901.1364, and λ is the wavelength in nanometers. The optical densities defined by Equation 1, before any shifts, peak at 528.2 nm. We emphasize that this photopigment spectrum was derived strictly for this analysis and is useful only in the range of measured wavelengths (400–700 nm). In a forthcoming publication we will use the spectral sensitivity data reported here to derive a new photopigment density spectrum and template. Although very similar to spectra measured by suction electrode recordings, those obtained from psychophysics are slightly steeper at longer wavelengths, probably because the suction electrode data are measured transversely through the photoreceptor outer segment rather than axially along it—as in normal vision. Axial beams are subject to waveguiding by the structure of the photoreceptor (for further discussion, see .
In deriving this template and analyzing the spectral sensitivity data, we made the simplifying assumption that the family of red, green, and red-green hybrid photopigment spectra are invariant in shape when plotted as a function of log(λ) (Mansfield and Levine, 1985;Stockman and Sharpe, MacNichol, 1986; Lamb, 1995). This simplification provides a straightforward means of analyzing the spectral sensitivity data, because the λmax of each photopigment can be estimated from a simple shift of the polynomial defined by Equation 1 (see alsoBaylor et al., 1987). A polynomial was used because we required a simple, continuous function that could be shifted along the log wavelength scale.
The primary fit of the photopigment template (Eq. 1) to the data was performed in the single fitting procedure defined by Equations 2 and3a:
| Equation 2 |
| Equation 3a |
In Equation 2, the template,POD(x), is shifted by Δxlog nanometers along the log wavelength scale and adjusted to a peak photopigment optical density of 0.5 to give the spectral sensitivity at the retinal level, Sretina(x). In Equation 3a, the spectral sensitivity at the retina,Sretina(x), then is corrected to the cornea, Scornea(x), by restoring the filtering effects of the lens pigment,alensOD(x), and macular pigment,bmacOD(x), two pigments that lie between the cornea and the photoreceptor. We assumed the lens pigment density spectrum [lensOD(x)] of van Norren and Vos (1974), slightly modified by Stockman et al. (1993a), and a macular pigment density spectrum [macOD(x)] based on the Vos (1972) estimate. Vertical shifts (cn) were estimated for each of then runs performed by each subject. Separate shifts for each run were required, because the short and long wavelength data sometimes were measured during separate runs. The lens density multiplier (a), the macular density multiplier (b), the template shift (Δx), and vertical shifts (cn) are all best-fitting values determined by the fit of the model defined by Equations 1, 2, and 3a to each set of data. The fit was performed with the Marquardt–Levenberg algorithm implemented in SigmaPlot (Jandel Scientific). In preliminary trials we also attempted to optimize the peak photopigment optical density (rather than assuming the fixed value of 0.5 in Eq. 2), but we found that it was too poorly constrained. We therefore fixed it at 0.5 for each observer (see .
In the primary analysis defined by Equations 1, 2, and 3a, which we refer to as the “full spectrum” fit, we fit the model at all measured wavelengths. In the secondary analysis, which we refer to as the “partial spectrum” fit, we performed the fit only for measurements made at wavelengths ≥520 nm. This simplifies the fitting procedure, because macular pigment plays little role, and the lens is relatively transparent (having an average optical density of only 0.10 log unit at 520 nm that declines with wavelength). Equation 3a was replaced by Equation 3b:
| Equation 3b |
The lens and macular densities were fixed at the mean population densities obtained in the full spectrum fit. Thus, lensOD(x) is 1.48 at 400 nm and macOD(x) is 0.37 at peak (see Table 5, and below). The average data for each subject were used for this fit so that there is only a single vertical shift (c). The template shift (Δx) and vertical shift (c) are best-fitting values determined by the fit of the model defined by Equations 1, 2, and 3b to the mean data (≥520 nm) for each subject. The partial spectrum fit served, in part, as a control for the full spectrum fit and in particular for the reliance (Eq. 3a) on best-fitting macular and lens densities, which could, in principle, distort the λmax estimates. Nonetheless, small differences between the λmax estimates obtained by the two methods should be expected, because the lens density assumed for the partial fit is the population mean density rather than the optimized individual density. We estimate that increasing and decreasing the lens density in Equation 3b by, for example, ±25% results in changes in λmax of approximately ± 0.5 nm. Given that small differences are expected between the two estimates, the agreement between the two is extremely good (see Tables3-5; Figs. 7-9).
Table 5.
Summary of results
| Genotype | n* | Age* | Full spectrum analysis | Partial spectrum analysis λmax | Slope (König–Helmholtz) | ||
|---|---|---|---|---|---|---|---|
| λmax | Lens density at 400 nm | Macular peak density | |||||
| R(A180) | 4 | 29.5 ± 4.6 | 557.88 ± 0.410 | 1.31 ± 0.16 | 0.39 ± 0.05 | 557.87 ± 0.34 | 0.459 ± 0.036 |
| R(G2;A180) | 1 | 24.0 | 556.88 | 1.31 | 0.35 | 556.17 | 0.417 |
| R(A180) & R(G2;A180) | 5 | 28.4 ± 3.7 | 557.68 ± 0.38 | 1.31 ± 0.11 | 0.38 ± 0.04 | 557.53 ± 0.43 | 0.451 ± 0.029 |
| R(S180) | 18 | 28.0 ± 1.5 | 560.26 ± 0.28 | 1.41 ± 0.08 | 0.39 ± 0.04 | 560.30 ± 0.35 | 0.393 ± 0.012 |
| R(G2;S180) | 1 | 22.0 | 558.52 | 1.66 | 0.29 | 558.78 | |
| R(S180) & R(G2;S180) | 19 | 27.7 ± 1.4 | 560.17 ± 0.28 | 1.43 ± 0.08 | 0.39 ± 0.04 | 560.22 ± 0.34 | 0.393 ± 0.012 |
| ALL R | 24 | 27.8 ± 1.3 | 559.65 ± 0.32 | 1.40 ± 0.07 | 0.39 ± 0.03 | 559.66 ± 0.36 | 0.408 ± 0.013 |
| R4G5(A180) | 1 | 23.0 | 535.45 | 1.60 | 0.25 | 535.36 | |
| R4G5(S180) | 1 | 29.0 | 534.18 | 1.25 | 0.34 | 533.91 | 0.662 |
| R3G4(S180) | 5 | 24.2 ± 2.5 | 531.49 ± 0.77 | 1.47 ± 0.14 | 0.43 ± 0.10 | 531.43 ± 0.86 | 0.721 ± 0.010 |
| R2G3(A180) | 3 | 28.0 ± 6.1 | 528.53 ± 0.72 | 1.77 ± 0.05 | 0.23 ± 0.11 | 528.78 ± 0.71 | 0.770 ± 0.005 |
| R1G2(A180) | 3 | 32.0 ± 6.8 | 527.76 ± 1.09 | 1.69 ± 0.07 | 0.24 ± 0.05 | 527.36 ± 0.93 | 0.801 ± 0.056 |
| R2G3+G(A180) | 2 | 28.0 ± 4.0 | 529.66 ± 0.07 | 1.41 ± 0.13 | 0.42 ± 0.14 | 529.67 ± 0.18 | 0.801 |
| R1G2+G(A180) | 1 | 45.0 | 529.69 | 1.83 | 0.61 | 529.99 | |
| R1G2, R2G3 & G(A180) | 9 | 31.2 ± 3.3 | 528.65 ± 0.47 | 1.67 ± 0.06 | 0.32 ± 0.06 | 528.64 ± 0.49 | 0.791 ± 0.026 |
| All | 40 | 28.1 ± 1.2 | 1.48 ± 0.05 | 0.37 ± 0.03 | |||
Summary of the mean results for different genotypes and groups: best-fitting photopigment λmax values, lens densities at 400 nm, and peak macular densities (at 460 nm) for the fit of the full spectrum model (Eqs. 1, 2, and 3a); the best-fitting photopigment λmax values and errors for the fit of the partial spectrum (≥520 nm) model (Eqs. 1, 2, and 3b); and the mean slopes of the regression lines fit to the Rayleigh matches. The SEMs are given. *The mean ages and the numbers of subjects (n) refer to the subjects who participated in the spectral sensitivity measurements.
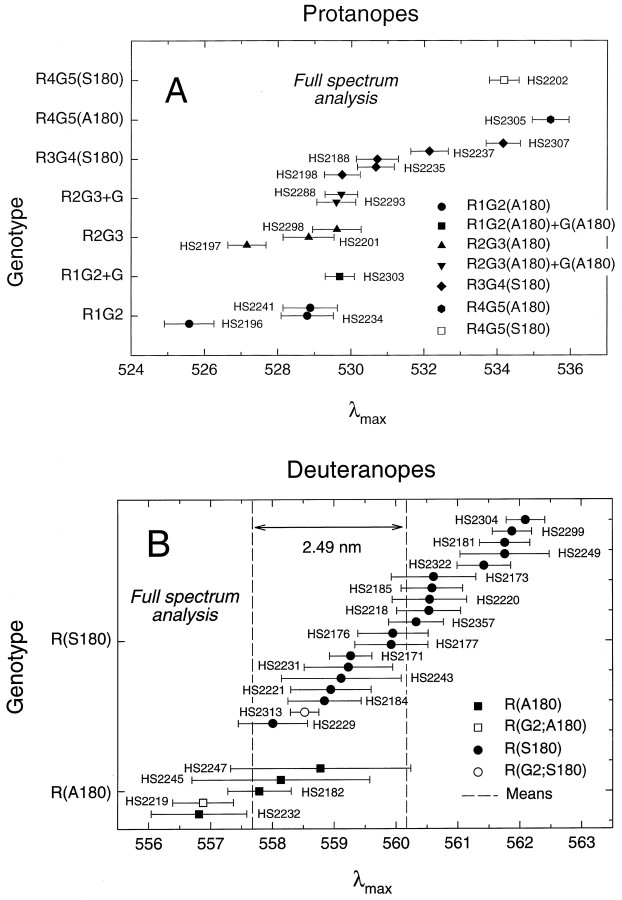
Fig. 7.
λmax values for red, green, or red-green hybrid pigments for individual dichromats grouped by genotypic class. The values shown here are from the full spectrum analysis with best-fitting lens and macular pigment corrections (see Table 3). The error bars indicate the SEs of the λmax estimates. A, Protanopes: R1G2(A180),filled circles; R1G2(A180)+G(A180), filled square; R2G3(A180), filled triangles; R2G3(A180)+G(A180), filled inverted triangles; R3G4(S180), filled diamonds; R4G5(A180) gene,filled hexagon; and R4G5(S180), open square. B, Deuteranopes: R(A180), filled squares; R(G2;A180), open square; R(S180),filled circles; and R(G2;S180), open circle. The vertical dashed lines represent the mean λmax estimates for the R(A180) and R(G2;A180) observers (left line, 557.68 nm) and R(S180) and R(G2;S180) observers (right line, 560.17 nm). The spectral difference between the two means is 2.49 nm.
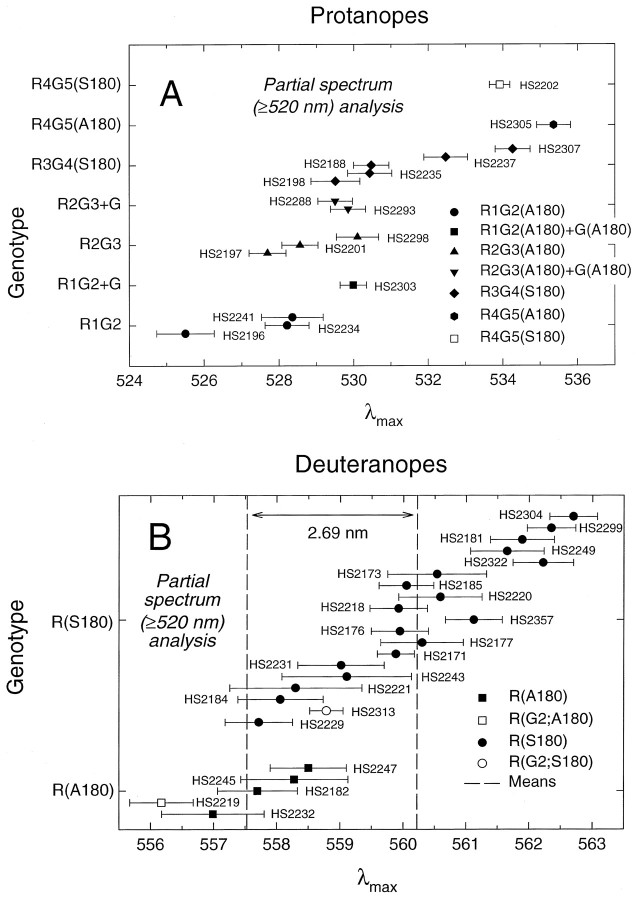
Fig. 8.
λmax values for red, green, or red-green hybrid pigments for individual dichromats grouped by genotypic class. The values shown here are from the partial spectrum (≥520 nm) analysis with mean lens pigment corrections (see Table 4). The error bars indicate SEs of the λmax estimates.A, Protanopes. B, Deuteranopes. The vertical position of each subject’s data point is the same as in Figure 7. Symbols are as in Figure 7. Thevertical dashed lines represent the mean λmax estimates for R(A180) and R(G2;A180) observers (left line, 557.53 nm) and R(S180) and R(G2;S180) observers (right line, 560.22 nm). The spectral difference between the two means is 2.69 nm.
Fig. 9.
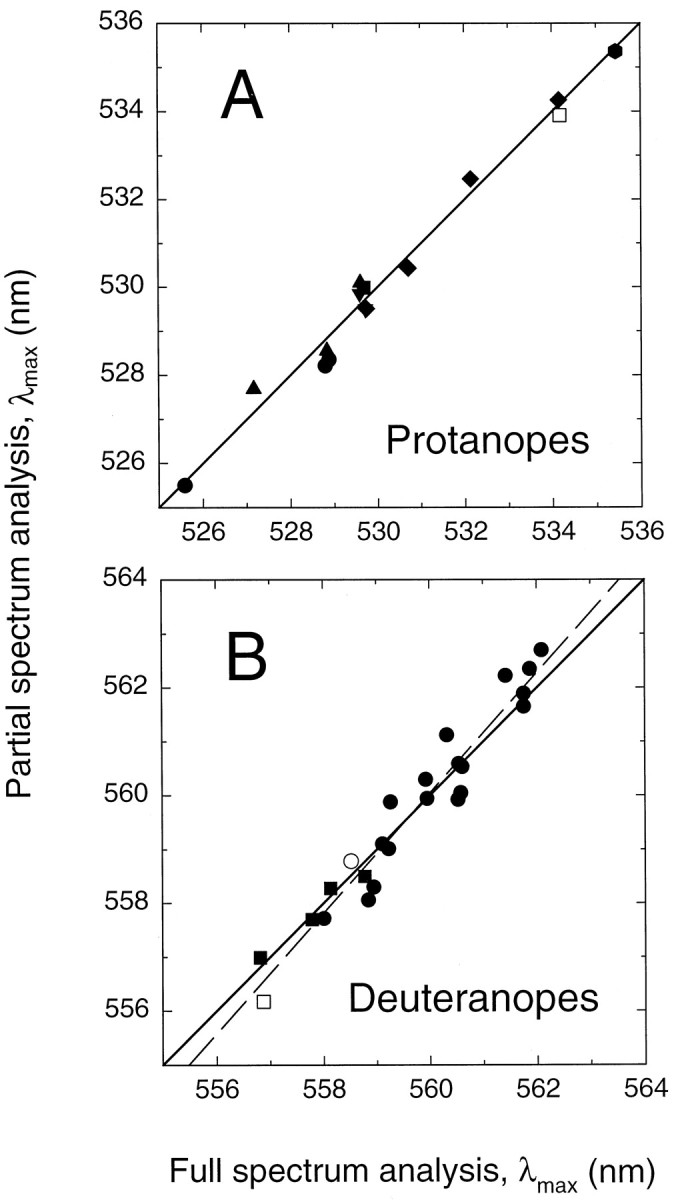
λmax values obtained from the full spectrum analysis plotted against those obtained from the partial spectrum (≥520 nm) analysis. The continuous diagonal lines (with slopes of 1) indicate perfect agreement between the two analyses. The regression line for protanopes (A) has a slope of 1.00 and is indistinguishable from the diagonal line, whereas that for deuteranopes (B) has a slope of 1.12, as shown by thedashed line. Symbols are as in Figures 9and 10. Perfect agreement should not be expected between the two analyses because mean, rather than individual, macular and lens densities are used in the partial spectrum, but not full spectrum, analysis.
We note that fits using just the long wavelength portion of the spectral sensitivity functions (Stockman and Sharpe, Baylor et al., 1987), in which individual differences in lens and photopigment density differences play little or no little role, are impracticable for our data. The problem is that we do not know precisely the absolute differences in sensitivity between the functions (i.e., their vertical positions relative to each other). We cannot tell—from just the long wavelength part of the spectrum—whether the difference between two functions is attributable to a difference in absolute sensitivity (vertical shift) or a difference in λmax (horizontal shift). We overcame this problem by fitting a photopigment template (Eq. 1) that includes, for both the partial spectrum and full spectrum fits, the peak of the photopigment curve as well as its long wavelength slope. Such fits allow us to determine both the vertical and the horizontal shifts together.
Below (see Tables 3-5), we use standard formats to define the results of the two fits. We tabulate (1) the position of the fitted photopigment template as its λmax in nanometers after the best-fitting shift (Δx), (2) the best-fitting lens density as the density at 400 nm, and (3) the macular density as the peak density of the pigment (at 460 nm). The SEs associated with each fit parameter also are given, along with the root mean-squared (rms) errors of the fits for each subject. The vertical shifts (c andcn), because they are of little interest in the context of this paper, are not tabulated individually. The mean value of cn is −8.17 log quanta · sec−1 · degree−2 (−8.10 for protanopes and −8.22 for deuteranopes) with a SE of 0.08, whereas the mean value of c is −8.17 log quanta · sec−1 · degree−2 (−8.14 for protanopes and −8.19 for deuteranopes) with a SE of 0.06. These values indicate the mean peak flicker photometric sensitivities on the 420 nm background field.
It is important to recognize that the spectral shifts (Δx) and the photopigment template [POD(x)] are calculated in log nanometers. Thus, the spectral shift in nanometers increaseswith wavelength. For example, a shift of 2.49 nm (full spectrum analysis; see Fig. 7B) or 2.69 nm (partial spectrum analysis; see Fig. 8B) between the R(S180) and R(A180) photopigments at their λmax becomes a shift of 3.08 nm and 3.33 nm, respectively, at 690 nm.
Systematic errors in both fits would be expected if different red-green photopigments are not shape-invariant when plotted against log10(λ) or if the peak photopigment optical density varies with genotype. Such errors would cause small shifts in the λmax estimates between genotypes but would have little effect on estimates within genotypes. Individual differences in photopigment optical density within a genotype will, of course, increase the variability of the λmax estimates within that group. As noted above, small errors in the secondary fit should be expected because of individual differences in lens pigment density.
RESULTS
Screening of dichromat subjects
Ninety-four dichromat males were recruited for this study. To qualify, they needed to produce a Rayleigh match by merely adjusting the intensity of the yellow light, regardless of the red/green ratio (Rayleigh, 1881). This is consistent with light absorption in this region of the spectrum being mediated by a single type of photoreceptor. To minimize confounding effects caused by age-related yellowing of the lens (Ruddock, 1965), we limited enrollment to young adults (average age, 28 years old; range 16–45). If we assume 25% efficient recruitment of subjects and a frequency of dichromacy among European males of ∼2% (Post, 1962), then we calculate that this study required a base population of 120,000 people.
Analysis of visual pigment gene structure
Dichromacy can arise from either intragenic or intergenic recombination within the tandem array of visual pigment genes (see Fig.1) (Nathans et al., 1986b; Deeb et al., 1992). In the simplest cases the product of recombination contains either a single red pigment gene as a result of intergenic recombination (producing deuteranopia, the loss of green cone sensitivity; abbreviated here G−R+), or a single 5′red-3′green hybrid gene as a result of intragenic recombination (producing protanopia, the loss of red cone sensitivity; abbreviated here G+R−). We will refer to subjects who carry such an arrangement as “single-gene dichromats.” As described more fully below, the heterogeneity in 5′red-3′green crossover points results in a variety of green-like pigments in G+R− dichromats, some of which differ from the green pigments of normal trichromats. In a few more complex cases the sequences suggest multiple recombination events. The location of the 5′ end of the red pigment gene at the edge of the visual pigment gene array precludes the production of the reciprocal hybrid—i.e., a 5′green-3′red gene—in an X chromosome that carries only a single gene. 5′green-3′red hybrid genes are found in place of, or in addition to, the normal green pigment gene(s) in G′R+ subjects (Nathans et al., 1986b; Deeb et al., 1992).
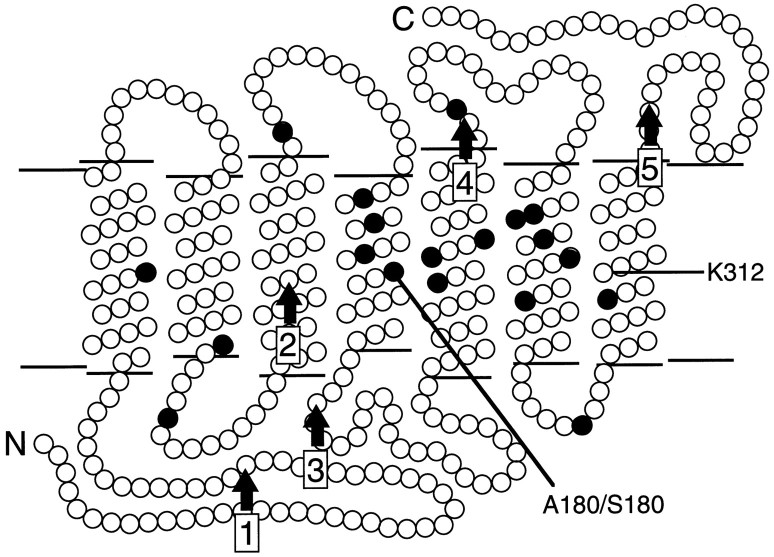
The red and green visual pigment genes each consist of six coding exons. Coding region sequence differences between these genes are confined to exons 2–5, which encode the seven membrane-embedded α-helices that together form the chromophore binding pocket. The present study confirms earlier observations that most intragenic recombination events between red and green pigment genes occur within intron sequences, presumably because of the ∼10-fold greater size of the introns as compared with the exons and the paradoxically greater DNA sequence similarity of the red and green pigment gene introns as compared with the exons (Shyue et al., 1994). Thus, in general, red-green hybrid genes contain some number of contiguous exons from one end of a red pigment gene joined to the remaining exons from the other end of a green pigment gene. In this paper, each hybrid pigment or the gene that encodes it is referred to by an abbreviation that reflects the origin of its exons and the identity of the polymorphic residue (alanine or serine) at position 180 in the third exon. For example, R4G5(A180) is a hybrid pigment encoded by a gene in which exons 1–4 are derived from a red pigment gene, exons 5 and 6 are derived from a green pigment gene, and position 180 is occupied by alanine. Red pigment genes are designated R(A180) or R(S180) to indicate the presence of alanine or serine, respectively, at position 180. Several of the pigment gene sequences reported here reveal a green pigment gene exon 2 embedded within a red pigment gene or a 5′red-3′green hybrid gene. In these cases the identity of the second exon is indicated in parentheses; for example R(G2;A180) represents a gene in which exons 1 and 3–6 are derived from a red pigment gene, exon 2 is derived from a green pigment gene, and position 180 is occupied by alanine. The fact that sequence differences between red and green pigment genes are confined to exons 2–5 implies that a R1G2 hybrid gene encodes a de facto green pigment.
Genomic DNA from each subject was analyzed first by Southern blot hybridization with a cDNA probe from exons 1 and 2 to determine the number of X chromosome visual pigment genes (see Fig. 1, Table1). As shown in Figure 1C, this probe reveals two fragments from the red pigment gene and two from the green pigment gene that differ as a result of sequence variation in the first intron and in 5′ flanking DNA. The 5′ hybridizing fragment from the red pigment gene, designated Br, is present as a single copy in all trichromat and dichromat X chromosomes (Nathans et al., 1986a,b). Intergenic or intragenic recombination events that reduce the array to a single gene result in the loss of fragment Bg and the loss of either Cr or Cg, depending on the exact location of the point of crossover.
Table 1.
Number of X-chromosome visual pigment genes among 94 dichromat males
| Phenotype | G−R+ | G+R− | all dichromats |
| Number of subjects with one gene | 28 | 13 | 41 |
| Number of subjects with >1 gene | 29 | 24 | 53 |
| Total number of subjects | 57 | 37 | 94 |
The assignment of dichromacy as G−R+(deuteranopia) or G+R− (protanopia) was made from the slope and intercept of the Rayleigh matches. The assignment of gene number was made by Southern blotting, as described in Figure 1 and in Materials and Methods, and was confirmed by sequencing for all single-gene dichromats.
As expected from previous studies (Nathans et al., 1986b; Deeb et al., 1992), multiple genotypes were found for both G+R− subjects and G−R+ subjects. In all, 41 of 94 dichromats (44%) were found to carry a single X-linked visual pigment gene. From these single-gene dichromats, DNA encompassing exons 2–5 was amplified and sequenced by using primers flanking each exon. In each of the 41 cases, a unique sequence was obtained for the four exons, consistent with the assignment of a single gene by Southern blotting. Exons 2–5 also were amplified and sequenced from four dichromats for which the Southern blot patterns predicted two or more genes (subject numbers 42–45 in Table2). Three of the four (HS2288, HS2293, and HS2312) show a superposition of different DNA sequences; the fourth (HS2303) carries two or more genes with identical coding region sequences.
A comparison of the deduced amino acid sequences from the 45 dichromats shows variations at 19 positions, most of which can be associated with either the normal green or the normal red pigment (Table 2). In each case the variable position encodes one of two possible amino acids, and most of these are located within the predicted transmembrane α-helices (Fig. 3). Previous work has implicated amino acid substitutions at three of these positions—180, 277, and 285—as major determinants of the difference in spectral sensitivity between red and green pigments (Neitz et al., 1991; Merbs and Nathans, 1992a,b, 1993; Asenjo et al., 1994). Amino acid substitutions at several other locations play a lesser role in spectral tuning (Merbs and Nathans, 1992b; Asenjo et al., 1994).
Fig. 3.
Schematic diagram of the human red and green pigments in the membrane. The N terminus (N) faces the extracellular space, and the C terminus (C) faces the cytosol. Eachfilled circle represents a position that differs between red and green pigments in the set of sequences studied here; these are positions 65, 111, 116, 153, 171, 174, 178, 180, 230, 233, 236, 249, 274, 275, 277, 279, 285, 298, and 309. The locations of lysine312 (K312), the site of covalent attachment of 11-cis retinal, and the polymorphic serine/alanine at position 180 (A180/S180) are indicated. The five intron positions are indicated byvertical arrows.
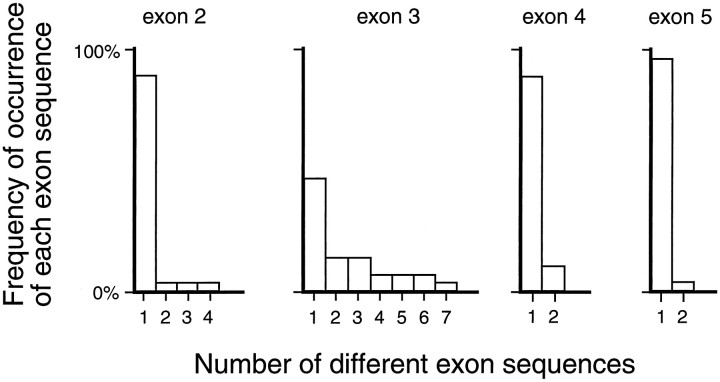
In accordance with the observations of Winderickx et al. (1993) on visual pigment gene arrays in normal trichromats, we find that exon 3 is more variable than exons 2, 4, or 5. Except for position 180, the highly polymorphic nature of the amino acids within exon 3 makes it impossible to discern whether any particular exon 3 is derived from a red or a green pigment gene, as can be seen by comparing the exon 3 sequences listed in Table 2. A quantitative representation of this variability among red pigment genes is shown in Figure4, in which the number and frequency of distinct amino acid sequences is plotted for each exon for those 28 G−R+ dichromats who carry a single red pigment gene. This group of subjects is presumed to carry a representative sample of normal red pigment genes, having lost their green pigment genes by intergenic recombination.
Fig. 4.
Distribution by exon of amino acid sequence variation in 28 red pigment genes (subjects 1–28 in Table 1). This tabulation includes all subjects with genotypes R(A180), R(S180), R(G2;A180), or R(G2;S180). Each bar in the histogram represents a distinct amino acid sequence for a given exon, and these are plotted on the horizontal axis for exons 2–5. The vertical axis shows the frequency of each type of sequence. Exon 3 shows greater polymorphism than exons 2, 4, or 5 do.
Current estimates suggest that among human red pigment genes ∼62% have serine and 38% have alanine at position 180, whereas among green pigment genes ∼9% have serine and 91% have alanine at position 180 (Winderickx et al., 1993). Our finding, among 28 G−R+ single-gene dichromats, of 21 R(S180) or R(G2;S180) genes and 7 R(A180) or R(G2;A180) genes is approximately consistent with these published allele frequencies for the red pigment gene. This distribution of the alanine/serine polymorphism is relevant to the assignment of the origin of exon 3 among red-green hybrid genes in which crossover occurred between exons 2 and 3 or between exons 3 and 4. Among red-green hybrid genes that carry serine at position 180, exon 3 (within which codon 180 resides) is more likely to derive from a red pigment gene, whereas among red-green hybrid genes that carry an alanine at position 180, exon 3 may have originated from either a green or a red pigment gene. Among subjects with R2G3 or R3G4 genotypes, we will, as a convention, refer to those genes that have serine at position 180 as R3G4 hybrids and refer to those genes that have alanine at position 180 as R2G3 hybrids.
Rayleigh matches
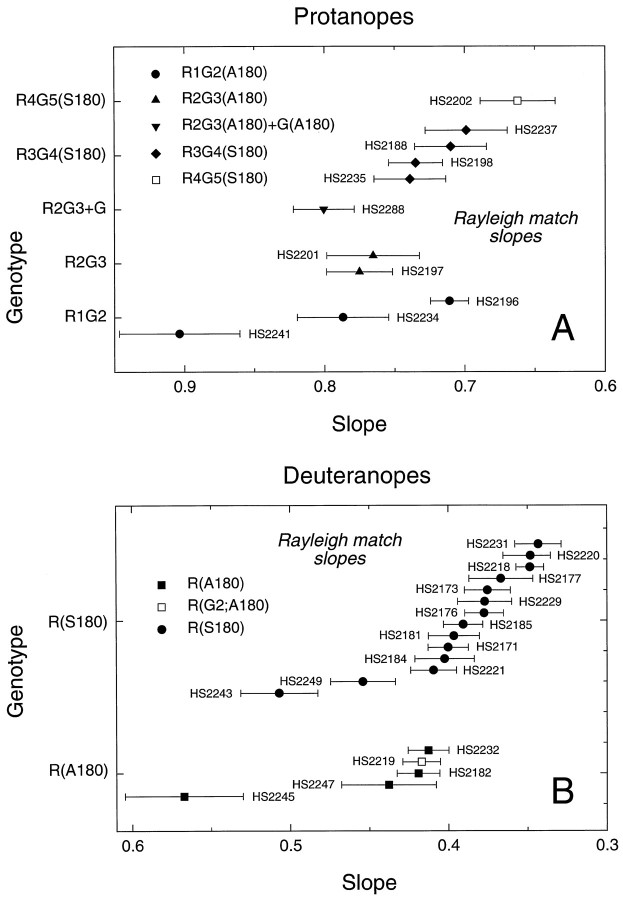
A comparison of the slopes of the regression lines fit to the Rayleigh matches for 29 single-gene dichromats tested with a König–Helmholtz colorimeter is shown in Figure5A for protanopes and Figure5B for deuteranopes. The values, which also are tabulated in Table 4, below, have not been corrected for absorption by the lens or macular pigment; the latter should be minimal at the wavelengths used (>546 nm; Wyszecki and Stiles, 1982). A progressive increase in the slopes of the regression lines fit to the Rayleigh matches, reflecting an increase in the ratio of red to green light required to match the yellow standard, is seen for subjects whose X-linked pigment contains progressively more green pigment-derived sequences. This is apparent in comparing the red pigments R(A180) and R(S180) and the 5′red-3′green hybrid pigments R4G5, R3G4, R2G3, and R1G2. The effect of alanine versus serine at position 180 is reflected in the differences in the slopes of the R(S180) and the R(A180) subjects. The former have a mean slope of 0.396, and the latter have a mean slope of 0.459, with some overlap in the distribution of individual data points. No significant difference is apparent between the mean slopes of the three R1G2(A180) subjects and the two R2G3(A180) subjects, nor are there significant differences between the slopes obtained for those single-gene dichromats who carry a red pigment gene versus those two subjects who carry a red pigment gene in which the second exon has been replaced by a second exon derived from a green pigment gene [i.e., R(G2;A180) and R(G2;S180)]. These data suggest that sequence differences in exon 2 play little or no role in determining the spectral differences among red and green pigments. In general, however, the Rayleigh matches provide limited information regarding spectral sensitivity because only three wavelengths are sampled.
Fig. 5.
Slopes of the regression lines fit to the Rayleigh matches obtained with a König–Helmholtz colorimeter for 29 single-gene dichromats. Data from protanopes are shown in A, and data from deuteranopes are shown in B. The green, yellow, and red primaries for this instrument were 546, 590, and 674 nm, respectively. The data have not been corrected for preretinal absorption. An increase in slope is apparent for subjects with pigments predicted to absorb at shorter wavelength. The mean values for the slopes of R(A180) and R(G2;A180) subjects are 0.452 ± 0.30, and for R(S180) subjects are 0.393 ± 0.012, a significant difference. The mean values for the slopes of R1G2(A180) and R2G3(A180) subjects, 0.801 ± 0.056 and 0.770 ± 0.05, respectively, are not significantly different (see Table 5).
Spectral sensitivity measurements
Foveal spectral sensitivities were determined in 40 subjects (37 single-gene and 3 multiple-gene dichromats) by heterochromatic flicker photometry. As noted in Materials and Methods, a reference light of 560 nm was alternated at a rate of 16 or 25 Hz with a superimposed test light, the wavelength of which was varied in 5 nm steps from 400 to 700 nm. Subjects found the radiance of the test light that eliminated or “nulled” the perception of flicker produced by the alternation of the two lights. Because the blue cones are desensitized by the background (and in any case make little contribution to flicker photometry under most conditions; Eisner and MacLeod, 1980;Stockman et al., 1991) and the rods are saturated, the nulled test and reference lights produce an equivalent level of activation of the remaining single class of red, green, or red-green hybrid cone in each single-gene dichromat. The radiance of the test light required to null the reference light as a function of wavelength is therefore an estimate of the spectral sensitivity of each subject’s single longer wavelength cone type.
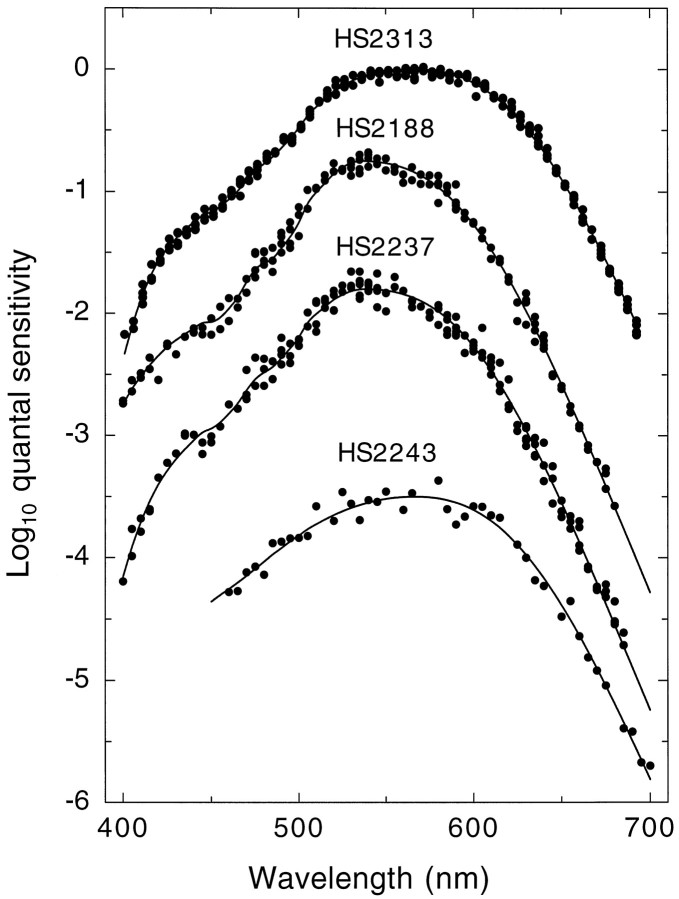
Figure 6 shows representative examples of the heterochromatic flicker data (symbols) for four subjects. The data sets for subjects HS2313, HS2188, and HS2237 are examples of the best, intermediate, and worst fits, respectively. The data set for subject HS2243 is an example of an intermediate fit, but with limited data. We have similarly limited data for 5 of the 40 subjects.
Fig. 6.
Representative examples of cone spectral sensitivity data (filled symbols) for four single-gene dichromats: HS2313 (top), HS2188 (upper middle), HS2237 (lower middle), and HS2243 (bottom), together with the fits of the visual pigment template (continuous lines). Data are from several single runs. The data sets for HS2313, HS2188, and HS2237 are examples of the best, intermediate, and worst fits, respectively, of the visual pigment templates to the data. The data set for subject HS2243 is an example of a subject for whom we have limited data. For clarity, the data sets have been displaced vertically.
We analyzed the spectral sensitivity measurements for each observer to estimate the wavelength of maximum sensitivity (λmax) of his underlying longer wavelength cone photopigment by using either the full range of measured wavelengths (400–700 nm), the “full spectrum” analysis (see Eqs. 1, 2, and 3a, below), or wavelengths ≥520 nm, the so-called “partial spectrum” analysis (see Eqs. 1, 2, and 3b, below).
In the full spectrum fit we determined the best-fitting macular and lens pigment densities as well as the spectral position of the underlying pigment template. The results are tabulated in Table3, along with each subject’s age and the rms error of the fit. Also tabulated are the SEs for each fitted parameter. A compilation of the λmax values and the SEs is shown in graphic form in Figure 7 for each protanope (Fig. 7A) and deuteranope (Fig.7B). The full spectrum fits to the data for subjects HS2313, HS2188, HS2237, and HS2243 are shown by the continuous lines in Figure6.
In the partial spectrum fit we fixed the macular and lens pigment densities at their mean best-fitting values and determined only the spectral position of the underlying pigment template. The results are tabulated in Table 4, and a compilation of the λmax values and the SEs is shown in Figure8. The partial spectrum fit also acts as a control for the full spectrum fit. Any distortion of the full spectrum λmax estimate caused by optimizing the lens and macular pigment densities should result in clear differences between the full spectrum and partial spectrum. These differences will be in addition to the small discrepancies that arise because mean rather than optimal lens and macular densities are assumed in the partial spectrum fit (see Materials and Methods).
Figure 9 presents a comparison of the λmax estimates obtained by the two methods. The estimates for the protanopes lie close to a straight line of unit slope (continuous line), which indicates good agreement between the two methods. The disagreements are <0.59 nm, with a mean of −0.06 nm (and a mean of 0.29 nm for the absolute differences). The estimates for deuteranopes fall along a slightly steeper line, with a best-fitting slope (Fig. 9B, dashed line) of 1.12, which suggests that the full spectrum fit, relative to the partial spectrum fit, slightly underestimates the λmax of longer wavelength red cone photopigments but overestimates that of shorter wavelength ones. In this case the discrepancies are <0.80 nm, with a mean of 0.00 nm (and a mean of 0.37 nm for the absolute differences).
We consider first the individual deuteranopic spectral sensitivities (see Figs. 7B, 8B, 9B, Tables3, 4). The estimated λmax values for four R(A180) subjects span a range of 2 nm, whereas those for 18 R(S180) subjects span a range of 4 nm with a partial overlap between the two populations. The λmax values of these various red pigments range from 556.8 to 562.1 nm (full spectrum analysis; Table 3) or from 556.2 to 562.7 (partial spectrum analysis; Table 4). Among the 10 R(S180) subjects with identical amino acid sequences (HS2173, HS2176, etc; see Table 2), the λmax values average 560.6 ± 0.4 with a range of 558.0 to 562.1 nm (full spectrum analysis; Table 3) and 560.4 ± 0.4 nm with a range of 557.7 to 562.7 nm (partial spectrum analysis; Table 4). Because this subject-to-subject variation occurs within the context of identical cone pigment sequences, it provides a measure of the precision with which these data and the analyses can be used to define the cone spectral sensitivity associated with a given pigment type. The λmax values of the two pigments that carry a green exon 2 embedded within an otherwise red pigment gene [R(G2;A180) and R(G2;S180)] are located on the short-wave limb of the corresponding R(A180) or R(S180) distributions, respectively. These data suggest that in this context a change in exon 2 from R2 to G2 shifts the λmax to slightly shorter wavelengths, but in terms of the distributions the effect is not significant. If we include the two types of R(G2) pigments together with their corresponding red pigments, then the means of the λmax values for the A180 and S180 red pigments have a separation of 2.49 nm (full spectrum analysis; Table 3) and 2.69 nm (partial spectrum analysis; Table 4). The variation in λmax among the 17 R(S180) subjects does not correlate in any simple way with the other polymorphic amino acids in exon 3.
The λmax values for 13 single-gene protanopes with 5′red-3′green hybrid pigments and three with two or more green or green-like pigments [i.e., R1G2+G(A180) and R2G3+G(A180)] are within the interval 525.6–535.5 nm (full spectrum analysis; Fig.7A, Table 3) or 525.5–535.4 nm (partial spectrum analysis; Fig. 8A, Table 4). These data confirm the major role of exon 5 sequence differences in determining the spectral separation between red and green pigments (Neitz et al., 1991; Merbs and Nathans, 1992b, 1993; Asenjo et al., 1994). Although the number of individuals with pigments of each type is small and there is variation of up to 3.5 nm in λmax values within a genotype, the spectral sensitivity measurements show a clear trend toward longer values of λmax for those pigments with a greater number of red pigment exons. In particular, the individual data indicate that there is, on average, a ∼1 nm spectral shift to longer wavelengths (range, −0.5 to 2.5 nm) associated with a change in exon 2 from green to red pigment sequences, a value similar to that described above for the red pigment—and not statistically significant. This value for the average spectral shift associated with differences in exon 2 is influenced significantly by one subject (HS2196) whose λmax (525.6 nm) is unusually short; without HS2196, the mean shift is <1 nm. For exon 4 the change from green to red pigment sequences is associated with, on average, a ∼2.8 nm spectral shift (range, −0.5 to 6 nm) to longer wavelengths. Finally, these data suggest that polymorphic amino acid sequence variations in exon 3 are associated with an average spectral shift of ∼3.3 nm, with a range from −0.5 to 7 nm.
To examine the effect of serine or alanine at position 180 among 5′red-3′green hybrid pigments, we can make a comparison between subjects who have either R2G3 or R3G4 genotypes. These genotypes can be considered together because, as noted above, except for position 180, the polymorphic nature of the other amino acids within exon 3 makes it impossible to discern whether it is derived from a red or a green pigment gene. A comparison between the five subjects with a R3G4(S180) pigment and the five subjects with R2G3(A180) or R2G3+G(A180) pigments shows a mean red shift of 2.5 nm (full spectrum analysis; Table 3) or 2.3 nm (partial spectrum analysis; Table 4) caused by the presence of serine at position 180. By contrast, the single subject with a R4G5(S180) pigment has a λmax that is 1.5 nm blue-shifted relative to the single subject with a R4G5(A180) pigment. Given the observations just noted with larger numbers of subjects that indicate a red-shifting effect of serine at position 180 and the several nanometer variability in λmax values among subjects with identical genotypes, we think that experimental variability and/or an unusual photopigment optical density may account for the unexpected relationship between this single pair of λmax values.
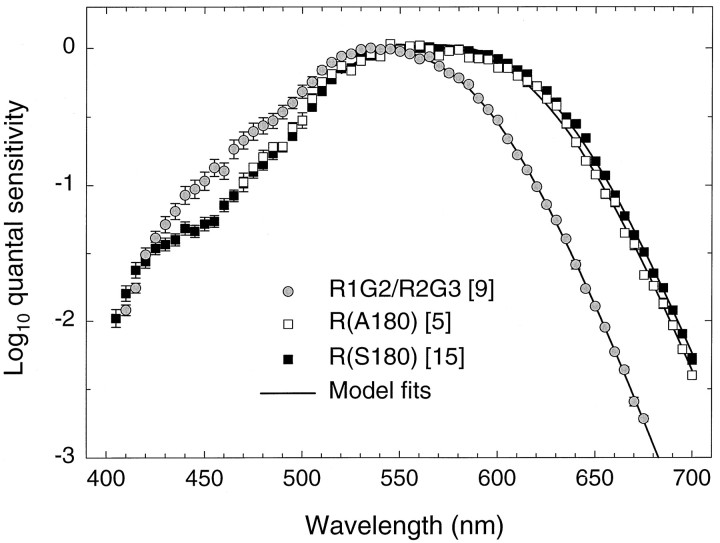
As well as individual data, we can consider mean spectral sensitivity data. Figure 10 shows data averaged across (1) five observers with a single R(A180) (n = 4) or R(A180;G2) gene (n = 1) (white squares); (2) 15 observers with a single R(G2;S180) (n = 1) or R(S180) (n = 14) gene (black squares); and (3) nine observers with a single R1G2 (n = 3) or R2G3 (n = 3) gene or with multiple R1G2+G (n = 1) or R2G3+G (n = 2) genes (gray circles). These mean analyses, with the exception of (1), include only subjects who made measurements throughout the spectrum. We have averaged together data from subjects whose genes differ only in their second exon (green vs red), because the substitution of a green for a red exon 2 does not alter spectral sensitivity significantly (Table5). Table 5 lists the results of the full spectrum and partial spectrum fits and the Rayleigh matches averaged across genotypes.
Fig. 10.
Averaged spectral sensitivity measurements for green, red(A180), and red(S180) pigments. Shown are mean data for nine observers with a single R1G2 (n = 3) or a single R2G3 (n = 3) gene or with multiple R1G2+G (n = 1) or R2G3+G (n = 2) genes (gray circles), five observers with a single R(G2;A180) (n = 1) or R(A180) (n = 4) gene (white squares), and 15 observers with a single R(G2;S180) (n = 1) or a single R(S180) (n = 14) gene (black squares). Three of the five R(A180) group made measurements only at wavelengths >470 nm, so the mean data are restricted to that region. The other means include data only from subjects who made measurements throughout the spectrum. Error bars indicate SE. Thecontinuous lines fit to each set of data ≥520 nm are fits of the partial spectrum model (Eqs. 1, 2, and 3b) to the mean data. See Materials and Methods for details.
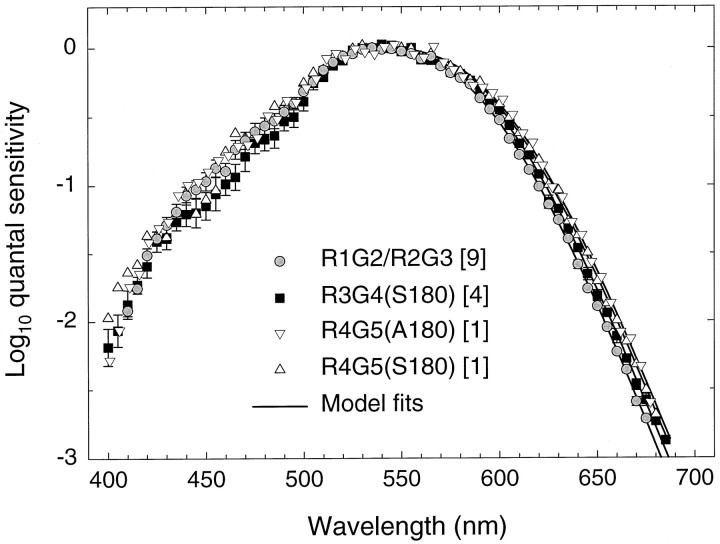
Figure 11 shows the mean spectral sensitivity data for protanopes with red-green hybrid photopigments. The gray circles are the same as in Figure 10; that is, they are the averages of data from the nine observers with single or multiple R1G2, R2G3, or green photopigment genes. The black circles denote the averages of data from the four observers with a single R3G4(S180) gene, whereas the white inverted triangles and white triangles are the individual data for the single observers with a R4G5(A180) and a R4G5(S180) gene, respectively.
Fig. 11.
Averaged spectral sensitivity measurements for red-green hybrid pigments. Shown are mean data for nine observers with R1G2 or R2G3 genes [a single R1G2 (n = 3) or a single R2G3 (n = 3) gene or multiple R1G2+G (n = 1) or R2G3+G (n = 2) genes] (gray circles), for four observers with a single R3G4(S180) gene (black squares), for one observer with a single R4G5(A180) gene (white inverted triangle), and for one observer with a single R4G5(S180) gene (white triangle). The mean data are only from subjects who made measurements throughout the spectrum. Of the protanopes, only one R3G4(S180) observer did not make complete measurements. Error bars indicate SE. The continuous lines fit to each set of data ≥520 nm are fits of the partial spectrum model to the mean or individual data. See Results for details.
The continuous lines fit to each set of data in Figures 10 and 11 are fits of the partial spectrum model (Eqs. 1, 2, and 3b) to the mean data. The λmax values of the fitted templates and the SEs of the fit are 557.5 ± 0.4 nm [R(A180)]; 560.4 ± 0.3 nm [R(S180)]; 528.56 ± 0.2 nm [R1G2/R2G3]; and 531.5 ± 0.3 nm [R3G4(S180)]. For the individual R4G5(A180) and R4G5(S180) data, the fits are as also given in Tables 4 and 5: 535.4 ± 0.4 nm [R4G5(A180)] and 533.9 ± 0.3 nm [R4G5(S180)]. The fits to the mean data agree well with the averages of the individual fits (see Table 5), although the R(S180) versus R(A180) difference of 2.9 nm is slightly larger than the differences estimated from the individual data (2.7 and 2.5 nm for the partial and full spectrum fits, respectively).
DISCUSSION
Genotype and phenotype among single-gene dichromats
By correlating the X-linked visual pigment sequences and color vision phenotypes of single-gene dichromats, we have characterized the spectral sensitivities in vivo of cones containing the normal human red and green pigments, as well as a series of 5′red-3′green hybrid pigments. We have determined psychophysically the spectral sensitivities of 37 single-gene dichromats and three dichromats who carry more than one X-linked visual pigment gene. Other investigators have reported DNA sequences and spectral sensitivities by using electroretinographic methods for small numbers of dichromats with one or more visual pigment genes (Neitz et al., 1995b), and our results are in approximate agreement with their data. The study of single-gene dichromats greatly simplifies the correlation of genotype and phenotype, and this approach should lead eventually to a comprehensive catalog of the spectral sensitivities of the various red, green, and 5′red-3′green hybrid pigments. The most significant limitation of using single-gene dichromats is that they do not carry 5′green-3′ red hybrid genes, which therefore can be studied only in G′R+anomalous trichromats.
Amino acid substitutions and spectral shifts
The psychophysical data reported here indicate that 5′red-3′green hybrid genes encode a range of pigments with altered spectral sensitivities and that in every case examined the spectral sensitivity of the hybrid is between those of the normal red and green pigments. The data further indicate that the spectral sensitivity of the hybrid depends on the position of the crossover and on the identity of other polymorphic amino acids, principally alanine or serine at position 180. For each exon the set of amino acids normally associated with the red or green pigments produces, respectively, spectral shifts to longer or shorter wavelengths, thus producing a monotonic relationship between the λmax and the fraction of the hybrid pigment derived from the red and green parental pigments.
The primary determinants of the spectral shift are located in exon 5, as seen by the clustering of the absorption maxima of all 5′red-3′green hybrids within 10 nm of the maxima of the normal green pigments (Tables3-5; Figs. 7, 8) (Merbs and Nathans, 1992b; Asenjo et al., 1994). A comparison of the spectral sensitivities of R(A180) and R(S180) with those of R4G5(A180) and R4G5(S180) indicates that red/green sequence differences in exon 5 result in a spectral shift of ∼25 nm (see Figs.7-10). The spectral sensitivity data further suggest that exon 2 contributes on average ∼1 nm (range, −0.5 to 2.5 nm), whereas exon 3 contributes on average ∼3.3 nm (range, −0.5 to 7 nm), exon 4 contributes on average ∼2.8 nm (range, −0.5 to 6 nm), and exon 5 contributes on average ∼24.9 nm (range, 22.2 to 27.6 nm). These results are in approximate agreement with results obtained from recombinant human cone pigments produced in transfected cells and studied in detergent solution (Merbs and Nathans, 1992b; Asenjo et al., 1994) and with inferences based on a comparison of primate visual pigment gene sequences and cone spectral sensitivity curves (Neitz et al., 1991; Ibbotson et al., 1992; Williams et al., 1992).
Over the past 6 years considerable attention has been paid to the importance of the serine/alanine polymorphism at position 180 (Merbs and Nathans, 1992a,b; Winderickx et al., 1992a; Sanocki et al., 1993,1994; Asenjo et al., 1994; Neitz et al., 1995b). All of the reported measurements have shown that serine-containing pigments are red-shifted with respect to alanine-containing pigments, but the magnitude of the shift has been controversial. Previous psychophysical and electroretinographic studies aimed at measuring the spectral shift produced by substitution at this position have been complicated by the presence of additional genes and/or additional amino acid substitutions within the same gene. On the basis of anomaloscope matches, Sanocki et al. (1993, 1994) estimated that the substitution of alanine by serine in the red pigment or within 5′red-3′green hybrid pigments results in a red shift of λmax by 4.3, 3.5, and 2.6–2.7 nm for G−R+ dichromats, G+R− dichromats, and normal trichromats, respectively. Neitz and Jacobs (1990) estimated a red shift of ∼3 nm from Rayleigh match data obtained from 60 normal trichromats. Red shift estimates of 5–7 nm were obtained from five dichromats by electroretinography, but this interpretation was complicated by additional amino acid differences between the pigments (Neitz et al., 1995b). In vitro measurements of recombinant pigments by Merbs and Nathans (1992a,b) showed a red shift of 4.3–4.4 nm in the red pigment and in 5′red-3′green and 5′green-3′red hybrid pigments. By contrast, Asenjo et al. (1994) found a range of red shifts depending on the parental pigment: a 2 nm shift in a 5′red-3′green hybrid and in the green pigment, a 4 nm shift in a 5′green-3′red hybrid, and a 7 nm shift in the red pigment. We do not have an explanation for the discrepancy between the 7 nm value reported by Asenjo et al. and the smaller values reported by Merbs and Nathans and obtained here. The spectral sensitivity curves reported here show a mean separation of 2.5–2.9 nm, depending on which of two methods is used to calculate the spectral sensitivity curve and whether individual or mean data are used. These results are consistent with other psychophysical measures of the variability of the red cone λmax in the normal population (Webster and MacLeod, 1988;Webster, 1992). The individual variation present in each genotypic group emphasizes the importance of averaging data from large numbers of subjects.
Implications for models of dichromacy and anomalous trichromacy
These results have significant implications for current theories of dichromacy. Most importantly, they require a qualification of the classical theory that all dichromats are merely missing one of the normal pigments and retain the others unchanged (the reduction model;Maxwell, 1860; König and Dieterici, 1886). Over one-half of the dichromats analyzed in this study (53/94) carry and presumably express more than one X-linked visual pigment gene. For G−R+ dichromats (deuteranopes) these larger arrays consist of a normal red pigment gene and a 5′green-3′red hybrid gene that encodes a pigment with a spectral sensitivity similar to that of the normal red pigment (Nathans et al., 1986b; Deeb et al., 1992). For G+R− dichromats (protanopes) these larger arrays consist of a 5′red-3′green hybrid gene that encodes a pigment with a spectral sensitivity similar to that of a green pigment and one or more normal green pigment genes (Nathans et al., 1986b; Deeb et al., 1992). Among single-gene dichromats those with G−R+ dichromacy constitute a true reduction type. By contrast, G+R− single-gene dichromats possess any one of several green-like hybrid pigments, and in the present study only 3 of 13 were found to possess a bona fide green pigment (i.e., a pigment encoded by a R1G2(A180) hybrid gene). An additional 3 of 13 possess a R2G3(A180) hybrid pigment, which has an absorbance spectrum that is identical or nearly identical to that of a normal green pigment. However, 7 of 13 G+R− single-gene dichromats reported here possess R3G4 or R4G5 hybrids, which would be predicted to produce G+R′ anomalous trichromacy if found in place of a red pigment gene and in conjunction with one or more normal green pigment genes. Thus, these subjects, who were found in this study to constitute 19% of all G+R−dichromats, can be thought of as “anomalous protanopes.”
The results described here are also relevant to contemporary theories of anomalous trichromacy. Although we have only studied anomalous pigments in dichromats, such pigments can, as noted above, be expressed from more complex gene arrays and, when paired with a normal pigment, produce anomalous trichromacy. Our finding of a diverse family of hybrid pigments is not consistent with the theory that all anomalous trichromats share a single anomalous pigment with a spectral sensitivity at a position intermediate between the normal red and green pigments (De Vries, 1948b; MacLeod and Hayhoe, 1974; Hayhoe and MacLeod, 1976), nor are the data consistent with the theory that there is one anomalous pigment common to all G′R+anomalous trichromats and another anomalous pigment common to all G+R′ anomalous trichromats (Rushton et al., 1973b;Pokorny et al., 1975; Pokorny and Smith, 1977; DeMarco et al., 1992). Finally, the data do not support, in its most extreme form, the theory that there are no distinct anomalous pigments but instead clusters of “normal” red and green pigments that differ in their peak sensitivities, and that anomalous trichromats draw their pigments from the same rather than different clusters (Alpern and Moeller, 1977;Alpern and Wake, 1977). Although this theory fails to account for the existence of bona fide hybrid pigments, it explains the phenotypes of that subset of anomalous trichromats in which the normal and anomalous pigments are distinguished only by the polymorphic alanine versus serine difference at position 180.
Any comprehensive model of anomalous trichromacy must account for the fact that anomalous trichromats differ greatly in the location and range of their Rayleigh matching points (for review, see Pokorny et al., 1979; Mollon, 1997). The data presented here strongly support a model of anomalous trichromacy in which any one of many green-like or red-like anomalous pigments can be paired with either of two major polymorphic versions of the more similar normal pigment (Merbs and Nathans, 1992b; Neitz et al., 1996). As the spectral sensitivities of the normal or the anomalous pigments shift, the midpoint of the Rayleigh match will shift, and as the separation between the spectral sensitivities of the normal and anomalous pigments increases or decreases, the better or poorer will be the subject’s chromatic discrimination.
Implications for the derivation of human cone fundamentals
The traditional method of estimating the red and green cone sensitivities is to use data obtained from G−R+ and G+R− dichromats. This approach is only valid, however, if those dichromacies are truly reduced forms of normal trichromacy in which one of the two normal X-linked pigments is absent and the other is identical in its spectral properties to that in the normal observer. As described above, the observations reported here underscore several important caveats that affect the validity of the reduction hypothesis. Moreover, the existence of polymorphism among normal red or green pigment genes, most especially the serine/alanine polymorphism at position 180, means that even true reduction dichromats constitute a heterogeneous group. It follows that a single set of cone fundamentals will describe accurately the color vision of only a subset of normal trichromats and that in the construction of an average set of fundamentals it is important that the ratio of polymorphic types within the test population match that in the general population. Thus, the most comprehensive approach to the construction of the cone fundamentals would be to determine spectral sensitivity curves for a large number of single-gene dichromats whose visual pigment gene sequences reveal which of the various possible pigments they possess. The averaged spectral sensitivity curves reported here for the R(A180), R(S180), and the green-like R1G2(A180) and R2G3(A180) pigments represent a first step toward this goal.
Footnotes
This work was supported by The Deutsche Forschungsgemeinschaft (Bonn; Grants SFB 430 Tp A6 and Sh23/5-1) and a Hermann und Lilly Schilling Professor award (L.T.S.), the Howard Hughes Medical Institute (J.N.), and the National Eye Institute (A.S.). We thank Drs. Donald MacLeod and Edward Famiglietti and two anonymous reviewers for helpful comments on this manuscript.
Correspondence should be addressed to Drs. Lindsay T. Sharpe, Andrew Stockman, or Jeremy Nathans at the above addresses.
aAmong visual psychophysicists the three cone classes now most often are referred to as long (L), middle (M), and short (S) wavelength-sensitive, according to the relative positions within the visible spectrum to which they are most sensitive. Among molecular biologists an older usage, in which the cones are referred to as red (R), green (G), and blue (B), is more prevalent. In this article, to be consistent and to avoid confusion caused by converting between terminologies, we adhere to the R, G, and B usage throughout. Hybrid visual pigment genes are denoted as 5′red-3′green or 5′green-3′red, to indicate which parental gene contributed sequences for the 5′ end and which contributed sequences for the 3′ end of the hybrid gene.
REFERENCES
- 1.Alpern M, Moeller J. The red and green cone visual pigments of deuteranomalous trichromacy. J Physiol (Lond) 1977;266:647–675. doi: 10.1113/jphysiol.1977.sp011786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alpern M, Wake T. Cone pigments in human deutan colour vision defects. J Physiol (Lond) 1977;266:595–612. doi: 10.1113/jphysiol.1977.sp011784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asenjo AB, Rim J, Oprian DD. Molecular determinants of human red/green color discrimination. Neuron. 1994;12:1131–1138. doi: 10.1016/0896-6273(94)90320-4. [DOI] [PubMed] [Google Scholar]
- 4.Baylor DA, Nunn BJ, Schnapf JL. Spectral sensitivity of the cones of the monkey Macaca fascicularis. J Physiol (Lond) 1987;390:145–160. doi: 10.1113/jphysiol.1987.sp016691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brindley GS. The effects on colour vision of adaptation to very bright lights. J Physiol (Lond) 1953;122:332–350. doi: 10.1113/jphysiol.1953.sp005003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dartnall HJA, Bowmaker JK, Mollon JD. Human visual pigments: microspectrophotometric results from eyes of seven persons. Proc R Soc Lond [Biol] 1983;220:115–130. doi: 10.1098/rspb.1983.0091. [DOI] [PubMed] [Google Scholar]
- 7.Deeb SS, Lindsey DT, Hibiya Y, Sanocki E, Winderickx J, Teller DY, Motulsky AG. Genotype–phenotype relationships in human red/green color vision defects: molecular and psychophysical studies. Am J Hum Genet. 1992;51:687–700. [PMC free article] [PubMed] [Google Scholar]
- 8.DeMarco P, Pokorny J, Smith VC. Full-spectrum cone sensitivity functions for X chromosome-linked anomalous trichromats. J Opt Soc Am [A] 1992;9:1465–1476. doi: 10.1364/josaa.9.001465. [DOI] [PubMed] [Google Scholar]
- 9.De Vries HL. The luminosity curve of the eye as determined by measurements with the flicker photometer. Physica. 1948a;14:319–348. [Google Scholar]
- 10.De Vries HL. The fundamental response curves of normal and abnormal dichromatic and trichromatic eyes. Physica. 1948b;14:367–380. [Google Scholar]
- 11.Drummond-Borg M, Deeb SS, Motulsky AG. Molecular patterns of X chromosome-linked color vision genes among 134 men of European ancestry. Proc Natl Acad Sci USA. 1989;86:983–987. doi: 10.1073/pnas.86.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisner A, MacLeod DIA. Blue sensitive cones do not contribute to luminance. J Opt Soc Am. 1980;70:121–123. doi: 10.1364/josa.70.000121. [DOI] [PubMed] [Google Scholar]
- 13.Enoch JM. Nature of the transmission of energy in the retinal receptors. J Opt Soc Am. 1961;51:1122–1126. doi: 10.1364/josa.51.001122. [DOI] [PubMed] [Google Scholar]
- 14.Enoch JM. Optical properties of the retinal receptors. J Opt Soc Am. 1963;53:71–85. [Google Scholar]
- 15.Hayhoe MM, MacLeod DIA. A single anomalous photopigment? J Opt Soc Am. 1976;66:276–277. doi: 10.1364/josa.66.000276. [DOI] [PubMed] [Google Scholar]
- 16.Hecht S. Brightness, visual acuity and color blindness. Doc Ophthalmol. 1949;3:289–306. doi: 10.1007/BF00162607. [DOI] [PubMed] [Google Scholar]
- 17.Hsia Y, Graham CH. Spectral luminosity curves for protanopic, deuteranopic, and normal subjects. Proc Natl Acad Sci USA. 1957;43:1011–1019. doi: 10.1073/pnas.43.11.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ibbotson RE, Hunt DM, Bowmaker JK, Mollon JD. Sequence divergence and copy number of the middle- and long-wave photopigment genes in Old World monkeys. Proc R Soc Lond [Biol] 1992;247:145–154. doi: 10.1098/rspb.1992.0021. [DOI] [PubMed] [Google Scholar]
- 19.König A, Dieterici C. Die Grundempfindungen und ihre Intensitäts-Vertheilung im Spektrum. Sitz Akad Wiss (Berl) 1886;1886:805–829. [Google Scholar]
- 20.Lamb TD. Photoreceptor spectral sensitivities: common shape in the long-wavelength spectral region. Vision Res. 1995;35:3083–3091. doi: 10.1016/0042-6989(95)00114-f. [DOI] [PubMed] [Google Scholar]
- 21.Macke JP, Nathans J. Individual variation in the size of the human red and green pigment gene array. Invest Ophthalmol Vis Sci. 1997;38:1040–1043. [PubMed] [Google Scholar]
- 22.MacLeod DIA, Hayhoe M. Three pigments in normal and anomalous color vision. J Opt Soc Am. 1974;64:92–96. doi: 10.1364/josa.64.000092. [DOI] [PubMed] [Google Scholar]
- 23.MacNichol EF. A unifying presentation of photopigment spectra. Vision Res. 1986;26:1543–1556. doi: 10.1016/0042-6989(86)90174-4. [DOI] [PubMed] [Google Scholar]
- 24.Mansfield RJW. Primate photopigments and cone mechanisms. In: Fein A, Levine JS, editors. The visual system. Liss; New York: 1985. pp. 89–106. [Google Scholar]
- 25.Maxwell JC. On the theory of compound colours and the relations of the colours of the spectrum. Philos Trans R Soc Lond [Biol] 1860;150:57–84. [Google Scholar]
- 26.Merbs SL, Nathans J. Absorption spectra of human cone pigments. Nature. 1992a;356:431–432. doi: 10.1038/356433a0. [DOI] [PubMed] [Google Scholar]
- 27.Merbs SL, Nathans J. Absorption spectra of the hybrid pigments responsible for anomalous color vision. Science. 1992b;258:464–466. doi: 10.1126/science.1411542. [DOI] [PubMed] [Google Scholar]
- 28.Merbs SL, Nathans J. Role of hydroxyl-bearing amino acids in differentially tuning the absorption spectra of the human red and green cone pigments. Photochem Photobiol. 1993;58:706–710. doi: 10.1111/j.1751-1097.1993.tb04956.x. [DOI] [PubMed] [Google Scholar]
- 29.Mitchell DE, Rushton WAH. Visual pigments in dichromats. Vision Res. 1971;11:1033–1043. doi: 10.1016/0042-6989(71)90110-6. [DOI] [PubMed] [Google Scholar]
- 30.Mollon JD. “… aus dreyerley Arten von Membranen oder Molekuelen”: George Palmer’s legacy. In: Cavonius CR, editor. Color vision deficiencies XIII. Kluwer; Dordrecht, the Netherlands: 1997. pp. 3–20. [Google Scholar]
- 31.Nathans J, Thomas D, Hogness DS. Molecular genetics of human color vision: the genes encoding blue, green, and red pigments. Science. 1986a;232:193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- 32.Nathans J, Piantandida TP, Eddy RL, Shows TB, Hogness DS. Molecular genetics of inherited variation in human color vision. Science. 1986b;232:202–210. doi: 10.1126/science.3485310. [DOI] [PubMed] [Google Scholar]
- 33.Nathans J, Merbs SL, Sung C-H, Weitz CJ, Wang Y. Molecular genetics of human visual pigments. Annu Rev Genet. 1992;26:401–422. doi: 10.1146/annurev.ge.26.120192.002155. [DOI] [PubMed] [Google Scholar]
- 34.Nathans J, Maumenee IH, Zrenner E, Sadowski B, Sharpe LT, Lewis RA, Hansen E, Rosenberg T, Schwartz M, Heckenlively JR, Traboulsi E, Klingaman R, Bech-Hansen NT, LaRoche GR, Pagon RA, Murphey WH, Weleber RG. Genetic heterogeneity among blue cone monochromats. Am J Hum Genet. 1993;53:987–1000. [PMC free article] [PubMed] [Google Scholar]
- 35.Neitz J, Jacobs GH. Polymorphism in normal color vision and its mechanism. Vision Res. 1990;30:621–636. doi: 10.1016/0042-6989(90)90073-t. [DOI] [PubMed] [Google Scholar]
- 36.Neitz J, Neitz M, Kainz P. Visual pigment gene structure and the severity of color vision defects. Science. 1996;274:801–804. doi: 10.1126/science.274.5288.801. [DOI] [PubMed] [Google Scholar]
- 37.Neitz M, Neitz J. Numbers and ratios of visual pigment genes for normal red-green color vision. Science. 1995;267:1013–1016. doi: 10.1126/science.7863325. [DOI] [PubMed] [Google Scholar]
- 38.Neitz M, Neitz J, Jacobs GH. Spectral tuning of pigments underlying red-green color vision. Science. 1991;252:971–974. doi: 10.1126/science.1903559. [DOI] [PubMed] [Google Scholar]
- 39.Neitz M, Neitz J, Grishok A. Polymorphism in the number of genes encoding long-wavelength-sensitive cone pigments among males with normal color vision. Vision Res. 1995a;35:2395–2407. [PubMed] [Google Scholar]
- 40.Neitz M, Neitz J, Jacobs GH. Genetic basis of photopigment variations in human dichromats. Vision Res. 1995b;35:2095–2103. doi: 10.1016/0042-6989(94)00306-8. [DOI] [PubMed] [Google Scholar]
- 41.Oprian DD, Asenjo AB, Lee N, Pelletier SL. Design, chemical synthesis, and expression of genes for the three human color visual pigments. Biochemistry. 1991;30:11367–11372. doi: 10.1021/bi00112a002. [DOI] [PubMed] [Google Scholar]
- 42.Piantandida TP, Sperling HG. Isolation of a third chromatic mechanism in the protanomalous observer. Vision Res. 1973a;13:2033–2047. doi: 10.1016/0042-6989(73)90180-6. [DOI] [PubMed] [Google Scholar]
- 43.Piantandida TP, Sperling HG. Isolation of a third chromatic mechanism in the deuteranomalous observer. Vision Res. 1973b;13:2049–2058. doi: 10.1016/0042-6989(73)90181-8. [DOI] [PubMed] [Google Scholar]
- 44.Pitt FHG. Characteristics of dichromatic vision. Medical Research Council special report series number 200. His Majesty’s Stationery Office; London: 1935. [Google Scholar]
- 45.Pokorny J, Smith VC. Evaluation of single-pigment shift model of anomalous trichromacy. J Opt Soc Am. 1977;67:1196–1209. doi: 10.1364/josa.67.001196. [DOI] [PubMed] [Google Scholar]
- 46.Pokorny J, Smith VC, Katz I. Derivation of the photopigment absorption spectra in anomalous trichromats. J Opt Soc Am. 1973;63:232–237. doi: 10.1364/josa.63.000232. [DOI] [PubMed] [Google Scholar]
- 47.Pokorny J, Moreland JD, Smith VC. Photopigments in anomalous trichromacy. J Opt Soc Am. 1975;65:1522–1524. [Google Scholar]
- 48.Pokorny J, Smith VC, Verriest G, Pinckers AJLG. Congenital and acquired color vision defects. Grune and Stratton; New York: 1979. [Google Scholar]
- 49.Post RH. Population differences in red and green color vision deficiency: a review, and a query on selection relaxation. Eugen Q. 1962;9:131–146. doi: 10.1080/19485565.1962.9987517. [DOI] [PubMed] [Google Scholar]
- 50.Rayleigh L. Experiments on color. Nature. 1881;25:64–66. [Google Scholar]
- 51.Ruddock KH. The effect of age upon colour vision. II. Changes with age in light transmission of the ocular media. Vision Res. 1965;5:47–58. doi: 10.1016/0042-6989(65)90074-x. [DOI] [PubMed] [Google Scholar]
- 52.Rushton WAH. A cone pigment in the protanope. J Physiol (Lond) 1963;168:345–359. doi: 10.1113/jphysiol.1963.sp007196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rushton WAH. A foveal pigment in the deuteranope. J Physiol (Lond) 1965;176:24–37. doi: 10.1113/jphysiol.1965.sp007532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rushton WAH, Powell DS, White KD. Exchange thresholds in dichromats. Vision Res. 1973a;13:1993–2002. doi: 10.1016/0042-6989(73)90177-6. [DOI] [PubMed] [Google Scholar]
- 55.Rushton WAH, Powell DS, White KD. Pigments in anomalous trichromats. Vision Res. 1973b;13:2017–2031. doi: 10.1016/0042-6989(73)90179-x. [DOI] [PubMed] [Google Scholar]
- 56.Sanocki E, Lindsey DT, Winderickx J, Teller DY, Deeb SS, Motulsky AG. Serine/alanine amino acid polymorphism of the L and M cone pigments: effects on Rayleigh matches among deuteranopes, protanopes, and color normal observers. Vision Res. 1993;33:2139–2152. doi: 10.1016/0042-6989(93)90012-l. [DOI] [PubMed] [Google Scholar]
- 57.Sanocki E, Shevell SK, Winderickx J. Serine/alanine amino acid polymorphism of the L-cone photopigment assessed by dual Rayleigh-type color matches. Vision Res. 1994;34:377–382. doi: 10.1016/0042-6989(94)90096-5. [DOI] [PubMed] [Google Scholar]
- 58.Schnapf JL, Kraft TW, Baylor DA. Spectral sensitivity of human cone photoreceptors. Nature. 1987;325:439–441. doi: 10.1038/325439a0. [DOI] [PubMed] [Google Scholar]
- 59.Shyue S-K, Li L, Chang BH-J, Li WH. Intronic conversion in the evolution of human X-linked color vision genes. Mol Biol Evol. 1994;11:548–551. doi: 10.1093/oxfordjournals.molbev.a040134. [DOI] [PubMed] [Google Scholar]
- 60.Smith VC, Pokorny J. Spectral sensitivity of the foveal cone photopigments between 400 and 500 nm. Vision Res. 1975;15:161–171. doi: 10.1016/0042-6989(75)90203-5. [DOI] [PubMed] [Google Scholar]
- 61.Stiles WS. The directional sensitivity of the retina and the spectral sensitivity of the rods and cones. Proc R Soc Lond [Biol] 1939;127:64–105. [Google Scholar]
- 62.Stiles WS. Further studies of visual mechanisms by the two-colour threshold technique. Coloq Probl Opt Vis (UIPAP, Madrid) 1953;1:65–103. [Google Scholar]
- 63.Stiles WS. Foveal threshold sensitivity on fields of different colors. Science. 1964;145:1016–1017. doi: 10.1126/science.145.3636.1016. [DOI] [PubMed] [Google Scholar]
- 64.Stockman A, MacLeod DIA, DePriest DD. The temporal properties of the human short-wave photoreceptors and their associated pathways. Vision Res. 1991;31:189–208. doi: 10.1016/0042-6989(91)90111-h. [DOI] [PubMed] [Google Scholar]
- 65.Stockman A, MacLeod DIA, Johnson NE. Spectral sensitivities of the human cones. J Opt Soc Am. 1993a;10:2491–2521. doi: 10.1364/josaa.10.002491. [DOI] [PubMed] [Google Scholar]
- 66.Stockman A, MacLeod DIA, Vivien JA. Isolation of the middle- and long-wavelength-sensitive cones in normal trichromats. J Opt Soc Am. 1993b;10:2471–2490. doi: 10.1364/josaa.10.002471. [DOI] [PubMed] [Google Scholar]
- 67. Stockman A, Sharpe LT. Cone spectral sensitivities and color matching. Color vision: from genes to perception Gegenfurtner K, Sharpe LT. 1999. Cambridge UP; Cambridge, UK, in press. [Google Scholar]
- 68.Stockman A, Sharpe LT, Fach CC (1999) The spectral sensitivity of the human short-wavelength sensitive cones derived from thresholds and color matches. Vision Res, in press. [DOI] [PubMed]
- 69.van Norren D, Vos JJ. Spectral transmission of the human ocular media. Vision Res. 1974;14:1237–1244. doi: 10.1016/0042-6989(74)90222-3. [DOI] [PubMed] [Google Scholar]
- 70.Vos JJ. Literature review of human macular absorption in the visible and its consequences for the cone receptor primaries. Netherlands Organization for Applied Scientific Research, Institute for Perception; Soesterberg, The Netherlands: 1972. [Google Scholar]
- 71.Wald G. The receptors of human color vision. Science. 1964;145:1007–1016. doi: 10.1126/science.145.3636.1007. [DOI] [PubMed] [Google Scholar]
- 72.Webster MA. A reanalysis of λmax variation in the Stiles and Burch 10 degree color matching functions. J Opt Soc Am [A] 1992;9:1419–1421. doi: 10.1364/josaa.9.001419. [DOI] [PubMed] [Google Scholar]
- 73.Webster MA, MacLeod DIA. Factors underlying individual differences in the color matches of normal observers. J Opt Soc Am [A] 1988;5:1722–1735. doi: 10.1364/josaa.5.001722. [DOI] [PubMed] [Google Scholar]
- 74.Williams AJ, Hunt DM, Bowmaker JK, Mollon JD. The polymorphic photopigments of the marmoset: spectral tuning and genetic basis. EMBO J. 1992;11:2039–2045. doi: 10.1002/j.1460-2075.1992.tb05261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Willmer EN. Further observations on the properties of the central fovea in colour-blind and normal subjects. J Physiol (Lond) 1950;110:422–446. doi: 10.1113/jphysiol.1949.sp004450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Winderickx J, Lindsey DT, Sanocki E, Teller DY, Motulsky AR, Deeb SS. Polymorphism in red photopigment underlies variation in colour matching. Nature. 1992a;356:431–433. doi: 10.1038/356431a0. [DOI] [PubMed] [Google Scholar]
- 77.Winderickx J, Battisti L, Motulsky AR, Deeb SS. Selective expression of the human X-linked green pigment genes. Proc Natl Acad Sci USA. 1992b;89:9710–9714. doi: 10.1073/pnas.89.20.9710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Winderickx J, Battisti L, Hibiya Y, Motulsky AR, Deeb SS. Haplotype diversity in the human red and green opsin genes: evidence for frequent sequence exchange in exon 3. Hum Mol Genet. 1993;2:1413–1421. doi: 10.1093/hmg/2.9.1413. [DOI] [PubMed] [Google Scholar]
- 79.Wyszecki G, Stiles WS. Color science, 2nd Ed. Wiley; New York: 1982. [Google Scholar]
- 80.Yamaguchi T, Motulsky AG, Deeb SS. Visual pigment gene structure and expression in human retinae. Hum Mol Genet. 1997;6:981–990. doi: 10.1093/hmg/6.7.981. [DOI] [PubMed] [Google Scholar]